Summary
Phosphorus, an essential mineral macronutrient, is a major constituent of fertilizers for maize (Zea mays L.) production. However, the molecular mechanisms of phosphate (Pi) acquisition in maize plants and its redistribution remain unclear. This study presents the functional characterization of ZmPT7 in Pi uptake and redistribution in maize. The ZmPT7 was expressed in roots and leaves, and induced during Pi starvation. The ZmPT7 complemented the Pi‐uptake deficiency of yeast mutant phoΔnull and Arabidopsis mutant pht1;1Δ4Δ, indicating that ZmPT7 functioned as a Pi transporter. We generated zmpt7 mutants by CRISPR/Cas9 and ZmPT7‐overexpressing lines. The zmpt7 mutants showed reduced, whereas the ZmPT7‐overexpressing lines displayed increased Pi‐uptake capacity and Pi redistribution from old to young leaves, demonstrating that ZmPT7 played central roles in Pi acquisition and Pi redistribution from old to young leaves. The ZmCK2 kinases phosphorylated ZmPT7 at Ser‐521 in old maize leaves, which enhanced transport activity of ZmPT7. The Ser‐520 of Arabidopsis AtPHT1;1, a conserved residue of ZmPT7 Ser‐521, was also phosphorylated by AtCK2 kinase, and the mutation of Ser‐520 to Glu (phosphorylation mimic) yielded enhanced transport activity of AtPHT1;1. Taken together, these results indicate that ZmPT7 plays important roles in Pi acquisition and redistribution, and its transport activity is modulated by phosphorylation.
Keywords: phosphate acquisition, phosphate redistribution, ZmPT7, phosphorylation modification, maize, Arabidopsis
Introduction
Phosphorus (P) is an essential macronutrient for plant growth and development. Inorganic phosphate (Pi) is the predominant form of P directly absorbed by plants. The soil Pi concentrations are often 10 μm or less (Schachtman et al., 1998), and Pi is one of the least available plant nutrients in soils (Raghothama and Karthikeyan, 2005).
The Pi is absorbed into plant cells through Pi transporters, which belong to the PHOSPHATE TRANSPORTER1 (PHT1) family (Młodzińska and Zboińska, 2016). The PHT1 transporters are only found in plants and fungi (Młodzińska and Zboińska, 2016), and their structures are conserved (Pedersen et al., 2013). The PHT1 transporters have been identified in many plant species (Loth‐Pereda et al., 2011), and there are nine and thirteen PHT1 members in Arabidopsis and rice, respectively (Liu et al., 2011; Mudge et al., 2002). Eight of nine Arabidopsis PHT1 genes and all rice PHT1 genes are expressed in root tissues (Liu et al., 2011; Mudge et al., 2002), consistent with their function in Pi uptake. Among nine Arabidopsis PHT1 transporters, AtPHT1;1 and AtPHT1;4 play major roles in Pi acquisition from the environment (Shin et al., 2004). Seven of thirteen rice PHT1 transporters are reported to participate in Pi uptake in roots (Ai et al., 2009; Jia et al., 2011; Sun et al., 2012; Wang et al., 2014b; Zhang et al., 2015); and two PHT1 transporters, AtPHT1;5 (Nagarajan et al., 2011) and OsPHT1;8 (also named OsPT8) (Li et al., 2015), are involved in Pi distribution from source to sink organs.
The PHT1 genes are precisely regulated at transcriptional level. Under Pi‐sufficient conditions, transcription factor WRKY42 positively regulates expression of AtPHT1;1 (Su et al., 2015); under Pi‐deficient conditions, the transcripts of AtPHT1;1 and AtPHT1;4 are up‐regulated by transcription factors WRKY75 (Devaiah et al., 2007), PHR1 (Bustos et al., 2010) and WRKY45 (Wang et al., 2014a). The PHT1 transporters are also modulated at post‐transcriptional level. The abundance of Arabidopsis PHT1 proteins can be modulated by an ubiquitin E3 ligase NLA (Lin et al., 2013; Park et al., 2014), and an ubiquitin–conjugation enzyme PHO2 (Huang et al., 2013). The PHOSPHATE TRANSPORTER TRAFFIC FACILITATOR1 (PHF1) proteins, AtPHF1 and OsPHF1, facilitate PHT1 transporters from the endoplasmic reticulum (ER) to the plasma membrane (Chen et al., 2011; González et al., 2005). Moreover, the phosphorylations of AtPHT1;1 Ser‐514 and OsPT8 Ser‐517 cause their retention in the ER (Bayle et al., 2011; Chen et al., 2015); AtPHT1;1 is also phosphorylated at Ser‐520 (Bayle et al., 2011; Nühse et al., 2004), but the role of Ser‐520 phosphorylation remains unknown.
Maize is one of the most important crops and is cultivated widely for staple food and industrial usage. Phosphorus is a major constituent of the fertilizers required to sustain high yields, whereas cultivated plants, including maize, use only approximately 20–30% of the applied phosphate (Chen and Liao, 2017; López‐Arredondo et al., 2014). Currently, the molecular mechanisms of Pi acquisition and distribution remain unclear in maize. Five PHT1 genes were reported in the maize genome (Nagy et al., 2006), and 13 ZmPHT1 genes were identified using bioinformatics (Liu et al., 2016; Sawers et al., 2017). Two mycorrhiza‐induced Pi transporters, ZmPHT1;6 and ZmPt9, were reported to affect maize growth and cob development or Pi uptake (Liu et al., 2018; Willmann et al., 2013).
In this study, we identified the function of ZmPT7 in maize Pi acquisition and redistribution. The ZmPT7 complemented the Pi‐uptake deficiency of yeast mutant phoΔnull and Arabidopsis mutant pht1;1Δ4Δ, indicating that ZmPT7 functioned as a Pi transporter. We generated the zmpt7 mutants and ZmPT7‐overexpressing lines, and found that ZmPT7 played central roles in Pi acquisition and Pi redistribution from old to young leaves. The ZmPT7 in old leaves was phosphorylated at Ser‐521, a conserved phosphorylation residue of AtPHT1;1 Ser‐520 (Bayle et al., 2011; Nühse et al., 2004). The phosphorylation modification at ZmPT7 Ser‐521 and AtPHT1;1 Ser‐520 enhanced transport activity of ZmPT7 and AtPHT1;1. Given this impact, altering the transcript and phosphorylation status of ZmPT7 might offer effective strategies to improve maize Pi acquisition and redistribution.
Results
Identification of maize Pi‐transporter genes and expression pattern of ZmPT7
The Arabidopsis AtPHT1;1 and rice OsPT8 are important Pi transporters participating in Pi acquisition from the environment (Jia et al., 2011; Shin et al., 2004). In order to identify Pi transporters in maize, BLAST searches were conducted using the amino acid sequences of AtPHT1;1 and OsPT8 against the maize B73 genome in the National Center for Biotechnology Information (NCBI; www.ncbi.nlm.nih.gov). To identify the sequences of putative maize PHT1 genes, the coding sequences of putative ZmPHT1 genes were amplified from the cDNA of maize inbred B73 and identified by direct sequencing of the diagnostic PCR product.
We obtained eight putative maize PHT1 transporters, named ZmPT1–ZmPT8 (Table S1), with nearly or over 50% sequence identities of AtPHT1;1 or OsPT8 (Table S2). Each putative ZmPHT1 protein contained 12 predicted transmembrane domains, six N‐terminal transmembrane domains and six C‐terminal transmembrane domains, separated by a central hydrophilic region (Table S1). The PHT1 conserved signature GGDYPLSATIxSE (Loth‐Pereda et al., 2011) was identified in these putative ZmPHT1 proteins (Table S1). The phylogenetic tree was constructed using the sequences of ZmPHT1 proteins and other PHT1 proteins from Arabidopsis thaliana, Oryza sativa and Glycine max. The phylogenetic analysis showed that the ZmPHT1 proteins were divided into two subgroups, the ZmPT2 and ZmPT7 were closely related to the OsPT8, and no ZmPHT1 protein was clustered with AtPHT1;1 (Figure 1a).
Figure 1.
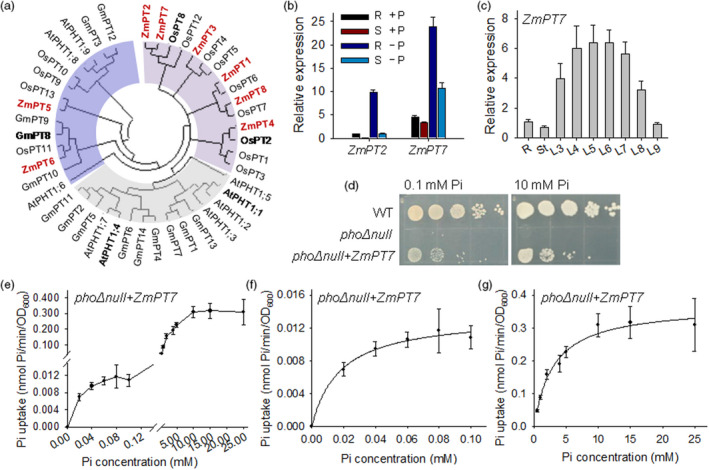
ZmPT7 is low‐Pi induced and complements yeast mutant pho∆null. (a) Neighbour‐joining tree analysis was conducted using MEGA4. Bootstrap values were calculated as a percentage of 1000 trials. The accession numbers of PHT1 proteins are listed in Table S1. (b) qRT‐PCR analysis of ZmPT2 and ZmPT7 in 7‐day‐old maize inbred B73 grown in hydroponic solution with 250 μm (+P) or 0 μm (−P) Pi for 5 days. S, shoot. R, root. Data are means ± standard error (SE), n = 4 biologically independent samples. (c) qRT‐PCR assay of ZmPT7 in various tissues of 40‐day‐old maize inbred B73. R, root. St, stalk. L, leaf. The leaves were numbered in ascending order according to their appearance as L3–L8. Data are means ± SE, n = 3 biologically independent samples. (d) Complementation of yeast mutant phoΔnull (pho84Δ pho87Δ pho89Δ pho90Δ pho91Δ) with maize ZmPT7 under different Pi concentrations. Equal volumes of tenfold serial dilutions applied for each yeast strain. (e‐g) 32Pi‐uptake rate of yeast phoΔnull + ZmPT7 transformants in the presence of different Pi concentrations. Data are means ± SE, n = 8 biologically independent samples.
The quantitative real‐time PCR (qRT‐PCR) analysis showed that the ZmPT2 was mainly expressed in roots, and ZmPT7 transcripts were abundant in roots and shoots (Figure 1b). Both ZmPT2 and ZmPT7 were up‐regulated during Pi starvation (Figure 1b). The transcript abundance of ZmPT7 was much higher than that of ZmPT2 under both Pi‐sufficient and Pi‐deficient conditions (Figure 1b), and the function of ZmPT7 was further analysed. Transcript levels of ZmPT7 were relatively high in roots at the early seedling stage (Figure 1b) and abundant in adult leaves at V6 stage (Figure 1c), suggesting that ZmPT7 had various functions in phosphate nutrition during maize development.
ZmPT7 complements the Pi‐uptake deficiency of yeast mutant phoΔnull and Arabidopsis mutant pht1;1Δ4Δ
To determine the role of ZmPT7 in Pi transport, we overexpressed ZmPT7 in a yeast mutant phoΔnull, in which five phosphate transporters were inactivated: PHO84, PHO87, PHO89, PHO90 and PHO91 (Wykoff and O'Shea, 2001; Popova et al., 2010). Similar to the previous report (Wykoff et al., 2001), the phoΔnull mutant failed to grow on standard synthetic medium (Figure 1d), and maize ZmPT7 complemented the synthetic lethality of the phoΔnull mutant (Figure 1d), indicating that ZmPT7 functioned as a Pi transporter in yeast. Then, we performed kinetic analysis of Pi uptake by yeast phoΔnull + ZmPT7 transformants using 32Pi as described before (Wykoff and O'Shea, 2001). The Pi‐uptake rate of ZmPT7 was Pi concentration‐dependent, revealing a biphasic pattern (Figure 1e). And the Km values of these two Pi‐uptake phases were obtained by fitting these measurements to the Michaelis–Menten equation, 17.8 ± 3.8 μm for high‐affinity phase and 3.02 ± 0.48 mm for low‐affinity phase (Figure 1f,g), suggesting that ZmPT7 was a dual‐affinity Pi transporter.
The AtPHT1;1 and AtPHT1;4 are two major Pi transporters for Arabidopsis Pi uptake from the environment, and the pht1;1Δ4Δ double mutant shows obvious deficits in growth and Pi uptake (Shin et al., 2004). To determine whether ZmPT7 functioned as a Pi transporter in plants, the coding sequence of ZmPT7 under the control of a Super promoter (Super:ZmPT7) was introduced into the pht1;1Δ4Δ mutant, and two homozygous pht1;1Δ4Δ/ZmPT7 transgenic lines were obtained (Figure 2a). The pht1;1Δ4Δ mutant was smaller than wild‐type plants under Pi‐sufficient (MS) or Pi‐deficient (LP) conditions, and the ZmPT7 (pht1;1Δ4Δ/ZmPT7) restored the growth deficit of the pht1;1Δ4Δ mutant (Figure 2b,c). The pht1;1Δ4Δ/ZmPT7 lines had greater biomass than wild‐type plants (Figure 2c).
Figure 2.
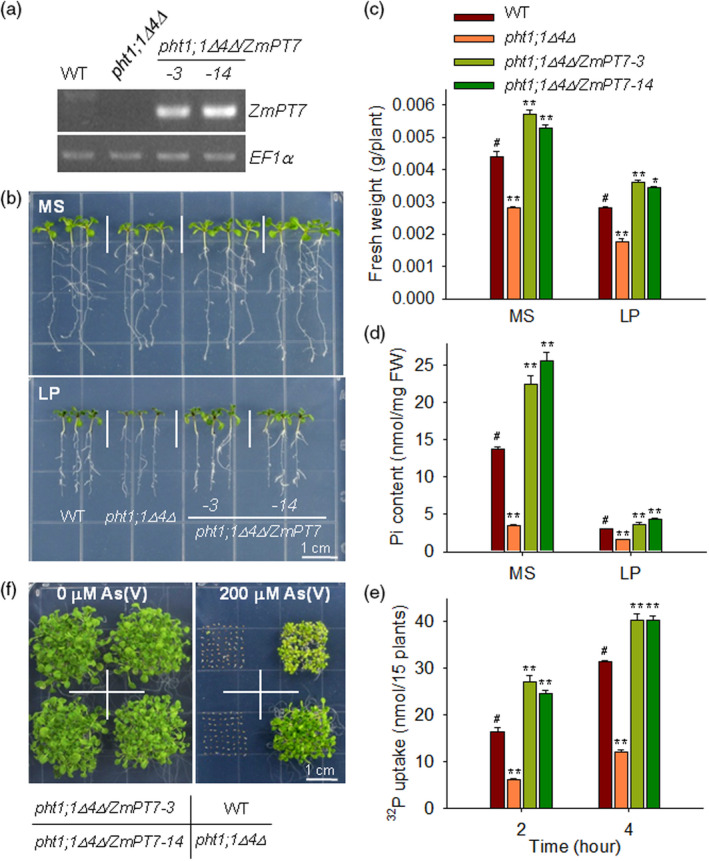
ZmPT7 complements Arabidopsis Pi‐uptake deficient mutant pht1;1Δ4Δ. (a) RT‐PCR analysis of ZmPT7 expression in the pht1;1Δ4Δ mutant, pht1;1Δ4Δ/ZmPT7 plants and wild‐type Arabidopsis plants (WT). EF1α was used as the control. (b) Phenotype comparison. Seven‐day‐old seedlings were transferred to Pi‐sufficient (MS) or Pi‐deficient (LP) medium for 7 days, and then, the photographs were taken. (c) Fresh weight of 7‐day‐old seedlings grown on MS or LP medium for 7 days. Data are means ± SE of 15 plants. (d) Pi contents of 7‐day‐old seedlings grown on MS or LP medium for 5 days. The experiments were done with three biological replicates, and a group of 15 seedlings was used as one biological sample. (e) Pi uptake was monitored over a 4‐h period in 7‐day‐old seedlings. The experiments were done with three biological replicates, and a group of 15 seedlings was used as one biological sample. Asterisks in c, d and e indicate significant differences compared with wild‐type plants (WT, #): *P < 0.05, **P < 0.01. (f) Arsenate‐tolerant phenotype comparison. All genotypes were germinated and grown on 1/2 MS medium without or with 200 μm arsenate [As(V)] for 14 days.
The Pi‐uptake capacities of the pht1;1Δ4Δ/ZmPT7 lines were also measured. Consistent with the previous report (Shin et al., 2004), the pht1;1Δ4Δ mutant showed reduced Pi content and Pi‐uptake rate compared with wild‐type plants (Figure 2d,e), whereas the Pi contents and Pi‐uptake rates of two pht1;1Δ4Δ/ZmPT7 lines were more elevated than those of pht1;1Δ4Δ mutant, and even of wild‐type plants (Figure 2d,e). Arsenate [As(V)], a toxic metalloid, is a structural analog of Pi and is transported into plant cells mainly via Pi transporters (Castrillo et al., 2013; Catarecha et al., 2007). When germinated and grown on medium in the presence of 200 μm As(V), the pht1;1Δ4Δ mutant displayed As(V)‐resistant phenotypes, similar to a previous report (Shin et al., 2004), whereas two pht1;1Δ4Δ/ZmPT7 lines were As(V)‐hypersensitive (Figure 2f). These data indicate that ZmPT7 has a Pi‐transporter activity in plants.
ZmPT7 modulates growth and phosphate uptake of maize
In an attempt to determine the roles of ZmPT7 in maize, we generated two maize zmpt7 mutants, zmpt7‐1 and zmpt7‐2, using the CRISPR/Cas9 technology (Figure S1). When germinated and grown for 40 days (V6 stage), both zmpt7‐1 and zmpt7‐2 mutants were smaller and showed reduced dry weights compared with wild‐type maize inbred B73 (Figure 3a,b). The leaves of zmpt7 mutants grew slowly, with length of 9th leaf (L9) reaching just a third of that of wild‐type plants (Figure 3c). These data indicate that disruption of ZmPT7 led to defects in maize vegetative growth.
Figure 3.
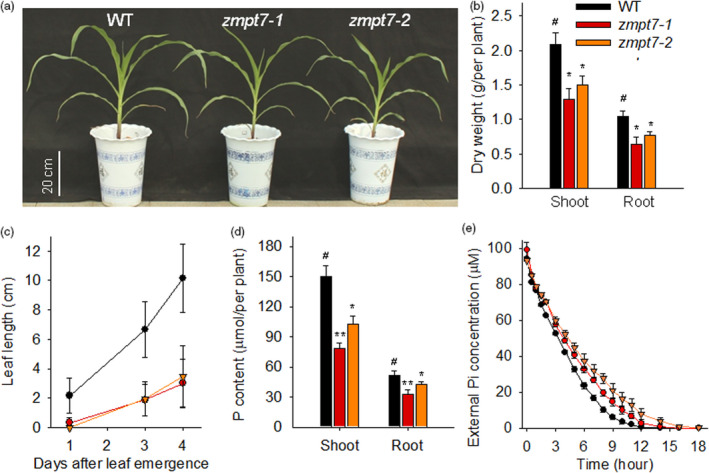
Disruption of ZmPT7 represses maize growth and Pi acquisition. (a) Phenotype comparison between the zmpt7 mutants and wild‐type maize, which germinated and grew for 40 days. (b) Dry weights of 40‐day‐old zmpt7 mutants and wild‐type maize plants. Data are means ± SE of six plants. (c) Leaf elongation measurement. Length of the ninth leaf was measured beginning at emergence. Data are means ± SE of six plants. (d) Total P contents of 40‐day‐old zmpt7 mutants and wild‐type maize. Data are means ± SE of six plants. (e) Pi‐uptake rate measurement. Seven‐day‐old seedlings were pretreated in Pi starvation solution for 3 days and then transferred to depletion solution with 100 μm Pi for Pi‐depletion experiment. Data are means ± SE of three biological repeats, each repeat contained two plants. Asterisks in b and d indicate significant differences compared with wild‐type plants (WT, #). *P < 0.05; **P < 0.01.
For ZmPT7 was a Pi transporter (Figures 1d and 2), the P content and Pi uptake were measured in zmpt7 mutants. The shoot and root P contents of two zmpt7 mutants were significantly lower than that of wild‐type maize (Figure 3d). And both zmpt7‐1 and zmpt7‐2 mutants exhibited obvious decreases in Pi‐uptake capacity compared with wild‐type plants (Figure 3e). These results demonstrated that disruption of ZmPT7 impaired Pi uptake of maize.
Transcripts of ZmPT7 were significantly induced in roots during Pi starvation (Figure 1b). To characterize the physiological function of transcriptional up‐regulation of ZmPT7, we generated the ZmPT7‐overexpressing lines, Ubi:ZmPT7‐1 and Ubi:ZmPT7‐2, with gradually increased ZmPT7 expression (Figure 4a). When grown for 40 days, the ZmPT7‐overexpressing lines were larger (Figure 4b), and with higher shoot dry weights (Figure 4c) than wild‐type maize. The ZmPT7‐overexpressing lines had a higher growth rate, with significantly longer L9 leaves than wild‐type plants (Figure 4d).
Figure 4.
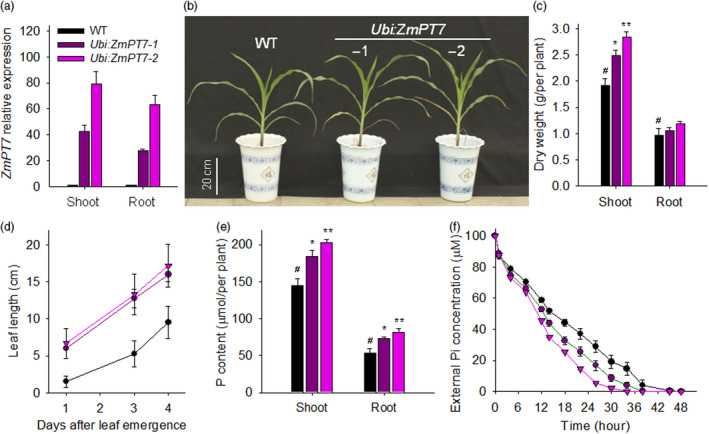
Overexpression of ZmPT7 increases maize Pi uptake. (a) qRT‐PCR analysis of ZmPT7 expression in maize ZmPT7‐overexpressing lines (Ubi:ZmPT7‐1 and Ubi:ZmPT7‐2). Data are means ± SE of three plants. (b) Phenotype comparison between the ZmPT7‐overexpressing lines and wild‐type maize, which germinated and grown for 40 days. (c) Dry weights of 40‐day‐old ZmPT7‐overexpressing lines and wild‐type maize. Data are means ± SE of six plants. (d) Leaf elongation of the ninth leaf. Data are means ± SE of six plants. (e) Total P contents of 40‐day‐old ZmPT7‐overexpressing lines and wild‐type maize. Data are means ± SE of six plants. (f) Pi‐uptake rate comparison among the 10‐day‐old ZmPT7‐overexpressing lines and wild‐type plants. Data are means ± SE of three biological repeats, each repeat contained two plants. Asterisks in c and e indicate significant differences compared with wild‐type plants (WT, #). *P < 0.05; **P < 0.01.
The P content was also measured. The two ZmPT7‐overexpressing lines, particularly the Ubi:ZmPT7‐2 line that had a higher transcript level of ZmPT7, showed elevated total P contents relative to wild‐type plants (Figure 4e). Consistent with P contents, the Pi‐uptake rates of the ZmPT7‐overexpressing lines were higher than that of wild‐type plants (Figure 4f). The increment of Pi‐uptake rate was related to the transcript level of ZmPT7, with more increased Pi‐uptake rate in the Ubi:ZmPT7‐2 line and less increment in the Ubi:ZmPT7‐1 line (Figure 4f), indicating that promotion of ZmPT7 expression enhanced Pi acquisition of maize.
ZmPT7 participates in P redistribution in maize
In addition to being expressed in roots, ZmPT7 was also expressed in leaves, particularly in mature leaves (Figure 1c), suggesting that ZmPT7 played a role in aerial parts of maize. When grown for 40 days (V6 stage), the old leaves rapidly senesced in ZmPT7‐overexpressing lines and stayed green in the zmpt7 mutants compared with wild‐type plants (Figure 5a). The ZmPT7‐overexpressing lines had larger young leaves, whereas zmpt7 mutants had smaller, compared to wild‐type plants (Figure 5a). Consistent with the phenotypes above, the dry weight of each leaf of zmpt7 mutants was lower than that of wild‐type plants, whereas the ZmPT7‐overexpressing lines showed similar dry weights in old leaves (L2–L5), and greater dry weight in young leaves (L6–L9) compared with wild‐type plants (Figure 5b).
Figure 5.
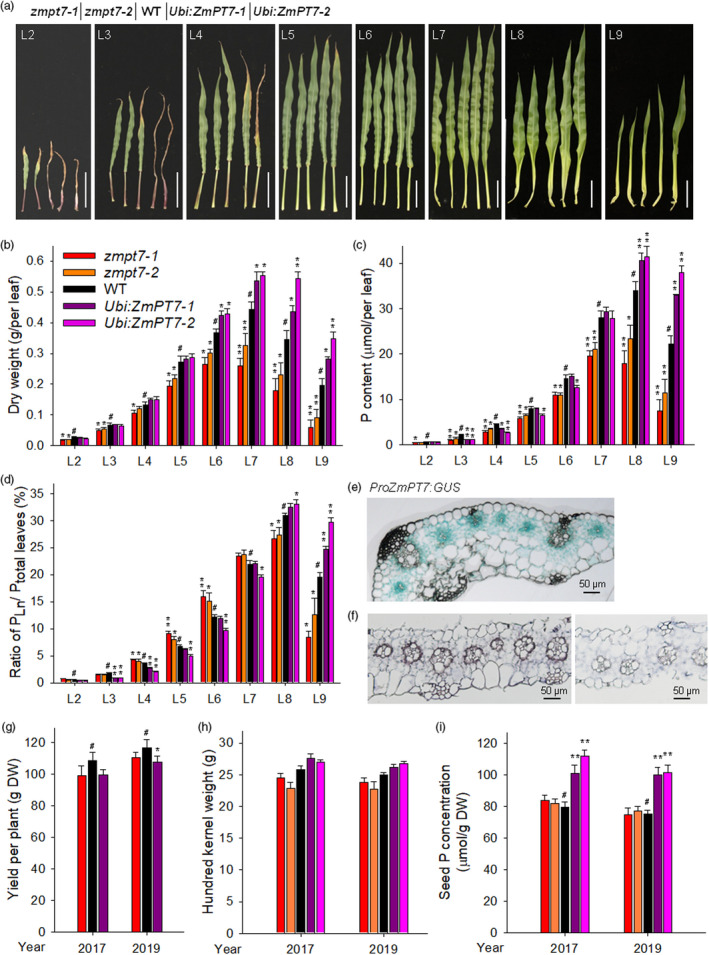
Characterization of leaf phenotypes between the zmpt7 mutants and ZmPT7‐overexpressing lines. (a) Leaf phenotypes. The zmpt7 mutants, ZmPT7‐overexpressing lines and wild‐type maize were germinated and grown for 40 days, and then, the leaves were harvested separately. The leaves (L2–L9) were numbered in ascending order according to their appearance. The leaves in each panel are displayed in the following order: zmpt7‐1, zmpt7‐2, WT, Ubi:ZmPT7‐1 and Ubi:ZmPT7‐2. Bars = 10 cm. (b) Leaf dry weight of 40‐day‐old maize plants. Data are means ± SE of six plants. (c) Leaf P content of 40‐day‐old plants. Data are means ± SE of six plants. (d) Ratio of PLn to Ptotal leaves. The ratio was calculated from the data presented in (c). Data are means ± SE of six plants. (e) GUS staining in leaf blade of ProZmPT7:GUS maize plant. (f) Expression of ZmPT7 in leaves determined by RNA in situ hybridization. The right panel is the negative control using the sense‐strand probe. (g) Yield per plant of zmpt7 mutant, ZmPT7‐overexpressing line and WT grown in the field. Data were obtained from at least 20 plants for each kind. (h) Hundred kernel weight of plants grown in the field. Data are means ± SE of 18 plants. (i) Seed P concentration of plants grown in the field. Data are means ± SE of 18 plants. Asterisks in b, c, d, g and i indicate significant differences compared with relative wild‐type plants (WT, #). *P < 0.05; **P < 0.01.
The P content was further measured. The leaf P contents were lower in zmpt7 mutants than wild‐type plants (Figure 5c). Interestingly, the ZmPT7‐overexpressing lines showed reduced P contents in old leaves and elevated P contents in young leaves, compared with wild‐type plants (Figure 5c). Then, the P‐distribution ratio among leaves (PLn/Ptotal leaves) was calculated using P content in one leaf (PLn) as a percentage of P content in all leaves (Ptotal leaves). The PLn/Ptotal leaves of zmpt7 mutants was elevated in bottom leaves (L4–L6) and reduced in top leaves (L8–L9), relative to wild‐type plants (Figure 5d). In contrast, the ZmPT7‐overexpressing lines exhibited lower PLn/Ptotal leaves in bottom leaves (L3–L5) and higher PLn/Ptotal leaves in top leaves (L8–L9) compared with wild‐type plants (Figure 5d), indicating that ZmPT7 modulated P redistribution from old to young leaves.
We then generated the ProZmPT7:GUS transgenic maize lines and found strong GUS staining in vascular bundles and bundle sheath cells of leaves (Figure 5e). A similar expression pattern of ZmPT7 in leaf blade was detected using an mRNA in situ hybridization assay (Figure 5f), consistent with the function of ZmPT7 in P redistribution among leaves.
We conducted field tests with ZmPT7‐overexpressing lines, zmpt7 mutants and wild‐type plants in Gongzhuling (Jilin, China) for two years. The zmpt7 mutant and ZmPT7‐overexpressing line showed slightly lower grain yields than wild‐type plants (Figure 5g). No significant differences of ZmPT7‐overexpressing lines or zmpt7 mutants compared with wild‐type plants were seen in terms of hundred kernel weight (Figure 5h). And the ZmPT7‐overexpressing lines exhibited higher seed P contents than wild‐type plants (Figure 5i). Collectively, these data indicated that increased expression of ZmPT7 benefited P redistribution to seeds.
ZmPT7 is phosphorylated by ZmCK2 at Ser‐521 in old maize leaves
The ZmPT7 was mainly expressed in mature leaves (Figure 1c), and we hypothesized that the increased Pi redistribution from old to young leaves of ZmPT7‐overexpressing lines was due to the elevated expression of ZmPT7 in old leaves. However, the transcript level of ZmPT7 was similar among all leaves of ZmPT7‐overexpressing lines (Figure S2). Another hypothesis we proposed was that ZmPT7 was post‐transcriptionally regulated in old leaves. Previous reports showed that Pi transporters can be phosphorylated at the hydrophilic C‐termini (CT) (Bayle et al., 2011; Chen et al., 2015), and some Ser residues were conserved among ZmPT7–CT, AtPHT1;1–CT, and OsPT8–CT (Figure 6a) (Bayle et al., 2011; Chen et al., 2015). A Phos‐tag mobility shift assay was conducted to determine whether ZmPT7 was phosphorylated in leaves. The recombinant ZmPT7–CT protein, fused with GST tag, was incubated with protein extracts from L3 or L8 of 40‐day‐old maize plants and subjected to Phos‐tag gel analysis. The ZmPT7–CT displayed a mobility shift in L3, and this lower mobility was abolished with a calf‐intestinal alkaline phosphatase (CIAP) treatment (Figure 6b). This slower migration was absent in L8 (Figure 6b). These data indicated that ZmPT7 was phosphorylated in old maize leaves.
Figure 6.
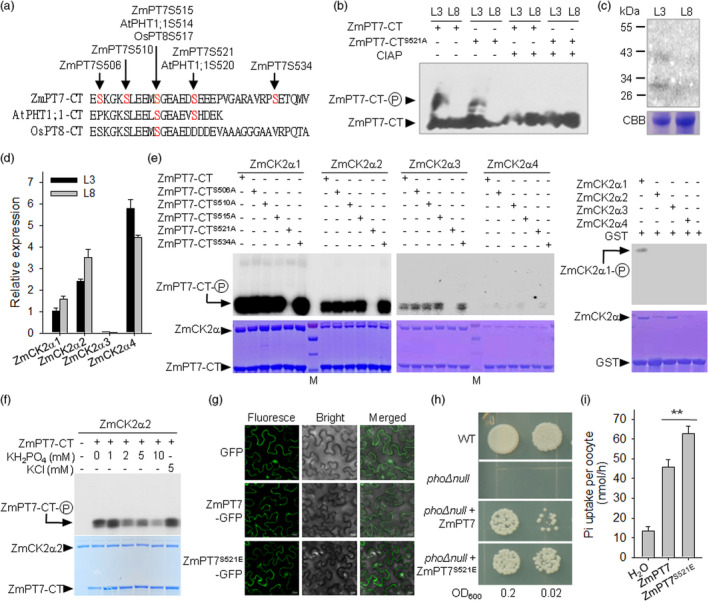
ZmPT7 is modulated by phosphorylation. (a) Sequence alignment of the hydrophilic C‐termini (CT) of ZmPT7, Arabidopsis PHT1;1 (AtPHT1;1) and rice PT8 (OsPT8). Arrow indicates the phosphorylated residues in the PHT1s. (b) Phosphorylation analysis of ZmPT7. The wild‐type maize plants were germinated and grown for 40 days, and then, the leaves were harvested individually for protein extraction. The recombinant GST‐ZmPT7–CT or GST‐ZmPT7–CTS521A was incubated with leaf protein extraction, with or without calf‐intestinal alkaline phosphatase (CIAP), and then, the mixtures were separated in a Phos‐tag SDS‐PAGE gel and immunoblotted with anti‐GST antibody. (c) In‐gel phosphorylation assay. Myelin basic protein (MBP) was used as a substrate for in‐gel phosphorylation assay with protein extracted from L3 or L8 leaf of 40‐day‐old maize B73 using GTP as a phosphate donor. Top, autoradiograph; bottom, Coomassie brilliant blue (CBB). (d) qRT‐PCR analysis of ZmCK2αs in L3 or L8 leaf of 40‐day‐old maize B73. Data are means ± SE of three plants. (e) In vitro phosphorylation of GST‐ZmPT7–CT, non‐phosphorylation mimicking GST‐ZmPT7–CTSA or GST alone by ZmCK2αs with corresponding CBB staining. (f) In vitro phosphorylation of GST‐ZmPT7–CT by ZmCK2α2 with different Pi concentration. (g) The location of wild‐type ZmPT7 (ZmPT7–GFP), Ser521Glu variant of ZmPT7 (ZmPT7S521E–GFP) and GFP alone in tobacco leaves. Bars = 20 μm. (h) Transport activity assay in yeast mutant pho∆null. ZmPT7 and Ser521Glu variant of ZmPT7 (ZmPT7S521E ) were expressed in pho∆null separately and incubated at 30 °C for 5 days. The initial OD600 is 0.2, equal volumes of tenfold serial dilutions applied for each yeast strain. (i) Transport activity assay in oocytes. The wild‐type ZmPT7 cRNA, Ser521Glu variant of ZmPT7 (ZmPT7S521E ) cRNA and water‐injected oocytes were incubated in ‐free ND96 solution for 36 h at 18 °C. Then, the oocytes were transferred into bath solution buffer containing 0.5 μm Pi with 32P (1 mCi/mL H3 32PO4) incubated for 2 h, pH 5.5. Data are means ± SE of n = 10. Asterisks indicate significant a difference between ZmPT7 and ZmPT7S521E cRNA‐injected oocytes, **P < 0.01.
A previous report showed that the hydrophilic CT of rice OsPT8 was phosphorylated at Ser‐517 by CK2 kinase (Chen et al., 2015). The CK2 is distinct from other kinases in that it can use GTP as a phosphoryl donor (Niefind et al., 1999), and the size of maize CK2 kinase is around 42 kDa (Vilela et al., 2015). We tested the kinase activity of maize CK2 kinase in old or young leaves using in‐gel kinase assay with GTP as a diagnostic phosphate donor. There was a near 42‐kDa band with CK2 activity in L3 and no obvious phosphorylation signal in L8 (Figure 6c). The CK2 kinase is a holoenzyme with two catalytic α subunits and two β subunits, and there are four α and four β subunit genes in the maize genome (Riera et al., 2011; Vilela et al., 2015). The qRT‐PCR results showed that the transcription accumulations were different among four ZmCK2αs (ZmCK2α1–ZmCK2α4), whereas the ZmCK2αs had similar transcriptional levels between L3 and L8 leaf (Figure 6d). The genes of four α subunits of maize CK2 were cloned and expressed in E. coli. In vitro phosphorylation assay showed that ZmPT7–CT was phosphorylated by ZmCK2α1, ZmCKα2 and ZmCKα3, but rarely by ZmCKα4 (Figure 6e).
Rice OsPT8, the closest rice Pi transporter to ZmPT7 (Figure 1a), was phosphorylated at Ser‐517 by rice CK2 kinase OsCK2α3 (Chen et al., 2015). The OsPT8 Ser‐517 residue was conserved with ZmPT7 Ser‐515 (ZmPT7S515) and AtPHT1;1 Ser‐514 (AtPHT1;1S514) (Figure 6a), and the Ser‐514 of AtPHT1;1 was phosphorylated in Arabidopsis cell suspensions (Bayle et al., 2011). To investigate whether ZmPT7 Ser‐515 was a putative phosphorylation site of ZmCK2 kinase, this residue was mutated to Ala (ZmPT7–CTS515A), mimicking the non‐phosphorylated ZmPT7–CT. The other four Ser residues in ZmPT7–CT (Figure 6a) were also separately replaced with Ala residues, for the Ser‐521 and Ser‐534 were phosphorylated in mature leaves of maize (Walley et al., 2016). In vitro phosphorylation assays showed that the phosphorylation signals of ZmPT7–CTS515A by ZmCK2αs were similar to that of wild‐type ZmPT7–CT, and ZmCK2αs‐mediated phosphorylation of ZmPT7–CT was almost abolished when only containing Ser‐521 point mutation (Figure 6e), indicating that ZmCK2αs phosphorylated ZmPT7 at residue Ser‐521 in vitro.
To determine whether ZmPT7 was phosphorylated at Ser‐521 in old maize leaves, the Phos‐tag mobility shift assay was also conducted using ZmPT7–CTS521A as a substrate. The phosphorylation signal of ZmPT7–CTS521A in old leaves (L3) was obviously reduced compared with wild‐type ZmPT7–CT (Figure 6b), suggesting that ZmPT7 can be phosphorylated at Ser‐521 residue in old maize leaves.
The ZmPT7–CT was phosphorylated in old maize leaves but not young leaves (Figure 6b), and the old leaves had lower P content than young leaves (Figure 5c); then, it was hypothesized that the phosphorylation of ZmPT7 was regulated by Pi. Then, we performed the in vitro phosphorylation assay using an increasing amount of Pi. As shown in Figure 6f, the phosphorylation of ZmPT7–CT by ZmCK2α2 was repressed in the presence of Pi, and this repression was a dose‐dependent response to Pi concentration, suggesting that the phosphorylation of ZmPT7 by ZmCK2 kinase was regulated by Pi level.
Phosphorylation of ZmPT7 at Ser‐521 affects its Pi‐transport activity
Previous reports demonstrate that phosphorylation of AtPHT1;1 at Ser‐514 or OsPT8 at Ser‐517 prevents AtPHT1;1 and OsPT8 exiting from the ER to the plasma membrane (Bayle et al., 2011; Chen et al., 2015). In order to investigate the role of Ser‐521 phosphorylation of ZmPT7, the Ser‐521 residue was changed by site‐directed mutagenesis to Glu (E) to mimic the phosphorylated form of ZmPT7. When transiently expressed in Nicotiana benthamiana leaves, the mutated ZmPT7 protein (ZmPT7S521E–GFP) showed a similar expression pattern to wild‐type ZmPT7 (Figure 6g), indicating that the phosphorylation modification at ZmPT7 Ser‐521 did not influence ZmPT7 subcellular trafficking.
We further hypothesized that this modification may be involved in regulation of ZmPT7 activity. To gain evidence for the capacity of phosphorylation to influence ZmPT7 activity, the wild‐type ZmPT7 and mutated ZmPT7 (ZmPT7S521E ) were separately overexpressed in the phoΔnull mutant. The wild‐type ZmPT7 complemented the growth deficiency of the phoΔnull mutant, and when the Ser521 was mutated to Glu, transformants with ZmPT7S521E grew much faster than those with wild‐type ZmPT7 (Figure 6h). To further confirm the role of phosphorylation modification at ZmPT7 Ser521, we expressed the ZmPT7 and ZmPT7S521E in oocytes by microinjecting and measured Pi transport. Oocytes expressing ZmPT7S521E showed significantly increased 32Pi‐uptake rate compared with those expressing ZmPT7 (Figure 6i). These data indicated that the phosphorylation of ZmPT7 at Ser521 enhanced the Pi‐transport activity of ZmPT7.
Phosphorylation modification of AtPHT1;1 at Ser‐520 enhances its Pi‐transport activity
The AtPHT1;1 is a main Pi transporter in Arabidopsis (Shin et al., 2004) and is phosphorylated at Ser‐520 (Bayle et al., 2011; Nühse et al., 2004), a conserved residue of ZmPT7 Ser521 (Figure 6a). Then, we assessed whether Arabidopsis AtPHT1;1 was phosphorylated at Ser‐520 by CK2 kinase. There were four catalytic α‐subunits of CK2α in Arabidopsis (Portolés and Más, 2010). Three of four Arabidopsis CK2α kinases, AtCK2α1, AtCK2α3 and AtCK2α4, phosphorylated the hydrophilic CT of PHT1;1 in vitro, and AtCK2α1/AtCK2α4‐mediated phosphorylation signals of AtPHT1;1–CT were abolished with the AtPHT1;1 Ser‐520 mutation to Ala (A) (Figure 7a), indicating that the AtPHT1;1 was phosphorylated at Ser‐520 by kinases AtCK2α1 and AtCK2α4 in vitro. When transiently expressed in N. benthamiana leaves, both wild‐type AtPHT1;1 and mutated AtPHT1;1 (AtPHT1;1S520E) were predominantly localized to the plasma membrane (Figure 7b), similar to the previous report (Bayle et al., 2011), suggesting that the phosphorylation modification of AtPHT1;1 Ser‐520 did not influence its subcellular localization. Then, the Pi‐transport activity of AtPHT1;1 was tested in the phoΔnull mutant. The wild‐type AtPHT1;1 and phosphorylated AtPHT1;1 (AtPHT1;1S520E) were separately transformed into the phoΔnull mutant. As a Pi transporter, AtPHT1;1 rescued the growth deficiency of the phoΔnull mutant (Figure 7c). The mimic phosphorylation form of AtPHT1;1 (AtPHT1;1S520E) displayed an increased growth rate compared with wild‐type AtPHT1;1 (Figure 7c). We also expressed the AtPHT1;1 and AtPHT1;1S520E in oocytes to measure their Pi transport. Oocytes expressing AtPHT1;1S520E showed a significantly increased 32Pi‐uptake rate compared with those expressing AtPHT1;1 (Figure 7d). These data indicated that phosphorylation modification of AtPHT1;1 Ser520 enhanced its Pi‐transport activity.
Figure 7.
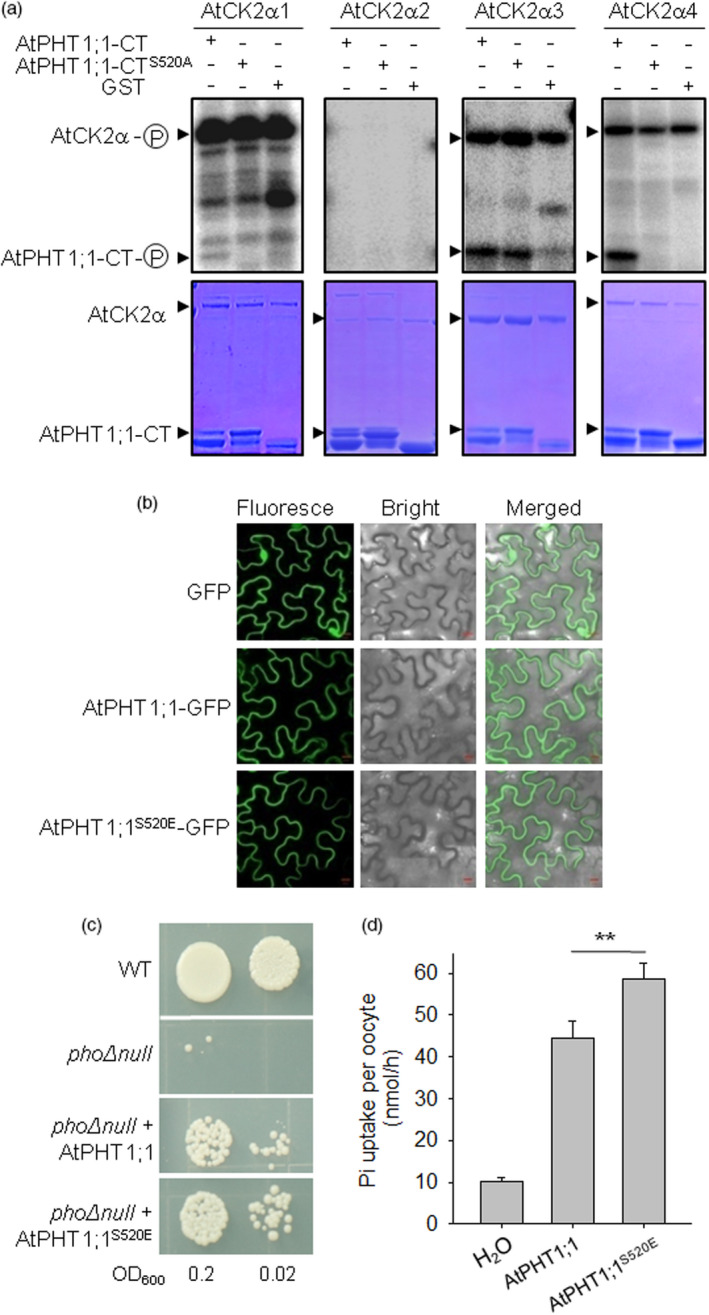
Arabidopsis AtPHT1;1 transport activity is modulated by phosphorylation at Ser‐520. (a) In vitro phosphorylation of GST‐AtPHT1;1–CT, non‐phosphorylation mimicking GST‐AtPHT1;1–CTS520A or GST alone by AtCK2αs with corresponding CBB staining. (b) Location of AtPHT1;1‐GFP, AtPHT1;1S520E‐GFP or GFP in tobacco leaves. Bars = 10 μm. (c) Transport activity test in yeast mutant pho∆null. Wild‐type AtPHT1;1 and AtPHT1;1S520E were expressed in pho∆null separately and incubated at 30 °C for 5 days. (d) Transport activity test in oocytes. Wild‐type AtPHT1;1 cRNA, AtPHT1;1S520E cRNA and water were injected into oocytes and then treated as the description of Figure 6i. Data are means ± SE of n = 6. Asterisks indicate a significant difference between AtPHT1;1 and AtPHT1;1S520E cRNA‐injected oocytes, **P < 0.01.
Discussion
ZmPT7 participates in Pi uptake and redistribution in maize
Maize takes up Pi directly through the PHT1 transporter or indirectly through mycorrhizal‐specific Pi transporter (Calderón‐Vázquez et al., 2011). Although several maize PHT1 genes were found by bioinformatics method and cloned (Liu et al., 2016; Nagy et al., 2006), it was not known whether these putative PHT1 proteins had Pi‐transport activity or which PHT1 transporter(s) participated in Pi acquisition in maize. The ZmPT7 can complement the Pi‐uptake and growth defects of yeast mutant phoΔnull and Arabidopsis mutant pht1;1Δ4Δ (Figures 1d and 2), indicating that ZmPT7 functioned as a Pi transporter. The CRISPR/Cas9 mutant of ZmPT7, zmpt7‐1 and zmpt7‐2 displayed obviously reduced P contents and Pi‐uptake rates compared with wild‐type plants (Figure 3), demonstrating that ZmPT7 played an important role in Pi acquisition in maize.
Transcripts of ZmPT7 accumulated in roots and shoots, and were obviously induced during Pi starvation (Figure 1b). The ZmPT7‐overexpressing lines displayed increased P contents and Pi‐uptake rates compared with wild‐type plants, and the increment was related to the transcript level of ZmPT7, with more increased P content and Pi‐uptake rate in the Ubi:ZmPT7‐2 line and less increment in the Ubi:ZmPT7‐1 line (Figure 4). These data suggest that the transcriptional regulation of ZmPT7 played a role in maize Pi acquisition. The transcript of Arabidopsis AtPHT1;1 is induced by low‐Pi stress (Shin et al., 2004) and is directly regulated by transcription factors AtPHR1 (Bustos et al., 2010), AtWRKY45 (Wang et al., 2014a) and AtWRKY42 (Su et al., 2015). Rice OsPHR2 is a homolog of AtPHR1 and positively regulates expression of OsPT2 (Liu et al., 2010). The W‐box and P1BS motifs are the binding sites of WRKY and PHR transcription factors, respectively (Bustos et al., 2010; Eulgem et al., 2000), and there are several W‐box and P1BS motifs within the 1.5‐kb ZmPT7 promoter (Figure S3), suggesting that ZmPT7 can be transcriptionally modulated by WRKY or PHR transcription factors.
The Pi distribution among plant organs and tissues is important for maintaining Pi homoeostasis. During growth, leaf senescence or Pi starvation, Pi is mobilized from old leaves and transported to the sink organs, such as young leaves. In Arabidopsis, about 78% of P was remobilized from senescing leaves (Shane et al., 2014), and Pi‐transporter AtPHT1;5 mobilized Pi between source and sink organs (Nagarajan et al., 2011). Rice Pi‐transporter OsPT8 was reported to be involved in Pi translocation from old leaves to sink organs, and knockdown of OsPT8 in shoots resulted in an increase in total P concentrations in old leaves (Li et al., 2015). Maize ZmPT7 was a close homolog of OsPT8 (Figure 1a), and ZmPT7 was mainly expressed in mature leaves (Figure 1c). The zmpt7 mutants showed reduced, whereas ZmPT7‐overexpressing lines displayed increased, Pi redistribution from old to young leaves (Figure 5a–d). The ZmPT7 was mainly expressed in bundle sheath cells of leaves (Figure 5e,f), which benefited Pi translocation into the phloem. The ZmPT7 was phosphorylated in old leaves, not in young leaves (Figure 6b), and this phosphorylation modulation enhanced the Pi‐transport capacity of ZmPT7 (Figure 6h,i). These data suggested that ZmPT7 modulated Pi redistribution from old to young leaves in a phosphorylation‐dependent way.
Phosphorylation is a main post‐transcriptional regulation for Pi transporters
Over the past decade, numerous studies revealed that Pi transporters were strongly regulated at the transcriptional level (Chiou and Lin, 2011; Liang et al., 2014; Rouached et al., 2010). Increasing numbers of reports showed that Pi transporters were also subjected to post‐transcriptional regulation. Arabidopsis ubiquitin E3 ligase NLA and ubiquitin–conjugation enzyme PHO2 modulated the abundances of Arabidopsis PHT1 proteins (Huang et al., 2013; Lin et al., 2013; Park et al., 2014), and Arabidopsis ALIX (ALG2‐interacting protein X) regulated vacuolar degradation of AtPHT1;1 (Cardona‐López et al., 2015). The plasma membrane location of Arabidopsis AtPHT1;1 and rice OsPT8 and OsPT2 was regulated by AtPHF1 and OsPHF1 (Chen et al., 2011; González et al., 2005). And the AtPHT1;1, AtPHT1;4 and OsPT8 were modulated by phosphorylation (Bayle et al., 2011; Chen et al., 2015; Nühse et al., 2004).
Protein phosphorylation is a well‐known type of post‐transcriptional modification and plays important roles in transporter function. Previous reports demonstrated that transporter AtNRT1.1 (AtNPF6.3) had at least two phosphorylated sites, Thr‐101 and His‐356, and the former site (NRT1.1 Thr‐101) was involved in nitrate sensing and switching the transport affinity of NRT1.1 (Liu and Tsay, 2003); the latter site (NRT1.1 His‐356) affected structural flexibility and in turn the transport rate of NRT1.1 (Parker and Newstead, 2014). The Arabidopsis ammonium transporter AMT1.1 exhibited active and inactive states which controlled by phosphorylation at Thr‐460 in the CT of AMT1.1 (Lanquar et al., 2009; Loqué et al., 2007). Similar to AMT1.1, the Pi transporters AtPHT1;1, ZmPT7 and OsPT8 were also phosphorylated in the CT (Bayle et al., 2011; Chen et al., 2015; Nühse et al., 2004; Walley et al., 2016). Arabidopsis AtPHT1;1 was phosphorylated at Ser‐514 and Ser‐520 (Bayle et al., 2011; Nühse et al., 2004). Phosphorylation of AtPHT1;1 Ser‐514 retains AtPHT1;1 in the ER retention of PTs (Bayle et al., 2011). In rice, the Ser‐517 of OsPT8 was a conserved serine residue of AtPHT1;1 Ser‐514 (Figure 6a) and was phosphorylated by rice CK2 kinase OsCK2α3 (Chen et al., 2015). The phosphorylation of OsPT8 Ser‐517 by OsCK2α3 inhibited the interaction of PT8 with OsPHF1 to retain OsPT8 in ER retention (Chen et al., 2015), similar to Arabidopsis PHT1;1 Ser‐514 (Bayle et al., 2011). ZmPT7 Ser‐515 was the conserved residue with AtPHT1;1 Ser‐514 and OsPT8 Ser‐517 (Figure 6a; Figure S4). Although the ZmPT7 Ser‐515 was not phosphorylated by kinase ZmCK2 in vitro (Figure 6e), the subcellular location of mimicking phosphorylated and non‐phosphorylated forms of ZmPT7 Ser‐515 (ZmPT7S515E and ZmPT7S515A) were tested in N. benthamiana leaves. To our surprise, ZmPT7S515A and ZmPT7S515E showed a similar expression pattern to the wild‐type ZmPT7 (Figure S5), suggesting that the putative phosphorylation modulation of ZmPT7 Ser‐515 did not affect the subcellular location of ZmPT7.
Previous reports showed that a general trend for the regulation of anion/cation uptake transporters, such as AKT1, NRT1.1 and AMT1.1, is preferentially linked to the phosphorylation (Liu and Tsay, 2003; Loqué et al., 2007; Parker and Newstead, 2014; Xu et al., 2006). Our results expand the understanding of phosphorylation modification in phosphate uptake transporters. Arabidopsis AtPHT1;1 was phosphorylated at Ser‐520 (Bayle et al., 2011; Chen et al., 2011), which was phosphorylated by kinases AtCK2α1 and AtCK2α4 in vitro (Figure 7a), and this phosphorylation modification at AtPHT1;1 Ser‐520 enhanced the Pi‐transport activity of AtPHT1;1 (Figure 7c,d). Maize Pi‐transporter ZmPT7 was phosphorylated by ZmCK2s in old leaves at ZmPT7 Ser‐521, a conserved residue of AtPHT1;1 Ser‐520 (Figure 6). This phosphorylation modification of ZmPT7 Ser‐521 enhanced the transporting function of ZmPT7 (Figure 6h,i). Together, these findings indicate that the regulation of uptake activity of transporters is preferentially linked to the phosphorylation of specific residues. In addition to ZmPT7 Ser‐521, the Ser‐534 of ZmPT7 was also phosphorylated (Walley et al., 2016). The ZmCK2 kinase cannot phosphorylate ZmPT7 at Ser‐534 in vitro (Figure 6e), suggesting that there was another kinase phosphorylating ZmPT7 Ser‐534. For PHT1 proteins, the serine residue of ZmPT7 Ser‐534 was not well conserved (Figure S4), indicating there was a different regulatory mechanism.
Experimental procedures
Plant material and growth conditions
The zmpt7 mutants were generated with CRISPR/Cas9 method. A sgRNA pair (C1, AACGTCGCGGCGGCGGTCAACGG; and C2, CGTGTACGGGATGACGCTCATGG) in ZmPT7 was designed and cloned into the pBUE411 vector (Xing et al., 2014). To obtain the ZmPT7‐overexpressing lines, the coding sequence of ZmPT7 amplified from maize inbred B73 was cloned into a modified pBCXUN, resulting in the Ubi:ZmPT7 construct. To generate the ProPT7:GUS line, the 2410‐bp promoter of ZmPT7 was cloned into pCM3300M‐GUS vector, resulting in the ProZmPT7:GUS construct. All these recombinant vectors were Agrobacterium‐transformed into maize inbred line B73. The T2 or T3 homozygous transgenic lines were used in this study.
The maize pot experiments were performed in solar greenhouse (Beijing) with a 14 h (28 ± 3 °C)/10 h (23 ± 3 °C) light/dark photoperiod, 400 μmol/m2/s irradiance and 45% relative humidity. The maize seeds were sterilized in 10% H2O2 for 30 min, washed with deionized water three times and then soaked in saturated CaSO4 solution overnight before germination. Each seed was germinated and grown in a pot with 5 kg of Turface® clay (Goron et al., 2015), premixed with 1.9 g of KH2PO4, 2.15 g of CO(NH2)2, 0.55 g of KCl, 1.25 g of MgSO4⋅7H2O, 472.3 μg of Ca(NO3)2⋅4H2O, 186 μg of Na2EDTA⋅2H2O, 139 μg of FeSO4⋅7H2O, 0.845 μg of MnSO4⋅H2O, 1.438 μg of ZnSO4⋅7H2O, 0.125 μg of CuSO4⋅5H2O, 0.309 μg of (NH4)6Mo7O24⋅4H2O and 0.309 μg of H3BO3. When grown to the V4 stage, a half amount of the premixed nutrients was further added.
For Arabidopsis experiments, the media [MS, LP, 0 μm As(V), and 200 μm As(V)] and growth conditions were conducted as described previously (Su et al., 2015). The Pi concentration was 1.25 mm in MS, 0 μm As(V) or 200 μm As(V) medium, and 10 μm in LP medium. For the pht1;1Δ4Δ/ZmPT7 lines, the coding sequence of ZmPT7 was cloned into the pCAMBIA1300‐ProSuper vector (Su et al., 2015), resulting in the Super:ZmPT7. The Super:ZmPT7 construct was transformed into the pht1;1Δ4Δ double mutant using the floral dip method (Clough and Bent, 1998), and the homozygous pht1;1Δ4Δ/ZmPT7 lines were obtained.
Construction of the phylogenetic tree
The maize PHT1 sequences were searched with the sequences of AtPHT1;1 and OsPT8 in the NCBI using tblastn. The putative homologs obtained were further characterized based on the identities, conserved domains and predicted transmembrane structures in comparison with Pi transporters in Arabidopsis and rice. To identify the sequences of putative maize PHT1 genes, total RNA of maize inbred B73 was extracted and treated with deoxyribonuclease I to eliminate genomic DNA contamination. The cDNA was synthesized from the treated RNA by reverse transcriptase using Oligo(dT)15 primer. The coding sequences of the putative ZmPHT1 genes were amplified from the cDNA of maize inbred B73 and identified by direct sequencing of the diagnostic PCR products.
For the phylogenetic analysis, the PHT1 amino acid sequences were aligned in ClustalX (version 2.0.12) with default parameters. The neighbour‐joining phylogenetic tree was conducted in MEGA4 (Tamura et al., 2007). Bootstrap values of the phylogenetic tree were calculated as a percentage of 1000 trials.
Yeast complementation assay and Yeast 32Pi‐uptake assay
The wild‐type and mutant coding sequences of ZmPT7 and AtPHT1;1 were introduced to pRS426 vector, respectively. The empty pRS426 and recombinant vectors were transformed into yeast mutant phoΔnull (Wykoff and O'Shea, 2001), respectively. The yeast complement experiments were conducted as described before (Wykoff and O'Shea, 2001). The primers used are listed in Table S3.
The yeast 32Pi‐uptake assay was conducted as described before (Wykoff and O'Shea, 2001). The yeast phoΔnull + ZmPT7 transformants were grown to log phase (OD600 = 0.8–1.0) in YPDA media and then transferred to SD media containing no phosphate (SD/‐Pi) for 3 h. Transformants were washed three times with SD/‐Pi media and resuspended in absorption solution (1.25 mm Tris–Base, 15 mm NaCl, 3% glucose, pH 5.5) with different concentration of KH2 32PO4 for 8 min, and stopped by Tris–succinate solution (25 mm, pH 6.0). And then transformants were washed for 3–5 times by 3% glucose before 32Pi measurement.
RT‐PCR and qRT‐PCR assays
The expression of ZmPT7 in the pht1;1Δ4Δ/ZmPT7, pht1;1Δ4Δ double mutant and Arabidopsis wild‐type plants (Ws genotype) was tested by RT‐PCR assay as described previously (Chen et al., 2009). Elongation Factor 1α (EF1α) was used as a quantitative control.
For qRT‐PCR assay, maize RNA was extracted with Trizol (Invitrogen), and transcript level of ZmPT7 was determined by qRT‐PCR method as described previously (Chen et al., 2009). Relative quantitative results were calculated by normalization to maize Ubiquitin (ZmUBQ) (GenBank accession number: BT018032). The primers used are listed in Table S3.
Pi‐depletion and 32Pi‐uptake assays
For the Pi‐depletion experiment, a group of two seedlings was transferred into a flask with 500 mL of depletion solution modified as previously reported (Liu et al., 2004), containing 100 μm KH2PO4, 325 μm MgSO4, 1 mm Ca(NO3)2, 375 μm K2SO4, 50 μm Fe‐EDTA, 0.5 μm H3BO3, 0.5 μm MnSO4, 0.5 μm ZnSO4, 0.05 μm CuSO4 and 0.025 μm (NH4)6Mo7O24. All flasks were kept on a shaking table at 130 r.p.m. A 500 μL volume of depletion solution was withdrawn at the indicated time, and the Pi concentration was measured.
The 32Pi‐uptake assay for Arabidopsis plants was conducted as described previously (Wang et al., 2014a).
Pi and total P content measurements
For Pi content measurement, the 7‐day‐old Arabidopsis seedlings were transferred to MS or LP medium for 5 days and then harvested for Pi content measurement as described before (Su et al., 2015).
For total P content measurement, tissues of 40‐day‐old maize plants were harvested, and the total P content was measured as described before (Chen et al., 2009).
Subcellular localization assay
The coding sequences of ZmPT7 and AtPHT1;1 were respectively cloned into pSuper1300:GFP vector, resulting in the ZmPT7–GFP and AtPHT1;1–GFP constructs. The constructs ZmPT7S521E–GFP and AtPHT1;1S520E–GFP were generated from ZmPT7–GFP or AtPHT1;1–GFP using site‐directed mutagenesis technology. The constructs were respectively transformed into N. benthamiana leaves, and after infiltration for 4 days, the GFP signal was observed using a confocal laser scanning microscope (LSM710, Carl Zeiss).
Phosphorylation assay
The sequence of hydrophilic CT of ZmPT7 or AtPHT1;1 was fused to pGEX‐4T‐1 vector and resulted in GST‐ZmPT7–CT and GST‐AtPHT1;1–CT constructs. The constructs GST‐ZmPT7–CTS506A , GST‐ZmPT7–CTS510A , GST‐ZmPT7–CTS515A , GST‐ZmPT7–CTS521A and GST‐ZmPT7–CTS534A were generated from GST‐ZmPT7–CT, and construct GST‐AtPHT1;1–CTS520A was generated from GST‐AtPHT1;1–CT, using site‐directed mutagenesis technology. The recombinant constructs were introduced into E. coli strain BL21. The E. coli cells were induced with 0.2 mm IPTG overnight at 18 °C. The fusion proteins were purified with glutathione–sepharose beads.
The in vitro phosphorylation assay used a 20‐μL kinase solution containing 25 mm Tris–HCl (pH 7.5), 10 mm MgCl2, 1 mm CaCl2, 1 mm DTT, 1 μm ATP, 5 μg of kinase (ZmCK2α1, ZmCK2α2, ZmCK2α3 or ZmCK2α4) and 5 μg of ZmPT7–CT protein. Phosphorylation was initiated by adding 1 μCi [γ‐32P] ATP. After incubation for 15 min at 30 °C, the reactions were stopped by adding 5 × loading buffer and incubated for 10 min at 95 °C. The reaction products were fractionated by SDS‐PAGE, and the phosphorylated proteins visualized by autoradiography.
The in vitro phosphorylation of GST‐ZmPT7–CT by ZmCK2α2 under different Pi concentration used a 20‐μL kinase solution containing 25 mm Tris–HCl (pH 7.5), 10 mm MgCl2, 1 mm DTT, 1 μm ATP, 5 μg of ZmCK2α2 protein, 5 μg of ZmPT7–CT protein, and 0, 1, 2, 5 or 10 mm KH2PO4, or 5 mm KCl. And the phosphorylation assay was done as above.
For the semi in vivo phosphorylation assay (Phos‐tag mobility shift assay), the wild‐type maize was germinated and grown for 40 days, and then, each leaf was harvested for protein extraction. To monitor the phosphorylation of recombinant GST‐ZmPT7–CT and GST‐ZmPT7–CTS521A, 1 μg of each purified recombinant protein was incubated with 300 μg of leaf total proteins at 28 °C for 1 h. The CIAP was used to dephosphorylate ZmPT7–CT as described before (Feng et al., 2014). The phosphorylated and dephosphorylated ZmPT7–CT peptides were distinguished using 8% Phos‐tag gel (NARD, AAL‐107) following the manufacturer’s protocol, and the ZmPT7–CT or ZmPT–CTS521A was detected by immunoblotting with anti‐GST antibody.
Transport activity assay in X. laevis oocytes
The coding sequence of ZmPT7 or AtPHT1;1 was cloned into pT7TS vector, resulting in pT7TS‐ZmPT7 or pT7TS‐AtPHT1;1. The constructs pT7TS‐ZmPT7S521E and pT7TS‐AtPHT1;1S520E were generated from ZmPT7–GFP or AtPHT1;1–GFP using site‐directed mutagenesis technology. After linearization of pT7TS plasmids with XbaI, RNA was transcribed in vitro using an mRNA synthesis kit (mMESSAGE mMACHINE T7 kit; Ambion). Oocytes were injected with 40 ng of RNA after recovery and were incubated in ‐free ND96 solution (98 mm NaCl, 2 mm KCl, 1 mm MgCl2, 1.8 mm CaCl2 and 5 mm HEPES, pH 7.5) for 36 h at 18 °C. The incubation solution was refreshed daily. Then, the oocytes were transferred into bath solution buffer containing 0.5 mm Pi with 32P (1 mCi/mL H3 32PO4, pH 5.5) for 2 h at 18 °C. The oocytes were washed five times, and radioactivity in the oocytes was measured.
Conflicts of interest
The authors declare no conflicts of interest related to this work.
Author contributions
Y.‐F. C. designed the research. P.‐J.C., F.W., Y.T., Y.H. and H.‐F.W. performed the research, and F.L. tested the gene expression in genetic materials. Y.‐F. C., F.W., P.‐J.C. and Y.T. analysed the data. Y.‐F. C., F.W. and P.‐J.C. wrote the paper.
Supporting information
Figure S1 Identification of zmpt7 mutants.
Figure S2 Analysis of transcript abundance of ZmPT7 in different leaves of 40‐day‐old Ubi:ZmPT7‐1 line by qRT‐PCR
Figure S3 In silico analysis of ZmPT promoter sequences
Figure S4 Sequence alignment of the hydrophilic C‐termini (CT) of the PHT1 transporters in maize, Arabidopsis and rice.
Figure S5 Location of ZmPT7–GFP and Ser515 variant of ZmPT7 in tobacco leaves.
Table S1 PHT1 transporters in maize, rice, soybean and Arabidopsis.
Table S2 Percentage of amino acid identity among maize PHT1 proteins, AtPHT1;1 and OsPT8.
Table S3 Primer sequences used in this study.
Acknowledgements
The transgenic maize lines and vectors (pBCXUN and pCM3300M‐GUS) used in this study were created by the Maize Functional Genomic Platform of China Agricultural University. We thank Erin K. O’Shea (Harvard University, USA) for phoΔnull mutant and Maria J. Harrison (Boyce Thompson Institute for Plant Research, USA) for Arabidopsis pht1;1∆4∆ double mutant. This work was supported by grants from the National Key Research and Development Program of China (No. 2016YFD0100707), the Ministry of Agriculture of China for transgenic research (No. 2016ZX08009002), the National Natural Science Foundation of China (Nos. 31670245 and 31970273), Chinese Universities Scientific Fund (2019TC122 and 2019TC228) and Beijing Outstanding University Discipline Program.
Wang, F. , Cui, P.‐J. , Tian, Y. , Huang, Y. , Wang, H.‐F. , Liu, F. and Chen, Y.‐F. (2020) Maize ZmPT7 regulates Pi uptake and redistribution which is modulated by phosphorylation. Plant Biotechnol. J., 10.1111/pbi.13414
References
- Ai, P. , Sun, S. , Zhao, J. , Fan, X. , Xin, W. , Guo, Q. , Yu, L. et al. (2009) Two rice phosphate transporters, OsPht1;2 and OsPht1;6, have different functions and kinetic properties in uptake and translocation. Plant J. 57, 798–809. [DOI] [PubMed] [Google Scholar]
- Bayle, V. , Arrighi, J.F. , Creff, A. , Nespoulous, C. , Vialaret, J. , Rossignol, M. , Gonzalez, E. et al. (2011) Arabidopsis thaliana high‐affinity phosphate transporters exhibit multiple levels of posttranslational regulation. Plant Cell, 23, 1523–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustos, R. , Castrillo, G. , Linhares, F. , Puga, M.I. , Rubio, V. , Pérez‐Pérez, J. , Solano, R. et al. (2010) A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet. 6, e1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderón‐Vázquez, C. , Sawers, R.J. and Herrera‐Estrella, L. (2011) Phosphate deprivation in maize: genetics and genomics. Plant Physiol. 156, 1067–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardona‐López, X. , Cuyas, L. , Marín, E. , Rajulu, C. , Irigoyen, M.L. , Gil, E. , Puga, M.I. et al. (2015) ESCRT‐III‐associated protein ALIX mediates high‐affinity phosphate transporter trafficking to maintain phosphate homeostasis in Arabidopsis. Plant Cell, 27, 2560–2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castrillo, G. , Sánchez‐Bermejo, E. , de Lorenzo, L. , Crevillén, P. , Fraile‐Escanciano, A. , Tc, M. , Mouriz, A. et al. (2013) WRKY6 transcription factor restricts arsenate uptake and transposon activation in Arabidopsis . Plant Cell, 25, 2944–2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catarecha, P. , Segura, M.D. , Franco‐Zorrilla, J.M. , García‐Ponce, B. , Lanza, M. , Solano, R. , Paz‐Ares, J. et al. (2007) A mutant of the Arabidopsis phosphate transporter PHT1;1 displays enhanced arsenic accumulation. Plant Cell, 19, 1123–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L. and Liao, H. (2017) Engineering crop nutrient efficiency for sustainable agriculture. J. Integr. Plant Biol. 59, 710–735. [DOI] [PubMed] [Google Scholar]
- Chen, Y.F. , Li, L.Q. , Xu, Q. , Kong, Y.H. , Wang, H. and Wu, W.H. (2009) The WRKY6 transcription factor modulates PHOSPHATE1 expression in response to low Pi stress in Arabidopsis . Plant Cell, 21, 3554–3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J. , Liu, Y. , Ni, J. , Wang, Y. , Bai, Y. , Shi, J. , Gan, J. et al. (2011) OsPHF1 regulates the plasma membrane localization of low‐ and high‐affinity inorganic phosphate transporters and determines inorganic phosphate uptake and translocation in rice. Plant Physiol. 157, 269–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J. , Wang, Y. , Wang, F. , Yang, J. , Gao, M. , Li, C. , Liu, Y. et al. (2015) The rice CK2 kinase regulates trafficking of phosphate transporters in response to phosphate levels. Plant Cell, 27, 711–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou, T.J. and Lin, S.I. (2011) Signaling network in sensing phosphate availability in plants. Annu. Rev. Plant Biol. 62, 185–206. [DOI] [PubMed] [Google Scholar]
- Clough, S.J. and Bent, A.F. (1998) Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana . Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Devaiah, B.N. , Karthikeyan, A.S. and Raghothama, K.G. (2007) WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol. 143, 1789–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem, T. , Rushton, P.J. , Robatzek, S. and Somssich, I.E. (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. [DOI] [PubMed] [Google Scholar]
- Feng, C.Z. , Chen, Y. , Wang, C. , Kong, Y.H. , Wu, W.H. and Chen, Y.F. (2014) Arabidopsis RAV1 transcription factor, phosphorylated by SnRK2 kinases, regulates the expression of ABI3, ABI4, and ABI5 during seed germination and early seedling development. Plant J. 80, 654–668. [DOI] [PubMed] [Google Scholar]
- González, E. , Solano, R. , Rubio, V. , Leyva, A. and Paz‐Ares, J. (2005) PHOSPHATE TRANSPORTER TRAFFIC FACILITATOR1 is a plant‐specific SEC12‐related protein that enables the endoplasmic reticulum exit of a high‐affinity phosphate transporter in Arabidopsis . Plant Cell, 17, 3500–3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goron, T.L. , Watts, S. , Shearer, C. and Raizada, M.N. (2015) Growth in Turface® clay permits root hair phenotyping along the entire crown root in cereal crops and demonstrates that root hair growth can extend well beyond the root hair zone. BMC Res. Notes. 8, 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, T.K. , Han, C.L. , Lin, S.I. , Chen, Y.J. , Tsai, Y.C. , Chen, Y.R. , Chen, J.W. et al. (2013) Identification of downstream components of ubiquitin‐conjugating enzyme PHOSPHATE2 by quantitative membrane proteomics in Arabidopsis roots. Plant Cell, 25, 4044–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, H. , Ren, H. , Gu, M. , Zhao, J. , Sun, S. , Zhang, X. , Chen, J. et al. (2011) The phosphate transporter gene OsPht1;8 is involved in phosphate homeostasis in rice. Plant J. 156, 1164–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanquar, V. , Loqué, D. , Hörmann, F. , Yuan, L. , Bohner, A. , Engelsberger, W.R. , Lalonde, S. et al. (2009) Feedback inhibition of ammonium uptake by a phospho‐dependent allosteric mechanism in Arabidopsis. Plant Cell, 21, 3610–3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Zhang, J. , Zhang, X. , Fan, H. , Gu, M. , Qu, H. and Xu, G. (2015) Phosphate transporter OsPht1;8 in rice plays an important role in phosphorus redistribution from source to sink organs and allocation between embryo and endosperm of seeds. Plant Sci. 230, 23–32. [DOI] [PubMed] [Google Scholar]
- Liang, C. , Wang, J. , Zhao, J. , Tian, J. and Liao, H. (2014) Control of phosphate homeostasis through gene regulation in crops. Curr. Opin. Plant Biol. 21, 59–66. [DOI] [PubMed] [Google Scholar]
- Lin, W.Y. , Huang, T.K. and Chiou, T.J. (2013) Nitrogen limitation adaptation, a target of microRNA827, mediates degradation of plasma membrane‐localized phosphate transporters to maintain phosphate homeostasis in Arabidopsis . Plant Cell, 25, 4061–4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, K.H. and Tsay, Y.F. (2003) Switching between the two action modes of the dual‐affinity nitrate transporter CHL1 by phosphorylation. EMBO J. 22, 1005–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. , Mi, G. , Chen, F. , Zhang, J. and Zhang, F. (2004) Rhizosphere effect and root growth of two maize (Zea mays L.) genotypes with contrasting P efficiency at low P availability. Plant Sci. 167, 217–223. [Google Scholar]
- Liu, F. , Wang, Z. , Ren, H. , Shen, C. , Li, Y. , Ling, H.Q. , Wu, C. et al. (2010) OsSPX1 suppresses the function of OsPHR2 in the regulation of expression of OsPT2 and phosphate homeostasis in shoots of rice. Plant J. 62, 508–517. [DOI] [PubMed] [Google Scholar]
- Liu, F. , Chang, X.J. , Ye, Y. , Xie, W.B. , Wu, P. and Lian, X.M. (2011) Comprehensive sequence and whole‐life‐cycle expression profile analysis of the phosphate transporter gene family in rice. Mol. Plant, 4, 1105–1122. [DOI] [PubMed] [Google Scholar]
- Liu, F. , Xu, Y. , Jiang, H. , Jiang, C. , Du, Y. , Gong, C. , Wang, W. et al. (2016) Systematic identification, evolution and expression analysis of the Zea mays PHT1 gene family reveals several new members involved in root colonization by arbuscular mycorrhizal fungi. Int. J. Mol. Sci. 17, 930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, F. , Xu, Y. , Han, G. , Wang, W. , Li, X. and Cheng, B. (2018) Identification and functional characterization of a maize phosphate transporter induced by mycorrhiza formation. Plant Cell Physiol. 59, 1683–1694. [DOI] [PubMed] [Google Scholar]
- López‐Arredondo, D.L. , Leyva‐González, M.A. , González‐Morales, S.I. , López‐Bucio, J. and Herrera‐Estrella, L. (2014) Phosphate nutrition: improving low‐phosphate tolerance in crops. Annu. Rev. Plant Biol. 65, 95–123. [DOI] [PubMed] [Google Scholar]
- Loqué, D. , Lalonde, S. , Looger, L.L. , von Wirén, N. and Frommer, W.B. (2007) A cytosolic trans‐activation domain essential for ammonium uptake. Nature, 446, 195–198. [DOI] [PubMed] [Google Scholar]
- Loth‐Pereda, V. , Orsini, E. , Courty, P.E. , Lota, F. , Kohler, A. , Diss, L. , Blaudez, D. et al. (2011) Structure and expression profile of the phosphate Pht1 transporter gene family in mycorrhizal Populus trichocarpa . Plant Physiol. 156, 2141–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Młodzińska, E. and Zboińska, M. (2016) Phosphate uptake and allocation ‐ A closer look at Arabidopsis thaliana L. and Oryza sativa L. Front. Plant Sci. 7, 1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudge, S.R. , Rae, A.L. , Diatloff, E. and Smith, F.W. (2002) Expression analysis suggests novel roles for members of the Pht1 family of phosphate transporters in Arabidopsis . Plant J. 31, 341–353. [DOI] [PubMed] [Google Scholar]
- Nagarajan, V.K. , Jain, A. , Poling, M.D. , Lewis, A.J. , Raghothama, K.G. and Smith, A.P. (2011) Arabidopsis Pht1;5 mobilizes phosphate between source and sink organs and influences the interaction between phosphate homeostasis and ethylene signaling. Plant Physiol. 156, 1149–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy, R. , Vasconcelos, M.J. , Zhao, S. , McElver, J. , Bruce, W. , Amrhein, N. , Raghothama, K.G. et al. (2006) Differential regulation of five Pht1 phosphate transporters from maize (Zea mays L.). Plant Biol. (Stuttg), 8, 186–197. [DOI] [PubMed] [Google Scholar]
- Niefind, K. , Pütter, M. , Guerra, B. , Issinger, O.G. and Schomburg, D. (1999) GTP plus water mimic ATP in the active site of protein kinase CK2. Nat. Struct. Biol. 6, 1100–1103. [DOI] [PubMed] [Google Scholar]
- Nühse, T.S. , Stensballe, A. , Jensen, O.N. and Peck, S.C. (2004) Phosphoproteomics of the Arabidopsis plasma membrane and a new phosphorylation site database. Plant Cell, 16, 2394–2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, B.S. , Seo, J.S. and Chua, N.H. (2014) Nitrogen limitation adaptation recruits phosphate2 to target the phosphate transporter PT2 for degradation during the regulation of Arabidopsis phosphate homeostasis. Plant Cell, 26, 454–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, J.L. and Newstead, S. (2014) Molecular basis of nitrate uptake by the plant nitrate transporter NRT1.1. Nature, 507, 68–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen, B.P. , Kumar, H. , Waight, A.B. , Risenmay, A.J. , Roe‐Zurz, Z. , Chau, B.H. , Schlessinger, A. et al. (2013) Crystal structure of a eukaryotic phosphate transporter. Nature, 496, 533–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popova, Y. , Thayumanavan, P. , Lonati, E. , Agrochão, M. and Thevelein, J.M. (2010) Transport and signaling through the phosphate‐binding site of the yeast Pho84 phosphate transceptor. Proc. Natl. Acad. Sci. USA, 107, 2890–2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portolés, S. and Más, P. (2010) The functional interplay between protein kinase CK2 and CCA1 transcriptional activity is essential for clock temperature compensation in Arabidopsis. PLoS Genet. 6, e1001201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghothama, K.G. and Karthikeyan, A.S. (2005) Phosphate acquisition. Plant Soil, 274, 37–49. [Google Scholar]
- Riera, M. , Irar, S. , Vélez‐Bermúdez, I.C. , Carretero‐Paulet, L. , Lumbreras, V. and Pagès, M. (2011) Role of plant‐specific N‐terminal domain of maize CK2β1 subunit in CK2β functions and holoenzyme regulation. PLoS One 6, e21909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouached, H. , Arpat, A.B. and Poirier, Y. (2010) Regulation of phosphate starvation responses in plants: signaling players and cross‐talks. Mol. Plant, 3, 288–299. [DOI] [PubMed] [Google Scholar]
- Sawers, R.J. , Svane, S.F. , Quan, C. , Grønlund, M. , Wozniak, B. , Gebreselassie, M.N. , González‐Muñoz, E. et al. (2017) Phosphorus acquisition efficiency in arbuscular mycorrhizal maize is correlated with the abundance of root‐external hyphae and the accumulation of transcripts encoding PHT1 phosphate transporters. New Phytol. 214, 632–643. [DOI] [PubMed] [Google Scholar]
- Schachtman, D.P. , Reid, R.J. and Ayling, S.M. (1998) Phosphorus uptake by plants: From soil to cell. Plant Physiol. 116, 447–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shane, M.W. , Stigter, K. , Fedosejevs, E.T. and Plaxton, W.C. (2014) Senescence‐inducible cell wall and intracellular purple acid phosphatases: implications for phosphorus remobilization in Hakea prostrata (Proteaceae) and Arabidopsis thaliana (Brassicaceae). J. Exp. Bot. 65, 6097–6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin, H. , Shin, H.S. , Dewbre, G.R. and Harrison, M.J. (2004) Phosphate transport in Arabidopsis: Pht1;1 and Pht1;4 play a major role in phosphate acquisition from both low‐ and high‐phosphate environments. Plant J. 39, 629–642. [DOI] [PubMed] [Google Scholar]
- Su, T. , Xu, Q. , Zhang, F.C. , Chen, Y. , Li, L.Q. , Wu, W.H. and Chen, Y.F. (2015) WRKY42 modulates phosphate homeostasis through regulating phosphate translocation and acquisition in Arabidopsis . Plant Physiol. 167, 1579–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, S. , Gu, M. , Cao, Y. , Huang, X. , Zhang, X. , Ai, P. , Zhao, J. et al. (2012) A constitutive expressed phosphate transporter, OsPht1;1, modulates phosphate uptake and translocation in phosphate‐replete rice. Plant Physiol. 159, 1571–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, K. , Dudley, J. , Nei, M. and Kumar, S. (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599. [DOI] [PubMed] [Google Scholar]
- Vilela, B. , Nájar, E. , Lumbreras, V. , Leung, J. and Pagès, M. (2015) Casein Kinase 2 negatively regulates abscisic acid‐activated SnRK2s in the core abscisic acid‐signaling module. Mol. Plant, 8, 709–721. [DOI] [PubMed] [Google Scholar]
- Walley, J.W. , Sartor, R.C. , Shen, Z. , Schmitz, R.J. , Wu, K.J. , Urich, M.A. , Nery, J.R. et al. (2016) Integration of omic networks in a developmental atlas of maize. Science, 353, 814–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H. , Xu, Q. , Kong, Y.H. , Chen, Y. , Duan, J.Y. , Wu, W.H. and Chen, Y.F. (2014a) Arabidopsis WRKY45 transcription factor activates PHOSPHATE TRANSPORTER1;1 expression in response to phosphate starvation. Plant Physiol. 164, 2020–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. , Wang, Y. , Piñeros, M.A. , Wang, Z. , Wang, W. , Li, C. , Wu, Z. et al. (2014b) Phosphate transporters OsPHT1;9 and OsPHT1;10 are involved in phosphate uptake in rice. Plant Cell Environ. 37, 1159–1170. [DOI] [PubMed] [Google Scholar]
- Willmann, M. , Gerlach, N. , Buer, B. , Polatajko, A. , Nagy, R. , Koebke, E. , Jansa, J. et al. (2013) Mycorrhizal phosphate uptake pathway in maize: vital for growth and cob development on nutrient poor agricultural and greenhouse soils. Front. Plant Sci. 4, 533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wykoff, D.D. and O'Shea, E.K. (2001) Phosphate transport and sensing in Saccharomyces cerevisiae. Genetics, 159, 1491–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing, H.L. , Dong, L. , Wang, Z.P. , Zhang, H.Y. , Han, C.Y. , Liu, B. , Wang, X.C. et al. (2014) A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 14, 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, J. , Li, H.D. , Chen, L.Q. , Wang, Y. , Liu, L.L. , He, L. and Wu, W.H. (2006) A protein kinase, interacting with two calcineurin B‐like proteins, regulates K+ transporter AKT1 in Arabidopsis . Cell, 125, 1347–1360. [DOI] [PubMed] [Google Scholar]
- Zhang, F. , Sun, Y. , Pei, W. , Jain, A. , Sun, R. , Cao, Y. , Wu, X. et al. (2015) Involvement of OsPht1;4 in phosphate acquisition and mobilization facilitates embryo development in rice. Plant J. 82, 556–569. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Identification of zmpt7 mutants.
Figure S2 Analysis of transcript abundance of ZmPT7 in different leaves of 40‐day‐old Ubi:ZmPT7‐1 line by qRT‐PCR
Figure S3 In silico analysis of ZmPT promoter sequences
Figure S4 Sequence alignment of the hydrophilic C‐termini (CT) of the PHT1 transporters in maize, Arabidopsis and rice.
Figure S5 Location of ZmPT7–GFP and Ser515 variant of ZmPT7 in tobacco leaves.
Table S1 PHT1 transporters in maize, rice, soybean and Arabidopsis.
Table S2 Percentage of amino acid identity among maize PHT1 proteins, AtPHT1;1 and OsPT8.
Table S3 Primer sequences used in this study.


