Abstract
Human ATP‐binding cassette transporter 6 of subfamily B (ABCB6) is an ABC transporter involved in the translocation toxic metals and anti‐cancer drugs. Using cryo‐electron microscopy, we determined the molecular structure of full‐length ABCB6 in an apo state. The structure of ABCB6 unravels the architecture of a full‐length ABCB transporter that harbors two N‐terminal transmembrane domains which is indispensable for its ATPase activity in our in vitro assay. A slit‐like substrate binding pocket of ABCB6 may accommodate the planar shape of porphyrins, and the existence of a secondary cavity near the mitochondrial intermembrane space side would further facilitate substrate release. Furthermore, the ATPase activity of ABCB6 stimulated with a variety of porphyrin substrates showed different profiles in the presence of glutathione (GSH), suggesting the action of a distinct substrate translocation mechanism depending on the use of GSH as a cofactor.
Keywords: ABCB6, hemin, porphyrins, transporter
1. INTRODUCTION
ATP‐binding cassette transporter 6 of subfamily B (ABCB6) is a widely expressed ABC transporter that has been found in many tissues, especially in liver, muscle, red blood cells (RBCs), and skin. 1 It was originally isolated from liver by screening P‐glycoprotein‐like genes related to drug resistance. 2 Sequence analysis indicates that ABCB6 is a human functional homologue of heavy metal tolerance factor 1. 2 , 3
ABCB6 was first reported to be located in the mitochondrial outer membrane as a porphyrin transporter which involved in the translocation of coproporphyrinogen III from the cytoplasm into the mitochondria for heme biosynthesis. 4 ATP‐driven translocation of porphyrin into mitochondria is severely impaired in the absence of ABCB6, 5 highlighting the role of ABCB6 in heme precursor translocation. Several studies have suggested that ABCB6 is also presented in other membrane systems like intracellular endo/lysosomal compartment 6 , 7 , 8 , 9 or the plasma membrane, 10 which is consistent with its ability to confer tolerance to toxic heavy metals. 1 , 3 ABCB6 has also been shown to protect cells from oxidative stress 11 and drug toxicity. 12 , 13 The overexpression of ABCB6 enhances drug resistance against the antineoplastic drug SN‐38 and vincristine, suggesting that the transporter is a potential anti‐cancer therapeutic target. 14 In addition to its role as a transporter, ABCB6 is also a Lan blood group antigen expressed on RBCs. The absence of ABCB6 protein expression on RBCs will lead to a rare blood phenotype Langereis (Lan). 15
Structurally, human ABCB6 is a homodimeric ABCB subfamily transporter, with each subunit containing 842 amino acid residues and 11 transmembrane (TMs) helices. Besides a canonical core domain structure formed by two TM domains (TMDs, i.e., TMs 6–11) and two cytoplasmic nucleotide‐binding domains (NBDs), ABCB6 was predicted to contain an additional N‐terminal TMD0 (i.e., TMs 1–5) in each subunit. Such TMD0 domains are only present in ABCB and ABCC subfamilies. Recently, the structures of two ABCC subfamily members, the multidrug resistant transporters MRP1 and SUR1, were determined to have a single TMD0. 16 , 17 However, no ABCB subfamily transporter with two TMD0s has yet been reported. To illustrate the porphyrin translocation mechanism and potential roles of TMD0 in ABCB6 function, we herein determined the 3D structure of human mitochondrial ABCB6 using single particle cryo‐electron microscopy (cryo‐EM). The obtained structure unravels the architecture of the first known full‐length ABCB subfamily that harbors two TMD0s and implies the existence of a distinct substrate transport mechanism of porphyrins.
2. RESULTS
2.1. Structural overview
The cryo‐EM sample was generated from full‐length ABCB6 proteins expressed in HEK293F cells. The proteins were extracted from the membrane and purified with lauryl maltose neopentyl glycol (LMNG). The purified protein sample was homogeneous and could bind porphyrin substrates which stimulated ATP hydrolysis (Figure S1). Eluted protein fractions were then immediately applied to supporting grids for the cryo‐EM study. 3D reconstitution yielded an overall resolution of full‐length ABCB6 of 5.2 Å (Figure S2, Table S1). Given the high flexibility of TMD0 domains, focused refinement on the transporter core region containing the two TMDs and two NBDs of ABCB6 was subsequently performed, which resulted in a better resolution of 4.0 Å at the core region (Figures S2 and S3, Table S1).
The ABCB6 protein was captured in an inward‐facing (i.e., facing the cytoplasmic side), open conformation that represents a substrate‐free apo state of the ABC exporter (Figure 1a,b). The two NBDs of ABCB6 separated approximately 22‐Å apart from each other (calculated with the Cα distance between two G827 residues at the C‐termini of NBDs) (Figure S4b), resulting in a large cytoplasmic accessible cavity in the membrane‐spanning region. TMs 9–10 of each subunit (Figure 1c) swapped with their counterparts and formed a canonical TMD structure of the Type‐I ABC exporter. 18 As suggested by the high similarity between their primary sequences (59% in core domain), the overall structure of the core domain of ABCB6 was very similar to the apo structure of yeast iron–sulfur cluster exporter Atm1, except that ABCB6 adopted a much more open conformation (Figure S4). This conformation discrepancy is putatively caused by the lack of interaction between the C‐terminal helices of NBDs in ABCB6, which is instead observed in Atm1 and locks the NBDs in a restricted inward‐facing conformation 19 (Figure S4). The N‐terminal TMD0 (Residues 27–206), which was predicted to be formed by TMs 1–5, symmetrically flanked on both sides of the core region, making a “frog”‐like structure for ABCB6 (Figure 1a,d).
FIGURE 1.

Overall structure of full‐length ATP‐binding cassette transporter 6 of subfamily B (ABCB6). (a) The overall structure of the full‐length human ABCB6 homodimer. The core domain of ABCB6 is shown as a ribbon presentation. TMD0s are shown as cylinders. Individual subunits are colored red and blue. (b) Ribbon presentation of the ABCB6 subunit colored as a rainbow spectrum ranging from blue to orange. (c) Schematic of the domain structure of ABCB6. Dashed lines represent interdomain regions that were not resolved in the cryo‐electron microscopy (EM) reconstructions and not included in the final model
2.2. The N‐terminal domain of TMD0
The presence of TMD0 was visible in the 2D classification (Figure 2a and S1). The two TMD0s of ABCB6 were observed apart from the core domain with no direct interactions in the final EM map. A structural comparison of ABCB6 with the ABCC subfamily transporter MRP1, which contains a single TMD0, revealed a quite different TMD0 position relative to the core domain (Figure 2b). In ABCB6, the two TMD0s resided in a line approximately 90° off the “TMD plane” that is perpendicular to the membrane and passes the two elbow helices. In comparison, the TMD0 domain of MRP1 is much closer to the TMD plane with an angle of about 60° (Figure 2c).
FIGURE 2.

TMD0 conformation of full‐length ATP‐binding cassette transporter 6 of subfamily B (ABCB6). (a) 2D classification of ABCB6 cryo‐electron microscopy (EM) particles with two TMD0s (marked by red arrows). (b,c) Structural alignment of human ABCB6 with bovine MRP1. Side view (b): the relative position of TMD0s of human ABCB6 is lower than that of bovine MRP1, moving toward the cytoplasmic side. Top view (c): the two TMD0s of human ABCB6 are perpendicular to the TMD axis. In comparison, the TMD0 in bovine MRP1 stays closer to the TMD axis with an angle of approximately 60°. (d) The BLI binding assay of full‐length and ΔTMD0 variants of the human ABCB6 protein with hemin (50 μM). The K D values of hemin toward the full‐length and ΔTMD0 samples were determined as 25.3 ± 0.7 μM and 15.3 ± 0.8 μM, respectively. (e) Normalized ATPase activity of full‐length and ΔTMD0 protein samples stimulated by different types of substrates. For the full‐length protein, coproporphyrin III, protoporphyrin IX, and chlorophyllin showed over twofold stimulated ATPase activity compared with apo protein. In comparison, the ΔTMD0 variant lost the substrate stimulation in the ATPase activity assay. Error bars represent SEM of technical replicates, n = 3
The observed conformational differences between TMD0 domains in ABCB6 and in MRP1 are possibly caused by a ~90 residue interfacial lasso motif that is specifically present in the ABCC subfamily but absent in ABCB6. In MRP1, the lasso motif interacts with and positions the TMD0 domain toward the TMD plane (Figure 2b). Lack of this lasso motif made the TMD0 of ABCB6 much more flexible and able to adopt a position in which its helices were nearly perpendicular to the nearby TMD helices in the detergent micelles. Accordingly, without the C‐terminal parts of lasso motifs lying near the cytoplasmic surface, the TMD0s of ABCB6 were located closer toward the NBDs compared with MRP1 (Figure 2b,c).
Because the core domain of the transporter already contains the minimal structural elements that are needed to perform substrate transport, 20 TMD0 domains are considered auxiliary docking sites that interact with other proteins. In the case of ABCB6, however, no protein partners have been identified to associate with TMD0. To test the role of TMD0 in ABCB6 function, we generated a core domain construct of ABCB6 (ΔTMD0) that is devoid of the N‐terminal 236 residues. This ΔTMD0 variant displayed a comparable hemin binding to the full‐length protein, indicating that TMD0 is not crucial for substrate binding (Figure 2d). However, the substrate‐stimulated ATPase activity was completely lost in ΔTMD0 (Figure 2e), whereas the basal activity of its apo protein increased, suggesting that the presence of TMD0 affects the apo state conformation(s) of the transporter.
2.3. Substrate binding and translocation pathway
Two potential substrate binding cavities were observed in ABCB6 (Figure 3a–c and S5). The canonical cavity (cavity‐1, ~1,351 Å3), which was surrounded by TM6, TM7, TM10, and TM11 of each subunit (Figure S5), opened to the cytoplasm and was extended deeply into the TM region. In particular, cavity‐1 was widely opened along the direction of NBD separation, but became narrowed at the cross direction because of the two TM11 helices that were in close proximity (Figure 3b and S5). Its slit shape enabled cavity‐1 to accommodate the polycyclic porphyrin substrates. Cavity‐1 mutants with increased pocket sizes resulting from the introduction of short chain residues at positions W546TM11 and M542TM11 at the pocket entrance (e.g., W546A, W546V, and M542A) resulted in the loss of substrate‐stimulated ATPase activity, whereas W546F maintained a similar ATPase activity to the wild‐type protein (Figures 3d and S6). Molecular docking of heme precursor protoporphyrin IX into ABCB6 showed that its porphyrin moiety could insert into the slit‐like pocket and form hydrophobic interactions with residues W546TM11 and M542TM11 (Figure S7). The binding mode of protoporphyrin IX to ABCB6 was similar to that of substrate LTC4 bound to MRP1. 16 In both cases, a hydrophobic moiety of the substrate binds at the central position of the substrate‐binding pocket, whereas hydrophilic moieties point to the lateral side of the pocket (Figure S7). These observations indicated that maintaining a narrow slit‐shaped pocket at the center of cavity‐1 is critical for translocation of porphyrin substrate. Unlike in P‐gp (also called ABCB1), the lateral membrane access route of ABCB6 was fully blocked by the TMD helices (Figure S8), suggesting that its substrates are bound from the cytoplasmic side. Consistently, despite its lipophilic nature, porphyrin contains negatively charged carboxylate side chains that are likely to prevent a substrate from diffusing through the membrane. 21 Indeed, the entrance of cavity‐1 was surrounded by basic amino acid residues (Figure S9), which may facilitate the initial recruitment of a substrate from the cytoplasm by interacting with negatively charged carboxylate groups of porphyrins.
FIGURE 3.

Substrate translocation pathway of ATP‐binding cassette transporter 6 of subfamily B (ABCB6). (a) Surface clip of ABCB6 protein, showing the two substrate‐binding cavities. (b) Cartoon representation of the residues forming the plug, lid, as well the cavities. Key residues in the entrance of cavity‐1 are shown as sticks. (c) Cartoon and surface presentation of the substrate pathway of human ABCB6 protein. Cavity‐1 and cavity‐2 are shown as a meshed surface. The plug is formed by Y286TM6 and V531TM11 of both subunits. The lid is formed by residues T294. (d) Substrate‐stimulated ATPase activity of the apo protein and each mutant. Error bars represent SEM of technical replicates, n = 3
At the top of cavity‐1, residues Y286 and V531 of the ABCB6 homodimer formed a “plug” that separated cavity‐1 from a secondary cavity (cavity‐2, ~31 Å3). This cavity‐2 was mainly made up of residues from TM6, TM11, and the extracellular loop‐3 (ECL3), and opened to an opposite direction (Figures 3a–c and S5). Both Y286A and V531A mutations abolish the substrate‐stimulated ATPase activity of ABCB6 and turn the substrates into inhibitor (Figures 3d and S6), suggesting that the plug residues are critical for substrate translocation. In the inward‐open apo state, cavity‐2 is much smaller than cavity‐1 and is sealed by a “lid” formed by two T294 residues at the N‐termini of TM6 helices (Figures 3b and S5), preventing solvent access from the outside. Destabilization of cavity‐2 by T294A mutation may lead to a further closure of the extracellular side of ABCB6, which resulted in a decreased basal activity (Figure 3d). To our knowledge, this cavity‐2 has only been observed in ABCB6 in this study and in ABCG2, 22 , 23 both of which are involved in the translocation of porphyrins. 24 Given the large size and rigidity of porphyrin moieties, the existence of cavity‐2 may facilitate substrate release by providing part of the transport path. Finally, the protein expression level of the key cavity mutants Y286A, T294A, V531A, W645A, and W645V are impaired compare to WT protein (Figure S6), indicating these cavity residues are critical for maintaining the structural stability of ABCB6.
2.4. Critical role of glutathione in ABCB6 substrate translocation
Glutathione (GSH) has been indicated in the function of several ABC transporters. To investigate possible functional roles of GSH in ABCB6‐mediated transport, we measured the ABCB6 ATPase activity stimulated by six of its potential substrates (Figure S10) in both the presence and absence of GSH (Figure 4a–e). All the porphyrins used in this paper, as well as GSH, showed a direct binding to ABCB6 in the biolayer interferometry (BLI) binding assay (Figure S11). For protoporphyrin IX, coproporphyrin III, and chlorophyllin, ATPase activity was stimulated by approximately twofold to threefold over the apo state without GSH (Figure 4e). In contrast, hemin, hematin, and Fe‐coproporphyrin III were found to exert no obvious effect on ATPase activity (Figure 4a–e). In the presence of GSH, however, hemin, hematin, and Fe‐coproporphyrin III stimulated the ATPase activity of ABCB6 by approximately twofold to threefold over the apo state (Figure 4e). In contrast with GSH, GSH analog ophthalmic acid, other reducing reagents, and component amino acids of GSH showed no stimulatory effect on the ATPase activity of ABCB6 (Figure 4d), indicating that it was the GSH chemical structure per se, rather than its reducing property, that increased the ATPase activity.
FIGURE 4.

Glutathione (GSH) plays a critical role in ATP‐binding cassette transporter 6 of subfamily B (ABCB6) substrate translocation and toxic metal resistance. (a) ATPase activity of full‐length human ABCB6 with different concentrations of GSH (0–5 mM) and hemin (10 μM). The dashed line represents the ATPase activity of ABCB6 with GSH only. The K M value determined for GSH in the presence of hemin (10 μM) was 51 ± 8 μM. The maximal ATPase activity was determined with GraphPad prism software to be 82.6 ± 1.6 nmol/mg/min. (b–d) The stimulated ATPase activity of ABCB6 of hemin (b), hematin (c), and Fe‐coproporphyrin III (d) in the presence or absence of GSH (1 mM). The K M values determined for hemin, hematin, and Fe‐coproporphyrin III in addition of GSH (1 mM) were 190 ± 23 nM, 600 ± 140 nM, and 148 ± 41 nM, respectively. The maximal ATPase activity was determined to be 79.6 ± 1.9 nmol/mg/min. (e) Stimulation of ATPase activity by different types of porphyrins with or without GSH (1 mM). (f) Hemin stimulated ATPase activity of ABCB6 in the presence of GSH, GSH derivatives, and other reducing agents. Among all these additives, only GSH could stimulate the ATPase activity of the protein when the same concentration (10 μM) of hemin was used. (g) GSH rescued the metal toxicity of Cu2+ to ABCB6. Without GSH, increased Cu2+ (0–300 μM) impaired the ATPase activity of ABCB6. In contrast, the addition of GSH (1 mM) protected ABCB6 from the metal toxicity of Cu2+ and displayed a stimulated ATPase activity. Error bars represent SEM of technical replicates, n = 3
Because GSH alone did not stimulate the ATPase activity of ABCB6 (Figure 4a,e), it may promote hemin binding and consequently the conformational change of ABCB6 through GSH conjugation with the ferric ion of hemin, which has been seen in other hemoprotein structures. 25 Such GSH conjugation may neutralize positive charges of the substrate and introduce additional substrate interactions with the protein, and thus stabilize a close NBD conformation which results in increased ATPase activity. In contrast, because protoporphyrin IX and coproporphyrin III do not chelate with any iron ion, the addition of GSH showed no extra stimulation on ATPase activity (Figure 4e). It is interesting to note that GSH also showed no improvement on chlorophyllin (Figure 4e). A possible explanation is that the Cu2+ cation is involved in an electrostatic interaction with the chlorophyll anion, so cannot conjugate with GSH. Furthermore, GSH was able to rescue and even enhance the ABCB6 ATPase activity impaired by Cu2+ (Figure 4g), suggesting that the metal toxicity resistance of ABCB6 is mediated by GSH conjugation.
3. DISCUSSION
Our three‐dimensional structure of ABCB6 together with functional data allows us to propose a transport mechanism for its substrates (Figure 5) based on the ATP‐driven alternating access. 26 In the absence of ATP, ABCB6 exists in an inward‐facing apo state with cavity‐1 ready to bind substrates from the cytoplasm. In this state, cavity‐2 is sealed by both the plug and lid and is in an occluded empty conformation. Porphyrin substrates without iron ions can bind directly to cavity‐1 from the cytoplasm. In contrast, for ion‐coordinated porphyrins such as hemin, and heavy metal ions, GSH conjugation is likely to be required for substrate binding possibly by shielding the protein from direct contact with the metal ion.
FIGURE 5.
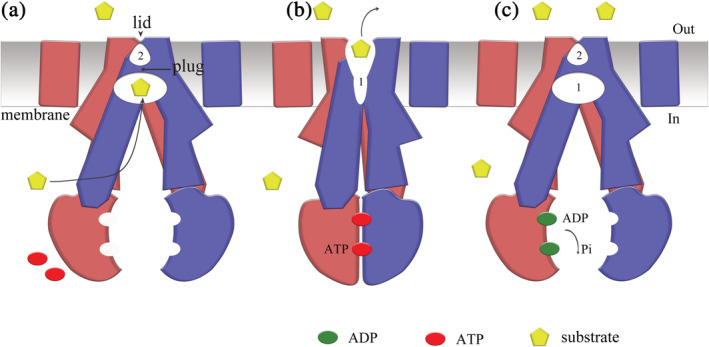
Proposed transport mechanism of ATP‐binding cassette transporter 6 of subfamily B (ABCB6). (a) In the apo state, human ABCB6 protein is inward‐facing, and cavity‐1 is ready to bind substrates from the cytoplasm. In comparison, cavity‐2 is much smaller than cavity‐1 and is sealed by the lid, so substrate binding from the opposite side is prevented. (b) Stimulated by substrate binding, two ATP molecules bind to the nucleotide‐binding domains (NBDs), drive the closing of the NBD dimer, and switch the conformation of ABCB6 to the outward‐facing state. The plug and lid are then opened, which allows the translocation of substrate from the reduced cavity‐1 to the enlarged cavity‐2. (c) Accompanied by ATP hydrolysis, ABCB6 releases substrate from cavity‐2 to cell exterior or into mitochondria. The NBDs release ADPs and inorganic phosphate (P i) into the cytoplasm. The protein is then recovered to the inner‐facing state and is ready for the next cycle of substrate transport
Upon substrate binding, the interaction of the substrate with ABCB6 drives the conformational change of cavity‐1. Such conformational change may propagate along the TM helices to the cytoplasmic NBDs, resulting in a close NBD interaction and promoting ATP binding. ATP binding between closed NBDs leads to a reduced size of cavity‐1 and increased size of cavity‐2. The plug is then opened to reach an outward‐facing conformation, such that the substrate can enter from cavity‐1 to cavity‐2. The presence of the substrate in cavity‐2 appears to further strengthen the interaction of NBDs, because an opened conformation of the NBD dimer would cause a steric clash inside cavity‐2 as what was proposed in the case of ABCG2. 22 With the substrate releasing though the opened lid, the catalytic sites formed by NBD dimerization hydrolyze ATP molecules to ADP and P i, which shifts the transporter back to the inward‐facing state ready for the next transport cycle. 27
The substrate dissociation constants (K Ds) toward the inward‐ and outward‐facing conformations of ABCB6 (K D,in and K D,out) are likely to be drastically different. The experimentally determined K D for the substrate (e.g., ~20 μM for hemin) is dominated by the affinity toward cavity‐1 in the inward‐facing conformation. Upon undergoing a conformational change, the substrate is squeezed from cavity‐1 into cavity‐2, and cavity‐1 itself diminishes. Because cavity‐2 is much smaller than cavity‐1, it only serves as part of the releasing path for the substrate. Therefore, the substrate affinity for ABCB6 in the outward‐facing conformation is reduced dramatically, corresponding to a change from the K D,in to a much higher K D,out. This change of the dissociation constant between the two conformations is associated with an endergonic energy term called differential binding energy, ΔG D (≡RTln(K D,out/K D,in), where R is the universal gas constant and T is the absolute temperature), which must be compensated for by the input energy from ATP binding as part of the ATP hydrolysis energy. 28 Another part of the ATP hydrolysis energy is likely to be converted to conformational energy (ΔG C) stored in the outward‐facing conformation. Upon substrate release and ATP hydrolysis, this conformational energy term is released to facilitate the conformational change of ABCB6 back to the lower‐energy, inward‐facing ground state. Moreover, the k cat of ABCB6 ATPase activity (in the presence of 10 μM hemin) determined in the in vitro assay (Figure 4a) is estimated to be ~10 per min. Although this is comparable with most reported values of ABC transporters, it appears rather low in a physiological context. Therefore, it is quite probable that in vitro experiments have missed some factors important for the in vivo ATPase activities of ABC transporters. One such factor may be the electrostatic potential across the cytoplasmic membrane. 28
TMD0 is a unique feature that is only present in certain transporters of both ABCB and ABCC subfamilies. Fundamental questions about these TMD0 domains include what their functions are and how TMD0 affects the activities of the transporter and the docking protein(s). In the heterodimeric TAP transporter (ABCB2/ABCB3), TMD0 is required for interacting with the tapasin protein to form a large peptide‐loading complex. 29 , 30 , 31 For SUR1 and SUR2, TMD0 interacts with potassium channel Kir6.2 to form a K ATP channel super complex. 17 In SUR1, TMD0 and the lasso motif mediate the cross talk between SUR1 and Kir6.2, and interactions between extracellular loops of TMD0 and the core domain were also observed. 32 However, the TMD0‐interacting protein(s) for ABCB6 currently remain elusive. Additionally, the loss of the lasso motif in ABCB6 results in increased flexibility of its two TMD0s in the absence of a docking protein. The communication between TMD0 and the core domain in ABCB6 is mediated by the TMD0‐core domain linker and possibly by loops from TMD0, which were not resolved in our cryo‐EM electron density map. We showed that the ΔTMD0 variant of ABCB6 lost substrate‐stimulated ATPase activity, despite possessing normal substrate binding. A possible interpretation of this is that interactions between the unresolved extracellular loops of TMD0 and loops from the core domain are important to transiently stabilize the outward‐facing conformation upon ATP hydrolysis. The existence of a lasso motif as a pivot also stabilizes the TMD0 conformation, and upon binding of Kir6.2 the TMD0 of SUR1 maintains a spatial position relative to the core domain similar to the TMD0 of the apo MRP1 structure. 16 , 17 It would be of interest to know whether TMD0 of ABCB6 remains close to the core domain when its docking protein is present, considering that no rigid structural element was observed between TMD0 and the core domain in our study. Previous work showed that some mutations in TMD0 of ABCB6 lead to loss of protein expression on the surface of RBCs, resulting in a “Lan−” blood group. 15 , 33 Thus, a properly folded TMD0 may be required for full‐length ABCB6 expression. 21 Several studies suggested that TMD0 of ABCB6 is important for ABCB6 subcellular targeting. 7 , 10 However, further studies are needed to elucidate the functional roles of TMD0.
In previous studies of certain ABC transporters, it was shown that GSH stimulates ATPase activity and is involved in the substrate transport process. 16 , 34 In such ABC transporters, for example MRP1, most of their substrates are GSH‐conjugated. 16 Direct binding of GSH to the transporter was observed in recently reported structures of NaAtm1 35 and yeast Atm1, 19 further demonstrating the potential roles of GSH in the substrate translocation of ABC transporters. In the current study, we showed that electronegative GSH(−1) can enhance the substrate‐stimulated ATPase activity of both hemin and hematin, most likely through conjugation with the coordinated ferric ion. Because free hemin and hematin are considered toxic in cells, 36 , 37 the GSH conjugation of hemin may naturally exist in vivo to protect cells from heme toxicity. In the ATPase hydrolysis assay, we observed stimulated ATPase activity in the presence of both Cu2+ and GSH. Previous studies have shown that ABCB6 is involved in cell resistance to arsenic and cadmium, 3 , 9 most likely through GSH conjugation. Although our data did not show direct binding or stimulated ATPase activity of GSH for ABCB6, it cannot be ruled out that GSH may bind to ABCB6 with low affinity. Moreover, given the relatively large substrate binding pocket of ABCB6, GSH alone is likely to be unable to interact with the transporter strongly enough to stabilize a close NBD conformation for ATP binding and hydrolysis. In future studies, an ABCB6 structure complexed with a GSH‐conjugated substrate will be helpful to identify the GSH binding site and to provide detailed structural information to more fully understand substrate binding.
4. MATERIALS AND METHODS
4.1. Cell culture
Mammalian cells (HEK293‐F) were recovered in Expi293 Medium (Thermo Fisher Scientific) for 3 days and subcultured in FreeStyle 293 Expression Medium (Thermo Fisher Scientific) at 37°C with 5% CO2. Insect cells (Sf9) were cultured in sf‐900 II SFM medium (Gibco) and maintained at 27°C.
4.2. Cloning, expression, and purification of human ABCB6
Human ABCB6 cDNA was kindly provided by Dr Jiahuai Han (Xiamen University, Xiamen, China) and cloned into the BacMam expression vector 38 with a GFP‐His10‐tag at the C‐terminal of the gene product. Recombinant baculovirus was obtained using the Bac‐to‐Bac Baculovirus Expression System (Invitrogen). Expression of human ABCB6 protein was performed through infection of HEK 293‐F cells at a density of 2 × 106 cells/ml with high‐titer virus stock at a multiplicity of infection of 5. Cells were harvested at 48 hr postinfection by centrifugation and stored at −80°C until use. Cells were suspended in lysis buffer containing 20 mM HEPES (pH 7.5), 150 mM NaCl, and a protease inhibitor cocktail tablet (Roche) 1:1,000; and were then gently broken with glass homogenizer. The cell membrane was collected by centrifugation at 100,000g for 1 hr at 4°C and solubilized in buffer containing 1% (wt/vol) 2,2‐didecylpropane‐1,3‐bis‐b‐d‐maltopyranoside (LMNG, Anatrace), 0.2% (wt/vol) cholesteryl hemisuccinate (CHS, Sigma) for 40 min at 4°C. ABCB6 protein was purified with cobalt resin (Thermo Fisher) followed by size‐exclusion chromatography in buffer containing 20 mM HEPES (pH 7.5), 150 mM NaCl, 2 mM Tris (2‐carboxyethyl) phosphine, 0.025% LMNG, and 0.005% CHS on a 10/300 Superose‐6 column (GE Healthcare).
4.3. Cryo‐EM sample preparation and data collection
The purified ABCB6 protein sample was collected at the peak of the size‐exclusion chromatography flow and diluted to a concentration of 1.5 mg/ml. The protein sample (4 μl) was then applied to a glow‐discharged Quantifoil holey gold grid (R1.2/1.3400 mesh), blotted using the Vitrobot Mark IV System (FEI, Thermo Fisher) for 2 s at blot force 2 after a waiting time of 5 s, then plunged into liquid ethane cooled by liquid nitrogen. A 300 kV Titan Krios (FEI, Thermo Fisher) equipped with a Gatan K2 Summit direct electron detector (Gatan) was used to image the grids. Images were recorded by serial EM 39 in super‐resolution mode with a pixel size of 1.04 Å for data collection. Each image was exposed for 6.4 s in 32 subframes with a dose rate of 10 electrons/pixel/s, resulting in a total dose of 60 electrons/Å2. Typical defocus values ranged from −1.5 to −2.5 μm.
4.4. Image processing
Motion‐corrected, dose‐weighted micrographs were directly imported to RELION‐2.1, 40 which was used for subsequent data processing except where specified. Contrast transfer function was estimated using CTFFIND4 software. 41 Approximately 10,000 bin2 particles were manually picked in Relion and used to generate 2D templates for automatic particle picking. A total of 869,750 particles were picked from 7,250 micrographs and subjected to several rounds of 2D classification to remove false particles and bad particles. A de novo 3D initial model of ABCB6 was generated by selected classes of 2D classification and used as a reference for further 3D classification. After several rounds of 3D classification, a class showing the visible density of two TMD0s was selected and re‐extracted, yielding 97,930 recentered raw particles. These particles were applied for autorefinement, yielding a 3D reconstruction of 7.5 Å. However, the TMD0 density was still poor and only a large micelle surrounded TMD0 was observed. Therefore, a skip alignment 3D classification with an adapted mask on the full‐length ABCB6 was performed, and 29,001 particles were selected. These particles were applied for further local refinement, resulting in a 3D reconstruction of 6.3 Å. After postprocessing, a map of full‐length ABCB6 with an overall resolution of 5.2 Å was obtained. To achieve a higher resolution of the core domain of ABCB6, a similar strategy was used with a core‐region soft mask. After skip alignment 3D classification with an adapted core domain mask, 74,135 particles from good classes were selected. These particles were subjected to autorefinement with a soft mask on the core domain of ABCB6, resulting in a 3D reconstruction of 5.3 Å. After sharpening using a core‐domain mask with a B‐factor of −260, a map of the ABCB6 core domain with an overall resolution of 4.0 Å was obtained. The overall resolution was estimated with the gold‐standard Fourier shell correlation criterion of 0.143. Local resolution variations were estimated using ResMap. 42
4.5. Model building and refinement
The core domain structure of ABCB6 was built first, then the nucleotide‐free TMD structure (PDB ID: 4MYC) of the yeast mitochondrial ABC transporter Atm1 was fitted into the B‐factor‐sharpened core domain map of ABCB6 and used as a template to generate the ABCB6 TMD model. Because the core domain of ABCB6 and Atm1 have high sequence identity (>40%), we used the conserved residues, bulky residues, and proline kinks as references to manually adjust the model in COOT software 43 according to the density. For the NBDs, the high‐resolution crystal structure of ABCB6 NBD (PDB ID: 3NH6) was modified to a ploy‐alanine model and docked into the EM map by Chimera software, 44 then connected with the TMD. The model was further refined by the software utility phenix.real_space_refine 45 with geometry and secondary structure restraints. For the cryo‐EM map of full‐length ABCB6, only cylindrical densities of TMD0s were observed. The TMD0 of ABCB6 was modeled with a homologous poly‐alanine structure of MRP1 TMD0 (PDB ID: 5UJA). The residues of five TM helices (Residues 27–206) were assigned according to the prediction by HMTMM software. 46 TMD0 was assigned to its adjacent subunit based on its position in MRP1. Because the poor TMD0 density did not allow for detailed modeling of its five helices and connectivity, we treated TM0 as a rigid body without further structural interpretation. All structural figures were prepared with either Pymol (Schrodinger, LLC.) or Chimera software.
4.6. BLI binding assay
The BLI binding assay was performed at room temperature on the Octet RED96 System (FortéBio) with running Buffer A (20 mM HEPES [pH 7.5], 150 mM NaCl, 0.025% LMNG, 0.005% CHS) and running Buffer B (20 mM HEPES [pH 7.5], 150 mM NaCl, 0.025% LMNG, 0.005% CHS, 1% dimethyl sulfoxide). Purified full‐length and core‐domain ABCB6 proteins were immobilized onto AR2G biosensors activated by EDC and Sulfo‐NHS mixtures. Then the AR2G biosensors with immobilized proteins were quenched with 1 M ethanolamine (pH 8.5) and equilibrated with running Buffer A for 3 min and then running Buffer B for 2 min. SN‐38, elacridar, hemin, and other porphyrins were diluted to suitable concentrations in Buffer B and incubated with the full‐length and core‐domain ABCB6 proteins for 2 min, then disassociated in running Buffer B for 2 min. AR2G biosensors which were immobilized with proteins were used for reference sensors. Data were analyzed using GraphPad Prism 5.0 software.
4.7. ATPase activity
The ATPase activity of purified ABCB6 was measured as the release of Pi from ATP in a colorimetric malachite green assay. The malachite green solution was prepared by mixing a solution of malachite green (58 mg/10 ml of ultrapure H2O) and ammonium molybdate tetrahydrate (562 mg/7 ml 4‐N HCl), followed by the addition of ultrapure H2O to a final volume to 50 ml. The solution was filtered over a 0.22‐μm‐pore size filter (Millipore) and stored at 4°C before use. Prior to the start of this assay, all mutant proteins were concentrated to the same concentration as WT ABCB6 (1.5 mg/ml). Purified protein (3.0 μg) in Buffer A supplemented with 5 mM MgCl2 and 2 mM Na‐ATP was incubated with potential substrates (see below) at 37°C in a 96‐well plate with a total reaction volume of 30 μl/well. After 10 min incubation, the ATPase reaction in each well was terminated by the addition of 150 μl malachite green solution and incubated for 5 min. Finally, 75 μl of 34% (wt/vol) citric acid was added to each well, and the plate was further incubated for 20 min in the dark to allow color development. A total of 30 μl drug buffer with different kinds of porphyrins or/and different concentrations of GSH (0–5 mM) was then added per well. Each sample well was accompanied by a reference well containing the same mixture but without the protein sample. The OD600 nm of the samples was measured in a Spectramax microplate reader (Molecular Devices) and compared with the OD600 nm of standards containing 0–4.2 nM P i. Data were analyzed by GraphPad Prism 5.0 software.
4.8. Molecular docking of protoporphyrin IX
To obtain a model of the ABCB6‐protoporphyrin IX complex, flexible dockings were performed with the Induced Fit Docking module in Maestro v11.1 (Schrodinger). SMILE string of protoporphyrin IX was imported to LigPrep to produce the corresponding molecular structure with proper ionization state (pH 7.0). The core domain structure of ABCB6 was used as protein template and prepared with the Protein Preparation Module. Hydrogen atoms and protein charges were added, followed by H‐bond assignment optimization and restrained minimization during this process. The grid file was generated with an enclosing box centered on cavity‐1 residue N545. Protoporphyrin IX was then docked into ABCB6 using the standard Glide‐SP mode. The output result was analyzed according to the favorable interaction energy and our visual inspection.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
Chunyu Wang: Performed the sample preparation and activity assay. Chunyu Wang and Can Cao: Performed cryo‐EM data collection. Chunyu Wang, Can Cao, and Nan Wang: Solved the structure of ABCB6. Can Cao and Chunyu Wang: Performed and analyzed molecular docking. Chunyu Wang, Can Cao, Xianping Wang, Xiangxi Wang, and Xuejun C. Zhang: Analyzed the data. Chunyu Wang, Can Cao, Nan Wang, and Xuejun C. Zhang: Wrote the manuscript.
Supporting information
Appendix S1. Figures.
TABLE S1 Cryo‐EM data collection, refinement, and validation statistics
ACKNOWLEDGMENTS
The authors would like to thank Prof J. Han of Xiamen University for providing human ABCB6 cDNA. The authors thank the Center for Biological Imaging (CBI), Institute of Biophysics, Chinese Academy of Science for our Cryo‐EM work, and are particularly grateful to X. Huang, B. Zhu, Z. Guo, and D. Fan for their help in taking cryo‐EM images. The authors also thank thank Y. Chen, Z. Yang, and X. Zhou for BLI technical assistance. The authors thank Dr S. Williams, from Liwen Bianji, Edanz Group China, for linguistic assistance during the preparation of this manuscript. This work was supported by the Chinese Academy of Sciences (XDB37030301 and XDB08020301), and National Natural Science Foundation of China (31971134).
Wang C, Cao C, Wang N, Wang X, Wang X, Zhang XC. Cryo‐electron microscopy structure of human ABCB6 transporter. Protein Science. 2020;29:2363–2374. 10.1002/pro.3960
Chunyu Wang, Can Cao, and Nan Wang contributed equally to this work.
Funding information National Natural Science Foundation of China, Grant/Award Number: 31971134; Chinese Academy of Sciences, Grant/Award Numbers: XDB08020301, XDB37030301
DATA AVAILABILITY STATEMENT
The coordinate of ABCB6 has been deposited into the PDB database with accession code: 7D7R (core‐domain) and 7D7N (full‐length). The cryo‐EM maps are accessible with EMD numbers: EMD‐30610 (core‐domain) and EMD‐30609 (full‐length).
REFERENCES
- 1. Rakvacs Z, Kucsma N, Gera M, et al. The human ABCB6 protein is the functional homologue of HMT‐1 proteins mediating cadmium detoxification. Cell Mol Life Sci. 2019;76:4131–4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Furuya KN, Bradley G, Sun D, Schuetz EG, Schuetz JD. Identification of a new P‐glycoprotein‐like ATP‐binding cassette transporter gene that is overexpressed during hepatocarcinogenesis. Cancer Res. 1997;57:3708–3716. [PubMed] [Google Scholar]
- 3. Chavan H, Oruganti M, Krishnamurthy P. The ATP‐binding cassette transporter ABCB6 is induced by arsenic and protects against arsenic cytotoxicity. Toxicol Sci. 2011;120:519–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Krishnamurthy PC, Du G, Fukuda Y, et al. Identification of a mammalian mitochondrial porphyrin transporter. Nature. 2006;443:586–589. [DOI] [PubMed] [Google Scholar]
- 5. Ulrich DL, Lynch J, Wang Y, et al. ATP‐dependent mitochondrial porphyrin importer ABCB6 protects against phenylhydrazine toxicity. J Biol Chem. 2012;287:12679–12690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kiss K, Brozik A, Kucsma N, et al. Shifting the paradigm: The putative mitochondrial protein ABCB6 resides in the lysosomes of cells and in the plasma membrane of erythrocytes. PLoS One. 2012;7:e37378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kiss K, Kucsma N, Brozik A, et al. Role of the N‐terminal transmembrane domain in the endo‐lysosomal targeting and function of the human ABCB6 protein. Biochem J. 2015;467:127–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bergam P, Reisecker JM, Rakvács Z, Kucsma N, Raposo G, van Szakacs NG. ABCB6 resides in melanosomes and regulates early steps of melanogenesis required for PMEL amyloid matrix formation. J Mol Biol. 2018;430:3802–3818. [DOI] [PubMed] [Google Scholar]
- 9. Kim S, Sharma AK, Vatamaniuk OK. N‐terminal extension and C‐terminal domains are required for ABCB6/HMT‐1 protein interactions, function in cadmium detoxification, and localization to the endosomal‐recycling system in Caenorhabditis elegans . Front Physiol. 2018;9:885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Paterson JK, Shukla S, Black CM, et al. Human ABCB6 localizes to both the outer mitochondrial membrane and the plasma membrane. Biochemistry. 2007;46:9443–9452. [DOI] [PubMed] [Google Scholar]
- 11. Lynch J, Fukuda Y, Krishnamurthy P, Du G, Schuetz JD. Cell survival under stress is enhanced by a mitochondrial ATP‐binding cassette transporter that regulates hemoproteins. Cancer Res. 2009;69:5560–5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Polireddy K, Chavan H, Abdulkarim BA, Krishnamurthy P. Functional significance of the ATP‐binding cassette transporter B6 in hepatocellular carcinoma. Mol Oncology. 2011;5:410–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Borel F, Han R, Visser A, et al. Adenosine triphosphate‐binding cassette transporter genes up‐regulation in untreated hepatocellular carcinoma is mediated by cellular microRNAs. Hepatology. 2012;55:821–832. [DOI] [PubMed] [Google Scholar]
- 14. Minami K, Kamijo Y, Nishizawa Y, et al. Expression of ABCB6 is related to resistance to 5‐FU, SN‐38 and vincristine. Anticancer Res. 2014;34:4767–4773. [PubMed] [Google Scholar]
- 15. Helias V, Saison C, Ballif BA, et al. ABCB6 is dispensable for erythropoiesis and specifies the new blood group system Langereis. Nat Genet. 2012;44:170–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Johnson ZL, Chen J. Structural basis of substrate recognition by the multidrug resistance protein MRP1. Cell. 2017;168:1075–1085. [DOI] [PubMed] [Google Scholar]
- 17. Li N, Wu J‐X, Ding D, Cheng J, Gao N, Chen L. Structure of a pancreatic ATP‐sensitive potassium channel. Cell. 2017;168:101–110. [DOI] [PubMed] [Google Scholar]
- 18. Wen PC, Verhalen B, Wilkens S, McHaourab HS, Tajkhorshid E. On the origin of large flexibility of P‐glycoprotein in the inward‐facing state. J Biol Chem. 2013;288:19211–19220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Srinivasan V, Pierik AJ, Lill R. Crystal structures of nucleotide‐free and glutathione‐bound mitochondrial ABC transporter Atm1. Science. 2014;343:1137–1140. [DOI] [PubMed] [Google Scholar]
- 20. Szollosi D, Rose‐Sperling D, Hellmich UA, Stockner T. Comparison of mechanistic transport cycle models of ABC exporters. Biochim Biophys Acta Biomembr. 2018;1860:818–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Boswell‐Casteel RC, Fukuda Y, Schuetz JD. ABCB6, an ABC transporter impacting drug response and disease. AAPS J. 2017;20:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Taylor NMI, Manolaridis I, Jackson SM, Kowal J, Stahlberg H, Locher KP. Structure of the human multidrug transporter ABCG2. Nature. 2017;546:504–509. [DOI] [PubMed] [Google Scholar]
- 23. Manolaridis I, Jackson SM, Taylor NMI, Kowal J, Stahlberg H, Locher KP. Cryo‐EM structures of a human ABCG2 mutant trapped in ATP‐bound and substrate‐bound states. Nature. 2018;563:426–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Krishnamurthy P, Schuetz JD. The role of ABCG2 and ABCB6 in porphyrin metabolism and cell survival. Curr Pharm Biotechnol. 2011;12:647–655. [DOI] [PubMed] [Google Scholar]
- 25. Yamada T, Takusagawa F. PGH2 degradation pathway catalyzed by GSH‐heme complex bound microsomal prostaglandin E2 synthase type 2: The first example of a dual‐function enzyme. Biochemistry. 2007;46:8414–8424. [DOI] [PubMed] [Google Scholar]
- 26. Locher KP. Mechanistic diversity in ATP‐binding cassette (ABC) transporters. Nat Struct Mol Biol. 2016;23:487–493. [DOI] [PubMed] [Google Scholar]
- 27. Higgins CF, Linton KJ. The ATP switch model for ABC transporters. Nat Struct Mol Biol. 2004;11:918–926. [DOI] [PubMed] [Google Scholar]
- 28. Zhang XC, Han L, Zhao Y. Thermodynamics of ABC transporters. Protein Cell. 2016;7:17–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Koch J, Guntrum R, Heintke S, Kyritsis C, Tampe R. Functional dissection of the transmembrane domains of the transporter associated with antigen processing (TAP). J Biol Chem. 2004;279:10142–10147. [DOI] [PubMed] [Google Scholar]
- 30. Leonhardt RM, Keusekotten K, Bekpen C, Knittler MR. Critical role for the tapasin‐docking site of TAP2 in the functional integrity of the MHC class I‐peptide‐loading complex. J Immunol. 2005;175:5104–5114. [DOI] [PubMed] [Google Scholar]
- 31. Oldham ML, Hite RK, Steffen AM, et al. A mechanism of viral immune evasion revealed by cryo‐EM analysis of the TAP transporter. Nature. 2016;529:537–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Martin GM, Yoshioka C, Rex EA, et al. Cryo‐EM structure of the ATP‐sensitive potassium channel illuminates mechanisms of assembly and gating. Elife. 2017;6:e24149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fukuda Y, Cheong PL, Lynch J, et al. The severity of hereditary porphyria is modulated by the porphyrin exporter and Lan antigen ABCB6. Nat Commun. 2016;7:12353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Brechbuhl HM, Gould N, Kachadourian R, Riekhof WR, Voelker DR, Day BJ. Glutathione transport is a unique function of the ATP‐binding cassette protein ABCG2. J Biol Chem. 2010;285:16582–16587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lee JY, Yang JG, Zhitnitsky D, Lewinson O, Rees DC. Structural basis for heavy metal detoxification by an Atm1‐type ABC exporter. Science. 2014;343:1133–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lisewski AM, Quiros JP, Ng CL, et al. Supergenomic network compression and the discovery of EXP1 as a glutathione transferase inhibited by artesunate. Cell. 2014;158:916–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Robinson SR, Dang TN, Dringen R, Bishop GM. Hemin toxicity: A preventable source of brain damage following hemorrhagic stroke. Redox Rep. 2009;14:228–235. [DOI] [PubMed] [Google Scholar]
- 38. Goehring A, Lee C‐H, Wang KH, et al. Screening and large‐scale expression of membrane proteins in mammalian cells for structural studies. Nat Protoc. 2014;9:2574–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. [DOI] [PubMed] [Google Scholar]
- 40. Scheres SH. RELION: Implementation of a Bayesian approach to cryo‐EM structure determination. J Struct Biol. 2012;180:519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rohou A. Grigorieff N (192) CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo‐EM density maps. Nat Methods. 2014;11:63–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Emsley P, Cowtan K. Coot: Model‐building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 44. Pettersen EF, Goddard TD, Huang CC, et al. UCSF chimera—A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. [DOI] [PubMed] [Google Scholar]
- 45. Adams PD, Afonine PV, Bunkoczi G, et al. PHENIX: A comprehensive Python‐based system for macromolecular structure solution. Acta Cryst. 2010;D66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J Mol Biol. 2001;305:567–580. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1. Figures.
TABLE S1 Cryo‐EM data collection, refinement, and validation statistics
Data Availability Statement
The coordinate of ABCB6 has been deposited into the PDB database with accession code: 7D7R (core‐domain) and 7D7N (full‐length). The cryo‐EM maps are accessible with EMD numbers: EMD‐30610 (core‐domain) and EMD‐30609 (full‐length).


