Abstract
Among the proteins encoded by the SARS-CoV-2 RNA, nsP3 (non-structural Protein3) is the largest multi-domain protein. Its role is multifaceted and important for the viral life cycle. Nonetheless, regarding the specific role of each domain there are many aspects of their function that have to be investigated. SARS Unique Domains (SUDs), constitute the nsP3c region of the nsP3, and were observed for the first time in SARS-CoV. Two of them, namely SUD-N (the first SUD) and the SUD-M (sequential to SUD-N), exhibit structural homology with nsP3b (“X” or macro domain); indeed all of them are folded in a three-layer α/β/α sandwich. On the contrary, they do not exhibit functional similarities, like ADP-ribose binding properties and ADP-ribose hydrolase activity. There are reports that suggest that these two SUDs may exhibit a binding selectivity towards G-oligonucleotides, a feature which may contribute to the characterization of their role in the formation of the replication/transcription viral complex (RTC) and of the interaction of various viral “components” with the host cell. While the structures of these domains of SARS-CoV-2 have not been determined yet, SUDs interaction with oligonucleotides and/or RNA molecules may provide a platform for drug discovery. Here, we report the almost complete NMR backbone and side-chain resonance assignment (1H,13C,15N) of SARS-CoV-2 SUD-N protein, and the NMR chemical shift-based prediction of the secondary structure elements. These data may be exploited for its 3D structure determination and the screening of chemical compounds libraries, which may alter SUD-N function.
Keywords: SARS-CoV-2, Non-structural protein, SARS unique domain (SUD), Solution NMR-spectroscopy, Protein druggability, Covid19-NMR
Biological context
Over the last 20 years three major human infections related to Coronaviruses attracted the attention of the global scientific community. The most recent infection, named COVID-19, spread rapidly worldwide and turned into a pandemic as declared by WHO. This disease is caused by Severe Acute Respiratory Syndrome coronavirus 2 (SARS-CoV-2), which along with SARS-CoV and MERS-CoV (Middle Eastern respiratory syndrome) coronaviruses, belongs to the beta Coronavirinae subfamily (i.e. 4 genera of Coronaviridae). All of the aforementioned members share a common morphology possessing a positive-sense single-stranded RNA (+ ssRNA) genome of up to 30 kb in length, encapsulated in a spherical virion (Burrell 2017). The 30 kb RNA of coronaviruses is the largest known RNA viral genome, containing multiple ORFs and possessing 5’ cap, 3’ poly-A tail, 5’ and 3’ UTRs with important roles in the regulations of replication and transcription (Snijder et al. 2016; Lei et al. 2018; Alhammad and Fehr 2020a).
One third of this genome (located at the proximal 3’ end) encodes genes that correspond to the Coronaviruses virion main structural proteins: the spike glycoprotein (S), the transmembrane protein (M) and the internal phosphorylated nucleocapsid protein (N); another minor protein is a small transmembrane protein (E). Furthermore, some coronaviruses exhibit a gene for an additional envelope protein with esterase and hemagglutination functions (HE), the latter is also found in influenza virus (Burrell et al. 2017; Alhammad and Fehr 2020a).
The two thirds at the proximal 5′ end of the genome consists of two ORF (ORF 1a and 1b). A ribosomal frame-shift leads to the translation of two polyproteins (pp1a and pp1ab) which are further processed to the necessary elements for the formation of the replication/transcription viral complex (RTC) (Perlman et al. 2009; V’kovski et al. 2020). In general, three virus-encoded proteases play a key role in the polyproteins cleavage. The papain-like protease 1(PL1pro), papain-like protease 2 (PL2pro) and the 3C-like main protease (Mpro). Among the resulting 16 proteins, generated by this cleavage, the multi-domain non-structural protein 3 (nsP3) is the largest and it is considered as a key component with various roles in different stages of the viral-life, from RTC formation to host proteins interactions. Nevertheless, there are still many undefined functions that have to be further investigated (Snijder et al. 2016; Burrell et al. 2017; Lei et al. 2017).
In contrast with the most of coronaviruses, SARS-CoV-2 genome encodes only two proteases (PLpro and Mpro). nsP3 contains PLpro and other globular domains such as the macro or “X” domain (Snijder et al. 2016). The macro domain is conserved in many viruses with a three-layer α/β/α sandwich fold, exhibiting ADP-ribose binding properties and ADP-ribose hydrolase activity and it is termed here nsP3b (Lei et al. 2018; Frick et al. 2020; Michalska et al. 2020; Alhammad et al. 2020b). nsP3b is sequentially followed by nsP3c the so-called SARS Unique Domain (SUD) (Tan et al. 2007; Tan et al. 2009; Chatterjee et al. 2009; Serrano et al. 2009; Johnson et al. 2010; Kusov et al. 2015). These domains occur mainly in SARS-CoV and they have been also related to viruses found in certain bats (Lei et al. 2017). SUD exhibits a three-domain architecture; SUD N, SUD M for middle domain, and SUD C.
Despite the similar fold-type compared to the macro domain of the two first SUDs, N and M, there is no sufficient experimental evidence for common functional properties with the macro domain. However, it is believed that the dual domain polypeptide of SARS-CoV, SUD-NM, binds oligo(G)-nucleotides capable of forming G-quadruplexes. Mutations either on SARS-CoV SUD-N or SUD-M reduce or abolish the G-quadruplex binding capacity and selectivity of these domains (Tan et al. 2009). According to in vitro experiments, it is now believed that due to this binding affinity of SUDs to G-oligonucleotides, SUDs may be involved in the virus’ RTC machinery and they may control the host-cell’s response to the viral infection (Kusov et al. 2015). Therefore, considering the functional role of both SARS-CoV macro and SUDs and the recent SARS-CoV-2 outbreak, the discovery of antiviral therapeutics that target these domains is of great interest. In the absence of the 3D structure of any of the SARS-CoV-2 SUDs, we report herein a preliminary NMR study and the complete backbone and side chains chemical shifts assignment of the SARS-CoV-2 SUD-N (spanning the residues 409–548; according to nsP3 numbering). These data provide the basis for drug-screening attempts and may be exploited in the identification of a lead compound with potential antiviral interest through the testing of a large chemical compound library.
Methods and experiments
Construct design
The coding sequence of the SUD-N domain (residues 409-548 of nsP3) was amplified using primers (forward: 5′ CGCGGATCCCAAGACGATAAG 3′ and reverse 5′ CCGCTCGAGTTATTATTCTTGTTTCTC 3′ designed on cDNA sequence encoding nsP3 residues 201-745 (GenBank entry: MT066156.1- nucleotide numbering of the whole genome 3319–4954- GenBank entry: QIA985533 orf1ab—protein numbering of 1019–1563) obtained as a codon optimized for expression in E. coli, synthetic gene from GenScript, (Piscataway, NJ). SARS-CoV-2 SUD-N coding sequence was cloned into pGex4T-1 expression vector, containing an N-terminal GST-tag followed by a thrombin cleavage site. The produced protein contained two artificial N-terminal residues (G-1, S-0) preceding the native protein sequence.
Protein expression and uniform 15N and 15N/13C labeling
An LB preculture was inoculated with BL21 (DE3) E.coli cells transformed with the above mentioned plasmid, and was grown overnight at 37 ˚C, 180 rpm with 0.1 mg/mL ampicillin. A culture of 0.5 L M9 medium (40 mM Na2HPO4, 22 mM KH2PO4, 8 mM NaCl) containing 0.5 g 15N labeled NH4Cl and 2 g unlabeled or 13C D-glucose, 1 mL from a stock solution containing 0.5 mg/mL biotin and 0.5 mg/mL thiamine, 0.5 mL 1 M Mg2SO4, 0.15 mL 1 M CaCl2, 1 mL solution Q (40 mM HCl, 50 mg/L FeCl2 . 4H2O, 184 mg/L CaCl2 · 2H2O, 64 mg/L H3BO3, 18 mg/L CoCl2 · 6H2O, 4 mg/L CuCl2 · 2H2O, 340 mg/L ZnCl2, 710 mg/L Na2MoO4 · 2H2O, 40 mg/L MnCl2 · 4H2O), and 0.1 mg/mL ampicillin was inoculated with the preculture. When the OD600 reached 0.6–0.8, IPTG was added to final concentration of 1 mM and the culture was incubated at 18 °C. Sixteen hours (16 h) after induction the cells were harvested by centrifugation.
Protein purification and sample preparation
The cell pellet was re-suspended with 25 mL lysis buffer containing 50 mM Tris-HCl pH 8, 300 mM NaCl, 2 mM DTT. Then protease inhibitor cocktail (Sigma Aldrich® P8849) and 25 μL DNase (1 mg/mL) were added to the re-suspended cells, and they were sonicated (PMisonix®, Sonicator 4000), after the sonication took place, the suspension was centrifuged at 4 °C and 20.000 rpm (Thermo Scientific®, Sorvall Lynx 6000) for 30 min. The soluble fraction containing the GST-tagged SARS-CoV2 SUD N was loaded onto a GSTrap HP affinity column (GE Healthcare) that had been previously equilibrated with 25 mL binding buffer (50 mM Tris-HCl pH 8, 300 mM NaCl). The column was washed with 50 mM Tris-HCl pH 8, 300 mM NaCl and then 100 μL thrombin (10 mg/mL) in 5 mL binding buffer were added to the column in order to remove the GST tag from the SUD-N. The column was incubated with thrombin for 16 h (overnight) at 4 °C. After the overnight incubation, the SUD-N was eluted with 50 mM Tris-HCl pH 8, 300 mM NaCl, as was verified with a 17% SDS-PAGE gel and finally GST-elution (50 mM Tris-HCl, pH 8, 10 mM Reduced Glutathione). With the use of an Amicon® Ultra 15 mL Centrifugal Filter membrane (nominal molecular weight cutoff 10 kDa) the protein was concentrated to final volume of 1 mL and buffer exchange was performed, to 50 mM NaPi, pH 7.2, 50 mM NaCl, 2mM EDTA, 2mM DTT. Finally, size exclusion chromatography was performed using Superdex 75 Increase 10/300 GE Healthcare column. The fractions containing the pure protein were pooled together and were concentrated to final volume of 500 μL to prepare the NMR sample.
Data acquisition, processing and assignment
Protein samples for NMR experiments were prepared in 500 μL of a mixed solvent of 90% H2O (50 mM NaPi, 50 mM NaCl, pH 7.2) and 10% D2O, 2 mM DTT, 2 mM EDTA, 2 mM NaN3, bacterial inhibitor cocktail (Sigma Aldrich®) and 0.25 mM DSS (4,4-dimethyl-4-silapentane-1-sulfonic acid) as internal 1H chemical shift standard. 13C and 15N chemical shifts were referenced indirectly to the 1H standard using a conversion factor derived from the ratio of NMR frequencies (Wishart et al. 1995). The protein concentration in the NMR sample was in the range of 0.9-1 mM. All NMR experiments were recorded at 298 K on a Bruker Avance III High-Definition four-channel 700 MHz NMR spectrometer equipped with a cryogenically cooled 5 mm 1H/13C/15N/D Z-gradient probe (TCI). The acquired NMR experiments used for sequence specific assignment are summarized in Table 1.
Table 1.
List of NMR experiments acquired to perform the sequence specific assignment of apo-nsP3c SUD-N domain and their main acquisition parameters
| Experiment for apo nsP3c SUD-N domain | Time domain data size (points) | Spectral width (ppm) | ns | Delay time (s) | ||||
|---|---|---|---|---|---|---|---|---|
| t1 | t2 | t3 | F1 | F2 | F3 | |||
| 1H-15N HSQC | 512 | 2048 | 44.0 (15N) | 16.0 (1H) | 8 | 1.0 | ||
| 1H-15N TROSY | 512 | 2048 | 40.0 (15N) | 14.0 (1H) | 2 | 1.0 | ||
| TROSY-HN(CO)CACB | 96 | 40 | 1024 | 72.0 (15N) | 44.0 (15N) | 14.0 (1H) | 16 | 1.0 |
| TROSY-HNCACB | 96 | 40 | 1024 | 72.0 (15N) | 44.0 (15N) | 14.0 (1H) | 32 | 1.0 |
| HN(CA)CO | 64 | 40 | 1024 | 18.0 (13C) | 44.0 (15N) | 14.0 (1H) | 8 | 1.0 |
| HNCO | 64 | 40 | 1024 | 18.0 (13C) | 44.0 (15N) | 14.0 (1H) | 8 | 1.0 |
| HNCA | 80 | 40 | 1024 | 42.0 (13C) | 44.0 (15N) | 14.0 (1H) | 8 | 1.0 |
| HN(CO)CA | 80 | 40 | 1024 | 42.0 (13C) | 44.0 (15N) | 14.0 (1H) | 8 | 1.0 |
| HBHA(CBCACO)NH | 112 | 40 | 1024 | 8.0 (1H) | 44.0 (15N) | 14.0 (1H) | 16 | 1.0 |
| hCCH-TOCSY | 128 | 48 | 1024 | 80.0 (13C) | 80.0 (13C) | 14.0 (1H) | 16 | 1.0 |
| 3D 1H-15N NOESY | 256 | 48 | 2024 | 14.0 (1H) | 40.0 (15N) | 14.0 (1H) | 8 | 1.0 |
| CBCACONH (sel no CG) | 64 | 44 | 1024 | 72.0 (13C) | 42.0 (15N) | 14.0 (1H) | 16 | 1.0 |
| CBCACONH (sel AlaCysSer) | 64 | 44 | 1024 | 72.0 (13C) | 38.0 (15N) | 14.0 (1H) | 16 | 1.0 |
Results
Assignments and data deposition
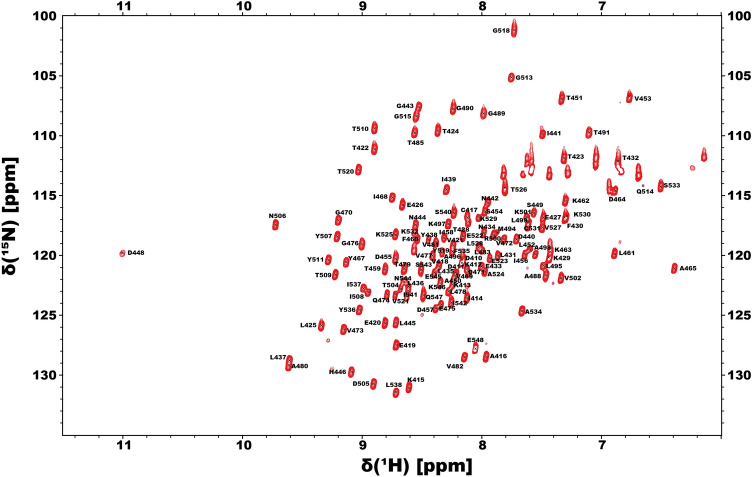
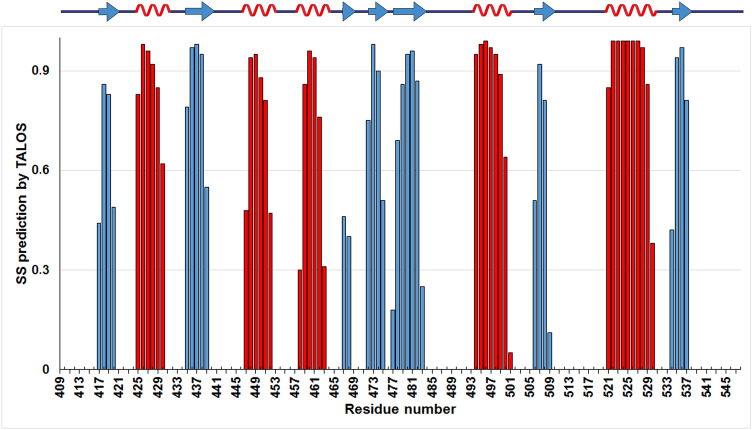
Backbone and sidechains assignments for the SARS-CoV-2 SUD-N were obtained from the following heteronuclear experiments: 2D [1H,15N]-HSQC and 2D [1H,15N]-TROSY, 3D HN(CO)CA, 3D HNCA, 3D TROSY HN(CO)CACB, 3D TROSY HNCACB, 3D HN(CA)CO, 3D HNCO, 3D HNHA, 3D HBHA(CO)NH, aliphatic 3D (H)CCH TOCSY, 2D [1H,13C]-HSQC and 3D 15N-edited NOESY (Table 1). To help the assignment process CBCA(CO)NH selective experiments have been performed in order to identify residues without CG and residues such as Ala, Cys and Ser (Table 1). All NMR data were processed with TOPSPIN 4.0.6 and analyzed with CARA 1.9.2a4 (Keller 2004). The 2D 1H,15N-HSQC spectrum shows well-dispersed amide signals (Fig. 1). For nsP3c SUD-N we assigned 95% of the resonances of the backbone atoms (HN, N, Hα, CO, Cα and Cβ) and about 80% of all the atom of side chains. The unassigned residues of nsP3c SUD-N are Gln409, Lys486, Lys487, Thr491, Glu492, Leu516 and Asn517 and are part of the loop regions as shown in Fig. 2.
Fig. 1.
700 MHz 1H,15N-HSQC assigned spectrum of the 13C,15N-labelled SARS-CoV-2 SUD-N nsP3c at 1 mM concentration in 50 mM NaPi pH 7.2, 50 mM NaCl, 2 mM EDTA, 2 mM DTT and 10% D2O acquired at 298 K. Amino acid numbering is according to the sequence of the multi-domain non-structural protein 3 (nsP3)
Fig. 2.
Predicted secondary structure of SARS-CoV-2 SUD-N nsP3c using TALOS+
Secondary structure prediction was performed using chemical shift assignments of five atoms (HN, Hα, Cα, Cβ, CO, N) for each residue in the sequence using TALOS+ (Shen et al. 2009). The secondary structure elements are in the following order from N-terminal to C-terminal: β/α/β/α/α/β/β/α. The order of the secondary structure segments is very similar to that of the nsP3b protein (Cantini et al 2020).
Chemical shift values for the 1H, 13C and 15N resonances of SARS-CoV-2 nsP3c SUD-N have been deposited at the BioMagResBank (https://www.bmrb.wisc.edu) under accession numbers 50448.
Acknowledgements
Work at BMRZ is supported by the state of Hesse. Work in COVID19-NMR was supported by the Goethe Corona Funds and the DFG in CRC902: “Molecular Principles of RNA-based regulation.” Work at CERM is supported by the Italian Ministry for University and Research (FOE funding) to the Italian Center (CERM, University of Florence) of Instruct-ERIC, a European Research Infrastructure, ESFRI Landmark. This work was also supported by the INSPIRED (MIS 5002550) which is implemented under the Action ‘Reinforcement of the Research and Innovation Infrastructure,’ funded by the Operational Program ‘Competitiveness, Entrepreneurship and Innovation’ (NSRF 2014–2020) and co-financed by Greece and the European Union (European Regional Development Fund). EU FP7 REGPOT CT-2011-285950–“SEE-DRUG” project is acknowledged for the purchase of UPAT’s 700 MHz NMR equipment.
Compliance with ethical standards
Conflict of interest
The authors declare no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Angelo Gallo, Aikaterini C. Tsika, and Nikolaos K. Fourkiotis have equally contributed.
Contributor Information
Nikolaos K. Fourkiotis, Email: covid19-nmr@dlist.server.uni-frankfurt.de
Lucia Banci, Email: banci@cerm.unifi.it.
Harald Schwalbe, Email: Schwalbe@nmr.uni-frankfurt.de.
Georgios A. Spyroulias, Email: G.A.Spyroulias@upatras.gr
References
- Alhammad YMO, Fehr AR. The viral macrodomain counters host antiviral ADP-ribosylation. Viruses. 2020;12(4):384. doi: 10.3390/v12040384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alhammad YMO, Kashipathy MM, Roy A, Johnson DK, McDonald P, Battaile KP, Gao P, Lovell S, Fehr AR (2020b) The SARS-CoV-2 conserved macrodomain is a highly efficient ADP-ribosylhydrolase enzyme. J Virol. 10.1128/JVI.01969-20 [DOI] [PMC free article] [PubMed]
- Burrell CJ, Howard CR, Murphy FA (2017) Chapter 31 – Coronaviruses In “Fenner and White's Medical Virology (5th Edition)” 437-446
- Cantini F, Banci L, Altincekic N, et al. 1H, 13C, and 15N backbone chemical shift assignments of the apo and the ADP-ribose bound forms of the macrodomain of SARS-CoV-2 non-structural protein 3b. Biomol NMR Assign. 2020;14:339–346. doi: 10.1007/s12104-020-09973-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A, Johnson MA, Serrano P, Pedrini B, Joseph JS, Neuman BW, Saikatendu K, Buchmeier MJ, Kuhn P, Wüthrich K. Nuclear magnetic resonance structure shows that the severe acute respiratory syndrome coronavirus-unique domain contains a macrodomain fold. J Virol. 2009;83:1823–36. doi: 10.1128/JVI.01781-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick DN, Virdi RS, Vuksanovic N, Dahal N, Silvaggi NR. Molecular Basis for ADP-Ribose Binding to the Mac1 Domain of SARS-CoV-2 nsP3. Biochemistry. 2020;59:2608–2615. doi: 10.1021/acs.biochem.0c00309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MA, Chatterjee A, Neuman BW, Wüthrich K. SARS coronavirus unique domain: three-domain molecular architecture in solution and RNA binding. J Mol Biol. 2010;400:724–42. doi: 10.1016/j.jmb.2010.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller, Rochus (2004). The Computer Aided Resonance Assignment Tutorial ISBN 3-85600-112-3, first edition.
- Kusov Y, Tan J, Alvarez E, Enjuanes L, Hilgenfeld R. A G-quadruplex-binding macrodomain within the "SARS-unique domain" is essential for the activity of the SARS-coronavirus replication-transcription complex. Virology. 2015;484:313–22. doi: 10.1016/j.virol.2015.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei J, Hilgenfeld R. RNA-virus proteases counteracting host innate immunity. FEBS Lett. 2017;591:3190–3210. doi: 10.1002/1873-3468.12827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei J, Kusov Y, Hilgenfeld R. NsP3 of coronaviruses: Structures and functions of a large multi-domain protein. Antiviral Res. 2018;149:58–74. doi: 10.1016/j.antiviral.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalska K, Kim Y, Jedrzejczak R, Maltseva NI, Stols L, Endres M, and Joachimiak A (2020) Crystal structures of SARS-CoV-2 ADP-ribose phosphatase (ADRP): from the apo form to ligand complexes. bioRxiv preprint. 10.1101/2020.05.14.096081. [DOI] [PMC free article] [PubMed]
- Perlman S, Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat Rev Microbiol. 2009;7(6):439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano P, Johnson MA, Chatterjee A, Neuman BW, Joseph JS, Buchmeier MJ, Kuhn P, Wüthrich K. Nuclear magnetic resonance structure of the nucleic acid-binding domain of severe acute respiratory syndrome coronavirus nonstructural protein 3. J Virol. 2009;83:12998–3008. doi: 10.1128/JVI.01253-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44(4):213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder EJ, Decroly E, Ziebuhr J. The Nonstructural proteins directing coronavirus RNA synthesis and processing. Adv Virus Res. 2016;96:59–126. doi: 10.1016/bs.aivir.2016.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J, Kusov Y, Mutschall D, Tech S, Nagarajan K, Hilgenfeld R, Schmidt CL. The "SARS-unique domain" (SUD) of SARS coronavirus is an oligo(G)-binding protein. Biochem Biophys Res Commun. 2007;364:877–82. doi: 10.1016/j.bbrc.2007.10.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J, Vonrhein C, Smart OS, Bricogne G, Bollati M, Kusov Y, Hansen G, Mesters JR, Schmidt CL, Hilgenfeld R. The SARS-unique domain (SUD) of SARS coronavirus contains two macrodomains that bind G-quadruplexes. PLoS Pathog. 2009;5:e1000428. doi: 10.1371/journal.ppat.1000428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- V’kovski P, Kratzel A, Steiner S, Stadler H, Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol. 2020 doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart DS, Bigam CG, Yao J, Abildgaard F, Dyson HJ, Oldfield E, Markley JL, Sykes BD. 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J Biomol NMR. 1995;6:135–140. doi: 10.1007/BF00211777. [DOI] [PubMed] [Google Scholar]