Abstract
It is unknown whether brain astrocytes and microglia have the capacity to present microbial antigens via the innate immune MR1/MAIT cell axis. We have detected MAIT cells in the normal mouse brain and found that both astrocytes and microglia are MR1+. When we stimulated brain astrocytes and microglia with E. coli, and then co-cultured them with MAIT cells, MR1 surface expression was upregulated and MAIT cells were activated in an antigen-dependent manner. Considering the association of MAIT cells with inflammatory conditions, including those in the CNS, the MR1/MAIT cell axis could be a novel therapeutic target in neuroinflammatory disorders.
Keywords: MR1, MAIT cells, Astrocytes, Microglia, Innate Immunity, Antigen Presentation
1. Introduction
The brain constitutes a unique immune-privileged organ. It is separated from the peripheral blood immune system by the presence of the blood brain barrier (BBB) that makes it “invisible” to immune cells (Larochelle et al. , 2011). However, the brain contains two unique resident cells, astrocytes and microglia, with the ability to activate immune responses via antigen presentation and hence, they constitute antigen presenting cells (APCs) in the brain (Aloisi et al. , 1998, Rostami et al. , 2020). These cells participate as first line defenders against inflammation and infection. In healthy conditions, their ability to communicate with immune cells is restricted by the presence of the BBB and low expression of antigen presenting molecules on their cell surface. However, an infection in the brain not only results in immune cells and microbial infiltration into the CNS by compromising the BBB, but it also up-regulates the expression of antigen presenting molecules on their surface, allowing them to efficiently stimulate various classes of T cells. Mucosal-associated invariant T (MAIT) cells are innate-like T cells that are highly enriched in the gut mucosa and constitute up to 10% of total T cells in adult human blood (Dusseaux et al. , 2011, Gherardin et al. , 2018). They recognize microbial vitamin B-derived metabolites (Corbett et al. , 2014) presented by MR1 (Huang et al. , 2005, Treiner et al. , 2003), a non-polymorphic major histocompatibility complex (MHC) class Ib molecule (Kjer-Nielsen et al. , 2012). MR1 surface expression is generally expressed at low levels on the cell surface under healthy conditions; it is subsequently upregulated in various disease states or cell exposure to microbial antigens (Huang, Gilfillan, 2005, Karamooz et al. , 2018, Kjer-Nielsen, Patel, 2012, Liu and Brutkiewicz, 2017, McWilliam and Villadangos, 2018, Ussher et al. , 2016).
When MAIT cells recognize the antigen/MR1 complex on APCs via their T cell receptor, they become activated, robustly secrete proinflammatory cytokines e.g., IFN-γ, TNF-α, IL-17 and GM-CSF (Dusseaux et al., 2011) and rapidly release cytotoxic molecules e.g., perforin and granzymes (Kurioka et al. , 2015, Le Bourhis et al. , 2013, Leeansyah et al. , 2015); this results in the lysis of the infected cells (Le Bourhis et al., 2013; Kurioka et al., 2015). This MR1-dependent mode of activation is operative in the case of infections with bacteria and fungi (Gold et al. , 2010, Le Bourhis et al. , 2010). Additionally, MAIT cells can be activated independently of MR1 recognition, in response to cytokines produced by infected cells, as observed previously in cases of several different virus infections (Hinks et al. , 2019, van Wilgenburg et al. , 2018); this mode of activation is termed,“MR1-independent activation” (Loh et al., 2016; Paquin-Proulx et al., 2018; (Suliman et al. , 2019, van Wilgenburg, Loh, 2018). Additionally, we and others have shown that Toll-like receptor signaling can modulate MR1 expression (Liu and Brutkiewicz, 2017, Ussher, van Wilgenburg, 2016).
Increasing evidence has highlighted the role of MAIT cells in neurodegenerative diseases such as multiple sclerosis (Held et al. , 2015, Willing et al. , 2014), brain tumors (Peterfalvi et al. , 2008) and other brain dysfunctions (Illés et al. , 2004).
In neurodegenerative diseases, MAIT cells infiltrate the CNS lesions, secrete higher amounts of pro-inflammatory cytokines, causing inflammation and thereby resulting in a poor outcome of the disease (Held, Bhonsle-Deeng, 2015, Willing, Leach, 2014). Considering that MAIT cells develop in the thymus (Legoux et al. , 2019a, Legoux et al. , 2019b, Salou et al. , 2019) and are highly abundant in the human gut mucosa (Giuffrida et al. , 2018, Toubal et al. , 2020), how they accumulate into the inflammatory lesions of MS and EAE (Van Kaer et al. , 2019) or brain tumors (Peterfalvi, Gomori, 2008) remains elusive. One hypothesis could be that activated MAIT cells acquire an effector-memory phenotype, express chemokine receptors and the very late antigen-4 (VLA-4) protein on the cell surface that helps their penetration into the brain, as it does in the liver (Jeffery et al. , 2016); this would be one way for MAIT cells to interact with other immune and non-immune brain-resident cells.
In the brain, astrocytes and microglia are known to maintain tissue homeostasis (Jha et al. , 2019). While the primary role of microglia is to protect the CNS from any infection (Chen et al. , 2019), astrocytes support the function of neurons by providing growth factors (Juaristi et al. , 2019). They also express several antigen presenting molecules (e.g., MHC-I, MHC-II and CD1d) on their surface (Bauer et al. , 2007, Busshoff et al. , 2001, Hoftberger et al. , 2004, Muir et al. , 2020, Rostami, Fotaki, 2020, Sheffield and Berman, 1998) and can therefore communicate with the immune cells that infiltrate the brain from the periphery during various environmental insults.
In terms of MR1, its mRNA expression in the brain has been reported in humans (Peterfalvi, Gomori, 2008), mice (Riegert et al. , 1998) and rats (Walter and Günther, 1998). However, these studies do not provide information at the protein level, which is critical for understanding the function of MR1-expressing cells. Also, which brain cell types express MR1 and how they interact with MAIT cells are equally important to know, in order to fully understand their function in neuropathogenesis. Chronic MAIT cell activation seems to be a common pathogenic mechanism in neurodegenerative and other inflammatory disorders (Giuffrida, Corazza, 2018, Hinks, 2016). Therefore, studies on the regulation of MAIT cell activation are important to better understand the mechanisms involved in neuroinflammatory disorders. Despite the information (albeit limited) implicating MAIT cells in neurological diseases, we decided to ask a fundamental question to help us begin to understand the role of the MR1/MAIT cell axis in the brain: is the MR1 present in the brain functional? That is, can it stimulate MAIT cells in an antigen-dependent manner? Here, we have assessed the functional expression of MR1 in brain astrocytes and microglia under normal conditions.
2. Materials and methods
2.1. Mice
Wildtype C57BL/6 mice were purchased from The Jackson Laboratory (Bar Harbor, ME). MR1-deficient (MR1 KO) mice (Treiner, Duban, 2003), were kindly provided by Dr. Daniel Hoft (St. Louis University, St. Louis, MO) and bred onsite. All procedures were approved by the Indiana University, School of Medicine’s Institutional Animal Care and Use Committee. Both male and female mice were used. In some experiments, they were used at an age of P3 (primary cultures of brain astrocytes and microglia) or 5 months (for isolation of brain resident MAIT cells).
2.2. Cell lines
The mouse brain cell line 8D1A (astrocyte-derived) was obtained from the American Type Culture collection (ATCC; Manassas, VA). The murine microglia-derived BV-2 cell line (Blasi et al. , 1990) was originally developed by Dr. Elisabetta Blasi (University of Perugia) and kindly provided by Dr. Linda Van Eldik (University of Kentucky, Lexington, KY).These cells were grown in DMEM (Lonza, Walkersville, MD) and DME:F12 medium (GE Healthcare Life Sciences, Logan UT), containing 10% FBS, 1% penicillin/streptomycin and 2 mM L-glutamine, respectively. The MAIT hybridoma cell 8D12 (Tilloy et al. , 1999, Treiner, Duban, 2003), was a kind gift from Dr. Shouxiong Huang (Cincinnati Children’s Hospital, Cincinnati, OH) and maintained in DMEM supplemented with 10% FBS.
2.3. Bacteria preparation
E. coli (DH5-α strain) was grown in LB media overnight and the colony forming units (CFU) as CFU/ml were determined. An E. coli bacterial pellet was washed in PBS and fixed in 1% paraformaldehyde (PFA) for 10 min at room temperature. After washing twice in PBS, the cell number was adjusted to 1×1010 CFU/ml in sterile PBS, aliquoted and stored at −80°C until further use.
2.4. Primary astrocytes and microglia culture generation
Primary brain astrocytes and microglia cultures were prepared as described previously with some modifications (McCarthy and de Vellis, 1980). Briefly, 3-day old C57BL/6 (P3) mice were euthanized by rapid decapitation with sharpened scissors. After euthanasia, the cortex was harvested and chopped finely with a razor blade. The cells were resuspended in 0.25% trypsin (HyClone, Logan, UT) and incubated for 20 min at 37°C in a water bath. The cells were triturated gently and centrifuged at 2,000 rpm for 5 min at 4°C.The supernatant was discarded and the cells were resuspended in fresh DMEM10 (DMEM containing 10 % FBS) media and seeded in poly-D lysine (PDL, 50μg/ml; Millipore Sigma, St. Louis, MO)-coated 75cm2 flasks. After 2 days, the medium was replaced with fresh DMEM10. After 7 days, when the astrocytes layer became confluent, the flasks were kept on an orbital shaker at 180 rpm for 30 min. The supernatants containing the microglia were harvested, centrifuged and transferred to a new PDL-coated flask containing DME:F12 complete media. The original astrocytes flask was replenished with 20 ml of fresh DMEM10 media and returned back to the orbital shaker for 6 h at 220 rpm to remove the oligodendrocytes. The supernatants containing oligodendrocytes were discarded and the remaining layer constituting astrocytes was trypsinized and incubated for an additional 14 days in DMEM10.
2.5. Immunofluorescence analyses
Astrocyte and microglia cell lines and primary purified cultures were seeded on PDL-coated cover glasses in 24-well plates at a density of 1×105cells/well. After overnight adherence, the cells were blocked with 5% goat serum for 30 min on ice. After washing with PBS, the cells were incubated either with an anti-MR1 mouse mAb hybridoma supernatant (26.5; (Huang, Gilfillan, 2005) or TW2.3 hybridoma supernatant (Yuwen et al. , 1993) followed by a 1h incubation with a Texas Red-conjugated goat anti-mouse antibody (Thermo Fisher Scientific, Eugene, OR). The cells were washed three times with cold PBS, fixed in cold 4 % PFA for 10 min at room temperature and permeabilized with 0.1% Triton X-100. The cells were blocked with mouse serum for 30 min on ice. The cells were then incubated with either a mouse Alexa-Fluor-488-conjugated anti-GFAP (Clone GA5; eBioscience) or FITC-labeled mouse anti-CD11b (Clone M1/70; BD Biosciences) for 1 h at room temperature in the dark. After washing three times with PBS, the cells were mounted on microscope slides using Vectashield antifade mounting medium (Vector Laboratories, Burlingame, CA) with 4′-diamidino-2-phenylindole (DAPI; Vector laboratories) and left to air dry. Images were captured on a Zeiss Axio Imager M2 microscope using a 20x objective.
2.6. Mouse brain single-cell homogenates
To prepare single-cell suspensions of whole brain cells, mice were sacrificed at 5 months of age. For MAIT cell detection in the brain, before perfusion, the mice were injected i.v. with 2 μg of CD45.2 mAb (PerCP/Cy5.5 labeled; Clone104; BioLegend) dissolved in 200 μl PBS, using a 1 ml insulin syringe as described previously (Anderson et al., 2014). After 3 min, the animals were perfused with 15 ml of PBS to wash out the peripheral blood. The brains were dissected out, the meninges layer was removed and both of the cerebral cortices were extracted. The cortices were cut into small pieces using a razor blade and homogenized thoroughly, yet gently, using a 15 ml dounce homogenizer. The homogenized cells were passed through a 40 μm filter and centrifuged at 2,000 rpm for 5 min at 4°C. The supernatant was discarded and the pellets were resuspended in 12 ml of 40% Percoll (GE Healthcare, Uppsala, Sweden). The cells were centrifuged at 2,200 rpm for 20 min at room temperature with the brake off. Cells were collected from the pellet and washed with ice cold FACS buffer (PBS containing 2% FBS). The cells were counted in a hemocytometer and the concentration was adjusted to 1×106 cells/ml. For MAIT cell detection, two or more mouse brains were pooled in order to obtain a sufficient number of cells for flow cytometric analysis.
2.7. Flow cytometry
In order to detect MR1 expression in brain cell suspensions and cell lines, isolated cells (1×105 cells/ml) were first incubated with anti-mouse CD16/32 mAb supernatant (the hybridoma was a kind gift from Dr. J. Yewdell, NIH) for 20 min on ice to block Fc receptors followed by staining with an APC-conjugated anti-MR1 mAb (Clone 26.5; BioLegend, San Diego, CA) for 30 min on ice in FACS buffer. An APC-labeled mouse IgG2a mAb (BioLegend) was used as an isotype control. After washing three times with PBS, the microglia cell line and primary microglia were stained with a FITC-conjugated anti-CD11b mAb (Clone M1/70; BD Biosciences) for 30 minutes on ice. After washing with PBS, the cells were fixed in 1% PFA for 10 min at room temperature, followed by permeabilization using 0.01% Triton X-100 for 5 min. Next, the cells were stained with an Alexa 488-labeled anti-GFAP mAb or a FITC-conjugated anti-IBA1 mAb (Abcam) for 30 min on ice and washed twice with FACS buffer.
For the detection of MAIT cells in the brain, the single cell suspensions generated above were first blocked with the anti-CD16/32 mAb followed by staining with anti-CD44 (Pacific Blue- labeled; Clone IM7, BioLegend), anti-F4/80 (FITC-labeled; Clone BM8, BioLegend), anti-CD45R/B220 (FITC-labeled; Clone RA3–6B2, BioLegend), anti-TCR-β (PE-labeled; Clone H57–597, BioLegend) mAbs, and a 5-OP-RU-loaded APC-conjugated MR1 tetramer (NIH Tetramer Core) for 30 min on ice. After washing three times with PBS, the cells were fixed and used for flow cytometry analysis. All samples were acquired on an LSRFortessa™cell analyzer (BD Biosciences). Cells that were stained with an isotype control mAb were used as negative controls. The gating strategy for the experiment shown in Fig. 1D was as follows: 1. Single cells (identified in the FSC-H vs. FSC-A profile) were then analyzed by a FSC vs SSC profile. 2. GFAP+ (astrocytes) and Iba1+ (microglia) cells were identified and gated. 3. These gated populations were then analyzed for their expression of MR1. The data were analyzed using FlowJo software (BD Biosciences).
Fig. 1.
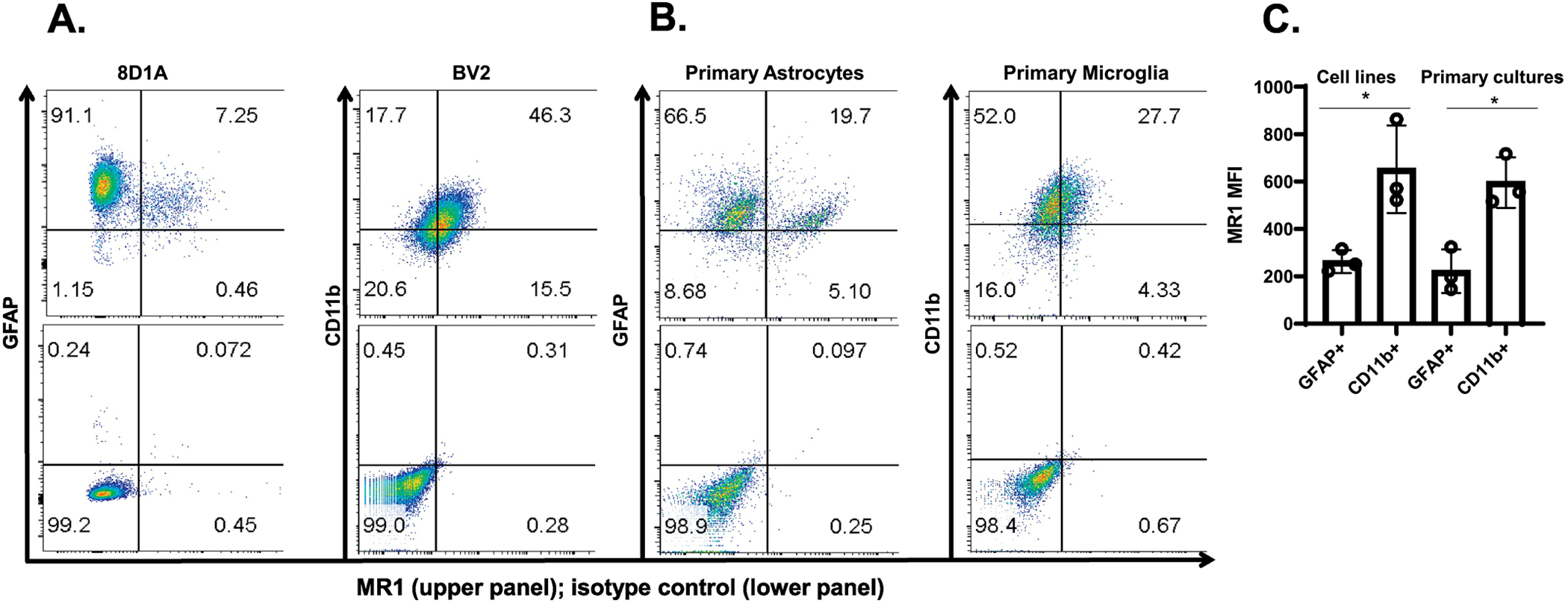
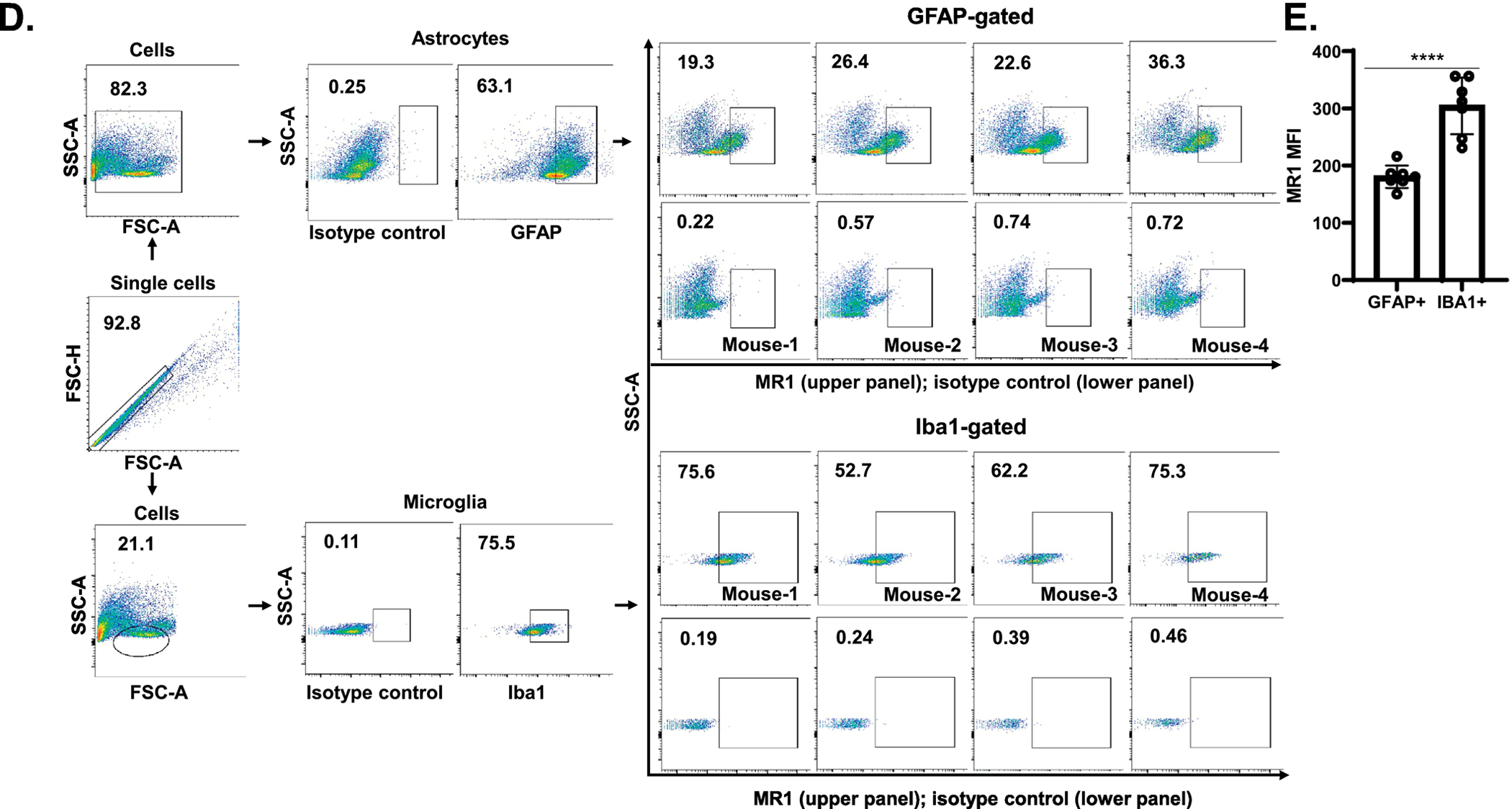
Analysis of MR1 surface expression by mouse brain astrocytes and microglia. A & B. An astrocyte cell line (8D1A), microglia cell line (BV2) and, primary astrocytes and primary microglia were double-stained using APC-labeled anti-MR1 and FITC-labeled anti-GFAP/CD11b mAbs and analyzed by flow cytometry. The percent of MR1+ cells is indicated in the upper panel of each quadrant, whereas the lower panel shows the isotype control for each group. A representative dot plot of one of the three experiments performed is shown. C. The MFI of MR1 in the GFAP+ and CD11b+ cells is shown and the data are presented as the mean ± SD from three independent experiments. D. Brain single cell suspensions from mice were double-stained with APC-labeled anti-MR1 and FITC-labeled anti-GFAP/anti-Iba1 mAbs and analyzed by flow cytometry. The GFAP+ and Iba1+ cells were first gated (the percentage of cells in the gate is indicated) and the percent of MR1+ cells was then analyzed in the population of GFAP+ and Iba1+ cells. The percent of MR1+ cells in GFAP+ and Iba1+ gated cells is indicated for each individual mouse (N=4) in the top row of the upper and lower panels, respectively, whereas the bottom row in both shows the isotype control for each group. The results show data from a representative experiment of two performed. E. MFI of MR1 in brain GFAP+ and Iba1+ cells are shown. The symbols represent individual mice and the data are shown for all analyzed mice (N=7) from two independent experiments. The data are shown as the mean ± SD. *, p<0.05, ****, p<0.0001, by an unpaired Student’s t-test.
2.8. Quantitative real-time PCR analysis
RNA was isolated from the brain-derived cell lines, purified primary cultures and mouse brain tissue using the RNeasy Mini Kit (Qiagen, Valencia, CA). As a calibrator, RNA from purified splenic B cells and intact spleen tissue was used as described previously (Livak and Schmittgen, 2001). The quality of RNA was assessed by spectrophotometry before analysis. A total of 1 μg of RNA was converted into cDNA using the Transcriptor First Strand cDNA synthesis kit (Roche Diagnostics, Mannheim, Germany) and random hexamer primers, according to the manufacturer’s instructions. Quantitative PCR was carried out in 10 μl reaction volumes containing 5 μl of 2X TaqMan Fast Advanced Master Mix (Applied Biosystems, Thermo Fisher Scientific), 1.0 μl cDNAs, 0.5 μl of each each primer (MR1 and GAPDH) and 3 μl of PCR grade water using the QuantStudio 6 Flex Real-Time PCR system (Applied Biosystems). The PCR conditions used were an initial hold stage at 95°C for 20 sec, followed by denaturing at 95°C for 1 sec, and annealing/extension stages at 60°C for 20 sec for 40 cycles. The probe for MR1 was labeled with the reporter dye FAM (Mm00468487_m1 Mr1) and the probe for GADPH was labeled with the reporter dye VIC (house-keeping gene, Mm 99999915_g1). Because of the different reporter dyes, amplification of MR1 and GAPDH was performed in the same well in three replicates. RNA from the spleen-derived B cells and spleen tissue was used as calibrators to determine the expression of MR1 in brain cell lines and whole brain tissue, respectively. The amount of MR1, normalized to GAPDH and relative to splenic B cells, was calculated by the 2−ΔΔ CT method as previously described (Livak and Schmittgen, 2001).
2.9. MR1 upregulation assay
Cell lines, as well as brain primary astrocytes and microglia, were cultured for 6 h in the presence or absence of PFA-fixed E. coli at a multiplicity of infection (MOI) of 300 in triplicate wells of 96-well U-bottom microtiter plates (1×105 cells/well). Then, both the E. coli-stimulated and control cells were stained with the anti-MR1 mAb and analyzed by flow cytometry as described above.
2.10. MAIT cell activation assay
To measure the ability to present E. coli-derived antigens to MAIT cells, primary brain astrocyte and microglia cultures, as well as the 8D1A and BV2 cell lines were mock-treated or treated with E. coli at an MOI of 300 for 6 h. To confirm our findings, we also included brain cells from MR1-deficient (MR1 KO) mice that are known to lack MAIT cells (Treiner, Duban, 2003), as a negative control. Next, the cells were washed three times with PBS and incubated with either a purified anti-MR1 blocking antibody (Clone 26.5; BioLegend, San Diego, CA) or purified mouse IgG2a isotype control (Clone MOPC-173; BioLegend) in 100μl of DMEM for 1 h at 37°C. These cells were co-cultured with the 8D12 mouse MAIT cell hybridoma in triplicate wells of 96-well U-bottom microtiter plates at a 1:1 MAIT:Target cell ratio (1 × 105 cells each / well / 200μl media). As controls, MAIT and target cells were cultured alone either in the presence of E. coli (MOI = 300) or PBS. After 48 hours of culture, cell supernatants were harvested, and IL-2 production was measured by ELISA.
2.11. Statistical analysis
The results were analyzed by an unpaired two-tailed Student’s t-test using Prism software (version 6.05 for Windows; GraphPad, San Diego, CA). The displayed error bars in the graphs indicate mean ± SD. A P value less than 0.05 was considered significant.
3. Results
3.1. Astrocytes and microglia express MR1 on the cell surface
To assess MR1 expression in an astrocyte and microglia cell line, as well as in primary cells, we first checked the purity of the primary cultures using cell-specific mAbs by flow cytometry. The purity level was 97 and 85 % in the primary cultures of astrocytes and microglia, respectively (data not shown). The flow cytometry analyses showed that both the brain cell lines (Fig. 1A) as well as the primary purified cultures of astrocytes and microglia (Fig. 1B) express MR1 on their surface. The expression level of MR1, as measured by mean fluorescent intensity (MFI) in the cell lines or primary cultures, was significantly higher overall in microglia compared to astrocytes (Fig. 1C). However, there were two populations of GFAP+ cells (astrocytes)—an MR1-negative/MR1lo population and one that was MR1hi (Fig. 1A & B). The level of MR1 expression on the small population was somewhat higher than that in the single CD11b+ cell population (microglia). This was the case in both the cell lines and primary cultures (Fig. 1A & B). Nonetheless, in freshly-isolated brain single cell suspensions, the level of MR1 expression was significantly higher in microglia than astrocytes (Fig. 1D & 1E).
We also performed immunofluorescent microscopy analysis of the astrocyte and microglia cell lines, as well as our cultures of primary astrocytes and microglia. In line with our flow cytometry results, all of these cells expressed MR1 (Fig. 2). The expression of Mr1 RNA in astrocyte and microglia cell lines, primary cultures (Fig. 3A), and in brain tissue from healthy mice (Fig. 3B), was detected via qPCR using GAPDH as an internal control and RNA from purified splenic B cells/intact spleen tissue as a calibrator. These cells were used as a calibrator because they are known to express Mr1. Mr1 mRNA was expressed in all the samples tested. The mRNA expression level of MR1 in both the microglia cell line (BV2) and in primary microglia was higher than that in the astrocyte cell line (8D1A) or in primary astrocytes (Fig. 3A). Moreover, Mr1 mRNA was also detected in mouse brain tissue (Fig. 3B). Therefore, these data demonstrate that populations of astrocytes and microglia in the brain, as well as transformed cell lines and primary cultures derived from them, express MR1.
Fig. 2.
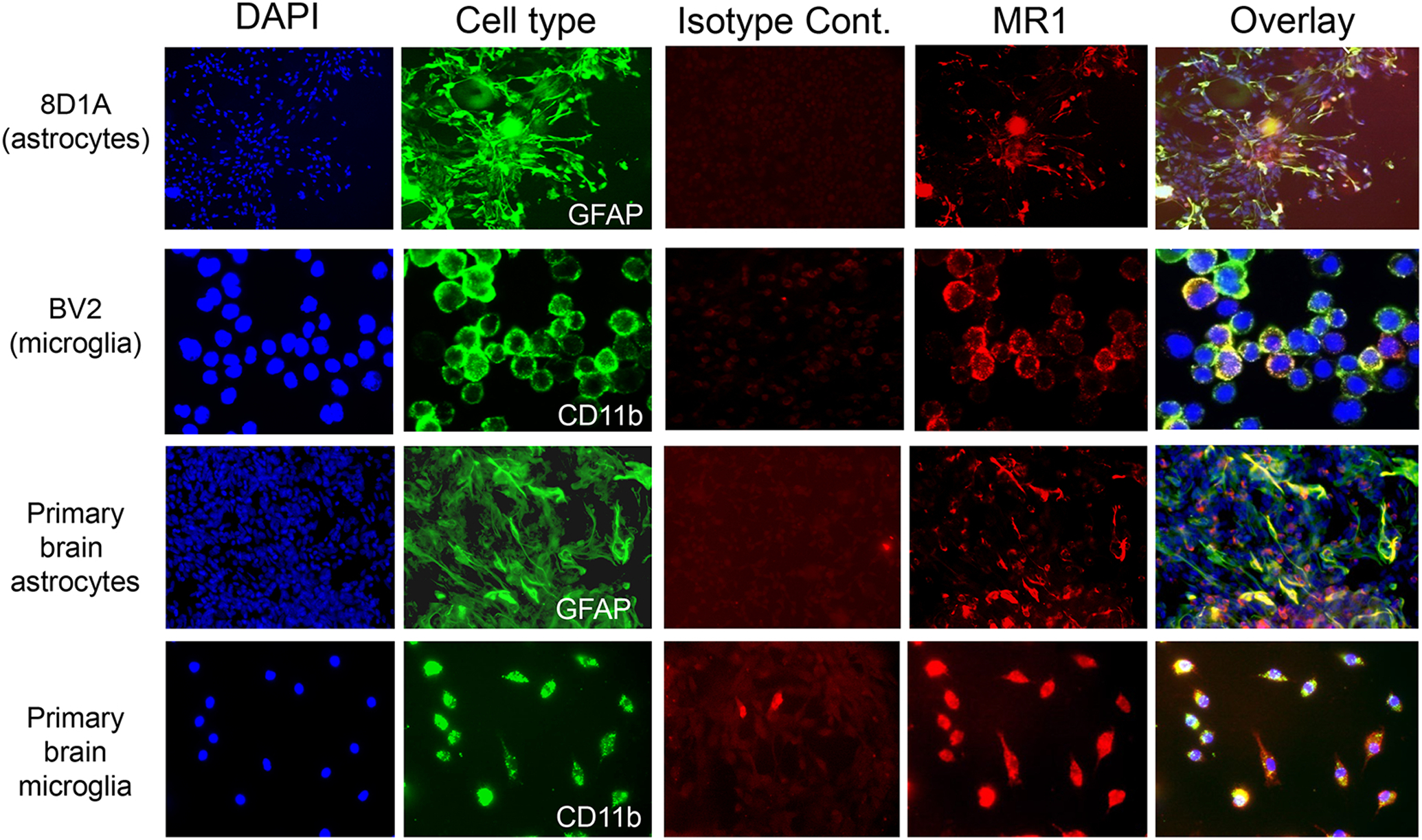
Immunofluorescent labelling of mouse brain astrocyte and microglia cell lines and primary cultures. The individual groups of cells were stained with the cell type-specific marker GFAP (astrocytes; green) or CD11b (microglia; green), as well as an anti-MR1 mAb (red) or an isotype control mAb (red). The nuclei were stained with DAPI (blue) and last column shows the overlay for each cell type. Magnification = 20X.
Fig. 3.
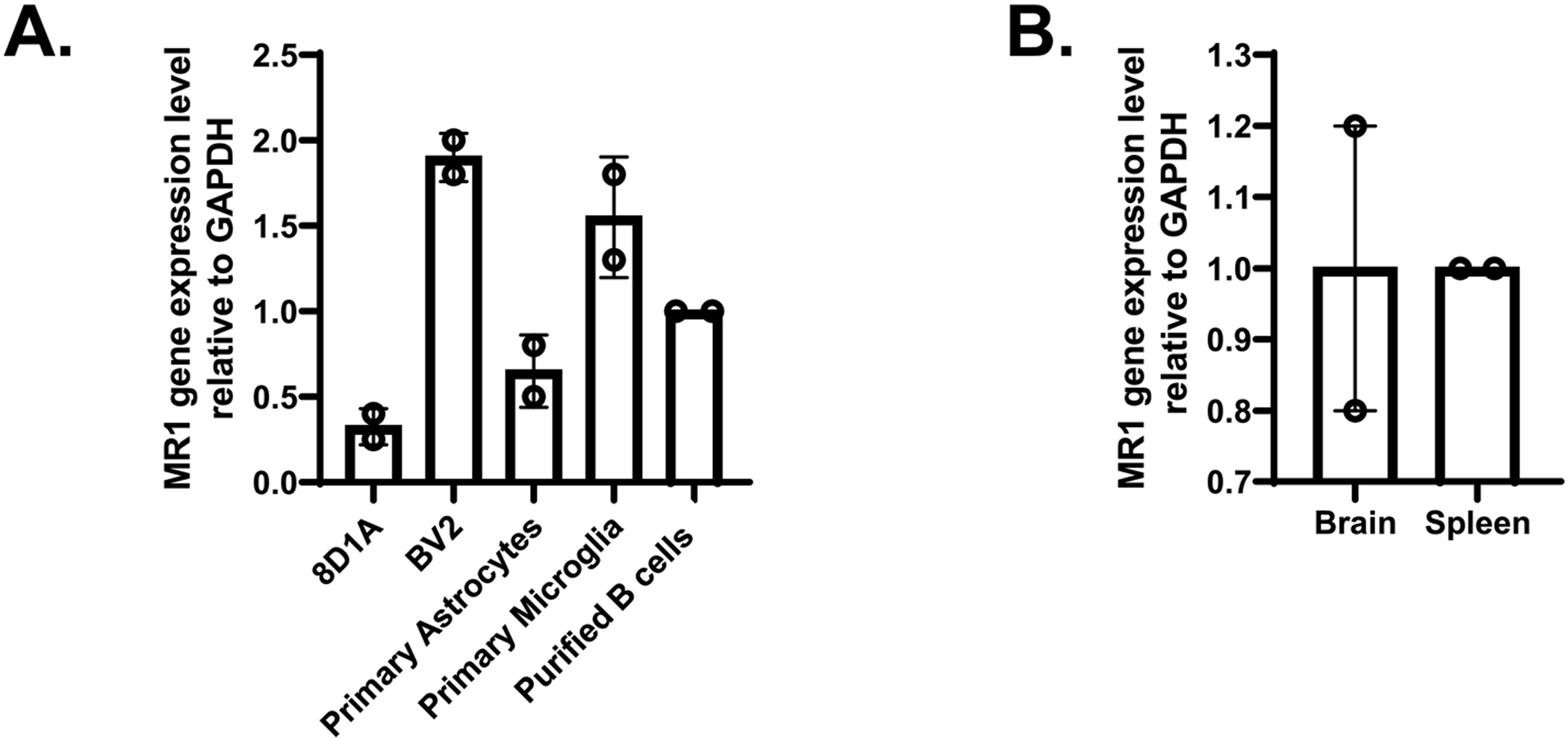
qPCR analysis of MR1 gene expression in A. 8D1A and BV2 cell lines, primary purified astrocyte and microglia cultures, and B. brain tissue. The housekeeping gene, GAPDH, was used as an internal control. RNA isolated from spleen-derived B cells and intact spleen tissue was used as a calibrator to determine the level of MR1 gene expression in A and B, respectively. The data are presented as the mean ± SD from two independent cDNA samples with three replicates from each sample.
3.2. Treatment with E. coli increases the surface expression of MR1 on brain cell lines and primary cells
We and others have shown that adding MR1 ligand-expressing bacteria to cells increases their levels of cell surface MR1 (Kjer-Nielsen, Patel, 2012, Liu and Brutkiewicz, 2017, Ussher, van Wilgenburg, 2016). To elucidate the effects of the presence of bacterial antigens on the surface expression of MR1 in brain cell lines and primary astrocytes and microglia cell cultures, the cells were stimulated for 6 hours with PBS or fixed E. coli at an MOI of 300 and subsequently stained for surface MR1 expression for flow cytometry analysis (Fig. 4A). An increase in cell surface expression of MR1 from 105 ± 14.7 in mock infected cells to 176.6 ± 16.9 in E. coli-stimulated cells was observed in 8D1A (astrocyte-derived) cells, whereas in the BV2 (microglia-derived) cell line, the increase was from 574 ± 9.2 to 907 ± 13. In primary cultures, MR1 expression in astrocytes increased from 135.3 ± 3.6 to 244.6 ± 20.4 and in microglia from 316 ± 43.15 to 489 ± 57.2 following stimulation with E. coli. A composite of the replicates of the MFI in this representative experiment is shown in Fig. 4B. Therefore, these data suggest that in these brain cells, expression of MR1 is upregulated in the presence of its bacterially-derived ligand.
Fig. 4.
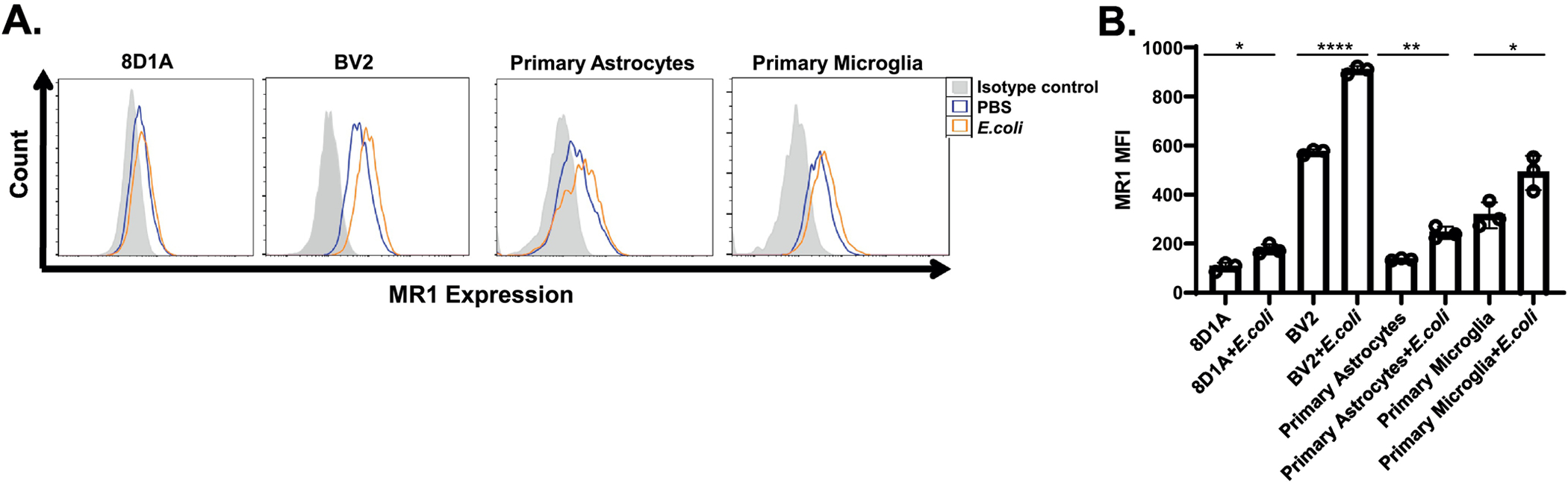
Upregulation of MR1 expression in brain astrocyte and microglia-derived cell lines and primary cultures by E. coli. A. Representative histogram overlays showing the cell surface staining of MR1 on the 8D12 and BV2 cell lines, as well as primary cultured astrocytes and microglia after 6 h of stimulation with PBS (negative control) or PFA-fixed E. coli at an MOI of 300. The grey shaded histogram is cells stained with the isotype control mAb, whereas the orange and blue line histograms are cells stained with an APC-labeled anti-MR1 antibody in the presence of E. coli or PBS, respectively. A representative experiment (out of three performed) is shown. B. The MFI levels of surface MR1 on the indicated cells after 6 h of stimulation with PBS or PFA-fixed E. coli at an MOI of 300 is shown. The data are representative of three independent experiments and expressed as the mean ± SD. *, p<0.05; **, p<0.01; ****, p<0.0001, by an unpaired Student’s t-test.
3.3. MAIT cells are detectable in the normal brain
MAIT cells have been reported to be present in the brain lesions of a variety of neurodegenerative diseases including multiple sclerosis (Held, Bhonsle-Deeng, 2015), found in the MS animal model, experimental allergic encephalomyelitis (Van Kaer, Postoak, 2019) and in brain tumors (Peterfalvi, Gomori, 2008), but it is not known if they are present in a healthy brain. To address this question, we performed flow cytometry on a single cell suspension generated from the brains of 5-month old mice using a mouse MR1 tetramer loaded with 5-OP-RU, that will specifically recognize MAIT cells (Corbett et al., 2014). As a negative control, we used a mouse MR1 tetramer loaded with 6-FP, that does not stain MAIT cells (Corbett et al., 2014). To discriminate vascular and tissue leukocytes, mice were injected i.v. with a PerCP/Cy5.5-labeled CD45.2-specific mAb. This approach allowed staining of all vascular leukocytes while excluding tissue resident leukocytes as described previously (Anderson et al. , 2014). MAIT cells in tissue were detected as a CD45.2-negative/B220-negative/F4/80-negative/TCR-β+/MR1Tetramer+/CD44+ cell population (Fig. 5A & 5B). As MAIT cells are present in mouse tissues at much lower levels than in humans (Godfrey et al. , 2019), we acquired a total of 10 million events for performing these experiments. To confirm our findings, we also included brain cells from MR1-deficient (MR1 KO) mice that are known to lack MAIT cells (Treiner, Duban, 2003), as a negative control. As a positive control, we stained MAIT cells in mouse liver and spleen, as these tissues are already known to constitute a percentage of total MAIT cells (Rahimpour et al. , 2015). Our results demonstrated that 1.07 ± 0.31 % of the gated cells were MAIT cells, which were clearly absent in MR1KO mice (Fig. 5C). The percentage of MAIT cells in liver and spleen was 0.62 ± .02 % and 0.11 ± .007 % respectively (Fig. 5C). Therefore, these data indicate that a small population of resident MAIT cells can be identified in the normal brains of wildtype mice.
Fig. 5.
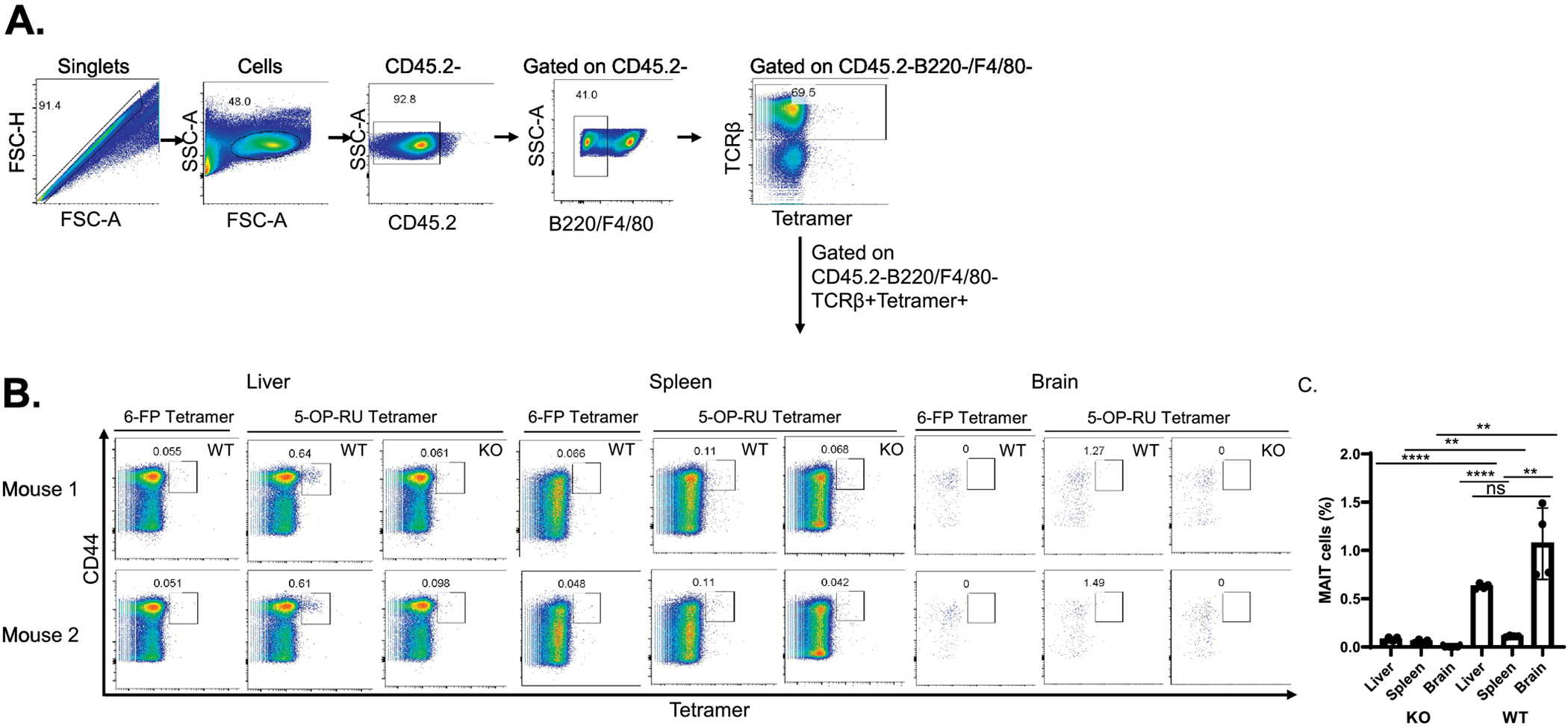
Detection of MAIT cells in the normal mouse brain. Five-month-old wildtype (WT) C57BL/6 and MR1 KO mice were injected i.v. with PerCP/Cy5.5-labeled anti-CD45.2 mAb. After 3 min, the animals were perfused with PBS to wash out the peripheral blood. A single cell suspension of brain cells from perfused mice was generated and stained with mAb against B220, F4/80, TCR-β, 5-OP-RU- or 6-FP (negative control)-loaded MR1 tetramers, and CD44. A. The gating strategy for the identification of MAIT cells is indicated. B. The identification of tissue-resident MAIT cells in the livers, spleens and brains of WT C57BL/6 mice and MR1 KO mice is shown. The left two-color plot in each group is the staining of WT cells with anti-CD44 and 6-FP-loaded MR1 tetramers, the latter serving as a negative control. The middle plot shows staining of WT cells with anti-CD44 and 5-OP-RU-loaded MR1 tetramers. The right panel in each group shows cells from MR1KO (i.e., MAIT cell-deficient) mice stained with anti-CD44 and 5-OP-RU-loaded MR1 tetramers. The percentage of the MAIT cells is shown in the plot. The results are representative of two independent experiments. (N=5 for wild type mice and N=4 for MR1 KO mice).
3.4. MR1-dependent activation of MAIT cells by brain APCs
Having demonstrated that MR1 is present in astrocytes and microglia, and that it can be upregulated upon exposure to its bacterial ligands, our final question was whether MR1 is actually functional on these cells. Thus, we performed an assay in which astrocytes and microglia (Fig. 6A), both cell lines and primary cell cultures (Fig. 6B), as well as freshly-isolated brain single cell suspensions from WT and MR1KO mice (Fig. 6C), were cocultured with MAIT cell hybridomas in the presence or absence of fixed E. coli. Forty-eight hours later, we harvested the supernatants and measured the levels of IL-2 by ELISA. E. coli-stimulated astrocytes and microglia induced a robust production of IL-2 by the MAIT cells. The activation of MAIT cells was dependent on MR1 expression, as blocking of MR1 with an MR1-specific mAb completely suppressed the secretion of IL-2 by the MAIT cells; this was not observed with the isotype control (Fig. 6A – C). As expected, the lack of E. coli in the co-culture assay resulted in no IL-2 release. Neither the MAIT cells nor the brain cells produced IL-2 when cultured alone; the addition of E. coli did not result in significant IL-2 release by brain cells, indicating that upon co-culture with E. coli-stimulated APCs, the MAIT cells (rather than the APCs) produced IL-2 (Fig. 6C). Therefore, these data demonstrate that brain astrocytes and microglia express functional MR1 that is capable of stimulating MAIT cells in the presence of bacteria-derived antigens.
Fig. 6.
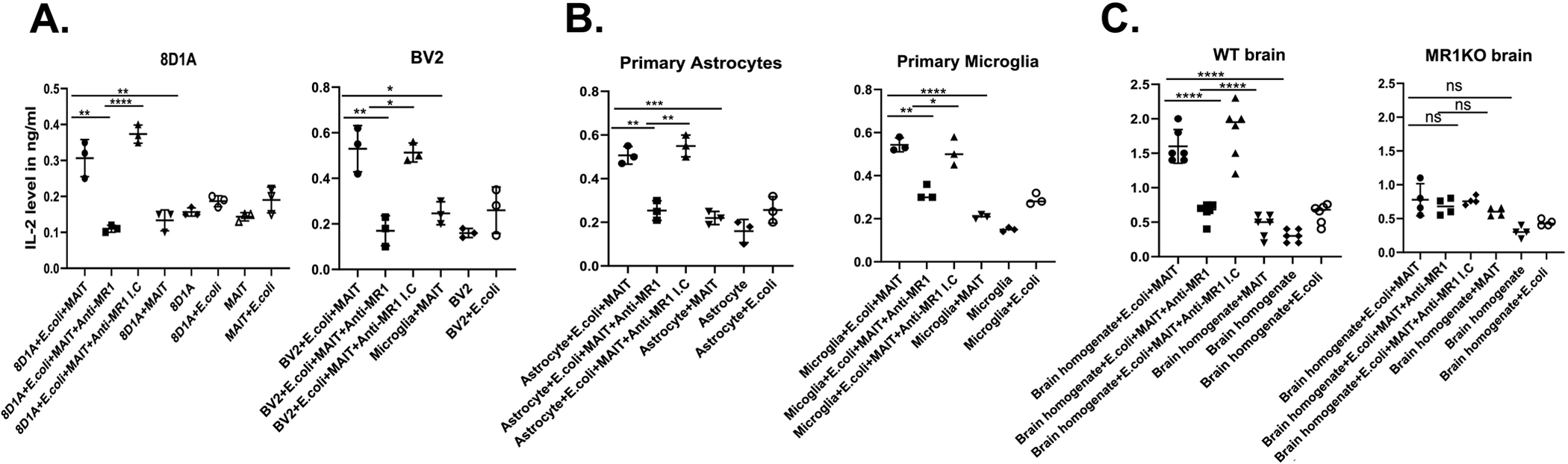
Activation of MAIT cells by E. coli-stimulated astrocytes and microglia. A. 8D1A and BV2 cell lines, B. Primary cultures of astrocytes and microglia, and C. Single cell suspensions from the brains of wild type (WT) and MR1 KO mice, were stimulated for 6 h with PBS or PFA-fixed E. coli at an MOI of 300. The cells were washed three times with PBS and incubated with either an MR1 blocking antibody or isotype control for 1 h, followed by the addition of the murine MAIT cell hybridoma, 8D12. As a control, the E. coli-stimulated target cells and MAIT cells were cultured alone. Following 48 h of culture, the supernatants were harvested and IL-2 production was measured by ELISA. The IL-2 level is expressed in ng/ml. A. & B. Each data point represents the mean of triplicate wells from one of three independent experiments. The data are inclusive of all three independent experiments and the error bars represent the mean ± SD. C. The data shown are from two combined, independent experiments with an N=6 for WT and N=4 for MR1KO mice. n.s, not significant; *, p< 0.05, **, p< 0.005, ***, p<0.0005; ****, p<0.0001; unpaired Student’s t-test.
4. Discussion
Major Histocompatibility Complex (MHC) class I molecules present peptide antigens to cytotoxic T cells (van de Weijer et al. , 2015). In general, cells in the CNS express relatively lower levels of MHC class I molecules under normal, steady state conditions (Corriveau et al. , 1998, Horwitz et al. , 1999, Johnson et al. , 2014). Upon an infection or following an injury/CNS disease, the MHC class I molecules are upregulated, most likely due to exposure of the cells to interferons and other pro-inflammatory cytokines (Horwitz, Evans, 1999, Johnson, Jin, 2014). As such, the peptides they present can be recognized by cytotoxic T cells and the cells subsequently lysed (van de Weijer, Luteijn, 2015). Type I interferons can also serve as co-stimulatory signals for MAIT cell activation (Lamichhane et al. , 2020). Obviously, this is especially problematic in the brain, where CNS cell death can result in functional and cognitive deficits.
There are also molecules related to the classical MHC class I glycoproteins, termed Class Ib or non-classical MHC class I molecules (Adams and Luoma, 2013, Brutkiewicz et al. , 2018, D’Souza et al. , 2019); many of these are also present in the CNS (Liu et al. , 2015, Tetruashvily et al. , 2016, Wiendl et al. , 2005, Wischhusen et al. , 2005), but they have not been extensively studied in that context. As a result, their functional capacity in terms of antigen presentation and their role in the CNS are poorly understood.
We study antigen presentation by the MHC class I-like molecule, MR1. Unlike classical MHC class I molecules, MR1 does not present peptides to the immune system; rather, it presents microbial-derived vitamin B metabolites to an innate T cell population called MAIT cells (Karamooz, Harriff, 2018, Keller et al. , 2017, Liu and Brutkiewicz, 2017, McWilliam and Villadangos, 2018). Many different cell types are MR1+ and can present antigen to MAIT cells (Karamooz, Harriff, 2018, Keller, Corbett, 2017, Liu and Brutkiewicz, 2017, McWilliam and Villadangos, 2018); these innate T cells mainly reside in human peripheral blood, liver and mucosal tissue (Godfrey, Koay, 2019). In the current report, we have demonstrated that MAIT cells are present in the normal brain as well.
Very little is known about the MR1/MAIT cell axis in the CNS. MAIT cells have recently been shown to be present in some neuroinflammatory disease states, such as in MS and the mouse model of MS, EAE (Van Kaer, Postoak, 2019). To the best of our knowledge, the functionality of this innate immune axis has not been studied in the normal CNS.
A number of CNS diseases or CNS injuries can result in a breaching of the BBB (Sweeney et al. , 2019). As a result, an influx of microbial pathogens can enter the CNS, inducing an inflammatory state due to pathogen-specific activation of Toll-like receptors (Kawai and Akira, 2011), upregulation of classical MHC molecules (Corriveau, Huh, 1998, Horwitz, Evans, 1999, Johnson, Jin, 2014, Liu, Shen, 2015) and the introduction of pro-inflammatory leukocytes and cytokines into the site of infection/injury (DiSabato et al. , 2016). We do not know the potential role of the MR1/MAIT cell axis in the neuroinflammation that develops in these disease states. In general, whether the site of inflammation is in the gut, such as with intestinal bowel disease (IBD) (Giuffrida, Corazza, 2018, Legoux, Bellet, 2019a, Ruijing et al. , 2012) or in the CNS with MS/EAE (Van Kaer, Postoak, 2019), MAIT cells are present. In the current report, we asked if, under normal conditions, CNS cell-specific MR1 had the ability to stimulate MAIT cells in a microbial antigen-specific manner.
Whether we used transformed cell lines or primary cultures of brain astrocytes and microglia, it was apparent that they were all MR1+ to various degrees. Furthermore, the addition of bacteria, E. coli in this case, caused an upregulation of MR1 in these cells, as we and others have shown in other cell type systems (Kjer-Nielsen, Patel, 2012, Le Bourhis, Dusseaux, 2013, Liu and Brutkiewicz, 2017). Each of these cell lines/primary cultured cells and freshly-isolated brain single cell suspensions demonstrated the effective capacity for MR1-mediated antigen presentation to MAIT cells. The presence of MAIT cells in the normal brain also leads one to speculate about the contribution of the MR1/MAIT cell axis in CNS diseases.
What does this mean as we think about the big picture? Is the influx of MAIT cells into sites of inflammation during disease good or bad for the patient? In IBD, this influx is associated with inflamed intestinal mucosa (Tominaga et al. , 2017). Moreover, it could be argued that in MS, with the neuroinflammation causing significant damage to the brain (Bjelobaba et al. , 2017), the presence of MAIT cells is also not helpful (Van Kaer, Postoak, 2019). We believe that the next steps ought to be investigations of MR1-deficient animal models in which MAIT cells are also absent (Treiner, Duban, 2003). It is already known that MAIT cells require an in vivo microbiome for their development (Legoux, Bellet, 2019a, Treiner, Duban, 2003); in fact, mice born into a germ-free environment do not have MAIT cells (Treiner, Duban, 2003). Thus, knowing that following BBB damage, microbes can enter the CNS with the ability to activate the MR1/MAIT cell axis, possibly contributing to the neuroinflammation observed in these disorders, may in the future lead to the therapeutic targeting of this innate immune axis in a variety of CNS diseases and injuries.
5. Conclusion
We have studied the functional expression of the MR1/MAIT cell axis using brain astrocytes and microglia, as a means to determine if they are capable of presenting bacteria-derived antigens to MAIT cells. Transformed cells lines, as well as primary cultures of brain astrocytes and microglia, stimulated MAIT cells when treated with E. coli, demonstrating that these cells can present microbial antigens via MR1. Moreover, the normal brain itself has a population of resident MAIT cells. As such, in various CNS diseases and injuries in which there could be a breach of the BBB, potentially allowing microbial pathogens to enter the brain, the MR1/MAIT cell axis could be an important underlying cause of neuroinflammation in these disorders. Further studies are needed to explore this hypothesis in depth.
Highlights.
The MR1 molecule that presents microbial-derived vitamin B metabolites to MAIT cells is expressed in astrocyte and microglia cell lines, as well as in primary cultures of brain astrocytes and microglia
MAIT cells are present in the normal mouse brain
Brain astrocyte and microglia MR1 can effectively process and present microbial antigens to MAIT cells, resulting in MAIT cell activation
The MR1/MAIT cell axis may play an important role in the CNS under both normal and disease conditions
Acknowledgements
We would like to thank Dr. Linda Van Eldik for the BV2 cells, Dr. J. Yewdell for the TW2.3 and 2.4G2 hybridomas, as well as the NIH Tetramer Core Facility for providing 6-FP and 5-OP-RU-loaded mouse MR1 tetramers. We would also like to acknowledge the Flow Cytometry Resource Facility, Indiana University School of Medicine, for their assistance.
This work was supported by National Institutes of Health grant R01 CA161178 and support from the Indiana Spinal Cord Injury and Brain Injury Grant Program (to R.R.B.), as well as U54 DK106846, supporting the Indiana University School of Medicine’s Cooperative Center of Excellence in Hematology.
Abbreviations:
- APC
allophycocyanin
- APCs
antigen presenting cells
- BBB
blood-brain barrier
- CFU
colony-forming units
- DAPI
4′-diamidino-2-phenylindole
- IBD
intestinal bowel disease
- MAIT cells
mucosal-associated invariant T cells
- MHC
major histocompatibility complex
- MOI
multiplicity of infection
- MR1 KO
MR1-deficient
- PDL
poly-D-lysine
- PFA
paraformaldehyde
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interest: Neither author declares any conflict of interest with this work
References
- Adams EJ, Luoma AM. The adaptable major histocompatibility complex (MHC) fold: structure and function of nonclassical and MHC class I-like molecules. Annu Rev Immunol. 2013;31:529–61. [DOI] [PubMed] [Google Scholar]
- Aloisi F, Ria F, Penna G, Adorini L. Microglia are more efficient than astrocytes in antigen processing and in Th1 but not Th2 cell activation. J Immunol. 1998;160:4671–80. [PubMed] [Google Scholar]
- Anderson KG, Mayer-Barber K, Sung H, Beura L, James BR, Taylor JJ, et al. Intravascular staining for discrimination of vascular and tissue leukocytes. Nat Protoc. 2014;9:209–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer J, Elger CE, Hans VH, Schramm J, Urbach H, Lassmann H, et al. Astrocytes are a specific immunological target in Rasmussen’s encephalitis. Ann Neurol. 2007;62:67–80. [DOI] [PubMed] [Google Scholar]
- Bjelobaba I, Savic D, Lavrnja I. Multiple Sclerosis and Neuroinflammation: The Overview of Current and Prospective Therapies. Curr Pharm Des. 2017;23:693–730. [DOI] [PubMed] [Google Scholar]
- Blasi E, Barluzzi R, Bocchini V, Mazzolla R, Bistoni F. Immortalization of murine microglial cells by a v-raf/v-myc carrying retrovirus. Journal of neuroimmunology. 1990;27:229–37. [DOI] [PubMed] [Google Scholar]
- Brutkiewicz RR, Yunes-Medina L, Liu J. Immune evasion of the CD1d/NKT cell axis. Current opinion in immunology. 2018;52:87–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busshoff U, Hein A, Iglesias A, Dörries R, Régnier-Vigouroux A. CD1 expression is differentially regulated by microglia, macrophages and T cells in the central nervous system upon inflammation and demyelination. J Neuroimmunol. 2001;113:220–30. [DOI] [PubMed] [Google Scholar]
- Chen Z, Zhong D, Li G. The role of microglia in viral encephalitis: a review. J Neuroinflammation. 2019;16:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett AJ, Eckle SB, Birkinshaw RW, Liu L, Patel O, Mahony J, et al. T-cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature. 2014;509:361–5. [DOI] [PubMed] [Google Scholar]
- Corriveau RA, Huh GS, Shatz CJ. Regulation of class I MHC gene expression in the developing and mature CNS by neural activity. Neuron. 1998;21:505–20. [DOI] [PubMed] [Google Scholar]
- D’Souza MP, Adams E, Altman JD, Birnbaum ME, Boggiano C, Casorati G, et al. Casting a wider net: Immunosurveillance by nonclassical MHC molecules. PLoS Pathog. 2019;15:e1007567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiSabato DJ, Quan N, Godbout JP. Neuroinflammation: the devil is in the details. J Neurochem. 2016;139 Suppl 2:136–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dusseaux M, Martin E, Serriari N, Péguillet I, Premel V, Louis D, et al. Human MAIT cells are xenobiotic-resistant, tissue-targeted, CD161hi IL-17-secreting T cells. Blood. 2011;117:1250–9. [DOI] [PubMed] [Google Scholar]
- Gherardin NA, Souter MN, Koay HF, Mangas KM, Seemann T, Stinear TP, et al. Human blood MAIT cell subsets defined using MR1 tetramers. Immunol Cell Biol. 2018;96:507–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuffrida P, Corazza GR, Di Sabatino A. Old and New Lymphocyte Players in Inflammatory Bowel Disease. Dig Dis Sci. 2018;63:277–88. [DOI] [PubMed] [Google Scholar]
- Godfrey DI, Koay HF, McCluskey J, Gherardin NA. The biology and functional importance of MAIT cells. Nat Immunol. 2019;20:1110–28. [DOI] [PubMed] [Google Scholar]
- Gold MC, Cerri S, Smyk-Pearson S, Cansler ME, Vogt TM, Delepine J, et al. Human mucosal associated invariant T cells detect bacterially infected cells. PLoS Biol. 2010;8:e1000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held K, Bhonsle-Deeng L, Siewert K, Sato W, Beltrán E, Schmidt S, et al. αβ T-cell receptors from multiple sclerosis brain lesions show MAIT cell-related features. Neurol Neuroimmunol Neuroinflamm. 2015;2:e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinks TS. Mucosal-associated invariant T cells in autoimmunity, immune-mediated diseases and airways disease. Immunology. 2016;148:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinks TSC, Marchi E, Jabeen M, Olshansky M, Kurioka A, Pediongco TJ, et al. Activation and In Vivo Evolution of the MAIT Cell Transcriptome in Mice and Humans Reveals Tissue Repair Functionality. Cell Rep. 2019;28:3249–62.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoftberger R, Aboul-Enein F, Brueck W, Lucchinetti C, Rodriguez M, Schmidbauer M, et al. Expression of major histocompatibility complex class I molecules on the different cell types in multiple sclerosis lesions. Brain pathology. 2004;14:43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz MS, Evans CF, Klier FG, Oldstone MB. Detailed in vivo analysis of interferon-gamma induced major histocompatibility complex expression in the the central nervous system: astrocytes fail to express major histocompatibility complex class I and II molecules. Lab Invest. 1999;79:235–42. [PubMed] [Google Scholar]
- Huang S, Gilfillan S, Cella M, Miley MJ, Lantz O, Lybarger L, et al. Evidence for MR1 antigen presentation to mucosal-associated invariant T cells. The Journal of biological chemistry. 2005;280:21183–93. [DOI] [PubMed] [Google Scholar]
- Illés Z, Shimamura M, Newcombe J, Oka N, Yamamura T. Accumulation of Valpha7.2-Jalpha33 invariant T cells in human autoimmune inflammatory lesions in the nervous system. Int Immunol. 2004;16:223–30. [DOI] [PubMed] [Google Scholar]
- Jeffery HC, van Wilgenburg B, Kurioka A, Parekh K, Stirling K, Roberts S, et al. Biliary epithelium and liver B cells exposed to bacteria activate intrahepatic MAIT cells through MR1. Journal of hepatology. 2016;64:1118–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha MK, Jo M, Kim JH, Suk K. Microglia-Astrocyte Crosstalk: An Intimate Molecular Conversation. Neuroscientist. 2019;25:227–40. [DOI] [PubMed] [Google Scholar]
- Johnson HL, Jin F, Pirko I, Johnson AJ. Theiler’s murine encephalomyelitis virus as an experimental model system to study the mechanism of blood-brain barrier disruption. J Neurovirol. 2014;20:107–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juaristi I, Contreras L, González-Sánchez P, Pérez-Liébana I, González-Moreno L, Pardo B, et al. The Response to Stimulation in Neurons and Astrocytes. Neurochem Res. 2019;44:2385–91. [DOI] [PubMed] [Google Scholar]
- Karamooz E, Harriff MJ, Lewinsohn DM. MR1-dependent antigen presentation. Semin Cell Dev Biol. 2018;84:58–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34:637–50. [DOI] [PubMed] [Google Scholar]
- Keller AN, Corbett AJ, Wubben JM, McCluskey J, Rossjohn J. MAIT cells and MR1-antigen recognition. Curr Opin Immunol. 2017;46:66–74. [DOI] [PubMed] [Google Scholar]
- Kjer-Nielsen L, Patel O, Corbett AJ, Le Nours J, Meehan B, Liu L, et al. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature. 2012;491:717–23. [DOI] [PubMed] [Google Scholar]
- Kurioka A, Ussher JE, Cosgrove C, Clough C, Fergusson JR, Smith K, et al. MAIT cells are licensed through granzyme exchange to kill bacterially sensitized targets. Mucosal Immunol. 2015;8:429–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamichhane R, Galvin H, Hannaway RF, de la Harpe SM, Munro F, Tyndall JD, et al. Type I interferons are important co-stimulatory signals during T cell receptor mediated human MAIT cell activation. Eur J Immunol. 2020;50:178–91. [DOI] [PubMed] [Google Scholar]
- Larochelle C, Alvarez JI, Prat A. How do immune cells overcome the blood-brain barrier in multiple sclerosis? FEBS Lett. 2011;585:3770–80. [DOI] [PubMed] [Google Scholar]
- Le Bourhis L, Dusseaux M, Bohineust A, Bessoles S, Martin E, Premel V, et al. MAIT cells detect and efficiently lyse bacterially-infected epithelial cells. PLoS pathogens. 2013;9:e1003681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bourhis L, Martin E, Peguillet I, Guihot A, Froux N, Core M, et al. Antimicrobial activity of mucosal-associated invariant T cells. Nature immunology. 2010;11:701–8. [DOI] [PubMed] [Google Scholar]
- Leeansyah E, Svard J, Dias J, Buggert M, Nystrom J, Quigley MF, et al. Arming of MAIT Cell Cytolytic Antimicrobial Activity Is Induced by IL-7 and Defective in HIV-1 Infection. PLoS pathogens. 2015;11:e1005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legoux F, Bellet D, Daviaud C, El Morr Y, Darbois A, Niort K, et al. Microbial metabolites control the thymic development of mucosal-associated invariant T cells. Science. 2019a;366:494–9. [DOI] [PubMed] [Google Scholar]
- Legoux F, Gilet J, Procopio E, Echasserieau K, Bernardeau K, Lantz O. Molecular mechanisms of lineage decisions in metabolite-specific T cells. Nat Immunol. 2019b;20:1244–55. [DOI] [PubMed] [Google Scholar]
- Liu J, Brutkiewicz RR. The Toll-like receptor 9 signalling pathway regulates MR1-mediated bacterial antigen presentation in B cells. Immunology. 2017;152:232–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Shen Y, Li M, Lv D, Zhang A, Peng Y, et al. Spatial-Temporal Expression of Non-classical MHC Class I Molecules in the C57 Mouse Brain. Neurochem Res. 2015;40:1487–96. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. [DOI] [PubMed] [Google Scholar]
- McCarthy KD, de Vellis J. Preparation of separate astroglial and oligodendroglial cell cultures from rat cerebral tissue. J Cell Biol. 1980;85:890–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWilliam HE, Villadangos JA. MR1 antigen presentation to MAIT cells: new ligands, diverse pathways? Curr Opin Immunol. 2018;52:108–13. [DOI] [PubMed] [Google Scholar]
- Muir FGW, Samadi-Bahrami Z, Moore GRW, Quandt JA. Expression of CD1d by astrocytes corresponds with relative activity in multiple sclerosis lesions. Brain Pathol. 2020;30:26–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterfalvi A, Gomori E, Magyarlaki T, Pal J, Banati M, Javorhazy A, et al. Invariant Valpha7.2-Jalpha33 TCR is expressed in human kidney and brain tumors indicating infiltration by mucosal-associated invariant T (MAIT) cells. Int Immunol. 2008;20:1517–25. [DOI] [PubMed] [Google Scholar]
- Rahimpour A, Koay HF, Enders A, Clanchy R, Eckle SB, Meehan B, et al. Identification of phenotypically and functionally heterogeneous mouse mucosal-associated invariant T cells using MR1 tetramers. J Exp Med. 2015;212:1095–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riegert P, Wanner V, Bahram S. Genomics, isoforms, expression, and phylogeny of the MHC class I-related MR1 gene. Journal of immunology. 1998;161:4066–77. [PubMed] [Google Scholar]
- Rostami J, Fotaki G, Sirois J, Mzezewa R, Bergström J, Essand M, et al. Astrocytes have the capacity to act as antigen-presenting cells in the Parkinson’s disease brain. J Neuroinflammation. 2020;17:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruijing X, Mengjun W, Xiaoling Z, Shu P, Mei W, Yingcheng Z, et al. Jα33+ MAIT cells play a protective role in TNBS induced intestinal inflammation. Hepatogastroenterology. 2012;59:762–7. [DOI] [PubMed] [Google Scholar]
- Salou M, Legoux F, Gilet J, Darbois A, du Halgouet A, Alonso R, et al. A common transcriptomic program acquired in the thymus defines tissue residency of MAIT and NKT subsets. J Exp Med. 2019;216:133–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield LG, Berman NE. Microglial expression of MHC class II increases in normal aging of nonhuman primates. Neurobiol Aging. 1998;19:47–55. [DOI] [PubMed] [Google Scholar]
- Suliman S, Murphy M, Musvosvi M, Gela A, Meermeier EW, Geldenhuys H, et al. MR1-Independent Activation of Human Mucosal-Associated Invariant T Cells by Mycobacteria. J Immunol. 2019;203:2917–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney MD, Zhao Z, Montagne A, Nelson AR, Zlokovic BV. Blood-Brain Barrier: From Physiology to Disease and Back. Physiol Rev. 2019;99:21–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetruashvily MM, Melson JW, Park JJ, Peng X, Boulanger LM. Expression and alternative splicing of classical and nonclassical MHCI genes in the hippocampus and neuromuscular junction. Mol Cell Neurosci. 2016;72:34–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilloy F, Treiner E, Park SH, Garcia C, Lemonnier F, de la Salle H, et al. An invariant T cell receptor alpha chain defines a novel TAP-independent major histocompatibility complex class Ib-restricted alpha/beta T cell subpopulation in mammals. J Exp Med. 1999;189:1907–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga K, Yamagiwa S, Setsu T, Kimura N, Honda H, Kamimura H, et al. Possible involvement of mucosal-associated invariant T cells in the progression of inflammatory bowel diseases. Biomed Res. 2017;38:111–21. [DOI] [PubMed] [Google Scholar]
- Toubal A, Kiaf B, Beaudoin L, Cagninacci L, Rhimi M, Fruchet B, et al. Mucosal-associated invariant T cells promote inflammation and intestinal dysbiosis leading to metabolic dysfunction during obesity. Nat Commun. 2020;11:3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treiner E, Duban L, Bahram S, Radosavljevic M, Wanner V, Tilloy F, et al. Selection of evolutionarily conserved mucosal-associated invariant T cells by MR1. Nature. 2003;422:164–9. [DOI] [PubMed] [Google Scholar]
- Ussher JE, van Wilgenburg B, Hannaway RF, Ruustal K, Phalora P, Kurioka A, et al. TLR signaling in human antigen-presenting cells regulates MR1-dependent activation of MAIT cells. Eur J Immunol. 2016;46:1600–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Weijer ML, Luteijn RD, Wiertz EJ. Viral immune evasion: Lessons in MHC class I antigen presentation. Semin Immunol. 2015;27:125–37. [DOI] [PubMed] [Google Scholar]
- Van Kaer L, Postoak JL, Wang C, Yang G, Wu L. Innate, innate-like and adaptive lymphocytes in the pathogenesis of MS and EAE. Cell Mol Immunol. 2019;16:531–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wilgenburg B, Loh L, Chen Z, Pediongco TJ, Wang H, Shi M, et al. MAIT cells contribute to protection against lethal influenza infection in vivo. Nat Commun. 2018;9:4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter L, Günther E. Isolation and molecular characterization of the rat MR1 homologue, a non-MHC-linked class I-related gene. Immunogenetics. 1998;47:477–82. [DOI] [PubMed] [Google Scholar]
- Wiendl H, Feger U, Mittelbronn M, Jack C, Schreiner B, Stadelmann C, et al. Expression of the immune-tolerogenic major histocompatibility molecule HLA-G in multiple sclerosis: implications for CNS immunity. Brain. 2005;128:2689–704. [DOI] [PubMed] [Google Scholar]
- Willing A, Leach OA, Ufer F, Attfield KE, Steinbach K, Kursawe N, et al. CD8+ MAIT cells infiltrate into the CNS and alterations in their blood frequencies correlate with IL-18 serum levels in multiple sclerosis. Eur J Immunol. 2014;44:3119–28. [DOI] [PubMed] [Google Scholar]
- Wischhusen J, Friese MA, Mittelbronn M, Meyermann R, Weller M. HLA-E protects glioma cells from NKG2D-mediated immune responses in vitro: implications for immune escape in vivo. J Neuropathol Exp Neurol. 2005;64:523–8. [DOI] [PubMed] [Google Scholar]
- Yuwen H, Cox JH, Yewdell JW, Bennink JR, Moss B. Nuclear localization of a double-stranded RNA-binding protein encoded by the vaccinia virus E3L gene. Virology. 1993;195:732–44. [DOI] [PubMed] [Google Scholar]


