Abstract
Structurally and functionally active synapses are essential for neurotransmission and for maintaining normal synaptic and cognitive functions. Researchers have found that synaptic dysfunction is associated with the onset and progression of neurodegenerative diseases, such as Alzheimer’s disease (AD), and synaptic dysfunction is even one of the main physiological hallmarks of AD. MiRNAs are present in small, subcellular compartments of the neuron such as neural dendrites, synaptic vesicles, and synaptosomes are known as synaptic miRNAs. Synaptic miRNAs involved in governing multiple synaptic functions that lead to healthy brain functioning and synaptic activity. However, the precise role of synaptic miRNAs has not been determined in AD progression. This review emphasizes the presence of miRNAs at the synapse, synaptic compartments and roles of miRNAs in multiple synaptic functions. We focused on synaptic miRNAs alteration in AD, and how the modulation of miRNAs effect the synaptic functions in AD. We also discussed the impact of synaptic miRNAs in AD progression concerning the synaptic ATP production, mitochondrial function, and synaptic activity.
Keywords: MicroRNAs, Synapse, synaptosome, Alzheimer’s disease, Synaptic activity
1. Introduction
Currently, 5.8 million people in the United States have been diagnosed with Alzheimer’s disease (AD)-related dementia; this number includes an estimated over 5.6 million people age 65 years and older (Alzheimer’s Disease Facts and Figures, 2019) [1]. In the United States, one in 10 people who are age 65 years and older has AD (Alzheimer’s Disease Facts and Figures, 2019) [1]. AD-related dementia has huge economic consequences, with the total medical costs of dementia worldwide for 2018 estimated at $1 trillion. In addition to dementia, AD is associated with the loss of synapses, synaptic dysfunction, mitochondrial structural and functional abnormalities, inflammatory responses, extracellular neuritic plaques, and intracellular neurofibrillary tangles (NFTs) [2-6]. Despite the tremendous progress that has been made in better understanding of synaptic abnormalities and AD pathogenesis, there are still no detectable markers, drugs, or agents that can prevent AD or slow its progression.
Increasingly severe synaptic dysfunction is the main cellular and physiological event in AD [5-7]. Synaptic dysfunction is an early cellular change in AD progression [5-7]. In a non-diseased state, synapses transmit neural impulses in neurons via neurotransmitters (e.g., acetylcholine or norepinephrine). Synapses and their components are critical for maintaining healthy synaptic function and neurotransmission in the brain [8]. Neurotransmitters are diffuse across the synaptic cleft and bind with specific receptors on the postsynaptic membrane of postsynaptic cells [9]. Synaptic dysfunction and reduced synaptic activity are caused by amyloid-beta (Aβ) and phosphorylated tau (p-tau) toxicities, which are the main pathophysiological factors involved in AD progression [10].
Researchers have been investigating different aspects of neurological pathology to prevent, delay, or cure neurological diseases. Recently, several groups are investigating the impact of functional and dysfunctional microRNAs (miRNAs) on neurons [11]. In particular, much miRNA research is attempting to block or to inhibit miRNA to determine the impact of the miRNA dysregulation on the expression of key genes (APP, BACE1, tau, PSEN1/2, and APOE) known to be associated with neurological diseases, such as AD [11,12]. Slowly, the research is revealing information about synapse-associated miRNAs and their roles in regulating local synaptic protein levels in AD progression [13].
It is known that miRNAs are important regulators of several synaptic genes (e.g., synapsin, synaptophysin, PSD95) [14], but the specific roles of miRNAs localized at synapse in AD neuron have not been identified. Identification of synaptosome-associated miRNAs in AD is very important in better understanding the role of synaptic miRNAs in AD development and progression. Besides, molecular connections of synaptosomal miRNAs need to be studied in Aβ and p-tau induced toxicities in synapses to determine synaptic functions of miRNAs. Because miRNAs are involved in several AD-related pathways and are known to regulate the AD pathogenesis [11-14]. MiRNAs research elucidating functions and dysfunctions in synapses will provide a valuable target for new therapeutics to prevent and/or delay AD symptoms.
Past studies have shown that synaptic plasticity, dendritic spine growth, and learning and memory are critically dependent upon the regulation of specific protein synthesis near or within the synapse [14-18]. The emerging evidence suggests that miRNAs play a critical role in regulating these as-yet-unidentified neuronal and synaptic proteins [14], given that many miRNAs and their precursors have been found to exist in synaptic fractions [15-18]. Studies have also shown that several specific miRNAs play important roles in regulating synaptic plasticity, including synaptogenesis, alteration of synaptic morphology, and modification of synaptic function [19-22]. MiRNA-mediated regulation of synaptic function, in particular, is not only responsible for synapse development and synaptic activity, but it also involved in the pathophysiology of plasticity-related diseases, such as Alzheimer’s, frontotemporal dementia (FTD) and other dementias.
The purpose of the article is to assess the role of functional/dysfunctional synapse localized miRNAs in AD progression. We begin our review, looking into the presence of small RNAs and miRNAs at the synapse, the synaptic vesicles, and synaptosomes and accessed their roles in regulating synaptic activity in neurodegenerative diseases and AD. In this article, we will also discuss research that has focused on synaptic miRNAs as possible therapeutic agents that capable of preventing or delaying AD onset or progression.
2. MicroRNAs
MiRNAs, small RNA molecules (about 22-25 nucleotide long), are expressed in humans, plants, fungi, bacteria, and viruses [23,24]. In humans, miRNAs are involved in all developmental and pathological processes via regulating the gene expression mainly by binding at 3’ untranslated region (UTR) of mRNA in a sequence-specific manner [25-29]. Some miRNA candidates are tissue-specific and are localized at certain cellular niches, while others are expressed throughout the human body [29,30]. MiRNAs are synthesized in the nucleus through the nuclear genome and modulate mRNA and protein levels of host cells. Under several circumstances, miRNAs (so-called circulating miRNAs) are released from cells and enter extracellular spaces where they participate in the governing of intra-cellular communication [31,32]. Research into miRNAs as potential biomarkers have focused on cancer, aging, diabetes, and such neurological conditions as Alzheimer’s, Huntington’s, Parkinson’s, Multiple sclerosis, and Ischemic stroke [13,25,33-42]. In AD patients, miRNA expression was found to be deregulated in the brain, extracellular fluid, such as cerebral spinal fluid, blood, plasma, and serum [23-25,27].
MiRNAs target approximately 30-60% of mammalian proteins and function in a variety of biological processes, including cell proliferation, differentiation, migration, and apoptosis [43-47]. The hundreds of miRNAs are expressed in the brain, which suggests that they could be an important regulator for the central nervous system (CNS) functioning [47].
Impairment of miRNA biogenesis processing is known to disrupt the development of the nervous system, in terms of the capability of the nervous system to assist in differentiation, learning, memory, and neuronal survival [43-47].
MiRNAs are present in small, subcellular compartments of the cell, including the rough endoplasmic reticulum, processing (P)-bodies, stress granules in the trans-Golgi network, early and late endosomes, multivesicular bodies, lysosomes, mitochondria, and synaptosomes, where they serve to micromanage the cells [28]. Each miRNA candidate usually interact/bind with a few hundred different mRNAs/genes [44] that it targets, although not every miRNA-mRNA interaction is necessarily of any physiological relevance [45]. Another feature of miRNAs is that not all miRNAs act as on-off switches, but rather they fine-tune the gene expression profiles [43], which usually occurs in the absence of mRNA degradation, a scenario that fits very well with the regulation of local dendritic mRNAs/proteins during storage and/or activation at the synapse [48,49].
In addition to miRNAs, circular RNAs, piwi-interacting RNAs (piRNAs), long non-coding RNAs (ncRNA) and other types of ncRNAs are also found to be implicated in AD [50]. Therefore, multiple forms of deregulated and multi-tasking ncRNAs are involved in the core pathophysiological mechanisms underlying AD. Altered expression of ncRNAs in the brain and peripheral circulation showed their biomarkers and therapeutic potential in AD [50].
3. Synapse
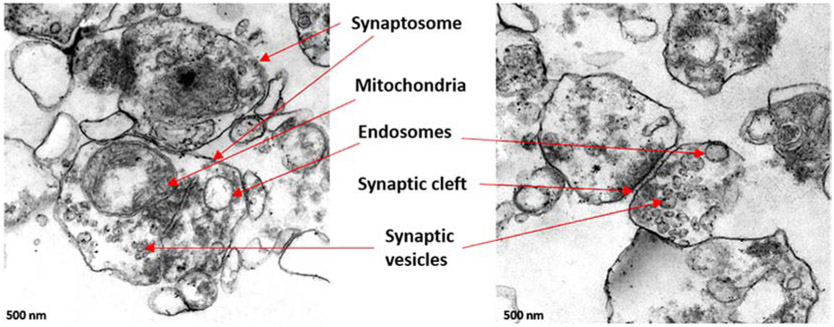
Synapse, the main compartments of the neuron, are essential and important to maintain healthy neurotransmission and cognitive function [51]. The synaptic dysfunction and poor prepost synaptic activities can lead to the progression of synaptic and neuron degeneration [52] in AD and neurological diseases [53]. Depending on signal transmission mechanisms, there are two basic types of synapses are chemical and electrical [54]. The human brain consists of hundreds of neuron types and possibly thousands of different kinds of the synapse. The total number of chemical synapses in the neocortex exceeds more than 100 trillion [54]. Therefore, it is a huge task to understand the molecular complexity and diversity of synaptic networks in the brain. The electron microscopic features of synapses are composed of synaptic vesicles, mitochondria, microsomes, lysosomes, postsynaptic density (PSD), and plasma membranes [52]. We extracted the synaptosomal fraction from the cortex of healthy postmortem brain tissue. The transmission electron microscopy showed the different components of the active synapse (Figure 1) (unpublished data).
Figure 1.
Synaptic organization in the human brain: Synaptosomes were extracted from the frontal cortex of postmortem brain. Transmission Electron Microscopy images showed the synapse and synaptosome components, the synaptic vesicles, mitochondria, endosomes and synaptic cleft.
4. Synaptosomes
Synaptosomes were first prepared in the late 1950s by Gray and Whittaker [55]. Gray and Whittaker (1962), studied the synaptic structure and functional components of the synapse and identified the major neurotransmitters and their uptake mechanisms [55,56]. Synaptosomes are predominately comprised of axon terminals with adherent postsynaptic densities, also called synaptoneurosomes, which are enriched with dendritic spines [15]. Synaptosomes are generated during the extraction process, which is the process of nerve terminals being torn from axons and dendrites and being resealed into spherical vesicles (around 0.5-1.5 μM in diameter) [57]. These detached and resealed synaptic terminals remain metabolically and enzymatically active. Synaptosomes have become important model systems for studying the functions of human synapses because they are a much more accessible system for in vivo than in ex vivo brain slices or primary neuronal cultures [52]. Human synaptosomes can be used to study neurotransmission mechanisms across synapses, metabolic changes within synapses, protein synthesis alterations in synapses, protein localization between synapses, and post-translational modification of neurons.
5. Messenger RNAs at dendrites
Messenger RNAs have been located on nerve terminals and dendrites and are associated with synapse-associated polyribosome complexes (SPRCs) in granule cells of the dentate gyrus [58]. These complexes and their associated membranous cisterns are localized in distal processes, beneath the postsynaptic sites on the dendrites of the CNS. The numerous research groups support the possibility that synaptic activity might be regulated by local protein synthesis and it has been confirmed in the presence of SPRCs [58,59]. These findings were further verified by in situ hybridization studies that showed most of the miRNA populations of the brain were present exclusively in the cell body [58]. However, a select population of miRNAs was also present in the dendritic compartment of vertebrate neurons. This population was found to be present in neuronal processes, suggesting that synaptic miRNAs play a critical role in synaptic functions [58].
6. Synaptic vesicles and small RNAs
The presence of small RNAs populations was discovered in the synaptic vesicles (SVs). SVs are small neuronal presynaptic organelles that carry the neurotransmitters and release them to the synaptic cleft. Before, it was presumed that synaptic vesicles contain the neurotransmitters only. In addition to the regular neurotransmitters, Li et al (2015), identified the presence of small ribonucleic acids (sRNAs), in the synaptic vesicles isolated from the electric organ of Torpedo californica [17]. Most of the sRNAs are transfer RNA molecules termed as tRNA fragments [17]. Not even Torpedo californica, but the SVs of the mouse brain contains abundant levels of sRNAs, including transfer RNA fragments (trfRNAs) and miRNAs [17]. These findings suggest that SV sRNAs are conserved and are possibly involved in transcriptional and translational regulation of local synaptic proteins. Besides, the release of neurotransmitters, now it is exciting to know whether SV contained sRNAs regulate the synaptic activity or change dendritic excitability. The SVs specific trfRNAs and miRNAs, both could directly regulate local protein synthesis and could have broad implications on neurotransmission and synaptic activity.
7. Neural microRNAs
Recent studies into neuronal miRNAs have identified a widespread role for miRNAs in developmental processes associated with the brain, such as synaptic development, dendritogenesis, synaptic formation, and synaptic maturation [60]. A key feature of synapse-associated miRNA includes an ability to regulate local mRNA translation in the synaptodendritic compartment and to modulate synaptic proteins and neuronal activity. Most studies of neuronal miRNAs were centered on neuronal development and differentiation [61]. However, the role of neural miRNAs in post-natal neuronal development is just beginning to emerge. A large pool of miRNA candidates is expressed in post-mitotic neurons during synapse development, and most of these miRNAs are part of translation regulatory complexes [62]. This type of complex function(s) to provide neural miRNA-regulated developmental pathways and to control the expression of several synapses -relevant proteins. The neuronal dendrites and synaptic fractions are enriched with neurotransmitter proteins, mature miRNAs, precursor miRNAs, and protein components of miRNA-mediated silencing complexes [15-18]. Several miRNAs have been classified as neural miRNAs based on their brain-specific origin and their significant role in brain development and function. MiR-134 and miR-138 are the two potent neural miRNAs that played important roles in synapse development and synaptic activity [60,63].
8. MicroRNAs and synaptic function
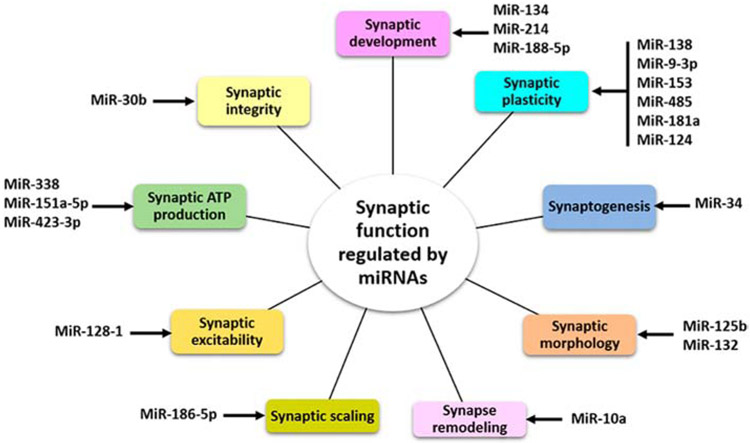
MiRNAs have been found to regulate the translation and degradation of target genes in the CNS. The increasing evidence suggests that quite a few specific synaptic miRNAs play important roles in various aspects of synaptic activity, including synaptic development, synaptic plasticity, synaptogenesis, synaptic morphology, synaptic remodeling, synaptic scaling, synaptic excitability, synaptic ATP production and synaptic integrity (Figure 2).
Figure 2.
Potential synaptic miRNAs and synaptic function. MiRNAs regulate the several process at synapse necessary for neural development and synaptic activity.
Synaptic plasticity is the biological process by which specific patterns of synaptic activity result in changes in synaptic strength and is thought to contribute to learning and memory. Both pre-synaptic and post-synaptic mechanisms can contribute to the expression of synaptic plasticity. MiRNA-mediated regulation of synaptic plasticity, as studied in miR-132, miR-134, miR-9-3p, miR-138, miR-125a, and miR-125b, is not only responsible for synaptic development and function but is also responsible for the pathophysiology of plasticity-related diseases, such as AD and FTD [14]. The synaptic dysfunction has been hypothesized to be involved in impaired synaptic plasticity, including synaptogenesis, synaptic morphology alteration, and synaptic functional modification of many neurological diseases including AD, Parkinson’s disease, and schizophrenia [60,61,64,65].
8.1. MicroRNA-134:
MiR-134 is a brain-specific miRNA, localized to the synaptodendritic compartment of rat’s hippocampal neurons. MiR-134 targets the mRNA, encoding a protein kinase Limk1 and negatively regulates the size of dendritic spines and in postsynaptic sites of excitatory synaptic transmission. The inhibition of Limk1 leads to control the spine development. The extracellular stimulus of neurons, such as via stimulation of a brain-derived neurotrophic factor, relieves miR-134 inhibition of Limk1 translation and induces synaptic development, maturation, and/or plasticity [60]. Thus, miR-134 seems to be very critical for synaptic development.
8.2. MicroRNA-138:
MiR-138 is another brain-specific miRNA that regulates the translation of local proteins in the brain by modulating Lypla1 [63]. Lypla1 is a kind of lysophospholipases enzyme that acts on the cell membranes to regulate the formation of multifunctional lysophospholipids. MiR-138 targets the 3’ UTR of Lypla1 through a conserved 8-mer seed match sequence and reduces the Lypla1 protein levels. However, endogenous inhibition of miR-138 by lock nucleic acid (LNA) in hippocampal neurons has led to a significant increase in endogenous Lypla1 levels. Interestingly, LNA-mediated inhibition of miR-138 did not change the endogenous Lypla1 mRNA levels suggesting that miR-138 regulates Lypla1 at the protein level rather than at the level of mRNA decay [63]. These results suggest a unifying mechanism for regulation of local protein translation by miRNA during synaptic plasticity.
8.3. MicroRNA-34:
The other conserved miRNA is miR-34, which regulates the synaptogenesis in both presynaptic and postsynaptic compartments [66]. The study conducted on Drosophila larval neuromuscular junction, to screen for novel regulators of synapse morphogenesis. The miR-34 mutant drosophila displays synaptic phenotypes and distinct downstream mechanisms in the presynaptic and postsynaptic compartments. The conserved downstream targets for miR-34 are junctional receptor CNTNAP4/Neurexin-IV (Nrx-IV) and the membrane cytoskeletal effector Adducin/Huli tai shao (Hts). MiR-34 restricted the synaptic expression of these proteins. Further, manipulation of miR-34, Nrx-IV or Hts-M alters function in motor neurons. Modulation of presynaptic miR-34 inhibits Nrx-IV, and modulation of postsynaptic miR-34 inhibits Hts proteins to regulate the initiation of bouton formation from presynaptic terminals [66]. MiR-34c, was also reported to be upregulated in the AD patients, AD mouse model and hippocampal neurons after Aβ treatment [67]. Hu et al (2015), unveiled the role of miR-34c in Aβ regulation and synaptic activity by targeting VAMP2 gene in AD. The suppression of VAMP2 level by miR-34c protects the neurons form Aβ induced synaptic failure as well as learning and memory deficits. In this way, miR-34c-VAMP2-Aβ pathway provides a novel underlying epigenetic mechanism to control Aβ toxicity in AD [67].
8.4. MicroRNA-125b:
Edbauer et al (2010), identified the roles of miR-125b and miR-132 in regulation of synaptic structure and function through interaction with fragile X mental retardation protein (FMRP) [68]. The action point of miR-125b and miR-132 are dendritic spine morphology and synaptic physiology in hippocampal neurons. MiR-125b target the NMDA receptor subunit NR2A, which is associated with FMRP in the brain. FMRP, miR-125b, and Argonaute 1 negatively regulate the NR2A expression and it mostly depends in part on miR-125b [68]. These observations have implications in miR-125b and NMDA levels in fragile X syndrome where plasticity is altered.
8.5. MicroRNA-9-3p:
MiR-9-3p is another critical miRNA that regulate synaptic plasticity, learning and memory [69]. MiR-9-3p is a brain-enriched miRNA known for brain developmental process and implicated in several neurological disorders. Inhibition of miR-9-3p, impaired long-term potentiation (LTP) in hippocampal neuron, resulting the deficits in learning and memory capacity. Furthermore, inhibition of miR-9-3p increased the expression of the LTP-related genes Dmd and SAP97, and expression of these LTP-related genes are negatively correlated with miR-9-3p levels [69]. These observations suggest that miR-9-3p plays important roles in regulation of synaptic plasticity and hippocampus-dependent memory.
8.6. MicroRNA-124:
MiR-124 also found to be required for maintaining homeostatic synaptic plasticity and neuronal activity [70]. GluA2-lacking, calcium permeable AMPARs (CP-AMPARs) biogenesis plays a crucial role in the homeostatic response and neuronal activity. Hou et al (2015), identified the role of miR-124, in biogenesis of CP-AMPARs and to maintain homeostatic plasticity [70]. MiR-124 was found to target 3’UTR of GluA2 and suppress their expression. Suppression of GluA2 was leads to the formation and induction of CP-AMPARs. Inhibition of miR-124 expression is stops the homeostatic response, whereas overexpression of miR-124 induces earlier homeostatic plasticity. Further, transcription of miR-124 was controlled by an inhibitory transcription factor EVI1, which work in association with the deacetylase HDAC1 [70]. This finding unveiled a cellular cascade in which EVI1/HDAC enhances miR-124 expression and high level of miR-124 induces the formation of CP-AMPARs and subsequently homeostatic synaptic plasticity.
8.7. MicroRNA-186-5p:
MiR-186-5p was also play a major role in regulation of GluA2 surface level and synaptic scaling in hippocampal neurons [71]. Synaptic scaling is a fundamental mechanism for neurons to maintain their normal activity within a dynamic range despite ongoing alterations in excitability and activity. The blockade of glutamate receptors- AMPA and NMDA induces changes in the neuronal mRNA and miRNA levels in the hippocampal neurons that leads to synaptic upscaling. Study by Silva et al (2019), found that miR-186-5p reduces the synaptic activity by targeting the 3′UTR of the AMPA receptor GluA2 subunit mRNA [71]. High level of miR-186 reduces GluA2 surface levels and induces synaptic expression of GluA2-lacking AMPA receptors, as a result synaptic scaling is blocked. However, by the inhibition of miR-186-5p, the surface levels of GluA2 is increased which leads to induction of AMPA receptor-mediated currents, and synaptic scaling induced by synaptic inactivity [71]. Therefore, several potential miRNAs have been explored that are involved and/or regulates crucial synaptic functions in disease process and healthy states.
9. Synaptosomal microRNAs
MiRNAs present throughout the cells, and some miRNAs are localized to cellular organelles. Subcellular compartmentalization and localization of miRNAs, miRNA induced silencing complex, and target mRNA have been observed to localize in multiple subcellular compartments, including rough endoplasmic reticulum, P-bodies, stress granules in the trans-Golgi network, early and late endosomes, multivesicular bodies, lysosomes, mitochondria, and the nucleus [28]. More recently, several studies identified miRNAs at the synapse and in synaptosomal fractions and their important roles in the regulation of local protein synthesis [15-18].
9.1. MicroRNA-146 and microRNA-125a.
Several miRNAs have been identified in the synaptoneurosome fractions of the cortex and hippocampus of C57B1/6 mice [15]. The top ones are- miR-429, miR-146, miR-100, miR-125a, miR-126, miR-153, and miR-301. The miR-146 and miR-125a were found predominantly in the synaptosome fraction. Precursor miRNAs (pre-miRNAs) and their corresponding mature miRNAs showed high synaptic enrichment. The mature miR-146 and miR-125a, exhibited significantly higher synaptic enrichment, compared to their pre-miRNAs [15]. These findings support the hypothesis that mature miRNAs expressed in synaptic fractions are likely to be generated from local processing of their precursors’ miRNAs.
Xu et al (2013), also found the several miRNAs that were secreted by synaptosomes via exocytosis. MiR-29a, miR-99a, and miR-125a were significantly elevated in synaptosome supernatants after membrane depolarization of neurons of C57Bl/6 mice [16]. Among them, miR- 125a was the most predominant. Within the synaptosomes, miR-125a is potentially bound to the Ago2 complex, target the key synaptic protein PSD95, and reduces their level in the synaptosome [16]. These findings suggest that the synapse miRNAs are assembled in the RNA-induced silencing complex, resulting in the suppression and/or degradation of target local mRNAs at synapse.
9.2. MicroRNA-128-1 and microRNA-99a.
The synaptic vesicles extracted from mouse CNS that contains several small RNAs, transfer-RNA, and miRNAs (miR-128-1, miR-99a, miR-100, miR-22, and miR-127) included high copy numbers of miR-128-1 and miR-99a relative to other miRNAs [17]. MiR-128 is highly enriched miRNA in the adult mouse and human brain that controls motor behavior and excitability [72]. MiR-128 regulate neuronal excitability by controlling the extracellular signal-regulated kinase ERK2 network [72]. The members of the miR-99 family (miR-99a) have been shown to coenrich with polyribosomes in mammalian neurons and to regulate the mammalian target of the rapamycin (mTOR) pathway [73]. Other than miR-128-1 and miR-99a, several other miRNAs (miR-124a-3p, miR-136-5p, and miR-376a-3p) were also abundantly expressed within synaptoneurosomes isolated from the prion-infected forebrain of animal model [18]. These synaptoneurosomes associated or synaptosome-specific miRNAs (termed synapto-miRs) could be very important in AD research and other neurodegenerative diseases. The potential synaptic miRNAs and their main functions are summarized in Table 1.
Table 1-.
Summary of synaptic miRNAs, their location, target genes and molecular functions.
| Synaptic miRNAs |
Location | Functions | Target genes | References |
|---|---|---|---|---|
| MiR-134 | Synaptodendritic compartments | Synaptic development, synaptic maturation, and synaptic plasticity | Limk1 | [60] |
| MiR-138 | Brain | Local protein translation at the synapse and synaptic plasticity. | Lypla1 | [63] |
| MiR-34 | Brain, Lungs | Synaptogenesis/Synapse morphology, dendritic growth | CNTNAP4/Neurexin- IV (Nrx-IV), VAMP2 | [66,67,92] |
| MiR-125b and MiR- 132 | Brain and other tissues | Synaptic structure and function | NMDA receptor subunit NR2A | [68] |
| MiR-9-3p | Brain | Synaptic plasticity, learning, memory, dendritic growth | LTP-related genes Dmd and SAP97 | [69,91] |
| MiR-124 | Brain and other tissues | Maintaining homeostatic synaptic plasticity and neuronal activity and memory loss | GluA2, PTPN1 | [19, 70] |
| MiR-186-5p | Brain and other tissues | Synaptic scaling | AMPA receptor GluA2 subunit | [71] |
| MiR-146 and MiR- 125a | Synaptic fraction Synaptosome | Synaptic plasticity, Suppression and/or degradation of target mRNAs at local synapses | PSD95 | [15,16] |
| MiR-128-1 | Neuron (Synaptic vesicles) | Regulates motor behavior by modulating neuronal signaling networks and excitability. | Extracellular signal-regulated kinase ERK2 network | [17,18,72] |
| MiR-99a | Polyribosomes in mammalian neurons | Regulate the mammalian target of the rapamycin (mTOR) pathway | [17,18,73] | |
| MiR-338 | Brain | Mitochondrial activity and ATP production | COXIV | [86] |
| MiR-151a- 5p | Brain and other tissues | Cellular respiration and ATP production | Cytochrome b (Cytb) | [88] |
| MiR-423-3p | Brain and other tissues | ATP production and energy metabolism | Cox6a2 | [89] |
| MiR-214 | Various tissues | Neuronal dendritic development | Qki | [90] |
| MiR-125b | Brain | Synaptic deficits, neurotrophic deficits, and astrogliosis | Synapsin-2 (SYN-2) and 15-lipoxygenase (15-LOX) | [94] |
| MiR-30b | Brain and other tissues | Synaptic integrity in AD | Ephrin type-B receptor 2 (ephB2), Sirtuin1 (sirt1), and Glutamate ionotropic receptor AMPA type subunit 2 (GluA2). | [20] |
| MiR-485 | Brain and other tissues | Homeostatic synaptic plasticity in AD | SV2A | [98] |
| MiR-153 | Brain and other tissues | Synaptic trafficking, presynaptic plasticity in AD | Stx1a, Snap25, Vamp2, Syt1 and synapsin 1 genes | [93, 99,100] |
| MiR-101b | Brain and other tissues | Cognitive impairment | AMPK, APP, RanBP9, Rab5 | [101,102] |
| MiR-132 | Brain | Neuron morphogenesis and plasticity | C1q, MeCP2 | [103,104] |
| MiR-181a | Brain | Synaptic plasticity and memory | Translin, GluA2 | [105] |
| MiR-10a | Brain and other tissues | Synapse remodeling | BDNF-TrkB | [106] |
| MiR-188-5p | Brain and other tissues | Dendritic spine density, Synaptic dysfunction | Nrp2 | [107] |
| MiR-574 | Brain and other tissues | Cognitive impairment | Nrn1 | [108] |
10. Synaptosome, microRNAs and Alzheimer’s disease
We have discussed the impact of functional and dysfunctional miRNAs in AD neurons [11-13]. Studies showed that miRNA plays crucial roles in synapse function and several aspects of synaptic activity in AD [19-22]. However, the function of synapse-enriched miRNAs is not determined in great detail in AD, although miRNAs, enriched at synaptic compartments, directly regulate local protein synthesis (synaptic proteins) and play a crucial role in neurotransmission [15-18]. For a long time, synaptosomes were prepared from the postmortem brains of AD patients and studied for AD-associated deficits such as neurotransmission, including acetylcholine, glutamate, and c-aminobutyric acid (GABA) systems [52,74,75]. These synaptosome studies identified decreased levels of neprilysin in AD patients [52]. In other studies, synaptosomes were prepared from the frontal cortical region of AD postmortem brains to study Aβ aggregates in a synaptosomal fraction [74,75]. Therefore, the study on the synaptosomal miRNAs is also needed in AD.
11. Alzheimer’s: a synaptic failure disease
It is well accepted that AD is a synaptic failure disease [76]. In addition, AD-related cognitive decline has been significantly associated with synapse loss [76,77]. A few losses of synapses, malformation of only a few synapses, or the dysfunction of just a few synapses could initiate AD onset [51,76]. We need to develop the innovative methods that could reveal brain structural changes alongside of brain functional changes.
Synapse as the initial target of synaptic degeneration in Alzheimer’s disease
Since the mid-1970s, neurochemical analyses of AD brain tissue revealed that the proteins that generate and metabolize acetylcholine neurotransmitters are depleted in AD [78]. The depleted level of acetylcholine was associated with the progression of AD and neuronal dysfunction [79]. Similarly, numerous other neurotransmitters (including corticotroping-releasing factor, somatostatin, GABA, and serotonin) were decreased during the AD progression. The dysfunction of cholinergic lesions and glutamatergic synapses are considered as early symptoms of AD onset [80,81]. In addition to neurotransmitter depletion, the alteration of other biochemical and morphological indicators also represents the initial attack on synapses in AD progression [80,81]. The study revealed a total of 25 to 35% decrease in the numerical density of synapses in AD cortexes, and a total of 15 to 35% decrease in the number of synapses per cortical neurons [82]. Further, postmortem examination of AD brains revealed that cognitive impairment correlates better with synaptic loss than with the number of extracellular neuritic plaques or NFTs in AD [83].
12. MicroRNAs and synaptic ATP production
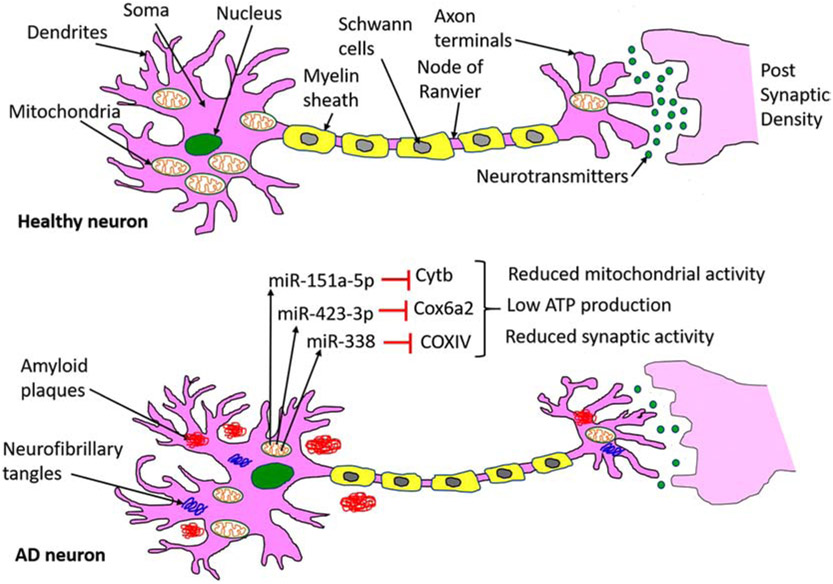
Synaptic energy or synaptic adenosine triphosphate (ATP) is critical to perform several synaptic functions, including synaptic vesicle formation, neurotransmitter release, neurite outgrowth in a healthy state, but it is defective in AD and other neurological diseases [51]. Synaptic mitochondria (mt) are dysfunctional due to increased oxidative stress that is caused by mtDNA alterations [84,85]. Defective and dysfunctional synaptic mitochondria produce low ATP, leading to synaptic degeneration in neurons [51]. Recent and emerging evidence suggests that mt miRNAs, particularly synaptic mt miRNAs play a large role in regulating synaptic ATP [62,86,87]. Figure 3 showed the action mechanism of mt miRNAs on the mitochondrial function, ATP production, and synaptic activity. A heterogeneous population of mRNAs is present in the axons and nerve terminals of primary sympathetic neurons including the nuclear-encoded mt gene COXIV. A brain-specific miRNA, miR-338 contains the putative binding site at the 3'UTR of COXIV mRNA. Over-expression of miR-338 into the axons of primary sympathetic neurons reduces the COXIV mRNA, protein levels as a result decrease in mitochondrial activity, and reduction of ATP occurs. Conversely, the reduction of miR-338 using synthetic anti-miR oligonucleotides increases COXIV levels and results in a significant increase in oxidative phosphorylation [86].
Figure 3.
Pictorial representation of miRNAs action on mitochondrial function and ATP production in the neuron. Healthy neurons showed the normal synaptic activity and neurotransmission, whereas in diseases state such as AD, Aβ, and tau toxicities cause miRNAs alteration in the neuron. Increased level of miRNAs reduces their target gene expression, those are important for mitochondrial function. Reduction of mitochondrial proteins leads to impaired mitochondrial function, reduced ATP, and poor synaptic activity.
Another miRNA that regulates cellular ATP production is miR-151a-5p. The level of miR-151a-5p was significantly increased in severe asthenozoospermia cases compared with healthy controls [88]. Overexpression of miR-151a-5p using mimics strategy decreases the mitochondrial respiratory activity and ATP level. Mechanistically, miR-151a-5p targets the Cytochrome b (Cytb) mRNA, and a high level of miR-151a-5p significantly reduced the Cytb expression at mRNA and protein levels. These findings indicate that miR-151a-5p plays a crucial role in cellular respiration and ATP production [88].
Siengdee et al, (2015) studied the correlation of mt miRNAs and mt gene during the differentiation of C2C12 myoblasts into myotubes [89]. Microarray expression profiles identified 14 miRNAs (miR-423-3p, miR-17, miR-130b, miR-301a/b, miR-345, miR-15a, miR-16a, miR-128, miR-615, miR-1968, miR-1a/b, and miR-194) as cellular ATP regulators and their targeting genes are (Cox4i2, Cox6a2, Ndufb7, Ndufs4, Ndufs5, and Ndufv1) which involved in mitochondrial energy metabolism during C2C12 differentiation. Among all, miR-423-3p showed a negative correlation with increasing ATP levels. MiR-423-3p target prediction analysis revealed Cox6a2 as a potential target, and a high level of miR-423-3p lead to the decreased cellular ATP level in C2C12 myogenic differentiation via suppressing Cox6a2 level [89]. These findings unveiled that miR-423-3p as a novel regulator of ATP production and energy metabolism [89]. Despite the finding of several miRNAs, more research is needed to better understand the involvement of miRNAs in cellular ATP production and energy metabolism in the neuron.
13. MicroRNAs and dendritic maturation
Dendritic spine maturation of neurons is critical for the formation of functional circuits during brain development. Any defects in dendrite morphogenesis are associated with neuropsychiatric disorders [90]. MiRNAs are found to be critical to maintain healthy dendritic spine maturation and development for e.g. miR-214 a key regulator essential for dendritic development. Irie at el (2016), reported that high level of miR-214 increased dendrite size and complexity in the hippocampal neurons of mice, whereas knockdown of endogenous miR-214, inhibited dendritic morphogenesis [90]. Author found that miR-214 targets quaking (Qki), an important gene implicated in psychiatric diseases such as schizophrenia. Downregulation of Qki by overexpressed miR-214 enhanced dendritic formation, dendritic growth and neuronal dendritic development [90]. MiR-9 is another crucial miRNA for dendritic maturation in the human brain. MiR-9 plays positive roles in neural progenitors, dendritic and synaptic development. Giusti et al (2014), developed a transgenic miRNA sponge mouse model for conditional inactivation of the miR-9 family in a spatio-temporal-controlled manner [91]. MiR-9 interacts and interferes with the genes that are needed to RE1 Silencing Transcription Factor (REST) protein translation. Hence, miR-9-mediated downregulation of the transcriptional repressor REST is essential for proper dendritic growth [91]. Another important miRNA is miR-34c, which expressed in mammalian hippocampi and alters the hippocampal dendritic spine density and induce memory impairment in AD mice. Cao et al (2018), showed that overexpression of miR-34c negatively regulated dendritic length and spine density in mice hippocampal neurons [92]. MiR-34c overexpressed hippocampal neurons showed shorter dendrites on average and fewer filopodia and spines relative to non-transfected control mice. Therefore, disruption of dendritic spine density by miR-34c influences synaptic plasticity and can contribute to AD pathogenesis [92].
14. MicroRNAs and synaptic receptors trafficking
Homeostatic plasticity is a compensatory regulation in neuronal activity and crucial for the maintenance of neuronal and neural circuit stability. Chronic deprivation of neuronal activity results in an increase in synaptic AMPARs and postsynaptic currents. The biogenesis of GluA2-lacking, calcium permeable AMPARs (CP-AMPARs) plays a crucial role in the neuronal homeostatic response. As we discussed previously, miR-124, is critical to maintain homeostatic plasticity by regulation of GluA2 and CP-AMPARs [70]. Another miRNA, miR-153 are known to control the synaptic vesicle trafficking in chronic cerebral hypoperfusion (CCH). MiR-153 level was upregulated in the hippocampus of 2VO rats [93]. In vitro study showed that miR-153 reduces the synapsin I protein level by targeting at the 3’UTR of synapsin I mRNA. Similarly, in vivo study replicated that upregulation of miR-153 reduced DRE phase and induces synapsin I deficiency as CCH. However, knockdown of miR-153 rescued the downregulation of synapsin I and shortened DRE phase in 2VO rats. These findings showed that CCH impairs hippocampal glutamatergic vesicle trafficking by upregulating miR-153 [93].
15. Synaptic microRNAs and Alzheimer’s disease
MiRNAs, as potential regulators of human genes, have been studied in AD [23]. MiRNAs have been widely accepted as critical in regulating synaptic activity. The elevated levels of miR-125b have been found to down-regulate synaptic protein synapsin-2 (SYN-2) and 15-lipoxygenase (15-LOX) which, in turn, possibly causes pathogenic effects associated with AD, such as synaptic deficits, neurotrophic deficits, and astrogliosis [94]. A novel miRNA-124/PTPN1 signal pathway mediates synaptic and memory deficits in AD [19]. Overexpression of miR-30b in the hippocampus of normal wild type (WT) mice impairs synaptic and cognitive functions, mimicking those seen in transgenic mice. Conversely, knockdown of endogenous miR-30b in transgenic mice was found to prevent synaptic functions associated with cognitive decline [20].
Even though this research has determined the significance of miRNAs, however, the role of synaptosomal-associated miRNAs has not been well-studied in AD progression. The identification of synaptosome-associated miRNAs in AD and their characterization are very important to better understand the biology of local miRNAs and synaptic activity. Also, molecular connections of synaptosomal miRNAs need to be studied in Aβ and p-tau induced toxicities at the synapse.
Synaptosomes prepared from human postmortem brains have been used to study AD-associated deficits in neurotransmission, including acetylcholine, glutamate, and c-aminobutyric acid (GABA) systems [95,96]. More recently, however, synaptosome studies have focused on studying the nature of Aβ and tau pathology in AD synapses and neprilysin in AD patients [74,97]. Likewise, the localization of huntingtin has been examined in the synaptosomes of Huntington’s disease patients [52]. These studies suggest that increasing progress is being made on synaptosomal miRNAs.
15.1. MicroRNA-124:
Wang et al, (2018) reported that miR-124 is an important modulator of synaptic function and memory, based on his finding that the level of miR-124 was found to be dramatically increased in the hippocampus of Tg2576 AD mice [19]. They also found that miR-124 levels elevated in specific brain regions affected by AD. Further, Wang et al, identified the Protein Tyrosine Phosphatase Non-Receptor Type 1 (PTPN1) gene as a direct target of miR-124, and induced levels of miR-124 or knockdown of PTPN1 recapitulating AD-like phenotypes in mice, which includes memory impairments and deficits in synaptic transmission and plasticity through glutamate receptor 2 membrane insertion. Most importantly, maintaining the balance of miR-124/PTPN1 levels by suppressing miR-124 through overexpression of PTPN1 could restore synaptic failure and memory deficits. These findings identified the miR-124/PTPN1 pathway as a critical mediator of synaptic dysfunction and memory loss in AD and could be a promising therapeutic target for AD patients [19].
15.2. MicroRNA-30b:
Song et al (2019), determined the role of miR-30b in AD and synaptic integrity [20]. MiR-30b targets several important genes that are important for maintaining healthy synaptic integrity, such as ephrin type-B receptor 2 (ephB2), sirtuin1 (sirt1), and glutamate ionotropic receptor AMPA type subunit 2 (GluA2). Levels of miR-30b were found to be significantly upregulated in the brains of both AD patients and APP transgenic mice, conversely, the expression of miR-30b targets was significantly downregulated. Induced expression of miR-30b in the hippocampus of normal WT mice impaired synaptic and cognitive functions, similarly to impairments of miR-30b in the APP mice [20]. However, the knockdown strategy of endogenous miR-30b in transgenic mice prevents synaptic and cognitive decline. Additionally, the levels of miR-30b were upregulated by pro-inflammatory cytokines and Aβ42 through NF-κβ signaling. These findings unveiled a previously undefined mechanism by which the deregulation of miR-30b causes synaptic and cognitive dysfunction in AD [20]. These findings suggest that the deregulation of miR-30b in the AD-affected brain may prevent or slow cognitive decline.
15.3. MicroRNA-485:
MiR-485 is another miRNA that could regulate the homeostatic synaptic plasticity in AD and HD. The miR-485 controls dendritic spine number and synapse formation in an activity dependent homeostatic manner in hippocampal neurons [98]. MiR-485 binds many plasticity-associated genes; however, the presynaptic protein SV2A was identified as a potential target of miR-485. By regulating the level of SV2A, miR-485 negatively regulate the dendritic spine density, postsynaptic density 95 clustering, and expression of GluR2. Furthermore, high level of miR-485 reduced spontaneous synaptic responses release of neurotransmitters [98]. This study revealed a role of miR-485 in regulation of SV2A protein, homeostatic plasticity and nervous system development with possible implications in AD.
15.4. MicroRNA-153:
Chronic brain hypoperfusion (CBH) pathology is closely associated with AD and vascular dementia (VaD). The levels of miR-153 was found to be upregulated in the plasma of dementia patients and in rats’ hippocampus following of bilateral common carotid artery ligation (2VO) surgery [99]. On the other side, levels of vesicle fusion-related proteins- SNAP-25, VAMP-2, syntaxin-1A and synaptotagmin-1 was significantly decrease. The expression of these fusion related protein was regulated by miR-153 through binding at the 3′ UTR of the Snap25, Vamp2, Stx1a and Syt1 genes. Therefore, high level of miR-153 controls CBH-induced presynaptic vesicle release impairment by regulating the level of four vesicle release-related proteins Stx1a, Snap25, Vamp2 and Syt1 genes [99]. This study identifies the role of miR-153 in presynaptic plasticity impairment, which could be a promising target for AD and VaD treatment.
Another study by Mathew et al, (2016) also unveiled the role of miR-153 in the SNARE-mediated vesicular transport pathway [100]. SNARE pathway is important in synaptic remodeling and long-term memories formation. Mathew et al (2016), detected the upregulation of miR-153 in the rodent hippocampus upon contextual fear conditioning [100]. The genes involved in vesicular transport and synaptogenesis pathways are the major targets of the fear-induced miRNAs e.g. miR-153. MiR-153 down-regulates Glutamate receptor A1 trafficking and neurotransmitter release as a result, the expression of key components of the vesicular transport machinery is reduce. Particularly, miR-153 level is induced during LTP induction and its knockdown enhanced fear memory in the hippocampus of adult mice [100]. These results suggest miR-153, a fear-induced miRNA, work as a negative feedback loop and blocks neuronal hyperactivity through inhibition of vesicular transport pathway.
15.5. MicroRNA-101b:
Histone deacetylase 2 (HDAC2) plays a major role in epigenetic regulation of gene expression and its expression is strongly increased in AD. HDAC2 overexpression induces AD-like tau hyperphosphorylation and aggregation, which is accompanied by the loss of dendritic complexity and spine density. MiR-101b regulates HDAC2 via targeting the 3’UTR of AMP-activated protein kinase (AMPK) mRNA. In vitro study showed that overexpression of miR-101b reduces AMPK levels and blocked HDAC2-induced tauopathy and dendritic impairments [101]. Similarly, in AD mice model, the induced level of miR-101b by mimics or AMPK siRNAs rescued tau pathology, dendritic abnormalities, and memory deficits [101]. This study unveiled complex relationship of HDAC2/miR-101/AMPK pathway in AD pathogenesis. Other study demonstrated that loss of miR-101 in hippocampal neurons induces cognitive decline and modulation of AD-related genes in mice [102]. Knockdown of endogenous miR-101 levels in hippocampus of C57BL/6J mice by using a miR-101 sponge (pLSyn-miR-101 sponge) showed cognitive impairment. AMPK hyperphosphorylation was observed in the hippocampus of miR-101 suppressed mice. The reduction of miR-101 caused cognitive impairment features and upregulation of miR-101 target genes associated with AD such as APP, RanBP9 and Rab5 and overproduction of Aβ42 levels [102].
15.6. MicroRNA-132:
As we discussed earlier miR-132 is another critical miRNA for synaptic function. MiR-132 is one of the most abundant brain-enriched miRNAs shown to regulate neuron morphogenesis and plasticity [103]. MiR-132 level was found to be is significantly reduced in the brains of AD patients and genetic deletion of miR-132 induces Aβ and p-tau deposition and memory impairment in mice [103]. MiR-132 target the C1q, a classical complement cascade protein, highly expressed in the synaptic regions in AD patients. The APP/PS1 transgenic mice transfected with miR-132 and given C1 inhibitors showed significantly increased expression of PSD95, Synapsin-1 and phosphorylated (p) Synapsin compared to non-transfected AD mice [103]. These results suggested that miR-132 might regulate synaptic proteins by controlling the C1q expression in AD. Xie at al (2019), further elaborate the role of miR-132 in tauopathies [104]. Overexpression of tau protein impairs the neuronal endocytosis by decreasing the GTPase dynamin 1 via miR- 132/ MeCP2 pathway in the mouse cortical neurons. The regulation of MeCP2 by miR-132 is very frequently reported in the brains of AD and hTau mice [104].
15.7. MicroRNA-181a:
Another hippocampal neuron abundant miRNA is miR-181a which controls the expression of key plasticity-related proteins at synapses. MiR-181a was found to be upregulated in the hippocampus of AD mouse model and it is correlated with reduced levels of plasticity-related proteins [105]. Rodriguez-Ortiz et al (2019), investigated the underlying mechanisms by which miR-181a negatively modulated synaptic plasticity and memory [105]. Reduced level of miR-181a in the primary hippocampal cultures correlated with upregulation of translin and GluA2 proteins, the predicted targets for miR-181a. Therefore, inhibition of miR-181a alleviated memory deficits and increased GluA2 and GluA1 levels, without restoring translin, in the AD mouse model [105].
15.8. MicroRNA-10a:
Wu et al (2018), unveiled the role of miR- 10a in synapse remodeling of neuronal cells in the rat model of AD through BDNF- TrkB signaling pathway [106]. The AD rat model showed the increased expression of miR-10a and reduced levels of p38, PSD95, BDNF, cAMP- response element- binding protein (CREB), and tropomyosin receptor kinase B (TrκB) proteins compared with the normal rats. In a similar way, neuronal cells transfected with miR- 10a mimics showed decreased levels of p38, PSD95, BDNF, CREB and TrκB, whereas miR- 10a inhibitors treated cells showed increased levels of these proteins [106]. These findings indicated that miR- 10a controls synapse remodeling and neuronal cell proliferation via inhibition of BDNF- TrkB signaling pathway in AD.
15.9. MicroRNA-188-5p:
Lee et al (2016), investigated the pathophysiological significance of miR-188-5p in AD [107]. The primary hippocampal neuron cultures treated with oligomeric Aβ1-42 showed reduced expression of miR-188-5p, while overexpression of miR-188-5p rescued cells from Aβ1-42-mediated synapse elimination and synaptic dysfunctions. MiR-188-5p level was found to be reduced in the AD postmortem brains and 5XFAD mice and it is regulated by CREB [107]. MiR-188-5p act through modulating the level of Nrp-2, a negative regulator of spine development and synaptic structure. The induced level of miR-188-5p reduces Nrp2 and rescued neuron from the Aβ toxicity; improve dendritic spine density and synaptic dysfunction [107].
15.10. MicroRNA-574:
Another study by Li et al, unveiled the involvement of miR-574 in cognitive impairment in APP/PS1 mice through modulation of neuritin protein [108]. The 5-month-old APP/PS1 mice were used for hippocampal miRNAs expression analysis. The microarray data and further validation analysis showed that miR-574 was significantly increased in the hippocampus of APP/PS1 mice relative to age matched wild type mice. Bioinformatic analysis showed that neuritin (Nrn1) mRNA as predicted target of miR-574 [108]. Induced expression of miR-574 in primary hippocampal neurons reduced the levels of neuritin and synaptic proteins. On the other side, suppression of miR-574 results a significant increased in the levels of neuritin and synaptic proteins [108]. All together results showed that miR-574 is involved in cognitive impairment in APP/PS1 mice through regulation of neuritin.
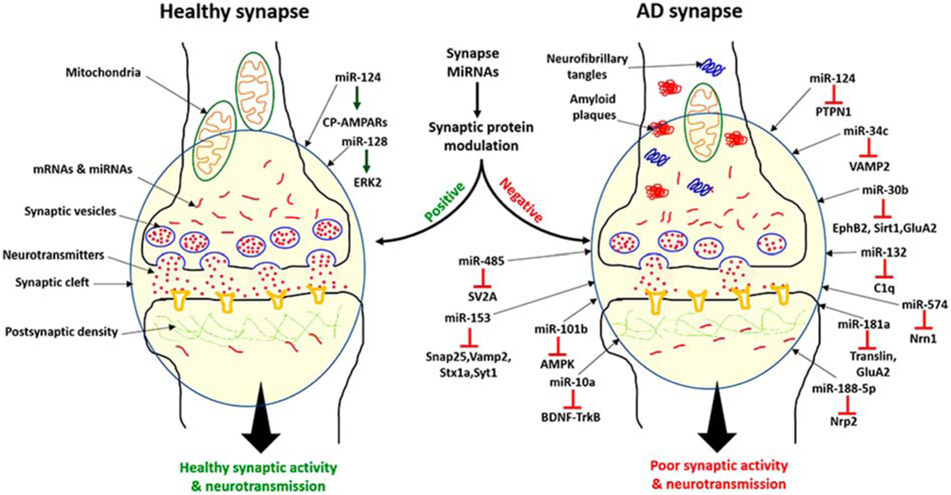
Although definitive proof that miRNAs function in a localized manner in the neurons is still lacking, it is quite possible that miRNAs localized in dendrites and axon terminals could modulate the expression of local mRNAs and proteins. As shown in Figure 4, the positive and negative regulation of proteins at the synapse by local miRNAs could alter synaptic functions. In a healthy state, cell homeostasis maintains balanced miRNAs levels, where miRNAs function in a positive way to maintain healthy cells functioning e.g. miR-124 and miR-128 regulates the CA-AMPARs and ERK2 pathway respectively [68,70]. Healthy neurons showed normal synaptic activity and neurotransmission. In AD state, the deregulation of synaptic miRNAs caused by the Aβ and tau toxicities. Increased level of miRNAs- miR-124, miR-34c, miR-30b, miR-485, miR-153, miR-101b, miR-132, miR-181a, miR-10a, miR-188-5p and miR-574 leads to the downregulation of their target gene/proteins PTPN1, VAMP2, EphB2, Sirt1, GluA2, SV2A, Snap25, Vamp2, Stx1a, Syt1, AMPK, C1q, GluA2, translin, Nrp2 and Nrn1) which are important for synaptic activity [19,20,67,70,72,98-108]. Suppression of synaptic and other proteins could impair synapse activity and neurotransmission (Figure 4).
Figure 4.
A working model of synaptic miRNAs: miRNAs localized in dendrites and axon terminals could modulate the expression of local mRNAs and proteins. Positive and negative regulation of synaptic proteins by local miRNAs at synapse could alter synaptic function. Neurons not affected by AD (healthy neuron) showed a balanced miRNAs levels, normal synaptic activity, and synaptic neurotransmission. Brain-specific miR-124 and 128 showed the positive modulation of synaptic activity via CA-AMPARs and ERK2 pathway. On the other hand in AD state, the deregulation of the brain-specific miRNAs (miR-124, miR-34c and miR-132) and other miR-485, miR-153, miR-30b, miR-188-5p, miR-132, miR-101b, miR-181a and miR-574 negatively modulate the synaptic activity via suppression of their target genes. The suppression of synaptic and other proteins leads to impaired synaptic activity and neurotransmission.
16. Conclusions and Future Directions
Synapse is thought to play important roles in the pathophysiology of many neurological diseases, including AD. In terms of synaptic components, synaptosomes contain synaptic vesicles, postsynaptic density, mitochondria, microsomes, lysosomes, plasma membranes, and miRNAs. Extensive research has been done on synaptosomes and synaptic plasticity in AD. However, we do not adequately understand the relationships across synaptosomes, miRNAs, and synaptic plasticity in AD and other neurological diseases. The current article sheds light on the status of synaptosomal miRNAs and their possible roles in maintaining healthy neurotransmission and synaptic activity. We summarized the molecular and physiological links among four different components- synaptosomes, synaptic plasticity, miRNAs, and AD.
Research has been lacking in the specific roles of synaptosomes and miRNAs in AD. Therefore, we attempted to summarize the current status of synaptosomal miRNAs in AD. Several key questions will need to be addressed: 1) Are miRNAs altered at the synaptosome in AD neurons with Aβ and p-tau induced toxicities? 2) Is miRNA deregulation at the synaptosome responsible for the poor synaptic activity and cognitive dysfunction in AD? 3) If yes, what could be the possible miRNAs-based research strategy to improve the synaptic function and cognitive behavior in AD and 4) what role synaptic mitochondrial miRNAs play in maintaining synaptic ATP? It is very important to understand the properties of synaptosomal miRNAs and their positive/negatives roles in AD progression.
Highlights.
MicroRNAs are present in small, subcellular compartments of the neuron, such as neural dendrites, synaptic vesicles, and synaptosomes.
MicroRNAs localize at or nearby synapse and regulate multiple synaptic functions
Synaptic miRNAs alteration(s) in Alzheimer’s disease, modulate the local protein levels at synapse.
Synaptic microRNAs are crucial to maintain healthy synaptic activity and synaptic function in Alzheimer’s disease.
Acknowledgments:
The research presented in this article was supported by NIH grants AG042178, AG047812, NS105473, AG060767, AG069333 (to PHR), and KAG065645 (to Subodh Kumar).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Statement
Authors declare that they do not have any conflict of interest
References
- 1.Alzheimer’s Disease Facts and Figures, 2019. (https://www.alz.org/media/documents/alzheimers-facts-and-figures-2019). [Google Scholar]
- 2.Selkoe DJ, Alzheimer's disease results from the cerebral accumulation and cytotoxicity of amyloid beta protein, J. Alzheimers Dis 3 (2001) 75–80. [DOI] [PubMed] [Google Scholar]
- 3.Mattson MP, Duan W, Wan R, Guo Z, Prophylactic activation of neuroprotective stress response pathways by dietary and behavioral manipulations, NeuroRx. 1 (2004) 111–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.LaFerla FM, Green KN, Oddo S, Intracellular amyloid-beta in Alzheimer's disease, Nat. Rev. Neurosci 8 (2007) 499–509. [DOI] [PubMed] [Google Scholar]
- 5.Reddy PH, Manczak M, Mao P, Calkins MJ, Reddy AP, Shirendeb U, Amyloid beta and mitochondria in aging and Alzheimer's disease: implications for synaptic damage and cognitive decline, J. Alzheimers Dis 20 (2010) S499–S512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reddy PH, Tripathi R, Troung Q, Tirumala K, Reddy TP, Anekonda V, Shirendeb UP, Calkins MJ, Reddy AP, Mao P, Manczak M, Abnormal mitochondrial dynamics and synaptic degeneration as early events in Alzheimer's disease: implications to mitochondria-targeted antioxidant therapeutics, Biochim. Biophys. Acta 1822 (2012) 639–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Criscuolo C, Fabiani C, Cerri E, Domenici L, Synaptic Dysfunction in Alzheimer's Disease and Glaucoma: From Common Degenerative Mechanisms Toward Neuroprotection, Front. Cell Neurosci 11 (2017) 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang DO, Kim SM, Zhao Y, Hwang H, Miura SK, Sossin WS, Martin KC, Synapse- and stimulus-specific local translation during long-term neuronal plasticity, Science. 324 (2009) 1536–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holt CE, Martin KC, Schuman EM, Local translation in neurons: visualization and function, Nat. Struct. Mol. Biol 26 (2019) 557–566. [DOI] [PubMed] [Google Scholar]
- 10.Marsh J, Alifragis P, Synaptic dysfunction in Alzheimer's disease: the effects of amyloid beta on synaptic vesicle dynamics as a novel target for therapeutic intervention, Neural Regen. Res 13 (2018) 616–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Silvestro S, Bramanti P, Mazzon E, Role of miRNAs in Alzheimer's Disease and Possible Fields of Application, Int. J. Mol. Sci 20 (2019) 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reddy PH, Tonk S, Kumar S, Vijayan M, Kandimalla R, Kuruva CS, P Reddy A, A critical evaluation of neuroprotective and neurodegenerative MicroRNAs in Alzheimer's disease, Biochem. Biophys. Res. Commun 483 (2017) 1156–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Siedlecki-Wullich D, Català-Solsona J, Fábregas C, Hernández I, Clarimon J, Lleó A, Boada M, Saura CA, Rodríguez-Álvarez J, Miñano-Molina AJ, Altered microRNAs related to synaptic function as potential plasma biomarkers for Alzheimer's disease, Alzheimers Res. Ther 15 (2019) 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ye Y, Xu H, Su X, He X, Role of MicroRNA in Governing Synaptic Plasticity, Neural Plast. 2016 (2016) 4959523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lugli G, Torvik VI, Larson J, Smalheiser NR, Expression of microRNAs and their precursors in synaptic fractions of adult mouse forebrain, J. Neurochem 106 (2008) 650–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu J, Chen Q, Zen K, Zhang C, Zhang Q, Synaptosomes secrete and uptake functionally active microRNAs via exocytosis and endocytosis pathways, J. Neurochem 124 (2013) 15–25. [DOI] [PubMed] [Google Scholar]
- 17.Li H, Wu C, Aramayo R, Sachs MS, Harlow ML, Synaptic vesicles contain small ribonucleic acids (sRNAs) including transfer RNA fragments (trfRNA) and microRNAs (miRNA), Sci. Rep 5 (2015) 14918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boese AS, Saba R, Campbell K, Majer A, Medina S, Burton L, Booth TF, Chong P, Westmacott G, Dutta SM, Saba JA, Booth SA, MicroRNA abundance is altered in synaptoneurosomes during prion disease, Mol. Cell. Neurosci 71 (2016) 13–24. [DOI] [PubMed] [Google Scholar]
- 19.Wang X, Liu D, Huang HZ, Wang ZH, Hou TY, Yang X, Pang P, Wei N, Zhou YF, Dupras MJ, Calon F, Wang YT, Man HY, Chen JG, Wang JZ, Hébert SS, Lu Y, Zhu LQ, Novel MicroRNA-124/PTPN1 Signal Pathway Mediates Synaptic and Memory Deficits in Alzheimer's Disease, Biol. Psychiatry 83 (2018) 395–405. [DOI] [PubMed] [Google Scholar]
- 20.Song Y, Hu M, Zhang J, Teng ZQ, Chen C, A novel mechanism of synaptic and cognitive impairments mediated via microRNA-30b in Alzheimer's disease, EBioMedicine. 39(2019) 409–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Long JM, Maloney B, Rogers JT, Lahiri DK, Novel upregulation of amyloid-β precursor protein (APP) by microRNA-346 via targeting of APP mRNA 5'-untranslated region: Implications in Alzheimer's disease, Mol. Psychiatry 24 (2019) 345–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chopra N, Wang R, Maloney B, Nho K, Beck JS, Pourshafie N, Niculescu A, Saykin AJ, Rinaldi C, Counts SE, Lahiri DK, MicroRNA-298 reduces levels of human amyloid-β precursor protein (APP), β-site APP-converting enzyme 1 (BACE1) and specific tau protein moieties, Mol. Psychiatry (2020) January 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar S, Reddy PH, Are circulating microRNAs peripheral biomarkers for Alzheimer's disease?, Biochim. Biophys. Acta 1862 (2016) 1617–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar S, Reddy PH, MicroRNA-455-3p as a Potential Biomarker for Alzheimer's Disease: An Update, Front. Aging Neurosci 10 (2018) 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar S, Chawla YK, Ghosh S, Chakraborti A, Severity of hepatitis C virus (genotype-3) infection positively correlates with circulating microRNA-122 in patients’ sera, Dis. Markers 2014 (2014) 35476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar S, Batra A, Kanthaje S, Ghosh S, Chakraborti A, Crosstalk between microRNA-122 and FOX family genes in HepG2 cells, Exp. Biol. Med 242 (2017) 436–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar S, Vijayan M, Reddy PH, MicroRNA-455-3p as a potential peripheral biomarker for Alzheimer's disease, Hum. Mol. Genet 26 (2017) 3808–3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Brien J, Hayder H, Zayed Y, Peng C, Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation, Front. Endocrinol 9 (2018) 402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar S, Reddy AP, Yin X, Reddy PH, Novel MicroRNA-455-3p and its Protective Effects Against Abnormal APP Processing and Amyloid Beta Toxicity in Alzheimer’s Disease, Biochim. Biophys. Acta. Mol. Basis Dis 1865 (2019) 2428–2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ludwig N, Leidinger P, Becker K, Backes C, Fehlmann T, Pallasch C, Rheinheimer S, Meder B, Stähler C, Meese E, Keller A, Distribution of miRNA expression across human tissues, Nucleic Acids Res. 44 (2016) 3865–3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turchinovich A, Samatov TR, Tonevitsky AG, Burwinkel B, Circulating miRNAs: cell-cell communication function?, Front. Genet 4 (2013) 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kosaka N, Yoshioka Y, Hagiwara K, Tominaga N, Ochiya T, Functional analysis of exosomal microRNA in cell-cell communication research, Methods Mol. Biol 1024 (2013) 1–10. [DOI] [PubMed] [Google Scholar]
- 33.Delay C, Mandemakers W, Hébert SS, MicroRNAs in Alzheimer's disease, Neurobiol. Dis 46 (2012) 285–290. [DOI] [PubMed] [Google Scholar]
- 34.Kumar S, Vijayan M, Bhatti JS, Reddy PH, MicroRNAs as Peripheral Biomarkers in Aging and Age-Related Diseases, Prog. Mol. Biol. Transl. Sci 146 (2017) 47–94. [DOI] [PubMed] [Google Scholar]
- 35.Vijayan M, Reddy PH, Peripheral biomarkers of stroke: Focus on circulatory microRNAs, Biochim. Biophys. Acta 1862 (2016) 1984–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williams J, Smith F, Kumar S, Vijayan M, Reddy PH, Are microRNAs true sensors of ageing and cellular senescence?, Ageing Res. Rev 35 (2017) 350–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vijayan M, Alamri FF, Al Shoyaib A, Karamyan VT, Reddy PH, Novel miRNA PC-5P-12969 in Ischemic Stroke, Mol. Neurobiol 56 (2019) 6976–6985. [DOI] [PubMed] [Google Scholar]
- 38.Condrat CE, Thompson DC, Barbu MG, Bugnar OL, Boboc A, Cretoiu D, Suciu M, Cretoiu SM, Voinea SC, miRNAs as Biomarkers in Disease: Latest Findings Regarding Their Role in Diagnosis and Prognosis, Cells. 9 (2020) 276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rupaimoole R, Slack FJ, MicroRNA therapeutics: towards a new era for the management of cancer and other diseases, Nat. Rev. Drug Discov 16 (2017) 203–222. [DOI] [PubMed] [Google Scholar]
- 40.Slack FJ, In this issue of Epigenetics: special focus on non-coding RNAs in epigenetic regulation, Epigenetics. 9 (2014) 1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Di Meco A, Praticò D, MicroRNAs as Therapeutic Targets for Alzheimer's Disease, J. Alzheimers Dis 53 (2016) 367–72. [DOI] [PubMed] [Google Scholar]
- 42.Lauretti E, Dincer O, Praticò D, Regional and temporal miRNAs expression profile in a transgenic mouse model of tauopathy: implication for its pathogenesis. Mol. Psychiatry (2020) January 27. [DOI] [PubMed] [Google Scholar]
- 43.Bartel DP, Chen CZ, Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs, Nat. Rev. Genet 5 (2004) 396–400. [DOI] [PubMed] [Google Scholar]
- 44.Bartel DP, MicroRNAs: target recognition and regulatory functions, Cell. 136 (2009) 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schratt G, microRNAs at the synapse, Nat. Rev. Neurosci 10 (2009) 842–849. [DOI] [PubMed] [Google Scholar]
- 46.Siegel G, Saba R, Schratt G, microRNAs in neurons: manifold regulatory roles at the synapse, Curr. Opin. Genet. Dev 21 (2011) 491–497. [DOI] [PubMed] [Google Scholar]
- 47.Rajman M, Schratt G, MicroRNAs in neural development: from master regulators to fine-tuners, Development. 144 (2017) 2310–2322. [DOI] [PubMed] [Google Scholar]
- 48.Song M, miRNAs-dependent regulation of synapse formation and function, Genes Genom. 42 (2020) 837–845. [DOI] [PubMed] [Google Scholar]
- 49.Hu Z, Li Z, miRNAs in synapse development and synaptic plasticity, Curr. Opin. Neurobiol 45 (2017) 24–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Millan MJ, Linking deregulation of non-coding RNA to the core pathophysiology of Alzheimer's disease: An integrative review, Prog. Neurobiol 156 (2017) 1–68. [DOI] [PubMed] [Google Scholar]
- 51.Reddy PH, Beal MF, Amyloid beta, mitochondrial dysfunction and synaptic damage: implications for cognitive decline in aging and Alzheimer's disease, Trends. Mol. Med 2 (2008) 45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jhou JF, Tai HC, The Study of Postmortem Human Synaptosomes for Understanding Alzheimer's Disease and Other Neurological Disorders: A Review, Neurol. Ther (Suppl 1) (2017) 57–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tannenberg RK, Scott HL, Tannenberg AE, Dodd PR, Selective loss of synaptic proteins in Alzheimer's disease: evidence for an increased severity with APOE varepsilon4, Neurochem. Int. 49 (2006) 631–639. [DOI] [PubMed] [Google Scholar]
- 54.Tang Y, Nyengaard JR, De Groot DM, Gundersen HJ, Total regional and global number of synapses in the human brain neocortex, Synapse. 41 (2001) 258–273. [DOI] [PubMed] [Google Scholar]
- 55.Gray EG, Whittaker VP, The isolation of nerve endings from brain: an electron microscopic study of cell fragments derived by homogenization and centrifugation, J. Anat 96 (1962) 79–88. [PMC free article] [PubMed] [Google Scholar]
- 56.Whittaker VP, Gray EG, The synapse: biology and morphology, Br. Med. Bull 18 (1962) 223–228. [DOI] [PubMed] [Google Scholar]
- 57.Whittaker VP, The morphology of fractions of rat forebrain synaptosomes separated on continuous sucrose density gradients, Biochem. J 106 (1968) 412–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Steward O, Wallace CS, Lyford GL, Worley PF, Synaptic activation causes the mRNA for the IEG Arc to localize selectively near activated postsynaptic sites on dendrites, Neuron. 21 (1998) 741–751. [DOI] [PubMed] [Google Scholar]
- 59.Gao FB, Messenger RNAs in dendrites: localization, stability, and implications for neuronal function. Bioessays. 20 (1998) 70–78. [DOI] [PubMed] [Google Scholar]
- 60.Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME, A brain-specific microRNA regulates dendritic spine development, Nature. 439 (2006) 283–289. [DOI] [PubMed] [Google Scholar]
- 61.Kosik KS, The neuronal microRNA system, Nat. Rev. Neurosci 12 (2006) 911–920. [DOI] [PubMed] [Google Scholar]
- 62.Kim J, Krichevsky A, Grad Y, Hayes GD, Kosik KS, Church GM, Ruvkun G, Identification of many microRNAs that copurify with polyribosomes in mammalian neurons, Proc. Natl. Acad. Sci. USA 101 (2004) 360–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Banerjee S, Neveu P, Kosik KS, A coordinated local translational control point at the synapse involving relief from silencing and MOV10 degradation, Neuron. 64 (2009) 871–884. [DOI] [PubMed] [Google Scholar]
- 64.Taoufik E, Kouroupi G, Zygogianni O, Matsas R, Synaptic dysfunction in neurodegenerative and neurodevelopmental diseases: an overview of induced pluripotent stem-cell-based disease models, Open Biol. 8 (2018) 180138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Aksoy-Aksel A, Zampa F, Schratt G, MicroRNAs and synaptic plasticity--a mutual relationship, Philos. Trans. R. Soc. Lond. B. Biol. Sci 369 (2014) 1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McNeill EM, Warinner C, Alkins S, Taylor A, Heggeness H, DeLuca TF, Fulga TA, Wall DP, Griffith LC, Vactor DV, The conserved microRNA miR-34 regulates synaptogenesis via coordination of distinct mechanisms in presynaptic and postsynaptic cells, Nat. Commun 11 (2020) 1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hu S, Wang H, Chen K, Cheng P, Gao S, Liu J, Li X, Sun X, MicroRNA-34c downregulation ameliorates amyloid-beta-induced synaptic failure and memory deficits by targeting VAMP2, J. Alzheimers Dis 48 (2015) 673–86. [DOI] [PubMed] [Google Scholar]
- 68.Edbauer D, Neilson JR, Foster KA, Wang CF, Seeburg DP, Batterton MN, Tada T, Dolan BM, Sharp PA, Shengal M, Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132, Neuron. 65 (2010) 373–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sim SE, Lim CS, Kim JI, Seo D, Chun H, Yu NK, Lee J, Kang SJ, Ko HG, Choi JH, Kim T, Jang EH, Han J, Bak MS, Park JE, Jang DJ, Baek D, Lee YS, Kaang BK, The Brain-Enriched MicroRNA miR-9-3p Regulates Synaptic Plasticity and Memory, The Journal of neuroscience : the official journal of the Society for Neuroscience, 36 (2016) 8641–8652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hou Q, Ruan H, Gilbert J, Wang G, Ma Q, Yao WD, Man HY, MicroRNA miR124 is required for the expression of homeostatic synaptic plasticity, Nature communications. 6 (2015)10045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Silva MM, Rodrigues B, Fernandes J, Santos SD, Carreto L, Santos MAS, Pinheiro P, Carvalho AL, MicroRNA-186-5p controls GluA2 surface expression and synaptic scaling in hippocampal neurons, Proc. Natl. Acad. Sci. USA 116 (2019) 5727–5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tan CL, Plotkin JL, Venø MT, von Schimmelmann M, Feinberg P, Mann S, Handler A, Kjems J, Surmeier DJ, O'Carroll D, Greengard P, Schaefer A, MicroRNA-128 governs neuronal excitability and motor behavior in mice, Science. 342 (2013) 1254–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jin Y, Tymen SD, Chen D, Fang ZJ, Zhao Y, Dragas D, Dai Y, Marucha PT, Zhou X, MicroRNA-99 family targets AKT/mTOR signaling pathway in dermal wound healing, PLoS One. 8 (2013) e64434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sokolow S, Henkins KM, Williams IA, Vinters HV, Schmid I, Cole GM, Gylys KH, Isolation of synaptic terminals from Alzheimer's disease cortex, Cytometry A. 81 (2012) 248–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Postupna NO, Keene CD, Latimer C, Sherfield EE, Van Gelder RD, Ojemann JG, Montine TJ, Darvas M, Flow cytometry analysis of synaptosomes from post-mortem human brain reveals changes specific to Lewy body and Alzheimer's disease, Lab Invest. 94 (2014) 1161–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Selkoe DJ, Alzheimer's disease is a synaptic failure, Science. 25 (2002) 789. [DOI] [PubMed] [Google Scholar]
- 77.Koffie RM, Hyman BT, Spires-Jones TL, Alzheimer's disease: synapses gone cold, Mol. Neurodegener 6 (2011) 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Davies P, Maloney AJ, Selective loss of central cholinergic neurons in Alzheimer's disease, Lancet. 2 (1976) 1403. [DOI] [PubMed] [Google Scholar]
- 79.Whitehouse PJ, Price DL, Struble RG, Clark AW, Coyle JT, Delon MR, Alzheimer's disease and senile dementia: loss of neurons in the basal forebrain, Science. 215 (1982) 1237–1239. [DOI] [PubMed] [Google Scholar]
- 80.Small DH, Mok SS, Bornstein JC, Alzheimer's disease and Abeta toxicity: from top to bottom, Nat. Rev. Neurosci 2 (2001) 595–598. [DOI] [PubMed] [Google Scholar]
- 81.Jackson J, Jambrina E, Li J, Marston H, Menzies F, Phillips K, Gilmour G, Targeting the Synapse in Alzheimer's Disease, Front. Neurosci 13 (2019) 735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Davies CA, Mann DM, Sumpter PQ, Yates PO, A quantitative morphometric analysis of the neuronal and synaptic content of the frontal and temporal cortex in patients with Alzheimer's disease, J. Neurol. Sci 78 (1987) 151–164. [DOI] [PubMed] [Google Scholar]
- 83.Terry RD, Masliah E, Salmon DP, Butters N, DeTeresa R, Hill R, Hansen LA, Katzman R, Physical basis of cognitive alterations in Alzheimer's disease: synapse loss is the major correlate of cognitive impairment, Ann. Neurol 30 (1991) 572–580. [DOI] [PubMed] [Google Scholar]
- 84.Swerdlow RH, Mitochondria and Mitochondrial Cascades in Alzheimer's Disease, J. Alzheimers Dis 62 (2018) 1403–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Swerdlow RH, Koppel S, Weidling I, Hayley C, Ji Y, Wilkins HM, Mitochondria, Cybrids, Aging, and Alzheimer's Disease, Prog. Mol. Biol. Transl. Sci 146 (2017) 259–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Aschrafi A, Schwechter AD, Mameza MG, Natera-Naranjo O, Gioio AE, Kaplan BB, MicroRNA-338 regulates local cytochrome c oxidase IV mRNA levels and oxidative phosphorylation in the axons of sympathetic neurons, J. Neurosci 28 (2008) 12581–12590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li P, Jiao J, Gao G, Prabhakar BS, Control of mitochondrial activity by miRNAs, J. Cell Biochem 113 (2012) 1104–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhou R, Wang R, Qin Y, Ji J, Xu M, Wu W, Chen M, Wu D, Song L, Shen H, Sha J, Miao D, Hu Z, Xia Y, Lu C, Wang X, Mitochondria-related miR-151a-5p reduces cellular ATP production by targeting CYTB in asthenozoospermia, Sci. Rep 2 (2015) 17743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Siengdee P, Trakooljul N, Murani E, Schwerin M, Wimmers K, Ponsuksili S, MicroRNAs Regulate Cellular ATP Levels by Targeting Mitochondrial Energy Metabolism Genes during C2C12 Myoblast Differentiation, PLoS One. 10 (2015) e0127850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Irie K, Tsujimura K, Nakashima H, Nakashima K, MicroRNA-214 Promotes Dendritic Development by Targeting the Schizophrenia-associated Gene Quaking (Qki), J. Biol. Chem 291 (2016) 13891–13904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Giusti SA, Vogl AM, Brockmann MM, Vercelli CA, Rein ML, Trümbach D, Wurst W, Cazalla D, Stein V, Deussing JM, Refojo D, MicroRNA-9 controls dendritic development by targeting REST, Elife. 18 (2014) e02755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kao YC, Wang IF, Tsai KJ, miRNA-34c Overexpression Causes Dendritic Loss and Memory Decline, Int. J. Mol. Sci 19 (2018) 2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang S, Yan ML, Yang L, An XB, Zhao HM, Xia SN, Jin Z, Huang SY, Qu Y, Ai J, MicroRNA-153 impairs hippocampal synaptic vesicle trafficking via downregulation of synapsin I in rats following chronic cerebral hypoperfusion, Exp. Neurol 332 (2020) 113389. [DOI] [PubMed] [Google Scholar]
- 94.Basavaraju M, de Lencastre A, Alzheimer's disease: presence and role of microRNAs, Biomol. Concepts 7 (2016) 241–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rylett RJ, Ball MJ, Colhoun EH, Evidence for high affinity choline transport in synaptosomes prepared from hippocampus and neocortex of patients with Alzheimer's disease, Brain Res. 289 (1983) 169–175. [DOI] [PubMed] [Google Scholar]
- 96.Hardy J, Cowburn R, Barton A, Reynolds G, Lofdahl E, O'Carroll AM, Wester P, Winblad B, Region-specific loss of glutamate innervation in Alzheimer's disease, Neurosci. Lett 73 (1987) 77–80. [DOI] [PubMed] [Google Scholar]
- 97.Wang DS, Lipton RB, Katz MJ, Davies P, Buschke H, Kuslansky G, Verghese J, Younkin SG, Eckman C, Dickson DW, Decreased neprilysin immunoreactivity in Alzheimer disease, but not in pathological aging, J. Neuropathol. Exp. Neurol 64 (2005) 378–385. [DOI] [PubMed] [Google Scholar]
- 98.Cohen JE, Lee PR, Chen S, Li W, Fields RD, MicroRNA regulation of homeostatic synaptic plasticity, Proc. Natl. Acad. Sci. USA 108 (2011) 11650–11655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yan ML, Zhang S, Zhao HM, Xia SN, Jin Z, Xu Y, Yang L, Qu Y, Huang SY, Duan MJ, Mao M, An XB, Mishra C, Zhang XY, Sun LH, Ai J, MicroRNA-153 impairs presynaptic plasticity by blocking vesicle release following chronic brain hypoperfusion, Cell Communication and Signaling. 18 (2020) 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mathew RS, Tatarakis A, Rudenko A, Johnson-Venkatesh EM, Yang YJ, Murphy EA, Todd TP, Schepers ST, Siuti N, Martorell AJ, Falls WA, Hammack SE, Walsh CA, Tsai LH, Umemori H, Bouton ME, Moazed D, A microRNA negative feedback loop downregulates vesicle transport and inhibits fear memory, eLife. 5 (2016) e22467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Liu D, Tang H, Li XY, Deng MF, Wei N, Wang X, Zhou YF, Wang DQ, Fu P, Wang JZ, Hébert SS, Chen JG, Lu Y, Zhu LQ, Targeting the HDAC2/HNF-4A/miR-101b/AMPK pathway rescues tauopathy and dendritic abnormalities in Alzheimer's Disease, Molecular therapy: the journal of the American Society of Gene Therapy, 25 (2017) 752–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Barbato C, Giacovazzo G, Albiero F, Scardigli R, Scopa C, Ciotti MT, Strimpakos G, Coccurello R, Ruberti F, Cognitive decline and modulation of Alzheimer's disease-related genes after inhibition of MicroRNA-101 in mouse hippocampal neurons, Molecular Neurobiology. 57 (2020) 3183–3194. [DOI] [PubMed] [Google Scholar]
- 103.Xu N, Li AD, Ji LL, Ye Y, Wang ZY, Tong L, miR-132 regulates the expression of synaptic proteins in APP/PS1 transgenic mice through C1q, Eur. J. Histochem 63 (2019) 3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Xie AJ, Hou TY, Xiong W, Huang HZ, Zheng J, Li K, Man HY, Hu YZ, Han ZT, Zhang HH, Wei N, Wang JZ, Liu D, Lu Y, Zhu LQ, Tau overexpression impairs neuronal endocytosis by decreasing the GTPase dynamin 1 through the miR-132/MeCP2 pathway, Aging cell. 18 (2019) e12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rodriguez-Ortiz CJ, Prieto GA, Martini AC, Forner S, Trujillo-Estrada L, LaFerla FM, Baglietto-Vargas D, Cotman CW, Kitazawa M, miR-181a negatively modulates synaptic plasticity in hippocampal cultures and its inhibition rescues memory deficits in a mouse model of Alzheimer's disease, Aging Cell. 3 (2020) e13118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wu BW, Wu MS, Guo JD, Effects of microRNA-10a on synapse remodeling in hippocampal neurons and neuronal cell proliferation and apoptosis through the BDNF-TrkB signaling pathway in a rat model of Alzheimer's disease, J. Cell Physiol 233 (2018) 5281–5292. [DOI] [PubMed] [Google Scholar]
- 107.Lee K, Kim H, An K, Kwon OB, Park S, Cha JH, Kim MH, Lee Y, Kim JH, Cho K, Kim HS, Replenishment of microRNA-188-5p restores the synaptic and cognitive deficits in 5XFAD Mouse Model of Alzheimer's Disease, Sci. Rep 6 (2016) 34433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Li F, Wei G, Bai Y, Li Y, Huang F, Lin J, Hou Q, Deng R, Zhou JH, Zhang SX, Chen DF, MicroRNA-574 is involved in cognitive impairment in 5-month-old APP/PS1 mice through regulation of neuritin, Brain Res. 1627 (2015) 177–188. [DOI] [PubMed] [Google Scholar]