Abstract
Chondrosarcoma is the second most common primary bone malignancy, representing one fourth of all primary bone sarcomas. It is typically resistant to radiation and chemotherapy treatments. However, the molecular mechanisms that contribute to cancer aggressiveness in chondrosarcomas remain poorly characterized. Here, we studied the role of mitochondrial transporters in chondrosarcoma aggressiveness including chemotherapy resistance. Histological grade along with stage are the most important prognostic biomarkers in chondrosarcoma. We found that high-grade human chondrosarcoma tumors have higher expression of the mitochondrial protein, translocase of the outer mitochondrial membrane complex subunit 20 (TOMM20), compared to low-grade tumors. TOMM20 overexpression in human chondrosarcoma cells induces chondrosarcoma tumor growth in vivo. TOMM20 drives proliferation, resistance to apoptosis and chemotherapy resistance. Also, TOMM20 induces markers of epithelial to mesenchymal transition (EMT) and metabolic reprogramming in these mesenchymal tumors. In conclusion, TOMM20 drives chondrosarcoma aggressiveness and resistance to chemotherapy.
Keywords: chondrosarcoma, mitochondria, proliferation, apoptosis, chemotherapy resistance
Introduction
Chondrosarcomas are malignant cartilaginous matrix-producing neoplasms [1] [2] [3]. They primarily affect adults and are the second most common type of bone malignancy after osteosarcoma [4]. Chondrosarcomas predominately arise in bone with endochondral ossification with the most common sites being the femur, pelvis, humerus, tibia and ribs [2]. Chondrosarcomas are classified based on morphology, whether they arise de novo (primary) or from a precursor benign lesion (secondary), or by location within the bone [1] [5] [6]. The majority of primary chondrosarcomas are classified as conventional and histological grading is the most important factor other than stage used to predict prognosis [7] [8] [9]. Chondrosarcomas are graded from 1 to 3 based on cellularity, nuclear size, nuclear atypia, mitotic activity, and matrix alterations [5]. Central atypical cartilaginous tumors (ACT)/chondrosarcoma grade 1 (CS1) neoplasms are low grade tumors, with a locally aggressive behavior. Tumors located in the appendicular skeleton are termed ACTs, whereas tumors located in the axial skeleton are termed CS1 [1] [3]. Chondrosarcoma grades 2 and 3 (CS2 and CS3) are high grade chondrosarcomas with high local recurrence and metastatic potential with a five-year survival rate of 74-99% for CS2 and 31-77% for CS3[1] [10]. Metastatic chondrosarcoma is incurable and leads to short overall survival [11]. Patients with low grade chondrosarcomas have less than 10% risk of metastasis, but metastatic risk increases to 50-70% in patients with high grade chondrosarcomas [2].
The histological grade and tumor location determine treatment of chondrosarcomas. Surgical excision is currently the primary treatment approach for non-metastatic chondrosarcomas because chemotherapy agents and radiation are not effective [2] [12]. The type of surgery can range from curettage for atypical chondromatous tumors (ACT) or CS1 to wide en-bloc excision for treatment of higher grade chondrosarcoma due to high risk of recurrence and metastasis [7]. However, en-bloc wide resection causes significant functional impairment [7]. Resistance to chemotherapy in chondrosarcoma was originally attributed to low proliferation rates and the hyaline cartilage and extracellular matrix, which restricts drug access to the cancerous cells [13] [14]. However, high grade lesions are also resistant to chemotherapy despite lower amounts of matrix, increased vascularization, and increased proliferation[15] and high expression of anti-apoptotic proteins is now recognized as a resistance contributing factor [16].
Mitochondrial metabolism is a driver of cancer aggressiveness in several human malignancies [17]. However, it is unknown if mitochondrial metabolism drives chondrosarcoma aggressiveness although isocitrate dehydrogenase (IDH) mutations, which modulate mitochondrial metabolism, are present in approximately 50% of chondrosarcomas and are the most common molecular event in this cancer subtype [1]. Also, glutaminolysis, which promotes mitochondrial metabolism, is a critical pathway in chondrosarcoma [17] [18]. Hence, we have studied the role of mitochondrial metabolism in chondrosarcoma aggressiveness.
The molecular mechanisms and signaling pathways that are contributing to drug resistance in chondrosarcoma remain poorly defined. Although development of metastatic disease is the most feared complication of chondrosarcoma, local recurrence is also a major issue [2]. Many patients with high grade chondrosarcomas undergoing surgery with curative intent will subsequently have a local recurrence and surgery can induce profound disability [6]. Consequently, chondrosarcomas are associated with poor clinical outcomes that have not improved despite rapid advances in chemotherapy for almost all other cancer subtypes [15]. Most studies on chemotherapy and resistance in chondrosarcoma have involved the use of anthracyclines such as Doxorubicin, since this is the most commonly employed drug in the management of sarcomas [11] [19]. Thus we have studied the role of mitochondrial metabolism in driving anthracycline resistance in chondrosarcoma.
Altered metabolism is a hallmark of cancer [20] and mitochondrial activity is increased in a subset of human cancers [17] [21] [22]. Most mitochondrial proteins are encoded by nuclear genes and are synthesized by cytosolic ribosomes with mitochondrial targeting signal sequences [23]. Translocase of the outer mitochondrial membrane complex subunit 20 (TOMM20) has been shown to be an important subunit of the translocase of the outer mitochondrial membrane complex that is responsible for recognizing and translocating mitochondrial proteins from the cytosol into the mitochondria [24] [25]. The components of the translocase of the outer mitochondrial membrane complex have been shown to aggregate into clusters at the membrane and the density of these clusters are tightly correlated with activity of the mitochondria [23]. The density of the TOMM20 clusters is higher in mitochondria with higher mitochondrial membrane potential [23]. Therefore, cells with greater mitochondrial mass and higher mitochondrial activity have more TOMM20 expression [23]. TOMM20 has previously been shown to be highly expressed in human epithelial and lymphoid cancers and is a prognostic biomarker [26] [27] [28] [29] [30].
In this study we set out to determine mechanisms of chemotherapy resistance in chondrosarcoma. Here, we determined that high grade human chondrosarcomas have high expression of TOMM20, TP53 Induced Glycolysis and Apoptosis Regulator (TIGAR), and Monocarboxylate Transporter 1 (MCT1) suggesting that aggressive chondrosarcomas have high mitochondrial oxidative phosphorylation (OXPHOS) metabolism. TOMM20 over-expression in chondrosarcoma cells promotes cancer aggressiveness by increasing tumor growth, invasiveness, proliferation, resistance to apoptosis, and resistance to chemotherapy.
Methods
Chemical Compounds.
Drug compounds were purchased from Sigma: Palbociclib (PZ0383, Millipore Sigma, Burlington, MA), Doxorubicin (D1515, Sigma Aldrich, St. Louis MO), Gemcitabine (G6423, Sigma Aldrich), Antimycin A (A8674, Sigma Aldrich), and Rotenone (45656, Sigma Aldrich).
Human Studies.
This study was approved by the institutional review board (IRB) at Thomas Jefferson University. Samples from low grade (ACT/CS1), and high grade (CS2 and CS3) chondrosarcomas were obtained from archived paraffin embedded tissue blocks for TOMM20, MCT1 and TIGAR histological analyses. The samples were scored by a bone and soft tissue tumor pathologist (W.J.) for each antibody. Percentage of tumor cells with positive staining was assessed. Intensity of staining within tumor cells was also assessed using a semiquantitative system with 0 having no staining, 1+ weak staining, 2+ moderate staining and 3+ high staining.
The relationship between MCT1 intensity and histological grade was analyzed by fitting a continuation ratio for ordinal responses with logit-link model using R [31] package VGAM [32]. The relationship between TOMM20 or TIGAR intensity and histological grade was analyzed by fitting a Firth’s bias-reduced logistic model using R package logistf [33] because of a lack of moderate and high intensity in the low-grade group. The relationship between percentage positive for TOMM20, MCT1 and TIGAR and intensity and histological grade was analyzed by fitting a multivariable linear mixed effects model with random effects of sample using R package nlme [34].
Immunohistochemistry.
Tumor samples of ACT/CS1, which are low grade chondrosarcomas (N=29) and CS2 and CS3, which are high grade chondrosarcomas (N=10) were stained by immunohistochemistry (IHC). Samples were stained with antibodies to TOMM20 (F-10) (1:5000; sc-17764, Santa Cruz Biotechnology, Dallas, TX), MCT1 (SLC16A1) 19-mer peptide (1:1000; YenZym Antibodies, South San Francisco, CA) and TIGAR (1:750; ab62533, Abcam, Cambridge, MA). Tissue sections were deparaffinized, rehydrated and antigen retrieval was performed on the Ventana Discovery ULTRA staining platform using Discovery CCI (Ventana #950-500, Roche Diagnostics, Indianapolis, IN) for a total application time of 64 minutes. Primary immunostaining was performed for 45 min followed by secondary immunostaining using a Horseradish Peroxidase (HRP) multimer cocktail (Ventana #760-500) and immune complexes were visualized using the ultraView Universal DAB (diaminobenzidine tetrahydrochloride) Detection Kit (Ventana #760-500). Slides were then washed with a Tris based reaction buffer (Ventana #950-300) and stained with Hematoxylin II (Ventana #790-2208) for 8 minutes. For mouse tumor staining, a manual 3-step HRP method was used. Four micron tumor xenograft paraffin sections were deparaffmized and rehydrated. Antigen retrieval was performed in lOmM citrate buffer pH 6.0 for 10 minutes with a pressure cooker followed by blocking for endogenous peroxidase with 3% H2O2 for 15 minutes. Blocking was also performed for endogenous biotin with an avidin-biotin blocking kit (Biocare Medical, Pacheco, CA). Samples were then incubated with 10% goat serum overnight at 4°C and the next day with anti-phospho-RB (1:500; #8516, Cell Signaling, Danvers, MA) for one hour at room temperature. After washing in PBS, the sections were incubated with a biotinylated goat-anti-rabbit IgG antibody followed by an avidin-biotinylated horseradish peroxidase complex (PK-6100, Vector Laboratories, Burlingame, CA). Immunoreactivity was determined using liquid DAB substrate chromogen (Agilent #K346811-2, Santa Clara, CA) and samples were counterstained with hematoxylin.
Phospho-retinoblastoma (phospho-RB) Quantification.
Phospho-RB staining in L2975 control and TOMM20 xenograft sections were quantified using Aperio software (Aperio, Nussloch, Germany). Briefly, digital images were captured with Leica and Aperio scanners under 3203 magnification with an average scan time of 120 seconds (compression quality, 70). A nuclear algorithm was used to identify phospho-RB positive nuclei and generate percentage positive nuclei in each of 9-11 areas per sample encompassing the majority of the viable tumor area for a total of 19 areas in the EV control and group and 29 areas in the TOMM20 group.
Cell culture.
The human chondrosarcoma cell line SW1353 was obtained from American TType Culture Collection (ATCC) and L2975 and CH2879 were a kind gift from Professor Judith Bovee at the Leiden University Medical Center, The Netherlands and Professor Antonio Llombart Universidad de Valencia, Spain. Cells were cultured in media containing 4.5 g/L glucose, ImM pyruvate, 10% FBS, 100 units/mL penicillin, and 100 units/mL streptomycin at 37°C, 5% CO2. RPMI cell culture media was used as the base medium for all cell lines.
Overexpression of TOMM20.
HA-TOMM20 (EX-G0283-Lvl20) and HA-control (EX-NEG-Lvl20) lentiviral vectors were purchased from GeneCopoeia (Rockville, MD). Lentiviruses were prepared according to the manufacturer’s protocol. Virus containing media was centrifuged, filtered with a 0.45 micron PES low protein filter, divided into 1 mL aliquots and stored at −80°C. Chondrosarcoma cells were seeded in 6-well culture dishes at 250,000 cells per well in growth media. At 24 hours post-seeding, the media was removed and replaced with lmL of media containing 5% FBS, 5ug/uL polybrene and 1 mL of virus containing media. Virus containing media was removed 24 hours post-infection and replaced with growth media. The cells were selected with puromycin (2ug/mL) for 3 days after infection.
Proteomic Analysis.
2D DIGE (two-dimensional difference gel electrophoresis) and mass spectrometry protein identification were run by Applied Biomics (Hayward, CA) as previously described [35], Briefly, image scans were performed immediately after SDS-PAGE using Typhoon TRIO. All of the images were analyzed using Image QuantTL software and subjected to in-gel and cross-gel analysis and protein differential expression was determined using DeCyder software. Image analysis spots were selected and subjected to in-gel trypsin digestion, peptide extraction, desalting, and MALDI-TOF/TOF (Applied Biomics, Hayward, CA) analysis to determine protein identity. All samples were prepared by washing cells 3X with cold PBS. Cells were scraped into 1.5mL micocentrifuge tubes and centrifuged at 13,000X g at 4°C for 15 minutes to generate a cell pellet. All samples were stored at −80°C until shipment. Proteins with 1.5 or greater fold change were considered to be upregulated in TOMM20 overexpressing cells compared to control.
Immunoblot Analysis.
Cell protein lysates were obtained by extraction with lysis buffer containing lOmM Tris, pH 7.5, 150mM NaCl, 1% Triton X-100, and 60mM n-ocytlglucoside containing protease and phosphatase inhibitors. Extracts were rotated at 4°C for 40 minutes and centrifuged at 13,000X g at 4°C for 15 minutes to remove the pellet. The protein concentration of all samples were determined by BCA reagent. Samples were separated by SDS PAGE and transferred to nitrocellulose membrane. Primary antibodies and dilutions are described in Table 1. Protein expression levels were determined by calculating densitometry and normalized to loading control.
Table 1.
Primary antibodies and their dilutions.
| Protein | Company | Reference | Dilution |
|---|---|---|---|
| N-Cadherin | BD Biosciences | BD-610820 | 1:1000 |
| Phosphorylated STAT3 Tyrosine705 | Cell Signaling Technology | CST9145 | 1:1000 |
| c-MYC | Cell Signaling Technology | CST5605 | 1:1000 |
| Cyclin D1 (DCS-6) | Invitrogen | MA5-12702 | 1:2000 |
| BCL2 | Abcam | ab181858 | 1:2000 |
| TIGAR | Abcam | ab31910 | 1:1000 |
| MCT1 | Santa Cruz Biotechnology | sc-365501 | 1:500 |
| MitoNEET (CDGSH iron sulfur domain 1) | Proteintech | 16006-1-AP | 1:10000 |
Apoptosis.
Apoptosis in culture was quantified by flow cytometry using PI and Annexin-V-APC. Cells were harvested and centrifuged and resuspended in Annexin-V binding buffer containing the Annexin V-APC conjugate (BDB550474, BD Biosciences, San Jose, CA) (8uL/mL) and DAPI (0.2-0.5ug/mL). Apoptotic cells (early, late, or dead) were identified using the BD LSR Fortessa cell analyzer. Statistical analysis was examined in GraphPad Prism using Student’s t-test. Values of p<0.05 were considered statistically significant.
Proliferation.
For DNA content and proliferation analyses, cells were incubated with lOuM EDU for 1 hour. Click-iT EdU Flow Cytometry Assay kit (C10418, Life Technologies) and FxCycle™ Far Red Stain (FI0348, Life Technologies, Carlsbad, CA) for cell cycle analysis was performed according to the standard protocol. Data was analyzed by Flowjo software. Statistical analysis was examined in GraphPad Prism using Student’s t test. Values of p<0.05 were considered significant.
Oxygen Consumption Rate Assessment.
A Seahorse Bioscience XF24 Extracellular Flux Analyzer was used. CH2879 and SW1353 cells were preseeded in the XF24 24-well plate with lOOuL of RPMI growth media containing 10% fetal bovine serum, 100 units/mL penicillin, 100 units/mL streptomycin in a 37°C, 5% CO2 incubator for one hour to allow for cell attachment. After one hour, an additional 200uL of growth media was added for a total volume of 300uL per well. On the second day, cells were incubated in non-buffered media containing 5mM glucose in a CO2 free incubator for one hour. Oxygen consumption measurements were obtained under basal conditions, after injection of lOmM lactate, and after injection of 0.5μM antimycin A and 0.5μM rotenone. Statistical analysis was performed using Student’s t-test. Values of p<0.05 were considered statistically significant. For CH2879 N=12 and for SW1353 N=6.
Animal Studies.
To evaluate the in vivo effects of TOMM20 overexpression on chondrosarcoma tumor growth, cells were injected into the flank of athymic NCr nude mice (Nu/Nu; Charles River at 6 weeks of age). All animals were maintained in a pathogen-free environment/barrier facility at the Sidney Kimmel Cancer Center at Thomas Jefferson University. The Institutional Animal Care and Use Committee (IACUC) approved all animal protocols and all experiments were performed in accordance with the National Institute of Health guidelines. U2975 cells were injected into the flank of male nude mice and CH2879 cells were injected into the flank of female nude mice following isoflurane anesthesia. For these studies, 2 million cancer cells were resuspended in 100μL of sterile PBS before injection. For each mouse experiment there were 5 mice injected bilaterally per experimental group for a total of 10 per group. Mice were sacrificed 4-12 weeks post-injection via CO2 inhalation and tumors were excised. Tumor volume was calculated using the formula V=X^2*Y/2 where V is the tumor volume, X is the length of the short axis and the Y is the length of the long axis measured using electronic calipers and tumor weight was measured in grams. Statistical analysis was performed using the Mann Whitney U Test in GraphPad Prism. Values of p<0.05 were considered statistically significant.
Results.
TOMM20, MCT1 and TIGAR expression in human chondrosarcoma
A cohort of 10 low grade chondrosarcomas (CS1) and 29 high grade chondrosarcomas (CS2 and CS3) were stained with antibodies to TOMM20, TIGAR, and MCT1 by immunohistochemistry (IHC) (Figure 1A). The samples were scored by a bone and soft tissue tumor pathologist for percentage of positive tumor cells and for staining intensity. Positive cells were defined as those that had at least weak expression. Also, the samples were scored for intensity semi-quantitatively using a 1+, 2+ and 3+ scoring system, where 1+ corresponded to weak expression, 2+ corresponded to moderate expression and 3+ corresponded to high expression.
Figure 1. TOMM20, MCT1 and TIGAR expression in human chondrosarcomas.

A, Representative images of immunohistochemistry for TOMM20, TIGAR, and MCT1 with CS1 and CS3 samples are shown. B, Percent positive cells and overall staining intensity were measured. Note that the percentage of tumor cells positive for TOMM20 and TIGAR is higher in high-grade chondrosarcoma (p<0.05) but not for MCT1 (p<0.23). C, Staining intensity for TOMM20, TIGAR, and MCT1 positively correlates with the percentage of tumor cells that are positive for TOMM20, TIGAR, and MCT1 respectively and higher grade tumors have higher staining intensity (p<0.05). *p<0.05
The percentage of tumor cells that were positive for TOMM20 and TIGAR was higher in high grade compared to low grade chondrosarcomas (Figure 1A) (p<0.05). There was a trend towards a higher percentage of tumor cells that were positive for MCT1 in high grade chondrosarcomas compared to low grade chondrosarcomas but this was not statistically significant (Figure 1A) (p<0.23). The intensity of TOMM20, TIGAR, and MCT1 on tumor cells was higher in high grade chondrosarcomas compared to low grade chondrosarcomas (Figure 1B) (p<0.05). The percentage of tumor cells that were positive for TOMM20, TIGAR, and MCT1 positively correlated with TOMM20, TIGAR, and MCT1intensity respectively (Figure 1C) (p<0.05). These data demonstrate that high grade chondrosarcomas have higher TOMM20 and TIGAR expression, which are markers of mitochondrial mass.
TOMM20 overexpression increases markers of cancer aggressiveness
To evaluate the effects of TOMM20 overexpression on chondrosarcoma aggressiveness, we overexpressed TOMM20 in the chondrosarcoma cells lines CH2879, L2975, and SW1353 (Figure 2A). Proteomic analysis was performed to determine the effect of TOMM20 overexpression on the chondrosarcoma cells and assess changes in protein levels and cellular processes in an unbiased fashion. The results of the proteomic analysis are summarized in Table 2. String DB was used with modifications to generate a network based on the proteins that increased with TOMM20 overexpression and on known and predicted protein-protein interactions (Figure 2B). The proteins and pathways that were found to be upregulated with TOMM20 overexpression by proteomic analysis were used to identify which pathways should be further studied. The most upregulated proteins with at least 1.5-fold increase in expression between the control and TOMM20 overexpressing cells were selected. We studied invasion, proliferation, apoptosis, mitochondrial metabolism, and markers of cancer aggressiveness by evaluating the expression of classical makers in the empty vector control (EV) and TOMM20 overexpressing cells. To study cancer aggressiveness and invasiveness, we analyzed expression of N-cadherin and phosphorylated STAT3, which are markers of an epithelial to mesenchymal transition (EMT) by immunoblot. TOMM20 overexpression increases N-cadherin and phospho-STAT3 compared to the EV control (Figure 2C). We evaluated the expression of cyclin D1 and c-MYC as markers of cell cycle progression and proliferation by immunoblot. TOMM20 overexpression increases cyclin D1 and c-MYC expression compared the EV control (Figure 2D–E). To study resistance to apoptosis with TOMM20 overexpression we evaluated the expression of the anti-apoptotic protein BCL2 in the EV control and TOMM20 overexpressing cells by immunoblot. We found that TOMM20 overexpression increases BCL2 expression compared to EV control (Figure 2F). TOMM20 overexpression increased the percentage of cells in S phase 1.3-fold compared to EV control cells (Figure 2G). Taken together, these data demonstrate that TOMM20 overexpression in chondrosarcoma cells may increase markers of aggressiveness by increasing the expression of proteins involved in invasion and metastasis, proliferation, and resistance to apoptosis (Figure 6F).
Figure 2. Effect of TOMM20 on markers of cancer aggressiveness.
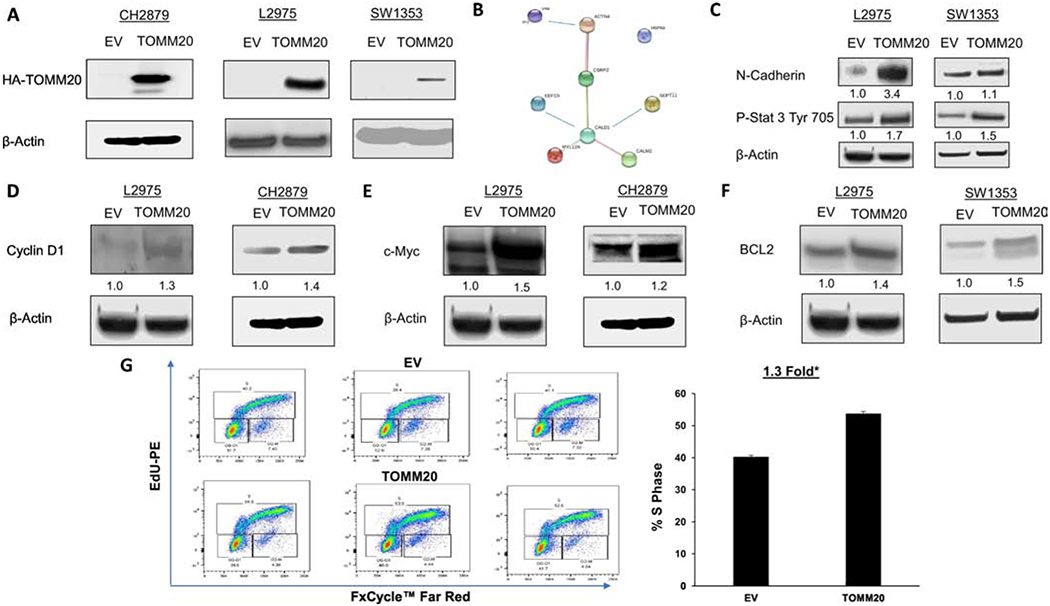
A, HA-TOMM20 immunoblot in CH2879, L2975, and SW1353 cells with TOMM20 overexpression compared to EV control. In CH2879 cells the two bands represent HA variants. B, The proteins identified from the proteomic analysis was subjected to String v11.0 analysis to identify functional interactions between the deregulated proteins. Each node represents a protein and each edge represents an interaction. C, Phospho-STAT3 tyr705 and N-cadherin immunoblot of EV control and TOMM20 overexpressing L2975 and SW1353 cells. D, Cyclin D1 immunoblot of EV control and TOMM20 overexpressing L2975 and CH2879 cells. E, C-MYC immunoblot of EV control and TOMM20 L2975 and CH2879 overexpressing cells. F, BCL2 immunoblot of EV control and TOMM20 overexpressing L2975 and SW1353 cells. G, Proliferation rates were measured with EdU incorporation and the cell cycle was assessed in carcinoma cells. DNA synthesis was measured with EdU-PE incorporation and ploidy was assessed with FxCycle™ Far Red Stain (N=3). All immunoblots are representative examples,*p<0.05.
Table 2.
Proteins that are upregulated with TOMM20 overexpression in chondrosarcoma.
| Protein name | Abbreviation | Pathway | Fold Change CTRL vs. TOMM20 |
|---|---|---|---|
| Heat shock cognate 71kDa protein | HSP7C | Mitochondrial metabolism | 1.5 |
| Elongation factor 1-delta | EF1D | Proliferation | 1.6 |
| Calmodulin-2 | CALM2 | Proliferation | 2.6 |
| Caldesmon | CALD1 | Proliferation | 1.5 |
| Septin-11 | SEP11 | Proliferation | 1.5 |
| Cysteine and glycine-rich protein 2 | CSRP2 | Resistance to cell death | 1.5 |
| Alpha-actinin-4 | ACTN4 | Dedifferentiation | 1.9 |
| Myosin regulatory light chain 12A | ML12A | Invasion | 1.5 |
Figure 6. Expression of mitochondrial markers in murine tumors.
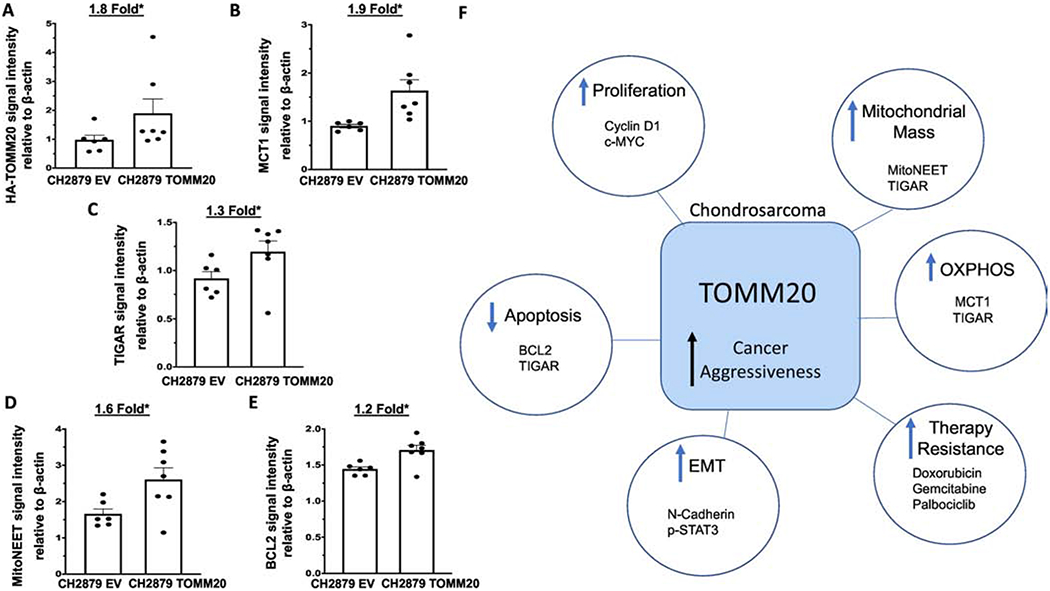
A, Quantification of TOMM20 protein levels normalized to β-actin control of tumors generated from EV control and TOMM20 overexpression. B, Quantification of MCT1 protein levels normalized to β-actin of tumors generated from control and EV TOMM20 overexpression. C, Quantification of TIGAR protein levels normalized to β-actin of tumors generated from EV control and TOMM20 overexpression. D, Quantification of mitoNEET protein levels normalized to β-actin of tumors generated from EV control and TOMM20 overexpression. E, Quantification of BCL2 protein levels normalized to β-actin of tumors generated from EV control and TOMM20 overexpression. F, Study schematic displaying the results of this study which include TOMM20 overexpression induces a more aggressive phenotype by increasing the expression of proteins involved in invasion and metastasis, proliferation, and resistance to apoptosis, promoting mitochondrial metabolism, and therapy resistance. EV control N=6, TOMM20 N=7, *p<0.05
TOMM20 overexpression increases mitochondrial metabolism
To assess the effect of TOMM20 overexpression on mitochondrial metabolism we studied the expression of TIGAR and MCT1 in EV control and TOMM20 overexpressing cells. The expression of TIGAR and MCT1 increases in TOMM20 overexpressing cells (Figure 3A–B). To further characterize the effects of TOMM20 on mitochondrial metabolism and OXPHOS, we studied oxygen consumption rates (OCR) in EV control and TOMM20 overexpressing chondrosarcoma cells. OCR was measured under basal conditions with media containing 5mM glucose, after exposure to 10mM lactate, and after treatment with 0.5μM Antimycin A and 0.5μM Rotenone. Antimycin A, a complex III inhibitor, and Rotenone, a complex I inhibitor, were used in combination to inhibit the mitochondrial electron transport chain in order to determine the contribution of non-mitochondrial sources to OCR [36] [37]. CH2879 TOMM20 overexpressing cells have 1.5-fold greater OCR than EV control cells at baseline and 1.4-fold greater OCR after lactate exposure (Figure 3C) (p<0.05). SW1353 TOMM20 overexpressing cells have 1.3-fold greater OCR than EV control cells at baseline and 1.4-fold greater OCR after lactate exposure (Figure 3D) (p<0.05). Treatment of both CH2879 and SW1353 cells with Antimycin A and Rotenone caused the expected reduction in oxygen consumption in both EV control and TOMM20 overexpressing cells. After subtraction of the non-mitochondrial oxygen consumption the increased OCR in TOMM20 overexpressing cells was maintained both at baseline and after lactate exposure. These data demonstrate that TOMM20 overexpression increases mitochondrial OCR and therefore increases mitochondrial OXPHOS metabolism in CH2879 and SW1353 cells. TOMM20 overexpressing cells also have increased OCR after lactate exposure suggesting that chondrosarcoma cells can utilize and catabolize lactate via mitochondrial OXPHOS metabolism. Taken together these data suggest that TOMM20 overexpression promotes mitochondrial metabolism (Figure 6F).
Figure 3. TOMM20 overexpression promotes mitochondrial metabolism.
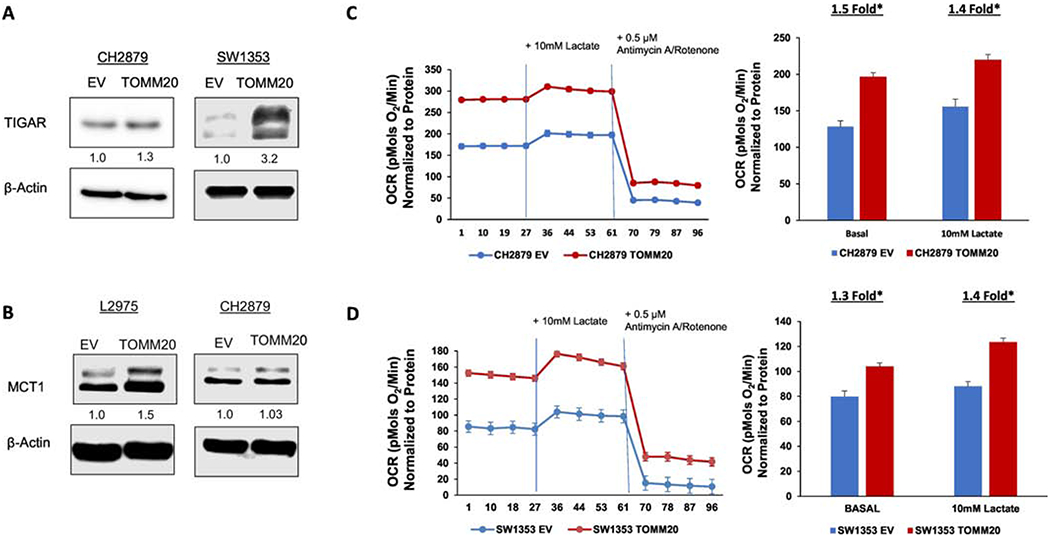
A, TIGAR immunoblot of EV control and TOMM20 overexpressing CH2879 and SW1353 cells. B, MCT1 immunoblot of EV control and TOMM20 overexpressing L2975 and CH2879 cells. C, Oxygen consumption was measured in EV control (blue) and TOMM20 overexpressing (red) CH2879 cells. Oxygen consumption was measured under baseline conditions with 5mM glucose, after injection of 10mM lactate, and after injection of 0.5μM Antimycin A and Rotenone. OCR values were normalized to protein concentration (N=12). D, Oxygen consumption was measured in EV control (blue) and TOMM20 overexpressing (red) SW1353 cells. Oxygen consumption was measured under baseline conditions with 5mM glucose, after injection of 10mM lactate, and after injection of 0.5μM Antimycin A and Rotenone. OCR values were normalized to protein concentration (N=6). *p<0.05
TOMM20 overexpression induces cancer chemotherapy resistance
Since TOMM20 overexpression increased markers of cancer aggressiveness including proliferation and resistance to apoptosis, we wanted to determine if TOMM20 overexpression can lead to resistance to anticancer drugs that target proliferating cells that have been studied in the context of chondrosarcoma such as Gemcitabine, Doxorubicin and Palbociclib. Gemcitabine and Doxorubicin are commonly used agents to treat sarcomas [11] [19] and recently the cyclin dependent kinase 4/6 inhibitor Palbociclib that targets proliferating cells has shown promise in chondrosarcoma preclinical models [38]. Chondrosarcoma cells were treated with Palbociclib and Gemcitabine alone or in combination, to determine the effects of these drugs on the proliferation of chondrosarcoma cells. Treatment with Palbociclib led to a 1.1-fold reduction, Gemcitabine led to a 3.2-fold reduction, and the combination of Palbociclib and Gemcitabine led to a 5.3-fold reduction in the percentage of cells in G2-M and S phase (Figure 4A) (p<0.05). We assessed the amount of cell death induced by the treatment of Palbociclib, Doxorubicin, and the combination. Treatment with Palbociclib alone induced a 1.2-fold increase, Doxorubicin induced a 2.6-fold increase, and the combination of Palbociclib and Doxorubicin induced a 5.1-fold increase in the percentage of apoptotic cells (Figure 4B) (p<0.05). Since treatment with these compounds led to reduced proliferation and increased cell death in chondrosarcoma cells, we wanted to determine the effects of TOMM20 overexpression on therapy efficacy in chondrosarcoma cells. EV control and TOMM20 overexpressing cells were exposed to Gemcitabine and then evaluated for apoptosis. The TOMM20 overexpressing cells had a 1.6-fold reduction in cell death compared to the EV control (Figure 4C) (p<0.05). Due to lack of activity of single chemotherapeutic agents in chondrosarcoma we investigated combination treatments in the context of TOMM20 overexpression. EV control and TOMM20 overexpressing cells were treated with Gemcitabine, Palbociclib, and the combination and proliferation rates were evaluated. At baseline, TOMM20 overexpressing cells had a 1.2-fold increase in the percentage of cells in G2-M and S phase compared to EV control cells (Figure 4D) (p<0.05). Treatment with Gemcitabine, Palbociclib, or the combination induced an 11-fold, 6-fold, or 12-fold reduction in proliferation in EV control cells compared to TOMM20 overexpressing cells (Figure 4D). EV control and TOMM20 cells were treated with Doxorubicin, Palbociclib, and the combination of Doxorubicin and Palbociclib and evaluated for proliferation rates as measured by the percentage of cells in G2-M, and S phase. Treatment with Doxorubicin, Palbociclib, or the combination induced a 4.2-fold, 7-fold, or 13-fold reduction in proliferation of EV control cells compared to TOMM20 overexpressing cells (Figure 4E) (p<0.05). TOMM20 overexpressing and EV control cells were also evaluated for apoptosis levels after treatment with Gemcitabine, Palbociclib and the combination. Under control conditions, TOMM20 overexpressing cells had a 1.4-fold reduction in apoptosis levels (Figure 4F) (p<0.05). After treatment with Gemcitabine, Palbociclib and the combination, TOMM20 cells had a 1.4, 1.3 and 1.6-fold reduction respectively in apoptosis rates compared to EV control cells (Figure 4F) (p<0.05). Taken together these data demonstrate that TOMM20 overexpression in chondrosarcoma induces resistance to commonly used chemotherapies, which is consistent with an aggressive phenotype (Figure 6F).
Figure 4. Effect of TOMM20 overexpression on therapy resistance.
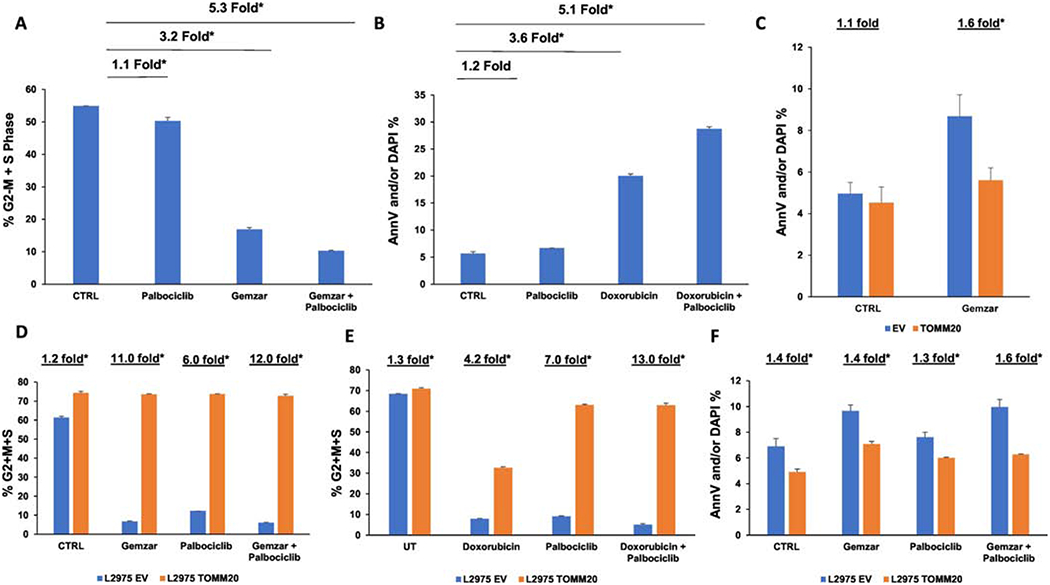
A Proliferation rates were measured with EdU incorporation and the cell cycle was assessed in carcinoma cells. DNA synthesis was measured with EdU-PE incorporation and ploidy was assessed with FxCycle™ Far Red Stain in L2975 chondrosarcoma cells after treatment with 20nM Palbociclib, 50nM Gemcitabine, and the combination of 20nM Palbociclib and 50nM Gemcitabine. B, Apoptosis levels were measured with Annexin-V (AnnV) and DAPI staining after treatment with 2μM Palbociclib, 2μM Doxorubicin, and the combination of 2μM Palbociclib and 2μM Doxorubicin. The percentage of apoptotic or dead L2975 cells (Annv-V-positive and/or DAPI positive) is shown. C, Apoptosis levels were measured in EV control and TOMM20 overexpressing L2975 cells after treatment with 5μM Gemcitabine (Gemzar). D, Proliferation rates were measured with EdU incorporation and the cell cycle was assessed in carcinoma cells. DNA synthesis was measured with EdU-PE incorporation and ploidy was assessed with FxCycle™ Far Red Stain in L2975 control and TOMM20 overexpressing cells after treatment with 0.5μM Palbociclib, 10μM Gemcitabine (Gemzar), and the combination of 0.5μM Palbociclib and 10μM Gemcitabine (Gemzar). E, Proliferation rates were measured with EdU incorporation and the cell cycle was assessed in carcinoma cells. DNA synthesis was measured with EdU-PE incorporation and ploidy was assessed with FxCycle™ Far Red Stainin L2975 control and TOMM20 overexpressing cells after treatment with 1μM Palbociclib, 1μM Doxorubicin, and the combination of 1μM Palbociclib and 1μM Doxorubicin. F, Apoptosis levels were measured with Annexin-V (AnnV) and DAPI staining after treatment with 2μM Palbociclib, 10μM Gemcitabine, and the combination of 2μM Palbociclib and 10μM Gemcitabine. The percentage of apoptotic or dead L2975 cells (Annv-V-positive and/or DAPI positive) is shown. N= 3-4, *p<0.05
TOMM20 overexpression induces larger tumors
Since TOMM20 overexpression induces a more aggressive cancer phenotype in chondrosarcoma cells, we wanted to determine if TOMM20 overexpression had an effect on tumor size in vivo. L2975 tumors with T0MM20 overexpression had 86.0-fold greater volume and 31.0-fold greater weight than EV control tumors (Figure 5A–B) (p<0.05). L2975 chondrosarcoma cells overexpressing TOMM20 had greater engraftment rates with 80 percent successful engraftment compared to the EV control cells that had only 20 percent successful engraftment (Figure 5C) (p<0.05). TOMM20 overexpressing L2975 tumors had 1.9-fold higher expression of phosphorylated Retinoblastoma (p-Rb) than EV control tumors (Figure 5D) (p<0.05). Tumor growth was also evaluated in CH2879 TOMM20 overexpressing and EV control cells. CH2879 TOMM20 overexpressing cells had a 5.2-fold greater volume and 2.8-fold greater weight than EV control tumors (Figure 5E–F) (p<0.05). Taken together, TOMM20 overexpression in chondrosarcoma cells promotes tumor growth.
Figure 5. Effect of TOMM20 overexpression on chondrosarcoma tumor growth.
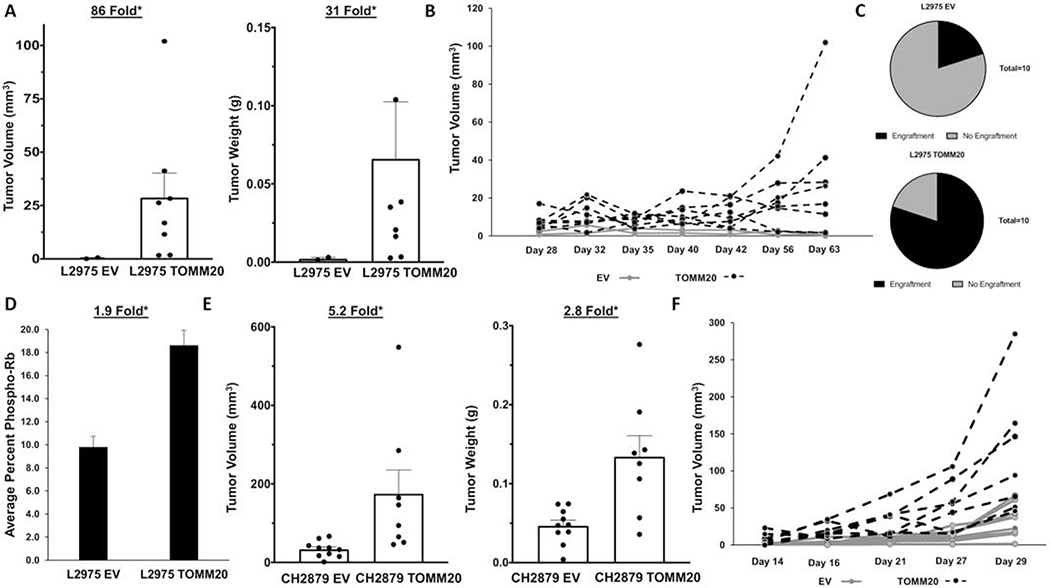
A, Tumor volume and weight measurement at the time of collection of L2975 EV and TOMM20 after injection into the flank of athymic nude mice at time of collection. B, Tumor volume growth chart of tumor volume started when the tumors were initially detected to endpoint. C, Relative engraftment of L2975 EV and TOMM20 tumors. D, Quantification of phospho-Rb staining measured by immunohistochemistry of the control and TOMM20 tumors and digitally quantified by Aperio software. E, Tumor volume and tumor weight measurement at the time of collection of CH2879 control and TOMM20 tumors injected into the flank of athymic nude mice. F, CH2879 tumor volume growth chart of tumor volume started when tumors were initially detected to endpoint. *p<0.05
TOMM20 overexpression induces expression of markers of mitochondrial reprogramming in tumors.
Tumors that were generated by injection into mice of cancer cells with overexpression of translocase of the outer mitochondrial membrane subunit 20 (TOMM20) or EV control tumors were analyzed for protein expression by immunoblot. Here we studied in vivo, the expression of several proteins associated with mitochondrial metabolic function by evaluating the expression of monocarboylate transporter 1 (MCT1), TP53-inducible glycolysis and apoptosis regulator (TIGAR) and mitoNEET. TOMM20 increased 1.8-fold in tumors overexpressing TOMM20 compared to EV control (Figure 6A) (p<0.05). The monocarboxylate transporters (MCTs) are a family of proton-linked membrane transporters that are responsible for the movement of single-carboxylate molecules such as lactate in and out of cells [39]. MCT1 has been found to be upregulated in cancer cells that have increased lactate uptake and oxidative phosphorylation (OXPHOS) metabolism. MCT1 increased 1.9-fold in TOMM20 overexpressing tumors compared to EV control (Figure 6B) (p<0.05). TIGAR has been described to be expressed in several solid tumor types and has been shown to induce OXPHOS in cancer cells [40]. TIGAR increased 1.3-fold in TOMM20 overexpressing tumors compared to EV control (Figure 6C) (p<0.05). To evaluate mitochondrial mass and energy regulation in tumors overexpressing TOMM20 we also studied mitoNEET expression. MitoNEET is an outer mitochondrial membrane protein containing an iron-sulfur cluster, which regulates oxidative capacity of the mitochondria [41]. MitoNEET expression increased 1.6-fold in tumors generated by TOMM20 overexpression compared to EV control (Figure 6D) (p<0.05). We also studied expression of BCL2 as a marker of mitochondrial anti-apoptotic function in chondrosarcoma tumors in mice. BCL2 expression increased 1.2-fold in tumors generated with TOMM20 overexpression compared to EV control (Figure 6E) (p<0.05). Therefore, MCT1, TIGAR, mitoNEET and BCL2 expression are increased upon overexpression of TOMM20.
Discussion
Chondrosarcomas are a clinically heterogeneous group of primary bone cancers that arise in bone with endochondral ossification [42]. Conventional chondrosarcomas are typically resistant to cytotoxic chemotherapy and therefore there are no effective therapies for patients with metastatic or inoperable chondrosarcomas [11]. It is important to study the biological events that drive chondrosarcoma aggressiveness to support the investigation of novel treatments for this complex disease.
Little is known about the metabolism of chondrocytes and the metabolism of chondrosarcomas. However, studies have suggested that mitochondrial metabolism may be important since it has been shown that isocitrate dehydrogenase mutations are common in chondrosarcoma and occur in approximately 50% of cases and there have also been reports of high glutaminolysis activity, high NAD flux, activation of the mTOR pathway as well as alterations in lipid metabolism in chondrosarcoma [43] [44] [45] [18] [46] [47]. However, other studies in chondrosarcomas based on expression analysis have suggested that glycolysis might be increased or OXPHOS reduced [48] [49]. Here, we studied mitochondrial protein import as a potential mechanism of chondrosarcoma aggressiveness and therapy resistance. We studied the expression of the translocase of the outer mitochondrial membrane subunit 20 (TOMM20) in human chondrosarcomas. High TOMM20 expression has been described in human epithelial and lymphoid cancers and is associated with poor outcomes [26] [27] [28] [29] [30]. We describe for the first time that TOMM20 is highly expressed in high grade compared to low grade human chondrosarcomas, suggesting that aggressive chondrosarcomas have increased mitochondrial metabolism. Future studies will be needed to further characterize the expression in normal human cartilage due to lack of access to these tissues for this study.
TOMM20 overexpressing chondrosarcoma cells were used to determine the effects of mitochondrial metabolism on cancer aggressiveness. From an unbiased proteomics analysis, we found that TOMM20 overexpression led to changes in protein expression associated with increased cancer aggressiveness including invasiveness, proliferation, resistance to cell death, and mitochondrial metabolism. Pathways and processes revealed by the proteomic analysis were then studied in TOMM20 overexpressing chondrosarcoma cell lines. N-cadherin and phosphorylated STAT3 both of which are involved in an epithelial to mesenchymal transition, invasion, and metastasis are increased in cells overexpressing TOMM20. Also, TOMM20 overexpressing cells have higher proliferation rates with a higher proportion of cells in S phase and increased expression of cyclin D1 and c-MYC. Lactate uptake and mitochondrial metabolism is upregulated by c-MYC, MCT1 and TIGAR[17] [40] [50] [51]. We demonstrate here that TOMM20 drives expression of c-MYC, MCT1 and TIGAR. Since MCT1 can be localized in the plasma membrane and in the mitochondrial membrane future studies will be required to further elucidate the localization of MCT1 and lactate dehydrogenase B (LDH-B) to differentiate between lactate uptake into the cell or into the mitochondria [52]. Resistance to apoptosis is another common feature of aggressive cancer including chondrosarcoma [53] and here we observed that TOMM20 overexpression leads to upregulation of the anti-apoptotic protein BCL2. Our data suggests that TOMM20 overexpression induces a more aggressive phenotype in chondrosarcoma.
To assess the effect of TOMM20 overexpression on mitochondrial metabolism, OCR was measured under baseline conditions with 5mM glucose, after exposure to 10mM lactate, and after addition of 0.5μM Antimycin A and 0.5μM Rotenone. We demonstrate that TOMM20 overexpression drives increased oxygen consumption rates and therefore mitochondrial metabolism. This increase in OCR is maintained after subtraction of non-mitochondrial oxygen consumption, which was determined by treatment with the mitochondrial electron transport chain inhibitors Antimycin A and Rotenone. TOMM20 overexpressing cells also have increased OCR after lactate exposure suggesting that chondrosarcoma cells can utilize and catabolize lactate via mitochondrial OXPHOS metabolism. Future studies will need to be performed to determine if in addition to glucose and lactate other substrates are utilized for mitochondrial oxygen consumption more efficiently by TOMM20 overexpressing cells. Here we have found a relationship between the expression of TOMM20 and oxygen consumption rates, but futures studies are needed to further determine the exact link between the expression of TOMM20 and OCR.
Chondrosarcoma is resistant to chemotherapy agents although Gemcitabine, Palbociclib or anthracyclines in combination with other agents appear promising [2] [11] [13] [19] [38] [54] [55]. TOMM20 overexpression in chondrosarcoma cells induced resistance to commonly used cell cycle specific chemotherapy agents. Treatment with Palbociclib, Doxorubicin, and Gemcitabine alone or in combination in parental chondrosarcoma cells led to a reduction in proliferation rates and increased apoptosis, demonstrating activity. However, we found that TOMM20 overexpressing chondrosarcoma cells were resistant to treatment of these agents alone or in combination. Therefore, we have discovered that TOMM20 in chondrosarcoma induces resistance to cell cycle specific chemotherapy agents. Future studies will need to determine if this is due to upregulation of BCL2 and/or other antiapoptotic proteins.
TOMM20 overexpression in cells provides a growth advantage in vivo. Chondrosarcoma cells overexpressing TOMM20 injected into mice generate larger tumors than injection of control cells. TOMM20 overexpressing L2975 cells also had more significant successful engraftment in mice than control cells. Tumors generated from L2975 cells overexpressing TOMM20, also had increased phospho-Rb levels compared to the control tumors, demonstrating that TOMM20 tumors had higher rates of proliferation in vivo similar to that observed with TOMM20 overexpressing cells in vitro.
In sum, this study demonstrates that TOMM20 overexpression leads to chondrosarcoma aggressiveness. We found that TOMM20 is expressed in human chondrosarcomas and when overexpressed in chondrosarcoma cells, increases expression of markers of cancer aggressiveness and promotes tumor growth. These data suggest that TOMM20 expression promotes tumor growth by promoting mitochondrial metabolism, increasing proliferation, and inducing markers of an epithelial to mesenchymal transition (EMT), which are consistent with other studies showing an association between TOMM20 expression and outcomes in human cancers [26] [27] [28] [29] [30]. These changes in chondrosarcoma cells induced by TOMM20 overexpression also induce resistance to cell cycle specific chemotherapy agents. Therefore, these data suggest that the expression of TOMM20 in chondrosarcomas could be a key driver of drug resistance and a potential mechanism of resistance to cell cycle specific chemotherapy agents. This is consistent with previous work demonstrating that the antiapoptotic protein BCL2 drives chemotherapy resistance [16]. Future studies will need to determine if agents targeting mitochondrial metabolism or apoptosis could be effective in treating chondrosarcomas.
Highlights.
High-grade human chondrosarcomas have higher expression of translocase of the outer mitochondrial membrane complex subunit 20 (TOMM20) than low-grade tumors.
TOMM20 overexpression in chondrosarcoma cells increases markers of mitochondrial reprogramming, proliferation, and resistance to apoptosis.
TOMM20 overexpression in chondrosarcoma induces resistance to anticancer drugs.
TOMM20 overexpression in chondrosarcoma cells induces larger tumors in vivo.
Acknowledgements:
This work was supported by the National Cancer Institute (NCI) of the National Institutes of Health (NIH), under Award Number NCI 5R37CA234239, NCI P30 CA056036.
We would like to thank Rachel and David Glyn for their longstanding support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest:
The authors disclose no conflict of interest.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:
Megan E. Roche
Zhao Lin
Diana Whitaker-Menezes
Ubaldo Martinez-Outschoorn*
Atrayee Basu-Mallick*
References:
- [1].World Health Organization., International Agency for Research on Cancer, WHO classification of tumours of soft tissue and bone, 5th ed., IARC Press, Lyon, 2020. [Google Scholar]
- [2].Nazeri E, Gouran Savadkoohi M, Majidzadeh AK, Esmaeili R, Chondrosarcoma: An overview of clinical behavior, molecular mechanisms mediated drug resistance and potential therapeutic targets, Crit Rev Oncol Hematol 131 (2018) 102–109. [DOI] [PubMed] [Google Scholar]
- [3].Roessner A, Smolle M, Schoeder V, Haybaeck J, [Cartilage tumors: morphology, genetics, and current aspects of target therapy], Pathologe 41(2) (2020) 143–152. [DOI] [PubMed] [Google Scholar]
- [4].Chow WA, Chondrosarcoma: biology, genetics, and epigenetics, F1000Res 7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Whelan JS, Davis LE, Osteosarcoma, Chondrosarcoma, and Chordoma, J Clin Oncol 36(2) (2018) 188–193. [DOI] [PubMed] [Google Scholar]
- [6].van Praag Veroniek VM, Rueten-Budde AJ, Ho V, Dijkstra PDS, B. Study group, t. Soft tissue, Fiocco M, van de Sande MAJ, Incidence, outcomes and prognostic factors during 25 years of treatment of chondrosarcomas, Surg Oncol 27(3) (2018) 402–408. [DOI] [PubMed] [Google Scholar]
- [7].Evola FR, Costarella L, Pavone V, Caff G, Cannavo L, Sessa A, Avondo S, Sessa G, Biomarkers of Osteosarcoma, Chondrosarcoma, and Ewing Sarcoma, Front Pharmacol 8 (2017) 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Shinohara N, Maeda S, Yahiro Y, Sakuma D, Matsuyama K, Imamura K, Kawamura I, Setoguchi T, Ishidou Y, Nagano S, Komiya S, TGF-beta signalling and PEG10 are mutually exclusive and inhibitory in chondrosarcoma cells, Sci Rep 7(1) (2017) 13494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Qasem SA, DeYoung BR, Cartilage-forming tumors, Semin Diagn Pathol 31(1) (2014) 10–20. [DOI] [PubMed] [Google Scholar]
- [10].Speetjens FM, de Jong Y, Gelderblom H, Bovee JV, Molecular oncogenesis of chondrosarcoma: impact for targeted treatment, Curr Opin Oncol 28(4) (2016) 314–22. [DOI] [PubMed] [Google Scholar]
- [11].Italiano A, Mir O, Cioffi A, Palmerini E, Piperno-Neumann S, Perrin C, Chaigneau L, Penel N, Duffaud F, Kurtz JE, Collard O, Bertucci F, Bompas E, Le Cesne A, Maki RG, Ray Coquard I, Blay JY, Advanced chondrosarcomas: role of chemotherapy and survival, Ann Oncol 24(11) (2013) 2916–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Rey V, Menendez ST, Estupinan O, Rodriguez A, Santos L, Tornin J, Martinez-Cruzado L, Castillo D, Ordonez GR, Costilla S, Alvarez-Fernandez C, Astudillo A, Brana A, Rodriguez R, New Chondrosarcoma Cell Lines with Preserved Stem Cell Properties to Study the Genomic Drift During In Vitro/In Vivo Growth, J Clin Med 8(4) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Polychronidou G, Karavasilis V, Pollack SM, Huang PH, Lee A, Jones RL, Novel therapeutic approaches in chondrosarcoma, Future Oncol 13(7) (2017) 637–648. [DOI] [PubMed] [Google Scholar]
- [14].Yahiro Y, Maeda S, Shinohara N, Jokoji G, Sakuma D, Setoguchi T, Ishidou Y, Nagano S, Komiya S, Taniguchi N, PEG10 counteracts signaling pathways of TGF-beta and BMP to regulate growth, motility and invasion of SW1353 chondrosarcoma cells, J Bone Miner Metab 37(3) (2019) 441–454. [DOI] [PubMed] [Google Scholar]
- [15].Mery B, Espenel S, Guy JB, Rancoule C, Vallard A, Aloy MT, Rodriguez-Lafrasse C, Magne N, Biological aspects of chondrosarcoma: Leaps and hurdles, Crit Rev Oncol Hematol 126 (2018) 32–36. [DOI] [PubMed] [Google Scholar]
- [16].van Oosterwijk JG, Herpers B, Meijer D, Briaire-de Bruijn IH, Cleton-Jansen AM, Gelderblom H, van de Water B, Bovee JV, Restoration of chemosensitivity for doxorubicin and cisplatin in chondrosarcoma in vitro: BCL-2 family members cause chemoresistance, Ann Oncol 23(6) (2012) 1617–26. [DOI] [PubMed] [Google Scholar]
- [17].Martinez-Outschoorn UE, Peiris-Pages M, Pestell RG, Sotgia F, Lisanti MP, Cancer metabolism: atherapeutic perspective, Nat Rev Clin Oncol 14(1) (2017) 11–31. [DOI] [PubMed] [Google Scholar]
- [18].Peterse EFP, Niessen B, Addie RD, de Jong Y, Cleven AHG, Kruisselbrink AB, van den Akker B, Molenaar RJ, Cleton-Jansen AM, Bovee J, Targeting glutaminolysis in chondrosarcoma in context of the IDH1/2 mutation, Br J Cancer 118(8) (2018) 1074–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Provenzano S, Hindi N, Morosi C, Ghilardi M, Collini P, Casali PG, Stacchiotti S, Response of conventional chondrosarcoma to gemcitabine alone: a case report, Clin Sarcoma Res 5 (2015) 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Hanahan D, Weinberg RA, Hallmarks of cancer: the next generation, Cell 144(5) (2011) 646–74. [DOI] [PubMed] [Google Scholar]
- [21].Wallace DC, Mitochondria and cancer, Nat Rev Cancer 12(10) (2012) 685–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ahn CS, Metallo CM, Mitochondria as biosynthetic factories for cancer proliferation, Cancer Metab 3(1) (2015) 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Wurm CA, Neumann D, Lauterbach MA, Harke B, Egner A, Hell SW, Jakobs S, Nanoscale distribution of mitochondrial import receptor Tom20 is adjusted to cellular conditions and exhibits an inner-cellular gradient, Proc Natl Acad Sci U S A 108(33) (2011) 13546–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Yano M, Terada K, Mori M, Mitochondrial import receptors Tom20 and Tom22 have chaperone-like activity, J Biol Chem 279(11) (2004) 10808–13. [DOI] [PubMed] [Google Scholar]
- [25].Yano M, Kanazawa M, Terada K, Takeya M, Hoogenraad N, Mori M, Functional analysis of human mitochondrial receptor Tom20 for protein import into mitochondria, J Biol Chem 273(41) (1998) 26844–51. [DOI] [PubMed] [Google Scholar]
- [26].Curry JM, Tuluc M, Whitaker-Menezes D, Ames JA, Anantharaman A, Butera A, Leiby B, Cognetti DM, Sotgia F, Lisanti MP, Martinez-Outschoorn UE, Cancer metabolism, sternness and tumor recurrence: MCT1 and MCT4 are functional biomarkers of metabolic symbiosis in head and neck cancer, Cell Cycle 12(9) (2013) 1371–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Zhao Z, Han F, He Y, Yang S, Hua L, Wu J, Zhan W, Stromal-epithelial metabolic coupling in gastric cancer: stromal MCT4 and mitochondrial TOMM20 as poor prognostic factors, Eur J Surg Oncol 40(10) (2014) 1361–8. [DOI] [PubMed] [Google Scholar]
- [28].Gooptu M, Whitaker-Menezes D, Sprandio J, Domingo-Vidal M, Lin Z, Uppal G, Gong J, Fratamico R, Leiby B, Dulau-Florea A, Caro J, Martinez-Outschoorn U, Mitochondrial and glycolytic metabolic compartmentalization in diffuse large B-cell lymphoma, Semin Oncol 44(3) (2017) 204–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Mikkilineni L, Whitaker-Menezes D, Domingo-Vidal M, Sprandio J, Avena P, Cotzia P, Dulau-Florea A, Gong J, Uppal G, Zhan T, Leiby B, Lin Z, Pro B, Sotgia F, Lisanti MP, Martinez-Outschoorn U, Hodgkin lymphoma: A complex metabolic ecosystem with glycolytic reprogramming of the tumor microenvironment, Semin Oncol 44(3) (2017) 218–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Park SH, Lee AR, Choi K, Joung S, Yoon JB, Kim S, TOMM20 as a potential therapeutic target of colorectal cancer, BMB Rep 52(12) (2019) 712–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].R. Core-Team, R: A language and environment for statistical computing, (2020) URL: https://www.R-project.org/. [Google Scholar]
- [32].Yee TY, Vector Generalized Linear and Additive Models: With an Implementation in R, Springer, NY: (2015). [Google Scholar]
- [33].Heinze GP, logistf M: Firth’s Bias-Reduced Logistic Regression, (2018) URL: https://CRAN.R-project.org/packageMogistif. [Google Scholar]
- [34].Pinheiro JB, D.; DebRoy S; Sarkar D; R Core Team, nlme: Linear and Nonlinear Mixed Effects Models, R package version 3.1-148 (2020) URL: https://CRAN.R-proiect.org/package=nlme. [Google Scholar]
- [35].Martinez-Outschoorn UE, Lin Z, Whitaker-Menezes D, Howell A, Sotgia F, Lisanti MP, Ketone body utilization drives tumor growth and metastasis, Cell Cycle 11(21) (2012) 3964–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].LeBleu VS, O’Connell JT, Gonzalez Herrera KN, Wikman H, Pantel K, Haigis MC, de Carvalho FM, Damascena A, Domingos Chinen LT, Rocha RM, Asara JM, Kalluri R, PGC-1alpha mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis, Nat Cell Biol 16(10) (2014) 992–1003, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Caro P, Kishan AU, Norberg E, Stanley IA, Chapuy B, Ficarro SB, Polak K, Tondera D, Gounarides J, Yin H, Zhou F, Green MR, Chen L, Monti S, Marto JA, Shipp MA, Danial NN, Metabolic signatures uncover distinct targets in molecular subsets of diffuse large B cell lymphoma, Cancer Cell 22(4) (2012) 547–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Ouyang Z, Wang S, Zeng M, Li Z, Zhang Q, Wang W, Liu T, Therapeutic effect of palbociclib in chondrosarcoma: implication of cyclin-dependent kinase 4 as a potential target, Cell Commun Signal 17(1) (2019) 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Brooks GA, The Science and Translation of Lactate Shuttle Theory, Cell Metab 27(4) (2018) 757–785. [DOI] [PubMed] [Google Scholar]
- [40].Ko YH, Domingo-Vidal M, Roche M, Lin Z, Whitaker-Menezes D, Seifert E, Capparelli C, Tuluc M, Birbe RC, Tassone P, Curry JM, Navarro-Sabate A, Manzano A, Bartrons R, Caro J, Martinez-Outschoorn U, TP53-inducible Glycolysis and Apoptosis Regulator (TIGAR) Metabolically Reprograms Carcinoma and Stromal Cells in Breast Cancer, J Biol Chem 291(51) (2016) 26291–26303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Wiley SE, Murphy AN, Ross SA, van der Geer P, Dixon JE, MitoNEET is an iron-containing outer mitochondrial membrane protein that regulates oxidative capacity, Proc Natl Acad Sci U S A 104(13) (2007) 5318–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].de Andrea CE, San-Julian M, Bovee J, Integrating Morphology and Genetics in the Diagnosis of Cartilage Tumors, Surg Pathol Clin 10(3) (2017) 537–552. [DOI] [PubMed] [Google Scholar]
- [43].Salinas D, Minor CA, Carlson RP, McCutchen CN, Mumey BM, June RK, Combining Targeted Metabolomic Data with a Model of Glucose Metabolism: Toward Progress in Chondrocyte Mechanotransduction, PLoS One 12(1) (2017) e0168326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Nakagawa M, Nakatani F, Matsunaga H, Seki T, Endo M, Ogawara Y, Machida Y, Katsumoto T, Yamagata K, Hattori A, Fujita S, Aikawa Y, Ishikawa T, Soga T, Kawai A, Chuman H, Yokoyama N, Fukushima S, Yahiro K, Kimura A, Shimada E, Hirose T, Fujiwara T, Setsu N, Matsumoto Y, Iwamoto Y, Nakashima Y, Kitabayashi I, Selective inhibition of mutant IDH1 by DS-1001b ameliorates aberrant histone modifications and impairs tumor activity in chondrosarcoma, Oncogene 38(42) (2019) 6835–6849. [DOI] [PubMed] [Google Scholar]
- [45].Peterse EFP, van den Akker B, Niessen B, Oosting J, Suijker J, de Jong Y, Danen EHJ, Cleton-Jansen AM, Bovee J, NAD Synthesis Pathway Interference Is a Viable Therapeutic Strategy for Chondrosarcoma, Mol Cancer Res 15(12) (2017) 1714–1721. [DOI] [PubMed] [Google Scholar]
- [46].Addie RD, de Jong Y, Alberti G, Kruisselbrink AB, Que I, Baelde H, Bovee J, Exploration of the chondrosarcoma metabolome; the mTOR pathway as an important pro-survival pathway, J Bone Oncol 15 (2019) 100222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Zhang H, Wei Q, Tsushima H, Puviindran V, Tang YJ, Pathmanapan S, Poon R, Ramu E, Al-Jazrawe M, Wunder J, Alman BA, Intracellular cholesterol biosynthesis in enchondroma and chondrosarcoma, JCI Insight 5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Rozeman LB, Hameetman L, van Wezel T, Taminiau AH, Cleton-Jansen AM, Hogendoorn PC, Bovee JV, cDNA expression profiling of chondrosarcomas: Ollier disease resembles solitary tumours and alteration in genes coding for components of energy metabolism occurs with increasing grade, J Pathol 207(1) (2005) 61–71. [DOI] [PubMed] [Google Scholar]
- [49].Hameetman L, Rozeman LB, Lombaerts M, Oosting J, Taminiau AH, Cleton-Jansen AM, Bovee JV, Hogendoorn PC, Peripheral chondrosarcoma progression is accompanied by decreased Indian Hedgehog signalling, J Pathol 209(4) (2006) 501–11. [DOI] [PubMed] [Google Scholar]
- [50].Doherty JR, Cleveland JL, Targeting lactate metabolism for cancer therapeutics, J Clin Invest 123(9) (2013) 3685–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Wanka C, Steinbach JP, Rieger J, Tp53-induced glycolysis and apoptosis regulator (TIGAR) protects glioma cells from starvation-induced cell death by up-regulating respiration and improving cellular redox homeostasis, J Biol Chem 287(40) (2012) 33436–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Hussien R, Brooks GA, Mitochondrial and plasma membrane lactate transporter and lactate dehydrogenase isoform expression in breast cancer cell lines, Physiol Genomics 43(5) (2011) 255–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].van Oosterwijk JG, Meijer D, van Ruler MA, van den Akker BE, Oosting J, Krenacs T, Picci P, Flanagan AM, Liegl-Atzwanger B, Leithner A, Athanasou N, Daugaard S, Hogendoorn PC, Bovee JV, Screening for potential targets for therapy in mesenchymal, clear cell, and dedifferentiated chondrosarcoma reveals Bcl-2 family members and TGFbeta as potential targets, Am J Pathol 182(4) (2013) 1347–56. [DOI] [PubMed] [Google Scholar]
- [54].Zhang YX, van Oosterwijk JG, Sicinska E, Moss S, Remillard SP, van Wezel T, Buhnemann C, Hassan AB, Demetri GD, Bovee JV, Wagner AJ, Functional profiling of receptor tyrosine kinases and downstream signaling in human chondrosarcomas identifies pathways for rational targeted therapy, Clin Cancer Res 19(14) (2013) 3796–807. [DOI] [PubMed] [Google Scholar]
- [55].Monderer D, Luseau A, Bellec A, David E, Ponsolle S, Saiagh S, Bercegeay S, Piloquet P, Denis MG, Lode L, Redini F, Biger M, Heymann D, Heymann MF, Le Bot R, Gouin F, Blanchard F, New chondrosarcoma cell lines and mouse models to study the link between chondrogenesis and chemoresistance, Lab Invest 93(10) (2013) 1100–14. [DOI] [PubMed] [Google Scholar]


