Abstract
Background
Coronavirus disease 2019 (COVID-19) caused by a novel betacoronavirus named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has attracted top health concerns worldwide within a few months after its appearance. Since viruses are highly dependent on the host small RNAs (microRNAs) for their replication and propagation, in this study, top miRNAs targeting SARS-CoV-2 genome and top miRNAs targeting differentially expressed genes (DEGs) in lungs of patients infected with SARS-CoV-2, were predicted.
Methods
All human mature miRNA sequences were acquired from miRBase database. MiRanda tool was used to predict the potential human miRNA binding sites on the SARS-CoV-2 genome. EdgeR identified differentially expressed genes (DEGs) in response to SARS-CoV-2 infection from GEO147507 data. Gene Set Enrichment Analysis (GSEA) and DEGs annotation analysis were performed using ToppGene and Metascape tools.
Results
160 miRNAs with a perfect matching in the seed region were identified. Among them, there was 15 miRNAs with more than three binding sites and 12 miRNAs with a free energy binding of −29 kCal/Mol. MiR-29 family had the most binding sites (11 sites) on the SARS-CoV-2 genome. MiR-21 occupied four binding sites and was among the top miRNAs that targeted up-regulated DEGs. In addition to miR-21, miR-16, let-7b, let-7e, and miR-146a were the top miRNAs targeting DEGs.
Conclusion
Collectively, more experimental studies especially miRNA-based studies are needed to explore detailed molecular mechanisms of SARS-CoV-2 infection. Moreover, the role of DEGs including STAT1, CCND1, CXCL-10, and MAPKAPK2 in SARS-CoV-2 should be investigated to identify the similarities and differences between SARS-CoV-2 and other respiratory viruses.
Keywords: COVID-19, SARS-CoV-2, microRNA, miR-29, miR-21
Highlights
-
•
miR-29 family had 11 binding site on the SARS-COV-2 genome.
-
•
miR-29a/b, 21, 761, 3130, 3167 and miR-3175 bound to the spike coding sequence.
-
•
miR-16, 146a, 21, 615 and let-7b/e targeted SARS-CoV-2 induced DEGs.
-
•
miR-146a, 203a, 24, 615 and miR-16 targeted DEGs involved in viral prosesess
1. Introduction
The COVID-19 disease, resulting from a novel coronavirus, is currently a global threat leading to considerable disease and mortality worldwide. Since November 1, 2020, a total of 46,493,580 confirmed cases, as well as 1,203,902 deaths from COVID-19 in 215 countries and territories have been reported [1]. The Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) is a close relative of SARS-CoV with 45–90% sequence similarity, which has resulted in the severe acute respiratory syndrome in over 8000 confirmed cases and about 800 deaths in 2003 [2,3].
Coronaviruses are a diverse family of viruses associated with multiple respiratory diseases with different severity, like common cold, pneumonia, and now COVID-19 [4]. With a single-stranded positive-sense RNA genome with genome sizes of 26–32 kilobases (kb) in length, they have the largest known genomes among all known RNA viruses [5]. The virus genome possesses a 5′ cap structure together with a 3′ poly(A) tail, like an mRNA to translate its proteins. About two-thirds of the genome at 5′ end is occupied by the replicase gene, which encoded two polyproteins, ORF1a and ORF1b. These polyproteins are further processed to generate the non-structural proteins (nsp). ORF1a is contributed to produce the nsp1-nsp11, while the rest of nsps (nsp12-nsp16) are originated from ORF1b [6]. Additionally, the viral structural proteins comprise surface (S), envelope (E), membrane (M), and nucleocapsid (N) proteins encoded by the one-third of genome at 3′ end [6,7].
A group of small non-coding RNAs, almost 19–24 base pairs in length, named microRNAs (miRNAs) plays a key role in the modulation of a wide range of biological processes, including development, immune system response, and cell death through gene expression regulation [8]. In addition, various aspects of the viral replication and proliferation, including host antiviral responses and viral pathogenesis can be influenced by miRNAs. MiRNAs mediate their regulatory function through direct binding to the target transcript. Perfect pairing in the seed region (position 2 to 8 from 5′ end) has an important impact on the regulatory function of a miRNA. MiRNAs play a negative or positive role in virus-related processes in three ways: direct binding to the viral genome, binding to the viral transcripts, or binding to the host transcripts [9]. Host miRNAs may promote viral RNA stability, replication, and infection or conversely, reinforce host antiviral responses against viruses.
However, the position, number, and distance between binding sites and point mutations in the seed region of a miRNA, can alter its target specificity and its subsequent impact [[9], [10], [11]]. It has been reported that in the samples infected with H5N1 influenza, miR-485 directly targets the viral PB1 gene coding an RNA-dependent RNA polymerase that is essential for virus replication [12]. Moreover, the induction of host immunity pathway like the interferon pathway upon viral infection can result in the enhanced expression of certain miRNA, including miR-155 to regulate the corresponding pathway [13]. For MERS-CoV genome, a total of 13 host miRNAs affecting the virus genome has been recognized, hence, their application as the appropriate therapeutics against viral infection appear promising as microRNAs are very specific in selecting the target regions [14]. Considering the current COVID-19 pandemic, it would be of great importance to investigate miRNAs involved in the host-SARS-CoV-2 interface. In this study, top miRNAs targeting SARS-CoV-2 genome and top miRNAs targeting differentially expressed genes (DEGs) in lungs of patients infected with SARS-CoV-2, were predicted.
2. Materials and methods
2.1. Prediction of the miRNA binding sites on the SARS-CoV-2 genome
The complete genome sequence of virus strain isolated in Wuhan, China (NC_045512.2) was downloaded from the National Center for Biotechnology Information (NCBI) database and considered as the reference viral sequence. In addition, the complete genome sequence of SARS-CoV-2 viruses isolated in the various geographical zones, including the United States (MT322413.1), Spain (MT359865.1), France (MT470137.1), Japan (LC529905.1), South Africa (MT324062.1), India (MT415321.1), Brazil (MT350282.1), Australia (MT007544.1), South Korea (MT304475.1), and Kazakhstan (MT428554.1) were obtained from the NCBI database. All human mature miRNA sequences were also acquired from miRBase database version 22.1. miRanda tool (version 3.3a) was used to predict the potential human miRNA binding sites on the SARS-CoV-2 genome sequence [15]. For this purpose, the thermodynamic folding energy and alignment score threshold values of −20 and 150 kcal/mol were set for miRanda tool; the strict alignment in the seed region was also considered with including the strict parameter.
2.2. Gene expression analysis
The gene count data derived from RNA sequencing in the lung tissue of COVID-19 patients (GSM4462415 and GSM4462416) and healthy individuals (GSM4462413 and GSM4462414) were obtained from the Gene Expression Omnibus (GEO) database with the accession number of GSE147507. Gene expression analysis was performed by edgeR package version 3.30.3 and genes with log2 fold change ≥ |1| and adjusted p-value (Benjamini-Hochberg procedure) threshold of 0.05 was considered as a significantly differentially expressed value.
2.3. Gene set enrichment analysis
Gene Set Enrichment Analysis (GSEA) is a useful approach for interpreting gene expression data based on the functional annotation of the differentially expressed genes (DEGs). In this study, ToppGene tool (https://toppgene.cchmc.org/) [16] was used to identify the enriched biological pathways, gene ontology (GO), and micRNAs that targeted DEGs. The FDR (Benjamini-Hochberg procedure) cutoff of 0.05 was considered significant. For annotation for DEGs, Metascape (https://metascape.org/gp/index.html#/main/step1), an online web tool to equip a comprehensive gene list annotation and analysis resource for experimental biologists, was used. Metascape simplifies and summarizes some results via bar graph colored according to p-values [17]. Finally, Cytoscape software version 3.7.2 was used to visualize miRNA-mRNA network and important GO term.
3. Results
3.1. Host human MiRNAs-SARS-CoV-2 genome interaction
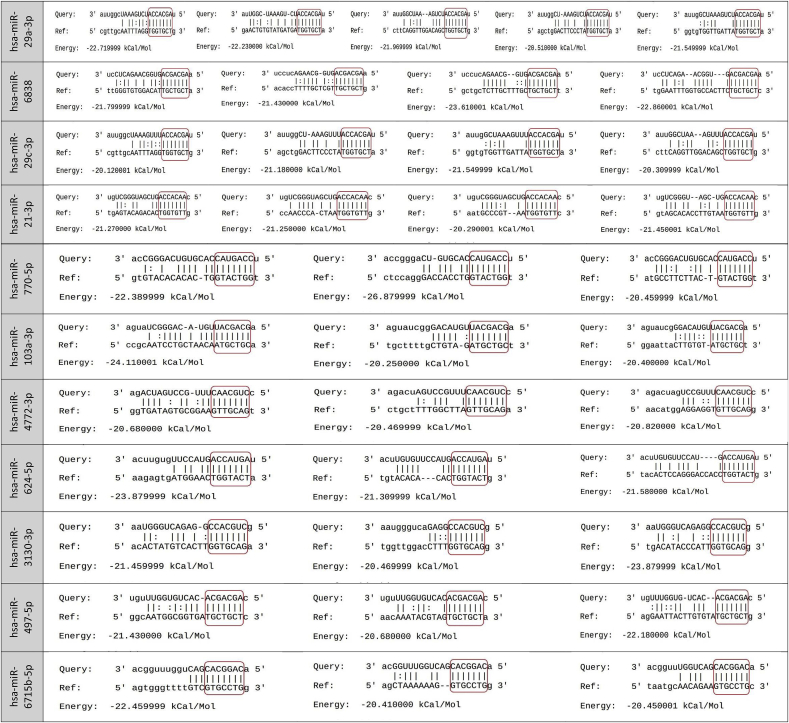
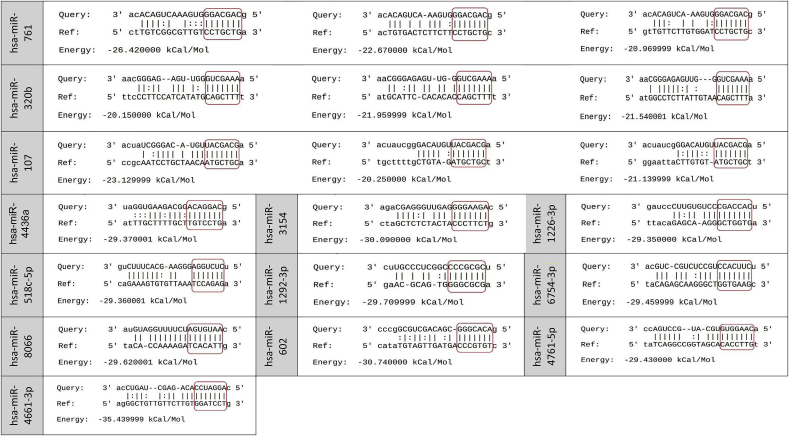
Among the 2654 human mature miRNAs, 444 miRNAs were identified with direct binding site on different positions along with the coronavirus 2 reference genome (Table S1). It was focused on the interactions with perfect matching in the seed region and 160 miRNAs were sorted out. Among them, there was 15 miRNAs with more than three binging sites (Table 1, Table 2) and 12 miRNAs bound to the coronavirus 2 reference genome with a free energy (ΔG) less than −29 kCal/Mol (Table 2, Table 3).
Table 1.
MiRNAs with more than three binding sites on the SARS-COV-2 reference genome.
| Three binding site |
|---|
| hsa-miR-770-5p, hsa-miR-103a-3p, hsa-miR-4772-3p, hsa-miR-624-5p, hsa-miR-3130-3p, hsa-miR-497-5p, hsa-miR-6715b-5p, hsa-miR-761, hsa-miR-320b & hsa-miR-107 |
| Four binding site |
| hsa-miR-6838, hsa-miR-29c-3p & hsa-miR-21-3p |
| Five binding site |
| hsa-miR-29a-3p |
Table 2.
MiRNAs that bind to the SARS-COV-2 reference genome with a free energy (ΔG) less than −29 kCal/Mol.
| One binding site |
|---|
| hsa-miR-4436a, hsa-miR-3154, hsa-miR-1226-3p, hsa-miR-518c-5p, hsa-miR-1292-3p, hsa-miR-6754-3p, hsa-miR-8066, hsa-miR-602, hsa-miR-4761-5p & hsa-miR-4661-3p. |
Table 3.
Pairing schemes of miRNA-target mRNA interactions are shown. The miRNA seed region is screened in the red box.
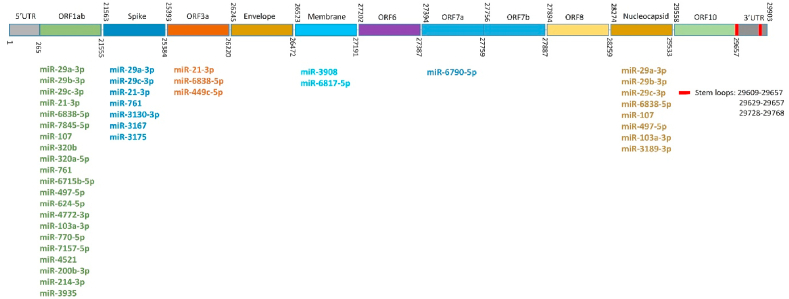
According to the results, miR-29 family (miR-29a, miR-29b, and miR-29c) had the most binding sites (11 sites) and miR-3175 had the least ΔG (−35 kCal/Mol). The position of binding sites on the SARS-CoV-2 for miRNAs with more than three binding sites or ΔG less than −29 kCal/Mol was also explored and ORF1ab, nucleocapsid, spike, ORF3a, membrane, and ORF7a coding regions with high capability for binding to host human miRNAs were found (Fig. 1). ORF1ab, nucleocapsid, and spike sequences had the most binding sites. Among the miRNAs, miR-29 exhibited various binding sites on ORF1ab, nucleocapsid, and spike sequences. MiR-21 had binding sites on ORF1ab, spike, and ORF3a. The spike region, which encodes the spike protein, is necessary for viral entry and is a promising target for antiviral therapy. Eight binding sites for miR-29a-3p, miR-29c-3p, miR-21-3p, miR-761, miR-3130-3p (2 sites), miR-3167, and miR-3175 were recognized on the spike coding region (Fig. 1). In particular, the binding pattern of miRNAs among genome sequences released from 10 different geographical locations was explored and no mutation and 100% similarity were found.
Fig. 1.
Schematic representation of human miRNA-binding sites on the SARS-CoV-2 genome (29903 bp). Different parts of the genome including untranslated regions (UTRs) (gray), coding sequences (different colors), stem loops (red), and miRNAs that bind to them are shown. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.2. Enrichment analysis of SARS-CoV-2 induced differentially expressed genes (DEGs)
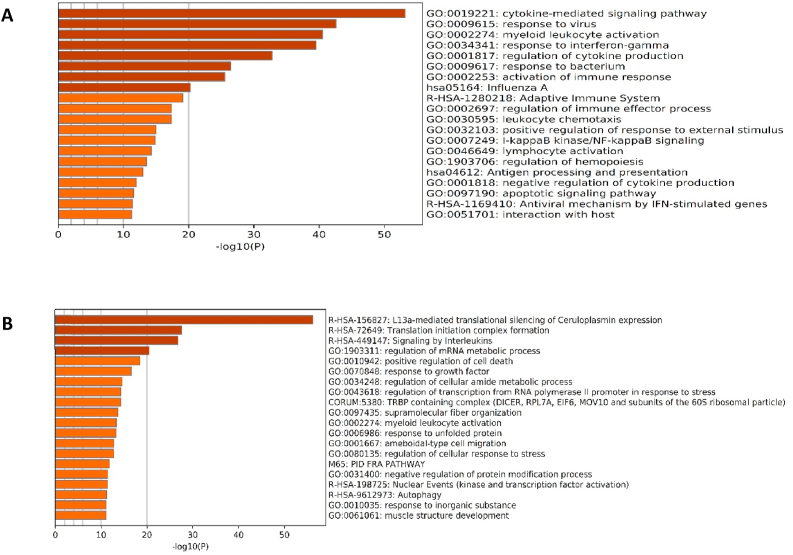
RNA-sequencing (RNA-seq) data (GSE 147507) of the lung tissues from two postmortem men infected with SARS-CoV-2 compared with those of two healthy controls were analyzed and 781 DEGs were found. 329 genes were up-regulated and 452 genes were down-regulated. The results of enrichment analysis of DEGs (Fig. 2) revealed that the genes involved in cytokine-mediated signaling pathway, response to virus, influenza A, NF-kB signaling, and antiviral mechanisms by INF- stimulated genes were significantly enriched and DEGs involved in L13a-mediated silencing of ceruloplasmin expression, translation initiation complex formation, and signaling by interleukins were significantly depleted in response to SARS-CoV-2 infection.
Fig. 2.
Gene set enrichment analyses. GO and KEGG pathway analysis of all up- (A) and down-regulated (B) genes in response to SARS-COV-2 infection.
3.3. miRNAs that target DEGs following SARS-CoV-2 infection
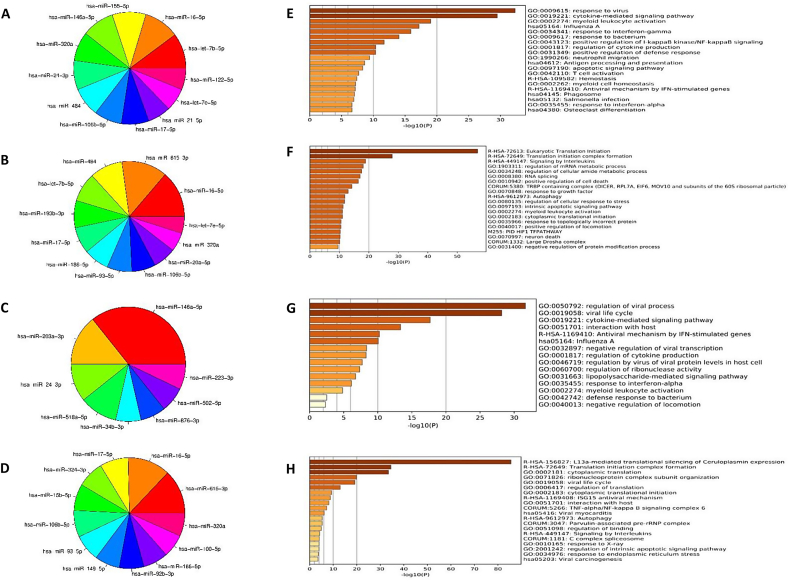
According to the results of enrichment analysis by ToppGene, top miRNAs targeted a large number of DEGs were identified (Fig. 3A and 3B). Among top miRNAs, miR-16 targeted 23 (10% of DEGs) up-regulated genes (Fig. 3A) as well as 100 (14% of the DEGs) down-regulated genes (Fig. 3B). MiR-615-5p (97 up-regulated DEGs, 13%), let-7b-5p (23 down-regulated genes, 10%), and miR-155 (22 down-regulated DEGs, 10%) occupied the next ranks for targeting SARS-CoV-2-induced DEGs (Fig. 3A and 3B). Among top miRNAs, miR-615-3p, miR-193b-3p, miR-186-5p, miR-93-5p, and miR-20a-5p were predicted to target only down-regulated DEGs (Fig. 3B), and miR-155-5p, miR-146a-5p, miR-24-3p, and miR-21-5p were predicted to target only up-regulated DEGs (Fig. 3B). MiR-16-5p, miR-484, let-7b-5p, miR-17-5p, miR-106b-5p, let-7e-5p, and miR-320a targets were from both up- and down-regulated DEGs. Analysis of DEGs targeted by miRNAs demonstrated that GO terms and biological pathways related to response to virus, influenza A, antiviral INF-stimulated genes, and positive regulator of NF-κB signaling were significantly enriched (Fig. 3E). However, the pathways and GO terms related to eukaryotic translation initiation and signaling by interleukins were significantly depleted (Fig. 3F). miRNA-mRNA network for up- and down-regulated DEGs in response to SARS-CoV-2 infection is illustrated in Fig. 4A and 4B, respectively. SARS-CoV-2-induced DEGs related to viral processes were also sorted out and top miRNAs, which target them were explored (Fig. 3C and D). According to the results, it was identified that 38 up-regulated (11%) and 77 (17%) down-regulated DEGs were enriched in viral processes. MiR-146a-5p, miR-203a-3p, and miR-24-3p, which were predicted to target 26%, 10%, and 10% of the up-regulated DEGs, were respectively involved in viral processes (Fig. 3C). Otherwise, 32% and 31% of the down-regulated DEGs involved in viral processes, were targeted by miR-615-3p and miR-16, respectively (Fig. 3D). MiRNA-mRNA network for miRNAs that target SARS-CoV-2-induced DEGs involved in viral processes are depicted in Fig. 5.
Fig. 3.
Pie chart of top miRNAs that target DEGs in response to SARS-CoV-2 infection. (A) and (B) refer to all the up- and down- regulated DEGs, (C) and (D) refer to up- down- regulated DEGs involved in viral biological processes, respectively. Enrichment analyses of GO and KEGG pathways for DEGs that are targeted by top miRNAs in response to SARS-CoV-2 infection. (E) and (F) refer to all the up- and down- regulated DEGs, (G) and (H) refer to the up- and down- regulated DEGs involved in viral biological processes, respectively.
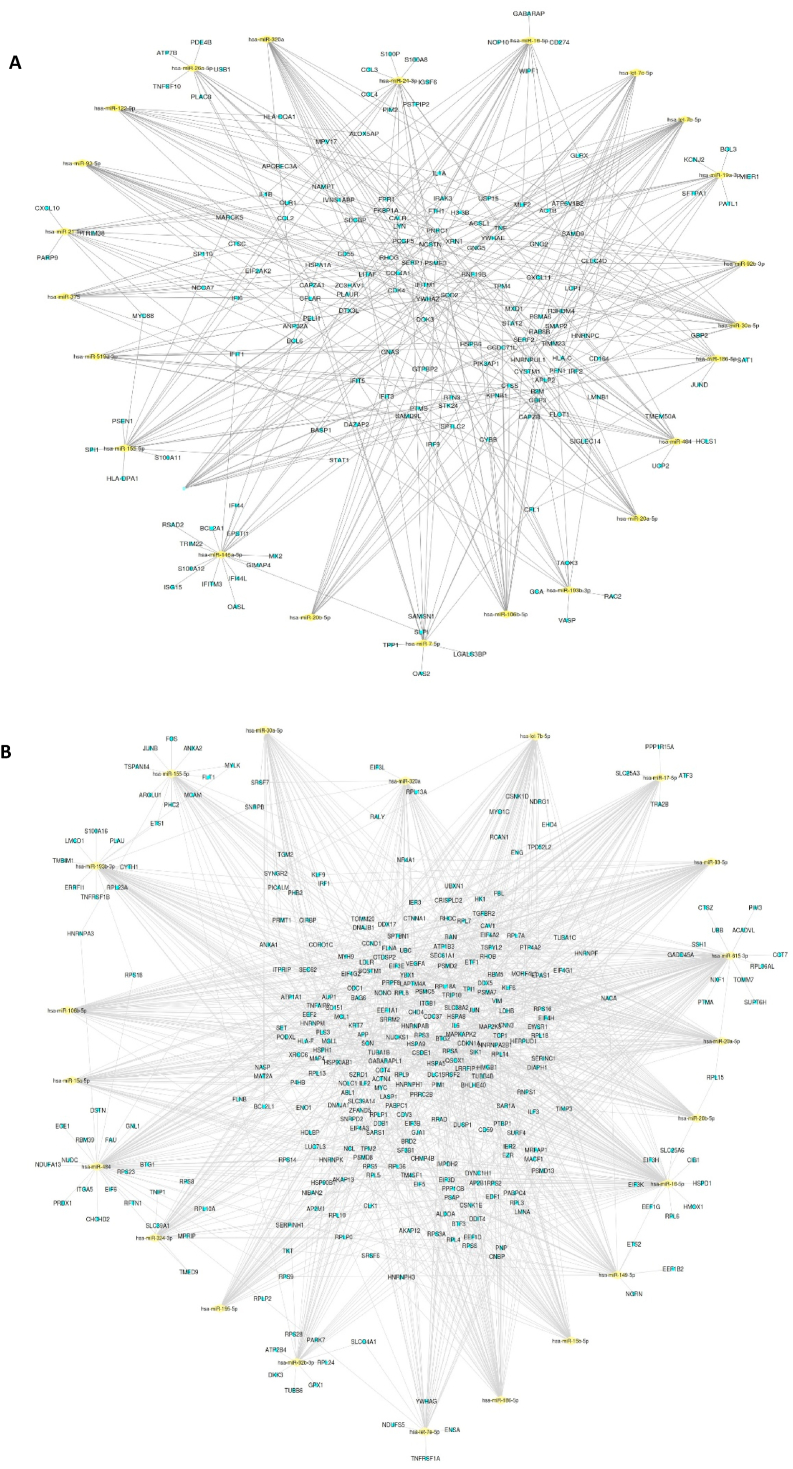
Fig. 4.
MiRNA-mRNA network analysis in response to SARS-CoV-2 infection. Networks constructed between up- (A) and down- (B) regulated mRNA and top miRNAs, which target them. Yellow diamonds represent miRNAs and small light blue dots represent mRNAs. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
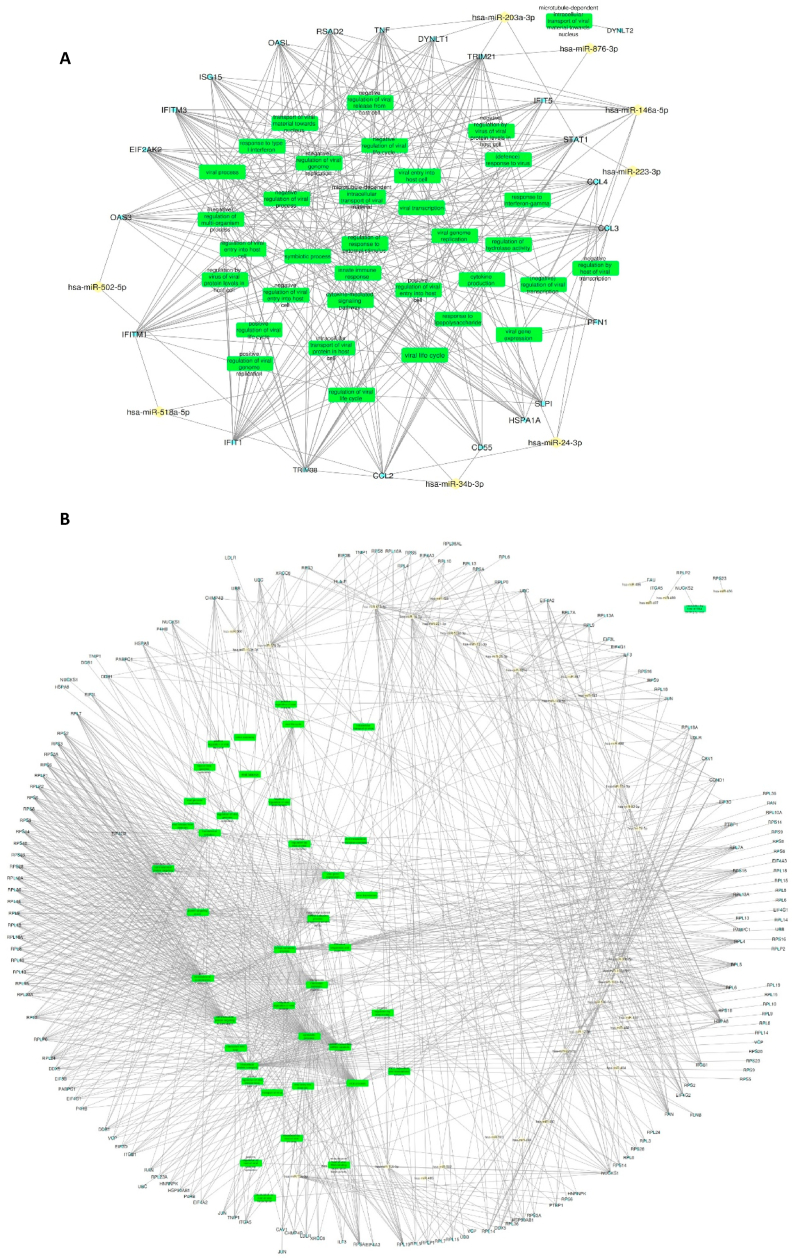
Fig. 5.
MiRNA-mRNA network analysis of DEGs involved in viral biological processes in response to SARS-CoV-2 infection. Networks constructed between up-(A) and down-(B) regulated mRNAs and top miRNAs, which target them. Yellow diamonds represent miRNAs, small light blue dots represent mRNAs, and green rectangles represent viral biological processes. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
Viral proteins have been broadly considered as targets for antiviral therapies, but the problem arises when the selective pressure results in the emergence of a new antiviral drug resistance lineage. Therefore, host-coded factors and particularly, microRNAs seem to be a better strategy [18]. miRNAs as antiviral targets is so important that the first clinical trial of miRNAs relates to their involvement in viral processes [19]. MiR-122, which is highly expressed in liver, binds directly to the 5′ untranslated region of the hepatitis C virus (HCV) and protects viral genome from pyrophosphatase and exonuclease activities and facilitates viral genome translation through structural modification [20,21].
Several studies have predicted possible binding sites for miRNAs on the SARS-CoV-2 genome from the beginning of the COVID-19 pandemic [[22], [23], [24], [25], [26]]. Arisan et al. (2020) identified seven miRNAs including miR-8066, miR-5197, miR-3611, miR-1307-3p, miR-3691-3p, miR-3934-3p, and miR-1468-5p, which bind to SARS-CoV-2 genome [25]. In another study by D. McLellan et al. (2020), ten microRNAs expressed in the SARS-CoV-2 target cells were filtered according to databases and published data and mutations within the binging sites of these miRNAs on the SARS-CoV-2 genome were investigated [26]. They identified eight mutations and hypothesized that these mutations might affect the host miRNA antiviral defenses. Eight of ten miRNAs (miR-197-5p, miR-3935-5p, miR-18b-5p, miR-3154, miR-4761-5p, miR-338-3p, miR-4661-3p, and miR-4436a) reported by them were found on the list (Table S1) reported in the present study with one binding site on the SARS-CoV-2 genome. However, in this study, it was focused on miRNAs with more than three binding sites on the SARS-COV-2 reference genome. Top miRNAs, which target host-related DEGs involved in viral processes in response to SARS-CoV-2 infection, were also predicted. In the present study, the number of binding sites, ΔG > −20 kCal/Mol, and perfect complementarity in the seed region were considered.
Among covid-19-binded miRNAs, miR-29 family had the greatest number of interactions (11 sites). miR-29 family consists of three members, namely, miR-29a, miR-29b, and miR-29c. In previous studies, the impact of host miR-29s on the regulation of viral processes depended on whether they directly bind to the viral genome or to the host transcripts [[27], [28], [29]].
Direct binding of miR-29a to the 3′ UTR region of the HIV genome, increased the transport of virus to p-bodies and reduction of HIV replication. Ahluwalia et al. (2008) also reported that the inhibitory impact of miR-29a on HIV infection is mediated through binding to the accessory viral protein negative factor (Nef), which is critical for viral persistence and release [30]. Therefore, miR-29a has been considered as a potential therapeutic target for HIV eradication [29]. According to the results of the present study, five miR-29s binding sites were predicted in the spike and nucleocapsid coding regions of SARS-CoV-2. Spike proteins protruded from the viral envelope are responsible and critical for host-receptor binding and viral entry. Nucleocapsid proteins specifically bind to the viral genome and facilitate viral entry, replication, and release. Both spike and nucleocapsid proteins were considered as targets for SARS-CoV-2 antiviral drug development. MiR-29s also targeted sequences in the ORF1ab region, which is the largest part of the genome and encoded for 16 nsps [31]. Despite having a large number of direct binding sites on the SARS-CoV-2, no miR-29s was found among the top miRNAs targeting host DEGs. However, in previous studies, direct binding of miR-29s to the host A20/TNFAIP3 transcript in response to influenza A and JEV infections, and subsequently, modulation of antiviral and pro-inflammatory responses have been reported [27]. Considering the high levels of miR-29s in the lungs of healthy adults and better response of these people to SARS CoV-2 compared to those with respiratory diseases with low levels of miR-29s, the probable role of miR-29s in modulating SARS-CoV-2 infection was suggested.
MiR-21, another SARs-CoV-2 binding microRNA, had four binding sites on the SARS-CoV-2 genome. miR-21 is one of the best known miRNAs whose expression increases in many pathological conditions including asthma, pulmonary fibrosis, and viral infection [32,33]. There is no report about direct binding of miR-21 on human viral genomes, and current reports about the involvement of miR-21 in viral infections are limited to modulating host transcripts. For example, the positive role of miR-21 in influenza A replication has been attributed to miR-21-host HDAC8 interaction [34]. In addition, it has been shown that miR-21 reduced the antiviral NF-KB pathway through binding to IRAK1 and TRAF6 transcripts in HIV and HCV infections [35,36]. According to the results of the present study, miR-21 had two binding sites on the spike coding regions. In addition, miR-21 was one of the top miRNAs which targeted up-regulated host-DEGs in response to SARS-CoV-2. One of the miR-21 targets is CXCL-10 that is a biomarker for viral, bacterial, fungal, and parasitic contamination [37]. In the present study, high levels of CXCL-10 in the lungs of COVID-19-infected patients compared to healthy ones were observed. Due to the fact that miR-21 targets are among up-regulated DEGs, the expression of miR-21 can be reduced in SARS-CoV-2 infection. However, experimental validation of miR-21 involvement in both binding to the SARS-CoV-2 genome and modulating host transcriptome are suggested.
Besides prediction of binding-miRNAs, top miRNAs including miR-16, let-7 family, and miR-146a, which were predicted to target host DEGs in response to the SARS-CoV-2, were also identified. MiR-16 was predicted to target the largest number of host DEGs in response to the SARS-CoV-2. Previously, RNA sequencing data from miR-16/15 deficient T cells revealed the inhibitory role of miR-16/15 family on T cell survival, differentiation, and proliferation, which is critical for host or mediated antiviral responses [38]. In addition, high levels of miR-16 expression have been reported in influenza A and RSV infections. Given that the majority of miR-16 targets were down-regulated in response to SARS-CoV-2, the expression of miR-16 might be increased in the lungs of COVID-19 patients. Zheng et al. (2017) showed that miR-16 suppressed cell cycle and prevented EV71 replication through decreasing CCND1 level, which is an important protein in G1 to S phase transition during cell cycle processes [39]. In the present study, decreased level of CCND1 in the lungs of patients infected with SARS-CoV-2 was observed, which seems to be not as effective as its reduction in EV1 infection, because the studied patients died from COVID-19.
Let-7e and let-7b, two let-7 family members, were also among the top miRNAs targeting host DEGs in SARS-CoV-19. The association between Let-7 family and several viral infections including RSV, influenza A, and hMPV has been demonstrated [40,41]. Similar to miR-16, Let-7 family also targets CCND1 [42]. In contrast to EV71 infection, the decreased level of CCND1 is an effective factor for influenza A replication. It seems that CCND1 reduction in the lungs of patients infected with SARS-CoV-2 has a positive effect on the virus replication, similar to what happens for influenza A.
MiR-146a is predicted to target 21 up-regulated genes in SARS-CoV-2 infection. The increased level of miR-146a in a wide range of viral infections including HCV, HBV, influenza A, HIV, H1N1, H3N2, and EBV has been previously reported [43]. MiR-146a protects the virus against the host through targeting and inhibiting STAT1 protein. In other words, miR-146a is a negative regulator of NF-kB signaling pathway. Considering the increased level of STAT1 and other targets of miR-146a in the lungs of patients infected with SARS-CoV-2, miR-146a can be decreased in SARS-CoV-2 infection. The preventive role of STAT1 against SARS-CoV replication has been previously reported [43]. In order to retaliate, SARS-CoV encodes a STAT1 antagonist (ORF6) to escape from eradication [44]. Although in this study, increased levels of STAT1 were observed in the lungs of patients infected with SARS-CoV-2 but it did not lead to an effective antiviral response. It may be, at least in part, due to an antagonizing strategy by SARS-CoV-2 similar to SARS-CoV.
In addition to STAT1, mitogen-activated protein kinase-activated protein kinase 2 (MAPKAPK2 or MK2) is the other altered gene which reveals the difference between SARS-CoV-2 and other viral infections. McCaskill et al. (2017) suggested that the reduction of MK2 by miR-24, miR-124, and miR-744 lead to a broad-spectrum inhibition of influenza and RSV infection [18]. According to the results of this study, MK2 which was predicted to be targeted by miR-16-5p, miR-615-3p, and miR-193b-3p, reduced in the lungs of SARS-CoV-2 patients. It seems that the reduction of MK2 in SARS-CoV-2 is not as effective as that in influenza and RSV infections.
Collectively, COVID-19 infection seems complicated and more experimental studies especially miRNA-based ones are needed to explore detailed molecular mechanisms of SARS-CoV-2 infection to clarify the similarities and differences between SARS-CoV-2 and other respiratory viruses.
CRediT authorship contribution statement
Saeideh Jafarinejad-Farsangi: Conceptualization, Supervision, Funding acquisition. Maryam Moazzam Jazi: Investigation, Methodology, Software. Farzaneh Rostamzadeh: Writing - original draft, Formal analysis. Morteza Hadizadeh: Investigation, Software, Methodology.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors would like to gratitude Kerman University of Medical Sciences, Kerman, Iran, for its financial support (Grant No: 99000343). This Study was approved by the Ethics Committee of Kerman University of Medical Sciences (IR.KMU.REC.1399.435). We are also grateful to Dorsay Hasani for her kind advice in reviewing the English text.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ncrna.2020.11.005.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.https://www.worldometers.info/coronavirus/11/9/2020
- 2.Lee N., Hui D., Wu A., Chan P., Cameron P., Joynt G.M., Ahuja A., Yung M.Y., Leung C. A major outbreak of severe acute respiratory syndrome in Hong Kong. KJNEJoM. 2003;348(20):1986–1994. doi: 10.1056/NEJMoa030685. [DOI] [PubMed] [Google Scholar]
- 3.Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26(4):450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lim Y.X., Ng Y.L., Tam J.P., Liu D.X.J.D. Human coronaviruses: a review of virus–host interactions. Diseases. 2016;4(3):26. doi: 10.3390/diseases4030026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mousavizadeh L., Ghasemi S. Genotype and Phenotype of COVID-19: Their Roles in Pathogenesis. Immunology & Infection. 2020 doi: 10.1016/j.jmii.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khailany R.A., Safdar M., Ozaslan M. Genomic characterization of a novel SARS-CoV-2. Gene Rep. 2020:100682. doi: 10.1016/j.genrep.2020.100682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Narayanan K., Ramirez S.I., Lokugamage K.G., Makino S. Coronavirus nonstructural protein 1: common and distinct functions in the regulation of host and viral gene expression. Virus Res. 2015;202:89–100. doi: 10.1016/j.virusres.2014.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Brien J., Hayder H., Zayed Y., Peng C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. 2018;9:402. doi: 10.3389/fendo.2018.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trobaugh D.W., Klimstra W.B. MicroRNA regulation of RNA virus replication and pathogenesis. Trends Mol. Med. 2017;23(1):80–93. doi: 10.1016/j.molmed.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Skalsky R.L., Cullen B. Viruses, microRNAs, and host interactions. Annu. Rev. Microbiol. 2010;64:123–141. doi: 10.1146/annurev.micro.112408.134243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sætrom P., Heale B.S., Snøve O., Jr., Aagaard L., Alluin J., Rossi J. Distance constraints between microRNA target sites dictate efficacy and cooperativity. Nucleic Acids Res. 2007;35(7):2333–2342. doi: 10.1093/nar/gkm133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ingle H., Kumar S., Raut A.A., Mishra A., Kulkarni D.D., Kameyama T., Takaoka A., Akira S., Kumar H. The microRNA miR-485 targets host and influenza virus transcripts to regulate antiviral immunity and restrict viral replication. Sci. Signal. 2015;8(406):ra126. doi: 10.1126/scisignal.aab3183. -ra126. [DOI] [PubMed] [Google Scholar]
- 13.Leon-Icaza S.A., Zeng M., Rosas-Taraco A. microRNAs in viral acute respiratory infections: immune regulation, biomarkers, therapy, and vaccines. ExRNA. 2019;1(1):1–7. doi: 10.1186/s41544-018-0004-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Canatan D., De Sanctis The impact of MicroRNAs (miRNAs) on the genotype of coronaviruses. Acta Biomed. 2020;91(2):195–198. doi: 10.23750/abm.v91i2.9534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.John B., Enright A.J., Aravin A., Tuschl T., Sander C., Marks DSJPb. Human microRNA targets. PLoS Biol. 2004;2(11):e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen J., Bardes E.E., Aronow B.J., Jegga A.G. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37(2):W305–W311. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O., Benner C., Chanda S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019;10(1):1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCaskill J.L., Ressel S., Alber A., Redford J., Power U.F., Schwarze J., Dutia B.M., Buck A.H. Broad-spectrum inhibition of respiratory virus infection by MicroRNA mimics targeting p38 MAPK signaling. Mol. Ther. Nucleic Acids. 2017;7:256–266. doi: 10.1016/j.omtn.2017.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van der Ree M.H., de Vree J.M., Stelma F., Willemse S., van der Valk M., Rietdijk S., Molenkamp R., Schinkel J., van Nuenen A.C., Beuers U. Safety, tolerability, and antiviral effect of RG-101 in patients with chronic hepatitis C: a phase 1B, double-blind, randomised controlled trial. Lancet (London, England) 2017;389(10070):709–717. doi: 10.1016/S0140-6736(16)31715-9. [DOI] [PubMed] [Google Scholar]
- 20.Jopling C.L., Yi M., Lancaster A.M., Lemon S.M., Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science (New York, NY) 2005;309(5740):1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 21.Bernier A., Sagan S.M. The diverse roles of microRNAs at the Host–Virus Interface. Viruses. 2018;10(8):440. doi: 10.3390/v10080440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Demirci M.D.S., Adan A.J.P. Computational analysis of microRNA-mediated interactions in SARS-CoV-2 infection. PeerJ. 2020;8:e9369. doi: 10.7717/peerj.9369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guterres A., Lima CHdA., Miranda R.L., Gadelha M.R. What is the potential function of microRNAs as biomarkers and therapeutic targets in COVID-19? Infect. Genet. Evol. 2020;85:104417. doi: 10.1016/j.meegid.2020.104417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ivashchenko A., Rakhmetullina A., Aisina D. SARS-CoV, and MERS-CoV; 2020. How miRNAs Can Protect Humans from Coronaviruses COVID-19. [Google Scholar]
- 25.Arisan E.D., Dart A., Grant G.H., Arisan S., Cuhadaroglu S., Lange S., Uysal-Onganer P.J.V. The prediction of miRNAs in SARS-CoV-2 genomes: hsa-miR databases identify 7 key miRs linked to host responses and virus pathogenicity-related KEGG pathways significant for comorbidities. Viruses. 2020;12(6):614. doi: 10.3390/v12060614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hosseini Rad Sm A., Ad M.L. Implications of SARS-CoV-2 mutations for genomic RNA structure and host microRNA targeting. Int. J. Mol. Sci. 2020;7(13):4807. doi: 10.3390/ijms21134807. 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang X., Dong C., Sun X., Li Z., Zhang M., Guan Z., Duan M. Induction of the cellular miR-29c by influenza virus inhibits the innate immune response through protection of A20 mRNA. Biochem. Biophys. Res. Commun. 2014;450(1):755–761. doi: 10.1016/j.bbrc.2014.06.059. [DOI] [PubMed] [Google Scholar]
- 28.Thounaojam M.C., Kaushik D.K., Kundu K., Basu A. Micro RNA‐29b modulates Japanese encephalitis virus‐induced microglia activation by targeting tumor necrosis factor alpha‐induced protein 3. J. Neurochem. 2014;129(1):143–154. doi: 10.1111/jnc.12609. [DOI] [PubMed] [Google Scholar]
- 29.Nathans R., Chu C.Y., Serquina A.K., Lu C.C., Cao H., Rana T.M. Cellular microRNA and P bodies modulate host-HIV-1 interactions. Mol. Cell. 2009;34(6):696–709. doi: 10.1016/j.molcel.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ahluwalia J.K., Khan S.Z., Soni K., Rawat P., Gupta A., Hariharan M., Scaria V., Lalwani M., Pillai B., Mitra D. Human cellular microRNA hsa-miR-29a interferes with viral nef protein expression and HIV-1 replication. Retrovirology. 2008;5(1):117. doi: 10.1186/1742-4690-5-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoshimoto F.K. The proteins of severe acute respiratory syndrome coronavirus-2 (SARS CoV-2 or n-COV19), the cause of COVID-19. Protein J. 2020;39(3):198–216. doi: 10.1007/s10930-020-09901-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu T.X., Munitz A., Rothenberg M. MicroRNA-21 is up-regulated in allergic airway inflammation and regulates IL-12p35 expression. J. Immunol. 2009;182(8):4994–5002. doi: 10.4049/jimmunol.0803560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu G., Friggeri A., Yang Y., Milosevic J., Ding Q., Thannickal V.J., Kaminski N., Abraham E. miR-21 mediates fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J. Exp. Med. 2010;207(8):1589–1597. doi: 10.1084/jem.20100035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xia B., Lu J., Wang R., Yang Z., Zhou X., Huang PJFic. miR-21-3p regulates influenza A virus replication by targeting histone deacetylase-8. Front Cell Infect Microbiol. 2018;8:175. doi: 10.3389/fcimb.2018.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Y., Chen J., Wang H., Shi J., Wu K., Liu S., Liu Y., Wu J.J.P.P. HCV-induced miR-21 contributes to evasion of host immune system by targeting MyD88 and IRAK1. PLoS Pathog. 2013;9(4) doi: 10.1371/journal.ppat.1003248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Houzet L., Yeung M.L., de Lame V., Desai D., Smith S.M., Jeang K.-T.J.R. MicroRNA profile changes in human immunodeficiency virus type 1 (HIV-1) seropositive individuals. Retrovirology. 2008;5(1):118. doi: 10.1186/1742-4690-5-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu M., Guo S., Hibbert J.M., Jain V., Singh N., Wilson N.O., Stiles J.K.J.C. CXCL10/IP-10 in infectious diseases pathogenesis and potential therapeutic implications. Cytokine Growth Factor Rev. 2011;22(3):121–130. doi: 10.1016/j.cytogfr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gagnon J.D., Kageyama R., Shehata H.M., Fassett M.S., Mar D.J., Wigton E.J., Johansson K., Litterman A.J., Odorizzi P., DJCr Simeonov. miR-15/16 restrain memory T cell differentiation, cell cycle, and survival. Cell Rep. 2019;28(8):2169–2181. doi: 10.1016/j.celrep.2019.07.064. e2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng C., Zheng Z., Sun J., Zhang Y., Wei C., Ke X., Liu Y., Deng L., Wang HJSr. MiR-16-5p mediates a positive feedback loop in EV71-induced apoptosis and suppresses virus replication. Sci. Rep. 2017;7(1):1–16. doi: 10.1038/s41598-017-16616-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu W., Choi E.-J., Lee I., Lee Y.S., Bao X.J.V. Non-coding RNAs and their role in respiratory syncytial virus (RSV) and human metapneumovirus (hMPV) infections. Viruses. 2020;12(3):345. doi: 10.3390/v12030345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma Y.J., Yang J., Fan X.L., Zhao H.B., Hu W., Li Z.P., Yu G.C., Ding X.R., Wang J.Z., XCJJoc Bo. Cellular micro RNA let‐7c inhibits M1 protein expression of the H1N1 influenza A virus in infected human lung epithelial cells. J. Cell Mol. Med. 2012;16(10):2539–2546. doi: 10.1111/j.1582-4934.2012.01572.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bakre A., Mitchell P., Coleman J.K., Jones L.P., Saavedra G., Teng M., Tompkins S.M. Tripp RAJTJogv, Respiratory syncytial virus modifies microRNAs regulating host genes that affect virus replication. 2012;93(Pt 11):2346. doi: 10.1099/vir.0.044255-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nahand J.S., Karimzadeh M.R., Nezamnia M., Fatemipour M., Khatami A., Jamshidi S., Moghoofei M., Taghizadieh M., Hajighadimi S., AJIl Shafiee. The role of miR‐146a in viral infection. J. Gen. Virol. 2020;72(3):343–360. doi: 10.1002/iub.2222. [DOI] [PubMed] [Google Scholar]
- 44.Frieman M., Yount B., Heise M., Kopecky-Bromberg S.A., Palese P., Baric R.S. Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane. J. Virol. 2007;81(18):9812–9824. doi: 10.1128/JVI.01012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.