Abstract
The coronavirus disease 2019 (COVID-19) pandemic, caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), poses an unprecedented global health crisis. It is particularly urgent to develop clinically effective therapies to contain the pandemic. The main protease (Mpro) and the RNA-dependent RNA polymerase (RdRP), which are responsible for the viral polyprotein proteolytic process and viral genome replication and transcription, respectively, are two attractive drug targets for SARS-CoV-2. This review summarizes up-to-date progress in the structural and pharmacological aspects of those two key targets above. Different classes of inhibitors individually targeting Mpro and RdRP are discussed, which could promote drug development to treat SARS-CoV-2 infection.
Keywords: Main protease, RNA-dependent RNA polymerase, SARS-CoV-2, Inhibitors
1. Introduction
Coronaviruses (CoVs) are enveloped, single-stranded positive-sense RNA (+ssRNA) viruses that feature a crown-like appearance under an electron microscope. CoVs belong to the subfamily Orthocoronavirinae, family Coronaviridae and the order Nidovirales. Members of the subfamily are classified into four genera according to their genome sequences and serological reactions: Alphacoronavirus, Be tacoronavirus, Gammacoronavirus, and Deltacoronavirus. Among them, Alphacoronavirus is further divided into subgroups 1a-1b, and Be tacoronavirus is divided into subgroups 2a-2d [1].
CoVs infect numerous species of vertebrates and can also be classified into animal and human CoVs according to their hosts. They are ecologically diverse, with enormous variation among bats, indicating that bats can serve as natural reservoirs for many CoVs [[2], [3], [4]]. Many diseases of domestic animals are associated with animal CoVs, such as porcine epidemic diarrhea virus (PEDV) [5], feline infectious peritonitis virus (FIPV) [6] and porcine deltacoronavirus (PDCoV) [7].
Some members of Alphacoronavirus and Betacoronavirus can infect humans and cause respiratory syndromes. Human coronaviruses (HCoVs) include 229 E (HCoV-229 E), OC43 (HCoV-OC43), NL63 (HCoV-NL63), and HKU1 (HCoV-HKU1), which are responsible for 10%–30% of upper respiratory tract infections in adults, characterized by a symptom of mild respiratory illness like common cold [8,9]. In the past two decades, there have been two severe respiratory syndrome epidemics caused by the transmission of an animal Betacoronavirus to humans [2]. In November 2002, the severe acute respiratory syndrome (SARS) first emerged in Guangdong Province, China [10], causing worldwide concern as it spread via international travel and trade to over 20 countries. Ultimately, measures of infection control rather than medical interventions contained the SARS pandemic. By July 2003, the SARS-CoV had resulted in 8096 reported cases and 774 deaths (mortality rate of approximately 10%) [11]. In June 2012, a decade after the emergence of SARS, another highly contagious CoV, Middle East respiratory syndrome coronavirus (MERS-CoV), was isolated from the sputum of a Saudi man who died from acute pneumonia and renal failure [12]. As of November 2019, there were 2494 confirmed cases of MERS and 858 deaths (mortality rate of approximately 35%) according to the World Health Organization (WHO), the majority of which were reported from Saudi Arabia [13].
In December 2019, the first clustered cases of a severe pneumonia were reported, and most of the patients are reported tohaving exposed to a large seafood market [[14], [15], [16], [17]]. On January 7th, the causative pathogen was isolated and identified as a novel member of Betacoronavirus that shares ∼96% sequence identity with bat RaTG13 CoV and ∼80% identity with SARS-CoV [16]. This virus was initially named 2019 novel coronavirus (2019-nCoV) and later renamed severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) according to its taxonomic and phylogenetic similarities with SARS-CoV [18]. Symptoms associated with COVID-19 include fever, cough, fatigue, nausea, and shortness of breath. The severity of the symptoms can range from mild to fatal, which poses a higher risk for older adults or people with medical conditions [19,20]. The WHO designated COVID-19 the name of this epidemic disease and subsequently declared it a pandemic. To date, there have been over 30 million confirmed COVID-19 cases and over 1 million related deaths worldwide across more than 180 countries [21]. The mortality rate of COVID-19 is estimated to be approximately 3%, which is lower than that of SARS and MERS. Although the remaining undetected and asymptomatic infections may affect the precise value [22], the basic reproduction number (R0) value of COVID-19 is estimated between 2–3 [23], which means it is highly contagious.
Unfortunately, no approved effective vaccines or specific antiviral drugs are currently available to prevent and treat COVID-19. Due to a stark warning over the spread of COVID-19, it is urgently required to identify and characterize drug targets for SARS-CoV-2 and to develop vaccines and effective drugs.
In this review, recent advances in structural and pharmacological studies of the SARS-CoV-2 main protease (Mpro) and RNA-dependent RNA polymerase (RdRP) will be summarized and discussed.
2. Structure and function of SARS-CoV-2 Mpro
2.1. Overview of SARS-CoV-2 Mpro
SARS-CoV-2 belongs to the Betacoronavirus genus, which also includes SARS-CoV and MERS-CoV [16,24]. The RNA genome of SARS-CoV-2 comprises ∼30,000 nucleotides with 14 open reading frames (ORFs). The replicase gene (ORF 1 ab) occupies two-thirds of the genome [16,24]. To improve the efficiency of their own replication and transcription, a number of positive-sense, single-stranded RNA viruses encode large polyproteins, which are further hydrolyzed to produce essential subunits required for replication. The production of functional elements is an important regulatory mechanism for RNA viruses.
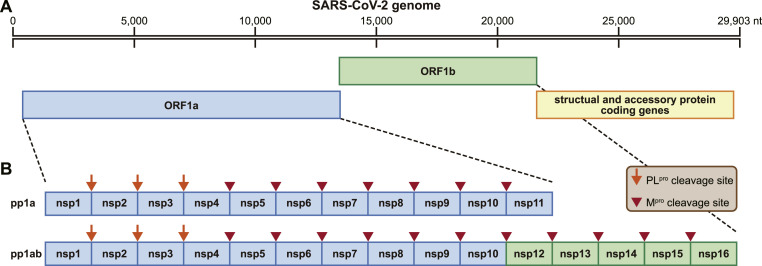
In the life cycle of SARS-CoV-2, ORF 1 ab encodes two long overlapping polyproteins, pp1a and pp1ab, which can be processed into 16 nonstructural proteins (nsps) required for RNA transcription and genome replication [16,25] (Fig. 1 ). This extensive proteolytic processing is achieved by two cysteine proteases, Mpro and papain-like protease (PLpro). PLpro is responsible for releasing nsp1-3 from the N-terminus of polyproteins. Mpro digests the polyprotein at the remaining 11 conserved cleavage sites (nsp 4/nsp5, nsp5/nsp6, nsp6/nsp7, nsp7/nsp8, nsp8/nsp9, nsp9/nsp10, nsp10/nsp12, nsp12/nsp13, nsp13/nsp14, and nsp14/nsp 15), starting with the autolytic cleavage of this enzyme (nsp5) from the polyproteins pp1a and pp1ab. It is called the main protease because it plays a major role in processing replicase polyproteins and thus facilitates viral gene expression and replication. Mpro is a chymotrypsin-like cysteine protease (∼33 kDa) [26]. Its alternative name is the 3C-like (3 C<superscript></superscript> L<superscript></superscript>) protease, which was assigned based on the picornavirus 3C protease because of the similar substrate specificity and the identification of cysteine as a catalytic residue in the context of two β-barrel structures [27,28].
Fig. 1.
The genomic organization of SARS-CoV-2. (A) The RNA genomic organization of SARS-CoV-2 (isolate Wuhan-Hu-1, NC_045,512). (B) Schematic representation of protease cleavage sites of the nonstructural protein (nsp) polyprotein. The orange arrows indicate papain-like protease (PLpro) cleavage sites, and the tailless red arrows indicate the main protease (Mpro) cleavage sites. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
2.2. Overall structure of SARS-CoV-2 main protease
On February 5th, 2020, the first crystal structure of SARS-CoV-2 Mpro in complex with an inhibitor N3 ((Protein Data Bank (PDB) 6LU7) was released to the public shortly after the outbreak began [26]. To date, hundreds of structures for SARS-CoV-2 Mpro have been reported for drug development [[29], [30], [31], [32], [33]].
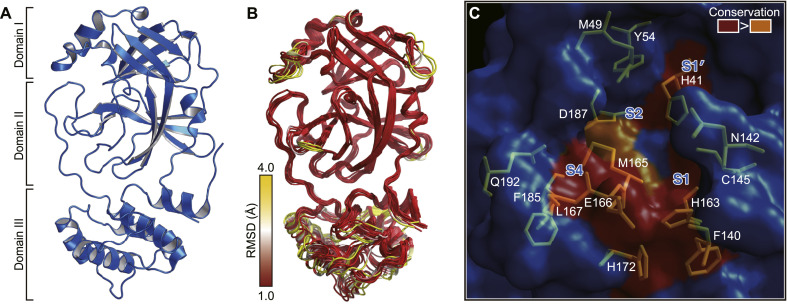
SARS-CoV-2 Mpro is composed of three domains [26] (Fig. 2 A). Domain I (residues 8–101) and domain II (residues 102–184) contain antiparallel β-barrel structures, which are reminiscent of the chymotrypsin family. SARS-CoV-2 Mpro possesses a catalytic dyad (Cys-His), and the substrate-binding site is located in a cleft between domain I and domain II. Domain III (residues 201–306) contains five α-helices that arrange into a large antiparallel globular cluster. Domain III has a unique topology in CoVs, which is required for homodimer formation. Domain III is connected to domain II through a long loop (residues 185–200).
Fig. 2.
The overall structure of SARS-CoV-2 Mpro and the substrate-binding pockets of Mpros across different species of coronavirus. (A) The structure of SARS-CoV-2 Mpro. (B) Superposition of structures of Mpros from 12 different species of coronaviruses. The color spectrum represents the root-mean-square deviation (r.m.s.d.) of the aligned Cα atoms. (C) Surface presentation of conserved substrate-binding pockets of Mpro from 12 coronaviruses. Red, residues are entirely identical among Mpros from all coronaviruses; orange, conserved substitution in Mpros of more than one of the coronaviruses. The S1, S2, S4, and S1’ subsites are indicated. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The crystal structures show that SARS-CoV-2 Mpro forms a homodimer with two protomers (denoted “A” and “B”) that are nearly perpendicular to each other, which is consistent with its active form in solution [34]. The dimer has a contact interface of ∼1394 Å2, predominantly between domain II of protomer A and the N-terminal residues of protomer B. In each protomer, the N-terminal finger (residues 1–7) inserts between domains II and III of its neighboring protomer, promoting the formation of the dimer and stabilizing the activity site of each protomer. Additionally, domain III regulates dimerization mainly through a salt-bridge interaction between Glu 290 of one protomer and Arg 4 from its neighbor [29].
2.3. Substrate-binding pocket and catalytic site
The substrate-binding pocket of SARS-CoV-2 Mpro is located in a cleft between domain I and domain II. According to the nomenclature of substrates for proteases [35], with the cleavage site as the center, the positions for the residues from the cleavage site to the N-terminus are named P1, P2, P3, P4, and P5 …, respectively. The positions for the residues from the cleavage site to the C-terminus are named P1’, P2’, P3’, P4’, and P5′ … respectively. The subsites in the substrate-binding pocket, which correspondingly accommodate each residue of the substrate, are named S1, S2, S3, S4, S5 … and S1’, S2’, S3’, S4’, S5′ …. The CoV Mpros are cysteine proteases, which are analogs of picornavirus 3 C<superscript></superscript> L<superscript></superscript> proteases. Although their structural similarities are limited, the two classes of viral proteases share common features in the specificity for their substrates. It has been reported that SARS-CoV Mpro prefers Leu-Gln-Ser or Leu-Gly-Ala at the P2–P1–P1’ sites [36]. Consistently, the SARS-CoV-2 Mpro cleaves polyproteins at least 11 sites that contain a conserved Leu-Gln↓(Ser, Ala, Asn) sequence.
In contrast with common serine and cysteine proteases, which adopt a catalytic triad of His-Asp(Glu)-Ser(Cys) [37,38], the CoV Mpros only possess only a catalytic dyad of His-Cys (His41-Cys145 for SARS-CoV-2) and completely lack a third catalytic residue at the active site. In the apo structure of SARS-CoV-2 Mpro, there is a stable water molecule in the oxyanion hole, corresponding to the position of the missing third catalytic residue [39]. This water molecule forms a hydrogen bond with the catalytic residue His41, indicating that its possible role is to protonate histamine during proteolytic cleavage, similar to the aspartic acid residue in classic serine proteases or glutamic acid in picornavirus 3 C<superscript></superscript> L<superscript></superscript> proteases [27,28,40]. It is believed that the proteolytic process follows a multistep mechanism. After the imidazole of histidine deprotonates the side chain of cysteine, the resulting nucleophilic thiolate attacks the carbonyl carbon of the substrate amide bond. The C-terminal portion of the peptide substrate is then released, and the histidine is deprotonated. Thereafter, the thioester is hydrolyzed to release the N-terminal portion of the peptide substrate, and the catalytic dyad is restored and ready for another round of catalytic reaction [36,41].
CoVs Mpros recognize mainly the P4 to P1’ positions of the substrate, and the substrate specificity is dominantly determined by S1, S2, S4 and S1’. Among them, the S1 subsite possesses the strongest selection and has an absolute requirement for glutamine at the P1 site of the substrate. The structure of SARS-CoV-2 Mpro revealed that the side chains of Phe140, Asn 142, His 163, Glu166, and His 172, along with the main chains of Phe140, and Leu141constitue the S1 subsite. Interestingly, the neighboring protomer also contributes to stabilizing the S1 site by extending its N-terminus close to the S1 site, allowing its Ser 1 to interact with Glu166, which helps maintain the conformation of the S1 subsite. The side chains of His41, Met 49, Tyr54, and Met165, as well as the alkyl portion of the side chain of Asp 187 are involved in the formation of the deep hydrophobic S2 subsite, which can accommodate the relatively bulky side chain of Leu or Phe at the P2 site of SARS-CoV-2 Mpro. The side chain of the P3 is solvent-exposed, which implies that this site could tolerate a wide range of functional groups. The side chains of Met165, Leu 167, Phe 185, and Gln 192, and the main chain of Gln 189 form a small hydrophobic S4 subsite of SARS-CoV-2 Mpro, which suggests that only small amino acid residues such as Ala, Val, Thr, or Pro, could be held at the P4 position. The S1′ of SARS-CoV-2 Mpro is a shallow subsite that can tolerate a small group of residues, such as Ser, Ala, or Asn.
3. Inhibitors of SARS-CoV-2 main protease
Previous studies indicate that Mpros have a highly conserved substrate-binding pocket across species, which serves as a drug target for the design of broad-spectrum inhibitors [42,43]. The superposition of the crystal structures of CoV Mpros, including those of SARS-CoV-2, SARS-CoV, MERS-CoV, HCoV-HKU1, HCoV-NL63, HCoV-229E, bat CoV-HKU4, mouse hepatitis virus (MHV), porcine epidemic diarrhea virus (PEDV), feline infectious peritonitis virus (FIPV), transmissible gastroenteritis virus (PDCoV), and infectious bronchitis virus (IBV) [26,29,39,[42], [43], [44], [45], [46], [47], [48], [49], [50], [51], [52]], shows that the most varied regions lie in the helical domain III and the surface loop (Fig. 2B). In contrast, the substrate-binding pocket is highly conserved among the Mpros of all the CoVs listed above (Fig. 2C). This finding suggests that antiviral inhibitors targeting this pocket should have broad-spectrum activity against various CoVs.
In particular, SARS-CoV-2 and SARS-CoV share 96% sequence identity in their Mpro sequences while their genomes only share only approximately 82% identity. Among the 12 different residues between SARS-CoV-2 Mpro and SARS-CoV Mpro, only residue Ser 46 in SARS-CoV-2 Mpro (Ala 46 in SARS-CoV Mpro) is located close to the entrance of the catalytic pocket. Hence inhibitors that can effectively inhibit SARS-CoV Mpro are expected to have a similar effect on SARS-CoV-2 Mpro. In the last few months, a series of inhibitors has been identified for SARS-CoV-2 Mpro (Fig. 3 ), and the structures of Mpro complexes have already been determined.
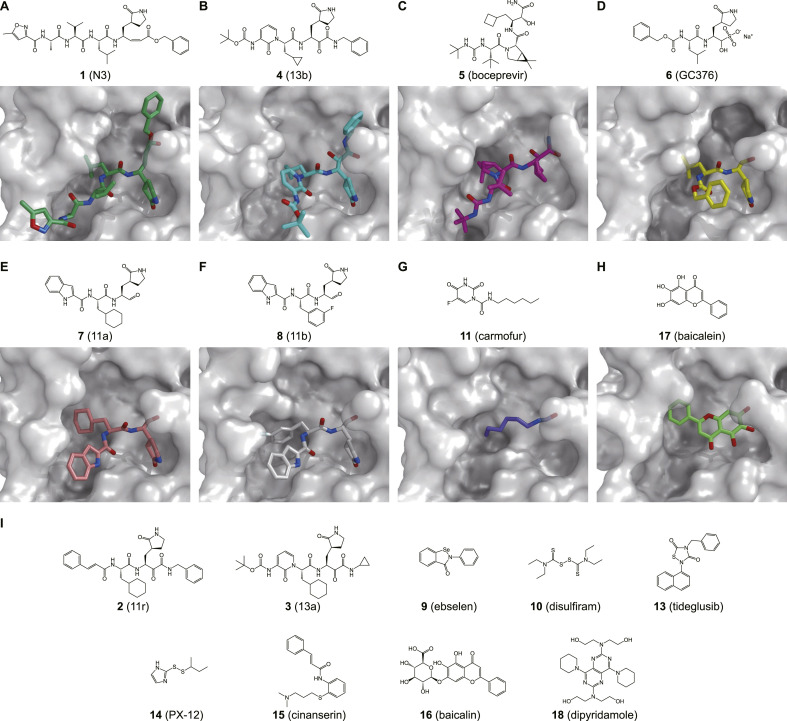
Fig. 3.
Inhibitors of SARS-CoV-2 Mpro and the crystal structures of Mpro in complex with individual inhibitors.
3.1. Peptidomimetic inhibitor with a Michael acceptor
Compound 1 (N3) is the first inhibitor whose structure in complex with SARS-CoV-2 Mpro has been deposited to the PDB [26]. This peptidomimetic inhibitor contains a Michael acceptor as a warhead that can irreversibly modify the catalytic residue Cys145. The strategy of designing this inhibitor involves replacing the amide bond at the cleavage site of the substrate with a Michael acceptor. The cysteine residue and the Michael acceptor group (warhead) of the inhibitor undergo 1,4-addition. In this process, the protonation of the α-carbanion from catalytic His-H+ results in the covalent bond formation. The structure shows that replacing the glutamine which is absolutely required at the P1 site, with a lactam group can result in strong binding for the inhibitor at the S1 subsite. Compound 1 inhibited SARS-CoV-2 with a half-maximal effective concentration (EC50) value of 16.77 μM in a plaque-reduction assay.
3.2. Peptidomimetic α-ketoamide inhibitors
Zhang et al. recently reported three α-ketoamides, including compound 2 (11r), 3 (13a), and 4 (13b), which can strongly inhibit SARS-CoV-2 Mpro [29]. Inhibitory activity assays and the crystal structure of compound 4 complexed with SARS-CoV-2 Mpro revealed that replacing the P2–P3 amide bond with a pyridone ring could improve the compound half-life in plasma. Additionally, compared with that of enterovirus 3C proteases, the S2 binding pocket of Mpro is smaller and less flexible.
α-Ketoamide is commonly used as a viral serine protease warhead in approved hepatitis C virus (HCV) drugs, such as boceprevir. Recently, researchers showed that compound 5 (boceprevir) and compound 6 (GC376), an inhibitor in preclinical studies against FIPV, could efficaciously inhibit SARS-CoV-2 in Vero cells by targeting Mpro [32,53,54].
3.3. Peptidomimetic inhibitor with an aldehyde
Compound 7 (11a) and 8 (11b) exhibited potent inhibition of SARS-CoV-2 Mpro with half-maximal inhibitory concentration (IC50) values of 0.053 μM and 0.040 μM, respectively [30]. Dai et al. designed these two peptidomimetic aldehydes with an indole moiety at the N-terminus (P3 site) and a C-terminal aldehyde warhead. The crystal structures show that these compounds can covalently bind to the catalytic site Cys145. Additionally, both peptidomimetic aldehyde compounds exhibited potent anti-SARS-CoV-2 infection activity in Vero cell-based assays and good pharmacokinetic and toxicity properties.
3.4. Inhibitors from drug repurposing
The repurposing of approved pharmaceutical drugs and drug candidates provides an alternative approach that allows the rapid identification of potential drug leads to addressing the current COVID-19 pandemic crisis. Jin et al. identified six hits, including ebselen (compound 9), disulfiram (compound 10), carmofur (compound 11), shikonin (compound 12), tideglusib (compound 13), and PX-12 (compound 14), via high-throughput screening (HTS). An in vitro enzymatic assay showed that the IC50 values of the six compounds ranged from 0.67 to 21.4 μM [26].
Among them, ebselen (9) exhibited promising antiviral activity in a plaque-reduction assay (EC50 = 4.67 μM). Ebselen (9) is an organoselenium compound that has previously been investigated for the treatment of multiple diseases, including bipolar disorders [55] and hearing loss [56]. Its low cytotoxicity in humans has been evaluated in clinical trials [[55], [56], [57]]. Ebselen (9) has been approved by the U.S. Food and Drug Administration (FDA) to enter phase II clinical trials (NCT04484025 and NCT04483973) to treat COVID-19. Carmofur (11) is a derivative of 5-fluorouracil (5-FU) and an approved antineoplastic agent. The X-ray crystal structure of SARS-CoV-2 Mpro in complex with carmofur (11) reveals that the carbonyl reactive group of carmofur (11) is covalently bound to the catalytic Cys145, whereas its fatty acid tail occupies the hydrophobic S2 subsite [31]. Carmofur (11) inhibits viral replication in cells (EC50 = 24.30 μM) and provides a basis for the rational design of analogs with enhanced inhibitory efficacy to treat COVID-19.
Virtual screening identified cinanserin (compound 15) as an inhibitor targeting SARS-CoV-2 Mpro. It is a well-characterized serotonin and a potential inhibitor [26]. It has previously been shown to inhibit SARS-CoV Mpro with an IC50 value of 5 μM [58]. It has displayed anti-SARS-CoV-2 activity with an EC50 value of 20.6 μM, which potential for further optimization as an antiviral drug lead.
Su et al. recently reported that baicalin (compound 16) and baicalein (compound 17), two natural products from Scutellaria baicalensis, are novel noncovalent, nonpeptidomimetic inhibitors for SARS-CoV-2 Mpro (IC50 = 6.41 μM and IC50 = 0.94 μM, respectively) [33]. Both baicalin (16) and baicalein (17) showed dose-dependent inhibition of SARS-CoV-2 replication, with EC50 values of 27.87 μM and 2.94 μM, respectively. The crystal structure of SARS-CoV-2 Mpro in complex with baicalein (17) revealed that this inhibitor noncovalently binds to the substrate-binding pocket of Mpro through interacting with the catalytic dyad His41-Cys145, the crucial S1/S2 subsites, and the oxyanion loop. It may act as a “shield” to effectively prevent the substrate from accessing the catalytic dyad [33].
By virtual screening of a U.S. FDA-approved drug library, Liu et al. identified an anticoagulation agent dipyridamole (compound 18), which may bind to SARS-CoV-2 Mpro through hydrogen bonds and hydrophobic interactions. It also showed an inhibitory effect by an in vitro assay (IC50 = 0.53 μM) [59]. Furthermore, it has been reported that dipyridamole (18) showed therapeutic improvement against COVID-19 in a small-scale clinical trial [59].
3.5. Fragment screening of inhibitors
To accelerate the development of antiviral drugs against COVID-19, Douangamath et al. performed large-scale fragment screening by combining mass spectrometry and X-ray approaches against SARS-CoV-2 Mpro [60]. The crystallographic screen identified 71 hits that bind to the substrate-binding pocket and 3 hits near the dimer interface. These structures provide information to develop more effective inhibitors against SARS-CoV-2 Mpro.
4. SARS-CoV-2 RdRP
4.1. Overview of SARS-CoV-2 RdRP
After entry into the cells, SARS-CoV-2 employs a multisubunit replication and transcription complex (RTC) to accomplish the replication and transcription of its RNA genome [61]. The RTC is assembled by an array of nsps released from polyprotein pp1ab by viral protease cleavage [62]. In this multisubunit machinery, the core component is nsp12, which is the catalytic subunit and is also called RdRP. Nsp12 is responsible for the catalysis of elongation of a new RNA chain from RNA templates [63]. However, nsp12 itself displays only little activity in RNA synthesis, and two other accessory factors, nsp7 and nsp8, are required to increase nsp12 binding to the template and subsequent RNA synthesis [64,65].
The RdRp core consists of the thumb, palm, and fingers subdomains forming an encircled human right-hand architecture, which is involved in template binding, polymerization, nucleoside triphosphate (NTP) entry, and other related functions [66,67]. In addition, SARS-CoV-2 nsp12 contains a nidovirus-specific N-terminal RdRp-associated nucleotidyltransferase (NiRAN) domain revealed by cryo-electron microscopy (cryo-EM) structures. The RdRP domain and NiRAN domain are bridged by an interface domain [67,68]. RdRP is an attractive antiviral drug target because of its essential role in the viral life cycle. Multiple nucleotide and nucleoside analogs targeting viral RdRP have been in clinical trials for the treatment of COVID-19 [[69], [70], [71]], such as remdesivir (RDV) and favipiravir. In comparison with other viral RdRPs, SARS-CoV-2 nsp12 shows structural similarity. Multiple key amino acid residues at the active site are conserved [72], indicating that antivirals targeting RdRPs may provide the first lines of defense against future CoV-associated diseases.
4.2. Structures of the SARS-CoV-2 RdRP complex
A series of cryo-EM structures has recently revealed that SARS-CoV-2 nsp12 (RdRP) forms a complex with nsp7 and nsp8, as previously observed for the RdRP complex from SARS-CoV. The viral RdRP interacts with an nsp7-nsp8 pair through its thumb subdomain and with another molecule of nsp8 through its finger subdomain [[66], [67], [68],[72], [73], [74], [75]]. These structures show that although the SARS-CoV-2 polymerase shares a similar structural architecture with that of SARS-CoV, an N-terminal β hairpin of SARS-CoV-2 nsp12 inserts into the groove clamped by the NiRAN domain and the palm subdomain, resulting in a cluster of tight contacts to stabilize the overall complex (Fig. 4 B). The most significant hit for the NiRAN domain identified by the DALI webserver search is P. syringae SelO [74], a pseudokinase harboring a kinase fold without key catalytic residues for canonical kinases [76]. Interestingly, the structural analysis shows that the residues involved in ADP-Mg2+ binding at the N-terminal domain of SARS-CoV nsp12 are very similar to those involved in AMP-PNP binding in SelO [74]. Although whether the NiRAN domain possesses nucleotidylation activity has yet to be determined, it has been found that the NiRAN domain plays an essential role in viral replication for SARS-CoV [77], suggesting that it may serve as a new druggable site.
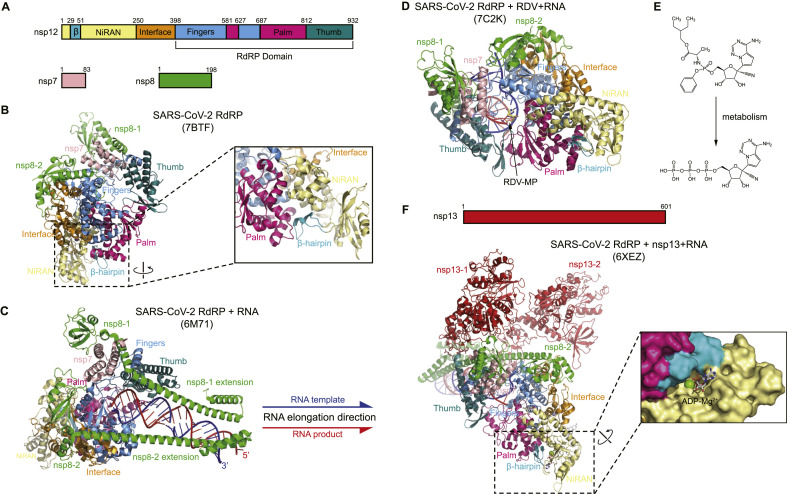
Fig. 4.
Structures of SARS-CoV-2 RdRP complex.
A) Schematic diagram illustrating the components of the SARS-CoV-2 RdRP complex, including nsp12, nsp8, and nsp7. The structural domains of nsp12 are labeled and divided by the residue numbers.
B) The overall structure of the apo form of the RdRP complex is displayed in the left panel (PDB: 7BTF). The right panel shows that the β hairpin is located in the groove clamped by the NiRAN domain and the palm subdomain. C) The overall structure of RdRP in complex with RNA (PDB: 6M71). The RNA contains a template chain (blue) and a product chain (red). The arrow represents the direction of the new RNA chain exit during viral replication.
D) The overall structure of SARS-CoV-2 RdRP-RNA in complex with a remdesivir monophosphate form (RDV-MP) (PDB: 7C2K). RDV-MP is showed as a yellow sticker model.
E) Chemical structure of the RDV and its active form of nucleoside triphosphate.
F) The overall structure of the RdRP complex consisting of RdRP, nsp7, nsp8 and nsp13 (helicase) is displayed in the left panel as a cartoon representation. Surface representation in the right panel shows the binding pocket of ADP in the NiRAN domain. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The structures of nsp12 in complex with nsp7, nsp8, and RNA template–product duplex provide detailed information for the interactions between RdRP and RNA. The cryo-EM structure of the RdRP complex shows that RdRP engages with over two turns of duplex RNA and identifies a long protruding RNA and extended protein regions in nsp8 [73]. The active site cleft of nsp12 binds to the first turn of RNA, and two copies of nsp8 bind to opposite sides of the cleft and the second turn of RNA (Fig. 4C).
It has been proposed that RdRP is able to cooperate with other nsp proteins for viral genomic replication and transcription. SARS-CoV-2 nsp13 belongs to a superfamily of 1B helicases that unwindRNA in an NTP-dependent manner [[78], [79], [80]]. Another cryo-EM structure of the RdRP complex demonstrates that nsp12, nsp8, nsp7, and nsp13 are able to form a larger complex [74]. This complex contains two molecules of nsp13, each of which binds to the N-terminal extension of each copy of nsp8. One nsp13 molecule also contacts the thumb subdomain of nsp12 (Fig. 4F). A possible backtracking mechanism was proposed for the subgenomic RNA transcription.
5. Inhibitors targeting SARS-CoV-2 RdRP
RDV, a nucleotide analog inhibitor that was developed targeting RdRP from Ebola virus, has demonstrated broad-spectrum inhibitory effects against a wide range of RNA viruses, including filoviruses, arenaviruses, and CoVs [[81], [82], [83]]. RDV is a prodrug that can be metabolized into its active triphosphate form in the cells (Fig. 4E), which could interfere with viral RNA synthesis and inhibit the elongation of the viral RNA product [81]. During the COVID-19 pandemic, clinical trials for RDV have been accelerated to determine its safety and efficacy in several countries. The structure of SARS-CoV-2 RdRP-RNA in complex with a monophosphate form of remdesivir (RDV-MP) reveals that incorporation of the active form of RDV may result in a stalled pre-translocated catalytic state (Fig. 4D), suggesting a delayed-chain-termination mechanism [72,84]. When RdRP synthesis proceeds to the i+3 position (where i corresponds to the position of the first incorporated RDV-MP), the first incorporated RDV-MP will be at position −3 or −4 for the pre- or post-translocated state, respectively. Nevertheless, the 1′-CN substituent of incorporated RDV-MP could encounter a steric clash with a serine side chain and probably lead to destabilization of the complex, hampering subsequent translocation. These structures provide a basis to develop antiviral drugs that interfere with this pivotal process of viral replication and transcription.
6. Concluding remarks
Following SARS-CoV and MERS-CoV, which emerged in 2003 and 2012, respectively, SARS-CoV-2 is the third CoV that crossed the species barrier to cause severe diseases in humans. Hence CoVs may still pose a threat to human health in the future. It is a plausible strategy to develop broad-spectrum antivirals as the first-line defense for future CoV-associated diseases. In the life cycle of CoVs, the Mpro and RdRP are prime drug targets that are well conserved among different CoVs. Great effort has been devoted to structural and functional studies of Mpro and RdRP, which can promote drug development against COVID-19 and other CoV-associated diseases.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
1. National Natural Science Foundation of China, China, 81520108019, 813300237;
2. Science and Technology Commission of Shanghai Municipality, China, 20431900200;
3. Natural Science Foundation of Tianjin, China, 18JCJQJC48000.
References
- 1.King A.M.Q., Adams M.J., Lefkowitz E., Carstens E.B. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier Science; 2011. [Google Scholar]
- 2.de Wit E., van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fan Y., Zhao K., Shi Z.-L., Zhou P. Bat coronaviruses in China. Viruses. 2019;11:210. doi: 10.3390/v11030210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woo P.C., Lau S.K., Huang Y., Yuen K.-Y. Coronavirus diversity, phylogeny and interspecies jumping. Exp. Biol. Med. 2009;234:1117–1127. doi: 10.3181/0903-MR-94. [DOI] [PubMed] [Google Scholar]
- 5.Jung K., Saif L.J. Porcine epidemic diarrhea virus infection: etiology, epidemiology, pathogenesis and immunoprophylaxis. Vet. J. 2015;204:134–143. doi: 10.1016/j.tvjl.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pedersen N.C. A review of feline infectious peritonitis virus infection. J. Feline Med. Surg. 2009;11:225–258. doi: 10.1016/j.jfms.2008.09.008. 1963–2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jung K., Hu H., Saif L.J. Porcine deltacoronavirus infection: etiology, cell culture for virus isolation and propagation, molecular epidemiology and pathogenesis. Virus Res. 2016;226:50–59. doi: 10.1016/j.virusres.2016.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paules C.I., Marston H.D., Fauci A.S. Coronavirus infections—more than just the common cold. J. Am. Med. Assoc. 2020;323:707–708. doi: 10.1001/jama.2020.0757. [DOI] [PubMed] [Google Scholar]
- 9.Fung T.S., Liu D.X. Human coronavirus: host-pathogen interaction. Annu. Rev. Microbiol. 2019;73:529–557. doi: 10.1146/annurev-micro-020518-115759. [DOI] [PubMed] [Google Scholar]
- 10.Zhong N., Zheng B., Li Y., Poon L., Xie Z., Chan K., Li P., Tan S., Chang Q., Xie J. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People’s Republic of China, in February, 2003. Lancet. 2003;362:1353–1358. doi: 10.1016/S0140-6736(03)14630-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.WHO Summary of probable SARS cases with onset of illness from 1 November 2002 to 31 July 2003. 2003. http://www.who.int/csr/sars/country/table2004_04_21/en/index.html
- 12.Zaki A.M., Van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 13.WHO Middle East respiratory syndrome coronavirus. 2019. https://www.who.int/emergencies/mers-cov/en/
- 14.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R. A novel coronavirus from patients with pneumonia in China, 2019. New England Journal of Medicine. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Qun, Guan Xuhua, Wu Peng, Wang Xiaoye, Zhou Lei, Tong Yeqing, Ren Ruiqi, Kathy S., Leung M., Eric H., Lau Y., Wong Jessica Y., Xing Xuesen, Xiang Nijuan, Wu Yang. Early transmission dynamics in wuhan, China, of novel coronavirus–infected pneumonia. N. Engl. J. Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., Yuan M.-L., Zhang Y.-L., Dai F.-H., Liu Y., Wang Q.-M., Zheng J.-J., Xu L., Holmes E.C., Zhang Y.-Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Viruses I.C.o.T.o. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nature Microbiology. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/s0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., Guan L., Wei Y., Li H., Wu X., Xu J., Tu S., Zhang Y., Chen H., Cao B. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/s0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gandhi M., Yokoe D., Havlir D. Asymptomatic transmission, the achilles’ heel of current strategies to control covid-19. N. Engl. J. Med. 2020;382:2158–2160. doi: 10.1056/NEJMe2009758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Y., Gayle A.A., Wilder-Smith A., Rocklöv J. The reproductive number of COVID-19 is higher compared to SARS coronavirus. J. Trav. Med. 2020;27 doi: 10.1093/jtm/taaa021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., Meng J., Zhu Z., Zhang Z., Wang J. Vol. 27. Cell host & microbe; 2020. Genome Composition and Divergence of the Novel Coronavirus (2019-nCoV) Originating in China; pp. 325–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O’Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., Tummino T.A., Hüttenhain R., Kaake R.M., Richards A.L., Tutuncuoglu B., Foussard H., Batra J., Haas K., Modak M., Kim M., Haas P., Polacco B.J., Braberg H., Fabius J.M., Eckhardt M., Soucheray M., Bennett M.J., Cakir M., McGregor M.J., Li Q., Meyer B., Roesch F., Vallet T., Mac Kain A., Miorin L., Moreno E., Naing Z.Z.C., Zhou Y., Peng S., Shi Y., Zhang Z., Shen W., Kirby I.T., Melnyk J.E., Chorba J.S., Lou K., Dai S.A., Barrio-Hernandez I., Memon D., Hernandez-Armenta C., Lyu J., Mathy C.J.P., Perica T., Pilla K.B., Ganesan S.J., Saltzberg D.J., Rakesh R., Liu X., Rosenthal S.B., Calviello L., Venkataramanan S., Liboy-Lugo J., Lin Y., Huang X.-P., Liu Y., Wankowicz S.A., Bohn M., Safari M., Ugur F.S., Koh C., Savar N.S., Tran Q.D., Shengjuler D., Fletcher S.J., O’Neal M.C., Cai Y., Chang J.C.J., Broadhurst D.J., Klippsten S., Sharp P.P., Wenzell N.A., Kuzuoglu-Ozturk D., Wang H.-Y., Trenker R., Young J.M., Cavero D.A., Hiatt J., Roth T.L., Rathore U., Subramanian A., Noack J., Hubert M., Stroud R.M., Frankel A.D., Rosenberg O.S., Verba K.A., Agard D.A., Ott M., Emerman M., Jura N., von Zastrow M., Verdin E., Ashworth A., Schwartz O., d’Enfert C., Mukherjee S., Jacobson M., Malik H.S., Fujimori D.G., Ideker T., Craik C.S., Floor S.N., Fraser J.S., Gross J.D., Sali A., Roth B.L., Ruggero D., Taunton J., Kortemme T., Beltrao P., Vignuzzi M., García-Sastre A., Shokat K.M., Shoichet B.K., Krogan N.J. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 27.Binford S., Maldonado F., Brothers M., Weady P., Zalman L., Meador J., Matthews D., Patick A. Conservation of amino acids in human rhinovirus 3C protease correlates with broad-spectrum antiviral activity of rupintrivir, a novel human rhinovirus 3C protease inhibitor. Antimicrob. Agents Chemother. 2005;49:619–626. doi: 10.1128/AAC.49.2.619-626.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cordingley M.G., Callahan P.L., Sardana V.V., Garsky V.M., Colonno R.J. Substrate requirements of human rhinovirus 3C protease for peptide cleavage in vitro. J. Biol. Chem. 1990;265:9062–9065. [PubMed] [Google Scholar]
- 29.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dai W., Zhang B., Jiang X.-M., Su H., Li J., Zhao Y., Xie X., Jin Z., Peng J., Liu F. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020;368:1331–1335. doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jin Z., Zhao Y., Sun Y., Zhang B., Wang H., Wu Y., Zhu Y., Zhu C., Hu T., Du X., Duan Y., Yu J., Yang X., Yang X., Yang K., Liu X., Guddat L.W., Xiao G., Zhang L., Yang H., Rao Z. Structural basis for the inhibition of SARS-CoV-2 main protease by antineoplastic drug carmofur. Nat. Struct. Mol. Biol. 2020;27:529–532. doi: 10.1038/s41594-020-0440-6. [DOI] [PubMed] [Google Scholar]
- 32.Fu L., Ye F., Feng Y., Yu F., Wang Q., Wu Y., Zhao C., Sun H., Huang B., Niu P. Both Boceprevir and GC376 efficaciously inhibit SARS-CoV-2 by targeting its main protease. Nat. Commun. 2020;11:1–8. doi: 10.1038/s41467-020-18233-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Su H.-x., Yao S., Zhao W.-f., Li M.-j., Liu J., Shang W.-j., Xie H., Ke C.-q., Hu H.-c., Gao M.-n. Anti-SARS-CoV-2 activities in vitro of Shuanghuanglian preparations and bioactive ingredients. Acta Pharmacol. Sin. 2020;41:1167–1177. doi: 10.1038/s41401-020-0483-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grum-Tokars V., Ratia K., Begaye A., Baker S.C., Mesecar A.D. Evaluating the 3C-like protease activity of SARS-Coronavirus: recommendations for standardized assays for drug discovery. Virus Res. 2008;133:63–73. doi: 10.1016/j.virusres.2007.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schechter I., Berger A. On the size of the active site in proteases. I. Papain. Biochem. Biophys. Res. Commun. 1967;27:157–162. doi: 10.1016/S0006-291X(67)80055-X. [DOI] [PubMed] [Google Scholar]
- 36.Pillaiyar T., Manickam M., Namasivayam V., Hayashi Y., Jung S.H. An overview of severe acute respiratory syndrome-coronavirus (SARS-CoV) 3CL protease inhibitors: peptidomimetics and small molecule chemotherapy. J. Med. Chem. 2016;59:6595–6628. doi: 10.1021/acs.jmedchem.5b01461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gorbalenya A.E., Donchenko A.P., Blinov V.M., Koonin E.V. Cysteine proteases of positive strand RNA viruses and chymotrypsin-like serine proteases: a distinct protein superfamily with a common structural fold. FEBS Lett. 1989;243:103–114. doi: 10.1016/0014-5793(89)80109-7. [DOI] [PubMed] [Google Scholar]
- 38.Bazan J.F., Fletterick R.J. Viral cysteine proteases are homologous to the trypsin-like family of serine proteases: structural and functional implications. Proc. Natl. Acad. Sci. Unit. States Am. 1988;85:7872–7876. doi: 10.1073/pnas.85.21.7872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou X., Zhong F., Lin C., Hu X., Zhang Y., Xiong B., Yin X., Fu J., He W., Duan J. Science China Life Sciences; 2020. Structure of SARS-CoV-2 main protease in the apo state; pp. 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matthews D.A., Smith W.W., Ferre R.A., Condon B., Budahazi G., Slsson W., Villafranca J., Janson C.A., McElroy H., Gribskov C. Structure of human rhinovirus 3C protease reveals a trypsin-like polypeptide fold, RNA-binding site, and means for cleaving precursor polyprotein. Cell. 1994;77:761–771. doi: 10.1016/0092-8674(94)90059-0. [DOI] [PubMed] [Google Scholar]
- 41.Huang C., Wei P., Fan K., Liu Y., Lai L. 3C-like proteinase from SARS coronavirus catalyzes substrate hydrolysis by a general base mechanism. Biochemistry. 2004;43:4568–4574. doi: 10.1021/bi036022q. [DOI] [PubMed] [Google Scholar]
- 42.Wang F., Chen C., Tan W., Yang K., Yang H. Structure of main protease from human coronavirus NL63: insights for wide spectrum anti-coronavirus drug design. Sci. Rep. 2016;6 doi: 10.1038/srep22677. 22677-22677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang H.T., Xie W.Q., Xue X.Y., Yang K.L., Ma J., Liang W.X., Zhao Q., Zhou Z., Pei D.Q., Ziebuhr J., Hilgenfeld R., Yuen K.Y., Wong L., Gao G.X., Chen S.J., Chen Z., Ma D.W., Bartlam M., Rao Z.H. Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biol. 2005;3 doi: 10.1371/journal.pbio.0030428. 2044-2044 e428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang H.T., Yang M.J., Ding Y., Liu Y.W., Lou Z.Y., Zhou Z., Sun L., Mo L.J., Ye S., Pang H., Gao G.F., Anand K., Bartlam M., Hilgenfeld R., Rao Z.H. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc. Natl. Acad. Sci. U. S. A. 2003;100:13190–13195. doi: 10.1073/pnas.1835675100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xue X.Y., Yu H.W., Yang H.T., Xue F., Wu Z.X., Shen W., Li J., Zhou Z., Ding Y., Zhao Q., Zhang X.J.C., Liao M., Bartlam M., Rao Z. Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design. J. Virol. 2008;82:2515–2527. doi: 10.1128/jvi.02114-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao Q., Li S., Xue F., Zou Y., Chen C., Bartlam M., Rao Z. Structure of the main protease from a global infectious human coronavirus, HCoV-HKU1. J. Virol. 2008;82:8647–8655. doi: 10.1128/JVI.00298-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee C.-C., Kuo C.-J., Ko T.-P., Hsu M.-F., Tsui Y.-C., Chang S.-C., Yang S., Chen S.-J., Chen H.-C., Hsu M.-C., Shih S.-R., Liang P.-H., Wang A.H.J. Structural basis of inhibition specificities of 3C and 3C-like proteases by zinc-coordinating and peptidomimetic compounds. J. Biol. Chem. 2009;284:7646–7655. doi: 10.1074/jbc.M807947200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ren Z.L., Yan L.M., Zhang N., Guo Y., Yang C., Lou Z.Y., Rao Z.H. The newly emerged SARS-Like coronavirus HCoV-EMC also has an "Achilles’ heel": current effective inhibitor targeting a 3C-like protease. Protein & Cell. 2013;4:248–250. doi: 10.1007/s13238-013-2841-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.St John S.E., Tomar S., Stauffer S.R., Mesecar A.D. Targeting zoonotic viruses: structure-based inhibition of the 3C-like protease from bat coronavirus HKU4-The likely reservoir host to the human coronavirus that causes Middle East Respiratory Syndrome (MERS) Bioorg. Med. Chem. 2015;23:6036–6048. doi: 10.1016/j.bmc.2015.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang F.H., Chen C., Liu X.M., Yang K.L., Xu X.L., Yang H.T. Crystal structure of feline infectious peritonitis virus main protease in complex with synergetic dual inhibitors. J. Virol. 2016;90:1910–1917. doi: 10.1128/jvi.02685-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang F.H., Chen C., Yang K.L., Xu Y., Liu X.M., Gao F., Liu H., Chen X., Zhao Q., Liu X., Cai Y., Yang H.T. Michael acceptor-based peptidomimetic inhibitor of main protease from porcine epidemic diarrhea virus. J. Med. Chem. 2017;60:3212–3216. doi: 10.1021/acs.jmedchem.7b00103. [DOI] [PubMed] [Google Scholar]
- 52.Cui W., Cui S.S., Chen C., Chen X., Wang Z.F., Yang H.T., Zhang L. The crystal structure of main protease from mouse hepatitis virus A59 in complex with an inhibitor. Biochem. Biophys. Res. Commun. 2019;511:794–799. doi: 10.1016/j.bbrc.2019.02.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ma C., Sacco M.D., Hurst B., Townsend J.A., Hu Y., Szeto T., Zhang X., Tarbet B., Marty M.T., Chen Y., Wang J. Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. Cell Res. 2020;30:678–692. doi: 10.1038/s41422-020-0356-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hung H.-C., Ke Y.-Y., Huang S.Y., Huang P.-N., Kung Y.-A., Chang T.-Y., Yen K.-J., Peng T.-T., Chang S.-E., Huang C.-T. Discovery of M Protease inhibitors encoded by SARS-CoV-2. Antimicrob. Agents Chemother. 2020;64 doi: 10.1128/AAC.00872-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Singh N., Halliday A.C., Thomas J.M., Kuznetsova O.V., Baldwin R., Woon E.C., Aley P.K., Antoniadou I., Sharp T., Vasudevan S.R. A safe lithium mimetic for bipolar disorder. Nat. Commun. 2013;4:1–7. doi: 10.1038/ncomms2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lynch E., Kil J. Development of ebselen, a glutathione peroxidase mimic, for the prevention and treatment of noise-induced hearing loss. Semin. Hear. 2009;30:47–55. [Google Scholar]
- 57.Kil J., Lobarinas E., Spankovich C., Griffiths S.K., Antonelli P.J., Lynch E.D., Le Prell C.G. Safety and efficacy of ebselen for the prevention of noise-induced hearing loss: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet. 2017;390:969–979. doi: 10.1016/S0140-6736(17)31791-9. [DOI] [PubMed] [Google Scholar]
- 58.Chen L., Gui C., Luo X., Yang Q., Günther S., Scandella E., Drosten C., Bai D., He X., Ludewig B. Cinanserin is an inhibitor of the 3C-like proteinase of severe acute respiratory syndrome coronavirus and strongly reduces virus replication in vitro. J. Virol. 2005;79:7095–7103. doi: 10.1128/JVI.79.11.7095-7103.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu X., Li Z., Liu S., Sun J., Chen Z., Jiang M., Zhang Q., Wei Y., Wang X., Huang Y.-Y. Potential therapeutic effects of dipyridamole in the severely ill patients with COVID-19. Acta Pharm. Sin. B. 2020;10:1205–1215. doi: 10.1016/j.apsb.2020.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Douangamath A., Fearon D., Gehrtz P., Krojer T., Lukacik P., Owen C.D., Resnick E., Strain-Damerell C., Ábrányi-Balogh P., Brandaõ-Neto J. bioRxiv; 2020. Crystallographic and Electrophilic Fragment Screening of the SARS-CoV-2 Main Protease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sola I., Almazan F., Zuniga S., Enjuanes L. Continuous and discontinuous RNA synthesis in coronaviruses. Annual review of virology. 2015;2:265–288. doi: 10.1146/annurev-virology-100114-055218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ziebuhr J., Snijder E.J., Gorbalenya A.E. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 2000;81:853–879. doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]
- 63.Te Velthuis A.J., Arnold J.J., Cameron C.E., van den Worm S.H., Snijder E.J. The RNA polymerase activity of SARS-coronavirus nsp12 is primer dependent. Nucleic Acids Res. 2010;38:203–214. doi: 10.1093/nar/gkp904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Subissi L., Posthuma C.C., Collet A., Zevenhoven-Dobbe J.C., Gorbalenya A.E., Decroly E., Snijder E.J., Canard B., Imbert I. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc. Natl. Acad. Sci. Unit. States Am. 2014;111:E3900–E3909. doi: 10.1073/pnas.1323705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., Wang T., Sun Q., Ming Z., Zhang L. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368:779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kirchdoerfer R.N., Ward A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 2019;10:2342. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., Wang T., Sun Q., Ming Z., Zhang L., Ge J., Zheng L., Zhang Y., Wang H., Zhu Y., Zhu C., Hu T., Hua T., Zhang B., Yang X., Li J., Yang H., Liu Z., Xu W., Guddat L.W., Wang Q., Lou Z., Rao Z. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368:779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yin W., Mao C., Luan X., Shen D.D., Shen Q., Su H., Wang X., Zhou F., Zhao W., Gao M., Chang S., Xie Y.C., Tian G., Jiang H.W., Tao S.C., Shen J., Jiang Y., Jiang H., Xu Y., Zhang S., Zhang Y., Xu H.E. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;368:1499–1504. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H., Spitters C., Ericson K., Wilkerson S., Tural A., Diaz G., Cohn A., Fox L., Patel A., Gerber S.I., Kim L., Tong S., Lu X., Lindstrom S., Pallansch M.A., Weldon W.C., Biggs H.M., Uyeki T.M., Pillai S.K., Washington State -nCo V.C.I.T. First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lai C.C., Shih T.P., Ko W.C., Tang H.J., Hsueh P.R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): the epidemic and the challenges. Int. J. Antimicrob. Agents. 2020;55:105924. doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang Q., Wu J., Wang H., Gao Y., Liu Q., Mu A., Ji W., Yan L., Zhu Y., Zhu C., Fang X., Yang X., Huang Y., Gao H., Liu F., Ge J., Sun Q., Yang X., Xu W., Liu Z., Yang H., Lou Z., Jiang B., Guddat L.W., Gong P., Rao Z. Structural basis for RNA replication by the SARS-CoV-2 polymerase. Cell. 2020;182:417–428. doi: 10.1016/j.cell.2020.05.034. e413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hillen H.S., Kokic G., Farnung L., Dienemann C., Tegunov D., Cramer P. Structure of replicating SARS-CoV-2 polymerase. Nature. 2020;584:154–156. doi: 10.1038/s41586-020-2368-8. [DOI] [PubMed] [Google Scholar]
- 74.Chen J., Malone B., Llewellyn E., Grasso M., Shelton P.M.M., Olinares P.D.B., Maruthi K., Eng E.T., Vatandaslar H., Chait B.T., Kapoor T.M., Darst S.A., Campbell E.A. Structural basis for helicase-polymerase coupling in the SARS-CoV-2 replication-transcription complex. Cell. 2020;182:1560–1573. doi: 10.1016/j.cell.2020.07.033. e1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Peng Q., Peng R., Yuan B., Zhao J., Wang M., Wang X., Wang Q., Sun Y., Fan Z., Qi J., Gao G.F., Shi Y. Structural and biochemical characterization of the nsp12-nsp7-nsp8 core polymerase complex from SARS-CoV-2. Cell Rep. 2020;31:107774. doi: 10.1016/j.celrep.2020.107774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dudkiewicz M., Szczepinska T., Grynberg M., Pawlowski K. A novel protein kinase-like domain in a selenoprotein, widespread in the tree of life. PloS One. 2012;7 doi: 10.1371/journal.pone.0032138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lehmann K.C., Gulyaeva A., Zevenhoven-Dobbe J.C., Janssen G.M., Ruben M., Overkleeft H.S., van Veelen P.A., Samborskiy D.V., Kravchenko A.A., Leontovich A.M., Sidorov I.A., Snijder E.J., Posthuma C.C., Gorbalenya A.E. Discovery of an essential nucleotidylating activity associated with a newly delineated conserved domain in the RNA polymerase-containing protein of all nidoviruses. Nucleic Acids Res. 2015;43:8416–8434. doi: 10.1093/nar/gkv838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lee N.R., Kwon H.M., Park K., Oh S., Jeong Y.J., Kim D.E. Cooperative translocation enhances the unwinding of duplex DNA by SARS coronavirus helicase nsP13. Nucleic Acids Res. 2010;38:7626–7636. doi: 10.1093/nar/gkq647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tanner J.A., Watt R.M., Chai Y.B., Lu L.Y., Lin M.C., Peiris J.S., Poon L.L., Kung H.F., Huang J.D. The severe acute respiratory syndrome (SARS) coronavirus NTPase/helicase belongs to a distinct class of 5’ to 3’ viral helicases. J. Biol. Chem. 2003;278:39578–39582. doi: 10.1074/jbc.C300328200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang R., Li Y., Cowley T.J., Steinbrenner A.D., Phillips J.M., Yount B.L., Baric R.S., Weiss S.R. The nsp1, nsp13, and M proteins contribute to the hepatotropism of murine coronavirus JHM.Wu. J. Virol. 2015;89:3598–3609. doi: 10.1128/JVI.03535-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Warren T.K., Jordan R., Lo M.K., Ray A.S., Mackman R.L., Soloveva V., Siegel D., Perron M., Bannister R., Hui H.C., Larson N., Strickley R., Wells J., Stuthman K.S., Van Tongeren S.A., Garza N.L., Donnelly G., Shurtleff A.C., Retterer C.J., Gharaibeh D., Zamani R., Kenny T., Eaton B.P., Grimes E., Welch L.S., Gomba L., Wilhelmsen C.L., Nichols D.K., Nuss J.E., Nagle E.R., Kugelman J.R., Palacios G., Doerffler E., Neville S., Carra E., Clarke M.O., Zhang L., Lew W., Ross B., Wang Q., Chun K., Wolfe L., Babusis D., Park Y., Stray K.M., Trancheva I., Feng J.Y., Barauskas O., Xu Y., Wong P., Braun M.R., Flint M., McMullan L.K., Chen S.S., Fearns R., Swaminathan S., Mayers D.L., Spiropoulou C.F., Lee W.A., Nichol S.T., Cihlar T., Bavari S. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature. 2016;531:381–385. doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.de Wit E., Feldmann F., Cronin J., Jordan R., Okumura A., Thomas T., Scott D., Cihlar T., Feldmann H. Prophylactic and therapeutic remdesivir (GS-5734) treatment in the rhesus macaque model of MERS-CoV infection. Proc. Natl. Acad. Sci. U. S. A. 2020;117:6771–6776. doi: 10.1073/pnas.1922083117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lo M.K., Jordan R., Arvey A., Sudhamsu J., Shrivastava-Ranjan P., Hotard A.L., Flint M., McMullan L.K., Siegel D., Clarke M.O., Mackman R.L., Hui H.C., Perron M., Ray A.S., Cihlar T., Nichol S.T., Spiropoulou C.F. GS-5734 and its parent nucleoside analog inhibit Filo-, Pneumo-, and Paramyxoviruses. Sci. Rep. 2017;7:43395. doi: 10.1038/srep43395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gordon C.J., Tchesnokov E.P., Woolner E., Perry J.K., Feng J.Y., Porter D.P., Gotte M. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 2020;295:6785–6797. doi: 10.1074/jbc.RA120.013679. [DOI] [PMC free article] [PubMed] [Google Scholar]