Abstract
Background
Osteosarcoma (OS) is one of the most common malignant bone tumors with a poor overall prognosis. MiR-1224-5p plays an important role in cancer, but its function and mechanism in OS have not been studied.
Materials and Methods
The expression of miR-1224-5p and PLK1 was detected by qRT-PCR in OS cells, adjacent tissues, and cell lines. Dual-luciferase reporter gene assay was used to verify the interaction between miR-1224-5p and PLK1. The expression of miR-1224-5p and PLK1 was intervened by transfection with miR-1224-5p mimic, NC mimic, pc-NC and PLK1, respectively. MTT, colony formation assay, Transwell and flow cytometry were used to observe the cell proliferation, invasion and apoptosis. Western blot was used to detect the expression levels of PLK1, PI3K/AKT/mTOR signaling pathway-related proteins, autophagy-related proteins, and epithelial-mesenchymal transition (EMT)-related proteins in the cells.
Results
We found that miR-1224-5p was down-regulated and PLK1 expression was up-regulated in OS tissues and cells. On the other hand, it is further confirmed that PLK1 was a target gene of miR-1224-5p. Overexpression of miR-1224-5p inhibited the proliferation, invasion while promoted the apoptosis of OS cells, whereas overexpression of PLK1 promoted the proliferation, invasion and inhibited the apoptosis of OS cells. In the miR-1224-5p group (overexpression of miR-1224-5p), PI3K, AKT, and mTOR protein phosphorylation levels were significantly reduced, while autophagic activity was significantly activated, and the degree of EMT was significantly reduced. But the results in the PLK1 group (overexpression of PLK1) were the opposite. In addition, overexpression of miR-1224-5p reversed the effect of PLK1 upregulation on OS cells.
Conclusion
MiR-1224-5p targets PLK1 to inhibit PI3K/AKT/mTOR signaling pathway, thus mediating the proliferation, invasion, apoptosis, autophagy and EMT in OS cells.
Keywords: osteosarcoma (OS), miR-1224-5p, PLK1, epithelial-mesenchymal transition (EMT), autophagy
Introduction
Osteosarcoma (OS) is a common malignant bone tumor, which often occurs in adolescents or children.1 Higher birth weight,2 adolescent hormone secretion3 and genetic variation4 are the risk factors affecting the occurrence of OS. Its current treatment is mainly based on neoadjuvant chemotherapy (NACT), surgical resection of the primary tumor and adjuvant chemotherapy.5 In the past 30 years, the occurrence and treatment of OS have not been improved. Adolescents or children with local non-invasive metastases have a 5-year survival rate of approximately 60–70%, whereas patients with metastatic OS have a poorer prognosis and a 5-year survival rate of approximately 23%.6 Among them, the lung is the most common site of OS metastasis.7 Therefore, to study the key protein molecules and their possible regulatory mechanisms in the occurrence, development and metastasis of OS is of great significance for its treatment.
Although researchers have explored and made some progress in the treatment of OS, the overall prognosis of OS patients is still poor, especially in patients with distant metastasis or recurrence.8 MicroRNAs (miRNAs) are endogenous non-coding single-stranded RNAs molecules. Based on the cutting or translational repression caused by sequence complementarity, miRNAs regulate the expression of gene after transcription in the cells by targeting and acting on the 3‘-UTR of mRNAs.9 As major members of the regulatory gene family in eukaryotic cells, miRNAs are involved in almost all life processes such as cell growth, development, senescence, and apoptosis.10,11 Currently, increasing reports show that different miRNAs influence the occurrence and development of OS by affecting multiple cellular signaling pathways to form a more complex regulatory network.12,13 For example, miR-497 activates P21 expression and promotes the apoptosis of OS cells by inhibiting the expression of MAPK/ERK signaling pathway.14 MiR-567 inhibits the proliferation, migration and invasion ability of OS cells by targeting fibroblast growth factor 5 (FGF5).15 MiR-493 inhibits the proliferation and invasion of OS cells by directly targeting SP1.16 MiR-30a inhibits the proliferation and metastasis of OS by down-regulating the expression of MEF2D.17 MiR-338-3p inhibits proliferation, migration, invasion and epithelial-mesenchymal transition (EMT) in OS cells by targeting heat shock protein HSP90.18 Thus, miRNAs are closely associated with the occurrence and development of OS.
As a microRNA, miR-1224-5p is closely associated with proliferation, drug resistance, invasion, and metastasis of tumor.19 For example, upregulation of miR-1224-5p can inhibit drug resistance of glioma to inhibit cancer.20 Meanwhile, miR-1224-5p inhibits the proliferation and invasion of ovarian cancer by targeting SND1.21 MiR-1224-5p/ELF3 signal axis mediates PI3K/AKT/Notch signal pathway to regulate the malignant biological behavior of pancreatic cancer.22 And miR-1224-5p inhibits the proliferation, apoptosis, and invasion of keloid through the TGF-β1/Smad3 signaling pathway.23 MiR-1224-5p also mediates mitochondrial damage to affect pulmonary fibrosis induced by silicon through targeting BECN1.24 These data prove that miR-1224-5p has a wide range of regulatory roles in a variety of tumors and it can regulate a variety of molecular pathways to affect the occurrence and development of tumors. Furthermore, miR-1224-5p is also involved in the EMT of tumor cells. It has been shown that miR-1224-5p inhibits the EMT in laryngeal papilloma cells and is strongly associated with patient prognosis.25 In addition, Hongkai Lian et al found that the Linc00460/miR-1224-5p/FADS1 axis is a potential therapeutic target for OS.26 However, the effect of miRNA-1224-5p on the EMT of OS cells needs to be explored.
Autophagy, a conserved catabolic process widely found in eukaryotes, is a biological pathway in which lysosomes are involved in the digestion and degradation of their own biomacromolecules or damaged organelles.27 Autophagy plays a dual role in the regulation of tumor. Autophagy can inhibit tumor formation, and can also help tumor cells against poor growth environments such as hypoxia and lack of nutritional factors, thus promoting the occurrence and development of tumors.28 Autophagy plays a dual role in the regulation of epithelial-mesenchymal transition (EMT) as well. For one thing, autophagy inhibits EMT to inhibit invasion of tumor cells; for another, autophagy can activate EMT to promote invasion of tumor cells.29 There is increasing evidence showing that autophagy is closely related to EMT, but the role of miR-1224-5p in EMT and autophagy in OS remains to be investigated. Therefore, we hypothesize that miR-1224-5p may affect the biological functions of OS cells such as proliferation, migration, invasion and EMT through autophagy, so as to provide new ideas. The present study will investigate this conjecture to provide new ideas for the treatment of OS.
Materials and Methods
Cell Culture
Human normal osteoblast line hFOB1.19, and OS cell lines MG-63, U2OS, SaOS2, HOS, 143B were purchased from the cell bank of Shanghai Institute of Cell Biology, Chinese Academy of Sciences. The cells were cultured in RPMI-1640 medium containing 10% fetal bovine serum (FBS), 100U/mL penicillin and 100μg/mL streptomycin in a saturated humidity incubator with 5% CO2 at 37°C. The cells were digested and passaged with 0.25% trypsin every 2–3 days, and the cells in logarithmic growth phase were taken for the subsequent experiments.
Cell Transfection and Vector Construction
MiR-1224-5p mimic, NC mimic, pcDNA3.1-NC and overexpression vector pcDNA3.1-PLK1 were designed and synthesized by RiboBio (Guangzhou, China). The cells were transfected using Lipofectamine 2000 in strict accordance with the instructions provided by the manufacturer. MG-63 cells and U2OS cells were divided into 6 groups: Control group, miR-1224-5p mimic group (miR-1224-5p group), miR-1224-5p mimic NC (NC mimic group), negative control group with PLK1 overexpression (pc-NC group), PLK1 overexpression group (PLK1 group) and miR-1224-5p overexpression + PLK1overexpression group (miR-1224-5p mimic+PLK1 group). The cells were seeded at 6-well plates with a density of 5×105 cells per well to 50–70% confluency. Subsequently, 2μL Lipofectamine2000 and 5nM miR-1224-5p mimic, NC mimic, pc-NC, PLK1 and miR-1224-5p mimic+PLK1 were mixed at 1:50 in volume for transfection. After 6 hours of transfection, the cells were rinsed with PBS for 3 times, the culture medium was replaced and then the cells were continuously cultured for 48 hours. The transfection efficiency was verified by qRT-PCR, and the cells with successful transfection were used in the follow-up experiments.
MTT Assay
The cells in logarithmic growth phase were seeded on 96-well plates at the concentration of 5.0 × 107/L (100 μL per well), and cultured in a saturated humidity incubator with 5% CO2 at 37°C for 24 hours. Following the cell attachment, the OS cells were transfected according to the method mentioned above. At 24, 48 and 72 hours after transfection, 20 μL MTT reaction solution (2 g/L) was added to each well and incubated for 4 hours, then the culture was terminated. Subsequently, the culture medium was aspirated and 150μL of DMSO was added to each well. The absorbance (A) value of each well was measured at 490nm using the microplate reader. Six parallel wells were set for each group, and the experiment was repeated 3 times.
Colony Formation Assay
At 48 hours after transfection, MG-63 and U2OS cells were digested with 0.25% pancreatin and centrifuged at 1000r/min for 5 min and then collected to resuspend and count. The cell suspension with gradient dilution was inoculated in the petri dish at appropriate cell density, and the cells were distributed evenly at the bottom as much as possible. The cells were continuously cultured in the incubator for 10 days until the colony was visible to the naked eye. Then the original culture medium was removed, PBS was used to rinse the cells once, and the cells were fixed for 15 min with 10% formaldehyde. Next, the cells were rinsed with PBS twice, stained with crystal violet for 30 min, rinsed with PBS again and dried, and finally photographed. The experiment was repeated 3 times.
Annexin V-PI Apoptosis Assay
The transfected cells were digested and then collected. A total of 5×104-10×104 cells were centrifuged at 1000r/min for 5 min, and the supernatant was aspirated. The cells were resuspended gently in 195μL binding buffer of Annexin V-FITC. Then, 5μL Annexin V-FITC reagent was added to the cell suspension and mixed gently. The cells were incubated at room temperature for 5 min in the dark. Subsequently, 10μL of propidium iodide (PI) staining solution was added and placed at room temperature for 10 min in the dark. Another 200μL of Annexin V-FITC binding buffer was added to gently resuspend the cells. Finally, apoptosis was detected using flow cytometry (FACSCanto TM II, BD Biosciences).
Transwell Invasion Assay
At 48 hours after transfection, MG-63 and U2OS cells were digested in the serum-free medium. Then, 200 μL cell suspension containing 8×107/L cells was added to the upper chamber of Transwell insert covered with matrix glue, while 600 μL medium containing 10% FBS was added to the lower chamber, and cultured with 5% CO2 at 37°C for 24 hours. Three parallel wells were set in each group. Transwell insert was taken out, Matrigel and non-invasive cells in the upper chamber were wiped off with cotton swab. And the invasive cells were fixed with methanol for 10 min, stained with 0.1% crystal violet for 40 min. Finally, five fields were randomly selected under a microscope (100 ×) to count the invasive cells, and the average value was taken.
Dual-Luciferase Reporter Gene Assay
PLK1 was predicted to be a target gene of miR-1224-5p using Starbase V2.0 software which was verified by luciferase reporter gene assay. The 3ʹuntranslated region (UTR) of the wild-type (WT) PLK1 gene was cloned into a luciferase vector plasmid, and the mutant (MUT) plasmid WAS obtained from the mutation of the binding domain of PLK1 and miR-1224-5p. MG-63 cells and U2OS cells were inoculated in a 24-well plate to 60% confluency. Then the cells were transfected with Lipofectamine 2000 according to the following groups: miR-1224-5p mimics and WT plasmid containing PLK1 3ʹ-UTR; NC mimics and WT plasmid containing PLK1 3ʹ-UTR; miR-1224-5p mimics and MUT plasmid containing PLK1 3ʹ-UTR; NC mimics and MUT plasmid containing PLK1 3ʹ-UTR. After 48 hours of co-transfection, the relative luciferase activity levels among different groups was detected by dual-luciferase assay system. The experiment was repeated 3 times.
Quantitative RT-PCR (qRT-PCR)
Total RNA was extracted from OS cell lines MG-63, U2OS, SaOS2, HOS, 143B, and normal osteoblast line hFOB1.19 and animal tumor tissue using Trizol reagent (Invitrogen) according to the instruction. A total of 500ng RNA was reversely transcribed to cDNA using the cDNA Transcription Kit (ABI). qRT-PCR reactions were performed with 40 cycles using the following parameters: 95°C for 2 min, 95°C for 15 seconds and 60°C for 15 seconds, respectively. All the results were standardized by the expression of GAPDH. Quantitative analysis was performed using the 2−ΔΔCt method. The primer sequences used were as follows: miR-1224-5p, Forward: 5ʹ-TGCGCGTGAGGACTCGGGA-3ʹ and Reverse: 5ʹ-CTCAAGTGTCGTGGAGTCGGCAA-3ʹ; U6, Forward: 5ʹ-GCTTCGGCAGCACATATACTAAAAT-3ʹ and Reverse: 5ʹ-CGCTTCACGAATTTGCGTGTCAT-3ʹ; PLK1, Forward: 5ʹ-ACCTGTTGTGTCTTCCCTCT-3ʹ and Reverse: 5ʹ-GTAGAACCCACACCCGAACA-3ʹ; GAPDH, Forward: 5ʹ-AACGGATTTGGTCGTATTG-3ʹ and Reverse: 5ʹ-GGAAGATGGTGATGGGATT-3ʹ.
Western Blot
At 48 hours after transfection, the cells were and lysed on ice for 30 min using RIPA buffer. After the protein content had been determined, 40 μg of protein was taken and separated using 10% SDS-PAGE. Subsequently, the proteins on the gels were transferred to PVDF membranes by wet transfer method. The PVDF membranes were blocked with blocking solution for 2 hours at room temperature. After that, the membranes were incubated with primary antibodies (Abcam, UK) overnight at 4°C. After rinsed with TBST 3 to 5 times (5–10 min/time), the membranes were incubated with the secondary antibodies at room temperature for 1 hour. Finally, the target proteins were developed with ECL solutions, and analyzed by gel imaging system. The images were quantified using Image J (National Institutes of Health).
Statistical Analysis
All the experiments were repeated 3 times, and the experimental data were expressed as mean ± standard deviation (SD). SPSS22.0 statistical software was used for data analysis. The comparisons of multiple groups were performed using one-way analysis of variance (ANOVA), and comparisons between groups using t-test. P<0.05 was considered significant.
Results
MiR-1224-5p Was Expressed at Low Levels in OS Tissues and Cell Lines
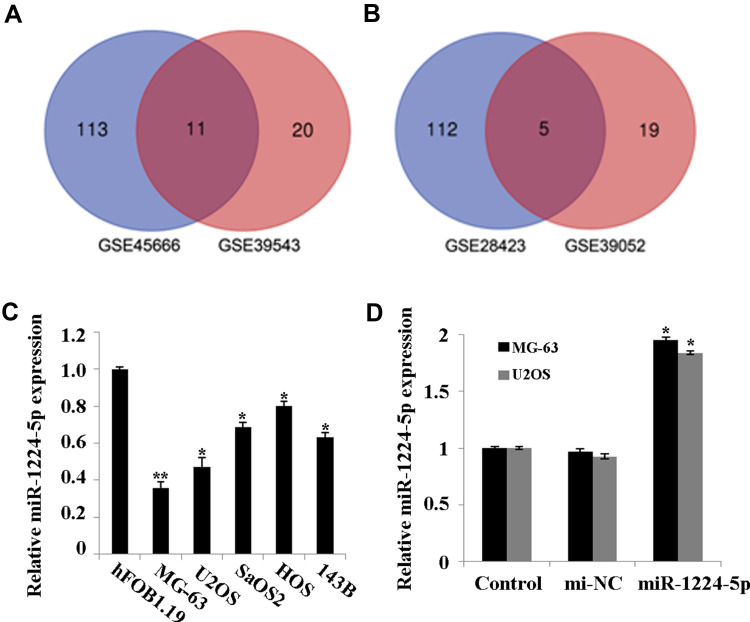
Firstly, GSE28423 and GSE39052 data sets were used to analyze the abnormally expressed miRNAs, and the intersection of the up-regulated miRNAs and the intersection of the down-regulated miRNAs were taken for the Venn diagram (Figure 1A and B). The results showed that a total of 11 miRNAs were co-up-regulated and 5 miRNAs were co-down-regulated. In addition, the expression of miR-1224-5p in human OS cell lines MG-63, U2OS, SaOS2, HOS and 143B was detected. Compared with human normal osteoblast line hFOB1.19, the levels of miR-1224-5p in MG-63, U2OS, SaOS2, HOS and 143B were significantly declined (Figure 1C). These results suggested that miR-1224-5p had the potential to be involved in the progression of OS. The expression of mir-1224-5p was lower in MG-63 cells and U2OS cells, so the two cells were selected for subsequent experiments. In order to further detect the effect of miR-1224-5p expression on OS progression, in MG-63 cells and U2OS cells were transfected with miR-1224-5p mimics to upregulate the expression of miR-1224-5p (Figure 1D).
Figure 1.
MiR-1224-5p is expressed at low levels in OS tissues and cell lines. (A) The Venn diagram of the intersection of up-regulated miRNAs in GSE28423 and GSE39052 data sets. (B) The Venn diagram of the intersection of down-regulated miRNAs intersection in GSE28423 and GSE39052 data sets. (C) QRT-PCR to detect the expression of miR-1224-5p in human osteoblast hFOB1.19 and OS cell lines MG-63, U2OS, SaOS2, HOS and 143B. *P<0.05 and **P<0.01 vs hFOB1.19. (D) QRT-PCR to detect the expression of miR-1224-5p in MG-63 cells and U2OS cells transfected with miR-1224-5p mimics or mimics NC. *P<0.05 vs mi-NC group.
Elevated miR-1224-5p Expression Inhibits Proliferation and Invasion While Promotes Apoptosis and Autophagy of OS Cells
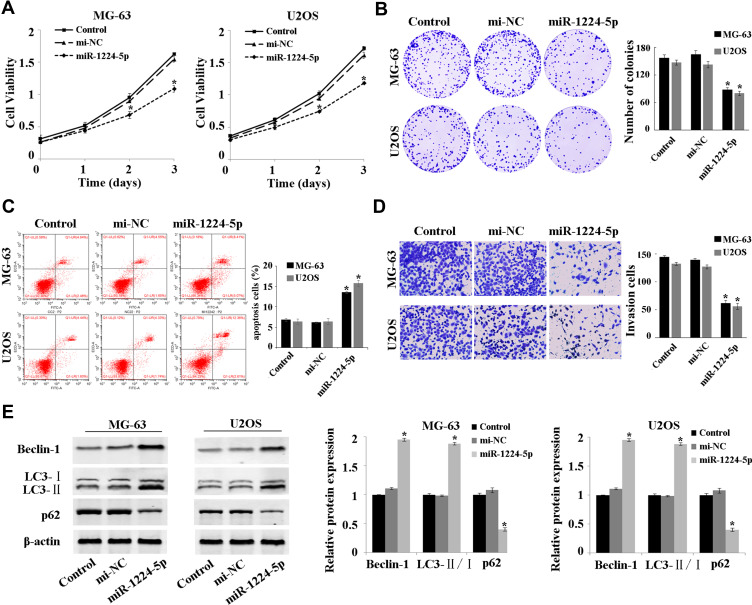
The expression of miR-1224-5p was overexpressed to investigate the effect of miR-1224-5p on the function of MG-63 and U2OS cells. The results of MTT assay (Figure 2A), colony formation assay (Figure 2B), flow cytometry (Figure 2C) showed that compared with mi-NC group, the proliferation of MG-63 cells and U2OS cells was inhibited and apoptosis was promoted in the miR-1224-5p group. The results of Transwell confirmed that overexpression of miR-1224-5p inhibited cell invasion ability (Figure 2D). Meanwhile, the results of Western blot revealed that overexpression of miR-1224-5p promoted the expression of Beclin-1 and LC3-Ⅱ/Ⅰ and inhibit the expression of p62 (Figure 2E). These results suggested that miR-1224-5p had the biological function to inhibit the proliferation, invasion and promote apoptosis and autophagy of OS cells.
Figure 2.
MiR-1224-5p inhibits proliferation and invasion while promotes the apoptosis and autophagy of OS cells. (A) MTT assay to detect the cell proliferation in MG-63 cells and U2OS cells after transfection with NC mimic and miR-1224-5p mimic. (B) Cell colony formation assay to detect the cell viability in MG-63 cells and U2OS cells after transfection with NC mimic and miR-1224-5p mimic. (C) Flow cytometry to detect the apoptosis rate of MG-63 cells and U2OS cells after transfected with NC mimic and miR-1224-5p mimic. (D) Transwell to detect the invasive ability of MG-63 cells and U2OS cells after transfection with NC mimic and miR-1224-5p mimic. (E) Western blot to detect the expression level of autophagy-related proteins Beclin-1, LC3-II/I and p62 in MG-63 cells and U2OS cells after transfection with NC mimic and miR-1224-5p mimic. *P<0.05 vs mi-NC group.
MiR-1224-5p Mediates PI3K/AKT/mTOR Pathway to Inhibit EMT of OS Cells
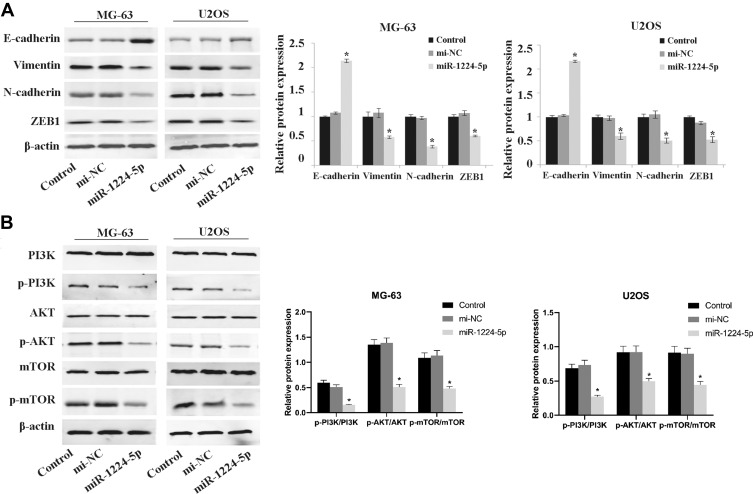
To further elucidate the mechanism of miR-1224-5p in the regulation of biological functions such as EMT of OS cells, Western blot was used to detect the expression of EMT-related proteins E-cadherin, Vimentin, N-cadherin, ZEB1 and the expression of related proteins of PI3K/AKT/mTOR signal pathway in MG-63 cells and U2OS cells after transfected with miR-1224-5p mimic and NC mimic. As shown in Figure 3A, compared with the mi-NC group, the protein expression level of E-cadherin in MG-63 cells and U2OS cells elevated significantly, while Vimentin, N-cadherin and ZEB1 declined obviously in MG-63 cells and U2OS cells in the miR-1224-5p mimic group. As shown in Figure 3B, the protein levels of p-PI3K, p-AKT and p-mTOR decreased significantly in MG-63 cells and U2OS cells in the miR-1224-5p group (P < 0.05). These results suggested that miR-1224-5p inhibited EMT of OS cells by mediating PI3K/AKT/mTOR signal pathway.
Figure 3.
Effect of miR-1224-5p on the expression of related proteins of PI3K/AKT/mTOR pathway and EMT. Western blot is applied to detect the expression levels of EMT-related proteins E-cadherin, Vimentin, N-cadherin, ZEB1 (A) and the expression levels of PI3K/AKT/mTOR signal pathway-related proteins p-PI3K, p-AKT, p-mTOR, PI3K, AKT and mTOR (B) in MG-63 cells and U2OS cells after transfection with miR-1224-5p mimic and NC mimic. *P<0.05 vs mi-NC group.
MiR-1224-5p Targets PLK1
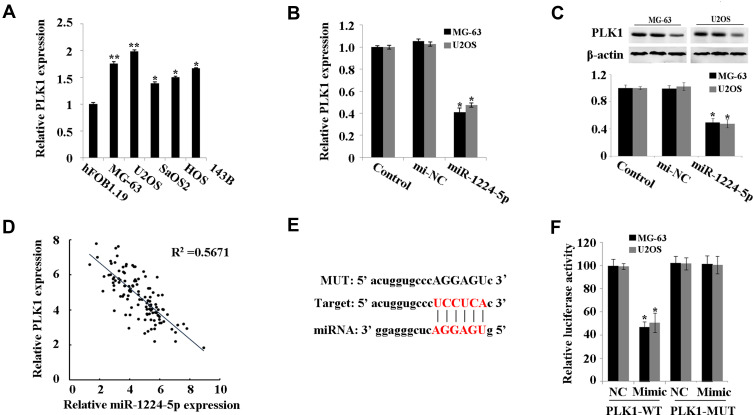
PLK1 was predicted to be a target gene of miR-1224-5p using Starbase V2.0. It was further found that compared with that in hFOB1.19 cells, PLK1 was expressed at low levels in MG-63, U2OS, SaOS2, HOS and 143B cells (Figure 4A). Meanwhile, the expression of PLK1 increased significantly in the miR-1224-5p group (Figure 4B and C). The results of correlation analysis showed a negative correlation between PLK1 and miR-1224-5p expression (Figure 4D). In order to further verify the targeting relationship between miR-1224-5p and PLK1, relative luciferase activity was measured and the results showed that the overexpression of miR-1224-5p significantly inhibited the luciferase reporter gene activity of the WT plasmid containing PLK1 3ʹ-UTR (Figure 4E and F). These results suggested that in OS cells, miR-1224-5p targeted and regulated the expression of PLK1 by binding to the PLK1 3ʹ-UTR.
Figure 4.
MiR-1224-5p targets PLK1. (A) qRT-PCR to detect the expression of PLK1 in hFOB1.19 and MG-63, U2OS, SaOS2, HOS and 143B. (B) qRT-PCR to detect the expression of PLK1 in MG-63 cells and U2OS cells after transfected with miR-1224-5p mimic and NC mimic. (C) Western blot to detect the protein expression of PLK1 in MG-63 cells and U2OS cells after transfected with miR-1224-5p mimic and NC mimic. *P<0.05 vs mi-NC group. (D) Correlation analysis between miR-1224-5p and PLK1 expression. (E) Starbase V2.0 to predict the binding site between miR-1224-5p and PLK1. (F) Dual-luciferase reporter gene assay to detect the targeting relationship between miR-1224-5p and PLK1. *P<0.05 vs NC group.
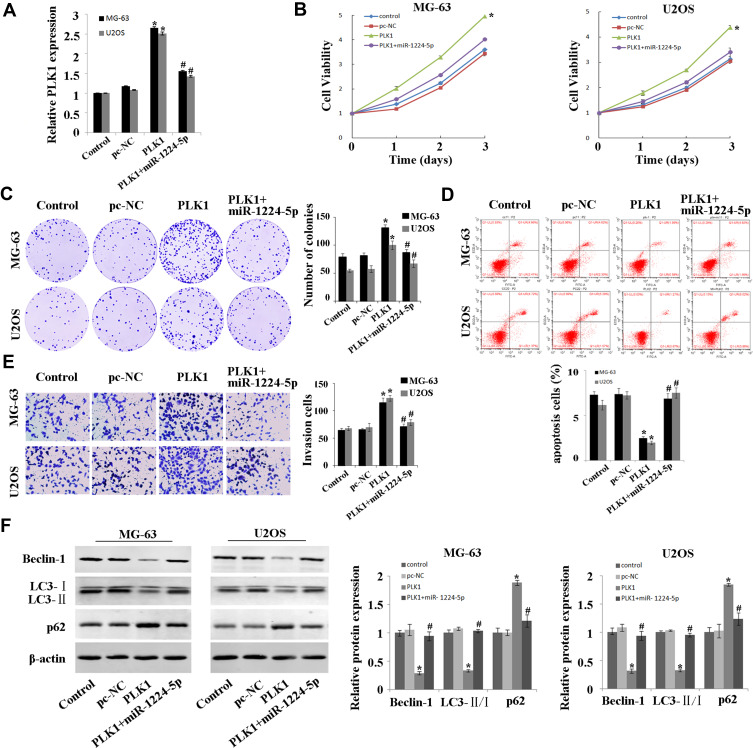
Overexpression of PLK1 Inhibits the Inhibitory Effect of miR-1224-5p on OS Cells
In order to explore whether miR-1224-5p can exert its biological function in OS cells by targeting PLK1, the rescue experiment was carried out. PLK1 overexpression plasmids was transfected into MG-63 cells and U2OS cells with overexpression of miR-1224-5p to detect cell function. After transfection, the expression of PLK1 in PLK1+miR-1224-5p group was significantly lower than that in PLK1 group (Figure 5A). The results of MTT, cell colony assay and flow cytometry (Figure 5B–D) showed that compared with pc-NC group, the proliferation of MG-63 cells and U2OS cells was promoted and the apoptosis was inhibited in the PLK1 group. The results of Transwell assay (Figure 5E) showed that overexpression of PLK1 promoted cell invasion, and inhibited the expression of autophagy-related proteins (Beclin-1 and LC3-Ⅱ/), while promoted the expression of P62 (Figure 5F). In addition, it was also found that compared with the pc-NC group, cell proliferation, apoptosis, invasion and autophagy in the PLK1+miR-1224-5p group were contrary to the above results. These results further suggested that miR-1224-5p inhibited the proliferation, invasion and promoted the apoptosis and autophagy of OS cells by regulating the expression of PLK1.
Figure 5.
MiR-1224-5p regulates the expression of PLK1 to inhibit the proliferation and invasion while promote the apoptosis and autophagy of OS cells. (A) QRT-PCR to detect the expression of PLK1 in MG-63 cells and U2OS cells after transfection with pc-NC, PLK1 and miR-1224-5p mimic+PLK1. (B) MTT assay to detect the cell proliferation in MG-63 cells and U2OS cells after transfection with pc-NC, PLK1 and miR-1224-5p mimic+PLK1. (C) Cell colony formation assay to detect the cell viability in MG-63 cells and U2OS cells after transfection with pc-NC, PLK1 and miR-1224-5p mimic+PLK1. (D) Flow cytometry to detect the apoptosis rate of MG-63 cells and U2OS cells after transfection with pc-NC, PLK1 and miR-1224-5p mimic+PLK1. (E) Transwell to detect the invasive ability of MG-63 cells and U2OS cells after transfection with pc-NC, PLK1 and miR-1224-5p mimic+PLK1. (F) Western blot to detect the expression levels of autophagy-related proteins Beclin-1, LC3-II/I and p62 in MG-63 cells and U2OS cells after transfection with pc-NC, PLK1 and miR-1224-5p mimic+PLK1. *P<0.01 vs pc-NC group; #P<0.01 vs PLK1 group.
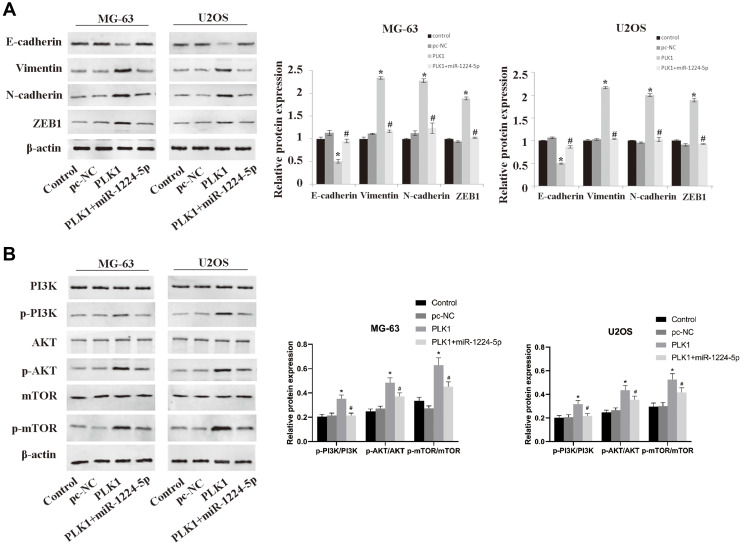
MiR-1224-5p Mediates PI3K/AKT/mTOR Pathway by Targeting PLK1 to Inhibit the EMT of OS Cells
In order to further investigate the molecular mechanism of miR-1224-5p in the regulation of EMT of OS cells by targeting PLK1, the expression of EMT-related proteins and PI3K/AKT/mTOR signal pathway-related proteins in MG-63 cells and U2OS cells after transfection with pc-NC, PLK1 and miR-1224-5p mimic+PLK1. The results of Western blot showed that compared with the pc-NC group, the protein expression level of E-cadherin in the PLK1 group was significantly lower, while Vimentin, N-cadherin and ZEB1 were significantly elevated (p < 0.05); and the protein levels of p-PI3K, p-AKT and p-mTOR also increased significantly in the PLK1 group. However, compared with the PLK1 group, the expression levels of these proteins in the PLK1+miR-1224-5p group were opposite (Figure 6A and B). These results suggested that miR-1224-5p mediated PI3K/AKT/mTOR signaling pathway by targeting PLK1 to inhibit the EMT of OS cells.
Figure 6.
Overexpression of PLK1 inhibits the effect of miR-1224-5p on the expression of related proteins of PI3K/AKT/mTOR and EMT. After transfection of pc-NC, PLK1 and miR-1224-5p mimic+PLK1, the expression levels of EMT-related proteins E-cadherin, Vimentin, N-cadherin, ZEB1 (A) and PI3K/AKT/mTOR signal pathway-related proteins p-PI3K, p-AKT, p-mTOR, PI3K, AKT and mTOR (B), in MG-63 cells and U2OS cells were detected by Western blot. *P < 0.05 vs mi-NC group; #P<0.01 vs PLK1 group.
Discussion
OS is a common primary malignant bone tumor in children and adolescents, with high metastasis rate, high malignant degree, high recurrence rate and high mortality rate. It causes serious impact on the physical and mental health of patients. Surgical amputation, radiotherapy and chemotherapy are commonly used in the clinical treatment of OS; although the survival time has been prolonged, the therapeutic effect still needs to be improved. Therefore, it is of great significance to find new therapeutic targets and effective drugs for the prognosis of OS. MiR-1224-5p is considered as a potential biomarker for tumor diagnosis, and this study provides a new direction for the treatment of OS by exploring the relationship between miR-1224-5p, EMT and autophagy.
Previous studies have found that miR-1224-5p plays an important role in tumorigenesis, regulating the proliferation, apoptosis and invasion of tumor cells. For example, based on the gene chip technology, Mosakhani et al studied the differences of miRNAs expression from 99 patients with metastatic colorectal cancer and found that there was a correlation between miR-1224-5p downregulation and survival rate.30 Della et al performed microarray analysis on the samples from 38 cases with rectal cancer who underwent surgery, and found that miR-1224-5p was significantly up-regulated.31 Jin et al found that miR-1224-5p was able to inhibit the occurrence and development of neuroglioma by targeting CREB1.32 However, the role of miR-1224-5p in OS is not clear currently. In this study, mir-1224-5p was down-regulated in OS cells; overexpression of miR-1224-5p inhibited the proliferation and invasion and promoted the apoptosis of osteosarcoma cells. These results confirmed that the inhibitory effect of miR-1224-5p on OS.
Furthermore, PLK1 gene was confirmed to be a direct target of miR-1224-5p. Then we found that overexpression of PLK1 gene promoted the proliferation and invasion and inhibit the apoptosis of OS cells. In addition, the results of rescue experiments confirmed that overexpression of PLK1 reversed the effect of miR-1224-5p on the function of OS tumor cells. PLK1 is a cell cycle kinase that participates in eukaryotes mitosis and can be used as a biomarker and potential therapeutic target in oncology.33 It has been reported that overexpression of PLK1 can promote the progression of breast cancer,33 renal cell carcinoma,34 gastric cancer.35 With regard to the study of PLK1 in OS, Zhu et al found that the proliferation of OS cells was inhibited with the intervention of PLK1 expression.36 Mo et al found that PLK1 could promote the proliferation of MYC-amplifying OS cells through autophagy.37 These results are consistent with our research results. It is suggested that miR-1224-5p regulates the effect of PLK1 expression on the occurrence and development in OS.
The occurrence and development of tumor is often accompanied by EMT which can promote tumor invasion and metastasis.38 The most important molecular markers in the process of EMT are the loss of epithelial marker E-cadherin and the gain of mesenchymal markers Vimentin, N-cadherin and ZEB1.39 Our study found that overexpression of miR-1224-5p could inhibit the expression of Vimentin, N-cadherin and ZEB1 in OS cells, and promote the expression of E-cadherin; overexpression of PLK1 promoted the occurrence of EMT in OS cells, while overexpression of miR-1224-5p could reverse the expression of the corresponding proteins.
Autophagy plays a dual role in the regulation of EMT. In human breast cancer cells, DEDD interacts with PI3KC3 to induce increase of autophagy, which in turn inhibits the occurrence of EMT, thus inhibiting the invasion of tumor cells.40 However, autophagy can also promote the invasion of hepatoma cells by activating EMT.41 Meanwhile, Lu et al have found that miR-517c inhibits autophagy and EMT of glioblastoma cells.42 Increasing evidence shows that autophagy is closely related to EMT, but autophagy inhibits or promotes EMT is not completely clear. We speculate that these different results may be caused by the inconsistency of microenvironment, cell type and experimental conditions. The molecular mechanisms by which autophagy inhibits or promotes EMT have not been found at present. It is well known that autophagy is regulated by autophagy-related molecules, and the expression levels of LC3-II/I, p62 and Beclin-1 can express autophagy activity.43 Our study found that through targeting PLK1, miR-1224-5p significantly activated the expression of autophagy-related proteins LC3II/I and Beclin-1 in OS, while decreased the level of p62. Other studies have confirmed that PI3K/AKT/mTOR pathway can reduce autophagy activity and promote EMT. This study further confirms that miR-1224-5p targets PLK1 to inhibit PI3K/AKT/mTOR pathway, thus activating autophagy activity to inhibit the occurrence and development of EMT.
In summary, miR-1224-5p is expressed at low levels in OS cells and tissues. MiR-1224-5p regulates the biological functions such as autophagy and EMT of OS through PLK1. The conclusions of this study still were required further animal experiments and clinical tissue samples to verify. This study suggests that miR-1224-5p is a potential target for the treatment of OS and has a good clinical application prospect for the prognosis of OS patients.
Acknowledgements
Thank you very much for the technical support of Guangzhou Yujia Biological Technology Co., Ltd.
Funding Statement
NHC Key Laboratory of Pulmonary Immune-related Diseases is supported by the non-profit central research Institute fund of Chinese academy of medical sciences (2019PT320003).
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Hendershot E, Volpe J, Taylor T, et al. Outpatient high-dose methotrexate for osteosarcoma: it’s safe and feasible, if you want it. J Pediatr Hematol Oncol. 2018;41(5):1. [DOI] [PubMed] [Google Scholar]
- 2.Tariq U, Sohail MS, Fatima Z, Khan A, Sheikh AB, Bhatti S. Simultaneous bilateral spontaneous pneumothorax: a rare complication of osteosarcoma. Cureus. 2018;10(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feng D, Liu T, Jin H, Wang W. Chimaphilin inhibits human osteosarcoma cell invasion and metastasis through suppressing the TGF-β1-induced epithelial-to-mesenchymal transition markers via PI-3K/Akt, ERK1/2, and smad signaling pathways. Can J Physiol Pharmacol. 2018;96. [DOI] [PubMed] [Google Scholar]
- 4.Pan Y, Lu L, Chen J, et al. Identification of potential crucial genes and construction of microRNA-mRNA negative regulatory networks in osteosarcoma. Hereditas. 2018;155(1):21. doi: 10.1186/s41065-018-0061-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paiva-Oliveira DI, Martins-Neves SR, Abrunhosa AJ, Fontes-Ribeiro C, Gomes CM. Therapeutic potential of the metabolic modulator metformin on osteosarcoma cancer stem-like cells. Cancer Chemother Pharmacol. 2018;81(1):49–63. doi: 10.1007/s00280-017-3467-6 [DOI] [PubMed] [Google Scholar]
- 6.Marko TA, Diessner BJ, Spector LG. Prevalence of metastasis at diagnosis of osteosarcoma: an international comparison. Pediatr Blood Cancer. 2016;63(6):1006–1011. doi: 10.1002/pbc.25963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zandueta C, Ormazábal C, Perurena N, et al. Matrix-Gla protein promotes osteosarcoma lung metastasis and associates with poor prognosis. J Pathol. 2016;239(4):438–449. doi: 10.1002/path.4740 [DOI] [PubMed] [Google Scholar]
- 8.Kunita A, Kashima TG, Ohazama A, et al. Podoplanin is regulated by AP-1 and promotes platelet aggregation and cell migration in osteosarcoma. Am J Pathol. 2011;179(2):1041–1049. doi: 10.1016/j.ajpath.2011.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Venkatadri R, Muni T, Iyer AKV, et al. Role of apoptosis-related miRNAs in resveratrol-induced breast cancer cell death. Cell Death Dis. 2016;7(2):e2104. doi: 10.1038/cddis.2016.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y, Xu B, Zhang XP. Effects of miRNAs on functions of breast cancer stem cells and treatment of breast cancer. Onco Targets Ther. 2018;11:4263–4270. doi: 10.2147/OTT.S165156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prada I, Gabrielli M, Turola E, et al. Glia-to-neuron transfer of miRNAs via extracellular vesicles: a new mechanism underlying inflammation-induced synaptic alterations. Acta Neuropathol. 2018;135(4):529–550. doi: 10.1007/s00401-017-1803-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thayanithy V, Sarver AL, Kartha RV, et al. Perturbation of 14q32 miRNAs-cMYC gene network in osteosarcoma. Bone. 2012;50(1):0–181. doi: 10.1016/j.bone.2011.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lauvrak SU, Munthe E, Kresse SH, et al. Functional characterisation of osteosarcoma cell lines and identification of mRNAs and miRNAs associated with aggressive cancer phenotypes. Br J Cancer. 2013;109(8):2228–2236. doi: 10.1038/bjc.2013.549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gui ZL, Wu TL, Zhao GC, Lin ZX, Xu HG. MicroRNA-497 suppress osteosarcoma by targeting MAPK/Erk pathway. Bratisl Lek Listy. 2017;118(08):449–452. [DOI] [PubMed] [Google Scholar]
- 15.Liu D, Zhang C, Li X, Zhang H, Pang Q, Wan A. MicroRNA-567 inhibits cell proliferation, migration and invasion by targeting FGF5 in osteosarcoma. EXCLI J. 2018;17:102–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qian M, Gong H, Yang X, et al. MicroRNA-493 inhibits the proliferation and invasion of osteosarcoma cells through directly targeting specificity protein 1. Oncol Lett. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Du L, Chen T, Zhao K, et al. miR-30a suppresses osteosarcoma proliferation and metastasis by downregulating MEF2D expression. Onco Targets Ther. 2018;11:2195–2202. doi: 10.2147/OTT.S102430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cao R, Shao J, Hu Y, et al. microRNA-338-3p inhibits proliferation, migration, invasion, and EMT in osteosarcoma cells by targeting activator of 90 kDa heat shock protein ATPase homolog 1. Cancer Cell Int. 2018;18(1):49. doi: 10.1186/s12935-018-0551-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu C, Cheng X, MingYu Q, Wang XB, Shen SQ. The effects of microRNA-1224-5p on hepatocellular carcinoma tumor endothelial cells. J Cancer Res Ther. 2019;15(2):329. [DOI] [PubMed] [Google Scholar]
- 20.Qian J, Li R, Wang -Y-Y, et al. MiR-1224-5p acts as a tumor suppressor by targeting CREB1 in malignant gliomas. Mol Cell Biochem. 2015;403(1):33–41. [DOI] [PubMed] [Google Scholar]
- 21.Wang J, Hu Y, Ye C, Liu J. miR-1224-5p inhibits the proliferation and invasion of ovarian cancer via targeting SND1. Hum Cell. 2020;1–10. [DOI] [PubMed] [Google Scholar]
- 22.Kong L, Liu P, Zheng M, et al. The miR-1224-5p/ELF3 axis regulates malignant behaviors of pancreatic cancer via PI3K/AKT/notch signaling pathways. Onco Targets Ther. 2020;13:3449–3466. doi: 10.2147/OTT.S248507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yao X, Cui X, Wu X, et al. Tumor suppressive role of miR-1224-5p in keloid proliferation, apoptosis and invasion via the TGF-β1/Smad3 signaling pathway. Biochem Biophys Res Commun. 2017;495(1):713. doi: 10.1016/j.bbrc.2017.10.070 [DOI] [PubMed] [Google Scholar]
- 24.Wu Q, Xu T, Liu Y, et al. miR-1224-5p mediates mitochondrial damage to affect silica-induced pulmonary fibrosis by targeting BECN1. Int J Mol Sci. 2017;18(11):2357. doi: 10.3390/ijms18112357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yin D, Wang Q, Wang S, Zhu G, Tang Q, Liu J. OGFOD1 negatively regulated by miR-1224-5p promotes proliferation in human papillomavirus-infected laryngeal papillomas. Mol Genet Genom. 2020;1–10. [DOI] [PubMed] [Google Scholar]
- 26.Lian H, Xie P, Yin N, et al. Linc00460 promotes osteosarcoma progression via miR-1224-5p/FADS1 axis. Life Sci. 2019;233:116757. doi: 10.1016/j.lfs.2019.116757 [DOI] [PubMed] [Google Scholar]
- 27.Yin Z, Pascual C, Klionsky D. Autophagy: machinery and regulation. Microbial Cell. 2016;3(12):588–596. doi: 10.15698/mic2016.12.546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garcia-Prat L, Martínez-Vicente M, Perdiguero E, et al. Autophagy maintains stemness by preventing senescence. Nature. 2016;534(7607):37–42. doi: 10.1038/nature16187 [DOI] [PubMed] [Google Scholar]
- 29.Zhang X, Cheng Q, Yin H, et al. Regulation of autophagy and EMT by the interplay between p53 and RAS during cancer progression (Review). Int J Oncol. 2017;51(1):18–24. doi: 10.3892/ijo.2017.4025 [DOI] [PubMed] [Google Scholar]
- 30.Mosakhani N, Lahti L, Borze I, et al. MicroRNA profiling predicts survival in anti-EGFR treated chemorefractory metastatic colorectal cancer patients with wild-type KRAS and BRAF. Cancer Genet Cytogenet. 2012;205(11):545–551. doi: 10.1016/j.cancergen.2012.08.003 [DOI] [PubMed] [Google Scholar]
- 31.Scarpati GDV, Falcetta F, Carlomagno C, et al. A specific miRNA signature correlates with complete pathological response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer. Int J Radiat Oncol Biol Phys. 2012;83(4):1113–1119. doi: 10.1016/j.ijrobp.2011.09.030 [DOI] [PubMed] [Google Scholar]
- 32.Jin Q, Li R, Wang -Y-Y, et al. MiR-1224-5p acts as a tumor suppressor by targeting CREB1 in malignant gliomas. Mol Cell Biochem. 2015;403(1–2):33–41. doi: 10.1007/s11010-015-2334-1 [DOI] [PubMed] [Google Scholar]
- 33.García IA, Garro C, Fernandez E, et al. Therapeutic opportunities for PLK1 inhibitors: spotlight on BRCA1-deficiency and triple negative breast cancers. Mutat Res. 2020;821:111693. doi: 10.1016/j.mrfmmm.2020.111693 [DOI] [PubMed] [Google Scholar]
- 34.Gao ZP, Man X, Li Z, et al. PLK1 promotes proliferation and suppresses apoptosis of renal cell carcinoma cells by phosphorylating MCM3. Cancer Gene Ther. 2020;27(6):412–423. doi: 10.1038/s41417-019-0094-x [DOI] [PubMed] [Google Scholar]
- 35.Chen ZH, Chai Y, Zhao T, et al. Effect of PLK1 inhibition on cisplatin-resistant gastric cancer cells. J Cell Physiol. 2019;234(5):5904–5914. doi: 10.1002/jcp.26777 [DOI] [PubMed] [Google Scholar]
- 36.Zhu JJ, Cui K, Cui Y, et al. PLK1 knockdown inhibits cell proliferation and cell apoptosis, and PLK1 is negatively regulated by miR-4779 in osteosarcoma cells. DNA Cell Biol. 2020;39(5):747–755. doi: 10.1089/dna.2019.5002 [DOI] [PubMed] [Google Scholar]
- 37.Mo H, He J, Yuan Z, et al. <p>PLK1 contributes to autophagy by regulating MYC stabilization in osteosarcoma cells. Onco Targets Ther. 2019;12:7527–7536. doi: 10.2147/OTT.S210575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Akalay I, Janji B, Hasmim M, et al. Epithelial-to-mesenchymal transition and autophagy induction in breast carcinoma promote escape from T-cell-mediated lysis. Cancer Res. 2013;73(8):2418–2427. doi: 10.1158/0008-5472.CAN-12-2432 [DOI] [PubMed] [Google Scholar]
- 39.Li J, Xia L, Zhou Z, et al. MiR-186-5p upregulation inhibits proliferation, metastasis and epithelial-to-mesenchymal transition of colorectal cancer cell by targeting ZEB1. Arch Biochem Biophys. 2018;640:53–60. doi: 10.1016/j.abb.2018.01.002 [DOI] [PubMed] [Google Scholar]
- 40.Lv Q, Wang W, Xue J, et al. DEDD interacts with PI3KC3 to activate autophagy and attenuate epithelial–mesenchymal transition in human breast cancer. Cancer Res. 2012;72(13):3238–3250. doi: 10.1158/0008-5472.CAN-11-3832 [DOI] [PubMed] [Google Scholar]
- 41.Grassi G, Di Caprio G, Santangelo L, et al. Autophagy regulates hepatocyte identity and epithelial-to-mesenchymal and mesenchymal-to-epithelial transitions promoting snail degradation. Cell Death Dis. 2015;6(9):e1880–e1880. doi: 10.1038/cddis.2015.249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu Y, Xiao L, Liu Y, et al. MIR517C inhibits autophagy and the epithelial-to-mesenchymal (-like) transition phenotype in human glioblastoma through KPNA2-dependent disruption of TP53 nuclear translocation. Autophagy. 2015;11(12):2213–2232. doi: 10.1080/15548627.2015.1108507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang J, Liu Y, Li X-H, et al. Curcumin protects neuronal cells against status-epilepticus-induced hippocampal damage through induction of autophagy and inhibition of necroptosis. Can J Physiol Pharmacol. 2017;95(5):501–509. doi: 10.1139/cjpp-2016-0154 [DOI] [PubMed] [Google Scholar]