Abstract
Despite the growing knowledge surrounding host–microbiome interactions, we are just beginning to understand how the gut microbiome influences—and is influenced by—host gene expression. Here, we review recent literature that intersects these two fields, summarizing themes across studies. Work in model organisms, human biopsies, and cell culture demonstrate that the gut microbiome is an important regulator of several host pathways relevant for disease, including immune development and energy metabolism, and vice versa. The gut microbiome remodels host chromatin, causes differential splicing, alters the epigenetic landscape, and directly interrupts host signaling cascades. Emerging techniques like single-cell RNA sequencing and organoid generation have the potential to refine our understanding of the relationship between the gut microbiome and host gene expression in the future. By intersecting microbiome and host gene expression, we gain a window into the physiological processes important for fostering the extensive cross-kingdom interactions and ultimately our health.
Electronic supplementary material
The online version of this article (10.1007/s00439-020-02237-0) contains supplementary material, which is available to authorized users.
Introduction
The human body plays host to large numbers of bacteria, fungi and other microorganisms—commonly referred to as the microbiota (Shreiner et al. 2015). These microbes are important to our health. They assist in establishing host immunity (Kamada et al. 2013), strengthen the gut barrier (Leclercq et al. 2014), and provide beneficial metabolites (Lukovac et al. 2014). Consequently, the microbiota is associated with a large number of complex diseases in humans, including inflammatory bowel disease (IBD) (Aleksandrova et al. 2017), cardiovascular disease (CVD) (Tang et al. 2017), and colon cancer (Nelson and Chia 2019). Whether shifts in the microbiota lead to or are the cause of disease remains largely unknown. Although, it has been demonstrated that the microbiota play causal roles in several diseases, including obesity (Ridaura et al. 2014; Turnbaugh et al. 2006) and diabetes (Wen et al. 2008). Given the importance for our health, it is necessary to characterize the physiological relationships between the microbiota and human host to understand disease etiology and design therapeutics involving the microbiome.
There is much to be learned about human–microbiome interactions by studying the genetic components of each. The human genome contains approximately 20,000 protein coding genes (Salzberg 2018). These genes are regulated in a tissue-specific manner by both intrinsic host factors as well as sensing environmental cues. Collectively, the microbial genomes within each of our microbiomes contain an estimated 100 times the gene content as our own genome (Nelson et al. 2010). This genetic material is sometimes referred to as our ‘second genome’, as the coding potential of the microbiome greatly expands upon the coding potential our own genome (Grice and Segre 2012). For example, genes unique to the microbiota create metabolites required by the human host (such as vitamin B12, biotin and folic acid) (Hooper et al. 2002) and allow for microbial survival (such as adhesion factors and transport systems) (Reidl et al. 2009). Understanding how the gene products of the host and microbiome interact can offer clues into what physiological processes are necessary for maintaining these complex cross-kingdom relationships.
In this review, we offer insights about the physiology of host–microbiota relationships gained by studying host gene expression jointly with the microbiome, specifically in the gut. We briefly describe the microbiome and how it is studied, synthesize themes gained through studies performed in powerful model systems and in humans, and highlight potential mechanisms through which host transcription–microbiota cross-talk occurs. Finally, we propose future directions, both experimental and analytical, that will further our understanding of how host transcription and the microbiome interact.
The human gut microbiome: a primer
The relationship between the host and the gut microbiome starts at birth when the microbiome of the newborn is seeded. Delivery mode (cesarean section or vaginal delivery) plays an important role in the establishment of the microbiome (Bäckhed et al. 2015; Dominguez-Bello et al. 2010; Montoya-Williams et al. 2018; Papathoma et al. 2016). Proper nutrition and the transition from breast feeding to more solid foods results in maturation of the infant microbiome (Bäckhed et al. 2015). The establishment of the microbiome in neonates works in concert with the establishment of innate mucosal immunity, but can be disrupted by early use of antibiotics (Russell et al. 2012). The composition of the microbiome rapidly diversifies up to the age of three, steadily increases until around the age of 40, and then remains fairly stable (de la Cuesta-Zuluaga et al. 2019; Yadav et al. 2016; Yatsunenko et al. 2012). Though highly unique between individuals, the microbiome is relatively resistant to long-term changes (Bäckhed et al. 2012). The microbiome will typically return to a state of equilibrium after a stress like a dietary change, a short term adult antibiotic treatment, or an acute invasion by a pathogenic bacterium (Bäckhed et al. 2012). However, short-term modulations of the gut microbiome can interrupt normal metabolite production (Yoon and Yoon 2018). This in turn may cause changes in host gene expression that could lead to more long-lasting effects in the host.
To investigate the microbiome, researchers employ a plethora of techniques, many involving next-generation sequencing technology. To explore the taxonomic makeup of the gut microbiome, sequencing a phylogenetic marker gene is easy and efficient. Most often the 16S rRNA gene is assayed, as it is universally present in archaea and bacteria. The gene contains alternating segments of high and low conservation, which can be used for PCR priming and taxonomic identification, respectively (Davidson and Epperson 2018).
To explore microbial genomes and functional capacity of the gut microbiome, shotgun metagenomics is used. This involves sequencing the total DNA composition of a sample. Several aspects of the microbial community can be assayed using metagenomics, including characterizing taxonomic composition [through programs like Kraken (Wood and Salzberg 2014) and MetaPhlAn2 (Truong et al. 2015)], functional capabilities [through the program HUMAnN2 (Franzosa et al. 2018)], and assembling individual microbial genomes [metagenome assembled genomes, or MAGs (Albertsen et al. 2013; Allen and Banfield 2005; Nielsen et al. 2014; Parks et al. 2017)]. While metagenomics has the potential to reveal additional functional information over phylogenetic marker gene sequencing, it is expensive and can be cost prohibitive in samples with a high host to bacterial biomass ratio.
Finally, to explore functional activity of the gut microbiome, RNA within a microbiome can be sequenced via RNA-seq (referred to as metatranscriptomics). Typically, researchers deplete ribosomal and transfer RNA experimentally to enrich for bacterial transcripts prior to sequencing, as ribosomal and transfer RNA make up approximately 95–97% of total RNA in a bacterial cell (Rosenow et al. 2001). Metatranscriptomics investigates bacterial gene expression (transcription) changes between conditions or individuals [analyzed with the program SAMSA2, for example (Westreich et al. 2018)]. 16S rRNA gene sequencing, metagenomics, and metatranscriptomic sequencing can be coupled with host transcriptomics to investigate how different aspects of the gut microbiome (composition, functional capacity, or functional activity) affects host gene expression and vice versa.
Relationship between microbiome and gene expression in model organisms
A powerful way to investigate both the microbiome and host transcriptomics is with model organisms, such as mice (Fig. 1), zebrafish, C. elegans or Drosophila melanogaster. With model organisms, researchers have control over the environment, complete control over diet, and, importantly, the ability to study all tissues. These aspects are either impossible or extremely difficult to do with human subjects. Organisms bred without a microbiome (e.g., germ-free mice) can be used to identify gut microbiome-mediated effects in the host organism. For example, if the effects of a treatment are lost in germ-free mice when compared to wild-type or conventionalized mice, then it can be inferred that gut microbiome plays a role in the specific effect. While powerful, there are caveats to consider when using germ-free mice. With no microbiome present, there is much thinner mucous layer in the gastrointestinal (GI) tract of a germ-free mouse compared to a conventional mouse (Miyakawa et al. 1971). Barrier function is worse, and there is likely innate inflammation present in the gut (Miyakawa et al. 1971). Germ-free mice also have reduced metabolic rates and enlarged ceca (Miyakawa et al. 1971). These innate functional differences must be noted when drawing conclusions with germ-free mice.
Fig. 1.
Using mice as a model organism. Mice are popular model organism for characterizing gut microbiome–host relationships. Researchers compare gnotobiotic (germ-free), mono-colonized, and/or conventionalized/wild-type mice to determine the role of the microbiome in their phenotype of interest. Orange elements represent host-derived tissues and techniques. Blue elements represent microbiome-derived samples and techniques. Both microbiome-derived and host-derived techniques can be categorized in three groups; sequencing techniques, other -omic techniques and gross tissue techniques. SHIME stands for the Simulator of the Human Intestinal Microbial Ecosystem (Molly et al. 1993)
Regardless, germ-free mouse experiments demonstrate that the microbiome plays a role in regulating host gene expression. For example, a murine model with a tissue-specific deletion of histone deacetylase 3 (HDAC3) results in the dysregulation of intestinal epithelial cell gene regulation dependent on the microbiome (Alenghant et al. 2013). Specifically, an intestinal epithelial cell specific HDAC3 knockout mouse line was bred (HDAC3ΔIEC) to investigate inflammatory bowel disease (IBD) progression. Conventionalized HDAC3ΔIEC mice show dysregulated host gene expression and disrupted homeostasis. Conversely, this dysregulation is not observed in a germ-free HDAC3ΔIEC mouse model, demonstrating that the effects were mediated by the microbiome. These results highlight one mechanism by which the microbiome is sensed and affects gene expression in the host: by influencing the expression of a host epigenetic modifying enzyme.
Modulation of the microbiome (either by probiotics or creating germ free mice) can have dramatic effects on host gene expression. In a study focusing on 303 xenobiotic processing genes expressed in the intestine, 116 genes were significantly differentially expressed between mice raised conventionally vs. germ-free (Fu et al. 2017). These genes include Phase I enzymes, Phase II enzymes, transporters, and transcription factors. Additionally, expression of drug-metabolizing enzymes in the liver are significantly altered between groups of mice raised conventionally, conventionally + probiotics (VSL3), germ-free, and germ-free + probiotics (Selwyn et al. 2016). These studies demonstrate the causal role the microbiome plays in altering host gene expression across the body.
Host energy metabolism is affected by the microbiome, as evidenced by changes in host gene expression. Short-chain fatty acids (SCFAs) are small molecules made solely by the microbiome via microbial fermentation. In animal guts, these molecules are generated by the breakdown of host-indigestible foods and serve as a major energy source for the host. For example, an abundant SCFA of major health importance is butyrate, which colonocytes use as an energy source (Donohoe et al. 2011). In germ-free mice, the TCA cycle is dysregulated in colonocytes, due to the lack of butyrate (Donohoe et al. 2011). However, upon supplementation of butyrate, isolated colonocytes from a germ-free mouse dramatically increased mitochondrial respiration. Additionally, the microbiome influences host hepatic lipogenesis in wild-type mice. Increased monosaccharide absorption in the mouse gut suppresses the expression of fasting-induced adipocyte factor (FAIF), which results in the deposit of triglycerides in adipocytes (Bäckhed et al. 2004). These are just two examples of how microbes in the gut regulate host energy metabolism, even in distant tissues.
Host immunity is also affected by the microbiome. In a mouse model comparing wild-type mice to mice lacking innate immunity (Myd88 knockout model) in both conventionally raised and germ-free conditions, over half of the expressed genes in the GI tract were regulated by the microbiome (2844 of 5652 genes) (Larsson et al. 2012). Gene Ontology (GO) analysis, showed the top differentially expressed genes were involved with host immune responses and host energy metabolism. Interestingly, the microbiomes involvement in both patterns of gene expression and dependency on MyD88 shifted along the GI tract. The composition, gene expression, and epigenetic profile of innate lymphoid cells (ILCs) are also shaped by the microbiome (Gury-BenAri et al. 2016). Specifically, single-cell RNA-seq, ATAC-seq, and iChIP-IVT on the intestinal lamina propria of conventional and antibiotic treated mice revealed hundreds of transcripts that were differentially regulated by the microbiome in several ILC clusters. These included genes related to cellular adhesion and interaction with the extracellular matrix, chemokine signaling, and MAPK signaling. In general, the transcriptomic profile of two ILC subsets (ILC1 and ILC2) shifted towards a third (ILC3). These results demonstrate the degree of plasticity host cells display in response to microbial stimuli and how that can affect downstream immune function. The relationship between the gut microbiome and host innate immunity has been extensively reviewed by Pott and Hornef (2012), Honda and Littman (2016), and Shi et al. (2017).
While we highlight mouse models above, many additional organisms serve as powerful laboratory systems to reveal insights into the relationship between the microbiome and host transcription. Studies conducted in zebrafish (Murdoch and Rawls 2019), C. elegans (Dirksen et al. 2020; Yang et al. 2019), and Drosophila melanogaster (Broderick et al. 2014; Douglas 2018; Elya et al. 2016) for example, all have revealed the impact of the microbiome on immunity, metabolism, and developmental gene regulation.
Gastric pathogens influence host transcription
Given the difficulties of studying human gene expression, far less is known about the relationship between the human microbiome and the human transcriptome. The area with the most research to date is the study of how gut pathogens influence host gene expression, either directly or indirectly through immune stimulation. One of the ways pathogens modulate host gene expression is indirectly through the activation of host microRNAs (miRNA), which are small RNA molecules (~ 20 nucleotides long) that repress transcribed mRNAs in human cells by targeting specific RNAs for degradation or inhibiting their translation (Maudet et al. 2014). Pathogenic bacteria like Listeria monocytogenes (Schnitger et al. 2011), Salmonella Typhimurium (Schulte et al. 2011) and Helicobacter pylori (Zhang et al. 2008) all stimulate host miRNAs that dampen the immune response, repress apoptotic signals, and increase autophagy to avoid host clearance (Maudet et al. 2014).
In addition to activating host miRNAs, pathogenic bacterial species release specific virulence factors called effector proteins which can either bind to host proteins to inhibit host cellular pathways or can act as enhancers or repressors for host genes (Shames and Finlay 2012). Pathogenic bacteria like Salmonella Typhimurium utilize their specific effector proteins to repress host innate immunity, leading to a successful invasion (Hausmann and Hardt 2019). In cases of Clostridium difficile infection (CDI), the two major toxins produced (TcdA and TcdB) increase expression of vascular endothelial growth factor A (VEGF-A) in gut epithelial cells (Huang et al. 2019). VEGF-A promotes angiogenesis and vasodilation and is upregulated in cases of IBD (Danese et al. 2006). Vasodilation exacerbates inflammation associated with IBD and promotes CDI pathogenesis (Huang et al. 2019). The toxins associated with CDI also downregulate the expression of aquaporins (specifically AQP1) in a human intestinal microvascular endothelial cell line (Hui et al. 2018). Irregular aquaporin activity results in diarrhea, due to disrupted osmosis (Hui et al. 2018).
Gut pathogens affect the host gene expression in a cell-type specific manner, and new techniques like dual RNA-seq and single cell RNAseq (scRNA-seq) can be used to investigate that relationship. Dual RNA-seq involves simultaneously sequencing both the pathogenic bacteria and the afflicted host cell (Westermann et al. 2012). This technique allows researchers to see the exact interactions that are occurring between the pathogen and the host cell (Westermann et al. 2016). The second technique mentioned, scRNA-seq, is a modified RNA-seq technique that involves sorting individual cells followed by RNA-seq to characterize the transcriptome on a cell-by-cell basis. ScRNA-seq allows for detection of gene expression differences both within and between different cell-types and characterization of cell-type proportions in a sample.
Currently, most scRNA-seq studies involving some microbiome component investigate the mechanisms underlying viral infection of host cells. For example, scRNA-seq revealed that there are highly heterogeneous infectivity and host transcriptional responses in a primary fibroblast model infected with herpes simplex virus 1 (HSV-1) (Drayman et al. 2019), identifiable transcriptomic signatures of SARS-CoV-2 infection in patients with severe disease (Bost et al. 2020), and no discernable differences between lytic and latent human cytomegalovirus (HCMV) transcriptomes in CD14 + monocytes and CD34 + HPCs (Shnayder et al. 2018). Though bacteria have different pathogenesis than viruses, scRNA-seq could be used to show how different microbial species of the microbiome affect the composition and transcriptional profiles of different host cells.
While exciting, technical limitations exist for scRNA-seq on host cells as well as for using scRNA-seq on a microbiome sample. The first hurdle of scRNA-seq is that the equipment needed for the experiments are expensive (Liu and Trapnell 2016). Additionally, the procedures used to isolate cells and their respective RNA are complicated and could introduce bias based on the enzyme treatments used. ScRNA-seq methods are most criticized for their low capture rates, meaning that sparse transcripts may be missed. When applying scRNA-seq to the microbiome, the above issues are compounded with the fact that there are thousands of species of bacteria in our gut. There has not been a method developed for using scRNA-seq specially on a microbiome sample. However, one group took existing scRNA-seq methods that address multiple species (Butler et al. 2017), zero inflation issues (Van den Berge et al. 2018) and issues with technical noise (Kharchenko et al. 2014), tested them on simulated and manually curated 16S and metagenomic microbiome data, and compared the results with existing metagenomic analysis techniques (Calgaro et al. 2020). This group concluded that there was no perfect method but existing microbiome methods like DESeq2, edgeR and corncob preformed the best when analyzing the data. The prospect of combining scRNA-seq with the microbiome is attractive, new analytical techniques will need to be implemented to deal with the complexity of both the host and the microbiome.
Relationship between the gut microbiome and host transcription in humans
When investigating microbiome–host interactions in humans, researchers employ various approaches, such as collecting biopsies (taken during colonoscopies, gastric surgeries, or colonic surgeries), generating organoids, the gut on a chip model (Kim et al. 2016) or using human cell cultures (Fig. 2, Supplemental Table 1). Biopsies provide the most biologically faithful representation of host gene expression–microbiome relationships at the interface of the intestinal lumen, but suffer from the drawback that they are invasive to obtain and are difficult to maintain the features of ‘normal’ human tissue. Consequently, many profiled biopsies originate from diseased tissue, and do not offer insight into host–microbiome interactions in healthy states. In some instances, matching control samples are collected from patients with diseased or inflamed tissue (Häsler et al. 2017; Lloyd-Price et al. 2019). This matched study design allows direct comparison between “normal” and inflamed tissue, which can be a more powerful approach that takes into account host-specific factors like diet and genetics versus comparing case and control samples collected in different individuals.
Fig. 2.
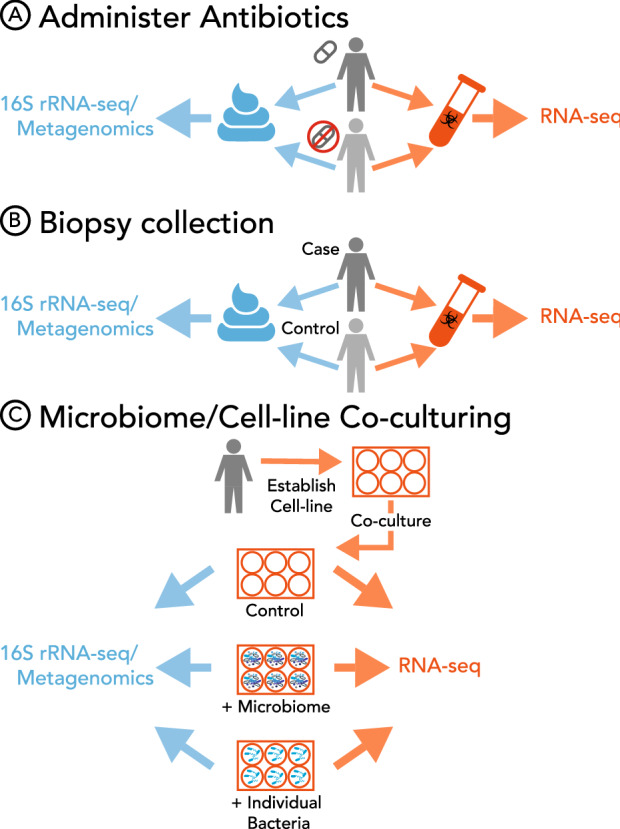
Techniques for examining microbiome—gene expression relationships in humans. Several different approaches are used to investigate the relationship between the microbiome and host transcriptome in humans. a Human subjects are split into two groups: one group is placed on antibiotics to temporally reduce the gut microbiome, while the other acts as a control. Researchers collect fecal and blood or tissue samples to study the microbiome and host transcriptome, respectively. b Tissue biopsies are collected during medical procedures from both inflamed and non-inflamed tissue, for example during colonoscopies. RNAseq data are collected from the host tissue, while the microbiome is characterized from either the associated mucosal layer or a separately collected fecal sample. c Human cell lines are established and co-cultured in the presence of a microbiome, individual bacterium, or vehicle control. Differences in gene expression between conditions can then be attributed to the microbiome
One such study used biopsies taken from the terminal ileum and sigmoidal colon from healthy individuals and those with IBD (inflamed and healthy intestinal tissue) to study IBD pathophysiology in humans (Häsler et al. 2017). Specifically, when comparing inflamed and non-inflamed tissues in the same individual, 13 pathways were differentially expressed in host tissue, including repression of citric acid cycle, bile acid synthesis and fatty acid oxidation, and induction of tryptophan, glycine, serine, alanine and threonine metabolism. Additionally, ~ 90% of the genes were differentially expressed between healthy and IBD individuals correlated with microbial taxa present in the healthy state and had almost no correlation to the microbial taxa present in the IBD gut. This “uncoupling” of mucosal gene regulation is hypothesized to be an important component of the environmental-host axis underlying IBD etiology.
A second study, also examining IBD progression used biopsies taken from patients during routine colonoscopies with both normal and inflamed intestinal tissue (4–14 biopsies per patient) coupled with stool and blood samples (Lloyd-Price et al. 2019). Interestingly, antimicrobial genes, including CXCL6, LCN2, DUOX2 and SAA2, showed increased expression in patients with IBD compared with controls. A separate study also saw increases in antimicrobial host genes in IBD [specifically those with ulcerative colitis (UC)] patients when compared to control (Bennet et al. 2018). The increase of antimicrobial genes in IBD patients could be one of the many reasons for the microbiome dysbiosis observed in the disease, due to the direct impact host antimicrobial genes would have on the members of the gut microbiome. This might also suggest that host gene expression plays a larger role in altering microbiome composition than the microbiome plays on altering host gene expression in cases of IBD. However, during outbreaks of pouchitis in patients who underwent ileal pouch-anal anastomosis as a treatment for IBD, the host transcriptome of ileal tissue drastically changed within the ileal pouch, while the taxonomic makeup of the microbiome remained stable between the ileal pouch and pre-pouch ileum (Morgan et al. 2015). This suggests that microbiome can act independently of host gene expression changes, and that the relationship between the two is highly dependent on disease context.
Biopsies are highly invasive and expensive, and alternative strategies exist for studying the relationship between the human microbiome and transcriptome. Cell cultures are cheap, relatively easy to maintain, and can be completely controlled by the researcher. To study how the host cells and microbes interact, co-culture techniques can be employed where intestinal cells are co-cultured with fecal isolates to simulate a host–microbiome environment (Richards et al. 2016). Although an imperfect proxy, colonocyte-microbiome co-culture experiments identify thousands of genes differentially regulated in the presence of a microbiome compared to control. The differentially expressed genes in the co-culture model are significantly enriched for genes identified as differentially expressed in murine colonic epithelial cells from conventional vs. germ-free mice, demonstrating the biological utility of the model.
The microbiome and host gene expression: a two-way conversation
It is clear from the many studies in model organisms and humans that there is an association between the gut microbiome and gene expression in the host. In many cases, it is unclear what the direction of causality is with these associations, however. Does a change in microbiome composition cause changes in host gene expression, or does a change in host gene expression change microbiome composition (Fig. 3)? Deciphering this relationship is important for understanding disease etiology and ultimately designing therapeutics that target the microbiome.
Fig. 3.
Direction of causality between microbiome and host gene expression associations. a When an association between the microbiome and host gene expression is identified, an open question is whether the microbiome is leading to the changes in gene expression or vice versa. b Here, differential expression of a host gene expression leads to changes in the microbiome. c Here, changes in the microbiome cause a change in host gene expression
Comparing microbiome-containing to germ-free system is one way to assess whether the microbiome plays a causative role in regulating gene expression. This type of approach can be done by comparing either germ-free to conventional organisms (Bäckhed et al. 2004; Fu et al. 2017; Larsson et al. 2012; Sayin et al. 2013; Selwyn et al. 2016) or by comparing cell cultures co-cultured with a microbiome or un-inoculated media (Richards et al. 2016). For example, expression of intestinal Cytochrome P450 (Cyp) 3a sub-family and transporter genes are significantly decreased in germ-free compared to conventionalized mice (Fu et al. 2017). Lower expression of these genes drastically reduces the detoxification capability of the host. The decrease in expression of these genes occurs in the germ-free host, demonstrating the causal role the microbiome plays in regulating detoxification pathways in the host.
In population samples, where germ-free conditions are unavailable, Mendelian Randomization (MR) offers a tool to assess whether the microbiome leads to a phenotype in the host or vice versa (Wade and Hall 2020). For example, MR analysis revealed that the gut microbiome casually effects metabolic traits related to type 2 diabetes and obesity (Sanna et al. 2019). Although not applied to studies of gene expression and the microbiome yet, this would be a powerful framework to examine direction of causality between the host transcriptome and microbiome.
Conversely, genome-wide association studies (GWAS) of the gut microbiota provide evidence that gene expression likely influences the abundance of certain bacteria in the gut (Blekhman et al. 2015; Bonder et al. 2016; Davenport 2016; Goodrich et al. 2016; Goodrich et al. 2017; Turpin et al. 2016; Wang et al. 2017). In particular, taxa such as Christensenellaceae, Akkermansia, and Bifidobacterium are either heritable or associated with genetic variation in the human genome. As it is expected that an individual’s genome sequence will not change in the presence of the microbiome, these studies demonstrate that host genetics, likely via gene regulation, modulates aspects of the gut microbiome.
Molecular mechanisms linking the microbiome to host gene expression
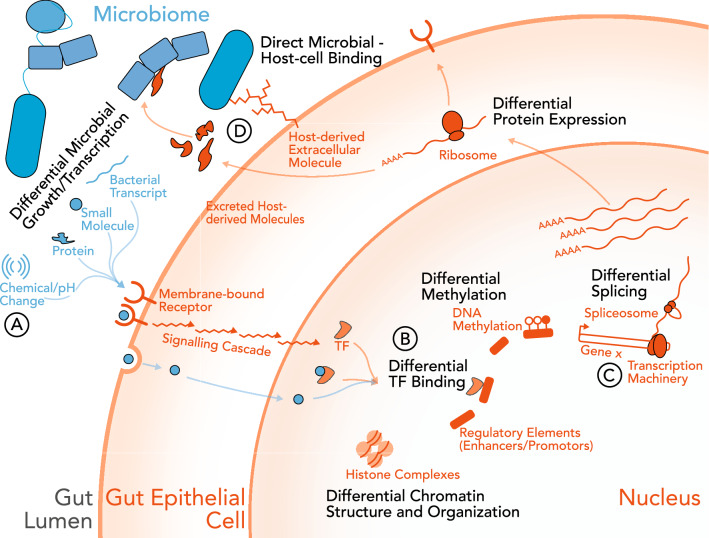
Many molecular mechanisms foster the cross-talk between the microbiome and host gene expression (Fig. 4). Transcription factors are host proteins that bind to DNA and regulate the transcription of genes. Elements of the microbiome bind directly to transcription factors (Davison et al. 2017; Krautkramer et al. 2017). In zebrafish, the microbiome suppresses the transcription factor hepatocyte nuclear factor 4A (HNF4α), preventing the regulation of host inflammatory pathways, potentially leading to an inflammatory state (Davison et al. 2017). Studies on the skin microbiome of mice showed that colonization of the skin microbiome regulates the expression of several key transcription factors (Klf4, AP-1 and SP-1), albeit via an unknown mechanism (Meisel et al. 2018).
Fig. 4.
Potential mechanisms underlying host gene expression–microbiome associations Interactions between the microbiome and microbiome-derived molecules and host can occur in either cellular or extra-cellular compartments and involve a variety of processes. a Microbiome derived stimuli are recognized by host cells, either via interaction with extracellular receptors or entering the cell. Stimuli include microbiome-derived transcripts, small molecules, proteins and enzymes, or pH changes. b Microbiome-derived molecules cause differential transcription factor binding, methylation, or chromatin availability. These changes lead to differential expression of host genes. c Microbiome-derived molecules can also lead to changes in transcription and alternative splicing, resulting in different host gene products. Both b and c result in differential protein expression, that are then released back into the gut lumen. d Host-derived proteins can also modulate the bacteria present in the gut microbiome, causing differential microbial growth and transcription. Microbiome-derived molecules illustrated in blue. Host-derived molecules illustrated in orange
The human microbiome also affects epigenetic modifications like DNA methylation and histone acetylation (Krautkramer et al. 2017; Yu et al. 2015) (Fig. 4b). For example, germ-free mice have lower genome-wide DNA methylation in colonic tissue compared to conventionally raised animals (Yu et al. 2015). A fecal transplant, however, drastically increases global DNA methylation in previously germ-free mice. Additionally, short-chain fatty acids (SCFAs) influence histone acetylation (Krautkramer et al. 2017). Even a small change in diet can cause major shifts in SCFAs and other bacterial metabolite levels in the host (David et al. 2014). Taken together, small dietary changes can potentially have major effects on host histone post-translational modifications, which then could change host gene expression for a variety of genes.
The microbiome also remodels the chromatin in host intestinal epithelial cells (Fig. 4b), albeit with conflicting evidence. ATAC-seq in a colonic epithelial cell co-culture model demonstrates that specific microbes regulate chromatin accessibility and transcription factor binding in host tissues (Richards et al. 2019). In mouse models, the presence of a microbiome results in more highly accessible chromatin in intestinal epithelial cells as compared to a gnotobiotic mouse (Semenkovich et al. 2016). The authors even speculate that the intestinal epithelium could have evolved to have a chromatin structure that requires a microbiome to activate appropriately. However, a separate study showed there are not significant changes in chromatin accessibility in intestinal epithelial cells between wild-type mice and germ-free mice (Camp et al. 2014). Instead microbial regulation of host gene expression most likely came from different transcription factor binding regions in the available chromatin.
Conversely, in a diet-induced obesity mouse experiment, the microbiome remodels chromatin in colonic cells (Qin et al. 2018). This results in an increase of accessible HNF4α binding sites, subsequently leading to downregulation of genes near those sites. One important gene that gains an HNF4α-binding site and is downregulated is Scd1 (Qin et al. 2018). Scd1 is an important regulator of the amount of free floating fatty acids and is responsible for combining them into triglyceride storage (Miyazaki et al. 2000). Downregulation of Scd1 has been seen in patients with nonalcoholic fatty liver disease (NAFLD) (Gornicka et al. 2011). Taken together it is possible that the microbiome in an obese state indirectly promotes the formation of NAFLD through modulation the structure of the chromatin in colonic cells, resulting in more binding sites for HNF4α.
Finally, the gut microbiome also has a role in the alternative splicing of host genes (Martínez-Montiel et al. 2016) (Fig. 4c). Products from Pseudomonas sp (FR901464) and Streptomyces sp (pladienolide) inhibit host splicing machinery (Fan et al. 2011). Additionally, 320 differential splicing events in intestinal tissue occurred between patients with IBD and healthy controls, after controlling for tissue type, inflammation status, and diagnosis. (Häsler et al. 2017). The alternative splicing events upregulated in IBD patients were mapped to the KEGG pathways for ‘bacterial invasion of epithelial cells’, ‘pathogenic E. coli infection’ and ‘allograft rejection’. It should be noted that there was only a weak correlation between host gene expression and the shared alternative splicing events. Research into the relationship between alternative splicing and the microbiome remains scarce and should be considered in future studies, considering the major role splicing plays in complex disease etiology (Li et al. 2016).
Future directions
The complexity of the interactions between the microbes in the gut and the host is immense. Although recent studies in model organisms and humans demonstrate that trans-kingdom crosstalk occurs, there are still many avenues to explore to gain further insight into host–microbiome interactions. Many existing studies identify individual bacteria associated with host gene expression. While this can occur, other effects are likely driven by a variety of taxa, due to functional redundancy. There are thousands of bacterial species present in the gut microbiome. Many different species share genes and pathways to produce the same metabolites (Moya and Ferrer 2016). Therefore, it is important to understand how the functional capability of the gut microbiome changes, in addition to shifts in taxonomy (Blakeley-Ruiz et al. 2019; Schirmer et al. 2019). Bacterial metagenomics, metatranscriptomics, and metaproteomics can be used to investigate the functional potential of the gut microbiome. Future studies that combine metatranscriptomics, metaproteomics, and 16S sequencing with metabolomics and RNAseq of host tissue may be able to reveal more insights about how microbiome functions interact with host gene expression.
As briefly mentioned above, scRNA-seq of both the microbiome and host tissue has the potential to reveal how the microbiome associates with cell-type composition and cell-specific gene expression. For example, using scRNA-seq, the transcriptome of only the first layer of intestinal epithelial cells that have direct contact with the microbiome could be assayed. This would limit the noise in gene expression measurements collected from multiple cell types and instead focus analysis on only the interactions happening directly at the interface of the host–gut axis.
Additionally, organoid systems can be utilized to create a better in vitro model of the human gut. Organoid systems for human colon and small intestinal tissue contain crypts and villi, which more accurately model an intestinal system versus cells adherent to a dish (Sato et al. 2011). Recently, intestinal organoids were used to model Clostridium difficile infections (CDI) (Leslie et al. 2015). The CDI organoid model more closely represented in vivo CDI when compared to CDI modeled with a cell culture (Leslie et al. 2015). The next step for these types of studies would be to move on to organoid studies and co-culture organoids with microbiome isolates. Recent methods involving microinjection could be employed to recapitulate the gut microbiome inside the organoids to provide researchers with a better model to study host–gut interactions (Williamson et al. 2018). Additionally, organoids created from patients with Crohn’s disease and healthy controls showed that the ex vivo organoid model could recapitulate the DNA methylation profiles seen in both host tissue (Howell et al. 2018). Organoid models have also been used to investigate intestinal cytokine secretion, independent of host immune functionality, to show that cytokine secretion is dependent on differentiation state (Lyons et al. 2018). Therefore, while we cannot make germ-free humans or knockout humans (like we can with mice), researchers can utilize 3D organoids with and without the microbiome instead.
Finally, novel computational methods need to be developed that integrate multiple highly complex datasets together to reveal biological insight, such as gene expression and taxonomic composition. Each type of high dimensional data has its own quirks, such as sparsity, compositionality, or overdispersion (Anders et al. 2010; Tsilimigras and Fodor 2016). Designing tools that specifically take these issues into account will allow researchers to identify novel and complex associations between the gut microbiome and host gene expression.
The study of the relationship between the microbiome and host gene expression has already yielded great insights into the physiological mechanisms that link a host with the microbes that live within it. With the exponential growth of sequencing techniques and technologies, new cell culture techniques, the growing popularity of scRNA-seq, and the advent of more advanced statistical methods, we are currently poised to gain even greater insight into the trans-kingdom relationship we have with our microbiome.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Author contributions
Performed literature search (RGN, ERD), wrote manuscript (RGN), revised manuscript (ERD).
Funding
The Pennsylvania State University.
Compliance with ethical standards
Conflict of interest
None to declare.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Robert G. Nichols, Email: rgn5011@psu.edu
Emily R. Davenport, Email: emily.davenport@psu.edu
References
- Albertsen M, Hugenholtz P, Skarshewsk A (2013) Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat Biotechnol 31 [DOI] [PubMed]
- Aleksandrova K, Romero-Mosquera B, Hernandez V. Diet, gut microbiome and epigenetics: Emerging links with inflammatory bowel diseases and prospects for management and prevention. Nutrients. 2017;9:1–13. doi: 10.3390/nu9090962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alenghant T, Osborne L, Saenz S, Kobuley D, Ziegler C, Mullican SE, Choi I, Grunberg S, Sinha R, Wynosky-Dolfi M, Snyder A, Gaicomin P, Joyce K, Hoang T, Bewtra M, Brodsky I, Sonnenberg G, Bushman F, Won K-J, Lazar M, Artis D. Histone Deacetylase 3 orchestrates commensal bacteria- dependent intestinal homeostasis. Nature. 2013;504:153–157. doi: 10.1038/nature12687.Histone. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen EE, Banfield JF. Community genomics in microbial ecology and evolution. Nat Rev Microbiol. 2005;3:489–498. doi: 10.1038/nrmicro1157. [DOI] [PubMed] [Google Scholar]
- Anders S, Huber W, Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR, Gu Z, Eils R, Schlesner M, Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, Bravo HC, Davis S, Gatto L, Girke T, Gottardo R, Hahne F, Hansen KD, Irizarry RA, Lawrence M, Love MI, MaCdonald J, Obenchain V, Oles̈ AK, Pagès H, Reyes A, Shannon P, Smyth GK, Tenenbaum D, Waldron L, Morgan M, Liao Y, Smyth GK, Shi W, Love MI, Huber W, Anders S, Narayana A, Mathew M, Tam M, Kannan R, Madden KM, Golfinos JG, Parker EC, Ott PA, Pavlick AC, Saeidipour B, Bakhshi S. Differential expression analysis for sequence count data via mixtures of negative binomials. Adv Environ Biol. 2010;7:2803–2809. [Google Scholar]
- Bäckhed F, Ding H, Wang T, Hooper LV, Gou YK, Nagy A, Semenkovich CF, Gordon JI. The gut microbiota as an environmental factor that regulates fat storage. PNAS. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckhed F, Fraser CM, Ringel Y, Sanders ME, Sartor RB, Sherman PM, Versalovic J, Young V, Finlay BB. Defining a healthy human gut microbiome: Current concepts, future directions, and clinical applications. Cell Host Microbe. 2012;12:611–622. doi: 10.1016/j.chom.2012.10.012. [DOI] [PubMed] [Google Scholar]
- Bäckhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, Li Y, Xia Y, Xie H, Zhong H, Khan MT, Zhang J, Li J, Xiao L, Al-Aama J, Zhang D, Lee YS, Kotowska D, Colding C, Tremaroli V, Yin Y, Bergman S, Xu X, Madsen L, Kristiansen K, Dahlgren J, Jun W. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. 2015;17:690–703. doi: 10.1016/j.chom.2015.04.004. [DOI] [PubMed] [Google Scholar]
- Bennet SMP, Sundin J, Magnusson MK, Strid H, Tap J, Derrien M, Le Nevé B, Doré J, Törnblom H, Simrén M, Öhman L. Altered intestinal antibacterial gene expression response profile in irritable bowel syndrome is linked to bacterial composition and immune activation. Neurogastroenterol Motil. 2018;30:1–15. doi: 10.1111/nmo.13468. [DOI] [PubMed] [Google Scholar]
- Blakeley-Ruiz JA, Erickson AR, Cantarel BL, Xiong W, Adams R, Jansson JK, Fraser CM, Hettich RL. Metaproteomics reveals persistent and phylum-redundant metabolic functional stability in adult human gut microbiomes of Crohn’s remission patients despite temporal variations in microbial taxa, genomes, and proteomes. Microbiome. 2019;7:1–15. doi: 10.1186/s40168-019-0631-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blekhman R, Goodrich JK, Huang K, Sun Q, Bukowski R, Bell JT, Spector TD, Keinan A, Ley RE, Gevers D, Clark AG. Host genetic variation impacts microbiome composition across human body sites. Genome Biol. 2015;16:1–12. doi: 10.1186/s13059-015-0759-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonder MJ, Kurilshikov A, Tigchelaar EF, Mujagic Z, Imhann F, Vila AV, Deelen P, Vatanen T, Schirmer M, Smeekens SP, Zhernakova DV, Jankipersadsing SA, Jaeger M, Oosting M, Cenit MC, Masclee AAM, Swertz MA, Li Y, Kumar V, Joosten L, Harmsen H, Weersma RK, Franke L, Hofker MH, Xavier RJ, Jonkers D, Netea MG, Wijmenga C, Fu J, Zhernakova A. The effect of host genetics on the gut microbiome. Nat Genet. 2016;48:1407–1412. doi: 10.1038/ng.3663. [DOI] [PubMed] [Google Scholar]
- Bost P, Giladi A, Liu Y, Bendjelal Y, Xu G, David E, Blecher-Gonen R, Cohen M, Medaglia C, Li H, Deczkowska A, Zhang S, Schwikowski B, Zhang Z, Amit I. Host-viral infection maps reveal signatures of severe COVID-19 patients. Cell. 2020 doi: 10.1016/j.cell.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broderick NA, Buchon N, Lemaitre B. Microbiota-induced changes in drosophila melanogaster host gene expression and gut morphology. mBio. 2014;5:1–13. doi: 10.1128/mBio.01117-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Physiol Behav. 2017;176:139–148. doi: 10.1016/j.physbeh.2017.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calgaro M, Romualdi C, Waldron L, Risso D, Vitulo N. Assessment of statistical methods from single cell, bulk RNA-seq, and metagenomics applied to microbiome data. Genome Biol. 2020;21:1–31. doi: 10.1186/s13059-020-02104-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp JG, Frank CL, Lickwar CR, Guturu H, Rube T, Wenger AM, Chen J, Bejerano G, Crawford GE, Rawls JF. Microbiota modulate transcription in the intestinal epithelium without remodeling the accessible chromatin landscape. Genome Res. 2014;24:1504–1516. doi: 10.1101/gr.165845.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danese S, Sans M, de la Motte C, Graziani C, West G, Phillips MH, Pola R, Rutella S, Willis J, Gasbarrini A, Fiocchi C. Angiogenesis as a novel component of inflammatory bowel disease pathogenesis. Gastroenterology. 2006;130:2060–2073. doi: 10.1053/j.gastro.2006.03.054. [DOI] [PubMed] [Google Scholar]
- Davenport ER. Elucidating the role of the host genome in shaping microbiome composition. Gut Microbes. 2016;7:178–184. doi: 10.1080/19490976.2016.1155022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson RM, Epperson LE. Microbiome sequencing methods for studying human diseases Rebecca. Dis Gene Identification: Methods Protocols. 2018;1706:77–90. doi: 10.1007/978-1-4939-7471-9_5. [DOI] [PubMed] [Google Scholar]
- Davison JM, Lickwar CR, Song L, Breton G, Crawford GE, Rawls JF. Microbiota regulate intestinal epithelial gene expression by suppressing the transcription factor Hepatocyte nuclear factor 4 alpha. Genome Res. 2017;27:1195–1206. doi: 10.1101/gr.220111.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cuesta-Zuluaga J, Kelley ST, Chen Y, Escobar JS, Mueller NT, Ley RE, McDonald D, Huang S, Swafford AD, Knight R, Thackray VG. Age- and sex-dependent patterns of gut microbial diversity in human adults. mSystems. 2019;4:1–12. doi: 10.1128/msystems.00261-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirksen P, Assié A, Zimmermann J, Zhang F, Tietje A-M, Marsh SA, Félix M-A, Shapira M, Kaleta C, Schulenburg H, Samuel B (2020) CeMbio—the Caenorhabditis elegans microbiome resource. bioRxiv, pp 1–48 [DOI] [PMC free article] [PubMed]
- Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, Knight R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. PNAS. 2010;107:11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohoe DR, Garge N, Zhang X, Sun W, O’Connell TM, Bunger MK, Bultman SJ. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab. 2011;13:517–526. doi: 10.1016/j.cmet.2011.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas A. The Drosophila model for microbiome research. Lab Anim. 2018;47:157–164. doi: 10.1038/s41684-018-0065-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drayman N, Patel P, Vistain L. HSV-1 single-cell analysis reveals the activation of anti-viral and developmental programs in distinct sub-populations. eLife. 2019;8:1–25. doi: 10.7554/eLife.46339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elya C, Zhang V, Ludington WB, Eisen MB. Stable host gene expression in the gut of adult Drosophila melanogaster with different bacterial mono-associations. PLoS ONE. 2016;11:1–23. doi: 10.1371/journal.pone.0167357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L, Lagisettia C, Edwards C, Webb T, Potter P. Sudemycins, novel small molecule analogues of FR901464, induce alternative gene splicing Liying. ACS Chem Biol. 2011;6:582–589. doi: 10.1038/jid.2014.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzosa EA, McIver LJ, Rahnavard G, Thompson LR, Schirmer M, Weingart G, Lipson KS, Knight R, Caporaso JG, Segata N, Huttenhower C. Species-level functional profiling of metagenomes and metatranscriptomes. Nat Methods. 2018;15:962–968. doi: 10.1038/s41592-018-0176-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu ZD, Selwyn FP, Cui JY, Klaassen CD. RNA-seq profiling of intestinal expression of xenobiotic processing genes in germ-free mice. Drug Metab Dispos. 2017;45:1225–1238. doi: 10.1124/dmd.117.077313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich J, Davenport E, Waters J, Clark A, Ley R. Cross-species comparisons of host genetic associations with the microbiome Julia. Science. 2016;352:532–535. doi: 10.1016/j.physbeh.2017.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich JK, Davenport ER, Clark AG, Ley RE. The relationship between the human genome and microbiome comes into view. Annu Rev Genet. 2017;51:413–433. doi: 10.1146/annurev-genet-110711-155532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gornicka A, Morris-Stiff G, Thapaliya S, Papouchado BG, Berk M, Feldstein AE. Transcriptional profile of genes involved in oxidative stress and antioxidant defense in a dietary murine model of steatohepatitis. Antioxid Redox Signal. 2011;15:437–445. doi: 10.1089/ars.2010.3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice E, Segre J. The human microbiome: our second genome. Annu Rev Genomics Hum Genet. 2012;13:151–170. doi: 10.1146/annurev-genom-090711-163814.The. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gury-BenAri M, Thaiss CA, Serafini N, Winter DR, Giladi A, Lara-Astiaso D, Levy M, Salame TM, Weiner A, David E, Shapiro H, Dori-Bachash M, Pevsner-Fischer M, Lorenzo-Vivas E, Keren-Shaul H, Paul F, Harmelin A, Eberl G, Itzkovitz S, Tanay A, Di Santo JP, Elinav E, Amit I. The spectrum and regulatory landscape of intestinal innate lymphoid cells are shaped by the microbiome. Cell. 2016;166:1231–1246.e13. doi: 10.1016/j.cell.2016.07.043. [DOI] [PubMed] [Google Scholar]
- Häsler R, Sheibani-Tezerji R, Sinha A, Barann M, Rehman A, Esser D, Aden K, Knecht C, Brandt B, Nikolaus S, Schäuble S, Kaleta C, Franke A, Fretter C, Müller W, Hütt MT, Krawczak M, Schreiber S, Rosenstiel P. Uncoupling of mucosal gene regulation, mRNA splicing and adherent microbiota signatures in inflammatory bowel disease. Gut. 2017;66:2087–2097. doi: 10.1136/gutjnl-2016-311651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausmann A, Hardt W-D. The interplay between salmonella enterica serovar typhimurium and the intestinal mucosa during oral infection. Microbiol Spectrum. 2019;7:1–16. doi: 10.1128/microbiolspec.bai-0004-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K, Littman DR. The microbiota in adaptive immune homeostasis and disease. Nature. 2016;535:75–84. doi: 10.1038/nature18848. [DOI] [PubMed] [Google Scholar]
- Hooper LV, Midtvedt T, Gordon JI. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu Rev Nutr. 2002;22:283–307. doi: 10.1146/annurev.nutr.22.011602.092259. [DOI] [PubMed] [Google Scholar]
- Howell KJ, Kraiczy J, Nayak KM, Gasparetto M, Ross A, Lee C, Mak TN, Koo BK, Kumar N, Lawley T, Sinha A, Rosenstiel P, Heuschkel R, Stegle O, Zilbauer M. DNA methylation and transcription patterns in intestinal epithelial cells from pediatric patients with inflammatory bowel diseases differentiate disease subtypes and associate with outcome. Gastroenterology. 2018;154:585–598. doi: 10.1053/j.gastro.2017.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Kelly C, Bakirtzi K, Villafuerte J, Lyras D, Mileto S, Larcombe S, Xu H, Yang X, Shields K, Zhu W, Zhang Y, Goldsmith J, Patel I, Hansen J, Huang M, Yla-Herttuala S, Moss A, Paredes-Sabja D, Pothoulakis C, Shah Y, Wang J, Chen X. Clostridium difficile toxins induce VEGF-A and vascular permeability to promote disease pathogenesis. Nat Microbiol. 2019;4:269–279. doi: 10.1097/CCM.0b013e31823da96d.Hydrogen. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui L, Zang K, Wang M, Shang F, Zhang G. Coculture with clostridium difficile promotes apoptosis of human intestinal microvascular endothelial cells. J Int Med Res. 2018;46:4631–4739. doi: 10.1177/0300060518799267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada N, Seo S-U, Chen GY, Núñez G. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol. 2013;13:321–335. doi: 10.1038/nri3430. [DOI] [PubMed] [Google Scholar]
- Kharchenko PV, Silberstein L, Scadden DT. Bayesian approach to single-cell differential expression analysis. Nat Methods. 2014;11:740–742. doi: 10.1038/nmeth.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Li H, Collins JJ, Ingber DE. Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip. Proc Natl Acad Sci USA. 2016;113:E7–E15. doi: 10.1073/pnas.1522193112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krautkramer KA, Kreznar JH, Romano KA, Vivas EI, Barrett-wilt GA, Rabaglia ME, Keller MP, Attie AD, Federico E, Denu JM. Diet-microbiota interactions mediate global epigenetic programming in multiple host tissues. Mol Cell. 2017;64:982–992. doi: 10.1016/j.molcel.2016.10.025.Diet-microbiota. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson E, Tremaroli V, Lee YS, Koren O, Nookaew I, Fricker A, Nielsen J, Ley RE, Bäckhed F. Analysis of gut microbial regulation of host gene expression along the length of the gut and regulation of gut microbial ecology through MyD88. Gut. 2012;61:1124–1131. doi: 10.1136/gutjnl-2011-301104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclercq S, Matamoros S, Cani PD, Neyrinck AM, Jamar F, Starkel P, Windey K, Tremaroli V, Backhed F, Verbeke K, de Timary P, Delzenne NM. Intestinal permeability, gut-bacterial dysbiosis, and behavioral markers of alcohol-dependence severity. Proc Natl Acad Sci U S A. 2014;111:E4485–E4493. doi: 10.1073/pnas.1415174111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie JL, Huang S, Opp JS, Nagy MS, Kobayashi M, Young VB, Spence JR. Persistence and toxin production by Clostridium difficile within human intestinal organoids result in disruption of epithelial paracellular barrier function. Infect Immun. 2015;83:138–145. doi: 10.1128/IAI.02561-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YI, Van De Geijn B, Raj A, Knowles DA, Petti AA, Golan D, Gilad Y, Pritchard JK. RNA splicing is a primary link between genetic variation and disease. Science. 2016;352:600–604. doi: 10.1126/science.aad9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Trapnell C. Single-cell transcriptome sequencing: recent advances and remaining challenges. F1000Research. 2016 doi: 10.12688/f1000research.7223.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd-Price J, Arze C, Ananthakrishnan AN, Schirmer M, Avila-Pacheco J, Poon TW, Andrews E, Ajami NJ, Bonham KS, Brislawn CJ, Casero D, Courtney H, Gonzalez A, Graeber TG, Hall AB, Lake K, Landers CJ, Mallick H, Plichta DR, Prasad M, Rahnavard G, Sauk J, Shungin D, Vázquez-Baeza Y, White RA, Bishai J, Bullock K, Deik A, Dennis C, Kaplan JL, Khalili H, McIver LJ, Moran CJ, Nguyen L, Pierce KA, Schwager R, Sirota-Madi A, Stevens BW, Tan W, ten Hoeve JJ, Weingart G, Wilson RG, Yajnik V, Braun J, Denson LA, Jansson JK, Knight R, Kugathasan S, McGovern DPB, Petrosino JF, Stappenbeck TS, Winter HS, Clish CB, Franzosa EA, Vlamakis H, Xavier RJ, Huttenhower C. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature. 2019;569:655–662. doi: 10.1038/s41586-019-1237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukovac S, Belzer C, Pellis L, Keijser BJ, de Vos WM, Montijn RC, Roeselers G. Differential modulation by akkermansia muciniphila and faecalibacterium prausnitzii of host peripheral lipid metabolism and histone acetylation in mouse gut organoids. mBio. 2014;5:1–10. doi: 10.1128/mBio.01438-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons J, Ghazi PC, Starchenko A, Tovaglieri A, Baldwin KR, Poulin EJ, Gierut JJ, Genetti C, Yajnik V, Breault DT, Lauffenburger DA, Haigis KM. The colonic epithelium plays an active role in promoting colitis by shaping the tissue cytokine profile. PLoS Biol. 2018;16:1–24. doi: 10.1371/journal.pbio.2002417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Montiel N, Rosas-Murrieta NH, Martínez-Montiel M, Gaspariano-Cholula MP, Martínez-Contreras RD. Microbial and natural metabolites that inhibit splicing: a powerful alternative for cancer treatment. Biomed Res Int. 2016 doi: 10.1155/2016/3681094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maudet C, Mano M, Eulalio A. MicroRNAs in the interaction between host and bacterial pathogens. FEBS Lett. 2014;588:4140–4147. doi: 10.1016/j.febslet.2014.08.002. [DOI] [PubMed] [Google Scholar]
- Meisel JS, Sfyroera G, Bartow-McKenney C, Gimblet C, Bugayev J, Horwinski J, Kim B, Brestoff JR, Tyldsley AS, Zheng Q, Hodkinson BP, Artis D, Grice EA. Commensal microbiota modulate gene expression in the skin. Microbiome. 2018;6:1–15. doi: 10.1186/s40168-018-0404-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyakawa M, Gordon HA, Wostmann BS. The gnotobiotic animal as a tool in the study of inflammation. Science. 1971;173:171–173. doi: 10.1126/science.173.3992.171. [DOI] [PubMed] [Google Scholar]
- Miyazaki M, Kim YC, Gray-Keller MP, Attie AD, Ntambi JM. The biosynthesis of hepatic cholesterol esters and triglycerides is impaired in mice with a disruption of the gene for stearoyl-CoA desaturase 1. J Biol Chem. 2000;275:30132–30138. doi: 10.1074/jbc.M005488200. [DOI] [PubMed] [Google Scholar]
- Molly K, Vande Woestyne M, Verstraete W. Development of a 5-step multi-chamber reactor as a simulation of the human intestinal microbial ecosystem. Appl Microbiol Biotechnol. 1993;39:254–258. doi: 10.1007/BF00228615. [DOI] [PubMed] [Google Scholar]
- Montoya-Williams D, Lemas DJ, Spiryda L, Patel K, Carney ONO, Neu J, Carson TL. The neonatal microbiome and its partial role in mediating the association between birth by cesarean section and adverse pediatric outcomes. Neonatology. 2018;114:103–111. doi: 10.1159/000487102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan XC, Kabakchiev B, Waldron L, Tyler AD, Tickle TL, Milgrom R, Stempak JM, Gevers D, Xavier RJ, Silverberg MS, Huttenhower C. Associations between host gene expression, the mucosal microbiome, and clinical outcome in the pelvic pouch of patients with inflammatory bowel disease. Genome Biol. 2015;16:67. doi: 10.1186/s13059-015-0637-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moya A, Ferrer M. Functional redundancy-induced stability of gut microbiota subjected to disturbance. Trends Microbiol. 2016;24:402–413. doi: 10.1016/j.tim.2016.02.002. [DOI] [PubMed] [Google Scholar]
- Murdoch CC, Rawls JF. Commensal microbiota regulate vertebrate innate immunity-insights from the zebrafish. Front Immunol. 2019;10:1–14. doi: 10.3389/fimmu.2019.02100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson H, Chia N. Gut microbiome and colon cancer: a plausible explanation for dietary contributions to cancer. J Am Coll Surg. 2019;229:231–235. doi: 10.1016/j.jamcollsurg.2019.05.003. [DOI] [PubMed] [Google Scholar]
- Nelson K, Weinstock G, Highlander S, Worley K, Creasy H, Wortman J, Rusch D, Mitreva M, Sodergren E, Chinwalla A, Feldgarden M, Gevers D, Haas B, Madupu R, Ward D, Birren B, Gibbs R, Methe B, Petrosino J, Strausberg R, Sutton G, White O. A catalog of reference genomes from the human microbiome. Science. 2010;328:994–999. doi: 10.1126/science.1183605.A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen B, Almeida M, Juncker A, Rasmussen S, Li J, Sunagawa S, Plichta D, Gautier L, Pedersen A, Chatelier E, Pelletier E, Bonde I, Nielsen T, Manichanah C, Arumugam M, Batto J-M, Quintanilha dos Santos M, Blom N, Borruel N, Burgdof K, Boumezbeur F, Casellas F, Dore J, Dworzynski P, Guarner F, Hansen T, Hildebrand F, Kass R, Kennedy S, Kristiansen K, Kultima JR, Leonard P, Levenez F, Lund O, Moumen B, Paslier D, Pons N, Pederson O, Prifti E, Qin J, Raes J, Sorensen S, Tap J, Tims S, Ussey D, Yamada T, Consortium M, Renault P, Sicheritz-Ponten T, Bork P, Wang J, Brunak S, Ehrlich D. Identification and assembly of genomes and genetic elements in complex metagenomic samples without using reference genomes. Nat Biotechnol. 2014;32:822–828. doi: 10.1038/nbt.2939. [DOI] [PubMed] [Google Scholar]
- Papathoma E, Triga M, Fouzas S, Dimitriou G. Cesarean section delivery and development of food allergy and atopic dermatitis in early childhood. Pediatr Allergy Immunol. 2016;27:419–424. doi: 10.1111/pai.12552. [DOI] [PubMed] [Google Scholar]
- Parks DH, Rinke C, Chuvochina M, Chaumeil PA, Woodcroft BJ, Evans PN, Hugenholtz P, Tyson GW. Recovery of nearly 8000 metagenome-assembled genomes substantially expands the tree of life. Nat Microbiol. 2017;2:1533–1542. doi: 10.1038/s41564-017-0012-7. [DOI] [PubMed] [Google Scholar]
- Pott J, Hornef M. Innate immune signalling at the intestinal epithelium in homeostasis and disease. EMBO Rep. 2012;13:684–698. doi: 10.1038/embor.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y, Roberts JD, Grimm SA, Lih FB, Deterding LJ, Li R, Chrysovergis K, Wade PA. An obesity-associated gut microbiome reprograms the intestinal epigenome and leads to altered colonic gene expression. Genome Biol. 2018;19:1–14. doi: 10.1186/s13059-018-1389-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reidl S, Lehmann A, Schiller R, Salam Khan A, Dobrindt U. Impact of O-glycosylation on the molecular and cellular adhesion properties of the Escherichia coli autotransporter protein Ag43. Int J Med Microbiol. 2009;299:389–401. doi: 10.1016/j.ijmm.2009.01.001. [DOI] [PubMed] [Google Scholar]
- Richards AL, Burns MB, Alazizi A, Barreiro LB, Pique-Regi R, Blekhman R, Luca F. Genetic and transcriptional analysis of human host response to healthy gut microbiota. mSystems. 2016;1:1–16. doi: 10.1128/msystems.00067-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards AL, Muehlbauer AL, Alazizi A, Burns MB, Findley A, Messina F, Gould TJ, Cascardo C, Pique-Regi R, Blekhman R, Luca F. Gut Microbiota Has a Widespread and Modifiable Effect on Host Gene Regulation. mSystems. 2019 doi: 10.1128/msystems.00323-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridaura VK, Faith JJ, Rey FE, Cheng J, Alexis E, Kau AL, Griffin NW, Lombard V, Henrissat B, Bain JR, Muehlbauer MJ, Ilkayeva O, Semenkovich CF, Funai K, Hayashi DK, Lyle BJ, Martini MC, Luke K, Clemente JC, Treuren WV, Wa W. Cultured gut microbiota from twins discordant for obesity modulate adiposity and metabolic phenotypes in mice. Science. 2014;341:1–22. doi: 10.1126/science.1241214.Cultured. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenow C, Saxena R, Durst M, Gingeras T. Prokaryotic RNA preparation methods useful for high density array analysis: comparison of two approaches. Nucleic Acids Res. 2001;29:112e–112. doi: 10.1093/nar/29.22.e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell SL, Gold MJ, Hartmann M, Willing BP, Thorson L, Wlodarska M, Gill N, Blanchet MR, Mohn WW, McNagny KM, Finlay BB. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep. 2012;13:440–447. doi: 10.1038/embor.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzberg SL. Open questions: how many genes do we have? BMC Biol. 2018;16:10–12. doi: 10.1186/s12915-018-0564-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanna S, Zuydam N, Mahajan A, Kurilshikov A, Vich Vila A, Vosa U, Mujagic Z, Masclee A, Jonkers D, Oosting M, Joosten L, Netea M, Franke L, Zhernakova A, Fu J, Wijmenga C, McCarthy M. Causal relationships between gut microbiome, short-chain fatty acids and metabolic diseases. Nat Genet. 2019;51:549–562. doi: 10.1097/CCM.0b013e31823da96d.Hydrogen. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Stange DE, Ferrante M, Vries RGJ, Van Es JH, Van Den Brink S, Van Houdt WJ, Pronk A, Van Gorp J, Siersema PD, Clevers H. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- Sayin SI, Wahlström A, Felin J, Jäntti S, Marschall HU, Bamberg K, Angelin B, Hyötyläinen T, Orešič M, Bäckhed F. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013;17:225–235. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- Schirmer M, Garner A, Vlamakis H, Xavier RJ. Microbial genes and pathways in inflammatory bowel disease. Nat Rev Microbiol. 2019;17:497–511. doi: 10.1038/s41579-019-0213-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnitger AKD, Machova A, Mueller RU, Androulidaki A, Schermer B, Pasparakis M, Krönke M, Papadopoulou N. Listeria monocytogenes infection in macrophages induces vacuolar-dependent host miRNA response. PLoS ONE. 2011 doi: 10.1371/journal.pone.0027435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte LN, Eulalio A, Mollenkopf HJ, Reinhardt R, Vogel J. Analysis of the host microRNA response to Salmonella uncovers the control of major cytokines by the let-7 family. EMBO J. 2011;30:1977–1989. doi: 10.1038/emboj.2011.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selwyn FP, Cheng SL, Klaassen CD, Cui JY. Regulation of hepatic drug-metabolizing enzymes in germ-free mice by conventionalization and probiotics. Drug Metab Dispos. 2016;44:262–274. doi: 10.1124/dmd.115.067504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenkovich NP, Planer JD, Ahern PP, Griffin NW, Lin CY, Gordon JI. Impact of the gut microbiota on enhancer accessibility in gut intraepithelial lymphocytes. Proc Natl Acad Sci USA. 2016;113:14805–14810. doi: 10.1073/pnas.1617793113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shames SR, Finlay BB. Bacterial effector interplay: a new way to view effector function. Trends Microbiol. 2012;20:214–219. doi: 10.1016/j.tim.2012.02.007. [DOI] [PubMed] [Google Scholar]
- Shi N, Li N, Duan X, Niu H. Interaction between the gut microbiome and mucosal immune system. Milit Med Res. 2017;4:1–7. doi: 10.1186/s40779-017-0122-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shnayder M, Nachshon A, Krishna B, Poole E, Boshkov A, Binyamin A, Maza I. Defining the transcriptional landscape during cytomegalovirus latency with single-cell RNA sequencing. MBio. 2018;9:1–17. doi: 10.1128/mBio.00013-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shreiner A, Kao J, Young V. The gut microbiome in health and in disease. Curr Opin Gastroenterol. 2015;31:69–75. doi: 10.1097/MOG.0000000000000139.The. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Kitai T, Hazen S. Gut microbiota in cardiovascular health and disease W.H. Circ Res. 2017;120:139–148. doi: 10.1016/j.physbeh.2017.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong DT, Franzosa EA, Tickle TL, Scholz M, Weingart G, Pasolli E, Tett A, Huttenhower C, Segata N. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat Methods. 2015;12:902–903. doi: 10.1038/nmeth.3589. [DOI] [PubMed] [Google Scholar]
- Tsilimigras MCB, Fodor AA (2016) Compositional data analysis of the microbiome: fundamentals, tools, and challenges. Ann Epidemiol 26. Elsevier Inc, pp 330–335 [DOI] [PubMed]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Turpin W, Espin-Garcia O, Xu W, Silverberg M, Kevans D, Smith M, Guttman D, Griffiths A, Panaccione R, Otley A, Xu L, Shestopaloff K, Moreno-Hagelsieb G, Consortium GPR, Paterson A, Croitoru K. Association of host genome with intestinal microbial composition in a large healthy cohort. Nat Genet. 2016;48:1413–1417. doi: 10.1038/ng.3693. [DOI] [PubMed] [Google Scholar]
- Van den Berge KVD, Perraudeau F, Soneson C, Love MI, Risso D, Vert J-p, Robinson MD, Dudoit S, Clement L (2018) Observation weights unlock bulk RNA-seq tools for zero inflation and single-cell applications. Genome Biology 1–17 [DOI] [PMC free article] [PubMed]
- Wade KH, Hall LJ (2020) Improving causality in microbiome research: Can human genetic epidemiology help? Wellcome Open Research 4 [DOI] [PMC free article] [PubMed]
- Wang J, Thingholm L, Skieceviciene J, Rausch P, Kummen M, Hov J, Degenhardt F, Heinsen F, Ruhlemann M, Szymczak S, Holm K, Esko T, Sun J, Pricop-Jeckstadt M, Al-Dury S, Bohov P, Bethune J, Sommer F, Ellinghaus D, Berge R, Hubenthal M, Koch M, Schwarz K, Rimbach G, Hubbe P, Oan W, Sheibani-Tezerji R, Hasler R, Rosensteil P, D'Amatto M, Cloppenborg-Schmidt K, Kunzel S, Laudes M, Marschall H, Leib W, Nothlings U, Karles T, Baines J, Franke A. Genome-wide association analysis identifies variation in vitamin D receptor and other host factors influencing the gut microbiota. Nat Genet. 2017;546:651–655. doi: 10.1038/nature22814.Trans-kingdom. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC, Hu C, Wong FS, Szot GL, Bluestone JA, Gordon JI, Chervonsky AV. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–1113. doi: 10.1016/j.physbeh.2017.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann AJ, Gorski SA, Vogel J. Dual RNA-seq of pathogen and host. Nat Rev Microbiol. 2012;10:618–630. doi: 10.1038/nrmicro2852. [DOI] [PubMed] [Google Scholar]
- Westermann AJ, Förstner KU, Amman F, Barquist L, Chao Y, Schulte LN, Müller L, Reinhardt R, Stadler PF, Vogel J. Dual RNA-seq unveils noncoding RNA functions in host-pathogen interactions. Nature. 2016;529:496–501. doi: 10.1038/nature16547. [DOI] [PubMed] [Google Scholar]
- Westreich ST, Treiber ML, Mills DA, Korf I, Lemay DG. SAMSA2: a standalone metatranscriptome analysis pipeline. BMC Bioinform. 2018;19:175. doi: 10.1186/s12859-018-2189-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson IA, Arnold JW, Samsa LA, Gaynor L, DiSalvo M, Cocchiaro JL, Carroll I, Azcarate-Peril MA, Rawls JF, Allbritton NL, Magness ST. A high-throughput organoid microinjection platform to study gastrointestinal microbiota and luminal physiology. Cmgh. 2018;6:301–319. doi: 10.1016/j.jcmgh.2018.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood DE, Salzberg SL. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15:R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav D, Ghosh TS, Mande SS. Global investigation of composition and interaction networks in gut microbiomes of individuals belonging to diverse geographies and age-groups. Gut Pathogens. 2016;8:17. doi: 10.1186/s13099-016-0099-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Petersen C, Pees B, Zimmermann J, Waschina S, Dirksen P, Rosenstiel P, Tholey A, Leippe M, Dierking K, Kaleta C, Schulenburg H. The inducible response of the nematode caenorhabditis elegans to members of its natural microbiota across development and adult life. Front Microbiol. 2019;10:1–16. doi: 10.3389/fmicb.2019.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsunenko T, Rey F, Manary M, Trehan I, Dominguez-Bello M, Contreras M, Magris M, Hidalgo G, Baldassano R, Anokhin A, Heath A, Warner B, Reeder J, Kuczynski J, Caporaso J, Lozupone C, Lauber C, Clemente J, Knights D, Knight R, Gordon J. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053.Human. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon MY, Yoon SS. Disruption of the gut ecosystem by antibiotics. Yonsei Med J. 2018;59:4–12. doi: 10.3349/ymj.2018.59.1.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu DH, Gadkari M, Zhou Q, Yu S, Gao N, Guan Y, Schady D, Roshan TN, Chen MH, Laritsky E, Ge Z, Wang H, Chen R, Westwater C, Bry L, Waterland RA, Moriarty C, Hwang C, Swennes AG, Moore SR, Shen L. Postnatal epigenetic regulation of intestinal stem cells requires DNA methylation and is guided by the microbiome. Genome Biol. 2015;16:211. doi: 10.1186/s13059-015-0763-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Li Z, Gao C, Chen P, Chen J, Liu W, Xiao S, Lu H. miR-21 plays a pivotal role in gastric cancer pathogenesis and progression. Lab Invest. 2008;88:1358–1366. doi: 10.1038/labinvest.2008.94. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.