Abstract
Mitochondria are highly dynamic organelles that continuously grow, divide, and fuse. The division of mitochondria is crucial for human health. During mitochondrial division, the mechano GTPase dynamin-related protein (Drp1) severs mitochondria at endoplasmic reticulum (ER)-mitochondria contact sites, where peripheral ER tubules interact with mitochondria. Here, we report that Drp1 directly shapes peripheral ER tubules in human and mouse cells. This ER-shaping activity is independent of GTP hydrolysis and located in a highly conserved peptide of 18 amino acids (termed D-octadecapeptide), which is predicted to form an amphipathic α-helix. Synthetic D-octadecapeptide tubulates liposomes in vitro and the ER in cells. ER tubules formed by Drp1 promote mitochondrial division by facilitating ER-mitochondria interactions. Thus, Drp1 functions as a two-in-one protein during mitochondrial division, with ER tubulation and mechano GTPase activities.
Keywords: Mitochondria, the endoplasmic reticulum, Drp1, Organelle contact sites
Graphical Abstract
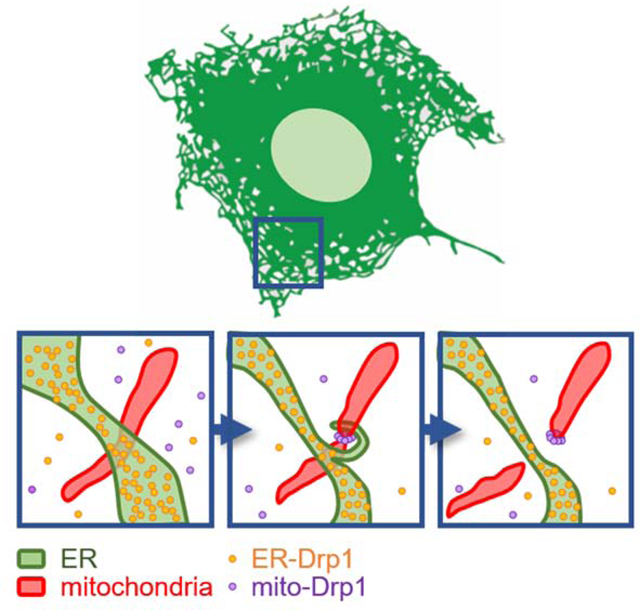
eTOC
Adachi et al. report that Drp1 shapes the ER into tubules independently of GTP hydrolysis. ER tubules formed by Drp1 promote mitochondrial division by facilitating ER-mitochondria interactions. Thus, Drp1 functions as a two-in-one protein during mitochondrial division, with ER tubulation and mechano GTPase activities.
Introduction
The endoplasmic reticulum (ER) consists of distinct sectors with distinct functions. Also called cisternae or matrices, ER sheets are contiguous to the nuclear membrane and associated with ribosomes. Furthermore, they create most of the rough ER for the biosynthesis of secreted and membrane proteins (Baumann and Walz, 2001; Nixon-Abell et al., 2016; Schwarz and Blower, 2016). In contrast, ER tubules functionally interact with other organelles, including mitochondria. Interactions between the ER and mitochondria play important roles in lipid biosynthesis, calcium signaling, cell death, and mtDNA replication (de Brito and Scorrano, 2008; Hirabayashi et al., 2017; Lewis et al., 2016; Prudent and McBride, 2017; Scorrano et al., 2019; Takeda and Yanagi, 2019).
The ratio of ER sheets and tubules varies depending on the type of cell. For example, cells that are specialized for secretion, such as pancreatic cells and B cells, contain large amounts of ER sheets (Baumann and Walz, 2001; Schwarz and Blower, 2016). In contrast, adrenal, liver, and muscle cells have more ER tubules. In cultures of most cell lines and fibroblasts, the ER sheets are present around the center of cells, while ER tubules are mainly located in the cell periphery (Chen et al., 2013; Goyal and Blackstone, 2013; Nixon-Abell et al., 2016; Terasaki, 2016; Zhang and Hu, 2016).
ER tubules frequently mark sites of mitochondrial division that are mediated by the dynamin-related GTPase Drp1 (Friedman et al., 2011; Guo et al., 2018; Korobova et al., 2013; Lewis et al., 2016; Liesa and Shirihai, 2013; Nagashima et al., 2020; Wong et al., 2018; Wu et al., 2018), a protein whose defects have been associated with many human diseases (Itoh et al., 2013; Liesa and Shirihai, 2013; Roy et al., 2015). In mitochondrial division, Drp1 forms high molecular weight oligomers that wrap around mitochondria, and subsequent oligomerization-induced GTP hydrolysis drives the conformational changes of the oligomers to sever mitochondria (Fonseca et al., 2019; Kalia et al., 2018; Kamerkar et al., 2018; Smirnova et al., 2001; Tamura et al., 2011). In this study, we report an unforeseen finding that Drp1 shapes peripheral ER tubules and promotes ER-mitochondria interactions independently of oligomerization and GTP hydrolysis.
Results
We visualized the morphology of the ER using its well-established marker GFP-Sec61β (Friedman et al., 2011), and quantified the relative amount of ER tubules and ER sheets in three types of Drp1-knockout (KO) cells (Materials and Methods). In wild-type (WT) mouse embryonic fibroblasts (MEFs), 40% of the ER was tubular, as shown by laser scanning confocal microscopy (Fig. 1A–1C) and structured illumination microscopy (Fig. S1A). The percentage of ER tubules decreased dramatically in Drp1-KO MEFs, and this ER phenotype was rescued by the re-expression of Drp1 (mouse isoform 3) (Fig. 1A–1C and S1A). The changes in ER morphology in Drp1-KO MEFs were confirmed using another ER marker, GFP-Cyb5 (Fig. S1B and S1C), and immunofluorescence microscopy with antibodies to an endogenous ER protein, BiP (Fig. 1D and 1E). Similar to KO, knockdown of Drp1 by shRNA also decreased levels of ER tubules in MEFs (Fig. 1F–1H). ER tubules were fully restored by shRNA-resistant Drp1 (Fig. 1F–1H). Similar to Drp1-KO MEFs, the number of ER tubules relative to ER sheets was decreased in both Drp1-KO Hela cells (Fig. 1I and 1J) (Otera et al., 2016) and hepatocytes isolated from liver-specific Drp1 knockout mice (Fig. S1D and S1E) (Yamada et al., 2018). As expected, mitochondria were elongated in all three types of Drp1-KO cells (Fig. 1A, 1C, 1I, 1J, S1D and S1E) and Drp1-knockdown cells (Fig. 1G and 1H). These data indicate that Drp1 regulates ER morphology in both mouse and human cells.
Fig. 1. Drp1 controls ER tubulation in cells.
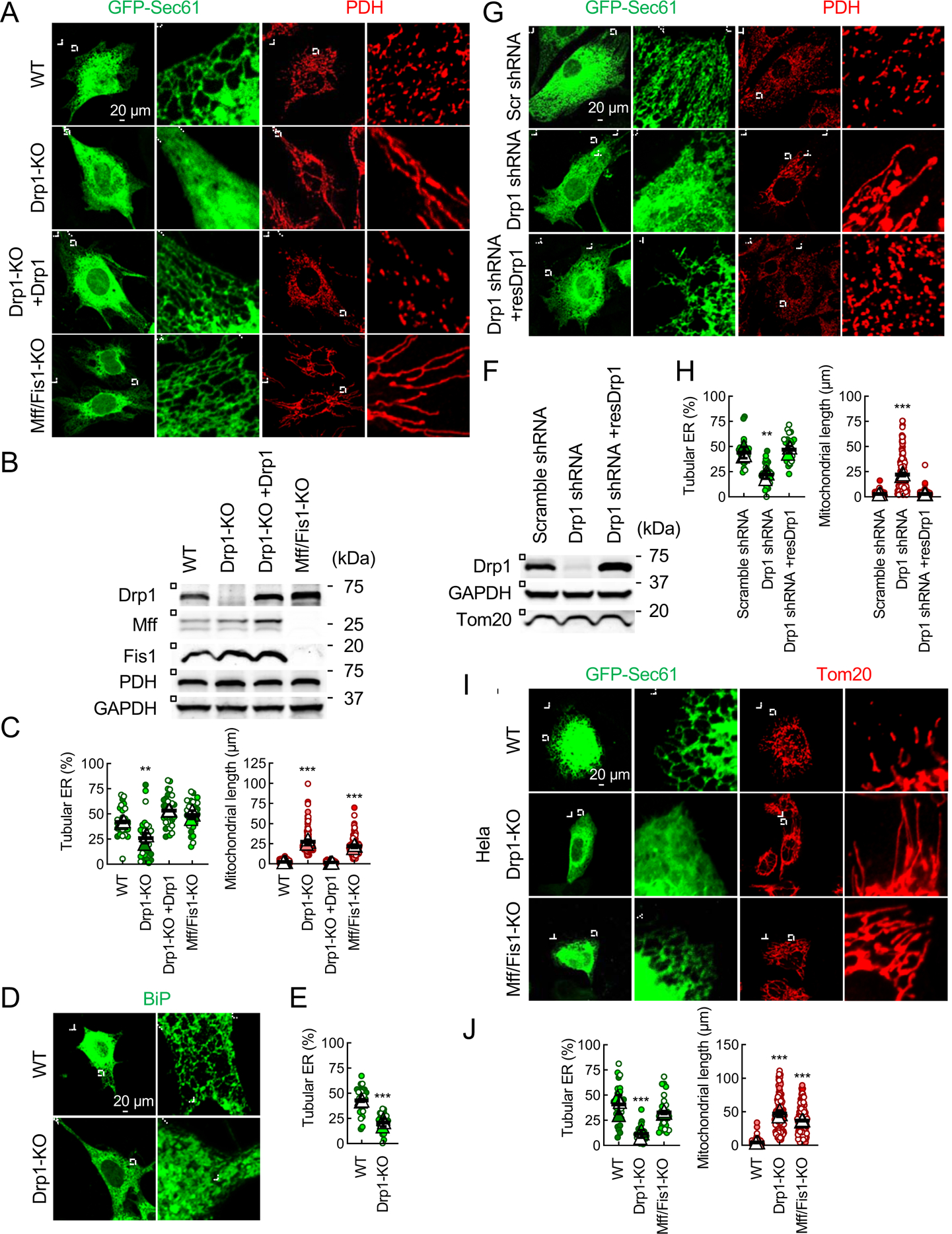
(A) WT, Drp1-KO, and Mff/Fis1 double-KO MEFs were transfected with an ER marker (GFP-Sec61β) and subjected to immunofluorescence confocal microscopy with antibodies to a mitochondrial protein, pyruvate dehydrogenase (PDH), and GFP. Boxed areas are enlarged. Cells are outlined by a dotted line. (B) The expression levels of Drp1, Mff, and Fis1 were analyzed by Western blotting. (C) Quantification of the morphology of the ER (green, n = 3 experiments, 10 cells were analyzed in each experiment) and mitochondria (red, n = 3 experiments, 50 mitochondria were examined in each experiment). Bars are average ± SD. (D) WT and Drp1-KO MEFs were subjected to immunofluorescence microscopy with antibodies to an endogenous ER protein, BiP. (E) Quantification of ER morphology. Bars are average ± SD (n = 3). (F, G and H) Knockdown of Drp1 decreases amounts of ER tubules in cells. WT MEFs were transduced with lentiviruses carrying scramble or Drp1-targeted shRNAs. (F) The expression of Drp1 was analyzed by Western blotting. (G) The ER and mitochondria were visualized by GFP-Sec61β and anti-PDH antibodies, respectively. (H) Quantification of the morphology of ER (green, n = 3) and mitochondria (red, n = 3). Bars are average ± SD. (I) The indicated Hela cells were transfected with the GFP-Sec61β plasmid and subjected to immunofluorescence microscopy with antibodies to a mitochondrial protein (Tom20) and GFP. (J) Quantification of the morphology of ER (green, n = 3) and mitochondria (red, n = 3) is shown. Bars are average ± SD. Statistical analysis was performed using One-way ANOVA with post-hoc Tukey (C, F and I), and Student’s t-test (E): **p<0.01, ***p<0.001. See also Figure S1.
To determine whether decreased percentages of ER tubules in Drp1-KO cells result from elongation of mitochondria, we analyzed the ER morphology in MEFs lacking Drp1 receptors Mff and Fis1 (Loson et al., 2013; Otera et al., 2016). As expected, the mitochondria became elongated in Mff/Fis1 double-KO MEFs (Fig. 1A–1C) and Hela cells (Fig. 1I and 1J). In contrast, the loss of Mff and Fis1 did not affect ER morphology in Mff/Fis1 double-KO MEFs (Fig. 1A and 1C) or Hela cells (Fig. 1I and 1J). Therefore, mitochondrial elongation is not the cause of decreases in ER tubules in Drp1-KO cells.
To test whether the loss of Drp1 affects the connectivity of the ER membrane and lumen, we performed fluorescence recovery after photobleaching (FRAP) experiments using GFP-Sec61β for the membrane and GFP-KDEL for the lumen. The results showed that the GFP fluorescence was similarly recovered after photobleaching for both the membrane and lumen markers in WT and Drp1-KO MEFs (Fig. 2).
Fig. 2. The loss of Drp1 does not affect the connectivity of the ER membrane and lumen.
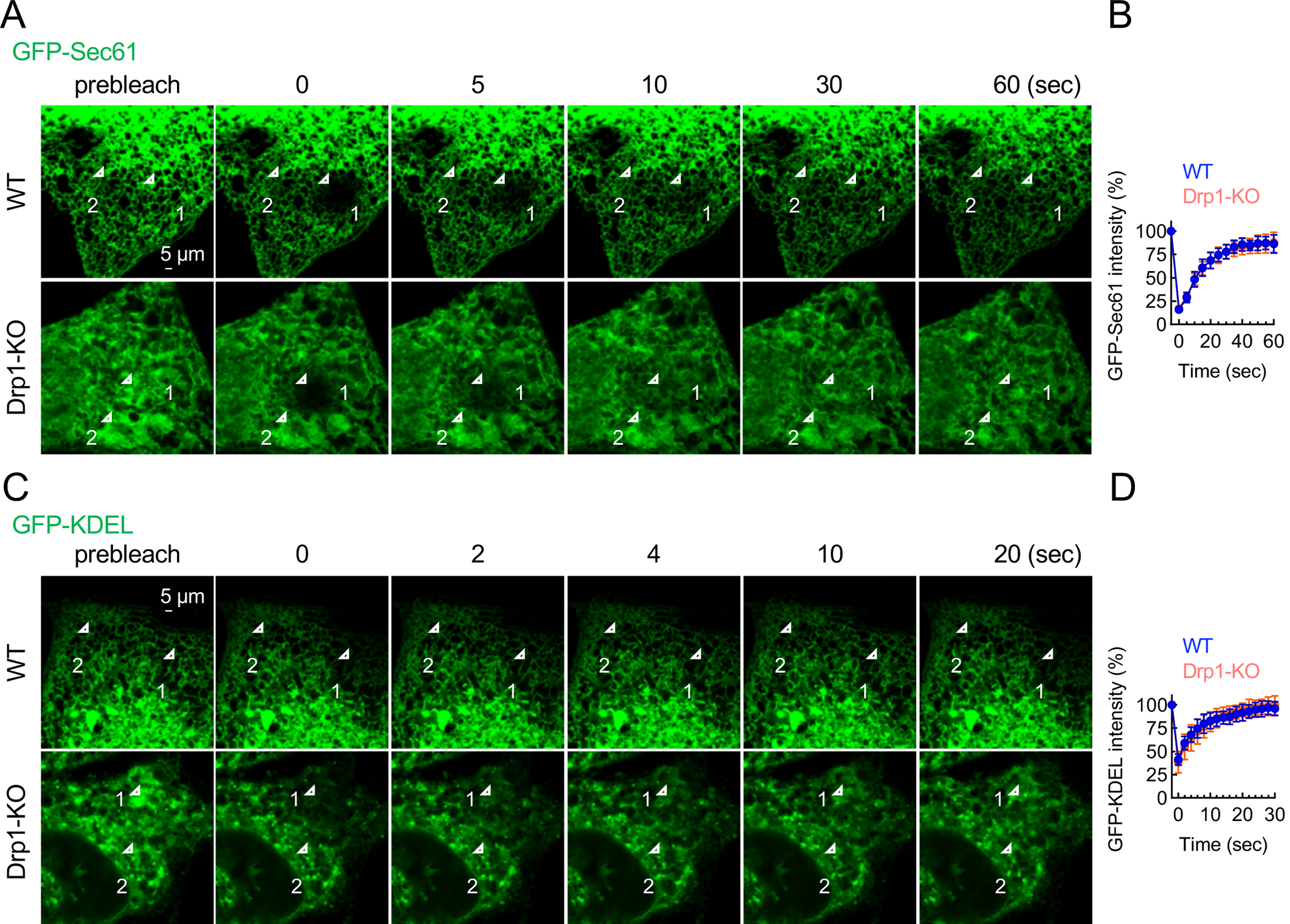
(A) FRAP analysis of GFP-Sec61β (an ER membrane probe) in WT and Drp1-KO MEFs. GFP-Sec61β in region #1 was photobleached at 0 sec. Images were taken at 5 sec intervals. (B) GFP intensity in region #1 was normalized to that of unbleached region #2 at each time point. Bars are the average ± SD (n = 10 cells). (C) FRAP analysis of GFP-KDEL (an ER luminal probe) in WT and Drp1-KO MEFs. Images were taken at 2-sec intervals. (D) Quantification of relative GFP intensity in region #1. Bars are the average ± SD (n = 10 cells).
It has been shown that the ER membrane proteins, reticulon 4a (Rtn4a) (Voeltz et al., 2006) and Climp63 (Shibata et al., 2010), form ER tubules and sheets, respectively. To test whether Drp1 regulates the peripheral ER morphology through these two proteins, we performed two sets of experiments. First, we created Rtn4a-KO MEFs in which the percentage of ER tubules were decreased (Fig. 3A–3C). We found similar decreases in the percentage of ER tubules in Rtn4a-KO and Drp1-KO MEFs (Fig. 1C and 3C). The ectopic expression of Drp1 restored ER tubules in Rtn4a-KO cells (Fig. 3A–3C). When we examined mitochondria in Rtn4a-KO MEFs, we found normal mitochondrial morphology (Fig. 3B and 3C), suggesting that mitochondrial morphology is independent of Rtn4a-mediated ER tubulation. Second, the overexpression of Climp63 produced more ER sheets and fewer ER tubules in WT MEFs (Fig. 3D and 3E) (Shibata et al., 2010). These ER sheets produced by Climp63 overexpression were converted to ER tubules by the ectopic expression of Drp1, similar to that of Rtn4a (Fig. 3D and 3E). Third, electron microscopy (EM) of ER sheets revealed that the luminal widths of ER sheets are comparable in WT and Drp1-KO hepatocytes (Fig. S1F and S1G). This is in sharp contrast with the decreased luminal widths of ER sheets in cells depleted for Climp63, which forms luminal bridges in ER sheets (Shibata et al., 2010). These findings suggest that Drp1 can convert ER sheets to tubules independently of Rtn4a and Climp63.
Fig. 3. Drp1 forms ER tubules independently of Rtn4a and Climp63.
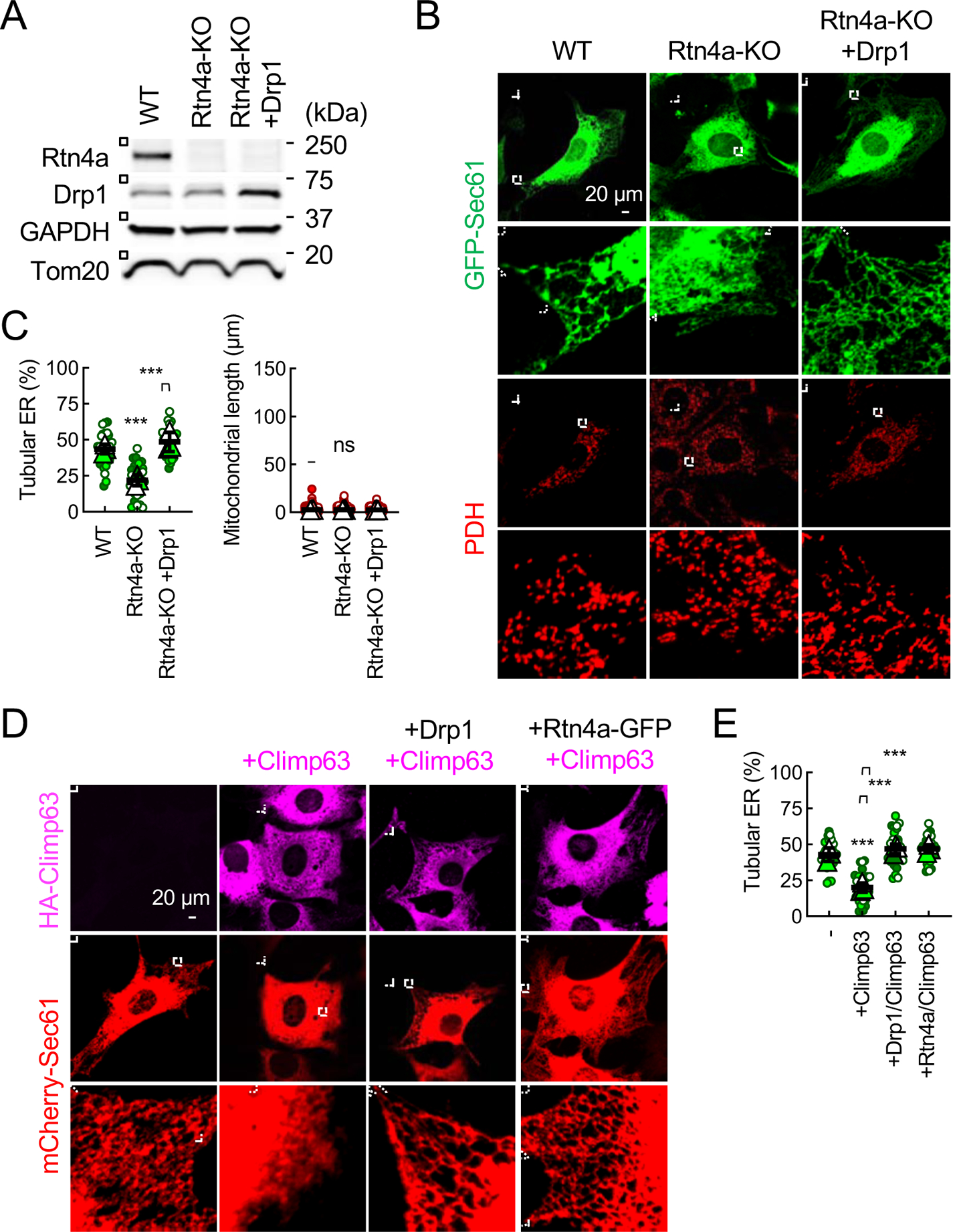
(A) The expression levels of Rtn4a and Drp1 were analyzed in WT MEFs, Rtn4a-KO MEFs, and Rtn4a-KO MEFs ectopically expressing Drp1 by Western blotting. (B) The ER and mitochondria in these MEFs were visualized. (C) Quantification of the morphology of the ER and mitochondria. Bars are average ± SD (n = 3 experiments). (D) WT MEF expressing mCherry-Sec61β were transduced with lentiviruses carrying HA-Climp63 along with Drp1 or Rtn4a-GFP. The efficiency of lentivirus transduction was essentially 100%. The MEFs were subjected to immunofluorescence microscopy with antibodies to HA and mCherry. (E) Quantification of ER morphology. Bars are average ± SD (n = 3). Statistical analysis was performed using One-way ANOVA with post-hoc Tukey (C and E): **p<0.01, ***p<0.001.
We found that a subset of endogenous Drp1 is associated with the ER when we performed four-color confocal microscopy for Drp1, the ER, mitochondria, and peroxisomes (Fig. 4A). WT and Mff/Fis1 double-KO MEFs were transduced with lentiviruses carrying mCherry-Sec61β and mitochondria-targeted Su9-HA-iRFP670, and then subjected to confocal immunofluorescence microscopy with antibodies against HA, mCherry, Drp1 and a peroxisomal protein, Pex14. Drp1 formed puncta on mitochondria in WT MEFs, and this mitochondrial localization of Drp1 was decreased in Mff/Fis1 double-KO MEFs (Fig. 4A, lower gain for Drp1) (Kameoka et al., 2018). We then examined the localization of Drp1 at the ER in regions where mitochondria and peroxisomes are absent: using confocal microscopy, the absence of mitochondria and peroxisomes was confirmed in both the X-Y axis and Z-axis. We detected significant amounts of Drp1 along ER tubules (Fig. 4A, higher gain for Drp1). Since the Drp1 signal at the ER tubules was weaker compared to that at mitochondria, it was critical to increase the detection gain on the confocal microscope to detect ER-located Drp1 signals (Fig. 4A). To ensure the specificity of these Drp1 signals, Drp1-KO MEFs were used as a negative control (Fig. 4A). A previous study has reported that a subset of Mff is located at the ER and promotes Drp1 oligomerization at the ER (Ji et al., 2017). We found that knockout of Mff does not grossly affect the localization of Drp1 on the ER (Fig. 4A). These data suggest that there are two populations of Drp1 in the ER. One population that is dependent on Mff, is recruited to the ER, oligomerized, and then transferred to mitochondria for the division, as reported (Ji et al., 2017). The other population of Drp1, which is independent of Mff, stays on the ER, and controls ER morphogenesis.
Fig. 4. Drp1 is located at the ER.
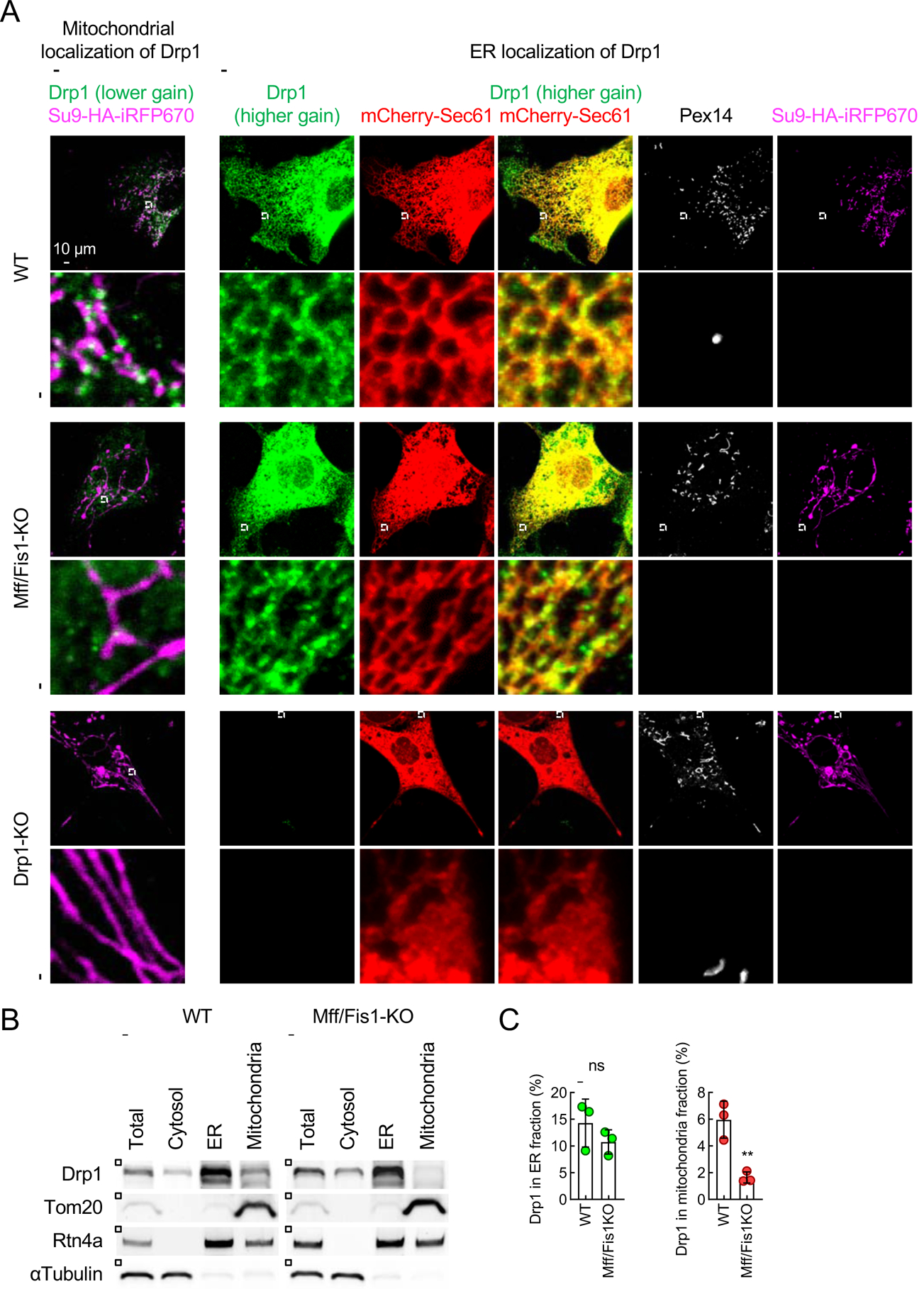
WT, Mff1/Fis1-KO, and Drp1-KO MEFs, all of which express mCherry-Sec61β and a mitochondrial marker (Su9-HA-iRFP670), were analyzed by immunofluorescence confocal microscopy with antibodies to Drp1, mCherry, a peroxisomal protein (Pex14), and HA. Drp1 signals were detected using two different gain settings (lower gain for mitochondrial localization and higher gain for ER localization). Regions where mitochondria and peroxisomes are absent are boxed and enlarged (higher gain). (B) The ER and mitochondrial fractions were obtained from WT and Mff/Fis1-KO MEFs using differential centrifugations. Each fraction was analyzed by western blotting using antibodies to Drp1, Tom20, Rtn4a, and α-Tubulin. (C) Relative amounts of Drp1 in the ER and mitochondrial fractions were quantified. Bars are the average ± SD (n = 3). Statistical analysis was performed using a student’s t-test: **p<0.01.
To confirm the ER localization of Drp1 biochemically, we performed subcellular fractionation of the ER and mitochondria in WT and Mff/Fis1-KO MEFs (Fig. 4B and 4C). After the cells were homogenized, the ER and mitochondria were separated by differential centrifugations (Sugiura et al., 2013). The results revealed a significant amount of Drp1 in the ER fraction (Fig. 4B and 4C). The amounts of Drp1 in the ER fractions were similar in WT and Mff/Fis1-KO MEFs (Fig. 4B and 4C). As expected, the amounts of Drp1 were lower in the mitochondrial fraction obtained from Mff/Fis1-KO MEFs (Fig. 4B and 4C). These data are consistent with the immunofluorescence microscopy results of Drp1 (Fig. 4A) and further support the localization of Drp1 at the ER.
For mitochondrial division, GTP hydrolysis by Drp1 and its oligomerization are essential (Fonseca et al., 2019; Kalia et al., 2018; Kamerkar et al., 2018; Smirnova et al., 2001; Tamura et al., 2011). To test whether these activities are required for the regulation of ER morphology, we expressed WT and three Drp1 mutants (Drp1K38A, which is defective in GTP hydrolysis; Drp1G350D, which is defective in oligomerization; and Drp1K38A,G350D, which is defective in both) in Drp1-KO MEFs (Fig. 5A–5C). Their expression levels were comparable (Fig. S2A). All three mutants failed to rescue mitochondrial morphology in Drp1-KO MEFs (Fig. 5B, 5C and S2C). Conversely, they all rescued ER morphology in Drp1-KO MEFs (Fig. 5B and 5C). Therefore, Drp1 regulates ER morphology independently of GTP hydrolysis and oligomerization. Drp1 consists of the GTPase, stalk, and variable domains (Fig. 5A and S3A). To determine which domain mediates ER morphogenesis, we expressed HA-tagged Drp1 constructs that contain different domains in Drp1-KO MEFs (Fig. S2B). The ER tubulation activity was found in the variable domain (Fig. 5B, 5C and S1A). In contrast, mitochondrial division required full-length Drp1 (Fig. 5C and S1A).
Fig. 5. D-octadecapeptide in the variable domain tubulates the ER in cells.
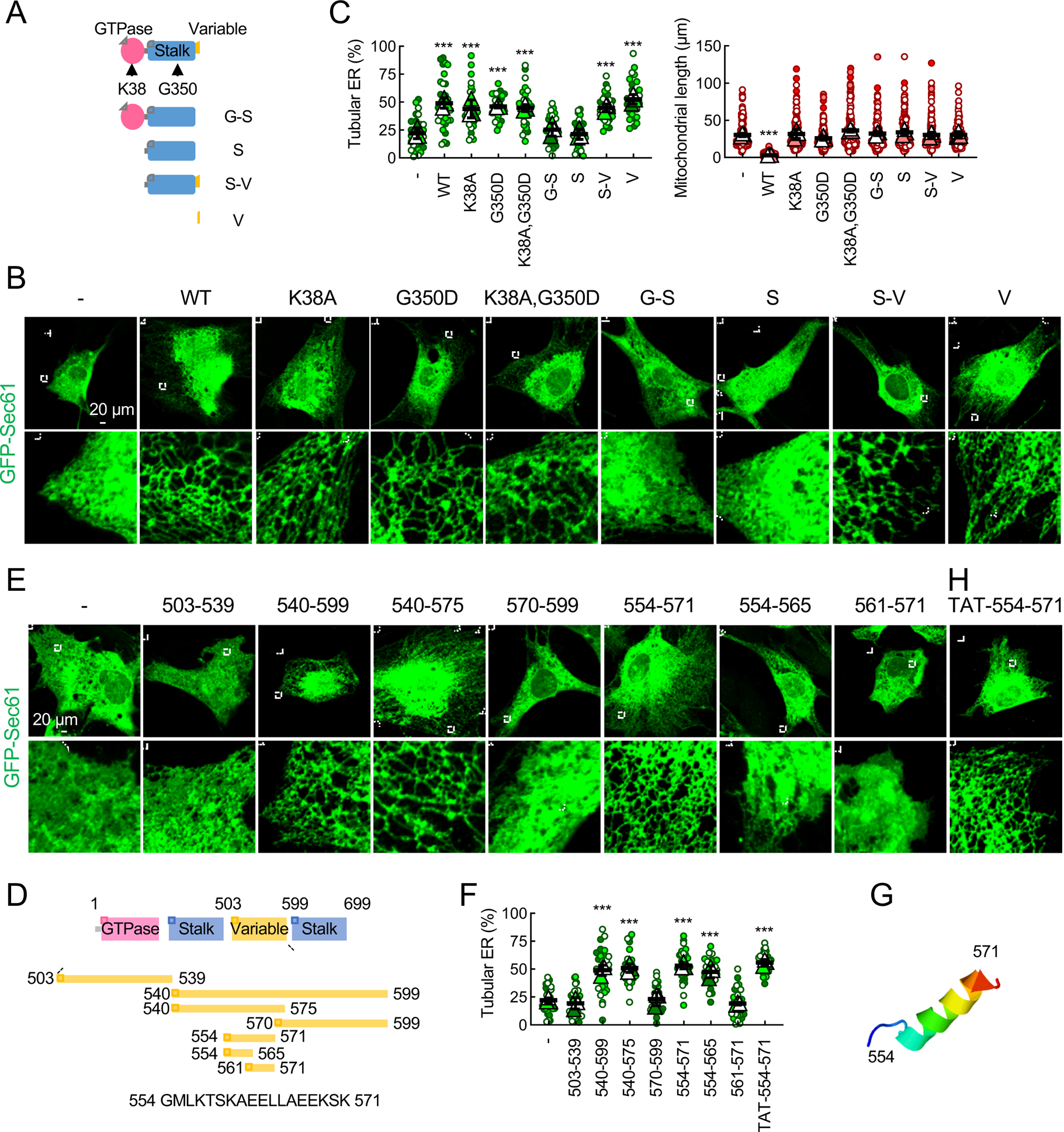
(A) Drp1-KO MEFs expressing the indicated Drp1 constructs (WT, mutants, or individual or combined domains) were transfected with GFP-Sec61β. (B) These MEFs were analyzed using immunofluorescence confocal microscopy with antibodies to GFP and Tom20 (see Fig. S2C). (C) Quantification of the morphology of ER (green, n = 3 experiments) and mitochondria (red, n = 3 experiments) is shown. Bars are average ± SD. (D) Truncations of the variable domain. (E) Drp1-KO MEFs carrying the truncations and GFP-Sec61β were subjected to immunofluorescence confocal microscopy with antibodies to GFP and PDH (see Fig. S2D). (F) Quantification of ER morphology. Bars are average ± SD (n = 3). (G) D-octadecapeptide is predicted to form an α-helix by ITASSER. (H) Drp1-KO MEFs expressing GFP-Sec61β were treated with synthetic D-octadecapeptide fused to a cell-permeable peptide TAT (50 μM) for 24 h and subjected to immunofluorescence microscopy with anti-GFP antibodies. Statistical analysis was performed using one-way ANOVA with the Tukey post-hoc test (C and F): ***p<0.001. See also Figure S2, S3 and S5.
To locate the ER morphogenesis activity within the variable domain with roughly 100 amino acids, the domain was truncated from the amino and carboxyl termini and expressed in Drp1-KO MEFs (Fig. 5D–5F). We found that a short sequence of 18 amino acids (termed D-octadecapeptide; 554 GMLKTSKAEELLAEEKSK 571 in mouse isoform 3) was sufficient to fully restore peripheral ER tubules (Fig. 5D–5F). Structural prediction programs suggested that D-octadecapeptide forms an amphipathic α-helix (Fig. 5G and S3B). The octadecapeptide sequence was unique to Drp1 and 100% conserved in human and mouse and present in all Drp1 isoforms (Fig. S3A). The sequence was slightly changed in zebrafish, and absent in fruit fly, worm, and yeast (Fig. S3A). Further truncations revealed that peptide554–565 only partially rescued ER morphology; peptide561–571 failed to do so (Fig. 5D–5F). Synthetic D-octadecapeptide fused to a cell-permeable, HIV-derived TAT peptide rescued ER morphology in Drp1-KO MEFs when added to the cell culture medium (Fig. 5F, 5H and S1A; TAT-554–571). These data suggest that D-octadecapeptide is a minimum region in Drp1 that controls ER morphology.
To ask if D-octadecapeptide directly binds and tubulates membranes, we incubated synthesized D-octadecapeptide with liposomes that mimic the ER membrane lipid composition in vitro (Fig. 6A and 6B). We have previously shown that Drp1 and the variable domain specifically binds phosphatidic acid (PA), but not other negatively charged phospholipids (Adachi et al., 2018; Adachi et al., 2017; Adachi et al., 2016). They also recognize saturated acyl chains of phospholipids for the additional specificity. We found that D-octadecapeptide specifically associates with liposomes containing saturated PA in a flotation assay (Fig. 6C). Mutations that change four positively charged lysines – which might interact with PA – to alanines (KA mutations), blocked the ability of D-octadecapeptide to bind saturated PA-containing liposomes (Fig. 6A and 6C). Surface plasmon resonance analysis further showed that D-octadecapeptide directly binds saturated PA, but not saturated PC, with an apparent Kd of 13 ± 3 μM (Fig. 6D). The KA mutations strongly inhibited the interactions of D-octadecapeptide with saturated PA in the surface plasmon resonance assay (Fig. 6D). Negative-stain EM showed that D-octadecapeptide tubulates the liposomes that contain saturated PA but not liposomes that lack saturated PA (Fig. 6E and 6F). The membrane-binding defective KA D-octadecapeptide failed to tubulate liposomes (Fig. 6E and 6F). The mean diameter of the membrane tubules induced by D-octadecapeptide was 18 ± 2 nm, which is similar to that of tubules generated by integral membrane proteins of the ER, reticulon and Yop1 (17 nm by negative-stain EM) (Hu et al., 2008). Two truncated peptides, peptide554–565 and peptide561–571, which exhibited decreased activities in ER tubulation in cells, also displayed decreased activities in liposome tubulation compared with D-octadecapeptide (Fig. S4A).
Fig. 6. Mechanism of ER tubulation by D-octadecapeptide.
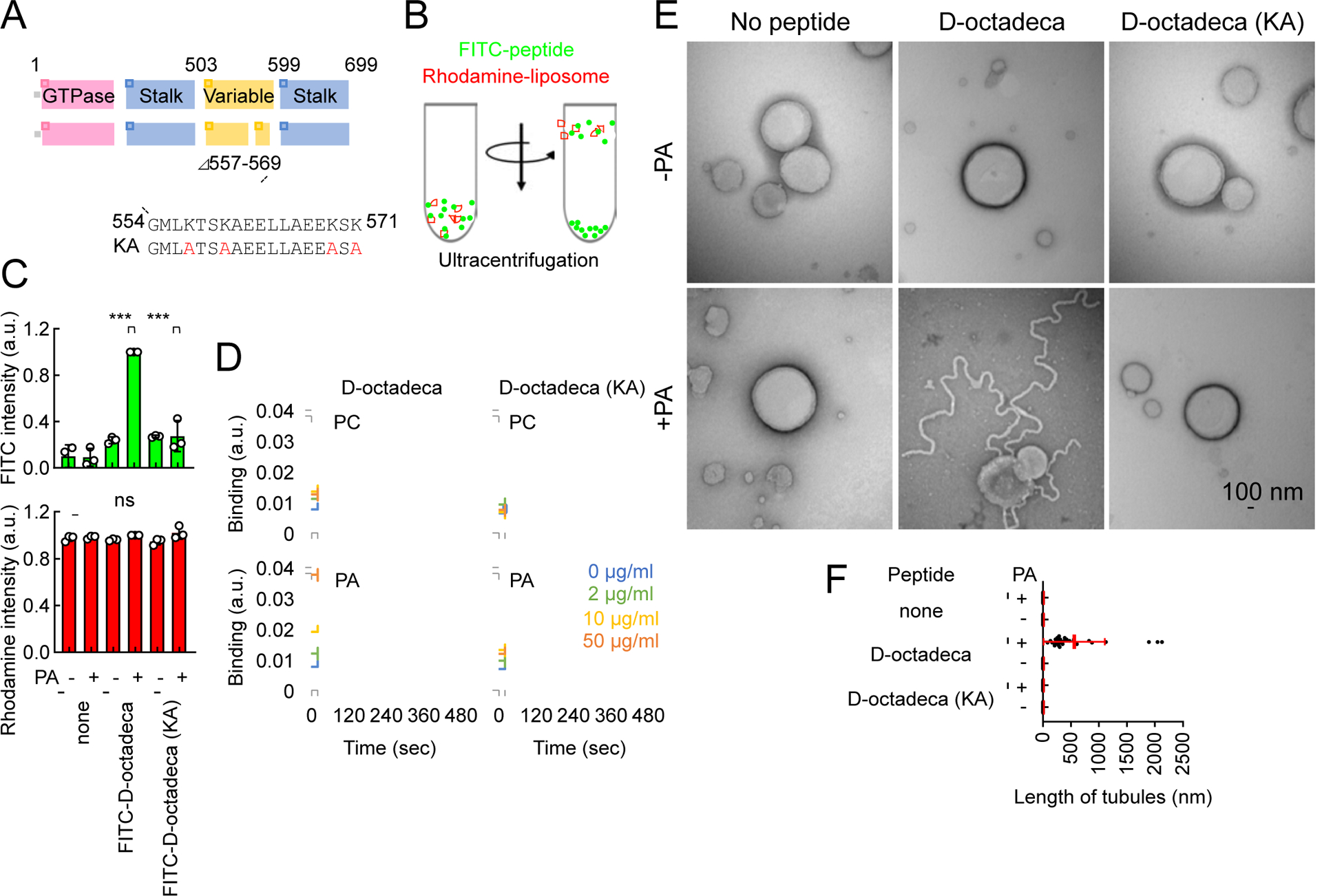
(A) The amino acid sequence of D-octadecapeptide is shown. The KA D-octadecapeptide mutant was created by changing four lysines to alanines (red). (B) Liposome flotation assay. Synthetic liposomes that mimic the ER membrane (which contains PC, PE, PI, and PS) carrying rhodamine-PE with or without saturated PA were incubated with FITC-labeled D-octadecapeptide or KA D-octadecapeptide and placed at the bottom of a sucrose gradient. (C) After ultracentrifugation, we collected four fractions from the top of the tube and measured FITC and rhodamine fluorescence. Almost 100% of the liposomes floated to the top fraction based on rhodamine fluorescence. Bars are average ± SD (n = 3 experiments). (D) Interactions of D-octadecapeptide and KA D-octadecapeptide with saturated PC or saturated PA were measured using surface plasmon resonance. Averages of triplicates for each lipid concentration are shown. (E) The same liposomes used in (B) were incubated with 1 μM D-octadecapeptide or KA D-octadecapeptide and viewed with negative-stain EM. (F) The length of the tubules that emerged from the liposomes was quantified. Bars are average ± SD (n = 20 tubules). Statistical analysis was performed using one-way ANOVA with the Tukey post-hoc test (C) and Kruskal-Wallis with Dunn’s post-hoc test (F): ***p<0.001. See also Figure S4.
We expressed a full-length Drp1 that lacks 14 amino acids (corresponding to the minimum sequence that forms the predicted α-helix) in the octadecapeptide (Drp1Δ557–569) and one that carries the KA mutations in Drp1-KO MEFs. We found that Drp1Δ557–569 and KA Drp1 are unable to rescue the ER morphology (Fig. 7A and 7B). Therefore, taken together with earlier data, we suggest that PA-binding D-octadecapeptide is critical for Drp1-mediated ER tubule formation. Remarkably, both Drp1 mutants defective in ER tubulation, Drp1Δ557–569 and KA Drp1, did not fully rescue mitochondrial morphology in Drp1-KO MEFs and left more mitochondria elongated compared with WT Drp1 (Fig. 7A and 7B). Furthermore, Drp1-KO MEFs expressing Drp1Δ557–569 failed to fragment mitochondria when mitochondrial division was induced by dissipating the membrane potential using carbonyl cyanide-4-phenylhydrazone (FCCP) (Fig. S5A–S5C). This inability of Drp1Δ557–569 to rescue mitochondrial division is not due to defects in the GTPase activity or mitochondrial localization since purified Drp1Δ557–569 normally hydrolyzed GTP (Fig. 7C) and HA-Drp1Δ557–569 was located at the mitochondria through Mff and Fis1 (Fig. S6). Instead, we suggest that Drp1Δ557–569 is unable to form ER-mitochondria contact sites for mitochondrial division. In support of this model, while the frequency of ER-mitochondria intersections was decreased after FCCP treatment in Drp1-KO expressing WT Drp1 due to mitochondrial division at the intersections, it remained unchanged in Drp1-KO MEFs expressing Drp1Δ557–569 (Fig. S5A and S5D).
Fig. 7. Function of ER tubulation by D-octadecapeptide.
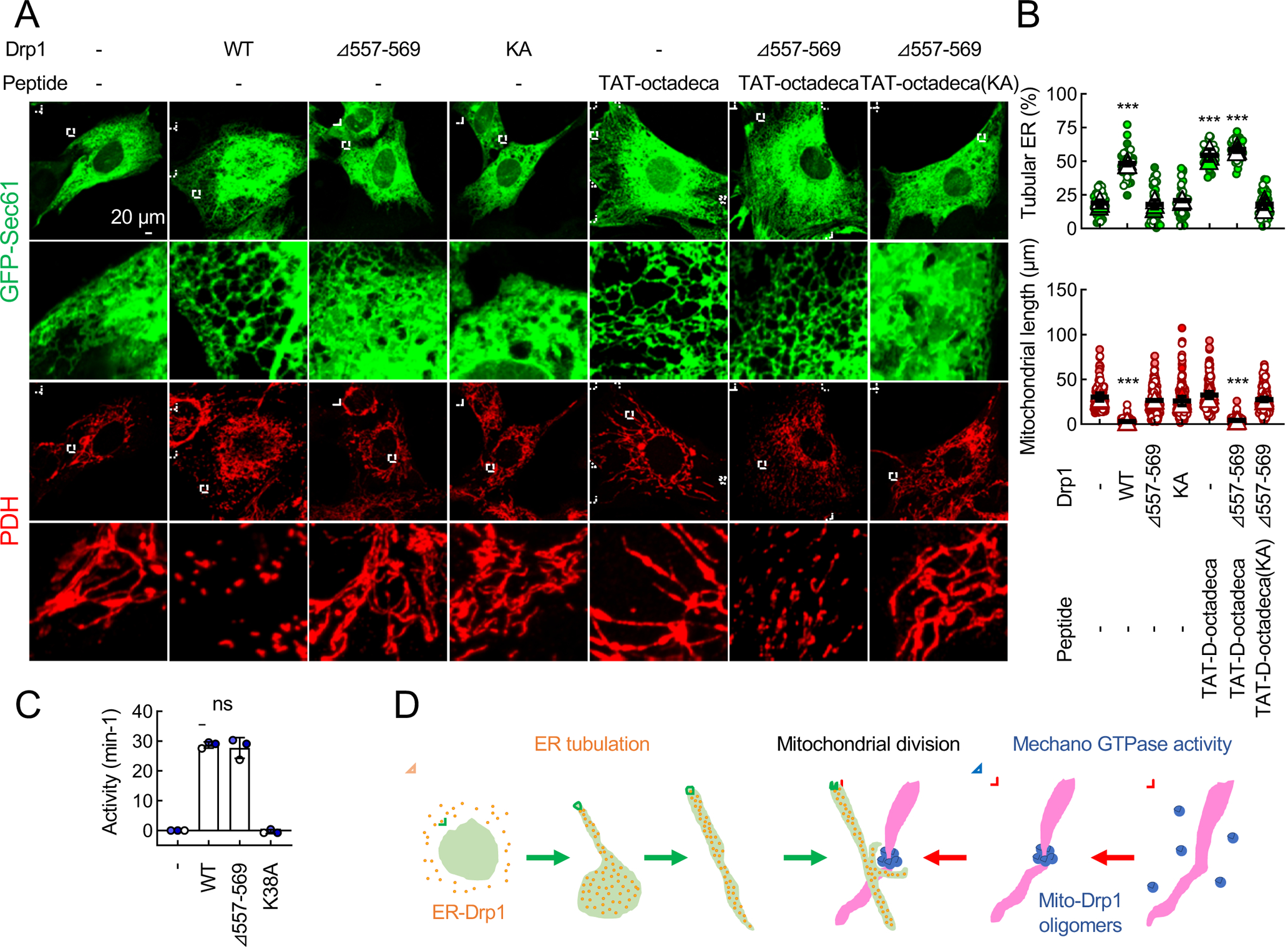
(A) Drp1-KO MEFs expressing the indicated Drp1 constructs were transfected with GFP-Sec61β. MEFs were then treated with 50 μM TAT-D-octadecapeptide or KA TAT-D-octadecapeptide for 24 h and analyzed using immunofluorescence confocal microscopy with antibodies to GFP and PDH. (B) Quantification of the morphology of ER (green, n = 3 experiments) and mitochondria (red, n = 3 experiments) is shown. Bars are average ± SD. (C) GTPase activity of purified His6-tagged WT Drp1, Drp1Δ557–569 and GTPase-dead Drp1K38A. Bars are average ± SD (n = 3). (D) Model for the role of Drp1 in ER tubule-associated mitochondrial division. Statistical analysis was performed using Kruskal-Wallis with Dunn’s post-hoc test (B and C): ***p<0.001. See also Figure S2 and S5. See also Figure S4, S5, S6 and S7.
Additional treatments of Drp1-KO MEFs expressing Drp1Δ557–569 with ER-tubulating, cell-permeable TAT-D-octadecapeptide fully rescued mitochondrial morphology (Fig. 7A and 7B). In contrast, TAT-D-octadecapeptide carrying the KA mutations that cannot rescue the ER morphology failed to rescue mitochondrial morphology (Fig. 7A and 7B). Finally, when we treated Drp1-KO MEFs with TAT-D-octadecapeptide this peptide increased mitochondrial constriction at ER-mitochondria intersections (Fig. S7). These data suggest that the ER tubules formed by D-octadecapeptide facilitates mitochondrial division by increasing ER-mitochondria interactions (Fig. 7D).
Discussion
This study reports that Drp1 is an ER-shaping protein that directly forms ER tubules. Unlike reticulon and DP1/Yop1, which contain essential transmembrane domains for ER tubulation (Hu et al., 2008; Powers et al., 2017; Voeltz et al., 2006), Drp1 generates ER tubules via peripheral membrane association of D-octadecapeptide. The role of Drp1 in ER morphogenesis is likely distinct from that of another dynamin-related protein, atlastin, because atlastin facilitates the ER network through GTP-dependent membrane fusion (Powers et al., 2017). We propose that Drp1 plays two distinct, critical roles in ER-assisted mitochondrial division. First, Drp1 generates ER tubules that facilitate the formation of ER-mitochondria contact sites, which likely occurs in collaboration with ER-mitochondria tethering proteins (Fig. 7D) (Giacomello et al., 2020). Second, Drp1 oligomers constrict mitochondria as a mechano GTPase at the ER-mitochondria contact sites (Fig. 7D). Therefore, these two activities of Drp1 integrate ER morphogenesis into mitochondrial dynamics. Since the loss of Rtn4a-mediated ER tubulation did not affect mitochondrial morphology, ER tubules formed by different mechanisms might be functionally different in mitochondrial division.
We propose that D-octadecapeptide forms an amphipathic α-helix and increases the curvature of the ER membrane (Fig. 5G and S3B). Previous studies have shown that protein-protein crowding on the surface of the membrane can induce the bending of the membrane (Day and Stachowiak, 2020). To deform membranes through this mechanism, high concentrations of proteins need to be present at the membrane. For example, 1 μM His-tagged GFP has been shown to tubulate liposomes containing 20% DOGS-NTA-Ni (Stachowiak et al., 2012). This occurs because the concentration of 1 μM His-GFP is ~100 times higher than the Kd value of DOGS-NTA-Ni (ñM). This condition creates a massive accumulation of His-GFP on the liposomes and induces crowding-dependent tubulation. In contrast, in the present work, we have shown that the Kd value of D-octadecapeptide for PA is 13 μM. Our liposome tubulation assay was performed using 1 μM of the peptide (10-times lower than Kd). Under this condition, only a small fraction of the peptide is associated with the liposomes and could not induce crowding. Therefore, crowding is not the mechanism for membrane tubulation by Drp1 and D-octadecapeptide.
The binding of Drp1 to PA plays important roles in the membrane tubulation activity of Drp1. We have previously reported that Drp1 separately recognizes the head group of PA and its saturated acyl chain. Although saturated PA could create phase separation in membranes (Shin and Brangwynne, 2017), Drp1 can bind PA independently of membranes. In supporting of this mechanism, we have shown that Drp1 interacts with saturated PA in a membrane-free dot blot assay and that Drp1 associates with saturated PA in a detergent solution (Adachi et al., 2016). These data support that Drp1 specifically binds saturated PA independently of the phase separation in membranes. However, these in vitro data do not rule out the possibility that the phase separation of saturated PA enhances its interaction with Drp1 at the ER in cells. It would be interesting to investigate how the biophysical characteristics of the ER membrane control its interactions with Drp1 and its morphology. We also predict that there are likely additional factors that control the function of Drp1 in ER tubulation along with PA to ensure the organelle specificity in cells.
Limitations
In the absence of Drp1, the amounts of peripheral ER sheets were increased. Since ER sheets could be flat sheets of the membrane or dense matrices of highly dynamic, intricate tubules (Nixon-Abell et al., 2016), it will be important to resolve the three-dimensional ultrastructure of ER sheets formed in Drp1-KO cells in future studies. We believe that such information would further advance our understanding of the mechanisms that control the morphogenesis, dynamics, and distribution of the ER membrane.
STAR Methods
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Hiromi Sesaki (hsesaki@jhmi.edu).
Materials Availability
All unique reagents generated in this study are available from the Lead Contact with a completed materials transfer agreement.
Data and Code Availability
This study did not generate any unique datasets or code.
Experimental Model and Subject Details
Cells
WT, Drp1-KO, and Mff/Fis1-KO MEFs (Loson et al., 2013; Wakabayashi et al., 2009) were grown in IMDM medium containing 10% FBS as previously described (Kageyama et al., 2014). WT, Drp1-KO, and Mff/Fis1-KO Hela cells (Otera et al., 2016) were grown in DMEM media supplemented with 10% FBS. Rtn4a-KO MEFs were generated using GeneArt CRISPR Nuclease Vector with OFP Reporter Kit (A21174, Invitrogen) following the manufacturer’s instructions. The target sequence (CCAGGTAACACTGTTTCGTC) was cloned into the vector and transfected into WT MEFs. Based on OFP fluorescent signal, transfected cells were sorted in 96-well plates as single cells at the Johns Hopkins Bloomberg Flow Cytometry and Immunology Core. The knockout of Rtn4a was confirmed by DNA sequencing and Western blotting with anti-Rtn4a antibodies. Rtn4-KO MEFs were grown in IMDM medium containing 10% FBS.
Animals
All animal work was performed according to guidelines established by the Johns Hopkins University Committee on Animal Care. Control and Alb-Drp1-KO mice were anesthetized by intraperitoneal injection of Avertin (Yamada et al., 2018). After the skin and abdominal muscle were incised, a cannula (25G) was inserted into the portal vein. The chest was incised and the thoracic inferior vena cava was cut. 50 ml of pre-warmed (37°C) HBSS supplemented with 0.5 mM EDTA was first perfused to confirm there was no leakage. Then, 30 ml of the DMEM medium containing 0.8 mg/ml collagenase I (17018029, Gibco) was perfused. The liver was harvested and further incubated in 20 ml of the collagenase solution at 37°C for 15 min. The liver was transferred to a plastic culture dish, finely chopped with a surgical scissor and pipetted up and down twice in 10 ml of HBSS supplemented with 0.5 mM EDTA. The chopped liver was filtered (70 μm pore size: 22363548, Thermo Fisher) and centrifuged at 50g for 3 minutes at 4°C to collect the hepatocytes. The hepatocytes were washed twice in the DMEM medium via centrifugation. The hepatocytes were resuspended in Williams’ medium E (A1217601, Gibco) supplemented with Hepatocyte Thawing and Plating Supplement Pack (CM3000, Gibco). 10,000 hepatocytes were plated on an 8-well chambered coverglass that had been coated with 0.3% collagen I (A1048301, Gibco) in 0.02 N acetic acid for 10 min and washed with PBS three times.
Method Details
Plasmids and lentiviruses
GFP-Sec61β plasmids were obtained from Addgene (15108). mCherry-Sec61β plasmids were constructed by replacing GFP with mCherry in the GFP-Sec61β plasmid. Rtn4a-GFP plasmids were obtained from Addgene (61807). Su9-HA-iRFP670 plasmids were generated by cloning the pre-sequence of Su9 (Yamada et al., 2018) fused to the HA epitope into piRFP670-N1 (Addgene, 45457). GFP-Cyb5 plasmids were made by cloning GFP and rat Cyb5 (100–134 amino acids) into the pcDNA3.1 plasmid.
Lentiviruses were produced as described previously (Kageyama et al., 2014). The pHR-SIN plasmids carrying full-length, mutant, or truncated Drp1 or N-terminally HA-tagged mouse Climp63 were cotransfected into HEK293T cells with two other plasmids, pHR-CMV8.2ΔR and pCMV-VSVG, using Lipofectamine 2000 (Invitrogen). Two days after transfection, the supernatant of the transfected cells containing released viruses was collected. The viruses were quick-frozen in liquid nitrogen and stored at −80°C. To generate shRNA lentiviruses, the following target sequences were cloned into pLKO.1: Scramble (CCTAAGGTTAAGTCGCCCTCGttcaagagaCGAGGGCGACTTAACCTTAGG) and Drp1 (GCTTCAGATCAGAGAACTTATttcaagagaATAAGTTCTCTGATCTGAAGC). To generate a knockdown-resistant Drp1, the target sequence for Drp1 was changed to GTTGCAAATTCGCGAGCTGAT in pHR-SIN. Lentiviruses were produced as described above.
Immunofluorescence microscopy
Cells were fixed with pre-warmed 4% paraformaldehyde in PBS for 20 min at room temperature in an 8-well chambered coverglasses (Adachi et al., 2016). The cells were permeabilized with PBS containing 0.1% Triton X-100 for 8 min at room temperature. The cells were incubated with primary antibodies in PBS containing 0.5% BSA, followed by the appropriate secondary antibodies. The primary antibodies were Drp1 (611113, BD Biosciences), Tom20 (sc-11415, Santa Cruz Biotechnology), PDH subunit E2/E3bp (ab110333, Abcam), Pex14 (10594–1-AP, Proteintech), BiP (ab21685, Abcam), GFP (Senoo et al., 2019) to enhance GFP-Sec61β, GFP-Cyb5 and Rtn4a-GFP signals, mCherry (M11217, invitrogen) to enhance mCherry-Sec61β, and HA (NB600–362, Novus Bio). The samples were viewed using a Zeiss LSM800 laser scanning confocal microscope. Image analysis was performed using NIH Image J software. The ER morphology was quantified by calculating the percentage of area that is covered by ER tubules relative to the total area covered by ER tubules or ER sheets. The morphology of mitochondria was quantified by measuring the lengths of mitochondria in a randomly selected region of cells in a circle with a diameter of 20 μm. We performed the experiments in triplicate, and in each one, 10 cells were analyzed for their ER morphology, while 50 mitochondria were measured for their length. The data are presented in Superplots (Lord et al., 2020).
For structured illumination microscopy, samples were viewed using a General Electric Deltavision OMX-SR super-resolution microscope at the Johns Hopkins University School of Medicine Microscope Facility. The data were acquired and processed using Delta Vision OMX Master Control software, SoftWoRx reconstruction and analysis software, and NIH Image J software.
Western blotting
Proteins were separated using SDS-PAGE and transferred onto PVDF membranes. The antibodies were Drp1, Fis1 (ALX-210-1037-0100, Enzo Life Sciences), Mff (Gandre-Babbe and van der Bliek, 2008), PDH, α-tubulin (#2125, Cell Signaling), Tom20, HA, Rtn4 (ab47085, Abcam) and GAPDH (MA5–15738, Thermo). Immunocomplexes were visualized using fluorescent-labeled secondary antibodies and detected using a PharosFX Plus molecular imager (Bio-Rad).
EM
Liver samples were prepared for EM as described previously (Yamada et al., 2018). Ultrathin sections were obtained using a Reichert-Jung ultracut E, stained with 2% uranyl acetate and lead citrate, and observed using a Hitachi 7600 transmission electron microscope equipped with dual CCD cameras (Advanced Microscopy Techniques). ImageJ software was used to measure the luminal widths of ER sheets.
FRAP
WT and Drp1-KO MEFs were plated on an 8-well chambered coverglass. On the next day, they were transfected with GFP-Sec61β and GFP-KDEL using Lipofectamin 3000 according to the manufacturer’s instructions. At 24 h after transfection, photobleaching and imaging were performed on a Zeiss LSM800 laser scanning confocal microscope equipped with an environmental chamber containing 5% CO2 at 37°C. The loss of GFP fluorescence and its subsequent recovery were monitored at intervals of 5 sec for GFP-Sec61β and 2 sec for GFPKDEL. Image analysis was performed using NIH Image J software and Zen 2012 SP1 black edition.
Subcellular fractionation
The ER and mitochondrial fractions were obtained as described with some modification (Sugiura et al., 2013). WT and Mff/Fis1-KO MEFs were cultured in 10-cm dishes. The plates were washed with ice-cold PBS and placed on ice. The cells were suspended in 1 ml of homogenization buffer [10 mM HEPES-KOH buffer (pH 7.4) containing 0.22 M mannitol, 0.07 M sucrose and protease inhibitors] and collected using a cell scraper. The cells were washed twice for 5 min at 4°C and homogenized with 10 strokes using a syringe with a 27-gauge needle. The postnuclear supernatant was obtained by centrifugation twice at 800 g for 5 and 10 min at 4°C. To obtain the ER and cytosolic fractions, after the clarification at 18,000 g for 20 min at 4°C, the postnuclear supernatant was further separated into the ER-enriched pellet and cytosolic supernatant by ultracentrifugation at 100,000 g for 1 h at 4°C. The fractions were analyzed by SDS-PAGE and Western blotting with antibodies to Drp1, Tom20, Rtn4a and α-Tubulin.
Peptides
D-Octadecapeptide (GMLKTSKAEELLAEEKSK), KA D-octadecapeptide (GMLATSAAEELLAEEASA), peptide554–565 (GMLKTSKAEELL), peptide561–571 (AEELLAEEKSK), biotin-conjugated D-octadecapeptide, biotin-conjugated KA D-octadecapeptide, TAT (GRKKRRQRRRPQ)-fused D-octadecapeptide, TAT-fused KA D-octadecapeptide, FITC-conjugated D-octadecapeptide and FITC-conjugated KA D-octadecapeptide, were synthesized at GenScript (Piscataway, NJ, USA) and Biomatik (Wilmington, DE, USA).
Phospholipids
POPC (850457), POPE (850757), Liver PI (840042), POPS (840034), DPPC (850355) and rhodamine-DPPE (810158) were purchased from Avanti Polar Lipids (Alabaster, AL, USA). DPPA was obtained from Avanti Polar Lipids (830855) and Echelon Biosciences (L-4116) (Salt Lake City, UT, USA)
Liposome tubulation assay
Liposomes were prepared as previously described (Adachi et al., 2016) with some modifications. The lipids were mixed in a POPC:POPE:Liver PI:POPS:DPPA:rhodamine-DPPE ratio of 44:25:10:5:15:1 (% mol). In control liposomes without DPPA, the amount of POPC was increased to 59%. The lipids were dried under a flow of nitrogen gas for 5 min and further dried in a SpeedVac overnight. The lipid film was resuspended at a concentration of 10 mM in 150 mM KCl and 20 mM MES (pH 7.0), voltexed for 1 hour, and subjected to freeze-thaw cycles 5 times using dry ice and a 42°C heat block. Unilamellar liposomes were generated via extrusion through a nanopore membrane with a pore size of 400 nm; this process was repeated 21 times. The liposomes (50 μM lipids) were incubated with peptides (0, 0.1, 0.3 and 1 μM) in 150 mM KCl and 20 mM MES (pH 7.0) at room temperature overnight. For negative-stain EM, the samples (20 μl) were placed on EM grids (CF400-CU-UL, Electron Microscopy Sciences, Hatfield, PA, USA) that have been glow discharged by Quorum GloQube-D (Laughton, East Sussex, UK). After being incubated for 2 min, the grids were washed with 150 mM KCl and 20 mM MES (pH 7.0) three times, stained with 0.1% uranyl acetate and 0.04% Tylose for 30 sec twice and then dried. The samples were observed using a Hitachi 7600 transmission electron microscope.
Liposome flotation assay
A liposome floatation assay was performed as previously described(Adachi et al., 2016) with some modifications. Liposomes were prepared in 150 mM KCl and 20 mM MES (pH 7.0), as described above in the liposome tubulation assay. The liposomes (50 μM lipids) were incubated with FITC-conjugated D-octadecapeptide or FITC-conjugated KA D-octadecapeptide (1 μM, final volume, 200 μl) in 150 mM KCl and 20 mM MES (pH 7.0) at room temperature with gentle mixing overnight. The peptide-liposome mixture was diluted in 1.728 M sucrose/20 mM MES (pH 7.0) (final volume, 1.25 ml) and placed at the bottom of tubes. We then overlaid 2.9 ml of 1.25 M sucrose/20 mM MES (pH 7.0) and 0.85 ml of 0.25 M sucrose/20 mM MES (pH 7.0). The sucrose gradient was centrifuged at 287,000 x g for 2 h at 4°C in an SW55Ti rotor (Beckman, Indianapolis, IN, USA). Four fractions (1.25 ml each) were collected from the top. The majority of liposomes were floated to the top fraction, and 125 μl of the top fraction was mixed with 125 μl of 1% TritonX-100. The fluorescence intensities of rhodamine-PE (Ex 550 nm/Em 605 nm) and FITC (Ex 488 nm/Em 535 nm) were measured using a CLARIOstar plate reader system (BMG Labtech, Cary, NC, USA).
Surface plasmon resonance
Surface plasmon resonance was measured using OpenSPR (Nicoya Lifesciences, Kitchener, ON, Canada). Biotin-conjugated D-octadecapeptide or D-octadecapeptide (KA) was resuspended at 100 μg/μl in 0.05% Tween 20, 100 mM NaCl and 20 mM MES (pH 7.0) and attached to a streptavidin sensor chip (SEN-AU-100–12-STRP) following the manufacturer’s instructions. DPPA and DPPC were dried in a SpeedVac overnight and resuspended at 2, 10 and 50 μg/ml in 0.05% Tween 20, 100 mM NaCl, 20 mM MES (pH 7.0). Interactions of the phospholipids with the peptides were monitored according to the manufacturer’s instructions. The apparent affinity of the DPPA for the biotin-conjugated D-octadecapeptide was estimated by taking the binding signal (a.u.) at saturation for each concentration of DPPA injected and plotting the saturation signal (a.u.)max versus the DPPA concentration injected. The apparent Kd was then estimated by fitting using the equation a.u. = (a.u.)max/(1 + Kd/C).
GTPase assay
GTP hydrolysis by purified His6-tagged Drp1 was measured as described previously (Adachi et a., 2016). 0.1 μM Drp1 was incubated in 150 mM KCl, 2 mM MgCl2, 1 mM DTT, 20 mM HEPES-KOH buffer (pH 7.4), and 1 mM GTP for 15 min at 37°C. Next, 25-μl samples were mixed with 100 μl of malachite green solution (Echelon, K-1501) at room temperature for 20 min, and the absorbance at OD620 was measured.
Quantification and Statistical Analysis
Statistical analysis
Unless otherwise noted, all data were quantified based on at least 3 independent experiments and presented as average ± SD. Statistical analysis was performed using One-way ANOVA with post-hoc Tukey and Student’s t-test: *p<0.05, **p<0.01, ***p<0.001.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-Drp1 | BD Biosciences | Cat#611113; RRID: AB_398424 |
| Mouse monoclonal anti-PDH | Abcam | Cat#ab110333; RRID: AB_10862029 |
| Rabbit polyclonal anti-Tom20 | Santa Cruz Biotechnology | Cat#sc-11415; RRID: AB_2207533 |
| Mouse monoclonal anti-GAPDH | Thermo Fisher Scientific | Cat#MA5–15738; RRID: AB_10977387 |
| Goat polyclonal anti-HA | Novus | Cat#NB600–362; RRID: AB_10124937 |
| Rat monoclonal anti-mCherry | Thermo Fisher Scientific | Cat#M11217; RRID: AB_2536611 |
| Rabbit polyclonal anti-Rtn4 (Nogo A+B) | Abcam | Cat#ab47085; RRID: AB_881718 |
| Rabbit polyclonal anti-Bip | Abcam | Cat#ab21685; RRID: AB_2119834 |
| Rabbit polyclonal anti-Fis1 | Enzo Life Sciences | Cat#ALX-210-1037; RRID: AB_2737586 |
| Rabbit polyclonal anti-PEX14 | Proteintech | Cat#10594-1-AP; RRID: AB_2252194 |
| Rabbit polyclonal anti-GFP | Senoo et al., Nat Cell Biol 2019 | N/A |
| Rabbit polyclonal anti-Mff | Gandre-Babbe and van der Bliek, Mol. Cell Biol 2008 | N/A |
| Alexa 405 anti-rabbit IgG | Abcam | Cat#ab175658; RRID: AB_2687445 |
| Alexa 488 anti-mouse IgG | Thermo Fisher Scientific | Cat#A21202; RRID: AB_141607 |
| Alexa 488 anti-rabbit IgG | Thermo Fisher Scientific | Cat#A21206; RRID: AB_2535792 |
| Alexa 488 anti-goat IgG | Thermo Fisher Scientific | Cat#A21467; RRID:AB_2535870 |
| Alexa 568 anti-mouse IgG | Thermo Fisher Scientific | Cat#A10037; RRID: AB_2534013 |
| Alexa 568 anti-rabbit IgG | Thermo Fisher Scientific | Cat#A10042; RRID: AB_2534017 |
| Alexa 568 anti-rat IgG | Abcam | Cat#AB175475; RRID: AB_2636887 |
| Alexa 647 anti-mouse IgG | Thermo Fisher Scientific | Cat#A31571; RRID: AB_162542 |
| Alexa 647 anti-goat IgG | Thermo Fisher Scientific | Cat#A21447; RRID: AB_2535864 |
| Bacterial and Virus Strains | ||
| Rosetta™ 2(DE3)pLysS competent cells | EMD Millipore | Cat#71403 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Chloroform | Sigma-Aldrich | Cat#C2432 |
| MES (2-morpholinoethanesulfonic acid, mono hydrate) | Sigma-Aldrich | Cat#M8250 |
| Sucrose | Thermo Fisher Scientific | Cat#S5–3 |
| beta-D-Galactopyranoside, Isopropyl-beta-D-thiogalactopyranoside (IPTG) | AMRESCO | Cat#0487 |
| HEPES | Thermo Fisher Scientific | Cat#BP310–1 |
| Imidazole | Sigma-Aldrich | Cat#I0125 |
| Coomassie Brilliant Blue R-250 | AMRESCO | Cat#M128 |
| 50% Ni-NTA beads | EMD Millipore | Cat#70666 |
| 4–20% Criterion TGX Precast Gel | Bio-Rad Laboratories | Cat#5671095 |
| DMSO | Sigma-Aldrich | Cat#D5879 |
| 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) | Avanti Polar Lipids | Cat#850457 |
| 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE) | Avanti Polar Lipids | Cat#850757 |
| L-α-phosphatidylinositol (Liver PI) | Avanti Polar Lipids | Cat#840042 |
| 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine (POPS) | Avanti Polar Lipids | Cat#840034 |
| 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC) | Avanti Polar Lipids | Cat#850355 |
| 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) (rhodamine-DPPE) | Avanti Polar Lipids | Cat#810158 |
| 1,2-dipalmitoyl-sn-glycero-3-phosphate (DPPA) | Avanti Polar Lipids | Cat#830855 |
| 1,2-dipalmitoyl-sn-glycero-3-phosphate (DPPA) | Echelon Biosciences | Cat#L-4116 |
| IMDM | Gibco | Cat#12440053 |
| Dulbecco’s Modified Eagle’s Medium - high glucose | Sigma-Aldrich | Cat#D5796 |
| Dulbecco’s Phosphate Buffered Saline | Sigma-Aldrich | Cat#D8537 |
| Fetal Bovine Serum | Sigma-Aldrich | Cat#F6178 |
| RIPA Buffer (10X) | Cell Signaling Technology | Cat#9806 |
| D-Mannitol | Sigma-Aldrich | Cat#M4125 |
| collagenase I | Gibco | Cat#17018029 |
| Williams’ medium E | Gibco | Cat#A1217601 |
| Collagen I | Gibco | Cat#A1048301 |
| Primary Hepatocyte Thawing and Plating Supplements | Gibco | Cat#CM3000 |
| Octadecapeptide554–571 (GMLKTSKAEELLAEEKSK) | This paper | N/A |
| KA octadecapeptide554–571 (GMLATSAAEELLAEEASA) | This paper | N/A |
| peptide554–565 (GMLKTSKAEELL) | This paper | N/A |
| peptide561–571 (AEELLAEEKSK) | This paper | N/A |
| biotin-conjugated octadecapeptide554–571 | This paper | N/A |
| biotin-conjugated KA octadecapeptide554–571 | This paper | N/A |
| TAT (GRKKRRQRRRPQ)-fused octadecapeptide554–571 | This paper | N/A |
| TAT-fused KA octadecapeptide554–571 | This paper | N/A |
| FITC-conjugated octadecapeptide554–571 | This paper | N/A |
| FITC-conjugated KA octadecapeptide554–571 | This paper | N/A |
| Uranyl Acetate 98%, ACS Reagent | Polysciences, Inc. | Cat#21447–25 |
| Tylose® MH 300 | Sigma-Aldrich | Cat#93800 |
| cOmplete™, Mini, EDTA-free Protease Inhibitor Cocktail | Roche | Cat#11836170001 |
| Critical Commercial Assays | ||
| Malachite Green Solution | Echelon Biosciences | Cat#K-1501 |
| Gibson Assembly® Master Mix | NEB | Cat#E2611 |
| Quick Ligation™ Kit | NEB | Cat#M2200 |
| Phusion® High-Fidelity DNA Polymerase | NEB | Cat#M0530 |
| Lipofectamine 2000 Transfection Reagent | Thermo Fisher Scientific | Cat#11668019 |
| Lipofectamine™ 3000 Transfection Reagent | Thermo Fisher Scientific | Cat#L3000150 |
| Biotin-Streptavidin Sensors Kit | Nicoya | Cat#NI SENAU100–10-STR |
| EM grids | Electron Microscopy Sciences | Cat#CF400-CU |
| Nuclepore™ Track-Etched Membranes | Whatman | Cat#800282 |
| GeneArt™ CRISPR Nuclease Vector with OFP Reporter Kit | Thermo Fisher Scientific | Cat#A21174 |
| Experimental Models: Cell Lines | ||
| Mouse embryonic fibroblasts (MEFs) | Wakabayashi et al., J. Cell Biol. 2009 | N/A |
| Drp1-KO MEFs | Wakabayashi et al., J. Cell Biol. 2009 | N/A |
| Mff/Fis1-KO MEFs | Loson et al., Mol Biol Cell 2013 | N/A |
| Hela cells | Otera et al., J Cell Biol. 2016 | N/A |
| Drp1-KO Hela cells | Otera et al., J Cell Biol. 2016 | N/A |
| Mff/Fis1-KO Hela cells | Otera et al., J Cell Biol. 2016 | N/A |
| Rtn4a-KO MEFs | This paper | N/A |
| HEK293T cells | ATCC | CRL-3216 |
| Experimental Models: Organisms/Strains | ||
| Mouse: Alb-Cre+/−::Drp1flox/flox | Yamada et al., Cell Metab 2018 | N/A |
| Oligonucleotides | ||
| CRISPR KO target sequence; Rtn4a CCAGGTAACACTGTTTCGTC |
This paper | N/A |
| shRNA target sequence; Scramble CCTAAGGTTAAGTCGCCCTCGttcaagagaCGAGGGCGACTTAACCTTAGG |
This paper | N/A |
| shRNA target sequence; Drp1 GCTTCAGATCAGAGAACTTATttcaagagaATAAGTTCTCTGATCTGAAGC |
This paper | N/A |
| Recombinant DNA | ||
| pHR-SIN Drp1 WT | This paper | N/A |
| pHR-SIN Drp1 K38A | This paper | N/A |
| pHR-SIN Drp1 G350D | This paper | N/A |
| pHR-SIN Drp1 K38, G350D | This paper | N/A |
| pHR-SIN Drp1 503–539 | This paper | N/A |
| pHR-SIN Drp1 540–599 | This paper | N/A |
| pHR-SIN Drp1 540–575 | This paper | N/A |
| pHR-SIN Drp1 570–599 | This paper | N/A |
| pHR-SIN Drp1 554–571 | This paper | N/A |
| pHR-SIN Drp1 554–565 | This paper | N/A |
| pHR-SIN Drp1 561–571 | This paper | N/A |
| pHR-SIN Drp1 Δ557–569 | This paper | N/A |
| pHR-SIN Drp1 KA | This paper | N/A |
| pHR-SIN HA-Drp1 S-V | This paper | N/A |
| pHR-SIN HA-Drp1 G-S | This paper | N/A |
| pHR-SIN HA-Drp1 V | This paper | N/A |
| pHR-SIN HA-Drp1 S | This paper | N/A |
| pHR-CMV8.2ΔR | Stewart et al., RNA 2003 | Addgene Plasmid #8455 |
| pCMV-VSVG | Stewart et al., RNA 2003 | Addgene Plasmid #8454 |
| pET15b His6-tagged Drp1 | Adachi et al., Mol Cell. 2016 | N/A |
| pET15b His6-tagged Drp1Δ557–569 | This paper | N/A |
| pET15b His6-tagged Drp1 K38A | This paper | N/A |
| GFP-Sec61β | Friedman et al., Science. 2011 | Addgene Plasmid #15108 |
| mCherry-Sec61β | This paper | N/A |
| Rtn4a-GFP | Shibata et al J Biol Chem. 2008 | Addgene Plasmid #61807 |
| HA-Climp63 | This paper | N/A |
| Su9-HA-iRFP670 | This paper | N/A |
| GFP-KDEL | This paper | N/A |
| GFP-Cyb5 | This paper | N/A |
| Software and Algorithms | ||
| Delta Vision OMX Master Control software | GE Healthcare Life Sciences | N/A |
| SoftWoRx reconstruction and analysis software | GE Healthcare Life Sciences | N/A |
| Image J | NIH | https://imagej.nih.gov/ij/index.html |
| Prism | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
Highlights.
Drp1 is associated with the ER
Drp1 shapes the ER into tubules independently of oligomerization and GTP hydrolysis
Octadecapeptide554–571 in the variable domain is sufficient for membrane tubulation
ER tubules formed by Drp1 promote mitochondrial division
Acknowledgments
We thank past and present members of the Iijima and Sesaki labs for helpful discussions and technical assistance. We are grateful to Dr. H. Otera for providing Drp1-KO and Mff/Fis1 double-KO Hela cells and Dr. A. van der Bliek for anti-Mff antibodies. This work was supported by NIH grants to MI (GM131768) and HS (GM123266 and GM130695).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
References
- Adachi Y, Iijima M, and Sesaki H (2018). An unstructured loop that is critical for interactions of the stalk domain of Drp1 with saturated phosphatidic acid. Small GTPases 9, 472–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adachi Y, Itoh K, Iijima M, and Sesaki H (2017). Assay to Measure Interactions between Purified Drp1 and Synthetic Liposomes. Bio Protocol 7, e2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adachi Y, Itoh K, Yamada T, Cerveny KL, Suzuki TL, Macdonald P, Frohman MA, Ramachandran R, Iijima M, and Sesaki H (2016). Coincident Phosphatidic Acid Interaction Restrains Drp1 in Mitochondrial Division. Mol Cell 63, 1034–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann O, and Walz B (2001). Endoplasmic reticulum of animal cells and its organization into structural and functional domains. Int Rev Cytol 205, 149–214. [DOI] [PubMed] [Google Scholar]
- Chen S, Novick P, and Ferro-Novick S (2013). ER structure and function. Curr Opin Cell Biol 25, 428–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day KJ, and Stachowiak JC (2020). Biophysical forces in membrane bending and traffic. Curr Opin Cell Biol 65, 72–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Brito OM, and Scorrano L (2008). Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature 456, 605–610. [DOI] [PubMed] [Google Scholar]
- Fonseca TB, Sanchez-Guerrero A, Milosevic I, and Raimundo N (2019). Mitochondrial fission requires DRP1 but not dynamins. Nature 570, E34–E42. [DOI] [PubMed] [Google Scholar]
- Friedman JR, Lackner LL, West M, DiBenedetto JR, Nunnari J, and Voeltz GK (2011). ER tubules mark sites of mitochondrial division. Science 334, 358–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandre-Babbe S, and van der Bliek AM (2008). The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol Biol Cell 19, 2402–2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomello M, Pyakurel A, Glytsou C, and Scorrano L (2020). The cell biology of mitochondrial membrane dynamics. Nat Rev Mol Cell Biol 21, 204–224. [DOI] [PubMed] [Google Scholar]
- Goyal U, and Blackstone C (2013). Untangling the web: mechanisms underlying ER network formation. Biochim Biophys Acta 1833, 2492–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Li D, Zhang S, Yang Y, Liu JJ, Wang X, Liu C, Milkie DE, Moore RP, Tulu US, et al. (2018). Visualizing Intracellular Organelle and Cytoskeletal Interactions at Nanoscale Resolution on Millisecond Timescales. Cell 175, 1430–1442 e1417. [DOI] [PubMed] [Google Scholar]
- Hirabayashi Y, Kwon SK, Paek H, Pernice WM, Paul MA, Lee J, Erfani P, Raczkowski A, Petrey DS, Pon LA, et al. (2017). ER-mitochondria tethering by PDZD8 regulates Ca(2+) dynamics in mammalian neurons. Science 358, 623–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Shibata Y, Voss C, Shemesh T, Li Z, Coughlin M, Kozlov MM, Rapoport TA, and Prinz WA (2008). Membrane proteins of the endoplasmic reticulum induce high-curvature tubules. Science 319, 1247–1250. [DOI] [PubMed] [Google Scholar]
- Itoh K, Nakamura K, Iijima M, and Sesaki H (2013). Mitochondrial dynamics in neurodegeneration. Trends in Cell Biology 23, 64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji WK, Chakrabarti R, Fan X, Schoenfeld L, Strack S, and Higgs HN (2017). Receptor- mediated Drp1 oligomerization on endoplasmic reticulum. J Cell Biol 216, 4123–4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama Y, Hoshijima M, Seo K, Bedja D, Sysa-Shah P, Andrabi SA, Chen W, Hoke A, Dawson VL, Dawson TM, et al. (2014). Parkin-independent mitophagy requires Drp1 and maintains the integrity of mammalian heart and brain. The EMBO journal 33, 2798–2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalia R, Wang RY, Yusuf A, Thomas PV, Agard DA, Shaw JM, and Frost A (2018). Structural basis of mitochondrial receptor binding and constriction by DRP1. Nature 558, 401–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kameoka S, Adachi Y, Okamoto K, Iijima M, and Sesaki H (2018). Phosphatidic Acid and Cardiolipin Coordinate Mitochondrial Dynamics. Trends Cell Biol 28, 67–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamerkar SC, Kraus F, Sharpe AJ, Pucadyil TJ, and Ryan MT (2018). Dynamin-related protein 1 has membrane constricting and severing abilities sufficient for mitochondrial and peroxisomal fission. Nat Commun 9, 5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korobova F, Ramabhadran V, and Higgs HN (2013). An actin-dependent step in mitochondrial fission mediated by the ER-associated formin INF2. Science 339, 464–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis SC, Uchiyama LF, and Nunnari J (2016). ER-mitochondria contacts couple mtDNA synthesis with mitochondrial division in human cells. Science 353, aaf5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liesa M, and Shirihai OS (2013). Mitochondrial dynamics in the regulation of nutrient utilization and energy expenditure. Cell metabolism 17, 491–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord SJ, Velle KB, Mullins RD, and Fritz-Laylin LK (2020). SuperPlots: Communicating reproducibility and variability in cell biology. J Cell Biol 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loson OC, Song Z, Chen H, and Chan DC (2013). Fis1, Mff, MiD49, and MiD51 mediate Drp1 recruitment in mitochondrial fission. Mol Biol Cell 24, 659–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagashima S, Tabara LC, Tilokani L, Paupe V, Anand H, Pogson JH, Zunino R, McBride HM, and Prudent J (2020). Golgi-derived PI(4)P-containing vesicles drive late steps of mitochondrial division. Science 367, 1366–1371. [DOI] [PubMed] [Google Scholar]
- Nixon-Abell J, Obara CJ, Weigel AV, Li D, Legant WR, Xu CS, Pasolli HA, Harvey K, Hess HF, Betzig E, et al. (2016). Increased spatiotemporal resolution reveals highly dynamic dense tubular matrices in the peripheral ER. Science 354, aaf3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otera H, Miyata N, Kuge O, and Mihara K (2016). Drp1-dependent mitochondrial fission via MiD49/51 is essential for apoptotic cristae remodeling. J Cell Biol 212, 531–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers RE, Wang S, Liu TY, and Rapoport TA (2017). Reconstitution of the tubular endoplasmic reticulum network with purified components. Nature 543, 257–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudent J, and McBride HM (2017). The mitochondria-endoplasmic reticulum contact sites: a signalling platform for cell death. Curr Opin Cell Biol 47, 52–63. [DOI] [PubMed] [Google Scholar]
- Roy M, Reddy PH, Iijima M, and Sesaki H (2015). Mitochondrial division and fusion in metabolism. Current opinion in cell biology 33C, 111–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DS, and Blower MD (2016). The endoplasmic reticulum: structure, function and response to cellular signaling. Cell Mol Life Sci 73, 79–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scorrano L, De Matteis MA, Emr S, Giordano F, Hajnoczky G, Kornmann B, Lackner LL, Levine TP, Pellegrini L, Reinisch K, et al. (2019). Coming together to define membrane contact sites. Nat Commun 10, 1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senoo H, Kamimura Y, Kimura R, Nakajima A, Sawai S, Sesaki H, and Iijima M (2019). Phosphorylated Rho-GDP directly activates mTORC2 kinase towards AKT through dimerization with Ras-GTP to regulate cell migration. Nat Cell Biol 21, 867–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata Y, Shemesh T, Prinz WA, Palazzo AF, Kozlov MM, and Rapoport TA (2010). Mechanisms determining the morphology of the peripheral ER. Cell 143, 774–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y, and Brangwynne CP (2017). Liquid phase condensation in cell physiology and disease. Science 357. [DOI] [PubMed] [Google Scholar]
- Smirnova E, Griparic L, Shurland DL, and van der Bliek AM (2001). Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol Biol Cell 12, 2245–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachowiak JC, Schmid EM, Ryan CJ, Ann HS, Sasaki DY, Sherman MB, Geissler PL, Fletcher DA, and Hayden CC (2012). Membrane bending by protein-protein crowding. Nature Cell Biology 14, 944–+. [DOI] [PubMed] [Google Scholar]
- Sugiura A, Nagashima S, Tokuyama T, Amo T, Matsuki Y, Ishido S, Kudo Y, McBride HM, Fukuda T, Matsushita N, et al. (2013). MITOL regulates endoplasmic reticulum mitochondria contacts via Mitofusin2. Molecular cell 51, 20–34. [DOI] [PubMed] [Google Scholar]
- Takeda K, and Yanagi S (2019). Mitochondrial retrograde signaling to the endoplasmic reticulum regulates unfolded protein responses. Mol Cell Oncol 6, e1659078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura Y, Itoh K, and Sesaki H (2011). SnapShot: Mitochondrial dynamics. Cell 145, 1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terasaki M (2016). A finer look at a fine cellular meshwork. Science 354, 415–416. [DOI] [PubMed] [Google Scholar]
- Voeltz GK, Prinz WA, Shibata Y, Rist JM, and Rapoport TA (2006). A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell 124, 573–586. [DOI] [PubMed] [Google Scholar]
- Wakabayashi J, Zhang Z, Wakabayashi N, Tamura Y, Fukaya M, Kensler TW, Iijima M, and Sesaki H (2009). The dynamin-related GTPase Drp1 is required for embryonic and brain development in mice. J Cell Biol 186, 805–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong YC, Ysselstein D, and Krainc D (2018). Mitochondria-lysosome contacts regulate mitochondrial fission via RAB7 GTP hydrolysis. Nature 554, 382–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Carvalho P, and Voeltz GK (2018). Here, there, and everywhere: The importance of ER membrane contact sites. Science 361, eaan5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Murata D, Adachi Y, Itoh K, Kameoka S, Igarashi A, Kato T, Araki Y, Huganir RL, Dawson TM, et al. (2018). Mitochondrial Stasis Reveals p62-Mediated Ubiquitination in Parkin-Independent Mitophagy and Mitigates Nonalcoholic Fatty Liver Disease. Cell Metab 28, 588–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, and Hu J (2016). Shaping the Endoplasmic Reticulum into a Social Network. Trends Cell Biol 26, 934–943. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate any unique datasets or code.


