Abstract
Background
Metastasis is the main cause of renal cell carcinoma (RCC) tumor death, and effective inhibition of RCC metastasis is an essential means to meliorate the prognosis of RCC patients. MicroRNAs (miRs) have been proved to be stable and important biomarkers for several malignancies. This study is therefore set out to explore the metastasis-related miR and its mechanism in RCC.
Methods
The expression of miR-26a-5p in RCC was analyzed using the expression profile in the Cancer Genome Atlas (TCGA). MiR-26a-5p and E2F transcription factor 7 (E2F7) in RCC patients were detected by qRT-PCR. Cell Counting Kit-8 (CCK-8) was adopted to assess cell proliferation, Transwell was utilized to evaluate migration and invasion, and flow cytometry (FC) was used to determine apoptosis. Mouse cell-derived and patient-derived xenotransplantation models were established to evaluate the effect of miR-26a-5p on tumor growth and metastasis in vivo. The molecular mechanism of miR-26a-5p was analyzed by dual-luciferase reporter (DLR) gene analysis, qRT-PCR, and Western blot (WB) both in vivo and in vitro.
Results
MiR-26a-5p was reduced in renal carcinoma cells and may serve as a biomarker for renal cancer metastasis and prognosis. MiR-26a-5p up-regulation inhibited migration and invasion in renal cell lines and tumor metastasis in vivo. Bioinformatics target prediction and RNA-seq results showed that E2F7 was among the targets of miR-26a-5p and was significantly inhibited by miR-26a-5p in vivo and in vitro.
Conclusion
MiR-26a-5p presents low expression in RCC and promotes RCC cell apoptosis and prevents cells from proliferating and invading by targeting E2F7, which is a promising therapeutic target for RCC.
Keywords: miR-26a-5p, E2F7, renal cell carcinoma, proliferation, invasion
Introduction
Renal carcinoma (RC), as the most lethal malignancy of the urinary system, accounts for 2% of all tumors globally.1 Among various tumors types, renal cell carcinoma (RCC) is the most pervasive one in RC, which is responsible for 80% of all the cases.2 According to a statistical survey,3 there are approximately 65,000 new cases inflicted by RC and about 13,000 deaths in the United States each year, with rising incidence at younger ages. Compared with other urinary tumors, chemotherapy has less benefit for the treatment of RCC.4 At present, the mainstay clinical treatment for RCC is surgical resection.5 However, the high occultation and invasiveness of RCC results in recurrence in 20% to 40% of patients after local nephrectomy.6 Therefore, it is pressing to explore new treatment options and to find potential therapeutic targets for RC.
As short non-coding RNA with a length between 17 and 21nt,7 microRNA (miR) targeting downstream genes through the 3-Untranslated Regions (UTR) terminal is reported to be effective in regulating cell proliferation, differentiation and programmed cell death.8 For example, Peng et al9 showed that by targeting ZFP91, miR-188-5p was able to inhibit the proliferation and invasion of gastric cancer cells. In addition, it was found that10 miR-216b down-regulated the oncogene KRAS and inhibited the proliferation and invasion of stem clear cell renal cell carcinoma (CCRCC) cells after transcription. MiR-26a-5p is a common tumor suppressor gene with low expression in a variety of tumors.11,12 Previously, serum miR-26a-5p was reported to be a feasible biomarker for RC,13 but the underlying mechanism remains elusive. In this study, we found a targeting relationship between E2F transcription factor 7 (E2F7) and miR-26a-5p through online prediction analysis. Besides, previous evidence also reported the high expression of E2F7 in RCC.
Therefore, in this study, we aim to explore the significance of miR-26a-5p and E2F7 in RCC to provide potential markers for clinical therapy.
Methods and Data
Clinical Data Collection
A total of 40 patients with RCC who received treatment in the People’s Hospital of Zhengzhou from January 2013 to January 2015 were enrolled. After intraoperative sampling, the tumor tissues and paracancerous tissues were transported with liquid nitrogen to the laboratory and stored at −80°C for later use. In this study, all the patients were first diagnosed with RCC and had not received any anti-tumor treatment, with a life expectancy of no less than 1 month. The Medical Ethics Committee of the People’s Hospital of Zhengzhou approved the study protocol, and all the enrolled subjects had provided the written informed consent to participate.
Cell Culture
Purchased from American Type Culture Collection (ATCC; Manassas, Virginia, United States), RCC cells (786-O, A498, ACHN) and CRL-2190 of healthy human renal proximal tubule epithelial cells (hRPTECs) were placed in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) for culture at 37°C with 5% CO2.
Cell Transfection
GeneCoposia TM (Guangzhou, China) was in charge of the synthesis of the vectors, including miR-26a-5p inhibitors (anti-miR-26a-5p), miR-26a-5p mimics, E2F7-specific inhibitors (si-E2F7) and all control oligonucleotides (miR-mimics, anti-miR and si-NC). Referring to the instructions, all oligonucleotides were transfected into A498, 786-O cells using Lipofectamine™ 2000 Transfection Reagent (Invitrogen, United States).
QRT-PCR Detection
Total RNA was extracted from patient tissues and tumor cells using TRIzol reagent (Invitrogen, United States). As instructed by the instructions provided with the kit, cDNA was synthesized from total RNA with a TaqMan MicroRNA reverse transcription kit (Applied Biosystems, United States). Subsequently, on ABI 7500 PCR system (ABI, United States), qRT-PCR was performed with the aid of TaqMan MicroRNA analysis kit (Invitrogen, United States) to measure the expression of miR-26a-5p and E2F7. Their levels were then calculated by,−ΔΔCt14 with GAPDH (mRNA) and U6 (miR) as the internal parameter of genes, respectively. miR-26a-5p upstream primer: 5ʹ-ACACTCCAGCTGGGTTCAAGTAATCCAGGA-3ʹ, downstream primer: 5ʹ-TGGTGTCGTGGAGTCG-3ʹ; E2F7 upstream primer: 5ʹ-AGGGATGGAGGTAAATTGTTTAACACT-3ʹ, downstream primer: 5ʹ-TTTCCCCATCTTCAACTAA-3ʹ; GAPDH upstream primer: 5ʹ-GATATTGTTGCCATCAATGAC-3ʹ, downstream primer: 5ʹ-TTGATTTTGGAGGGATCTCG-3ʹ; U6 stream primer: 5ʹ-CTCGCTTCGGCAGCACA-3ʹ, downstream primer: 5ʹ-ACGCTTCACGAATTTGCGT-3ʹ.
Western Blot (WB)
The collected cells were lysed using RIPA buffer, and the protein concentration was measured with the BCA. Having electrophoresed on 10% SDS-PAGE, an equivalent amount of the extracted protein (50 µg) was then transferred to a PVDF membrane, and sealed with phosphate-buffered saline with skim milk (5%) and Tween-20 (0.1%) for 1 h. Thereafter, the membrane was incubated with rabbit anti-E2F7, Bax, Bcl-2, cle-caspase3 and anti-β-actin (Abcam, United States) at 4°C overnight. After incubating the membrane with a secondary antibody conjugated with horseradish peroxidase for 30 min, the protein bands were visualized using the Pierce ECL substrate WB detection system (Bio-Rad Laboratories).
CCK-8 Detection
Cell proliferation was determined using Cell Counting Kit-8 (Dojindo, Kumamoto, Japan), strictly following the kit instructions. The specific steps were as follows. The transfected cells were prepared as 5×103 cells/well and seeded in 96-well plates in the growth medium. After 0, 24, 48, and 72 hours of incubation, each well was added with CCK-8 solution (20 μL) for 2 hours at 37°C. Then, the optical density (OD) at 450 nm was determined by a microplate reader (Thermo Fisher Scientific, United States).
Transwell Experiment
RCC cell migration and invasion cells were measured using 24-well Transwell plates (8 μm; BD Biosciences, San Jose, California). The migration and invasion of RCC cells were measured in test chambers with or without Matrigel (BD Biosciences, San Jose, CA, USA). The serum-free medium containing 2×104 transfected cells was added to the upper chamber, and the medium containing 10% FBS was added to the lower chamber as chemotactic agent. After 24 hours culture at 37°C, cells migrating to or invading the basal chamber were immobilized, dyed and finally calculated in five fields at random under the microscope (Olympus, Japan).
Flow Cytometry (FC)
In this study, cell apoptosis was tested by Annexin V-FITC/PI staining, and the operating procedures were strictly in accordance with the kit instructions. The specific protocol was as follows. After adjusting the transfected cells to 1×105 cells/mL, they were collected in 6-well plates, immersed in Annexin V-FITC and PI with 5 µL each, and incubated in darkness for 15 min for detection with a Flow Cytometer (BD LSRII; BD Pharmingen).
Dual-Luciferase Report
Firstly, online target gene prediction sites including TargetScan,15 starBase,16 miRDB17 and miRTarBase18 were used to predict downstream target genes of mir-26a-5p. The 3ʹ-UTR of E2F7 containing the putative miR-26a-5p binding site was subsequently amplified by PCR. Then, pmirGLO-E2F7-WT-3ʹUTR plasmid was established by cloning the wild-type (WT) 3ʹ-UTR into the downstream of the pmirGLO vector firefly luciferase gene. While pmirGLO-E2F7-MT-3ʹUTR was generated by mutating (MT), the plasmid at miR-26a-5p’s binding sites. All the constructs were validated by sequencing. 786-O and A498 cells were planted in 24-well plates and co-transfected with miR-26a-5p-mimics, interfered mimics and WT/MT 3ʹUTR plasmids. Luciferase activity was measured 48 h after transfection.
Nude Mouse Tumorigenicity Assay
Ten BALB/c-nu nude mice (6 weeks, 16–18 g) were purchased from Beijing Charles River Company. 786-O cells (6×106 cells) with lentivirus overexpressed miR-26a-5p (LV-miR-26a-5p) were injected into the axilla of the right forelimb of BALB/c-nu nude mice. On the 0th, 7th, 14th, 21st, and 28th days after implantation, the tumor volume and tumor size [formula: (length×width2)/2 (mm3)] were measured. Twenty-eight days later, the animals were euthanized, and the tumor was excised and weighed. The animal experiment was ratified by the Animal Ethics Committee, of the People’s Hospital of Zhengzhou, and was conducted in accordance with the Guidelines for the Care and Use of Experimental Animals of the United States National Institutes of Health.19
Statistical Analysis
In this study, GraphPad 7 software package was employed for data analysis and image rendering. The measurement data were recorded as mean±standard deviation (mean±SD) and compared by the independent sample t-test between groups. The counting data were recorded as percentage (%), and verified by chi-square test (denoted by χ2). One-way analysis of variance (ANOVA) was employed for intra-group comparisons (recorded as F), and LSD-t was adopted for post hoc pairwise comparisons. Multiple time point expressions were analyzed by repeated measure ANOVA (denoted by F), and tested by Bonferroni. Correlation analysis of each gene was performed by Pearson test. The plotting and analysis of patient overall survival (OS) were done by Kaplan-Meier (K-M) survival curves and Log rank test, respectively, and the independent factors affecting the prognosis were performed by Cox regression. When P<0.05, there was a statistical difference.
Results
Reduced miR-26a-5p in RCC and Indicated Poor Prognosis
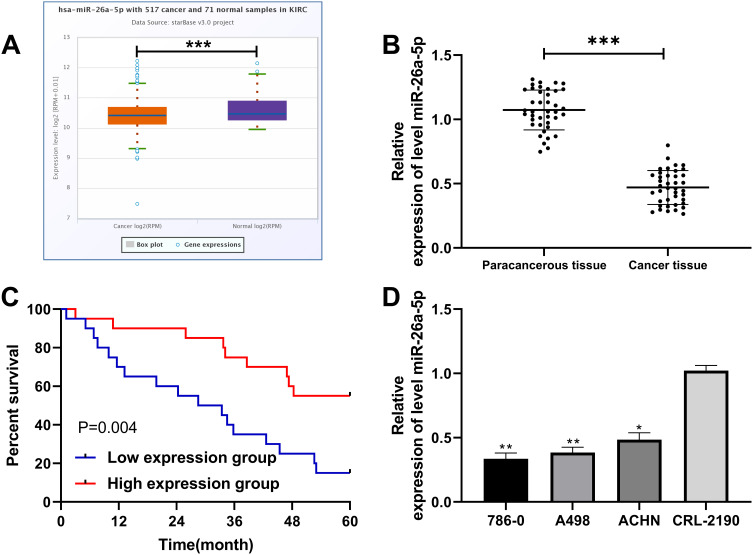
First of all, in order to verify the expression of miR-26a-5p in RCC, we analyzed its profile in RCC in the TCGA database through the starBase online analysis website. It was found that miR-26a-5p was under-expressed in RCC (Figure 1A). QRT-PCR detection of tumor tissues in RCC patients also identified lower miR-26a-5p in tumor tissues than in paracancerous tissues (Figure 1B). In addition, patients were classified into high- and low- miR-26a-5pexpression groups with the median value as the boundary, and the correlation between miR-26a-5p expression and patient pathological data were further observed. The results showed that patients with low miR-26a-5p expression had remarkably increased odds of high TNM stage and lymphatic metastasis (Table 1). Moreover, K-M survival analysis found a dramatically lower five-year survival rate of patients with low expression compared with those with high expression (Figure 1C). According to Cox regression analysis, miR-26a-5p was an independent risk factor affecting patient outcomes (Table 2). We also detected the expression of miR-26a-5p in RCC cells and found its markedly reduced expression in RCC cells compared with normal renal cells (Figure 1D).
Figure 1.
MiR-26a-5p expression and survival in RCC patients. (A) StarBase online website analysis of the expression of miR-26a-5pin RCC patients in TCGA database. ***Indicates P<0.001. (B) qRT-PCR detection of the relative expression of miR-26a-5p in tumor tissue of RCC patients. ***Indicates P<0.001. (C) K-M survival analysis of 5-year survival of patients with high and low expression of miR-26a-5p. (D) qRT-PCR detection of the relative expression of miR-26a-5p in RCC cells. *Indicates P<0.05 vs CRL-2190, and **Indicates P<0.01 vs CRL-2190.
Table 1.
Relationship Between miR-26a-5p and Pathological Data of RCC Patients
| Factors | miR-26a-5p | P value | ||
|---|---|---|---|---|
| Low Expression (n=20) | High Expression (n=20) | |||
| Gender | 0.507 | |||
| Male (n=26) | 14 | 12 | ||
| Female (n=14) | 6 | 8 | ||
| Age | 0.327 | |||
| ≥60 years old (n=25) | 11 | 14 | ||
| <60 years old (n=15) | 9 | 6 | ||
| Tumor size | 0.057 | |||
| ≥7cm (n=22) | 8 | 14 | ||
| <7cm (n=28) | 12 | 6 | ||
| Fuhrman grade | 0.749 | |||
| I+II (n=23) | 11 | 12 | ||
| III+IV (n=17) | 9 | 8 | ||
| Lymph node metastasis | 0.011 | |||
| Negative (n=18) | 5 | 13 | ||
| Positive (n=22) | 15 | 7 | ||
| TNM stage | 0.004 | |||
| I+II (n=19) | 5 | 14 | ||
| III+IV (n=21) | 15 | 6 | ||
Abbreviation: TNM, tumor node metastasis.
Table 2.
Cox Regression Analysis
| Factors | Univariate | Multivariate | ||||
|---|---|---|---|---|---|---|
| P value | HR | 95CI% | P value | HR | 95CI% | |
| Gender | 0.283 | 1.578 | 0.685–3.634 | |||
| Age | 0.632 | 0.827 | 0.379–1.802 | |||
| Tumor size | 0.650 | 1.197 | 0.550–2.608 | |||
| Fuhrman grade | 0.886 | 0.944 | 0.433–2.060 | |||
| Lymph node metastasis | 0.028 | 0.408 | 0.184–0.906 | 0.045 | 0.436 | 0.194–0.980 |
| TNM stage | 0.049 | 0.443 | 0.196–0.998 | 0.066 | 0.428 | 0.173–1.058 |
| miR-26a-5p | 0.007 | 0.322 | 0.142–0.728 | 0.010 | 0.338 | 0.148–0.772 |
Abbreviation: TNM, tumor node metastasis.
MiR-26a-5p Up-Regulation Inhibited RCC Cell Growth and Enhanced Apoptosis
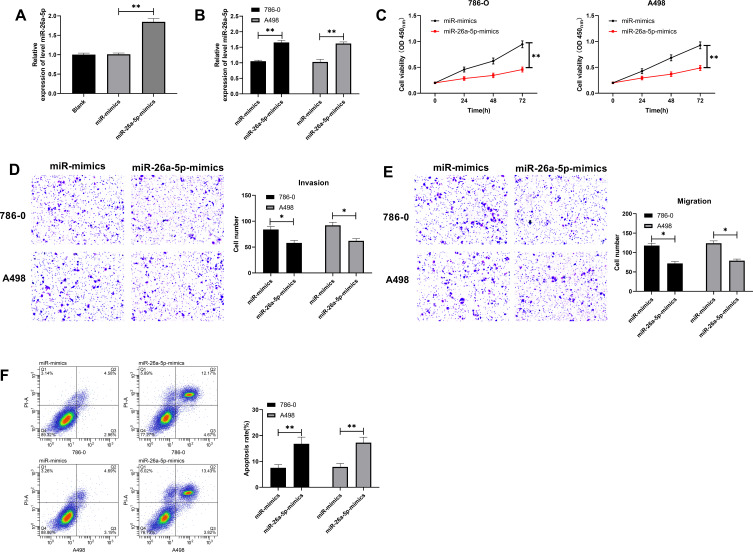
To further explore the effect of miR-26a-5pon RCC cell growth, we constructed miR-26a-5p-mimics (Figure 2A) and transfected it into 786–0 and A498 cells to observe the cell changes (Figure 2B). CCK-8 assay found that the cell proliferation ability was significantly weakened after miR-26a-5p-mimics transfection compared with miR-mimics (Figure 2C). Transwell detection showed that the number of cell invasion and migration statistically decreased after miR-26a-5p-mimics transfection compared with miR-mimics (Figure 2D–E). In addition, FC exhibited that miR-26a-5p-mimics transfection markedly induced RCC cell apoptosis (Figure 2F), suggesting that miR-26a-5p may be a feasible therapeutic marker for RCC.
Figure 2.
Effects of up-regulating miR-26a-5p on the growth of RCC cells. (A) qRT-PCR detection of the relative expression of miR-26a-5p after transfection with miR-26a-5p-mimics. **Indicates P<0.01. (B) qRT-PCR detection of the relative expression of miR-26a-5p in RCC cells after transfection with miR-26a-5p-mimics. **Indicates P<0.01. (C) CCK-8 assay of the proliferation of RCC cells after transfection with miR-26a-5p-mimics. **Indicates P<0.01. (D–E) Transwell detection of the changes of invasion and migration of RCC cells after transfection with miR-26a-5p-mimics. *Indicates P<0.05. (F) Flow cytometry of the change of RCC cell apoptosis rate after transfection with miR-26a-5p-mimics. **Indicates P<0.01.
MiR-26a-5p Could Target E2F7
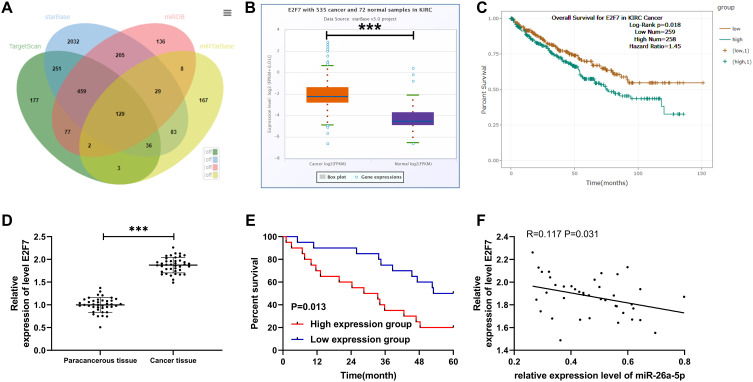
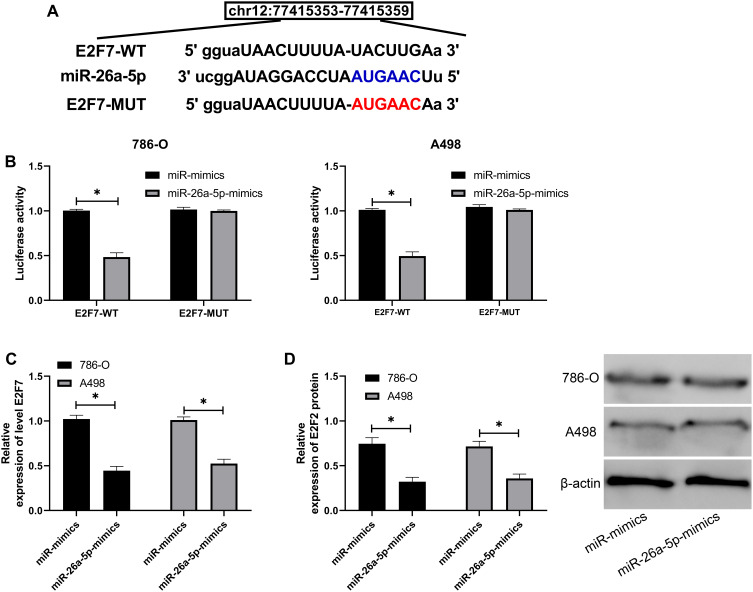
It has been reported that miR is involved in the growth of tumor cells by targeting downstream target genes. We predicted the downstream target genes of miR-26a-5p through TargetScan, starBase, miRDB, and miRTarBase (Figure 3A), and found a targeted binding relationship between miR-26a-5p and E2F7 in all the above four online analysis sites (Figure 4A). In order to determine the expression of E2F7 in RCC, we used GEPIA2 to analyze the expression and survival of E2F7in RCC in TCGA data. We found that E2F7increased in RCC (Figure 3B), and the OS rate of patients with high E2F7decreased notably (Figure 3C). Through qRT-PCR analysis, we also found that E2F7was increased in RCC patients, and K-M survival analysis revealed a decreased five-year survival rate in patients with high E2F7(Figure 3D–E). Pearson correlation test identified that miR-26a-5p was in negative association with E2F7 in RCC patients (Figure 3F). Therefore, we speculated the existence of a regulatory relationship between the two, which was later confirmed by DLR (Figure 4B). Also, we found significant inhibition of E2F7protein and mRNA after miR-26a-5p-mimics transfection by qRT-PCR and WB (Figure 4C and D). All these demonstrated that miR-26a-5p targeted E2F7and they two may be potential targets for RCC.
Figure 3.
E2F7was highly expressed in RCC and was negatively correlated with miR-26a-5p. (A) Venn diagram was used to map the online tools of TargetScan, starBase, miRDB and miRTarBase to predict the common target gene of miR-26a-5p. (B–C) StarBase online site analysis of the relative expression of E2F7and the survival of RCC patients. (D) qRT-PCR detection of the relative expression of E2F7in RCC patients. ***Indicates P<0.001. (E) K-M test analysis of the survival of RCC patients with high and low E2F7expression. (F) The relationship between miR-26a-5p and E2F7tested by Pearson.
Figure 4.
MiR-26a-5p can target E2F7. (A) The specific binding site of miR-26a-5p and E2F7. (B) Dual-luciferase reporter detected that miR-26a-5ptargeted E2F7. *Indicates P<0.05. (C and D) qRT-PCR and Western blot detection of the relative mRNA and protein expression of E2F7in cells transfected with miR-26a-5p-mimics. *Indicates P<0.05.
MiR-26a-5p Down-Regulation Reversed E2F7 Knockdown-Induced Inhibition on RCC Cell Growth and Metastasis
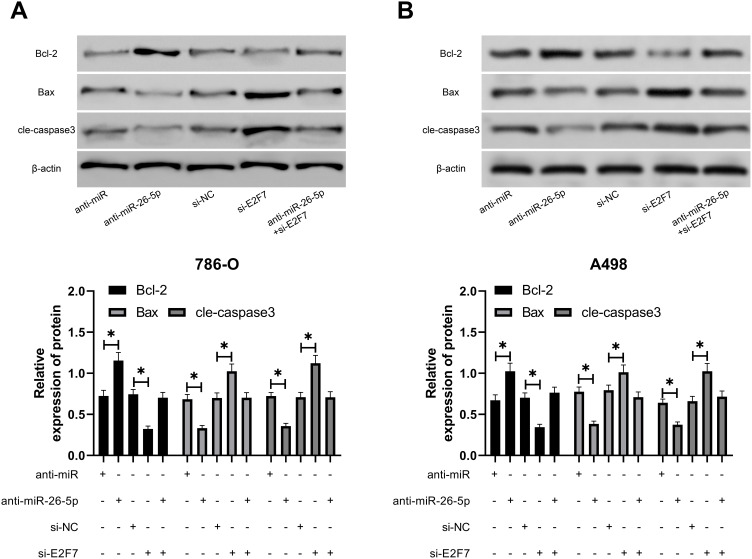
We have determined that miR-26a-5p targets E2F7in the preceding study, but to check out whether there is a regulatory relationship between the two, we conducted a rescue experiment. After transfection of si-E2F7into RCC cells, the proliferation capacity of si-E2F7transfected cells as assessed by CCK-8 assay was found to be remarkably reduced compared with that of si-NC cells (Figure 5A), while the cell invasion and migration as detected by Transwell assay was observed to be decreased notably (Figure 5B and C). FC analysis revealed that cell apoptosis was enhanced after transfection of si-E2F7(Figure 5D). WB detection also found that Bcl-2 protein expression in si-E2F7 transfected cells declined dramatically, while Bax and cle-caspase3 elevated notably (Figure 5E and F). However, the above results were reversed after anti-miR-26a-5p and si-E2F7 co-transfection indicating that miR-26a-5p regulated E2F7to participate in the progression of RCC cells.
Figure 5.
MiR-26a-5p regulated E2F7to participate in the occurrence of RCC. (A) CCK-8 assay of the changes of proliferation ability of RCC cells transfected with anti-miR-26a-5p and si-E2F7. *Indicates P<0.05, and **Indicates P<0.01. (B and C) Transwell assay showed that the number of invasion and migration of RCC cells increased after transfection of miR-26a-5p-mimics. *Indicates P<0.05. (D) Flow cytometry of the change of RCC cell apoptosis rate after transfection with miR-26a-5p-mimics. *Indicates P<0.05, and **Indicates P<0.01. *Indicates P<0.05.
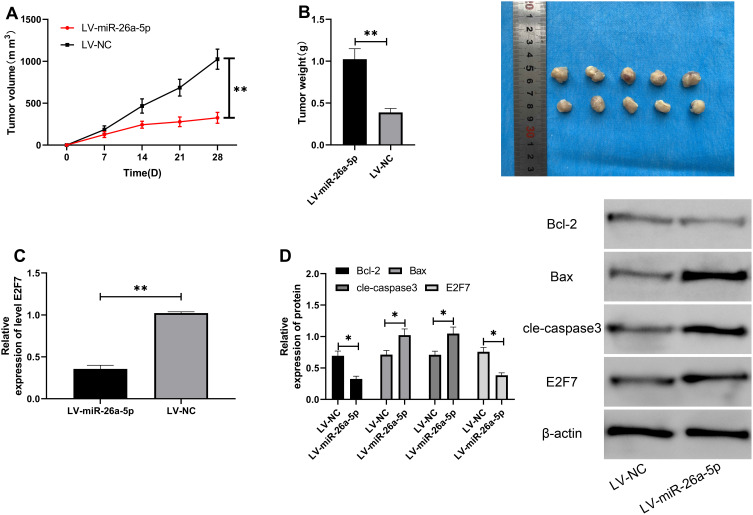
MiR-26a-5p Up-Regulation Inhibited Tumor Volume Growth in Nude Mice
At the end of the study, we conducted in vivo experiments. Through the establishment of nude mouse model, we found that the tumor volume of nude mice injected with LV-miR-26a-5p was evidently reduced compared with LV-NC (Figure 6A), and the tumor mass was also noticeably reduced after the execution (Figure 6B). Furthermore, we detected the tumor tissue by qRT-PCR and found that the E2F7was inhibited in the tumor tissue of nude mice intervened by LV-miR-26a-5p (Figure 7A and B). WB detection showed that E2F7and Bcl-2 protein levels were evidently decreased, and Bax, Cle-caspase3 protein levels were noticeably increased in nude mouse tumor tissues injected with LV-miR-26a-5p (Figure 7C–D), suggesting that miR-26a-5p could be used as a potential therapeutic target for RCC.
Figure 6.
Effect of MiR-26a-5p regulated E2F7on apoptotic protein of RCC cells. (A and B) Western blot of the changes of apoptosis-related proteins in RCC cells transfected with anti-miR-26a-5p and si-E2F7. *Indicates P<0.05.
Figure 7.
Up-regulation of miR-26a-5p inhibited the growth of tumor volume in nude mice. (A) Changes of tumor volume in nude mice. **Indicates P<0.01. (B) Changes of tumor weight in nude mice within 28 days. **Indicates P<0.01. (C) qRT-PCR detection of the expression of E2F7in tumor tissues of nude mice. **Indicates P<0.01. (D) Western blot detection of the relative expression of apoptosis-related proteins and E2F7in tumor tissues of nude mice. *Indicates P<0.05.
Discussion
RCC is highly latent. It is shown that20 RCC is not sensitive to chemoradiotherapy, which leads to the lack of effective treatment options for patients with advanced stage. Therefore, it is particularly important to probe into the potential therapeutic targets. In the present study, we find that miR-26a-5p was underexpressed in RCC patients and could inhibit the growth and metastasis of RCC cells by targeting E2F7, which is a promising therapeutic target.
Located on the human 12q14.1 chromosome, miR-26a-5p shows different levels in lung cancer,21 liver cancer22 and gastric cancer.23 However, little research has been done on the role of miR-26a-5p in RCC. In our research, we first found that miR-26a-5p was highly expressed in RCC through TCGA database analysis, and then verified it by detecting tumor tissues of patients with RCC. In addition, the results identified that the five-year survival rate was statistically reduced in patients with low miR-26a-5p expression, indicating that miR-26a-5p was an independent prognostic indicator of RCC. These findings indicate that miR-26a-5p is involveon of RCC and is a potential indicator to observe the prognosis of RCC patients, but whether it is a feasible treatment target for RCC remains unclear.
Effectively inhibiting the tumorigenic phenotype of tumor cells and promoting apoptosis is a vital means to inhibit tumor growth and metastasis.24 To confirm that miR-26a-5p regulates RCC growth and metastasis, we examined its expression to found its low expression in RCC cells. Then, miR-26a-5p-mimics was transfected into RCC cells to further observe the functional changes. It turned out that miR-26a-5p up-regulation resulted in dramatically suppressed malignant phenotype of cells and induced apoptosis, indicating its role as a possible therapeutic marker for RCC. MiRs have been reported to be implicated in the development of RCC by targeting downstream target genes. For example, Xu et al25 showed that miR-143 inhibited the transfer potential of RCC cells by inhibiting ABL2. Another research identified that26 miR-124 and miR-203 co-inactivated the EMT pathway in CCRCC by co-regulating ZEB2. Aiming to further inquire into the in-depth mechanism of miR-26a-5p, we predicted its downstream target genes and found the shared binding sites with E2F7. E2F7, highly expressed in a variety of tumors, is an initiator involved in cell cycle regulation, apoptosis and differentiation.27 However, previous studies did not find any link between E2F7and RCC. In this study, we foud increased E2F7expression in tissues of RCC patients, and the 5-year survival rate of patients with high E2F7levels was dramatically decreased, which accorded with the results in TCGA database. Later, we identified a negative association between E2F7and miR-26a-5p in RCC patients, which suggested that there might be a targeted binding relationship between the two. Therefore, we carried out DLR assay for verification. It was found that miR-26a-5p-mimics inhibited the fluorescence activity of E2F7-WT, and the expression of E2F7as detected by qRT-PCR and WB was found to be remarkably suppressed in RCC cells after miR-26a-5p-mimics transfection, demonstrating that miR-26a-5p targeted to regulate E2F7. To verify that the two have the same regulatory role in RCC cells, we carried out rescue experiments at the end of the study. We found that after knocking down E2F7, cell proliferation, invasion and migration were notably inhibited, and apoptosis was induced. Moreover, through WB assay, we observed that Bcl-2 protein expression decreased while Bax and cle-caspase3 protein expression increased, which suggested that inhibition of E2F7could promote cell apoptosis, thereby inhibiting cell proliferation, invasion and migration. However, the above results were reversed after anti-miR-26a-5p and si-E2F7 co-transfection, indicating that miR-26a-5p regulated E2F7to participate in the occurrence of RCC. What is more, in vivo experiment revealed that the tumor volume and mass decreased in nude mice injected with LV-miR-26a-5p. WB detection exhibited that E2F7 and Bcl-2 protein expression in tumor tissues of nude mice injected with LV-miR-26a-5p decreased evidently, while Bax and cle-caspase3 protein expression increased notably. These studies confirm that by regulating E2F7, miR-26a-5p promotes apoptosis of RCC cells, thus preventing cell proliferation and invasion.
All in all, our study confirms that miR-26a-5p is under-expressed in RCC, and by targeting E2F7, it can enhance apoptosis and inhibit proliferation and invasion of RCC cells, which may be a feasible therapeutic target for RCC.
Disclosure
The authors report no conflicts of interest for this work.
References
- 1.arcinoma: standards and controversies. World J Urol. 2018;36:1889–1890. [PMID: 30209553]. doi: 10.1007/s00345-018-2490-5 [DOI] [PubMed] [Google Scholar]
- 2.Molina AMD. A multidisciplinary approach for the management of earlier stage renal cell carcinoma. Urol Oncol. 2018;36:15–16. [PMID: 29198585]. doi: 10.1016/j.urolonc.2017.09.020 [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. [PMID: 30620402]. doi: 10.3322/caac.21551 [DOI] [PubMed] [Google Scholar]
- 4.De Meerleer G, Khoo V, Escudier B, et al. Radiotherapy for renal-cell carcinoma. Lancet Oncol. 2014;15:e170–e177. [PMID: 24694640]. doi: 10.1016/S1470-2045(13)70569-2 [DOI] [PubMed] [Google Scholar]
- 5.Roos FC, Rubben H, Stief C, Stockle M, Thuroff JW. [Surgical treatment for renal cell carcinoma]. Aktuelle Urol. 2010;41(4):252–256. German. doi: 10.1055/s-0030-1247480 [DOI] [PubMed] [Google Scholar]
- 6.Sun M, Choueiri TK. Kidney cancer: recurrence in renal cell carcinoma: the work is not done. Nat Rev Urol. 2016;13:246–247. [PMID: 27030528]. doi: 10.1038/nrurol.2016.57 [DOI] [PubMed] [Google Scholar]
- 7.Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol. 2018;141:1202–1207. [PMID: 29074454 PMCID: PMC5889965]. doi: 10.1016/j.jaci.2017.08.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gebert LFR, MacRae IJ. Regulation of microRNA function in animals. Nat Rev Mol Cell Biol. 2019;20:21–37. [PMID: 30108335 PMCID: PMC6546304]. doi: 10.1038/s41580-018-0045-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peng Y, Shen X, Jiang H, et al. miR-188-5p suppresses gastric cancer cell proliferation and invasion via targeting ZFP91. Oncol Res. 2018;27:65–71. [PMID: 29471891]. doi: 10.3727/096504018X15191223015016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y, Dong D, Jiang S, et al. miR-216b post-transcriptionally downregulates oncogene KRAS and inhibits cell proliferation and invasion in clear cell renal cell carcinoma. Cell Physiol Biochem. 2018;49:1755–1765. [PMID: 30231239]. doi: 10.1159/000493621 [DOI] [PubMed] [Google Scholar]
- 11.Wang H, Hu Z, Chen L. Enhanced plasma miR-26a-5p promotes the progression of bladder cancer via targeting PTEN. Oncol Lett. 2018;16:4223–4228. [PMID: 30197668 PMCID: PMC6126335]. doi: 10.3892/ol.2018.9163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song Q, Liu B, Li X, et al. MiR-26a-5p potentiates metastasis of human lung cancer cells by regulating ITGbeta8- JAK2/STAT3 axis. Biochem Biophys Res Commun. 2018;501:494–500. [PMID: 29746867]. doi: 10.1016/j.bbrc.2018.05.020 [DOI] [PubMed] [Google Scholar]
- 13.Hauser S, Wulfken LM, Holdenrieder S, et al. Analysis of serum microRNAs (miR-26a-2*, miR-191, miR-337-3p and miR-378) as potential biomarkers in renal cell carcinoma. Cancer Epidemiol. 2012;36:391–394. [PMID: 22542158]. doi: 10.1016/j.canep.2012.04.001 [DOI] [PubMed] [Google Scholar]
- 14.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta delta C(T)) method. Methods. 2001;25:402–408. [PMID: 11846609]. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 15.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015:4 [PMID: 26267216 PMCID: PMC4532895]. doi: 10.7554/eLife.05005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(Database issue):D92–D97. [PMID: 24297251 PMCID: PMC3964941]. doi: 10.1093/nar/gkt1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu W, Wang X. Prediction of functional microRNA targets by integrative modeling of microRNA binding and target expression data. Genome Biol. 2019;20:18 [PMID: 30670076 PMCID: PMC6341724]. doi: 10.1186/s13059-019-1629-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chou CH, Shrestha S, Yang CD, et al. miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2018;46:D296–D302. [PMID: 29126174 PMCID: PMC5753222]. doi: 10.1093/nar/gkx1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.National Research Council. In the Editor Guide for the Care and Use of Laboratory Animals. Washington (DC); 2011. [Google Scholar]
- 20.Mery B, Rancoule C, Rowinski E, et al. Cancer du rein et radiothérapie: radiorésistance et au-delà. Bull Cancer. 2018;105(Suppl 3):S280–S285. [PMID: 30595157]. doi: 10.1016/S0007-4551(18)30383-7 [DOI] [PubMed] [Google Scholar]
- 21.Liang R, Xiao G, Wang M, et al. SNHG6 functions as a competing endogenous RNA to regulate E2F7 expression by sponging miR-26a-5p in lung adenocarcinoma. Biomed Pharmacother. 2018;107:1434–1446. [PMID: 30257360]. doi: 10.1016/j.biopha.2018.08.099 [DOI] [PubMed] [Google Scholar]
- 22.Chang L, Li K, Guo T. miR-26a-5p suppresses tumor metastasis by regulating EMT and is associated with prognosis in HCC. Clin Transl Oncol. 2017;19:695–703. [PMID: 27864783]. doi: 10.1007/s12094-016-1582-1 [DOI] [PubMed] [Google Scholar]
- 23.Li Y, Wang P, Wu LL, Yan J, Pang XY, Liu SJ. miR-26a-5p inhibit gastric cancer cell proliferation and invasion through mediated Wnt5a. Onco Targets Ther. 2020;13:2537–2550. [PMID: 32273724 PMCID: PMC7108881]. doi: 10.2147/OTT.S241199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiao Y, Zhao Q, Du B, Chen HY, Zhou DZ. MicroRNA-187 inhibits growth and metastasis of osteosarcoma by downregulating S100A4. Cancer Invest. 2018;36:1–9. [PMID: 29303365]. doi: 10.1080/07357907.2017.1415348 [DOI] [PubMed] [Google Scholar]
- 25.Xu B, Wang C, Wang YL, et al. miR-143 inhibits renal cell carcinoma cells metastatic potential by suppressing ABL2. Kaohsiung J Med Sci. 2020;36(8):592–598. [PMID: 32196963]. doi: 10.1002/kjm2.12207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen J, Zhong Y, Li L. miR-124 and miR-203 synergistically inactivate EMT pathway via coregulation of ZEB2 in clear cell renal cell carcinoma (ccRCC). J Transl Med. 2020;18:69 [PMID: 32046742 PMCID: PMC7014595]. doi: 10.1186/s12967-020-02242-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitxelena J, Apraiz A, Vallejo-Rodriguez J, et al. An E2F7-dependent transcriptional program modulates DNA damage repair and genomic stability. Nucleic Acids Res. 2018;46:4546–4559. [PMID: 29590434 PMCID: PMC5961008]. doi: 10.1093/nar/gky218 [DOI] [PMC free article] [PubMed] [Google Scholar]