Abstract
Background and objectives
Chest X-ray data have been found to be very promising for assessing COVID-19 patients, especially for resolving emergency-department and urgent-care-center overcapacity. Deep-learning (DL) methods in artificial intelligence (AI) play a dominant role as high-performance classifiers in the detection of the disease using chest X-rays. Given many new DL models have been being developed for this purpose, the objective of this study is to investigate the fine tuning of pretrained convolutional neural networks (CNNs) for the classification of COVID-19 using chest X-rays. If fine-tuned pre-trained CNNs can provide equivalent or better classification results than other more sophisticated CNNs, then the deployment of AI-based tools for detecting COVID-19 using chest X-ray data can be more rapid and cost-effective.
Methods
Three pretrained CNNs, which are AlexNet, GoogleNet, and SqueezeNet, were selected and fine-tuned without data augmentation to carry out 2-class and 3-class classification tasks using 3 public chest X-ray databases.
Results
In comparison with other recently developed DL models, the 3 pretrained CNNs achieved very high classification results in terms of accuracy, sensitivity, specificity, precision, score, and area under the receiver-operating-characteristic curve.
Conclusion
AlexNet, GoogleNet, and SqueezeNet require the least training time among pretrained DL models, but with suitable selection of training parameters, excellent classification results can be achieved without data augmentation by these networks. The findings contribute to the urgent need for harnessing the pandemic by facilitating the deployment of AI tools that are fully automated and readily available in the public domain for rapid implementation.
Keywords: COVID-19, Chest X-rays, Artificial intelligence, Deep learning, Classification
Introduction
COVID-19 (coronavirus disease 2019) is an infectious disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which is a strain of coronavirus. The disease was officially announced as a pandemic by the World Health Organisation (WHO) on 11 March 2020. Given spikes in new COVID-19 cases and the re-opening of daily activities around the world, the demand for curbing the pandemic is to be more emphasized.
Medical images and artificial intelligence (AI) have been found useful for rapid assessment to provide treatment of COVID-19 infected patients. Therefore, the design and deployment of AI tools for image classification of COVID-19 in a short period of time with limited data have been an urgent need for fighting the current pandemic. Radiologists have recently found that deep learning (DL) developed in AI, which was able to detect tuberculosis in chest X-rays, could be useful for identifying lung abnormalities related to COVID-19 and help clinicians in deciding the order of treatment of high-risk COVID-19 patients [1]. The role of medical imaging has also been confirmed by others as playing an important source of information to enable the fast diagnosis of COVID-19 [2], and the coupling of AI and chest imaging can help explain the complications of COVID-19 [3].
Regarding the image analysis of COVID-19, chest X-ray is an imaging method to diagnose COVID-19 infection adopted by hospitals, particularly the first image-based approach used in Spain [4]. The protocol is that if a clinical suspicion about the infection remains after the examination of a patient, a sample of nasopharyngeal exudate is obtained to test the reverse-transcription polymerase chain reaction (RT-PCR) and the taking of a chest X-ray film follows. Because the results of the PCR test may take several hours to become available, information revealed from the chest X-ray plays an important role for a rapid clinical assessment. This means if the clinical condition and the chest X-ray are normal, the patient is sent home while awaiting the results of the etiological test. But if the X-ray shows pathological findings, the suspected patient will be admitted to the hospital for close monitoring. In general, the absence or presence of pathological findings on the chest X-ray is the basis for making a clinical decision in sending the patient home or keeping the patient in the hospital for further observation.
While radiography in medical examinations can be quickly performed and become widely available with the prevalence of chest radiology imaging systems in healthcare systems, the interpretation of radiography images by radiologists is limited due to the human capacity in detecting the subtle visual features present in the images. Because AI can discover patterns in chest X-rays that normally would not be recognized by radiologists [5–8], there have been many studies reported in literature about new developments of DL models using convolutional neural networks (CNNs) for differentiating COVID-19 from non-COVID-19 using public databases of chest X-rays (related works are presented in the next section).
This study attempted to investigate the potential of the parameter adjustments in the transfer learning of three popular pretrained CNNs: AlexNet, GoogLeNet, and SqueezeNet, which are known to have least prediction and training iteration times among other pretrained CNNs reported from the ImageNet Large-Scale Visual Recognition Challenge [9]. If these fine-tuned networks can achieve desired performance in the classification of COVID-19 chest X-ray images by a configuration in such a way to highly perform the task, then the contribution of the findings to the coronavirus pandemic relief would be significant. This is because it can facilitate the urgent need for rapidly deploying AI tools to assist clinicians in making optimal clinical decisions by saving time, resources, and technical efforts in developing models that may result in the same or lower performance.
The new contribution of this study is the finding of the relatively simple yet powerful performance of several fine-tuned pretrained CNNs that can produce better accuracy in classifying COVID-19 chest X-ray data with less training effort than other existing deep-learning models.
Related works
Peer-reviewed works that are related to the study presented herein are described as follows.
The Bayes-SqueezeNet [10] was introduced for detecting the COVID-19 using chest X-rays. The proposed net consists of the offline augmentation of the raw dataset and model training using the Bayesian optimization. The Bayes-SqueezeNet was applied for classifying X-ray images labeled in 3 classes as normal, viral pneumonia, and COVID-19. Using the data augmentation, the net claimed to overcome the problem of imbalanced data obtained from the public databases.
As another CNN, the CoroNet [11] was developed for detecting COVID-19 infection from chest X-ray images. This model was based on the pretrained CNN known as the Xception [12]. CoroNet adopted the Xception as base model with a dropout layer and two fully-connected layers added at the end. As a result, CoroNet has 33,969,964 parameters in total out of which 33,969,964 trainable and 54,528 are non-trainable parameters. The net was applied for 3-class classification (COVID-19, pneumonia, and normal) as well as 4-class classification (COVID-19, pneumonia bacterial, pneumonia viral, and normal).
The CovidGAN [13] was proposed as an auxiliary classifier generative adversarial network based on GAN (generative adversarial network) [15] for the detection of COVID-19. The architecture of the CovidGan was built on the pretrained VGG-16 [14], which is connected with four custom layers at the end with a global average pooling layer followed by a 64 units dense layer and a dropout layer with 0.5 probability. The net further utilized the GAN approach for generating synthetic chest X-ray images to improve the classification performance.
The DarkCovidNet [16], which was built on the DarkNet model [17], is another CNN model proposed for COVID-19 detection using chest X-rays. The DarkCovidNet consists of fewer layers and (gradually increased) filters than the original DarkNet. This model was tested for a 2-class classification (COVID-19 and no-findings) and 3-class classification (COVID-19 no-findings, and pneumonia).
The work reported in [18] implemented VGG-19 [14], MobileNet-v2 [19], Inception [20], Xception [12], and Inception ResNet-v2 [20] as pretrained CNNs for the detection of COVID-19 from X-ray images. These pretrained CNNs were applied to 2-class and 3-class classification cases using 2 datasets consisting of images of COVID-19, bacterial pneumonia, viral pneumonia, and healthy conditions.
Pretrained CNNs and training parameters for transfer learning
Three prerained CNNs, which are AlexNet [21], GoogLeNet [22], and SqueezeNet [23], were selected in this study. The reason for selecting these CNNs was that these three models require the least training time among other pretrained CNNs. The architectures and specification of training parameters for transfer learning of AlexNet, GoogLeNet, and SqueezeNet are described as follows.
First, the layer graph from the pretrained network was extracted. If the network was a SeriesNetwork object, such as AlexNet, then the list of layers was converted to a layer graph. In the pretrained networks, the last layer with learnable weights is a fully connected layer. This fully connected layer was replaced with a new fully connected layer with the number of outputs being equal to the number of classes in the new data set, which is 2 or 3, in this study. In the pretrained SqueezeNet, the last learnable layer is a 1-by-1 convolutional layer instead. In this case, the convolutional layer was replaced with a new convolutional layer with the number of filters equal to the number of classes.
The original chest X-ray images were converted into RGB images and resized to fit into the input image size of each pretrained CNN. For the training options, the stochastic gradient descent with momentum optimizer was used, where the momentum value = 0.9000; gradient threshold method = norm; minimum batch size = 10; maximum number of epochs = 10; initial learning rate= 0.0003; the learning rate remained constant throughout training; the training data were shuffled before each training epoch, and the validation data were shuffled before each network validation; and factor for regularization (weight decay) = 0.0001.
Basic properties of the three networks in terms of depth, size, numbers of parameters, and input image size are given in Table 1. Other hyperparameters of the three networks can be found in [21] (AlexNet), [22] (GoogLeNet), and [23] (SqueezeNet).
Table 1.
Basic properties of 3 pretrained convolutional neural networks
| Network | Depth | Size (MB) | Parameters (millions) | Input image size |
|---|---|---|---|---|
| AlexNet | 8 | 227 | 61.0 | |
| GoogLeNet | 22 | 27 | 7.0 | |
| SqueezeNet | 18 | 5.2 | 1.24 |
Chest X-ray databases
This study used 3 public databases of COVID-19 chest X-rays: (1) COVID-19 Radiography Database [24], (2) COVID-19 Chest X-Ray Dataset Initiative [25], and (3) IEEE8023/Covid Chest X-Ray Dataset [26].
The COVID-19 Radiography Database consists of chest X-rays of 219 COVID-19 positive images, 1341 normal images, and 1345 viral pneumonia images. The COVID-19 Chest X-Ray Dataset Initiative has 55 COVID-19 positive images. IEEE8023/Covid Chest X-Ray Dataset is part of the COVID-19 Image Data Collection of chest X-ray and CT images of patients which are positive or suspected of COVID-19 or other viral and bacterial pneumonias, in which 706 images are chest X-rays. The numbers of images in these databases, which are expected to increase over time with more available data, were reported on the date of access.
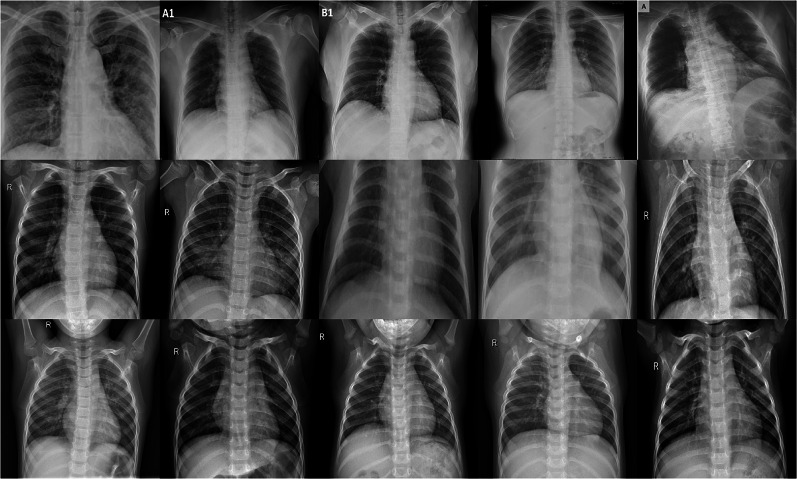
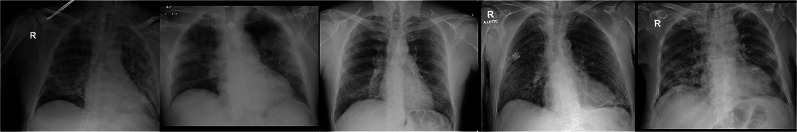
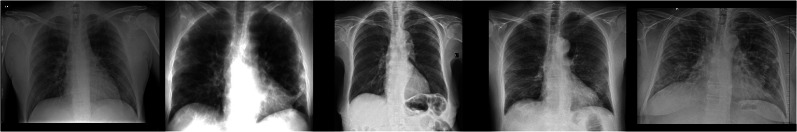
Figure 1 shows some chest X-ray images of COVID-19, viral pneumonia, and normal subjects provided by the COVID-19 Radiography Database. Figures 2 and 3 show some chest X-ray images of COVID-19 obtained from the COVID-19 Chest X-Ray Dataset Initiative and IEEE8023/Covid Chest X-Ray Dataset, respectively.
Fig. 1.
Chest X-rays from COVID-19 Radiography Database: COVID-19 (Row 1), viral pneumonia (Row 2), and normal (Row 3)
Fig. 2.
Chest X-rays of COVID-19 from the Chest X-Ray Dataset Initiative
Fig. 3.
Chest X-rays of COVID-19 from the IEEE8023/Covid Chest X-Ray Dataset
Design of chest X-ray subsets
Six subsets of chest X-ray data were constructed out of the COVID-19 Radiography Database (Kaggle), COVID-19 Chest X-Ray Dataset Initiative, and IEEE8023/Covid Chest X-Ray Dataset to test and compare the performance of the pretrained CNNs. These 6 subsets are described as follows.
Dataset 1
This dataset includes 403 chest X-rays of COVID-19 and 721 chest X-rays of healthy subjects . All images of the healthy subjects were taken from the COVID-19 Radiography Database. This dataset was designed for a two-class classification to compare with the study reported in [13].
Dataset 2
This chest X-ray dataset has 438 images of COVID-19 and 438 images of healthy subjects. All images of the healthy subjects were taken from the COVID-19 Radiography Database. This balanced dataset was designed for a two-class classification with more COVID-19 images.
Dataset 3
This chest X-ray dataset has 438 images of COVID-19 and 876 images of healthy and viral pneumonia subjects (438 healthy and 438 viral pneumonia) cases. All images of the healthy and viral pneumonia subjects were taken from the COVID-19 Radiography Database. This dataset was designed for a two-class classification.
Dataset 4
To carry out a three-class classification, this chest X-ray dataset has 438 images of COVID-19, 438 images of viral pneumonia, and 438 images of healthy subjects. All images of the healthy and viral pneumonia subjects were taken from the COVID-19 Radiography Database.
Dataset 5
This two-class dataset consists of all images of the COVID-19 (class 1), and healthy and viral pneumonia subjects (class 2) of the COVID-19 Radiography Database.
Dataset 6
This three-class dataset consists of all images of the COVID-19 (class 1), viral pneumonia (class 2), and healthy subjects (class 3) of the COVID-19 Radiography Database.
Performance metrics
Six metrics used for evaluating the performance of the CNNs are accuracy, sensitivity, specificity, precision, score, and area under a receiver operating characteristic curve (AUC).
The sensitivity (SEN) is defined as the percentage of COVID-19 patients who are correctly identified as having the infection, and expressed as
| 1 |
where TP is called true positive, denoting the number of COVID-19 patients who are correctly identified as having the infection, FN false negative, denoting the number of COVID-19 patients who are misclassified as having no infection of COVID-19, and P the total number of COVID-19 patients.
The specificity (SPE) is defined as the percentage of non-COVID-19 subjects who are correctly classified as having no infection of COVID-19:
| 2 |
where TN is called true negative and denotes the number of non-COVID-19 subjects who are correctly identified as having no infection of COVID-19, FP false positive, denoting the number of non-COVID-19 subjects who are misclassified as having the infection, and N the total number of non-COVID-19 subjects.
The accuracy (ACC) of the classification is defined as
| 3 |
The precision (PRE) is also known as the percentage of positive predictive value and defined as:
| 4 |
The score is defined as the harmonic mean of precision and sensitivity:
| 5 |
The receiver operating characteristic (ROC) is a probability curve created by plotting the TP rate against the FP rate at various threshold settings, and the AUC represents the measure of performance of a classifier. The higher the AUC is, the better the model at distinguishing between COVID-19 and non-COVID-19 cases. For a perfect classifier, AUC = 1, and an AUC = 0.5 indicates a classifier that randomly assigns observations to classes. The AUC is calculated using the trapezoidal integration to estimate the area under the ROC curve.
Results
All results are reported as the average values and standard deviations of 3 executions of randomly selected ratios of training and testing data.
Table 2 shows the classification results obtained from the transfer learning of AlexNet, GoogLeNet, and SqueezeNet, using Dataset 1 with two different training and testing data ratios. The 3 pretrained CNNs achieved very high accuracy, sensitivity, specificity, precision, score, and AUC in all cases. Particularly, GoogLeNet and SqueezeNet had almost 100% accuracy with 80% training and 20% testing data. The AUCs were almost perfect in all cases for all three CNNs.
Table 2.
Two-class classification results: Dataset 1.
| CNN | Accuracy (%) | Sensitivity (%) | Specificity (%) | Precision (%) | score | AUC |
|---|---|---|---|---|---|---|
| 80% training and 20% testing | ||||||
| AlexNet | 99.14 ± 0.62 | 98.44 ± 1.19 | 99.51 ± 0.66 | 99.10 ± 1.21 | 0.988 ± 0.009 | 0.999 ± 0.002 |
| GoogLeNet | 99.70 ± 0.52 | 100 ± 0.00 | 99.54 ± 0.80 | 99.16 ± 1.46 | 0.996 ± 0.007 | 0.999 ± 0.000 |
| SqueezeNet | 99.85 ± 0.26 | 100 ± 0.00 | 99.77 ± 0.40 | 99.57 ± 0.74 | 0.998 ± 0.004 | 0.999 ± 0.000 |
| 50% training and 50% testing | ||||||
| AlexNet | 99.19 ± 0.23 | 98.32 ± 0.65 | 99.65 ± 0.14 | 99.35 ± 0.26 | 0.988 ± 0.003 | 0.999 ± 0.000 |
| GoogLeNet | 99.22 ± 0.46 | 99.14 ± 0.60 | 99.26 ± 0.42 | 98.63 ± 0.79 | 0.989 ± 0.007 | 0.999 ± 0.002 |
| SqueezeNet | 98.43 ± 2.10 | 95.85 ± 6.28 | 99.81 ± 0.32 | 99.66 ± 0.032 | 0.976 ± 0.000 | 0.999 ± 0.000 |
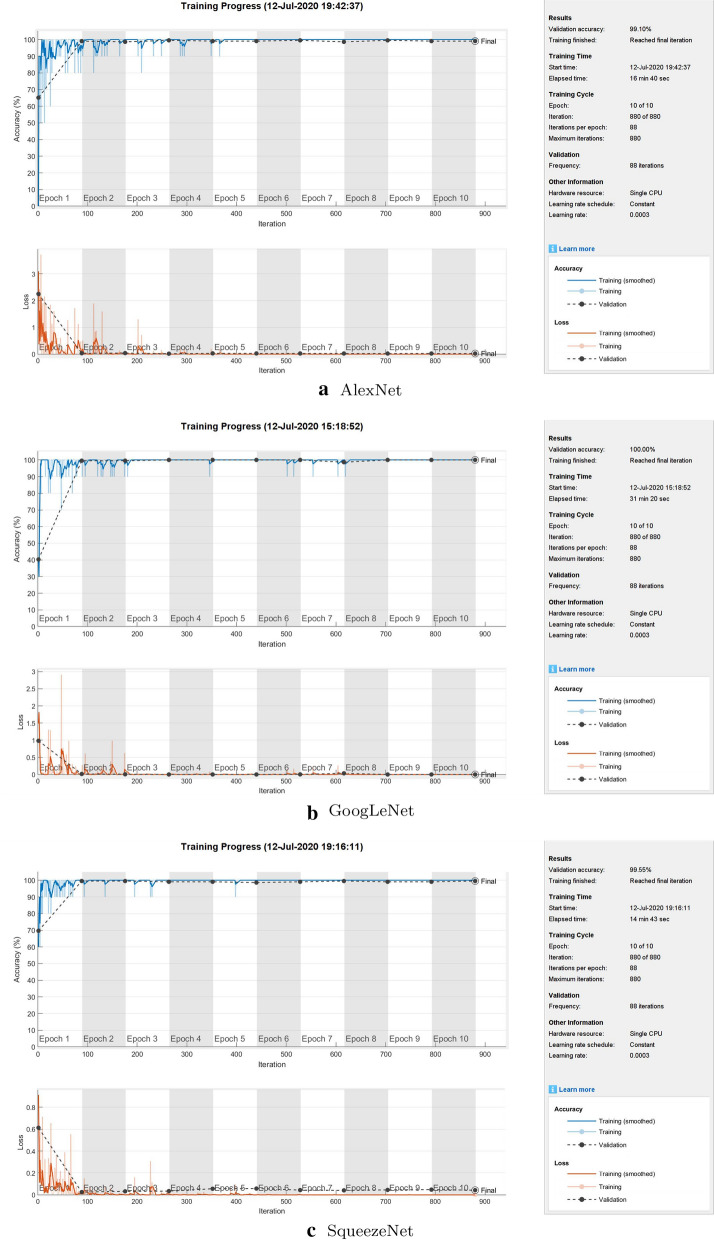
Figure 4 shows the training processes of the transfer learning of the three CNNs, and Fig. 5 shows the features at the fully connected layers extracted from transfer learning of the three CNNs, all using Dataset 1 with 80% training and 20% testing.
Fig. 4.
Transfer-learning processes of the pretrained convolutional neural networks for the 2-class classification using Dataset 1
Fig. 5.
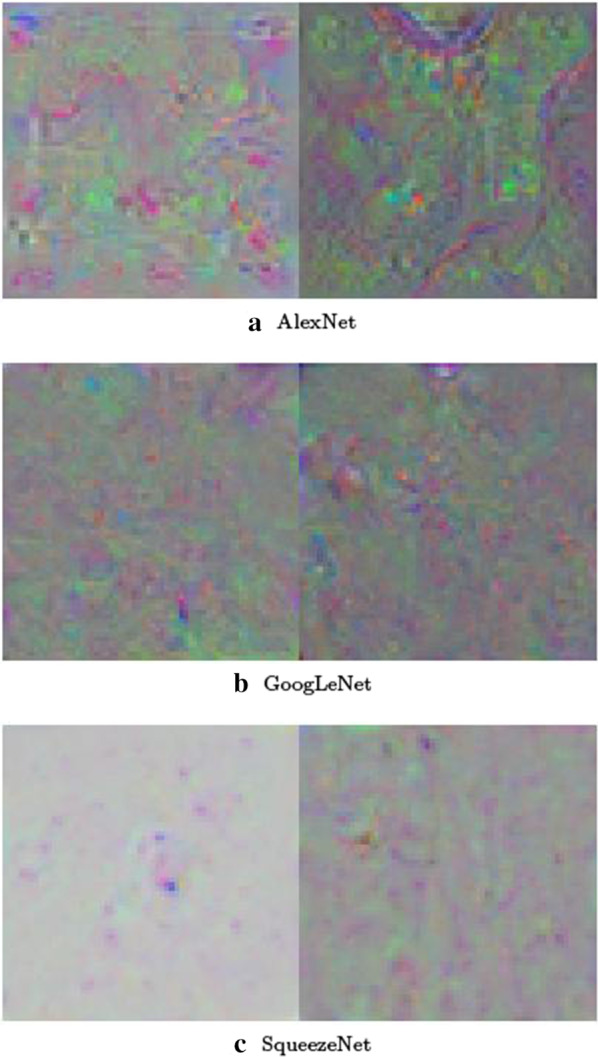
Features at the fully connected layers of the pretrained convolutional neural networks for the 2-class classification using Dataset 1: This feature visualization provides insights into the performance of the convolutional neural networks for differentiating COVID-19 (left images) from normal (right images) conditions. The networks first learned simple edges and texture, then more abstract properties of the two classes in higher layers, resulting in distinctive features for effective pattern classification
Tables 3 and 4 show the classification results obtained from the AlexNet, GoogLeNet, and SqueezeNet for a 2-class classification of COVID-19 and normal cases (Dataset 2), and COVID-19 and both normal and viral pneumonia (Dataset 3) with 50% of the data for training and the other 50% for testing, respectively. For Dataset 2, all classifiers achieved accuracy, sensitivity, specificity, and precision > 99%, and score > 0.990, and AUC almost being 1. For Dataset 3, all achieved > 98% in accuracy, > 97% in sensitivity, > 98% in specificity and precision, > 0.975 for score, and almost 1 for AUC.
Table 3.
Two-class classification results: Dataset 2 (50% training and 50% testing)
| CNN | Accuracy (%) | Sensitivity (%) | Specificity (%) | Precision (%) | score | AUC |
|---|---|---|---|---|---|---|
| AlexNet | 99.40 ± 0.31 | 99.34 ± 0.54 | 99.45 ± 0.75 | 99.44 ± 0.76 | 0.994 ± 0.003 | 1.00 ± 0.000 |
| GoogLeNet | 99.61 ± 0.13 | 99.21 ± 0.27 | 100 ± 0.00 | 100 ± 0.00 | 0.996 ± 0.001 | 0.999 ± 0.000 |
| SqueezeNet | 99.69 ± 0.13 | 99.53 ± 0.47 | 99.85 ± 0.26 | 99.84 ± 0.27 | 0.997 ± 0.001 | 1 ± 0.000 |
Table 4.
Two-class classification results: Dataset 3 (50% training and 50% testing)
| CNN | Accuracy (%) | Sensitivity (%) | Specificity (%) | Precision (%) | score | AUC |
|---|---|---|---|---|---|---|
| AlexNet | 98.43 ± 0.86 | 97.35 ± 1.14 | 98.95 ± 0.86 | 97.83 ± 1.77 | 0.976 ± 0.013 | 0.998 ± 0.002 |
| GoogLeNet | 98.82 ± 0.54 | 97.79 ± 2.24 | 99.32 ± 0.46 | 98.58 ± 0.93 | 0.982 ± 0.009 | 0.998 ± 0.002 |
| SqueezeNet | 98.77 ± 0.31 | 97.79 ± 1.80 | 99.24 ± 0.48 | 98.43 ± 0.96 | 0.981 ± 0.005 | 0.999 ± 0.000 |
Table 5 shows the 3-class classification (COVID-19, viral pneumonia, and normal) results obtained from the transfer learning of three pretrained CNNs using Dataset 4 with 50% of the data for training and the other 50% for testing. All the three CNNs achieved accuracies > 96%, sensitivity > 97%, specificity > 95%, precision > 96%, score 0.970, and AUC = 0.998.
Table 5.
Three-class classification results: Dataset 4 (50% training and 50% testing)
| CNN | Accuracy (%) | Sensitivity (%) | Specificity (%) | Precision (%) | score | AUC |
|---|---|---|---|---|---|---|
| AlexNet | 96.46 ± 1.36 | 97.35 ± 1.37 | 96.03 ± 1.44 | 98.11 ± 1.93 | 0.977 ± 0.015 | 0.998 ± 0.001 |
| GoogLeNet | 96.20 ± 0.44 | 97.79 ± 2.24 | 95.43 ± 0.46 | 98.44 ± 1.15 | 0.981 ± 0.007 | 0.998 ± 0.002 |
| SqueezeNet | 96.25 ± 0.90 | 98.10 ± 1.71 | 95.36 ± 0.53 | 96.01 ± 1.75 | 0.970 ± 0.010 | 0.998 ± 0.001 |
Table 6 shows the results obtained from the three CNNs using Dataset 5, of which accuracies (> 99%), AUCs (= 0.999), and specificity (> 99%) are similar for both cases of (1) 90% training and 10% testing data, and (2) 50% training and 50% testing data. The AlexNet achieved the best average sensitivity (98.48%) using 90% training and 10% testing data, and the SqueezeNet achieved the best average sensitivity (98.47%) for 50% training and 50% testing data.
Table 6.
Two-class classification results: Dataset 5
| CNN | Accuracy (%) | Sensitivity (%) | Specificity (%) | Precision (%) | score | AUC |
|---|---|---|---|---|---|---|
| 90% training and 10% testing | ||||||
| AlexNet | 99.77 ± 0.20 | 98.48 ± 2.62 | 99.88 ± 0.21 | 98.55 ± 2.51 | 0.985 ± 0.013 | 0.999 ± 0.008 |
| GoogLeNet | 99.31 ± 0.34 | 95.45 ± 4.55 | 99.63 ± 0.37 | 95.63 ± 4.18 | 0.955 ± 0.023 | 0.999 ± 0.001 |
| SqueezeNet | 99.54 ± 0.37 | 96.97 ± 5.25 | 99.75 ± 0.43 | 97.22 ± 4.81 | 0.970 ± 0.026 | 0.999 ± 0.002 |
| 50% training and 50% testing | ||||||
| AlexNet | 99.13 ± 0.42 | 91.93 ± 3.69 | 99.72 ± 0.22 | 96.37 ± 2.79 | 0.941 ± 0.029 | 0.998 ± 0.001 |
| GoogLeNet | 99.47 ± 0.14 | 96.94 ± 1.40 | 99.68 ± 0.09 | 96.07 ± 1.03 | 0.965 ± 0.010 | 0.999 ± 0.000 |
| SqueezeNet | 99.38 ± 0.42 | 98.47 ± 1.06 | 99.45 ± 0.50 | 93.83 ± 5.23 | 0.960 ± 0.026 | 0.999 ± 0.000 |
For the 3-class classification using Dataset 6, the results as shown in Table 7 are still very high but slightly lower than those obtained using Dataset 5 for the 2-class classification. For both cases of 1) 90% training and 10% testing data, and 2) 50% training and 50% testing data, all the accuracies are 96%, specificity > 96%, and AUC > 0.998. The SqueezeNet has the highest sensitivity (98.48%) for 90% training and 10% testing data, while the GoogLeNet achieved the highest sensitivity (95.23%) for 50% training and 50% testing data. The precision (98.48%) is highest for the GoogLeNet using 90% training and 10% testing data, and highest (96.75%) for the SqueezeNet using 50% training and 50% testing data. The GoogLeNet achieved the highest scores as 0.977 and 0.952 for both 90% training and 10% testing, and 50% training and 50% testing, respectively.
Table 7.
Three-class classification results: Dataset 6
| CNN | Accuracy (%) | Sensitivity (%) | Specificity (%) | Precision (%) | score | AUC |
|---|---|---|---|---|---|---|
| 90% training and 10% testing | ||||||
| AlexNet | 97.59 ± 0.60 | 95.45 ± 4.55 | 97.76 ± 0.37 | 98.55 ± 2.51 | 0.969 ± 0.014 | 0.998 ± 0.004 |
| GoogLeNet | 96.09 ± 2.30 | 96.97 ± 2.62 | 96.02 ± 2.28 | 98.48 ± 2.62 | 0.977 ± 0.023 | 0.999 ± 0.004 |
| SqueezeNet | 97.47 ± 1.31 | 98.48 ± 2.62 | 97.39 ± 1.30 | 94.20 ± 2.51 | 0.963 ± 0.026 | 0.999 ± 0.009 |
| 50% training and 50% testing | ||||||
| AlexNet | 95.89 ± 0.42 | 92.11 ± 6.31 | 96.20 ± 0.66 | 96.66 ± 2.30 | 0.942 ± 0.029 | 0.998 ± 0.000 |
| GoogLeNet | 96.07 ± 0.63 | 95.23 ± 1.51 | 96.14 ± 0.72 | 95.24 ± 1.72 | 0.952 ± 0.015 | 0.999 ± 0.001 |
| SqueezeNet | 95.95 ± 0.52 | 92.11 ± 3.02 | 96.26 ± 0.63 | 96.75 ± 1.20 | 0.943 ± 0.014 | 0.998 ± 0.001 |
Comparions with related works
The CovidGAN [13] aimed to generate synthetic chest X-ray images using the principle of GAN for the classification. Using the combination of three databases (COVID-19 Radiography Database, COVID-19 Chest X-Ray Dataset Initiative, and IEEE8023/Covid Chest X-Ray Dataset) with about 80% training and 20% testing data, this network achieved 95% in accuracy, 90% in sensitivity, and 97% in specificity. Using the same database combination with 80% training and 20% testing data without data augmentation, all three fine-tuned CNNs reported in the present study (Table 2) achieved accuracy > 99%, sensitivity from 98% (AlexNet) to 100% (GoogLeNet and SqueezeNet), and specificity > 99%.
The Bayes-SqueezeNet [10] carried out the 3-class classification of X-ray images labeled as normal, bacterial pneumonia, and COVID-19. The Bayes-SqueezeNet used the COVID-19 Radiography Database (Kaggle) and IEEE8023/Covid Chest X-Ray Dataset, with approximately 89% (3697 images) training and validation (462 images) and 11% (459 images) testing data, and applied data augmentation for the network training. The accuracy, specificity, and scores were 98%, 99%, and 0.98. The current study used the updated COVID-19 Radiography Database with 90% training and 10% testing, resulting in similar 3-class classification results obtained from the fine-tuned AlexNet without data augmentation (Table 7: 98% in accuracy, 98% in specificity, and 0.97 for score.
To avoid the unbalanced data problem, the CoroNet [11] randomly selected 310 normal, 330 bacterial pneumonia, and 327 COVID-19 X-ray images from the COVID-19 Radiography Database. The 4-fold cross-validation (75% training and 25% testing) results for the 2-class classification were: accuracy = 99%, sensitivity = 99%, specificity = 99%, precision = 98%, score = 0.99. The 4-fold cross-validation results for the 3-class classification were: accuracy = 95%, sensitivity = 97%, specificity = 98%, precision = 95%, score = 0.96. Using the whole updated COVID-19 Radiography Database with 90% for training and 10% for testing, the 2-class classification results obtained from the fined-tuned AlexNet were (Table 6): accuracy = 100%, sensitivity = 98%, specificity = 100%, precision = 99%, score = 1.00. With 50% for training and 50% for testing, the 2-class classification results obtained from the fined-tuned GoogLeNet were (Table 6): accuracy = 99%, sensitivity = 97%, specificity = 100%, precision = 96%, score = 0.97. Using the whole updated COVID-19 Radiography Database with 90% for training and 10% for testing, the 3-class classification results obtained from the fined-tuned AlexNet were (Table 7): accuracy = 98%, sensitivity = 95%, specificity = 98%, precision = 99%, score = 0.97. With 50% for training and 50% for testing, the 3-class classification results obtained from the fined-tuned GoogLeNet were (Table 7): accuracy = 96%, sensitivity = 95%, specificity = 96%, precision = 95%, score = 0.95.
The DarkCovidNet [16] used the IEEE8023/Covid Chest X-Ray Dataset and Chestx-ray8 Database [27] to perform 2-class classification (COVID-19 and no-findings) and 3-class classification (COVID-19, no-findings, and pneumonia). Using the 5-fold cross validation (80% training and 20% testing) for the 2-class classification, the DarkCovidNet achieved accuracy = 98%, sensitivity = 95%, specificity = 95%, precision = 98%, and score = 0.97. Using the 5-fold cross validation for the 3-class classification, the DarkCovidNet achieved accuracy = 87%, sensitivity = 85%, specificity = 92%, precision = 90%, and score = 0.87. For the 2-class classification (Table 3), the three fine-tuned CNNs reported in this study, which used the Dataset 2 with 50% training and 50% testing , achieved > 99% in accuracy, sensitivity, specificity, and precision, and score > 0.99; and for the 3-class classification (Table 5), > 98% in accuracy, > 97% in sensitivity, 99% in specificity, 98% in precision, and score = 0.98.
The VGG-19, MobileNet-v2, Inception, Xception, and Inception ResNet-v2 were implemented for the classification of COVID-19 chest X-ray images [18]. Those networks were trained and tested using the IEEE8023/Covid Chest X-Ray Dataset and other chest X-rays collected on the internet. The best 10-cross-validation (90% training and 10% testing) results obtained from the VGG-19 were: accuracy = 98.75% for the 2-class classification and 93.48% for the 3-class classification. Using the COVID-19 Radiography Database, the fine-tuned AlexNet, GoogLeNet, and SqueezeNet achieved 2-class classification accuracies > 99% for both 90% training-10% testing and 50% training-50% testing (Table 6), and 3-class classification accuracies > 96% for 90% training-10% testing (Table 7).
Discussions
The results obtained from the transfer learning of the fine-tuned AlexNet, GoogLeNet, and SqueezeNet illustrate the high accomplishment of the pretrained models for the classification of COVID-19. Due to the database updates over time and the public availability of other data collections, it is impossible to carry out exact comparisons of the results reported herein and many other works. Comparisons with base-line works using the same datasets as shown in Table 8 strongly suggest that the fine-tuned pretrained networks achieved better performance than several other base-line methods in terms of classification accuracy, and partitions of training and testing data.
Table 8.
Comparisons of classification accuracy and partitions of training and testing data between (proposed) fine-tuned and other deep-learning methods
| Methods | Training (%) | Testing (%) | Accuracy (%) |
|---|---|---|---|
| Dataset 1, number of classes = 2 | |||
| CovidGAN [13] | 80 | 20 | 95 |
| Fine-tuned AlexNet | 80 | 20 | 99 |
| Fine-tuned GoogLeNet | 80 | 20 | 100 |
| Fine-tuned SqueezeNet | 80 | 20 | 100 |
| Fine-tuned AlexNet | 50 | 50 | 99 |
| Fine-tuned GoogLeNet | 50 | 50 | 99 |
| Fine-tuned SqueezeNet | 50 | 50 | 98 |
| Dataset 2, number of classes = 2 | |||
| DarkCovidNet [16] | 80 | 20 | 99 |
| Fine-tuned AlexNet | 50 | 50 | 100 |
| Fine-tuned GoogLeNet | 50 | 50 | 100 |
| Fine-tuned SqueezeNet | 50 | 50 | 100 |
| Dataset 4, number of classes = 3 | |||
| DarkCovidNet [16] | 80 | 20 | 87 |
| Fine-tuned AlexNet | 50 | 50 | 96 |
| Fine-tuned GoogLeNet | 50 | 50 | 96 |
| Fine-tuned SqueezeNet | 50 | 50 | 96 |
| Dataset 5, number of classes = 2 | |||
| CoroNet [11] | 75 | 25 | 99 |
| VGG-19 [18] | 90 | 10 | 99 |
| Fine-tuned AlexNet | 50 | 50 | 99 |
| Fine-tuned GoogLeNet | 50 | 50 | 99 |
| Fine-tuned SqueezeNet | 50 | 50 | 99 |
| Dataset 6, number of classes = 3 | |||
| VGG-19 [18] | 90 | 10 | 93 |
| Fine-tuned AlexNet | 90 | 10 | 98 |
| Fine-tuned GoogLeNet | 90 | 10 | 96 |
| Fine-tuned SqueezeNet | 90 | 10 | 97 |
| CoroNet [11] | 75 | 25 | 95 |
| Fine-tuned AlexNet | 50 | 50 | 96 |
| Fine-tuned GoogLeNet | 50 | 50 | 96 |
| Fine-tuned SqueezeNet | 50 | 50 | 96 |
Both AlexNet and SqueezeNet take the least training and prediction time among many other pretrained CNNs. In this study, data augmentation was not applied to the transfer learning of the three networks. However, very high classification results could be obtained by using suitable parameters for the transfer learning of new data. This finding emphasizes the role of fine tuning of pretrained CNNs for handling new data before adding more complex architectures to the networks. The finding in this study can be useful for the rapid deployment of available AI models for the fast, reliable, and cost-effective detection of COVID-19 infection.
Conclusion
The transfer learning of three popular pretrained CNNs for the classification of chest X-ray images of COVID19, viral pneumonia, and normal conditions using several subsets of three publicly available databases have been presented and discussed. The performance metrics obtained from different settings of training and testing data have demonstrated the effectiveness of these three networks. The present results suggest the fine tuning of the network learning parameters is important as it can help avoid making efforts in developing more complex models when existing ones can result in the same or better performance.
Due to limited data labeling, this study did not consider the sub-classification of COVID-19 into mild, moderate, or severe disease. Another issue is that only a single chest X-ray series was obtained for each patient. This data limitation has an implication that it is not possible to determine if patients developed radiographic findings as the illness progressed [28]. Nevertheless, hospitals and institutions across continents have been trying to rapidly develop AI-based solutions for solving the time-sensitive COVID-19 crisis. The findings reported in this study can facilitate the free availability of AI models to all participants for clinical validations.
Funding
The author received no funding for this work.
Availability of data
Six data subsets designed in this study were obtained from three third-party databases, which are publicly available. The data links for the 1) COVID-19 Radiography Database (Kaggle), 2) COVID-19 Chest X-Ray Dataset Initiative, and 3) IEEE8023/Covid Chest X-Ray Dataset are given at [24, 25], and [26], respectively.
Compliance with ethical standards
Conflicts of interest
The author declares no conflict of interest.
Code availability
The MATLAB code used in this study is available at the author’s personal homepage: https://sites.google.com/view/tuan-d-pham/.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Yi PH, Kim TK, Lin CT. Generalizability of deep learning tuberculosis classifier to COVID-19 chest radiographs. J Thorac Imaging. 2020;35:W102–W104. doi: 10.1097/RTI.0000000000000532. [DOI] [PubMed] [Google Scholar]
- 2.Yang W, Sirajuddin A, Zhang X, Liu G, Teng Z, Zhao S, Lu M. The role of imaging in 2019 novel coronavirus pneumonia (COVID-19) Eur Radiol. 2020;15:1–9. doi: 10.1007/s00330-020-06827-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kundu S, Elhalawani H, Gichoya JW, Kahn CE. How might AI and chest imaging help unravel COVID-19’s mysteries? Radiol: Artif Intell. 2020;2:3. doi: 10.1148/ryai.2020200053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Imaging the Coronavirus Disease COVID-19. https://healthcare-in-europe.com/en/news/imaging-the-coronavirus-disease-covid-19.html.
- 5.Kim TK, Yi PH, Hager GD, Lin CT. Refining dataset curation methods for deep learning-based automated tuberculosis screening. J Thorac Dis. 2019;12:2. doi: 10.21037/jtd.2019.08.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong HYF, Lam HYS, Fong AH-T, Leung ST, Chin TWY, et al. Frequency and distribution of chest radiographic findings in COVID-19 positive patients. Radiology. 2020 doi: 10.1148/radiol.2020201160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zech JR, Badgeley MA, Liu M, Costa AB, Titano JJ, Oermann EK. Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: a cross-sectional study. PLoS Med. 2018;15:e1002683. doi: 10.1371/journal.pmed.1002683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hurt B, Kligerman S, Hsiao A. Deep learning localization of pneumonia. J Thorac Imaging. 2020;35:W87–W89. doi: 10.1097/RTI.0000000000000512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Russakovsky O, Deng J, Su H, Krause J, Satheesh S, et al. ImageNet LargeScale visual recognition challenge. Int J Comput Vis. 2015;115:211–252. doi: 10.1007/s11263-015-0816-y. [DOI] [Google Scholar]
- 10.Ucar F, Korkmaz D. COVIDiagnosis-net: deep bayes-SqueezeNet based diagnosis of the coronavirus disease 2019 (COVID-19) from X-ray images. Med Hypotheses. 2020;140:109761. doi: 10.1016/j.mehy.2020.109761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khan AI, Shah JL, Bhat MM. CoroNet: a deep neural network for detection and diagnosis of COVID-19 from chest x-ray images. Comput Methods Programs Biomed. 2020;196:105581. doi: 10.1016/j.cmpb.2020.105581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chollet F. Xception: deep learning with depthwise separable convolutions. arXiv 2017; 161002357.
- 13.Waheed A, Goyal M, Gupta D, Khanna A, Al-Turjman F, Pinheiro PR. CovidGAN: data augmentation using auxiliary classifier GAN for improved Covid-19 detection. IEEE Access. 2020;8:91916–91923. doi: 10.1109/ACCESS.2020.2994762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. arXiv 2015; 1409.1556.
- 15.Goodfellow I, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, et al. Generative adversarial nets. In: Proceedings of advances in neural information processing systems; 2014. p. 2672–2680.
- 16.Ozturk T, Talo M, Yildirim EA, Baloglu UB, Yildirim O, Acharya UR. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput Biol Med. 2020;121:103792. doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Redmon J, Farhadi A. Yolo9000: better, faster, stronger. arXiv 2016; 1612.08242
- 18.Apostolopoulos ID, Mpesiana TA. Covid. 19: automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Phys Eng Sci Med. 2020;43:635–640. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Howard AG, Zhu M, Chen B, Kalenichenko D, Wang W, et al. MobileNets: efficient convolutional neural networks for mobile vision applications. arXiv 2017; 170404861.
- 20.Szegedy C, Ioffe S, Vanhoucke V, Alemi AA. Inception-v4, inception-resnet and the impact of residual connections on learning. In: Thirty-first AAAI conference on artificial intelligence; 2017. p. 4278–4284.
- 21.Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. Commun. ACM. 2017;60:84–90. doi: 10.1145/3065386. [DOI] [Google Scholar]
- 22.Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, et al. Going deeper with convolutions. In: Proceedings of the IEEE conference on computer vision and pattern recognition; 2015. p. 1-9.
- 23.Iandola FN, Han S, Moskewicz MW, Ashraf K, Dally WJ, Keutzer K. SqueezeNet: AlexNet-level accuracy with 50x fewer parameters and 0.5MB model size. arXiv 2016; 1602.07360.
- 24.COVID-19 Radiography Database. https://www.kaggle.com/tawsifurrahman/covid19-radiography-database. Accessed 2 July 2020.
- 25.COVID-19 Chest X-Ray Dataset Initiative. https://github.com/agchung/Figure1-COVID-chestxray-dataset. Accessed 2 July 2020.
- 26.IEEE8023/Covid Chest X-Ray Dataset. https://github.com/ieee8023/covid-chestxray-dataset. Accessed 2 July 2020.
- 27.Wang X, Peng Y, Lu L, Lu Z, Bagheri M, Summers RM. Chestx-ray8: hospitalscale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In: Proceedings of the IEEE conference on computer vision and pattern recognition; 2017. p. 2097–2106.
- 28.Weinstock MB, et al. Chest X-ray findings in 636 ambulatory patients with COVID-19 presenting to an urgent care center: a normal chest X-ray is no guarantee. J Urgent Care Med. 2020;14:13–18. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Six data subsets designed in this study were obtained from three third-party databases, which are publicly available. The data links for the 1) COVID-19 Radiography Database (Kaggle), 2) COVID-19 Chest X-Ray Dataset Initiative, and 3) IEEE8023/Covid Chest X-Ray Dataset are given at [24, 25], and [26], respectively.