Abstract
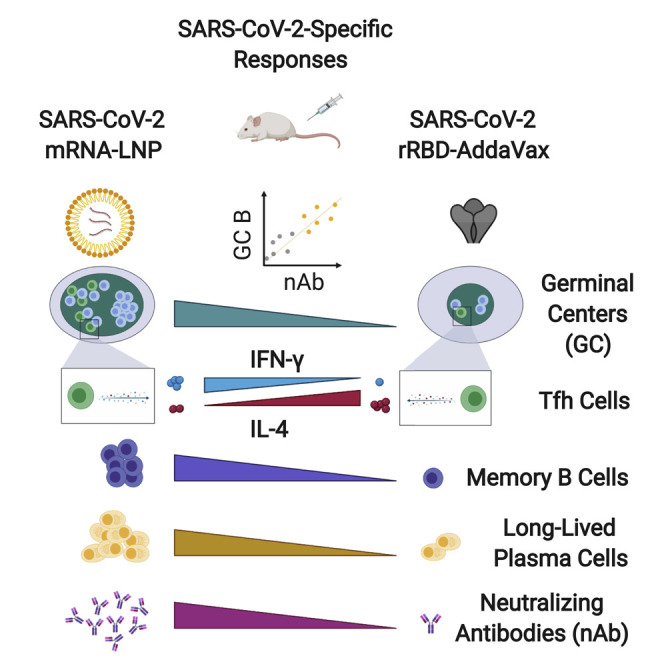
The deployment of effective vaccines against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is critical to eradicate the coronavirus disease 2019 (COVID-19) pandemic. Many licensed vaccines confer protection by inducing long-lived plasma cells (LLPCs) and memory B cells (MBCs), cell types canonically generated during germinal center (GC) reactions. Here, we directly compared two vaccine platforms—mRNA vaccines and a recombinant protein formulated with an MF59-like adjuvant—looking for their abilities to quantitatively and qualitatively shape SARS-CoV-2-specific primary GC responses over time. We demonstrated that a single immunization with SARS-CoV-2 mRNA, but not with the recombinant protein vaccine, elicited potent SARS-CoV-2-specific GC B and T follicular helper (Tfh) cell responses as well as LLPCs and MBCs. Importantly, GC responses strongly correlated with neutralizing antibody production. mRNA vaccines more efficiently induced key regulators of the Tfh cell program and influenced the functional properties of Tfh cells. Overall, this study identifies SARS-CoV-2 mRNA vaccines as strong candidates for promoting robust GC-derived immune responses.
Keywords: germinal centers, germinal center B cells, T follicular helper cells, SARS-CoV-2, COVID-19, mRNA vaccines, neutralizing antibodies
Graphical Abstract
Highlights
-
•
SARS-CoV-2 mRNA vaccines elicit potent GC B cell responses
-
•
GC responses are associated with a robust development of neutralizing antibodies
-
•
SARS-CoV-2 mRNA vaccines promote antigen-specific Tfh cells
-
•
Key elements of the Tfh cell program are modulated by SARS-CoV-2 mRNA vaccines
Herein, Lederer et al. show a nucleic-acid-based vaccine platform for SARS-CoV-2 that potently induces germinal center (GC) responses. GCs are microanatomical sites harboring the formation of high-quality, protective antibody responses. Such vaccine platforms can be promising candidates to mitigate the COVID-19 pandemic.
Introduction
The coronavirus disease 2019 (COVID-19) outbreak was declared a pandemic in March 2020 (Cucinotta and Vanelli, 2020) and has led to a worldwide health emergency, calling for rapid and efficient countermeasures. Vaccination is one of the most cost-effective and successful public health interventions to prevent infections and save millions of lives (Orenstein and Ahmed, 2017). Hence, developing efficacious vaccine approaches for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of COVID-19, is of paramount importance (Graham, 2020; Sempowski et al., 2020).
Most efficient vaccines generate prolonged immunity by eliciting long-lived plasma cells (LLPCs) and memory B cells (MBCs) (Sallusto et al., 2010). LLPCs and MBCs with high affinity for the pathogens are formed during germinal center (GC) reactions. GCs are microanatomical structures in secondary lymphoid organs where antigen-activated B cells undergo the Darwinian process of affinity maturation (Allen et al., 2007; Mesin et al., 2016). In GCs, somatic hypermutation (SHM), which introduces random mutations in the immunoglobulin genes, is followed by positive selection of high-affinity GC B cells. After multiple rounds of this iterative process, GC B cells differentiate into MBCs or antibody (Ab)-secreting LLPCs. The GC reaction is tightly regulated by specialized B cell helpers named T follicular helper (Tfh) cells, which enable proliferation, survival, and differentiation of GC B cells through the delivery of costimulatory molecules and cytokines (Crotty, 2019; Vinuesa et al., 2016). Of note, Tfh cells can display a heterogeneous functional profile that influences B cell responses (Schmitt et al., 2014; Weinstein et al., 2016). Overall, GC reactions are fundamental for the induction of high-quality long-lasting B cell responses.
A large number of different vaccine strategies for SARS-CoV-2 are being simultaneously pursued by multiple pharmaceutical companies and laboratories around the world (Amanat and Krammer, 2020; Graham, 2020; Sempowski et al., 2020). The long list of SARS-CoV-2 vaccine candidates includes newer vaccine approaches such as nucleic-acid-based vaccines as well as more traditional vaccine platforms. Nucleic-acid-based vaccines like nucleoside-modified mRNA vaccines encapsulated in lipid nanoparticles (LNPs) can elicit powerful, durable protective Ab responses after a single immunization (Pardi et al., 2018a; 2017) and have attracted much interest in recent years (Pardi et al., 2018b). The promise of SARS-CoV-2 mRNA-LNP vaccine candidates is substantiated by our recent study showing that mRNA vaccines induce elevated levels of neutralizing Abs (nAbs) (Laczkó et al., 2020), as well as by similar findings on SARS-CoV-2 mRNA vaccine candidates currently in clinical trials (Anderson et al., 2020; Corbett et al., 2020a; 2020b; Jackson et al., 2020; Mulligan et al., 2020a; Sahin et al., 2020; Walsh et al., 2020; Zhang et al., 2020). Along with the newer platforms, more traditional vaccine approaches are currently being evaluated in clinical trials (WHO, 2020). For instance, SARS-CoV-2 recombinant protein vaccines are being tested in combination with FDA-approved adjuvants such as MF59 (O’Hagan, 2007), an oil-in-water emulsion licensed for use in seasonal influenza vaccines. The SARS-CoV-2 vaccine field is moving at a fast pace, and several preclinical and clinical studies have been published thus far reporting a limited characterization of the immune responses induced by the SARS-CoV-2 vaccine candidates (Corbett et al., 2020b; 2020a; Gao et al., 2020; Jackson et al., 2020; Laczkó et al., 2020; Mulligan et al., 2020b; Sahin et al., 2020; Smith et al., 2020; Yu et al., 2020; Zhang et al., 2020). However, the lack of a direct comparison and universal metrics for the assessment of the immune responses elicited by the different vaccines make it challenging to conclude what vaccines can induce superior immune responses. Additionally, no study has deeply investigated the capacity of the SARS-CoV-2 vaccines to elicit GC responses and the ensuing quality of Ab responses. This is a fundamental gap in our knowledge given the crucial role of GCs in regulating B cell responses and high-quality Ab generation.
Herein, we systematically compared SARS-CoV-2 mRNA vaccines encoding the receptor-binding domain (RBD) and full-length spike protein of SARS-CoV-2 with recombinant SARS-CoV-2 RBD protein (rRBD) formulated with AddaVax, an MF59-like adjuvant. Our in-depth analysis was aimed at shedding light on the ability of the two different vaccine platforms to mold GC responses and the functional properties of Tfh cells. Overall, our study revealed a superior ability of the SARS-CoV-2 mRNA vaccines to elicit SARS-CoV-2-specific GC B cell responses, which were associated with an efficient production of nAbs after a single or a booster immunization. Moreover, it proved that the mRNA vaccines could efficiently promote the induction of the Tfh cell program and shape the functional properties of Tfh cells. Taken together, our data highlight the importance of GC responses in SARS-CoV-2 vaccination for the generation of protective Abs and point toward mRNA vaccines as promising candidates to elicit potent and high-quality adaptive immune responses.
Results
Nucleoside-Modified SARS-CoV-2 mRNA Vaccines Promote Potent GC Formation, whereas rRBD Vaccine Formulated in AddaVax Fails to Trigger Robust Primary GC Responses
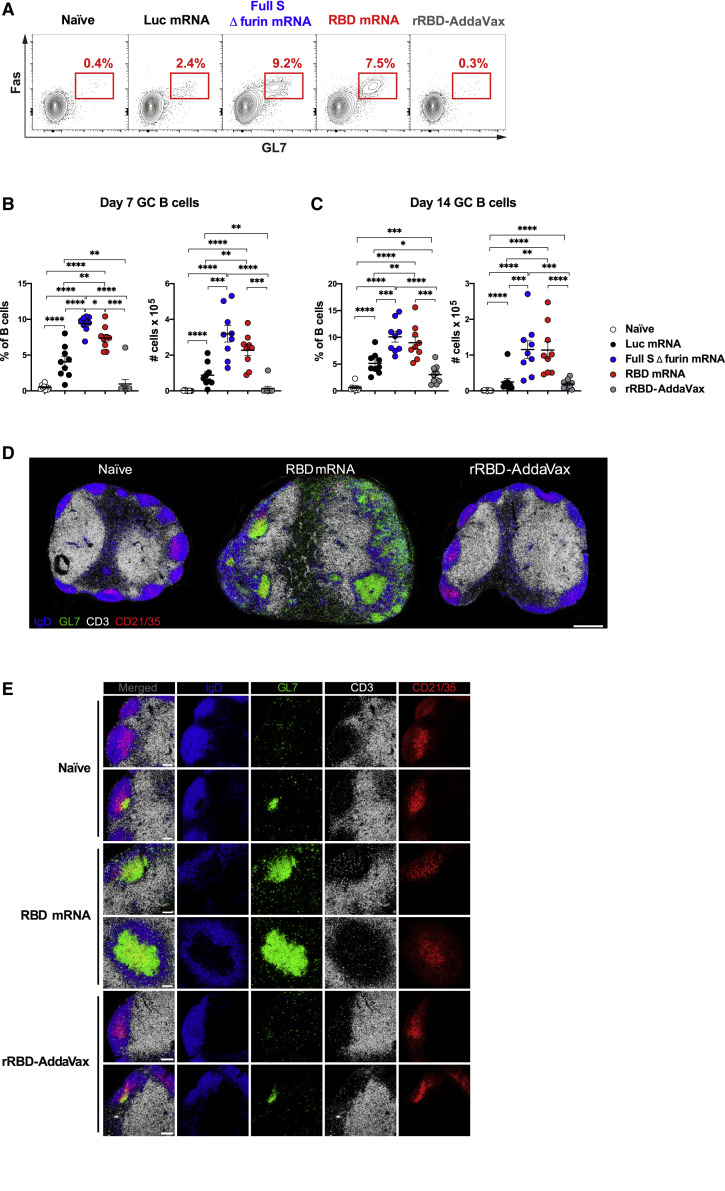
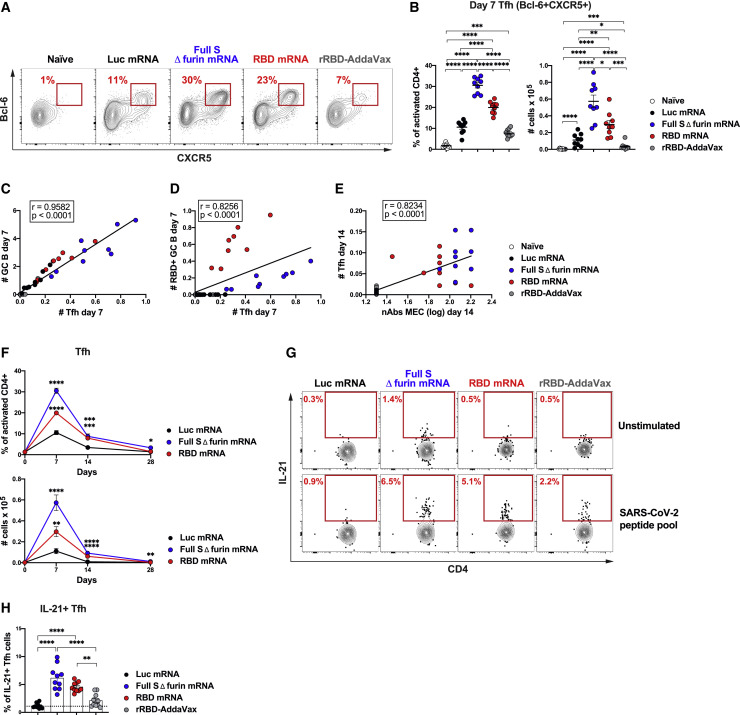
Hundreds of millions of people will have to be vaccinated to combat the COVID-19 pandemic, and vaccination strategies deploying one or few immunizations are an attractive option if capable of eliciting efficient immune responses. mRNA vaccines have been previously shown to promote potent or protective Ab responses upon a single immunization (Laczkó et al., 2020; Pardi et al., 2018b). Conversely, purified/recombinant proteins formulated with adjuvants often require multiple immunizations to achieve sufficient titers of protective Abs. In light of the importance of GCs for the generation of high-quality persistent Ab responses, we hypothesized that a single immunization with SARS-CoV-2 mRNA vaccines (Laczkó et al., 2020) can result in superior GC formation in comparison to a SARS-CoV-2 recombinant protein formulated with the MF59-like adjuvant AddaVax. To test our hypothesis, BALB/c mice were immunized once intramuscularly (i.m.) with 30 μg of an mRNA-LNP vaccine encoding a SARS-CoV-2 stabilized full-length spike glycoprotein with deleted furin cleavage site (full S Δ furin mRNA), or with a second mRNA-LNP encoding the SARS-CoV-2 RBD (RBD mRNA), a prominent target for nAbs on the full S protein (Laczkó et al., 2020). A third and a fourth group of mice were immunized i.m. with a control mRNA-LNP vaccine encoding firefly luciferase (Luc mRNA) or with 10 μg of recombinant SARS-CoV-2 RBD adjuvanted in AddaVax (rRBD-AddaVax), respectively. GC B cells were evaluated in draining inguinal lymph nodes (LNs) 7 days post-immunization by flow cytometry (Figures 1A and 1B). After a single immunization, no detectable GC B cells (Fas+GL7+ B cells) were induced by rRBD-AddaVax when compared to unimmunized naive animals. By contrast, a robust population of GC B cells was elicited by the two SARS-CoV-2 mRNA vaccines. At this early time point, SARS-CoV-2 mRNA vaccines, particularly the full S Δ furin mRNA vaccine, were also superior at inducing short-lived plasma cells (PCs) (Figures S1A and S1B). To test whether rRBD-AddaVax induced elevated GC responses that were delayed in comparison to SARS-CoV-2 mRNA vaccines, frequencies and absolute numbers of GC B cells were evaluated 14 days after immunization. In line with day 7 post-immunization results, rRBD-AddaVax induced minimal GC responses, whereas a sizable GC B cell population was present in SARS-CoV-2-mRNA-immunized animals at this later time point (Figure 1C). Similarly, GC B cell counts were still significantly higher in animals immunized with SARS-CoV-2 mRNA than in rRBD-AddaVax-immunized mice at day 21 (Figure S1C). An extended analysis of different draining LNs (popliteal) failed to show differences in drainage pattern of the two vaccine platforms, as only SARS-CoV-2-mRNA-immunized mice displayed elevated GC B cells and short-lived PCs at this site (Figures S1D and S1E).
Figure 1.
A Single Immunization with SARS-CoV-2 mRNA Vaccines Results in Robust GC B Cell Responses
Mice were immunized i.m. with Luc, full S Δ furin or RBD mRNA, or recombinant RBD protein adjuvanted with AddaVax (rRBD-AddaVax). Naive mice were also included. Inguinal LNs were analyzed 7 days ([A], [B], [D], and [E]) and 14 days (C) later.
(A) Representative flow cytometry contour plots of GC B cells, defined as live dump− CD19+GL7+Fas+ cells.
(B and C) Frequency (left) and absolute counts (right) of total GC B cells (defined as in [A]).
(D) Representative confocal images of whole LN sections. LN sections were stained with monoclonal Abs against IgD (blue), GL7 (green), CD3 (white), and CD21/35 (red); bar, 400 μm.
(E) Selected magnified images of B cell follicles. Images display merged signals and each channel separately; bar, 100 μm.
In (A)–(C), n = 9 mice per group were analyzed. Data are combined from three independent experiments. Mean ± SEM is shown, and each data point represents an individual mouse. One-way-ANOVA with Bonferroni correction or unpaired two-tailed Mann-Whitney U tests were conducted according to the distribution of the data. In (D) and (E), n = 4 mice per group from two independent experiments were analyzed, and representative samples were displayed. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001. See also Figure S1 and Tables S2 and S5.
To confirm the superior ability of SARS-CoV-2 mRNA vaccines to foster the formation of GCs after a single immunization, we performed microscopy studies on inguinal LNs from mice immunized 7 days earlier with RBD mRNA or rRBD-AddaVax (Figures 1D and 1E). GCs are defined by microscopy as clusters of GL7+ cells forming around a network of follicular dendritic cells (FDCs, CD21/35hi cells) (Heesters et al., 2014) and are surrounded by a mantle of IgD+ naive B cells (De Silva and Klein, 2015). GCs are anatomically segregated from T cell areas (CD3+). A global view of LN sections revealed a robust induction of GCs by the RBD mRNA vaccine as opposed to minimal GC formation in rRBD-AddaVax-immunized mice that resembled the one observed in naive mice (Figure 1D). A previously published study also described small GC formation by microscopy after one immunization with a protein antigen in MF59 (Liang et al., 2017). The high level of colocalization of GL7+ cells and CD21/35hi FDCs, together with the presence of infiltrating CD3+ cells, suggested that the structures observed in the LNs of RBD-mRNA-immunized mice were bona fide GCs (Figure 1E). Therefore, SARS-CoV-2 mRNA vaccines, but not rRBD-AddaVax, elicited potent GC formation upon a single immunization.
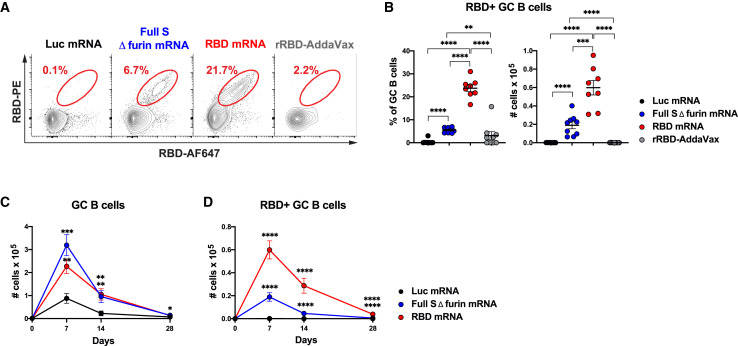
GC B Cells Elicited by SARS-CoV-2 mRNA Vaccines Are Enriched in RBD-Specific B Cells and Contract by Day 28
We sought to determine whether GC B cells induced by the SARS-CoV-2 mRNA vaccines were antigen specific. To this aim, we generated two fluorescently labelled rRBD probes to track RBD-specific GC B cells induced by the different vaccines via flow cytometry. A large fraction of the GC B cell response was RBD specific in RBD-mRNA-immunized mice 7 days post-immunization (Figures 2A and 2B). RBD-specific GC B cells were also induced by full S Δ furin mRNA, although at significantly lower levels than in RBD-mRNA-immunized mice (Figures 2A and 2B). Because the full S protein includes additional epitopes besides the ones contained in RBD, we measured full S-specific GC B cells by taking advantage of two fluorescently labelled full S probes (Figures S2A and S2B). This analysis revealed a similar number of RBD-specific (Figure 2B, mean 0.19 × 105 cells) and full S-specific GC B cells (Figure S2B, mean 0.23 × 105 cells) in mice that received full S Δ furin mRNA vaccine, suggesting that GC B cell responses induced by full S might be focused on epitopes included in the RBD domain. The mice immunized with the control vaccine (Luc mRNA) did not mount SARS-CoV-2-specific GC B cell responses (Figures 2A, 2B, S2A, and S2B). In agreement with the poor induction of GC responses, mice that received the rRBD-AddaVax vaccine did not form SARS-CoV-2-specific GC B cells 7 days after a single vaccine administration (Figures 2A, 2B, S2A, and S2B). Similar findings emerged from the analysis of popliteal draining LNs or when a higher dose of rRBD (20 μg) in AddaVax was used as immunogen along with a lower SARS-CoV-2 mRNA vaccine dose (20 μg) (Figures S2C–S2F).
Figure 2.
SARS-CoV-2 mRNA Vaccines Elicit Strong Antigen-Specific GC B Cell Responses.
Mice were i.m. immunized with SARS-CoV-2 mRNA vaccines, Luc mRNA, or rRBD-AddaVax. GC B cells in inguinal LNs were measured 7, 14, or 28 days post-immunization.
(A) Representative flow cytometry contour plots showing RBD-specific GC B cells, defined as live dump− CD19+Fas+GL7+RBD-PE+RBD-AF647+ cells.
(B) Frequency (left) and absolute counts (right) of RBD-specific GC B cells (defined as in [A]) at 7 days post-immunization.
(C) Kinetics of absolute numbers of total GC B cells.
(D) RBD-specific GC B cell absolute numbers over time. In (C) and (D), day 0 represents the average of 18 naive animals.
In (A) and (B), n = 8 mice for RBD-mRNA immunization, and n = 9 mice per all other groups were analyzed. Data are combined from three independent experiments. Mean ± SEM is shown, and each data point represents an individual mouse. In (C) and (D), the same number of mice and experiments were analyzed as in (A) and (B) for days 7 and 14. n = 10 mice per group were analyzed, and data are combined from two independent experiments at day 28. Mean ± SEM is graphed. One-way-ANOVA with Bonferroni correction or unpaired two-tailed Mann-Whitney U tests were conducted according to the distribution of the data. In (C) and (D), statistics were calculated versus Luc mRNA group. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001. See also Figure S2 and Table S2.
Next, we interrogated the capacity of SARS-CoV-2 mRNA vaccines to regulate the longevity of GC responses over time. Published work has shown that mRNA vaccines can induce GC B cells (Lindgren et al., 2017; Pardi et al., 2018a), but the kinetics of GC responses upon immunization with mRNA vaccines was not previously investigated. During vaccination with mRNA, host cells take up the mRNA and translate it into proteins (Pardi et al., 2018b), raising the possibility that this in situ protein production results in prolonged antigen availability and extended GC reactions. Immunization studies with model antigens have taught us that fully formed GCs can be found 7 days post-immunization (De Silva and Klein, 2015) and suggested that GCs have a 2–3 week duration (Allen et al., 2007). A quantitative evaluation of primary GC kinetics was performed by evaluating frequencies and absolute numbers of total and SARS-CoV-2-specific GC B cells over time. Our analysis indicated that GC B cell responses elicited by SARS-CoV-2 mRNA vaccines quantitatively peaked between days 7 and 14, then underwent a contraction phase and were deeply blunted by day 28 post-immunization (Figures 2C and 2D). Total GC B cells and SARS-CoV-2-specific GC B cells followed similar kinetics (Figures 2C, 2D, and S2G). In stark contrast with the absolute counts, the frequencies of total and SARS-CoV-2-specific GC B cells persisted at elevated levels until day 28 after immunization (Figures S2H and S2I). The numerical decrease of GC B cells was driven by a reduction in total LN cellularity (Figure S2J) that at this time point went back to baseline values. GC B cell and RBD-specific GC B cell responses following rRBD-AddaVax immunization were only evaluated up to 21 days and revealed modest and delayed GC B cell formation in comparison to immunization with SARS-CoV-2 mRNA (Figures S2K and S2L).
Overall, our data indicated that SARS-CoV-2 mRNA vaccines can potently induce SARS-CoV-2-specific GC B cells responses that are mostly resolved within 4 weeks after the immunization. Conversely, poor SARS-CoV-2-specific GC B cell responses were elicited by rRBD formulated with AddaVax after a single immunization.
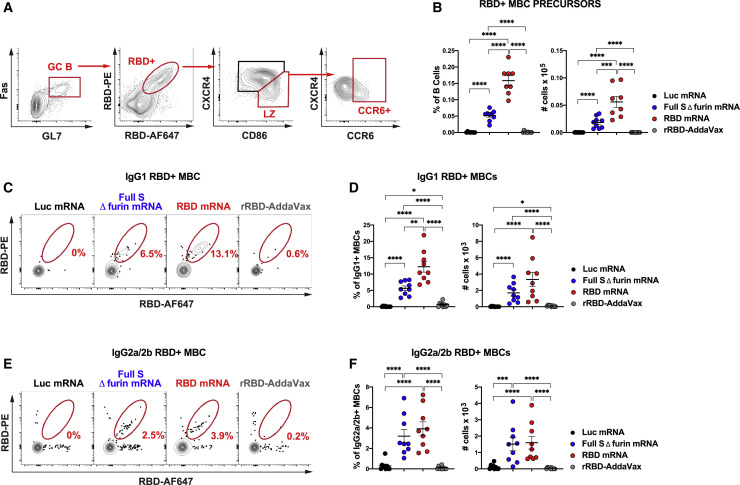
SARS-CoV-2-Specific GC MBC Precursors and MBCs Are Induced by SARS-CoV-2 mRNA Vaccines, but Not by rRBD-AddaVax
MBCs are powerful mediators of secondary immune responses, and their activation can lead to a rapid burst of PCs and serum Abs (Sallusto et al., 2010). Precursors of MBCs can be identified within the light zone (LZ) of mouse and human GCs (Suan et al., 2017). We hypothesized that, by virtue of its capacity to foster robust antigen-specific GC responses, a SARS-CoV-2 mRNA vaccine would be superior at inducing SARS-CoV-2-specific MBC precursors compared to rRBD-AddaVax. Thus, we immunized mice with one dose of the various vaccines and measured MBC precursors (as defined in Figure 3 A) in the inguinal LNs 7 days post-immunization. Consistent with our hypothesis, we found a substantial population of RBD-specific MBC precursors only in mice immunized with SARS-CoV-2 mRNA vaccines (Figure 3B). Neither rRBD-AddaVax nor Luc mRNA vaccines generated significant RBD+ MBC precursors (Figure 3B). Similarly, a considerable population of full S+ MBC precursors was induced by SARS-CoV-2 mRNA vaccines, but not by rRBD-AddaVax nor Luc mRNA vaccines (Figure S3A).
Figure 3.
Antigen-Specific MBC Precursors and Bona Fide MBCs are Induced by SARS-CoV-2 mRNA Vaccination
Mice were i.m. immunized with SARS-CoV-2 mRNA vaccines, Luc mRNA control or rRBD-AddaVax.
(A and B) Inguinal LNs were analyzed 7 days post-immunization.
(A) Representative gating strategy for RBD-specific MBC precursors (CCR6+ cells) in the LZ of the GCs (CXCR4Lo/−CD86+). Plots shown were pre-gated on live dump− CD19+ cells.
(B) Frequency (left) and absolute counts (right) of RBD-specific MBC precursors.
(C–F) Spleens were assessed 60 days post-immunization.
(C) Representative contour plots of RBD-specific IgG1+ MBCs. Cells were pre-gated on live dump− CD19+B220+IgD−Fas−CD38+IgG1+ B cells.
(D) Frequency (left) and absolute numbers (right) of RBD-specific IgG1+ MBCs.
(E) Representative contour plots of RBD-specific IgG2a/2b+ MBCs, pre-gated on live dump− CD19+B220+IgD−Fas−CD38+IgG2a/2b+ B cells.
(F) Frequency (left) and absolute numbers (right) of RBD-specific IgG2a/2b+ MBCs.
In (A) and (B), n = 8 mice for RBD-mRNA immunization, and n = 9 mice per group for other immunization conditions. In (C)–(F), n = 9 mice per group were analyzed. In (A)–(F), data were combined from three independent experiments. Mean ± SEM is shown, and each data point represents an individual mouse. One-way ANOVA with Bonferroni correction or unpaired two-tailed Mann-Whitney U tests were conducted according to the distribution of the data. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001. See also Figure S3 and Tables S2 and S3.
To assess whether the MBC precursor induction by the SARS-CoV-2 mRNA vaccines was predictive of bona fide SARS-CoV-2-specific MBC generation at a memory time point, SARS-CoV-2-specific MBCs were measured in spleen of animals 60 days after vaccination. While SARS-CoV-2 mRNA vaccines promoted the generation of class-switched RBD-specific and full S-specific MBCs expressing immunoglobulin (Ig)G1 (Figures 3C, 3D, S3B, and S3C) and IgG2a/2b (Figures 3E, 3F, S3D, and S3E), rRBD-AddaVax and the control Luc mRNA vaccine failed to induce sizable SARS-CoV-2-specific class-switched MBC populations. None of the vaccines elicited SARS-CoV-2-specific MBCs expressing IgM (Figures S3F–S3I).
Thus, only SARS-CoV-2 mRNA, but not the rRBD-AddaVax vaccine, efficiently generated antigen-specific MBC precursors in GCs during the primary response and also formed class-switched MBCs that persisted at least 2 months post-immunization.
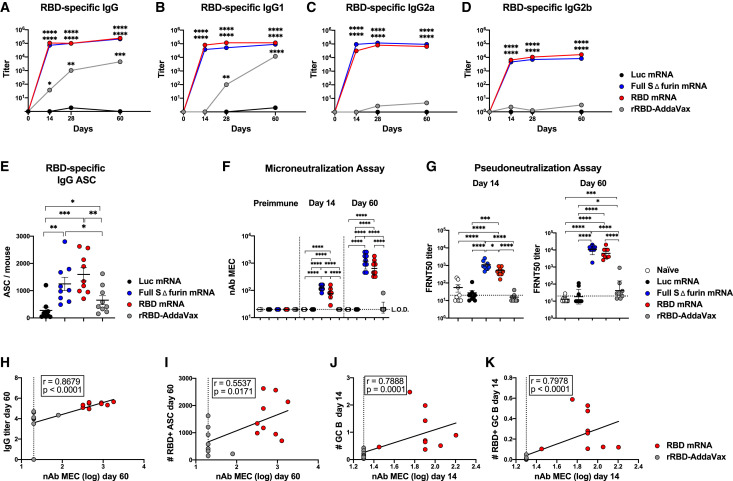
Primary GC Responses Are Predictive of Neutralizing Ab Formation in SARS-CoV-2 Immunization
Building on our recent findings (Laczkó et al., 2020), we next sought to compare the ability of SARS-CoV-2 mRNA vaccines to the rRBD-AddaVax vaccine to induce SARS-CoV-2-specific Abs, SARS-CoV-2-specific bone marrow LLPCs, and SARS-CoV-2 nAbs.
We first performed a longitudinal analysis of the SARS-CoV-2-specific serum IgG titers induced by a single dose of the different vaccines under investigation (Figures 4 A and S4A). Both SARS-CoV-2 mRNA vaccines elicited elevated titers of SARS-CoV-2-specific IgG as early as day 14 post-immunization, and the titers remained high through at least day 60 post-vaccination. Despite of an almost complete lack of GC induction, mice receiving rRBD-AddaVax generated rRBD-specific Ab responses, although SARS-CoV-2-specific IgG titers rose more slowly than they did after mRNA vaccination and remained lower in rRBD-AddaVax at day 60 post-immunization (Figures 4A and S4A). This observation confirmed that rRBD-AddaVax preparation was not inactive and induced a delayed antigen-specific immune response. SARS-CoV-2-specific IgG1 accounted for the majority of the antigen-specific IgG responses in the mice immunized with rRBD-AddaVax, most of which had negligible IgG2a and IgG2b responses (Figures 4B–4D and S4B–S4D). In contrast, SARS-CoV-2-mRNA-immunized mice displayed high levels of SARS-CoV-2-specific IgG2a and IgG2b titers, together with IgG1 at all time points (Figures 4B–4D and S4B–S4D).
Figure 4.
Elevated IgG Titers and nAbs Are Driven by SARS-CoV-2 mRNA Immunization
Mice were i.m. immunized with SARS-CoV-2 mRNA vaccines, Luc mRNA control, or rRBD-AddaVax.
(A–D) Serum was collected at various time points for analysis. SARS-CoV-2-specific Ab titers were determined by ELISA. Kinetics of RBD-specific IgG (A), IgG1 (B), IgG2a (C), and IgG2b (D) titers are shown.
(E) Bone marrow (BM) was collected 60 days post-immunization. Quantification of RBD-specific IgG+ ASC was determined by ELISPOT.
(F) nAbs were measured by microneutralization assays at pre-immune conditions and day 14 and day 60 post-immunization in serum samples. Minimal effective concentration (MEC) is shown.
(G) Levels of nAbs at 14 (left) and 60 (right) days post-immunization were confirmed by pseudoneutralization assay.
(H–K) For all correlations, nAbs from microneutralization assays were used. Data from day 60 ([H] and [I]) and day 14 ([J] and [K]) were included. Spearman correlations of (H) RBD-specific IgG titers and nAbs, (I) BM RBD-specific IgG+ ASC and nAbs, (J) inguinal LN GC B cells (cells × 105) and nAb, and (K) inguinal LN RBD-specific GC B cells (cells × 105) and nAb levels are shown.
In (A)–(K), n = 9 mice per group were analyzed. Data are combined from three independent experiments. In (A)–(D), serological data are represented as geometric mean. In (A)–(D), day 0 represents the average of 36 naive mice, and statistics were calculated versus Luc mRNA. In (E), mean ± SEM is shown, and each data point represents an individual mouse. In (F) and (G), geometric mean ± geometric SD is shown, and each data point represents an individual mouse. L.O.D., limit of detection (dotted line). One-way-ANOVA with Bonferroni correction or unpaired two-tailed Mann-Whitney U tests were conducted according to the distribution of the data. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001. See also Figure S4.
LLPCs persistently secrete Abs without the need for antigen re-exposure and are responsible for maintaining protective levels of circulating Abs that can prevent or hinder a re-infection by pathogens (Sallusto et al., 2010). We compared bone marrow SARS-CoV-2-specific Ab-secreting cells (ASCs) elicited by the different vaccines 60 days post-immunization. Consistent with the serum IgG data above, mice immunized with rRBD-AddaVax generated RBD-specific IgG+ LLPCs, despite minimal GC responses. However, mice immunized with mRNA vaccines had substantially higher RBD-specific IgG+ LLPC numbers (Figures 4E and S4E), suggesting more efficient generation of multiple facets of long-term humoral immunity by these vaccines.
To investigate the apparent discrepancy between the inability to form GCs and the presence of detectable Ab and LLPC responses of rRBD-AddaVax-immunized mice, we performed a qualitative analysis of nAb responses. We tested nAbs in sera from animals immunized 14 or 60 days earlier with the various SARS-CoV-2 vaccines by performing in vitro microneutralization assays with authentic SARS-CoV-2. Remarkably, in contrast to mice immunized with SARS-CoV-2 mRNA vaccines where nAbs were present in the sera at both time points tested, no detectable nAbs were induced by rRBD-AddaVax after a single immunization (Figure 4F). Comparable data were obtained with an in vitro pseudoneutralization assay (Figure 4G). The level of nAbs detected by the microneutralization assay was associated with SARS-CoV-2-specific IgG responses, but it showed a weaker correlation with RBD-specific LLPCs (Figures 4H, 4I, S4F, and S4G), suggesting that some of the ASCs detected at this time point could reflect lower-quality or non-neutralizing responses. Because SARS-CoV-2-mRNA-immunized mice generated both elevated nAbs and GC B cells, we hypothesized the existence of a strong association between these two parameters. As anticipated, total and SARS-CoV-2-specific GC B cells strongly correlated with the levels of nAbs at day 14 post-immunization (Figures 4J, 4K, and S4H–S4J).
Altogether, these data strongly indicate that efficient GC responses are fundamental to obtain high-quality SARS-CoV-2 nAbs and that a potent GC B cell induction in mice immunized once with SARS-CoV-2 mRNA vaccine, but not with rRBD-Addavax, is connected to efficient SARS-CoV-2 nAb production.
SARS-CoV-2 mRNA Vaccines Are Superior in Comparison to rRBD-AddaVax Vaccine at Inducing SARS-CoV-2-Specific Tfh Cells
Tfh cells are key regulators of GC responses (Crotty, 2019; Vinuesa et al., 2016). Knowing that Tfh cell responses normally peak between days 7 and 9 in mice (Baumjohann et al., 2011; Botta et al., 2017), we determined the frequencies and absolute numbers of Tfh cells induced by the mRNA vaccines and rRBD-AddaVax in draining LNs 7 days post-immunization. Tfh cells were measured by flow cytometry as CD4+CD44hiCD62L− cells expressing the Tfh cell signature markers CXCR5 and Bcl-6 (Figure 5 A) or CXCR5 and PD-1 (Figure S5A). In line with the potent GC B cell induction driven by the SARS-CoV-2 mRNA vaccines (Figures 1A and 1B), a robust generation of Tfh cells was promoted by full S Δ furin mRNA and RBD mRNA vaccines (Figures 5A, 5B, S5A, and S5B). Conversely, rRBD-AddaVax elicited only a modest Tfh cell population at day 7 (Figures 5A, 5B, S5A, and S5B), which was still significantly lower by absolute numbers than what was seen in the SARS-CoV-2 mRNA vaccine group at day 21 post-immunization (Figure S5C) (closer to the GC peak for this vaccine group). This was an anticipated outcome based on the poor GC B cell induction measured in response to rRBD-AddaVax (Figures 1A and 1B). Additionally, we found a robust correlation between the numbers of Tfh cells, defined either as CXCR5+Bcl-6+ or CXCR5+PD-1+ cells, and total or RBD-specific GC B cells (Figures 5C, 5D, and S5D). Of note, Tfh cell absolute numbers were significantly correlated with nAbs after a single-dose immunization (Figures 5E and S5E).
Figure 5.
SARS-CoV-2 mRNA Vaccines Elicit a Robust Differentiation of Antigen-Specific Tfh Cells
Mice were i.m. immunized with SARS-CoV-2 mRNA vaccines, Luc mRNA control, or rRBD-AddaVax. Naive mice were also used as control. Tfh cells were from inguinal ([A]–[F]) or pooled inguinal and popliteal LNs ([G] and [H]). (A)–(D) and (G) and (H) refer to data analyzed at 7 days post-immunization.
(A) Representative analysis of Tfh cells (Bcl-6+CXCR5+). Cells were pre-gated on live CD4+B220−CD44hiCD62L− populations.
(B) Frequency (left) and absolute counts (right) of Tfh cells as detailed in (A).
(C–E) Spearman correlations of (C) GC B cells (cells × 105) and Bcl-6+CXCR5+ Tfh cells (cells × 105), (D) RBD-specific GC B cells (cells × 105) and Bcl-6+CXCR5+ Tfh cells (cells × 105), and (E) Bcl-6+CXCR5+ Tfh cells (cells × 105) and nAb levels (MEC) at 14 days post-immunization.
(F) Kinetics of Bcl-6+CXCR5+ Tfh cell frequency (top) and absolute counts (bottom).
(G) Representative analysis of antigen-specific Tfh cells. Lymphocytes were stimulated with a SARS-CoV-2 peptide pool or were left unstimulated. SARS-CoV-2-specific Tfh cells were identified as IL-21+PD-1+CXCR5+ cells. Cells were pre-gated on live CD4+B220−CD44hi populations.
(H) Frequency of SARS-CoV-2-specific IL-21+ Tfh cells as detailed in (G). Dotted line represents the average of 45 unstimulated samples.
In (A)–(E), n = 9 mice per group were analyzed. Data are combined from three independent experiments. In (F), n = 9 mice per group were analyzed at day 7 and day 14. Data are combined from three independent experiments. n = 10 mice per group were analyzed at day 28, and data are combined from two independent experiments. In (G) and (H), n = 10 mice per group were analyzed. Data are combined from four independent experiments. In (B), and (F)–(H), data were graphed as mean ± SEM. One-way-ANOVA with Bonferroni correction or unpaired two-tailed Mann-Whitney U tests were conducted according to the distribution of the data. For kinetics in (F), day 0 represents the average of 18 naive animals, and statistics were calculated versus Luc mRNA group. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001. See also Figure S5 and Table S1.
To determine whether the kinetic of Tfh cell induction mirrored GC B cell responses, we evaluated Tfh cell frequencies and absolute numbers over time following immunization with SARS-CoV-2 mRNA vaccines or Luc mRNA vaccine. We found that Tfh responses driven by SARS-CoV-2 mRNA vaccines peaked at day 7 post-immunization and then waned over time (Figure 5F). At day 28 post-immunization, elevated Tfh cell responses were no longer detectable.
The correlation between Tfh cells and RBD+ GC B cells suggested that the Tfh cells induced by the SARS-CoV-2 mRNA vaccines were SARS-CoV-2 specific. To directly measure antigen-specific polyclonal Tfh cell populations, we successfully established a robust in vitro assay for the detection of SARS-CoV-2-specific Tfh cells. In this assay, the stimulation with a SARS-CoV-2 peptide pool, aided by CD28 costimulation, was able to induce a detectable production of IL-21 (a canonical Tfh cytokine) by CXCR5+PD-1+ Tfh cells, measured via flow cytometry intracellular staining (ICS) (Figures 5G, 5H, and S5F). Under these conditions, we efficiently identified SARS-CoV-2-specific Tfh cells producing IL-21 after peptide stimulation only in SARS-CoV-2-mRNA-immunized mice, but not Luc-mRNA-immunized mice (Figures 5G and 5H). By comparison, rRBD-AddaVax immunization resulted in considerably weaker IL-21+ Tfh responses, with many mice indistinguishable from control Luc-mRNA-immunized mice (Figures 5G and 5H).
Collectively, our study directly shows that, even when analyzed in an antigen-specific fashion, SARS-CoV-2-mRNA-immunized mice presented a more robust generation of Tfh cells than did rRBD-AddaVax.
Tfh Cells Induced by SARS-CoV-2 mRNA and rRBD-AddaVax Vaccines Are Endowed with Diverse Functional Profiles and Maturation Status
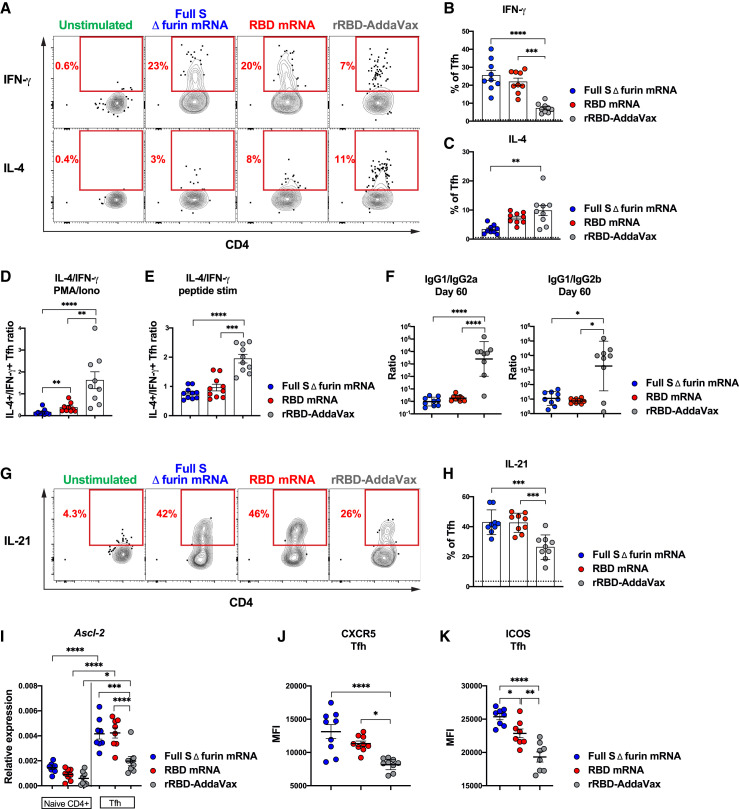
Tfh cells are a functionally heterogeneous population (Morita et al., 2011; Locci et al., 2013; Weinstein et al., 2016). To shed light on the functional characteristics of the Tfh cells induced by the vaccines under investigation, we evaluated the capacity of the mRNA vaccines and rRBD-AddaVax to shape the Tfh cell potential to produce interferon (IFN)-γ and interleukin (IL)-4 upon in vitro stimulation. To this aim, mice were immunized with rRBD-AddaVax or with SARS-CoV-2 mRNA vaccines. Seven days after immunization, lymphocytes from draining LNs were stimulated in vitro with PMA and ionomycin, and IFN-γ and IL-4 production by Tfh cells was measured by ICS (Figures 6 A–6C and S6A). This analysis revealed a clear skew toward a Th2 functional profile of the Tfh cells induced by rRBD-AddaVax, characterized by elevated IL-4 production along with a significantly reduced IFN-γ production in comparison to the Tfh cells induced by mRNA vaccines (Figures 6A–6C). The Th2 bias of the rRBD-AddaVax-driven Tfh cells was also indicated by the higher Th2/Th1 cytokine ratio upon PMA/ionomycin (Figure 6D) and SARS-CoV-2 peptide pool (Figures 6E and S6B–S6E) in vitro stimulations in comparison to the Tfh cells induced by mRNA vaccines. RBD mRNA vaccines instead induced Tfh cells with a mixed Th1-Th2 functional profile capable of producing IFN-γ and IL-4 (Figures 6A–6E and S6B–S6E). Tfh cells from mice immunized with full S Δ furin mRNA showed a significant trend for a more pronounced Th1 polarization. A similar polarization was found also in C57BL/6 mice (Figures S6F–S6H), which also presented more robust induction of GC B cells, RBD-specific GC B cells, and Tfh cells in response to RBD mRNA vaccine than in response to rRBD-AddaVax (Figures S6I–S6K).
Figure 6.
SARS-CoV-2 mRNA Vaccines Drive High Expression of Tfh Signature Molecules
Mice were i.m. immunized with SARS-CoV-2 mRNA vaccines or rRBD-AddaVax. For (A)–(D), (G), (H), and (J), inguinal LNs and pooled inguinal and popliteal LNs ([E], [I], and [K]) were analyzed. In (A)–(E) and (G)–(K), data were analyzed at 7 days and in (F) at 60 days post-immunization.
(A) Representative analysis of IFN-γ+ and IL-4+ Tfh cells (PD-1+CXCR5+) measured by ICS after PMA/ionomycin stimulation. Cells were pre-gated on live CD4+B220−CD44hi populations.
(B) Frequency of IFN-γ+ Tfh cells as described in (A). In (B), (C), and (H), dotted line represents an average of 35 unstimulated samples.
(C) Frequency of IL-4+ Tfh cells as described in (A).
(D and E) Ratio of IL-4+/IFN-γ+ Tfh cells measured after (D) PMA/ionomycin stimulation or (E) SARS-CoV-2 peptide stimulation. Data used for (E) are detailed in Figure S6.
(F) Ratio of IgG1/IgG2a (left) and IgG1/IgG2b titers (right). Data used for this ratio is detailed in Figure 4.
(G) Representative analysis of IL-21+ Tfh cells (PD-1+CXCR5+) measured by ICS after PMA/ionomycin stimulation. Cells were pre-gated on live CD4+B220−CD44hi populations.
(H) Frequency of IL-21+ Tfh cells as described in (G).
(I) Relative expression of Ascl-2 on purified naive CD4 and Tfh cells (PD-1+CXCR5+) measured by qPCR.
(J and K) Mean fluorescence intensity (MFI) of (J) CXCR5 or (K) ICOS on Tfh cells (CXCR5+Bcl-6+).
In (A)–(D) (F), (G), (H), and (J), n = 9 mice per group were analyzed. Data were combined from three independent experiments. In (I) and (K), n = 8 mice per group were analyzed. Data were combined from two independent experiments. In (B)–(E) and (H)–(K), mean ± SEM is shown. In (F), ratios of Ab titers are shown as geometric mean ± geometric SD. Each data point represents an individual mouse. One-way-ANOVA with Bonferroni correction or unpaired two-tailed Mann-Whitney U tests were conducted according to the distribution of the data. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001 See also Figure S6 and Table S1.
To better understand the biological consequences of the observed Tfh functional polarization, we calculated the ratio of IgG1 to IgG2a and IgG1 to IgG2b SARS-CoV-2-specific Abs in the sera of mice immunized 60 days earlier with rRBD-AddaVax or the SARS-CoV-2 mRNA vaccines (Figure 6F). In mice, Th2-biased responses are associated with IgG1 production. Conversely, Th1 polarized responses are linked to IgG2 Ab production (Reinhardt et al., 2009; Snapper and Paul, 1987; Stevens et al., 1988). In agreement with the functional polarization of Tfh cells, we found higher IgG1/IgG2 Ab ratios in mice immunized with rRBD-AddaVax in comparison to the mRNA vaccines (Figure 6F).
To gain insights on the mechanisms by which mRNA vaccines foster Tfh cell responses, we evaluated the expression of distinctive transcription factors and key molecules associated with the follicular program and function of Tfh cells. We evaluated IL-21+ Tfh cells by ICS after PMA/ionomycin in vitro stimulation. IL-21 is a cytokine produced by Tfh cells that regulates proliferation and differentiation of GC B cells into PCs (Crotty, 2019; Vinuesa et al., 2016). Our analysis revealed a higher frequency of IL-21+ Tfh cells driven by SARS-CoV-2 mRNA vaccine in comparison to rRBD-AddaVax (Figures 6G and 6H). Ascl-2 is a transcription factor (TF) regulating the initiation of Tfh cell development (Liu et al., 2014) and has been shown to promote the expression of CXCR5, a chemokine receptor involved in Tfh homing to B cell follicles (Crotty, 2019; Vinuesa et al., 2016). qPCR analysis of Tfh cells isolated from mice immunized 7 days earlier with SARS-CoV-2 mRNA vaccines revealed a higher expression of Ascl-2 in comparison to Tfh cells from rRBD-AddaVax-immunized mice (Figure 6I). Consistent with the heightened expression of Ascl-2, CXCR5 was expressed at higher levels in Tfh cells from SARS-CoV-2-mRNA-immunized mice when compared to Tfh cells induced by rRBD-AddaVax (Figure 6J). Differently, the Tfh signature TF Bcl-6 (Crotty, 2019; Vinuesa et al., 2016), which is not influenced by Ascl-2 (Liu et al., 2014), was expressed at comparable levels on the Tfh cells driven by the two different vaccine platforms (Figure S6L). The co-stimulatory molecule ICOS plays a central role in establishing the Tfh program and serves as migration receptor to promote the follicular homing of Tfh cells (Crotty, 2019; Xu et al., 2013). At the protein level, ICOS expression was enhanced in Tfh cells of mRNA-vaccinated mice (Figure 6K) and positively correlated with the absolute numbers of vaccine elicited Tfh cells (Figure S6M).
Overall, SARS-CoV-2 mRNA vaccines appear to mediate superior Tfh cell responses when compared to rRDB-AddaVax by modulating molecules that are important for the initiation of the Tfh gene program as well as for the follicular homing and B cell helper function of Tfh cells.
A Booster Immunization with SARS-CoV-2 mRNA Induces a Second Wave of GC Responses and Further Enhances nAb Production
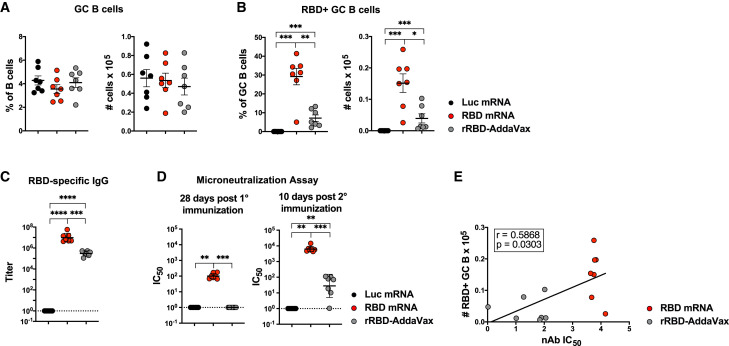
Since repeated immunizations can enhance antibody responses, we sought to investigate secondary GC formation and SARS-CoV-2 RBD-specific Ab responses after a booster immunization. To this aim, we immunized mice twice, 28 days apart, with RBD or Luc mRNA, or rRBD-AddaVax. At the time of the boost, primary GC response has almost completely waned (Figure 2). We investigated secondary GC B cell responses in the draining LNs at 10 days post-boost, a time point chosen based on published prime-boost GC studies with MF59 (Lofano et al., 2015). Differently from the primary response, we detected a robust induction of GC B cells in all groups (Figure 7 A), including rRBD-AddaVax-immunized mice, upon the second immunization. In line with the GC B cell findings, Tfh cell responses were not statistically different across the immunization groups (Figure S7A). Of note, despite the similar magnitude of GC formation, mice immunized with rRBD-AddaVax failed to mount robust RBD-specific GC B cell responses 10 days post-boost (Figure 7B), as opposed to the relevant formation of SARS-CoV-2-specific GC B cells in the RBD mRNA group.
Figure 7.
A Booster Immunization with a SARS-CoV-2 mRNA Vaccine Induces Robust Secondary GCs and nAbs
Mice were i.m. immunized twice, 28 days apart, with Luc mRNA, RBD mRNA, or rRBD-AddaVax. Serum was collected before boost and 10 days post-boost. Inguinal LNs were analyzed 10 days after the second immunization.
(A and B) Frequencies (left) and absolute numbers (right) of (A) total GC B cells defined as live dump− CD19+Fas+GL7+ and (B) RBD-specific GC B cells defined as live dump− CD19+Fas+GL7+RBD-PE+RBD-AF647+ cells.
(C) RBD-specific IgG titers measured by ELISA.
(D) nAbs measured by microneutralization assays at 28 days post-first (left) and post-second (right) immunization.
(E) Spearman correlation of RBD-specific GC B cells and nAbs at day 10 post-boost.
In (A)–(C), n = 7 mice per group were analyzed. In (D) and (E), n = 6 mice per group for Luc mRNA and n = 7 mice per group for all other groups were analyzed. Data are combined from two independent experiments. In (A) and (B), data were graphed as mean ± SEM. In (C) and (D), geometric mean ± geometric SD is shown. In (A)–(E), each data point represents an individual mouse. One-way-ANOVA with Bonferroni correction or unpaired two-tailed Mann-Whitney U tests were conducted according to the distribution of the data. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001. See also Figure S7.
We next questioned whether the difference in the magnitude of secondary RBD-specific GC responses between groups was paralleled by quantitative and qualitative differences in Ab responses post-boost. Although both rRBD-AddaVax and RBD mRNA groups had detectable levels of RBD-specific IgG titers after the second immunization, the RBD mRNA group had a superior induction of RBD-specific IgG in comparison with the rRBD-AddaVax group (Figure 7C). Next, we measured nAbs by SARS-CoV-2 microneutralization assays before and after the booster immunization. In line with the data shown in Figure 4, only RBD mRNA induced substantial nAb responses 4 weeks after a single immunization (Figure 7D). Differently, after a booster immunization both RBD-mRNA- and rRBD-AddaVax-immunized animals were able to produce detectable nAb levels. There was, however, a marked difference in the magnitude of nAb responses that mirrored RBD-specific GC B cell responses, with the group of animals immunized with RBD mRNA presenting nAb responses two-logs higher than the rRBD-AddaVax group (Figure 7D). Consistent with these findings, secondary RBD-specific GC B cell responses correlated with nAb responses at day 10 post-boost (Figure 7E). No correlation was observed between total numbers of GC B cells and nAbs upon secondary immunization (Figure S7B).
Altogether, these data indicate that rRBD-AddaVax induces negligible GC formation and nAb production after a single-dose immunization, along with larger but mostly non-RBD-specific GC B cell responses that are associated with low levels of nAb production after a booster immunization. Conversely, SARS-CoV-2 mRNA vaccines elicit powerful SARS-CoV-2-specific GC responses as well as a robust nAb production that can be further enhanced by a booster immunization.
DISCUSSION
Recent studies have revealed that multiple SARS-CoV-2 vaccine approaches are capable, to varying extent, of mediating the production of nAbs that can neutralize SARS-CoV-2 in vivo and/or in vitro (Anderson et al., 2020; Corbett et al., 2020b; 2020a; Gao et al., 2020; Jackson et al., 2020; Laczkó et al., 2020; Mulligan et al., 2020a; Sahin et al., 2020; Smith et al., 2020; Yu et al., 2020; Zhang et al., 2020). Outstanding questions were left unanswered thus far in the SARS-CoV-2 vaccine field: what is a good metric to predict nAb formation? Are GCs important for nAb formation? And how do different vaccine platforms compare to each other? Our study, built on a systematic comparison between two vaccine platforms, nucleoside-modified mRNA-LNP and recombinant protein formulated with the MF59-like adjuvant AddaVax (rRBD-AddaVax), evaluated quantitatively and qualitatively the GC responses to the novel virus SARS-CoV-2 upon immunization. We found that SARS-CoV-2 mRNA vaccines had a superior capacity, in comparison to rRBD-AddaVax, to elicit potent SARS-CoV-2-specific GC B cell responses after the administration of a single vaccine dose. Importantly, we demonstrate here that GC B cells and Tfh cells strongly correlated with the production of nAbs. In fact, mice immunized with SARS-CoV-2 mRNA vaccines had robust GC responses coupled with in vitro neutralization of SARS-CoV2 virus. Conversely, a single immunization with rRBD-AddaVax resulted in minimal GC responses and a lack of nAb production over time. The notion that effective nAb formation deeply relies on GC reactions has been widely studied in the HIV field (Havenar-Daughton et al., 2017). It is well established that the generation of broadly neutralizing Abs (bnAbs) capable of neutralizing most HIV strains depend on a hardcore GC-dependent affinity maturation process lasting several years. The generation of HIV bnAbs is an extreme example for many reasons, including the elevated rate of mutation of HIV that forces bnAbs to acquire an unusually high degree of SHM and to target less immunodominant-conserved epitopes (Havenar-Daughton et al., 2017). Many viruses do not require such drastic GC responses to confer protection. Nevertheless, studies on other viruses such as influenza virus revealed that most human Abs to influenza are heavily mutated, and these mutations are likely critical to mediate broad protection against the virus (Victora and Wilson, 2015). Hence, it is not unexpected to observe here a strong connection between GC responses, which harbor SHM and the affinity maturation process, and the capacity of the SARS-CoV-2 mRNA vaccines to foster a high level of nAbs after a single immunization. Future studies will be required to establish the degree of SHM and affinity maturation resulting from immunization with SARS-CoV-2 mRNA vaccines and how SHM translates to in vivo protection.
The correlation between nAbs and Tfh cells after a single immunization also hints at the importance of T cell help for the generation of protective SARS-CoV-2-specific Ab responses. This finding is in line with recently reported correlations of antigen-specific CD4 T cells (Grifoni et al., 2020) and circulating Tfh cells (Mathew et al., 2020) with SARS-CoV-2-specific Abs in COVID-19 donors. Of note, increased frequencies of Th1- and Th2-biased circulating Tfh cells were found in humans to be associated with the highest plasma-neutralizing activity in COVID-19 patients (Juno et al., 2020). This is an important finding that strengthens the idea that the functional profile of Tfh cells might be relevant in shaping the quality of neutralizing response to SARS-CoV-2. Our study demonstrated that the bona fide Tfh cells induced by SARS-CoV-2 mRNA vaccines had a mixed Th1-Th2 functional profile (especially clear in RBD-mRNA-immunized BALB/c mice) that was characterized by the production of IFN-γ and IL-4. This is opposed to the Th2-skewed Tfh cell profile triggered by rRBD-AddaVax. In agreement with these data, we found that the rRBD-AddaVax vaccine favors Th2-type Ab responses (high RBD-IgG1 titers, low RBD-IgG2 titers), whereas mRNA vaccines are superior at molding mixed Th1-Th2 type Ab responses (high RBD-IgG1 and RBD-IgG2 titers). Multiple studies have demonstrated that mRNA vaccines for SARS-CoV-2 trigger CD4 T cells responses that are dominated by Th1 cytokine production (Corbett et al., 2020b; 2020a; Laczkó et al., 2020; Zhang et al., 2020). In light of the importance of IL-4 in maintaining the survival of GC B cells and regulating GC reactions (Crotty, 2019; Vinuesa et al., 2016), it is expected that Tfh cells, differently from conventional CD4 T cells, might exhibit some degree of IL-4 production even in a Th1-inducing milieu. This differential behavior of conventional CD4 T cells and Tfh cells can be explained by alternative transcriptional requirement for the regulation of IL-4 production in these different cell types. Indeed, Gata3 regulates IL-4 production in conventional CD4 T cell, whereas Tfh cell biology does not depend on Gata3 (Nurieva et al., 2008), and the SLAM-Sap pathway is instead responsible for the modulation of IL-4 production by Tfh cells (Yusuf et al., 2010). Overall, the fact that mRNA vaccines generate Tfh cells with a mixed Th1/Th2 profile along with Th1-biased conventional CD4 T cells might be a desirable feature to potentially avoid vaccine-associated enhanced respiratory disease (VAERD) (Graham, 2020). In fact, VAERD has been connected to immune responses skewed toward a Th2 profile in other preclinical and clinical studies on different respiratory viruses.
At least two reasons could explain the superior ability of SARS-CoV-2 mRNA vaccines to promote GC responses. First, mRNA-encoding SARS-CoV-2 antigens are translated into proteins directly in the host cells (Pardi et al., 2018b), and it was shown for the Luc mRNA control vaccine that Luciferase expression is detectable for at least 10 days after injection (Pardi et al., 2018a; 2015). This could lead to a more prolonged antigen availability along with continuous presentation of antigens via major histocompatibility complex class II molecules in comparison to the administration of a recombinant protein, which in turn could foster greater GC responses. In keeping with this hypothesis, prolonged antigen administration via osmotic pumps results in improved GC B cell and Tfh generation in non-human primate and murine HIV vaccine models (Cirelli et al., 2019; Tam et al., 2016). It is worth highlighting that a possible extended protein production driven by mRNA vaccines did not cause the formation of overt overextended GC reactions. Our analysis of GC responses over time demonstrated a deep quantitative reduction of Tfh and GC B cells after 4 weeks post-immunization. However, it is interesting to notice that the frequency of SARS-CoV-2-specific GC B cells was still elevated at this time point, possibly suggesting that low-level GC activity might still be occurring and contributing to the elevated SARS-CoV-2-specific IgG titers and nAbs found 60 days post-immunization. In various experimental models, sensitive techniques have indeed shown that antigen-specific GCs might be present for a prolonged time and that late GC responses could contribute to the clonal diversity of B cell responses (Allen et al., 2007). A second less-explored alternative/complementary explanation for the superior GC activity of mRNA vaccines could be that a component of this vaccine platform is endowed with an intrinsic Tfh cell adjuvanticity and/or is more efficient at priming naive CD4 T cells.
Overall, our study provides a detailed quantitative and qualitative overview of the GC responses induced by two types of SARS-CoV-2 vaccines and uncovers a strong connection between SARS-CoV-2 nAb generation and SARS-CoV-2-specific GC reactions. With multiple vaccines currently being considered as candidates to fight the COVID-19 pandemic, this study sets a benchmark that can improve the evaluation of the immune responses induced by SARS-CoV-2 vaccine candidates in future pre-clinical and clinical studies.
Limitations of Study
Although the points discussed above could explain the different performance of the two vaccine platforms in regulating GC primary responses upon a single immunization, we do not rule out the possibility that sequential doses of booster immunizations with rRBD-AddaVax might give rise to improved GC responses and nAb production, as indicated by our analysis of secondary GC B cell and Tfh cell responses at 10 days post-boost. A late nAb response to rRBD-AddaVax following booster immunizations, which might be suggested by the observed delayed kinetics of GC B and Tfh cells after a single immunization, could also be plausible and cannot be excluded by our investigation. Additionally, as suggested by a recently reported Phase 1 clinical trial (Keech et al., 2020), the usage of SARS-CoV-2 full S as immunogen, which is a much longer protein containing a higher number of epitopes, combined with stronger adjuvants such as Matrix-M1, might likely lead to superior GC responses and nAb formation in comparison to rRBD-AddaVax, even during primary responses. Hence, it is possible that repeated immunizations with different recombinant SARS-CoV-2 protein/adjuvant combinations might represent a successful approach to elicit GC responses and nAbs in humans. Future studies will be needed to address all the points discussed above.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-mouse CD16/CD32 Purified | BioXCell | Cat#BE0307; RRID: AB_2736987 |
| anti-mouse CD28 purified | Tonobo | Cat#40-0281-M001; RRID: AB_2621445 |
| anti-mouse Bcl-6 AF 647 | BD Biosciences | Cat#561525; RRID: AB_10898007 |
| anti-mouse CCR6 BV785 | Biolegend | Cat#129823; RRID: AB_2715923 |
| anti-mouse CD3 AF 594 | Biolegend | Cat#100240; RRID: AB_2563427 |
| anti-mouse CD3 APC-Fire750 | Biolegend | Cat#100248; RRID: AB_2572118 |
| anti-mouse CD4 PerCP-Cy5.5 | Biolegend | Cat#100540; RRID: AB_893326 |
| anti-mouse CD19 BV605 | Biolegend | Cat#115540; RRID: AB_2563067 |
| anti-mouse CD21/CD35 (CR2/CR1) BV421 | Biolegend | Cat#123422; RRID: AB_2650891 |
| anti-mouse CD38 PE-Cy7 | Biolegend | Cat#102718; RRID: AB_2275531 |
| anti-mouse CD44 BV605 | Biolegend | Cat#103047; RRID: AB_2562451 |
| anti-mouse CD44 FITC | Biolegend | Cat#103006; RRID: AB_312957 |
| anti-mouse/human CD45R/B220 BV650 | Biolegend | Cat#103241; RRID: AB_11204069 |
| anti-mouse/human CD45R/B220 AF 700 | eBioScience | Cat#56-0452-82; RRID: AB_891458 |
| anti-mouse CD62L BUV395 | BD Biosciences | Cat#740218; RRID: AB_2739966 |
| anti-mouse CD86 BV421 | Biolegend | Cat#105032; RRID: AB_2650895 |
| anti-mouse CD138 BV650 | BD Biosciences | Cat#564068; RRID: AB_2738574 |
| anti-human/mouse/rat CD278 (ICOS) AF 488 | Biolegend | Cat#313514; RRID: AB_2122584 |
| anti-mouse CXCR4 Biotin | eBioScience | Cat#13-9991-82; RRID: AB_10609202 |
| anti-mouse CXCR5 Biotin | eBioScience | Cat#13-7185-82; RRID: AB_2572800 |
| anti-mouse CXCR5 BV421 | Biolegend | Cat#145512; RRID: AB_2562128 |
| anti-mouse FAS BV510 | BD Biosciences | Cat#563646; RRID: AB_2738345 |
| anti-mouse IgD PE-Cy7 | Biolegend | Cat#405720; RRID: AB_2561876 |
| Anti-mouse IgD AF 647 | Biolegend | Cat#405708; RRID: AB_893528 |
| anti-mouse IgG1 V450 | BD Bioscience | Cat#562107; RRID: AB_10894002 |
| anti-mouse IgG2a/2b BB700 | BD Bioscience | Cat#745969; RRID: AB_2743378 |
| anti-mouse IgM FITC | Jackson ImmunoResearch | Cat#115-095-020; RRID: AB_2338592 |
| anti-mouse IFN-g BV650 | Biolegend | Cat#505832; RRID: AB_2734492 |
| anti-mouse IL-4 AF 647 | Biolegend | Cat#504110; RRID: AB_493322 |
| anti-mouse GL7 PerCP-Cy5.5 | Biolegend | Cat#144610; RRID: AB_2562979 |
| anti-mouse PD-1 PE-Cy7 | Biolegend | Cat#109110; RRID: AB_572017 |
| anti-mouse Ter119 APC-Fire750 | Biolegend | Cat#116250; RRID: AB_2819833 |
| Goat anti-Human IgG Fc Secondary Antibody, PE | eBioScience | Cat#12-4998-82; RRID: AB_465926 |
| goat anti-mouse IgG-HRP | Jackson ImmunoResearch | Cat#115-035-003; RRID: AB_10015289 |
| rat anti-mouse IgG1-HRP | Southern Biotech | Cat#1144-05; RRID: AB_2734757 |
| rat anti-mouse IgG2a-HRP | Southern Biotech | Cat#1155-05; RRID: AB_2794648 |
| rat anti-mouse IgG2b-HRP | Southern Biotech | Cat#1186-05; RRID: AB_2794687 |
| anti-VSV Indiana G | Absolute Antibody | Cat#Ab01401-2.0; RRID: AB_2883992 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| ACK Lysing Buffer | Lonza | Cat#10-548E |
| Cytofix/Cytoperm | BD Biosciences | Cat#554714; RRID: AB_2869008 |
| ELISPOT AEC Substrate Set | BD Biosciences | Cat#551951; RRID: AB_2868954 |
| Expifectamine 293 transfection kit | Gibco | Cat#A14525 |
| Fixable Viability Dye eFluor780 | eBioScience | Cat#115-095-020 |
| GolgiPlug | BD Biosciences | Cat#555029; RRID: AB_2869014 |
| Lightning-Link(R) Phycoerythrin | Expedeon | Cat#703-0010 |
| Lightning-Link(R) Alexa Fluor 647 | Expedeon | Cat#336-0005 |
| Pierce TMB Substrate Kit | Thermo Scientific | Cat#34021 |
| Recombinant Mouse IL-21R Fc Chimera Protein | R&D Systems | Cat#596-MR-100 |
| Recombinant RBD and and Full Spike | Florian Krammer | N/A |
| SARS-CoV-2 spike peptide pools | JPT | Cat#PM-WCPV-S |
| Streptavidin-BV421 | Biolegend | Cat#405225 |
| Streptavidin-AF488 | Biolegend | Cat#405235 |
| Experimental Models: Cell Lines | ||
| HEK293T | ATCC | Cat#CRL11268; RRID: CVCL_1926 |
| FreeStyle 293 (293F) cells | Gibco | Cat#R79007 |
| Experimental Models: Organisms/Strains | ||
| Balb/c mouse | The Jackson Laboratory | Stock#000651 |
| C57BL/6J mouse | The Jackson Laboratory | Stock#000664 |
| Balb/c mouse | Charles River Laboratories | N/A |
| Experimental Models: Bacterial and Virus Strains | ||
| SARS-CoV-2 (USA/WA1/2020) | BEI Resources | NR-52281 |
| Oligonucleotides | ||
| Actb | Integrated DNA Technologies | Forward 5’- GAGGTATCCTGACCCTGA AGTAReverse 5’- GCTCGAAGTCTAGAG CAACATAG; |
| Ascl2 | Integrated DNA Technologies | Forward 5’-GAGAGCTAAGCCCGATG GAReverse 5’-AGGTCCACCAGGAGT CACC |
| Software and Algorithms | ||
| FlowJo v10 | FlowJo LLC | N/A |
| S6 Fluorospot Analyzer | CTL | N/A |
| Graphpad Prism v8 | Graphpad | N/A |
| Volocity 6.3 | Perkin Elmer | N/A |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Michela Locci (michela.locci@pennmedicine.upenn.edu).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
This study did not generate/analyze datasets/code. Supplementary material can be located at: https://doi.org/10.17632/96m659kfrp.1.
Experimental Model and Subject Details
Mice
BALB/cJ or C57BL/6J mice were purchased from The Jackson Laboratories or Charles River and housed in an Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC)-accredited facility. Mice between 6-8-weeks of age were used for experiments. All studies were conducted under protocols approved by the University of Pennsylvania’s Institutional Animal Care and Use Committee (IACUC).
Method Details
Immunogen preparation and immunization
SARS-CoV-2 RBD protein (rRBD) was produced in 293F cells, as described previously (Amanat et al., 2020; Stadlbauer et al., 2020). Briefly, 600 million cells were transfected with 200 μg of purified DNA encoding codon-optimized RBD of SARS-CoV-2 using ExpiFectamine 293 transfection kit (Gibco, A14525). The manufacturer’s protocol was followed and cells were harvest on day 3. Cells were spun at 4000 x g for 10 minutes and sterile-filtered with a 0.22 μm filter. Supernatant was incubated with Ni-NTA resin (Qiagen, 30230) for 2 hours. Then, this mixture was loaded onto columns and the protein was eluted using elution buffer with high amounts of imidazole. Protein was concentrated using 10 kDa Amicon centrifugal units (Millipore Sigma cat#UFC901024) and re-constituted in PBS. Concentration was measured using Bradford reagent (Bio-Rad, 5000201) and a reducing sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) was run to check the integrity of the protein. SARS-CoV-2 full length S (full S) protein was prepared as described above for SARS-CoV-2 rRBD.
Full length spike Δ furin (full S Δfurin, RRAR furin cleavage site abolished) and RBD mRNA vaccines were designed based on the full spike (S) protein sequence of SARS-CoV-2 (Wuhan-Hu-1, GenBank: MN908947.3) (Laczkó et al., 2020). mRNA coding sequences of RBD, full S Δfurin and firefly luciferase (Luc) were codon-optimized, synthesized, produced and encapsulated in lipid nanoparticles (LNP) as previously reported (Freyn et al., 2020).
rRBD was diluted in phosphate buffered saline (PBS, Corning, 21-031-CV) and combined in a 1:1 ratio with AddaVax (InVivoGen, vac-adx-10). mRNA-LNP were diluted in PBS prior to inoculation. Vaccines were injected into the gastrocnemius muscle using a 0.5 mL 28 G x 1/2” insulin syringe (BD Biosciences, 329461). For booster immunizations, the same dose of the respective vaccine was injected into the same site as the primary immunization.
Sample collection and processing
Sample collection: Seven to sixty days post-immunization, mice were anesthetized with isoflurane and blood was collected from the orbital sinus using capillary tubes without anticoagulant for sera separation. Inguinal and popliteal lymph nodes (LNs), spleens and both tibias and femurs were harvested and placed in cold complete Dulbecco’s Modified Eagle’s Medium [(DMEM, Corning, T10014CV) containing 10% heat inactivated Fetal Bovine Serum (FBS, Corning, 35-015), 1% Glutamax (Gibco, 35050-061) and 1% Penicillin/Streptomycin (Gibco, 5070063)]. All organs were kept on ice and immediately processed after collection.
LNs and spleen processing: Organs were homogenized with a syringe plunger and filtered through a 40 μm cell strainer on ice. Red blood cells (RBCs) of spleens were lysed using ACK lysing buffer (Lonza, 10-548E) for 5-8 minutes on ice and the reaction was stopped with cold PBS.
Bone Marrow (BM) processing: BM was flushed from both femurs and tibias from each mouse using a 1 mL 25 G x 5/8” syringe (BD Biosciences, 30962). RBCs were lysed as described above.
Cells from all tissues were resuspended in ice cold complete DMEM and immediately used for counting, culture, or staining.
Serum processing. Following collection, blood was centrifuged at 14,000 x g (maximum speed) for 30 minutes, 4°C. Serum was recovered and stored at -20°C for neutralization assays and ELISAs.
SARS-CoV-2 fluorescent probe generation
rRBD or full S proteins used for flow cytometry experiments were independently conjugated to both R-PE and Alexa Fluor 647 using Lightning-Link® R-Phycoerythrin (R-PE) (Expedeon, 336-0005) and Lightning-Link (R) Rapid Alexa Fluor 647 (Expedeon, 703-0010) kits, following manufacturer’s instructions.
Immunophenotyping by flow cytometry
All staining steps were carried out at 4°C in FACS buffer (PBS with 2% heat inactivated FBS). Single cell suspensions were Fc blocked with anti-CD16/CD32 monoclonal antibody (mAb) prior to staining.
Tfh cell: Cells were incubated with CXCR5-biotin for 1 hour, washed and then incubated with a cocktail of fluorescently labeled anti-mouse mAbs, streptavidin, and Fixable Viability dye eFluor780 for 30 minutes. Cells were washed with FACS buffer, then fixed and permeabilized in FoxP3/Transcription Factor Staining Buffer Set (eBioScience, 00-5523-00) according to manufacturer’s instructions before intranuclear staining with mAbs (Table S1) for 30 minutes.
GC B cell: Cells were incubated with anti-CXCR4-biotin for 1 hour and subsequently washed. Then, they were stained with a cocktail of fluorescently labeled anti-mouse mAbs containing streptavidin, Fixable Viability dye eFluor780, RBD- or full S-PE, and RBD- or full S-Alexa Fluor 647 (Table S2) for 30 minutes. Excess antibodies were washed away, and cells were fixed with 1% PFA for 30 minutes prior to acquisition.
Memory B cell: Cells were stained with a cocktail of fluorescently labeled anti-mouse mAbs containing Fixable Viability Dye eFluor780, RBD- or full S-PE, and RBD- or full S-Alexa Fluor 647 (Table S3) for 30 minutes. Cells were washed and fixed with 1% paraformaldehyde (PFA) for 30 minutes prior to acquisition.
All samples were acquired on a 5 laser Cytoflex LX (Beckman Coulter) or Aurora (Cytek) and data analyzed in FlowJo v10 (Treestar).
Tfh cell intracellular cytokine assay
Antigen-specific stimulation: one million cells from LNs (pool of inguinal and popliteal) were stimulated for 6 hours with 1.5 μg/mL SARS-CoV-2 spike peptide pool (JPT PM-WCPV-S1, N-terminal dissolved in DMSO) and 2.5 μg/mL anti-CD28 mAb (Tonbo #40-0281-M001) in a U-bottom plate at 37°C, 5% CO2. Cultures were performed in complete Iscove’s Modified Dulbecco’s Medium [(IMDM, Gibco, 12440-053) containing 10% heat inactivated FBS, 1% Penicillin/Streptomycin, 2 mM L-glutamine, 1 mM sodium pyruvate (Gibco, 11360-070), and 55 μM 2-mercaptoethanol (Gibco, 21985-023)] in the presence of GolgiPlug (BD Biosciences, 555029). Cells incubated with DMSO for 6 hours served as a negative control and cells incubated with 50 ng/mL phorbol 12-myristate 13-acetate (PMA, Sigma Aldrich, P1585) and 1 μg/mL ionomycin (Sigma Aldrich, I0634) for 5 hours were used as a positive control. Flow cytometry staining is described in detail below.
PMA/onomycin stimulation: cells from inguinal LNs were stimulated and stained as described previously (Gowthaman et al., 2019) with minor modifications. Briefly, 0.5-1 million cells were incubated for 2 hours at 37°C in complete IMDM with 50 ng/mL PMA and 1 μg/mL ionomycin. After 2 hours, GolgiPlug was added to the culture at a final concentration of 1:1,000 and cells were further incubated for 3 additional hours. Unstimulated cell controls were cultured without PMA and ionomycin in the presence of GolgiPlug.
Upon stimulation, cells were washed with cold PBS and kept at 4°C for the entirety of the staining process. Following Fc block for 20 minutes and the surface staining with PD-1 PE-Cy7 and Fixable Viability Dye eFluor780 for 25 minutes, cells were fixed and permeabilized with Cytofix/Cytoperm - Fixation/Permeabilization Solution Kit (BD Biosciences, 554715), according to manufacturer’s instructions. Cells were incubated with IL-21R-Fc chimera protein (R&D Systems, 596MR100) for 30 minutes in permeabilization solution, washed and then incubated with anti-human IgG Fc PE mAb (Invitrogen, 12499882) for 20 minutes. Samples were extensively washed and incubated overnight (ON) with permeabilization solution containing the mAb cocktail described in Table S4. The following day, samples were washed and acquired on a FACSymphony A3 or LSRFortessa (BD Biosciences).
Immunofluorescence stain and microscopy
Inguinal LN were harvested into PBS and fixed with 4% PFA plus 10% Sucrose for 30 minutes at room temperature (RT). Following three washes with PBS, samples were equilibrated in 30% sucrose for 1 hour to ON. Samples were washed three times with PBS and snap frozen in cryomolds using Optimal Cutting Temperature compound (OCT, Fisher Healthcare 23-730-571) in a dry ice-cooled bath of 2-methylbutane. 10 μm tissue sections were blocked for 30 minutes at room temperature in staining solution (5% donkey serum in PBS-Tween) in the presence of FcR block reagent, and subsequently stained ON at 4°C in staining solution with the mAb cocktail described in Table S5. After washing in PBS, slides were mounted using ProLong™Diamond antifade (Invitrogen), and imaged using a Zeiss LSM710 confocal microscope. Tile scan images were taken using a 10X magnification lens with 10% overlap. Enlarged images were taken using a 20X magnification lens. Images were processed and analyzed using Zeiss Blue 3.1 (Zeiss) and Volocity 6.3 (Perkin Elmer) software.
ELISAs
Nunc Maxisorp flat-bottom 96 well plates (Invitrogen, 44-2404-21) were coated with 1 μg/mL SARS-CoV-2 rRBD protein in bicarbonate buffer ON at 4°C. Plates were washed four times with wash buffer [0.05% Tween-20 (Sigma Aldrich, P1379) in PBS] and blocked with 2% bovine serum albumin (BSA, Fisher BioReagents, Lot 170763A) in PBS for 1 hour at RT. Serum samples were serially diluted in blocking buffer and incubated for 2 hours at RT followed by five washes. HRP-conjugated IgG1 (Southern Biotech, 1144-05), IgG2a (Southern Biotech, 1155-05), or IgG2b (Southern Biotech, 1186-05) were diluted 1:5,000 in blocking buffer and incubated for 1 hour, then washed seven times. Plates were developed with Pierce TMB Substrate (Thermo Scientific, 34021), and the reaction was stopped with 2 N sulfuric acid. Absorbance was measured at 450 nm using a SpectraMax microplate reader. RBD-endpoint antibody titers were calculated as reciprocal dilutions giving OD signals > average of blanks plus three times the standard deviation using GraphPad Prism. An arbitrary value of 1 was assigned to the samples with OD values below the limit of detection for which it was not possible to interpolate the titer.
ELISPOT
A detailed protocol was previously published (Laczkó et al., 2020). Briefly, multiScreen HTS IP filter plates of 0.45 μm (Millipore Sigma, MSIPS4W10) were coated with 2.5 μg/mL SARS-CoV-2 rRBD or full S proteins in bicarbonate buffer (35 mM NaHCO3 and 15 mM Na2CO3) ON at 4 °C. Plates were washed three times with PBS and blocked with complete DMEM for at least 1 hour at 37 °C. Single cell suspensions of BM cells were serially diluted in complete DMEM with halving dilutions starting at one million cells. Following ON incubation at 37 °C and 5% CO2, plates were washed three times with 0.05% Tween-20 in PBS. Membranes were incubated with anti-IgG-HRP (Jackson ImmunoResearch, 115-035-003) detection antibody diluted in complete DMEM for 2 hours at RT. Plates were washed three times with 0.05% Tween-20 in PBS and spots corresponding to antigen-specific antibody-secreting cells (ASC) were developed using BD ELISPOT AEC Substrate Set (551951). Membranes were dried ON and counted using a CTL Immunospot analyzer (Shaker Heights, OH).
Microneutralization Assay
Vero E6 cells (2 x 104) were added to flat-bottom 96 well tissue culture plates and allowed to adhere ON. Serum samples were heat inactivated and diluted two-fold in virus diluent (MEM containing Earl’s salts and L-glutamine supplemented with 100 U/mL Penicillin, 100 μg/mL Streptomycin, 2% FBS, 1 mM sodium pyruvate, and 1X MEM nonessential amino acids). Assay controls were treated and diluted in the same manner as experimental samples. 100 TCID50 of authentic SARS-CoV-2 (isolate USA-WA1/2020, BEI Resources NR-52281) was added to each sample and incubated for 1 hour. Cells were washed 1 time in PBS. Sample/virus (100 μL) was added to cells and incubated for 4 days. Cells were fixed in 10% Neutral-buffered formalin (VWR 16004-128) and stained with 0.1% Crystal Violet (Sigma C38886-500g). Neutralization is demonstrated by absence of cytopathic effect in Vero E6 monolayers. Data is reported as the geometric mean of the reciprocal dilution factor of two replicates and defined as Minimal effective concentration (MEC). An improved fluorescent assay was performed to generate the neutralization data in Figures 7 and S7 with the following modifications to the original assay. Vero E6 cells (4 x 104) were used to coat the plates. Each serum sample, prepared as described above, was incubated with 100 TCID50 fluorescent SARS-CoV-2 (Xie et al., 2020), strain USA/WA1/2020, for 1 hour at 37°C. Following incubation, the serum/virus solution was added to Vero E6 cells and incubated at 37°C for 24 hours. The cells were fixed using 10% neutral-buffered formalin, and washed with PBS. Fluorescent signal was measured using a BioTek Synergy H1 microplate reader. Percent inhibition was calculated and used to determine IC50 values with non-linear regression analysis tools from GraphPad Prism. In each assay, samples were measured in duplicate.
Pseudovirus Neutralization Assay
Production of VSV pseudotypes with SARS-CoV-2 S: 293T cells plated 24 hours previously at 5 x 106 cells per 10 cm dish were transfected using calcium phosphate with 35 μg of pCG1 SARS-CoV S delta18 expression plasmid encoding a codon optimized SARS-CoV S gene with an 18 residue truncation in the cytoplasmic tail (kindly provided by Stefan Pohlmann). Twelve hours post transfection the cells were fed with fresh DMEM (Corning, 10-013-CV) containing 10% heat inactivated FBS (Corning, 35-010-CV) and 5 mM sodium butyrate (Alfa Aesar, A11079) to increase expression of the transfected DNA. Thirty hours after transfection, the SARS-CoV-2 S expressing cells were infected for 2-4 hours with VSV-G pseudotyped VSVΔG-RFP at a MOI of ∼1-3; cells were washed twice with media to remove unbound virus. Supernatants containing the VSVΔG-RFP SARS-CoV-2 pseudotypes were harvested 28-30 hours after infection and clarified by centrifugation twice at 6000 x g, aliquoted and stored at -80 °C until used for antibody neutralization analysis.
Antibody neutralization assay using VSVΔG-RFP SARS-CoV-2: all mouse sera were heat-inactivated for 30 minutes at 55 °C prior to use in neutralization assay. Vero E6 cells stably expressing TMPRSS2 were seeded in 100 μL at 2 x 104 cells/well in a 96-well collagen coated plate. The next day, 2-fold serially diluted serum samples were mixed with VSVΔG-RFP SARS-CoV-2 pseudotype virus (100-400 focus forming units/well) and incubated for 1 hour at 37 °C. The mouse anti-VSV Indiana G 1E9F9 was also included in this mixture to neutralize any potential VSV-G carryover virus at a concentration of 600 ng/mL (Absolute Antibody, Ab01402-2.0). Cells were infected with the respective serum-virus mixture for 23-24 hours. Then, cells were washed and fixed with 4% PFA before visualization on an S6 FluoroSpot Analyzer (CTL). Individual infected foci were enumerated, and the values compared to control wells without serum. The focus reduction neutralization titer 50% (FRNT50) was measured as the greatest serum dilution at which focus count was reduced by at least 50% relative to control cells that were infected with pseudotype virus in the absence of mouse serum. FRNT50 titers for each sample were measured in at least two technical replicates performed on separate days.
Tfh cell sorting and qPCR
Both inguinal and popliteal LNs were harvested, pooled per mice, and processed as described above. Cells were incubated with anti-mouse CD16/CD32 and CXCR5-biotin antibodies as outlined in the Tfh staining. The following cocktail of anti-mouse mAbs was used to stain the cells and identify Tfh and naive CD4 T cells: B220 Alexa Fluor 700, CD4 PerCP-Cy5.5, CD44 FITC, and PD-1 PE-Cy7; together with Streptavidin BV421 and Fixable Viability dye eFluor780. Cells were washed and resuspended in FACS buffer and naive CD4 T (live B220-CD4+CD44-) and Tfh (live B220-CD4+CD44hiCXCR5+PD-1hi) cell populations were sorted on FACSAria Fusion (BD Biosciences). The purity of the sorted cells was immediately checked using the same instrument and was 97% or higher. Total RNA was extracted with RNeasy plus micro kit (Qiagen, 74034) following manufacturer’s instructions.
Total RNA was retrotranscribed into cDNA with SuperScript II reverse transcriptase (Invitrogen, 18064014). Quantitative real-time PCRs (qPCR) for Ascl2 and Actb were performed using the following primer pairs: Actb: Forward 5’- GAGGTATCCTGACCCTGAAGTA and reverse 5’- GCTCGAAGTCTAGAGCAACATAG; and Ascl2: Forward 5’-GAGAGCTAAGCCCGATGGA and reverse 5’-AGGTCCACCAGGAGTCACC (Zhan et al., 2019). These primers were synthesized by Integrated DNA Technology (IDT, Coralville, IA). qPCRs were set up using SYBR Green PowerUp Master Mix (Applied Biosystems, A25742) and ran on QuantStudio 6 PCR System (Applied Biosystems). Relative mRNA levels were calculated by 2-ΔCt according to Actb gene abundance.
Quantification and Statistical Analysis
GraphPad Prism version 8 was used to conduct all statistical analysis. Shapiro−Wilk and Kolmogorov−Smirnov tests were performed to establish the normal distribution of the data. Significance of differences among groups were calculated according to the distribution of the data using unpaired one-way ANOVA with Bonferroni correction or two-tailed Mann−Whitney U tests. Correlations were determined using the Spearman’s rank coefficient with a 95% confidence interval. Rare samples that presented technical issues in the flow cytometry staining were excluded from the analysis. The precise number of samples analyzed in each graph is reported in figure legends. Statistical significance was set at the critical values of p < 0.05 (∗), p < 0.01 (∗∗), p < 0.001 (∗∗∗), and p < 0.0001 (∗∗∗∗).
Acknowledgements
This work was funded by NIH NIAID grant R01 AI123738 to M.L. and R01 AI091627 to I.M. N.P. was supported by NIH NIAID 1R01AI146101. T.B.M was supported by NIH T32AI007324. P.B. was supported by a Peer Reviewed Medical Research Program award PR182551 and NIH R21AI129531 and R21AI142638. M.J.H. is a Cancer Research Institute Irvington Fellow supported by the Cancer Research Institute. Live SARS-CoV-2 microneutralization assays were performed in the Virology Unit of the Duke Regional Biocontainment Laboratory, which received partial support for construction from the NIH NIAD (UC6AI058607; G.D.S). Graphical abstract was created with BioRender.com. We thank the Cell and Developmental Biology Microcopy core at the University of Pennsylvania for their assistance with microscopy imaging. We thank Dr. Florin Tuluc and Jennifer Murray of the CHOP Flow Cytometry core facility and Dr. Derek D. Jones of the Penn Cytomics and Cell Sorting Resource Laboratory at the University of Pennsylvania for technical assistance. We thank Dr. E. John Wherry and Dr. Shane Crotty for providing valuable scientific feedback on the manuscript.
Author Contributions
K.L., D.C., and M.L. designed and/or performed experiments and analyzed data. D.G.A. and I.M. generated and analyzed microscopy data. T.H.O and G.D.S performed and analyzed microneutralization assays. S.W. performed ELISA. T.B.M., P.C., K.A.L., and P.B. performed and analyzed pseudoneutralization assays. M.J.H and L.C.E. shared expertise and reagents for antigen-specific stimulation. Y.K.T. and S.H.Y.F. provided lipid nanoparticles. F.A. and F.K. provided rRBD and full S proteins. H.M. prepared mRNA-LNP vaccines. N.P. and D.W. designed and provided mRNA vaccines. M.L. wrote the manuscript with help from K.L and D.C. and input from the other authors. M.L. conceived and supervised the study.
Declaration of Interests
In accordance with the University of Pennsylvania policies and procedures and our ethical obligations as researchers, we report that Drew Weissman is named on patents that describe the use of nucleoside-modified mRNA as a platform to deliver therapeutic proteins. Drew Weissman and Norbert Pardi are also named on a patent describing the use of nucleoside-modified mRNA in lipid nanoparticles (LNPs) as a vaccine platform. We have disclosed those interests fully to the University of Pennsylvania, and we have in place an approved plan for managing any potential conflicts arising from licensing of our patents. Ying K. Tam and Steven H.Y. Fan are employees of Acuitas Therapeutics, a company involved in the development of mRNA-LNP therapeutics. Ying Tam is named on patents that describe LNPs for delivery of nucleic acid therapeutics including mRNA and the use of modified mRNA in LNPs as a vaccine platform.
Published: November 21, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.immuni.2020.11.009.
Supplemental Information
References
- Allen C.D.C., Okada T., Cyster J.G. Germinal-center organization and cellular dynamics. Immunity. 2007;27:190–202. doi: 10.1016/j.immuni.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amanat F., Krammer F. SARS-CoV-2 Vaccines: Status Report. Immunity. 2020;52:583–589. doi: 10.1016/j.immuni.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amanat F., Stadlbauer D., Strohmeier S., Nguyen T.H.O., Chromikova V., McMahon M., Jiang K., Arunkumar G.A., Jurczyszak D., Polanco J. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020;26:1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson E.J., Rouphael N.G., Widge A.T., Jackson L.A., Roberts P.C., Makhene M., Chappell J.D., Denison M.R., Stevens L.J., Pruijssers A.J., mRNA-1273 Study Group Safety and Immunogenicity of SARS-CoV-2 mRNA-1273 Vaccine in Older Adults. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2028436. NEJMoa2028436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumjohann D., Okada T., Ansel K.M. Cutting Edge: Distinct waves of BCL6 expression during T follicular helper cell development. J. Immunol. 2011;187:2089–2092. doi: 10.4049/jimmunol.1101393. [DOI] [PubMed] [Google Scholar]
- Botta D., Fuller M.J., Marquez-Lago T.T., Bachus H., Bradley J.E., Weinmann A.S., Zajac A.J., Randall T.D., Lund F.E., León B., Ballesteros-Tato A. Dynamic regulation of T follicular regulatory cell responses by interleukin 2 during influenza infection. Nat. Immunol. 2017;18:1249–1260. doi: 10.1038/ni.3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirelli K.M., Carnathan D.G., Nogal B., Martin J.T., Rodriguez O.L., Upadhyay A.A., Enemuo C.A., Gebru E.H., Choe Y., Viviano F. Slow Delivery Immunization Enhances HIV Neutralizing Antibody and Germinal Center Responses via Modulation of Immunodominance. Cell. 2019;177:1153–1171.e28. doi: 10.1016/j.cell.2019.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett K.S., Edwards D., Leist S.R., Abiona O.M., Boyoglu-Barnum S., Gillespie R.A., Himansu S., Schäfer A., Ziwawo C.T., DiPiazza A.T. SARS-CoV-2 mRNA Vaccine Development Enabled by Prototype Pathogen Preparedness. bioRxiv. 2020 doi: 10.1101/2020.06.11.145920. 2020.06.11.145920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett K.S., Flynn B., Foulds K.E., Francica J.R., Boyoglu-Barnum S., Werner A.P., Flach B., O’Connell S., Bock K.W., Minai M. Evaluation of the mRNA-1273 Vaccine against SARS-CoV-2 in Nonhuman Primates. N. Engl. J. Med. 2020;383:1544–1555. doi: 10.1056/NEJMoa2024671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S. T Follicular Helper Cell Biology: A Decade of Discovery and Diseases. Immunity. 2019;50:1132–1148. doi: 10.1016/j.immuni.2019.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cucinotta D., Vanelli M. WHO Declares COVID-19 a Pandemic. Acta Biomed. 2020;91:157–160. doi: 10.23750/abm.v91i1.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva N.S., Klein U. Dynamics of B cells in germinal centres. Nat. Rev. Immunol. 2015;15:137–148. doi: 10.1038/nri3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freyn A.W., Ramos da Silva J., Rosado V.C., Bliss C.M., Pine M., Mui B.L., Tam Y.K., Madden T.D., de Souza Ferreira L.C., Weissman D. A Multi-Targeting, Nucleoside-Modified mRNA Influenza Virus Vaccine Provides Broad Protection in Mice. Mol. Ther. 2020;28:1569–1584. doi: 10.1016/j.ymthe.2020.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q., Bao L., Mao H., Wang L., Xu K., Yang M., Li Y., Zhu L., Wang N., Lv Z. Development of an inactivated vaccine candidate for SARS-CoV-2. Science. 2020;369:77–81. doi: 10.1126/science.abc1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowthaman U., Chen J.S., Zhang B., Flynn W.F., Lu Y., Song W., Joseph J., Gertie J.A., Xu L., Collet M.A. Identification of a T follicular helper cell subset that drives anaphylactic IgE. Science. 2019;365:eaaw6433. doi: 10.1126/science.aaw6433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham B.S. Rapid COVID-19 vaccine development. Science. 2020;368:945–946. doi: 10.1126/science.abb8923. [DOI] [PubMed] [Google Scholar]
- Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havenar-Daughton C., Lee J.H., Crotty S. Tfh cells and HIV bnAbs, an immunodominance model of the HIV neutralizing antibody generation problem. Immunol. Rev. 2017;275:49–61. doi: 10.1111/imr.12512. [DOI] [PubMed] [Google Scholar]
- Heesters B.A., Myers R.C., Carroll M.C. Follicular dendritic cells: dynamic antigen libraries. Nat. Rev. Immunol. 2014;14:495–504. doi: 10.1038/nri3689. [DOI] [PubMed] [Google Scholar]
- Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N., McCullough M.P., Chappell J.D., Denison M.R., Stevens L.J., mRNA-1273 Study Group An mRNA Vaccine against SARS-CoV-2 - Preliminary Report. N. Engl. J. Med. 2020;383:1920–1931, a2022489. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juno J.A., Tan H.-X., Lee W.S., Reynaldi A., Kelly H.G., Wragg K., Esterbauer R., Kent H.E., Batten C.J., Mordant F.L. Humoral and circulating follicular helper T cell responses in recovered patients with COVID-19. Nat. Med. 2020;26:1428–1434. doi: 10.1038/s41591-020-0995-0. [DOI] [PubMed] [Google Scholar]
- Keech C., Albert G., Cho I., Robertson A., Reed P., Neal S., Plested J.S., Zhu M., Cloney-Clark S., Zhou H. Phase 1-2 Trial of a SARS-CoV-2 Recombinant Spike Protein Nanoparticle Vaccine. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2026920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laczkó D., Hogan M.J., Toulmin S.A., Hicks P., Lederer K., Gaudette B.T., Castaño D., Amanat F., Muramatsu H., Oguin T.H., 3rd A single immunization with nucleoside-modified mRNA vaccines elicits strong cellular and humoral immune responses against SARS-CoV-2 in mice. Immunity. 2020;53:724–732.e7. doi: 10.1016/j.immuni.2020.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang F., Lindgren G., Sandgren K.J., Thompson E.A., Francica J.R., Seubert A., De Gregorio E., Barnett S., O’Hagan D.T., Sullivan N.J. Vaccine priming is restricted to draining lymph nodes and controlled by adjuvant-mediated antigen uptake. Sci. Transl. Med. 2017;9:eaal2094. doi: 10.1126/scitranslmed.aal2094. [DOI] [PubMed] [Google Scholar]
- Lindgren G., Ols S., Liang F., Thompson E.A., Lin A., Hellgren F., Bahl K., John S., Yuzhakov O., Hassett K.J. Induction of Robust B Cell Responses after Influenza mRNA Vaccination Is Accompanied by Circulating Hemagglutinin-Specific ICOS+ PD-1+ CXCR3+ T Follicular Helper Cells. Front. Immunol. 2017;8:1539. doi: 10.3389/fimmu.2017.01539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Chen X., Zhong B., Wang A., Wang X., Chu F., Nurieva R.I., Yan X., Chen P., van der Flier L.G. Transcription factor achaete-scute homologue 2 initiates follicular T-helper-cell development. Nature. 2014;507:513–518. doi: 10.1038/nature12910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locci M., Havenar-Daughton C., Landais E., Wu J., Kroenke M.A., Arlehamn C.L., Su L.F., Cubas R., Davis M.M., Sette A., International AIDS Vaccine Initiative Protocol C Principal Investigators Human circulating PD-1+CXCR3-CXCR5+ memory Tfh cells are highly functional and correlate with broadly neutralizing HIV antibody responses. Immunity. 2013;39:758–769. doi: 10.1016/j.immuni.2013.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lofano G., Mancini F., Salvatore G., Cantisani R., Monaci E., Carrisi C., Tavarini S., Sammicheli C., Rossi Paccani S., Soldaini E. Oil-in-Water Emulsion MF59 Increases Germinal Center B Cell Differentiation and Persistence in Response to Vaccination. J. Immunol. 2015;195:1617–1627. doi: 10.4049/jimmunol.1402604. [DOI] [PubMed] [Google Scholar]
- Mathew D., Giles J.R., Baxter A.E., Oldridge D.A., Greenplate A.R., Wu J.E., Alanio C., Kuri-Cervantes L., Pampena M.B., D’Andrea K., UPenn COVID Processing Unit Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science. 2020;369 doi: 10.1126/science.abc8511. eabc8511, c8529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesin L., Ersching J., Victora G.D. Germinal Center B Cell Dynamics. Immunity. 2016;45:471–482. doi: 10.1016/j.immuni.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita R., Schmitt N., Bentebibel S.-E., Ranganathan R., Bourdery L., Zurawski G., Foucat E., Dullaers M., Oh S., Sabzghabaei N. Human blood CXCR5(+)CD4(+) T cells are counterparts of T follicular cells and contain specific subsets that differentially support antibody secretion. Immunity. 2011;34:108–121. doi: 10.1016/j.immuni.2010.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan M.J., Lyke K.E., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Raabe V., Bailey R., Swanson K.A. Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature. 2020;586:589–593. doi: 10.1038/s41586-020-2639-4. [DOI] [PubMed] [Google Scholar]
- Mulligan M.J., Lyke K.E., Kitchin N., Absalon J., Gurtman A., Lockhart S.P., Neuzil K., Raabe V., Bailey R., Swanson K.A. Phase 1/2 Study to Describe the Safety and Immunogenicity of a COVID-19 RNA Vaccine Candidate (BNT162b1) in Adults 18 to 55 Years of Age: Interim Report. medRxiv. 2020:1–16. doi: 10.1101/2020.06.30.20142570. [DOI] [Google Scholar]
- Nurieva R.I., Chung Y., Hwang D., Yang X.O., Kang H.S., Ma L., Wang Y.-H., Watowich S.S., Jetten A.M., Tian Q., Dong C. Generation of T follicular helper cells is mediated by interleukin-21 but independent of T helper 1, 2, or 17 cell lineages. Immunity. 2008;29:138–149. doi: 10.1016/j.immuni.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hagan D.T. MF59 is a safe and potent vaccine adjuvant that enhances protection against influenza virus infection. Expert Rev. Vaccines. 2007;6:699–710. doi: 10.1586/14760584.6.5.699. [DOI] [PubMed] [Google Scholar]
- Orenstein W.A., Ahmed R. Simply put: Vaccination saves lives. Proc. Natl. Acad. Sci. USA. 2017;114:4031–4033. doi: 10.1073/pnas.1704507114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardi N., Tuyishime S., Muramatsu H., Karikó K., Mui B.L., Tam Y.K., Madden T.D., Hope M.J., Weissman D. Expression kinetics of nucleoside-modified mRNA delivered in lipid nanoparticles to mice by various routes. J. Control. Release. 2015;217:345–351. doi: 10.1016/j.jconrel.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardi N., Hogan M.J., Pelc R.S., Muramatsu H., Andersen H., DeMaso C.R., Dowd K.A., Sutherland L.L., Scearce R.M., Parks R. Zika virus protection by a single low-dose nucleoside-modified mRNA vaccination. Nature. 2017;543:248–251. doi: 10.1038/nature21428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardi N., Hogan M.J., Naradikian M.S., Parkhouse K., Cain D.W., Jones L., Moody M.A., Verkerke H.P., Myles A., Willis E. Nucleoside-modified mRNA vaccines induce potent T follicular helper and germinal center B cell responses. J. Exp. Med. 2018;215:1571–1588. doi: 10.1084/jem.20171450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardi N., Hogan M.J., Porter F.W., Weissman D. mRNA vaccines - a new era in vaccinology. Nat. Rev. Drug Discov. 2018;17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt R.L., Liang H.-E., Locksley R.M. Cytokine-secreting follicular T cells shape the antibody repertoire. Nat. Immunol. 2009;10:385–393. doi: 10.1038/ni.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin U., Muik A., Derhovanessian E., Vogler I., Kranz L.M., Vormehr M., Baum A., Pascal K., Quandt J., Maurus D. COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T cell responses. Nature. 2020;586:594–599. doi: 10.1038/s41586-020-2814-7. [DOI] [PubMed] [Google Scholar]
- Sallusto F., Lanzavecchia A., Araki K., Ahmed R. From vaccines to memory and back. Immunity. 2010;33:451–463. doi: 10.1016/j.immuni.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt N., Bentebibel S.-E., Ueno H. Phenotype and functions of memory Tfh cells in human blood. Trends Immunol. 2014;35:436–442. doi: 10.1016/j.it.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sempowski G.D., Saunders K.O., Acharya P., Wiehe K.J., Haynes B.F. Pandemic Preparedness: Developing Vaccines and Therapeutic Antibodies For COVID-19. Cell. 2020;181:1458–1463. doi: 10.1016/j.cell.2020.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T.R.F., Patel A., Ramos S., Elwood D., Zhu X., Yan J., Gary E.N., Walker S.N., Schultheis K., Purwar M. Immunogenicity of a DNA vaccine candidate for COVID-19. Nat. Commun. 2020;11:2601–2613. doi: 10.1038/s41467-020-16505-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snapper C.M., Paul W.E. Interferon-gamma and B cell stimulatory factor-1 reciprocally regulate Ig isotype production. Science. 1987;236:944–947. doi: 10.1126/science.3107127. [DOI] [PubMed] [Google Scholar]
- Stadlbauer D., Amanat F., Chromikova V., Jiang K., Strohmeier S., Arunkumar G.A., Tan J., Bhavsar D., Capuano C., Kirkpatrick E. SARS-CoV-2 Seroconversion in Humans: A Detailed Protocol for a Serological Assay, Antigen Production, and Test Setup. Curr. Protoc. Microbiol. 2020;57:e100. doi: 10.1002/cpmc.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens T.L., Bossie A., Sanders V.M., Fernandez-Botran R., Coffman R.L., Mosmann T.R., Vitetta E.S. Regulation of antibody isotype secretion by subsets of antigen-specific helper T cells. Nature. 1988;334:255–258. doi: 10.1038/334255a0. [DOI] [PubMed] [Google Scholar]
- Suan D., Kräutler N.J., Maag J.L.V., Butt D., Bourne K., Hermes J.R., Avery D.T., Young C., Statham A., Elliott M. CCR6 Defines Memory B Cell Precursors in Mouse and Human Germinal Centers, Revealing Light-Zone Location and Predominant Low Antigen Affinity. Immunity. 2017;47:1142–1153.e4. doi: 10.1016/j.immuni.2017.11.022. [DOI] [PubMed] [Google Scholar]
- Tam H.H., Melo M.B., Kang M., Pelet J.M., Ruda V.M., Foley M.H., Hu J.K., Kumari S., Crampton J., Baldeon A.D. Sustained antigen availability during germinal center initiation enhances antibody responses to vaccination. Proc. Natl. Acad. Sci. USA. 2016;113:E6639–E6648. doi: 10.1073/pnas.1606050113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victora G.D., Wilson P.C. Germinal center selection and the antibody response to influenza. Cell. 2015;163:545–548. doi: 10.1016/j.cell.2015.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinuesa C.G., Linterman M.A., Yu D., MacLennan I.C.M. Follicular Helper T Cells. Annu. Rev. Immunol. 2016;34:335–368. doi: 10.1146/annurev-immunol-041015-055605. [DOI] [PubMed] [Google Scholar]
- Walsh E.E., Frenck R., Falsey A.R., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Mulligan M.J., Bailey R. RNA-Based COVID-19 Vaccine BNT162b2 Selected for a Pivotal Efficacy Study. medRxiv. 2020 doi: 10.1101/2020.08.17.20176651. 2020.08.17.20176651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein J.S., Herman E.I., Lainez B., Licona-Limón P., Esplugues E., Flavell R., Craft J. TFH cells progressively differentiate to regulate the germinal center response. Nat. Immunol. 2016;17:1197–1205. doi: 10.1038/ni.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (2020). DRAFT landscape of COVID-19 candidate vaccines – 20 April 2020. Available at: https://www.who.int/blueprint/priority-diseases/key-action/novel-coronavirus-landscape-ncov.pdf. Accessed: August 7, 2020.
- Xie X., Muruato A., Lokugamage K.G., Narayanan K., Zhang X., Zou J., Liu J., Schindewolf C., Bopp N.E., Aguilar P.V. An Infectious cDNA Clone of SARS-CoV-2. Cell Host Microbe. 2020;27:841–848.e3. doi: 10.1016/j.chom.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Li X., Liu D., Li J., Zhang X., Chen X., Hou S., Peng L., Xu C., Liu W. Follicular T-helper cell recruitment governed by bystander B cells and ICOS-driven motility. Nature. 2013;496:523–527. doi: 10.1038/nature12058. [DOI] [PubMed] [Google Scholar]
- Yu J., Tostanoski L.H., Peter L., Mercado N.B., McMahan K., Mahrokhian S.H., Nkolola J.P., Liu J., Li Z., Chandrashekar A. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science. 2020;369:806–811. doi: 10.1126/science.abc6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusuf I., Kageyama R., Monticelli L., Johnston R.J., Ditoro D., Hansen K., Barnett B., Crotty S. Germinal center T follicular helper cell IL-4 production is dependent on signaling lymphocytic activation molecule receptor (CD150) J. Immunol. 2010;185:190–202. doi: 10.4049/jimmunol.0903505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan T., Ambrosi G., Wandmacher A.M., Rauscher B., Betge J., Rindtorff N., Häussler R.S., Hinsenkamp I., Bamberg L., Hessling B. MEK inhibitors activate Wnt signalling and induce stem cell plasticity in colorectal cancer. Nat. Commun. 2019;10:2197–2217. doi: 10.1038/s41467-019-09898-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N.-N., Li X.-F., Deng Y.-Q., Zhao H., Huang Y.-J., Yang G., Huang W.-J., Gao P., Zhou C., Zhang R.-R. A thermostable mRNA vaccine against COVID-19. Cell. 2020;182:1271–1283.e16. doi: 10.1016/j.cell.2020.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate/analyze datasets/code. Supplementary material can be located at: https://doi.org/10.17632/96m659kfrp.1.