Abstract
The morphology of bulliform cells located on the upper epidermis of leaves is one of the most important cell structures affecting leaf shape. Although many mechanisms regulating the development of bulliform cells have been reported, the fine regulatory mechanisms governing this process have rarely been described. To identify novel components regulating rice leaf morphology, a mutant showing a constitutively rolling phenotype from the seedling stage to flowering, known as crm1‐D, was selected for further analysis. Anatomical analyses in crm1‐D were attributable to the size reduction of bulliform cells. The crm1‐D was controlled by a single dominant nuclear gene. Map‐based cloning revealed that Roc8, an HD zipper class IV family member, was responsible for the crm1‐D phenotype. Notably, the 50‐bp sequence in the 3′‐untranslated region (3′‐UTR) of the Roc8 gene represses Roc8 at the translational level. Moreover, the roc8 knockdown lines notably increased the size of bulliform cells. A series of assays revealed that Roc8 negatively regulates the size of bulliform cells. Unexpectedly, Roc8 was also observed to positively mediate lignin biosynthesis without incurring a production penalty. The above results show that Roc8 may have a practical application in cultivating materials with high photosynthetic efficiency and low lignin content.
Keywords: rice, Roc8, 3′‐UTR, bulliform cells, lignin, photosynthesis
Introduction
Leaf morphology is an important part of the ideal plant architecture of rice (Zhang et al., 2009). Moderate leaf curling helps to keep the leaves upright, increase the light‐receiving space at the base of the middle and late stages of population, improve the utilization rate of light energy and enhance the light quantity and intensity at the base of the canopy (Xu et al., 2018). Moderate leaf curling also helps to improve root system activity and strengthen lodging resistance (Zhang et al., 2009; Zou et al., 2011). Therefore, the study of leaf morphology has become the focus of basic theory and practical breeding applications.
Considerable progress has been made in genetic studies of leaf morphogenesis in rice (Xu et al., 2018). Leaf curling was primarily caused by bulliform cell changes in monocotyledons. Bulliform cells, also called motor cells, are among the most important cell structures and are located on the upper epidermis of leaves. The osmotic pressure change of bulliform cells leads to leaf curling. Physiological studies showed that under water stress, bulliform cells shrank, and the leaves rolled. When the stress was removed, the bulliform cells swelled and absorbed water, and the leaves returned to their original state (Sylvester et al., 2001; Zou et al., 2011). To date, at least 10 genes controlling bulliform cell development have been reported in rice (Xu et al., 2018). These genes mainly included BRD1 (Hong et al., 2002), NRL1 (Hu et al., 2010), RL14 (Fang et al., 2012), SRL1 (Xiang et al., 2012), LC2 (Zhao et al., 2010), Roc5 (Zou et al., 2011), ACL1/ACL2 (Li et al., 2010) and OsLBD3‐7 (Li et al., 2016). These genes played an important role in controlling the number and size of bulliform cells. NRL1, RL14 and ACL1/ACL2 positively regulated the development of bulliform cells, while SRL1, BRD1, LC2, Roc5 and OsLBD3‐7 negatively regulated the development of bulliform cells. Most of the genes encoded oxidative, reductive or synthetase and were located at the lowest level of the signalling pathway. Despite several findings, the molecular mechanism of leaf rolling has not been fully elucidated. Few detailed studies have been conducted on directly regulating the development of bulliform cells by transcription factors.
Notably, the transcription factor Roc5 encodes an HD zipper class IV family member that negatively regulates bulliform cell development in combination with downstream target gene protodermal factor‐like (Zou et al., 2011). Whether other Roc families in rice have similar mechanism remains to be explored. In this study, a crm1‐D mutant showing a rolling phenotype was selected for analysis. Roc8, an HD zipper class IV family member, was responsible for the crm1‐D phenotype. We verified that the 50‐bp sequence in the Roc8 3′‐UTR represses Roc8 at the translational level. Roc8 was observed to negatively regulate the size of bulliform cells. In particular, Roc8 was determined to positively modulate leaf lignin contents in rice leaves. The results of this study may be helpful to understand the function of the rice outermost cell‐specific gene (Roc) family and provide a potential theoretical basis for cultivating materials with high photosynthesis or low lignin contents in rice or forage grass.
Results
Phenotypic characterization of the crm1‐D mutant
To identify novel components regulating rice leaf morphology, a mutant named crm1‐D showed a constitutively inward rolling phenotype from the seedling to flowering stage (Figure 1a, b, c, d), which was selected for analysis. The rolling degree in crm1‐D adds up to ~0.9 compared with the wild type (Figure 1i). F1 crosses of crm1‐D with the wild type suggested that the recovered allele was dominant (Figure 1a). Genetic analyses of heterozygous F1 progeny showed that the phenotype in crm1‐D segregated in a 3:1 ratio (χ 2 = 1.1145, χ2 < 0.05, 1) of wild‐type (45) and mutant‐like (156) plants, indicating that the crm1‐D phenotype was caused by a single dominant mutation.
Figure 1.

Phenotypic comparison of crm1‐D and wild‐type plants. (a) Plant stature of Wt, crm1‐D and F1 plants (bar = 5 cm). (b) Leaf phenotypes of Wt (left) and crm1‐D mutant (right) at the tiller stage (bar = 1 cm). (c‐d) Transverse leaf sections of Wt (c) and crm1‐D mutants (d) at the tiller stage (bar = 0.5 cm). (e) Composite image of mature leaf blade of Wt from midvein to leaf margin. (f) Composite image of the mature leaf blade of the crm1‐D mutant. In order to better display the paraffin section results of crm1‐D, f is generated by crm1‐D multiple field photographs (using the Photoshop6 software overlap function). Red boxes represent the overlapping position of crm1‐D photographs with different vision fields. (g) Wild‐type leaf showing the placement of bulliform cells (BCs) between a major and minor vein (bar = 20 μm). (h) crm1‐D mutant leaf showing reduced size of bulliform cells (bar = 20 μm). (i‐k): (i) Leaf rolling index (n = 10), (j) bulliform cell area and (k) bulliform cell number of wild‐type and mutant plants (n = 10). Bars represent the SD of measurements. Asterisks denote significant differences based on Student’s t‐test (**P < 0.01).
A detailed histological examination of the crm1‐D mutant leaves identified no gross abnormalities in leaf ultra‐structure with the exception of bulliform cells that appeared smaller in the mutant relative to the wild type (Figure 1e, f). At the mature stage, the number of bulliform cells in crm1‐D was equivalent to that in wild type (Figure 1k), but the size of bulliform cells in crm1‐D was considerably smaller than that in wild type (Figure 1g, h, j). The reduced size of bulliform cells in crm1‐D was correlated with increased stomatal conductance (Figure S1b, Method S17), increased transpiration rates (Figure S1c), reduced photosynthetic capacity (Figure S1a) and higher internal CO2 level of the leaf (Figure S1d) relative to the wild type. Collectively, these results suggest that plant morphological changes in crm1‐D plants caused by the size of bulliform cells led to a reduction in photosynthetic efficiency.
Bulliform cells can be stained by toluidine blue O (Hernandez et al., 1999). The adaxial leaf crm1‐D and wild‐type plants at the 10th leaf stage were peeled, and bulliform cells and epidermal cells were stained purple and blue, respectively, with toluidine blue O (Figure S2a). The cross section of seedling leaves of crm1‐D and wild‐type plants were observed by scanning electron microscopy (Figure S2b). The size and area of bulliform cells in crm1‐D were significantly reduced compared with wild type (Figure S2a, b), again demonstrating decreased size of bulliform cells in crm1‐D compared with the wild type.
Map‐based cloning of CRM1
The crm1‐D mutant was coarsely mapped in an F2 population to the short arm of rice chromosome 6 between markers Indel6‐3 and Indel6‐5 (Figure 2a). To fine‐map the CRM1, we screened 890 individuals using markers designed to the region between Indel6‐3 and Indel6‐5 (Figure 2a). The CRM1 locus was further delimited to a 30‐kb region, which contained five putative open reading frames located between markers Indel1‐6 and Indel1‐7 (Figure 2a).
Figure 2.

Map‐based cloning of the CRM1 gene. (a) The CRM1 locus was mapped to a 5‐cM interval on chromosome 6 between Indel markers Indel6‐3 and Indel6‐5. The location of the CRM1 locus was fine‐mapped to a 30‐kb region between InDel markers 1–6 and 1–7 on BAC clone P0664C05 using 890 F2 homozygous plants. There were five open reading frames (ORF1‐ORF5) as potential candidate genes in this interval. (b) The structure of CRM1 (ORF3). Putative ATG start and TAA stop codons are shown. A 50‐nt deletion 658 bp downstream of TAA was detected in crm1‐D. (c) P1 and P2 primers shown in (b) were used to distinguish the CRM1 deletion region from wild type and crm1‐D in genomic DNA (left). PCR amplification of products from RNA (no bands) and cDNA confirmed the presence of a 50‐nt deletion present in crm1‐D transcripts. (d) Western blot analysis was performed using a Roc8 polyclonal antibody generated against the full‐length Roc8 fusion protein. (e) Comparison of transcript abundance of Roc8 in wild‐type (Wt) and crm1‐D mutant by qRT–PCR. (f) Comparison of protein abundance of Roc8 in wild‐type (Wt) and crm1‐D mutants by quantification of signal from Western blot using ImageJ analysis. Bars represent the SD of measurements. Asterisks denote significant differences based on Student’s t‐test (**P < 0.01).
To identify the candidate gene in this interval, quantitative PCR was used to measure transcript levels of each of the five ORFs (LOC_Os06g10580, LOC_Os06g10590, LOC_Os06g10600, LOC_Os06g10610 and LOC_Os06g10620) in the crm1‐D mutant and wild type. Surprisingly, no significant differences in gene expression in any of the genes were detected between wild‐type and crm1‐D plants in tissues from the 3rd leaf stage (Figure 2e, Figure S3a, b, c, d and Method S4). However, sequence analysis of the ORFs revealed a 50‐bp deletion located 650 bp downstream of the stop codon of LOC_Os06g10600 (Figure 2b). This gene was predicted to encode Roc8, a putative homeodomain leucine zipper class IV gene (Ito et al., 2002), closely related to Roc5, a gene previously shown to regulate leaf rolling in rice (Zou et al., 2011). The Roc8 gene may be an optimum candidate gene. Roc8 is predicted to encode a transcript of 3070 nucleotides with an open reading frame of 2094 nucleotides that direct the synthesis of a 697 amino acid protein of predicted molecular mass of 75 kD. The coding region of Roc8 consists of nine exons interrupted by eight introns (Figure 2b). The Roc8 protein is predicted to encode two conserved domains, a DNA‐binding HD‐ZIP at the N‐terminus and a putative lipid‐interacting START domain at the C‐terminus (Figure S5a). Roc8 shares 76.52% protein identity with the maize homolog OCL1, which is highly expressed in immature tassels, including meiotic tissues and anthers (Depege‐Fargeix et al., 2011). Roc8 also shares extensive sequence homology with GL2, previously shown to regulate trichome differentiation in Arabidopsis thaliana (Khosla et al., 2014).
Roc8 transcription profiling
Data submitted to the microarray transcript profiling database indicated that Roc8 was widely expressed in rice with strong expression in leaf tissue (Zimmermann et al., 2004). To validate these data, we conducted an expression analysis by qRT‐PCR using RNA samples prepared from wild‐type tissues at the seedling and mature stages. Roc8 transcript abundance was highest in young leaf blade and sheath tissues and lowest in root and culm tissues (Figure S4).
An in situ hybridization assay was used to detect the expression pattern of the Roc8 gene. Shoot apexes of cross section and longitudinal cutting of six‐day seedlings were probed with digoxigenin‐labelled sense or antisense strand of Roc8 RNA. The results showed that in the sense probe, there were no visible hybridization signals, whereas strong signals were expressed in the epidermal cell and vascular bundle using antisense probe hybridization (Figure 3a, b, c, d). This result indicated that Roc8 may function in epidermal cell differentiation and vascular bundle development.
Figure 3.

In situ hybridization and subcellular localization of Roc8. (a‐d) In situ hybridization using fragments from the partial coding sequence of the Roc8 gene. (a) Transverse or (c) longitudinal sections through the shoot apex of six‐day‐old seedlings were hybridized with a digoxigenin‐labelled (a, c) antisense probe or (b, d) sense strand probe of Roc8. Arrows denote both adaxial and abaxial expression of Roc8 restricted to the L1 layer of developing leaves. c, d: bar = 100 μm. a, b: bar = 20 μm. (e)–(h) A 35S::Roc8:GFP vector was transformed into rice protoplasts, and green fluorescent signals were visualized using confocal microscopy. DAPI staining of nuclei revealed that the GFP signal colocalized with DAPI fluorescence, indicating that Roc8 was exclusively targeted to the rice nucleus. e‐h: bar: 100 µm.
3′RACE (Rodriguez‐Cazorla et al., 2015) was conducted to define the 3′ end of the transcriptional unit of Roc8 in wild type. The results indicated that the crm1‐D mutant carried a 50‐bp deletion immediately upstream of the poly(A) signal in the 3′UTR of Roc8 (Figure 2c). The dominant nature of the crm1‐D mutant phenotype suggested that the sequences within the 50‐bp deletion might negatively regulate Roc8 protein expression in the wild type and might thus result in increased or ectopic expression of the Roc8 protein when deleted. To test this hypothesis, a polyclonal antibody was generated to the Roc8 peptide and used in Western blot analysis (Method S5). As shown (Figure 2d, f), Roc8 protein accumulated to approximately sevenfold higher levels in mutant leaf tissue relative to the wild type. To confirm that the overexpression of Roc8 was responsible for the mutant phenotype, we introduced a 4.8‐kb BAC genomic fragment containing the full‐length coding and regulatory sequences of the wild‐type CRM1 locus into Nipponbare callus (Figure 4a). A total of 50 independent transgenic lines were generated, and all phenocopied the original crm1‐D mutant in terms of plant stature (Figure 4b), leaf shape (Figure 4c, d, e), bulliform cell size (Figure 4f, g, h, j, k) and leaf rolling index (Figure 4i). The results strongly suggested that misexpression of Roc8 resulted in the crm1‐D phenotype. To facilitate the description, we used Roc8 to replace CRM1 in subsequent studies.
Figure 4.

Overexpression of Roc8 recapitulates the crm1‐D Phenotype. (a) Schematic of the pCRM1 expression vector containing the full Roc8 open reading frame derived by Pst1/EcoRI digestion of BAC P0664C05. (b) Plant stature of Wt and Roc8 overexpression in T1 plants (OE1, OE2) at maturity. (c‐e) Leaf phenotypes of (c) Wt, (d) OE1 and (e) OE2 at maturity (ab: abaxial side; ad: adaxial side, bar = 5 cm). (f) Transverse section of Wt leaf showing the placement of bulliform cells between a major and minor vein (bar = 20 μm). (g‐h) Transverse sections of (g) OE1 and (h) OE2 leaves showing a reduced size of bulliform cells (bar = 20 μm). (i‐j) Quantitative analysis of leaf traits in the Wt, OE1 and OE2 lines (i, j, k: n = 10). Bars represent the SD of measurements. Student’s t‐test was performed to determine significance: ** represents P < 0.01.
Roc8 is a transcriptional activator
As Roc8 belongs to an HD‐ZIP IV family member, it is predicted to encode a transcriptional activator but does not contain a classical nuclear localization signal. To test for subcellular localization of Roc8, green fluorescent signals were visualized using confocal microscopy (Chen et al., 2006). As expected, the GFP signal colocalized with DAPI fluorescence, indicating that Roc8 was exclusively targeted to the rice nucleus (Figure 3e, f, g, h, Method S7).
To decide whether Roc8 has transcriptional activation activity, the Roc8 full‐length coding sequence was fused in frame with the pGBKT7 vector, and pGBKT7 was regarded as a negative control. Yeast cells containing pGBKT7‐Roc8 and the control were added to selective medium lacking Trp, His and Leu. As shown in Figure S5b, the clones (negative control) could not grow at all on medium lacking Trp, His and Leu. In contrast, the pGBKT7‐Roc8 strain exhibited gradual growth at 10‐1, 10−2 and even 10−3 dilutions and showed blue colour plus X‐α‐gal chromogenic reagent (Figure S5b). These results indicate that Roc8 has transcriptional activation activity.
Roc8 had three main motifs, containing the N‐terminus HD‐Zip domain (amino acids 1 to 200), START domain (amino acids 201 to 460) and C‐terminus (Figure S5a). To determine whether the N‐terminus, START domain or C‐terminus is responsible for transcription activation, Roc8 deletion vectors (Roc8‐A, B, C) fused with pGBKT7 were constructed. Among the three constructs, yeast cells containing the START domain only grew well (Figure S5b), indicating that the START domain of Roc8 may be responsible for its transcriptional activation activity.
Roc8 3′‐UTR functions as a translational repressor
Three experiments were designed to verify Roc8 3′‐UTR function. First, GFP constructs containing different Roc8 3′‐UTRs were transiently transformed into rice protoplasts. GFP fluorescence, as well as GFP protein and mRNA levels, was tracked and compared to the control GFP‐Roc8 lacking the 3′‐UTR (Method S9). All the constructs localized to the nucleus, indicating that the Roc8 3′‐UTR did not influence Roc8 cellular localization. As expected, control GFP‐Roc8 was strongly expressed in the nucleus, whereas the constructs (GFP‐Roc8‐3′UTR‐FL and GFP‐Roc8‐50b) could significantly inhibit GFP fluorescence signals. Construct (GFP‐Roc8‐3′UTR‐△M) did not repress the GFP fluorescence signals in protoplasts under the same settings (Figure S6a). The reduction in GFP fluorescence was correlated with a reduction in GFP protein but not in GFP mRNA levels, as shown by Western blot and RT‐PCR analyses, respectively. The RT‐PCR experiment showed that the reduced GFP proteins were not caused by GFP mRNA levels (Figure S6b, c). The results indicate that Roc8 3′‐UTR 50‐bp sequence may have a post‐transcriptional regulation function.
Second, to explore whether the 3′‐UTR mediated Roc8 translation directly, we used the dual‐luciferase reporter assay in rice protoplasts to judge the Roc8 3′‐UTR effect (Method S10). The luciferase gene fused vectors bearing the Roc8 3′‐UTR deletion versions or the 35S terminator (35S Ter) were selected as reporters. A Renilla luciferase gene with a 35S terminator in its 3′‐UTR was cotransfected to normalize the transfection efficiency (Figure S7a). The ratio of relative luminescence/mRNA level was considered to estimate the relative translational efficiency. The results showed that the complete Roc8 3′‐UTR and the partial 3′‐UTR containing the 50‐bp sequence remarkably repressed luciferase activity, but the Roc8 3′‐UTR lacking the 50‐bp sequence had no obvious repression effect (Figure S7b). Furthermore, we replaced the 3′‐UTR 50‐bp sequence with an unrelated sequence (3′‐UTR‐50‐R1 or 3′‐UTR‐50‐R2). Compared to the complete 3′‐UTR (3′‐UTR‐FL), the ratio of the relative luminescence/mRNA level in the 3′‐UTR‐50‐R1 or 3′‐UTR‐50‐R2 constructs increased approximately threefold (Figure S7a, b). These results indicate that the Roc8 3′‐UTR 50‐bp sequence is a functional translational repression factor.
Third, a stable transformation experiment was used to test the Roc8 3′‐UTR functions (Method S15). Transgenic studies suggested that the 50‐bp deletion of the Roc8 3′UTR in crm1‐D resulted in the overproduction of Roc8‐encoded protein. The 50‐bp sequence comparisons suggested that 20 of 50 base pairs were conserved across multiple monocot and dicot species (Figure S8a, b). Furthermore, the deletion spanned a region required for a predicted stem–loop structure (Figure S8c). We speculated that a secondary stem–loop structure present in the 3′‐UTR of Roc8 was required for efficient target repression and that deletion of this sequence results in ectopic expression of the Roc8 transcript. To explore this hypothesis, we overexpressed the full‐length wild‐type Roc8 gene with its native 3′‐UTR (Roc8‐FL), the full‐length Roc8 gene including its 3′‐UTR (deletion 50 bp) (Roc8‐FL‐△M) and the complete coding sequence without the 3′‐UTR (Roc8‐FL‐△U) in the Nipponbare background (Figure 5a). As expected, the Roc8‐FL overexpression lines showed a semi‐rolled leaf phenotype (LRI: 0.72 ± 0.01; Figure 5b, c). However, the same transcript carrying a 50‐bp deletion in the 3′UTR (Roc8‐FL‐△M) resulted in a much stronger rolling phenotype (LRI: 0.82 ± 0.05), which was similar to plants carrying a construct that completely lacked the 3′ UTR region (Roc8‐FL‐△U) (LRI: 0.84 ± 0.06) (Figure 5b, c). Interestingly, equivalent levels of Roc8 mRNA were observed in the Roc8‐FL and Roc8‐FL‐△M, Roc8‐FL‐△U overexpression lines (Figure 5d), but the Roc8 protein level was higher in the Roc8‐FL‐△M, Roc8‐FL‐△U (threefold increase compared to the wild type; Figure 5e). The above findings strongly suggest that the 50‐bp region in the Roc8 3′ UTR negatively regulates the expression of Roc8 activity and that the deletion of this sequence mediates the enhanced accumulation of Roc8 protein.
Figure 5.

Features in the Roc8 3′‐UTR function as translational repressors. (a) Roc8 expression constructs. Transgenic expression of full‐length and deletion derivatives of the Roc8 gene are shown; the predicted CDS is in black, 3′‐UTRs are in white, and the 50 bp deletion found in the crm1‐D allele is shown in blue. All constructs were driven by the OsActin promoter as detailed in the Method S15. (b, c) Transgenic plants showing (b) representative T1 plants at maturity and (c) close‐up images of leaves and leaf rolling index of plant expression Roc8 vectors detailed above images (ab: abaxial side; ad: adaxial side). The nontransformed wild‐type (Wt1.1) control is shown at an equivalent developmental stage (bar = 5 cm). LRI: leaf erect index, n = 10. (d) Quantitative PCR assays of Roc8 mRNA levels in T1 transgenic and corresponding near‐isogenic null segregants (n = 3). Bars show the mean expression ± SD (**P < 0.01, Student's t‐test). (e) Semiquantitative analysis of Roc8 protein levels as measured by Western blotting using the Roc8 antibody in T1 transgenic lines and corresponding near‐isogenic null segregants. Actin was used as a loading control, and band intensities were quantified using ImageJ analysis of Western blots. The fold change shown above the blot is relative to Wt controls normalized by actin in each lane.
Roc8 knockdown lines increased the size of bulliform cells and caused rolling outward
Since crm1‐D is a gain‐of‐function mutant, a Roc8 deletion mutation was created using CRISPR/Cas9 technology for further analysis. Two lines, roc8‐m1 and roc8‐m2, contained a 10‐bp deletion in the 2nd exon (from 123‐132 bp) and a 5‐bp deletion in the 2nd exon of Roc8 (from 138‐142 bp), respectively, causing early translation termination (Figure 6a, b). Quantitative PCR analysis in roc8‐m1 and roc8‐m2 plants determined that the Roc8 mRNA expression level was reduced significantly in the roc8‐m1 and roc8‐m2 plants, which indicated that roc8‐m1 and roc8‐m2 plants were knockdown mutants (Figure 6c, d). Furthermore, roc8‐m1 and roc8‐m2 plants showed an obvious rolling outward phenotype (Figure 6e, f, g), accompanied by increased bulliform cell size and leaf rolling index (Figure 6h, i, j, l, m) and normal bulliform cell numbers. The physiological measurements showed that roc8‐m1 and roc8‐m2 had higher photosynthesis, lower stomatal conductance, lower transpiration rate and lower Ci than the wild type (Figure S9a, b, c, d; Method S17). These results further suggest that Roc8 negatively regulates the size of bulliform cells.
Figure 6.
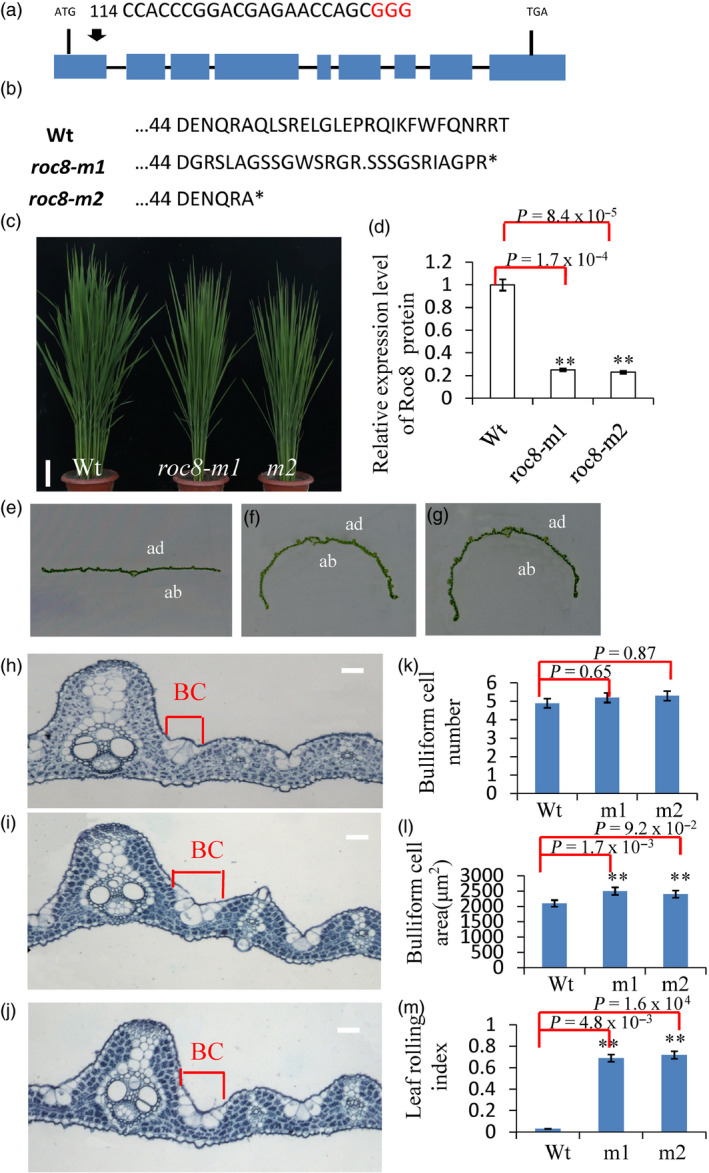
CRISPR/Cas9‐induced mutations in Roc8. (a) Schematic map of the genomic region of Roc8 and the sgRNA target site; arrow shows the sgRNA target site on the Roc8 genomic sequence; the PAM motif (NGG) is shown in red. Blue boxes represent Roc8 exons, and the black lines indicate intron sequences. (b) Amino acid alignment surrounding the sgRNA target region showing the predicted peptide sequence of wild‐type and mutant alleles. The site of the predicted frame‐shifted sequence is underlined, and new stop codons are shown by asterisks. (c) The phenotype of Wt, roc8‐m1 and roc8‐m2 homozygous mutants at the tillering stage (bar = 5 cm). (d) Quantitative PCR analysis showing Roc8 expression levels in Wt, roc8‐m1 and roc8‐m2 homozygous mutant plants. Actin was selected as an internal reference. (e‐g) Leaf phenotypes of (e) Wt, (f) roc8‐m1 and (g) roc8‐m2 homozygous mutants at maturity (ab: abaxial side; ad: adaxial side). (h‐j) Mature leaf showing the extent of bulliform cell area between a major and minor vein in (h) Wt, roc8‐m1 (i) and roc8‐m2 (j) homozygous mutants (bar = 20 μm). (k‐m) Bulliform cell characteristics in Wt and mutant plants, including (k) cell number, (l) area and (m) leaf rolling index (n = 10). Bars represent the SD of measurements. Student’s t‐test was performed to determine significance: ** represents P < 0.01.
Double mutant analysis between Roc8 knockdown and Roc5 knockout mutants
Rice outermost cell‐specific genes (Roc) are transcription factors that play an important role in epidermal cell fate. Roc5 negatively regulated bulliform cell fate and leaf development (Zou et al., 2011). Ito reported that the Roc family could form heterodimers to act (Ito et al., 2002). The knockdown mutants of Roc5 and Roc8 had similar leaf rolling towards the abaxial surface. We investigated whether Roc5 and Roc8 proteins interact in vitro. A yeast two‐hybrid assay verified that Roc5–AD could interact with Roc8–BD (Figure S9, Method S8). The double mutant was created by crossing roc8‐m1 and roc5 knockout mutant (Zou et al., 2011). F1 progeny showed a normal leaf shape phenotype. The double mutant was obtained by PCR amplification and DNA sequencing in F2 progeny. The leaf rolling degree in the double mutant was more serious than that in the single mutant (Figure S10a, b, c, d, e). The anatomical analyses in the double mutant (roc5/roc8‐m1) revealed that the number and size of the bulliform cells increased considerably compared to a single mutant (Figure S10f‐l). The expression analysis of the Roc5 gene in the crm1‐D mutant or the expression analysis of the Roc8 gene in roc5 mutants showed no significant difference compared with wild type (Figures S14, S15). The above results indicated that Roc8 and Roc5 may function redundantly for the leaf rolling phenotype.
Identification of Roc8 downstream pathways
Transcriptome analysis was used to determine Roc8 downstream gene expression. Because Roc8 expression was the highest at the 3rd leaf stage, triplicate mRNA samples of the crm1‐D plants and wild‐type plants were harvested (Method S6). Transcriptome data analysis indicated that 317 genes were up‐regulated and 251 genes were down‐regulated in crm1‐D plants (differentially expressed genes with more than twofold change (P < 0.01); Dataset S1). The differentially expressed genes mainly involved photosynthesis, organ compound metabolic process, generation of precursor metabolites and energy. Careful transcriptome data analysis identified that some genes related to the development of bulliform cells were inhibited in the crm1–D mutant (Table S2). Therefore, we selected five vacuolar cell developmental marker genes for qPCR experiments. The vacuolar cell development genes were generally inhibited (Figure S11). Additionally, some genes controlling bulliform cell development, such as Roc5, NRL1, RL14, SRL1, ZHD1, OsMYB103L, AGO7 and OsLBD3‐7, were selected for qRT‐PCR detection in the crm1‐D mutant. The results showed that except for the SRL1, OsMYB103L and OsLBD3‐7 genes, the expression levels of other genes were not significantly different in the crm1‐D mutant compared to the wild type (Figure S12). The above experiments indicated that Roc8 may be involved independent of or partially dependent on the changed genes to regulate the development of bulliform cells.
The L1 box (AACATTTA) is well conserved within promoter regions of the regulated target genes of all L1‐specific HD‐Zip IV genes analysed to date (Ohashi., 2004; Zhang et al., 2010; Zou et al., 2011). Of all bulliform development‐related genes, only the promoter of the OsLBD3‐7 gene (LOC_Os03g57670) contains an L1 box located approximately –487 to –494 bp from the transcription start site. The OsLBD3‐7 expression level in crm1‐D increased more than threefold compared to the wild type (Figure S12).
The OsLBD3‐7 gene was reported to regulate the size of bulliform cells. Overexpression of OsLBD3‐7 could induce adaxially rolled leaves in rice (Li et al., 2016). We sought to determine whether the Roc8 gene could bind to the promoter of the OsLBD3‐7 gene. First, vectors containing 3XL1 boxes and Roc8‐AD were constructed and imported into yeast. The strains could grow in SD medium (‐TLH + 3‐AT), but the control did not survive in SD medium (‐TLH + 3‐AT; Figure 7. b). Second, a luciferase activity assay was used to detect whether Roc8 can activate OsLBD3‐7. In the rice protoplasts cotransfected with the effector and reporter vectors, the ratio between LUC (firefly luciferase) and Ren (Renilla luciferase) of the effector OsLBD3‐7pro‐LUC was threefold higher than that of the empty vector control (Figure S13, Method S13). Third, a chromatin immunoprecipitation (ChIP) assay was performed using the Roc8 antibody (Method S14). As shown in Figure S14, the P3 region of the OsLBD3‐7 promoter was more enriched than other regions using Roc8 antibody precipitation (Figure S14). The results showed that the OsLBD3‐7 gene was one of the candidate targets of Roc8 and Roc8 as a transcriptional activator that positively regulated the size of bulliform cells by directly activating the OsLBD3‐7 gene.
Figure 7.

Roc8 regulated lignin content in rice leaves. (a) Schematic of the OsLAC17 promoter region showing regions of the promoter used for PCR amplification. The L1 box is located approximately −679— −617 base pairs upstream of the OsLAC17 start codon. (b) A yeast one‐hybrid assay was used to test Roc8 binding to the L1 box. The AD‐only vector was used as a control. (c) Anti‐Roc8 ChIP assays were performed on etiolated seedlings at the 3rd leaf stage. (d‐f) Histochemical analysis of lignin in cross sections of leaf vascular bundle using phloroglucinol staining in the (d) homozygous crm1‐D (e) Wt and (f) roc8‐m1 mutant. Bar: 25 μm. (g) Lignin contents were measured in mature leaf blades and sheath tissues (n = 3).
Roc8 knockdown mutant reduced lignin contents without production penalty
Notably, many genes involved in cell wall lignin formation metabolism and cell wall cellulose formation also displayed a marked change in crm1‐D mutants in differentially expressed data (Table S2). Several celluloses and lignin‐related genes were selected for qPCR tests. The results further verified that cellulose formation and lignin synthesis genes were greatly increased in the crm1‐D mutant (Figure S16a, b). The promoters of these genes were used to determine whether they contain the L1 box. Among these genes, only the OsLAC17 and OsLAC22 promoters contained the L1 box, ranging from −679 to −671 bp and from −1178 to −1170 bp from the transcription start site, respectively. We speculated that the OsLAC17 and OsLAC22 genes may be potential target genes of Roc8. Li reported that OsLAC17 was responsible for lignin biosynthesis (Li et al., 2017); thus, OsLAC17 was selected as a candidate gene for subsequent analysis. We further tested whether Roc8 could bind to OsLAC17 chromatin to mediate lignin biosynthesis. First, a luciferase activity assay was used to detect whether Roc8 can activate OsLAC17. In the rice protoplasts cotransfected with the effector and reporter vectors, the ratio between LUC (firefly luciferase) and Ren (Renilla luciferase) of the effector OsLAC17pro‐LUC was significantly threefold higher than that of the empty vector control (Figure S13). Second, a chromatin immunoprecipitation (ChIP) qPCR assay was performed using the Roc8 antibody. As shown in Figure 7a, c, the P3 region of the OsLAC17 promoter was more enriched than other regions using Roc8 antibody precipitation. The results showed that the OsLAC17 gene was also one of the candidate targets of Roc8 and was activated by Roc8.
To further confirm whether the lignin changed in the mutant, the phloroglucinol dye assay was used to display lignin changes. The phloroglucinol–HCl staining assay showed stronger red staining of the leaf vascular bundle in the crm1‐D mutant and weak red staining of the leaf vascular bundle in roc8 knockdown lines compared to the wild type (Figure 7d, e, f). Next, leaf blade/sheath lignin contents in the crm1‐D mutant and the wild type were measured (Method S12). The crm1‐D mutant retains a significantly higher lignin content in leaves (Figure 7g). In roc8 knockdown lines, the lignin contents of the leaf blade and sheath were reduced to 75%–85% of the wild type. Notably, roc8 knockdown lines m1 and m2 showed excellent agronomic traits, such as normal height, normal seed set rate (Figure 6, Table S1), higher photosynthetic efficiency, lower transpiration rate and higher 1000‐grain weight (Figure S17, Table S1, Method S16). Taken together, the results of this study indicate that Roc8 also positively mediated lignin biosynthesis without the production penalty. This result indicated that knocking down the Roc8 homologous gene in forage grass may have strong practical application prospects in cultivating low lignin materials without a production penalty.
Discussion
The epidermis is the outermost cell layer of a plant leaf and provides a barrier that allows maintenance of leaf water homeostasis (Martin and Glover, 2007). Epidermal cells are specially differentiated cells and have different cell types, such as bulliform cells, cuticular trichome structures and stomata. Extensive studies have established regulatory mechanisms controlling epidermal cell fate in Arabidopsis and other species (Cheng et al., 2014; Le et al., 2014; Takada and Iida, 2014; Takada et al., 2013; Wang and Chen, 2014). Bulliform cells belong to the monocotyledon‐specific epidermal cell structure. Only by studying monocotyledon model plant leaf morphology can we elucidate this kind of cell regulation mechanism. At present, much progress has been made in regulating the development of bulliform cells (Xu et al., 2018). However, the fine regulatory network governing this process has not been elucidated. In this study, a constitutively rolling leaf mutant, crm1‐D, was obtained from our previous rice mutant population (Wan et al., 2009; Zou et al., 2014), and the corresponding cloned gene was Roc8 (Figure 2), a member of the HD‐Zip IV family. Sequence analysis revealed that the 50‐bp deletion occurred in the Roc8 3′‐UTR in crm1–D (Figure 2). A series of experiments showed that the 50‐bp sequence in the Roc8 3′‐UTR represses Roc8 at the translational level in vitro and in vivo (Figures 5, S6, S7). The size of bulliform cells was increased in Roc8 knockdown lines (Figure 6). Moreover, a series of assays, including the transcriptome profile, qPCR analysis, yeast two‐hybrid assays, luciferase activity detection and ChIP experiments, confirmed that Roc8 acted as a transcriptional activator and bound to the OsLBD3‐7 promoter, regulating the size of bulliform cells (Figures S12,S13,S14). Therefore, our work showed that Roc8 plays an essential role in the formation and development of epidermal bulliform cells in rice. HD‐Zip IV family genes seem to have a conserved function in regulating epidermal cell fate both in dicots and monocots.
Roc8 and Roc5 play a redundant role in regulating the size of bulliform cells in rice
Leaf rolling is a simple and general phenotype that is easy to observe by the naked eye and thus has received more attention from traditional breeders. However, this phenotype is not due to a simple development process and is regulated by multiple tissue cells. At present, there are many factors that affect leaf rolling in rice, including changes in the number and size of bulliform cells, programmed cell death of sclerenchyma cells on the adaxial/abaxial surface (Zhang et al., 2009) and cuticle development (Wu et al., 2011). Bulliform cells generally occur intercostally as long strips several cells wide. Bulliform cells aligned at the abaxial side with linear patterning by positional information in maize, similar to the trichome pattern in Arabidopsis (Hernandez et al., 1999). In Arabidopsis, trichomes are single cells and never linked to each other; bulliform cells are arranged in clusters of several cells, and the clusters are not adjacent to each other in rice. Our study revealed that Roc8, a member of the rice outermost cell‐specific gene (Roc) family, plays an important role in epidermal cell fate. At present, Roc1, Roc4 and Roc5 have been clarified in detail. The expression of Roc1 in the outermost cells may be dependent on the positional information of cells in the embryo or meristemoid cells prior to cell fate determination of the protoderm (epidermis; Ito et al., 2002). Roc4 negatively regulates cuticular wax biosynthesis by E3 ligase degradation in rice (Wang et al., 2018). In contrast to Roc1 and Roc4, Roc8 and Roc5 play a redundant role in regulating the size of bulliform cells in rice. First, overexpression in Roc8 and Roc5 caused leaf rolling with adaxially curved leaves. Suppression in Roc8 and Roc5 caused leaf rolling with abaxially curved leaves. Second, the size of bulliform cells increased in Roc8 knockdown and Roc5 knockout plants but was reduced in Roc5‐overexpressing lines and crm1‐D lines. Third, Roc5 and Roc8 have similar expression patterns. Roc5 is mainly expressed in the L1 layer of the meristem but not in mature leaves (Ito et al., 2002). In situ hybridization assays showed that Roc8 was expressed in epidermal cells and vascular bundles (Figure 3), suggesting that Roc5 or Roc8 may participate in the establishment of bulliform cells, rather than their maintenance in the adaxial epidermis. Fourth, the yeast two‐hybrid assays showed that Roc5 interacted with Roc8 in vitro, which was consistent with Roc family members acting as heterodimers (Ito et al., 2002). Fifth, cytology analysis showed that the size of bulliform cells in the double mutant was significantly increased compared with that of the single mutant.
Although the two genes Roc5 and Roc8 are functionally redundant in controlling the development of bulliform cells, the developmental processes controlling cell expansion and division may be slightly different. First, the Roc8 expression pattern was slightly different from the Roc5 expression pattern. From the Roc8 in situ hybridization pattern (Figure 3) and Roc5 GUS analysis (Zou et al., 2011), Roc8 expression began earlier than Roc5 expression. Roc8 expression began before the 3rd stage, whereas Roc5 expression began at the 3rd stage. This finding is consistent with the period of the respective phenotype. Second, Roc8 mainly controls the size of bulliform cells, while Roc5 primarily controls the number and size of bulliform cells. We speculate that Roc8 may be the main regulator of bulliform cell expansion, while Roc5 may be the regulator of bulliform cell division. Quantitative PCR analysis showed that in the roc8‐m1 and roc8‐m2 lines (knockdown mutants), the expression of some genes controlling cell expansion, such as OsXTH9, OsXTH11, OsEXPA2, OsEXPA4, OsEXPA5 and OsEXPB11, increased significantly (Figure S17a), while in the roc5 mutant, some genes controlling the cell cycle, such as the cell cycle proteins OsCycA1;2 and OsCycA2;1, increased significantly (Figure S17c). Additionally, the expansion genes, such as OsEXPA2, OsEXPA4, OsEXPB11 and OsXTH11, increased slightly in roc5 (Figure S17a), which also indicated that Roc8 and Roc5 were partially redundant in controlling the size of bulliform cells.
Potential regulation mechanism of bulliform cell expansion
Although we have found two direct target genes, how could Roc8 regulate the cell expansion of bulliform cells in rice? We think that Roc8 can regulate the expansion of bulliform cells by the following possible mechanisms. First, the expansion of bulliform cells needs to change the composition of the cell wall. The nrl1 mutant showed reduced leaf width, half‐rolled leaves and dwarfing. The nrl1 mutant had a smaller bulliform cell size. The Nrl1 gene encodes the cellulose synthetase gene (Hu et al., 2010). Roc8 can induce the expression of xyloglucan endotransglucosylase/hydrolases (XTHs) and expansion genes, and whether Roc8 directly binds with its promoter needs to be further verified. Second, gibberellin biosynthesis and auxin biosynthesis are involved in the developmental process of bulliform cells. YAB1 was involved in feedback regulation of GA3 biosynthesis in rice. The YAB1 RNAi plants rolled abaxially to form a cylinder‐like structure (Dai et al., 2007). Nal7 encodes a riboflavin monooxygenase of the YUCCA family member. The nal7 mutant showed narrowed leaves and reduced auxin contents. The number and size of bulliform cells in nal7 were reduced, resulting in the leaf rolling phenotype (Fujino et al., 2008). The cow1 mutant, an allele of Nal7, results in rupture of the largest bulliform cell and consequently rolled leaves (Woo et al., 2007). In the transcriptome data and quantitative PCR analysis, auxin synthesis‐related genes showed significant changes in the mutant. Roc8 may regulate the size of bulliform cells by affecting auxin synthesis genes (Figure S18b) but is not involved in the gibberellin biosynthesis pathway (Figure S19). Third, the acid growth theory was used to explain cell expansion (Moloney et al., 1981). Roc8 activated OsLBD3‐7. Lateral organ boundary domain families (LBDs) and auxin‐responsive genes (ARFs) are often involved in auxin signalling pathways (Kim and Lee, 2013). Deregulation of the OsmiR160 target gene OsARF18 leads to leaf rolling in rice by affecting auxin signalling (Huang et al., 2016). Auxin activated the PMAs and acidified the intercellular space. Changes in apoplastic pH activity result in higher turgor pressure between the outer and inner membranes, thereby enabling cell expansion (Hager., 2003). Roc8 may affect cell expansion in all the above ways, and the identification of direct Roc8 target genes should help to clarify this question.
Roc8 3′‐UTR functions as a translational repressor
The 3′‐UTR plays an important role in the growth and development of animals and plants. At present, the mechanism of this region primarily affects transcription level, post‐transcriptional modification and protein translation (Li et al., 2015; Shen et al., 2017). Extensive evidence supports a 3′‐UTR as a regulatory factor that could influence the expression of adjacent genes (Kumar, 2014; Silveyra et al., 2014). The 3′‐UTR could alter adjacent gene transcription through epigenetic modifications (Mao et al., 2015; Wei et al., 2014). We first analysed the effect of crm1‐D on Roc8 DNA methylation using bisulphite genomic sequencing. The CG, CHG and CHH methylation levels were unchanged (Figure S20 and Method S11). This finding supported the conclusion that the mRNA level of Roc8 in crm1‐D did not vary and that the 3′‐UTR did not regulate Roc8 gene expression at the transcriptional level. Shen reported a translational repression mechanism mediated by a stowaway‐like MITE (sMITE) site embedded in the 3′‐UTR of Ghd2 (Shen et al., 2017). Klein‐Cosson provided evidence for the existence of a 24‐nt small RNA, which was complementary to the 3′‐UTR of OCL1 (Outer Cell Layer1; Klein‐Cosson et al., 2015). These researchers argued that it was possible for a secondary stem–loop structure present in the 3′ UTR of OCL1 to be an efficient target for repression. The 3′‐UTR of OCL1 not only influences the OCL1 mRNA level but also affects the translational level (Klein‐Cosson et al., 2015). However, this conclusion lacked more genetic data to support this hypothesis. In this study, using a series of in vitro and in vivo experiments, we verified that the Roc8 3′‐UTR does not only affect the expression of RNA but also affects the protein level. 3′‐UTR analyses of Roc8, Roc5, Roc4, Roc6, ZmOCL1, ZmOCL4, AtANL2 and AtHDG1 (Figure S8) showed that a conserved 20‐bp sequence exists in HD‐Zip IV families, indicating that some gene duplications in dicots and monocots occurred before their divergence.
We further analysed the possible regulation mechanism of the 50‐bp sequence as follows. (i) This region may be siRNA or miRNA target sites, similar to OCL1 in rice (Klein‐Cosson et al., 2015). We predicted that the 3′ UTR of Roc8 contained a miRNA‐ or siRNA‐binding region, but there were no relevant miRNAs or siRNAs complementary to the 3′ UTR of Roc8 in the current database. The search for miRNA or small RNA precursors targeting the 3′ UTR of Roc8 would be a target for future research. (ii) This region may be a transposon regulatory region, and some of the smaller transposons are small reverse repeat transposon elements whose transcription instinct forms a stable stem–loop structure. (iii) The stem–loop structure may serve as a termination element, which may be similar to poly(A) as a termination signal (Wang et al., 2019). Roc8, as a transcription factor, may regulate multiple physiological processes of plant development. Finely controlling the Roc8 mRNA or protein level is necessary to improve plant adaptability. The next step is to determine which factors affect the binding of this target region and whether other RNA‐binding proteins (RBPs) or regulatory factors are needed to participate in this process of translation repression.
Roc8 positively regulated leaf lignin biosynthesis
Lignin is a complex organic polymer that forms important structural materials in vascular plants and some algae supporting tissues (Vanholme et al., 2012). Lignin plays an important role in the structure and repair mechanism of the plant cell wall, and scientists hope to modulate lignin contents by genetic engineering (Halpin, 2019). In this study, by analysing the transcriptome data, some controlling lignin biosynthesis genes increased in crm1‐D. Five additional experiments confirmed the hypothesis that Roc8 positively regulated leaf lignin contents. First, qPCR verified that the OsLAC17 expression level was significantly increased in the crm1‐D mutant. Second, a chromatin immunoprecipitation (ChIP) assay using the Roc8 antibody in wild‐type protoplasts confirmed that Roc8 could directly bind to the promoter L1 region of the OsLAC17 gene. Third, in the rice protoplasts cotransfected with the effector and reporter vectors, the ratio between LUC (firefly luciferase) and Ren (Renilla luciferase) of the effector OsLAC17pro‐LUC was significantly (threefold) higher than that of the empty vector control. Fourth, the phloroglucinol histochemical analysis in leaf vascular bundle cross sections showed that there was a higher signal in the walls of the xylem cells in the crm1‐D mutant and a lower signal in the Roc8 knockdown lines compared to wild‐type plants. Fifth, the lignin content measurements showed that there were higher lignin contents in crm1‐D and lower lignin contents in Roc8 knockdown lines. The above five experiments strongly verified that Roc8 positively regulated the lignin contents in rice.
Many transcription factors, such as OsTF1L, OsIDD2 and OsMYB103L, participate in the regulation of lignin biosynthesis in rice. Overexpression of OsTF1L, a rice HD‐Zip transcription factor, promoted lignin biosynthesis (Bang et al., 2019). OsIDD2, a zinc finger and indeterminate domain protein, regulates secondary cell wall formation (Huang et al., 2018). OsMYB103L, an R2R3‐MYB transcription factor, influenced leaf rolling and mechanical strength in rice (Yang et al., 2014). Simultaneously, overexpression of these genes often caused unfavourable developmental defects, such as stunted growth, dwarf, brittle culms and decreased fertility, which limited their application prospects. Notably, the lignin contents decreased slightly in Roc8 knockdown lines (75%–85%). These lines not only did not exhibit adverse agronomic traits but also showed high photosynthetic efficiency due to plant keeping moderate rolling, making the leaves upright, more leaves exposed to light and enhancing the light quantity and intensity at the base of the canopy, suggesting that the use of Roc8 or homologous genes in the production of materials with lower lignin content has important practical application value.
The results of this study support a proposed model illustrating the Roc8 mechanism (Figure S21). We verified the regulatory function of the 3′‐UTR of Roc8 by forward genetics and provided substantial evidence to clarify Roc8 function by regulating the size of bulliform cells by activating the OsLBD3‐7 gene. In addition, Roc8 bound to the promoter of the OsLAC17 gene, which could be involved in modulating lignin biosynthesis. By reverse genetics, we created Roc8 knockdown lines using gene editing technology and revealed that the Roc8 knockdown lines not only improved the photosynthetic efficiency because of leaf morphology change from the large size of bulliform cells but also had lower lignin contents without the production penalty. Future research may obtain functional clues for combining other Roc homologous genes to cultivate high photosynthetic efficiency crops in rice or low lignin contents in forage grass.
Materials and methods
Material screening and growth condition
In previous research, a rice mutant population was constructed using the Japonica cultivar Nipponbare as the recipient (Wan et al., 2009). The crm1‐D mutant was screened from this population. Genetic analysis was conducted by cross test between crm1‐D and Nipponbare (wild type). A map of the F2 population was created from a cross between crm1‐D and Dular (Indica). The materials were grown under natural growth conditions in a paddy field (Zou et al., 2014).
Positional cloning of Roc8
An initial set of crm1‐D/Dular F2 progeny was used for coarse linkage analysis; fine mapping was then performed based on 890 homozygous mutant F2 segregants. The polymorphic InDel markers used to genotype these progenies are presented in Table S3. The positional cloning strategy was previously described (Zhang et al., 1994). The amplicons were separated on 6% polyacrylamide gels and visualized by silver staining. Finally, products in the candidate region were amplified, sequenced and aligned. The mutant region was verified using primer pairs p1/p2 between wild‐type and crm1‐D DNA or cDNA.
Recapturing the crm1‐D phenotype
BAC clone P0664C05 containing the Roc8 gene was first cut by PstI and EcoRI restriction enzymes. The product carrying a 1.8 kb Roc8 native promoter, full‐length Roc8 coding region and 1 kb 3′‐UTR were ligated into pCambia2300 with the selectable marker NPTII. The expression vector was introduced into the Agrobacterium tumefaciens strain AGL1 using the heat‐shock method and then transformed into Nipponbare calli. The regenerated plant leaves in T1 progeny were harvested, and histological observations were conducted. The areas and numbers of bulliform cells were numbered as mentioned above. Ten independent visual fields were counted (n = 10).
Creation of the Roc8 knockdown lines
Roc8 knockdown plants were generated using CRISPR/Cas9. The sgRNAs were designed to target Roc8 locations. The single sgRNA was created in the BGK03 vector containing Cas9, which was introduced into Agrobacterium strain EHA105 and transformed into Nipponbare. Eight independent lines of sgRNA were obtained. To examine the function of CRISPR/Cas9 in vivo, genomic DNA was extracted from transgenic plants, and primer pairs flanking the designed target site were used for PCR amplification (Table S3). Sequence alignment revealed that Roc8 knockdown lines were obtained.
Phloroglucinol–HCl staining test
Fresh mature leaves were collected and carefully cut with razor blades. The leaves were moved to the slide and stained with phloroglucinol (1%) staining liquid for two minutes. After staining, concentrated hydrochloric acid was added to the slide (Srivastava, 1966). The photographs were taken with a light microscope (Zeiss). (n = 3).
Histology and cytology observation
Histology, cytology, TEM analysis and in situ hybridization were described in Methods S1, S2 and S3.
Statistical analysis
Bars show the mean value ± s.d. Student’s t‐test analysis was used to test for significant differences between the above lines (*P < 0.05, **P < 0.01).
Primer sequences
The primers used in this study are listed in Table S3.
Conflicts of interest
The authors declare no conflicts of interest.
Author Contribution
Z.G.Z. and L.T.G designed the experiments and wrote the manuscript. S.J, C.X.A and T.S.Z performed map‐based cloning of Roc8. Z.K.L performed the gene expression analysis. W.Y.W performed ChIP experiment and Western blotting test. C.Z.H performed subcellular localization assays. W.J.X, S.X.H and A.P.F conducted the transgenic experiment. W.P.Q gave critical advice. Z.G.Z and T.G.L designed the total experiments.
Supporting information
Figure S1 Photosynthesis parameter measurements from the crm1‐D plant and wild type at the flowering stage.
Figure S2 Toluidine blue O staining and scanning electron microscopy observations of cross‐sections of seedling leaves in Oryza sativa.
Figure S3 Comparison of transcript abundance of candidate genes in wild‐type (Wt) and crm1‐D mutant by qRT–PCR.
Figure S4 Expression pattern analysis of Roc8 at the seedling stage and mature stage.
Figure S5 Protein structure, transactivation analysis of Roc8.
Figure S6 Fluorescence detection of GFP‐Roc8 3′‐UTR with different fusion constructs using a rice protoplast transient transformation system.
Figure S7 Roc8 3′‐UTR regulates luciferase activity.
Figure S8 Roc8 3‐UTR sequence analysis.
Figure S9 A yeast two‐hybrid interaction assay was performed with Roc5 and Roc8.
Figure S10 Double mutant analysis of loss‐of‐function alleles of roc5/roc8.
Figure S11 Expression analysis of vacuolar development‐related genes.
Figure S12 Bulliform cell development‐related gene expression analysis in rice.
Figure S13 LUC transient transactivation assay in rice protoplasts.
Figure S14 Chip experiment for Roc8 binding to the promoter of the OsLBD3‐7 gene.
Figure S15 Expression analysis of Roc8 in the Roc5 mutant and wild type at the 3rd leaf stage.
Figure S16 Expression analysis of cellulose‐related genes and lignin biosynthesis genes in rice.
Figure S17 Photosynthesis parameter measurements from the roc8‐m1 plant and wild‐type plant at the late tillering stage.
Figure S18 RT‐qPCR expression analysis of cell expansion and cell division genes.
Figure S19 RT‐qPCR expression analysis of the gibberellin biosynthesis pathway genes.
Figure S20 Distribution of three cytosine contexts and methylation patterns in the ∼800‐bp promoter region of Roc8 between wild type and crm1‐D.
Figure S21 Schematic of the hypothesis illustrating the possible mechanism underlying Roc8 in this study.
Table S1 Agronomic trait comparison between wild type, crm1‐D and Roc8‐m1 mutants at the mature stage.
Table S2 Differentially expressed proteins associated with cellulose, lignin, epidermis development in leaves.
Table S3 Primers used for fine mapping, qRT‐PCR, vector construction and etc.
Methods S1 Transmission Electron Microscopy (TEM) Analysis.
Methods S2 Histology and cytology observation.
Methods S3 In situ hybridization.
Methods S4 RNA preparation and quantitative real‐time (qRT)‐PCR analysis.
Methods S5 Protein extraction, sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS‐PAGE), and Western blotting.
Methods S6 Transcriptome data analysis.
Methods S7 Subcellular localization of Roc8 in rice.
Methods S8 Yeast two‐Hybrid assay and Yeast one‐hybrid assay.
Methods S9 Rice protoplast transformation assay.
Methods S10 Luciferase assay to detect Roc8 3′‐UTR function using a rice protoplast transient transformation system.
Methods S11 Bisulfite sequencing analysis.
Methods S12 Lignin content measurement.
Methods S13 Luciferase transient transcriptional activity assay in rice protoplasts.
Methods S14 Chromatin immunoprecipitation (ChIP) and quantitative PCR analysis.
Methods S15 Roc8 3′‐UTR deletion construction.
Methods S16 Field evaluation for agronomic traits.
Methods S17 Photosynthetic parameters measurement.
Dataset S1 Differentially expressed genes in crm1‐D and wild type.
Acknowledgements
This work was supported in part by NSFC‐CGIAR (31861143006) and Innovation Project of Chinese Academy of Agricultural Science (CAAS‐ZDXT2019003) to Tiegang Lu and Zhiguo Zhang.
Sun, J. , Cui, X. , Teng, S. , Kunnong, Z. , Wang, Y. , Chen, Z. , Sun, X. , Wu, J. , Ai, P. , Quick, W. P. , Lu, T. and Zhang, Z. (2020) HD‐ZIP IV gene Roc8 regulates the size of bulliform cells and lignin content in rice. Plant Biotechnol. J., 10.1111/pbi.13435
†These authors contributed equally to this work.
Contributor Information
Xuean Cui, Email: cuixuean@caas.cn.
Tiegang Lu, Email: lutiegang@caas.cn.
Zhiguo Zhang, Email: zhangzhiguo@caas.cn.
References
- Bang, S.W. , Lee, D.K. , Jung, H. , Chung, P.J. , Kim, Y.S. , Choi, Y.D. , Suh, J.W. et al. (2019) Overexpression of OsTF1L, a rice HD‐Zip transcription factor, promotes lignin biosynthesis and stomatal closure that improves drought tolerance. Plant Biotechnol. J. 17, 118–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S. , Tao, L. , Zeng, L. , Vega‐Sanchez, M.E. , Umemura, K. and Wang, G.L. (2006) A highly efficient transient protoplast system for analyzing defence gene expression and protein‐protein interactions in rice. Mol. Plant Pathol. 7, 417–427. [DOI] [PubMed] [Google Scholar]
- Cheng, Y. , Zhu, W. , Chen, Y. , Ito, S. , Asami, T. and Wang, X. (2014) Brassinosteroids control root epidermal cell fate via direct regulation of a MYB‐bHLH‐WD40 complex by GSK3‐like kinases. eLife, 3, e02525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, M. , Zhao, Y. , Ma, Q. , Hu, Y. , Hedden, P. , Zhang, Q. and Zhou, D.X. (2007) The rice YABBY1 gene is involved in the feedback regulation of gibberellin metabolism. Plant Physiol. 144, 121–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depege‐Fargeix, N. , Javelle, M. , Chambrier, P. , Frangne, N. , Gerentes, D. , Perez, P. , Rogowsky, P.M. et al. (2011) Functional characterization of the HD‐ZIP IV transcription factor OCL1 from maize. J. Exp. Botany, 62, 293–305. [DOI] [PubMed] [Google Scholar]
- Fang, L. , Zhao, F. , Cong, Y. , Sang, X. , Du, Q. , Wang, D. , Li, Y. et al. (2012) Rolling‐leaf14 is a 2OG‐Fe (II) oxygenase family protein that modulates rice leaf rolling by affecting secondary cell wall formation in leaves. Plant Biotechnol. J. 10, 524–532. [DOI] [PubMed] [Google Scholar]
- Fujino, K. , Matsuda, Y. , Ozawa, K. , Nishimura, T. , Koshiba, T. , Fraaije, M.W. and Sekiguchi, H. (2008) NARROW LEAF 7 controls leaf shape mediated by auxin in rice. Mol. Genet. Genom. 279, 499–507. [DOI] [PubMed] [Google Scholar]
- Hager, A. (2003) Role of the plasma membrane H+‐ATPase in auxin‐induced elongation growth: historical and new aspects. J. Plant Res. 116, 483–505. [DOI] [PubMed] [Google Scholar]
- Halpin, C. (2019) Lignin engineering to improve saccharification and digestibility in grasses. Curr. Opin. Biotechnol. 56, 223–229. [DOI] [PubMed] [Google Scholar]
- Hernandez, M.L. , Passas, H.J. and Smith, L.G. (1999) Clonal analysis of epidermal patterning during maize leaf development. Develop. Biol. 216, 646–658. [DOI] [PubMed] [Google Scholar]
- Hong, Z. , Ueguchi‐Tanaka, M. , Shimizu‐Sato, S. , Inukai, Y. , Fujioka, S. , Shimada, Y. , Takatsuto, S. et al. (2002) Loss‐of‐function of a rice brassinosteroid biosynthetic enzyme, C‐6 oxidase, prevents the organized arrangement and polar elongation of cells in the leaves and stem. Plant J. 32, 495–508. [DOI] [PubMed] [Google Scholar]
- Hu, J. , Zhu, L. , Zeng, D. , Gao, Z. , Guo, L. , Fang, Y. , Zhang, G. et al. (2010) Identification and characterization of NARROW AND ROLLED LEAF 1, a novel gene regulating leaf morphology and plant architecture in rice. Plant Mol. Biol. 73, 283–292. [DOI] [PubMed] [Google Scholar]
- Huang, J. , Li, Z. and Zhao, D. (2016) Deregulation of the OsmiR160 target gene OsARF18 causes growth and developmental defects with an alteration of auxin signaling in rice. Sci. Rep. 6, 29938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, P. , Yoshida, H. , Yano, K. , Kinoshita, S. , Kawai, K. , Koketsu, E. , Hattori, M. et al. (2018) OsIDD2, a zinc finger and INDETERMINATE DOMAIN protein, regulates secondary cell wall formation. J. Integr. Plant Biol. 60, 130–143. [DOI] [PubMed] [Google Scholar]
- Ito, M. , Sentoku, N. , Nishimura, A. , Hong, S.K. , Sato, Y. and Matsuoka, M. (2002) Position dependent expression of GL2‐type homeobox gene, Roc1: significance for protoderm differentiation and radial pattern formation in early rice embryogenesis. Plant J. 29, 497–507. [DOI] [PubMed] [Google Scholar]
- Khosla, A. , Paper, J.M. , Boehler, A.P. , Bradley, A.M. , Neumann, T.R. and Schrick, K. (2014) HD‐Zip proteins GL2 and HDG11 have redundant functions in arabidopsis trichomes, and GL2 activates a positive feedback loop via MYB23. Plant Cell, 26, 2184–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J. and Lee, H.W. (2013) Direct activation of EXPANSIN14 by LBD18 in the gene regulatory network of lateral root formation in Arabidopsis. Plant Signal Behav. 8, e22979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein‐Cosson, C. , Chambrier, P. , Rogowsky, P.M. and Vernoud, V. (2015) Regulation of a maize HD‐ZIP IV transcription factor by a non‐conventional RDR2‐dependent small RNA. Plant J. 81, 747–758. [DOI] [PubMed] [Google Scholar]
- Kumar, R. (2014) Role of microRNAs in biotic and abiotic stress responses in crop plants. Appl. Biochem. Biotechnol. 174, 93–115. [DOI] [PubMed] [Google Scholar]
- Le, J. , Zou, J. , Yang, K. and Wang, M. (2014) Signaling to stomatal initiation and cell division. Front. Plant Sci. 5, 297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L. , Shi, Z.Y. , Li, L. , Shen, G.Z. , Wang, X.Q. , An, L.S. and Zhang, J.L. (2010) Overexpression of ACL1 (abaxially curled leaf 1) increased Bulliform cells and induced Abaxial curling of leaf blades in rice. Mol. Plant, 3, 807–817. [DOI] [PubMed] [Google Scholar]
- Li, W. , Ma, M. , Feng, Y. , Li, H. , Wang, Y. , Ma, Y. , Li, M. et al. (2015) EIN2‐directed translational regulation of ethylene signaling in Arabidopsis. Cell, 163, 670–683. [DOI] [PubMed] [Google Scholar]
- Li, C. , Zou, X. , Zhang, C. , Shao, Q. , Liu, J. , Liu, B. , Li, H. et al. (2016) OsLBD3‐7 overexpression induced adaxially rolled leaves in rice. PLoS One, 11, e156413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W.Q. , Zhang, M.J. , Gan, P.F. , Qiao, L. , Yang, S.Q. , Miao, H. , Wang, G.F. et al. (2017) CLD1/SRL1 modulates leaf rolling by affecting cell wall formation, epidermis integrity and water homeostasis in rice. Plant J. 92, 904–923. [DOI] [PubMed] [Google Scholar]
- Mao, H. , Wang, H. , Liu, S. , Li, Z. , Yang, X. , Yan, J. , Li, J. et al. (2015) A transposable element in a NAC gene is associated with drought tolerance in maize seedlings. Nat. Commun. 6, 8326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, C. and Glover, B.J. (2007) Functional aspects of cell patterning in aerial epidermis. Curr. Opin. Plant Biol. 10, 70–82. [DOI] [PubMed] [Google Scholar]
- Moloney, M.M. , Elliott, M.C. and Cleland, R.E. (1981) Acid growth effects in maize roots: Evidence for a link between auxin‐economy and proton extrusion in the control of root growth. Planta, 152, 285–291. [DOI] [PubMed] [Google Scholar]
- Ohashi, Y. (2004) Roles and regulations of an Arabidopsis homeobox gene, GL2 in epidermal cell development. Tanpakushitsu Kakusan Koso, 49, 148–154. [PubMed] [Google Scholar]
- Rodriguez‐Cazorla, E. , Andujar, A. , Ripoll, J.J. , Bailey, L.J. , Martinez‐Laborda, A. , Yanofsky, M.F. and Vera, A. (2015) 3′ rapid amplification of cDNA Ends (3′ RACE) using Arabidopsis samples. Bio. Protoc. 5, e1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, J. , Liu, J. , Xie, K. , Xing, F. , Xiong, F. , Xiao, J. , Li, X. et al. (2017) Translational repression by a miniature inverted‐repeat transposable element in the 3' untranslated region. Nat. Commun. 8, 14651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silveyra, P. , DiAngelo, S.L. and Floros, J. (2014) An 11‐nt sequence polymorphism at the 3'UTR of human SFTPA1 and SFTPA2 gene variants differentially affect gene expression levels and miRNA regulation in cell culture. Am. J. Physiol. Lung Cell Mol. Physiol. 307, L106–L119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava, L.M. (1966) Histochemical studies on lignin. Tappi, 49, 173–183. [Google Scholar]
- Sylvester, A.W. , Parker‐Clark, V. and Murray, G.A. (2001) Leaf shape and anatomy as indicators of phase change in the grasses: comparison of maize, rice, and bluegrass. Am. J. Botany, 88, 2157–2167. [PubMed] [Google Scholar]
- Takada, S. and Iida, H. (2014) Specification of epidermal cell fate in plant shoots. Front. Plant Sci. 5, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada, S. , Takada, N. and Yoshida, A. (2013) Induction of epidermal cell fate in Arabidopsis shoots. Plant Signal Behav. 8, e26236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme, R. , Morreel, K. , Darrah, C. , Oyarce, P. , Grabber, J.H. , Ralph, J. and Boerjan, W. (2012) Metabolic engineering of novel lignin in biomass crops. New Phytol. 196, 978–1000. [DOI] [PubMed] [Google Scholar]
- Wan, S. , Wu, J. , Zhang, Z. , Sun, X. , Lv, Y. , Gao, C. , Ning, Y. et al. (2009) Activation tagging, an efficient tool for functional analysis of the rice genome. Plant Mol. Biology, 69, 69–80. [DOI] [PubMed] [Google Scholar]
- Wang, S. and Chen, J.G. (2014) Regulation of cell fate determination by single‐repeat R3 MYB transcription factors in Arabidopsis. Front. Plant Sci. 5, 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z. , Tian, X. , Zhao, Q. , Liu, Z. , Li, X. , Ren, Y. , Tang, J. et al. (2018) The E3 Ligase DROUGHT HYPERSENSITIVE negatively regulates cuticular wax biosynthesis by promoting the degradation of transcription factor ROC4 in rice. Plant Cell, 30, 228–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. , Monford Paul Abishek, N. Jeon, H.J. , Lee, Y. , He, J. , Adhya, S. and Lim, H.M . (2019) Processing generates 3′ ends of RNA masking transcription termination events in prokaryotes. Proc. Natl. Acad. Sci. USA, 116, 4440–4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, L. , Gu, L. , Song, X. , Cui, X. , Lu, Z. , Zhou, M. , Wang, L. et al. (2014) Dicer‐like 3 produces transposable element‐associated 24‐nt siRNAs that control agricultural traits in rice. Proc. Natl. Acad. Sci. USA, 111, 3877–3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo, Y.M. , Park, H.J. , Su'Udi, M. , Yang, J.I. , Park, J.J. , Back, K. , Park, Y.M. et al. (2007) Constitutively wilted 1, a member of the rice YUCCA gene family, is required for maintaining water homeostasis and an appropriate root to shoot ratio. Plant Mol. Biol. 65, 125–136. [DOI] [PubMed] [Google Scholar]
- Wu, R. , Li, S. , He, S. , Wassmann, F. , Yu, C. , Qin, G. , Schreiber, L. et al. (2011) CFL1, a WW domain protein, regulates cuticle development by modulating the function of HDG1, a class IV homeodomain transcription factor, in rice and Arabidopsis. Plant Cell, 23, 3392–3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang, J.J. , Zhang, G.H. , Qian, Q. and Xue, H.W. (2012) Semi‐rolled leaf1 encodes a putative glycosylphosphatidylinositol‐anchored protein and modulates rice leaf rolling by regulating the formation of bulliform cells. Plant Physiol. 159, 1488–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, P. , Ali, A. , Han, B. and Wu, X. (2018) Current advances in molecular basis and mechanisms regulating leaf morphology in rice. Front. Plant Sci. 9, 1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, C. , Li, D. , Liu, X. , Ji, C. , Hao, L. , Zhao, X. , Li, X. et al. (2014) OsMYB103L, an R2R3‐MYB transcription factor, influences leaf rolling and mechanical strength in rice (Oryza sativa L.). BMC Plant Biol. 14, 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Q. , Shen, B.Z. , Dai, X.K. , Mei, M.H. , Saghai, M.M. and Li, Z.B. (1994) Using bulked extremes and recessive class to map genes for photoperiod‐sensitive genic male sterility in rice. Proc. Natl. Acad. Sci. USA, 91, 8675–8679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, G.H. , Xu, Q. , Zhu, X.D. , Qian, Q. and Xue, H.W. (2009) SHALLOT‐LIKE1 is a KANADI transcription factor that modulates rice leaf rolling by regulating leaf abaxial cell development. Plant Cell, 21, 719–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, F. , Zuo, K. , Zhang, J. , Liu, X. , Zhang, L. , Sun, X. and Tang, K. (2010) An L1 box binding protein, GbML1, interacts with GbMYB25 to control cotton fibre development. J. Exp. Botany, 61, 3599–3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, S.Q. , Hu, J. , Guo, L.B. , Qian, Q. and Xue, H.W. (2010) Rice leaf inclination2, a VIN3‐like protein, regulates leaf angle through modulating cell division of the collar. Cell Res. 20, 935–947. [DOI] [PubMed] [Google Scholar]
- Zimmermann, P. , Hirsch‐Hoffmann, M. , Hennig, L. and Gruissem, W. (2004) GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136, 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, L.P. , Sun, X.H. , Zhang, Z.G. , Liu, P. , Wu, J.X. , Tian, C.J. , Qiu, J.L. et al. (2011) Leaf rolling controlled by the homeodomain leucine zipper class IV gene Roc5 in rice. Plant Physiol. 156, 1589–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, L.P. , Zhang, Z.G. , Qi, D.F. , Peng, M. and Lu, T.G. (2014) Cytological mechanisms of leaf rolling in rice. Crop Sci. 54, 1–12. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Photosynthesis parameter measurements from the crm1‐D plant and wild type at the flowering stage.
Figure S2 Toluidine blue O staining and scanning electron microscopy observations of cross‐sections of seedling leaves in Oryza sativa.
Figure S3 Comparison of transcript abundance of candidate genes in wild‐type (Wt) and crm1‐D mutant by qRT–PCR.
Figure S4 Expression pattern analysis of Roc8 at the seedling stage and mature stage.
Figure S5 Protein structure, transactivation analysis of Roc8.
Figure S6 Fluorescence detection of GFP‐Roc8 3′‐UTR with different fusion constructs using a rice protoplast transient transformation system.
Figure S7 Roc8 3′‐UTR regulates luciferase activity.
Figure S8 Roc8 3‐UTR sequence analysis.
Figure S9 A yeast two‐hybrid interaction assay was performed with Roc5 and Roc8.
Figure S10 Double mutant analysis of loss‐of‐function alleles of roc5/roc8.
Figure S11 Expression analysis of vacuolar development‐related genes.
Figure S12 Bulliform cell development‐related gene expression analysis in rice.
Figure S13 LUC transient transactivation assay in rice protoplasts.
Figure S14 Chip experiment for Roc8 binding to the promoter of the OsLBD3‐7 gene.
Figure S15 Expression analysis of Roc8 in the Roc5 mutant and wild type at the 3rd leaf stage.
Figure S16 Expression analysis of cellulose‐related genes and lignin biosynthesis genes in rice.
Figure S17 Photosynthesis parameter measurements from the roc8‐m1 plant and wild‐type plant at the late tillering stage.
Figure S18 RT‐qPCR expression analysis of cell expansion and cell division genes.
Figure S19 RT‐qPCR expression analysis of the gibberellin biosynthesis pathway genes.
Figure S20 Distribution of three cytosine contexts and methylation patterns in the ∼800‐bp promoter region of Roc8 between wild type and crm1‐D.
Figure S21 Schematic of the hypothesis illustrating the possible mechanism underlying Roc8 in this study.
Table S1 Agronomic trait comparison between wild type, crm1‐D and Roc8‐m1 mutants at the mature stage.
Table S2 Differentially expressed proteins associated with cellulose, lignin, epidermis development in leaves.
Table S3 Primers used for fine mapping, qRT‐PCR, vector construction and etc.
Methods S1 Transmission Electron Microscopy (TEM) Analysis.
Methods S2 Histology and cytology observation.
Methods S3 In situ hybridization.
Methods S4 RNA preparation and quantitative real‐time (qRT)‐PCR analysis.
Methods S5 Protein extraction, sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS‐PAGE), and Western blotting.
Methods S6 Transcriptome data analysis.
Methods S7 Subcellular localization of Roc8 in rice.
Methods S8 Yeast two‐Hybrid assay and Yeast one‐hybrid assay.
Methods S9 Rice protoplast transformation assay.
Methods S10 Luciferase assay to detect Roc8 3′‐UTR function using a rice protoplast transient transformation system.
Methods S11 Bisulfite sequencing analysis.
Methods S12 Lignin content measurement.
Methods S13 Luciferase transient transcriptional activity assay in rice protoplasts.
Methods S14 Chromatin immunoprecipitation (ChIP) and quantitative PCR analysis.
Methods S15 Roc8 3′‐UTR deletion construction.
Methods S16 Field evaluation for agronomic traits.
Methods S17 Photosynthetic parameters measurement.
Dataset S1 Differentially expressed genes in crm1‐D and wild type.


