Abstract
Mitochondria accumulate copper in their matrix for the eventual maturation of the cuproenzymes cytochrome c oxidase and superoxide dismutase. Transport into the matrix is achieved by mitochondrial carrier family (MCF) proteins. The major copper transporting MCF described to date in yeast is Pic2, which imports the metal ion into the matrix. Pic2 is one of ~30 MCFs that move numerous metabolites, nucleotides and co-factors across the inner membrane for use in the matrix. Genetic and biochemical experiments showed that Pic2 is required for cytochrome c oxidase activity under copper stress, and that it is capable of transporting ionic and complexed forms of copper. The Pic2 ortholog SLC25A3, one of 53 mammalian MCFs, functions as both a copper and a phosphate transporter. Depletion of SLC25A3 results in decreased accumulation of copper in the matrix, a cytochrome c oxidase defect and a modulation of cytosolic superoxide dismutase abundance. The regulatory roles for copper and cuproproteins resident to the mitochondrion continue to expand beyond the organelle. Mitochondrial copper chaperones have been linked to the modulation of cellular copper uptake and export and the facilitation of inter-organ communication. Recently, a role for matrix copper has also been proposed in a novel cell death pathway termed cuproptosis. This review will detail our understanding of the maturation of mitochondrial copper enzymes, the roles of mitochondrial signals in regulating cellular copper content, the proposed mechanisms of copper transport into the organelle and explore the evolutionary origins of copper homeostasis pathways.
Keywords: Copper, mitochondria, mitochondrial carrier family, cytochrome c oxidase, superoxide dismutase
1.0. Introduction
Copper (Cu) is required in eukaryotes for oxygen metabolism, oxygen radical detoxification and iron (Fe) uptake [1]. More recently, a role for Cu in the regulation of numerous other cellular processes via metalloallostery has been described. Metalloallostery refers to the ability of Cu to bind to previously unknown sites in proteins to regulate their activity [2]. This newly discovered regulatory modality has expanded our view of Cu beyond that of a static co-factor. In fact, there is now considerable evidence that Cu is also a dynamic signaling element that has considerable influence over a diverse list of processes that includes lipolysis, cellular proliferation, autophagy and neural activity [3–9]. The expanding role of Cu in maintaining or restoring homeostasis reiterates how important it is to control its biological availability inside and outside of the cell [2, 8, 10]. Various biological ligands and proteins are used to achieve the coordinated regulation of Cu distribution within the cell. Protein-mediated delivery of Cu occurs in the cytosolic compartment, with copper chaperone for superoxide dismutase (CCS) delivering Cu to superoxide dismutase (SOD1) and antioxidant 1 copper chaperone (ATOX1) trafficking Cu to the ATPases ATP7A and ATP7B, which then transport Cu into the trans-Golgi cisternae for the metallation of various enzymes, Figure 1 [1]. ATOX1 interacts with the amino-terminus of these Cu-ATPases to regulate their activity by modulating the rate of ATP hydrolysis and, in turn, Cu transport [11]. ATP7A and ATP7B are homologous ATPases that, when mutated, result in the inherited Cu transport disorders Menkes and Wilson disease, respectively [12]. The mechanisms underlying disease etiology and tissue-specific expression of these transporters are extensively reviewed elsewhere [13, 14]. However, it is important to note here that when cytosolic Cu reaches excess, ATP7A re-localizes to the plasma membrane to facilitate Cu export from the cell while ATP7B re-localizes to vesicles that mediate export of Cu into the bile [13, 14]. This defense mechanism is complemented by binding of Cu into inert complexes with metallothionein and/or a range of molecules that chelate Cu with varying affinity and efficiency [15]. Genetic studies have clearly established that export is the major form of protection against Cu toxicity, as cells lacking ATP7A are significantly more sensitive to excess Cu when compared to those that lack metallothioneins [16]. Membrane-bound compartments within the cell further contribute to limiting cytosolic Cu and minimizing toxicity by serving as sites of sequestration to reduce damage to lipids and proteins and prevent the inappropriate mis-metallation of co-factor binding sites of cytosolic enzymes. In so doing, however, organelles assume the risk of Cu-mediated damage.
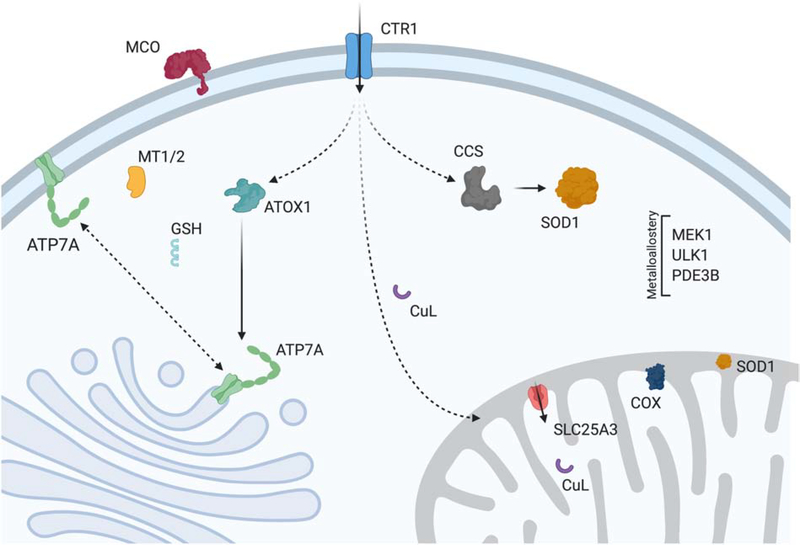
Figure 1: Cu homeostasis in mammals.
Cu enters the cell via the high affinity plasma membrane transporter CTR1 and then is distributed to various targets via metallochaperones (ATOX1, CCS) or alternative ligands (CuL, GSH). Cu enters the trans-Golgi network through an interaction between ATOX1 and ATP7A. Once the Cu enters the cisternae, it can be incorporated into numerous enzymes including the multi-Cu oxidases (MCO). In mitochondria, Cu is transported into the matrix by SLC25A3 for its storage. Upon being triggered by an unknown mechanism Cu is translocated back across the IM to IMS-localized metallochaperones which use it to facilitate assembly of cytochrome c oxidase (COX) and maturation of superoxide dismutase (SOD1). Cu in the cytosol is used as a co-factor by SOD1 for protection against oxidative stress, and CCS is required for metallation of this site and the formation of an essential disulfide bond. When Cu reaches excess levels, it is bound by metallothionein (MT1/2) and ATP7A relocalizes to the PM where it acts as a Cu exporter. In specific cell types, ATP7A is replaced by ATP7B which relocalizes to exocytic vesicles during Cu stress to promote its excretion via the bile (not shown). Dynamic Cu pools within the cell bind at allosteric sites that regulate the activity of the kinases MEK1 and ULK1, and the phosphodiesterase PD3EB in adipocytes.
The major Cu enzymes in mitochondria are cytochrome c oxidase (COX) which is found in the mitochondrial inner membrane, and Cu, Zn superoxide dismutase (SOD1) which is localized to the intermembrane space (IMS) [17, 18]. The organelle has a suite of protein chaperones dedicated to Cu delivery to both COX and SOD1; the associated Cu transfer reactions occur in the IMS, the compartment between the outer and inner membrane (IM), Figure 2 [19]. The protein import pathways in mitochondria require that the proteins enter the IMS in an apo, non-metallated form and therefore a Cu source is required. Mitochondria also have mechanisms to sequester Cu, transporting it to the matrix compartment for storage [20, 21]. While matrix Cu content correlates with the enzymatic activities of both COX and SOD1, this sequestration strategy poses a challenge with respect to utilization because the Cu prosthetic groups are matured in the IM or IMS.
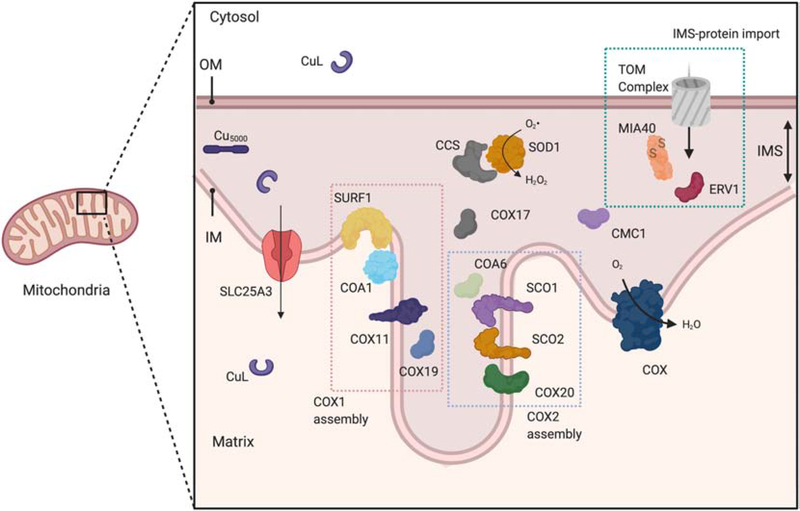
Figure 2: Assembly of COX and SOD1 in mitochondria.
Mitochondrial IM and IMS enzymes are assembled as a result of the concerted actions of multiple metallochaperones. COX is assembled in modules that are subsequently combined to yield the mature holoenzyme complex. COX1 which contains the CuB site and heme co-factors is assembled as a large, modular complex that include SURF1, COA1 and COX11. COX19 is involved in the reduction of the Cu-binding cysteines and activation of COX11. The CuA site is assembled by a complex suite of redox-dependent interactions that ultimately allow SCO1 to insert Cu into apo-COX2. COX20 physically interacts with apo-COX2 upon its insertion into the IM, and COA6 and SCO2 work in concert to ensure that the active site cysteines in SCO1 and COX2 are maintained in the reduced state during CuA site maturation. COX17 supplies the Cu necessary to assemble both the COX1 and COX2 modules. CMC1 is also involved in COX assembly, and genetic studies in yeast support an additional role for CMC1 in promoting SOD1 activity. CCS delivers Cu to SOD1 and forms the disulfide bond required for its activity. The Cu used in these reactions is proposed to come from the mitochondrial matrix. The Cu is imported by SLC25A3 after being delivered across the cytosol by an unknown non-proteinaceous ligand (CuL). An alternate hypothesis is that Cu is stored in an IMS complex (Cu5000) that interacts directly with the metallochaperone proteins resident to this compartment. The cysteine containing IMS chaperones (e.g. COA6, CMC1, COX17, COX19, CCS, SOD1) are imported into the IMS via the TOM complex of the outer membrane and the MIA4/ERV1 disulfide relay machinery.
In the Long-Evans Cinnamon rat, an animal model of Wilson disease associated with Cu overload high levels of Cu accumulate in mitochondria, disrupting the integrity of mitochondrial membranes, depleting glutathione stores and increasing oxidative stress related damage within the organelle [22, 23]. Highlighting the fact that this damage is a consequence of Cu reaching a threshold where it is associated with a variety of weaker affinity ligands in the organelle, these defects can be partially rescued by Cu chelation with methanobactin, a peptide produced by Methylosinus trichosporium OB3b that has a high affinity for Cu [24]. Methanobactin treatment prevented hepatocyte death, subsequent liver failure and morbidity in this animal model. Importantly, from the perspective of mitochondria, Cu toxicity is also linked to the disruption of iron-sulfur (FeS) containing enzymes. In Saccharomyces cerevisiae Cu-mediated damage to the accessible, labile FeS of mitochondrial ferredoxin leads to downstream growth-inhibitory effects [25]. In vitro Cu can block FeS cluster formation by inhibiting the activity of relevant mitochondrial assembly proteins [26]. However, it should be noted that FeS targets outside mitochondria are also an important facet of the observed Cu toxicity. Depletion of the mitochondrial ABC-type transporter Atm1 that transports intermediates required for cytosolic FeS cluster formation exacerbated the Cu-toxicity phenotype in Cryptococcus neoformans [27].
The discovery that ionophores such as elesclomol can be used to deliver Cu and restore function in numerous animal and cell models of Cu-related disorders has been a major advance in the field [28]. While formerly tested at much higher concentrations as a cancer therapeutic, the application of exceptionally low concentrations of elesclomol was found to enhance Cu uptake and liberation of internal Cu stores, allowing for the rescue of Cu associated defects in a number of subcellular compartments in several yeast and mammalian models [28–31]. In addition, direct injection of the Cu-binding ionophore ATSM is in a Phase2/3 clinical trial (clincaltrials.gov: NCT04082832) to partially correct disease symptoms in patients with amyotrophic lateral sclerosis (ALS) [32]. However, all ionophores and related compounds are toxic when used at high concentrations. In yeast, compounds like 2-(6-benzyl-2-pyridyl)quinazoline cause significant cellular and mitochondrial accumulation of Cu resulting in enhanced toxicity [33]. Cu-elesclomol can induce cell death in mammalian systems, and a genome wide CRISPR screen for suppressors resistant to this ionophore identified mitochondrial ferredoxin as a target that can modulate toxicity [34]. This discovery led to the description of a Cu-elesclomol triggered, ferredoxin-dependent form of cell death termed cuproptosis, which occurs independently of known markers of apoptosis and ferroptosis [34]. The dramatic difference in outcomes, from striking phenotypic rescue of fatal Cu handling disorders to potentiation of cell death, emphasizes that regulation of Cu in mitochondria is critical to normal cellular physiology, Figure 3. This review therefore focuses on mitochondrial Cu homeostasis and how the organelle impinges upon other Cu handling pathways in the cell.
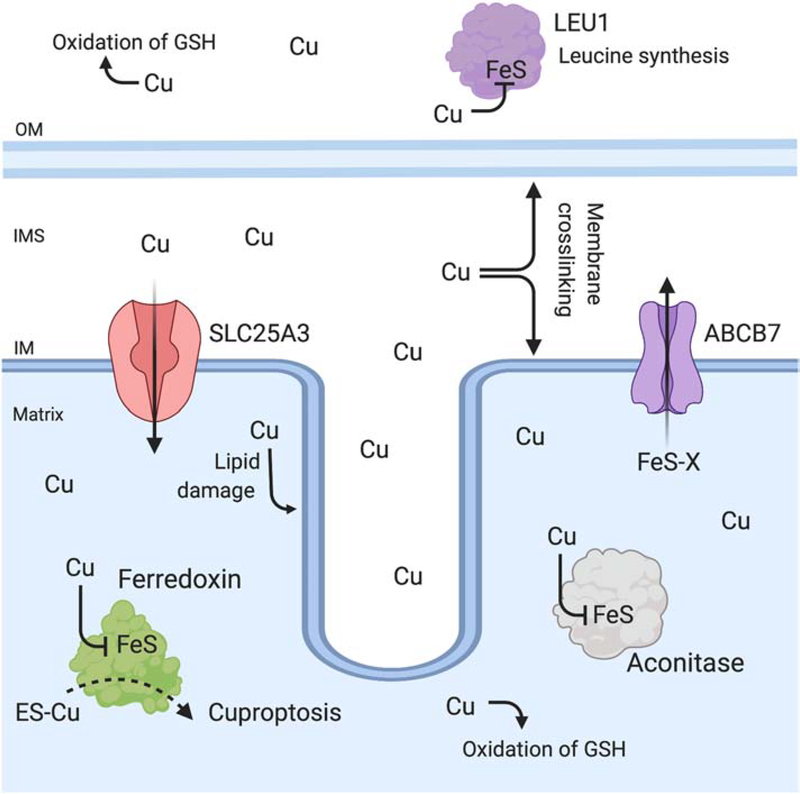
Figure 3: Cu toxicity in mitochondria.
Cu is essential yet becomes toxic when its content exceeds cellular capacity for binding Cu in inert complexes. In mitochondria, Cu toxicity results in lipid damage, protein oxidation, redox imbalance due to inappropriate binding to cysteine-rich sites and depletion of reduced glutathione (GSH). In addition, Cu disrupts and displaces Fe from exposed FeS cluster containing enzymes. Cu has been shown to inactivate ferredoxin and aconitase, which are both localized to the matrix. Cu also inactivates cytosolic enzymes required for leucine synthesis (LEU1). In fungal models, deletion of the ABCB7-homolog (Atm1) responsible for transporting a FeS intermediate of unknown identity (FeS-X) increases Cu toxicity, and emphasizes that cytosolic targets are also an important aspect of Cu toxicity. The ionophore elesclomol (ES-Cu) increases Cu accumulation in mitochondria, and at high concentrations induces cell death via a ferredoxin-dependent mechanism named cuproptosis.
2.0. Mitochondrial Cu enzymes
2.1. Cytochrome c oxidase: COX
COX is the terminal complex of the electron transport chain and accepts the electrons from cytochrome c that are required to convert oxygen to water [35]. Mammalian COX contains 14 subunits, two of which bind the 3 redox centers required for electron transfer and proton pumping, Figure 4. The catalytic core consists of the mitochondrially-encoded subunits COX1, COX2 and COX3. COX2 binds the binuclear CuA site required for accepting electrons from cytochrome c. These electrons are then transferred to the co-factors of COX1, first to heme a and then to the heme a3-CuB site where oxygen is bound. While COX3 does not contain co-factors, it contributes to the minimal core of the enzyme that is conserved in the homologous bacterial complexes. The Cu and heme co-factors of COX1 are buried within the core structure of the enzyme that is embedded in the IM, while the CuA site of COX2 is localized to the IMS and is solvent exposed. Insertion of the co-factors is required for timely progression of COX assembly and ultimately the stability of the mature protein complex itself [19].
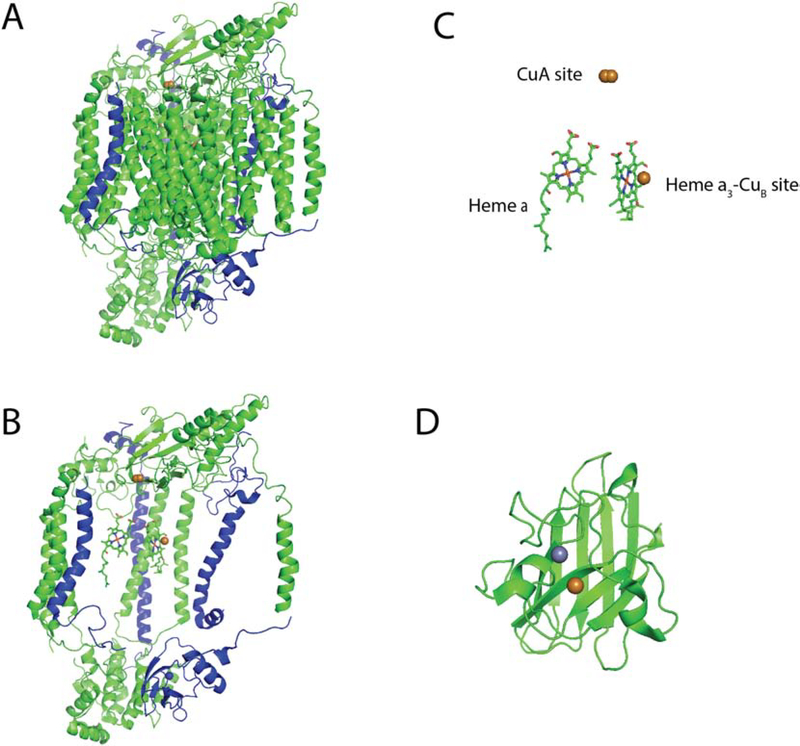
Figure 4: Structure of COX and SOD1.
A) The intact, 14-subunit structure of human cytochrome c oxidase (PDB: 5z62) represented as a cartoon backbone with the 3 mitochondrially-encoded subunits highlighted in blue. B) COX structure with the mitochondrial subunits removed to reveal the heme and Cu co-factors. C) Enlarged rendering of the bi-nuclear CuA site, the heme a and the mixed metal heme a3-CuB site. D) Cartoon depiction of one half of the SOD1 dimer (PDB: 2c9v) showing its Cu and zinc co-factors.
COX biogenesis requires >25 accessory proteins known as COX assembly factors [35]. At least 9 of these factors facilitate the insertion of the two Cu co-factors that are essential for its catalytic activity, Figure 2. Formation of COX requires that the expression of the three mitochondrially-encoded subunits be coordinated with that of the remaining nuclear-encoded subunits which must be imported into the organelle. To coordinate and shepherd the process of COX biogenesis, assembly factors residing within the matrix, IM or IMS work in concert to construct multiple protein modules and assembly intermediates that are eventually combined into the final enzyme complex [36, 37]. Different COX assembly factors exchange into and out of these relatively transient complexes as protein modules are assembled and matured, to promote the incorporation of structural subunits into the IM and the insertion of the heme and Cu co-factors into COX1 and COX2 [38]. Individual modules formed during the assembly process are based around each of the mitochondrially-encoded subunits. Failure to completely assemble any of these modules results in turnover of the intermediate complex by quality control proteases. Although this turnover is relatively rapid, stalled or incompletely assembled intermediates have been detected by blue-native page and their accumulation has been used as a diagnostic tool to define the deficient stage of COX assembly in a number of disease states [35].
COX1 translation and insertion into the IM is mediated by a suite of specific translational activators and chaperones that form a complex known as MITRAC (mitochondrial translation regulation assembly intermediate of COX) [39]. Once COX1 is inserted into the IM, SURF1 adds the heme co-factor and COX11 facilitates the formation of the CuB site. The Cu required for CuB site maturation is donated to COX11 in the IMS by COX17 after the cysteinyl sulfurs of the COX11 Cu-binding site have been reduced by COX19 [18, 40–42]. Like COX1, specific factors also activate COX2 translation and membrane insertion. COX2 is then ultimately bound by a complex composed minimally of COX20, SCO1, SCO2 and COA6 that facilitates the maturation of the binuclear CuA site [43–48]. CuA site formation requires that Cu is transferred from COX17 to SCO1 [41, 49], which then catalyzes its insertion into COX2. SCO2 appears to be essential for this latter Cu transfer step because it acts as an oxidoreductase on the cysteinyl thiolates of COX2 [50]. These reactions are coupled in vivo with COA6 which is also an essential disulfide reductase that reduces COX2 and SCO1, thus allowing for Cu-binding in the oxidizing environment of the IMS [46]. While some of the translational activators and membrane insertion components differ between yeast and humans, the Cu insertion machinery is fully conserved in both species.
2.2. Superoxide dismutase: SOD1
SOD1 catalyzes the conversion of the radical in molecular oxygen to hydrogen peroxide (H2O2) through the alternate reduction and oxidation of Cu, Figure 4 [51]. H2O2 is then detoxified independently by catalase and glutathione peroxidase. While >90% of total cellular SOD1 is found in the cytosol, about 5% is localized to the IMS [52].
The mitochondrial localization of SOD1 appears to be dependent on a “fold-and-trap”-like mechanism, as retention of the protein upon import into the IMS requires that SOD1 co-localizes with its cognate chaperone CCS and that its Cu co-factor is available [51–53]. SOD1 lacks a traditional mitochondrial targeting sequence at its amino terminus or an internal amino acid motif that targets it to the organelle. Rather, mitochondrial import of SOD1 has been shown to depend on the activity of the MIA40/ERV1 pathway, which is also responsible for CCS import, Figure 2. To function properly, this pathway requires that MIA40 form a transient disulfide bonded intermediate with CCS before its subsequent resolution and release of the substrate. ERV1 then oxidizes MIA40 so it can catalyze additional import reactions [54]. SOD1 must be metal free and have its essential disulfide bond reduced to translocate across the outer membrane (OM) and interact with the MIA40/ERV1 relay system. Once unfolded apo-SOD1 enters the IMS, the metallochaperone CCS facilitates insertion of its Cu co-factor and formation of the disulfide bond, effectively trapping SOD1 in the IMS [55–58].
The relative prioritization and distribution of Cu to COX and SOD1 within the IMS appears to be regulated at least in part by the COX assembly factor CMC1, because manipulating CMC1 expression levels affects the amount of enzymatically active SOD1 [59, 60]. However, the mechanisms that distribute Cu within the IMS and prioritize it for COX assembly or SOD1 maturation are unknown.
3.0. Mitochondrial Cu Availability
3.1. Matrix Cu
Cu delivery to, and accumulation within, the mitochondrion is independent of the presence and activity of COX or SOD1 [61]. The total Cu concentration in purified mitochondria can be compared to estimates of heme A concentrations. This highly modified heme is only found in fully assembled COX, and therefore can be used as a proxy of holoenzyme abundance. Such estimates suggest mitochondrial Cu levels are in a 5 to 10-fold excess of COX content, depending on the growth conditions [61, 62]. Evidence derived from the use of biochemical fractionation, chemical probes specific for Cu and phenotypes associated with the expression of heterologous Cu-binding proteins in the matrix all further suggest that the majority of mitochondrial Cu is found in the matrix in an exchangeable, non-proteinaceous form [53, 61, 63]. However, the exact identify of the matrix Cu complex or complexes remains unsolved. The initial purification scheme used mitochondria isolated under normal atmospheric conditions that were then fractionated aerobically by sonication, anion exchange, size exclusion and reverse phase chromatography to yield a fluorescent complex that was termed CuL [61]. While the CuL complex has been recalcitrant to analysis by mass spectroscopy and therefore to identification, its levels positively correlate with cellular Cu concentration and the ability to assemble COX [20, 53]. Further, it was established that the matrix Cu pool is also present in mitochondria isolated from mouse liver and that the soluble Cu has the same retention index characteristics and fluorescent signature of the yeast CuL complex [53]. Subsequent direct measurements of the Cu content of highly purified mitochondria isolated from patient fibroblast lines with isolated COX deficiencies ranging from 10–50% of age-matched control lines showed that this same matrix Cu pool is present, and that it is maintained at a level in excess of the requirements for COX assembly [63]. Cu-responsive probes targeted to the mitochondrial matrix in intact cells have been used to demonstrate that this pool responds dynamically to both genetic and pharmacological stimuli that either increase or decrease the total Cu content of the cell [63–65]. The presence of a Cu complex(es) in the matrix is congruent with the mechanism of Cu toxicity that is linked to impaired FeS synthesis in matrix-localized proteins, and the fact that Cu toxicity can be delayed by the deletion of the matrix-localized FeS protein ferredoxin, Figure 3 [25, 33]. Subsequently, a soluble Cu complex of 5000 Da was isolated from brain mitochondrial extracts prepared under anaerobic conditions and fractionated based on a gel filtration and size exclusion system that allowed for simultaneous detection of Cu content, Figure 3 [66]. It should be noted that this 5000 Da Cu complex is proposed to be IMS-localized and has been cited to challenge the presence of a Cu pool within the matrix [67].
Biochemical depletion of the matrix Cu pool by ectopic expression of Cu-binding proteins localized to this mitochondrial compartment results in a defect in COX and IMS-SOD1 activity in yeast, and a failure to grow on non-fermentable carbon sources [53]. The expression of matrix-localized, Cu-binding proteins also inhibits the activity of a heterologously expressed version of SOD1 that is physically tethered to the IM on the IMS side of the leaflet. By artificially tethering SOD1 to the IM, it no longer requires Cu availability to localize to mitochondria and therefore can be used as biomarker of labile Cu in the IMS. Importantly, rescue of the SOD1-specific lysine auxotrophy phenotype in SOD1 or CCS1 mutants is lost in this biomarker strain when a matrix targeted Cu-binding protein is expressed, strongly suggesting that the matrix Cu pool is redistributed to the IMS for maturation of both COX and SOD1 [53]. An evolutionary rationale and biological argument for why the matrix pool functions in this manner is outlined later in this review in more detail. However, the basic premise is that by maintaining a pool of Cu in the matrix the original endosymbiont would have control over Cu availability. An alternative model based on the 5000 Da Cu species discovered in brain mitochondria proposes that the IMS localization of this complex would provide a Cu source for COX and SOD1 without the need for additional transporters, Figure 5.
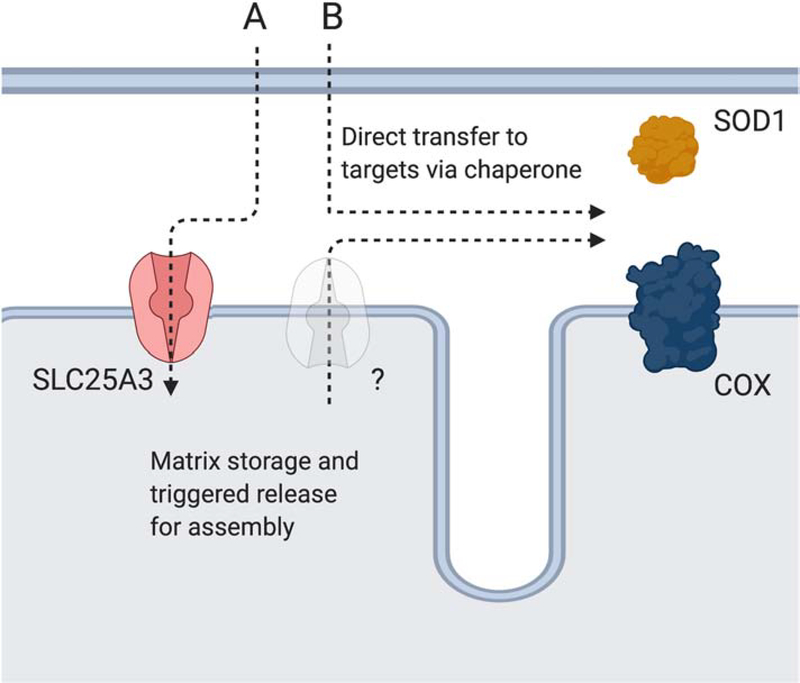
Figure 5: Cu recruitment from the cytosol for COX and IMS-SOD1 maturation.
Two alternative pathways may exist for Cu delivery to COX and IMS-SOD1. Pathway A depicts the matrix storage pathway where Cu that was recruited to the matrix by SLC25A3 for storage is transported back to the IMS by an unidentified transporter for enzyme maturation. The existence of this pathway is supported by multiple studies under Cu stress. Pathway B depicts the direct access pathway where chaperones bind Cu as it enters the IMS or access it after storage in this compartment to support enzyme maturation. Storage in the IMS could be facilitated by the Cu5000 complex that was originally isolated from brain mitochondria.
3.2. Cytosolic Cu transport to mitochondria
The transit of Cu through the cytosol to mitochondria is achieved via an unknown mechanism. To date, a cytosolic protein that can cause a specific decrease in total mitochondrial Cu has not been identified. If in fact such a protein exists, it may be responsible for chaperoning Cu to the OM where it would then transfer the metal ion to an OM pore to facilitate its transport into the IMS. This model predicts that one would observe a decrease in total mitochondrial Cu when said gene is deleted. An alternative model is that Cu is delivered to the OM bound by CuL, the 5000 Da Cu complex or other non-proteinaceous ligands. These complexes could pass through outer membrane porins into the IMS for immediate use in COX assembly or SOD1 maturation or for uptake into the matrix. The use of non-proteinaceous chelating molecules for metal transport is prevalent in multiple biological systems, including the siderophore uptake system for Fe or the chalkophore methanobactin. Methanobactin is a modified peptide produced by M. trichosporium OB3b that is exported into the environment when the organism switches from the soluble, non-Cu requiring methane monooxygenase to the particulate, Cu-requiring methane monooxygenase [68–70]. Using the biophysical characteristics of the hypothetical CuL complex from the matrix, it was proposed that the potential ligand was present in the cytosol in an apo- form where it would act as a Cu chelator for Cu destined for mitochondria. Chelator challenge and spectroscopy studies suggest that this CuL complex is stable in the cuprous redox state and can then be transported to the matrix for storage [53]. Alternatively, the ligand may release the Cu for its subsequent transport across the IM or utilization by IMS chaperones.
4.0. Identifying a mitochondrial Cu transporter
Mitochondria have four major classes of transporters in the IM that include the mitochondrial carrier family (MCF/SLC25), ATP-binding cassette (ABC) transporters, mitochondrial pyruvate carrier (MPC) and sideroflexins [71]. MCF proteins form the largest family in the IM with 53 members in humans and 30 members in yeast that are collectively responsible for the transport of numerous substrates including various Krebs cycle intermediates, nucleoside di- and triphosphates for energy metabolism and nucleotide replication, and amino acids for degradation or maintenance of the urea cycle [72]. MCF transporters have a conserved fold consisting of three repeats of approximately 100-amino acids that contain two TM helices connected by a loop with a short α-helix, Figure 6 [73, 74]. In general, this fold allows for transport of a substrate without leakage of protons to avoid dissipation of membrane potential. The ability of this fold to control proton leakage is highlighted by the fact that the MCF family also includes the uncoupling proteins, which are required under different conditions and in various cell types for proton leakage to maintain normal physiology [75]. The MCF fold is stabilized by salt bridges that form at the closed end of the channel on both the matrix and IMS side of the IM, depending on which conformational state the protein adopts. The two states are known as the cytoplasmic state (c-state) which is open to the IMS, and the matrix state (m-state) which is open to the matrix side [76, 77]. The c-state is so called because the IMS is contiguous with the cytoplasm with respect to access to metabolites and other substrates, given the porous nature of the OM. Each odd-number TM helices contains a conserved PX(D/E)XX(R/K) motif that is a signature of all MCF proteins, Figure 6 [73]. The salt bridges form on the matrix side of the protein in the c-state. MCF proteins also have a complementary (Y/F)(D/E)XX(R/K) motif in the even-numbered helices that form salt bridge and hydrogen bonding contacts on the IMS side of the protein in the m-state [74]. The strength of these salt bridge interactions is an important predictor of the requirement for counter or co-substrates and the directionality of transport [73]. Sixteen of the 53 human MCFs have no known substrate(s) as of yet, and the established promiscuity of some of the characterized family members raises the possibility that even those MCFs may have multiple substrates [71].
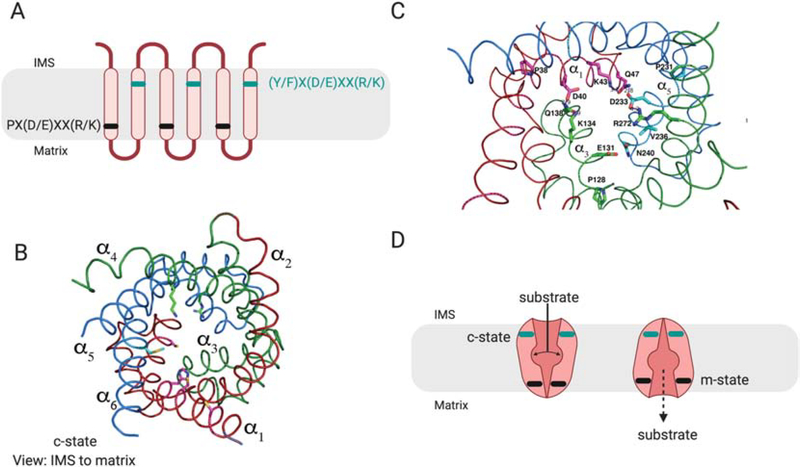
Figure 6: Schematic of MCF structure.
MCF proteins have a conserved, repeated three TM helices structure. A) Within the odd numbered helices a PX(D/E)XX(R/K) motif, shown in black, forms a matrix salt bridge that prevents entry of substrates to the matrix and blocks proton leak. A complementary (Y/F)X(D/E)XX(R/K) motif shown in teal is found in the even numbered helices, and forms salt bridges on the IMS side of the protein. B) The cartoon backbone of the modelled structure of yeast Pic2 in the c-state based on the structure of the ADP/ATP exchanger (PDB: 4C9G). The conserved repeated structure is highlighted in red for helices 1 and 2 (α1, α2), green for helices 3 and 4 (α3, α4), blue for helices 5 and 6 (α5, α6). The structure is shown from the IMS side. C) The salt bridge interactions poise the MCF in either the c-state open to the IMS, or the m-state open to the matrix. The transition between these two states requires substrate binding, and the relative strength of the salt bridge interactions determines whether the transporter acts as a uniporter or as an exchanger. Strong IMS and matrix salt bridges would require exchange of a substrate to reset to the opposite state. Any weaker salt bridges would allow the transporter to be unidirectional.
There are 4 mitochondrially-localized ABC-transporters (ABCB6, ABCB7, ABCB8 and ABCB10) which are part of a large superfamily of proteins that mediate nucleotide-dependent transport [78]. All four of these ABC-transporters have been linked to Fe homeostasis, moving FeS and heme related metabolites into and out of mitochondria [71]. One of the five sideroflexin proteins has been shown to have a role in serine transport in mitochondria [79]. Finally, there is the MPC which is a heterodimeric complex of MPC1 and MPC2 that is responsible for transporting cytosolic pyruvate into the organelle [80, 81]. The MPC is an example of a near 40-year search for a transporter central to carbohydrate, fatty acid and amino acid metabolism [82] whose identification relied on evidence gathered using four distinct experimental approaches; 1) phenotypic characterization of null mutants, 2) detection of a defect in substrate (i.e. pyruvate) accumulation, 3) the use of a specific inhibitor of transport and 4) heterologous expression studies in Lactococcus lactis. We co-opted the suite of techniques used to identify the MPC to uncover possible mitochondrial Cu transporters.
Over the years, multiple genetic screens focused on cellular Cu availability have been attempted using S. cerevisiae. These screens led to the identification of gene products linked to the role of Cu as a catalytic co-factor in Fe uptake and oxygen metabolism, and included multiple COX assembly factors [17, 19, 40, 83–86]. The COX assembly factor Cox17 was originally identified as having a potential role in delivering Cu to mitochondria based on genetic analyses and the ability to bypass its deletion with high Cu in the medium [87]; however, Cox17 was subsequently shown to only be required for Cu delivery to COX within the IMS [41, 49, 88–90]. A critical aspect of the defining investigations that reassigned the role of Cox17 was its ability to function when tethered to the IM, and the fact that there was no change in mitochondrial Cu levels when the gene was deleted [61, 90]. Subsequently, mitochondrial Cu defects were identified in yeast mutants lacking the COX assembly factors Coa1, Shy1 and Coa6, Figure 2 [43, 91–93]. Coa1 and Shy1 are IM proteins that exchange into and out of multiple complexes that chaperone COX1 from translation through to its insertion into the IM and addition of its heme and Cu co-factors [92, 94, 95]. Coa6 is responsible for controlling the redox status of Cu-binding thiolates of Sco1, Sco2 and Cox2 [43–47, 93]. The localization and topology of all three proteins make it unlikely that they are direct transporters of Cu; however, these findings do strongly suggest that Cu availability in mitochondria and COX assembly are tightly coordinated processes.
Vest et al showed that silver (Ag) effectively inhibited Cu uptake into mitochondria by acting as a direct competitor [20] and then used a genetic screen to identify yeast mutants that had glycerol growth defects that were induced by Ag and rescued by Cu. The guiding hypothesis was that upon deletion of the gene encoding the highest affinity Cu transporter Ag would more easily inhibit the “lower” affinity transporters, Figure 7. This screening strategy yielded the MCF protein Pic2 as a candidate mitochondrial Cu transporter [20]. Strains with Pic2 deleted were COX deficient, were directly affected by the inhibitor Ag and had mitochondria with lower Cu levels. Moreover, purified mitochondria from pic2Δ yeast had a defect in the rate of Cu uptake. Therefore, Pic2 was assigned as a transporter required for Cu import into the mitochondrial matrix. Cu loading of an IM-tethered SOD1 was also dependent on Pic2, providing further evidence that Cu cycles through the matrix before being delivered to the IMS for utilization, at least under conditions of stress and/or competition [20].
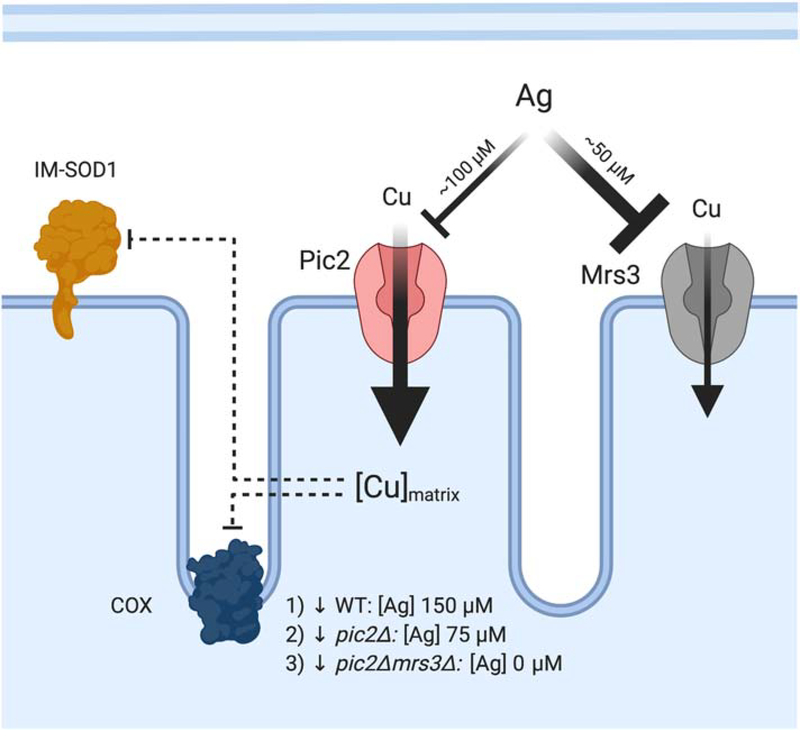
Figure 7: Phenotypic analysis of mitochondrial Cu transporters.
Cu-transporting MCF proteins were identified using genetic screens based on limiting Cu availability with the addition of Ag as a competitor. At concentrations of 150 μM, Ag inhibits Cu uptake and results in decreased COX activity. To elicit the same defect in cells lacking PIC2 only 75–100 μM Ag is required, suggesting a loss of high-affinity transport. Mrs3 was identified as a lower affinity Cu transporter based on additive phenotypes. Yeast cells lacking PIC2 and MRS3 grown under mild Cu chelation without the addition of Ag had decreased COX, decreased IM-SOD1 activity and lower mitochondrial Cu uptake and accumulation.
5.0. Mitochondrial Carrier Family
5.1. Cu transport into the matrix
MCF proteins are known to insert into the cytoplasmic membrane of L. lactis. This experimental system has been used to demonstrate the ability of MCFs to actively transport a range of metabolites [96]. Pic2 expressed in L. lactis mediates Cu-uptake into the cells, consistent with the idea that Pic2 is an importer of Cu in mitochondria [20]. In addition, L. lactis expressing Pic2 exhibit increased sensitivity to the toxic Cu mimetic Ag relative to cells expressing an empty vector. Finally, purified Pic2 transports Cu when reconstituted into proteoliposomes [20]. However, it cannot be the only protein involved in mitochondrial Cu import since deletion of the PIC2 gene only causes a growth defect under Cu-limiting conditions, and mitochondria from pic2Δ yeast still harbor residual Cu and are able to import Cu, albeit to a lesser degree than wild-type organelles. Using the MCF family, a directed screen was performed by crossing pic2Δ into the remaining viable single deletion MCF strains under more permissive conditions. This screen and subsequent validation experiments showed that MRS3 contributes as a secondary importer to mitochondrial Cu transport [97]. This conclusion was based on the additive growth phenotypes, biochemical defects in total Cu, further impairment of Cu uptake in the double deletion strain and the ability of MRS3 to partially suppress pic2Δ phenotypes. Mrs3 is a recognized Fe transporter and this finding therefore represents an additional overlap between Cu and Fe homeostasis in yeast. The transport of Cu by Mrs3 has been reported in studies using mitochondrially derived vesicles from yeast and plants and has been observed for its mammalian homolog Mitoferrin (MFRN1) in a reconstituted assay [98–101].
The homolog of Pic2 in mammals is SLC25A3 [72, 102]. To investigate a role for SLC25A3 in mitochondrial Cu homeostasis, we focused on COX assembly as a marker of Cu availability [21]. Cells lacking SLC25A3 have a COX deficiency that can be rescued by adding Cu but not phosphate to the culture medium. Mitochondrial Cu levels are also reduced in these cells based on in vivo detection of Cu using chemical probes. These phenotypic data were supported by direct measurement of SLC25A3 Cu transport activity after its purification from E. coli and reconstitution into liposomes [21]. Consistent with these observations, expression of SLC25A3 in L. lactis results in time-dependent Cu uptake and enhanced Ag toxicity. A modified L. lactis assay in which Ag was substituted with the phosphate mimic arsenate confirmed that SLC25A3 retained its phosphate transport ability in this expression system [21].
5.2. MCF: Mechanism of transport
Insight into the transport mechanism of MCFs comes largely from pivotal structural studies of the ADP/ATP exchanger [76, 77]. Based on this elegant work, MCFs undergo significant conformational transition from the c-state to the m-state during the transport of large biochemical substrates, while preventing proton leak across the IM. The structure of the ADP/ATP exchanger has been solved in the c- and m-states using specific inhibitors to lock the individual conformations [76, 77]. The c-state, trapped with the inhibitor and nucleotide analog carboxyatractyloside, has the channel open to the IMS side allowing for substrate entry. Substrate binding triggers a conformational change that then switches the protein to the m-state, which is open to the matrix side. The m-state structure was determined in the presence of bongkrekic acid. The transition between the two states has large relative movements of the transmembrane helices, especially helices 2, 4 and 6, and results in a disruption of the matrix salt bridges and the concomitant formation of the IMS side salt bridges. Structural analysis of both the c- and m-states of the ADP/ATP exchanger provides critical information for expanding our understanding of how the TM helices pack to prevent substrate and proton leakage [74]. The proline residue within the signature Px(D/E)xx(R/K)xxx(Q/N) motif results in a kink in the helix that allows it to partially bend, thereby blocking access to the channel. Other residues from this motif interact with the adjacent glutamine residue to form what has been coined the Q-brace to stabilize the structure via hydrogen bonding and prevent uncontrolled movement of the substrate across the IM. In the m-state, conserved tyrosine or phenylalanine residues of the (Y/F)(D/E)XX(R/K) motif on the even numbered helices interact to perform a similar function, forming what is called a Y-brace. During the transport of substrates, structural changes cause the close packing of the helices. The close packing of helices is achieved due to two additional, conserved motifs: πGπxπGπ in the odd-numbered helices and πxxxxπ motif in the even-numbered helices, where π is any amino acid with a small side chain [74]. The knowledge of the substrate-binding sites is less well developed since the structures were determined with substrate inhibitors that lock the structure into one of the two bound states. However, extrapolations from computational models and alignments in conjunction with mutational studies suggest that three contact sites are required for a substrate to bind and trigger the transition between c- and m-states [73, 74]. For the ADP/ATP exchanger, contact site I (Arg, Thr, Asn) on helix 2 mediates electrostatic interactions with the substrate, contact site II (Gly, Ile) on helix 4 discriminates between substrates while contact site III (Arg) is a positively charged residue on helix 6 that is required for activity but is not involved in substrate recognition [73]. A combination of charged residues in the channel neutralizes the charge of the anionic substrate to allow for its transport into the matrix.
Recruitment of Cu, Fe, zinc and manganese to the matrix is required for optimal mitochondrial function; however, the transporters that move these ions across the IM are largely unknown and the function of those that have been identified remains poorly understood. The MCF proteins MITOFERRIN1 (MFRN1/SLC25A37) and MITOFERRIN2 (MFRN2/SLC25A28) are responsible for high-affinity Fe transport across the IM [99, 103, 104]. Fe transport is critical to the essential process of FeS assembly that takes place in the mitochondrial matrix in eukaryotes, and to the synthesis of heme that is critical to the viability of aerobic eukaryotes [105–107]. The transport of Fe was phenotypically demonstrated by deletion of MFRN1 in zebrafish and mouse, which gives rise to severe anemias in each model system due to the lack of heme production in developing erythroid cells. MFRN2 function partially overlaps with MFRN1 in that it is able to transport Fe, but its low abundance in erythroid cells prevents it from suppressing the anemia phenotype [103].
The ability of MFRN1/Mrs3 to transport Fe has been shown biochemically in IM-derived vesicles and proteoliposomes [98–100]. Purified MFRN1 can transport Fe, Cu, manganese and zinc but not nickel [100]. The transport of Fe is dependent on residues across four of the TM helices (His55, Cys119, Met156, Met202, His210, and Cys253) and the amino-terminal domain of the protein. Interestingly, the residues required for Fe transport are found outside of the substrate binding site predicted from the structures of the ADP/ATP exchanger. The transport of Fe by MFRN1 in proteoliposomes was independent of proton translocation and presumably uses histidine, methionine and cysteine residues in the TM helices as ligands to allow for the transport of Fe [100]. No specific ligands are required for Fe transport suggesting that any Fe-complexes present in the IMS could be substrates. As techniques are refined and new chemical probes are developed, we continue to gain a better understanding of the dynamic nature of the redox states of these essential metals in cells and their potential to exchange with multiple buffering components. Numerous Fe-related diseases have large deposits of “insoluble” Fe in mitochondria. If all transport was ligand-mediated, it is unclear why the metal complex would dissociate in the matrix, allowing for inappropriate Fe interactions [108, 109]. Therefore, a potential role for a stable endogenous Fe ligand(s) may be to maintain soluble matrix metal pools and prevent initiation of inappropriate reactions. The identity of such Fe ligands has yet to be revealed.
To gain a greater understanding of a possible transport pathway for Cu, we created two structural models of Pic2 based on the c-state and m-state structures of the ADP/ATP exchanger, Figure 8. A surface rendering of the c-state model of Pic2 shows a series of three cysteine (Cys21, Cys29, Cys44), one histidine (His33) and one methionine (Met275) residues that align in the channel. These residues, like those mutated in MFRN1, may be involved in forming transient sites that bind Cu directly as it moves through the IM. Their relative spacing in the channel likely limits them from forming a stable Cu-binding site but may allow for Cu to walk from ligand to ligand, thereby directing transport into the matrix. In our model, the Cys29(SG)-His33(NE2) side chain atoms has a distance of 4.5 Å that could be the major Cu-binding site in the channel. Cys21 could help to recruit Cu from the IMS and present it to residues Cys29-His33. Based on sequence alignments, His33 is equivalent to His55 of MFRN1/Mrs3 which is important for transport of Fe both in vitro and in vivo [98, 100]. Met275 could form a bridge to Cys44 (d=8.3 Å) which sits on the matrix side of the channel below the matrix salt bridges. In the model of the m-state, the proximity of Cys29-His33 is maintained and Cys44 is exposed; however, Cu would need to relocate by 6.6 Å to Met275 then 8.3 Å to Cys44 making it likely that there are additional ligands required for Cu translocation through the channel.
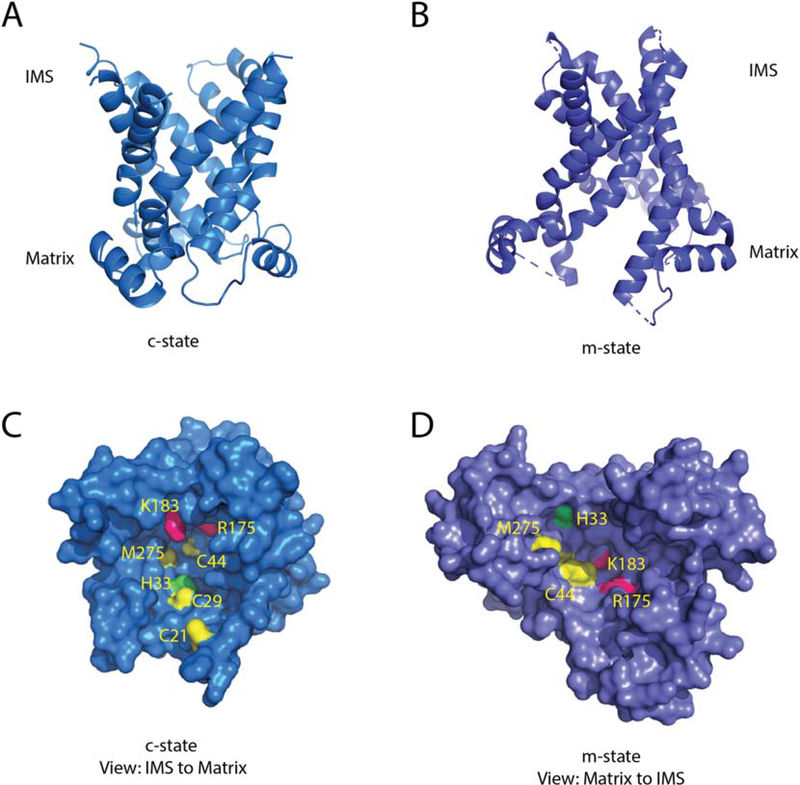
Figure 8: Structural model of Pic2.
A) A cartoon backbone representation of Pic2 modeled onto the c-state of the ADP/ATP exchanger (PDB:4C9G) B) A cartoon backbone representation of Pic2 modeled onto the m-state of the ADP/ATP exchanger (PDB: 6GCI). C) Surface rendering of the Pic2 model in the c-state model looking down the channel from the IMS side of the IM highlighting the positioning of Cys21, Cys29, Cys44, His33, Met275, Arg175 and Lys183 residues in yellow, green and pink. D) Surface rendering of the Pic2 model in the m-state model highlighting the residues listed in C).
Biochemically, Pic2 is able to transport ionic Cu and Cu in the ligated CuL form [20]. Fluorescence anisotropy measurements show that the CuL complex binds to both Pic2 and SLC25A3. The CuL complex is negatively charged based on anion exchange chromatography used in the purification scheme, so positively charged residues in the channel may stabilize this interaction [61]. Pic2 has two positively charged residues, Arg175 and Lys183, on the opposite side of the channel relative to the Cys29-His33 site that are at an equivalent position to Met202 and His210 in MFRN1, which are important for Fe transport. Based on our computational analysis, the proposed contact sites for an anionic substrate in Pic2 are contact site I (Tyr83, Gln86, Lys90), contact site II (Arg175, Gln176) and contact site III (Met275) [73]. The spatial arrangement of these residues may allow for CuL binding and subsequent release of Cu to the Cys29-His33 site, or for a conformational change that facilitates transport of the intact CuL complex.
In yeast, it is important to note that simultaneous deletion of the MFRN1/2 homologs MRS3 and MRS4 only causes a growth defect under severe Fe-depleted conditions, suggesting that additional proteins also function in Fe transport across the IM. RIM2 was identified as one of those transporters. Originally characterized as a transporter of pyrimidine nucleotides, subsequent studies demonstrated that RIM2 also transports Fe-nucleotides and finally that the Fe and pyrimidine nucleotide transporting functions can be separated [110–112]. Mutation of Glu248 specifically blocked mitochondrial Fe transport activity while mutation of Lys299 specifically disrupted pyrimidine nucleotide transport [112]. Sequence alignments of PIC2 and RIM2 introduce very large gaps owing to low identity between these two proteins, which makes it difficult to place the position of Glu248 or Lys299 in the correct context. However, these experiments reinforce the idea that even MCFs with defined substrates can contribute to metal transport, and that metals may be more promiscuous in usage of MCFs than other substrates tested to date.
6.0. Mitochondrial regulation of cellular Cu homeostasis and associated processes
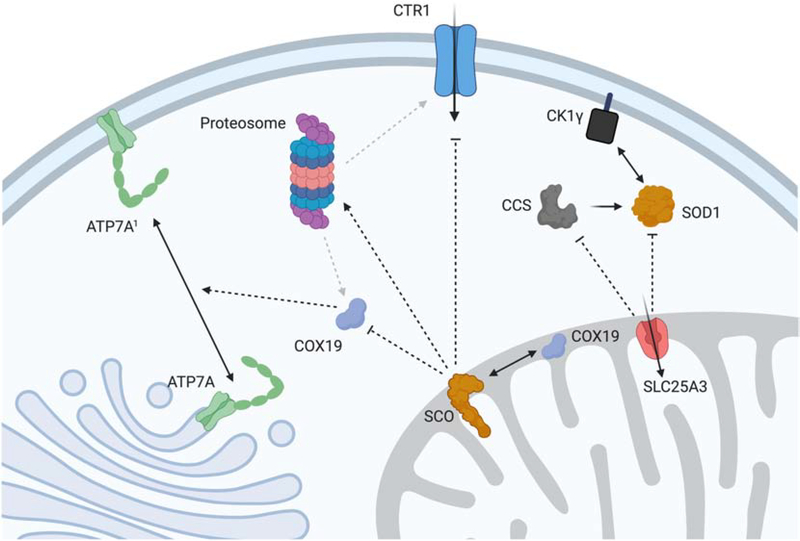
Mitochondria generate and transduce a redox signal that can regulate the activity of the cellular Cu import and export machinery and, therefore, total Cu concentrations, Figure 9. The integrity of this signaling pathway is compromised by mutations in the metallochaperones SCO1 and SCO2 [113–117]. Patients with pathogenic mutations in SCO1 or SCO2 present with clinically heterogeneous forms of fatal disease that typically affect liver, heart or brain function. Affected tissues in SCO patients are both COX and Cu deficient, and both deficiencies appear to be key components of the underlying tissue-specificity of disease [115–117]. In cultured fibroblasts harboring the SCO1 P174L allele, a constitutive signal is generated that leads to a sustained increase in Cu export via ATP7A. The Cx9C protein COX19 is distributed between the IMS and the cytosol, and its partitioning appears to relate to the functional status of SCO1 and the transduction of a redox-related signal outside of the organelle, Figure 9 [114]. More specifically, results from biochemical fractionation and knockdown experiments are consistent with a model whereby COX19 accumulation in the cytosol indicates a state of Cu-overload that stimulates the relocalization and export activity of ATP7A, effectively rendering SCO patient fibroblasts Cu
Figure 9: Mitochondrial Cu-associated regulation of cellular physiology.
Genetic and pharmacological manipulation of mitochondrial Cu in mammalian models results in dramatic remodeling of cellular Cu homeostasis. SCO proteins generate a redox signal that is transduced to the cytosol in part by COX19 to trigger the relocalization of a fraction of the ATP7A pool to the PM to facilitate Cu export. In addition, the SCO-mediated signal(s) affects Cu import via CTR1 by promoting its proteasomal degradation or preventing its localization to the PM. While the combination of these two events causes SCO mutant cells to be profoundly Cu deficient, their mitochondrial Cu pool is preserved. In contrast, deletion of SLC25A3 causes a mitochondrial Cu defect that triggers a signal that results in decreased CCS levels and cytosolic SOD1 activity. We speculate that this would affect the known interaction between SOD1 and casein kinase 1γ (CK1γ), thereby regulating glucose utilization in this mutant cell line.
deficient. However, it is unclear if COX19 fulfills the same function in other cell types and if it interacts directly with ATP7A to regulate its trafficking. The mechanistic details of how the SCO1 redox signal is triggered also remain under investigation. In mouse models of disease, deletion of Sco1 in the liver or heart instead results in a profound deficit in Cu import, with the high affinity plasma membrane transporter CTR1 being mislocalized in Sco1 null hearts and rapidly degraded by the proteasome in Sco1 null livers [115, 116]. These findings argue that a mitochondrial redox signaling pathway emanating from SCO1 is critical to the activity of both the Cu efflux and import machinery. However, further investigations are required to define the mechanisms that affect CTR1 function in each of these Sco1 animal models.
While SCO-deficient cells preserve their mitochondrial Cu pool even in the face of a severe global Cu deficiency, cultured mouse fibroblasts lacking Slc25a3 exhibit a significant depletion in the matrix Cu pool and only a modest decrease in total cellular Cu levels [21]. However, both the activity and abundance of cytosolic SOD1 are reduced in Slc25a3 null fibroblasts, suggesting a novel regulatory role for the mitochondrial Cu pool in some aspect of SOD1 activation. This may be due to a heretofore unappreciated role for the organelle in regulating Cu that is bioavailable for CCS-mediated delivery to cytosolic SOD1. Alternatively, mitochondria may somehow stimulate the degradation of CCS as its steady-state levels are lower in Slc25a3 null fibroblasts. Intriguingly, cytosolic SOD1 is known to transmit signals from oxygen and glucose to affect respiration rates via an interaction with casein kinase 1-γ [118]. This protein-protein interaction promotes the stability of the kinase and requires the activity of SOD1 to convert superoxide to hydrogen peroxide. Therefore, when mitochondria are Cu deficient, the turnover or lack of activity of SOD1 may be a mechanism to adjust glucose metabolism to compensate for the decrease in COX.
Given the fundamental role mitochondria play in metabolism and metabolic regulation it is tempting to speculate that mechanisms analogous to those that regulate SOD1 activity may also impinge upon other, newly defined Cu regulated processes linked to metabolism such as Cu availability for PDE3 and lipolysis, MEK1 kinase and cell proliferation or ULK1/2 kinases and autophagy. This possibility is further bolstered by the identification of CCS as the cytosolic chaperone that delivers Cu to MEK1, and the observation that CCS stability is at least in part regulated by SLC25A3 [21, 119]. Whether the exchangeable nature of the matrix Cu pool allows it to participate in additional metalloallosteric regulation of flux through a variety of metabolic pathways remains unknown. However, the connection between matrix Cu and the regulation of transporters and cytosolic SOD1 activation opens up such opportunities and suggests a potential for mitochondria to function upstream of many Cu related processes.
7.0. Mitochondria as the central hub for Cu metabolism: An endosymbiotic hypothesis
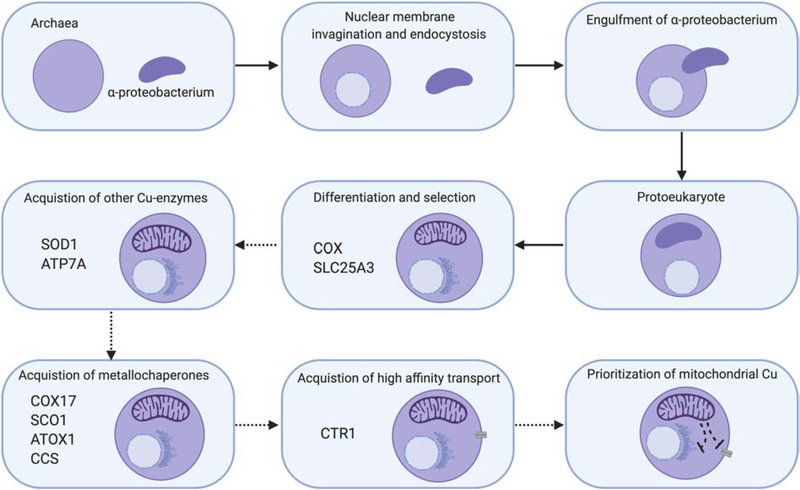
Eukaryogenesis was a multi-step process that combined a host cell of archaeal origin with a symbiont cell of bacterial origin [120]. The basic steps can be simplified as the host archaea 1) underwent changes resulting in membrane invaginations to make a primitive nucleus, 2) acquired phagocytosis-related machinery, and 3) engulfed of free-living bacteria destined to become semi-autonomous organelles such as mitochondria and chloroplasts, Figure 10. The resultant protoeukaryotes then underwent selection before a final set of internal structures were fully differentiated. During these selection steps, a series of niche adaptation pressures made the initial symbiotic relationship a permanent innovation.
Figure 10: Proposed model for Cu homeostasis during eukaryogenesis.
Eukaryotes are thought to have evolved from ancient archaea that had initially differentiated to gain a nuclear membrane and acquire endocytic machinery. Thereafter, the endocytosis of an α-proteobacterium started a symbiotic relationship between the two organisms that resulted in permanent innovation, with retention of the α-proteobacterium as a mitochondrion and differentiation into a multi-organelle protoeukaryote. During its adaptation and development, we speculate that COX was one of the major Cu requirements for the protoeukaryote given its role as a co-factor in the primitive enzyme. As its evolution continued, numerous intermediate steps selected for other Cu-requiring processes and the metallochaperones required to enhance their activity and Cu recruitment. The exact order of gains and losses has not been investigated and therefore is shown here with dashed arrows. One of the final steps based on preliminary investigation of diverse genomes from the tree of life was the acquisition of a high affinity Cu transport system at the plasma membrane to fulfill these Cu needs. We propose that to maintain prioritization of Cu for mitochondria, the protoeukaryote then refined the hierarchical regulation of Cu handling around signals originating from the organelle.
Mitochondrial endosymbiosis was initiated when an archaeal host phagocytosed an α-proteobacterium [121]. The exact nature of the original free-living organism is debated, but the origin of several mitochondrial functions including aerobic metabolism, FeS cluster assembly, heme biosynthesis, β-oxidation of fatty acids, cristae formation and organelle maintenance and replication can be traced to an α-proteobacterium [120]. While mitochondria of many eukaryotes are capable of aerobic metabolism, some are not. Some Oxymonads, for example, have lost mitochondria and have the bacterial-like cluster of genes to allow cytosolic FeS assembly or retained a mitosome with minimal Fe-S machinery, arguing that at least in the niche inhabited by these organisms Fe-S assembly is the only essential process afforded by the ancestral organelle [122–124].
While aerobic electron transport has not been retained by all niches of eukaryotes, it is essential for humans. Therefore, we predict that for the original endosymbiont, oxidative phosphorylation was advantageous for the protoeukaryotic cell as atmospheric oxygen levels rose. The cells that had this terminal electron acceptor complex may have been able to produce increased levels of ATP or partially remove oxygen from the local environment and this may have helped them gain traction in the developing niche. The initial electron transport chain must have contained a primitive relative of COX since the central hydrophobic core of this enzyme is still encoded by the mitochondrial genome and no nuclear to mitochondrial gene translocation has occurred, meaning that it was part of the ancient α-proteobacterium genome.
We hypothesize that during eukaryogenesis the advantage gained by retaining a symbiont with a Cu-oxidase meant that selective pressure was placed on the maintenance and recruitment of Cu to mitochondria as this was key to remaining viable in an oxygen-rich environment. As part of this speculation, we assume that the cytosol of the protoeukaryote became the environment from which mitochondria had to recruit Cu. A “selfish” motive to remain productive in the symbiosis would have driven accumulation of a dedicated pool of Cu that was primarily accessible to the symbiont. We believe this pool is housed in the mitochondrial matrix, making it inaccessible to other Cu-dependent processes or binding partners. This selection process predicts that cellular Cu homeostasis and signaling should be mediated through mitochondria to guarantee the maintenance of this Cu pool. Presumably as oxygen levels rose the need for detoxification of reactive oxygen species increased and availability of iron decreased in the environment, leading to higher Cu requirements for the cell as the enzymes that gave a competitive advantage to these pathways were cuproenzymes (e.g. SOD1 and multi-copper oxidases). Therefore, with higher demands cells with a high-affinity Cu transport system and Cu chaperones that optimized availability were favored. Eukaryotes employ a series of thiol-oxidoreductase steps essential for the maturation of the Cu sites that are not always present in the bacterium. These reactions are catalyzed by proteins that use one or more Cx9C motifs (e.g. COA6, COX19) to play important roles in controlling the accessibility of the Cu sites by reducing relevant ligands in targets to ensure insertion when Cu is available. The duplication of multiple Cx9C motif-containing proteins with these redox properties must have provided a selective advantage for these organisms. Interestingly, the Monoceromonoides eukaryotes that lack mitochondria also lack all Cu homeostasis proteins, suggesting that they either lost all other Cu-related machinery or never acquired it.
Our thesis is that initially, mitochondria ensured prioritization of Cu to the organelle and maintenance of a matrix pool by transducing signals that regulate its global homeostasis, Figure 10. The ability of Cu-handling COX assembly factors to regulate the Cu export and import machinery, the maintenance of an intact mitochondrial Cu pool in Cu-deficient SCO1 cells, and the cytosolic SOD1 defect in cells lacking SLC25A3 are consistent with the evolutionary conservation of this signaling circuitry.
8.0. Conclusions and Future Directions
Existing data suggest that mitochondria retain a dedicated pool of Cu for eventual use in the assembly of COX and SOD1 in the IMS. Matrix storage prevents allocation to other Cu-dependent processes and the importance of maintaining this pool may have led to the evolution of a signaling cascade that make mitochondria a priority for Cu homeostasis. The identification of Pic2 and SLC25A3 as dual function Cu and phosphate carriers opens up many opportunities to investigate how cellular Cu homeostasis is affected by specific depletion of this pool. Chelators are a valuable tool to investigate these pathways; however, they simultaneously limit multiple pathways and this may complicate the interpretation of Cu-dependent signaling cascades. Therefore, it is reasonable to expect that cell lines depleted of individual Cu-handling proteins like SLC25A3 will be more informative with respect to uncovering new pathways regulated by Cu. In addition, by identifying additional MCF family members that transport Cu it will be possible to further flesh out how the metal ion is moved between various organellar compartments.
Several major outstanding questions remain with respect to mitochondrial Cu handling and its influence over Cu homeostasis in healthy and diseased states. How is Cu translocated back to the IMS? How is that export regulated? It can be speculated that a reason genetic screens have failed to identify additional Cu transporters is that perhaps Pic2/SLC25A3 can reverse the direction of transport with this change in directionality dependent on interacting partners. In this model, interactions with IMS chaperones could regulate the release of Cu back to the IMS for assembly of COX and SOD1. To address this question a comprehensive catalog of the proteins that interact with Pic2/SLC25A3 is required. However, these efforts have been challenged by the fact that SLC25A3 is a frequent contaminant in complex protein mixtures generated using physical pulldown methods [125]. Can phosphate and Cu transport be functionally separated in SLC25A3? How does the mitochondrial signal get transduced to the cytosol to coordinately regulate the activity of various cellular Cu homeostasis pathways? At present, it appears that the regulation often includes degradation of the targets. So, how does the proteasome or other proteases receive the signal to selectively degrade specific Cu proteins? What other physiological functions are disturbed by manipulating mitochondrial Cu pools? How is Cu toxicity managed in the mitochondrial matrix relative to FeS, especially in the light of cuproptosis? Answering these questions as well as expanding our knowledge of metalloallostery of mitochondrial targets will be the major advances made around mitochondrial Cu over the next few years.
Highlights.
Mitochondria accumulate copper in their matrix via the mitochondrial carrier family proteins, Pic2/SLC25A3, for the eventual maturation of the cuproenzymes cytochrome c oxidase and superoxide dismutase.
The regulatory roles for copper and cuproproteins resident to the mitochondrion continue to expand beyond the organelle with links to the modulation of cellular copper uptake and export and the facilitation of inter-organ communication.
The roles of mitochondrial signals in regulating cellular copper content, the proposed mechanisms of copper transport into the organelle are explored the context of evolutionary origins of copper homeostasis pathways.
Acknowledgements
We thank Dr. Katherine Buckley (Auburn University) for her suggestions on the text of this article. Figures were created using BioRender.com and exported under a paid subscription. Funding: This work is supported by a grant from National Institutes of Health [R01GM120211 to PAC and SCL]. PAC is supported by the Alabama Agricultural Experiment Station.
Footnotes
Declaration of competing interest
Authors declare that they have no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
9.0 References
- [1].Robinson NJ, Winge DR, Copper metallochaperones, Annu Rev Biochem 79 (2010) 537–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Chang CJ, Searching for harmony in transition-metal signaling, Nat Chem Biol 11(10) (2015) 744–7. [DOI] [PubMed] [Google Scholar]
- [3].Krishnamoorthy L, Cotruvo JA Jr., Chan J, Kaluarachchi H, Muchenditsi A, Pendyala VS, Jia S, Aron AT, Ackerman CM, Wal MN, Guan T, Smaga LP, Farhi SL, New EJ, Lutsenko S, Chang CJ, Copper regulates cyclic-AMP-dependent lipolysis, Nat Chem Biol 12(8) (2016) 586–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Brady DC, Crowe MS, Turski ML, Hobbs GA, Yao X, Chaikuad A, Knapp S, Xiao K, Campbell SL, Thiele DJ, Counter CM, Copper is required for oncogenic BRAF signalling and tumorigenesis, Nature 509(7501) (2014) 492–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Tsang T, Posimo JM, Gudiel AA, Cicchini M, Feldser DM, Brady DC, Copper is an essential regulator of the autophagic kinases ULK1/2 to drive lung adenocarcinoma, Nat Cell Biol 22(4) (2020) 412–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Turski ML, Brady DC, Kim HJ, Kim BE, Nose Y, Counter CM, Winge DR, Thiele DJ, A novel role for copper in Ras/mitogen-activated protein kinase signaling, Mol Cell Biol 32(7) (2012) 1284–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Dodani SC, Domaille DW, Nam CI, Miller EW, Finney LA, Vogt S, Chang CJ, Calcium-dependent copper redistributions in neuronal cells revealed by a fluorescent copper sensor and X-ray fluorescence microscopy, Proc Natl Acad Sci U S A 108(15) (2011) 5980–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Dodani SC, Firl A, Chan J, Nam CI, Aron AT, Onak CS, Ramos-Torres KM, Paek J, Webster CM, Feller MB, Chang CJ, Copper is an endogenous modulator of neural circuit spontaneous activity, Proc Natl Acad Sci U S A 111(46) (2014) 16280–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Xiao T, Ackerman CM, Carroll EC, Jia S, Hoagland A, Chan J, Thai B, Liu CS, Isacoff EY, Chang CJ, Copper regulates rest-activity cycles through the locus coeruleus-norepinephrine system, Nat Chem Biol 14(7) (2018) 655–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Ackerman CM, Chang CJ, Copper signaling in the brain and beyond, J Biol Chem 293(13) (2018) 4628–4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Yu CH, Yang N, Bothe J, Tonelli M, Nokhrin S, Dolgova NV, Braiterman L, Lutsenko S, Dmitriev OY, The metal chaperone Atox1 regulates the activity of the human copper transporter ATP7B by modulating domain dynamics, J Biol Chem 292(44) (2017) 18169–18177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kaler SG, Inborn errors of copper metabolism, Handb Clin Neurol 113 (2013) 1745–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hartwig C, Zlatic SA, Wallin M, Vrailas-Mortimer A, Fahrni CJ, Faundez V, Trafficking mechanisms of P-type ATPase copper transporters, Curr Opin Cell Biol 59 (2019) 24–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Polishchuk R, Lutsenko S, Golgi in copper homeostasis: a view from the membrane trafficking field, Histochem Cell Biol 140(3) (2013) 285–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Morgan MT, Bourassa D, Harankhedkar S, McCallum AM, Zlatic SA, Calvo JS, Meloni G, Faundez V, Fahrni CJ, Ratiometric two-photon microscopy reveals attomolar copper buffering in normal and Menkes mutant cells, Proc Natl Acad Sci U S A 116(25) (2019) 12167–12172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Gudekar N, Shanbhag V, Wang Y, Ralle M, Weisman GA, Petris MJ, Metallothioneins regulate ATP7A trafficking and control cell viability during copper deficiency and excess, Scientific Reports 10(1) (2020) 7856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Baker ZN, Cobine PA, Leary SC, The mitochondrion: a central architect of copper homeostasis, Metallomics 9(11) (2017) 1501–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Leary SC, Winge DR, Cobine PA, “Pulling the plug” on cellular copper: the role of mitochondria in copper export, Biochim Biophys Acta 1793(1) (2009) 146–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Jett KA, Leary SC, Building the CuA site of cytochrome c oxidase: a complicated, redox-dependent process driven by a surprisingly large complement of accessory proteins, J Biol Chem (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Vest KE, Leary SC, Winge DR, Cobine PA, Copper Import into the Mitochondrial Matrix in Saccharomyces cerevisiae is Mediated by Pic2, a Mitochondrial Carrier Family Protein, J Biol Chem 288 (33) (2013) 23884–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Boulet A, Vest KE, Maynard MK, Gammon MG, Russell AC, Mathews AT, Cole SE, Zhu X, Phillips CB, Kwong JQ, Dodani SC, Leary SC, Cobine PA, The mammalian phosphate carrier SLC25A3 is a mitochondrial copper transporter required for cytochrome c oxidase biogenesis, J Biol Chem 293(6) (2018) 1887–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Zischka H, Lichtmannegger J, Pathological mitochondrial copper overload in livers of Wilson’s disease patients and related animal models, Ann N Y Acad Sci 1315 (2014) 6–15. [DOI] [PubMed] [Google Scholar]
- [23].Zischka H, Lichtmannegger J, Schmitt S, Jagemann N, Schulz S, Wartini D, Jennen L, Rust C, Larochette N, Galluzzi L, Chajes V, Bandow N, Gilles VS, DiSpirito AA, Esposito I, Goettlicher M, Summer KH, Kroemer G, Liver mitochondrial membrane crosslinking and destruction in a rat model of Wilson disease, J Clin Invest 121(4) (2011) 1508–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Lichtmannegger J, Leitzinger C, Wimmer R, Schmitt S, Schulz S, Kabiri Y, Eberhagen C, Rieder T, Janik D, Neff F, Straub BK, Schirmacher P, DiSpirito AA, Bandow N, Baral BS, Flatley A, Kremmer E, Denk G, Reiter FP, Hohenester S, Eckardt-Schupp F, Dencher NA, Adamski J, Sauer V, Niemietz C, Schmidt HH, Merle U, Gotthardt DN, Kroemer G, Weiss KH, Zischka H, Methanobactin reverses acute liver failure in a rat model of Wilson disease, J Clin Invest 126(7) (2016) 2721–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Vallieres C, Holland SL, Avery SV, Mitochondrial Ferredoxin Determines Vulnerability of Cells to Copper Excess, Cell Chem Biol 24(10) (2017) 1228–1237 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Brancaccio D, Gallo A, Piccioli M, Novellino E, Ciofi-Baffoni S, Banci L, [4Fe-4S] Cluster Assembly in Mitochondria and Its Impairment by Copper, J Am Chem Soc 139(2) (2017) 719–730. [DOI] [PubMed] [Google Scholar]
- [27].Garcia-Santamarina S, Uzarska MA, Festa RA, Lill R, Thiele DJ, Cryptococcus neoformans Iron-Sulfur Protein Biogenesis Machinery Is a Novel Layer of Protection against Cu Stress, mBio 8(5) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Soma S, Latimer AJ, Chun H, Vicary AC, Timbalia SA, Boulet A, Rahn JJ, Chan SSL, Leary SC, Kim BE, Gitlin JD, Gohil VM, Elesclomol restores mitochondrial function in genetic models of copper deficiency, Proc Natl Acad Sci U S A 115(32) (2018) 8161–8166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Hasinoff BB, Yadav AA, Patel D, Wu X, The cytotoxicity of the anticancer drug elesclomol is due to oxidative stress indirectly mediated through its complex with Cu(II), J Inorg Biochem 137 (2014) 22–30. [DOI] [PubMed] [Google Scholar]
- [30].Nagai M, Vo NH, Shin Ogawa L, Chimmanamada D, Inoue T, Chu J, Beaudette-Zlatanova BC, Lu R, Blackman RK, Barsoum J, Koya K, Wada Y, The oncology drug elesclomol selectively transports copper to the mitochondria to induce oxidative stress in cancer cells, Free Radic Biol Med 52(10) (2012) 2142–50. [DOI] [PubMed] [Google Scholar]
- [31].Guthrie LM, Soma S, Yuan S, Silva A, Zulkifli M, Snavely TC, Greene HF, Nunez E, Lynch B, De Ville C, Shanbhag V, Lopez FR, Acharya A, Petris MJ, Kim B-E, Gohil VM, Sacchettini JC, Elesclomol alleviates Menkes pathology and mortality by escorting Cu to cuproenzymes in mice, Science 368(6491) (2020) 620–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Williams JR, Trias E, Beilby PR, Lopez NI, Labut EM, Bradford CS, Roberts BR, McAllum EJ, Crouch PJ, Rhoads TW, Pereira C, Son M, Elliott JL, Franco MC, Estevez AG, Barbeito L, Beckman JS, Copper delivery to the CNS by CuATSM effectively treats motor neuron disease in SOD(G93A) mice co-expressing the Copper-Chaperone-for-SOD, Neurobiol Dis 89 (2016) 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Foster AW, Dainty SJ, Patterson CJ, Pohl E, Blackburn H, Wilson C, Hess CR, Rutherford JC, Quaranta L, Corran A, Robinson NJ, A chemical potentiator of copper-accumulation used to investigate the iron-regulons of Saccharomyces cerevisiae, Mol Microbiol 93(2) (2014) 317–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Tsvetkov P, Detappe A, Cai K, Keys HR, Brune Z, Ying W, Thiru P, Reidy M, Kugener G, Rossen J, Kocak M, Kory N, Tsherniak A, Santagata S, Whitesell L, Ghobrial IM, Markley JL, Lindquist S, Golub TR, Mitochondrial metabolism promotes adaptation to proteotoxic stress, Nat Chem Biol 15(7) (2019) 681–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Diaz F, Cytochrome c oxidase deficiency: patients and animal models, Biochim Biophys Acta 1802(1) (2010) 100–10. [DOI] [PubMed] [Google Scholar]
- [36].Barros MH, McStay GP, Modular biogenesis of mitochondrial respiratory complexes, Mitochondrion 50 (2020) 94–114. [DOI] [PubMed] [Google Scholar]
- [37].McStay GP, Su CH, Tzagoloff A, Modular assembly of yeast cytochrome oxidase, Mol Biol Cell 24(4) (2013) 440–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Soto IC, Fontanesi F, Liu J, Barrientos A, Biogenesis and assembly of eukaryotic cytochrome c oxidase catalytic core, Biochim Biophys Acta 1817(6) (2012) 883–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Dennerlein S, Rehling P, Human mitochondrial COX1 assembly into cytochrome c oxidase at a glance, J Cell Sci 128(5) (2015) 833–7. [DOI] [PubMed] [Google Scholar]
- [40].Vest KE, Hashemi HF, Cobine PA, The copper metallome in eukaryotic cells, Met Ions Life Sci 12 (2013) 451–78. [DOI] [PubMed] [Google Scholar]
- [41].Banci L, Bertini I, Ciofi-Baffoni S, Hadjiloi T, Martinelli M, Palumaa P, Mitochondrial copper(I) transfer from Cox17 to Sco1 is coupled to electron transfer, Proc Natl Acad Sci U S A 105(19) (2008) 6803–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Bode M, Woellhaf MW, Bohnert M, van der Laan M, Sommer F, Jung M, Zimmermann R, Schroda M, Herrmann JM, Redox-regulated dynamic interplay between Cox19 and the copper-binding protein Cox11 in the intermembrane space of mitochondria facilitates biogenesis of cytochrome c oxidase, Mol Biol Cell 26(13) (2015) 2385–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Ghosh A, Pratt AT, Soma S, Theriault SG, Griffin AT, Trivedi PP, Gohil VM, Mitochondrial disease genes COA6, COX6B and SCO2 have overlapping roles in COX2 biogenesis, Hum Mol Genet 25(4) (2016) 660–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Pacheu-Grau D, Bareth B, Dudek J, Juris L, Vogtle FN, Wissel M, Leary SC, Dennerlein S, Rehling P, Deckers M, Cooperation between COA6 and SCO2 in COX2 maturation during cytochrome c oxidase assembly links two mitochondrial cardiomyopathies, Cell Metab 21(6) (2015) 823–33. [DOI] [PubMed] [Google Scholar]
- [45].Pacheu-Grau D, Wasilewski M, Oeljeklaus S, Gibhardt CS, Aich A, Chudenkova M, Dennerlein S, Deckers M, Bogeski I, Warscheid B, Chacinska A, Rehling P, COA6 Facilitates Cytochrome c Oxidase Biogenesis as Thiol-reductase for Copper Metallochaperones in Mitochondria, J Mol Biol (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Soma S, Morgada MN, Naik MT, Boulet A, Roesler AA, Dziuba N, Ghosh A, Yu Q, Lindahl PA, Ames JB, Leary SC, Vila AJ, Gohil VM, COA6 Is Structurally Tuned to Function as a Thiol-Disulfide Oxidoreductase in Copper Delivery to Mitochondrial Cytochrome c Oxidase, Cell Rep 29(12) (2019) 4114–4126 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Stroud DA, Maher MJ, Lindau C, Vogtle FN, Frazier AE, Surgenor E, Mountford H, Singh AP, Bonas M, Oeljeklaus S, Warscheid B, Meisinger C, Thorburn DR, Ryan MT, COA6 is a mitochondrial complex IV assembly factor critical for biogenesis of mtDNA-encoded COX2, Hum Mol Genet 24(19) (2015) 5404–15. [DOI] [PubMed] [Google Scholar]
- [48].Leary SC, Sasarman F, Nishimura T, Shoubridge EA, Human SCO2 is required for the synthesis of CO II and as a thiol-disulphide oxidoreductase for SCO1, Human Molecular Genetics 18(12) (2009) 2230–40. [DOI] [PubMed] [Google Scholar]
- [49].Horng YC, Cobine PA, Maxfield AB, Carr HS, Winge DR, Specific copper transfer from the Cox17 metallochaperone to both Sco1 and Cox11 in the assembly of yeast cytochrome C oxidase, J Biol Chem 279(34) (2004) 35334–40. [DOI] [PubMed] [Google Scholar]
- [50].Morgada MN, Abriata LA, Cefaro C, Gajda K, Banci L, Vila AJ, Loop recognition and copper-mediated disulfide reduction underpin metal site assembly of CuA in human cytochrome oxidase, Proc Natl Acad Sci U S A 112(38) (2015) 11771–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Kawamata H, Manfredi G, Import, maturation, and function of SOD1 and its copper chaperone CCS in the mitochondrial intermembrane space, Antioxid Redox Signal 13(9) (2010) 1375–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Sturtz LA, Diekert K, Jensen LT, Lill R, Culotta VC, A fraction of yeast Cu,Zn-superoxide dismutase and its metallochaperone, CCS, localize to the intermembrane space of mitochondria. A physiological role for SOD1 in guarding against mitochondrial oxidative damage, The Journal of Biological Chemistry 276(41) (2001) 38084–9. [DOI] [PubMed] [Google Scholar]
- [53].Cobine PA, Pierrel F, Bestwick ML, Winge DR, Mitochondrial matrix copper complex used in metallation of cytochrome oxidase and superoxide dismutase, J Biol Chem 281(48) (2006) 36552–9. [DOI] [PubMed] [Google Scholar]
- [54].Neal SE, Dabir DV, Tienson HL, Horn DM, Glaeser K, Ogozalek Loo RR, Barrientos A, Koehler CM, Mia40 Protein Serves as an Electron Sink in the Mia40-Erv1 Import Pathway, J Biol Chem 290(34) (2015) 20804–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Gross DP, Burgard CA, Reddehase S, Leitch JM, Culotta VC, Hell K, Mitochondrial Ccs1 contains a structural disulfide bond crucial for the import of this unconventional substrate by the disulfide relay system, Mol Biol Cell 22(20) (2011) 3758–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Kloppel C, Suzuki Y, Kojer K, Petrungaro C, Longen S, Fiedler S, Keller S, Riemer J, Mia40-dependent oxidation of cysteines in domain I of Ccs1 controls its distribution between mitochondria and the cytosol, Mol Biol Cell 22(20) (2011) 3749–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Reddehase S, Grumbt B, Neupert W, Hell K, The disulfide relay system of mitochondria is required for the biogenesis of mitochondrial Ccs1 and Sod1, J Mol Biol 385(2) (2009) 331–8. [DOI] [PubMed] [Google Scholar]
- [58].Varabyova A, Topf U, Kwiatkowska P, Wrobel L, Kaus-Drobek M, Chacinska A, Mia40 and MINOS act in parallel with Ccs1 in the biogenesis of mitochondrial Sod1, FEBS J 280(20) (2013) 4943–59. [DOI] [PubMed] [Google Scholar]
- [59].Horn D, Al-Ali H, Barrientos A, Cmc1p is a conserved mitochondrial twin CX9C protein involved in cytochrome c oxidase biogenesis, Mol Cell Biol 28(13) (2008) 4354–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Horn D, Zhou W, Trevisson E, Al-Ali H, Harris TK, Salviati L, Barrientos A, The conserved mitochondrial twin Cx9C protein Cmc2 Is a Cmc1 homologue essential for cytochrome c oxidase biogenesis, J Biol Chem 285(20) (2010) 15088–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Cobine PA, Ojeda LD, Rigby KM, Winge DR, Yeast contain a non-proteinaceous pool of copper in the mitochondrial matrix, J Biol Chem 279(14) (2004) 14447–55. [DOI] [PubMed] [Google Scholar]
- [62].Garber Morales J, Holmes-Hampton GP, Miao R, Guo Y, Munck E, Lindahl PA, Biophysical characterization of iron in mitochondria isolated from respiring and fermenting yeast, Biochemistry 49(26) (2010) 5436–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Dodani SC, Leary SC, Cobine PA, Winge DR, Chang CJ, A targetable fluorescent sensor reveals that copper-deficient SCO1 and SCO2 patient cells prioritize mitochondrial copper homeostasis, J Am Chem Soc 133(22) (2011) 8606–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Shen C, Kolanowski JL, Tran CM, Kaur A, Akerfeldt MC, Rahme MS, Hambley TW, New EJ, A ratiometric fluorescent sensor for the mitochondrial copper pool, Metallomics 8(9) (2016) 915–9. [DOI] [PubMed] [Google Scholar]
- [65].Yang L, McRae R, Henary MM, Patel R, Lai B, Vogt S, Fahrni CJ, Imaging of the intracellular topography of copper with a fluorescent sensor and by synchrotron x-ray fluorescence microscopy, Proc Natl Acad Sci U S A 102(32) (2005) 11179–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].McCormick SP, Moore MJ, Lindahl PA, Detection of Labile Low-Molecular-Mass Transition Metal Complexes in Mitochondria, Biochemistry 54(22) (2015) 3442–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Lindahl PA, Moore MJ, Labile Low-Molecular-Mass Metal Complexes in Mitochondria: Trials and Tribulations of a Burgeoning Field, Biochemistry 55(30) (2016) 4140–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Kenney GE, Rosenzweig AC, Chalkophores, Annu Rev Biochem 87 (2018) 645–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Kim HJ, Graham DW, DiSpirito AA, Alterman MA, Galeva N, Larive CK, Asunskis D, Sherwood PM, Methanobactin, a copper-acquisition compound from methane-oxidizing bacteria, Science 305(5690) (2004) 1612–5. [DOI] [PubMed] [Google Scholar]
- [70].Semrau JD, DiSpirito AA, Gu W, Yoon S, Metals and Methanotrophy, Appl Environ Microbiol 84(6) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Cunningham CN, Rutter J, 20,000 picometers under the OMM: diving into the vastness of mitochondrial metabolite transport, EMBO Rep (2020) e50071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Palmieri F, Scarcia P, Monne M, Diseases Caused by Mutations in Mitochondrial Carrier Genes SLC25: A Review, Biomolecules 10(4) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Robinson AJ, Overy C, Kunji ER, The mechanism of transport by mitochondrial carriers based on analysis of symmetry, Proc Natl Acad Sci U S A 105(46) (2008) 17766–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Ruprecht JJ, Kunji ERS, The SLC25 Mitochondrial Carrier Family: Structure and Mechanism, Trends Biochem Sci 45(3) (2020) 244–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Crichton PG, Lee Y, Kunji ER, The molecular features of uncoupling protein 1 support a conventional mitochondrial carrier-like mechanism, Biochimie 134 (2017) 35–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Ruprecht JJ, King MS, Zogg T, Aleksandrova AA, Pardon E, Crichton PG, Steyaert J, Kunji ERS, The Molecular Mechanism of Transport by the Mitochondrial ADP/ATP Carrier, Cell 176(3) (2019) 435–447 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Pebay-Peyroula E, Dahout-Gonzalez C, Kahn R, Trezeguet V, Lauquin GJ, Brandolin G, Structure of mitochondrial ADP/ATP carrier in complex with carboxyatractyloside, Nature 426(6962) (2003) 39–44. [DOI] [PubMed] [Google Scholar]
- [78].Rees DC, Johnson E, Lewinson O, ABC transporters: the power to change, Nat Rev Mol Cell Biol 10(3) (2009) 218–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Kory N, Wyant GA, Prakash G, Uit de Bos J, Bottanelli F, Pacold ME, Chan SH, Lewis CA, Wang T, Keys HR, Guo YE, Sabatini DM, SFXN1 is a mitochondrial serine transporter required for one-carbon metabolism, Science 362(6416) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Bricker DK, Taylor EB, Schell JC, Orsak T, Boutron A, Chen YC, Cox JE, Cardon CM, Van Vranken JG, Dephoure N, Redin C, Boudina S, Gygi SP, Brivet M, Thummel CS, Rutter J, A mitochondrial pyruvate carrier required for pyruvate uptake in yeast, Drosophila, and humans, Science 337(6090) (2012) 96–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Herzig S, Raemy E, Montessuit S, Veuthey JL, Zamboni N, Westermann B, Kunji ER, Martinou JC, Identification and functional expression of the mitochondrial pyruvate carrier, Science 337(6090) (2012) 93–6. [DOI] [PubMed] [Google Scholar]
- [82].Halestrap AP, The mitochondrial pyruvate carrier: has it been unearthed at last?, Cell Metab 16(2) (2012) 141–3. [DOI] [PubMed] [Google Scholar]
- [83].Vest KE, Cobine PA, Copper in Mitochondria, Encyclopedia of Inorganic and Bioinorganic Chemistry (2011) 1–12. [Google Scholar]
- [84].Vest KE, Zhu X, Cobine PA, Copper Disposition in Yeast, Clinical and Translational Perspectives on WILSON DISEASE (2019) 115–126. [Google Scholar]
- [85].Horn D, Barrientos A, Mitochondrial copper metabolism and delivery to cytochrome c oxidase, IUBMB Life 60(7) (2008) 421–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Timon-Gomez A, Nyvltova E, Abriata LA, Vila AJ, Hosler J, Barrientos A, Mitochondrial cytochrome c oxidase biogenesis: Recent developments, Semin Cell Dev Biol 76 (2018) 163–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Glerum DM, Shtanko A, Tzagoloff A, SCO1 and SCO2 act as high copy suppressors of a mitochondrial copper recruitment defect in Saccharomyces cerevisiae, J Biol Chem 271(34) (1996) 20531–5. [DOI] [PubMed] [Google Scholar]
- [88].Beers J, Glerum DM, Tzagoloff A, Purification, characterization, and localization of yeast Cox17p, a mitochondrial copper shuttle, J Biol Chem 272(52) (1997) 33191–6. [DOI] [PubMed] [Google Scholar]
- [89].Horng YC, Cobine PA, Maxfield AB, Carr HS, Winge DR, Specific copper transfer from the Cox17 metallochaperone to both Sco1 and Cox11 in the assembly of yeast cytochrome C oxidase, J Biol Chem 279(34) (2004) 35334–40. [DOI] [PubMed] [Google Scholar]
- [90].Maxfield AB, Heaton DN, Winge DR, Cox17 is functional when tethered to the mitochondrial inner membrane, J Biol Chem 279(7) (2004) 5072–80. [DOI] [PubMed] [Google Scholar]
- [91].Bestwick M, Jeong MY, Khalimonchuk O, Kim H, Winge DR, Analysis of Leigh syndrome mutations in the yeast SURF1 homolog reveals a new member of the cytochrome oxidase assembly factor family, Mol Cell Biol 30(18) (2010) 4480–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Pierrel F, Bestwick ML, Cobine PA, Khalimonchuk O, Cricco JA, Winge DR, Coa1 links the Mss51 post-translational function to Cox1 cofactor insertion in cytochrome c oxidase assembly, EMBO J 26(20) (2007) 4335–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Ghosh A, Trivedi PP, Timbalia SA, Griffin AT, Rahn JJ, Chan SS, Gohil VM, Copper supplementation restores cytochrome c oxidase assembly defect in a mitochondrial disease model of COA6 deficiency, Hum Mol Genet 23(13) (2014) 3596–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Mick DU, Wagner K, van der Laan M, Frazier AE, Perschil I, Pawlas M, Meyer HE, Warscheid B, Rehling P, Shy1 couples Cox1 translational regulation to cytochrome c oxidase assembly, EMBO J 26(20) (2007) 4347–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Wang C, Richter-Dennerlein R, Pacheu-Grau D, Liu F, Zhu Y, Dennerlein S, Rehling P, MITRAC15/COA1 promotes mitochondrial translation in a ND2 ribosome-nascent chain complex, EMBO Rep 21(1) (2020) e48833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Monne M, Chan KW, Slotboom DJ, Kunji ER, Functional expression of eukaryotic membrane proteins in Lactococcus lactis, Protein Sci 14(12) (2005) 3048–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Vest KE, Wang J, Gammon MG, Maynard MK, White OL, Cobine JA, Mahone WK, Cobine PA, Overlap of copper and iron uptake systems in mitochondria in Saccharomyces cerevisiae, Open Biol 6(1) (2016) 150223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Brazzolotto X, Pierrel F, Pelosi L, Three conserved histidine residues contribute to mitochondrial iron transport through mitoferrins, Biochem J 460(1) (2014) 79–89. [DOI] [PubMed] [Google Scholar]
- [99].Froschauer EM, Schweyen RJ, Wiesenberger G, The yeast mitochondrial carrier proteins Mrs3p/Mrs4p mediate iron transport across the inner mitochondrial membrane, Biochim Biophys Acta 1788(5) (2009) 1044–50. [DOI] [PubMed] [Google Scholar]
- [100].Christenson ET, Gallegos AS, Banerjee A, In vitro reconstitution, functional dissection, and mutational analysis of metal ion transport by mitoferrin-1, J Biol Chem 293(10) (2018) 3819–3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Jain A, Dashner ZS, Connolly EL, Mitochondrial Iron Transporters (MIT1 and MIT2) Are Essential for Iron Homeostasis and Embryogenesis in Arabidopsis thaliana, Front Plant Sci 10 (2019) 1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Palmieri F, The mitochondrial transporter family (SLC25): physiological and pathological implications, Pflugers Arch 447(5) (2004) 689–709. [DOI] [PubMed] [Google Scholar]
- [103].Paradkar PN, Zumbrennen KB, Paw BH, Ward DM, Kaplan J, Regulation of mitochondrial iron import through differential turnover of mitoferrin 1 and mitoferrin 2, Mol Cell Biol 29(4) (2009) 1007–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Shaw GC, Cope JJ, Li L, Corson K, Hersey C, Ackermann GE, Gwynn B, Lambert AJ, Wingert RA, Traver D, Trede NS, Barut BA, Zhou Y, Minet E, Donovan A, Brownlie A, Balzan R, Weiss MJ, Peters LL, Kaplan J, Zon LI, Paw BH, Mitoferrin is essential for erythroid iron assimilation, Nature 440(7080) (2006) 96–100. [DOI] [PubMed] [Google Scholar]
- [105].Braymer JJ, Lill R, Iron-sulfur cluster biogenesis and trafficking in mitochondria, J Biol Chem 292(31) (2017) 12754–12763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Lill R, From the discovery to molecular understanding of cellular iron-sulfur protein biogenesis, Biol Chem (2020). [DOI] [PubMed] [Google Scholar]
- [107].Lill R, Freibert SA, Mechanisms of Mitochondrial Iron-Sulfur Protein Biogenesis, Annu Rev Biochem (2020). [DOI] [PubMed] [Google Scholar]
- [108].Hadzhieva M, Kirches E, Mawrin C, Review: iron metabolism and the role of iron in neurodegenerative disorders, Neuropathol Appl Neurobiol 40(3) (2014) 240–57. [DOI] [PubMed] [Google Scholar]
- [109].Llorens JV, Soriano S, Calap-Quintana P, Gonzalez-Cabo P, Molto MD, The Role of Iron in Friedreich’s Ataxia: Insights From Studies in Human Tissues and Cellular and Animal Models, Front Neurosci 13 (2019) 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Yoon H, Zhang Y, Pain J, Lyver ER, Lesuisse E, Pain D, Dancis A, Rim2, a pyrimidine nucleotide exchanger, is needed for iron utilization in mitochondria, Biochem J 440(1) (2011) 137–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Froschauer EM, Rietzschel N, Hassler MR, Binder M, Schweyen RJ, Lill R, Muhlenhoff U, Wiesenberger G, The mitochondrial carrier Rim2 co-imports pyrimidine nucleotides and iron, Biochem J 455(1) (2013) 57–65. [DOI] [PubMed] [Google Scholar]
- [112].Knight SAB, Yoon H, Pandey AK, Pain J, Pain D, Dancis A, Splitting the functions of Rim2, a mitochondrial iron/pyrimidine carrier, Mitochondrion 47 (2019) 256–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Leary SC, Antonicka H, Sasarman F, Weraarpachai W, Cobine PA, Pan M, Brown GK, Brown R, Majewski J, Ha KC, Rahman S, Shoubridge EA, Novel mutations in SCO1 as a cause of fatal infantile encephalopathy and lactic acidosis, Hum Mutat 34(10) (2013) 1366–70. [DOI] [PubMed] [Google Scholar]
- [114].Leary SC, Cobine PA, Nishimura T, Verdijk RM, de Krijger R, de Coo R, Tarnopolsky MA, Winge DR, Shoubridge EA, COX19 mediates the transduction of a mitochondrial redox signal from SCO1 that regulates ATP7A-mediated cellular copper efflux, Mol Biol Cell 24(6) (2013) 683–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Hlynialuk CJ, Ling B, Baker ZN, Cobine PA, Yu LD, Boulet A, Wai T, Hossain A, El Zawily AM, McFie PJ, Stone SJ, Diaz F, Moraes CT, Viswanathan D, Petris MJ, Leary SC, The Mitochondrial Metallochaperone SCO1 Is Required to Sustain Expression of the High-Affinity Copper Transporter CTR1 and Preserve Copper Homeostasis, Cell Rep (2015) pii: S2211-1247(15)00032-7. [DOI] [PubMed] [Google Scholar]
- [116].Baker ZN, Jett K, Boulet A, Hossain A, Cobine PA, Kim B-E, Zawily AME, Lee L, Tibbits GF, Petris MJ, Leary SC, The mitochondrial metallochaperone SCO1 maintains CTR1 at the plasma membrane to preserve copper homeostasis in the murine heart, Human Molecular Genetics Advance Article: ddx344 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Leary SC, Cobine PA, Kaufman BA, Guercin GH, Mattman A, Palaty J, Lockitch G, Winge DR, Rustin P, Horvath R, Shoubridge EA, The human cytochrome c oxidase assembly factors SCO1 and SCO2 have regulatory roles in the maintenance of cellular copper homeostasis (vol 5, pg 9, 2007), Cell Metabolism 5(5) (2007) 403–403. [DOI] [PubMed] [Google Scholar]
- [118].Reddi AR, Culotta VC, SOD1 integrates signals from oxygen and glucose to repress respiration, Cell 152(1–2) (2013) 224–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Grasso M, Bond GJ, Kim Y-J, Alwan KB, Boyd S, Dzebo MM, Valenzuela S, Tsang T, Schibrowsky NA, Matthews ML, Burslem GM, Wittung-Stafshede P, Winkler DD, Blackburn NJ, Marmorstein R, Brady DC, Structural and molecular determinants of CCS-mediated copper activation of MEK1/2, bioRxiv (2020) 2020.05.01.072124. [Google Scholar]
- [120].Roger AJ, Munoz-Gomez SA, Kamikawa R, The Origin and Diversification of Mitochondria, Curr Biol 27(21) (2017) R1177–R1192. [DOI] [PubMed] [Google Scholar]
- [121].Wang Z, Wu M, An integrated phylogenomic approach toward pinpointing the origin of mitochondria, Sci Rep 5 (2015) 7949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Karnkowska A, Vacek V, Zubacova Z, Treitli SC, Petrzelkova R, Eme L, Novak L, Zarsky V, Barlow LD, Herman EK, Soukal P, Hroudova M, Dolezal P, Stairs CW, Roger AJ, Elias M, Dacks JB, Vlcek C, Hampl V, A Eukaryote without a Mitochondrial Organelle, Curr Biol 26(10) (2016) 1274–84. [DOI] [PubMed] [Google Scholar]
- [123].Vacek V, Novak LVF, Treitli SC, Taborsky P, Cepicka I, Kolisko M, Keeling PJ, Hampl V, Fe-S Cluster Assembly in Oxymonads and Related Protists, Mol Biol Evol 35(11) (2018) 2712–2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Karnkowska A, Treitli SC, Brzon O, Novak L, Vacek V, Soukal P, Barlow LD, Herman EK, Pipaliya SV, Panek T, Zihala D, Petrzelkova R, Butenko A, Eme L, Stairs CW, Roger AJ, Elias M, Dacks JB, Hampl V, The Oxymonad Genome Displays Canonical Eukaryotic Complexity in the Absence of a Mitochondrion, Mol Biol Evol 36(10) (2019) 2292–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Mellacheruvu D, Wright Z, Couzens AL, Lambert JP, St-Denis NA, Li T, Miteva YV, Hauri S, Sardiu ME, Low TY, Halim VA, Bagshaw RD, Hubner NC, Al-Hakim A, Bouchard A, Faubert D, Fermin D, Dunham WH, Goudreault M, Lin ZY, Badillo BG, Pawson T, Durocher D, Coulombe B, Aebersold R, Superti-Furga G, Colinge J, Heck AJ, Choi H, Gstaiger M, Mohammed S, Cristea IM, Bennett KL, Washburn MP, Raught B, Ewing RM, Gingras AC, Nesvizhskii AI, The CRAPome: a contaminant repository for affinity purification-mass spectrometry data, Nat Methods 10(8) (2013) 730–6. [DOI] [PMC free article] [PubMed] [Google Scholar]