Abstract
Introduction
The association between genetic background and the risk of invasive aspergillosis (IA) has not been addressed in Thailand. We conducted genetic risk surveillance for IA among Thai hematologic patients.
Methods
We conducted a prospective observational cohort study including moderate- to high-risk hematology patients at Ramathibodi Hospital. IA occurrence, relevant clinical data, and genetic analyses were assessed. Odds ratios (ORs) of IA were assessed for the presence of the selected single nucleotide polymorphism genotype using logistic regression.
Results
A total of 357 patients were enrolled. The most common hematologic disease was non-Hodgkin lymphoma (45.1%). IA was diagnosed in 36 patients (10.10%). The C allele of IL10rs1800896 was associated with an increased risk of IA (adjusted OR 5.297; 95% confidence interval [CI] 2.032–13.809, p = 0.001). In multivariate Cox regression analysis, prolonged neutropenia and the C allele of IL10rs1800896 were associated with IA (hazard ratio [HR] 12.585; 95% CI 3.866–40.967, p < 0.001 and HR 2.449; 95% CI 1.097–5.468, p = 0.042, respectively).
Conclusions
Carrying the C allele of IL10rs1800896 was associated with an increased risk of IA among moderate- to high-risk Thai patients with hematologic diseases. This finding can potentially lead to a novel risk stratification scheme to further prevent IA in resource-limited settings.
Electronic supplementary material
The online version of this article (10.1007/s40121-020-00331-4) contains supplementary material, which is available to authorized users.
Keywords: Antifungal prophylaxis, Genetic susceptibility, Hematology patients, Invasive aspergillosis
Key Summary Points
| Why carry out this study? |
| Hematology patients are at high risk for invasive aspergillosis. |
| Universal antifungal prophylaxis is not feasible in a resource-limited setting even in high-risk patients. |
| Genetic risk surveillance may assist in risk stratification among this population. |
| What was learned from the study? |
| Carrying the C allele of IL10rs1800896 was associated with an increased risk of IA among Thai patients with moderate- to high-risk hematologic diseases. |
| This finding potentially leads to a novel risk stratification scheme to further prevent IA in a resource-limited setting. |
Digital Features
This article is published with digital features to facilitate understanding of the article. You can access the digital features on the article’s associated Figshare page. To view digital features for this article go to 10.6084/m9.figshare.12794684.
Introduction
Aspergillus is a ubiquitous fungus with a wide range of clinical features, depending on the degree of host immunity [1]. Invasive aspergillosis (IA) is a major cause of morbidity and mortality in immunosuppressed patients. IA has been estimated to occur in 5–40% of hematology patients, especially in acute myeloid leukemia (AML) and allogeneic hematopoietic stem cell transplant (HSCT) [2–6], with a mortality rate attributable to IA of 25–50% [2, 3, 7, 8]. Additionally, patients with IA usually require high-cost treatment, either antifungal therapy or radiologic investigations [7, 9].
Although exposure to Aspergillus conidia through inhalation is common, only a minority of those exposed will develop lung disease [10]. Several factors have been shown to affect the risk of IA development [5, 11–13]; however, the factors determining IA susceptibility have not been completely discovered. Innate immunity and genetic susceptibility have been proposed among several other important factors determining disease occurrence [14–16].
The physical barrier of the respiratory tract is the first line of resistance against inhaled conidia of Aspergillus [17]. If passed successfully through the ciliated epithelium, conidia are then challenged by alveolar macrophages, dendritic cells, and polymorphonuclear cells [15]. In the early stages of infection, conidia are destroyed by local alveolar macrophages, and extracellular killing of germinating hyphae is mediated by recruited polymorphonuclear cells. Conidia and hyphae are recognized via soluble and membrane-bound pattern recognition receptors [18]. Pattern recognition receptors (PRRs) sense pathogen-associated molecular patterns (PAMPs) and drive the secretion of proinflammatory cytokines and chemokines [18–20].
Genetic variation within key innate immune response genes could influence disease susceptibility and the outcome of this infection [21–23]. Defects in Toll-like receptors (TLR)-2, TLR-3, TLR-4, TLR-9, dendritic cell-specific intercellular adhesion molecule-3-grabbing nonintegrin (DC-SIGN; also known as CD209 antigen), dendritic cell-associated C-type lectin 1 (DECTIN-1), pentraxin-3 (PTX-3), and interleukin (IL)-10 have been proposed to affect IA pathogenesis [15]. However, only certain single nucleotide polymorphisms have been documented in Asian populations, according to national and global databases. Of these, a single polymorphism on DECTIN-1 is the most frequent genotype identified in Asia, followed by PTX-3. Although positive associations between genetic variants in cytokine genes and vulnerability to IA have been reported [24–36], the lack of functional validation and underpowered design of most studies preclude definite conclusions [15]. Furthermore, most previous studies were conducted among white participants with HSCT status only; therefore, studies among Asian populations are needed.
Genetic surveillance for risk stratification of IA is a logical step to identify patients at risk and potentially assist clinicians to design more intensive monitoring and to more specifically select anti-mold prophylaxis [11, 23]. We analyzed the association of selected single nucleotide polymorphisms (SNPs) and the risk of IA among moderate- to high-risk hematology patients.
The primary objective of this study was to analyze the potential genetic predisposition to IA among moderate- to high-risk hematology patients. The secondary objectives were to study the epidemiology of IA and the potential impact of genetic predisposition on clinical outcomes related to IA.
Methods
Definitions
Confirmed and probable IA were specified according to the revised criteria of the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group and the National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) Consensus Group in the year of 2008 [37].
Moderate-risk hematologic diseases were defined as any of the following conditions: acute lymphoblastic leukemia (ALL), chronic lymphocytic leukemia (CLL), non-Hodgkin lymphoma (NHL), myelodysplastic syndromes (MDS), severe aplastic anemia (AA), multiple myeloma, and graft-versus-host disease (GVHD) after HSCT [13, 14].
High-risk hematologic diseases were defined as diagnoses of AML and allogeneic HSCT [13, 14].
Prior history of proven or probable invasive mold disease was defined as evidence of IA within 1 year at study entry [11].
High-dose corticosteroid treatment was defined as receipt of corticosteroids, at least 0.3 mg/kg/day of prednisolone equivalent for longer than 3 weeks [37].
Hospital admission for high-risk chemotherapy was defined as receipt of chemotherapy for AML and conditioning chemotherapy for allogeneic HSCT [38].
Prolonged neutropenia was defined as absolute neutrophil count of less than 500 cells/mm3 for longer than 10 days within 30 days before admission or following chemotherapy [11].
Lymphocytopenia and probable impaired lymphocyte function at the time of admission were defined as an absolute lymphocyte count of less than 1000 cells/mm3 or CD4 count of less than 50 cells/mm3, or any allogeneic HSCT patient receiving cyclosporine, tacrolimus, or anti-thymocyte globulin [11].
Positive serum galactomannan was defined as an optical density ratio of at least 0.5 in two consecutive positive samples or at least 0.7 in a serum sample [39].
Positive bronchoalveolar lavage (BAL) fluid galactomannan was defined as an optical density ratio at least 0.8 in a fluid sample [40].
Patients
We conducted this prospective observational study at Ramathibodi Hospital, Mahidol University, an academic tertiary care medical center in Bangkok, Thailand between January 2015 and December 2016. All Thai patient of all ages with moderate- to high-risk hematologic disease and no known diagnosis of ongoing IA were prospectively screened, and 357 patients were enrolled. Blood samples were obtained for genetic analyses. Patients were excluded if palliative care had been administered and the chemotherapy plan was discontinued. This study was conducted in accordance with the Declaration of Helsinki of 1964 and its later amendments and was approved by the Institutional Review Board of Ramathibodi Hospital (approval no. MURA 2015/18). The committee approved the use of patient samples and data for the publication of this study. Written consent was obtained from all participants.
Data Collection
We retrieved information of patient demographics including age, sex, smoking history, medical condition, prior diagnosis of IA, diagnosis of hematologic disease, type, and intensity of immunosuppressive agents received, relevant clinical data during hospitalization, detection of cytomegalovirus (CMV) DNAemia or infection, medical ward assignment (high-efficiency particulate air filter equipped or non-high-efficiency particulate air filter equipped), type of antifungal prophylaxis if one was to be received, site of infection in individuals with IA. Galactomannan assay and radiological findings were recorded. Treatment outcome was also collected, to compare patients.
DNA Extraction and SNP Genotyping
A 5- to 10-mL sample of whole blood was obtained from each patient. Genomic DNA was extracted using the FavorPrep™ Blood Genomic DNA extraction kit (Favorgen Biotech Corp., Taiwan) and stored at 4 °C. Samples were subsequently stored at − 20 °C for possible future analyses. In individuals who underwent stem cell transplantation, donor DNA was isolated from post-engraftment blood samples. The genotyping process was performed using the TaqMan SNP genotyping assay (Thermo Fisher Scientific, USA) according to the manufacturer’s instructions in a ViiA™ 7 system (Applied Biosystems, California, USA). Primer and probe sequences used in this study are shown in Table 1 in the supplementary material. Laboratory personnel were blinded to patients’ history and conditions.
Serum and BAL fluid galactomannan were tested using a Platelia galactomannan assay (Bio-Rad Laboratories, California, USA), according to the manufacturer’s instructions. Factors potentially associated with false positive galactomannan status were recorded and were carefully reviewed, namely concomitant use of piperacillin–tazobactam and amoxicillin–clavulanic acid, diagnoses of other fungal infection such as histoplasmosis, and certain types of dietary consumption [41–43].
Study Endpoint
The study endpoint was documentation of proven or probable IA, the need for second stem cell transplantation, death, or at 12 months after diagnosis of the hematologic disease. For patients with IA, the clinical outcome at 90 days after IA diagnosis was indicated.
Selection of Genetic Polymorphisms
A total of five SNPs in five genes of interest were selected for genotyping analyses in this study, namely rs4804800 in DC-SIGN [32], rs1800896 in IL10 [24, 25], rs7309123 in DECTIN-1 [32], rs4986790 in TLR4 [26, 31, 33], and rs2305619 in PTX3 [34], on the basis of previous clinical studies identifying important SNPs and the frequency of gene identification among Asian populations. The analysis was performed in all allele types for every gene of interest. According to the global Single Nucleotide Polymorphism Database (https://www.ncbi.nlm.nih.gov/SNP/) and accessed national data on ThaiSNP2 (https://www4a.biotec.or.th/thaisnp2/), the frequency of the five high-risk genotypes in Asian populations was 37.2%, 4.3%, 46.5%, 1.3%, and 44.2%, respectively. There was only one study from Asia by Seo et al. that reported an association of IL10 polymorphisms with a higher risk of IA among Korean patients who underwent allogeneic HSCT [24].
Statistical Analysis
Genotypes and allele frequencies were calculated and compared between patients with and without IA using Pearson–chi-square and Fisher’s exact tests. Odds ratios (ORs) and 95% confidence intervals (CIs) were assessed for the presence (homozygous or heterozygous) or absence (homozygous wild-type allele) of the selected SNPs using logistic regression analysis. All polymorphisms were tested for the Hardy–Weinberg equilibrium to evaluate genetic variation and distribution patterns. Demographics in patients with and without IA were compared using a two-tailed t test or Mann–Whitney test for continuous data, and a chi-square test or Fisher’s exact test for categorical data. Significant variables (p < 0.05) were included in multivariate Cox regression analysis. The log-rank test was used to compare the time to IA and survival time according to different baseline characteristics and polymorphisms of interest.
Results
Between January 2015 and December 2016, a total of 410 patients were screened. Fifty-three patients were excluded from the study because they did not meet the criteria for moderate- to high-risk hematologic malignancy or had not received any treatment for such hematologic conditions at the time. The patients’ characteristics are shown in Table 1. Among 357 patients, 188 were male patients (52.7%). The median age was 57 years old (interquartile range [IQR] 44.0–64.0). The most common hematologic disease was NHL, documented in 161/357 patients (45.1%), followed by AML and multiple myeloma (60/357, 16.8% each). The median age of patients with AML and allogeneic HSCT was 37 years (IQR 31.0–57.5 years). Twenty-eight patients among the total had diabetes mellitus, with median glycated hemoglobin (HbA1C) of 7.0% (IQR 6.0–7.5%). Five patients had poorly controlled diabetes with HbA1C at enrollment over 8.0%. Eight patients with NHL were coinfected with HIV. Only 25/357 patients (7.0%) received antifungal prophylaxis; among these, 3 patients were given itraconazole, 21 patients received fluconazole, and 1 patient received micafungin. No patients received more than one agent during their clinical course. However, complete data on prophylaxis were missing in 45 patients.
Table 1.
Characteristics of patients
| Characteristics | Patients, n = 357 (%) | No IA, n = 321 (%) | IA, n = 36 (%) | IA rate, % | P value |
|---|---|---|---|---|---|
| Median age (IQR) | 57.0 (44.0–64.0) | 57.0 (46.0–65.0) | 43.5 (32.5–58.0) | – | 0.001 |
| Age, years | < 0.001 | ||||
| < 40 | 77 (21.6) | 60 (18.7) | 17 (47.2) | 22.1 | |
| 41–60 | 147 (41.2) | 135 (42.1) | 12 (33.3) | 8.1 | |
| > 60 | 133 (37.3) | 126 (39.3) | 7 (19.4) | 5.3 | |
| Male gender | 188 (52.7) | 172 (53.6) | 16 (44.4) | 8.5 (male), 11.8 (female) | 0.298 |
| Underlying hematologic disease | < 0.001 | ||||
| Allogeneic HSCT | 21 (5.9) | 19 (5.9) | 2 (5.6) | 9.5 | |
| Acute myeloid leukemia | 60 (16.8) | 40 (12.5) | 20 (55.6) | 33.3 | |
| Acute lymphoblastic leukemia | 28 (7.8) | 24 (7.5) | 4 (11.1) | 14.3 | |
| Chronic lymphocytic leukemia | 9 (2.5) | 8 (2.5) | 1 (2.8) | 11.1 | |
| Non-Hodgkin’s lymphoma | 161 (45.1) | 156 (48.6) | 5 (13.9) | 3.1 | |
| Myelodysplastic syndrome | 13 (3.6) | 12 (3.7) | 1 (2.8) | 7.7 | |
| Severe aplastic anemia | 4 (1.1) | 3 (0.9) | 1 (2.8) | 25.0 | |
| Multiple myeloma | 60 (16.8) | 59 (18.4) | 1 (2.8) | 1.7 | |
| Severe GVHD after HSCT | 1 (0.3) | 0 (0) | 1 (2.8) | 100.0 | |
| Other underlying disease | |||||
| Diabetes mellitus | 28 (14.8) | 26 (17.0) | 2 (5.6) | 7.1 | 0.082 |
| HIV | 8 (2.3) | 8 (2.5) | 0 (0) | 0 | 1.000 |
IA invasive aspergillosis, IQR interquartile range, HSCT hematopoietic stem cell transplant, GVHD graft versus host disease
IA was diagnosed in 36/357 patients (10.1%), characterized as proven IA in 6 patients; the remainder were probable cases. The median time from enrollment to IA diagnosis was 56.5 days (IQR 26.0–150.5) days. IA was mostly diagnosed in patients with AML, followed by those with NHL receiving salvage therapy. No patients with poorly controlled diabetes were found to have developed IA (Table 1). The median age of patients with IA was 43.5 years (IQR 32.5–58.0 years). Thirty-three of 36 patients had single organ involvement; 30 patients had pulmonary involvement, and three patients had paranasal sinus infections. Three patients had multiple organ infections involving the respiratory and central nervous systems. Aspergillus species were identified in five patients, among which three patients were found to have A. flavus, one patient had combined A. flavus and A. fumigatus infection, and one patient had A. terreus infection. The 90-day IA-related mortality rate was 36.1% (13/36 patients). The median hospital length of stay among patients with IA was 34.0 days (IQR 25.5–58.5 days) compared with 16.0 days (IQR 7–27 days) in patients without IA (p < 0.001). There was no significant difference between probable and proven IA in terms of length of stay, duration of treatment, and survival. One patient each developed IA while taking fluconazole and itraconazole prophylaxis, but therapeutic drug monitoring was not performed in either patient when IA was diagnosed.
The genotypes of interest, DC-SIGNrs4804800, IL10rs1800896, DECTIN1rs7309123, TLR4rs4986790, and PTX3rs2305619, were identified in 47 patients (14.0%), 3 patients (0.9%), 230 patients (68.2%), 0 (0%), and 141 patients (42.0%), respectively; Table 2 displays the alleles of interest according to proportion. Individuals carrying the C/C and T/C genotypes of the IL10rs1800896 SNP had a higher incidence of IA diagnosis (adjusted OR 5.297; 95% CI 2.032–13.809, p = 0.001). The associated risk factors of IA are listed in Table 3. In the multivariate Cox proportional hazards model, only prolonged neutropenia and the C allele of IL10rs1800896 SNP were independent factors contributing to IA development (HR 12.585; 95% CI 3.866–40.967, p < 0.001 and HR 2.449; 95% CI 1.097–5.468, p = 0.029, respectively). Figure 1 shows the cumulative risk of IA in association with individuals carrying C/C and T/C genotypes of IL10rs1800896 SNP, who tended to develop IA earlier in their clinical course compared with other patients. A combination of the SNPs of interest did not increase the risk of IA development in this cohort. There was no significant association between the tested polymorphisms and galactomannan levels in either serum or BAL fluid.
Table 2.
Association of invasive aspergillosis and selected polymorphisms
| SNP | Genotype | No IA, n (%) | IA, n (%) | Adjusted OR (95% CI)a | P value |
|---|---|---|---|---|---|
| DC-SIGNrs4804800 | A/A | 123 (41.1) | 10 (30.3) | 1.000 | – |
| A/G | 135 (45.2) | 17 (51.5) | 1.499 (0.622–3.616) | 0.367 | |
| G/G | 41 (13.7) | 6 (18.2) | 1.568 (0.485–5.069) | 0.453 | |
| G/G + A/G | 176 (58.9) | 23 (69.7) | 1.516 (0.656–3.501) | 0.330 | |
| IL10rs1800896 | T/T | 263 (88.9) | 24 (70.6) | 1.000 | – |
| T/C | 31 (10.5) | 9 (26.5) | 5.517 (2.043–14.896) | 0.001 | |
| C/C | 2 (0.7) | 1 (2.9) | 3.538 (0.194–64.546) | 0.394 | |
| C/C + T/C | 33 (11.1) | 10 (29.4) | 5.297 (2.032–13.809) | 0.001 | |
| DECTIN1rs7309123 | C/C | 8 (2.7) | 2 (6.1) | 1.000 | – |
| C/G | 83 (28.0) | 8 (24.2) | 0.311 (0.046–2.098) | 0.230 | |
| G/G | 205 (69.3) | 23 (69.7) | 0.449 (0.074–2.725) | 0.384 | |
| G/G + C/G | 288 (97.3) | 31 (93.9) | 0.404 (0.068–2.404) | 0.319 | |
| TLR4rs4986790 | A/A | 284 (96.3) | 33 (100.0) | 1.000 | – |
| A/G | 11 (3.7) | 0 (0.0) | – | – | |
| G/G | 0 (0.0) | 0 (0.0) | – | – | |
| PTX3rs2305619 | A/A | 34 (11.4) | 2 (5.9) | 1.000 | – |
| A/G | 140 (47.0) | 15 (44.1) | 1.130 (0.228–5.598) | 0.881 | |
| G/G | 124 (41.6) | 17 (50.0) | 1.398 (0.283–6.909) | 0.681 | |
| G/G + A/G | 264 (88.6) | 32 (94.1) | 1.253 (0.267–5.893) | 0.775 |
SNP single nucleotide polymorphism, IA invasive aspergillosis, OR odds ratio, DC-SIGN dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin, IL10 interleukin-10, DECTIN1 dendritic cell-associated c-type lectin 1, TLR4 toll like receptor 4, PTX3 pentraxin-3
aLogistic regression analysis with model adjusted for age group and high-risk hematologic disease
Table 3.
Associated risk factors of invasive aspergillosis in this cohort
| Risk factors | No IA, n = 380 admission episodes (%) | IA, n = 36 admission episodes (%) | Univariate Cox regression HR (95% CI), P value | Multivariate Cox regression HR (95% CI), P value |
|---|---|---|---|---|
| Female | 163 (42.9) | 20 (55.6) | 1.681 (0.871–3.245), 0.121 | |
| Age > 40 years | 272 (71.6) | 19 (52.8) | 0.504 (0.262–0.970), 0.040 | 0.956 (0.443–2.063), 0.909 |
| At-risk occupationa | 27 (8.8) | 1 (2.8) | 0.281 (0.038–2.053), 0.252 | |
| Current smoking | 12 (3.9) | 4 (11.1) | 2.309 (0.816–6.534), 0.115 | |
| Prior invasive mold disease | 7 (2.3) | 1 (2.8) | 1.177 (0.161–8.594), 0.873 | |
| Diabetes mellitus | 40 (12.7) | 2 (5.6) | 0.388 (0.093–1.615), 0.193 | |
| Corticosteroids | 72 (23.8) | 2 (5.6) | 0.215 (0.052–0.894), 0.035 | 0.424 (0.089–2.021), 0.282 |
| Lymphocytopenia or anti-T cell use | 52 (17.1) | 5 (13.9) | 0.813 (0.316–2.094), 0.668 | |
| Prolonged neutropenia | 37 (12.1) | 26 (74.3) | 12.672 (5.930–27.081), < 0.001 | 12.585 (3.866–40.967), < 0.001 |
| Mucositis, grade 3–4 | 9 (3.0) | 4 (11.1) | 3.068 (1.084–8.681), 0.035 | 1.672 (0.460–6.086), 0.435 |
| CMV DNAemia | 13 (4.3) | 3 (8.3) | 1.675 (0.512–5.471), 0.393 | |
| Admitted in non-HEPA room | 168 (62.9) | 25 (75.8) | 1.951 (0.877–4.338), 0.101 | |
| High-risk malignancyb | 87 (23.0) | 23 (65.7) | 5.001 (2.488–10.052), < 0.001 | 1.074 (0.420–2.743), 0.882 |
| Uncontrolled malignancyc | 213 (69.2) | 31 (86.1) | 2.986 (1.160–7.688), 0.023 | 2.835 (0.813–9.892), 0.102 |
| High-risk chemotherapyd | 156 (49.4) | 27 (75.0) | 2.453 (1.153–5.219), 0.020 | 0.609 (0.237–1.569), 0.305 |
| Antifungal prophylaxis | 34 (11.2) | 2 (5.6) | 0.474 (0.114–1.973), 0.305 | |
| DC-SIGNrs4804800 G/G genotype | 48 (13.4) | 6 (18.2) | 1.445 (0.596–3.500), 0.415 | |
| DC-SIGNrs4804800 G carriage | 195 (54.5) | 23 (69.7) | 1.761 (0.838–3.699), 0.135 | |
| IL10rs1800896 C/C genotype | 0 (0) | 1 (2.9) | – | |
| IL10rs1800896 C carriage | 36 (10.1) | 10 (29.4) | 3.436 (1.641–7.196), 0.001 | 2.449 (1.097–5.468), 0.029 |
IA invasive aspergillosis, HR hazard ratio, CMV cytomegalovirus, HEPA high efficiency particulate air filter, DC-SIGN dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin, IL10 interleukin 10, DECTIN1 dendritic cell-associated c-type lectin 1, TLR4 Toll-like receptor 4, PTX3 pentraxin-3
aPatient works as a farmer, mason, carpenter/construction, or has outdoor work with likely spore exposures
bHigh-risk malignancy was defined as diagnosis of acute myeloid leukemia or allogeneic HSCT
cUncontrolled malignancy was defined as malignancy status that was not in partial or complete remission
dHigh-risk chemotherapy was defined as chemotherapy for AML and conditioning chemotherapy for allogeneic HSCT
Fig. 1.
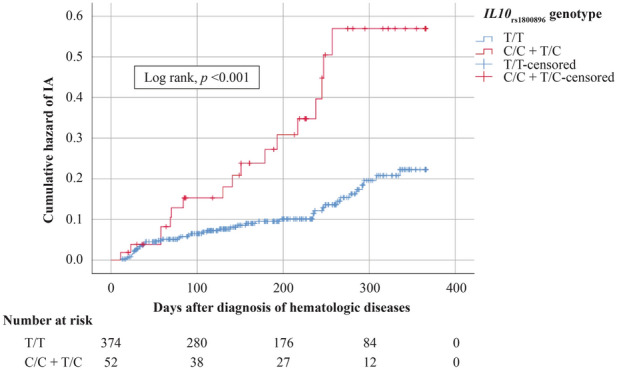
Kaplan–Meier analysis shows association of IL10rs1800896 genotype and cumulative risk of IA
All-cause 1-year mortality in this cohort was 14.8% (53/357 patients). Age over 60 years old, prolonged neutropenia, and uncontrolled malignancy were associated with increased all-cause mortality (HR 2.313, 95% CI 1.209–4.426, p = 0.003; HR 3.240, 95% CI 1.663–6.312, p = 0.001; HR 2.661, 95% CI 1.039–6.813, p = 0.041, respectively). Individuals carrying the C allele on IL10rs1800896 tended to have increased all-cause 1-year mortality (HR 2.275; 95% CI 1.166–4.441, p = 0.016) in the univariate Cox regression model, but this did not retain statistical significance in the multivariate Cox proportional hazards model. The presence of the other tested SNPs did not influence hospital length of stay and overall mortality, regardless of IA diagnosis. A complete list of associated risk factors affecting all-cause 1-year mortality is given in Table 4.
Table 4.
Risk factors of all-cause one-year mortality
| Risk factors | Survived, n = 363 admission episodes (%) | Dead, n = 53 admission episodes (%) | Univariate Cox regression HR (95% CI), P value | Multivariate Cox regression HR (95% CI), P value |
|---|---|---|---|---|
| Female | 159 (43.8) | 24 (45.3) | 1.110 (0.647–1.907), 0.704 | |
| > 60 years old | 100 (27.5) | 24 (45.3) | 1.972 (1.148–3.388), 0.014 | 2.313 (1.209–4.426), 0.011 |
| At risk occupationa | 25 (8.5) | 3 (6.4) | 0.667 (0.207–2.151), 0.498 | |
| Current smoking | 11 (3.7) | 5 (10.4) | 2.153 (0.852–5.437), 0.105 | |
| Prior invasive mold disease | 7 (2.3) | 1 (2.1) | 0.933 (0.129–6.768), 0.945 | |
| Diabetes | 34 (11.2) | 8 (16.3) | 1.264 (0.592–2.701), 0.545 | |
| Corticosteroids | 66 (22.5) | 8 (17.4) | 0.763 (0.356–1.636), 0.487 | |
| Lymphocytopenia or anti-T cell use | 44 (15.1) | 13 (27.1) | 1.859 (0.982–3.520), 0.057 | |
| Prolonged neutropenia | 44 (15.1) | 19 (39.6) | 2.904 (1.626–5.187), < 0.001 | 3.240 (1.663–6.312), 0.001 |
| Mucositis, grade 3–4 | 9 (3.1) | 4 (8.3) | 2.335 (0.839–6.501), 0.105 | |
| CMV infection | 10 (3.4) | 6 (12.5) | 2.624 (1.113–6.187), 0.028 | 2.077 (0.777–5.552), 0.145 |
| Admitted in non-HEPA room | 164 (64.3) | 29 (64.4) | 1.125 (0.609–2.076), 0.707 | |
| High-risk malignancyb | 91 (25.2) | 19 (35.8) | 1.460 (0.833–2.561), 0.186 | |
| Uncontrolled malignancyc | 202 (68.2) | 42 (87.5) | 3.374 (1.433–7.944), 0.005 | 2.661 (1.039–6.813), 0.041 |
| High-risk chemotherapyd | 63 (21.1) | 14 (29.2) | 0.879 (0.499–1.550), 0.656 | |
| Antifungal prophylaxis | 35 (11.9) | 1 (2.1) | 0.178 (0.025–1.290), 0.088 | |
| DC-SIGNrs4804800 G/G genotype | 49 (14.4) | 5 (9.8) | 0.718 (0.285–1.808), 0.482 | |
| DC-SIGNrs4804800 G carriage | 186 (54.7) | 32 (62.7) | 1.277 (0.724–2.253), 0.399 | |
| IL10rs1800896 C/C genotype | 0 (0) | 1 (2.0) | – | – |
| IL10rs1800896 C carriage | 35 (10.4) | 11 (21.6) | 2.275 (1.166–4.441), 0.016 | 1.770 (0.881–3.554), 0.109 |
HR hazard ratio, CMV cytomegalovirus, HEPA high efficiency particulate air filter, DC-SIGN dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin, IL10 interleukin-10, DECTIN1 dendritic cell-associated c-type lectin 1, TLR4 Toll-like receptor 4, PTX3 pentraxin-3
aPatient works as a farmer, mason, carpenter/construction, or has outdoor work with likely spore exposures
bHigh-risk malignancy defined as diagnosis of acute myeloid leukemia or allogeneic HSCT
cUncontrolled malignancy defined as malignancy status that is not in partial or complete remission
dHigh-risk chemotherapy was defined as chemotherapy for AML and conditioning chemotherapy for allogeneic HSCT
Discussion
A variety of associated risk factors for IA have been identified, e.g., prolonged neutropenia with absolute neutrophil count of less than 500/mm3 for longer than 10 days, uncontrolled hematologic malignancy, poorly controlled diabetes mellitus particularly among Asians, and the influence of genetic components in patients at risk, according to various clinical studies [44–46]. In this study, most patients at risk of having IA were individuals with AML because they tended to have prolonged neutropenia following chemotherapy, with a prevalence rate of IA 33.3%. Most of these patients were diagnosed with AML and IA at a young age. Despite the high prevalence rates of IA among patients with ALL, CLL, and severe AA, these were not considered to represent a large proportion of our study population.
Prior history of invasive mold infection and lymphopenia have been reported to be independent risk factors of IA according to Stanzani et al. [11]. However, neither factors were good predictors for IA development in this study. This was potentially owing to a low prevalence of previous IA diagnosis in three patients. Uncontrolled diabetes mellitus has been reported more frequently as a comorbid condition in IA; more recently, a study in five Asian countries emphasized this unique associated risk factor [46]. The precise mechanism is still unknown, but one possibility is impaired neutrophil function causing compromised oxidative and non-oxidative killing mechanisms, which are crucial for hyphal and germinating conidia control [14, 47]. However, diabetes mellitus was not a significant contributing factor to IA in this cohort. Garcia-Vidal et al. described that CMV infection has a strong relationship with invasive fungal infection diagnosis, especially in individuals with lymphoproliferative malignancies [48]. The exact mechanism to illustrate this finding is not fully understood but is potentially owing to the immunomodulatory effects of CMV itself [49]. However, CMV DNAemia did not predict an increased IA risk in this cohort, likely secondary to the low prevalence of CMV DNAemia (14 patients). Therefore, an impact of CMV was not noted in this study. A higher-risk trend of IA development was observed among individuals with high-risk malignancy, uncontrolled malignancy, and receipt of high-risk chemotherapy. Nevertheless, none of these showed significant effects in multivariate Cox regression. Among all well-established risk factors mentioned, the only two associated factors that remained significant regarding increased IA risk were prolonged neutropenia and IL10rs1800896 C carriage (Table 3).
Genetic predisposition to IA has been increasingly studied over the past decade, mostly in North America and Europe. Some studies have demonstrated an increased risk of IA with the presence of polymorphisms of DC-SIGN, DECTIN1, IL10, TLR4, and PTX3 [24–26, 32, 34], given that the corresponding proteins have been discovered to be involved in IA pathogenesis. Moreover, some SNPs have been identified to be associated with elevated serum galactomannan levels, without any significant correlation to IA development [32]. IA has long been documented to be a fatal infectious complication among immunocompromised patients, particularly hematology patients [50]. Data on the epidemiology of IA in Thailand are limited, with only a few retrospective studies conducted but none in hematology patients [8, 48]. In the modern medicine era, antifungal prophylaxis has been prescribed widely in at-risk patients, to minimize invasive fungal infection occurrence. This schematic approach is considered a high-cost treatment and generally applies in high-income countries only. Therefore, applying a personalized medicine strategy by screening genetic predisposition to IA may be worthwhile, to identify patients truly in need of antifungal prophylaxis.
Among our study population, the prevalence rate of IA was highest in individuals with AML, followed by those with severe AA, ALL, CLL, and allogeneic HSCT recipients. Patients with AML accounted for 16.8% of all patients, second only to those with NHL. This reflects that patients with AML undergoing chemotherapy may be an appropriate group in which to consider antifungal prophylaxis when the cost of the antifungal agent is a concern. However, prescribing universal antifungal prophylaxis to all patients with AML undergoing chemotherapy may not be a feasible approach in low-income countries.
Regarding human immune response, macrophages and dendritic cells are responsible for the initial step in pathogen recognition, particularly fungi including Aspergillus spp., via various types of receptors on their surface, e.g., DC-SIGN, TLR-4, PTX-3, and so on. A complex interplay between immune cells is then triggered to control the invading microorganisms. Hence, any change in these genes could impair normal host immune response, leading to devastating fungal infection [21, 51]. Several genetic polymorphisms have been found to be associated with increased risk for IA, e.g., DECTIN- 1, DC-SIGN, TLR4, IL10, S100B, PTX3, PLG, IFNG, and TNFR1 [15, 35, 52]. However, these proteins have mainly been identified in North America and Europe, except for a single study in South Korea [24]. On the basis of national data of ThaiSNP2 (https://www.4a.biotec.or.th/thaisnp2/) and the global Single Nucleotide Polymorphism Database (https://www.ncbi.nlm.nih.gov/SNP/), only five SNPs of interest in this study have been identified in the Thai population.
In this study, we identified a strong association between the C allele of IL10rs1800896 and increased risk of IA, regardless of a diagnosis of hematologic disease; the same was not true for the other tested SNPs. The influence of this finding is second only to a well-known risk factor, namely prolonged neutropenia affecting IA development in moderate- to high-risk patients, despite the fact that there was no trend of increased mortality among individuals carrying such an SNP. Carrying the IL10rs1800896 C/C genotype, however, showed less effect on IA diagnosis in comparison with other genotypes. This finding may be explained by the relatively small number of individuals with a homozygous C genotype. The overall mortality of IA in our study was 14.8%; this was relatively lower than in previous studies and had no significant correlation with any tested SNPs [2, 3, 6].
Among 21 cases of HSCT in our study, only two patients developed IA. This prevalence rate was much lower than AML and severe AA [53]. One reason that could explain this relatively low prevalence could be the stringent pre-transplant scheme; complete remission of patients’ hematologic condition at transplant may reduce the risk of IA development at our institution.
The limitations of this study are as follows. (1) This was a single-center study, and therefore the data may not represent the characteristics of the entire region; (2) missing data in some patients made a complete genetic analysis impossible; (3) there was a lack of data on environmental exposure after hospitalization, which may affect IA development. The strength of this study is that we included a relatively large number of participants and gained new insight into IA development among hematology patients in Thailand. Genetic surveillance of patients at risk, particularly those with AML, and considering antifungal prophylaxis with IL10rs1800896 C carriage is recommended, especially when universal antifungal prophylaxis is not an option. Finally, a multicenter study is encouraged, to further clarify the effect of genetics on IA susceptibility.
Conclusion
Apart from traditional clinical risk factors, polymorphisms of immunoregulatory gene IL10rs1800896 may be an important risk factor to consider in risk stratification of IA development, particularly in patients with AML. This could potentially be a new strategy in hematology patients to lessen the risk of IA, especially in a resource-limited setting where universal antifungal prophylaxis is not possible in real-life practice.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank all participants for their contribution and all staff for help with the enrollment of patients in this study. We thank Analisa Avila, ELS, of Edanz Group (https://en-author-services.edanzgroup.com/ac) for editing a draft of this manuscript.
Funding
This study was supported by the Faculty of Medicine Ramathibodi Hospital, Mahidol University (CF_61004). The funder had no role in the study design, data collection, and analysis, decision to publish, or preparation of the manuscript. The Rapid Service Fee was funded by the authors.
Authorship
All named authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this article, take responsibility for the integrity of the work as a whole, and have given their approval for this version to be published.
Prior Presentation
Part of this study was presented by the authors at ID Week 2016.
Disclosures
Tananun Tanpaibule, Natini Jinawath, Aruchalean Taweewongsounton, Pimjai Niparuck and Porpon Rotjanapan have nothing to declare.
Compliance with Ethics Guidelines
This study was conducted in accordance with the Declaration of Helsinki of 1964 and its later amendments and was approved by the Institutional Review Board of Ramathibodi Hospital (approval no. MURA 2015/18). The committee approved the use of patient samples and data for the publication of this study. Written consent was obtained from all participants.
Data Availability
The datasets generated during and/or analyzed in the current study are not publicly available but are available from the corresponding author on reasonable request.
Footnotes
Digital Features
To view digital features for this article go to 10.6084/m9.figshare.12794684.
References
- 1.Kosmidis C, Denning DW. The clinical spectrum of pulmonary aspergillosis. Thorax. 2015;70:270–277. doi: 10.1136/thoraxjnl-2014-206291. [DOI] [PubMed] [Google Scholar]
- 2.Kontoyiannis DP, Marr KA, Park BJ, et al. Prospective surveillance for invasive fungal infections in hematopoietic stem cell transplant recipients, 2001–2006: overview of the Transplant-Associated Infection Surveillance Network (TRANSNET) Database. Clin Infect Dis. 2010;50:1091–1100. doi: 10.1086/651263. [DOI] [PubMed] [Google Scholar]
- 3.Pagano L, Caira M, Candoni A, et al. Invasive aspergillosis in patients with acute myeloid leukemia: a SEIFEM-2008 registry study. Haematologica. 2010;95:644–650. doi: 10.3324/haematol.2009.012054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pagano L, Caira M, Candoni A, et al. The epidemiology of fungal infections in patients with hematologic malignancies: the SEIFEM-2004 study. Haematologica. 2006;91:1068–1075. [PubMed] [Google Scholar]
- 5.Girmenia C, Ferretti A, Barberi W. Epidemiology and risk factors for invasive fungal diseases in hematopoietic stem cell transplantation. Curr Opin Hematol. 2014;21:459–465. doi: 10.1097/MOH.0000000000000086. [DOI] [PubMed] [Google Scholar]
- 6.Azie N, Neofytos D, Pfaller M, Meier-Kriesche HU, Quan SP, Horn D. The PATH (Prospective Antifungal Therapy) Alliance® registry and invasive fungal infections: update 2012. Diagn Microbiol Infect Dis. 2012;73:293–300. doi: 10.1016/j.diagmicrobio.2012.06.012. [DOI] [PubMed] [Google Scholar]
- 7.Slobbe L, Polinder S, Doorduijn JK, et al. Outcome and medical costs of patients with invasive aspergillosis and acute myelogenous leukemia-myelodysplastic syndrome treated with intensive chemotherapy: an observational study. Clin Infect Dis. 2008;47:1507–1512. doi: 10.1086/591531. [DOI] [PubMed] [Google Scholar]
- 8.Kiertiburanakul S, Thibbadee C, Santanirand P. Invasive aspergillosis in a tertiary-care hospital in Thailand. J Med Assoc Thail. 2007;90:895–902. [PubMed] [Google Scholar]
- 9.Ananda-Rajah MR, Cheng A, Morrissey CO, et al. Attributable hospital cost and antifungal treatment of invasive fungal diseases in high-risk hematology patients: an economic modeling approach. Antimicrob Agents Chemother. 2011;55:1953–1960. doi: 10.1128/AAC.01423-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwon-Chung KJ, Sugui JA. Aspergillus fumigatus–what makes the species a ubiquitous human fungal pathogen? PLoS Pathog. 2013;9:e1003743. doi: 10.1371/journal.ppat.1003743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stanzani M, Lewis RE, Fiacchini M, et al. A risk prediction score for invasive mold disease in patients with hematological malignancies. PLoS One. 2013;8:e75531. doi: 10.1371/journal.pone.0075531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baddley JW. Clinical risk factors for invasive aspergillosis. Med Mycol. 2011;49:S7–S12. doi: 10.3109/13693786.2010.505204. [DOI] [PubMed] [Google Scholar]
- 13.Pagano L, Akova M, Dimopoulos G, Herbrecht R, Drgona L, Blijlevens N. Risk assessment and prognostic factors for mould-related diseases in immunocompromised patients. J Antimicrob Chemother. 2011;66:i5–i14. doi: 10.1093/jac/dkq437. [DOI] [PubMed] [Google Scholar]
- 14.Herbrecht R, Bories P, Moulin JC, Ledoux MP, Letscher-Bru V. Risk stratification for invasive aspergillosis in immunocompromised patients. Ann NY Acad Sci. 2012;1272:23–30. doi: 10.1111/j.1749-6632.2012.06829.x. [DOI] [PubMed] [Google Scholar]
- 15.Cunha C, Aversa F, Romani L, Carvalho A. Human genetic susceptibility to invasive aspergillosis. PLoS Pathog. 2013;9:e1003434. doi: 10.1371/journal.ppat.1003434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wojtowicz A, Bochud PY. Host genetics of invasive Aspergillus and Candida infections. Semin Immunopathol. 2015;37:173–186. doi: 10.1007/s00281-014-0468-y. [DOI] [PubMed] [Google Scholar]
- 17.Whitsett JA, Alenghat T. Respiratory epithelial cells orchestrate pulmonary innate immunity. Nat Immunol. 2014;16:27–35. doi: 10.1038/ni.3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Camargo JF, Husain S. Immune correlates of protection in human invasive aspergillosis. Clin Infect Dis. 2014;59:569–577. doi: 10.1093/cid/ciu337. [DOI] [PubMed] [Google Scholar]
- 19.Romani L. Immunity to fungal infections. Nat Rev Immunol. 2011;11:275–288. doi: 10.1038/nri2939. [DOI] [PubMed] [Google Scholar]
- 20.Lass-Florl C, Roilides E, Loffler J, Wilflingseder D, Romani L. Minireview: host defence in invasive aspergillosis. Mycoses. 2013;56:403–413. doi: 10.1111/myc.12052. [DOI] [PubMed] [Google Scholar]
- 21.Park SJ, Mehrad B. Innate immunity to Aspergillus species. Clin Microbiol Rev. 2009;22:535–551. doi: 10.1128/CMR.00014-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carvalho A, Cunha C, Bozza S, et al. Immunity and tolerance to fungi in hematopoietic transplantation: principles and perspectives. Front Immunol. 2012;3:156. doi: 10.3389/fimmu.2012.00156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oliveira-Coelho A, Rodrigues F, Campos A, Jr, Lacerda JF, Carvalho A, Cunha C. Paving the way for predictive diagnostics and personalized treatment of invasive aspergillosis. Front Microbiol. 2015;6:411. doi: 10.3389/fmicb.2015.00411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seo KW, Kim DH, Sohn SK, et al. Protective role of interleukin-10 promoter gene polymorphism in the pathogenesis of invasive pulmonary aspergillosis after allogeneic stem cell transplantation. Bone Marrow Transpl. 2005;36:1089–1095. doi: 10.1038/sj.bmt.1705181. [DOI] [PubMed] [Google Scholar]
- 25.Sainz J, Hassan L, Perez E, et al. Interleukin-10 promoter polymorphism as risk factor to develop invasive pulmonary aspergillosis. Immunol Lett. 2007;109:76–82. doi: 10.1016/j.imlet.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 26.Bochud PY, Chien JW, Marr KA, et al. Toll-like receptor 4 polymorphisms and aspergillosis in stem-cell transplantation. N Engl J Med. 2008;359:1766–1777. doi: 10.1056/NEJMoa0802629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mezger M, Steffens M, Beyer M, et al. Polymorphisms in the chemokine (C-X-C motif) ligand 10 are associated with invasive aspergillosis after allogeneic stem-cell transplantation and influence CXCL10 expression in monocyte-derived dendritic cells. Blood. 2008;111:534–536. doi: 10.1182/blood-2007-05-090928. [DOI] [PubMed] [Google Scholar]
- 28.Zaas AK, Liao G, Chien JW, et al. Plasminogen alleles influence susceptibility to invasive aspergillosis. PLoS Genet. 2008;4:e1000101. doi: 10.1371/journal.pgen.1000101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sainz J, Salas-Alvarado I, Lopez-Fernandez E, et al. TNFR1 mRNA expression level and TNFR1 gene polymorphisms are predictive markers for susceptibility to develop invasive pulmonary aspergillosis. Int J Immunopathol Pharmacol. 2010;23:423–436. doi: 10.1177/039463201002300205. [DOI] [PubMed] [Google Scholar]
- 30.Cunha C, Giovannini G, Pierini A, et al. Genetically-determined hyperfunction of the S100B/RAGE axis is a risk factor for aspergillosis in stem cell transplant recipients. PLoS One. 2011;6:e27962. doi: 10.1371/journal.pone.0027962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Boer MG, Jolink H, Halkes CJ, et al. Influence of polymorphisms in innate immunity genes on susceptibility to invasive aspergillosis after stem cell transplantation. PLoS One. 2011;6:e18403. doi: 10.1371/journal.pone.0018403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sainz J, Lupianez CB, Segura-Catena J, et al. Dectin-1 and DC-SIGN polymorphisms associated with invasive pulmonary aspergillosis infection. PLoS One. 2012;7:e32273. doi: 10.1371/journal.pone.0032273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koldehoff M, Beelen DW, Elmaagacli AH. Increased susceptibility for aspergillosis and post-transplant immune deficiency in patients with gene variants of TLR4 after stem cell transplantation. Transpl Infect Dis. 2013;15:533–539. doi: 10.1111/tid.12115. [DOI] [PubMed] [Google Scholar]
- 34.Cunha C, Aversa F, Lacerda JF, et al. Genetic PTX3 deficiency and aspergillosis in stem-cell transplantation. N Engl J Med. 2014;370:421–432. doi: 10.1056/NEJMoa1211161. [DOI] [PubMed] [Google Scholar]
- 35.Lupianez CB, Canet LM, Carvalho A, et al. Polymorphisms in host immunity-modulating genes and risk of invasive aspergillosis: results from the AspBIOmics consortium. Infect Immun. 2015;84:643–657. doi: 10.1128/IAI.01359-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fischer M, Spies-Weisshart B, Schrenk K, et al. Polymorphisms of dectin-1 and TLR2 predispose to invasive fungal disease in patients with acute myeloid leukemia. PLoS One. 2016;11:e0150632. doi: 10.1371/journal.pone.0150632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De Pauw B, Walsh TJ, Donnelly JP, et al. Revised definitions of invasive fungal disease from the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group and the National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) Consensus Group. Clin Infect Dis. 2008;46:1813–1821. doi: 10.1086/588660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martino R, Parody R, Fukuda T, et al. Impact of the intensity of the pretransplantation conditioning regimen in patients with prior invasive aspergillosis undergoing allogeneic hematopoietic stem cell transplantation: a retrospective survey of the Infectious Diseases Working Party of the European Group for Blood and Marrow Transplantation. Blood. 2006;108:2928–2936. doi: 10.1182/blood-2006-03-008706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maertens J, Theunissen K, Verbeken E, et al. Prospective clinical evaluation of lower cut-offs for galactomannan detection in adult neutropenic cancer patients and haematological stem cell transplant recipients. Br J Haematol. 2004;126:852–860. doi: 10.1111/j.1365-2141.2004.05140.x. [DOI] [PubMed] [Google Scholar]
- 40.Lamoth F. Galactomannan and 1,3-beta-d-glucan testing for the diagnosis of invasive aspergillosis. J Fungi (Basel) 2016;2:22. doi: 10.3390/jof2030022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aubry A, Porcher R, Bottero J, et al. Occurrence and kinetics of false-positive Aspergillus galactomannan test results following treatment with beta-lactam antibiotics in patients with hematological disorders. J Clin Microbiol. 2006;44:389–394. doi: 10.1128/JCM.44.2.389-394.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mattei D, Rapezzi D, Mordini N, et al. False-positive Aspergillus galactomannan enzyme-linked immunosorbent assay results in vivo during amoxicillin-clavulanic acid treatment. J Clin Microbiol. 2004;42:5362–5363. doi: 10.1128/JCM.42.11.5362-5363.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barton RC. Laboratory diagnosis of invasive aspergillosis: from diagnosis to prediction of outcome. Scientifica. 2013;2013:459405. doi: 10.1155/2013/459405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muhlemann K, Wenger C, Zenhausern R, Tauber MG. Risk factors for invasive aspergillosis in neutropenic patients with hematologic malignancies. Leukemia. 2005;19:545–550. doi: 10.1038/sj.leu.2403674. [DOI] [PubMed] [Google Scholar]
- 45.Atalla A, Garnica M, Maiolino A, Nucci M. Risk factors for invasive mold diseases in allogeneic hematopoietic cell transplant recipients. Transpl Infect Dis. 2015;17:7–13. doi: 10.1111/tid.12328. [DOI] [PubMed] [Google Scholar]
- 46.Rotjanapan P, Chen YC, Chakrabarti A, et al. Epidemiology and clinical characteristics of invasive mould infections: a multicenter, retrospective analysis in five Asian countries. Med Mycol. 2018;56:186–196. doi: 10.1093/mmy/myx029. [DOI] [PubMed] [Google Scholar]
- 47.Latge JP. Aspergillus fumigatus and aspergillosis. Clin Microbiol Rev. 1999;12:310–350. doi: 10.1128/cmr.12.2.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Garcia-Vidal C, Upton A, Kirby KA, Marr KA. Epidemiology of invasive mold infections in allogeneic stem cell transplant recipients: biological risk factors for infection according to time after transplantation. Clin Infect Dis. 2008;47:1041–1050. doi: 10.1086/591969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marchesi F, Pimpinelli F, Di Domenico EG, et al. Association between CMV and invasive fungal infections after autologous stem cell transplant in lymphoproliferative malignancies: opportunistic partnership or cause-effect relationship? Int J Mol Sci. 2019;20:1373. doi: 10.3390/ijms20061373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.D'Haese J, Theunissen K, Vermeulen E, et al. Detection of galactomannan in bronchoalveolar lavage fluid samples of patients at risk for invasive pulmonary aspergillosis: analytical and clinical validity. J Clin Microbiol. 2012;50:1258–1263. doi: 10.1128/JCM.06423-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vautier S, MacCallum DM, Brown GD. C-type lectin receptors and cytokines in fungal immunity. Cytokine. 2012;58:89–99. doi: 10.1016/j.cyto.2011.08.031. [DOI] [PubMed] [Google Scholar]
- 52.Cunha C, Carvalho A. Genetic defects in fungal recognition and susceptibility to invasive pulmonary aspergillosis. Med Mycol. 2019;57:S211–S218. doi: 10.1093/mmy/myy057. [DOI] [PubMed] [Google Scholar]
- 53.Omrani AS, Almaghrabi RS. Complications of hematopoietic stem transplantation: fungal infections. Hematol Oncol Stem Cell Ther. 2017;10:239–244. doi: 10.1016/j.hemonc.2017.05.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed in the current study are not publicly available but are available from the corresponding author on reasonable request.


