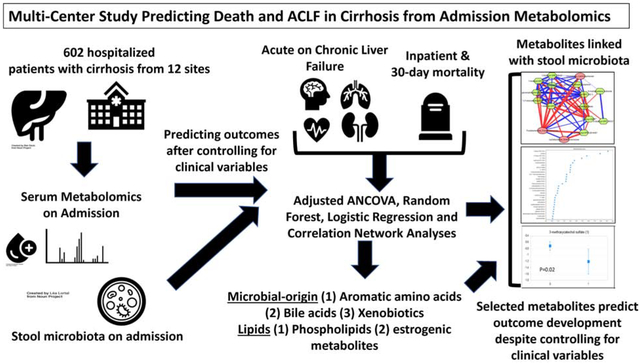
Abstract
Background & Aims:
Inpatients with cirrhosis have high rates of acute on chronic failure (ACLF) development and high mortality within 30 days of admission to the hospital. Better biomarkers are needed to predict these outcomes. We performed metabolomic analyses of serum samples from patients with cirrhosis at multiple centers to determine whether metabolite profiles might identify patients at high risk for ACLF and death.
Methods:
We performed metabolomic analyses, using liquid chromatography, of serum samples collected at time of admission to 12 North American tertiary hepatology centers from 602 patients in the NACSELD consortium sites from 2015 through 2017 (mean age, 56 years; 61% men; mean model for end-stage liver disease score, 19.5). We performed analysis of covariance, adjusted for model for end-stage liver disease at time of hospital admission, serum levels of albumin and sodium, and white blood cell count, to identify metabolites that differed between patients who did vs did not develop ACLF and patients who did vs did not die during hospitalization and within 30 days. We performed random forest analysis to identify specific metabolite(s) that were associated with outcomes and area under the curve (AUC) analyses to analyze them in context of clinical parameters. We analyzed microbiomes of stool samples collected from 133 patients collected at the same time and examined associations with serum metabolites.
Results:
Of the 602 patients analyzed, 88 developed ACLF (15%), 43 died in the hospital (7%), and 72 died within 30 days (12%). Increased levels of compounds of microbial origin (aromatic compounds, secondary or sulfated bile acids, and benzoate) and estrogen metabolites, as well as decreased levels of phospholipids, were associated with development of ACLF, inpatient, and 30-day mortality and were also associated with fecal microbiomes. Random forest analysis and logistic regression showed that levels of specific microbially produced metabolites identified patients who developed ACLF with an AUC of 0.84 (95% CI, 0.78–0.88; P=.001), patients who died while in the hospital with an AUC of 0.81 (95% CI, 0.74–0.85, P=.002), and patients who died within 30 days with an AUC of 0.77 (95% CI, 0.73–0.81; P=.02). The metabolites were significantly additive to clinical parameters for predicting these outcomes. Metabolites associated with outcomes were also correlated with microbiomes of stool samples.
Conclusions:
In an analysis of serum metabolites and fecal microbiomes of patients hospitalized with cirrhosis at multiple centers, we associated metabolites of microbial origin and lipid moieties with development of ACLF and death as an inpatient or within 30 days, after controlling for clinical features.
Keywords: tryptophan, estrone, sepsis
Graphical Abstract
LAY SUMMARY
Hospitalized patients with cirrhosis often develop poor outcomes. We found that gut microbes and their circulating products collected on admission can predict death and acute-on-chronic liver failure.
INTRODUCTION:
Cirrhosis is the end stage of any chronic liver disease and is a leading cause of death worldwide1, 2. Mortality from cirrhosis is linked with hospitalizations and the development of organ failures3, that define Acute-on-Chronic Liver Failure (ACLF), an entity with a high 30-day mortality that can occur in 20–45% of hospitalized patients with cirrhosis4. ACLF poses a major healthcare burden and its prevalence is steadily increasing5. However, current definitions of ACLF rely on the identification of organ failures and its severity is dependent on the number of organ failures6, 7. These definitions have been bolstered using systemic inflammatory markers and metabolomics, which demonstrate energy failure and distinguish ACLF from decompensated cirrhosis8, 9. However, there are no recognized parameters that will predict the development of ACLF in hospitalized patients.
Promising biomarkers in this regard are microbiota and their functional alterations tested through untargeted and targeted metabolomics10, 11. Single and multiple-center studies have demonstrated that alterations of stool microbiota on admission, can independently predict ACLF development and mortality12–14. These microbial changes were more advanced on admission in those who ultimately had negative outcomes and were much more altered in patients with renal injury12, 15. However, the relative numbers are limited and functional aspects of microbiota analysis, through which their impact is exerted, need to be explored. Metabolomics has been used in prior studies to predict outcomes and provide insights into pathophysiology of disease processes in patients with cirrhosis and can have a human, microbial or a shared human-microbial source16–21. Prominent metabolites in tryptophan and phenylalanine metabolism, bile acid metabolism, lyso-phospholipid, benzoate, and choline metabolism have a predominant microbial origin22. Therefore, a multi-center approach to determining the role of metabolomics, with a focus on microbial metabolism, in predicting organ failures, ACLF and in-hospital mortality is required.
Our aim was to evaluate admission serum metabolomics in the multi-center NACSELD (North American Consortium for the Study of End-Stage Liver Disease) cohort of hospitalized patients with cirrhosis in order to determine whether they can predict inpatient outcomes independent of clinical variables.
METHODS
The NACSELD consortium consists of 14 North American tertiary hepatology centers that have collected prospective data from patients with cirrhosis hospitalized non-electively, without HIV infection or prior organ transplants. Data was collected after informed consent and entered in a REDCAP database. The study was approved by institutional review boards at all sites and all participants gave informed consent. For this study, we only included the subset that was (a) admitted without organ failures and (b) who consented to providing serum samples within 12 hours of admission. Patients with pre-existing ACLF on admission or those who were unable or unwilling to provide samples were excluded from this sub-study. All sites were instructed on uniform sample collection practices before study initiation and samples were stored in −80 degrees centigrade freezers until the time for analysis.
Data pertaining to demographics, cirrhosis details, medications, reasons for admission and hospital course were recorded. Organ failures were defined according to the NACSELD-ACLF criteria23, 24. These criteria are 2 of the following: brain failure [grade III/IV hepatic encephalopathy (HE)], renal failure (patients on renal replacement therapy), circulatory failure (shock and requirement for vasopressors) and respiratory failure (BiPAP/mechanical ventilation). Outcomes determined were the development of ACLF, in-hospital mortality and 30-day mortality.
Analyses were performed at Metabolon Inc. using validated Ultrahigh Performance Liquid Chromatography-Tandem Mass Spectroscopy. Data analyzed pertains to all metabolites including amino acids, carbohydrates, lipids, nucleotides, microbiota metabolism, energy, cofactors & vitamins, xenobiotics, and novel metabolites (supplementary data). To account for variability related to patient-level variables, ANCOVA analyses were performed adjusting for age, gender, alcohol-related etiology, admission values of MELD, WBC, serum sodium and serum albumin using false discovery rate (FDR) adjustment, represented by the q-value generated by the method of Storey and Tibshirani. Following log transformation and imputation of missing values, if any, with the minimum observed value for each compound, ANOVA contrasts and Welch’s two-sample t-test were used to identify biochemicals that differed significantly between groups. Finally, ANCOVA was performed. An estimate of the FDR was calculated to consider the multiple comparisons that normally occur in metabolomic-based studies11. Instrument variability was determined by calculating the median relative standard deviation (RSD) for the internal standards that were added to each sample prior to injection into the mass spectrometers. Overall process variability was determined by calculating the median RSD for all endogenous metabolites (i.e., non-instrument standards) present in 100% of the Client Matrix samples, which are technical replicates of pooled client samples. Overall process variability was determined by calculating the median RSD for all endogenous metabolites (i.e., non-instrument standards) present in the technical replicates.
Metabolites that were independently associated with the outcomes of interest (ACLF, inpatient mortality and 30-day mortality) and infected patients on ANCOVA were considered predictive of such outcomes. The ANCOVA tables were ranked according to p-values, false discovery rates and pathways found to be consistently involved in protection from or associated with the outcomes were then explored deeper for each outcome.
Then a random forest analysis (RFA) was performed, which is a supervised classification technique based on an ensemble of decision trees25. For a given decision tree, a random subset of the data with identifying true class information is selected to build the tree without replacement and sample the same number from each group. The in-bag samples are different for each tree. Then after the forest is constructed, the predictions are made for the out-of-bag (OOB) samples for each tree. For each tree, only a subset of variables is considered as determined by the mtry parameter (“bootstrap sample” or “training set”). The final classification of each sample is determined by computing the class prediction frequency (“votes”) for the OOB samples over the whole forest. This method is unbiased since the prediction for each sample is based on trees built from a subset of samples that do not include that sample. To determine which metabolites, make the largest contribution to the classification, a “variable importance” measure called the “Mean Decrease Accuracy” (MDA) was computed. The MDA is determined by randomly permuting a variable, running the observed values through the trees, and then reassessing the prediction accuracy. If a variable is not important, then this procedure will have little change in the accuracy of the class prediction (permuting random noise will give random noise). By contrast, if a variable is important to the classification, the prediction accuracy will drop after such a permutation, which we record as the MDA. Thus, the random forest analysis provides an “importance” rank ordering of metabolites and the first 30 for each outcome are displayed. AUCs were calculated for the ANCOVA-adjusted models for each category.
Then, in order to determine the specific metabolites that were relevant, we performed a backwards elimination binary logistic regression analysis was performed to develop a baseline model for each of the three outcomes (ACLF, in-patient and 30-day mortality). The base model variables considered were: admission Albumin, MELD, Na, and WBC and patient age, gender, and alcohol-related etiology, which have affected these outcomes in prior NACSELD studies24. The predictive ability of the final base model was assessed using the area under the curve (AUC) for the receiver operator characteristic curve. Next, for each of the outcomes, a Lasso regression (penalized) was utilized on thirty-six metabolites that were found to be significant on the ANCOVA-adjusted analyses to find those that were most predictive of the outcome. Then, microbially-derived metabolites identified as having significant association with the outcome were added to the base model and then a backwards elimination procedure was again performed focusing on the metabolite variables until all metabolites were significant at the p = 0.05 level of significance when assessed by the likelihood ratio Chi-Square test. The predictive ability of the base model + metabolites was assessed using the AUC and the difference between the AUC of the Base Model and the Base+Metabolite model was assessed using the DeLong test26 for comparing the AUCs under two correlated ROC curves.
Finally, we correlated the serum metabolites that were significant on ANCOVA with stool microbiota analyzed using 16srRNA sequencing collected in the subset of patients who gave both samples on admission using published techniques in R27. Microbial taxa linked with metabolites that were significant at r=>0.7/−0.7 and p<0.001 were visualized in Cytoscape between patients who developed outcomes versus those who did not. Microbiota collection, processing and composition findings have already been published28.
RESULTS:
Study participants:
We enrolled 602 participants with cirrhosis from 12 sites (Table S1). The mean age was 56.0±9.6 years, with 367 (61%) men. The MELD on admission was 19.5±7.5, serum sodium 133.8±6.0 mEq/L, admission WBC count 7.9±4.9 /ml and admission albumin was 2.8±0.7 g/dl. The major etiology was alcohol-related (n=178) followed by HCV only (n=155), NASH (n=105), HCV+alcohol (n=94) and others (n=69). Of the 178 patients with alcohol-related cirrhosis, only 15 were actively drinking among whom 10 had alcohol associated hepatitis. 54 patients were on SBP prophylaxis with ciprofloxacin and 265 patients were on rifaximin for HE on admission. None were on urso-deoxycholic acid (UDCA). On admission, infections were found in 237 (39%), of which SBP was the most common (n=83), followed by UTI (n=73), spontaneous bacteremia (n=38), skin/soft-tissue (n=31) and others. Of the remaining 365 patients, 89 were admitted for HE, 65 for GI bleeding, 61 for renal dysfunction, 57 for anasarca, 21 for electrolyte-related and 72 had liver-unrelated admissions. 133 subjects had also provided stool.
Clinical Outcomes:
144 patients (24%) required intensive care unit (ICU) transfer related to organ failures. Brain failure was the most common (n=137), then renal (n=81), respiratory (n=74) and circulatory failure (n=71). Eighty-eight patients developed ACLF, 43 patients died during the hospitalization while a total of 72 patients died within 30 days. A significantly higher number of patients with ACLF died during the inpatient (79% vs 10%, p<0.0001) and 30-day (60% vs 8%, p<0.0001) period. Most patients who died as an inpatient (35 vs 8, p<0.0001 had ACLF; the 8 who died as inpatients without ACLF died as an inpatient due to cirrhosis-unrelated reasons or procedural complications. Rifaximin or SBP prophylaxis on admission did not significantly impact outcomes.
Metabolomics:
Metabolite overview:
1464 metabolites were identified, of which 268 were unknown. Most of the named metabolites belonged to lipids (n=459) followed by xenobiotics (n=329) and amino acids (n=223). Remaining were nucleotides (n=43), peptides (n=40), cofactors and vitamins (n=36), partially characterized metabolites (n=27) and carbohydrates (n=26).
1060 metabolites correlated with MELD score with p-values and q-values (FDR rate) <0.05. The 30 most positive and negative correlations with MELD score are shown in Table S2. The majority positively linked to MELD score were related to bilirubin and aromatic amino acid metabolites while those negatively linked to MELD score were related to lipid metabolism. Table 2 shows microbially-derived metabolites and their ANCOVA-adjusted direction of change for all outcomes and infected patients. Tables S3–6 show all ANCOVA adjusted metabolites that are p<0.05 and q<0.05, while Table S10 shows these grouped according to metabolic pathways with significance indicated by red (higher) and blue (lower) in those who developed the three outcomes.
Table 2:
Microbially derived metabolites that were significantly different between patients who developed outcomes
| ACLF vs no-ACLF | Infection or no | In-patient death vs. no | 30-day death vs no | |
|---|---|---|---|---|
| Choline metabolism | Choline↓ Betaine↓ Tri-methyl amine oxide↓ |
Tri-methyl amine oxide↓ | - | - |
| Bile acid | glycocholenate sulfate↑ taurocholenate sulfate↑ ursodeoxycholate↓ 7-ketodeoxycholate↓ |
Isoursodeoxycholic acid↓ ursodeoxycholate↓ deoxycholic acid glucuronide↑ |
glycochenodeoxycholate glucuronide↓ glycochenodeoxycholate 3-sulfate↓ 7-ketodeoxycholate↓ glycoursodeoxycholate↓ glycohyocholate↓ taurohyocholate↓ hyocholate↓ glycocholenate sulfate↑ taurocholenate sulfate↑ |
glycocholenate sulfate↑ taurocholenate sulfate↑ 7-ketodeoxycholate↓ |
| Aromatic amino acid metabolism | 3-indoxyl sulfate↓ 3-(3-hydroxyphenyl) propionate↓ 3-(4-hydroxyphenyl) propionate↓ 5-hydroxy indoleacetate↑ indolepropionate↓ phenylacetylglutamine↓ 3-methoxycatechol sulfate↓ N-acetyltryptophan↑ Tyramine sulfate↓ |
3-indoxyl sulfate↓ indolepropionate↓ serotonin↓ |
3-methoxycatechol sulfate↓ 4-hydroxy phenylacetate↑ phenol sulfate↑ phenol glucuronide↑ |
3-methoxycatechol sulfate↓ 5-hydroxy indoleacetate↑ phenyllactate↑ hydroxyphenylacetate↑ 4-hydroxy phenylacetate↑ phenylacetylglutamine↑ phenol sulfate↑ indolelactatet phenol glucuronide↑ |
| Xenobiotic | 3-phenylpropionate ↓ 3-(4-hydroxyphenyl) propionate↓ p-cresol sulfate↓ p-cresol glucuronide↓ o-cresol sulfate↑ methyl-4-hydroxybenzoate↑ |
3-phenylpropionate ↓ 3-(4-hydroxy phenyl)propionate↓ p-cresol sulfate↓ p-cresol glucuronide↓ methyl-4-hydroxybenzoate↑ |
daidzein sulfate ↑ methyl-4-hydroxybenzoate↑ 4-hydroxyhippurate↑ 4-hydroxybenzoate↑ genistein sulfate↑ |
3-phenylpropionate ↓ 3-(4-hydroxyphenyl) propionate↓ 4-hydroxyhippurate↑ 4-hydroxybenzoate↑ daidzein sulfate ↑ genistein sulfate↑ methyl-4-hydroxy benzoate↑ hippurate↑ |
| Polyamines | Spermidine↓ | |||
| Lipid metabolism | Lyso-Phosphocholine↓ Lyso-Phospho ethanolamine↓ |
Lyso-Phosphocholine↓ Lyso-Phospho ethanolamine↓ |
Lyso-Phosphocholine↓ Lyso-Phospho ethanolamine↓ |
Lyso-Phosphocholine↓ Lyso-Phospho ethanolamine↓ |
Fold-change compared to no-outcomes are presented with arrows: ↓: Lower in those who developed outcome vs those who did not, ↑: Higher in those who developed outcome vs not. All data are ANCOVA adjusted for age, gender, alcohol etiology, admission MELD, albumin and sodium. Details of all other metabolites are in supplementary table 4. ACLF: acute on chronic liver failure
ACLF development:
Clinical variables:
Patients who developed ACLF were younger, more likely with alcohol-related and more advanced cirrhosis and admitted with infection compared to the rest. Patients with ACLF had a longer LOS and experienced higher rates of ICU transfer, inpatient and 30-day mortality.
ANCOVA and FDR-adjusted analysis:
The 117 metabolites associated with ACLF development with the lowest p- and significant q-values were selected. Of these 47 were lipids, 19 were amino acids and xenobiotics respectively, while 32 were not characterized. The top 30 named metabolites are shown in table 2, which demonstrate relatively higher aromatic amino acids, and lower phospholipids in those who developed ACLF compared to those who did not. Several microbially-related metabolites were also involved shown in figure 1 and supplementary figure 1. Prominent among these were metabolites related to tryptophan, bile acid and benzoate metabolism. Other metabolites of amino acids, such as arginine metabolites including asymmetric and symmetric dimethylarginine (ADMA and SDMA) were higher in those who developed ACLF. Some lipid moieties were largely lower in those who developed ACLF: specifically, lower choline, plasmalogens, phospholipids and glyceryl moieties and their degradation products hexosylceramides. Choline products betaine and TMAO were also lower in those who developed ACLF. No changes in long or medium chain saturated/unsaturated fatty acids were identified. In contrast, dicarboxylic fatty acids, and sex steroids that were glucuronidated and sulfated in the liver and their estrogenic metabolite, estrone-3-sulfate were higher in patients who developed ACLF. The uremic toxin 3-carboxy-4-methyl-5-propyl-2-furanpropanoate was also higher in patients who developed ACLF. In patients with infection on admission, there was also a lower TMAO but higher spermidine and again lower phospholipids. Importantly estrone-3-sulfate levels did not differentiate between groups with/without infection.
Figure 1: ACLF-Related Serum Metabolites.
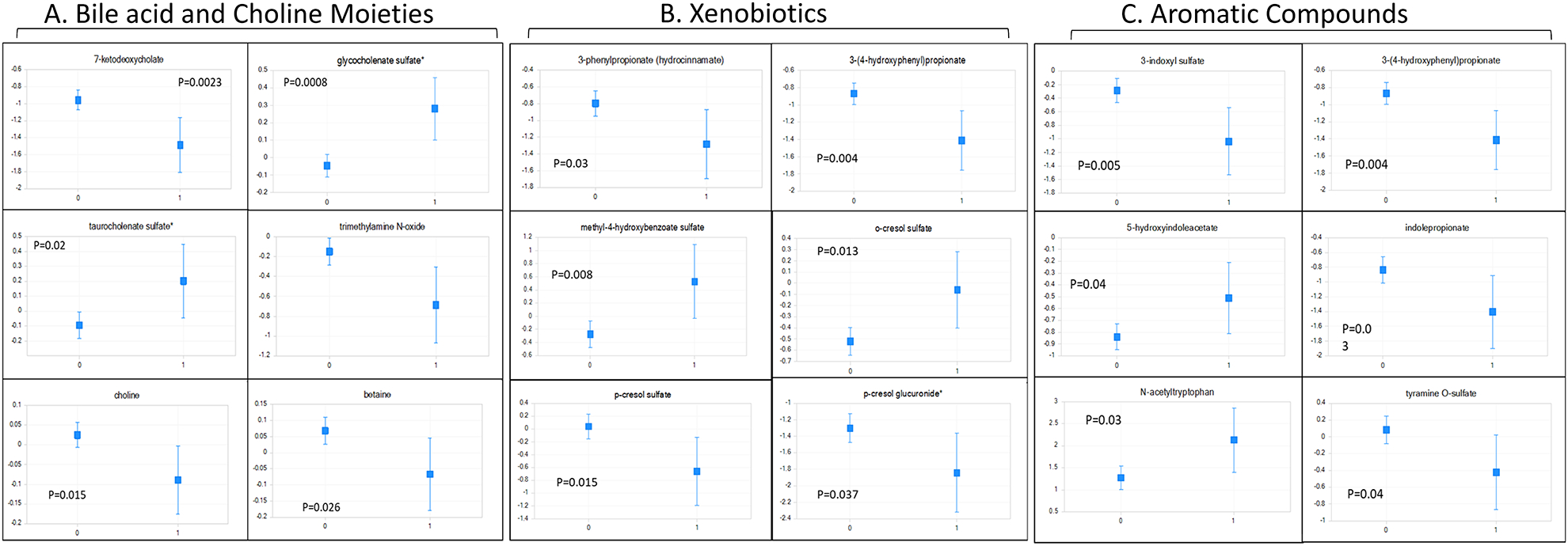
Least square means of ANCOVA-corrected microbial-origin metabolites presented as mean and 95% CI between patients who did (marked as 1) and did not develop ACLF (marked as 0). A: Bile acids and Choline moieties, B: Aromatic compounds and C: Xenobiotics. Y axis: scaled intensity. FDR-corrected p values are shown for each metabolite.
Random forest analysis:
ANCOVA adjusted metabolites were used to determine outcomes with the OOB rate 0.30, meaning that the ANCOVA model predicted the development at 0.70 level (Range 0–1). The top 30 metabolites in order of decreasing MDA are shown in figure 4A, the majority of which belonged to phospholipids, xenobiotics, estrone-3-sulfate and bacterial metabolites of aromatic amino acid metabolism.
Figure 4: Mean Decrease Accuracy on Random Forest Analysis.
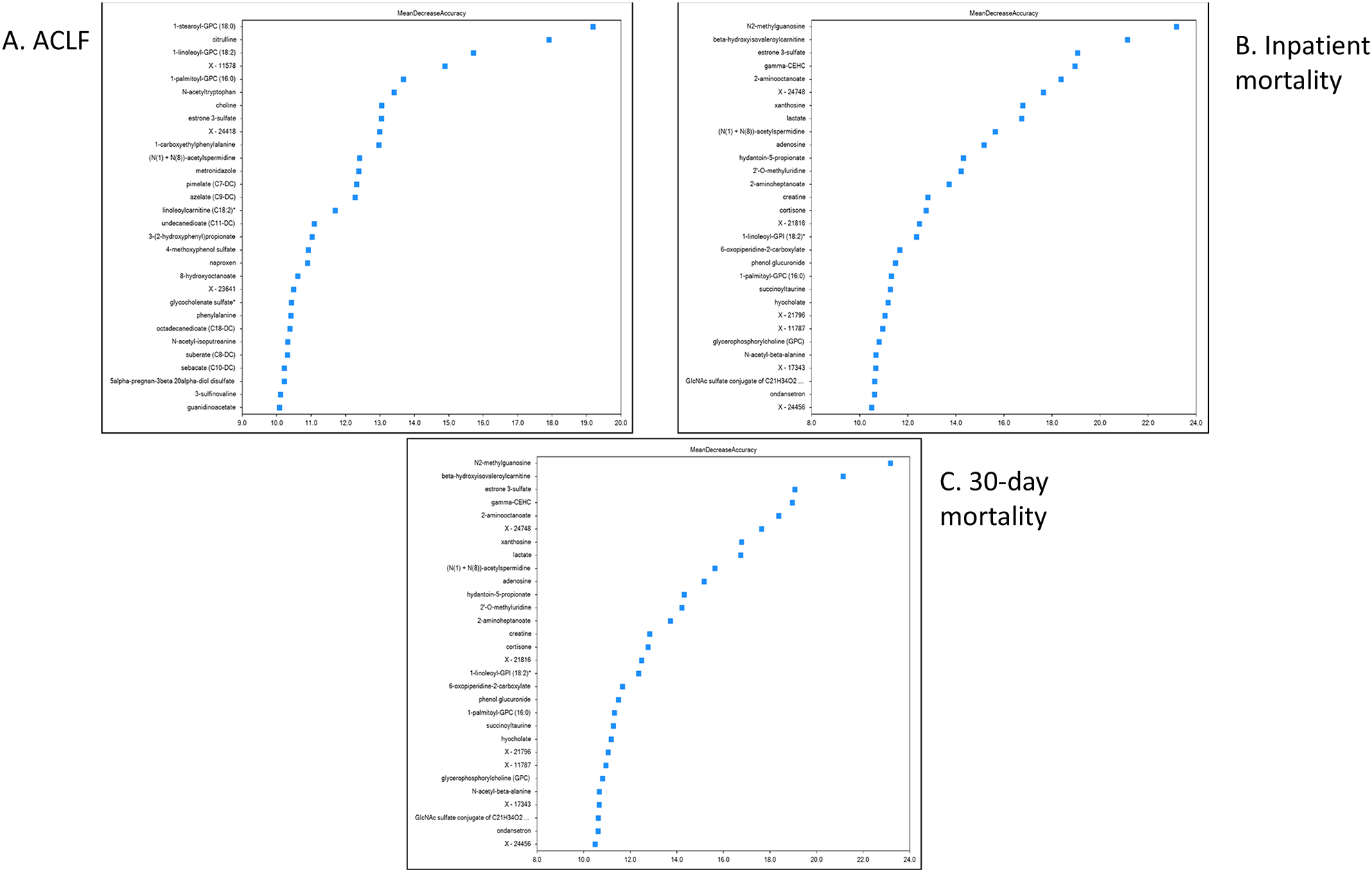
Mean Decrease Accuracy of Top 30 Metabolites in the Random Forest Analysis. Metabolites that caused the greatest change in the random forest analysis are positioned from top to bottom with percentage change in the X axis.
4A: Mean decrease accuracy in those who developed ACLF
4B: Mean decrease accuracy in those who developed inpatient death
4C: Mean decrease accuracy in those who died within 30 days
In-hospital mortality:
Clinical variables:
Age, gender, and etiology of cirrhosis were similar between patients who died during hospitalization (n=43) compared to those who survived hospitalization (N=559). However, cirrhosis severity on admission (MELD, WBC, albumin and Na) was worse in those who died during the hospitalization. Patients who died during hospitalization were also more likely to be infected, have higher LOS, have higher likelihood of ICU transfer and to develop ACLF during the admission.
ANCOVA and FDR-adjusted analysis:
119 metabolites were significantly associated with inpatient mortality after FDR adjustment. Of these 65 were lipids, 18 were amino acids and 14 were xenobiotics. These again demonstrated that aromatic amino acid metabolites were higher while lipids were lower in those who died compared to survivors. Again, tryptophan, bile acid, ADMA/SDMA and lipid moieties including estrone-3-sulfate were different between groups.
Random forest analysis:
OOB rates were 0.27. Figure 4B shows the 30 metabolites with highest MDA. These were also related to the microbial products of amino acids, bile acids, lipid metabolites with cortisone and estrone sulfate and markers of energy failure such as lactate.
30-day mortality:
Clinical variables:
Patients who died within 30 days had similar demographics and etiology distribution but had a higher MELD, lower serum albumin and Na and higher admission WBC count compared to those who survived. These patients were more likely to have had infections on admission, ICU transfer, longer LOS and ACLF development.
ANCOVA and FDR-adjusted analysis:
427 metabolites were significant after FDR adjustment, of which 178 were lipids, 106 were amino acids, 57 were xenobiotics and 86 were uncharacterized. As found in other outcomes, metabolites related to aromatic amino acids, ADMA/SDMA, bile acids and steroids were associated with 30-day mortality.
Random forest analysis:
OOB rates were 0.26. The top 30 metabolites ranked by MDA are found in Figure 4C which were related to lipid metabolism, cortisone hormones, amino acids including their products like dimethylarginine, bilirubin and lipid metabolites.
Logistic regression:
The baseline clinical model for ACLF showed AUC of 0.78 (0.72–0.83) which significantly increased to 0.84 (0.78–0.88, p=0.001) with 3-pheynlpropinoate,4-hydroxybenzoate, 5-hydroxyindoleacetate, Choline, Glycohyocholate, o-cresol sulfate and Trimethylamine N-oxide (Table S7). The clinical model for inpatient mortality showed an AUC of 0.73 (0.66–0.80), which was significantly enhanced to 0.81 (0.74–0.85, p=0.002) with 3-methoxycatechol sulfate, 4-hydroxybenzoate, Choline, Glycohyocholate and o-cresol sulate (Table S8). For 30-day mortality, the AUC for the clinical model was 0.73 (0.69–0.78), which increased to AUC 0.77(0.73–0.81, p=0.02) with Daidzein sulfate, Methyl-4-hydroxybenzoate sulfate, N-acetyltryptophan, p-cresol glucuronide and phenol sulfate being independently contributory (Table S9).
Microbiota correlation network changes:
We only found significant correlations in groups with outcomes at the cut-off. As shown in figure 5, Clostridium cluster XI, Enterococcaceae and Fusobacteriaceae were linked with compounds and higher in those who developed these outcomes while autochthonous taxa such as Lachnospiraceae and Ruminococcaceae were the reverse.
Figure 5: Correlation networks between metabolites and stool mtcrobiota.
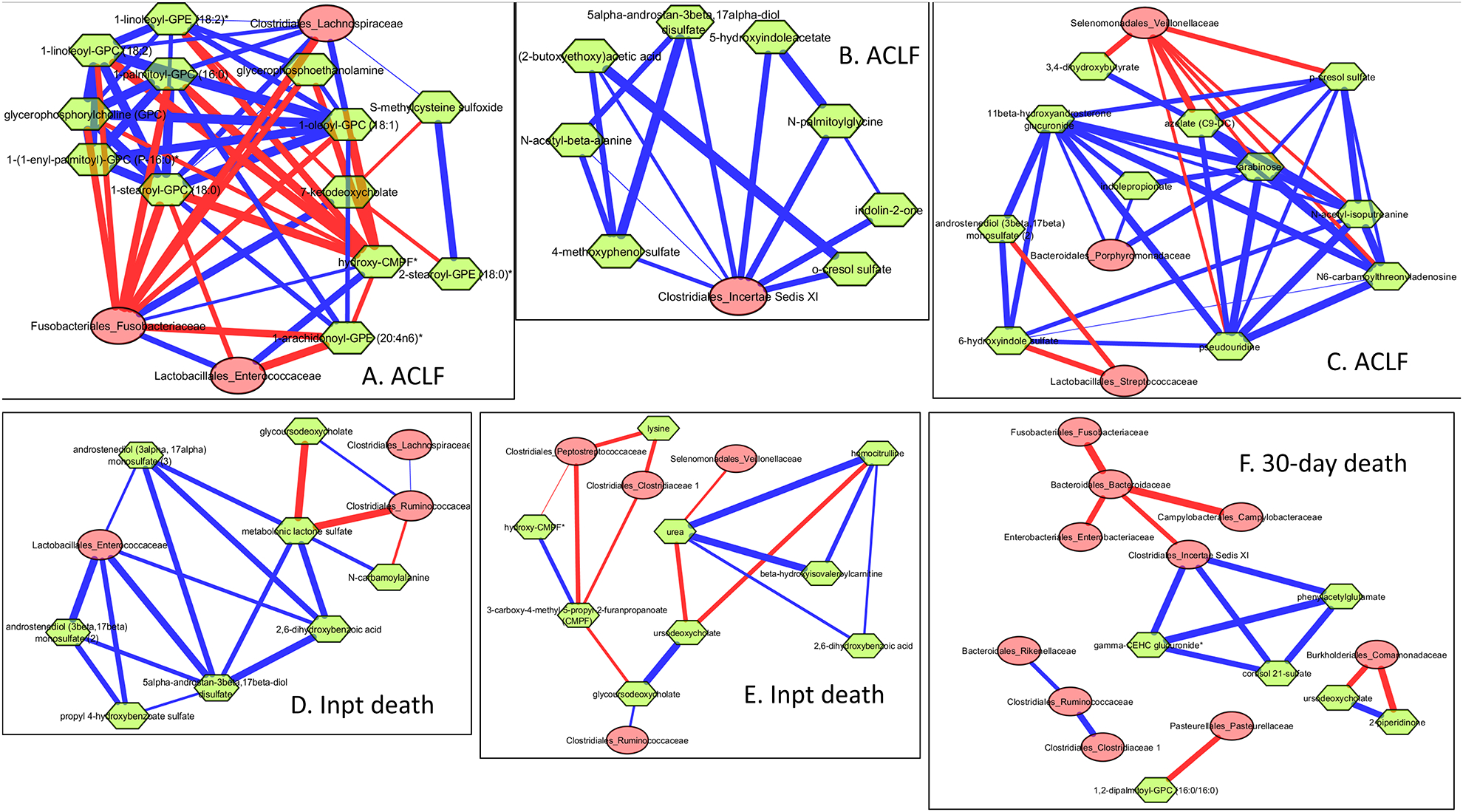
Correlation networks in patients reaching outcomes filtered at r>0.7/r<−0.7 and p<0.001 between microbiota (red nodes) and metabolites (green nodes). Red edges indicate negative while blue indicate positive correlation.
5A-C: Sub-networks of important taxa in those who developed ACLF
5A: Fusobacteriaceae and Enterococcaceae showed positive linkages with CMPF and negative with phospholipids, while the reverse was seen with Lachnospiraceae
5B: Clostridium cluster XI showed positive linkages with aromatic compounds
5C. Veillonellaceae, Porphyromonadaceae linked to compounds protective against ACLF
5D-E: Sub-networks of important taxa in those who died as an inpatient.
5D: Enterococcaceae associated with sex steroids and benzoate compounds associated with death, while Ruminococcaceae and Lachnospiraceaeae are positively linked with glycoursodeoxycholate, which is protective.
5E: Clostridiaceae and Peptostreptococcaceae are negatively linked with CMPF/hydoxy-CMPF, which in turn are negatively linked with protective glycoursodeoxycholate
5F: 30-day death sub-network showed Clostridium cluster XI linked positively with phenylacetylglutamine, which is associated with death and negatively with commensal Bacteroidaceae, which in turn are negatively correlated with pathobionts. Pasteurellaceae are negatively linked with phospholipids and Commamondaceae negatively with protective ursodeoxycholate.
DISCUSSION:
Patients with cirrhosis are prone to poor inpatient outcomes such as the development of ACLF or mortality. Prediction of these outcomes in hospitalized patients with cirrhosis is not only critical for adequate prognostication and, to judge futility but, importantly, to determine a pathogenic link that would identify possible prevention strategies. In addition, since most accepted definitions of ACLF require the presence of major organ failures, novel means of identifying patients who will develop ACLF before organ failure development are urgently needed. The current multi-center metabolomic experience demonstrates a serum signature on the day of hospital admission in patients with cirrhosis who will subsequently develop ACLF or mortality centered on microbially-derived metabolites and xenobiotics.
ACLF marks a sentinel event in the natural history of cirrhosis and can lead to high mortality unless liver transplant can be performed. The definitions of ACLF vary worldwide but by and large focus on organ failures late in the disease process, especially the NACSELD definition29. Prediction of ACLF development, which could potentially identify a high-risk subgroup before advanced organ failure has set in, is important to test and later implement potential preventative strategies. Prior studies have focused on using specialized markers, including metabolomics, systemic inflammation and kyunerine to differentiate ACLF from acute decompensation8, 30, 31. These results clearly demonstrate that there is a bio-energetics failure coupled with systemic inflammatory response in ACLF patients compared to those with decompensated cirrhosis. However, the prediction of who will develop ACLF is an open question with markedly important consequences.
Single center studies from USA and China have shown that stool microbial composition on admission can predict inpatient death and organ failures13, 14. These results demonstrated higher gram-negative taxa and lower autochthonous beneficial taxa belonging to Lachnospiraceae, Ruminococcaceaeae, and Clostridia. This was further extended by a multi-center study with stool microbiota collected on admission28. The results demonstrated the admission stool microbiota composition can independently predict the development of ACLF, organ failures and mortality. Stool microbiota composition in those who developed ACLF and individual organ failures was characterized by higher relative abundance of gram-negative bacteria belonging to Proteobacteria. When 30-day mortality was considered, there were also higher Enterococcaceae in addition to Proteobacteria. Correlation networks found that metabolites associated with negative outcomes were more likely linked with pathobionts such as Clostridium Cluster XI, which includes C.difficile, Enterococcaceae, Fusobacteriaceae and Commanomadaceae, while the reverse was found with autochthonous, beneficial taxa such as Lachnospiraceae and Ruminococcaceae32. While we were able to determine the linkage with composition in a subset, these metabolites represent a major means of human-microbial interactions10, 33, 34.
These microbially-derived metabolites can influence several important metabolic pathways that are critical for cell homeostasis, optimal organ function and circulation and are related to bile acid, xenobiotic and aromatic amino acid metabolism35. Overall, we found that serum metabolites related to microbial metabolism such as bile acids (lower conjugated, secondary, and sulfated bile acids), aromatic amino acid metabolites (lower indoxyl-sulfate and indole propionate and higher indoleacetate), xenobiotics (higher o-cresol and phenol and lower p-cresol sulfates and glucuronides), choline metabolism (lower betaine and TMAO) and those linked with lipid metabolism (lower phospholipids and higher estrogenic metabolites) were associated with ACLF and death.
We found a consistent increase in serum tauro and glyco-cholenate in their sulfated form in patients who developed ACLF or died. Cholenate is formed as a result of the acidic or non-traditional pathway of bile acid synthesis from cholesterol in the liver36. Usually it is a minor part of the bile acid profile, but serum levels increase in patients with cholestasis37. Glycine and taurine conjugation and finally sulfation are performed to make these moieties more hydrophilic and able to be excreted38. Microbiota are involved in deconjugation of the glycine and taurine conjugates and desulfation39. Therefore, the relative increase in these moieties reflect cholestasis along with a potential reduction in the activity of gut microbiota belonging to Clostridium, Lactobacillus, Bacteroides and Fusobacterium that can de-sulfate and deconjugate these bile acids38, 39. This is interesting because these were significant despite adjusting for serum bilirubin levels in MELD score. UDCA, which was lower in those with outcomes, is produced by bacterial epimerization of the primary bile acid chenodeoxycholic acid and signifies the presence of specific beneficial Clostridium and Ruminococcus species40; none of the patients were on UDCA treatment. Similar beneficial species also can form 7-ketodeoxycholate or 7-oxo-deoxycholate and secondary bile acids and their relative reduction likely represents a bacterial functional alteration that predisposes patients to mortality, and ACLF41.
In addition to bile acids, several metabolites that are part of the aromatic amino acid, arginine and benzoate metabolism were significantly associated with the development of ACLF and death. The arginine metabolites ADMA and SDMA, which adversely impact vascular reactivity and brain function, were higher in patients who developed poor outcomes compared to those who did not42, 43. Tryptophan metabolites have the potential to engage the aryl-hydrocarbon (AhR) receptor and can cause several downstream positive effects on the systemic circulation, brain and liver34, 44. Indoxyl-sulfate and indole propionate were lower in those who developed ACLF. Indolepropionic acid is associated with stabilization of the intestinal barrier and its relative absence could potentially predispose to increased intestinal permeability, bacterial translocation and disease progression33. Indoxyl-sulfate and p-cresol sulfate and glucuronides are microbial-mammalian co-metabolites, which require conversion of aromatic amino acids by microbiota followed by sulfation in the liver45, 46. While some of these metabolites were higher in patients with chronic kidney disease, they also require intact liver function to be synthesized46. Therefore, their reduction in those who develop ACLF could reflect a combination of impaired liver sulfation and microbial dysbiosis. Other microbial metabolites of phenylalanine and tyrosine that engage the AhR receptor and promote IL-22 secretion and promote local immune efficacy were lower in those who developed ACLF, showing a relative immunodeficiency47, 48. We also extended prior studies of increased kyunerine and kyunerate, and quinolinate, which are endogenous metabolites of tryptophan that can be induced by systemic inflammation and macrophage activation, into this North American cohort in the context of other aromatic acid metabolites30, 49. 5-hydroxy indoleacetate was higher in those who developed ACLF and death. This is a stable breakdown product of the relatively unstable serotonin that can modulate microbial growth and intestinal motility, which is often impaired in cirrhosis47 and may be related to the dysbiosis and small intestinal bacterial overgrowth known to be more frequent in patients with more advanced liver disease.
In addition, to the microbial metabolites, lipids and hormonal biochemical entities were important prognosticators. Daidzein and genistein are isoflavones found in food that are modified by specific microbiota and sulfated in the liver. They have estrogenic properties and are linked with estrone and estrone-3-sulfate. In addition, the estrogenic activity of the other xenobiotic intermediate, 4-hydroxybenzoate, could be contributory50. The estrone-3-sulfate increase was accompanied by an increase in androgenic steroid precursors. Estrogen increase and conversion of androgens into estrogenic compounds are known to occur to a greater extent as liver disease progresses and could be harbingers of ACLF and death regardless of MELD score and other clinical variables adjusted for on ANCOVA. Therefore, the concomitant contribution of gut microbiota-associated generation of metabolites with estrogenic activity could also contribute towards the prediction of negative outcomes in patients with cirrhosis.
Another group of metabolites associated with a reduction in development of negative outcomes was related to phosphatidylethanolamines, phosphatidylserines, phosphatidylcholines and their lyso- conjugated biochemicals. These are associated with numerous bacterial and human metabolic pathways. Choline deficiency was associated with increased gram-negative taxa in prior studies51–53. Choline is metabolized by microbiota into methylamines that are oxidized by the liver to trimethylamine oxide, a product that indicates healthy liver function. In patients who develop ACLF, choline, betaine and TMAO were lower. Phosphatidylethanolamine, phosphatidylinositol and phosphatidycholine were lower in patients who developed all negative outcomes, including in infected patients. These findings extend prior single-center studies into this multi-center experience and in the context of ACLF in the Western world11, 44, 53. These molecules are essential components of the cell membrane, with PE being more focused on the inner leaflet and the mitochondrial membranes. Phosphatidylethanolamine and phosphatidycholine have been associated with protection in human liver disease in prior single center studies and our study extends that into a multi-center context54. We found that some changes were more likely in infected patients; lower spermidine, a polyamine that is utilized by intestinal microbiota, lower bile acid moieties and similar estrone-3-sulfate levels unlike other outcomes55. It is likely that lower bile acid and estrone-3-sulfate are associated with underlying liver disease rather than infection. As found in other studies of infected patients regardless of cirrhosis. There continued to be lower phospholipids in infected patients56.
We did not find significant changes in bioenergetics or long/medium-chain fatty acids in our study. Fatty acid changes and some features of altered bioenergetics are found in metabolomic studies of stable patients with cirrhosis before/after therapy and could have been obscured by the phospholipid release through cell membrane stress in inpatients in our study21, 57. Regarding bio-energetics, a recent study showed significant energy failure using blood metabolomics, however those patients had already developed ACLF, while ours were taken before that occurrence9. In addition, since several of the energy metabolites can be collinear with disease severity, our adjustment for clinical and demographic factors may have also reduced their relative significance in our cohort. While we determined the change in microbial functional metabolites, these were linked to microbial composition only in the subset of subjects who provided stool. However, the functional changes are more relevant in affecting changes in the host and indeed several metabolites that affected prognosis were related to the microbial taxa that are changed during admission in patients with cirrhosis58. ACLF can often lead to death, therefore, there was an overlap between the metabolites were found in all outcomes.
These results need validation in independent samples. A small minority had alcoholic hepatitis or were actively drinking so that group could not be tested separately. Potential translation of these findings could be creation of serum panels focused on these specific metabolites that could be drawn on admission to prognosticate patients and analyzed in hospital laboratories if validated. These findings can also form the basis of future clinical trials focused on favorably altering gut microbiota such as the use of prebiotics, probiotics and even potentially microbiota transplantation in hospitalized patients with cirrhosis.
We conclude that in this multi-center, prospective study of inpatients with cirrhosis, serum metabolomic profiles focused on gut microbial metabolites, hormonal and phospholipid moieties can predict the development of ACLF, inpatient and 30-day death. These predictions remain significant despite controlling for clinically significant variables and likely reflect the alteration in systemic milieu and hepatic functional capacity that is not fully captured by current clinical prognostic scales. In a subset of patients who also provided stool, these were linked to gut microbial composition. Specific microbially-derived metabolites are associated with a significantly higher AUC for ACLF, inpatient and 30-day mortality compared to purely clinical factors. Serum panels focused on metabolomics on admission in addition to our current clinical parameters have the potential to refine prognostication of inpatients with cirrhosis in order to develop pro-active rather than reactive strategies to prevent ACLF.
Supplementary Material
Figure 2: Inpatient Mortality-Related Serum Metabolites.
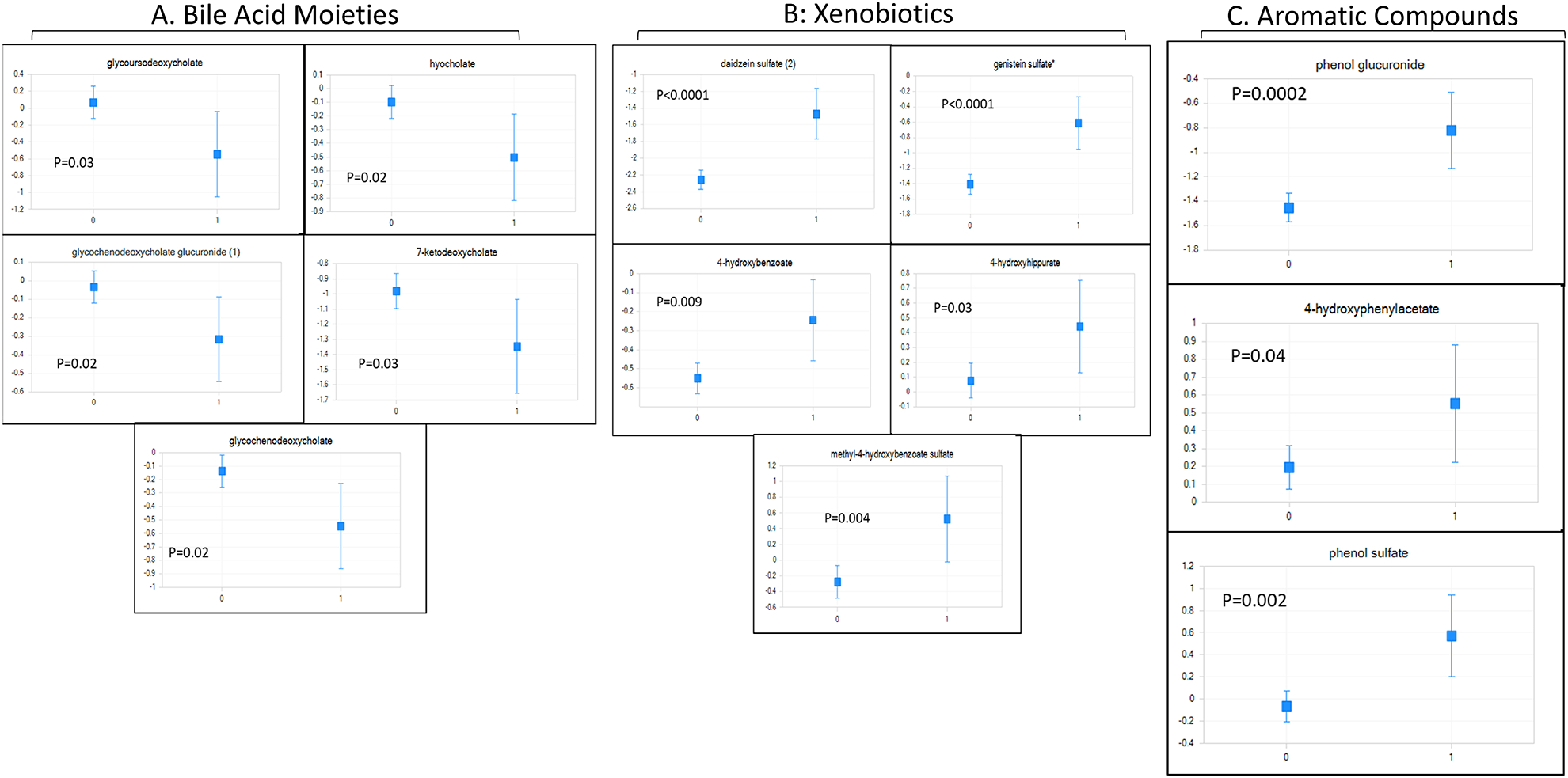
Least square means of ANCOVA-corrected microbial-origin metabolites presented as mean and 95% CI between patients who did (marked as 1) and did not develop inpatient mortality (marked as 0). Y axis: scaled intensity. A: Bile acids moieties, B: Aromatic compounds and C: Xenobiotics FDR-corrected p values are shown for each metabolite.
Figure 3: 30-day Mortality-Related Serum Metabolites.
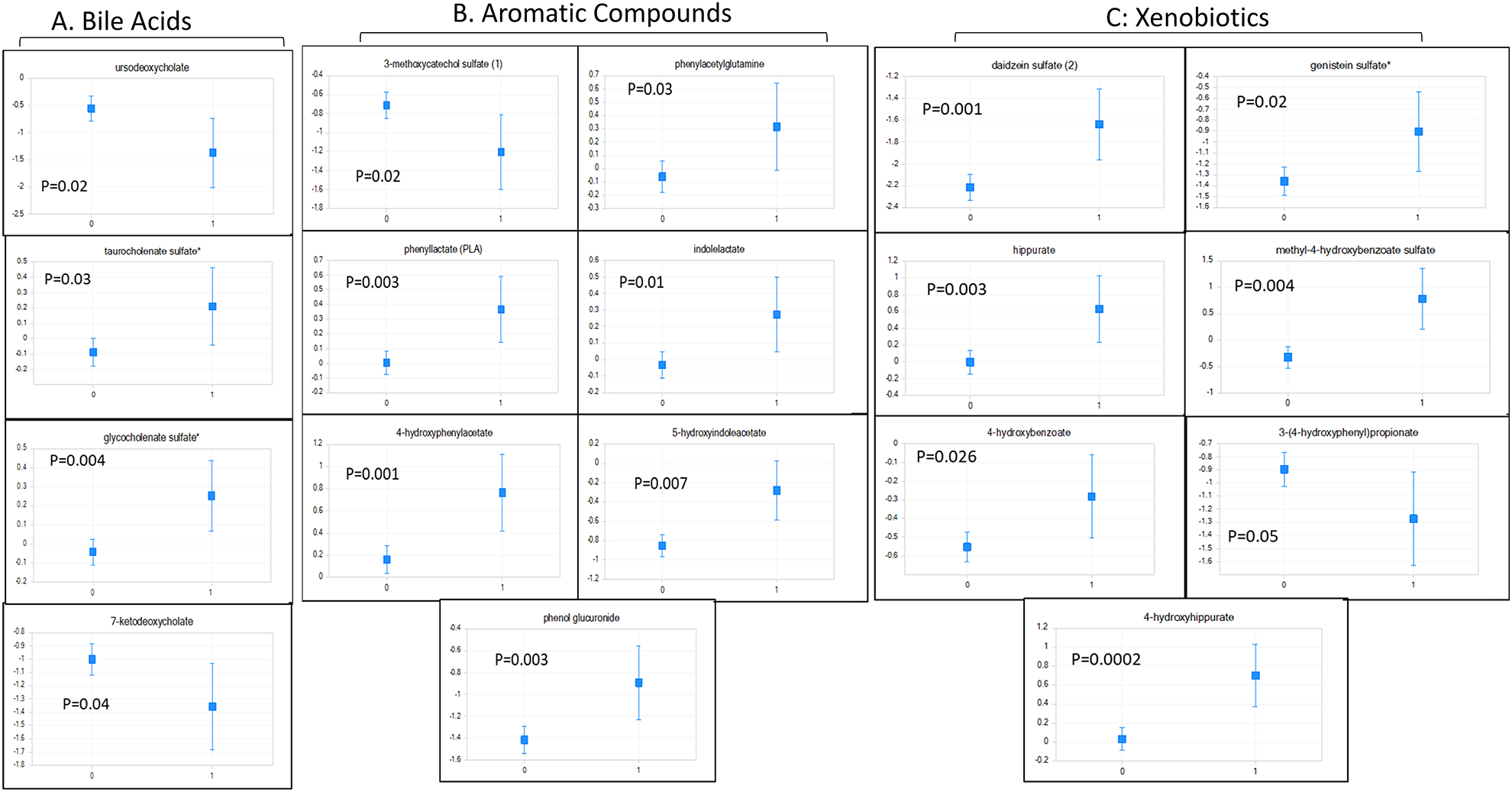
Least square means of ANCOVA-corrected microbial-origin metabolites presented as mean and 95% CI between patients who did (marked as 1) and did not develop 30-day mortality (marked as 0). Y axis: scaled intensity. A: Bile acids moieties, B: Aromatic compounds and C: Xenobiotics. FDR-corrected p values are shown for each metabolite.
Table 1:
Clinical comparisons between patients who developed ACLF, inpatient death and 30-day mortality
| Variable | Developed ACLF | Died in the hospital | Died within 30-days | ||||||
|---|---|---|---|---|---|---|---|---|---|
| No (n=514) | Yes (n=88) | P value | No (n=559) | Yes (n=43) | P value | No (n=530) | Yes (n=72) | P value | |
| Age (years) | 56.4±9.4 | 53.2±10.0 | 0.008 | 56.2±9.6 | 53.6±9.6 | 0.11 | 56.2±9.5 | 54.7±10.2 | 0.26 |
| Men | 317 (62%) | 45 (51%) | 0.18 | 337 (60%) | 24 (56%) | 0.95 | 319 (60%) | 46 (64%) | 0.37 |
| Etiology (see below) | 136/146/82/93/52 | 18/32/9/13/17 | 0.03 | 143/167/85/98/59 | 9/11/6/9/8 | 0.58 | 139/154/84/95/5 | 16/25/7/12/12 | 0.33 |
| MELD score | 18.3±7.3 | 25.8±7.4 | <0.0001 | 18.9±7.5 | 26.0±7.6 | <0.0001 | 18.6±7.4 | 25.9±7.3 | <0.0001 |
| Serum Na (meq/L) | 134.2±5.7 | 130.9±7.0 | <0.0001 | 134.0±5.9 | 130.9±6.7 | 0.007 | 13.42±5.8 | 130.8±6.8 | <0.0001 |
| Serum Albumin (g/dl) | 2.83±0.65 | 2.73±0.76 | 0.25 | 2.84±0.66 | 2.57±0.66 | 0.01 | 2.85±0.66 | 2.62±0.67 | 0.01 |
| WBC count (X103/ml | 7.5±4.7 | 10.1±5.6 | <0.0001 | 7.8±4.8 | 10.4±5.6 | 0.005 | 7.6±4.8 | 10.1 ±5.6 | 0.001 |
| Admission infection | 175 (34%) | 59 (67%) | <0.0001 | 205 (37%) | 30 (70%) | <0.0001 | 193 (36%) | 44 (61%) | <0.0001 |
| Admission rifaximin (N=265) | 39 (44%) | 226 (44%) | 0.92 | 251 (44%) | 14 (49%) | 0.12 | 237 (45%) | 28 (39%) | 0.34 |
| Admission SBP prophylaxis | 44 (9%) | 10 (12%) | 0.38 | 51 (9%) | 3 (7%22) | 0.79 | 49 (9%) | 5 (7%) | 0.52 |
| ICU transfer | 74 (14%) | 70 (80%) | <0.0001 | 107 (19%) | 34 (79%) | <0.0001 | 99 (19%) | 45 (62%) | <0.0001 |
| LOS | 10.2±35.9 | 26.9±27.8 | <0.0001 | 11.7±35.6 | 26.1 ±31.0 | 0.01 | 11.4±36.1 | 22.3±35.8 | 0.003 |
Data is presented as mean ± SD or in raw numbers (%). Comparisons were performed using unpaired t-tests or Mann-Whitney tests as appropriate. All laboratory values are on admission to hospital. Etiology categories are Hepatitis C/Alcohol/Both hepatitis C and alcohol/non-alcoholic fatty liver or cryptogenic/Other, ACLF: acute on chronic liver failure, ICU: intensive care unit.
WHAT YOU NEED TO KNOW.
Background and Context: Inpatients with cirrhosis have high rates of acute on chronic failure (ACLF) development and death within 30 days of admission to the hospital. Better biomarkers are needed to predict these outcomes
New Findings: In an analysis of serum metabolites and fecal microbiomes of patients hospitalized with cirrhosis at multiple centers, we associated metabolites of microbial origin and lipid moieties with development of ACLF and death as an inpatient or within 30 days, after controlling for clinical features
Limitations: Studies are needed to validate these findings.
Impact: Serum metabolite and fecal microbiome analyses might be used to determine prognoses of patients with cirrhosis and to identify mechanism of disease progression.
Acknowledgements:
We would like to thank Kari Wong and Matthew Mitchell from Metabolon Inc.
Grant Support: Supported by an investigator-initiated grant by Mallinckrodt Pharmaceuticals for metabolomics analysis.
NACSELD database and sample collection was supported by investigator-initiated grant from Grifols Pharmaceuticals and partly through VA Merit review 2I0CX001076.
Abbreviations:
- NACSELD
North American Consortium for the Study of End-Stage Liver Disease
- ACLF
Acute-on-Chronic Liver Failure
- HE
hepatic encephalopathy
- FDR
false discovery rate
- RFA
random forest analysis
- MDA
Mean decrease accuracy
- OOB
out of bag
- RSD
relative standard deviation
- ICU
intensive care unit
- ADMA
asymmetric dimethyl arginine
- SDMA
symmetric dimethyl arginine
- UDCA
ursodeoxycholic acid
- AUC
area under the curve
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: none for any author
References:
- 1.Kim WR, Brown RS Jr., Terrault NA, et al. Burden of liver disease in the United States: summary of a workshop. Hepatology 2002;36:227–42. [DOI] [PubMed] [Google Scholar]
- 2.Asrani SK, Devarbhavi H, Eaton J, et al. Burden of liver diseases in the world. J Hepatol 2019;70:151–171. [DOI] [PubMed] [Google Scholar]
- 3.Jalan R, Moreau R, Kamath PS, et al. Acute-on-Chronic Liver Failure: A Distinct Clinical Condition. Semin Liver Dis 2016;36:107–8. [DOI] [PubMed] [Google Scholar]
- 4.Bajaj JS, Moreau R, Kamath PS, et al. Acute-on-Chronic Liver Failure: Getting ready for prime-time. Hepatology 2018. [DOI] [PubMed] [Google Scholar]
- 5.Allen AM, Kim WR, Moriarty JP, et al. Time trends in the health care burden and mortality of acute on chronic liver failure in the United States. Hepatology 2016;64:2165–2172. [DOI] [PubMed] [Google Scholar]
- 6.Hernaez R, Sola E, Moreau R, et al. Acute-on-chronic liver failure: an update. Gut 2017;66:541–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bajaj JS, Moreau R, Kamath PS, et al. Acute-on-Chronic Liver Failure: Getting Ready for Prime Time? Hepatology 2018;68:1621–1632. [DOI] [PubMed] [Google Scholar]
- 8.Claria J, Stauber RE, Coenraad MJ, et al. Systemic inflammation in decompensated cirrhosis: Characterization and role in acute-on-chronic liver failure. Hepatology 2016;64:1249–64. [DOI] [PubMed] [Google Scholar]
- 9.Moreau R, Claria J, Aguilar F, et al. Blood metabolomics uncovers inflammation-associated mitochondrial dysfunction as a potential mechanism underlying ACLF. J Hepatol 2019. [DOI] [PubMed] [Google Scholar]
- 10.Bajaj JS, Khoruts A. Microbiota changes and Intestinal Microbiota Transplantation in Liver Diseases and Cirrhosis. J Hepatol 2020. [DOI] [PubMed] [Google Scholar]
- 11.McPhail MJW, Shawcross DL, Lewis MR, et al. Multivariate metabotyping of plasma predicts survival in patients with decompensated cirrhosis. J Hepatol 2016;64:1058–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bajaj JS, Vargas HE, Reddy KR, et al. Association Between Intestinal Microbiota Collected at Hospital Admission and Outcomes of Patients with Cirrhosis. Clin Gastroenterol Hepatol 2018. [DOI] [PubMed] [Google Scholar]
- 13.Chen Y, Guo J, Qian G, et al. Gut dysbiosis in acute-on-chronic liver failure and its predictive value for mortality. J Gastroenterol Hepatol 2015. [DOI] [PubMed] [Google Scholar]
- 14.Bajaj JS, Heuman DM, Hylemon PB, et al. Altered profile of human gut microbiome is associated with cirrhosis and its complications. J Hepatol 2014;60:940–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bajaj JS, Thacker LR, Fagan A, et al. Gut microbial RNA and DNA analysis predicts hospitalizations in cirrhosis. JCI Insight 2018;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams HR, Cox IJ, Walker DG, et al. Differences in gut microbial metabolism are responsible for reduced hippurate synthesis in Crohn’s disease. BMC Gastroenterol 2010;10:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kang DJ, Kakiyama G, Betrapally NS, et al. Rifaximin Exerts Beneficial Effects Independent of its Ability to Alter Microbiota Composition. Clin Transl Gastroenterol 2016;7:e187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bajaj JS, Gillevet PM, Patel NR, et al. A longitudinal systems biology analysis of lactulose withdrawal in hepatic encephalopathy. Metab Brain Dis 2012;27:205–15. [DOI] [PubMed] [Google Scholar]
- 19.Bajaj JS, Kakiyama G, Zhao D, et al. Continued Alcohol Misuse in Human Cirrhosis is Associated with an Impaired Gut-Liver Axis. Alcohol Clin Exp Res 2017;41:1857–1865. [DOI] [PubMed] [Google Scholar]
- 20.Bajaj JS, Kakiyama G, Cox IJ, et al. Alterations in gut microbial function following liver transplant. Liver Transpl 2018;24:752–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bajaj JS, Fan S, Thacker LR, et al. Serum and urinary metabolomics and outcomes in cirrhosis. PLoS One 2019;14:e0223061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bajaj JS, O’Leary JG, Reddy KR, et al. Second infections independently increase mortality in hospitalized patients with cirrhosis: the North American consortium for the study of end-stage liver disease (NACSELD) experience. Hepatology 2012;56:2328–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bajaj JS, O’Leary JG, Reddy KR, et al. Survival in infection-related acute-on-chronic liver failure is defined by extrahepatic organ failures. Hepatology 2014;60:250–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O’Leary JG, Reddy KR, Garcia-Tsao G, et al. NACSELD Acute-on-Chronic Liver Failure (NACSELD-ACLF) Score Predicts 30-Day Survival in Hospitalized Patients with Cirrhosis. Hepatology 2018. [DOI] [PubMed] [Google Scholar]
- 25.Breiman L Random Forests In: Schapire RE, ed. Machine Learning. Volume 45 Netherlands: Kluwer Academic Publishers, 2001:5–32. [Google Scholar]
- 26.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 1988;44:837–845. [PubMed] [Google Scholar]
- 27.Naqvi A, Rangwala H, Keshavarzian A, et al. Network-based modeling of the human gut microbiome. Chem Biodivers 2010;7:1040–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bajaj JS, Vargas HE, Reddy KR, et al. Association Between Intestinal Microbiota Collected at Hospital Admission and Outcomes of Patients With Cirrhosis. Clin Gastroenterol Hepatol 2019;17:756–765 e3. [DOI] [PubMed] [Google Scholar]
- 29.Rosenblatt R, Shen N, Tafesh Z, et al. The NACSELD-ACLF Score Accurately Predicts Survival: An External Validation Using a National Cohort. Liver Transpl 2019. [DOI] [PubMed] [Google Scholar]
- 30.Claria J, Moreau R, Fenaille F, et al. Orchestration of Tryptophan-Kynurenine Pathway, Acute Decompensation, and Acute-on-Chronic Liver Failure in Cirrhosis. Hepatology 2019;69:1686–1701. [DOI] [PubMed] [Google Scholar]
- 31.Moreau R, Claria J, Aguilar F, et al. Blood metabolomics uncovers inflammation-associated mitochondrial dysfunction as a potential mechanism underlying ACLF. J Hepatol 2019. [DOI] [PubMed] [Google Scholar]
- 32.Ohashi Y, Fujisawa T. Analysis of Clostridium cluster XI bacteria in human feces. Biosci Microbiota Food Health 2019;38:65–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roager HM, Licht TR. Microbial tryptophan catabolites in health and disease. Nat Commun 2018;9:3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holmes E, Li JV, Athanasiou T, et al. Understanding the role of gut microbiome-host metabolic signal disruption in health and disease. Trends Microbiol 2011;19:349–59. [DOI] [PubMed] [Google Scholar]
- 35.Wikoff WR, Anfora AT, Liu J, et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A 2009;106:3698–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ridlon JM, Harris SC, Bhowmik S, et al. Consequences of bile salt biotransformations by intestinal bacteria. Gut Microbes 2016;7:22–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Minder EI, Karlaganis G, Paumgartner G. Radioimmunological determination of serum 3 beta-hydroxy-5-cholenoic acid in normal subjects and patients with liver disease. J Lipid Res 1979;20:986–93. [PubMed] [Google Scholar]
- 38.Ridlon JM, Bajaj JS. The human gut sterolbiome: bile acid-microbiome endocrine aspects and therapeutics. Acta Pharm Sin B 2015;5:99–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gerard P Metabolism of cholesterol and bile acids by the gut microbiota. Pathogens 2013;3:14–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Winston JA, Theriot CM. Diversification of host bile acids by members of the gut microbiota. Gut Microbes 2019:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Doden H, Sallam LA, Devendran S, et al. Metabolism of Oxo-Bile Acids and Characterization of Recombinant 12alpha-Hydroxysteroid Dehydrogenases from Bile Acid 7alpha-Dehydroxylating Human Gut Bacteria. Appl Environ Microbiol 2018;84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bajaj JS, Ahluwalia V, Wade JB, et al. Asymmetric dimethylarginine is strongly associated with cognitive dysfunction and brain MR spectroscopic abnormalities in cirrhosis. J Hepatol 2013;58:38–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Langer DA, Shah VH. A gas, an amino acid, and an imposter: the story of nitric oxide, L-arginine, and ADMA in portal hypertension. Hepatology 2005;42:1255–7. [DOI] [PubMed] [Google Scholar]
- 44.Amathieu R, Nahon P, Triba M, et al. Metabolomic approach by 1H NMR spectroscopy of serum for the assessment of chronic liver failure in patients with cirrhosis. J Proteome Res 2011;10:3239–45. [DOI] [PubMed] [Google Scholar]
- 45.Oliphant K, Allen-Vercoe E. Macronutrient metabolism by the human gut microbiome: major fermentation by-products and their impact on host health. Microbiome 2019;7:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin CJ, Liou TC, Pan CF, et al. The Role of Liver in Determining Serum Colon-Derived Uremic Solutes. PLoS One 2015;10:e0134590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao J, Xu K, Liu H, et al. Impact of the Gut Microbiota on Intestinal Immunity Mediated by Tryptophan Metabolism. Front Cell Infect Microbiol 2018;8:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rowland I, Gibson G, Heinken A, et al. Gut microbiota functions: metabolism of nutrients and other food components. Eur J Nutr 2018;57:1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lahdou I, Sadeghi M, Oweira H, et al. Increased serum levels of quinolinic acid indicate enhanced severity of hepatic dysfunction in patients with liver cirrhosis. Hum Immunol 2013;74:60–6. [DOI] [PubMed] [Google Scholar]
- 50.Khetan SK. Endocrine Disruptors in the Environment: Wiley, 2014. [Google Scholar]
- 51.Sherriff JL, O’Sullivan TA, Properzi C, et al. Choline, Its Potential Role in Nonalcoholic Fatty Liver Disease, and the Case for Human and Bacterial Genes. Adv Nutr 2016;7:5–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spencer MD, Hamp TJ, Reid RW, et al. Association between composition of the human gastrointestinal microbiome and development of fatty liver with choline deficiency. Gastroenterology 2011;140:976–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lieber CS, Robins SJ, Li J, et al. Phosphatidylcholine protects against fibrosis and cirrhosis in the baboon. Gastroenterology 1994;106:152–9. [DOI] [PubMed] [Google Scholar]
- 54.McPhail MJ, Shawcross DL, Lewis MR, et al. Multivariate metabotyping of plasma predicts survival in patients with decompensated cirrhosis. J Hepatol 2016;64:1058–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shah P, Swiatlo E. A multifaceted role for polyamines in bacterial pathogens. Mol Microbiol 2008;68:4–16. [DOI] [PubMed] [Google Scholar]
- 56.Cambiaghi A, Pinto BB, Brunelli L, et al. Characterization of a metabolomic profile associated with responsiveness to therapy in the acute phase of septic shock. Sci Rep 2017;7:9748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bajaj JS, Heuman DM, Sanyal AJ, et al. Modulation of the metabiome by rifaximin in patients with cirrhosis and minimal hepatic encephalopathy. PLoS One 2013;8:e60042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Allaband C, McDonald D, Vazquez-Baeza Y, et al. Microbiome 101: Studying, Analyzing, and Interpreting Gut Microbiome Data for Clinicians. Clin Gastroenterol Hepatol 2019;17:218–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


