Abstract
Background
Cardiovascular disease is one of the most common causes of morbidity and mortality worldwide. Several cardiovascular diseases require therapy with warfarin, an anticoagulant with large interindividual variability resulting in dosing difficulties. The selected genes and their polymorphisms have been implicated in several Genome-Wide Association Study (GWAS) to be associated with cardiovascular disease.
Objective
The goal of this study is to discover if there are any associations between rs646776 of PSRC1, rs660240 and rs12740374 of CELSR2, and rs602633 of SORT1 to coronary heart disease (CHD) and warfarin dose variability in patients diagnosed with cardiovascular disease undergoing warfarin therapy.
Methods
The study was directed at the Queen Alia Hospital Anticoagulation Clinic in Amman, Jordan. DNA was extracted and genotyped using the Mass ARRAY™ system, statistical analysis was done using SPSS.
Results
The study found several associations between the selected SNPs with warfarin, but none with cardiovascular disease. All 4 studied SNPs were found to be correlated to warfarin sensitivity during the stabilization phase except rs602633 and with warfarin dose variability at the initiation phase. CELSR2 SNPs also showed association with dose variability during the stabilization phase. Also, rs646776 and rs12740374 were linked to warfarin sensitivity over the initiation phase. Only rs602633 was associated with INR treatment outcomes.
Conclusion
The findings presented in this study found new pharmacogenomic associations for warfarin, that warrant further research in the field of genotype-guided warfarin dosing.
Keywords: warfarin, SNPs, pharmacogenetics, Jordan
Introduction
Cardiovascular disease, namely heart artery disease (CHD) or coronary artery disease (CAD) is one of the main causes of global morbidity and mortality, with the World Health Organization (WHO) predicting that it will become the leading cause of death by 2030.1–4 Risk factors for the development of CAD are divided into modifiable (hypertension, diabetes, obesity, hypercholesteremia, smoking, and sedentary lifestyles) and non-modifiable (age, gender, genetic). 40–50% of CAD risk is attributable to non-modifiable hereditary factors.5,6 Recent GWAS have identified several genetic risk factors to be associated with CAD, including mutations in PSRC1, CELSR2, SORT1 genes on chromosome 1p13.3.4,7 Since then, several studies have linked polymorphisms in these candidate genes to the development of CAD through their effect on hypercholesteremia and elevated levels of circulating LDL-C, known contributors to atherosclerosis.8,9 Being a multifactorial disease with non-modifiable genetic aspects, the early detection of significant mutations through genetic analysis and genotyping could potentially improve outcomes in patients if treatment is initiated early. However, further exploration is necessary.
The Cadherin EGF LAG Seven Pass Type Receptor 2 gene (CELSR2) encodes a Cadherin responsible for receptor-ligand-based cellular interactions and contact-mediated cellular adhesion. The Proline-Serine Rich Coiled Coil (PSRC1) gene is responsible partly for microtubule destabilization. The Sortilin 1 (SORT1) gene encodes the Sortilin 1 protein, which responsible for a significant role in lipid uptake. In addition to being linked with CAD, studies have found several other associations to the aforementioned genes. PSRC1 gene was found to be associated with atrial fibrillation and hypertension.10 SORT1 gene was also associated with early-onset myocardial infarction, abdominal aortic aneurysms, and aortic valve calcification.11,12 CELSR2 gene has been linked with lower protein C levels, a vitamin K-dependent glycoprotein synthesized by the liver that has anticoagulant effect.13 Warfarin is the most commonly used anticoagulant medication worldwide. It is used in the treatment of a variety of pathologies including, but not limited to atrial fibrillation and deep venous thrombosis.14–16
Warfarin is a highly water-soluble oral medication, consisting of R and S isomers in an equal distribution. The more potent s-warfarin is metabolized in the liver by CYP2C9 gene. It is absorbed through the gastrointestinal system and has a very high bioavailability.17
Warfarin exerts an anticoagulation effect through binding non-competitively to vitamin K epoxide reductase subunit 1 (VKORC1), an enzyme needed for the clotting factors II, VII, IX, and X activation.18 The International Normalized Ration (INR) is a tool used to monitor and measure the effect of anticoagulants status. Careful monitoring of the INR is a must in order to achieve the desired anticoagulatory effect and avoid the risk of over-anticoagulation.19 Patients being treated with warfarin are typically expected to maintain an INR of 2–3.14 Large interindividual variation exists in response to warfarin; this is partly due to genetic variability. VKORC1 and CYP2C9 polymorphisms are responsible for a significant portion of the variation on warfarin dosage.20 Warfarin dosing is divided into an initiation phase and a stabilization phase. The initiation phase consists of a period when INR levels are fluctuating, and dose modifications are necessary to achieve the desired INR. Once a patient has a therapeutic INR for two consecutive visits, the patient is considered to be in the stabilization phase of therapy.15
Owing to its slow onset of action, wide interindividual variability, multiple drug–drug interactions (rifampin, phenobarbital, quinidine), and narrow therapeutic index, warfarin is one of the drugs that most frequently cause adverse effects.17–19 Warfarin major adverse effect is bleeding, which usually occurs during the first 3 months of treatment onset. Therefore, reaching the maintenance phase of therapy earlier is necessary. Warfarin dosing poses a great challenge for physicians prescribing the drug. As a solution to this issue, several studies and trials have been conducted to assess the best method for predicting the necessary warfarin dose. The two methods most heavily compared in literature are genotype-guided dosing vs dosing using clinical algorithms. Clinical algorithms rely on clinical factors known to affect bleeding or clotting tendencies such as age, diabetes, smoking, history of thromboembolic events, and concurrent use of medications known to interact with warfarin.19,20 The goal of the current study is to assess for genotypic associations between selected SNPs in PSRC1, CELSR2, and SORT1 genes and susceptibility to cardiovascular disease and warfarin dose variability in Jordanian patients.
Patients and Methods
Study Design and Patients
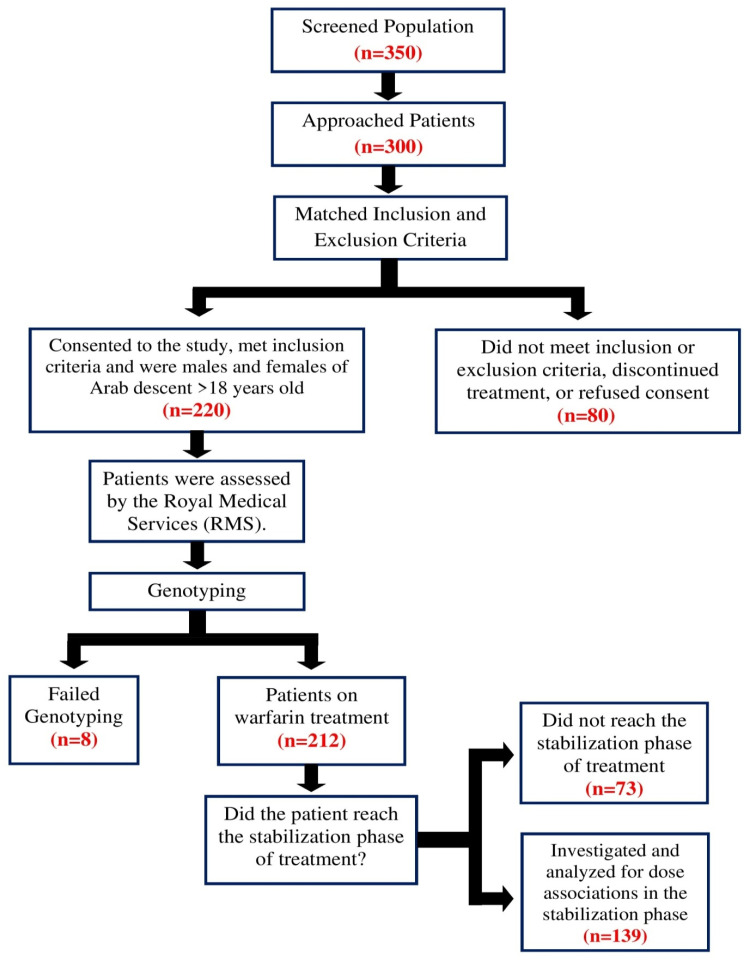
The study populations consist of 220 healthy control subjects were defined as 18 years of age or older and without CHD and 212 patients from a Jordanian Arab population who have cardiovascular disease and are being treated with warfarin from the anticoagulation clinic at Queen Alia Hospital in Amman, Jordan. Informed consent was obtained from patients who participated in the study, and ethical approval was obtained from the Ethical Committees at the Jordan University of Science and Technology and the Royal Medical Services. The inclusion criteria were patients >18 years old who have been on warfarin treatment for at least 3 months and have had regular visits to the anticoagulation clinic. The exclusion criteria were patients with lost clinical data, consent refusal, discontinuation of warfarin treatment or those taking medications that had pharmacokinetic interactions with warfarin according to the Dutch standards (Standaard Afhandeling Cumarine Interacties, 2003) . The included patients were assessed by the Royal Medical Services and samples were sent for genotyping. Eight samples failed to genotype, either due to low DNA content or degradation, leaving 212 patients from whom data for the initiation phase of therapy was obtained. 139 of these patients reached the stabilization phase, whose data were used for analysis during the stabilization phase. The study design is summarized in (Figure 1).
Figure 1.
A flow chart outlining the study design.
Data Collection and Follow-up Time
Demographical data were collected about each patient including their age, gender, body mass index (BMI), smoking status, and eating habits. In addition to information about the indication for anticoagulation, therapeutic INR range, INR measurements, warfarin dosage, comorbidities, and lipid profile (total cholesterol, LDL, and HDL). Our previous study on the same patient pool found no significant association between the collected data and warfarin metabolizer status.15
SNP Selection and Genotyping
Candidate SNPs relating to warfarin pharmacokinetics and pharmacodynamics were chosen from public databases. Four SNPs were selected from the 1p13.3 locus containing 3 genes: PSRC1, CELSR2, and SORT1. Table 1 lists the chosen genes, the SNP IDs, and location. DNA extraction was done using Wizard Genomic DNA Purification Kit (Promega), quantitative samples were sent to the Australian Genome Research Facility (AGRF) who use Mass ARRAY™ System (iPLEX GOLD) (Sequenom, San Diego, CA, USA). Mass ARRAY™ protocols and primers information for the selected SNPs can be provided upon request.
Table 1.
Genes and SNPs Characteristics
| Gene | SNP ID | Chr Positiona | SNP | SNP Type |
|---|---|---|---|---|
| PSRC1 | rs646776 | 1:109,275,908 | C/T | Downstream Variant |
| CELSR2 | rs660240 | 1:109,275,216 | T/C | 3 Prime UTR Variant |
| rs12740374 | 1:109,274,968 | G/T | 3 Prime UTR Variant | |
| SORT1 | rs602633 | 1:109,278,889 | T/G | N/Ab |
Notes: aChromosome positions are based on NCBI Human Genome Assembly Build. b.N/A: not available.
Outcome Measurements
Our study focuses on finding associations for the selected SNPs for their effect on warfarin sensitivity, responsiveness, and dosage variability. We used Gordon (2009)21 to divide patients into 3 categories for measuring sensitivity:
Extensive Metabolizes/Warfarin Resistance: Patients in this group require higher doses of high doses >49mg/week.
Moderate Metabolizes/Warfarin Response: Patients in this group require doses between 21–49mg/week.
Poor Metabolizes/Warfarin Sensitive: Patients in this group require doses <21mg/week.
We used Higashi et al (2012)22 for classifying patients according to response:
Good Responders: Patients with INR values within the therapeutic range.
Poor Responders: Patients with INR values that failed to reach the target INR.
Ultra-Responders: Patients with INR values that surpassed the target INR.
The maintenance dose was determined by the average of all administered doses during the stabilization phase of warfarin therapy, which in turn is defined as the calculated dose that is unchanged for two consecutive visits with INR values within the therapeutic range.
Statistical Analysis
The statistical package for social sciences (SPSS) v21.0 was used for all statistical analysis done. The p-value was calculated for Minor Allele Frequency (MAF) and Hardy Weinberg Equilibrium (HWE). To evaluate for HWE deviation, the Pearson x2 test was used. To test for association between selected SNPs and warfarin response, multiple analysis tests were performed including Chi-Square, Non-parametric correlation Krusak Wallis, Tukey HSD post hoc multiple comparisons, and unidirectional analysis of variance.
Results
The demographical data published in Al-Eitan et.al (2018) previous study showed no significance between the data collected and warfarin metabolizer status.15 The mean age and standard deviation for patients in the good metabolizers group, who were the majority was 53.2 [15.3]. The mean ages for patients in the extensive metabolizers and poor metabolizers groups were 47.6 [17.7] and 54.2 [18.4], respectively. The most abundant comorbidity is hypertension and Aspirin is the most common concurrent medication. 41.7% of patients had a target INR of 2–3, while the remaining 58.3% had a target INR of 2.5–3.5. The mean and standard deviation for controls age was 37.5 [13.7] and (BMI) it was 26.9 [4.9]. And 53.6% of them were male, 46.3% were female and 32.3% of the cohort was smokers. Match the demographics in patients with a mean age of 54.1 [15.3] and a mean BMI of 26.9 [4.9] with 55.2% male and 44.8% female and 34.4% of patients were smokers. Patient and control demographics data published in a previous study by Al-Eitan et al.15,23
Association of Selection SNPs to CHD
The MAF of the selected SNPs listed in Table 2. No significant associations were found between the studied polymorphisms and cardiovascular disease when comparing allele frequencies of the selected SNPs in patients with cardiovascular and their frequencies in healthy controls Table 3. The CELSR2 haplotypes also failed to reach significance with cardiovascular disease, as depicted in Table 4.
Table 2.
List of SNPs, Their Minor Allele Frequencies, and HWE p-values
| Gene | SNP ID | MAa | Patients MAFb | Controls MAFb | HWEc P-value |
|---|---|---|---|---|---|
| PSRC1 | rs646776 | C | 0.2 | 0.18 | 0.53 |
| CELSR2 | rs660240 | C | 0.19 | 0.18 | 0.26 |
| rs12740374 | T | 0.18 | 0.16 | 0.49 | |
| SORT1 | rs602633 | T | 0.23 | 0.22 | 0.2 |
Abbreviations: aMA, minor allele; bMAF, minor allele frequency; cHWE, Hardy–Weinberg equilibrium.
Table 3.
The Distributions of PSRC1, CELSR2, and SORT1 SNPs in 212 Cardiovascular Patients and 213 Healthy Controls
| Gene | SNP ID | Model | Patients % | Controls % | P-value* |
|---|---|---|---|---|---|
| PSRC1 | rs646776 | TT/CT/CC | 66/28.8/5.2 | 66.7/30.5/2.8 | 0.44 |
| TT/(CT+CC) | 66/34 | 66.7/33.3 | 0.89 | ||
| (TT+CT)/CC | 94.8/5.2 | 97.2/2.8 | 0.21 | ||
| CELSR2 | rs660240 | CC/TC/TT | 66.8/27.9/5.3 | 67/29.7/3.3 | 0.58 |
| CC(TC+TT) | 66.8/33.2 | 67/33 | 0.97 | ||
| (CC+TC)/TT | 94.7/5.3 | 96.7/3.3 | 0.31 | ||
| rs12740374 | GG/GT/TT | 68.7/27/4.3 | 70/27.7/2.4 | 0.54 | |
| GG/(GT+TT) | 68.7/31.3 | 70/30.1 | 0.78 | ||
| (G+GT)/TT | 95.7/4.3 | 97.7/2.4 | 0.27 | ||
| SORT1 | rs602633 | GG/GT/TT | 62.1/30.8/7.1 | 61.8/33.5/4.7 | 0.53 |
| GG/(GT+TT) | 62.1/37.9 | 61.8/38.2 | 0.95 | ||
| (GG+GT)/TT | 92.9/7.1 | 95.3/4.7 | 0.3 |
Note: *Chi-square test with p< 0.05 is considered significant.
Table 4.
The Distributions of CELSR2 Gene Haplotypes and 211 Cardiovascular Patients Compared to 213 Healthy Controls
| Gene | Haplotypes | Patients (%) | Controls (%) | Odds Ratio (95% CI) | P-value* |
|---|---|---|---|---|---|
| CELSR2 | CG | 0.807 | 0.812 | 1 | — |
| TT | 0.175 | 0.157 | 1.12 (0.78–1.60) | 0.53 | |
| TG | 0.015 | 0.026 | 0.55 (0.20–1.51) | 0.25 |
Notes: *Chi–Square test with p<0.05 is considered significant.
Impact of SNPs to Warfarin Sensitivity During the Initiation Phase of Therapy
Upon studying the relationship between the selected SNPs and their effect on warfarin sensitivity during the initiation phase of therapy, it was noted that major allele homozygotes (11.4%) and heterozygotes (26.6%) for PSRC1 rs646776 had significant associations with being sensitive to warfarin, with the SNP as a whole having an overall significant p-value of 0.028 (p<0.05). Patients homozygous for the minor allele of rs660240 at the CELSR2 gene showed no significant association with sensitivity to the drug during the initiation phase. In addition, patients with the (GG) and (TT) genotypes for rs12740374 exhibited associations with warfarin sensitivity and warfarin moderate response, respectively. Both these SNPs for the CELSR2 gene reached an overall significant p-value. SORT1 gene, on the other hand, did not reach an overall gene association. However, patients heterozygous for rs602633 demonstrated an association with warfarin sensitivity Table 5. Haplotypes distribution for the (TG) and (TT) genotypes at the CELSR2 gene showed significant distribution associations with warfarin sensitivity with the homozygous haploid eliciting a stronger association with a p-value <0.001 Table 6.
Table 5.
Association of PSRC1, CELSR2, and SORT1 SNPs with Warfarin Sensitivity During the Initiation Phase of Therapy of 212 Cardiovascular Patients Treated with Warfarin
| Gene | SNP ID | Genotype | Sensitivea (n= 32) | Moderateb (n= 146) | Resistancec (n= 34) | Overall P-value* |
|---|---|---|---|---|---|---|
| PSRC1 | rs646776 | CC | (0/11) 0.0% | (8/11) 72.7% | (3/11) 27.3% | 0.028 |
| P-value* | 0.15 | 0.78 | 0.3 | |||
| CT | (16/61) 26.6% | (39/61) 63.9% | (6/61) 9.8% | |||
| P-value* | 0.004 | 0.32 | 0.12 | |||
| TT | (16/140) 11.4% | (99/140) 70.7% | (25/140) 17.9% | |||
| P-value* | 0.04 | 0.42 | 0.31 | |||
| CELSR2 | rs660240 | CC | (17/139) 12.2% | (97/139) 69.8% | (25/139) 18% | 0.062 |
| P-value* | 0.12 | 0.02 | 0.15 | |||
| TC | (14/58) 24.1% | (39/58) 67.2% | (5/58) 8.6% | |||
| P-value* | 0.81 | 0.7 | 0.8 | |||
| TT | (0/11) 0.0% | (8/11) 72.7% | (3/11) 27.3% | |||
| P-value* | 0.23 | 0.08 | 0.29 | |||
| rs12740374 | GG | (16/145) 11% | (102/145) 70.3% | (27/145) 18.6% | 0.013 | |
| P-value* | 0.03 | 0.004 | 0.2 | |||
| GT | (15/57) 26.3% | (38/57) 66.7% | (4/57) 7% | |||
| P-value* | 0.59 | 0.63 | 0.86 | |||
| TT | (0/9) 0.0% | (6/9) 66.7% | (3/9) 33.3% | |||
| P-value* | 0.14 | 0.03 | 0.15 | |||
| SORT1 | rs602633 | GG | (16/131) 12.2% | (93/131) 71% | (22/131) 16.8% | 0.29 |
| P-value* | 0.13 | 0.37 | 0.75 | |||
| GT | (15/65) 23.1% | (41/65) 63.1% | (9/65) 13.8% | |||
| P-value* | 0.04 | 0.24 | 0.53 | |||
| TT | (1/15) 6.7% | (11/15) 73.3% | (3/15) 20% | |||
| P-value* | 0.32 | 0.65 | 0.65 |
Notes: *Chi-square test with p< 0.05 is considered significant. aWarfarin sensitive group (required minimum warfarin dose < 21 mg/week). b.Warfarin moderate response group (required average warfarin dose between 21 and 49 mg/week). cWarfarin resistance group (required high warfarin dose > 49 mg/week).
Table 6.
The Distributions of CELSR2 Haplotypes Among 212 Warfarin Sensitive Patients
| Gene | Haplotypes | Frequency (%) | Odds Ratio (95% CI) | P-value* |
|---|---|---|---|---|
| CELSR2 | CG | 0.6835 | 0.00 | — |
| TG | 0.1388 | 0.12 (−0.01–0.26) | 0.07 | |
| TC | 0.1243 | 0.11 (−0.03–0.25) | 0.14 | |
| TT | 0.0534 | −0.63 (−0.9 - −0.37) | <0.0001 |
Notes: *Chi–Square test with p<0.05 is considered significant.
Impact of SNPs to Warfarin Sensitivity During the Stabilization Phase of Therapy
The major allele homozygous and heterozygous genotypes for rs646776 were significantly associated with warfarin sensitivity, with the polymorphism as a whole reaching a significant overall p-value of 0.031. Minor allele homozygous and heterozygous genotypes of rs660240 and rs12740374 both elicited significant associations with warfarin sensitivity during the stabilization phase. Both rs660240 and rs12740374 also showed overall associations with warfarin sensitivity with p-values of 0.038 and 0.026, respectively. SORT1 SNP rs602633 minor allele homozygotes and heterozygotes displayed an association with warfarin sensitivity during the stabilization, yet the polymorphism failed to reach overall significance Table 7.
Table 7.
Association of PSRC1, CELSR2, and SORT1 SNPs with Warfarin Sensitivity During the Stabilization Phase of Therapy of 139 Cardiovascular Patients Treated with Warfarin
| Gene | SNP ID | Genotype | Sensitivea (n= 20) | Moderateb (n= 86) | Resistancec (n= 33) | Overall P-value* |
|---|---|---|---|---|---|---|
| PSRC1 | rs646776 | CC | (0/6) 0.0% | (4/6) 66.7% | (2/6) 33.3% | 0.031 |
| P-value* | 0.32 | 0.84 | 0.55 | |||
| CT | (13/48) 27.1% | (27/48) 56.3% | (8/48) 16.7% | |||
| P-value* | 0.002 | 0.32 | 0.16 | |||
| TT | (7/85) 8.2% | (55/85) 64.7% | (23/85) 27.1% | |||
| P-value* | 0.01 | 0.37 | 0.23 | |||
| CELSR2 | rs660240 | CC | (7/83) 8.4% | (52/83) 62.7% | (24/83) 28.9% | 0.038 |
| P-value* | 0.02 | 0.69 | 0.13 | |||
| TC | (12/46) 26.1% | (27/46) 58.7% | (7/46) 15.2% | |||
| P-value* | 0.004 | 0.62 | 0.07 | |||
| TT | (0/6) 0.0% | (4/6) 66.7% | (2/6) 33.3% | |||
| P-value* | 0.32 | 0.76 | 0.62 | |||
| rs12740374 | GG | (7/89) 7.9% | (58/89) 65.2% | (24/89) 27% | 0.026 | |
| P-value* | 0.01 | 0.37 | 0.27 | |||
| GT | (12/44) 27.3% | (25/44) 56.8% | (7/44) 15.9% | |||
| P-value* | 0.002 | 0.37 | 0.13 | |||
| TT | (0/5) 0.0% | (3/5) 60% | (2/5) 40% | |||
| P-value* | 0.37 | 0.92 | 0.37 | |||
| SORT1 | rs602633 | GG | (6/79) 7.6% | (52/79) 65.8% | (21/79) 26.6% | 0.071 |
| P-value* | 0.01 | 0.23 | 0.37 | |||
| GT | (13/50) 26% | (27/50) 54% | (10/50) 20% | |||
| P-value* | 0.004 | 0.16 | 0.42 | |||
| TT | (1/9) 11.1% | (6/9) 66.7% | (2/9) 22.2% | |||
| P-value* | 0.76 | 0.76 | 0.92 |
Notes: *Chi-square test with p< 0.05 is considered significant. aWarfarin sensitive group (required minimum warfarin dose < 21 mg/week). bWarfarin moderate response group (required average warfarin dose between 21 and 49 mg/week). cWarfarin resistance group (required high warfarin dose > 49 mg/week).
Impact of SNPs to Warfarin Dosage Variability
One-way ANOVA analysis on the four studied SNPs reached overall significance with regards to initiation dosage (p<0.05). rs646776, rs660240, and rs12740374 all had p-values <0.0001 and rs602633 had a p-value of 0.002. As for variability of dosage during the maintenance phase, only rs66024 and rs12740374 of the CELSR2 gene were shown to have a significant association with p-values of 0.036 and 0.031, respectively Table 8. Post-Hock multiple comparisons test; compare the means of the initiation and maintenance dose among all genotypes exhibited on Table 9.
Table 8.
Association of PSRC1, CELSR2, and SORT1 SNPs with Variability on Warfarin Required Doses
| Gene | SNP ID | Genotype | Initiation Dose | Overall P-value* | Maintenance Dose | Overall P-value* |
|---|---|---|---|---|---|---|
| PSRC1 | rs646776 | CC | 66.92 [78.79] | <0.0001 | 48.55 [29.53] | 0.107 |
| CT | 33.74 [16.20] | 34.85 [17.20] | ||||
| TT | 37.67 [13.19] | 39.95 [17.03] | ||||
| CELSR2 | rs660240 | CC | 38.03 [14.42] | <0.0001 | 41.00 [18.32] | 0.036 |
| TC | 33.07 [13.30] | 33.78 [14.43] | ||||
| TT | 66.92 [78.79] | 48.55 [29.53] | ||||
| rs12740374 | GG | 38.17 [14.25] | <0.0001 | 40.48 [17.89] | 0.031 | |
| GT | 32.22 [13.15] | 33.67 [14.63] | ||||
| TT | 75.59 [85.41] | 51.26 [32.17] | ||||
| SORT1 | rs602633 | GG | 37.06 [13.05] | 0.002 | 40.02 [17.23] | 0.33 |
| GT | 35.57 [16.70] | 35.69 [17.31] | ||||
| TT | 57.79 [68.63] | 42.37 [26.07] |
Notes: *One-way ANOVA test with p< 0.05 is considered significant, Mean Standard deviation in square brackets.
Table 9.
Post Hoc Tests for the Association of PSRC1, CELSR2, and SORT1 SNPs with Variability on Warfarin Required Doses
| Gene | SNP ID | Genotype | Initiation Dose P-value* | Maintenance Dose P-value* | |
|---|---|---|---|---|---|
| PSRC1 | rs646776 | CC | CT | <0.0001 | 0.18 |
| TT | <0.0001 | 0.49 | |||
| CT | CC | <0.0001 | 0.18 | ||
| TT | 0.479 | 0.26 | |||
| TT | CC | <0.0001 | 0.49 0.26 |
||
| CT | 0.479 | ||||
| CELSR2 | rs660240 | CC | TC | 0.326 | 0.07 |
| TT | 0.0001 | 0.57 | |||
| TC | CC | 0.326 | 0.07 | ||
| TT | <0.0001 | 0.14 | |||
| TT | CC | 0.0001 | 0.57 0.14 |
||
| TC | <0.0001 | ||||
| rs12740374 | GG | GT | 0.186 | 0.09 0.38 |
|
| TT | <0.0001 | ||||
| GT | GG | 0.186 | 0.09 0.09 |
||
| TT | <0.0001 | ||||
| TT | GG | <0.0001 | 0.38 | ||
| GT | <0.0001 | 0.09 | |||
| SORT1 | rs602633 | GG | GT | 0.902 | 0.38 0.93 |
| TT | 0.003 | ||||
| GT | GG | 0.902 | 0.38 | ||
| TT | 0.002 | 0.56 | |||
| TT | GG | 0.003 | 0.93 | ||
| GT | 0.002 | 0.56 | |||
Note: *Post-Hoc multiple comparisons test with p< 0.05 is considered significant.
Impact of SNPs to Warfarin Responsiveness During the Initiation and Stabilization Phase of Therapy
No significant associations were found between any of the studied SNPs and warfarin responsiveness during both the initiation and maintenance phases of warfarin treatment in patients with cardiovascular disease Tables 10 and 11. Nevertheless, haplotype distribution frequencies for 3 CELSR2 haploid genotypes (TG, TC, and TT) reached significant associations (p<0.05) with warfarin response Table 12.
Table 10.
Association of PSRC1, CELSR2, and SORT1 SNPs with Warfarin Responsiveness During the Initiation Phase of Therapy of 212 Cardiovascular Patients Treated with Warfarin
| Gene | SNP ID | Genotype | Poora (n= 39) | Goodb (n= 162) | Ultrac (n= 11) | Overall P-value* |
|---|---|---|---|---|---|---|
| PSRC1 | rs646776 | CC | (4/11) 36.4% | (7/11) 63.6% | (0/11) 0.0% | 0.264 |
| P-value* | 0.11 | 0.31 | 0.43 | |||
| CT | (7/61) 11.5% | (51/61) 83.6% | (3/61) 4.9% | |||
| P-value* | 0.10 | 0.12 | 0.92 | |||
| TT | (28/140) 20% | (104/140) 74.3% | (8/140) 5.7% | |||
| P-value* | 0.40 | 0.31 | 0.63 | |||
| CELSR2 | rs660240 | CC | (28/139) 20.1% | (103/139) 94.1% | (8/139) 5.8% | 0.323 |
| P-value* | 0.48 | 0.37 | 0.69 | |||
| TC | (7/58) 12% | (48/58) 82.8% | (3/58) 5.2% | |||
| P-value* | 0.13 | 0.16 | 1 | |||
| TT | (4/11) 36.4% | (7/11) 63.6% | (0/11) 0.0% | |||
| P-value* | 0.13 | 0.32 | 0.42 | |||
| rs12740374 |
GG | (28/145) 19.3% | (109/145) 75.2% | (8/145) 5.5% | 0.646 | |
| P-value* | 0.65 | 0.31 | 0.24 | |||
| GT | (8/57) 14% | (46/57) 80.7% | (3/57) 5.3% | |||
| P-value* | 0.57 | 0.36 | 0.49 | |||
| TT | (3/9) 33.3% | (6/9) 66.7% | (0/9) 0.0% | |||
| P-value* | 0.76 | 1 | 0.47 | |||
| SORT1 | rs602633 | GG | (27/131) 20.6% | (97/131) 74% | (7/131) 5.3% | 0.196 |
| P-value* | 0.32 | 0.23 | 0.62 | |||
| GT | (7/65) 10.8% | (55/65) 84.6% | (3/65) 4.6% | |||
| P-value* | 0.06 | 0.07 | 0.32 | |||
| TT | (5/15) 33.3% | (10/15) 66.7% | (0/15) 0.0% | |||
| P-value* | 0.13 | 0.32 | 0.37 |
Notes: *Chi-square test with p< 0.05 is considered significant. a Poor responders (international normalized ratio (INR) value below target). b Good responders who have an INR in the target range (therapeutic range). cUltra-responders (INR over target).
Table 11.
Association of PSRC1, CELSR2, and SORT1 SNPs with Warfarin Responsiveness During the Stabilization Phase of Therapy of 139 Cardiovascular Patients Treated with Warfarin
| Gene | SNP ID | Genotype | Poora (n= 9) | Goodb (n= 124) | Ultrac (n= 6) | Overall P-value* |
|---|---|---|---|---|---|---|
| PSRC1 | rs646776 | CC | (0/6) 0.0% | (6/6) 100% | (0/6) 0.0% | 0.715 |
| P-value* | 0.48 | 0.37 | 0.62 | |||
| CT | (2/48) 4.2% | (43/48) 89.6% | (3/48) 6.3% | |||
| P-value* | 0.42 | 0.92 | 0.42 | |||
| TT | (7/85) 8.2% | (75/85) 88.2% | (3/85) 3.5% | |||
| P-value* | 0.27 | 0.62 | 0.55 | |||
| CELSR2 | rs660240 | CC | (7/83) 8.4% | (73/83) 88% | (3/83) 3.6% | 0.718 |
| P-value* | 0.32 | 0.69 | 0.55 | |||
| TC | (2/46) 4.3% | (41/46) 89.1% | (3/46) 6.5% | |||
| P-value* | 0.42 | 0.92 | 0.42 | |||
| TT | (0/6) 0.0% | (6/6) 100% | (0/6) 0.0% | |||
| P-value* | 0.48 | 0.37 | 0.62 | |||
| rs12740374 | GG | (7/89) 7.9% | (79/89) 88.8% | (3/89) 3.4% | 0.766 | |
| P-value* | 0.37 | 0.84 | 0.42 | |||
| GT | (2/44) 4.5% | (39/44) 88.6% | (3/44) 6.8% | |||
| P-value* | 0.55 | 0.92 | 0.32 | |||
| TT | (0/5) 0.0% | (5/5) 100% | (0/5) 0.0% | |||
| P-value* | 0.55 | 0.42 | 0.62 | |||
| SORT1 | rs602633 | GG | (7/79) 8.9% | (69/79) 87.3% | (3/79) 3.8% | 0.691 |
| P-value* | 0.19 | 0.27 | 0.92 | |||
| GT | (2/50) 4% | (46/50) 92% | (2/50) 4% | |||
| P-value* | 0.37 | 0.55 | 0.84 | |||
| TT | (0/9) 0.0% | (9/9) 100% | (0/9) 0.0% | |||
| P-value* | 0.42 | 0.32 | 0.55 |
Notes: *Chi-square test with p< 0.05 is considered significant. a Poor responders (international normalized ratio (INR) value below target). b Good responders who have an INR in the target range (therapeutic range). c Ultra-responders (INR over target).
Table 12.
The Distributions of CELSR2 Haplotypes Among 212 Warfarin Responsiveness Patients
| Gene | Haplotypes | Frequency (%) | Odds Ratio (95% CI) | P-value* |
|---|---|---|---|---|
| CELSR2 | CG | 0.6755 | 0.00 | – |
| TG | 0.1468 | 0.16 (0.05–0.27) | 0.006 | |
| TC | 0.1324 | 0.19 (0.07–0.31) | 0.0027 | |
| TT | 0.0453 | −0.42 (−0.62 - −0.21) | <0.0001 |
Notes: *Chi–Square test with p<0.05 is considered significant.
Impact of SNPs INR Treatment Outcomes
Associations between the selected polymorphisms were found when studied and analyzed for an effect on treatment outcomes based on INR values. Only rs602633 of the SORT1 gene showed overall significance in relation to maintenance INR while none of the studied SNPs revealed any association with initiation phase INR Table 13. Post-Hock multiple comparisons test; compare the means of the initiation and maintenance INR values among all genotypes exhibited on Table 14.
Table 13.
Association of PSRC1, CELSR2, and SORT1 SNPs with INR Treatment Outcome
| Gene | SNP ID | Genotype | Initiation INR | Overall P-value* | Maintenance INR | Overall P-value* |
|---|---|---|---|---|---|---|
| PSRC1 | rs646776 | CC | 2.00 [0.74] | 0.087 | 2.38 [0.34] | 0.109 |
| CT | 2.56 [0.61] | 2.67 [0.41] | ||||
| TT | 2.45 [0.83] | 2.72 [0.38] | ||||
| CELSR2 | rs660240 | CC | 2.44 [0.83] | 0.09 | 2.72 [0.38] | 0.109 |
| TC | 2.56 [0.62] | 2.67 [0.39] | ||||
| TT | 2.00 [0.74] | 2.38 [0.34] | ||||
| rs12740374 | GG | 2.44 [0.81] | 0.344 | 2.72 [0.37] | 0.160 | |
| GT | 2.53 [0.66] | 2.66 [0.40] | ||||
| TT | 2.13 [0.76] | 2.40 [0.38] | ||||
| SORT1 | rs602633 | GG | 2.65 [0.84] | 0.047 | 2.73 [0.38] | 0.021 |
| GT | 2.53 [0.59] | 2.67 [0.38] | ||||
| TT | 1.99 [0.67] | 2.37 [0.32] |
Notes: *One-way ANOVA test with p< 0.05 is considered significant, Mean Standard deviation in square brackets.
Table 14.
Post Hoc Tests for the Association of PSRC1, CELSR2, and SORT1 SNPs with INR Treatment Outcome
| Gene | SNP ID | Genotype | Initiation INR P-value* | Maintenance INR P-value* | |
|---|---|---|---|---|---|
| PSRC1 | rs646776 | CC | CT | <0.0001 | 0.18 |
| TT | <0.0001 | 0.09 | |||
| CT | CC | <0.0001 | 0.18 0.78 |
||
| TT | 0.479 | ||||
| TT | CC | <0.0001 | 0.09 0.78 |
||
| CT | 0.479 | ||||
| CELSR2 | rs660240 | CC | TC | 0.326 | 0.77 |
| TT | 0.0001 | 0.09 | |||
| TC | CC | 0.326 | 0.77 | ||
| TT | <0.0001 | 0.19 | |||
| TT | CC | 0.0001 | 0.09 | ||
| TC | <0.0001 | 0.19 | |||
| rs12740374 | GG | GT | 0.186 | 0.65 | |
| TT | <0.0001 | 0.17 | |||
| GT | GG | 0.186 | 0.65 | ||
| TT | <0.0001 | 0.33 | |||
| TT | GG | <0.0001 | 0.17 0.33 |
||
| GT | <0.0001 | ||||
| SORT1 | rs602633 | GG | GT | 0.902 | 0.58 |
| TT | 0.003 | 0.02 | |||
| GT | GG | 0.902 | 0.58 | ||
| TT | 0.002 | 0.07 | |||
| TT | GG | 0.003 | 0.02 | ||
| GT | 0.002 | 0.07 | |||
Notes: *Post-Hoc multiple comparisons test with p<0.05 is considered significant. Compare initiation and maintenance dose among all genotypes.
Discussion
The findings of the current study failed to find any significant associations between rs646776, rs660240, rs12740374, and rs602633 and cardiovascular disease in Jordanian population, although some of them are involved as a candidates based on GWAS. Arvind et al, found that rs646776 was associated with a protective effect against CAD and that PSRC1 gene expression levels were significantly lower in cases vs controls. However, they found no such association with CELSR2 and SORT1 genes in their study of Asian-Indian populations.1 Guo et al, had similar findings. They found that high levels of PSRC1 gene expression were linked with a decrease in circulating LDL-C levels and an increase in HDL levels, therefore decreasing the risk of CAD.10 However, the disagreement between the results of our study and previous studies could be due to differences in ethnicity, population stratification, and sample size. The frequency of the C allele of rs646776 in our population (0.20) differs from that reported in African Americans and Asians by (0.36) (0.06) respectively (http://www.hapmap.org/). Corresponding to our result, a study conducted on the Lebanese population found that rs646776 was not associated with CAD.24 In addition, Rizk et al, studied a population similar to the one in our study on an Arab population. Their study found no correlation between the rs646776 polymorphism and coronary artery disease categorized by angiography. However, they found an association between this SNP and a decrease in LDL-C levels, such a finding possibly due to gene–gene interaction especially since the PSRC1, CELSR2, and SORT1 genes are found in a highly linked disequilibrium.6 (Samani et al, Zhou et al, and Musunuru et al,) found that expression of a minor protective allele of rs646776 in the human liver was associated with an increase in hepatic expression of CELSR2, PRSC1, and SORT1 genes which correlated negatively with plasma LDL-C levels.25–27 A GWAS conducted by O’donnel et al, showed that rs12740374 may act as a proxy for rs646776, a finding consistent with a genetic component associated with LDL-C levels, CAD, and myocardial infarction.28 Kjolby et al, conducted an experiment to study for the effect of the sortilin protein on LDL-C levels. The absence of sortilin in double knockout mice resulted in a 30% decrease in cholesterol levels and upon over expression of the gene a 42% increase in cholesterol levels was noted.12 Qin et al, studies the effect of rs646776 and rs12740374 on peripheral arterial disease (PAD) in a Chinese population with type 2 diabetes mellitus. Associations were found between the minor (G) allele of rs646776 and the minor (T) allele of rs12740374 and a decrease risk for PAD in the studies population.7 Consistent with our result, it was also found that rs602633 in the SORT1 gene was not associated with CAD in Japanese Americans (p = 0.88), but was associated with CAD in African Americans and Latinos (p = 0.004) and (p = 0.04).29 The variation on these results it could be due to differences in allele frequency and sample size between different ethnic populations and it is possible that this SNP is present in high linkage disequilibrium with additional variants in genes involved in the lipid metabolism pathway.
To the best of our knowledge, the present study was the first to explore the association between rs646776 of PSRC1 gene, rs660240 and rs12740374 of CELSR2 gene, and rs602633 of SORT1 gene on warfarin sensitivity and responsiveness. The associations found between rs646776 and rs12740374 with warfarin sensitivity during the initiation phase and those found between all the studied SNPs with warfarin sensitivity during the stabilization phase indicated that patients with these polymorphisms required lower doses of the drug to achieve its therapeutic effect. Their associations with the variability of dosage showed significance during the initiation phase, however, only the CELSR2 gene showed significance with maintenance dose variability as well. Interestingly, a study by Pankow et al, found an association between rs12740374 and decreased levels of protein C in whites and blacks, that when combined reached a genome-wide significance.13 Protein C, like clotting factors II, VII, IX, and X, also requires vitamin K to undergo carboxylation.18 Being an anticoagulant, the association between our results and those of Pankow et al, might seem paradoxical. Even though warfarin suppresses the function of protein C, its anticoagulatory effects are superior to its prothrombotic properties. Since little is known about the pathway in which rs12740374 produces these effects on warfarin and protein C, more research on the subject matter is necessary to explain these novel findings.
SORT1 SNP rs602633 did not display an association with warfarin sensitivity during both phases of therapy. But after applying the multiple post-hock comparison tests, a significant association was found only with the heterozygous genotype. Supposedly the small sample size influenced the statistical power (only 32 sensitive patients, 15 of them were GT carriers and only one was a carrier of TT), a study on larger sample size is required to confirm our result.
However, further study is required to validate the result of current study, especially with these patients carry additional mutations in VKORC1 and CYP2C9 genes which was previously reported by our team in 2019, indicating that the carrying VKORC1 or CYP2C9 polymorphisms is associated with warfarin sensitivity and warfarin dose variability during the initiation and stabilization phases of treatment.14,15 Therefore, to eliminate genetic cofounding variables an additional study is required in wild-type genotyped subjects for the above polymorphism.
Two previous studies by Al-Eitan et al, have explored the associations between VKORC1 and CYP2C9 genes and warfarin sensitivity and responsiveness in a Jordanian population. Both studies were in accordance with GWAS findings and have shown to be associated with warfarin sensitivity, with the exception of those that failed to correlate with drug responsiveness in the stabilization phase of therapy.14,15 There were several large trials conducted to tackle the issue of genotype-guided warfarin dosing vs dosing using clinical algorithms. Cavallari et al, published two review articles summarizing the latest evidence from the trials conducted in the field of warfarin pharmacogenomics. The trials focused on studying CYP2C9 and VKORC1 genes, were multicenter, with large study samples, and multiethnic.20 The European Pharmacogenetics of Anticoagulant Therapy (EU-PACT) trial found that patients treated with a genotype guided approach have spent more time in the therapeutic range (TTR), had fewer medication side effects (bleeding), and required less time to reach the therapeutic INR and stabilization phase of treatment.20 The Clarification of Optimal Anticoagulation through Genetics (COAG) trial found no better outcomes using genotype-guided warfarin dosing in comparison to a clinical algorithmic approach.31–34
However, a large proportion of the study population was African Americans, whose warfarin sensitivity polymorphisms were not included in genotyping.35 The second review by Cavallari et al, published in 2017, revisited the debate with new evidence from the Genetics Informatics Trial (GIFT), which found that a 27% reduction in venous thromboembolism, hemorrhage, INR values >4, and death in the group that was doused with a genotype-guided approach.35 The Clinical Pharmacogenetic Implementation Consortium (CPIC) guidelines recommend a genotype-guided dosing approach when strong genotype-associated evidence is available for the population in terms of warfarin dosing requirements.36,37
Limitation
This study is the first study on the pharmacogenetic association of the PSRC1, CELSR2 and SORT1 polymorphisms in Jordanian cardiovascular patients treated with warfarin. However, the present study has several limitations. The number of subjects enrolled in the study was relatively small and larger population studies are needed to identify and clarify our findings. Furthermore, a larger sample would allow an analysis of the frequencies of deficient alleles and genotypes in the present study. Furthermore, gene–gene and gene–environment interactions cannot be ruled out. Therefore, more studies with larger sample sizes and including multivariate analyses are needed.
Future Research Directions
Further studies are needed to confirm our results with a larger sample size and more specific clinical phenotypic data. Future studies will also be needed to examine the combined effects of these SNPs and the contribution of multiple genes to susceptibility to heart disease and the efficacy of warfarin treatment. Therefore, the future could be towards multivariate and haplotype analysis.
In addition, there is a need to develop a more effective and safe CV therapy, especially with warfarin, which has a narrow therapeutic index and which has been reported that most of the variability in its required dose explained by genetic together with the clinic factors.16,19 Many investigators have developed pharmacogenetic dosing algorithms that estimate the warfarin dose based on genetic and clinical factors. In fact, current warfarin dosing algorithms do not take into account the Arab population. Previous studies in our cohort have shown that the association between CYP2C9 and VKORC1 SNPs and warfarin sensitivity and variability in the required dose.14,15 However, examination of other genetic factors in combination with the clinical factor will help clinicians prescribe warfarin more efficient. However, the algorithms that are needed in our population are still in the early stages of development and replication in a larger sample, and a different cohort is needed for validation.
Conclusion
In conclusion, the present study revealed novel associations between PSRC1, CELSR2, and SORT1 genes with warfarin sensitivity and INR treatment outcomes in Jordanian patients with cardiovascular disease, with the CELSR2 gene haplotype frequencies exhibiting an association as well. However, no associations were found between the studied polymorphisms and CHD or warfarin responsiveness. Looking at the bigger picture, the associations found in this study warrant further research into the associations of these genes with warfarin. This will build a stronger pharmacogenomic database for potentially strengthening the argument of genotype-guided dosing and hopefully making personalized medicine the standard of care for patients in the future.
Funding Statement
Appreciation is due to Queen Alia Hospital, who was supportive of this project. This work was funded by the Deanship of Scientific Research at the Jordan University of Science and Technology under grant number (203/2014).
Disclosure
The authors of this study report no conflicts of interest.
References
- 1.Arvind P, Nair J, Jambunathan S, et al. CELSR2–PSRC1–SORT1 gene expression and association with coronary artery disease and plasma lipid levels in an Asian Indian cohort. J Cardiol. 2014;64(5):339–346. doi: 10.1016/j.jjcc.2014.02.012 [DOI] [PubMed] [Google Scholar]
- 2.Below JE, Parra EJ, Gamazon ER, et al. Meta-analysis of lipid-traits in Hispanics identifies novel loci, population-specific effects and tissue-specific enrichment of eQTLs. Sci Rep. 2016;6(1):19429. doi: 10.1038/srep19429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kjolby M, Andersen OM, Breiderhoff T, et al. Sort1, Encoded by the Cardiovascular Risk Locus 1p13.3, Is a Regulator of Hepatic Lipoprotein Export. Cell Metab. 2010;12(3):213–223. doi: 10.1016/j.cmet.2010.08.006 [DOI] [PubMed] [Google Scholar]
- 4.Zhou L, Ding H, Zhang X, et al. Genetic Variants at Newly Identified Lipid Loci Are Associated with Coronary Heart Disease in a Chinese Han Population. PLoS One. 2011;6(11):e27481. doi: 10.1371/journal.pone.0027481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deloukas P, Kanoni S, Willenborg C, et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet. 2013;45(1):25–33. doi: 10.1038/ng.2480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rizk NM, El-Menyar A, Egue H, et al. The Association between Serum LDL Cholesterol and Genetic Variation in Chromosomal Locus 1p13.3 among Coronary Artery Disease Patients. Biomed Res Int. 2015;2015:1–12. doi: 10.1155/2015/678924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qin J, Tian J, Liu G, et al. Association between 1p13 polymorphisms and peripheral arterial disease in a Chinese population with diabetes. J Diabetes Investig. 2018;9(5):1189–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robinson JR, Denny JC, Roden DM, et al. Genome-wide and Phenome-wide Approaches to Understand Variable Drug Actions in Electronic Health Records. Clin Transl Sci. 2018;11(2):112–122. doi: 10.1111/cts.12522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holdt LM, Teupser D. From genotype to phenotype in human atherosclerosis - recent findings. Curr Opin Lipidol. 2013;24(5):410–418. doi: 10.1097/MOL.0b013e3283654e7c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo K, Hu L, Xi D, et al. PSRC1 overexpression attenuates atherosclerosis progression in apoE −/− mice by modulating cholesterol transportation and inflammation. J Mol Cell Cardiol. 2018;116:69–80. doi: 10.1016/j.yjmcc.2018.01.013 [DOI] [PubMed] [Google Scholar]
- 11.Goettsch C, Kjolby M, Sortilin AE. Its Multiple Roles in Cardiovascular and Metabolic Diseases. Arterioscler Thromb Vasc Biol. 2018;38(1):19–25. doi: 10.1161/ATVBAHA.117.310292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kjolby M, Nielsen MS, Petersen CM. Sortilin, Encoded by the Cardiovascular Risk Gene SORT1, and Its Suggested Functions in Cardiovascular Disease. Curr Atheroscler Rep. 2015;17(4):18. [DOI] [PubMed] [Google Scholar]
- 13.Pankow JS, Tang W, Pankratz N, et al. Identification of Genetic Variants Linking Protein C and Lipoprotein metabolism: the ARIC study (Atherosclerosis Risk in Communities). Arterioscler Thromb Vasc Biol. 2017;37(3):589–597. doi: 10.1161/ATVBAHA.116.308109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Al-Eitan LN, Almasri AY, Khasawneh RH. Effects of CYP2C9 and VKORC1 polymorphisms on warfarin sensitivity and responsiveness during the stabilization phase of therapy. Saudi Pharm J. 2019;27(4):484–490. doi: 10.1016/j.jsps.2019.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Al-Eitan L, Almasri A, Khasawneh R. Impact of CYP2C9 and VKORC1 Polymorphisms on Warfarin Sensitivity and Responsiveness in Jordanian Cardiovascular Patients during the Initiation Therapy. Genes. 2018;9(12):578. doi: 10.3390/genes9120578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Emery JD. Pharmacogenomic testing and warfarin: what evidence has the GIFT trial provided? JAMA. 2017;318(12):1110–1112. doi: 10.1001/jama.2017.11465 [DOI] [PubMed] [Google Scholar]
- 17.Fawzy AM, Lip GY. Pharmacokinetics and pharmacodynamics of oral anticoagulants used in atrial fibrillation. Expert Opin Drug Metab Toxicol. 2019;15(5):381–398. doi: 10.1080/17425255.2019.1604686 [DOI] [PubMed] [Google Scholar]
- 18.Johnson JA, Cavallari LH. Warfarin pharmacogenetics. Trends Cardiovasc Med. 2015;25(1):33–41. doi: 10.1016/j.tcm.2014.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kasner SE, Wang L, French B, et al. Warfarin Dosing Algorithms and the Need for Human Intervention. Am J Med. 2016;129(4):431–437. doi: 10.1016/j.amjmed.2015.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cavallari LH. Time to revisit warfarin pharmacogenetics. Future Cardiol. 2017;13(6):511–513. doi: 10.2217/fca-2017-0061 [DOI] [PubMed] [Google Scholar]
- 21.Consortium IWP. Estimation of the warfarin dose with clinical and pharmacogenetic data. N Eng J Med. 2009;360(8):753–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Higashi MK, Veenstra DL, Kondo LM, et al. Association between CYP2C9 genetic variants and anticoagulation-related outcomes during warfarin therapy. JAMA. 2002;287(13):1690–1698. doi: 10.1001/jama.287.13.1690 [DOI] [PubMed] [Google Scholar]
- 23.Al-Eitan LN, Almasri AY, Al-Habahbeh SO. Effects of coagulation factor VII polymorphisms on warfarin sensitivity and responsiveness in Jordanian cardiovascular patients during the initiation and maintenance phases of warfarin therapy. Pharmgenomics Pers Med. 2019;Volume 12:1–8. doi: 10.2147/PGPM.S189458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Large-Scale Association MN. Analysis Identifies 13 New Susceptibility Loci for Coronary Artery Disease. Circ Cardiovasc Genet. 2011;4(3):327–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Samani NJ, Braund PS, Erdmann J, et al. The novel genetic variant predisposing to coronary artery disease in the region of the PSRC1 and CELSR2 genes on chromosome 1 associates with serum cholesterol. J Mol Med. 2008;86(11):1233–1241. doi: 10.1007/s00109-008-0387-2 [DOI] [PubMed] [Google Scholar]
- 26.Zhou Y, Yang Q, Yin R. GW26-e2113 Polymorphisms in the CELSR2-PSRC1-SORT1 are associated with serum lipid traits, the risk of coronary artery disease and ischemic stroke. J Am Coll Cardiol. 2015;66(16):C104–C105. doi: 10.1016/j.jacc.2015.06.406 [DOI] [Google Scholar]
- 27.Musunuru K, Strong A, Frank-Kamenetsky M, et al. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature. 2010;466(7307):714–719. doi: 10.1038/nature09266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Donnell CJ, Kavousi M, Smith AV, et al. Genome-Wide Association Study for Coronary Artery Calcification With Follow-Up in Myocardial Infarction. Circulation. 2011;124(25):2855–2864. doi: 10.1161/CIRCULATIONAHA.110.974899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ke W, Rand K, Conti D, et al. Evaluation of 71 Coronary Artery Disease Risk Variants in a Multiethnic Cohort. Fron Cardiovasc Med. 2018;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Al-Eitan L, Haddad Y. Emergence of Pharmacogenomics in Academic Medicine and Public Health in Jordan: history, Present State and Prospects. Curr Pharmacogenomics Person Med. 2014;12(3):167–175. doi: 10.2174/1875692113666150115221210 [DOI] [Google Scholar]
- 31.Xu H, Xie X, Wang B, et al. Meta-analysis of efficacy and safety of genotype-guided pharmacogenetic dosing of warfarin. Int J Cardiol. 2014;177(2):654–657. doi: 10.1016/j.ijcard.2014.09.174 [DOI] [PubMed] [Google Scholar]
- 32.Wu AH. Pharmacogenomic-guided dosing for warfarin: too little too late? Per Med. 2018;15(2):71–73. doi: 10.2217/pme-2017-0080 [DOI] [PubMed] [Google Scholar]
- 33.Maluso A. Pharmacogenomic Testing and Warfarin Management. Oncol Nurs Forum. 2015;42(5):563–565. doi: 10.1188/15.ONF.563-565 [DOI] [PubMed] [Google Scholar]
- 34.Tavares LC, Marcatto LR, Santos PC. Genotype-guided warfarin therapy: current status. Pharmacogenomics. 2018;19(7):667–685. doi: 10.2217/pgs-2017-0207 [DOI] [PubMed] [Google Scholar]
- 35.Cavallari LH, Nutescu EA. Warfarin Pharmacogenetics: to Genotype or Not to Genotype, That Is the Question. Clin Pharmacol Ther. 2014;96(1):22–24. doi: 10.1038/clpt.2014.78 [DOI] [PubMed] [Google Scholar]
- 36.Johnson J, Caudle K, Gong L, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for Pharmacogenetics-Guided Warfarin Dosing: 2017 Update. Clin Pharmacol Ther. 2017;102(3):397–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Al-Eitan L, Practical Challenges TA. Translational Issues in Pharmacogenomics and Personalized Medicine from 2010 Onwards. Curr Pharmacogenomics Person Med. 2016;14(1):7–17. doi: 10.2174/1875692115666161215103842 [DOI] [Google Scholar]