Abstract
Numerous studies have reported the association of long non-coding RNAs (lncRNAs) in cancers, yet the function of lncRNA high expressed in hepatocellular carcinoma (HEIH) in esophageal carcinoma (EC) has seldom been explored. Here, we aimed to explore the mechanism of HEIH on EC via microRNA-185 (miR-185)/kallikrein-related peptidase 5 (KLK5) modulation. Cancer and non-tumoral tissues were collected, in which HEIH, miR-185 and KLK5 expression were detected, as well as their correlations. Also, the relation between the prognosis of EC patients and HEIH/miR-185/KLK5 expression was clarified. EC cells (KYSE-30 and TE-1) were screened for subsequent gain- and loss-of-function assays and their biological functions were further monitored. Tumor volume and weight in EC mice were also measured. Results from this study indicated that HEIH and KLK5 were elevated and miR-185 was declined in EC. The positive correlation was seen in HEIH and KLK5 expression, while the negative correlation was observed in HEIH or KLK5 and miR-185 expression. High HEIH and KLK5 indicated worse prognosis and high miR-185 suggested better prognosis of EC patients. Depleting HEIH or restoring miR-185 suppressed the malignant phenotypes of EC cells, and delayed tumor growth in EC mice. HEIH was found to bind with miR-185 to regulate KLK5 expression. Overexpressing KLK5 alone promoted EC cell progression while up-regulating miR-185 reversed such effects on EC cells. Collectively, we reveal that HEIH depletion dampens EC progression by upregulating miR-185 and downregulating KLK5, which provides novel treatments for EC.
Subject terms: Cancer, Diseases
Introduction
Esophageal carcinoma (EC) is the eighth most common cancer over the world, and almost four to five cases arise in nonindustrialized countries, with the highest rates in Asia and Africa1. There are a wide range of risk factors for EC, such as smoking, alcohol intake, obesity, gastroesophageal reflux disease, and genetic factors2. Preoperative neoadjuvant therapy is increasingly applied to treat locally advanced EC to improve survival3. However, both intrinsic or acquired drug resistance generally contributes to the treatment failure of EC4. Therefore, it is imperative to seek for new effective approaches to EC treatment.
Long noncoding RNAs (lncRNAs) are implicated in various biological processes and usually abnormally expressed in cancers5. A recent study has reported that as one of the lncRNAs, lncRNA SNHG7 is elevated in EC, and is capable of facilitating proliferation and limits apoptosis of EC cells6. It has also been suggested that lncRNA high expressed in hepatocellular carcinoma (HEIH) is of great significance in boosting colorectal cancer (CRC) tumorigenesis7. Another article has indicated that lncRNA HEIH results in induced melanoma cell growth8. HEIH is suggested to involve the pathogenesis of EC9. MicroRNAs (miRNAs) are small endogenous ncRNAs, which modulate both intracellular pathologic and physiologic processes, such as cell differentiation, proliferation, and apoptosis10. Literatures have recorded that several miRNAs, such as miR-186 and miR-1207-5p, works as a tumor inhibitor in EC11,12. It is evidenced that miR-185 is lowly expressed in esophageal squamous cell carcinoma (ESCC) patients and is likely to dampen ESCC migration and invasion13. Kallikrein-related peptidase 5 (KLK5), which is encoded by KLK5 gene, is originally confirmed as a trypsin-like enzyme overexpressed in human epidermis and a critical player in skin desquamation14. Researchers have demonstrated that KLK5 is upregulated in CRC, and its upregulation is connected with the malignant behaviors of CRC15. Another study has suggested that elevated KLK10 could suppress EC cell proliferation and induce cisplatin sensitivity in vitro16. Nevertheless, there is almost no study exploring the impacts of HEIH/miR-185/KLK5 axis in EC development. Hence, we made this research to understanding the mechanism of HEIH on EC development through modulation of miR-185 and KLK5.
Materials and methods
Ethics statement
Written informed consents were obtained from all patients before the study. The protocols of this study were approved by the Ethic Committee of Chinese Academy of Medical Sciences and Peking Union Medical College. All animal experiments were in line with the Guide for the Care and Use of Laboratory Animal by International Committees.
Subjects
A total of 56 ESCC patients (39 males, 17 females, 46–73 years old, median age: 61 years old) who underwent surgery in Chinese Academy of Medical Sciences and Peking Union Medical College from 2014 to 2016 were enrolled in this study. Inclusion criteria: patients confirmed as EC by gastroscopy; preoperative chest enhancement computed tomography (CT) and upper abdominal B-ultrasound showed no visible cancer metastasis; and preoperative transesophageal endoscopic ultrasonography suggested no obvious swollen lymph nodes in the mediastina, and the esophageal infiltration was within T2. Exclusion criteria: patients with history of cervical spondylosis and not able to recline their head; patients with history of myocardial infarction, cerebral infarction, or pulmonary infarction; and chest CT or endoscopic ultrasonography showed swollen lymph nodes in the mediastina (>1 cm). None patients received any radiotherapy or chemotherapy before operation. Referred to Union for International Cancer Control, it was clarified that two cases were in stage I, 22 cases in stage II, 24 cases in stage III, and 8 cases in stage IV of tumor-node-metastasis (TNM) staging. There were 13 cases with high differentiation, 16 cases with moderate differentiation, and 27 cases with poor differentiation; 31 cases with lymph node metastasis (LNM) and 25 cases without LNM. EC tissues (cancer tissue) and non-tumoral tissues 3–5 cm away from the carcer tissues were collected. Each tissue specimen was divided into two parts. One part was fixed in 10% neutral formalin solution, then routinely made into wax masses and preserved for hematoxylin–eosin (HE) staining, immunohistochemistry, and in situ hybridization. The other part was stored at −80 °C for RNA and protein extraction. Patients were followed up by outpatient service or telephone for 30 months (until December 2018).
HE staining
The tissues were routinely dehydrated, embedded, cut into sections, dewaxed in xylene I and II, dehydrated in descending concentrations of alcohol, and stained with hematoxylin, followed by differentiation with hydrochloric acid alcohol. Subsequently, the sections were immersed in ammonia water for 10–30 s to return to the blue, stained with eosin solution, and dehydrated with ascending concentrations of alcohol. After that, xylene permeabilization and neutral gum sealing were performed before observation under a microscope.
Immunohistochemistry
The tumor paraffin sections were routinely deparaffinized, dehydrated, and immersed in 3% methanol H2O2. Rinsed sequentially with distilled water and 0.1 M PBS, sections were treated with antigen retrieval in water bath, sealed with normal goat serum blocking solution (ZSGB-Bio, Beijing, China), and probed with the primary antibody KLK5 (1:200, Proteintech Group Inc., Wuhan, China) overnight. After that, the sections were added with secondary antibody and incubated with horseradish peroxidase-labeled streptomyces working solution (ZSGB-Bio). Finally, the sections were developed by diaminobenzidine (ZSGB-Bio), followed by counterstaining, dehydration, permeabilization, and sealing. Immunohistochemistry of tumor tissues was independently evaluated by two pathologists17.
In situ hybridization
Cancer tissue and non-tumoral tissue sections were deparaffinized and dehydrated. The sections on the hybridization oven were reacted with 200 μL of pre-hybridization solution at 65 °C, and incubated with the probe hybridization solution of HEIH and miR-185 (500 ng/mL, 200 μL, DAKO, USA) at 55 °C. Then, the sections were developed by NBT/BCIP without light exposure and terminated with pure water. After that, the sections were stained by 200 μL nuclear fast red staining solution, counterstained, dehydrated (50%, 75%, and 100% alcohol), and mounted with neutral gum. BCIP/NBT staining was blue, while nuclear fast red color staining was red. The results were independently evaluated by two pathologists, and the cytoplasmic blue staining represented the positive cells18. Each section was randomly observed from five fields of view under a 200-fold microscope to calculate the percentage of positive cells. Negative: positive cells <5%; positive: positive cells ≥5%.
Cell selection and culture
Human normal esophageal epithelial cells Het-1A and human EC cell lines (KYSE-30, TE-1, Eca-109, EC9706, and KYSE-150), available from American type culture collection (ATCC, Rockefeller, Maryland, USA), were cultured in RPMI 1640 (Gibco, Carlsbad, California, USA) with 10% fetal bovine serum (FBS) and 8% NaHCO3, and the medium was renewed every 2 days. The cells reached 80–90% confluence were passaged. After two to three stable passages, the cells were taken for reverse transcription quantitative polymerase chain reaction (RT-qPCR) and western blot analysis. The cell lines with the largest and smallest HEIH, miR-185, and KLK5 expression difference from human normal esophageal epithelial cells Het-1A were screened for subsequent cell experiments.
Cell transfection
TE-1 and KYSE-30 cells of passages 3–6 were transfected with corresponding siRNAs, mimic, inhibitor, and plasmids, respectively, via Lipofectamine 2000 (Invitrogen, USA), as per instructions provided by the manufacturer.
KYSE-30 cells were transfected with HEIH overexpression vector, HEIH overexpression vector NC, miR-185 inhibitor, miR-185 inhibitor NC, HEIH overexpression vector and miR-185 mimic NC, HEIH overexpression vector and miR-185 mimic, KLK5 overexpression vector NC and miR-185 mimic NC, KLK5 overexpression vector and miR-185 mimic NC, or KLK5 overexpression vector and miR-185 mimic.
TE-1 cells were transfected with HEIH low expression vector, HEIH low expression vector NC, miR-185 mimic NC, miR-185 mimic, HEIH low expression vector (sh-HEIH) and miR-185 inhibitor NC, HEIH low expression vector and miR-185 inhibitor, KLK5 low expression vector NC and miR-185 inhibitor NC, KLK5 low expression vector and miR-185 inhibitor NC, or KLK5 low expression vector and miR-185 inhibitor.
The transfection plasmids were supplied by GenePharma (Shanghai, China).
MTS assay
3-(4,5-Dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) kit (Promega, Madison, WI, USA) was used to detect cell proliferation. The transfected cell were cultured for 24 h, detached by 0.2% trypsin, and suspended in medium containing 10% FBS to prepare cell suspension of 1 × 104 cells/mL. Then cell solution (100 μL) was seeded into 96-well plates, and appended to each well at 0, 24, 48, or 72 h after cell adherence. After 4-h incubation, the optical density value at 492 nm was measured by a 2010-type microplate reader (Anthos Labtec Instruments, Salzburg, Austria).
EdU assay
TE-1 and KYSE-30 cell DNA replication ability was measured with 5-ethynyl-2′-deoxyuridine (EdU) kit (KeyGen BioTECH, Jiangsu, China). Cells were resuspended and counted after routinely detached, and seeded in 96-well plates at 1 × 104 cells/well (five parallel wells set up in every group). After incubation with 100 μL EdU solution (50 μmol/L), the cells were fixed with 4% paraformaldehyde incubated with 2% glycine, and permeabilized with 150 μL penetrant. Five random fields were screened to capture images under a DWI40CCB fluorescence inverted microscope (Leica, Wetzlar, Germany). Blue fluorescence stood for all cells, and red fluorescence for cells being replicating that had been infiltrated by EdU. The percentage of EdU-positive cells was calculated.
PI single staining
When cell confluence reached 80%, TE-1 and KYSE-30 cells were trypsinized with 0.25% trypsin, seeded in 12-well plates at 1 × 105 cells/well, adhered, and starved with serum-free medium for 12 h. The cells were trypsinized again, fixed in 70% ethanol, and then incubated with RNaseA for 0.5 h (20 mg/L), prepared with PBS with 0.2% Triton-X100. After incubation with 50 mg/L propidium iodide (PI), 2 × 104 cells were collected and analyzed by ModFit software for cell cycle detection.
AnnexinV FITC/PI double staining
TE-1 and KYSE-30 cells were detached, centrifuged at 1000 r/min, resuspended in precooled PBS, centrifuged again to remove the supernatant, suspended with 5 mL binding buffer, and dyed with 5 μL AnnexinV-fluorescein isothiocyanate (FITC) and PI staining solution for 10 min in the dark. Lastly, cell apoptosis was detected by the flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA).
Hoechst33342 fluorescent staining
Cells were seeded in six-well plates at 1 × 105 cells/well. When cell confluence reached 70%, the cells were incubated in serum-free medium, fixed with 1 mL 4% paraformaldehyde, and dyed with 1 mL Heochst33342 staining solution. Finally, cells were viewed under the inverted fluorescence microscope and photographed.
Transwell assay
Cell invasion
Matrigel-coated transwell chambers (Corning, NY, USA; polycarbonate membrane pore size: 0.8 μm) were seeded in the 24-well culture plate. The upper chamber was separated from the lower chamber by a polycarbonate membrane. Suspensions of TE-1 and KYSE-30 cells at 3 × 105 cells/mL were prepared by serum-free medium, and appended to the upper chamber at 200 μL/well containing 0.5% bovine serum albumin (BSA). Next, the culture medium containing 10% serum was appended to the lower chamber at 600 μL/well and cultured for 48 h. Cells in the lower chamber were fixed in 4% paraformaldehyde, dyed in 0.1% crystal violet dying solution, and decolorized (the residual cells were removed by a cotton swab). Five random fields were selected for photography. Cells were counted and analyzed by ImageJ software.
Cell migration
The other steps were the same as the invasion assay except that Matrigel was not added and the incubation time was shortened to 24 h.
RT-qPCR
HEIH, miR-185, and KLK5 primer sequences (Supplementary Table 1) were entrusted to Genechem Co., Ltd. (Shanghai, China) for synthesis. Total RNA of EC and non-tumoral tissues, and TE-1 and KYSE-30 cells was extracted by Trizol (Thermo Fisher Scientific, Waltham, MA, USA), and the concentration and purity of RNA were determined. RNA reverse transcription into cDNA was conducted by PrimeScript RT Master Mix (Takara, Dalian, China), and RT-qPCR was carried out by PrimeScript RT reagent Kit and SYBR Premix Ex Taq (Takara), as per the manufacturer’s instructions. RT-qPCR was conducted in the ABI 7500 real-time PCR system (Applied Biosystems, CA, USA). MiR-185 was expressed relative to U6, while HEIH and KLK5 to glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Gene expression was calculated by 2−ΔΔCT method.
Western blot analysis
Total protein of EC and non-tumoral tissues, and TE-1 and KYSE-30 cells was extracted, and the protein concentration was measured by bicinchoninic acid kit (Boster, Hubei, China). After that, the extracted protein was supplemented to the loading buffer, and each well loaded with 30 μg sample. Then, the protein was separated by electrophoresis with 10% polyacrylamide gel (Boster), electroblotted onto the polyvinylidene fluoride membrane, and sealed with 5% BSA. Then primary antibody KLK5 (1:1000, Abcam, Cambridge, UK) and GAPDH (1:2000, Jackson Immuno Research, Grove, Pennsylvania, USA), as well as horseradish peroxidase-labeled secondary antibody (1:500, Jackson Immuno Research) were added for incubation. Images were obtained with Odyssey two-color infrared fluorescence scanning imaging system, and the gray values of target bands were determined by Quantity One image analysis software.
RNA-FISH assay
Subcellular localization of HEIH in TE-1 and KYSE-30 cells was predicted at http://lncatlas.crg.eu/ and then identified by fluorescence in situ hybridization (FISH) assay with FISH kit (Guangzhou RiboBio Co., Ltd., Guangdong, China). The Cy3-labeled HEIH probe was compounded by GenePharma. Cells were fixed with 4% paraformaldehyde, permeabilized, and reacted with specific probes overnight. The nucleus was stained with 4′,6-diamidino-2-phenylindole (Sigma-Aldrich, St. Louis, MO, USA) and observed by a fluorescence microscope (Nikon, Tokyo, Japan).
Dual luciferase reporter gene assay
The biological prediction website https://cm.jefferson.edu/rna22/Precomputed/ predicted the potential binding sites between HEIH and miR-185. HEIH containing wild-type (WT) or mutant (MUT) miR-185 response elements were cloned into the pmirGLO vector (Promega) to produce HEIH-WT and HEIH-MUT. HEIH-WT and HEIH-MUT were supplied by Wuhan GeneCreate Biological Engineering Co., Ltd. (Hubei, China). HEIH-WT and HEIH-MUT with correct sequencing were co-transfected with miR-185 mimic or mimic-NC into TE-1 and KYSE-30 cells, respectively, for 36–48 h. Luciferase activity was measured by the dual luciferase assay system (Promega).
The binding site between miR-185 and KLK5 was predicted at http://www.targetscan.org/vert_72/. KLK5 3′-untranslated region embodying the miR-135a binding site was inserted into a pmirGLO vector to construct KLK5-WT and KLK5-MUT. The correctly sequenced KLK5-WT and KLK5-MUT were co-transfected with miR-185 mimic or mimic-NC into TE-1 and KYSE-30 cells, respectively, for 36–48 h, after which luciferase activities were determined by dual luciferase assay system (Promega).
RNA pull-down assay
Biotin-labeled HEIH was synthetized as the probe for RNA pull-down assay, and the binding of Ago2 was determined by western blot analysis and RT-qPCR. The DNA fragment of HEIH sequence was amplified with a primer containing a T7 promoter and cloned into a pCR8 vector (Invitrogen). Subsequently, the plasmid was treated with the restriction enzyme NotI. Biotin-labeled RNAs were reversely transcribed with Biotin RNA Labeling Mix (Roche Diagnostics, NJ, USA) together with T7 RNA polymerase (Roche, Basel, Switzerland). The product was purified with RNeasy Mini Kit (Qiagen, Valencia, CA, USA) after treated with Rnase-free DNase I (Roche, IN, USA). After biotin-labeled HEIH was incubated with TE-1 and KYSE-30 cell lysate overnight, biotin-labeled HEIH was captured using streptavidin magnetic beads, and incubated for 1 h. Western blot analysis was adopted to detect biotin-labeled HEIH level, and RT-qPCR to determine miR-185 level in RNA precipitates.
Tumor xenografts in nude mice
Seventy BALB/c nude mice (4–5 w, the Institute of Laboratory Animal Science, Chinese Academy of Medical Science and Peking Union Medical College) were raised under specific pathogen-free conditions. Nude mice were adjusted to the new environment in the animal room for a week. The condition of mice was observed every day, and food and water supplies were ensured (all were strictly disinfected). TE-1 and KYSE-30 cells were detached with 0.25% trypsin and centrifuged at 1000 r/min to remove the supernatant. Next, cells were resuspended in normal saline and then centrifuged again at 1000 r/min. TE-1 and KYSE-30 cells (1 × 107 cell, 200 mL) were injected into the unilateral armpit of nude mice with a 1-mL syringe. After successful tumor xenografts, mice were observed regularly. After nude mice were disinfected by ultraviolet irradiation, 200 μL 1% pentobarbital sodium solution was injected into mouse abdominal cavity, and mice were photographed to monitor the tumor size (tumor volume = (length × width)/2. The mean tumor volume of five nude mice was counted to plot the tumor growth curves. Immediately after imaging, the nude mice were sent to the breeding room to prevent death from hypothermia caused by anesthesia, observed every 4 h until the nude mice got back to normal, and then reared conventionally. The nude mice were euthanized with carbon dioxide, and their subcutaneous tumors were completely removed and weighed.
Statistical analysis
All data were statistically analyzed with SPSS 21.0 (IBM Corp., Armonk, NY, USA). Mean ± standard deviation were the standard form for measurement data. The data between two groups were compared by t test. Data among multiple groups were compared by one-way analysis of variance (ANOVA), followed by pairwise comparison by Tukey’s multiple comparison test. The relationship between HEIH expression and the clinicopathological features of EC patients was determined by chi-square test. The prognosis of EC patients were analyzed by Kaplan–Meier analysis. P < 0.05 was considered statistically significance.
Results
HEIH and KLK5 are upregulated, and miR-185 is downregulated in EC tissues
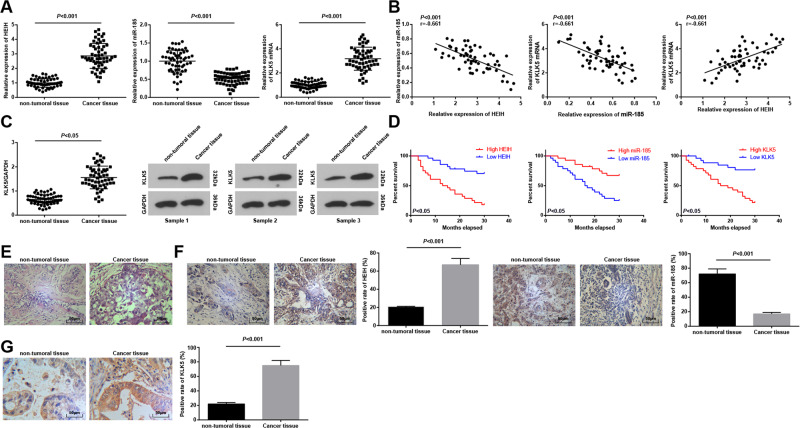
EC is a common cancer worldwide. There is more evidence emphasizes the key roles of lncRNAs in tumorigenesis. HEIH was differentially expressed in EC, and participated in the occurrence and development of EC9, suggesting that HEIH may be a possible therapeutic target for the treatment of EC. miR-185-5p overexpression was able to inhibit the invasion and migration of ESCC cells19, and KLF5 was the core regulatory factor for ESCC cells20. Given that, we detected HEIH, miR-185, and KLK5 expression by RT-qPCR and western blot assay in cancer and non-tumoral tissues of EC patients, and found that HEIH and KLK5 were elevated, and miR-185 was declined in EC tissues vs non-tumoral tissues (Fig. 1A, C). The relationship between HEIH, miR-185, and KLK5 levels was assessed by Pearson test, and the results revealed that HEIH and miR-185, and miR-185 and KLK5 were negatively correlated, while HEIH and KLK5 were positively correlated (Fig. 1B).
Fig. 1. HEIH and KLK5 are upregulated, and miR-185 is downregulated in EC tissues.
A Detection of HEIH, miR-185, and KLK5 expression in EC tissues by RT-qPCR (n = 56). B The relationship between HEIH, miR-185, and KLK5 analyzed by Pearson test (n = 56). C Detection of KLK5 protein expression in EC tissues by western blot analysis (n = 56). D The influence of HEIH on prognosis of patients with EC analyzed by Kaplan–Meier (n = 56). E Observation of pathological changes of EC tissues by HE staining. F HEIH and miR-185 expression in EC tissues detected by in situ hybridization (n = 56). G KLK5 expression in EC tissues detected by immunohistochemstry (n = 56); data in the figure were all measurement data expressed as mean ± standard deviation; comparison between two groups were analyzed by t test.
Patients were divided into low expression group (n = 27) and high expression group (n = 29) in the light of the median value of HEIH, miR-185, and KLK5 relative expression, and the effects of HEIH, miR-185, and KLK5 expression on survival and prognosis of EC patients were analyzed by Kaplan–Meier analysis. The results revealed that worse prognosis was found in EC patients with high HEIH or KLK5 expression, while better prognosis was observed in EC patients with high miR-185 expression (Fig. 1D).
Cancer tissues and non-tumoral tissues were sectioned and stained with HE. Under the microscope, the cells in non-tumoral tissues were arranged orderly with intact structure and uniform staining, and cells in cancer tissues were damaged with obvious vacuoles and inflammatory infiltration (Fig. 1E).
In situ hybridization detected HEIH and miR-185 expression in cancer tissues and non-tumoral tissues. It was manifested that HIEH expression was increased, while miR-185 expression was decreased in cancer tissues (Fig. 1F). Also, immunohistochemistry detected that KLK5 was mainly located in the cytoplasm and its expression was raised in cancer tissues (Fig. 1G).
The relationship between HEIH expression and clinicopathological features of EC patients was assessed. The results mirrored that EC patients with large tumor, great infiltration depth, and advanced TNM stage had increased proportion of high HEIH expression, indicating that tumor size, infiltration depth, and TNM staging were correlated with HEIH expression, but not with age, gender, and invasion of lymph (Table 1).
Table 1.
Relationship between HEIH expression and clinicopathological features of patients with esophageal carcinoma.
| Clinicopathological data | n | HEIH expression | P | |
|---|---|---|---|---|
| Low (n = 28) | High (n = 28) | |||
| Age (years old) | ||||
| ≥60 | 34 | 16 | 18 | 0.785 |
| <60 | 22 | 12 | 10 | |
| Gender | ||||
| Male | 39 | 18 | 21 | 0.562 |
| Female | 17 | 10 | 7 | |
| Tumor diameter (cm) | ||||
| <5 | 36 | 24 | 12 | 0.002 |
| ≥5 | 20 | 4 | 16 | |
| Infiltration depth | ||||
| pT1–pT2 | 25 | 17 | 8 | 0.031 |
| pT3–pT4 | 31 | 11 | 20 | |
| TNM staging | ||||
| I–II | 31 | 21 | 10 | 0.007 |
| III–IV | 25 | 7 | 18 | |
| Invasion of lymph | ||||
| Yes | 28 | 13 | 15 | 0.137 |
| No | 38 | 25 | 13 | |
The data in this table were measurement data analyzed by chi-square test.
HEIH and KLK5 are upregulated, and miR-185 is downregulated in EC cells
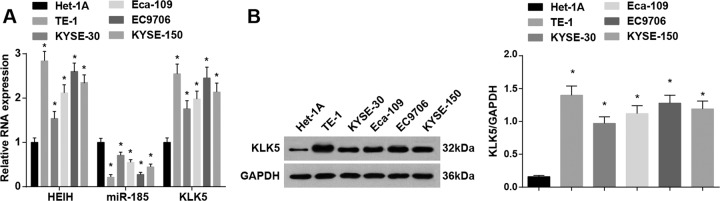
HEIH, miR-185, and KLK5 expression in Het-1A and human EC cells (KYSE-30, TE-1, Eca-109, EC9706, and KYSE-150) were detected. The results suggested that HEIH and KLK5 were upregulated, and miR-185 was downregulated in KYSE-30, TE-1, Eca-109, EC9706, and KYSE-150 cells. TE-1 cells showed the highest HEIH and KLK5 expression and the lowest miR-185 expression, which suggested the most difference from Het-1A cells, and KYSE-30 cells showed the lowest HEIH and KLK5 expression and the highest miR-185 expression, which suggested the least difference from Het-1A cells (Fig. 2A, B). Thus, TE-1 and KYSE-30 cells were selected for subsequent assays.
Fig. 2. HEIH and KLK5 are upregulated, and miR-185 is downregulated in EC cells.
A Detection of HEIH, miR-185, and KLK5 expression in Het-1A, KYSE-30, TE-1, Eca-109, EC9706, and KYSE-150 cells by RT-qPCR. B Detection of KLK5 protein expression in Het-1A, KYSE-30, TE-1, Eca-109, EC9706, and KYSE-150 cells by western blot analysis. *P < 0.05 vs the normal esophageal epithelial cell Het-1A; data in the figure were expressed as mean ± standard deviation; comparisons among multiple groups were analyzed by ANOVA, followed by pairwise comparison by Tukey’s multiple comparison test.
HEIH downregulation and miR-185 upregulation dampen proliferation and suppress cell cycle progression of EC cells
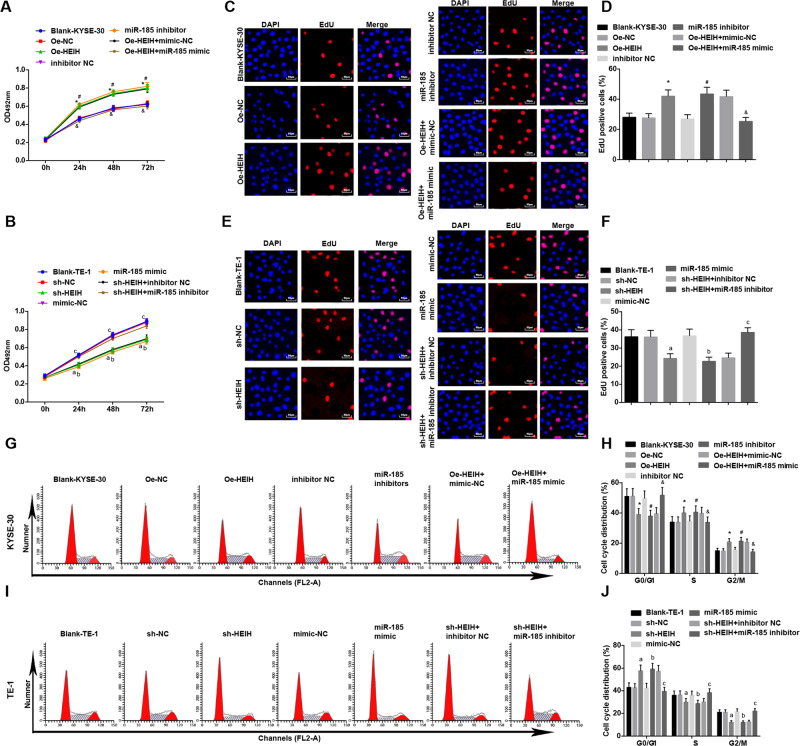
Cell proliferative capacity was detected by MTS and EdU assays, while cell cycle by flow cytometry. The results indicated that HEIH overexpression or miR-185 inhibition strengthened KYSE-30 cell proliferation ability, decreased cells in G0/G1, and increased cells in S and G2/M phases. miR-185 upregulation reversed HEIH overexpression-mediated effects on KYSE-30 cell proliferation and cell cycle.
As to TE-1 cells, it was discovered that the cell proliferative capacity was impaired and cells arrested in G0/G1 phase in response to HEIH downregulation or miR-185 upregulation. However, silencing miR-185 weakened the impacts of HEIH downregulation on cell proliferation and cell cycle distribution (Fig. 3A–J).
Fig. 3. HEIH downregulation and miR-185 upregulation dampen proliferation and cell cycle progression of EC cells.
A Detection of of KYSE-30 cell proliferation by MTS assay. B Detection of TE-1 cell proliferation by MTS assay. C Detection of KYSE-30 cell proliferation by EdU assay. D EdU-positive cell ratio of KYSE-30 cells. E Detection of TE-1 cell proliferation by EdU assay. F EdU-positive cell ratio of TE-1 cells. G Detection of KYSE-30 cell cycle distribution by flow cytometry. H Quantitative results of KYSE-30 cell cycle changes. I Detection of TE-1 cell cycle distribution by flow cytometry. J Quantitative results of TE-1 cell cycle changes. *, P < 0.05 vs the Oe-NC group; #, P < 0.05 vs the inhibitor NC group; &, P < 0.05 vs the Oe-HEIH + mimic NC group; a, P < 0.05 vs the sh-NC group; b, P < 0.05, the mimic NC group; c, P < 0.05 vs the sh-HEIH + inhibitor NC group; data in the figure were expressed as mean ± standard deviation; comparisons among multiple groups were analyzed by ANOVA, followed by pairwise comparison by Tukey’s multiple comparison test.
HEIH downregulation and miR-185 upregulation promote EC cell apoptosis
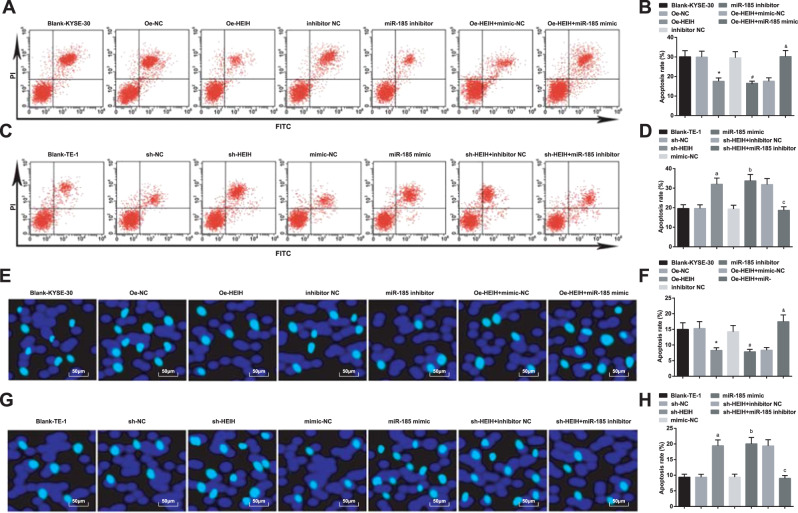
KYSE-30 and TE-1 cell apoptosis rates were detected by flow cytometry and Hoechst33342 fluorescent staining (Fig. 4A–H). The results revealed that KYSE-30 cell apoptosis rate lowered by overexpressing HEIH or inhibiting miR-185. Elevating miR-185 reversed the effect of HEIH overexpression on KYSE-30 cell apoptosis. TE-1 cell apoptosis rate was increased by decreasing HEIH or increasing miR-185. Suppressing miR-185 reversed the influence of HEIH decrease on TE-1 cell apoptosis.
Fig. 4. HEIH downregulation and miR-185 upregulation promote EC cell apoptosis.
A Detection of KYSE-30 cell apoptosis by flow cytometry. B Quantitative results of KYSE-30 cell apoptosis rate by flow cytometry. C Detection of TE-1 cell apoptosis by flow cytometry. D Quantitative results of TE-1 cell apoptosis rate by flow cytometry. E Detection of KYSE-30 cell apoptosis rate by Hoechst33342 fluorescent staining. F Quantitative results of KYSE-30 cell apoptosis rate by Hoechst33342 fluorescent staining. G Detection of TE-1 cell apoptosis rate by Hoechst33342 fluorescent staining. H Quantitative results of TE-1 cell apoptosis rate by Hoechst33342 fluorescent staining. *, P < 0.05 vs the Oe-NC group; #, P < 0.05 vs the inhibitor NC group; &, P < 0.05 vs the Oe-HEIH + mimic NC group; a, P < 0.05 vs the sh-NC group; b, P < 0.05, the mimic NC group; c, P < 0.05 vs the sh-HEIH + inhibitor NC group; data in the figure were expressed as mean ± standard deviation; comparisons among multiple groups were analyzed by ANOVA, followed by pairwise comparison by Tukey’s multiple comparison test.
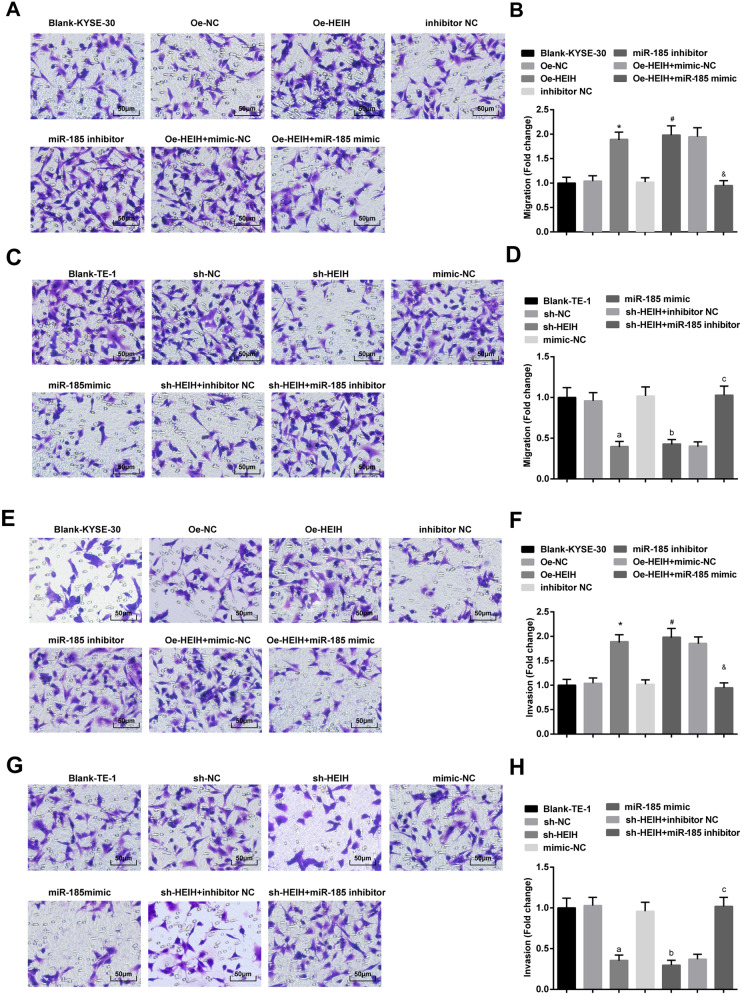
HEIH downregulation and miR-185 upregulation weaken EC cell migration and invasion ability
KYSE-30 and TE-1 cell migration and invasion ability was detected by transwell assay (Fig. 5A–H). It was indicated that HEIH elevation or miR-185 suppression enhanced KYSE-30 cell migration and invasion ability. The promoting effects of HEIH elevation on KYSE-30 cell migration and invasion were mitigated by upregulating miR-185. TE-1 cells treated with lowly expressed HEIH or overexpressed miR-185 showed impaired cell migration and invasion. However, the impaired migration of invasion induced by HEIH low expression was reversed through downregulating miR-185.
Fig. 5. HEIH downregulation and miR-185 upregulation weaken EC cell migration and invasion ability.
A Detection of KYSE-30 cell migration ability by transwell assay. B Quantitative results of KYSE-30 cell migration ability by transwell assay. C Detection of TE-1 cell migration ability by transwell assay. D Quantitative results of TE-1 cell migration ability by transwell assay. E Detection of KYSE-30 cell invasion ability by transwell assay. F Quantitative results of KYSE-30 cell invasion ability by transwell assay. G Detection of TE-1 cell invasion ability by transwell assay. H Quantitative results of TE-1 cell invasion ability by transwell assay. *, P < 0.05 vs the Oe-NC group; #, P < 0.05 vs the inhibitor NC group; &, P < 0.05 vs the Oe-HEIH + mimic NC group; a, P < 0.05 vs the sh-NC group; b, P < 0.05, the mimic NC group; c, P < 0.05 vs the sh-HEIH + inhibitor NC group; data in the figure were expressed as mean ± standard deviation; comparisons among multiple groups were analyzed by ANOVA, followed by pairwise comparison by Tukey’s multiple comparison test.
LncRNA HEIH binds with miR-185 to regulate KLK5 expression
To explore the mechanism of HEIH, we first analyzed it at http://lncatlas.crg.eu/ and found that HEIH was mainly positioned in the cytoplasm (Supplementary Fig.1A), and verification by RNA-FISH assay suggested that HEIH was concentrated in the cytoplasm in KYSE-30 and TE-1 cells (Supplementary Fig. 1B), implying that HEIH functioned in the cytoplasm.
Online analysis software predicted a binding region between HEIH and miR-185 (Supplementary Fig. 1C), which was further confirmed by dual luciferase reporter gene assay. The findings revealed that the luciferase activity (Supplementary Fig. 1D, F) suggested great abatement in KYSE-30 and TE-1 cells co-transfected with HEIH-WT and miR-185 mimic, while that in cells co-transfected with HEIH-MUT and miR-185 mimic showed no alterations, indicating that miR-185 mimic specifically bound to HEIH. RNA pull-down assay verified that miR-185 specifically indeed bound to HEIH (Supplementary Fig. 1E, G).
A targeted relationship between miR-185 and KLK5 was predicted at http://www.targetscan.org (Supplementary Fig. 1H). Dual luciferase reporter gene assay further suggested that the relative luciferase activity in KYK5-WT, and miR-185 mimic co-transfected KYSE-30 and TE-1 cells showed great diminution, while that in KLK5-MUT and miR-185 mimic co-transfected KYSE-30 and TE-1 cells suggested no alterations (Supplementary Fig. 1I, J), indicating that KLK5 was a direct target gene of miR-185.
HEIH and KLK5 were elevated, while miR-185 was diminished in KYSE-30 cells transfected with HEIH overexpression vector. miR-185 was downregulated, while KLK5 was upregulated in KYSE-30 cells transfected with miR-185 inhibitor. miR-185 was upregulated, while KLK5 was downregulated in KYSE-30 cells co-transfected with HEIH overexpression vector and miR-185 mimic (Supplementary Fig. 1K, L).
HEIH and KLK5 were declined, while miR-185 was enhanced in TE-1 cells transfected with HEIH low expression vector. miR-185 was upregulated, while KLK5 was downregulated in TE-1 cells transfected with miR-185 mimic. miR-185 was diminished, while KLK5 was elevated in TE-1 cells co-transfected with HEIH low expression vector and miR-185 inhibitor (Supplementary Fig. 1M, N).
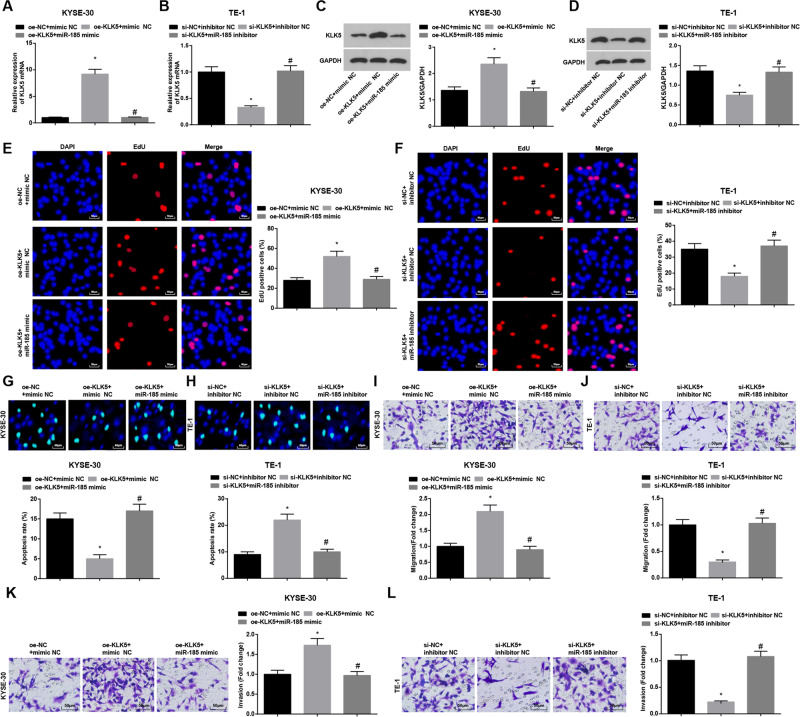
KLK5 upregulation promotes the progression of EC cells
To further explore the roles of KLK5 and miR-185 in the biological functions of EC cells, KYSE-30 cells were spontaneously transfected with KLK5 overexpression vector and miR-185 mimic, as well as their corresponding NCs, while TE-1 with KLK5 low expression vector and miR-185 inhibitor, as well as their corresponding NCs. The transfection efficacy was confirmed by RT-qPCR and western blot (Fig. 6A–D). Then, functional experiments verified that KLK5 overexpression enhanced KYSE-30 cell proliferation, invasion, and migration and depressed cell apoptosis. Moreover, upregulating miR-185 impaired KLK5 overexpression-induced KYSE-30 cell growth promotion. As to TE-1 cells, downregulating KLK5 repressed cell progression. In addition, lowering miR-185 expression could weaken KLK5 inhibition-induced effects on the biological functions of EC cells (Fig. 6E–L).
Fig. 6. miR-185 overexpression induce the progression of EC cells.
A–D Detection of KLK5 expession in KYSE-30 and TE-1 cells by RT-qPCR and western blot analysis. E, F Detection of KYSE-30 and TE-1 cell proliferation by EdU assay. G, H Detection of KYSE-30 and TE-1 cell apoptosis rate by Hoechst33342 fluorescent staining. I–L Detection of KYSE-30 and TE-1 cell migration and invasion by transwell assay; *, P < 0.05 vs the oe-NC + mimic NC group; #, P < 0.05 vs the oe-KLK5 + mimic NC group; &, P < 0.05 vs the si-NC + inhibitor NC group; @, P < 0.05 vs the si-KLK5 + inhibitor NC group; data in the figure were expressed as mean ± standard deviation; comparison among multiple groups by ANOVA, followed by pairwise comparison by Tukey’s multiple comparison test.
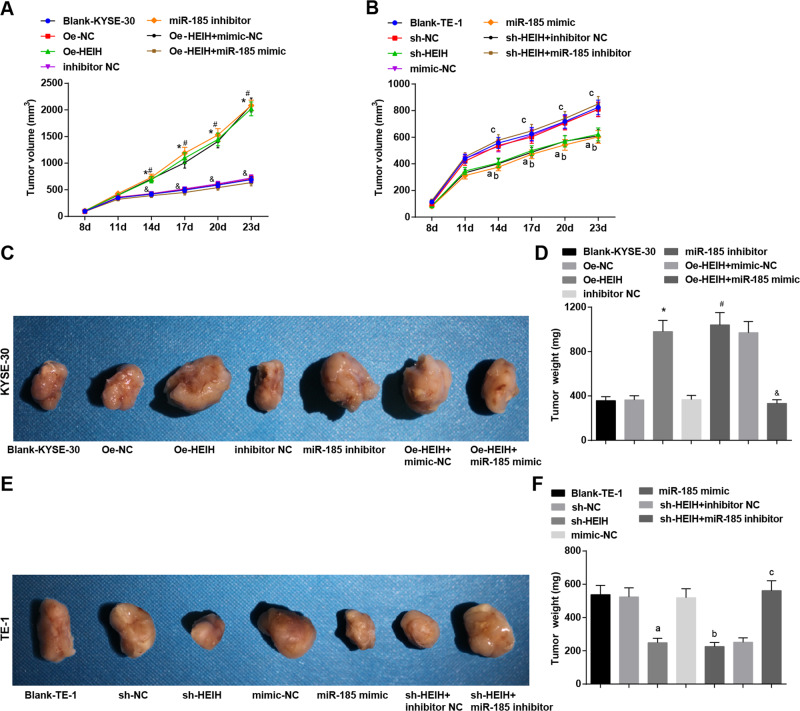
HEIH downregulation and miR-185 upregulation decline tumor volume and weight of EC mice in vivo
Eight days after tumor xenografts of KYSE-30 and TE-1 cells, the tumors of nude mice grew to varying degrees in a time-dependent manner. Fourteen days after tumor xenografts, the tumor volume were detected and 28 days after tumor xenografts, mice were euthanized to obtain and weigh the tumors. It was discovered that the tumor volume and weight increased in mice injected with KYSE-30 cells that had been transfected with HEIH overexpression vector or miR-185 inhibitor. miR-185 mimic in KYSE-30 cells reversed the function of HEIH overexpression vector on tumor volume and weight. In mice injected with TE-1 cells that had been transfected with HEIH low expression vector or miR-185 mimic, the tumor volume and weight were decreased. miR-185 inhibitor in TE-1 cells reversed HEIH low expression vector-induced effects on tumor growth and weight (Fig. 7A–F).
Fig. 7. HEIH downregulation and miR-185 upregulation decline tumor volume and weight of EC mice in vivo.
A Tumor growth curve of nude mice injected with KYSE-30 cells. B Tumor growth curve of nude mice injected with TE-1 cells. C Representative figure of nude mice and the subcutaneous tumors on the 23rd d after xenografts of KYSE-30 cells. D Quantitative results of nude mice and the subcutaneous tumors on the 23rd d after xenografts of KYSE-30 cells. E Representative figure of nude mice and the subcutaneous tumors on the 23rd d after xenografts of TE-1 cells in each group. F Quantitative results of nude mice and the subcutaneous tumors on the 23rd d after xenografts of TE-1 cells in each group. *, P < 0.05 vs the Oe-NC group; #, P < 0.05 vs the inhibitor NC group; &, P < 0.05 vs the Oe-HEIH + mimic NC group; a, P < 0.05 vs the sh-NC group; b, P < 0.05, the mimic NC group; c, P < 0.05 vs the sh-HEIH + inhibitor NC group; data in the figure were expressed as mean ± standard deviation; comparisons among multiple groups were analyzed by ANOVA, followed by pairwise comparison by Tukey’s multiple comparison test.
Discussion
EC is one of the most fatal cancers all over the world, as a result of its extremely aggressive nature and unfavorable survival rate21. Lately, the levels of ncRNAs (lncRNAs and miRNAs) in tissue and plasma from EC patients have been proposed to closely related to the survival and EC development22. Thus, in our study, we are meant to probe into the function of HEIH in EC development via modulation of miR-185 and KLK5. Altogether, we demonstrate that HEIH diminution dampens EC progression by upregulating miR-185 and downregulating KLK5.
To begin with, we found that HEIH was downregulated in EC tissues and cells, and functional knockout of HEIH impaired EC cell proliferation, invasion, and migration, and elevated cell apoptosis rate in vitro, as well as delayed tumor growth in mice in vivo. In compliance with our findings, the overexpressed HEIH was presented in ESCC, and depleting HEIH depressed the viability and invasion of ESCC cells9. Also, it has been reported that HEIH was highly expressed in hepatocellular carcinoma (HCC) cells, which strengthened cancer cell proliferation and invasion23. A report has shown that overexpressed HEIH in triple-negative breast cancer had oncogenic potential, therefore, facilitating proliferation and damping apoptosis of cancer cells24. A similar result has proposed the high expression of HEIH in melanoma, and knocking down HEIH greatly prevented melanoma cells from proliferation, migration, and invasion through binding to miR-200b/a/429 (ref. 8). Another study by Ma et al. has proffered that knockdown of HEIH exerted negative impacts on liver cancer cell development and metastasis by modulating miR‐199a‐3p (ref. 25). Furthermore, it has been suggested that HEIH overexpression catalyzed non-small cell lung cancer (NSCLC) proliferation and metastasis26. Anyway, it was proved that HEIH was a pro-tumor actor in cancers, and its suppression could be repressive for cancer development.
Then, we discovered that HEIH bound with miR-185 which was downregulated in EC. Moreover, we uncovered that miR-185 upregulation owned the similar effects of HEIH knockdown on EC cell progression and tumor growth. In a late research by Jing et al., miR-185 expression was low in ESCC and restoring miR-185-disabled ESCC cells to migrate, invade, and develop distal pulmonary metastases13. Not only in ESCC, the low expression of miR-185 also showed in gastric cancer and elevating miR-185 could restrain cancer progression27. A recent study has revealed that elevated miR-185 blocked NSCLC cell proliferation and invasion via modulating KLF7 (ref. 28). Literature has also recorded that enhanced miR-185 suppressed CRC cancer cell colony formation ability and enhanced apoptosis29. A similar study by Zhang et al. has reported that miR-185 was downregulated in HCC, and enhancing miR-185 could dampen HCC cell migration and invasion30. Anyhow, miR-185 was an-tumor in cancers and its upregulation served to slow down tumor progression.
Followed by that, we explored that miR-185 targeted KLK5, the upregulated gene to function in EC development. A prior research has revealed that KLK5 was elevated in CRC, and upregulated KLK5 was correlated with the malignant behavior of CRC15. Impressively, downregulating KLK5 in orthotopic oral tumors suppressed the development of aggressive lesions and inflammatory infiltrate31. In oral squamous cell carcinoma, KLK5 level was investigated to elevate and silencing KLK5 restricted the metastatic dissemination of cancer cells32.
To conclude, we reveal that HEIH depletion depresses growth of EC cells by upregulating miR-185 and downregulating KLK5 (Supplementary Fig. 2). Our study facilitates the understanding of the role of HEIH in EC development, and suggests HEIH as a novel target for EC treatment. However, more research has to be done give in-depth descriptions of the inner mechanism.
Supplementary information
Acknowledgements
We would like to acknowledge the reviewers for their helpful comments on this paper. This work was supported by National key research and development plan (No. 2016YFC0901401).
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by E. Candi
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article has been retracted. Please see the retraction notice for more detail: https://doi.org/10.1038/s41419-023-06365-z
These authors contributed equally: Bing Wang, Xuezhi Hao
Change history
12/18/2023
This article has been retracted. Please see the Retraction Notice for more detail: 10.1038/s41419-023-06365-z
Contributor Information
Fang Lv, Email: 13681130082@139.com.
Yushun Gao, Email: GGGaoyushun@163.com.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41419-020-03170-w).
References
- 1.Short MW, Burgers KG, Fry VT. Esophageal cancer. Am. Fam. Physician. 2017;95:22–28. [PubMed] [Google Scholar]
- 2.Huang FL, Yu SJ. Esophageal cancer: risk factors, genetic association, and treatment. Asian J. Surg. 2018;41:210–215. doi: 10.1016/j.asjsur.2016.10.005. [DOI] [PubMed] [Google Scholar]
- 3.Zhang, X. & Jain, D. Updates in staging and pathologic evaluation of esophageal carcinoma following neoadjuvant therapy. Ann. N. Y. Acad. Sci.10.1111/nyas.14462 (2020). [DOI] [PubMed]
- 4.Duan L, et al. EI24 inhibits cell proliferation and drug resistance of esophageal squamous cell carcinoma. Front. Oncol. 2020;10:1570. doi: 10.3389/fonc.2020.01570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duguang L, et al. The involvement of lncRNAs in the development and progression of pancreatic cancer. Cancer Biol. Ther. 2017;18:927–936. doi: 10.1080/15384047.2017.1385682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu LJ, et al. LncRNA SNHG7 promotes the proliferation of esophageal cancer cells and inhibits its apoptosis. Eur. Rev. Med. Pharm. Sci. 2018;22:2653–2661. doi: 10.26355/eurrev_201805_14961. [DOI] [PubMed] [Google Scholar]
- 7.Cui C, et al. Long noncoding RNA HEIH promotes colorectal cancer tumorigenesis via counteracting miR-939-mediated transcriptional repression of Bcl-xL. Cancer Res Treat. 2018;50:992–1008. doi: 10.4143/crt.2017.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao, H. et al. Long noncoding RNA HEIH promotes melanoma cell proliferation, migration and invasion via inhibition of miR-200b/a/429. Biosci. Rep.37, BSR20170682, 2017. [DOI] [PMC free article] [PubMed]
- 9.Wang D, You D, Pan Y, Liu P. Downregulation of lncRNA-HEIH curbs esophageal squamous cell carcinoma progression by modulating miR-4458/PBX3. Thorac. Cancer. 2020;11:1963–1971. doi: 10.1111/1759-7714.13489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sethi S, Ali S, Sethi S, Sarkar FH. MicroRNAs in personalized cancer therapy. Clin. Genet. 2014;86:68–73. doi: 10.1111/cge.12362. [DOI] [PubMed] [Google Scholar]
- 11.Yang X, et al. miRNA-1207-5p is associated with cancer progression by targeting stomatin-like protein 2 in esophageal carcinoma. Int. J. Oncol. 2015;46:2163–71.. doi: 10.3892/ijo.2015.2900. [DOI] [PubMed] [Google Scholar]
- 12.Lang B, Zhao S. miR-486 functions as a tumor suppressor in esophageal cancer by targeting CDK4/BCAS2. Oncol. Rep. 2018;39:71–80. doi: 10.3892/or.2017.6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jing R, et al. Plasma miR-185 is decreased in patients with esophageal squamous cell carcinoma and might suppress tumor migration and invasion by targeting RAGE. Am. J. Physiol. Gastrointest. Liver Physiol. 2015;309:G719–G729. doi: 10.1152/ajpgi.00078.2015. [DOI] [PubMed] [Google Scholar]
- 14.Pampalakis G, Zingkou E, Sotiropoulou G. KLK5, a novel potential suppressor of vaginal carcinogenesis. Biol. Chem. 2018;399:1107–1111. doi: 10.1515/hsz-2017-0302. [DOI] [PubMed] [Google Scholar]
- 15.Wu Y, et al. Upregulation of kallikreinrelated peptidase 5 is associated with the malignant behavior of colorectal cancer. Mol. Med. Rep. 2016;14:2164–70.. doi: 10.3892/mmr.2016.5516. [DOI] [PubMed] [Google Scholar]
- 16.Li L, et al. Upregulated KLK10 inhibits esophageal cancer proliferation and enhances cisplatin sensitivity in vitro. Oncol. Rep. 2015;34:2325–32.. doi: 10.3892/or.2015.4211. [DOI] [PubMed] [Google Scholar]
- 17.Zanatta A, et al. The relationship among HOXA10, estrogen receptor alpha, progesterone receptor, and progesterone receptor B proteins in rectosigmoid endometriosis: a tissue microarray study. Reprod. Sci. 2015;22:31–37. doi: 10.1177/1933719114549846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo L, et al. Decreased miR-320 expression is associated with breast cancer progression, cell migration, and invasiveness via targeting Aquaporin 1. Acta Biochim. Biophys. Sin. 2018;50:473–480. doi: 10.1093/abbs/gmy023. [DOI] [PubMed] [Google Scholar]
- 19.Liu JQ, et al. lncRNA KLF3-AS1 suppresses cell migration and invasion in ESCC by impairing miR-185-5p-targeted KLF3 inhibition. Mol. Ther. Nucleic Acids. 2020;20:231–241. doi: 10.1016/j.omtn.2020.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang, Y. Y. et al. TP63, SOX2, and KLF5 establish a core regulatory circuitry that controls epigenetic and transcription patterns in esophageal squamous cell carcinoma cell lines. Gastroenterology159, 1311–1327 (2020). [DOI] [PubMed]
- 21.Zhang Y. Epidemiology of esophageal cancer. World J. Gastroenterol. 2013;19:5598–5606. doi: 10.3748/wjg.v19.i34.5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hou X, Wen J, Ren Z, Zhang G. Non-coding RNAs: new biomarkers and therapeutic targets for esophageal cancer. Oncotarget. 2017;8:43571–43578. doi: 10.18632/oncotarget.16721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y, et al. Molecular mechanism of HEIH and HULC in the proliferation and invasion of hepatoma cells. Int. J. Clin. Exp. Med. 2015;8:12956–12962. [PMC free article] [PubMed] [Google Scholar]
- 24.Li, P., Zhou, B., Lv, Y. & Qian, Q. LncRNA HEIH regulates cell proliferation and apoptosis through miR-4458/SOCS1 axis in triple-negative breast cancer. Hum. Cell32, 522–528 (2019). [DOI] [PubMed]
- 25.Ma Y, et al. Silence of lncRNA HEIH suppressed liver cancer cell growth and metastasis through miR-199a-3p/mTOR axis. J. Cell Biochem. 2019;120:17757–17766. doi: 10.1002/jcb.29041. [DOI] [PubMed] [Google Scholar]
- 26.Jia K, Chen F, Xu L. Long noncoding RNA HEIH promotes the proliferation and metastasis of non-small cell lung cancer. J. Cell Biochem. 2019;120:3529–3538. doi: 10.1002/jcb.27629. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Q, Chen B, Liu P, Yang J. XIST promotes gastric cancer (GC) progression through TGF-beta1 via targeting miR-185. J. Cell Biochem. 2018;119:2787–2796. doi: 10.1002/jcb.26447. [DOI] [PubMed] [Google Scholar]
- 28.Zhao, L. et al. MiR-185 inhibits cell proliferation and invasion of non-small cell lung cancer by targeting KLF7. Oncol. Res.27, 1015–1023 (2018). [DOI] [PMC free article] [PubMed]
- 29.Afshar S, et al. MiR-185 enhances radiosensitivity of colorectal cancer cells by targeting IGF1R and IGF2. Biomed. Pharmacother. 2018;106:763–769. doi: 10.1016/j.biopha.2018.07.002. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Q, Chen Y, Liu K. miR-185 inhibits cell migration and invasion of hepatocellular carcinoma through CDC42. Oncol. Lett. 2018;16:3101–3107.. doi: 10.3892/ol.2018.8971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson JJ, et al. Protease-activated receptor-2 (PAR-2)-mediated Nf-kappaB activation suppresses inflammation-associated tumor suppressor microRNAs in oral squamous cell carcinoma. J. Biol. Chem. 2016;291:6936–45.. doi: 10.1074/jbc.M115.692640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang R, Shi Z, Johnson JJ, Liu Y, Stack MS. Kallikrein-5 promotes cleavage of desmoglein-1 and loss of cell-cell cohesion in oral squamous cell carcinoma. J. Biol. Chem. 2011;286:9127–35.. doi: 10.1074/jbc.M110.191361. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.