Highlights
-
•
There were multiple introduction events of SARS-CoV-2 into Thailand.
-
•
One lineage, designated A/Thai-1, has expanded and has become a predominant and unique lineage in Thailand.
-
•
A major frame-shift mutation was found at the gene encoding ORF7a, a putative host antagonizing factor of the virus.
Keywords: COVID-19, Genomic surveillance, SARS-CoV-2, Thailand
Abstract
Coronavirus Disease 2019 (COVID-19) is a global public health threat. Genomic surveillance of SARS-CoV-2 was implemented in March of 2020 at a major diagnostic hub in Bangkok, Thailand. Several virus lineages supposedly originated in many countries were found, and a Thai-specific lineage, designated A/Thai-1, has expanded to be predominant in Thailand. A virus sample in the SARS-CoV-2 A/Thai-1 lineage contains a frame-shift deletion at ORF7a, encoding a putative host antagonizing factor of the virus.
1. Introduction
Coronavirus Disease 2019 (COVID-19) has reached the status of global pandemic. Genomic surveillance of its etiological virus, SARS-CoV-2, plays an important role in epidemiological investigations and transmission control strategies (Gudbjartsson et al., 2020). Genetic variation data of the virus could reveal transmission chains between infected individuals and could even map the connection between outbreak cohorts. Thailand has suffered from the spread of COVID-19 with the total number of confirmed cases over 3000 and with more than 120,000 individuals screened as of May 2020. Since January 2020, when both imported and locally-transmitted COVID-19 cases were reported in Thailand, the country implemented several measures to combat COVID-19 on a national scale (Okada et al., 2020; Pongpirul et al., 2020; Hinjoy et al., 2020).
Genomic surveillance could be a powerful tool in the implementation of the national COVID-19 control strategy in Thailand. ARTIC multiplex tiling PCR allows whole-genome sequencing with minuscule amount of material by generating genome-wide overlapping amplicons, which has led to its success during the Zika virus outbreak investigation in Brazil (Quick et al., 2017; Giovanetti et al., 2020). Using leftover RNA samples from a standard RT-PCR diagnosis, the genomic information of SARS-CoV-2 can be decoded in less than a week. The data presented here provide an insight into the genetic repertoire, origins and viral lineages of SARS-CoV-2 in Thailand. The information is particularly important given the multiple introduction events into the country and the local expansion of the Thai-specific SARS-CoV-2 lineages.
2. Genomic surveillance of SARS-CoV-2 populations in Thailand
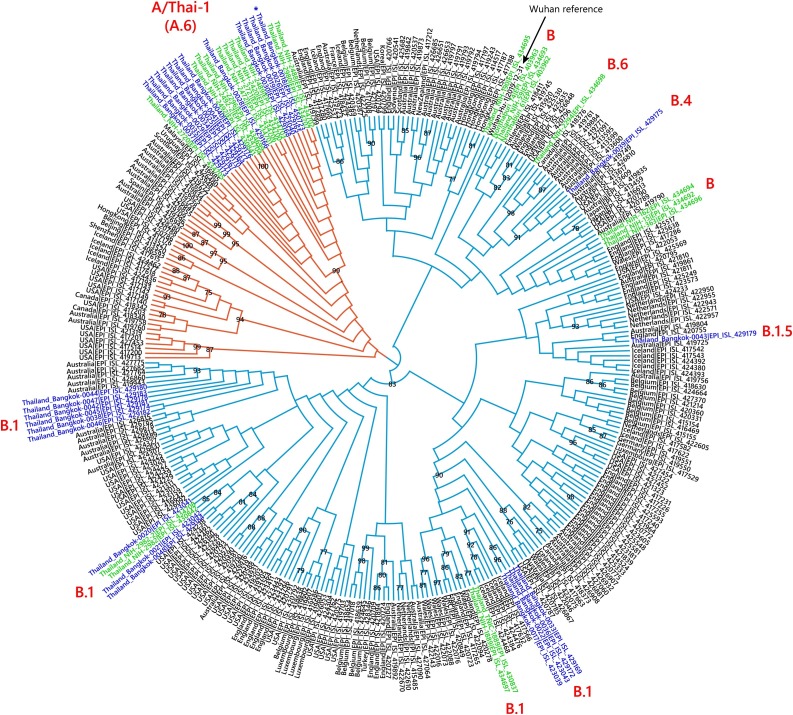
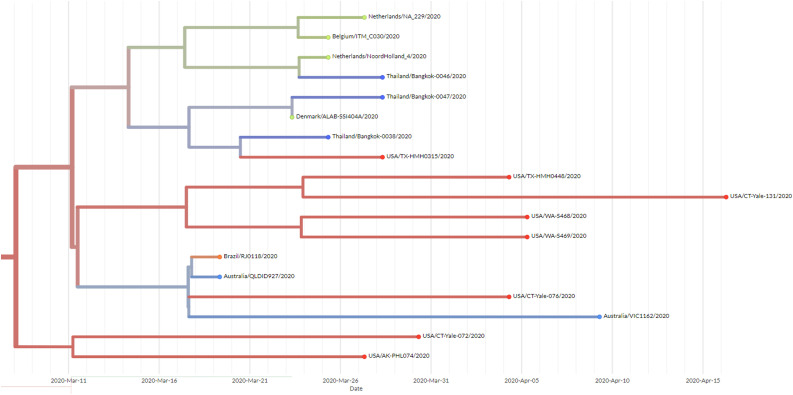
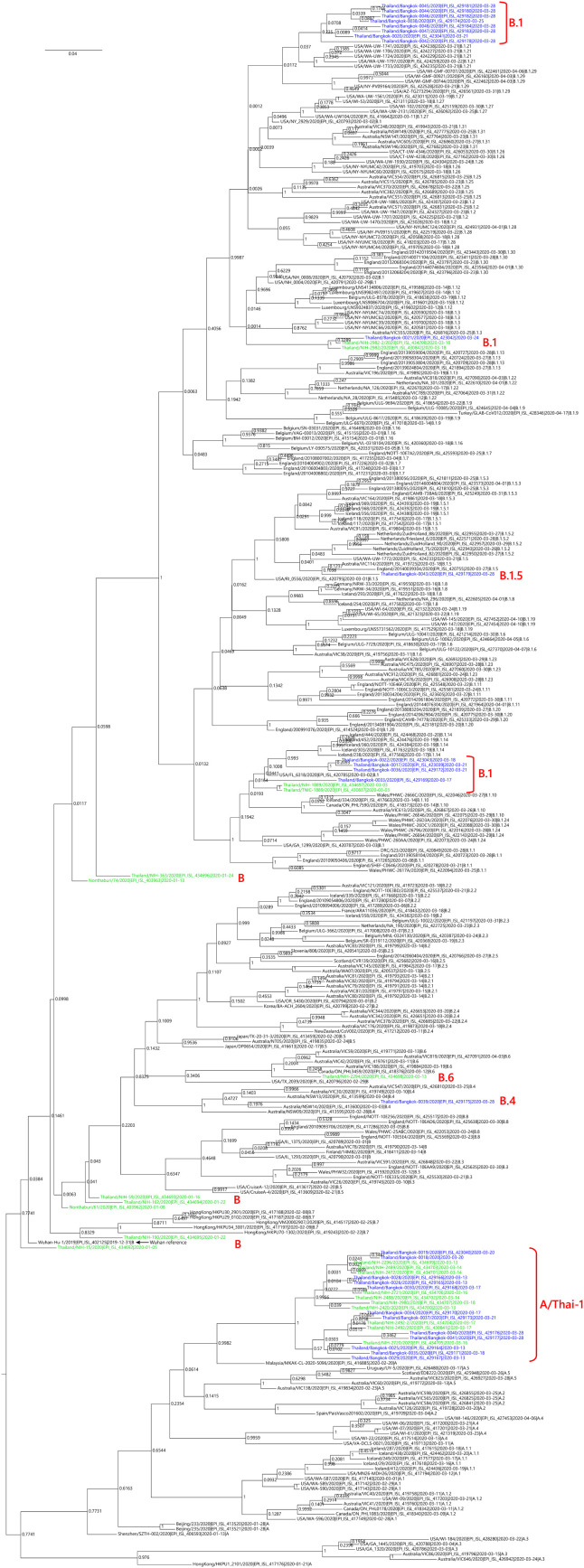
We sequenced 27 anonymized RT-qPCR positive virus transport media samples containing nasopharyngeal/oropharyngeal swabs from Ramathibodi Hospital in Bangkok from March 13, 2020 to March 28, 2020 (Supplementary Table 1) [EC approval number: MURA2020/676]. The hospital acted as one of the major diagnostic hubs for COVID-19 in Bangkok during the study period. Enrichment and amplification steps were done according to the ARTIC Network protocol with ARTIC primer version 2 (Quick, 2020). The libraries were prepared using KAPA HyperPrep and KAPA Library Amplification kits and subsequently sequenced using with a MiSeq Reagent Kit v2 according to the manufactures’ protocols. Variant calling was performed using the ncov2019-artic-nf pipeline (https://github.com/connor-lab/ncov2019-artic-nf) using the default Illumina parameters (variants called by iVar v1.11 with minimum frequency threshold 0.75 and minimum depth 10) (Grubaugh et al., 2019). Consensus sequences were used to construct the maximum-likelihood and Bayesian phylogenetic trees with recommended representatives from various lineages worldwide utilizing IQ-TREE 2.0 and BEAST v1.10.4, respectively (Supplementary Table 2) (Drummond et al., 2012; Minh et al., 2020; Rambaut et al., 2020a). Interestingly, Thailand appears to have had multiple introduction events of SARS-CoV-2 into the country, as evidenced by at least six separate clusters in the maximum-likelihood tree (Fig. 1 and Supplementary Fig. 1) (Hadfield et al., 2018). Based on a Pangolin classification system (Database version 27 April 2020), they are grouped into A (12 samples), B.1 (13 samples), B.1.5 (1 sample) and B.4 (1 sample) lineages (Fig. 1, Supplementary Fig. 1 and Supplementary Table 1) (Rambaut et al., 2020b; O’Toole, 2020). The relationship and the origin of these lineages were described in (Rambaut et al., 2020a). Considering the origins and lineage branches, these SARS-CoV-2 lineages are likely to have recent ancestors outside Thailand. For example, six of the samples in the B.1 lineage in our collection are grouped tightly with virus samples commonly found in the United States of America and Europe and collected during the same period (March 2020) as visualized by Nextstrain Timetree (Supplementary Fig. 2 ). The constructed Bayesian tree displayed the similar structure to that of the maximum-likelihood approach (Supplementary Fig. 3).
Fig. 1.
Cladogram based on a maximum-likelihood tree showing Thai populations of SARS-CoV-2. The cladogram represents a matching maximum-likelihood tree (1,000 bootstrap replicates) shown in Supplementary Fig. 1. Genomes generated in this study are labelled in blue with remaining lineage representatives labelled in black. Bootstrap values (≥ 75) are shown at the nodes. Thai virus genomes independently generated by other groups and deposited in GISAID are labelled in green. Genomes collected from January 2020 are similar to that of reference Wuhan-Hu-1. Our data showed at least four independently lineages, and two additional events of lineage B from January 2020 and lineage B.6 (shown in green), could be recognized as potential introduction events. The A and B lineages are coloured in orange and cyan, respectively. * indicates the sample with the ORF7a deletion.
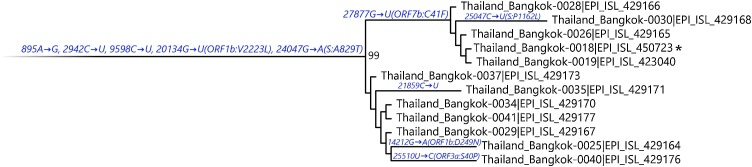
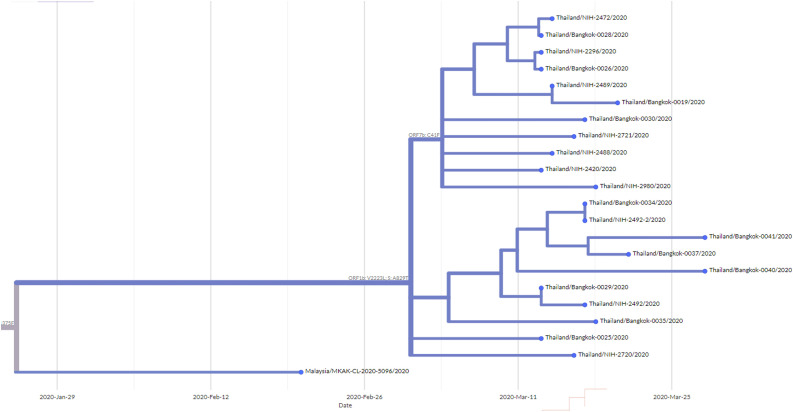
Fig. 2.
Maximum-likelihood tree containing A/Thai-1 subset. For visualization of A/Thai-1 lineage, the A/Thai-1 genomes from Fig. 1 generated in this study are presented. The lineage defining node has a bootstrap value of 99 (1,000 replicates) as shown in Supplementary Fig. 1. Mutations that define each branch point are shown. 20,134G→U in Thailand_Bangkok-0030 could not be called. * indicates the sample with the ORF7a deletion.
It is worth noting the local expansion of a putative Thai specific lineage (Fig. 1 and Fig. 2). This cluster of viruses has passed the criteria of a novel lineage as follows: (a) exhibits two shared nucleotide differences from the ancestral lineage [cut point ≥ 1], (b) contains 11 genomes with > 95 % of the genome sequenced [cut point ≥ 5], (c) exhibits one shared nucleotide change (27,877G→U) among the ongoing transmission groups from March 2020 [cut point ≥ 1], and (d) has 99 % bootstrap value for the lineage defining node [cut point > 70 %] (Rambaut et al., 2020b). This lineage, designated A/Thai-1 (Fig. 1, Fig. 2), descended from the original A lineage (based on the maximum-likelihood based classification system), which was first reported in China before expanding into various countries in Asia, Europe, North America, South America and Australia. This A/Thai-1 branch (designated A.6 in the Pangolin system (Rambaut et al., 2020a)) is separated from the rest of the original A lineage and subgroups. Upon visual inspection in Nextstrain, only one Malaysian sample (MKAK-CL-2020-5096) is the closest to A/Thai-1, but only with 63 % bootstrap value by the maximum-likelihood approach and one shared lineage-specific nucleotide substitution (4,390G→U) (Supplementary Figure 4). Non-synonymous mutations unique to A/Thai-1 are 20,134G→U (ORF1b) and 24,047G→A (Spike protein) (the full mutation list is shown in Table 1 ). Among the changes, 20,134G→U mutation has been independently found in two samples in lineage B.1 from the Netherlands and USA. It remains to be determined with a larger sample size whether this is the result of convergent evolution or genetic recombination. This pattern of homoplasy was also hypothetically linked to putative RNA editing (Simmonds, 2020).
Table 1.
Mutations present in A/Thai-1 in comparison with Wuhan-Hu-1.
| Position | Ref | Alt | Syn/Non-syn | Gene | Samples | Note |
|---|---|---|---|---|---|---|
| 895 | A | G | S | ORF1a | 0018, 0019, 0025, 0026, 0028, 0029, 0030, 0034, 0035, 0037, 0040, 0041 | Unique to A/Thai-1 |
| 2942 | C | U | S | ORF1a | 0018, 0019, 0025, 0026, 0028, 0029, 0030, 0034, 0035, 0037, 0040, 0041 | Unique to A/Thai-1 |
| 4390 | G | U | NS | ORF1a | 0018, 0019, 0025, 0026, 0028, 0029, 0030, 0034, 0035, 0037, 0040, 0041 | Shared with Malaysia/MKAK-CL-2020-2096 (A lineage) |
| 8782 | C | U | S | ORF1a | 0018, 0019, 0025, 0026, 0028, 0029, 0030, 0034, 0035, 0037, 0040, 0041 | Shared among ancestral A lineages |
| 9598 | C | U | S | ORF1a | 0018, 0019, 0025, 0026, 0028, 0029, 0030, 0034, 0035, 0037, 0040, 0041 | Shared with Scotland/EDB2081 (B.1 lineage) |
| 14212 | G | A | NS | ORF1b | 0025 | – |
| 20134 | G | U | NS | ORF1b | 0018, 0019, 0025, 0026, 0028, 0029, 0034, 0035, 0037, 0040, 0041 | Cannot be called in 0030; Shared with Netherlands/NA_300 and USA/UN-UW-5172 (B.1 lineage) |
| 21859 | C | U | S | S | 0035 | – |
| 24047 | G | A | NS | S | 0018, 0019, 0025, 0026, 0028, 0029, 0030, 0034, 0035, 0037, 0040, 0041 | Unique to A/Thai-1 |
| 25047 | C | U | NS | S | 0030 | – |
| 25510 | U | C | NS | ORF3a | 0040 | – |
| 27694-27697 | TTTC | – | Frame-shift deletion | ORF7a | 0018 | – |
| 27877 | G | U | NS | ORF7b | 0018, 0019, 0026, 0028, 0030 | Shared with Taiwan/170 and USA/UT-00514 (B.1 lineage) |
| 28144 | U | C | NS | ORF8 | 0018, 0019, 0025, 0026, 0028, 0029, 0030, 0034, 0035, 0037, 0040, 0041 | Shared among ancestral A lineages |
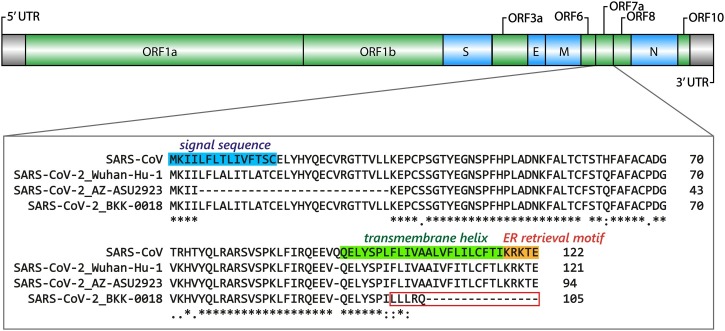
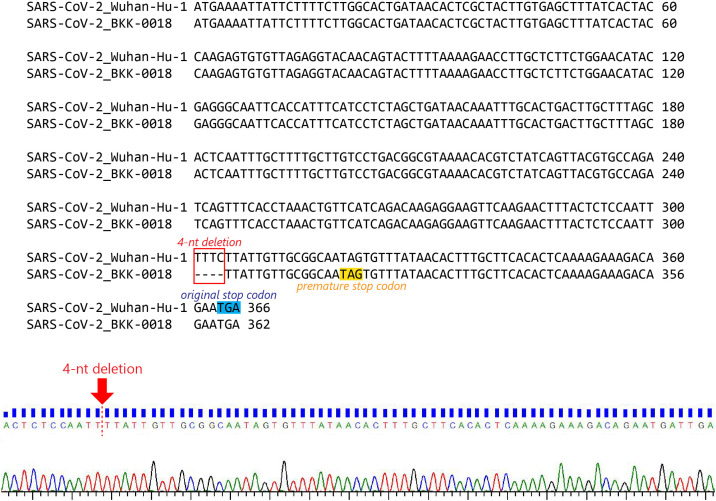
Thailand/Bangkok-0018, a sample in the A/Thai-1 lineage, contains a 4-nt frame-shift deletion at position 27,694-27,697, causing a premature truncation in ORF7a, which now contains five altered amino acid residues and loses the 16 original C-terminal residues (Fig. 3 ). The deletion was confirmed by Sanger sequencing twice using two independent RT-PCR reactions (Supplementary Figure 5). The frame-shift mutation alters approximately one-sixth (21/121 residues) of the ORF7a protein. Based on protein homology to SARS-CoV, the missing region corresponds to a transmembrane helix and an ER retrieval motif, required for antagonizing a host antiviral factor (Taylor et al., 2015; Fielding et al., 2004). One sample from Arizona, USA also contains an 81-nt in-frame deletion in the ORF7a gene (Holland et al., 2020). So far, only one sample in A/Thai-1 appears to have this frame-shift deletion. It is tempting to speculate on the relationship between ORF7a deletions and virus attenuation. However, further investigations by laboratory-based functional experiments are needed before reaching any conclusion on their biological and clinical significance.
Fig. 3.
Diagram depicting Thailand/Bangkok-0018 frame-shift deletion in ORF7a. The upper diagram shows the gene organization of the SARS-CoV-2 genome. The functional domains of ORF7a are indicated based on SARS-CoV. Sequences from SARS-CoV, the SARS-CoV-2 Wuhan-Hu-1 reference, the Arizona sample with an 81-nucleotide deletion (EPI_ISL_424669) and Thailand/Bangkok-0018 are aligned to demonstrate the altered region in red box.
When the analysis was extended to an additional set of 22 genomes, independently deposited in GISAID by the Thai Red Cross and the Thai National Institute of Health, samples collected in January 2020 are grouped closely with the B lineage from China including the Wuhan-Hu-1 reference (Supplementary Table 3). The genetic repertoires from this additional collection also support the notion of multiple virus lineages introduced into Thailand. A/Thai-1 was the largest lineage in Thailand during the period of March 2020, with the total of 22 virus samples designated to A/Thai-1 (12 from our sequencing work and 10 from other independent genomic sequencing projects) from the total of 49 genomes available.
3. Implication of the findings
Genomic surveillance is likely to be pivotal in the identification and the elimination of transmission cohorts and chains (Kumpornsin et al., 2019; Gardy and Loman, 2018). The genetic composition presented here suggests the necessity for screening and monitoring international travelers during the period of COVID-19 pandemic. The local expansion of A/Thai-1 strongly indicates a series of local transmission events, allowing an evolutionary branch unique to Thailand. This lineage needs to be investigated further for its compatibility to diagnosis and vaccine tools under development.
CRediT authorship contribution statement
Khajohn Joonlasak: Investigation. Elizabeth M Batty: Formal analysis, Software. Theerarat Kochakarn: Formal analysis, Software. Bhakbhoom Panthan: Investigation. Krittikorn Kümpornsin: Investigation, Methodology, Writing - review & editing. Poramate Jiaranai: Investigation. Arporn Wangwiwatsin: Investigation, Methodology. Angkana Huang: Formal analysis, Software. Namfon Kotanan: Investigation. Peera Jaru-Ampornpan: Formal analysis. Wudtichai Manasatienkij: Formal analysis. Treewat Watthanachockchai: Resources. Kingkan Rakmanee: Resources. Anthony R. Jones: Supervision, Writing - review & editing. Stefan Fernandez: Supervision, Writing - review & editing. Insee Sensorn: Project administration. Somnuek Sungkanuparph: Resources, Writing - review & editing. Ekawat Pasomsub: Resources, Formal analysis. Chonticha Klungthong: Supervision, Formal analysis, Writing - review & editing. Thanat Chookajorn: Formal analysis, Supervision, Project administration, Writing - original draft, Funding acquisition, Writing - review & editing. Wasun Chantratita: Formal analysis, Supervision, Project administration, Funding acquisition, Writing - review & editing.
Declaration of Competing Interest
None.
Acknowledgements
The work here was supported by Ramathibodi Foundation, Thailand Center of Excellence for Life Sciences (TCELS), the National Research Council of Thailand (NRCT) and Mahidol University. The authors acknowledge NSTDA Supercomputer Center (ThaiSC) for providing computing resources for this work. We are grateful for the comment and suggestion from P.Wilairat and Y. Yuthavong. The computational aspects of this research were supported by the Wellcome Trust Core Award Grant Number 203141/Z/16/Z and the NIHR Oxford BRC. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health. The original surveillance study was funded by the Armed Forces Health Surveillance Branch and its Global Emerging Infections Surveillance branch (P0128_20_AF_13) under fiscal year 2020. Material has been reviewed by the Walter Reed Army Institute of Research. There is no objection to its presentation and/or publication. The opinions or assertions contained herein are the private views of the author, and are not to be construed as official, or as reflecting true views of the Department of the Army or the Department of Defense.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.virusres.2020.198233.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- Drummond A.J. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012;29(8):1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielding B.C. Characterization of a unique group-specific protein (U122) of the severe acute respiratory syndrome coronavirus. J. Virol. 2004;78(14):7311–7318. doi: 10.1128/JVI.78.14.7311-7318.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardy J.L., Loman N.J. Towards a genomics-informed, real-time, global pathogen surveillance system. Nat. Rev. Genet. 2018;19(1):9–20. doi: 10.1038/nrg.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovanetti M. Genomic and epidemiological surveillance of zika virus in the Amazon region. Cell Rep. 2020;30(7):2275–2283. doi: 10.1016/j.celrep.2020.01.085. e7. [DOI] [PubMed] [Google Scholar]
- Grubaugh N.D. An amplicon-based sequencing framework for accurately measuring intrahost virus diversity using PrimalSeq and iVar. Genome Biol. 2019;20(1):8. doi: 10.1186/s13059-018-1618-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudbjartsson D.F. Spread of SARS-CoV-2 in the icelandic population. N. Engl. J. Med. 2020 doi: 10.1056/NEJMc2027653. [DOI] [PubMed] [Google Scholar]
- Hadfield J. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34(23):4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinjoy S. A self-assessment of the Thai Department of Disease Control’s communication for international response at early phase to the COVID-19. Int. J. Infect. Dis. 2020 doi: 10.1016/j.ijid.2020.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland L.A. An 81 nucleotide deletion in SARS-CoV-2 ORF7a identified from sentinel surveillance in Arizona (jan-Mar 2020) J. Virol. 2020 doi: 10.1128/JVI.00711-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumpornsin K., Kochakarn T., Chookajorn T. The resistome and genomic reconnaissance in the age of malaria elimination. Dis. Model. Mech. 2019;12(12) doi: 10.1242/dmm.040717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh B.Q. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020;37(5):1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole, A.M., (2020) JT. Phylogenetic Assignment of Named Global Outbreak LINeages. [cited 2020 6 May]; Available from: https://github.com/hCoV-2019/pangolin.
- Okada P. Early transmission patterns of coronavirus disease 2019 (COVID-19) in travellers from Wuhan to Thailand, January 2020. Euro Surveill. 2020;25(8) doi: 10.2807/1560-7917.ES.2020.25.8.2000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pongpirul W.A. Journey of a thai taxi driver and novel coronavirus. N. Engl. J. Med. 2020;382(11):1067–1068. doi: 10.1056/NEJMc2001621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quick J. 2020. nCoV-2019 Sequencing Protocol.https://www.protocols.io/view/ncov-2019-sequencing-protocol-bbmuik6w Available from: [Google Scholar]
- Quick J. Multiplex PCR method for MinION and Illumina sequencing of Zika and other virus genomes directly from clinical samples. Nat. Protoc. 2017;12(6):1261–1276. doi: 10.1038/nprot.2017.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020 doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A., Holmes E.C., Pybus O.G. 2020. A Dynamic Nomenclature for SARS-CoV-2 to Assist Genomic Epidemiology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmonds P. Rampant C-->U hypermutation in the genomes of SARS-CoV-2 and other coronaviruses: causes and consequences for their short- and long-term evolutionary trajectories. mSphere. 2020;5(3) doi: 10.1128/mSphere.00408-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J.K. Severe acute respiratory syndrome coronavirus ORF7a inhibits bone marrow stromal antigen 2 virion tethering through a novel mechanism of glycosylation interference. J. Virol. 2015;89(23):11820–11833. doi: 10.1128/JVI.02274-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.