Abstract
Patient-derived xenograft (PDX) tumor models represent a valuable platform for identifying new biomarkers and novel targets, to evaluate therapy response and resistance mechanisms. This study aimed at establishment, characterization and therapy testing of colorectal carcinoma-derived PDX. We generated 49 PDX and validated identity between patient tumor and corresponding PDX. Sensitivity of PDX toward conventional and targeted drugs revealed that 92% of PDX responded toward irinotecan, 45% toward 5-FU, 65% toward bevacizumab, and 61% toward cetuximab. Expression of epidermal growth factor receptor (EGFR) ligands correlated to the sensitivity toward cetuximab. Proto-oncogene B-RAF, EGFR, Kirsten rat sarcoma virus oncogene homolog gene copy number correlated positively with cetuximab and erlotinib sensitivity. The mutational analyses revealed an individual mutational profile of PDX and mainly identical profiles of PDX from primary tumor vs corresponding metastasis. Mutation in PIK3CA was a determinant of accelerated tumor doubling time. PDX with wildtype Kirsten rat sarcoma virus oncogene homolog, proto-oncogene B-RAF, and phosphatidylinositol-4,5-bisphosphate 3-kinaseM catalytic subunit alfa showed higher sensitivity toward cetuximab and erlotinib. To study the molecular mechanism of cetuximab resistance, cetuximab resistant PDX models were generated, and changes in HER2, HER3, betacellulin, transforming growth factor alfa were observed. Global proteome and phosphoproteome profiling showed a reduction in canonical EGFR-mediated signaling via PTPN11 (SHP2) and AKT1S1 (PRAS40) and an increase in anti-apoptotic signaling as a consequence of acquired cetuximab resistance. This demonstrates that PDX models provide a multitude of possibilities to identify and validate biomarkers, signaling pathways and resistance mechanisms for clinically relevant improvement in cancer therapy.
Keywords: Colorectal carcinoma, Patient-derived xenograft models, In vivo drug testing, Personalized medicine
Abbreviations: AKT, Serine threonine protein kinase AKT; AREG, Amphiregulin; APC, adenomatous polyposis coli; BTC, betacellulin; CRC, colorectal cancer; c-MET, tyrosine protein kinase MET; BRAF, proto-oncogene B-RAF; EGF, epidermal growth factor; EGFR, epidermal growth factor receptor; EpCAM, epithelial cell adhesion molecule; EREG, epiregulin; 5-FU, 5-fluorouracil; FFPE, formalin fixed paraffin embedded; HER2,3,4, human epidermal growth factor receptor 2,3,4; GCN, gene copy number; KRAS, Kirsten rat sarcoma virus oncogene homolog; IHC, imunno histochemistry; MAPK, mitogen activated protein kinase; mTOR, mammalian target of rapamycin; NSG mice, NOD scid gamma mice; PDX, patient derived xenograft; PIK3CA, phosphatidylinositol-4,5-bisphosphate 3-kinaseM catalytic subunit alfa; T/C, treated to control value; TDT, tumor doubling time; TGFα, transforming growth factor alfa; TNM, tumor nodule metastasis classification; TP53, tumor protein p53
Introduction
Colorectal cancer (CRC) is the third most diagnosed cancer affecting about 10% of men and 9% of women worldwide. CRC represents the fourth most common cause of death due to cancer worldwide and the second in Europe [1]. The inherent complexity of the disease, characterized by multiple genetic aberrations in interconnected signaling cascades has impact on therapy outcome and also on drug discovery [2,3].
CRC is treated by surgery, combined targeted and chemotherapy and radiation [4]. However, therapeutic success is highly dependent on choosing the right therapeutic modality for the right patient, who will benefit most from an appropriate and effective treatment. Regarding chemotherapy of CRC patients, 5-fluorouracil (5-FU), modulated by leucovorin (LV), was for decades the only drugs available for CRC and metastatic CRC. Currently, 5-FU is combined with oxaliplatin or irinotecan as first-line treatment. The introduction of targeted anti-cancer drugs such as bevacizumab and cetuximab prolonged the survival of CRC patients [5].
However, identification of resistance signatures and stratifying responsive patients is a current challenge in clinical management of CRC. This demands appropriate and reliable preclinical models that reproduce cancer pathway dynamics and closely resemble the clinical situation. In this context, appropriate clinically relevant in vivo models are required, such as patient-derived xenograft models (PDX). PDX were demonstrated to maintain the morphological and molecular characteristics of the original heterogeneous patient tumor and have been identified as a superior model system for translational research [6]. Several studies have demonstrated that PDX models can be used for the correct prediction for sensitivity or resistance of the tumor to better guide therapies for the patient [7], [8], [9], [10], [11], [12], [13]. Analysis of clinically validated biomarkers such as Kirsten rat sarcoma virus oncogene homolog (KRAS) mutations and resistance to epidermal growth factor receptor (EGFR) inhibitors in CRC PDX studies led to the same conclusions as clinical trials and explored resistance signatures for improved therapy in CRC [14,15].
In this study, we established 49 CRC PDX models, performed thorough characterization of molecular features such as gene copy number, mutational status, gene expression as well as proteome profiles. We characterized tumor responsiveness toward anti-cancer drugs and generated in vivo resistance models to be used for molecular analysis of key features associated with cetuximab-induced resistance. Our study strongly supports the power and value of PDX models as a platform for biomarker identification and verification and for more pre-clinical testing of individualized cancer therapies.
Material and methods
Patient samples
Approval of the local ethical committees was given and informed consent was obtained from all patients prior to sample acquisition and experimentation. All patient data were used in an anonymized fashion according to the ethical guidelines.
Establishment of PDX
All animal experiments were carried out in accordance to the German Animal Welfare Act as well as the UKCCCR (United Kingdom Coordinating Committee on Cancer Research).
Fresh tumor fragments were transplanted subcutaneously (s.c.) into the left flank of anaesthetized NOD scid gamma mice. Mice were observed for maximum 120 d and maintained under sterile and controlled conditions (22 °C, 50% relative humidity, 12 h light–dark cycle, autoclaved food and bedding, acidified drinking water). Tumor growth was measured in 2 dimensions with a caliper. Tumor volumes (TV) were determined by the formula: TV = (width² x length) x 0.5. Tumors were routinely passaged at TV = 1 cm³. Xenograft material was snap frozen and stored at -80 °C or processed to formalin fixed, paraffin embedded (FFPE) blocks.
Chemosensitivity testing of the PDX
Groups of 5 mice were randomized to receive either solvent as control or one of the respective drugs. Treatment was started at tumor size of approx. 0.1 cm³. For evaluation of therapeutic efficacy, the ratio of the mean TV of the treated group (T) and the control group (C) was expressed as the T/C-value in percentage. Antitumor activity was also defined based on the Response Evaluation Criteria in Solid Tumors. A relative tumor volume (RTV, normalized to the TV on the first treatment day) greater than 1.2 was classified as progressive disease (PD), an RTV of 0.7 to 1.2 as stable disease (SD), an RTV lower than 0.7 as partial response (PR), and a complete disappearance of the tumor was classified as complete response (CR).
Generation of cetuximab‐resistant PDX sublines
To generate resistant PDX isogenic models, 2 cetuximab sensitive PDX models were continuously treated with cetuximab for 10 in vivo passages (weekly treated with increasing doses of cetuximab: 50, 75, 100 mg/kg). PDX with highest RTV was selected for further passages. The corresponding untreated PDX was passaged in parallel as corresponding sensitive model.
HE‐staining of tumor samples
FFPE blocks were sectioned (5 to 8 µm), deparaffinized. Shock frozen tissues were cut (4 to 5 µm) and fixed in 96% ethanol for 5 min. Specimens were stained according to standard hematoxylin eosin protocol for histopathological evaluation.
Immunohistochemistry and immunofluorescence
For immunohistochemistry frozen sections were fixed with 4% formalin at RT, blocked with peroxidase-block, washed in phosphate buffered saline (PBS) and blocked with a Streptavidin-Biotin block (both from Vector Laboratories), according to manufacturer's instructions. Then sections were blocked with 20% goat serum for 30 min at RT, incubated with the biotinylated anti-EpCAM antibody (using the Animal Research Kit (Dako), 1:200, 2 h, RT (Enzo Life Sciences), and horse radish peroxidase-labeled streptavidin (1:800, 20 min, RT).
For immunofluorescence FFPE samples were washed in PBS and permeabilized in 0.1% Triton-X, incubated with Cy3-labeled human anti-nuclei antibody (Merck Millipore, dilution 1:300) 1 h at 37 °C, washed in PBS and mounted in glycerol and PBS (1:1) containing 5 µg/mL 4′,6-Diamidin-2-phenylindole (DAPI) (Roth). Staining was visualized with 3,3′-Diaminobenzidine substrate (Dako) and counterstained with hematoxylin, then mounted with VectaMount Aqueous Mounting Medium.
Total DNA extraction from tissues
Isolation of total DNA from tissues was carried out using the DNeasy Blood and Tissue kit (QIAGEN) following manufacturer's instructions. The DNA was eluted in 200 µL of distilled H2O and used for further analysis.
DNA sequencing
DNA samples from the 49 PDX were analyzed with the Illumina TruSeq Amplicon Cancer Panel (Illumina), targeting 212 amplicons from 48 oncogenes. Paired patient tumor and normal tissue from 3 samples were analyzed and compared to their respective PDX in order to corroborate the conservation of the original genetic profile in the xenografts. All reagents were supplied in the TruSeq Amplicon Cancer Panel Kit (Illumina). MiSeq sequencing was carried out on Illumina MiSeq Desktop Sequencer. Illumina Variant Studio 2.1 was used for sample analysis. For correlation analysis, known SNPs were excluded and only somatic mutations with an allelic frequency > 5% were considered.
Total RNA extraction from tissues
The total RNA from xenograft tissue samples was isolated by using the RNeasy Mini Kit (QIAGEN) following the manufacturer's instructions. The RNA was eluted in RNAse-free 50 µL H2O and used for further analyses.
Gene expression analysis
The global mRNA expression was compared between sensitive PDX and its cetuximab resistant counterparts. Transcriptome analysis of 12 CRC PDX samples was performed using Illumina Beadchips (HumanHT-12, V4). Sample preparation was done according to the ``Whole-Genome Gene Expression Direct Hybridization Assay GuidePart # 11322355 Rev. A'' from Illumina. For analysis, the Illumina HumanHT-12 array data were quantile normalized on probe level (47,323 probes) and on gene level without background correction using Illumina GenomeStudio V2011.1. cDNA synthesis
The Reverse-Transcriptase-Kit (Applied Biosystems) was used according to manufacturer's instructions to generate a master mix of 1 x RT-buffer, containing 500 µM dNTP's, 5.5 mM MgCl2, 2.5 µM random hexamers, 0.4 U/µL RNase inhibitor und 1.25 U/µL MultiScribe-Reverse-Transkriptase. Then, 200 ng of RNA were diluted in 10 µl RT-buffer and the RT-PCR reaction was conducted at 25 °C, 10 min; 48 °C, 30 min, and 95 °C, 5 min.
Gene expression analysis by quantitative real‐time PCR
200 ng of cDNA, TaqMan Fast Master Mix and Gene Expression Assay kit were combined in a total volume of 20 µL according to manufacturer's instructions. The real-time polymerase chain reaction (PCR) was carried out on a StepOnePlus System and was conducted at 95 °C for 20 s, followed by 40 cycles of 95 °C, 1 s and at 60 °C, 20 s. The amplification plots were evaluated with the StepOne Software Version 2.3. The threshold cycle (CT) of the gene of interest was normalized to the CT of β-actin and the ΔCT-values were used to compare the expression between samples.
Enzyme‐linked immunosorbent assay
Protein lysates were generated for EGFR and its ligand analysis. Tumor tissues were homogenized in 200 µL T-PER Tissue Protein Extraction Reagent (Life Technologies), supplemented with protease and phosphatase inhibitors (Sigma-Aldrich). Then, samples were freeze-thawed and centrifuged 15 min, 4 °C at 13,000 x g. Protein concentration was measured with a Protein Assay (Bio-Rad), using a bovine serum albumine (BSA) standard curve and adjusted to 4 µg/µL. DuoSet enzyme-linked immunosorbent assays were used to measure the concentration of EGFR, Amphiregulin (AREG), epiregulin (EREG), betacellulin (BTC), EGF, and transforming growth factor alfa (TGFα) according to manufacturer's instructions (USCN Life Science). The concentrations were normalized to the total protein concentration.
Protein extraction and digestion
The tissues were cryo-fractured on dry ice using a Covaris CP02 cryoPREP Automated Dry Pulverizer. Proteins were extracted with an 8 M Urea-based extraction buffer and reduced with 5 mM DTT (Thermo Fisher Pierce) for 1 h at 37 °C. Cysteine residues were alkylated by adding IAA (Sigma-Aldrich) to a final concentration of 10 mM, followed by incubation for 45 min in the dark at 25°C. Samples were diluted 1:4 with 50 mM Tris-HCl (pH 8.0). LysC (Wako) was added in an enzyme/substrate ratio of 1:50 followed by incubation for 2 h at 25 °C. Trypsin (Promega, enzyme/substrate ratio of 1:50) was added for overnight digestion at 25 °C, then quenched by acidifying the mixture to a final concentration of 1% FA (Sigma-Aldrich). The peptide samples were desalted using tC18 SepPak cartridges with a vacuum manifold.
Tandem Mass Tag labeling and fractionation
For multiplexing, 200 µg peptide samples were labeled with 400 µg tandem mass tag (TMT)-10 reagents (Thermo Scientific). The samples were separated by high-pH reversed-phase liquid chromatography using Agilent 1290 Infinity II LC system into 96 fractions that were combined in a step-wise manner, into 28 fractions for proteome and 14 fractions for phosphoproteome analysis. Early-, middle-, and late-eluting peptides were combined by mixing every 28th original fraction for the proteome and every 12th original fraction for the phosphoproteome analysis. A total of 10% by volume of the material was used for proteome analysis. The remaining 90% of each sample was enriched for phosphopeptides by immobilized metal affinity chromatography using high capacity Fe (III)-NTA cartridges on Agilent Bravo automated liquid handling platform.
LC‐MS/MS analysis
Tryptic peptides were analyzed on an EASY-nLC 1200 system coupled to a Q-Exactive HF-X (Thermo Fisher Scientific). The EASY-nLC system was equipped with a 75 µm x 20 cm column (packed in-house with 1.9 um C18 resin; Reprosil Gold, Dr. Maisch) and operated at a flow rate of 250 nL/min applying a 110 min linear gradient from 2 to 90% solvent B (90% ACN, 0.1% FA) in solvent A (3% ACN, 0.1% FA). MS measurements were performed on Q Exactive HF-X with the following modifications: MS1 spectra were recorded at a resolution of 60k using a maxIT of 10 ms. Fragment spectra were acquired at 45k resolution using a maxIT of 86 ms for global proteome measurements and a maxIT of 120 ms for phosphoproteome measurements.
Data analysis
For TMT experiments, peptide identification and quantification were performed using MaxQuant (version 1.6.0.1) [16]. Tandem mass spectra were searched against human and mouse reference proteome (Uniprot fasta, downloaded on 09.01.2017) supplemented with common contaminants. For all searches carbamidomethylated cysteine was set as fixed modification and oxidation of methionine, N-terminal protein acetylation, and for immobilized metal affinity chromatography data also phosphorylation on serine, threonine, and tyrosine residues as variable modifications. Trypsin/P was specified as the proteolytic enzyme with up to 2 missed cleavage sites allowed. Results were adjusted to 1% false discovery rate. The reporter-ion intensities were corrected for isotopic impurities before using the reporter-ion signals in each MS/MS spectrum for quantitative calculation s [17].
Statistical analysis
All statistical analyses were performed with Graph Pad Prism 5. For the response evaluation in the sensitivity characterization, two-tailed ANOVA-test was used. Correlation analysis was performed as Spearman rank-order correlation with a two-tailed Pvalue, and Spearman Rho (rS) were calculated. Mann-Whitney U tests were performed to compare the generated cetuximab resistant PDX sublines with their sensitive counterparts. A P value of < 0.05 was considered as statistically significant.
Statistical analysis of proteome and phosphoproteome
The corrected reporter ion intensities obtained from the mass-spectrometric measurements were divided by the internal standard mix reporter ion intensities and log-transformed using the R-statistical software package [18]. The ratios for the resistant vs control were calculated and used for the one-sample moderated t test with Proteomics Toolset for Integrative Data Analysis (Protigy; https://github.com/broadinstitute/protigy). All identifications were considered significant with adj. P< 0.1 and reproducibility filter alfa = 0.01 in. Significantly regulated proteins were used for the Gene Ontology analysis using the DAVID online tool [19]. Heatmaps were created using pheatmap package in R.
Results
Histological characteristics of CRC PDX models
For establishment of the 49 CRC PDX, 87 surgical tumor samples (patient characteristics Table 1) were used, reflecting an engraftment rate of 56%. The average time to initial engraftment in NOD scid gamma mice was 49.4 ± 19.5 d. Of these 49 PDX, 27 (55%) were derived from colon and 22 (45%) from rectum (Table 2).
Table 1.
Key clinical data of patients from whom PDX models were established.
| PDX | Patient characteristics |
Tumor sample characteristics |
||||||
|---|---|---|---|---|---|---|---|---|
| Gender | Age | TNM-status | Grading | Classification | Primary site | Metastasis |
||
| Type | Site | |||||||
| Co 5676 | f | 67 | T3aN0M0 | G2L0V1R0 | primary | rectum | ||
| Co 5677 | m | 71 | T4bN1M0 | G2L0V0R0 | primary | colon | ||
| Co 5679 | f | 51 | T3N2M1 | G3L1V1R0 | primary | colon | ||
| Co 5682 | f | 78 | T4bN2M0 | G2L0V1R0 | primary | rectum | ||
| Co 5734 | m | 79 | T3bN0M0 | G2L0V0R0 | primary | colon | ||
| Co 5735 | m | 84 | T3N0M1 | G2 | metastasis | rectum | met | liver |
| Co 5736 | f | 77 | T4N0M0 | G2L0V0R0 | primary | rectum | ||
| Co 5771 | m | 49 | T3N1M0 | G2LxV0R0 | primary | rectum | ||
| Co 5776 | m | 55 | yT3N1M1 | G3L1V1R0 | metastasis | rectum | syn | liver |
| Co 5841 | m | 55 | T3N2M1 | G3L1V1R0 | primary | rectum | ||
| Co 5854 | f | 52 | T4N2M0 | G3L1V1R0 | primary | colon | ||
| Co 5896 | m | 71 | T2N0M0 | G2L0V0R0 | primary | rectum | ||
| Co 6044 | m | 53 | T4N2M1 | G2L0V1R2 | metastasis | rectum | syn | lymph node |
| Co 6228 | f | 60 | rpTxNxM1 | GxLxVxR0 | metastasis | colon | met | liver |
| Co 72711 | f | 66 | pT2pN1 | G3L0V1R0 | metastasis | colon | syn | lung |
| Co 7475 | f | 68 | pT2pN0 | G2 | metastasis | rectum | met | lung |
| Co 7515 | m | 53 | pT3 pN0M0 | L0V0 R0 | metastasis | rectum | met | lung |
| Co 7523 | f | 64 | pT3pN1cM0 | n.a. | metastasis | colon | met | lung |
| Co 7553A2 | m | 77 | T3pN1M0 | G3 | metastasis | colon | met | lung |
| Co 7553B2 | m | 77 | T3pN1M0 | G3 | metastasis | colon | met | lung |
| Co 7567 | m | 75 | pT3pN0M1 | R0 | metastasis | colon | syn | lung |
| Co 7596 | f | 72 | pT3NxcM0C2 | n.a. | metastasis | rectum | met | lung |
| Co 76601 | f | 67 | pT2pN1 | G3L0V1R0 | metastasis | colon | met | lung |
| Co 7689 | f | 58 | pT3N2M0 | G2c | metastasis | rectum | met | lung |
| Co 7809 | m | 67 | yrp TxNxM1 | R0LxVxG3 | metastasis | colon | syn | liver |
| Co 7818 | m | 68 | rpTxNxM1 | R0LxVxG2 | metastasis | colon | syn | lung |
| Co 7835 | m | 75 | rpTxNxM1 | R0LxVxG2 | metastasis | colon | syn | lung |
| Co 7888 | m | 73 | yrepTxNxM1 | R0LxVxGx | metastasis | rectum | syn | liver |
| Co 7935 | m | 65 | rpTxNxM1 | R0LxVxG2 | metastasis | rectum | syn | liver |
| Co 9587 | f | 80 | pT3pN0(0/16) | G2R0L1V0 | primary | colon | ||
| Co 9634 | f | 61 | pT3C4pN1C4cM1 | L0V0 | metastasis | colon | met | liver |
| Co 9689A3 | m | 53 | pT3pN2cM1 | L1V1 | metastasis | rectum | met | liver |
| Co 9689B3 | m | 53 | pT3pN2cM1 | L1V1 | metastasis | rectum | met | liver |
| Co 9729 | m | 68 | pT3pN1c | G3L1V0 | primary | rectum | ||
| Co 9775 | m | 67 | pT3pN2apM1 | G2R0L0V0 | primary | colon | ||
| Co 9946 | m | 81 | pT4bpN0 | G2R0 | primary | colon | ||
| Co 9978 | f | 50 | pT4apN1bpM1 | G2R0L1V1 | primary | rectum | ||
| Co 99974 | f | 27 | pT4apN2b | G3R0L1V1 | primary | rectum | ||
| Co 10,158 | m | 67 | pT4apN0 | G2R0L0V0 | primary | colon | ||
| Co 10,194 | m | 78 | pT3pN0 | G2R0L0V0 | primary | colon | ||
| Co 103004 | f | 27 | pT4apN2b | G3R0L1V1 | metastasis | rectum | syn | liver |
| Co10302A5 | m | 64 | pT4b(m)pN1b | G2RXL1V0 | primary | colon | ||
| Co10302B5 | m | 64 | pT4b(m)pN1b | G2RXL1V0 | primary | colon | ||
| Co 10,377 | m | 50 | ypT3ypN0 | RxL0V1G2 | primary | colon | ||
| Co 10,383 | m | 51 | pT2pN0pM1 | G3R0L0V0 | primary | rectum | ||
| Co 10,588 | m | 72 | pT3pN0 | G2R0 | primary | colon | ||
| Co 10,764 | m | 62 | yrpT4bpN1apM1 | n.a. | primary | colon | ||
| Co 10,925 | m | 73 | pT3pN2bcM0 | R0 | primary | colon | ||
| Co 11,061 | f | 81 | pT3pN0 | G2R0L0V0 | primary | colon | ||
1-5PDX models were derived from the same patient; syn = synchronous metastasis; met = metachronous metastasis.
Table 2.
Summary of key parameters of the established 49 PDX.
| Number | Ratio of PDX [%] | |
|---|---|---|
| Patients | 44 | |
| Male | 28 | |
| Female | 16 | |
| Generated PDX | 49 | |
| PDX derived from colon carcinoma | 27 | 55 |
| PDX derived from rectum carcinoma | 22 | 45 |
| PDX derived from primary tumors | 26 | 53 |
| PDX derived from metastases | 23 | 47 |
| Liver | 11 | 22 |
| Lung | 11 | 22 |
| Lymph node | 1 | 2 |
The number of PDX derived from primary tumors was balanced to those PDX derived from metastases (53% vs 47% respectively). The metastasis-derived PDX were evenly distributed between metastasis from lung or liver. Histology of primary CRC patient tumor tissue was compared with corresponding PDX tissue (Figure 1).
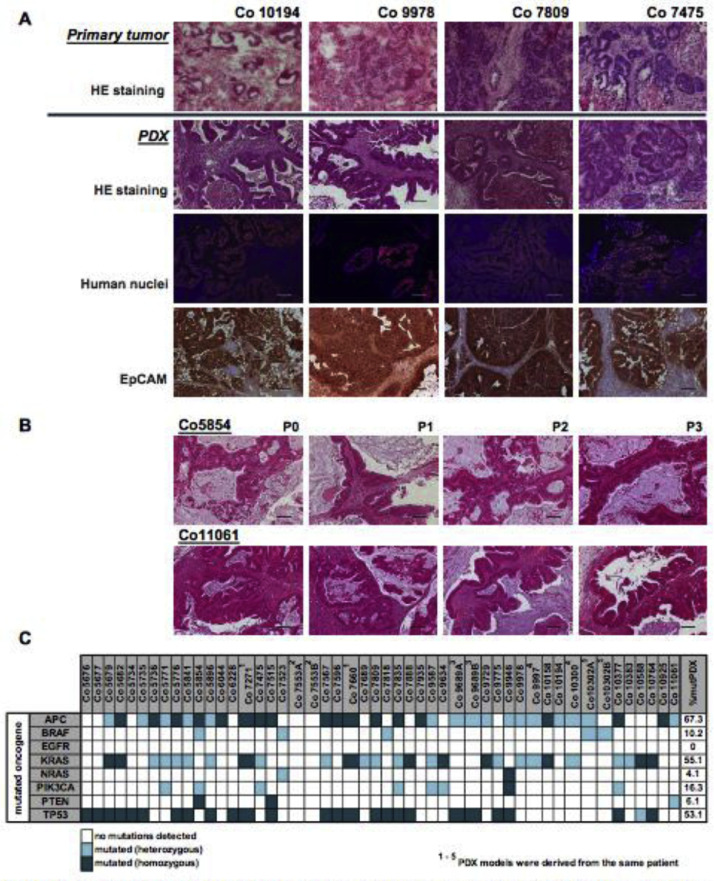
Figure 1.
Histological and mutational status of CRC PDX. (A) Staining of the original patient tissue paired to the corresponding PDX tissue. Representative patient/PDX pairs were chosen: Co10194 is derived from a primary colon carcinoma sample, Co9978 from a rectum sample, Co7809 was derived from a liver metastasis and Co7475 is derived from a lung metastasis. Upper panel, original patient tissue: HE staining; panel 2 to 4, PDX tissue: HE staining; IF staining for human nuclei, DAPI = blue; human nuclei = orange (Cy3); IHC staining for EpCAM. (B) Representative HE staining of consecutive passages of PDX Co5854 and Co11061 demonstrate characteristic phenotype of human colorectal carcinoma. Scale bar represents 100 µm. (C) Summary of mutational status of all PDX models.
All PDX showed similar characteristics of adenocarcinomas and were therefore well comparable with the initial patient tumor sample (Figure 1A). This adenocarcinoma histology was maintained through serial passages (Figure 1B). The tumor cells in the PDX expressed the human epithelial marker EpCAM. Further, they were positive for human nuclei antibody staining, whereas the surrounding stroma was negative, indicating murine origin of the stroma in the stably engrafted PDX (Figure 1A), indicating a replacement of human by murine stroma during in vivo passaging of the PDX.
Mutational status and genetic stability of CRC PDX models
Mutation analysis in the PDX revealed frequent mutations in the prominent oncogenes, such as adenomatous polyposis coli (APC), KRAS, and P53, as well-known key players in CRC (Figure 1C; Suppl. Table 1). Highest mutational frequency of 67.3% was detected for APC, followed by 55.1% for KRAS and 53.1% for P53 in the 49 CRC PDX. A lower percentage of PDX possessed mutated NRAS, PTEN, BRAF or PIK3CA (4.1%, 6.1%, 10.2%, or 16.3%, respectively).
To evaluate whether the PDX retain the genetic profile, mutational analyses were performed for all models regarding normal-, primary tumor- and PDX tissues. The mutation analysis of the original patient tumor after establishment and also over serial passages in 3 representative PDX revealed mutations in the genes APC, KRAS, PIK3CA, and TP53 of the respective patient tumor tissues. These were also present in all analyzed passages of the matched PDX. No mutations were detected in the corresponding normal tissues. Comparison of primary tumor tissue vs PDX indicated no gains or losses of mutations throughout the xenografting and passaging process (Table 3) supporting stability of the mutational status within driver oncogenes and suppressor genes in the PDX.
Table 3.
Patient tumor and matched normal tissue, paired with tissue from different, sequential passages of the corresponding PDX models was sequenced using the TruSeq Amplicon – Cancer Panel. 212 amplicons in 48 oncogenes were targeted. The results obtained for the main oncogenes are summarized in the table (AA = amino acid, n.a. = not analyzed).
| Mutation detected, (AA mutation) |
|||||||
|---|---|---|---|---|---|---|---|
| Patient or PDX, passage | APC | BRAF | EGFR | KRAS | MET | PIK3CA | TP53 |
| Co9587, normal tissue | - | - | - | - | - | - | - |
| Co9587, patient tumor | n.a. | n.a. | n.a. | G12D | n.a. | n.a. | n.a. |
| Co9587, P0 | ins1554;R876 | - | - | G12D | - | del104 | - |
| Co9587, P1 | ins1554;R876X | - | - | G12D | - | del104 | - |
| Co9587, P2 | ins1554;R876X | - | - | G12D | - | del104 | - |
| Co9587, P3 | ins1554;R876X | - | - | G12D | - | del104 | - |
| Co9587, P4 | ins1554;R876X | - | - | G12D | - | del104 | - |
| Co9775 normal tissue | - | - | - | - | - | - | - |
| Co9775, patient tumor | - | - | - | G12D | - | - | G245S |
| Co9775, P0 | - | - | - | G12D | - | - | G245S |
| Co9775, P1 | - | - | - | G12D | - | - | G245S |
| Co9775, P3 | - | - | - | G12D | - | - | G245S |
| Co9775, P4 | - | - | - | G12D | - | - | G245S |
| Co10925 normal tissue | - | - | - | - | - | - | - |
| Co10925,patient tumor | E1379X | - | - | - | - | - | - |
| Co10925, P1 | E1379X | - | - | - | - | - | - |
| Co10925, P2 | E1379X | - | - | - | - | - | - |
| Co10925, P4 | E1379X | - | - | - | - | - | - |
Growth characteristics of the PDX models
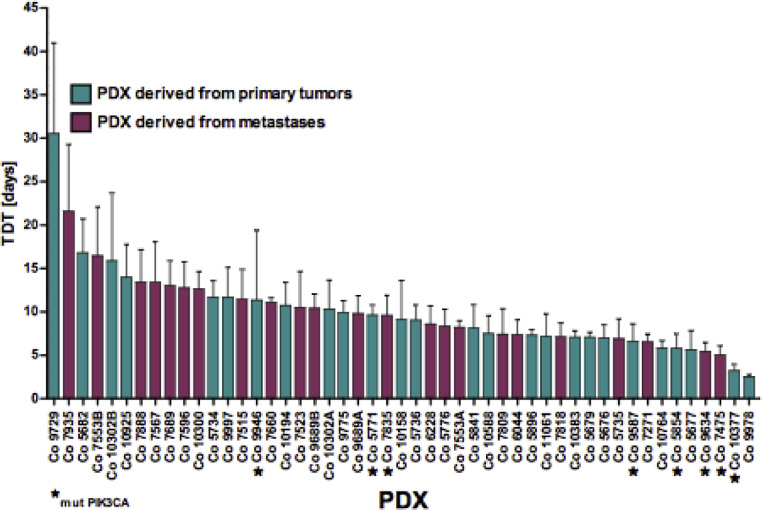
Tumor doubling time (TDT) was determined as one parameter for PDX growth (Figure 2). Mean TDT of the 49 CRC PDX was 9.96 ± 4.73 d, with a broad range between 2.5 and 30.5 d. TDT of 50% of PDX models clustered in a rather narrow range of 7.1 to 11.7 d. PDX derived from the colon or rectum were not different in their TDT. Similarly, TDT of PDX derived from primary tumors and PDX derived from metastasis did not show differences. Analysis of the mutational status of the PDX regarding TDT showed no correlation between mutations in APC, KRAS, TP53, PTEN, BRAF, or NRAS. Interestingly, mutated PIK3CA (n = 8 PDX) had a significant impact (P= 0.019) on TDT (7.1 ± 2.8 d in mutated vs 10.5 ± 4.8 d in wild type PIK3CA; n = 41).
Figure 2.
Tumor doubling times (TDT) of the PXD models. The tumor doubling time was measured once the models were considered as stably established. The PDX with mutated PIK3CA are marked by an “*.”
Sensitivity of the PDX models toward drug treatment
One key parameter of PDX models is responsiveness toward conventional and targeted drugs.
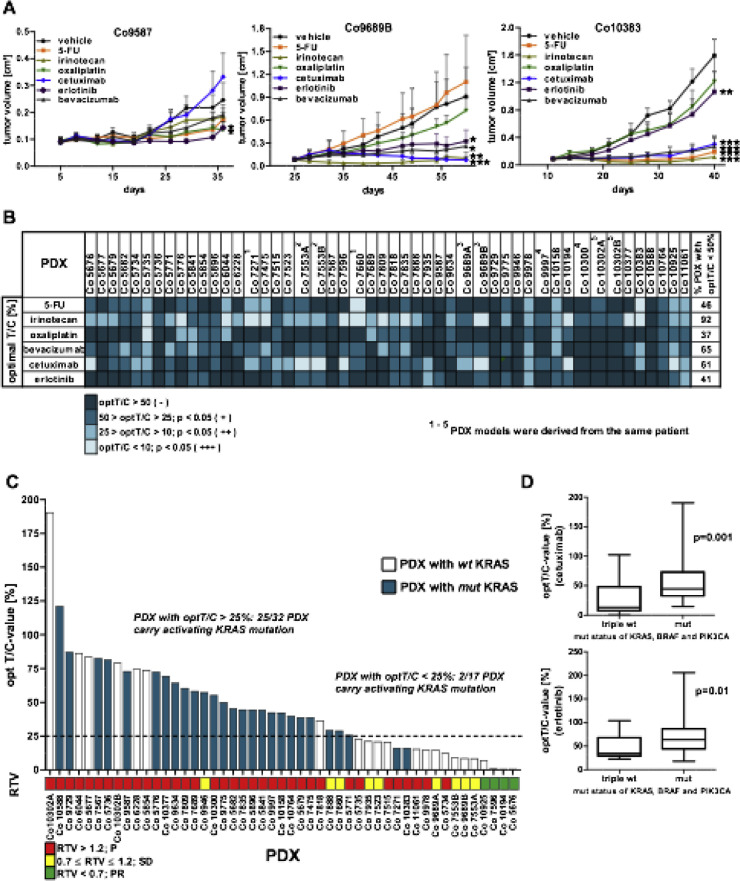
The sensitivity of PDX toward chemotherapeutics including 5-FU, oxaliplatin and irinotecan as well as toward targeted drugs bevacizumab, cetuximab and erlotinib were tested as monotherapy (Figure 3A and B and Suppl. Tables 2 and 3). The criterion for response was fulfilled, if the treated to control value (T/C) was <50%. By this, 44% of the 49 PDX responded to 5-FU and 37% to oxaliplatin treatment. Best responses were achieved for irinotecan in 92% of the PDX, which can be attributed to the specific activating metabolism in mice [20]. For the targeted agents, best response rates were obtained with cetuximab (61% of PDX) and bevacizumab (65% of PDX). Erlotinib showed a response in 41% of the PDX. Interestingly, comparison of PDX models derived from tumor samples of the same patient showed almost identical sensitivity profiles (Figure 3B, Suppl. Tables 2 and 3).
Figure 3.
Chemosensitivity of the PDX models. (A) Three representative chemosensitivity curves of PDX Co9587, Co9689B, and Co10383 are shown. Groups of 5 tumor-bearing mice were treated either with vehicle (control group) or specific drug as monotherapy. (*P< 0.05, **P < 0.01, ***P < 0.001). (B) Summary of sensitivity characteristics of the PDX models toward standard of care cytostatic and targeted drugs based on the optimal treated to control (optT/C) values, expressed in percent. (C) Sensitivity of PDX models toward cetuximab: the 49 PDX models are arranged according to their optT/C-value for cetuximab and are correlated to the KRAS mutational status. The bar below represents the relative tumor volume (RTV) values. The bar colors indicate the respective mutational status of KRAS. (D) Comparison of optT/C-values for EGFR-inhibitors cetuximab and erlotinib between PDX with wildtype KRAS, BRAF, and PIK3CA (triple wt) and PDX with an activating mutation in one of these genes (mut).
Since mutational status of KRAS is recognized as an important biomarker for resistance toward EGFR-targeted therapies, the influence of KRAS, BRAF and PIK3CA was correlated to PDX sensitivity toward cetuximab and erlotinib. Among the sensitive PDX with T/C-value <25%, only 2/17 models (12%) have an activating KRAS mutation. By contrast, 25/32 (78%) of resistant PDX carried a KRAS mutation (Figure 3C). Of 5 PDX with a BRAF mutation, 3 carrying the V600E mutation (Co5854, Co10302A, and Co10302B) were resistant to cetuximab. The T/C-values for cetuximab and erlotinib were not significantly different between PDX with a mutated or a wild type PIK3CA or BRAF. However, more importantly, triple wild-type PDX for BRAF, KRAS, and PIK3CA were significantly more sensitive toward cetuximab or erlotinib (significantly lower T/C-values; P= 0.001 or P = 0.01 respectively) than PDX with an activating mutation in 1 or more of these 3 genes (Figure 3D).
Influence of gene copy numbers on PDX sensitivity toward targeted drugs
As for gene mutational status, we also analyzed if alterations in gene copy numbers (GCN) have an impact on responsiveness of the PDX toward targeted drugs and analyzed EGFR, KRAS, NRAS, BRAF, and MET GCN alterations (Figure 4).
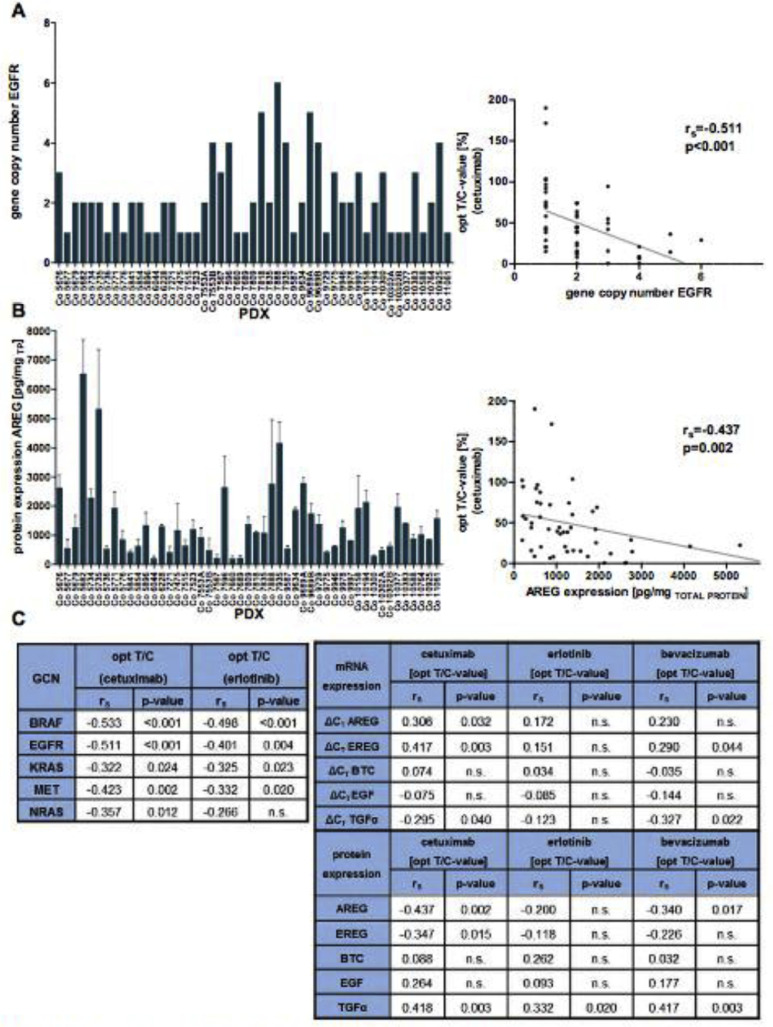
Figure 4.
Molecular characterization of PDX regarding GCN and AREG/EREG protein expression and chemosensitivity toward targeted drugs. (A) GCN of EGFR in the 49 CRC PDX models, determined by a real-time PCR (left panel) and correlation to cetuximab response (right panel). (B) AREG expression at protein level (by ELISA) in the PDX models and correlation to cetuximab response (right panel). The protein concentrations have been normalized to the total protein content in the sample. Three PDX samples from different passages were measured (n = 3). (C) Summary of correlation analysis (spearman coefficients rs and their corresponding P values) of the GCN of the key molecules of the EGFR network (left table) and the sensitivity of the PDX toward cetuximab and erlotinib, as well as ligand expression at mRNA and protein level (right table).
This analysis indicated, that 14/49 PDX harbor increased GCN (>2) for EGFR (Figure 4A, Suppl. Table 4). For KRAS, 3/49 PDX had 5 to 9 copies of the gene. BRAF amplifications were detected in 9/49 PDX (Co7596 had a BRAF copy number of 23). The correlation analysis revealed better response to cetuximab or erlotinib in the PDX with high GCN for EGFR, KRAS, and BRAF (Figure 4C). This indicates that a potentially higher activity of EGFR/MAPK signaling increases the tumors’ vulnerability toward cetuximab or erlotinib therapies.
Impact of EGFR ligand and receptor expression on PDX sensitivity toward targeted drugs
Regarding targeted drug action, we also analyzed expression of EGFR ligands AREG, EREG, EGF, BTC, and TGFα in association with responsiveness toward cetuximab or erlotinib. This analysis revealed a significant (P< 0.05) correlation for expression of AREG, EREG and TGFα with cetuximab response of the PDX. No such correlation was detected regarding erlotinib response (Figure 4B and C). This reflects the close molecular interplay of cetuximab and the respective natural ligands as determinant of drug efficacy. By contrast, no significant correlation was found between sensitivity toward cytostatic drugs and the expression of EGFR and its ligands.
Generation and characterization of cetuximab resistant CRC PDX
We generated isogenic PDX pairs of cetuximab responsive and resistant models by the continuous treatment of PDX with cetuximab (Figure 5).
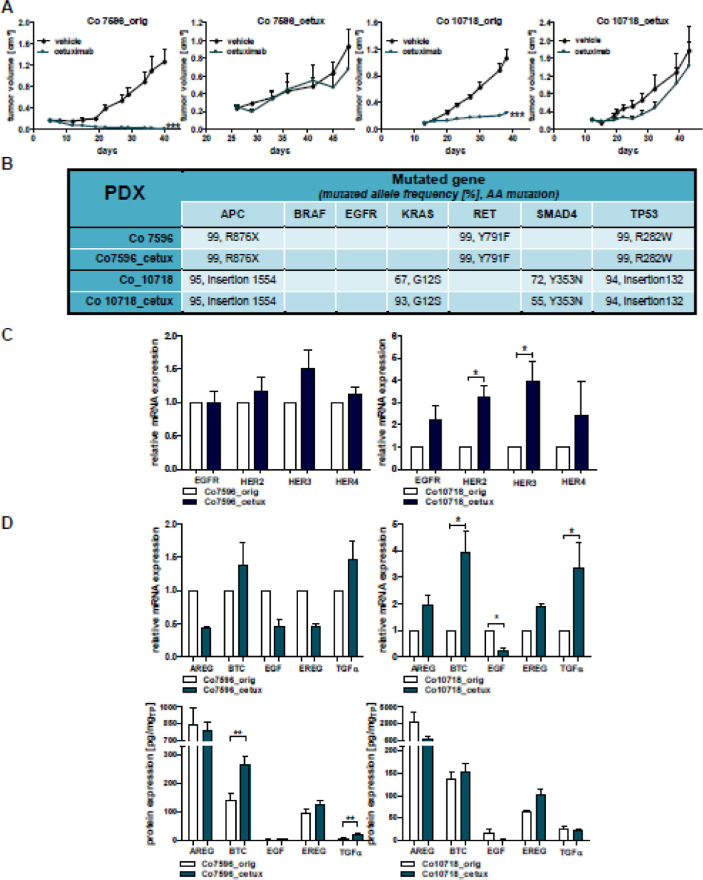
Figure 5.
Analysis of newly generated cetuximab resistant PDX models. (A) The PDX models Co7596 and Co10718 were initially cetuximab sensitive (left panels). After continued cetuximab treatment the cetuximab resistant models Co7596-cetux and Co19718_cetux were generated (right panels). (B) Comparison of genetic profiles in the 2 PDX pairs by the Illumina TruSeq Amplicon Cancer Panel reflected by mutated allele frequency and specific amino acid (AA) mutations. (C) Analysis of relative mRNA expression of EGFR, HER2, HER3, and HER4 in the PDX pairs. (D) Analysis of relative mRNA and protein expression of the EGFR ligands AREG, BTC, EGF, EREG, and TGFα in the PDX pairs. (*P< 0.05, **P < 0.01, ***P < 0.001).
After 10 passages of the PDX with cetuximab, resistance had developed, reflected by increase in T/C values for the Co7596 model (Co7596_orig vs Co7596_cetux) from 0.7% up to 74% and for the Co10718 model (Co10718_orig vs Co10718_cetux) from 23% to 46% (Figure 5A).
For these pairs we determined if emergence of cetuximab resistance is associated with alterations in mutational status. The analysis revealed no alterations for the mutational status and mutated allele frequency (Figure 5B) of APC, BRAF, KRAS, RET, SMAD4, and TP53 in Co7596_orig vs Co7596_cetux. In Co10718_orig vs Co10718_cetux, cetuximab resistance was correlated with an increased frequency of mutated KRAS from 67% to 93% and decreased frequency for SMAD4 from 72% to 55%. This indicates the emergence of rather individual alterations during the emergence of cetuximab resistance.
Since we had observed a significant correlation between GCN of BRAF and EGFR and response toward targeted drugs, we also analyzed this correlation regarding cetuximab resistance. However, we did not detect significant alterations in GCN during emergence of resistance in the 2 models (Suppl. Table 5).
Expression alteration of EGFR family members and their ligands during cetuximab resistance in PDX
Since targeted therapies aim at EGFR signaling, the expression of the EGFR receptor family and its ligands was compared in original PDX and their resistant counterparts. Expression analysis for EGFR, HER2, HER3 and HER4 revealed no significant changes in Co7596_orig vs Co7596_cetux (Figure 5C). By contrast, in the Co10718_cetux model, a statistically significant increase in HER2 and HER3 (P< 0.05) was detected, pointing to the diverging response of the CRC tumors toward cetuximab treatment.
As expression of EGFR ligands correlated with sensitivity to cetuximab in the 49 PDX, expression of these molecules was also analyzed in the cetuximab resistant PDX to evaluate potential changes of AREG, EREG, BTC, EGF, or TGFα. In this regard elevated expression only of BTC and TGFα was detected at protein level in the Co7596_cetux, and at mRNA-level in Co10718_cetux PDX (Figure 5D). The concentration of BTC in PDX was almost 2-fold in the Co7596_cetux (from 141.95 pg/mg protein to 263.36 pg/mg protein) when compared to the original PDX. Also, in Co10718_orig vs Co10718_cetux, a statistically significant difference in ΔCT-values was detected for BTC, EGF, and TGFα. The expression levels of BTC and TGFα were elevated and the level of EGF was lowered.
Molecular analysis of cetuximab resistant CRC PDX
To obtain a deeper insight into potential alteration mediating resistance toward cetuximab, differential gene expression of Co7596_orig and Co7596_cetux, as well as Co10718_orig and Co10718_cetux, was performed (Figure 6A).
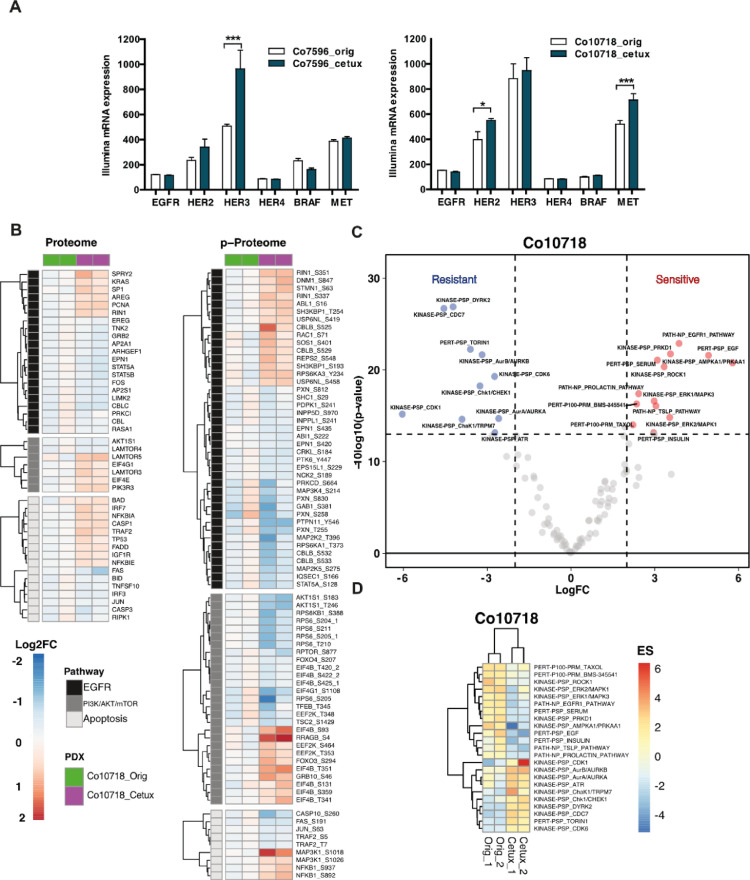
Figure 6.
Gene expression and proteome, phosphoproteome analysis of cetuximab sensitive vs resistant PDX. (A) Comparison of the gene expression of EGFR receptors and molecules involved in EGFR signal transduction. (B) Heatmaps of significantly changed proteins and phosphosites (One-sample t test, adj. P value < 0.1, reproducibility filter = 0.01) within the EGFR, PI3K/AKT/Mtor, and apoptosis pathways (Wiki Pathway annotations) for the Co10718 model. P values are calculated with data of 2 replicates for the Co10718_cetux models normalized against Co10718_orig models. The annotation column shows 2 lanes for 2 replicates of sensitive (Co10718_orig) and cetuximab resistant (Co10718_cetux) models with green and magenta colors, respectively. On the heatmap, blue color indicates down-regulation whereas red color corresponds to proteins and phosphosites up-regulated in the cetuximab resistant comparing to sensitive pair. (C) Volcano plot of the P values vs the logFC (fold change) of enrichment scores of PTM-SEA (Ref DOI: 10.1074/mcp.TIR118.000943). Two-sample t test is performed between resistant and sensitive replicates of the Co10718 model. Proteins crossing the significance lines (|logFC| ≥ 2, Pvalue ≤ 0.05) are colored in blue or red. (D) Heatmap depicting enrichment scores of significantly regulated pathways in the PTM-SEA analysis.
The differential gene expression analysis revealed decreased EGFR expression in Co10718_cetux compared to Co10718_orig (Differential score = –2.55). HER2 and HER3 were upregulated in Co7596_cetux and Co10718_cetux, respectively. Interestingly, an up-regulation of c-MET was seen in Co10718_cetux, and BRAF was down-regulated (not significantly) in Co7596_cetux, similarly to its CGN. (*P < 0.05, **P < 0.01, ***P < 0.001)
Proteomics and phosphoproteomic analysis of cetuximab resistant CRC PDX
To characterize proteomic and phosphoproteomic changes involved in the acquisition of cetuximab resistance in CRC PDX models, we performed TMT labeling-based global proteome and phosphoproteome analysis of Co10718 and Co7596 resistant models and their sensitive counterparts (Figure 6B, Suppl. Figure 1; Suppl. Table 6). Among the 15,167 proteins and 24,890 phosphosites that were quantified without missing values, we detected 9095 proteins, 15,237 phosphosites that could be matched to the human proteome. Of these, 1335 proteins and 1259 phosphosites were significantly regulated in the Co10718_cetux model (adj. P value < 0.05, one-sample t test), whereas no significant changes were observed for the Co7596_cetux model due to sample related variability between replicates (Suppl. Figures 2 and 3).
We performed pathway enrichment analyses for the significantly up and down regulated proteins and phosphosites in Co10718 model. Top 20 significantly up and down regulated GO terms and KEGG pathways are shown in the bar graphs (Suppl. Figure 4). Here, endocytosis, cell-cell adhesion, tight and adherence junctions related terms were enriched in the downregulated proteins and phosphosites population of the resistant model.
We also examined proteomic and phosphoproteomic profiles of proteins belonging to EGFR and downstream signaling cascades including MAPK, PI3K/AKT/mTOR and apoptosis pathways (significance cut-off adj. P value <0.1, one-sample t test; Figure 6B). The results on protein level indicated no significant change on the EGFR itself, while KRAS was slightly upregulated in the cetuximab resistant model. Notably, among EGFR ligands, AREG level was increased whereas EREG level was slightly decreased in the Co10718_cetux model (Figure 6B). Also, we found increased levels of RIN1, a RAS effector protein known to compete with RAF for RAS interaction.
Deep phosphoproteome analysis showed increased phosphorylation levels of RIN1 (Ser 351) and its downstream effector ABL. Both proteins have previously been shown to be involved in EGFR stabilization and inhibition of macropinocytosis (PMID: 22,976,291). In addition, increased phosphorylation of DNM1 (dynamin) and RAC1, proteins involved in endocytosis and re-organization of the actin cytoskeleton was observed. We detected increased phosphorylation on MAP3K1 (MEKK1) (Figure 6B), known to have unique structural characteristics that mediate its specific activities including regulation of cell survival and apoptosis [56].
Metabolic pathways, cell cycle and mRNA processing, and transport-related terms were enriched for the upregulated proteins and phosphosites. In order to further elucidate pathways which are activated or shut down upon acquired cetuximab resistance, we performed phosphosite-centric PTM-SEA (Post Translational Modifications Set Enrichment Analysis) analysis using individual quantified phosphosites (n = 11,917 phosphosites) of the Co10718 PDX model and queried against PTMsigDB. PTM-SEA resulted in 102 enriched pathway signatures which includes at least 5 matched phosphosites. Enrichment scores were calculated for each sample and individual pathway signatures. To obtain differentially enriched pathway scores between cetuximab resistant and sensitive models, we applied two-sample moderated t test to the replicates of the Co10718 model. In total, we detected 23 significantly enriched signatures between resistant and sensitive models (Figure6C and D). We observed positive enrichment of EGFR and EGF pathway signatures in the cetuximab sensitive models. Among the signatures that showed positive enrichment upon acquired cetuximab resistance were proliferative kinase signatures such as CDK1 and CDK6, Aurora kinases and DYRK2.
Discussion
This study described the successful establishment of 49 CRC PDX and their thorough characterization. We were able to stably generate PDX with cetuximab resistance to further explore the molecular features associated with emergence of this resistance. We employed these models to explore in detail the impact of the EGFR signaling network in the context of chemosensitivity.
Histology, stability, and sensitivity of the PDX
All 49 engrafted PDX displayed the characteristic adenocarcinoma architecture with intense uniform membranous and cytoplasmic staining for EpCAM [21]. Human nuclei staining confirmed that CRC tumor cells reconstructed structures of primary tumors by replacing human stroma through several passages of PDX. In the vast majority of studies, as well as in our set of PDX, only murine stroma was detected in stably engrafted PDX [22], [23], [24]. Regarding growth characteristics, the mean TDT for the 49 PDX was 10 d. This indicates that cells growing in a PDX undergo much less divisions compared to xenografts derived from established tumor cell lines, preventing genetic drift [25].
Mutation profiling showed that the key genetic elements driving tumor growth (e.g., p53, KRAS) in patients were maintained in PDX [26,27]. Frequency of mutations in CRC PDX closely reflects frequency in human primary tumor samples [22,28]. The frequency of 55.1% we describe for KRAS in the PDX is rather at the upper level of the overall described frequencies for CRC. However, for CRC in patient tumors and PDX models frequencies ranging from 35% to 51% were described [15,29]. Therefore, these models are well suited for sensitivity testing [7,28,30,31].
Regarding sensitivity toward drugs our set of PDX reflects the heterogeneity known for CRC. The T/C-values for the cytostatic drugs 5-FU and oxaliplatin reflect well the clinical response of CRC to the drugs. Irinotecan reached highest response among all cytostatics due to the more efficient drug metabolism in mice [20]. Lowest response was seen for oxaliplatin. For targeted drugs, T/C-values for erlotinib correlated strongly with those for cetuximab underlining their interference with same signaling pathway.
We showed, that GCNs of BRAF, EGFR, and KRAS, correlated to drug response to EGFR-inhibitors of the PDX. Amplification of EGFR has been associated with sensitivity to cetuximab by several studies, which is in accordance with our own results [32], [33], [34], [35]. Thus, PDX models can be considered representative of the patient tissue regarding GCN.
EGFR pathway and targeted drug response in CRC PDX
The expression of EGFR and its ligands has been linked to a more aggressive disease or a poor prognosis in several cancers, including CRC [35,36]. Regarding EGFR ligands, highest and most differential expression was found for AREG and EREG, indicating their biological relevance in the PDX. Another analysis of 144 CRC tumors also determined that AREG and EREG, are tightly co-expressed in primary tumors as well as in liver metastases [37,38]. We showed that this link extends to BTC. Expression of EGFR ligands can be upregulated upon activation of the receptor by the ligand itself (auto-induction), as well as by other members of the EGFR ligand family [39]. The correlation pattern of ligands and receptors in this study confirms the redundant EGFR signaling in the PDX.
Higher expression of AREG and EREG correlated to better tumor growth inhibition by cetuximab, which is well documented in CRC patients [40,41]. These correlations were not observed for erlotinib, underlining different mechanisms of action of the 2 drugs. TGFα behaved inversely. Since TGFα is an epithelial-specific autocrine mitogen, but also acts in a paracrine manner to modulate the tumor microenvironment, its mitogenic action might protect the PDX from tumor growth inhibition. In a study of 62 CRC patients, AREG and EREG were elevated in sensitive tumors, whereas TGFα behaved inversely [42].
Comparison of the genetic profile between original PDX and cetuximab resistant sub‐lines
We generated models of acquired cetuximab resistance to examine relevant mechanisms of acquired cetuximab-resistance at molecular level. Efforts to generate preclinical models of cetuximab-resistance from xenograft tumors have been limited to date [43], [44], [45].
The genetic profile and expression of EGFR-related molecules were analyzed in 2 original PDX and their cetuximab-resistant counterparts. The mRNA analysis both, by qRT-PCR and also by Illumina, showed a downregulation of EGFR expression in Co10718_cetux, as well as an increase of HER2 and HER3. Such upregulation of the HER2 and HER3 receptors or their increased activation as response to EGFR inhibition has been reported in preclinical models and cancer patients [44,46,47]. An increased ligand level (e.g., TGFα) was observed in both resistant models, indicating that ligand production can be a mechanism of resistance to EGFR blockade. Redundant EGFR signaling and use of alternative receptors to activate downstream mitogenic cascades are crucial in resistance to cetuximab. Increased expression of HER2 and HER3 can open novel treatments that target HER2 and/or HER3.
Comparison of the proteomic and phosphoproteomic profile between original PDX and cetuximab‐resistant sub‐lines
Proteome and phosphoproteome analysis revealed that acquired resistance to cetuximab in CRC PDX affects multiple pathways downstream of the EGFR. Increased phosphosites within the EGFR pathway belong to proteins associated with the regulation of EGFR trafficking, ubiquitination and proteasomal degradation (RIN1, ABL1, CBLB, and SH3KBP1). It was demonstrated before that increased EGFR degradation is associated with acquired cetuximab resistance in metastatic CRC cells [48]. Additional proteins known to play a role in endocytosis and the regulation of cytoskeletal components include dynamin, ABL and the src family kinase-binding protein SH3KBP1. These phosphorylation events indicate that alterations in EGFR and growth factor receptor internalization and trafficking are associated with resistance to cetuximab and warrant further investigation.
Deep phosphoproteome profile analysis showed that canonical MAPK and PI3K/AKT/mTOR pathways have major dephosphorylation events, such as reduced phosphorylation of pTyr546 of PTPN11 (SHP2), pSer381 of GAB1, pSer183 and pThr246 of AKT1S1 (PRAS40), pSer388 of RPS6KB1 (p70S6K) kinase, and several phosphosites on its substrate RPS6. In contrast to downregulated phosphorylation profiles of several MAPK pathway kinases such as MAP3K4, MAP2K2, or MAP2K5, and other kinases such as PDK1 and PTK6 in the Co10718_cetux model, we detected increased phosphorylation on MAP3K1 (MEKK1) (Figure 6B). MEKK1 is known to have unique structural and functional characteristics compared to other MAPK pathway proteins, that mediate its specific activity including regulation of cell survival and apoptosis [56].
Among the EGFR ligands, AREG level was increased whereas EREG level was slightly decreased in the Co10718_cetux model. Regulation of AREG and EREG expression has been reported to be involved in cancer metastasis and drug resistance in various cancers [49], [50], [51], [52].
Even though significant downregulation in the phosphorylation profile of several MAPK pathway proteins was observed, we detected increased phospho-MAP3K1 in the Co10718_cetux model. Several studies demonstrated that MAP3K1 promotes cell survival or apoptosis depending on the cell type, genetic alteration or stimulus [52], [53], [54]. In this regard, decreased Casp3 protein level with increased phosphorylated MAP3K1 and NF-κB point toward a MAP3K1-mediated cell survival mechanism in this cetuximab resistance model [55], [56], [57]. Thus, tumor profiling on the genome and proteome level after different treatments can determine possible changes in the molecular drivers and signaling pathways and elucidate resistance mechanisms in vivo.
In summary, a set of CRC PDX was established and extensively characterized. The genetic and sensitivity profile of the PDX reflects the heterogeneity of CRC. Correlation analysis of molecules involved in the EGFR pathway, as well as targeted therapy addressing this pathway, reflected the dynamics of the EGFR pathway regulation in the PDX models, confirming them as a tool for preclinical cancer research.
CRediT authorship contribution statement
Maria Rivera: Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing - original draft. Iduna Fichtner: Conceptualization, Funding acquisition, Supervision. Annika Wulf-Goldenberg: Methodology. Christine Sers: Funding acquisition, Writing - review & editing. Johannes Merk: Resources. Giannino Patone: Methodology, Software. Keziban M. Alp: Investigation, Data curation, Visualization, Writing - review & editing. Tamara Kanashova: Investigation, Writing - review & editing. Philipp Mertins: Investigation, Methodology, Resources, Writing - review & editing. Jens Hoffmann: Project administration, Resources, Writing - review & editing. Ulrike Stein: Conceptualization, Writing - original draft, Writing - review & editing. Wolfgang Walther: Conceptualization, Project administration, Supervision, Writing - original draft, Writing - review & editing.
Acknowledgments
We would like to thank Carsta Werner and Stefanie Tannert for their technical assistance in animal care and for the performance of the molecular biological analyses.
Footnotes
Funding: This project was funded by the OncoPath consortium of the Federal Ministry of Education and Research, Germany (Projektträger Jülich, OncoPath 031618F, project J).
Declaration of competing interest: The authors declare that they have no competing financial interests of personal relationships that could influence the work reported in this paper.
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.neo.2020.11.005.
Appendix. Supplementary materials
References
- 1.Siegel R.L., Miller K.D., Goding Sauer A., Fedewa S.A., Butterly L.F., Anderson J.C., Cercek A., Smith R.A., Jemal A. Colorectal cancer statistics. CA Cancer J Clin. 2020;70:145–164. doi: 10.3322/caac.21601. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Stratton M.R., Campbell P.J., Futreal P.A. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Santis C.E., Lin C.C., Mariotto A.B., Siegel R.L., Stein K.D., Kramer J.L., Alteri R., Robbins A.S., Jemal A. Cancer treatment and survivorship statistics. CA Cancer J Clin. 2014;64:252–271. doi: 10.3322/caac.21235. [DOI] [PubMed] [Google Scholar]
- 5.Saltz L.B., Meropol N.J., Loehrer P.J.S., Needle M.N., Kopit J., Mayer R.J. Phase II trial of cetuximab in patients with refractory colorectal cancer that expresses the epidermal growth factor receptor. J Clin Oncol. 2004;22:1201–1208. doi: 10.1200/JCO.2004.10.182. [DOI] [PubMed] [Google Scholar]
- 6.Siolas D., Hannon G.J. Patient-derived tumor xenografts: transforming clinical samples into mouse models. Cancer Res. 2013;73:5315–5319. doi: 10.1158/0008-5472.CAN-13-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fiebig H.H., Maier A., Burger A.M. Clonogenic assay with established human tumour xenografts: correlation of in vitro to in vivo activity as a basis for anticancer drug discovery. Eur J Cancer. 2004;40:802–820. doi: 10.1016/j.ejca.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 8.Guenot D., Guérin E., Aguillon-Romain S., Pencreach E., Schneider A., Neuville A., Chenard M.P., Duluc I., Du Manoir S., Brigand C. Primary tumour genetic alterations and intra-tumoral heterogeneity are maintained in xenografts of human colon cancers showing chromosome instability. J Pathol. 2006;208:643–652. doi: 10.1002/path.1936. [DOI] [PubMed] [Google Scholar]
- 9.Merk J., Rolff J., Becker M., Leschber G., Fichtner I. Patient-derived xenografts of non-small-cell lung cancer: a pre-clinical model to evaluate adjuvant chemotherapy? Eur J Cardiothorac Surg. 2009;36:454–459. doi: 10.1016/j.ejcts.2009.03.054. [DOI] [PubMed] [Google Scholar]
- 10.Zhang X., Claerhout S., Prat A., Dobrolecki L.E., Petrovic I., Lai Q., Landis M.D., Wiechmann L., Schiff R., Giuliano M. A renewable tissue resource of phenotypically stable, biologically and ethnically diverse, patient-derived human breast cancer xenograft models. Cancer Res. 2013;73:4885–4897. doi: 10.1158/0008-5472.CAN-12-4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho Y.B., Hong H.K., Choi Y.-L., Oh E., Joo K.M., Jin J., Nam D.H., Ko Y.H., Lee W.Y. Colorectal cancer patient-derived xenografted tumors maintain characteristic features of the original tumors. J Surg Res. 2014;187:502–509. doi: 10.1016/j.jss.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 12.Izumchenko E., Paz K., Ciznadija D., Sloma I., Katz A., Vasquez-Dunddel D., Ben-Zvi I., Stebbing J., McGuire W., Harris W. Patient-derived xenografts effectively capture responses to oncology therapy in a heterogeneous cohort of patients with solid tumors. Ann Oncol. 2017;28:2595–2605. doi: 10.1093/annonc/mdx416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klinghammer K., Walther W., Hoffmann J. Choosing wisely – preclinical test models in the era of precision medicine. Cancer Treat Rev. 2017;55:36–45. doi: 10.1016/j.ctrv.2017.02.009. [DOI] [PubMed] [Google Scholar]
- 14.Krumbach R., Schüler J., Hofmann M., Giesemann T., Fiebig H.H., Beckers T. Primary resistance to cetuximab in a panel of patient-derived tumour xenograft models: activation of MET as one mechanism for drug resistance. Eur J Cancer. 2011;47:1231–1243. doi: 10.1016/j.ejca.2010.12.019. [DOI] [PubMed] [Google Scholar]
- 15.Schütte M., Risch T., Abdavi-Azar N., Boehnke K., Schumacher D., Keil M., Yildirimman R., Jandrasits C., Borodina T., Amstislavskiy V. Molecular dissection of colorectal cancer in pre-clinical models identifies biomarkers predicting sensitivity to EGFR inhibitors. Nat Comm. 2017;8:14262. doi: 10.1038/ncomms14262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cox J J., Mann M. MaxQuant enables high peptide identification rates, individualized ppb-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 17.Shadforth I.P., Dunkley T.P., Lilley K.S., Bessant C. i-Tracker: for quantitative proteomics using iTRAQ. BMC Genomics. 2005;6:145–150. doi: 10.1186/1471-2164-6-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.R Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2014. R: A language and environment for statistical computing.http://www.R-project.org/ URL. [Google Scholar]
- 19.Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 20.Morton C.L., Iacono L., Hyatt J.L., Tylor K.R., Cheshire P.J., Houghton P.J., Danks M.K., Stewart C.F., Potter P.M. Activation and antitumor activity of CPT-11 in plasma esterase-deficient mice. Cancer Chemother Pharmacol. 2005;56:629–636. doi: 10.1007/s00280-005-1027-y. [DOI] [PubMed] [Google Scholar]
- 21.Zhou F.Q., Qi Y.M., Xu H., Wang Q.Y., Gao X.S., Guo H.G. Expression of EpCAM and Wnt/ β-catenin in human colon cancer. Genet Mol Res. 2015;14:4485–4494. doi: 10.4238/2015.May.4.6. [DOI] [PubMed] [Google Scholar]
- 22.Dangles-Marie V., Pocard M., Richon S., Weiswald L.B., Assayag F., Saulnier P., Judde J.G., Janneau J.L., Auger N., Validire P. Establishment of human colon cancer cell lines from fresh tumors versus xenografts: comparison of success rate and cell line features. Cancer Res. 2007;67:398–407. doi: 10.1158/0008-5472.CAN-06-0594. [DOI] [PubMed] [Google Scholar]
- 23.Julien S., Merino-Trigo A., Lacroix L., Pocard M., Goere D., Mariani P., Landron S., Bigot L., Nemati F., Dartigues P. Characterization of a large panel of patient-derived tumor xenografts representing the clinical heterogeneity of human colorectal cancer. Clin Cancer Res. 2012;18:5314–5328. doi: 10.1158/1078-0432.CCR-12-0372. [DOI] [PubMed] [Google Scholar]
- 24.Maykel J., Liu J.H., Li H., Schultz L.D., Greiner D.L., Houghton J.M. NOD-scidIl2rg (tm1Wjl) and NOD-Rag1 (null) Il2rg (tm1Wjl) : a model for stromal cell-tumor cell interaction for human colon cancer. Dig Dis Sci. 2014;59:1169–1179. doi: 10.1007/s10620-014-3168-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hlatky L., Olesiak M., Hahnfeldt P. Measurement of potential doubling time for human tumor xenografts using the cytokinesis-block method. Cancer Res. 1996;56:1660–1663. [PubMed] [Google Scholar]
- 26.Fearon E.R. Molecular Genetics of Colorectal Cancer. Annu Rev Pathol. 2011;6:479–507. doi: 10.1146/annurev-pathol-011110-130235. [DOI] [PubMed] [Google Scholar]
- 27.Hidalgo M., Amant F., Biankin A.V., Budinská E., Byrne A.T., Caldas C., Clarke R.B., de Jong S., Jonkers J., Mælandsmo G.M. Patient-Derived Xenograft Models: An Emerging Platform for Translational Cancer Research. Cancer Discov. 2014;4:998–1013. doi: 10.1158/2159-8290.CD-14-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seol H.S., Kang H.J., Lee S.I., Kim N.E., Kim T.I., Chun S.M., Kim T.W., Yu C.S., Suh Y.A., Singh S.R. Development and characterization of a colon PDX model that reproduces drug responsiveness and the mutation profiles of its original tumor. Cancer Lett. 2014;345:56–64. doi: 10.1016/j.canlet.2013.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tignanelli C.J., Loeza S.G.H., Yeh J.J. KRAS and PIK3CA mutation frequencies in patient derived xenograft (PDX) models of pacreatic and colorectal cancer are reflective of patient tumors and stable across passages. Am Surg. 2014;80:873–877. [PMC free article] [PubMed] [Google Scholar]
- 30.Fichtner I., Slisow W., Gill J., Becker M., Elbe B., Hillebrand T., Bibby M. Anticancer drug response and expression of molecular markers in early-passage xenotransplanted colon carcinomas. Eur J Cancer. 2004;40:298–307. doi: 10.1016/j.ejca.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 31.Fichtner I., Rolff J., Soong R., Hoffmann J., Hammer S., Sommer A., Becker M., Merk J. Establishment of patient-derived non-small cell lung cancer xenografts as models for the identification of predictive biomarkers. Clin Cancer Res. 2008;14:6456–6468. doi: 10.1158/1078-0432.CCR-08-0138. [DOI] [PubMed] [Google Scholar]
- 32.Moroni M., Veronese S., Benvenuti S., Marrapese G., Sartore-Bianchi A., Di Nicolantonio F., Gambacorta M., Siena S., Bardelli A. Gene copy number for epidermal growth factor receptor (EGFR) and clinical response to antiEGFR treatment in colorectal cancer: a cohort study. Lancet Oncol. 2005;6:279–286. doi: 10.1016/S1470-2045(05)70102-9. [DOI] [PubMed] [Google Scholar]
- 33.Cappuzzo F., Finocchiaro G., Rossi E., Jänne P.A., Carnaghi C., Calandri C., Bencardino K., Ligorio C., Ciardiello F., Pressiani T. EGFR FISH assay predicts for response to cetuximab in chemotherapy refractory colorectal cancer patients. Ann Oncol. 2008;19:717–723. doi: 10.1093/annonc/mdm492. [DOI] [PubMed] [Google Scholar]
- 34.Campanella C., Mottolese M., Cianciulli A., Torsello A., Merola R., Sperduti I., Melucci E., Conti S., Diodoro M.G., Zeuli M. Epidermal growth factor receptor gene copy number in 101 advanced colorectal cancer patients treated with chemotherapy plus cetuximab. J Transl Med. 2010;8:36. doi: 10.1186/1479-5876-8-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shigeishi H., Higashikawa K., Hiraoka M., Fujimoto S., Mitani Y., Ohta K., Takechi M., Kamata N. Expression of epiregulin, a novel epidermal growth factor ligand associated with prognosis in human oral squamous cell carcinomas. Oncol Rep. 2008;19:1557–1564. [PubMed] [Google Scholar]
- 36.Zhang J., Iwanaga K., Choi K.C., Wislez M., Raso M.G., Wei W., Wistuba I.I., Kurie J.M. Intratumoral epiregulin is a marker of advanced disease in non-small cell lung cancer patients and confers invasive properties on EGFR-mutant cells. Cancer Prev Res. 2008;1:201–207. doi: 10.1158/1940-6207.CAPR-08-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuramochi H., Nakajima G., Kaneko Y., Nakamura A., Inoue Y., Yamamoto M., Hayashi K. Amphiregulin and Epiregulin mRNA expression in primary colorectal cancer and corresponding liver metastases. BMC Cancer. 2012;12:88. doi: 10.1186/1471-2407-12-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pentheroudakis G., Kotoula V., De Roock W., Kouvatseas G., Papakostas P., Makatsoris T., Papamichael D., Xanthakis I., Sgouros J., Televantou D. Biomarkers of benefit from cetuximab-based therapy in metastatic colorectal cancer: interaction of EGFR ligand expression with RAS/RAF, PIK3CA genotypes. BMC Cancer. 2013;13:49. doi: 10.1186/1471-2407-13-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dahlhoff M., Wolf E., Schneider M.R. The ABC of BTC: structural properties and biological roles of betacellulin. Semin Cell Dev Biol. 2014;28:42–48. doi: 10.1016/j.semcdb.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 40.Jacobs B., De Roock W., Piessevaux H., Van Oirbeek R., Biesmans B., De Schutter J., Fieuws S., Vandesompele J., Peeters M., Van Laethem J.L. Amphiregulin and epiregulin mRNA expression in primary tumors predicts outcome in metastatic colorectal cancer treated with cetuximab. J Clin Oncol. 2009;27:5068–5074. doi: 10.1200/JCO.2008.21.3744. [DOI] [PubMed] [Google Scholar]
- 41.Yoshida M., Shimura T., Sato M., Ebi M., Nakazawa T., Takeyama H., Joh T. A novel predictive strategy by immunohistochemical analysis of four EGFR ligands in metastatic colorectal cancer treated with anti-EGFR antibodies. J Cancer Res Clin Oncol. 2013;139:367–378. doi: 10.1007/s00432-012-1340-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tabernero J., Cervantes A., Rivera F., Martinelli E., Rojo F., von Heydebreck A., Macarulla T., Rodriguez-Braun E., Eugenia Vega-Villegas M., Senger S. Pharmacogenomic and pharmacoproteomic studies of cetuximab in metastatic colorectal cancer: biomarker analysis of a phase I dose-escalation study. J Clin Oncol. 2010;28:1181–1189. doi: 10.1200/JCO.2009.22.6043. [DOI] [PubMed] [Google Scholar]
- 43.Ciardiello F., Bianco R., Caputo R., Caputo R., Damiano V., Troiani T., Melisi D., De Vita F., De Placido S., Bianco A.R. Antitumor activity of ZD6474, a vascular endothelial growth factor receptor tyrosine kinase inhibitor, in human cancer cells with acquired resistance to antiepidermal growth factor receptor therapy. Clin Cancer Res. 2004;10:784–793. doi: 10.1158/1078-0432.ccr-1100-03. [DOI] [PubMed] [Google Scholar]
- 44.Quesnelle K.M., Grandis J.R. Dual kinase inhibition of EGFR and HER2 overcomes resistance to cetuximab in a novel in vivo model of acquired cetuximab resistance. Clin Cancer Res. 2011;17:5935–5944. doi: 10.1158/1078-0432.CCR-11-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quesnelle K.M., Wheeler S.E., Ratay M.K., Grandis J.R. Preclinical modeling of EGFR inhibitor resistance in head and neck cancer. Cancer Biol Ther. 2012;13:935–945. doi: 10.4161/cbt.20846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bertotti A., Migliardi G., Galimi F., Sassi F., Torti D., Isella C., Corà D., Di Nicolantonio F., Buscarino M., Petti C. A molecularly annotated platform of patient-derived xenografts (“xenopatients”) identifies HER2 as an effective therapeutic target in cetuximab-resistant colorectal cancer. Cancer Discov. 2011;1:508–523. doi: 10.1158/2159-8290.CD-11-0109. [DOI] [PubMed] [Google Scholar]
- 47.Yonesaka K., Zejnullahu K., Okamoto I., Satoh T., Cappuzzo F., Souglakos J., Ercan D., Rogers A., Roncalli M., Takeda M. Activation of ERBB2 signaling causes resistance to the EGFR-directed therapeutic antibody cetuximab. Sci Transl Med. 2011;3:99ra86. doi: 10.1126/scitranslmed.3002442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu Y., Li X., Liang K., Luwor R., Siddik Z.H., Mills G.B., Mendelsohn J., Fan Z. Epidermal growth factor receptor (EGFR) ubiquitination as a mechanism of acquired resistance escaping treatment by the anti-EGFR monoclonal antibody cetuximab. Cancer Res. 2007;67:8240–8247. doi: 10.1158/0008-5472.CAN-07-0589. [DOI] [PubMed] [Google Scholar]
- 49.Khambata-Ford S., Garrett C.R., Meropol N.J., Basik M., Harbison C.T., Wu S., Wong T.W., Huang X., Takimoto C.H., Godwin A.K. Expression of epiregulin and amphiregulin and K-ras mutation status predict disease control in metastatic colorectal cancer patients treated with cetuximab. Journal Clin Oncol. 2007;25:3230–3237. doi: 10.1200/JCO.2006.10.5437. [DOI] [PubMed] [Google Scholar]
- 50.Jonker D.J., Karapetis C.S., Harbison C., O'Callaghan C.J., Tu D., Simes R.J., Malone D.P., Langer C., Tebbutt N., Price T.J. Epiregulin gene expression as a biomarker of benefit from cetuximab in the treatment of advanced colorectal cancer. Brit J Cancer. 2014;110:648–655. doi: 10.1038/bjc.2013.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tepper S.R., Zuo Z., Khattri A., Heß J., Seiwert T.Y. Growth factor expression mediates resistance to EGFR inhibitors in head and neck squamous cell carcinomas. Oral Oncol. 2016;56:62–70. doi: 10.1016/j.oraloncology.2016.03.008. [DOI] [PubMed] [Google Scholar]
- 52.Tokunaga S., Nagano T., Kobayashi K., Katsurada M., Nakata K., Yamamoto M., Tachihara M., Kamiryo H., Yokozaki H., Nishimura Y. Amphiregulin as a Novel Resistance Factor for Amrubicin in Lung Cancer Cells. Anticancer Res. 2017;37:2225–2231. doi: 10.21873/anticanres.11558. [DOI] [PubMed] [Google Scholar]
- 53.Bonvin C., Guillon A., van Bemmelen M.X., Gerwins P., Johnson G.L., Widman C. Role of the amino-terminal domains of MEKKs in the activation of NFκB and MAPK pathways and in the regulation of cell proliferation and apoptosis. Cell Signal. 2002;14:123–131. doi: 10.1016/s0898-6568(01)00219-4. [DOI] [PubMed] [Google Scholar]
- 54.Pham T.T., Angus S.P., Johnson G.L. MAP3K1: genomic alterations in cancer and function in promoting cell survival or apoptosis. Genes Cancer. 2013;4:419–426. doi: 10.1177/1947601913513950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Widmann C., Gerwins P., Johnson N.L., Jarpe M.B., Johnson G.L. MEK kinase 1, a substrate for DEVD-directed caspases, is involved in genotoxin-induced apoptosis. Mol Cell Biol. 1998;18:2416–2429. doi: 10.1128/mcb.18.4.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schlesinger T.K., Bonvin C., Jarpe M.B., Fanger G.R., Cardinaux J.R., Johnson G.L., Widman C. Apoptosis stimulated by the 91-kDa caspase cleavage MEKK1 fragment requires translocation to soluble cellular compartments. J Biol Chem. 2002;277:10283–10291. doi: 10.1074/jbc.M106885200. [DOI] [PubMed] [Google Scholar]
- 57.Gebauer G., Mrakhur B., Nguyen Q., Shore S.K., Simpkins H., Dhanasekaran N. Cisplatin-resistance involves the defective processing of MEKK1 in human ovarian adenocarcinoma 2008/C13 cells. Int J Oncol. 2000;16:321–326. doi: 10.3892/ijo.16.2.321. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.