Abstract
In this study, 6 infants with type-1 retinopathy of prematurity (ROP) were compared with 4 high-risk preterm neonates without any ROP but similar baseline neonatal comorbidities. The infants with type-1 ROP showed significant enrichment of Enterobacteriaceae at 28 weeks’ postmenstrual age. Several metabolic pathways, including several amino acid metabolism pathways, were enriched in gut microbiota of infants without ROP. Based on these findings, we posit a possible association between early gut microbiome profile and ROP pathogenesis. Furthermore, it is possible that absence of Enterobacteriaceae overabundance, in addition to enrichment of amino acid biosynthesis pathways, may protect against severe ROP in high-risk preterm infants.
Gut microbiota may play a role in several ocular diseases, but their role in retinopathy of prematurity (ROP) has not been explored.1 In addition to traditional risk factors for ROP, such as low birth weight (BW) and gestational age (GA), early-onset sepsis2 and poor postnatal weight gain (as a marker of insulinlike growth factor [IGF-1] deficiency) may also play a role.2 We have previously demonstrated the association of gut microbiota and brain development through the IGF-1 pathway in premature infants.3 Based on the relationship of microbiota to the IGF-1 pathway, which also plays a key role in ROP, we aimed in this study to determine whether a possible association exists between early gut microbiome composition and ROP development by comparing the gut microbiome in high-risk preterm infants with type-1 ROP and high-risk preterm neonates without any ROP.
Subjects and Methods
This study was approved by the University of Chicago Institutional Review Board and conformed to requirements of the US Health Insurance Portability and Accountability Act of 1996 and the tenets of the Declaration of Helsinki. Written consent was obtained for all patients.
As part of a larger neonatal intensive care unit study by the senior author (ECC), fecal samples were collected weekly, from birth through discharge, from high-risk preterm neonates (≤27 weeks’ GA or BW of ≤750 g) who underwent ROP screening at a single level III neonatal intensive care unit. All patients born before 37 weeks were offered inclusion in the larger microbiome study. Subjects were excluded if they had severe genetic or congenital anomalies, such as major congenital heart disease, major kidney disease, gastrointestinal, and lung or brain malformations. Subjects were stratified into two groups: (1) high-risk preterm infants with type 1 ROP and (2) high-risk preterm infants without any ROP. The following data were collected: BW, GA, delivery mode, postnatal weight gain rate, days of antibiotics, days on mechanical ventilation, severe intraventricular hemorrhage, necrotizing enterocolitis, and bronchopulmonary dysplasia. Bacterial DNA was extracted from fecal samples followed by amplification of the 16S rRNA V4 region using MiSeq sequencing (Illumina Inc, San Diego, CA). For 16S rRNA analysis, 16 million paired-end reads were joined and demultiplexed with QIIME 1.9.1, and exact sequence variants (ESV) were selected using the Deblur pipeline. Alpha and beta diversity were analyzed using QIIME 1.9.1 and phyloseq. Bacteria-encoded pathways were predicted using PICRUSt (PICRUSt- version 1.1.4; https://picrust.github.io/picrust/install.html). Significance was determined using permutational multivariate analysis of variance for alpha diversity, t tests for beta diversity, analysis of composition of microbiomes (http://scikitbio.org/docs/0.4.2/generated/generated/skbio.stats.composition.ancom.html) for ESV differences, and the Mann-Whitney U test for pathway level differences.
Results
A total of 6 infants with type 1 ROP and 4 infants without any ROP were included. Subject characteristics are described in Table 1. There was a trend toward older GA among high-risk preterm infants without ROP; otherwise neonatal comorbidities were similar at baseline.
Table 1.
Baseline characteristics of subjects without retinopathy of prematurity (ROP) and with type 1 ROP
| Factor | No ROP (n = 4) | Type 1 ROP (n = 6) | P valuea |
|---|---|---|---|
| GA at birth, weeks, median (IQR) | 25.6 (25.2–26.5) | 24.1 (24.0–24.6) | 0.052 |
| BW, g, median (IQR) | 653 (593 – 678) | 700 (560 – 794) | 0.593 |
| C-section, no. (%) | 4 (100%) | 3 (50%) | 0.200 |
| Bronchopulmonary dysplasia, no. (%) | 4 (100%) | 5 (83%) | 1.000 |
| Necrotizing enterocolitis, no. (%) | 1 (25%) | 1 (17%) | 1.000 |
| Severe intraventricular hemorrhage,b no. (%) | 0 (0%) | 2 (33%) | 0.467 |
| Thrombocytopenia,c no. (%) | 2 (67%) | 5 (83%) | 1.000 |
| Infection,d no. (%) | 1 (25%) | 3 (50%) | 0.571 |
| Post-natal weight gain (g/day), median (IQR) | 11.1 (8.8 – 13.8) | 11.4 (10.0–22.4) | 0.784 |
| Days on antibiotics before 29 weeks’ PMA, median (IQR) | 5.5 (4.0–7.5) | 12.5 (3.0–17.0) | 0.336 |
| Days on ventilator, median (IQR) | 55.5 (38–68.5) | 64.0 (55.0–85.0) | 0.336 |
BW, birth weight; GA, gestational age; IQR, interquartile range; PMA, postmenstrual age.
Fisher exact test for proportions; rank sum for difference in mean.
Grade 3 or 4.
<150 k platelets before 30 weeks’ PMA.
Culture proven or culture negative with presumed sepsis.
Neonatal fecal samples showed divergence in microbiome composition; infants with type-1 ROP showed significant enrichment of Enterobacteriaceae at 28 weeks’ postmenstrual age (PMA; P < 0.05). See Figure 1A. Pathway abundance analysis showed several metabolic pathways enriched in infants without ROP, including arginine and proline metabolism pathways, histidine metabolism pathways, valine, leucine and isoleucine degradation pathways, oxidative phosphorylation, benzoate degradation pathways, and propanoate metabolism and aminobenzoate degradation pathways (Figure 1B).
FIG 1.
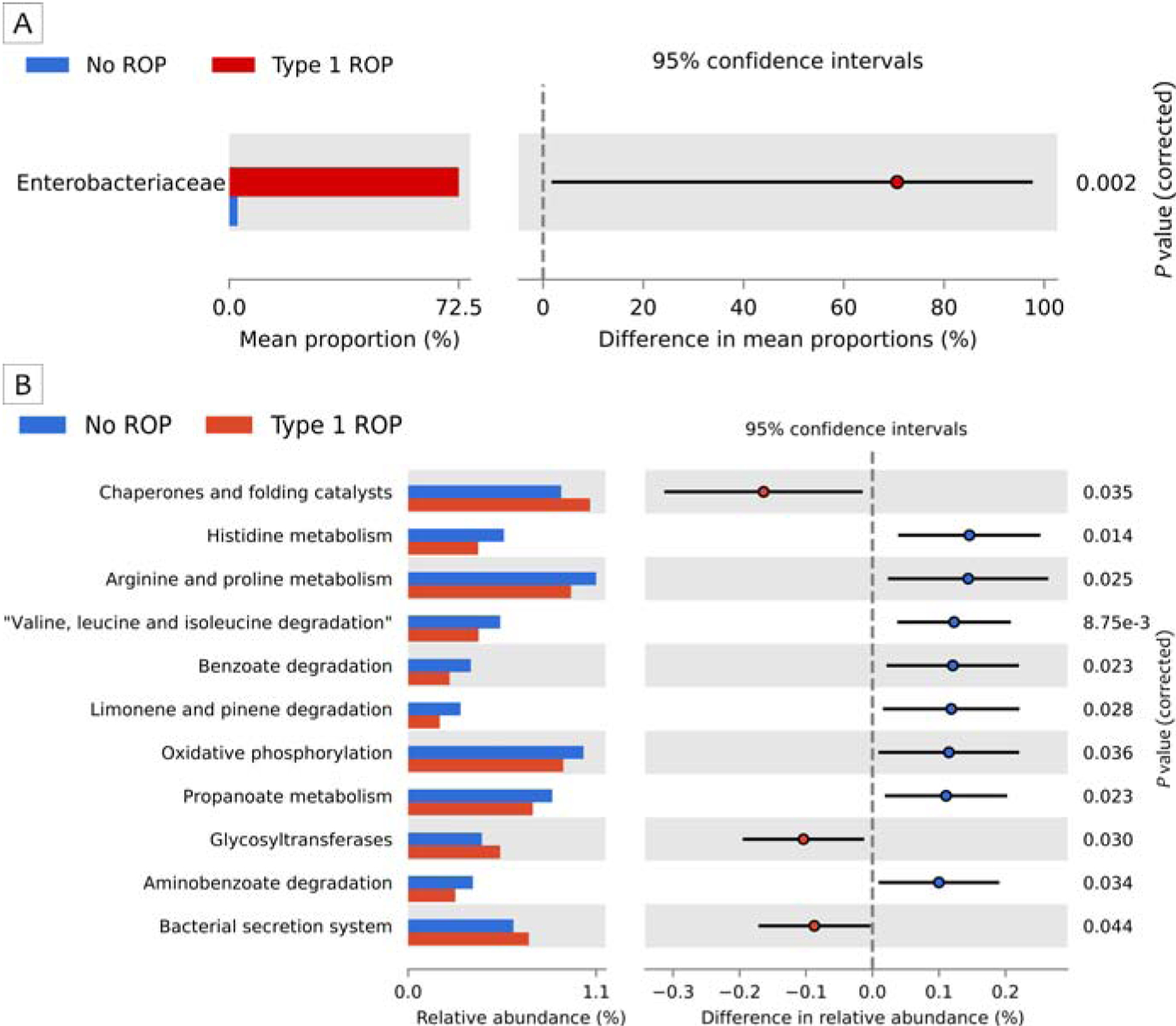
A, Significant enrichment of Enterobacteriaceae at 28 weeks’ postmenstrual age (P < 0.05; Benjamini-Hochberg [BH] FDR corrected) was noted in fecal samples of premature infants that developed type-1 retinopathy of prematurity (ROP). B, Pathway-level comparisons between the two groups using Mann-Whitney U test. BH FDR corrected P values are listed for each differentially abundant pathway.
Discussion
To our knowledge, this is the first report showing a difference in early gut microbiome composition in severe ROP and indicates metabolic pathways that could be protective. Overabundance of Enterobacteriaceae as well as arginine biosynthesis pathways have been associated with macular degeneration, another neovascular retinal disease driven by vascular endothelial growth factor (VEGF).4 Because microbiota have been shown to regulate IGF-1 expression,3 we hypothesize that early gut microbiome composition could affect retinal vascular development during phase 1 of ROP marked by IGF-1 deficiency, which plays a key role in VEGF-driven angiogenesis. Of note, Enterobacteriaceea species, such as Escherichia Coli, have been shown to affect VEGF expression.5 High-throughput RNA sequencing has revealed that mouse retina IGF-1, IGF-1 receptor, VEGF, and VEGF-receptor gene expression may be regulated by microbiome status (unpublished data, D. Skondra). The microbiome could possibly affect ROP via dysregulation of IGF or VEGF. The fact that amino acids such as argininin/proline have been shown to control IGF-1 expression could explain the protective role of these pathways in high-risk infants without ROP.6 Disruption of early gut microbiome composition in bacteremia or early-onset sepsis may also contribute to increased ROP risk.2
Although there was a statistically significant difference in GA at baseline (25.6 vs 24.1 months), both groups remained very high risk for severe ROP. Because gestational age may also affect the microbiome, this represents an important study limitation, and larger studies are needed to support our results. Nonetheless, it is possible that absence of Enterobacteriaceae overabundance in addition to enrichment of amino acid biosynthesis pathways may protect against severe ROP in high-risk preterm infants. Early gut microbiome profiles may play a role in ROP pathogenesis and could be target for future intervention, but further studies are needed to elucidate their role in ROP and the pathways involved.
Funding support:
National Institutes of Health grants HD083481 (EC) and UG3OD0232281 (EC); Duchossois Family Institute (EC).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: Bree Andrews is a cofounder of Preeme+You.
References
- 1.Baim A, Movahedah A, Farooq A, Skondra D. The microbiome and ophthalmic disease. Exp Biol Med (Maywood) 2019;244:419–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim SJ, Port AD, Swan R, Campbell JP, Chan RVP, Chiang MF. Retinopathy of prematurity: a review of risk factors and their clinical significance. Surv Ophthalmol 2018;63:618–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu J, Lu L, Yu Y, Cluette-Brown J, Martin CR, Claud EC. Effects of intestinal microbiota on brain development in humanized gnotobiotic mice. Sci Rep 2018;8:5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ziknernager MS, Zysset-Burri DC, Keller I, et al. Association of the intestinal microbiome with the development of neovascular age-related macular degeneration. Sci Rep 2017;7:40826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cane G, Moal VL, Pagès G, Servin AL, Hofman P, Vouret-Craviari V. Up-regulation of intestinal vascular endothelial growth factor by afa/dr diffusely adhering Escherichia coli. PLoS One 2007;2:e1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brameld JM, Gilour RS, Buttery PJ. Glucose and amino acids interact with hormones to control expression of insulin-like growth factor-i and growth hormone receptor mRNA in cultured pig hepatocytes J. Nutr 1999;129:1298–306. [DOI] [PubMed] [Google Scholar]


