Abstract
With the goal of achieving carbon sequestration, emission reduction and cleaner production, biological methods have been employed to convert carbon dioxide (CO2) into fuels and chemicals. However, natural autotrophic organisms are not suitable cell factories due to their poor carbon fixation efficiency and poor growth rate. Heterotrophic microorganisms are promising candidates, since they have been proven to be efficient biofuel and chemical production chassis. This review first briefly summarizes six naturally occurring CO2 fixation pathways, and then focuses on recent advances in artificially designing efficient CO2 fixation pathways. Moreover, this review discusses the transformation of heterotrophic microorganisms into hemiautotrophic microorganisms and delves further into fully autotrophic microorganisms (artificial autotrophy) by use of synthetic biological tools and strategies. Rapid developments in artificial autotrophy have laid a solid foundation for the development of efficient carbon fixation cell factories. Finally, this review highlights future directions toward large-scale applications. Artificial autotrophic microbial cell factories need further improvements in terms of CO2 fixation pathways, reducing power supply, compartmentalization and host selection.
Keywords: CO2 fixation, autotrophy, heterotrophy, synthetic biology, reducing power, cell factory
Introduction
The carbon element is the most important component in all types of living organic matter, accounting for approximately 50% of the dry weight of the organics. In nature, elemental carbon exists in many forms, including carbon dioxide (CO2) in the atmosphere, CO2 dissolved in water (H2CO3, HCO, and CO) and carbon in organics, as well as carbon in rocks and fossil fuels. Currently, combustion of fossil fuels leads to considerable emission of greenhouse CO2 and causes global warming, which is a major concern for all societies (Irfan et al., 2019). Thus, it is urgent to minimize CO2 emissions by reducing the consumption of fossil fuels and reinforcing CO2 fixation (Pacala and Socolow, 2004). Compared to chemical methods, biological carbon sequestration is an attractive option for CO2 fixation, as it has several advantages, including mild reaction conditions and an eco-friendly approach (Li et al., 2012). Organisms that fix carbon include plants and autotrophic microorganisms. In green plants, carbon flux occurs primarily from CO2 to biomass driven by solar energy. Autotrophic microorganisms exist in certain special conditions on Earth in which plants cannot live, and they assimilate CO2 into biomass driven by solar energy or chemical energy produced by oxidizing inorganic substance in a more direct and rapid way. It is supposed that CO2 fixation might be more economic and efficient when sustainable bioprocesses producing biofuel and valuable chemicals directly from CO2 are realized. In this respect, autotrophic microorganisms can be regarded as the most suitable cell factories (Liu et al., 2020).
Autotrophic microorganisms are capable of incorporating CO2 into biomass via six natural carbon fixation pathways (Bar-Even et al., 2012). Since the discovery of the Calvin-Benson-Bassham (CBB) cycle in the 1940s and 1950s, another five CO2 assimilation mechanisms have been elucidated, namely, the reductive citric acid cycle (rTCA), the reductive acetyl-CoA pathway (Wood-Ljungdahl pathway), the 3-hydroxypropionate bicycle (3HP bicycle), the 3-hydroxypropionate/4-hydroxybutyrate cycle (3HP/4HB cycle), and dicarboxylate/4-hydroxybutyrate cycle (DC/HB cycle) (Berg, 2011). The CBB cycle is ubiquitous in autotrophic microorganisms, primarily occurring in cyanobacteria. Remarkable progress has been made in producing biofuel and chemicals from CO2 and solar energy by engineering the native CBB cycle in cyanobacteria, including ethanol, butanol, lactic acid, acetone, isobutyraldehyde, isoprene, and biodiesel (Savakis and Hellingwerf, 2015; Veaudor et al., 2020). However, the titer of target products is unsatisfactory (Hagemann and Hesse, 2018; Luan et al., 2020). The CO2 capturing rate of ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) (EC 4.1.1.39) in the CBB cycle is an order of magnitude lower than the average of central metabolic enzymes, and efforts to improve Rubisco’s kinetic properties have attained only limited success so far (Antonovsky et al., 2017; Liang et al., 2018). Moreover, low levels of growth and CO2 fixation efficiency, as well as deficient genetic operating platforms have largely limited industrial applications of cyanobacteria (Blankenship et al., 2011; Kushwaha et al., 2018). Hence, there is a need for faster and more efficient bioprocesses for the conversion of CO2 into the desired products. Generally, heterotrophic microorganisms have the advantage that growth and production yields are typically superior compared to the autotrophic life cycle. Massive and mature genetic manipulation tools make heterotrophy more accessible to a brand-new metabolism and proliferation mode (Nielsen and Keasling, 2016). The considerable advances in synthetic biology allow the engineering of novel functions and de novo metabolism networks in heterotrophic microorganisms for industrial biotechnology applications (Chen Y. et al., 2020; Clarke and Kitney, 2020). Previous developments in engineering of CO2-fixing pathways for improving the efficiency of CO2 fixation in autotrophic and heterotrophic microorganisms have been addressed in reported reviews (Claassens et al., 2016; Antonovsky et al., 2017; Hu et al., 2018).
In this review, we discuss recent advances in developing artificial autotrophic microorganisms for reinforcing CO2 fixation (Figure 1). Starting from heterotrophs to hemiautotrophs, and finally to complete autotrophs, a variety of natural and artificial synthetic CO2 fixation pathways have been developed and introduced into heterotrophic chassis. These engineered artificial autotrophs represent very promising candidates for highly efficient CO2 fixation and sustainable production of biofuels and value-added chemicals.
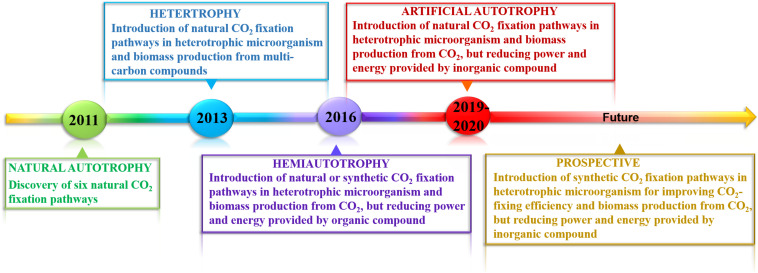
FIGURE 1.
Milestone in developing artificial autotrophy in the last decade. Until 2011, six naturally occurring CO2 fixation pathways have been identified, some of which were introduced into heterotrophic microorganisms. However, all of biomass was derived from additional multi-carbon compounds, such as glucose. The strong advances in synthetic biology enable the engineering of synthetic CO2 fixation pathways for improving carbon assimilation efficiency. Since 2016, hemiautotrophy has been successfully constructed by integrating natural or synthetic CO2 fixation pathways in heterotrophic hosts using organic compound as reducing power and energy source, in which biomass was completely derived from CO2. In 2019 and 2020, the conversion of heterotrophy to fully autotrophy was realized though integration of natural CO2 fixation pathways to support cell growth and inorganic compound’s oxidation to provide reducing power and energy.
Natural Co2 Fixation Pathways
Carbon dioxide assimilation is the process of reducing CO2 into cellular carbon which requires reducing equivalents and energy provided by ATP hydrolysis. The reductant varies by different microorganisms. In anaerobes, low-potential electron donors bearing more energy are responsible for offering reducing equivalents, such as ferredoxin E0’ ≈ −400 mV. In contrast, aerobic microbial organisms require more ATP equivalents since high-potential electron donors with less energy are used as electron donors, such as NADPH E0’ ≈ −320 mV (Berg, 2011). The sensitivity of enzymes toward molecular oxygen decides the distribution of CO2 fixation pathways in both anaerobic and aerobic microorganisms.
Currently, six autotrophic CO2-fixing pathways in microorganisms have been elucidated in detail. These pathways are divided into two groups according to the tolerance of key enzymes toward oxygen. The aerobic pathways include the CBB cycle, 3HP bicycle and 3HP/4HB cycle, while the rTCA cycle, Wood-Ljungdahl pathway and DC/HB cycle belong to the anaerobic pathways group since strictly anaerobic enzymes operate in these routes.
For common microorganisms, the CBB cycle is the most important mechanism of autotrophic CO2 fixation (Bar-Even et al., 2012). The entire cycle is composed of 13 steps and three stages, consisting of carboxylation, reduction and regeneration (Bassham and Calvin, 1962). First, the key enzyme Rubisco catalyzes the carboxylation of CO2 and ribulose-1,5-bisphosphate (RuBP) to generate 3-phosphoglycerate (3-PGA). Free energy change (ΔrG) is −37.8 kJ/mol. Then this product is reduced to glyceraldehyde-3-phosphate (GAP) consuming ATP and NADPH by 3-phosphoglycerate kinase and GAP dehydrogenase, in which the reaction catalyzed by 3-phosphoglycerate kinase is thermodynamically challenging (ΔrG = +18.7 kJ/mol). Next, 5-bisphosphate (Ru5P) is regenerated through conversion between C3, C4, C5, C6, and C7 sugar. Finally, Ru5P is phosphorylated by phosphoribulokinase to regenerate RuBP (ΔrG = −24.2 kJ/mol). After one cycle, the CBB cycle can fix three CO2 molecules and produce one GAP molecule at the cost of nine molecules of ATP and six molecules of NADPH reducing equivalents. The regeneration of energy carrier (ATP) and reducing equivalents (NADPH) in living microbes can be realized by photosystems. Despite the CBB cycle being the most prevalent CO2 fixation mechanism on Earth, the efficiency of carbon assimilation is unsatisfactory. First, the final product of this cycle is a C3 compound that is inefficient for the synthesis of acetyl-CoA since CO2 is inevitably lost during the conversion of GAP to acetyl-CoA. Acetyl-CoA is the central precursor for producing value-added multicarbon compounds, such as fatty acids (Blatti et al., 2013). Furthermore, the strong side reaction of Rubisco with oxygen can incur loss of fixed carbon, and the side product 2-phosphoglycolate is toxic to cell (Hagemann and Bauwe, 2016). In addition, large amounts of ATP and NADPH are consumed in this cycle (Berg, 2011).
As the key enzyme in the CBB cycle, Rubisco is the most abundant protein in the biosphere, which fixes ∼1011 tons of CO2 from the atmosphere per year (Hayer-Hartl and Hartl, 2020). Rubisco is a well-studied enzyme in photosynthetic organism since it provides organic carbon for life (Iniguez et al., 2020). Crystal structures of Rubisco from several origins have been analyzed by X-ray, including Rhodospirillum rubrum, Synechococcus PCC6301, Chlorobium tepidum, Thermococcus kodakaraensis KOD1, and so on (Iniguez et al., 2020). Two forms of Rubisco participate in the CBB cycle. Form I exists in all photosynthetic organisms, consisting of eight large-subunits (50–60 kDa) and eight small-subunits (12–18 kDa) that form an L8S8 structure. The tertiary structure of Form I is similar to a barrel, in which the L8 core is formed through the tetramerization of four interactional L2 dimers. Eight small subunits equally divided into two sets separately assemble the L8 core upside and downside. The large subunit harbors catalytic sites, especially amino acid sequences between 169 and 220, as well as 321 and 340. Compared to the small subunits, homology of the large subunits among different species is high. Small subunits have regulatory functions. Form II Rubisco is found in purple nonsulfur photosynthetic bacteria, such as Rhodospirillum rubrum and some marine dinoflagellates. This type is composed of two large-subunits (L2). Form I and Form II Rubisco are simultaneously found in several nonsulfur phototropic bacteria, such as Rhodobacter sphaeroides and R. capsulatus, as well as Thiobacillus sp. and Hydrogenovibrio marinus. Although Rubisco plays a pivotal role in global carbon fixation, its extremely poor carboxylation activity and competing oxygenase activity greatly hinder high carbon-fixing efficiency in the CBB cycle. CO2 and O2 competitively combine in the active site of Rubisco. The competitive efficiency for CO2 and O2 is defined as the specificity factor (Ω = VcKo/VoKc), which is the ratio of the catalytic efficiency (Vmax/KM) for the carboxylation and oxygenation reactions. The specificity factor Ω of Form II (Ω = 10 to15) is much lower than that of Form I (Ω = 25 to 75 in bacteria). Significant efforts have been made to improve the value of specificity factor Ω and the efficiency of carbon capturing. In recent years, remarkably improved heterologous expression levels have been realized. However, molecular modification of Rubisco has been challenging owing to its complicated protein structure and intricate nature. Escherichia coli has long been the preferred host for the directed evolution of Rubisco for the sake of enhancing carboxylation activity toward CO2 due to its high transformation efficiency and simple genetic tools (Mueller-Cajar and Whitney, 2008a). Several screening platforms for directed evolution of Rubisco have been developed by coupling cell growth to Rubisco activity (Parikh et al., 2006; Mueller-Cajar et al., 2007; Mueller-Cajar and Whitney, 2008b; Cai et al., 2014; Wilson et al., 2016; Zhou and Whitney, 2019). Unfortunately, affinity and catalytic efficiency toward CO2 failed to synchronously increase (Kubis and Bar-Even, 2019). Moreover, remarkable kinetic enhancement was not obtained, likely due to an improper library screening platform, such as the host. In addition to model heterotrophs, Rhodobacter capsulatus has been used as an autotrophic host for directed evolution and rational design of Rubisco (Satagopan et al., 2019). In the R. capsulatus Rubisco deletion strain, complementation of a foreign Rubisco gene allowed the selection of substitutions. These screened mutants exhibited an improved affinity toward CO2 rather than improved carboxylation activity. For example, the V186I/T327AL mutant showed a 42% decreased Kc and a 41% increased Ko. These mutant sites are located in the hemiconserved hydrophobic region adjacent to the active site and connect the large-small interface. The above studies related to Rubisco engineering are limited to bacterial Rubisco since prokaryotic hosts cannot form the eukaryotic holoenzyme. By swapping certain residues of the large-subunit’s primary structure that are responsible for preventing heterologous eukaryotic holoenzyme formation, Koay et al. (2016) created bacterial and eukaryotic Rubisco hybrids that could express and assemble in E. coli. These chimeric Rubisco proteins serve as targets for directed evolution and rational design in the future work.
The 3HP bicycle was first discovered in 1986 by Helge Holo and primarily exists in photosynthetic green non-sulfur bacteria, such as Chloroflexus (Mattozzi et al., 2013). In the first cycle, three HCO molecules and one acetyl-CoA molecule are converted to glyoxylate via ten steps catalyzed by ten enzymes (ΔrG = −109.4 kJ/mol). In the second cycle, glyoxylate and propionyl-CoA synthesize acetyl-CoA and pyruvate through five steps (ΔrG = −55.4 kJ/mol). In addition to the number of steps in this pathway, the energy costs are higher than in the other four natural pathways. 3HP bicycle fixes three CO2 molecules and produces one pyruvate molecule with the consumption of seven ATP molecules and five molecules of reducing equivalents. A “chimeric” photosynthetic system is used to regenerate ATP and reducing equivalents (Xin et al., 2018).
The 3HP/4HB cycle was first identified in archaea (Berg et al., 2007). In this pathway, acetyl-CoA/propionyl-CoA carboxylase fixes two molecules of HCO to produce succinyl-CoA (ΔrG = −61.9 kJ/mol). To regenerate acetyl-CoA, succinyl-CoA is reduced to 4-hydroxybutyrate, which is then activated to 4-hydroxybutyryl-CoA (ΔrG = −17.0 kJ/mol). Crotonyl-CoA is synthesized by the key enzyme 4-hydroxybutyryl-CoA dehydratase from 4-hydroxybutyryl-CoA (ΔrG = −7.7 kJ/mol). Finally, crotonyl-CoA is oxidized and subsequently cleaved into acetyl-CoA (ΔrG = −16.5 kJ/mol). The 3HP/4HB cycle fixes two molecules of HCO to produce one molecule of acetyl-CoA at the expense of six ATPs and four NAD(P)H reducing equivalents after 16 steps. In this cycle, the hydrolysis of pyrophosphate may provide energy (Berg, 2011).
The rTCA cycle exists in photosynthetic green sulfur bacteria and anaerobic bacteria (Buchanan and Arnon, 1990). This cycle reverses the reactions of the oxidative citric acid cycle (TCA cycle) and forms acetyl-CoA from two molecules of CO2 at the cost of two molecules of ATP, which is far less than the above three pathways. The regeneration of ATP and reducing equivalents in living microbes can be realized by photosynthetic reaction center type I (Berg, 2011). To reverse the TCA cycle, three irreversible enzymes are required, including fumarate reductase, ferredoxin-dependent 2-oxoglutarate synthase and ATP-citrate lyase, among which ferredoxin-dependent 2-oxoglutarate synthase is strictly anaerobic. Thermodynamically challenging reactions (ΔrG > 10 kJ/mol) are catalyzed by ATP-citrate lyase, 2-ketoglutarate synthase and isocitrate dehydrogenase in this cycle.
The Wood-Ljungdahl pathway is characterized by high energetic efficiency because in this route two CO2 molecules are fixed to produce acetyl-CoA with the consumption of only one ATP (Ljungdahl, 1969). In detail, this pathway begins with one CO2 molecule reduction to formic acid by highly oxygen sensitive formate dehydrogenase (FDH), which is thermodynamically challenging (ΔrG = +18.0 kJ/mol). Another CO2 molecule is reduced to carbon monoxide by a nickel atom in the active center of another highly oxygen sensitive CO dehydrogenase, which is also thermodynamically challenging (ΔrG = +32.6 kJ/mol). Then formic acid is attached to tetrahydrofolate (THF) for further reduction. Finally, the one-carbon unit transfers to nickel-bound CO and forms acetyl-CoA. In acetogens, CO2 is reduced to acetic acid with H2 via the Wood-Ljungdahl pathway, in which the ATP required for formate activation is regenerated in the acetate kinase reaction (Mock et al., 2015; Lemaire et al., 2020).
The DC/HB cycle is a another strictly anaerobic CO2 fixation pathway, in which two molecules of HCO and acetyl-CoA are converted to succinyl-CoA by two carboxylase pyruvate synthase and phosphoenolpyruvate (PEP) carboxylase (Huber et al., 2008). The regeneration of acetyl-CoA occurs through the same route as in the 3HP/4HB cycle. However, some enzymes and electron carriers, such as pyruvate synthase and ferredoxin, are inactivated by oxygen. Within this cycle, one molecule of HCO and one molecule of CO2 are fixed to generate one molecule of acetyl-CoA via 13 steps catalyzed by 10 enzymes with the consumption of five molecules of ATP.
Introduction of Natural Co2 Fixation Pathways Into Heterotrophic Microorganisms
Although autotrophic organisms are the sponsors to fix CO2 in nature, low activity of carbon capturing dramatically influences the efficient conversion of CO2 to biomass. Many efforts have been made to improve CO2 carboxylation rate during the CBB cycle since Rubisco exhibits an extremely low affinity toward CO2 and obvious oxygenation side-reactions in conditions of low levels of CO2 (Ducat and Silver, 2012; Lin et al., 2014). Unfortunately, limited success has been achieved through engineering Rubisco or other related enzymes. Moreover, it is challenging to attempt genetic manipulation of autotrophic microorganisms owing to their unclear genetic background and the absence of genetic tools. Meanwhile, overexpressing new genes responsible for foreign CO2 fixation pathway in autotrophic microorganisms may exert negative influences on regulatory networks in cells, such as carbon/nitrogen metabolism (Song et al., 2020). As model heterotrophs, E. coli and Saccharomyces cerevisiae have been widely used to serve as chassis to express natural CO2 fixation routes (Antonovsky et al., 2017).
E. coli
The 3-HP bi-cycle from Chloroflexus aurantiacus has been successfully integrated into E. coli. To heterologously express the entire bicycle, a grouping strategy was employed, by which a 16-step enzymatic reaction was divided into four functional subpathways, and each group was independently expressed in a synthetic operon. This study provides evidence for the evolution of bacteria by horizontal gene transfer (Mattozzi et al., 2013). However, the functional co-expression of the overall bicycle in E. coli failed to support autotrophic growth.
The possibility of producing sugar for cell proliferation was explored by switching off the inflow of organic carbon into the CBB cycle. In this pioneer work, reconstruction of the fully functional CBB cycle conferred in E. coli the capacity to synthesize sugar and other biomass components from CO2 by implementing a comprehensive approach that involved rational metabolic network construction, heterologous recombination expression and adaptive laboratory evolution (ALE). To cut off gluconeogenesis, central metabolism was divided into two independent modules by introducing Rubisco from Synechococcus sp. PCC 7002 and phosphoribosyl kinase (PRK) from S. elongatus PCC 7942, simultaneously deleting the phosphoglycerol mutase gene. The first module included upstream glycolysis, pentose phosphate pathway (PPP) and recombinant CBB cycle enzymes (Rubisco and PRK). Meanwhile, the second module was an energy portion containing downstream glycolysis and TCA cycle, providing ATP and reducing power for carbon fixation of module one. However, the resultant strain could not grow in the presence of CO2 and pyruvate (In this study, energy and reducing power were supplied by the oxidation of pyruvate). Only addition of a second sugar could support cell growth. The ALE strategy has advanced our ability to rapidly obtain a desired phenotype (Lee and Kim, 2020). The engineered strain was initially grown in a xylose-restricted chemostat, and at last, the evolved strain could grow with pyruvate as the sole carbon source at high CO2 concentrations. Further mass spectrometry analysis showed that CO2 was the only carbon source for phospho-sugars synthesis in the evolved strain, including ribose-5 phosphate, sedoheptulose-7 phosphate, fructose-6-phosphate, and glucose-6 phosphate, indicating that the fully functional CBB cycle in E. coli can directly synthesize sugar from CO2. Four molecules of pyruvate were consumed per sugar. After genome-wide sequencing, the prs gene, encoding ribose phosphate pyrophosphate kinase, was the only common mutant gene found in three chemostat experiments, which is the primary branching enzyme of the CBB cycle module. This study shows that the functionality of the CBB cycle depends not only on heterologous enzymes (Rubisco and PRK) but also on the endogenous components that interact with them, particularly metabolic enzymes in the circulating carbon pool (Antonovsky et al., 2016). Proper balance of kinetic properties in enzymes at branch point is imperative to maintain a stable metabolism in vivo (Barenholz et al., 2017). In this work, three molecules of CO2 were fixed to one molecule of pyruvate via CBB cycle. However, three molecules of CO2 were produced during the TCA cycle using pyruvate as substrate. Therefore, the net CO2 fixation was zero.
S. cerevisiae
Biofuel has been widely regarded as a promising alternative to fossil fuels concerning energy security, renewability, and global warming (Caspeta et al., 2013). Bioethanol is currently the most highly produced biofuel on an industrial scale (Nielsen et al., 2013). S. cerevisiae is the most commonly used host for bioethanol fermentation due to its high ethanol productivity and strong ethanol tolerance (Alper et al., 2006). However, during the process of yeast fermentation to produce ethanol, the production of ethanol is accompanied by CO2 release, causing carbon loss and greenhouse gas emissions. In addition, excessive NADH causes excessive accumulation of the byproduct glycerol. To recycle CO2, heterologous PRK and Rubisco derived from various origins were introduced into S. cerevisiae to construct the CO2-fixing route. The product of CO2 fixing by Rubisco is glycerate 3-phosphate (G3P). Ethanol is produced from G3P. At the same time, glycerol can also be formed using GAP as substrate by the actions of glycerol-3-phosphate dehydrogenase and glycerol-3-phosphatase. GAP is produced during xylose fermentation via non-oxidative PPP. This approach increased ethanol production and reduced the accumulation of the byproduct glycerol. For example, researchers co-expressed PRK from spinach and Rubisco from Thiobacillus denitrificans in S. cerevisiae. T. denitrificans Rubisco belongs to type II, composed of 8 large subunits. Its active expression requires the assistance of E. coli chaperons GroEL and GroES. Compared to the original strain, 90% in the reduction of byproduct glycerol and 10% increase of the production of ethanol were realized when the media was supplemented with glucose and galactose (Guadalupe-Medina et al., 2013). The use of sugarcane and corn starch as raw materials for the production of ethanol has the problem of “competing with people for food” (Tilman et al., 2009). As the second most abundant sugar in hydrolysis products from lignocellulose, xylose is an ideal feedstock to yield bioethanol via the xylose reductase (XR)/xylitol dehydrogenase (XDH) pathway in S. cerevisiae (Nogue and Karhumaa, 2015). CO2 fixation has been achieved by introducing PRK from Spinacia oleracea and Rubisco from Rhodospirillum rubrum using xylose as substrate. Three molecules of CO2 were fixed into one molecule of GAP by introducing PRK and Rubisco into yeast. Then, one molecule of ethanol was produced from one molecule of GAP with releasing one molecule of CO2. And pyruvate was formed from GAP to produce acetyl-CoA for other metabolism to support cell growth with inevitably releasing CO2. Therefore, the net production of CO2 was observed in this engineered strain. Note that, results showed that the reduced release of CO2 was observed during xylose fermentation, suggesting that the CO2 generated by pyruvate decarboxylase was partially re-assimilated through the synthetic reductive PPP (Xia et al., 2017). Results demonstrated that the net ethanol production of engineered yeast had increased 10%, and the byproducts decreased 11%, demonstrating that the introduction of the PRK-Rubisco route achieved CO2 recycling (Xia et al., 2017). An improved dual-module system was constructed in S. cerevisiae by employing mutants with alternate cofactor preference. Moreover, PRK and Rubisco from Ralstonia eutropha H16 were co-expressed in yeast (Li et al., 2017). R. eutropha H16 Rubisco belongs to type I, which is composed of 8 large subunits and 8 small subunits. To assist with proper folding of proteins, the endogenous chaperones of S. cerevisiae (Hsp60-HSP10) were co-expressed. Results showed that the productivity of ethanol was 15% higher than in the control strain, and the CO2 fixation rate reached 336.6–436.3 mg CO2/(L h) with consuming 3.1 g/L total sugar, which was significantly higher than previous natural or engineered microorganisms [5.8–147.0 mg CO2/(L h)]. Of note, this experiment proves that type I Rubisco has higher carboxylation activity than type II, likely because the small subunit has the ability to enrich CO2, which increases the concentration of CO2 at the active site of the enzyme (Li et al., 2017). In the above studies, the introduction of Rubisco and PRK into brewer’s yeast to achieve in situ fixation of CO2 in the process of bioethanol production and increase the output of the target product bioethanol has laid the foundation for the production of other fuels and chemicals from lignocellulose.
Introduction of Synthetic Co2 Fixation Pathways Into Heterotrophic Microorganism
Despite their naturally existing diversity, the application of CO2 fixation pathways for biomanufacturing valuable compounds directly from CO2 has been limited so far. These naturally occurring CO2 assimilation mechanisms contribute to cell growth rather than to products of interest. Moreover, CO2-fixing efficiency of these natural or ALE routes is still unsatisfactory and most of them are inactivated in the presence of oxygen. The emerging field of synthetic biology facilitates to creation of artificial synthetic CO2 fixation pathways.
With the goal of enhancing CO2 assimilation efficiency, much interest has been devoted to creating stoichiometrically and thermodynamically feasible routes. Starting with CO2 capturing enzymes, oxygen-insensitive and kinetically superior carboxylases or reductases have been identified, for instance, PEP carboxylase (EC 4.1.1.31) (Durall et al., 2020), pyruvate decarboxylase (EC 6.4.1.1) (Bar-Even et al., 2010), CoA-dependent carboxylase and metal-dependent FDH (Schwander et al., 2016; Cotton et al., 2018). Next, a series of CO2 fixation pathways were predicted in silico by evaluating the stoichiometric and thermodynamic feasibility of theoretical pathways, from which to recruit a route characterized by high energy-efficiency. To sustain the proposed draft, potential enzymes were selected from the natural enzyme pool and then engineered to improve their kinetic profiles through directed evolution or rational design. Finally, a cocktail containing all of these enzymes was developed to investigate the feasibility of the proposed pathways in vitro, and further, all of these enzymes were introduced into heterotrophic model microorganisms to create artificial autotrophy. 13C-labeling method is a powerful tool to pinpoint carbon flow. The ultimate aim is to produce biofuels and value-added compounds directed from CO2 through the developed artificial autotrophy.
The first attempt of de novo design of a CO2 fixation pathway in vitro was the crotonyl–coenzyme A (CoA)/ethylmalonyl-CoA/hydroxybutyryl-CoA (CETCH) cycle. Under aerobic condition, one molecule of glyoxylate was generated from two molecules of CO2 consuming two molecules of ATP and three molecules of NADPH though this completely artificial carbon fixation pathway with 12 core reactions (Schwander et al., 2016). ECRs with high catalytic efficiency toward CO2 were used as the initial enzyme of the entire cycle (ΔrG = −26.1 kJ/mol). The methylsuccinyl-CoA dehydrogenase (Mcd) was a rate-limiting enzyme, and a protein engineering approach was used to convert Mcd into a methylsuccinyl-CoA oxidase (Mco). A thermodynamically challenging reaction (ΔrG ≥ 10 kJ/mol) is catalyzed by 4-hydroxybutyryl-CoA synthetase in this cycle. Compared to other aerobic naturally occurring CO2 fixation flux, the CETCH process consumed the least amount of ATP. The feasibility of the CETCH cycle in vitro is a big breakthrough, demonstrating that more efficient CO2 assimilation can be realized by rewiring natural elements. However, the final product of this cycle is glyoxylate rather than acetyl-CoA, which hinders the efficient connection between the CETCH cycle and central metabolism. Moreover, it is not economical to employ a complex enzyme assembly to synthesize useful products from CO2 on a large scale. The challenges of introducing this artificial synthetic pathway into a heterotrophic host involve the complex interplay among these enzymes used in the CETCH cycle and aboriginal enzymes, as well as this route and endogenous metabolism network. Metabolic regulation also influences expression levels of each enzyme. Last but not least, it is crucial to maintain an efficient supply of reducing power for CO2 fixation.
There are several different strategies to activate CO2 with specific enzymes in nature. In addition to carboxylation, reduction can also be used to convert CO2 to formate by another kind of carbon capturing enzyme, which is FDH. It catalyzes the reversible reaction between CO2 and formate, taking part in various metabolic pathways with a variety of redox partners in different subcellular locations (Maia et al., 2017). Roughly, FDH can be divided into two classes, including NAD-independent and NAD-dependent (Jormakka et al., 2003). NAD-independent FDH is characterized by high activity toward CO2 and sensitivity to oxygen (Maia et al., 2016; Yu et al., 2017). This class of FDH contains complex redox-active centers harboring different transition metals, such as molybdenum (Mo), tungsten and nonhaemiron, molybdopterin guanine dinucleotide (MGD) and selenocysterine (Niks and Hille, 2019; Lemaire et al., 2020). It has been identified only in prokaryotic organisms. In contrast, NAD-dependent FDH has relatively low activity toward CO2 and is insensitive to oxygen (Hartmann and Leimkuhler, 2013; Choe et al., 2014; Altas et al., 2017). This kind of FDH has no metal ions or other redox-active centers and is widely distributed in bacteria, yeasts, fungi, and plants (Maia et al., 2017). However, the conversion of CO2 to formate is unfavorable (ΔrG = +18.0 kJ/mol) (Liu et al., 2020).
After reducing CO2 to formate, the next stage of carbon fixation can be implemented via natural or synthetic formate assimilation pathways (Bar-Even, 2016). Two well-known natural enzymes are capable of activating formate, oxygen-tolerant formate-tetrahydrofolate ligase (FTL) and oxygen-sensitive pyruvate formate-lyase (PFL) (Zelcbuch et al., 2016; Cotton et al., 2018). Starting from these entry-points, two types of formate assimilation pathways proceed, termed the reductive glycine (rGly) pathway and the PFL-PKT cycle (Cotton et al., 2018). Within the aerobic rGly process, formate is attached to THF to form formyl-THF, which can be subsequently reduced to methylene-THF. Glycine is formed by attaching another CO2 molecule to methylene-THF via glycine cleavage/synthase system (GCS). Next, serine is generated by adding another methylene-THF molecule. Last, serine is reduced to pyruvate for biomass production. The overall thermodynamics of the pathway starting from formate to serine favor the reductive direction with ΔrG = −6 kJ/mol (Yishai et al., 2018). During the anaerobic PFL-PKT cycle, PFL catalyzes the reaction of acetyl-CoA and formate to yield pyruvate. This intermediate is transformed to sugar-phosphates though gluconeogenic and pentose-phosphate pathways. Phosphoketolase (PKT) catalyzes the generation of acetyl-phosphate (AcP) from xylulose 5-phosphate. Finally, acetyl-CoA is regenerated from AcP by the action of phosphate acetyltransferase (PTA, EC 2.3.1.8), and the cycle is closed.
As mentioned above, the rGly pathway is an efficient synthetic route for aerobic assimilation of formate. In E coli, foreign enzymes required for the rGly process were expressed, including formate-THF ligase, 5,10-methenyl-THF cyclohydrolase (Fch) and 5,10-methylene-THF dehydrogenase (MtdA) from the Methylobacterium extorquens GCS. The resultant strain could biosynthesize cellular glycine and C1 compounds derived from formate and CO2 when cells grown under heterotrophic condition (Yishai et al., 2018). Unlike the CBB cycle, the rGly process has few overlaps with cellular central metabolism, reducing the influence of metabolic regulations. Bang and Lee (2018) introduced this pathway in E. coli to realize one-carbon assimilation in vivo by overexpressing enzymes related to the rGly pathways as well as NAD-dependent FDH for producing reducing power from formate. After glucose depletion, the engineered strain maintained a slight growth using only formate and CO2 as substrates, confirming its feasibility for supplying energy and reducing power via NAD-dependent FDH oxidative activity. Based on these pioneer studies, an outstanding work made an attempted to endow E. coli grown on formate or methanol and CO2 without any other organics via the rGly route. Enzymes in rGly pathways, as well as NAD-dependent FDH, were overexpressed in the strain, in which formate was the only source for cell growth. To improve growth rate, a short-term laboratory evolution was performed. After 40 generations of culture, the doubling time of cells was reduced by 6–8-fold. Genome sequence analysis of the initial and evolved strains revealed two mutations involving energy and reducing power supply (Kim et al., 2020). In another study, cell growth was achieved when integrating the formate assimilation pathway and subsequent ALE without FDH supplying NADH. Genomic sequence analysis showed that mutations covered rGly pathway related genes, biofilm formation genes and formate-utilizing genes (Kim et al., 2019). However, in above studies, the net CO2 emission was inevitable since one molecule of CO2 was lost during the process of oxidizing formate to supply NADH.
In addition to ethanol, S. cerevisiae also has the ability to tolerate high levels of formate in the environment. Moreover, this host harbors necessary genes for the rGly route. Given these unique features, overexpression of rGly route native enzymes in S. cerevisiae resulted in net production of glycine using formate as feedstock. However, the addition of sugar instead of formate supported cell growth (de la Cruz et al., 2019). Engineering downstream biomass formation from glycine and subsequent ALE holds promise for use in the development of yeast that are fully autotrophic.
Compared to formate, the more active feature allows formaldehyde easier access to the central metabolism. Therefore, the reduction of formate to formaldehyde coupled with subsequent the natural or synthetic formaldehyde assimilation pathway is an attractive bypass to fix one-carbon compounds. With the goal of shorting the process of one-carbon utilization and accelerating growth starting from formate, an artificial synthetic route was computationally designed, the formolase (FLS) pathway. In this linear route, two natural enzymes with considerable activity toward substrate analogs were identified. Acetyl-CoA synthase (ACS) derived from E. coli catalyzes the ATP-dependent conversion of formate into formyl-CoA, and then putative acylating aldehyde dehydrogenase (ACDH) from Listeria monocytogenes catalyzes the NADH-dependent reduction of formyl-CoA to formaldehyde. Next, a novel enzyme FLS, which catalyzes the carboligation of three molecules of formaldehyde into one molecule of dihydroxyacetone was created through rational protein design and site-directed mutagenesis based on benzaldehydelyase (BAL) from Pseudomonas fluorescens biovar I. Finally, this C3 product can flow into central metabolism. It is speculated by in silicon calculations that this completely new synthetic pathway is superior to any natural one-carbon utilization starting from formate pathways due to having the fewest steps and the highest chemical driving force under fully aerobic conditions (the total Gibbs energy change ΔrG from formaldehyde to acetyl-CoA is −110.2 kJ/mol). However, the low enzymatic activity of FLS resulted in undetectable cell growth with formate as a substrate (Siegel et al., 2015), suggesting the biotransformation of C1 directly to C3 is difficult to achieve. Given these findings, investigators wondered whether it was possible to produce C2 directly from C1. Recently, a synthetic aerobic acetyl-CoA (SACA) pathway was designed and constructed wherein two molecules of formaldehyde were transferred into one molecule of acetyl-CoA through only three steps (ΔrG is −96.7 kJ/mol). First, formaldehyde was condensed into glycolaldehyde (GALD) by glycolaldehyde synthase (GALS). Then, GALD was converted into AcP by acetyl-phosphate synthase (A EC 4.1.2.9). GALS and ACPS were screened and engineered to enhance their catalytic efficiency toward their new substrates, respectively. Finally, the phosphate group of AcP was replaced with CoA catalyzed by PTA, and acetyl-CoA was successfully produced both in vitro and in vivo (Bar-Even et al., 2010; Li et al., 2019). Although the SACA process is characterized by carbon-conserved and ATP-independent processes, achieving high-efficiency of the pathway has been challenging, likely due to the toxicity of formaldehyde to cells and kinetic bottlenecks of enzymes. To address this issue, renovating the host and implementing further protein engineering may represent promise solutions.
Reduction of Co2 Loss in Microorganisms
In heterotrophic hosts, fixed carbon flows into the central metabolism for synthesis of metabolites for cell growth, during which carbon inevitably suffers great or small loss. For example, the product dihydroxyacetone of the FLS pathway can be converted into acetyl-CoA via the glycolysis pathway with carbon loss and a theoretical carbon yield of 66.7%. To address this issue, significant efforts have been devoted to designing an artificial bypass to reduce the loss of carbon during acetyl-CoA formation. The nonoxidative glycolysis (NOG) pathway achieved biosynthesis of acetyl-CoA from sugar without carbon loss by rewiring the known carbon rearrangement pathway (Bogorad et al., 2013). In this pathway, bifunctional phosphoketolase (Fxpk) from Bifidobacterium adolescentis breaks down three molecules of fructose 6-phosphate (F6P) into three molecules of AcP and three molecules of erythrose 4-phosphate (E4P). The three molecules of E4P regenerate two molecules of F6P through carbon rearrangement. The net reaction includes one molecule of F6P that produces three molecules of AcP without carbon loss. By overexpressing Fxpk and removing succinic acid, lactic acid, ethanol, formic acid, and other competitive pathways, the engineered E. coli strain produces acetic acid with a yield of 2.2 mol/mol from xylose, close to the theoretical maximum yield (2.5 mol/mol) and exceeding the maximum theoretical value of producing acetic acid from xylose though the EMP pathway (1.67 mol/mol). However, this pathway itself cannot support the growth of cells in a minimal medium that uses sugar as a carbon source, and requires the assistance of reducing equivalents and metabolites in the EMP pathway. To overcome this challenge, researchers further constructed an E. coli strain that does not use EMP for glycocatabolism. The engineered strain, which contained 11 overexpressed genes, 10 deleted genes, and more than 50 gene mutations, including three overall regulatory factors, was obtained through directed evolution. This strain can grow in the medium containing glucose, and the carbon conversion rate of anaerobic fermentation of glucose to acetic acid is close to 100% (Lin et al., 2018). By combining the ribulose monophosphate (RuMP) and NOG pathways, acetyl-CoA was produced from methanol through the methanol condensation cycle (MCC) pathway (Bogorad et al., 2014). The first step in MCC is the oxidation of methanol to formaldehyde. This C1 compound and Ru5P were converted to F6P via upstream of the RuMP pathway. Next, acetyl-CoA is produced from F6P by downstream of NOG bypass. Meanwhile, Ru5P was regenerated though the RuMP pathway. Throughout the entire pathway, phosphates were conserved, and C1 compounds were assimilated into acetyl-CoA in an ATP-independent manner. However, construction of MCC in vivo has not been implemented, and protein engineering of key enzymes is expected to accelerate carbon flux.
In addition to the EMP pathway, a high level of carbon loss also occurs within TCA cycle. In glyoxysome of plants, the glyoxylate shunt (GS) shares several common intermediates with the TCA cycle. GS involves the conversion of fatty acid to sugar, and the net reaction includes two molecules of acetyl-CoA producesing one molecule of succinic acid without carbon loss. It can be assumed that the reverse version of the glyoxylate shunt (rGS) might be used to generate acetyl-CoA to circumvent loss of carbon. Based on the rGS, a synthetic pathway was designed, in which malate and succinate were converted to oxaloacetate and two molecules of acetyl-CoA. However, the driving force of key steps relies on the hydrolysis of ATP, and the growth rate of the resultant strain was relatively slow (Mainguet et al., 2013). Like the abovementioned C4 metabolites, acetyl-CoA can also be produced from C3 metabolites. A synthetic malonyl-CoA-glycerate (MCG) pathway has been demonstrated to be an efficient way to produce acetyl-CoA (Yu et al., 2018). First, two molecules of PEP were carboxylated into two molecules of oxaloacetate by inputting two molecules of bicarbonate. This reaction was catalyzed by PEP carboxylase, an attractive CO2 fixing enzyme regarding its robustness and activity. Oxaloacetate was reduced to malate and then activated to malyl-CoA with ATP consumption. Two molecules of malyl-CoA were split into two acetyl-CoAs and two glyoxylates. The latter products were regenerated to PEP via the glyoxylate assimilation route, during which one CO2 was released. Thus, coupled with the CBB cycle, the MCG pathway can fix two molecules of CO2 to produce one molecule of acetyl-CoA with the consumption of 5.5 molecules of ATP equivalents and 4 molecules of reducing equivalents. The feasibility of the MCG pathway has been demonstrated both in vitro and in vivo. Moreover, when coupled with the CBB cycle in cyanobacteria, both acetyl-CoA level and bicarbonate assimilation rate increased one-fold.
Therefore, the novel synthetic pathway can be designed by coupling CO2-fixing reactions with the abovementioned carbon loss-reducing pathways to improve CO2 fixation efficiency (Figure 2 and Table 1). Once a source of reducing power is provided, this pathway could theoretically allow growth with CO2 as the sole carbon source.
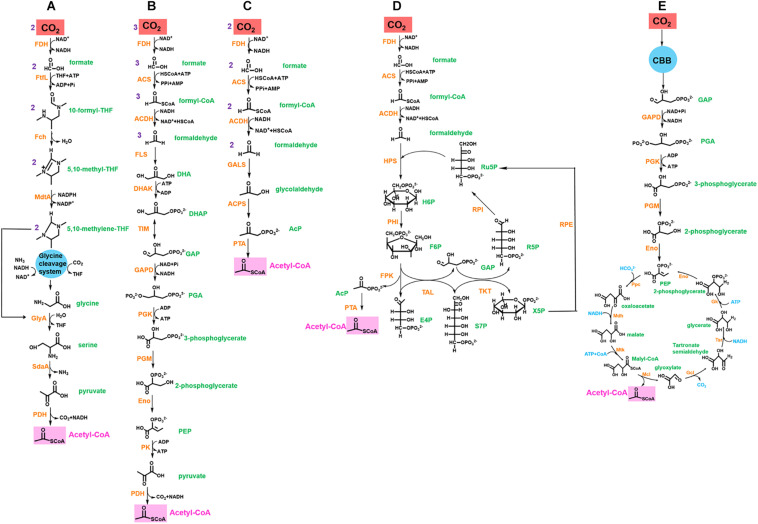
FIGURE 2.
Synthetic CO2 fixation pathways for acetyl-CoA synthesis. (A) rGly pathway coupled FDH. (B) Formolase pathway coupled FDH. (C) SACA pathway coupled FDH and partial formolase path. (D) MCC pathway coupled FDH and partial formolase path. (E) MCG cycle coupled CBB cycle. Abbreviation for enzymes (orange in color): FDH, formate dehydrogenase; FtfL, formate-THF ligase; FchA, 5,10-methenyl-THF cyclohydrolase; MtdA, 5,10-methenyl-THF dehydrogenase; GlyA, serine hydroxymethyl-transferase; SdaA, serine deaminase; PDH, pyruvate dehydrogenase; ACS, acetyl-CoA synthase; ACDH, acetaldehyde dehydrogenase; FLS, formolase; DHAK, dihydroxyacetone kinase; TIM, triose phosphate isomerase; GAPD, glyceraldehyde-3-phosphate dehydrogenase; PGK, 3-phosphoglycerate kinase; PGM, phosphoglycerate mutase; Eno, enolase; PK, pyruvate kinase; GLAS, glycolaldehyde synthase; ACPS, acetyl-phosphate synthase; PTA, phosphate acetyltransferase; HPS, hexulose-6-phosphate synthase; PHI, phosphohexulose isomerase; FPK, fructose-6-phosphate phosphoketolase; TAL, transaldolase; TKT, transketolase; RPE, ribulose 5-phosphate epimerase; RPI, ribose-5 phosphate isomerase; Ppc, phosphoenolpyruvate carboxylase; Mdh, malate dehydrogenase; Mtk, malate thiokinase; Mcl, malyl-CoA lyase; Gcl, glyoxylate carboligase; Tsr, tartronate semialdehyde reductase; Gk, glycerate kinase. Abbreviation for metabolites (green in color): DHA, dihydroxyacetone; DHAP, dihydroxyacetone phosphate; GAP, glyceraldehyde 3-phosphate; PGA, 1,3-biphosphoglycerate; PEP, phosphoenolpyruvate; AcP, acetyl phosphate; H6P, hexulose-6-phosphate; Ru5P, ribulose-5 phosphate; F6P, fructose-6-phosphate; R5P, ribose-5 phosphate; E4P, erythrose-4 phosphate; S7P, sedoheptulose-7 phosphate; X5P, xylose-5 phosphate.
TABLE 1.
Comparison of synthetic CO2 fixation pathways with natural ones.
| Pathway | Status | Fixed CO2 or HCO | Product | ATP consumption | NAD(P)H consumption | CO2 capturing enzyme | Specific activity μ mol/min/mg | References |
| CBB | Natural | 3 | Glyceraldehyde-3-phosphate | 9 | 6 | Ribulose-1,5-bisphosphate, Carboxylase/oxygenase | 3.5 | Bar-Even et al., 2010 |
| 3HP bicycle | Natural | 3 | Pyruvate | 7 | 5 | Acetyl-CoA carboxylase, propionyl-CoA carboxylase | 18, 30 | Herter et al., 2001; Bar-Even et al., 2010 |
| 3HP/4HB cycle | Natural | 2 | Acetyl-CoA | 6 | 4 | Acetyl-CoA carboxylase, propionyl-CoA carboxylase | 18, 30 | Berg et al., 2007; Bar-Even et al., 2010 |
| rTCA | Natural | 2 | Pyruvate | 2 | 5 | 2-oxoglutarate synthase, isocitrate dehydrogenase | –, 53 | Kim et al., 1992; Martinez et al., 2007 |
| Wood-Ljungdahl | Natural | 2 | Acetyl-CoA | 1 | 4 | NAD-independent formate dehydrogenase, CO dehydrogenase/acetyl-CoA synthase | 2.34, 0.46 | Ragsdale, 1997 |
| DC/HB cycle | Natural | 2 | Acetyl-CoA | 5 | 4 | Pyruvate synthase, PEP carboxylase | −, 35 | Huber et al., 2008; Garcia-Gonzalez and De Wever, 2017 |
| CETCH | Synthetic | 2 | Glyoxylate | 1 | 4 | CoA-dependent carboxylase | ||
| rGly | Synthetic | 3 | Pyruvate | 2 | 3 | Glycine cleavage system | ||
| Formolase path | Synthetic | 3 | Dihydroxyacetone phosphate | 4 | 3 | NAD-independent formate dehydrogenase | ||
| Partial formolase path + SACA path | Synthetic | 2 | Acetyl-CoA | 2 | 2 | NAD-independent formate dehydrogenase | ||
| partial formolase path + MCC path | Synthetic | 1 | Acetyl-CoA | 1 | 1 | NAD-independent formate dehydrogenase | ||
| CBB + MCG path | Natural + synthetic | 2 | Acetyl-CoA | 5.5 | 4 | PEP carboxylase, ribulose-1,5-bisphosphate, carboxylase/oxygenase |
The Conversion of Microorganisms From Hemiautotrophy Into Fully Autotrophs
Challenges in rewiring hemiautotrophy to complete autotrophy for steady-state growth on CO2 as the sole source of carbon include (1) replacement of the native sugar transport system with a foreign CO2 transport system, (2) deletion of part of the sugar metabolism pathway, (3) integration of systems for CO2 fixation and central metabolism, (4) supply of reducing power, and (5) adaption of the strain to grow under this rewired metabolism. Fully considering these criteria, completely autotrophic microorganisms have been developed (Gleizer et al., 2019; Gassler et al., 2020) (Figure 3). For E. coli, carbonic anhydrase (CA, which catalyzes the reversible reaction between CO2) was used to concentrate and transport CO2 into cells. Key enzymes in the CBB cycle were heterologously expressed (Rubisco and PRK), whereas genes related to glycolysis and the oxidative pentose-phosphate pathway were knocked out. Importantly, NAD-dependent FDH was introduced into cell to generate NADH from formate. NADH provides the reducing power to drive carbon fixation and serves as the substrate for ATP generation via oxidative phosphorylation. Although three necessary enzymes were introduced into E. coli, these efforts failed to generate a complete autotrophy due to the complexity of native metabolism and regulation networks in cells. Thus, long-time ALE was performed to redistribute the central metabolic flux. Finally, a complete autotrophic strain was obtained after 350 days of evolving. Genomic sequence analysis showed that CBB cycle-related genes, genetic selective pressure-related genes and nonfunctional genes were mutated (Erb et al., 2019; Gleizer et al., 2019). However, one molecule of CO2 was inevitably lost during the process of oxidizing formate to supply NADH, resulting in a net CO2 emission under autotrophic conditions.
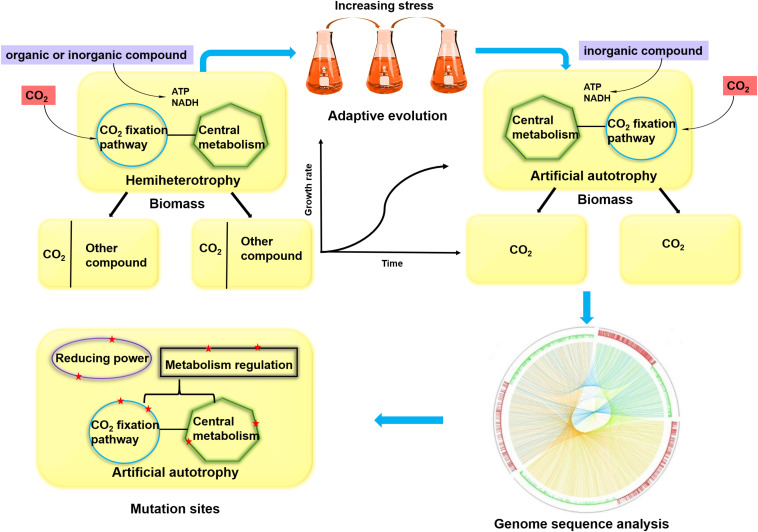
FIGURE 3.
The conversion process of hemiheterotrophy to artificial autotrophy. In hemiheterotrophic microorganism, CO2 was fixed into organic carbon via introducing CO2 fixation pathway and organic or inorganic compound provided reducing power and energy. Biomass was derived from CO2 and another compound. The artificial autotrophic microorganism can be obtained through ALE strategy, in which inorganic compound provided reducing power and energy and biomass was completely derived from CO2. To understand the genetic basis for adaptation to autotrophy, genomic analysis can be carried out, and mutation sites were related to CO2 fixation pathway, central metabolism, reducing power and metabolism regulation (depicted by red star).
As a model eukaryotic microbe, Pichia pastoris is also regarded as a proper chassis to construct autotrophs. The CBB cycle was introduced into cells though rewriting the xylulose monophosphate cycle and deleting methanol assimilation genes in peroxisomes. Energy and reducing power were provided by methanol oxidation. The maximum specific growth rate on CO2 increased more than two-fold via adaptive evolution. Genomic sequence analysis showed that mutations were involved in CBB cycle related genes, NADH and ATP synthesis genes. These results indicated the importance of carbon fixation pathways, energy and reducing power supply on cell growth using CO2 as a sole carbon source (Gassler et al., 2020). However, similar with employing formate as energy and reducing power source, the oxidation of methanol also resulted in the net emission of CO2.
Conclusion and Perspectives
Efforts to implement CO2 fixation metabolism into heterotrophic microorganisms have not only focused on the well-known CBB cycle. Other naturally occurring pathways, as well as synthetic routes, have also been successfully introduced into heterotrophic hosts. Recent advances in the conversion of heterotrophic metabolism to fully autotrophic growth marked a new period of “artificial autotrophy.” However, net CO2 assimilation has not yet been demonstrated since fixed CO2 is lost again during reducing power and energy generation. Sustainable production of biofuels and value-added compounds will become possible only if artificial autotrophy can efficiently fix CO2 without carbon loss. Future efforts to develop artificial autotrophy are discussed in this section.
First of all, to achieve autotrophic growth, sustainable sources of energy and reducing equivalents are vital. Introducing light-cycle reactions into heterotrophic hosts remains challenging due to their extremely complex nature. Artificial photoautotrophy might be realized by introducing a fully functional proteorhodopsin photosystem into model hosts or hybridizing light-capturing nanoparticles with cells (Martinez et al., 2007; Guo et al., 2018). However, these photosynthetic biological hybridization systems are still in the early stages of development. Other challenges also require continuous research and exploration, such as biocompatibility of materials, selection of light collection devices, and seamless coupling of biological and nonbiological components (Cestellos-Blanco et al., 2020). In addition to light energy, electrical energy can also be used to produce energy carriers, such as formate, hydrogen, carbon monoxide, methanol, methane, and so on (Chen H. et al., 2020). In a pioneer study, a genetically engineered autotrophic microorganism, Ralstonia eutropha H16, produced higher alcohol levels in an electric bioreactor that used CO2 as the sole carbon source and electricity as the sole energy input. The CO2 on the cathode driven by electricity was reduced to formic acid, which was then converted into isobutanol and 3-methyl-1-butanol. This process integrated CO2 fixation, electrochemical formic acid formation, and higher alcohol synthesis, opening up possibilities for electrically driving CO2 to commercial chemicals (Li et al., 2012). Although this work proves the feasibility of electrochemical fixation of CO2, the lack of a full understanding of the host and the production of hydrogen peroxide during the production process affected cell growth, limiting continuous improvement of the system. Enzymatic catalysis is a powerful platform to drive CO2 fixation, such as NAD-dependent FDH and hydrogenase (Antonovsky et al., 2017). The former has been successfully applied in construction of artificial autotrophy via the CBB cycle as described above. However, carbon loss is inevitable during the generation of NADH. Therefore, hydrogenase is a promising option for providing energy and reducing equivalents without carbon loss (Yu, 2018). In this context, the (an)aerobic fermentation of synthesis gas (syngas) (H2/CO/CO2) and industrial off-gases has been proven to be an attractive platform for fixing CO2 to produce a variety of chemicals in some natural autotrophic microorganisms, including acetogenic bacteria (Clostridium autoethanogenum and Clostridium ljungdahlii) (Mock et al., 2015; Emerson et al., 2019), Cupriavidus necator (Garcia-Gonzalez and De Wever, 2017) and Oligotropha carboxidovorans (Siebert et al., 2020), in which carbon can be fixed via CBB cycle or the Wood-Ljungdahl pathway with hydrogen as energy source. Similarly, Keller et al. (2013) expressed five genes of subpathway 1 in 3-HP/4-HB cycle from the thermophilic bacteria Metallosphaera sedula in Pyrococcus furiosus. The engineered strain used hydrogen as the electron donor to convert CO2 and acetyl-CoA into the valuable chemical 3-hydroxypropionic acid (Keller et al., 2013).
Second, CO2 fixation pathways are the key component of artificial autotrophy. As described above, although the CBB cycle is ubiquitous in the biosphere, carbon yield is lower and energy consumption is higher. Moreover, other natural aerobic CO2 assimilation routes involve too many enzymes, for instance 3-HP bicycle. Hence, a synthetic pathway with higher carbon yield, reduced energy cost and relatively fewer enzymes is a more attractive candidate for reconstruction of artificial autotrophy. In this respect, strong advances in synthetic biology have enabled the rational design of a novel CO2 fixation pathway based on a variety of established routes reported over the last decade. For example, the formate assimilation pathway has been engineered in model microorganisms, and an entire metabolic pathway can be developed by coupling upstream CO2 capturing and downstream production of acetyl-CoA and other metabolites for cell growth and product biosynthesis. CO2 can be reduced to formate by the action of NAD-independent FDH or electrodes, and the recruitment of SACA bypasses downstream pathways and can efficiently produce acetyl-CoA without any carbon loss. Moreover, the influence of carbon flux regulation on CO2 assimilation metabolism will be dramatically minimized due to few overlaps between exogenous metabolism and the endogenous cellular metabolic network. In addition to well-studied CO2 capturing enzymes, such as Rubisco or NAD-independent FDH, more novel and efficient enzyme candidates in the CO2 fixation pathway might be identified using advanced technologies for DNA sequencing, bioinformatics and structure-function predictions. Recently, a CO2-reducing formate dehydrogenase complex (FdhAB) was identified from environmental samples by genome-resolved metagenomics (Figueroa et al., 2018). Moreover, the rate-limiting enzyme in CO2 fixation pathways may have to be evolved or engineered with new features or improved kinetic properties.
Third, in most reactions, increased substrate concentrations can improve thermodynamics and enzyme conversion efficiency, as well as reducing enzyme side reaction activity. CO2 capture mechanisms can be used to boost CO2 concentration, including transmembrane bicarbonate pumps, transport proteins, carbonic anhydrase and microcompartments (Kerfeld and Erbilgin, 2015). Functional expression of foreign carboxysome in E. coli laid the groundwork for CO2 condensation and fixation (Bonacci et al., 2012; Gong et al., 2015). Moreover, this spatial organization provides more stable enzyme structures, facilitates substrate channeling between active sites, and promotes carbon flux in a desirable direction (Siu et al., 2015).
Last but not least, the selection of a proper host plays a crucial role in the development of artificial autotrophy. A variety of criteria should be taken into account, including tolerance of feedstock, culture conditions, products of interest, cell growth rate, robustness, feasibility and stability of genetic manipulation. Compared to E. coli, S. cerevisiae has a higher tolerance for formate or other toxic substrates, as well as more endogenous carbon anabolic enzymes. From a biotechnology perspective, these features make S. cerevisiae a very promising chassis for the CO2-fixing bio-industry.
Although all of abovementioned essential components for autotrophy are introduced into a suitable host, the constructed artificial autotrophic microorganism cannot grow solely on CO2 since it is “accustomed” to heterotrophy. Therefore, this engineered strain should be adapted to be “fed” with CO2. In recent studies, autotrophic growth was achieved by use of an ALE approach, and many essential mutation sites were identified, involving the CO2 fixation pathway, central metabolism, reducing power and metabolic regulation (Figure 3 and Table 2). These sites can be used for modified targets in future work.
TABLE 2.
Essential mutations for adaptation to autotrophy.
| Overexpression enzymes | Energy source and reducing power | Mutated genes | Functions | Host | References |
| Ribulose-1,5-bisphosphate, carboxylase/oxygenase, phosphoribosyl kinase, formate dehydrogenase | Pyruvate | prs (ribose-phosphate diphoskinase), pgi (glucosephosphate isomerase), serA (3-phosphoglycerate dehydrogenase) | CBB cycle | E. coli | Herz et al., 2017 |
| crp (cAMP receptor protein), ppsR (PEP synthetase regulatory protein) | Metabolism regulation | ||||
| Formate-THF ligase, methenyl-THF cyclohydrolase, methylene-THF dehydrogenase, serine glyoxylate transaminase, serine hydroxymethyltransferase hydroxypyruvate reductase, glycerate kinase | Formate | metF (methylenetetrahydrofolate reductase) purU (formyltetrahydrofolate deformylase) | Folate metabolism | E. coli | Kim et al., 2019 |
| purT/purN (phosphoribosylglycinamide formyltransferase) hycA/fnr (formate hydrogenlyase) | Formate hydrogen lyase regulation | ||||
| Ribulose-1,5-bisphosphate, carboxylase/oxygenase, phosphoribosyl kinase, formate dehydrogenase, carbonic anhydrase | Formate | prs (ribose-phosphate diphoskinase), pgi (glucosephosphate isomerase), aroH (2-dehydro-3-deoxyphosphoheptonate aldolase) | CBB cycle | E. coli | Gleizer et al., 2019 |
| fdh (formate dehydrogenase) | Reducing power | ||||
| Phosphoribosyl kinase phosphoglycerate kinase glyceraldehyde-3-phosphate dehydrogenase triosephosphate isomerase transketolase | Methanol | Prk (phosphoribosyl kinase) | CBB cycle | P. pastoris | Gassler et al., 2020 |
| NMA1 (nicotinic acid mononucleotide adenylyltransferase) | Reducing power | ||||
| Formate-THF ligase, methenyl-THF cyclohydrolase, methylene-THF dehydrogenase, glycine cleavage/synthase system, serine hydroxymethyltransferase, serine deaminase, formate dehydrogenase | Formate | fdh (formate dehydrogenase), pntAB (membrane-bound transhydrogenase) | Reducing power | E. coli | Kim et al., 2020 |
In conclusion, the development of synthetic biology has provided the possibility of designing efficient biological carbon-fixing processes through understanding the diversity of carbon-fixing organisms and their metabolic pathways in nature, as well as the exploration of efficient carbon-fixing elements. We believe that CO2 fixation by microbial organisms may make a significant contribution to building a sustainable society.
Author Contributions
BL and JY designed the manuscript. BL and YZ prepared the manuscript. JY polished the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Caroline S. Harwood for useful suggestions which helped to improve the quality of the manuscript.
Footnotes
Funding. This work was supported by grants from Key Laboratory of Biofuels, Qingdao Institute of Bioenergy and Bioprocess Technology, Chinese Academy of Sciences (CASKLB201805), the “First class grassland science discipline” program in Shandong Province, the National Natural Science Foundation of China (31860011), and the Talents of High Level Scientific Research Foundation (Grants 6651117005 and 6651119011) of Qingdao Agricultural University.
References
- Alper H., Moxley J., Nevoigt E., Fink G. R., Stephanopoulos G. (2006). Engineering yeast transcription machinery for improved ethanol tolerance and production. Science 314 1565–1568. 10.1126/science.1131969 [DOI] [PubMed] [Google Scholar]
- Altas N., Aslan A. S., Karatas E., Chronopoulou E., Labrou N. E., Binay B. (2017). Heterologous production of extreme alkaline thermostable NAD(+)-dependent formate dehydrogenase with wide-range pH activity from Myceliophthora thermophila. Process. Biochem. 61 110–118. 10.1016/j.procbio.2017.06.017 [DOI] [Google Scholar]
- Antonovsky N., Gleizer S., Milo R. (2017). Engineering carbon fixation in E. coli: from heterologous RuBisCO expression to the Calvin-Benson-Bassham cycle. Curr. Opin. Biotechnol. 47 83–91. 10.1016/j.copbio.2017.06.006 [DOI] [PubMed] [Google Scholar]
- Antonovsky N., Gleizer S., Noor E., Zohar Y., Herz E., Barenholz U., et al. (2016). Sugar synthesis from CO2 in Escherichia coli. Cell 115–125. 10.1016/j.cell.2016.05.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bang J., Lee S. Y. (2018). Assimilation of formic acid and CO2 by engineered Escherichia coli equipped with reconstructed one-carbon assimilation pathways. Proc. Natl. Acad. Sci. U.S.A. E9271–E9279. 10.1073/pnas.1810386115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barenholz U., Davidi D., Reznik E., Bar-On Y., Antonovsky N., Noor E., et al. (2017). Design principles of autocatalytic cycles constrain enzyme kinetics and force low substrate saturation at flux branch points. eLife 6:e20667. 10.7554/eLife.20667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Even A. (2016). Formate assimilation: the metabolic architecture of natural and synthetic pathways. Biochemistry 55 3851–3863. 10.1021/acs.biochem.6b00495 [DOI] [PubMed] [Google Scholar]
- Bar-Even A., Noor E., Lewis N. E., Milo R. (2010). Design and analysis of synthetic carbon fixation pathways. Proc. Natl. Acad. Sci. U.S.A. 107 8889–8894. 10.1073/pnas.0907176107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Even A., Noor E., Milo R. (2012). A survey of carbon fixation pathways through a quantitative lens. J. Exp. Bot. 63 2325–2342. 10.1093/jxb/err417 [DOI] [PubMed] [Google Scholar]
- Bassham J. A., Calvin M. (1962). The way of CO2 in plant photosynthesis. Comp. Biochem. Physiol. 4 187–204. 10.1016/0010-406x(62)90004-x [DOI] [PubMed] [Google Scholar]
- Berg I. A. (2011). Ecological aspects of the distribution of different autotrophic CO2 fixation pathways. Appl. Environ. Microbiol. 77 1925–1936. 10.1128/AEM.02473-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg I. A., Kockelkorn D., Buckel W., Fuchs G. (2007). A 3-hydroxypropionate/4-hydroxybutyrate autotrophic carbon dioxide assimilation pathway in Archaea. Science 318 1782–1786. 10.1126/science.1149976 [DOI] [PubMed] [Google Scholar]
- Blankenship R. E., Tiede D. M., Barber J., Brudvig G. W., Fleming G., Ghirardi M., et al. (2011). Comparing photosynthetic and photovoltaic efficiencies and recognizing the potential for improvement. Science 332 805–809. 10.1126/science.1200165 [DOI] [PubMed] [Google Scholar]
- Blatti J. L., Michaud J., Burkart M. D. (2013). Engineering fatty acid biosynthesis in microalgae for sustainable biodiesel. Curr. Opin. Chem. Biol. 17 496–505. 10.1016/j.cbpa.2013.04.007 [DOI] [PubMed] [Google Scholar]
- Bogorad I. W., Chen C. T., Theisen M. K., Wu T. Y., Schlenz A. R., Lam A. T., et al. (2014). Building carbon-carbon bonds using a biocatalytic methanol condensation cycle. Proc. Natl. Acad. Sci. U.S.A. 111 15928–15933. 10.1073/pnas.1413470111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogorad I. W., Lin T. S., Liao J. C. (2013). Synthetic non-oxidative glycolysis enables complete carbon conservation. Nature 502 693–697. 10.1038/nature12575 [DOI] [PubMed] [Google Scholar]
- Bonacci W., Teng P. K., Afonso B., Niederholtmeyer H., Grob P., Silver P. A., et al. (2012). Modularity of a carbon-fixing protein organelle. Proc. Natl. Acad. Sci. U.S.A. 109 478–483. 10.1073/pnas.1108557109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan B. B., Arnon D. I. (1990). A reverse KREBS cycle in photosynthesis: consensus at last. Photosynth. Res. 24 47–53. 10.1007/BF00032643 [DOI] [PubMed] [Google Scholar]
- Cai Z., Liu G. X., Zhang J. L., Li Y. (2014). Development of an activity-directed selection system enabled significant improvement of the carboxylation efficiency of Rubisco. Protein Cell 5 552–562. 10.1007/s13238-014-0072-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspeta L., Buijs N. A. A., Nielsen J. (2013). The role of biofuels in the future energy supply. Energ. Environ. Sci. 6 1077–1082. 10.1039/c3ee24403b [DOI] [Google Scholar]
- Cestellos-Blanco S., Zhang H., Kim J. M., Shen Y. X., Yang P. D. (2020). Photosynthetic semiconductor biohybrids for solar-driven biocatalysis. Nat. Catal. 3 245–255. 10.1038/s41929-020-0428-y [DOI] [Google Scholar]
- Chen H., Dong F. Y., Minteer S. D. (2020). The progress and outlook of bioelectrocatalysis for the production of chemicals, fuels and materials. Nat. Catal. 3 225–244. 10.1038/s41929-019-0408-2 [DOI] [Google Scholar]
- Chen Y., Banerjee D., Mukhopadhyay A., Petzold C. J. (2020). Systems and synthetic biology tools for advanced bioproduction hosts. Curr. Opin. Biotechnol. 64 101–109. 10.1016/j.copbio.2019.12.007 [DOI] [PubMed] [Google Scholar]
- Choe H., Joo J. C., Cho D. H., Kim M. H., Lee S. H., Jung K. D., et al. (2014). Efficient CO2-reducing activity of NAD-dependent formate dehydrogenase from Thiobacillus sp KNK65MA for formate production from CO2 gas. PLoS One 9:e103111. 10.1371/journal.pone.0103111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claassens N. J., Sousa D. Z., dos Santos V. A. P. M., de Vos W. M., van der Oost J. (2016). Harnessing the power of microbial autotrophy. Nat. Rev. Microbiol. 14 692–706. 10.1038/nrmicro.2016.130 [DOI] [PubMed] [Google Scholar]
- Clarke L., Kitney R. (2020). Developing synthetic biology for industrial biotechnology applications. Biochem. Soc. Trans. 48 113–122. 10.1042/BST20190349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotton C. A. R., Edlich-Muth C., Bar-Even A. (2018). Reinforcing carbon fixation: CO2 reduction replacing and supporting carboxylation. Curr. Opin. Biotech. 49 49–56. 10.1016/j.copbio.2017.07.014 [DOI] [PubMed] [Google Scholar]
- de la Cruz J. G., Machens F., Messerschmidt K., Bar-Even A. (2019). Core catalysis of the reductive glycine pathway demonstrated in yeast. Acs Synthet. Biol. 8 911–917. 10.1021/acssynbio.8b00464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducat D. C., Silver P. A. (2012). Improving carbon fixation pathways. Curr. Opin. Chem. Biol. 16 337–344. 10.1016/j.cbpa.2012.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durall C., Lindberg P., Yu J. P., Lindblad P. (2020). Increased ethylene production by overexpressing phosphoenolpyruvate carboxylase in the cyanobacterium Synechocystis PCC 6803. Biotechnol. Biofuels 13:16. 10.1186/s13068-020-1653-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson D. F., Woolston B. M., Liu N., Donnelly M., Currie D. H., Stephanopoulos G. (2019). Enhancinghydrogen-dependent growth of and carbon dioxide fixation by Clostridium ljungdahlii through nitrate supplementation. Biotechnol. Bioeng. 116 294–306. 10.1002/bit.26847 [DOI] [PubMed] [Google Scholar]
- Erb T. J., Keller P., Vorholt J. A. (2019). Escherichia coli in auto(trophic) mode. Cell 1244–1245. 10.1016/j.cell.2019.10.040 [DOI] [PubMed] [Google Scholar]
- Figueroa I. A., Barnum T. P., Somasekhar P. Y., Carlstrom C. I., Engelbrektson A. L., Coates J. D. (2018). Metagenomics-guided analysis of microbial chemolithoautotrophic phosphite oxidation yields evidence of a seventh natural CO2 fixation pathway. Proc. Natl. Acad. Sci. U.S.A. 115 E92–E101. 10.1073/pnas.1715549114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gonzalez L., De Wever H. (2017). Valorisation of CO2 rich off gases to biopolymers through biotechnological process. FEMS Microbiol. Lett. 364 1–8. 10.1093/femsle/fnx196 [DOI] [PubMed] [Google Scholar]
- Gassler T., Sauer M., Gasser B., Egermeier M., Troyer C., Causon T., et al. (2020). The industrial yeast Pichia pastoris is converted from a heterotroph into an autotroph capable of growth on CO2. Nat. Biotechnol. 38 210–218. 10.1038/s41587-019-0363-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleizer S., Ben-Nissan R., Bar-On Y. M., Antonovsky N., Noor E., Zohar Y., et al. (2019). Conversion of Escherichia coli to generate all biomass carbon from CO2. Cell 179 1255–1263. 10.1016/j.cell.2019.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong F., Liu G., Zhai X., Zhou J., Cai Z., Li Y. (2015). Quantitative analysis of an engineered CO2-fixing Escherichia coli reveals great potential of heterotrophic CO2 fixation. Biotechnol. Biofuels 8:86. 10.1186/s13068-015-0268-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guadalupe-Medina V., Wisselink H. W., Luttik M. A. H., de Hulster E., Daran J. M., Pronk J. T., et al. (2013). Carbon dioxide fixation by Calvin-Cycle enzymes improves ethanol yield in yeast. Biotechnol. Biofuels 6:125. 10.1186/1754-6834-6-125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J., Suastegui M., Sakimoto K. K., Moody V. M., Xiao G., Nocera D. G., et al. (2018). Light-driven fine chemical production in yeast biohybrids. Science 362 813–816. 10.1126/science.aat9777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemann M., Bauwe H. (2016). Photorespiration and the potential to improve photosynthesis. Curr. Opin. Chem. Biol. 35 109–116. 10.1016/j.cbpa.2016.09.014 [DOI] [PubMed] [Google Scholar]
- Hagemann M., Hesse W. R. (2018). Systems and synthetic biology for the biotechnological application of cyanobacteria. Curr. Opin. Biotech. 49 94–99. 10.1016/j.copbio.2017.07.008 [DOI] [PubMed] [Google Scholar]
- Hartmann T., Leimkuhler S. (2013). The oxygen-tolerant and NAD(+)-dependent formate dehydrogenase from Rhodobacter capsulatus is able to catalyze the reduction of CO2 to formate. FEBS J. 280 6083–6096. 10.1111/febs.12528 [DOI] [PubMed] [Google Scholar]
- Hayer-Hartl M., Hartl F. U. (2020). Chaperone machineries of Rubisco-The most abundant enzyme. Trends Biochem. Sci. 20:30119. 10.1016/j.tibs.2020.05.001 [DOI] [PubMed] [Google Scholar]
- Herter S., Farfsing J., Gad’On N., Rieder C., Eisenreich W., Bacher A., et al. (2001). Autotrophic CO2 fixation by Chloroflexus aurantiacus: study of glyoxylate formation and assimilation via the 3-hydroxypropionate cycle. J. Bacteriol. 183 4305–4316. 10.1128/JB.183.14.4305-4316.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herz E., Antonovsky N., Bar-On Y., Davidi D., Gleizer S., Prywes N., et al. (2017). The genetic basis for the adaptation of E-coli to sugar synthesis from CO2. Nat. Commun. 8:1705. 10.1038/S41467-017-01835-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G. P., Li Y., Ye C., Liu L. M., Chen X. L. (2018). Engineering microorganisms for enhanced CO2 sequestration. Trends Biotechnol. 37 532–547. 10.1016/j.tibtech.2018.10.008 [DOI] [PubMed] [Google Scholar]
- Huber H., Gallenberger M., Jahn U., Eylert E., Berg I. A., Kockelkorn D., et al. (2008). A dicarboxylate/4-hydroxybutyrate autotrophic carbon assimilation cycle in the hyperthermophilic Archaeum Ignicoccus hospitalis. Proc. Natl. Acad. Sci. U.S.A. 105 7851–7856. 10.1073/pnas.0801043105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iniguez C., Capo-Bauca S., Niinemets U., Stoll H., Aguilo-Nicolau P., Galmes J. (2020). Evolutionary trends in RuBisCO kinetics and their co-evolution with CO2 concentrating mechanisms. Plant J. 101 897–918. 10.1111/tpj.14643 [DOI] [PubMed] [Google Scholar]
- Irfan M., Bai Y., Zhou L., Kazmi M., Yuan S., Mbadinga S. M., et al. (2019). Direct microbial transformation of carbon dioxide to value-added chemicals: a comprehensive analysis and application potentials. Bioresource Technol. 288:121401. 10.1016/j.biortech.2019.121401 [DOI] [PubMed] [Google Scholar]
- Jormakka M., Byrne B., Iwata S. (2003). Formate dehydrogenase–a versatile enzyme in changing environments. Curr. Opin. Struct. Biol. 13 418–423. 10.1016/s0959-440x(03)00098-8 [DOI] [PubMed] [Google Scholar]
- Keller M. W., Schut G. J., Lipscomb G. L., Menon A. L., Iwuchukwu I. J., Leuko T. T., et al. (2013). Exploiting microbial hyperthermophilicity to produce an industrial chemical, using hydrogen and carbon dioxide. Proc. Natl. Acad. Sci. U.S.A. 110 5840–5845. 10.1073/pnas.1222607110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerfeld C. A., Erbilgin O. (2015). Bacterial microcompartments and the modular construction of microbial metabolism. Trends Microbiol. 23 22–34. 10.1016/j.tim.2014.10.003 [DOI] [PubMed] [Google Scholar]
- Kim B. W., Chang H. N., Kim I. K., Lee K. S. (1992). Growth kinetics of the photosynthetic bacterium Chlorobium thiosulfatophilum in a fed-batch reactor. Biotechnol. Bioeng. 40 583–592. 10.1002/bit.260400505 [DOI] [PubMed] [Google Scholar]
- Kim S., Lindner S. N., Aslan S., Yishai O., Wenk S., Schann K., et al. (2020). Growth of E. coli on formate and methanol via the reductive glycine pathway. Nat. Chem. Biol. 16 538–545. 10.1038/s41589-020-0473-5 [DOI] [PubMed] [Google Scholar]
- Kim S. J., Yoon J., Im D. K., Kim Y. H., Oh M. K. (2019). Adaptively evolved Escherichia coli for improved ability of formate utilization as a carbon source in sugar-free conditions. Biotechnol. Biofuels 12:207. 10.1186/S13068-019-1547-Z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koay T. W., Wong H. L., Lim B. H. (2016). Engineering of chimeric eukaryotic/bacterial Rubisco large subunits in Escherichia coli. Genes Genet. Syst. 139–150. 10.1266/ggs.15-00054 [DOI] [PubMed] [Google Scholar]
- Kubis A., Bar-Even A. (2019). Synthetic biology approaches for improving photosynthesis. J. Exp. Bot. 70 1425–1433. 10.1093/jxb/erz029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushwaha D., Upadhyay S. N., Mishra P. K. (2018). Growth of Cyanobacteria: optimization for increased carbohydrate content. Appl. Biochem. Biotechnol. 184 1247–1262. 10.1007/s12010-017-2620-3 [DOI] [PubMed] [Google Scholar]
- Lee S., Kim P. (2020). Current status and applications of adaptive laboratory evolution in industrial microorganisms. J. Microbiol. Biotechnol. 30 793–803. 10.4014/jmb.2003.03072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire O. N., Jespersen M., Wagner T. (2020). CO2-fixation strategies in energy extremophiles: what can we learn from acetogens? Front. Microbiol. 11:486. 10.3389/Fmicb.2020.00486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Huang L., Liu T., Liu J., Zhen L., Wu J., et al. (2019). Electrochemical reduction of carbon dioxide to formate via nano-prism assembled CuO microspheres. Chemosphere 237:124527. 10.1016/j.chemosphere.2019.124527 [DOI] [PubMed] [Google Scholar]
- Li H., Opgenorth P. H., Wernick D. G., Rogers S., Wu T. Y., Higashide W., et al. (2012). Integrated electromicrobial conversion of CO2 to higher alcohols. Science 335 1596–1596. 10.1126/science.1217643 [DOI] [PubMed] [Google Scholar]
- Li Y. J., Wang M. M., Chen Y. W., Wang M., Fan L. H., Tan T. W. (2017). Engineered yeast with a CO2-fixation pathway to improve the bio-ethanol production from xylose-mixed sugars. Sci. Rep. 7:43875. 10.1038/srep43875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang F. Y., Lindberg P., Lindblad P. (2018). Engineering photoautotrophic carbon fixation for enhanced growth and productivity. Sustain. Energ. Fuels 2 2583–2600. 10.1039/c8se00281a [DOI] [Google Scholar]
- Lin M. T., Occhialini A., Andralojc P. J., Parry M. A. J., Hanson M. R. (2014). A faster Rubisco with potential to increase photosynthesis in crops. Nature 513 547–550. 10.1038/nature13776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin P. P., Jaeger A. J., Wu T. Y., Xu S. C., Lee A. S., Gao F. K., et al. (2018). Construction and evolution of an Escherichia coli strain relying on nonoxidative glycolysis for sugar catabolism. Proc. Natl. Acad. Sci. U.S.A. 115 3538–3546. 10.1073/pnas.1802191115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z. H., Wang K., Chen Y., Tan T. W., Nielsen J. (2020). Third-generation biorefineries as the means to produce fuels and chemicals from CO2. Nat. Catal. 3 274–288. 10.1038/s41929-019-0421-5 [DOI] [Google Scholar]
- Ljungdahl L. G. (1969). Total synthesis of acetate from CO2 by heterotrophic bacteria. Annu. Rev. Microbiol. 23 515–538. 10.1146/annurev.mi.23.100169.002503 [DOI] [PubMed] [Google Scholar]
- Luan G., Zhang S., Lu X. (2020). Engineering cyanobacteria chassis cells toward more efficient photosynthesis. Curr. Opin. Biotechnol. 62 1–6. 10.1016/j.copbio.2019.07.004 [DOI] [PubMed] [Google Scholar]
- Maia L. B., Fonseca L., Moura I., Moura J. J. (2016). Reduction of carbon dioxide by a molybdenum-containing formate dehydrogenase: a kinetic and mechanistic study. J. Am. Chem. Soc. 138 8834–8846. 10.1021/jacs.6b03941 [DOI] [PubMed] [Google Scholar]
- Maia L. B., Moura I., Moura J. J. G. (2017). Molybdenum and tungsten-containing formate dehydrogenases: aiming to inspire a catalyst for carbon dioxide utilization. Inorg. Chim. Acta 455 350–363. 10.1016/j.ica.2016.07.010 [DOI] [Google Scholar]
- Mainguet S. E., Gronenberg L. S., Wong S. S., Liao J. C. (2013). A reverse glyoxylate shunt to build a non-native route from C4 to C2 in Escherichia coli. Metab. Eng. 19 116–127. 10.1016/j.ymben.2013.06.004 [DOI] [PubMed] [Google Scholar]
- Martinez A., Bradley A. S., Waldbauer J. R., Summons R. E., DeLong E. F. (2007). Proteorhodopsin photosystem gene expression enables photophosphorylation in a heterologous host. Proc. Natl. Acad. Sci. U.S.A. 104 5590–5595. 10.1073/pnas.0611470104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattozzi M., Ziesack M., Voges M. J., Silver P. A., Way J. C. (2013). Expression of the sub-pathways of the Chloroflexus aurantiacus 3-hydroxypropionate carbon fixation bicycle in E. coli: toward horizontal transfer of autotrophic growth. Metab. Eng. 16 130–139. 10.1016/j.ymben.2013.01.005 [DOI] [PubMed] [Google Scholar]
- Mock J., Zheng Y. N., Mueller A. P., Ly S., Tran L., Segovia S., et al. (2015). Energy conservation associated with ethanol formation from H2 and CO2 in Clostridium autoethanogenum involving electron bifurcation. J. Bacteriol. 197 2965–2980. 10.1128/Jb.00399-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller-Cajar O., Morell M., Whitney S. M. (2007). Directed evolution of rubisco in Escherichia coli reveals a specificity-determining hydrogen bond in the form II enzyme. Biochemistry 16 14067–14074. 10.1021/bi700820a [DOI] [PubMed] [Google Scholar]
- Mueller-Cajar O., Whitney S. M. (2008a). Directing the evolution of Rubisco and Rubisco activase: first impressions of a new tool for photosynthesis research. Photosynth. Res. 98 667–675. 10.1007/s11120-008-9324-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller-Cajar O., Whitney S. M. (2008b). Evolving improved Synechococcus Rubisco functional expression in Escherichia coli. Biochem. J. 414 205–214. 10.1042/BJ20080668 [DOI] [PubMed] [Google Scholar]
- Nielsen J., Keasling J. D. (2016). Engineering cellular metabolism. Cell 164 1185–1197. 10.1016/j.cell.2016.02.004 [DOI] [PubMed] [Google Scholar]
- Nielsen J., Larsson C., van Maris A., Pronk J. (2013). Metabolic engineering of yeast for production of fuels and chemicals. Curr. Opin. Biotech. 24 398–404. 10.1016/j.copbio.2013.03.023 [DOI] [PubMed] [Google Scholar]
- Niks D., Hille R. (2019). Molybdenum- and tungsten-containing formate dehydrogenases and formylmethanofuran dehydrogenases: structure, mechanism, and cofactor insertion. Protein Sci. 28 111–122. 10.1002/pro.3498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogue V. S., Karhumaa K. (2015). Xylose fermentation as a challenge for commercialization of lignocellulosic fuels and chemicals. Biotechnol. Lett. 37 761–772. 10.1007/s10529-014-1756-2 [DOI] [PubMed] [Google Scholar]
- Pacala S., Socolow R. (2004). Stabilization wedges: solving the climate problem for the next 50 years with current technologies. Science 305 968–972. 10.1126/science.1100103 [DOI] [PubMed] [Google Scholar]
- Parikh M. R., Greene D. N., Woods K. K., Matsumura I. (2006). Directed evolution of RuBisCO hypermorphs through genetic selection in engineered E.coli. Protein Eng. Des. Sel. 19 113–119. 10.1093/protein/gzj010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragsdale S. W. (1997). The eastern and western branches of the Wood/Ljungdahl pathway: how the east and west were won. Biofactors 6 3–11. 10.1002/biof.5520060102 [DOI] [PubMed] [Google Scholar]
- Satagopan S., Huening K. A., Tabita F. R. (2019). Selection of cyanobacterial (Synechococcus sp. Strain PCC 6301) RubisCO variants with improved functional properties that confer enhanced CO2-dependent growth of Rhodobacter capsulatus, a photosynthetic bacterium. mBio 10:e01537-19. 10.1128/mBio.01537-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savakis P., Hellingwerf K. J. (2015). Engineering cyanobacteria for direct biofuel production from CO2. Curr. Opin. Biotech. 33 8–14. 10.1016/j.copbio.2014.09.007 [DOI] [PubMed] [Google Scholar]
- Schwander T., von Borzyskowski L. S., Burgener S., Cortina N. S., Erb T. J. (2016). A synthetic pathway for the fixation of carbon dioxide in vitro. Science 354 900–904. 10.1126/science.aah5237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebert D., Busche T., Metz A. Y., Smaili M., Queck B. A. W., Kalinowski J., et al. (2020). Genetic engineering of Oligotropha carboxidovorans strain OM5-A promising candidate for the aerobic utilization of synthesis gas. Acs Synth. Biol. 9 1426–1440. 10.1021/acssynbio.0c00098 [DOI] [PubMed] [Google Scholar]
- Siegel J. B., Smith A. L., Poust S., Wargacki A. J., Bar-Even A., Louw C., et al. (2015). Computational protein design enables a novel one-carbon assimilation pathway. Proc. Natl. Acad. Sci. U.S.A. 112 3704–3709. 10.1073/pnas.1500545112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu K. H., Chen R. P., Sun Q., Chen L., Tsai S. L., Chen W. (2015). Synthetic scaffolds for pathway enhancement. Curr. Opin. Biotech. 36 98–106. 10.1016/j.copbio.2015.08.009 [DOI] [PubMed] [Google Scholar]
- Song S., Timm S., Lindner S. N., Reimann V., Hess W. R., Hagemann M., et al. (2020). Expression of formate-tetrahydrofolate ligase did not improve growth but interferes with nitrogen and carbon metabolism of Synechocystis sp. PCC 6803. Front. Microbiol. 11:1650 10.3389/fmicb.2020.01650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilman D., Socolow R., Foley J. A., Hill J., Larson E., Lynd L., et al. (2009). Energy. Beneficial biofuels–the food, energy, and environment trilemma. Science 325 270–271. 10.1126/science.1177970 [DOI] [PubMed] [Google Scholar]
- Veaudor T., Blanc-Garin V., Chenebault C., Diaz-Santos E., Sassi J. F., Cassier-Chauvat C., et al. (2020). Recent advances in the photoautotrophic metabolism of Cyanobacteria: biotechnological implications. Life 10:71. 10.3390/life10050071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R. H., Alonso H., Whitney S. M. (2016). Evolving Methanococcoides burtonii archaeal Rubisco for improved photosynthesis and plant growth. Sci. Rep. 6:22284. 10.1038/srep22284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia P. F., Zhang G. C., Walker B., Seo S. O., Kwak S., Liu J. J., et al. (2017). Recycling carbon dioxide during xylose fermentation by engineered Saccharomyces cerevisiae. ACS Synth. Biol. 6 276–283. 10.1021/acssynbio.6b00167 [DOI] [PubMed] [Google Scholar]
- Xin Y., Shi Y., Niu T., Wang Q., Niu W., Huang X., et al. (2018). Cryo-EM structure of the RC-LH core complex from an early branching photosynthetic prokaryote. Nat. Commun. 9:1568. 10.1038/s41467-018-03881-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yishai O., Bouzon M., Doring V., Bar-Even A. (2018). In vivo assimilation of one-carbon via a synthetic reductive glycine pathway in Escherichia coli. Acs Synth. Biol. 2023–2028. 10.1021/acssynbio.8b00131 [DOI] [PubMed] [Google Scholar]
- Yu H., Li X., Duchoud F., Chuang D. S., Liao J. C. (2018). Augmenting the Calvin-Benson-Bassham cycle by a synthetic malyl-CoA-glycerate carbon fixation pathway. Nat. Commun. 9:2008. 10.1038/s41467-018-04417-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J. (2018). Fixation of carbon dioxide by a hydrogen-oxidizing bacterium for value-added products. World J. Microb. Biot. 34:89. 10.1007/s11274-018-2473-0 [DOI] [PubMed] [Google Scholar]
- Yu X., Niks D., Mulchandani A., Hille R. (2017). Efficient reduction of CO2 by the molybdenum-containing formate dehydrogenase from Cupriavidus necator (Ralstonia eutropha). J. Biol. Chem. 292 16872–16879. 10.1074/jbc.M117.785576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelcbuch L., Lindner S. N., Zegman Y., Slutskin I. V., Antonovsky N., Gleizer S., et al. (2016). Pyruvate formate-lyase enables efficient growth of Escherichia coli on acetate and formate. Biochemistry 55 2423–2426. 10.1021/acs.biochem.6b00184 [DOI] [PubMed] [Google Scholar]
- Zhou Y., Whitney S. (2019). Directed evolution of an improved Rubisco; In vitro analyses to decipher fact from fiction. Int. J. Mol. Sci. 20:5019. 10.3390/ijms20205019 [DOI] [PMC free article] [PubMed] [Google Scholar]