Abstract
Angular leaf spot (ALS), caused by Pseudocercospora griseola, is one of the most devastating diseases of common bean (Phaseolus vulgaris L.) in tropical and subtropical production areas. Breeding for ALS resistance is difficult due to the extensive virulence diversity of P. griseola and the recurrent appearance of new virulent races. Five major loci, Phg-1 to Phg-5, conferring ALS resistance have been named, and markers tightly linked to these loci have been reported. Quantitative trait loci (QTLs) have also been described, but the validation of some QTLs is still pending. The Phg-1, Phg-4, and Phg-5 loci are from common bean cultivars of the Andean gene pool, whereas Phg-2 and Phg-3 are from beans of the Mesoamerican gene pool. The reference genome of common bean and high-throughput sequencing technologies are enabling the development of molecular markers closely linked to the Phg loci, more accurate mapping of the resistance loci, and the comparison of their genomic positions. The objective of this report is to provide a comprehensive review of ALS resistance in common bean. Furthermore, we are reporting three case studies of ALS resistance breeding in Latin America and Africa. This review will serve as a reference for future resistance mapping studies and as a guide for the selection of resistance loci in breeding programs aiming to develop common bean cultivars with durable ALS resistance.
Common bean (Phaseolus vulgaris L.), which includes dry and snap beans, is the world’s most important grain legume for direct human consumption and is also an important source of protein, fiber, calories, and vital micronutrients, particularly for millions of people in Latin America and eastern and southern Africa (Singh, 1999; Broughton et al., 2003). Frequent consumption of dry seeds of common bean combined with cereals ensures a balanced diet of essential amino acids and other nutrients that contribute to alleviating malnutrition and preventing cardiovascular disease, diabetes, and certain types of cancer (Broughton et al., 2003; Thompson et al., 2017; Viguiliouk et al., 2017).
The Americas are the largest common bean-producing region, and Brazil is the world’s largest producer and consumer (Singh, 1999; Beebe, 2012). Africa, where common bean was introduced after the discovery of the Americas, is second in production. Moreover, consumption in several African countries, up to 66 kg person−1 yr−1, is greater than that in Latin America (Wortmann et al., 1998; Singh, 1999; Broughton et al., 2003; Beebe, 2012).
Numerous infectious diseases caused by fungi, viruses, bacteria, and nematodes represent major limitations to common bean production throughout the world (Schwartz and Pastor-Corrales, 1989; Schwartz et al., 2005). Angular leaf spot (ALS), a disease caused by Pseudocercospora griseola (Sacc.) Crous & Braun [previously referred to as Phaeoisariopsis griseola (Sacc.) Ferrari], was until the 1980s considered to be of minor importance in Latin America, particularly in Brazil (Rava Seijas et al., 1985). However, in the mid-1980s, ALS began to be considered a significant constraint to dry bean production in Brazil, Central America, and eastern and southern Africa (Rava Seijas et al., 1985; Liebenberg and Pretorius, 1997; Pastor-Corrales et al., 1998; Aggarwal et al., 2004). Angular leaf spot is currently one of the most recurring and devastating diseases of dry beans in Latin America and Africa, the most important production areas of the world (Correa-Victoria. et al., 1989; Liebenberg and Pretorius, 1997; Wortmann et al., 1998; Stenglein et al., 2003; Sartorato, 2004; Crous et al., 2006). Angular leaf spot has also been reported to occur sporadically in countries of the temperate climate zone, including the United States and Canada (Correa and Saettler, 1987; Melzer and Boland, 2001), and ALS was recently reported for the first time in northern Spain (Landeras et al., 2017).
Yield losses caused by ALS can reach up to 80% (Schwartz et al., 1981; Rava Seijas et al., 1985; de Jesus Junior et al., 2001). Although fungicides are an option for the control of ALS, they are often expensive or not readily available to smallholder farmers, the predominant producers of dry beans in the tropics. Cultivars with resistance to P. griseola offer a cost-effective, easy-to-use, and environmentally friendly management strategy (Pastor-Corrales et al., 1998), and several sources of ALS resistance have been identified among primary and secondary gene pools of P. vulgaris (CIAT, 1996, 1999; Pastor-Corrales et al., 1998; Busogoro et al., 1999a; Mahuku et al., 2003, 2009, 2011). However, development of common bean cultivars with durable ALS resistance is difficult due to the broad and changing virulence diversity of the ALS pathogen that renders varieties that are resistant in one year or location susceptible in another (Pastor-Corrales et al., 1998; Mahuku et al., 2002b).
Resistance to the ALS pathogen is largely conferred by single dominant resistance genes, hereafter also referred to as loci. Recent studies also show a more quantitative nature of resistance that includes quantitative trait loci (QTLs). To date, five ALS resistance loci have been approved by the Bean Improvement Cooperative (BIC) Genetics Committee (http://arsftfbean.uprm.edu/bic/wp-content/ uploads/2018/04/Bean_Genes_List_2017.pdf) that maintains the guidelines for the nomenclature of disease resistance genes in common bean. These include three dominant and independent Phg loci named Phg-1, Phg-2, and Phg-3 and two major QTLs named Phg-4 and Phg-5 (de Carvalho et al., 1998; Sartorato et al., 1999a; Corrêa et al., 2001; Namayanja et al., 2006; Gonçalves-Vidigal et al., 2011, 2013; Oblessuc et al., 2012, 2013; Keller et al., 2015).
New resources, including the reference genome of common bean (Schmutz et al., 2014) and high-throughput sequencing, facilitate the development of different types of molecular markers that are tightly linked to these loci and provide new insight into the relationship between existing and newly discovered disease resistance loci. This review aims to (i) discuss the virulence diversity of P. griseola and its impact on disease resistance breeding, (ii) discuss the current knowledge of ALS resistance, (iii) comment on how new genomic resources might facilitate and accelerate ALS research, including gene discovery and the development of highly accurate molecular markers, and (iv) present three case studies of ALS resistance breeding in Brazil, Colombia, and Uganda.
PSEUDOCERCOSPORA GRISEOLA, THE CAUSAL AGENT OF ANGULAR LEAF SPOT DISEASE
The ALS pathogen belongs to the class Dothideomycetes, the largest and most diverse class of ascomycete fungi, which contains many important plant pathogens, endophytes, and saprobes (Crous et al., 2006; Schoch et al., 2009). Although P. griseola can be transmitted through seeds, the most frequent source of primary inoculum to initiate ALS disease under natural conditions is the presence of plant debris infected with the pathogen (Correa-Victoria. et al., 1989). Pseudocercospora griseola is considered a fastidious pathogen (Correa-Victoria et al., 1989), yet it grows and produces spores on artificial culture media. Lyophilization has been successfully used for the long-term storage of spores (Castellanos et al., 2016). The response of common bean germplasm to P. griseola is usually evaluated using a disease severity scale ranging from 1 to 9, where scores of 1 to 3 are considered resistant, 4 to 6 are intermediate, and 6 to 9 are susceptible (van Schoonhoven and Pastor-Corrales, 1987).
The ALS pathogen is known for its extensive virulence diversity (Guzman et al., 1995; Chacón et al., 1997; Pastor-Corrales et al., 1998; Busogoro et al., 1999b; Mahuku et al., 2002b; Aggarwal et al., 2004; Sartorato, 2004). In the early 1980s, while working in the Bean Program of CIAT, Cali, Colombia, the corresponding author of this manuscript developed a set of 12 common bean differential cultivars: six Andean and six Mesoamerican. To standardize races of P. griseola (Table 1), a binary code was implemented and launched in 1995 during the first ALS workshop (CIAT, 1995).
Table 1.
Set of 12 common bean differential cultivars and binary code used to designate races of Pseudocercospora griseola. Each race is assigned two numbers based on the summation of the binary numbers of the susceptible Andean and Mesoamerican cultivars, respectively. The example in the table shows how a Mesoamerican isolate of P. griseola was characterized as race 25-39.
| Differential cultivars | Seed size | Common bean race | Resistance gene/ linkage group | Binary value | Reaction and binary value of susceptible cultivars |
|---|---|---|---|---|---|
| Andean cultivars† | |||||
| Don Timoteo | Large | Chile | Unknown | 1 | Susceptible: 1 |
| G11796 | Large | Peru | Unknown | 2 | Resistant |
| Bolon Bayo | Large | Peru | Unknown | 4 | Resistant |
| Montcalm | Large | Nueva Granada | Unknown | 8 | Susceptible: 8 |
| Amendoim | Large | Nueva Granada | Unknown | 16 | Susceptible: 16 |
| G5686 | Large | Nueva Granada | Phg-4/Pv04; Phg-5/Pv10 | 32 | Resistant |
| Mesoamerican cultivars‡ | |||||
| PAN 72 | Small | Mesoamerica | Unknown | 1 | Susceptible: 1 |
| G2858 | Medium | Durango | Unknown | 2 | Susceptible: 2 |
| Flor De Mayo | Small | Jalisco | Unknown | 4 | Susceptible: 4 |
| Mexico 54 | Medium | Jalisco | Phg-2/Pv08 | 8 | Resistant |
| BAT 332 | Small | Mesoamerica | Phg-22/Pv08 | 16 | Resistant |
| Cornell 49-242 | Small | Mesoamerica | 32 | Susceptible: 32 | |
Andean binary value (1 + 8 + 16) = 25.
Mesoamerican binary value (1 + 2 + 4 + 32) = 39 (race 25–39).
Characterization of the virulence phenotype (known as races) of isolates of P. griseola on Andean and Mesoamerican common bean differential cultivars has resulted in the separation of these isolates into two distinct virulence groups (Pastor-Corrales and Jara, 1995). Isolates obtained from large-seeded bean cultivars of the Andean gene pool from Ecuador, Colombia, and Argentina were virulent only on Andean differential bean cultivars, and these races were referred to as Andean. Isolates from small- and medium-seeded Mesoamerican differential cultivars from Central America, Brazil, Bolivia, and Argentina were virulent on both Mesoamerican and Andean ALS differential cultivars and were referred to as Mesoamerican (Beebe and Pastor-Corrales, 1991; Guzman et al., 1995; Pastor-Corrales and Jara, 1995; Pastor-Corrales et al., 1998; Mahuku et al., 2002b; Stenglein and Balatti, 2006).
Similar studies using differential cultivars and molecular techniques have revealed that the virulence and genetic diversity of two other pathogens that cause anthracnose (Colletotrichum lindemuthianum) and rust (Uromyces appendiculatus) also segregate into two distinct groups that mirror the diversity of their common bean host (Beebe and Pastor-Corrales, 1991; Pastor-Corrales, 1996; Pastor-Corrales and Aime, 2004).
The set of differential cultivars has been extensively used throughout the world and has permitted the comparison of races of the ALS pathogen between continents, countries, and locations. In general, races of P. griseola differ between regions, countries, and even continents.
Some of the most aggressive isolates of P. griseola, belonging to race 63-63, that are virulent on all Andean and Mesoamerican differential cultivars were first observed in Latin America. These races have been recurrently found in Brazil, Argentina, and Central America (Mahuku et al., 2002b; Sartorato, 2004; Stenglein and Balatti, 2006; Silva et al., 2008). Later, these races were infrequently found in Africa (Mahuku et al., 2002a; Wagara et al., 2004, 2011). Although Andean and Mesoamerican races have been found in the Americas and Africa, their predominance on these continents differ. Initially, Andean races infecting only Andean differential cultivars were the predominant races in Africa (Guzman et al., 1995; Aggarwal et al., 2004). Shortly afterward, the first reports of races that infected mainly Andean differentials but also a few Mesoamerican differential cultivars appeared. These races were termed Afro-Andean (CIAT, 1997; Mahuku et al., 2002a; Wagara et al., 2004, 2011). Using molecular markers to analyze the diversity of Afro-Andean races, Mahuku et al. (2002b) suggested that the Afro-Andean races belonged to the Andean group, disputing the existence of the Afro-Andean group.
Recently, Serrato-Diaz et al. (personal communication, 2018) further investigated the population structure of P. griseola by sequencing four nuclear genes to construct a phylogenetic tree. These authors found that isolates from Puerto Rico, Honduras, and Guatemala clustered in the Mesoamerican clade, whereas the Tanzanian isolates clustered into three clades: Mesoamerican, Andean, and Afro-Andean. These findings suggest a more diverse population structure than the previously reported Mesoamerican and Andean groups, but additional research is needed to resolve the extent of the population structure of this pathogen.
Although differential cultivars provide a simple and effective method for studying pathogen virulence diversity, some or all differential cultivars may become susceptible to new virulent isolates due to changes in the virulence spectra of pathogens over time and space. In fact, the virulence diversity of P. griseola appears to have changed. Many new isolates belonging to race 63-63 that are virulent on all differential cultivars have been found in Latin America and Africa (Sartorato, 2004; Wagara et al., 2004, 2011; Stenglein and Balatti, 2006; Silva et al., 2008; Ddamulira et al., 2014b; Pereira et al., 2015). Because of the increased occurrence of isolates of race 63-63, efforts are underway to develop a new set of differential cultivars, and in 2015, new candidates were proposed during the Common Bean Disease Workshop in Skukuza, South Africa. However, before a new set of differential cultivars can become available, they need to be tested for their reaction to P. griseola isolates from different countries and, in particular, for reactions to isolates of race 63-63.
Because of the high virulence diversity of P. griseola and the great potential for overcoming resistance, a successful resistance breeding strategy requires a sound understanding of the virulence diversity and evolution of this pathogen. Based on what is known about the parallel evolution between gene pools of the common bean host and pathogen, proposed strategies for durable ALS resistance include pyramiding of Andean and Mesoamerican resistance genes or using Mesoamerican resistance sources in areas where predominantly Andean isolates exist, and vice versa (Guzman et al., 1995; Pastor-Corrales et al., 1998).
OVERVIEW OF MAJOR LOCI CONDITIONING RESISTANCE TO PSEUDOCERCOSPORA GRISEOLA
Since the beginning of the 1980s, many studies have reported new sources and genes conferring resistance to ALS, as well as molecular markers linked to ALS resistance genes (Schwartz et al., 1982). However, many of these publications did not include appropriate physical linkage information or allelism tests and thus could not be validated. Only well-characterized loci or QTLs for which linked molecular markers are available can be submitted for acceptance to the BIC Genetics Committee. The currently approved ALS resistance loci include three dominant and independent loci, Phg-1, Phg-2, and Phg-3, as well as two major QTLs, Phg-4 and Phg-5 (Table 2).
Table 2.
Named and mapped angular leaf spot resistance genes in common bean. Resistance loci are stated with their new name accepted by the Bean Improvement Cooperative Genetics Committee and their originally published name. Table modified from Souza et al. (2016).
| Locus symbol |
Resistance source | Gene pool† | Chromosome | Pathogen race | Reference | |
|---|---|---|---|---|---|---|
| New | Original | |||||
| Phg-1 | Phg-1 | AND 277 | A | Pv01 | 63-23 | de Carvalho et al. (1998), Gonçalves-Vidigal et al. (2011) |
| Phg-2 | Phg-2 | Mexico 54 | MA | Pv08 | 63-19 | Sartorato et al. (1999a), Sartorato et al. (2000) |
| Phg-22 | – | BAT 332 | MA | Pv08 | 63-39 | Namayanja et al. (2006) |
| Phg-3 | Phg-ON | Ouro Negro | MA | Pv04 | 63-39 | Corrêa et al. (2001), Gonçalves-Vidigal et al. (2013) |
| Phg-4 | PhgG5686A, ALS4.1GS,UC | G5686 | A | Pv04 | 31-0, 31-0 | Mahuku et al. (2009), Keller et al. (2015) |
| Phg-5 | ALS10.1DG,UC | CAL 143 | A | Pv10 | 0-39, field | Oblessuc et al. (2012, 2013) |
| Phg-5 | ALS10.1DG,UC,GS | G5686 | A | Pv10 | 31-0 | Keller et al. (2015) |
A, Andean; MA, Mesoamerican.
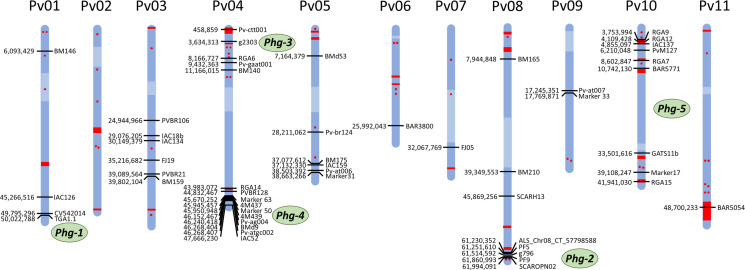
Methods of characterizing ALS resistance loci have changed over time. Initial work with random amplification of polymorphic DNA (RAPD), amplified fragment length polymorphism (AFLP), and restriction fragment length polymorphism (RFLP) markers was followed by sequence characterized amplified region (SCAR), simple sequence repeat (SSR), and single-nucleotide polymorphism (SNP) marker systems (Semagn et al., 2006; Jiang, 2013; Fang, 2015). The publication of the common bean reference genome (Schmutz et al., 2014) has permitted the mapping and comparison of the positions of most SCAR, SSR, and SNP markers (Fig. 1). In this section, we discuss the progress in ALS resistance characterization, focusing mainly on genetic mapping studies and remapping molecular markers linked to ALS resistance genes on the reference genome of common bean. For details on markers, see Supplemental Table S1.
Fig.1.
Genetic map showing positions of reported markers tagging angular leaf spot (ALS) resistance loci. Markers linked to ALS resistance loci are shown with their location mapped on the Phaseolus vulgaris reference genome version 2.1 (available at https://phytozome.jgi.doe.gov/pz/portal.html). The resistance genes approved by the Bean Improvement Cooperative Genetics Committee and their approximate positions are marked in green on the right side of the chromosome. Centromere regions are shown in light blue as reported in the reference genome (Schmutz et al., 2014). Resistance genes, containing an ARC domain (PF00931) are marked in red, with points if there are less than three loci and with bands if there are three or more loci. A summary of markers linked to resistance loci and their primer sequences is given in the Supplemental Table S1.
Phg-1
Origin
The Phg-1 locus was reported on chromosome Pv01, and it is tightly linked to the anthracnose resistance locus Co-14 in the Andean cultivar AND 277 (Gonçalves-Vidigal et al., 2011).
Molecular Markers
The Phg-1 and Co-14 loci are tightly linked (0.0 cM) to each other on chromosome Pv01 (Gonçalves-Vidigal et al., 2011). Two molecular markers, CV542014450 and TGA1.1570, flanking the Co-14/Phg-1 loci were identified as linked at 0.7 and 1.3 cM, respectively.
Alleles
No alleles were reported.
Breeding Value
The cultivar AND 277, which was obtained from the cross of Andean cultivars G 21720 ´ BAT 1386, is an important ALS resistance source that has been used in breeding programs in Brazil and southern Africa (de Carvalho et al., 1998; Aggarwal et al., 2004; Arruda et al., 2008). AND 277 was reported to be resistant to P. griseola and C. lindemuthianum under field conditions during 2 yr of evaluations in Malawi (Aggarwal et al., 2004). The ALS-resistant Andean cultivar CAL143, derived from a cross G12229 ´ AND 277, may carry Phg-1 present in the AND 277 parent; however, there are no studies supporting this assertion (Aggarwal et al., 2004). CAL 143 is a high-yielding variety that has a strong level of resistance to ALS, rust, and halo blight (caused by Pseudomonas syringae pv. phaseolicola) under field and greenhouse conditions (Chataika et al., 2010; Oblessuc et al., 2012; Cichy et al., 2015).
Phg-2
Origin
The Phg-2 locus was discovered in Mesoamerican cultivar Mexico 54 as a single dominant resistance locus on chromosome Pv 08 (Sartorato et al., 1999a).
Molecular Markers
Sartorato et al. (1999a) reported that the RAPD markers OPN 02, OPN 14, and OPE 04 were linked to Phg-2 in Mexico 54 at 5.9, 6.6, and 11.8 cM, respectively. The SCAR marker N02, which was developed based on the SN02890 fragment, showed polymorphisms identical to the original RAPD OPN 02 marker (Sartorato et al., 1999b, Sartorato et al., 2000). However, the N02 marker was not polymorphic in other evaluated populations using Mexico 54 as the resistant parent (Namayanja et al., 2006). The polymerase chain reaction (PCR) marker g796, which is highly specific for Mexico 54, was linked to the Phg-2 locus at 3 cM distance (Miller et al., 2018).
Alleles
In addition to Mexico 54, other Mesoamerican cultivars including Cornell 49-242, MAR 2, G10474, BAT 332, and G10909 contain ALS resistance loci that map to the lower end of chromosome Pv08. . Physical position analysis using the common bean reference genome sequence (Schmutz et al., 2014) indicated that the ALS resistance in these cultivars may be conferred by alleles of Phg-2 (Souza et al., 2016). However, this information requires verification.
A single dominant resistance locus was found in cultivar Cornell 49-242 controlling resistance to race 31-17 (Nietsche et al., 2000; Caixeta et al., 2005). The OPN02890 and OPE04650 RAPD markers linked to Phg-2 in Mexico 54 were also found to be linked to the resistance locus in Cornell 49-242 at 3.2 and 12.5 cM, respectively (Nietsche et al., 2000). Given that the same markers are linked to ALS resistance in Mexico 54 and Cornell 49-242, these loci might be allelic (Nietsche et al., 2000). OPE04650 was also found to be linked at 5.8 cM to a single dominant resistance locus in MAR 2 conferring resistance to race 63-39 (Ferreira et al., 2000).
Two genes in bean genotype G10909, on chromosomes Pv04 and Pv08 confer resistance to the highly virulent race 63-63 (Mahuku et al., 2011). The resistance gene PhgG10909B on chromosome Pv08 was found to cosegregate with SCAR markers PF13310, PF9260, and OPE04709 at 4.9, 7.4, and 9.9 cM, respectively (Mahuku et al., 2011).
A single and dominant gene in cultivar BAT 332 linked to RAPD markers OPAA07950 and OPAO12950 at 5.10 and 5.83 cM, respectively, confers resistance to race 61-41 (Caixeta et al., 2003). Additionally, an allelism test between Mexico 54 and BAT 332 inoculated with race 63-39 showed no segregation, which is an indication that the loci conferring ALS resistance in Mexico 54 and BAT 332 are allelic (Namayanja et al., 2006). Angular leaf spot resistance in BAT 332 has been designated as the Phg-22 allele. At present, Phg-22 is the only allele of Phg-2 officially accepted by the BIC Genetics Committee.
A single dominant resistance gene in Mesoamerican common bean accession G10474 was linked to the codominant SCAR marker PF5, positioned 5.0 cM from the resistance locus. The gene-pool-specific PF5 marker can be used to transfer resistance from G10474 to Andean common bean cultivars (Mahuku et al., 2004). In addition, another highly specific marker, ALS_Chr08_ CT_57798588, found through whole-genome sequencing of G10474, can be used in marker-assisted selection (MAS) (Lobaton et al., 2018).
In summary, the known physical positions of linked markers suggest that the ALS resistance genes in MAR 2, Cornell 49-242, G10474, and G10909 are either alleles of Phg-2 from Mexico 54, or they may represent different loci within a resistance gene cluster. Allelism studies have only confirmed that the resistance loci in BAT 332 and MAR 2 are allelic to Phg-2 (Caixeta et al., 2005; Namayanja et al., 2006). Further genomic characterization of the Phg-2 locus is necessary to clarify allelic relationships or the presence of potentially different genes within the locus.
Breeding Value
The Phg-2 locus found in Mexico 54 and its potential alleles reported in various Mesoamerican cultivars confer the broadest known resistance. These alleles are present in several bean cultivars used in ALS resistance breeding. Mexico 54 has been extensively used in breeding and research, and it is very resistant to African isolates of P. griseola, though this cultivar was resistant to only 5 of 19 Colombian races used in one study (CIAT, 2005; Chilagane et al., 2013; Ng’ayu-Wanjau, 2013; Ddamulira et al., 2015). The putative Phg-2 allele of G10474 confers broad resistance to most races of P. griseola, and it has been extensively used in breeding at CIAT. G10474 was found to be resistant to most races screened under greenhouse conditions. Only races from Haiti and South Africa caused disease in this cultivar (Mahuku et al., 2004, CIAT, 2005).
Phg-3
Origin
The Phg-3 ALS resistance locus was first reported in the Mesoamerican common bean cultivar Ouro Negro (Corrêa et al., 2001). A later study reported that Phg-3 cosegregates with the anthracnose resistance locus Co-34 (previously named Co-10), and that Phg-3/Co-34 alleles were tightly linked to each other (0.0 cM) on chromosome Pv04 (Gonçalves-Vidigal et al., 2013).
Molecular Markers
Gonçalves-Vidigal et al. (2013) reported that the Phg-3/ Co-34 resistance alleles were tightly linked to the marker g2303, at 0.0 cM on Pv04, enabling the use of MAS to transfer the ALS and anthracnose resistance cluster to commercial bean cultivars.
Alleles
The resistance locus PhgG10909A in the Mesoamerican cultivar G10909 is located on chromosome Pv04, 13 cM from microsatellite marker Pv-gaat001 (Mahuku et al., 2011). PhgG10909A and Phg-3 are in the same region, though no allelism tests between these loci have been conducted.
Breeding Value
Ouro Negro is a highly productive black-seeded Mesoamerican cultivar with desirable agronomic and cooking characteristics that was selected from CIAT accession G3680, also known as Honduras 35 (Alzate-Marin et al., 2003). The Phg-3 ALS, Co-34 anthracnose, and Ur-14 rust resistance alleles present in Ouro Negro are very important for common bean breeding programs in Brazil (Alzate-Marin et al., 2003; Souza et al., 2011; Gonçalves-Vidigal et al., 2013), conferring resistance to at least 21 C. lindemuthianum races and seven P. griseola races, including highly virulent race 63-63 (Faleiro et al., 2001, 2003; Gonçalves-Vidigal and Kelly, 2006; Gonçalves-Vidigal et al., 2009, 2013; Ragagnin et al., 2009).
Phg-4
Origin
The Phg-4 locus, previously named PhgG5686A, was discovered in the Andean cultivar G5686 inoculated with P. griseola race 31-0 (Mahuku et al., 2009). Keller et al. (2015) used a fine-mapping approach to characterize and delimit the QTL on chromosome Pv-04 in G5686 and named it ALS4.1GS, UC. Because of the consistent and significant effects of this major locus across different environments and populations (Mahuku et al., 2009; Keller et al., 2015), the BIC Genetics Committee has approved the name Phg-4 for the ALS4.1 QTL (Souza et al., 2016).
Molecular Markers
The Phg-4 locus was found to be linked, at 0.0 cM, to the microsatellite marker Pv-ag004 from G5686 (Mahuku et al., 2009). Keller et al. (2015) used fine mapping to investigate the Phg-4 locus in detail, delimiting it to a 418-kb genomic region between markers Marker63 and 4M439. The delimited region includes 36 candidate genes, including 11 serine/ threonine protein kinases arranged in a repetitive array, which are promising candidate genes for ALS resistance. Single nucleotide polymorphism-based markers highly specific to Phg-4 in G5686 are available on several genotyping platforms (Keller et al., 2015; Lobaton et al., 2018)
Alleles
Using a CAL 143 (resistant) ´ IAC-UNA (susceptible) cross, Oblessuc et al. (2012) reported two adjacent QTLs, ALS4.1GS, UC delimited by markers IAC52 and BMd9, and ALS4.2GS, UC delimited by markers PVBR92 and Pv-gaat001. Although these two QTLs were reported as being close to the Phg-4 locus, they have not been approved by the BIC Genetics Committee as alleles of Phg-4.
Breeding Value
G5686 has been inoculated with >500 isolates of P. griseola from 27 countries and found to be one of the most resistant genotypes. G5686 is currently being used in breeding line development at CIAT.
Phg-5
Origin
The resistance locus Phg-5 on chromosome Pv10 has been found in two different Andean common bean cultivars: CAL 143 and G5686. The Phg-5 locus was first reported in a CAL 143 ´ IAC-UNA recombinant inbred line (RIL) population evaluated in the field under natural infection and inoculated under greenhouse conditions with P. griseola race 0-39 (Oblessuc et al., 2012). Phg-5, previously named QTL ALS10.1, exhibits a strong effect in booth, field, and greenhouse environments. Keller et al. (2015) confirmed the presence of Phg-5 in common bean accession G5686. Besides a rough positional analysis, there is no evidence that Phg-5 in CAL 143 and G5686 is the same allele or that they are different genes.
Molecular Markers
The closely linked markers GATS11b and IAC137 flank the Phg-5 locus in CAL 143 (Oblessuc et al., 2012). Although the two markers are closely linked, the physical positions of GATS11b (33.50 Mb) and IAC137 (4.86 Mb) are very far apart; this range coincides with the centromeric region of chromosome Pv10, which shows little recombination (Schmutz et al., 2014). Oblessuc et al. (2013) increased the marker density around the Phg-5 locus and identified ATA220 marker, which coincided with the peak LOD score, but ATA220 did not map to the common bean genome reference. Investigation of the transcriptional modulation of the Phg-5 region revealed an enrichment of genes involved in plant–pathogen interactions, and seven of the 323 genes located in the core region were found to be differentially regulated after infection (Oblessuc et al., 2015). Keller et al. (2015) reported a minor QTL linked to Marker17 on chromosome Pv10 in G5686, which was designated as Phg-5.
Alleles
López et al. (2003) reported four resistance gene analogs on chromosome Pv10 linked to ALS resistance in a RIL population from the G19833 ´ DOR364 cross. Another minor QTL was found to be associated with marker BAR5771 in AND 277 on chromosome Pv10, conferring resistance to race 1-21 (Bassi et al., 2017).
Breeding Value
CAL 143 has been used extensively in breeding, and it is a popular variety that has been released in several African countries, including under the name of Lyambai in Zambia. G5686 is one of the most resistant Andean genotypes known, and it carries the resistance locus Phg-5, but it also contains Phg-4 (Mahuku et al., 2009, Keller et al., 2015).
OTHER REPORTED ANGULAR LEAF SPOT RESISTANCE LOCI TAGGED BY MARKERS
Besides the well-characterized five ALS resistance loci (Phg-1 to Phg-5), other loci have been reported, but they showed either a weak effect on resistance or there was not sufficient evidence to validate their existence and to assign them a Phg symbol.
Chromosome Pv01
In phenotypic evaluations under field conditions of a Jalo EEP 558 (resistant) ´ Small White (susceptible) cross, Teixeira et al. (2005) found an ALS QTL linked to marker BM146 of Jalo EEP 558, which they reported to be on chromosome Pv05. However, this marker has been mapped to the upper end of chromosome Pv01 on the reference genome, with Phg-1 positioned at the lower end of the same chromosome (Fig. 1).
Chromosome Pv03
Using an RIL population from the CAL 143 ´ IAC-UNA cross, Oblessuc et al. (2012) found a minor QTL on chromosome 3 flanked by markers PVBR21 and FJ19. These authors also reported an ALS resistance locus on chromosome Pv02 flanked by markers IAC134 and IAC18b. However, these markers have been mapped to chromosome Pv03 of the reference genome, in proximity to other reported markers (Fig. 1). The resistant allele at this locus is derived from CAL 143.
Chromosome Pv05
Oblessuc et al. (2012) also discovered two additional QTLs on chromosome Pv05 in the CAL 143 ´ IAC-UNA cross. The QTL ALS5.1UC was flanked by markers BMd53 and FJ05, and QTL ALS5.2UC, which exhibited a strong effect under greenhouse conditions, was flanked by markers BM175 and IAC261. By mapping the above mentioned markers onto the reference genome, BMd53 and BM175 were confirmed to be located on chromosome Pv05, however; FJ05 and IAC261 were mapped to chromosomes Pv07 and Pv01, respectively (Fig. 1). In another study using RILs from the AND 277 (resistant) ´ SEA 5 cross (susceptible) inoculated with race 1-21, a minor QTL was found associated with marker IAC159 on chromosome Pv05 (Bassi et al., 2017). Quantitative trait locus mapping in G5686 ´ Sprite revealed a minor QTL in the same genomic region that explained 3.7% of the variance associated with Marker 31 (Keller et al., 2015).
Chromosome Pv06
In the AND 277 ´ SEA 5 RIL population, another minor QTL associated with marker BAR3800 was found on chromosome 6 (Bassi et al., 2017).
Chromosome Pv08
An ALS resistance QTL was found on the upper arm of chromosome Pv08, opposite to Phg-2. Teixeira et al. (2005) found markers BM210 and BM165 to be linked to ALS resistance in Jalo EEP 558. Marker BM165 was initially reported to be on chromosome Pv05, though it maps to chromosome 8 (Fig. 1).
Chromosome Pv09
A study of G5686 (resistant) ´ Sprite (susceptible) cross revealed a locus named PhgG5686C on chromosome Pv09 linked, at 12.1 cM, to marker Pv-at007 (Mahuku et al., 2009). This locus was confirmed as a minor QTL explaining 1.7% of the variance linked to Marker 33 (Keller et al., 2015).
Chromosome Pv11
Bassi et al. (2017) reported a major QTL conferring ALS resistance on chromosome Pv-11, explaining 26.5% of the observed phenotypic variance using RIL of AND 277 ´ SEA 5. Marker BAR5054 was associated with ALS resistance in AND 277, and also with susceptibility to powdery mildew (caused by Erysiphe poligoni DC) in AND 277 (Bassi et al., 2017).
SEGREGATION AND ALLELISM STUDIES
In addition to the abovementioned studies reporting molecular markers linked to ALS resistance loci, numerous other segregation studies have been conducted. In these studies, resistance sources were crossed to susceptible varieties or to other resistance sources, and segregation ratios were analyzed to draw conclusions about the inheritance or allelism of resistance loci (Caixeta et al., 2005; CIAT, 2006; Namayanja et al., 2006; Mahuku et al., 2011). However, these studies were often deficient and did not always support their conclusions. First, ALS resistance is often quantitative; thus, the classification of ALS responses into resistant and susceptible categories is not suitable because the score distribution of resistant and susceptible plants does not segregate into two well-defined groups (Teixeira et al., 2005; Chataika et al., 2010; Oblessuc et al., 2012). Second, when evaluations are conducted using single F2 plants, errors can be introduced during phenotyping and due to hybridization problems. Finally, distinct interactions between the resistance loci in common bean and isolates of P. griseola exist; thus, the type of resistance loci observed are contingent on the isolate of P. griseola that is used. Taken together, the diversity of isolates of the ALS pathogen and technical and statistical issues of most published allelism studies render these results difficult to interpret, thus limiting the knowledge gained from such studies.
OUTLOOK ON GENETIC CHARACTERIZATION OF ANGULAR LEAF SPOT RESISTANCE
Methods to genetically characterize ALS resistance in common bean have changed substantially in recent years. For instance, the publication of the reference genome of common bean has allowed for the assignment of positions for most SCAR, SSR, and SNP markers and the comparison of loci obtained from different studies. For several genotypes, resistance loci have been mapped to similar positions on the genome. Although mapping studies are often complemented by allelism and segregation analyses, some allelism studies are difficult to interpret and frequently report more and different genes involved in resistance. Overall, additional studies are needed to resolve these discrepancies.
The identification of resistance genes will be a major goal for geneticists in order to understand the nature of defense genes and to define haplotypes for marker design to aid in breeding. In this respect, the locus best characterized to date is Phg-5 where the expression of its candidate genes has been investigated (Oblessuc et al., 2015). However, fine mapping of Phg-5 has been hindered by the partial localization of the core QTL region to a pericentromeric region that spans several megabases and where little recombination occurs. In contrast, the Phg-4 locus has been mapped to a much smaller region in which 36 candidate genes have been annotated (Keller et al., 2015). Although the whole genome sequencing data of G5686 is available, the repetitive nature of the Phg-4 region poses a barrier to de novo assembly and to the correct identification of all copies of potential resistance genes and polymorphisms. Other technologies, such as 10´ sequencing (www.10xgenomics.com), PacBio (www.pacb.com), and Oxford Nanopore (www.nanoporetech.com), are promising for overcoming the issues of short read assemblies and will allow researchers to assemble repetitive genomic regions more accurately.
Next-generation sequencing technologies have reduced sequencing costs, provided opportunities for additional genotyping, and allowed agricultural research scientists to perform a wide variety of applications, such as high-throughput genotyping by sequencing (GBS), whole-genome sequencing (WGS), genome-wide association studies (GWAS), and genomic selection. So far, most ALS resistance studies published have been conducted on biparental mapping populations with associated SNPs that often are polymorphic only in segregating populations from crosses between Andean and Mesoamerican cultivars. Genome-wide association studies will allow for finding resistance loci in a more diverse genetic background than biparental mapping, where allelic diversity is limited.
Genome-wide association studies have been used to explore the genetic basis of disease resistance, to identify new genomic regions controlling resistance, and to find molecular markers associated with ALS resistance in common bean (Perseguini et al., 2016; Zuiderveen et al., 2016; Tock et al., 2017). Perseguini et al. (2016) using GWAS detected 17 and 11 significant marker–trait associations, on a 0.05 significance level, for ALS and anthracnose resistance loci, respectively. Significantly, associated markers were distributed on most chromosomes of the genome. These authors reported that their results indicated a quantitative and complex inheritance pattern for the ALS and anthracnose diseases of common bean. Conversely, Nay et al. (2018) conducted GWAS in a large common bean panel and tested it with a mixture of five races of P. griseola in the field in Darien, Colombia, and with one race (63-47) in the greenhouse. A single ALS resistance locus on chromosome Pv08 was conferring resistance in the field and greenhouse trials. The GWAS results in this study suggested a qualitative nature of the ALS resistance, and a SNP that clearly distinguished resistant from susceptible bean lines was reported (Nay et al., 2018). More GWAS studies are underway to evaluate the pathotype-specific effect of resistance against prevalent races of P. griseola in Colombia and Uganda (M.M. Nay, unpublished data, 2018). These studies will give insights into the effectiveness of ALS resistance loci on different continents. High-density marker data obtained using GBS or WGS will enable the selection of markers that specifically tag ALS resistance loci, unlike previously developed markers that were effective in two-parental study populations but have been ineffective when used in breeding populations.
BREEDING OF ANGULAR LEAF SPOT-RESISTANT CULTIVARS
Conventional ALS resistance breeding is based on the selection of resistant lines under field conditions using natural or artificial inoculation. Characterization of ALS resistance loci has advanced in recent years, and molecular markers linked to resistance alleles in some of the most resistant donor genotypes are available. Moreover, MAS allows for the selection of resistant lines from segregating populations using genetic analysis instead of phenotypic screening (reviewed in Miklas et al., 2006). Conversely, DNA-based selection techniques, such as marker-assisted and genomic selection, have not been routinely applied in common bean breeding, which still depends heavily on traditional techniques. In the next sections, we review some of the ALS resistance breeding strategies implemented in different breeding programs in the tropics, where ALS is a recurrent and severe disease.
BREEDING FOR ANGULAR LEAF SPOT RESISTANCE AT CIAT HEADQUARTERS IN CALI, COLOMBIA
Breeding for resistance to ALS has long been a major objective of the bean program at CIAT in Cali, Colombia. Large collections of 22,832 wild and cultivated common beans have been screened in the field using local pathogen isolates, and highly resistant sources such as G10909, G10474, and G5686 have been identified (Pastor-Corrales et al., 1998; Mahuku et al., 2003). Pre-breeding lines of the Mesoamerican small red-seeded grain class were created by crosses and backcrosses with identified resistance sources, and ALS resistance was introgressed into elite breeding germplasm by subsequent crossing and selection under field conditions.
In the Andean breeding program, resistance sources AND 277 and CAL 143 of the Andean gene pool were used for introgressing ALS resistance. Recently, transfer of the ALS resistance of Mesoamerican origin into Andean bush types was attempted through selection by crossing Andean elite lines with ALS-resistant Mesoamerican pre-breeding lines (Solarte, 2014). A new, highly specific marker (ALS_Chr08_CT_57798588) based on the whole genome sequence of the Mesoamerican bean G10474 is now available, and it has been validated to tag Phg2 specifically in G10474 and another Mesoamerican resistant accession G4691, but not in Mesoamerican breeding lines (Lobaton et al., 2018). Another project at CIAT is aimed at pyramiding five QTLs from the Andean (AND 277 and G5686) and Mesoamerican (G10474 and G10909) ALS resistance sources and to combine them with the well-accepted grain quality of CAL143 and KAT B1 cultivars. Single nucleotide polymorphism markers that tag ALS resistance genes have also been established at a commercial genotyping service, allowing for high-throughput screening of the progeny of crosses.
Taken together, efforts to generate ALS-resistant breeding lines at CIAT, Colombia, are ongoing due to the persistent need in Africa and other regions. Better molecular markers that are more specific, easier to use on a large scale, and tag different ALS resistance loci are becoming available and should increase the efficiency of ALS resistance breeding. Despite the availability of molecular tools, phenotypic evaluations under field conditions will remain an important method and the final quality check for selecting new varieties.
BREEDING FOR ANGULAR LEAF SPOT RESISTANCE IN BRAZIL: RECURRENT SELECTION STRATEGY
With the expansion of irrigated areas in Brazil, common bean has become a highly valuable crop that can be grown all year. Brazil is the largest producer and the largest consumer of common bean in the world (Beebe, 2012). The increase in production has also resulted in an increase in the incidence of common bean diseases, with ALS causing significant crop damage (Singh and Schwartz, 2010; de Paula Júnior et al., 2015). As the P. griseola races occurring in Brazil are some of the most aggressive, it is difficult to obtain common bean lines with durable resistance (Nietsche et al., 2001; Sartorato and Alzate-Marin, 2004; Silva et al., 2008; Balbi et al., 2009; Pereira et al., 2013; Sanglard et al., 2013).
One of the methods used for breeding durable ALS-resistant cultivars in Brazil is recurrent selection (Ramalho et al., 2012). In 1998, a phenotypic recurrent selection program was initiated in the state of Minas Gerais with the goal of obtaining elite lines that accumulate important ALS resistance alleles combined with high seed yield and the carioca grain type, the market class preferred by Brazilian consumers (Amaro et al., 2007; Arantes et al., 2010). The breeding program involved a circulating diallel cross of seven carioca-seeded lines and 10 sources of ALS resistance from different market classes including Andean and Mesoamerican common bean cultivars (AN 512561, AND 277, Ouro Negro, Compuesto Negro Chimaltenango, CAL 143, MAR 2, MAR 1, G5686, MA 4.137, and Jalo). In 2011, the progress of the breeding program was evaluated, and the five best lines from Cycles I to VII were evaluated in three locations in Minas Gerais (T.I.P.O. Souza, unpublished data, 2018). The mean ALS severity scores of the lines from each recurrent selection cycle ranged from 6.2 in Cycle I in Lambari to 3.2 in Cycle III in Patos de Minas. In addition, the interactions of lines ´ locations were significant for ALS severity, which suggested that the prevalent races of P. griseola differ among locations and/or that environmental conditions favor the development of the disease differently among locations. On average, in the three environments, the genetic progress for ALS severity estimated by the cycle of selection was −2.9% and for seed yield was 1.8%, confirming the efficiency of a recurrent selection program. The chosen strategy, therefore, appears to have been successful in accumulating alleles for ALS resistance and for high seed yield.
Furthermore, the 27 highest performing lines from different cycles were tested in different years in the final field trials for agronomic performance evaluation of elite common bean lines conducted by Universidade Federal de Lavras–EMBRAPA in Minas Gerais. The lines MAIV 18-529 and MAIV 18-524 were selected as the most promising, and they have been used as parents (ALS elite resistance sources) in elite crosses in different breeding programs in Brazil.
BREEDING FOR ANGULAR LEAF SPOT RESISTANCE IN EAST AFRICA
Angular leaf spot is one of the most important diseases of common bean in East Africa (Pastor-Corrales et al., 1998), with annual yield losses in Africa up to 384,200 metric tons, as estimated by Wortmann et al. (1998). In fact, this disease has been associated with up to 50% yield losses among released bean varieties in Uganda (Pastor-Corrales et al., 1998; Opio et al., 2001; Namayanja et al., 2006). Moreover, progress in breeding for ALS resistance has been slow, mainly because of the high diversity of ALS pathogen races found in East Africa (Wagara et al., 2011; Ddamulira et al., 2014b; Leitich, 2016; Kijana et al., 2017). Research on ALS in East Africa has focused on the identification and genetic characterization of new sources of resistance in local landraces and released varieties (Ng’ayu-Wanjau, 2013; Ddamulira et al., 2014a; Leitich, 2016; Kijana et al., 2017). In these studies, Mexico 54 was found to be resistant to most races of P. griseola, and this cultivar is the most common source of ALS resistance in breeding in Uganda and East Africa (Namayanja et al., 2006; Chilagane et al., 2013; Ng’ayu-Wanjau, 2013; Ddamulira et al., 2015; Miller et al., 2018).
Marker-assisted selection can improve breeding efficiency by facilitating introgression of resistance loci into elite cultivars, and it allows pyramiding of resistance loci for more durable resistance. In the 2000 to 2010 period, the National Agricultural Research System (NARS) common bean breeding programs in East and Central Africa had little access to MAS facilities and hence collaborated with CIAT, which had established a simple but fully functional MAS laboratory at the National Agricultural Research Laboratories (NARKL) at Kawanda, Uganda. This laboratory was and is still used for MAS projects by CIAT in collaboration with NARS for targeting disease resistance, including resistance to ALS. For example, the SCAR marker OPE4709, developed by Mahuku et al. (2011), and which tags Phg-2 from Mexico 54, was used to select resistant progeny containing Phg-2. Using this marker, the prevalence of the Phg-2 locus in advanced lines was assessed, and it was found that 60% of lines from Rwanda and 13% of lines from Uganda harbor the resistance allele. The lines from Rwanda had previously undergone selection at an ALS hotspot in Rubona, which explains the relatively high frequency of lines carrying the Phg-2 resistance locus. Given the high diversity of P. griseola races found in Africa (Wagara et al., 2011; Ddamulira et al., 2014b; Leitich, 2016; Kijana et al., 2017), more than one resistance gene will be required to confer durable ALS resistance to a wide range of races. Ddamulira et al. (2015) pyramided resistance genes from AND 277 (Phg-1), Mexico 54 (Phg-2), and G5686 (Phg-4, Phg-5, and PhgG5686C) and introgressed the resistance into the susceptible cultivars Kanyebwa (local landrace) and CAL 96. The presence of the resistance loci was verified by molecular markers, and the resulting population showed reduced ALS symptom severity compared with single crosses when inoculated with race 61-63 (Ddamulira et al., 2015). Supported by MAS, breeding at CIAT Uganda focuses on combining ALS resistance with resistance to other stresses, and in one study, six genes for resistance to anthracnose, Pythium, viruses, and ALS were pyramided into four genotypes (Okii et al., 2017).
Angular leaf spot studies at CIAT Uganda have heavily relied on student projects, but the obtained knowledge about genes, markers, and new resistant germplasm is used in systematic breeding, supporting phenotypic selection in germplasm development. However, reliance on Mexico 54 as the major source of resistance has resulted in less-than-adequate progress in the desired market classes. This might be caused by the undesired characteristics introduced through crossing the small-seeded Mesoamerican cultivar Mexico 54 with large-seeded Andean beans, which are the preferred grain type in East Africa. Identification of additional effective ALS resistance sources from the Andean gene pool for use in breeding for resistance in East Africa will reduce the linkage drag and accelerate breeding progress.
OUTLOOK ON BREEDING FOR RESISTANCE TO ANGULAR LEAF SPOT
Breeding common bean cultivars with durable resistance to P. griseola will continue to be a high priority in many countries of Latin America and Africa, where ALS is a recurring disease. However, durable resistance is difficult to achieve due to the extensive virulence diversity of the ALS pathogen and its capacity to produce new virulent strains. In this review, we examined the components of a durable strategy for resistance to ALS, which includes a review of the virulence diversity of the ALS pathogen, the existing situation of ALS resistance loci, and molecular markers linked to these loci for use in resistance breeding. We also consider how new technologies could facilitate and accelerate ALS resistance breeding but also disease resistance breeding in common bean in general. Different populations of the ALS pathogen have co-evolved separately with Andean and Mesoamerican common beans, such that there are distinct Andean and Mesoamerican races of the ALS pathogen. A similar pattern was observed in the common bean diseases anthracnose and rust. In rust, combining Andean and Mesoamerican resistance genes in single cultivars has resulted in resistance to all known races of the rust pathogen (Pastor-Corrales et al., 2007).
These results have important implications for the development and deployment of ALS-resistant varieties. Even though there are three Andean and two Mesoamerican ALS resistance loci, there are no common bean cultivars grown by farmers that combine Andean and Mesoamerican resistance loci that would allow evaluating whether these combinations of genes confer broad and durable resistance. The extensive virulence diversity of the ALS pathogen suggests that common bean cultivars with single genes for resistance to ALS will likely succumb to new virulent races of the ALS pathogen in the future. This has frequently been observed in bean cultivars harboring single genes for resistance to the rust and anthracnose pathogens (Kelly et al., 1994; del Río et al., 2003; Pastor-Corrales et al., 2010). In addition, there is ample evidence that the virulence diversity of the ALS pathogen is changing. New races that overcome the resistance of all differential cultivars have been found in Brazil, Central America, and, more recently, in Africa. This situation calls for a breeding strategy based on a broad diversity of quantitative and qualitative resistance, which may confer broad-spectrum and durable resistance (St. Clair, 2010). Breeding, however, is currently based on a few well-characterized resistance loci: the Mesoamerican Mexico 54 with Phg-2 in Africa, the Mesoamerican Ouro Negro with Phg-3 in Brazil, the Andean G5686 with Phg-4, and the Mesoamerican G10474 likely with Phg-2 in Colombia. These single resistance genes are easy to transfer to new commercial cultivars, but they are also at risk of losing their resistance to new virulence races of the ALS pathogen. Thus, there is a need to discover new Andean and Mesoamerican ALS resistance genes to broaden the genetic base of common bean against the highly virulent ALS pathogen. There is also the need to learn about the spectrum of resistance of each of the ALS resistance genes by challenging them with a broad diversity of Andean and Mesoamerican races of the pathogen, and to combine (pyramid) various effective genes into commercial cultivars. Releasing and disseminating new superior varieties will stabilize bean productivity in vulnerable populations and will positively affect the livelihoods of the producers that depend on this crop for nutrition and income.
Supplementary Material
Acknowledgments
The authors thank the organizers (M.A. Pastor-Corrales, P.N. Miklas, T. Porch, and D. Fourie) of the Common Bean Disease Workshop on Angular Leaf Spot and Root Rot for offering us the opportunity to write this review. M.M. Nay is supported by ETH Global, the Sawiris Foundation for Social Development, and the Molecular Plant Breeding Group of ETH Zürich. T.L.P.O. Souza, M.C. Gonçalves-Vidigal, Â.F.B. Abreu, and L.C. Melo are supported by CNPq, the Brazilian Council for Scientific and Technological Development. B. Raatz and C.M. Mukankusi would like to acknowledge funding support from the Tropical Legumes III—Improving Livelihoods for Smallholder Farmers: Enhanced Grain Legume Productivity and Production in Sub-Saharan Africa and South Asia (OPP1114827) projects funded by the Bill & Melinda Gates Foundation. M.A. Pastor-Corrales was supported, in part, by funding from the Norman Borlaug Commemorative Research Initiative of the US Agency for International Development (Project no. 0210-22310-004-96R), and by the USDA-ARS (Project no. 8042-22000-286-00D).
Supplemental Material Available
Supplemental material is available online for this article.
Conflict of Interest
The authors declare that there is no conflict of interest.
References
- Aggarwal V.D., Pastor-Corrales M.A., Chirwa R.M., and Buruchara R.A.. 2004. Andean beans (Phaseolus vulgaris L.) with resistance to the angular leaf spot pathogen (Phaeoisariopsis griseola; ) in southern and eastern Africa. Euphytica: 136:201–210. doi: 10.1023/B:EUPH.0000030678.12073.a9 [DOI] [Google Scholar]
- Alzate-Marin A.L., Costa M.R., Arruda K.M., De Barros E.G., and Moreira M.A.. 2003. Characterization of the anthracnose resistance gene present in Ouro Negro (Honduras 35) common bean cultivar. Euphytica 133:165–169. doi: 10.1023/A:1025551006444 [DOI] [Google Scholar]
- Amaro G.B., Abreu Â.F.B., Ramalho M.A.P., and Silva F.B.. 2007. Phenotypic recurrent selection in the common bean (Phaseolus vulgaris L.) with carioca-type grains for resistance to the fungi Phaeoisariopsis griseola. Genet. Mol. Biol. 30:584–588. doi: 10.1590/S1415-47572007000400014 [DOI] [Google Scholar]
- Arantes L.D.O., Abreu Â.F.B., and Ramalho M.A.P.. 2010. Eight cycles of recurrent selection for resistance to angular leaf spot in common bean. Crop Breed. Appl. Biotechnol. 10:232–237. doi: 10.1590/S1984-70332010000300008 [DOI] [Google Scholar]
- Arruda K.M.A., Alzate-Marin A.L., Gomes Oliveira M.S., de Barros E.G., and Moreira M.. 2008. Inheritance studies for anthracnose resistance genes of common bean cultivar AND 277. Annu. Rep. Bean Improv. Coop. 51:170–171. [Google Scholar]
- Balbi B.P., Sanglard D.A., Arruda K.M.A., Costa M.R., Piovesan N.D., de Barros E.G., and Moreira M.A.. 2009. Characterization of Pseudocercospora griseolaisolates collected in the state of Minas Gerais, Brazil. Annu. Rep. Bean Improv. Coop. 52:56–57. [Google Scholar]
- Bassi D., Briñez B., Rosa J.S., Oblessuc P.R., De Almeida C.P., Nucci S.M., et al. 2017. Linkage and mapping of quantitative trait loci associated with angular leaf spot and powdery mildew resistance in common beans. Genet. Mol. Biol. 40:109–122. doi: 10.1590/1678-4685-gmb-2015-0314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beebe S.E. 2012. Common bean breeding in the tropics. In: Janick J, editor, Plant breeding reviews. Vol. 36 Wiley-Blackwell, Hoboken, NJ. doi: 10.1002/9781118358566.ch5 [DOI] [Google Scholar]
- Beebe S.E., and Pastor-Corrales. M.A.. 1991. Breeding for disease resistance. In: van Schoonhoven A. and Voysest O., editors, Common beans: Research for crop improvement. CAB Int., Wallingford, UK. [Google Scholar]
- Broughton W.J., Hernández G., Blair M., Beebe S., Gepts P., and Vanderleyden J.. 2003. Beans (Phaseolus spp.); model food legumes. Plant Soil 252:55–128. doi: 10.1023/A:1024146710611 [DOI] [Google Scholar]
- Busogoro J.P., Jijakli M.H., and Lepoivre P.. 1999a. Identification of a novel source of resistance to angular leaf spot disease of common bean within the secondary gene pool. Plant Breed. 118:417–423. doi: 10.1046/j.1439-0523.1999.00413.x [DOI] [Google Scholar]
- Busogoro J.P., Jijakli M.H., and Lepoivre P.. 1999b. Virulence variation and RAPD polymorphism in African isolates of Phaeoisariospis griseola (Sacc.) Ferr., the causal agent of angular leaf spot of common bean. Eur. J. Plant Pathol. 105:559–569. doi: 10.1023/A:1008707101645 [DOI] [Google Scholar]
- Caixeta E.T., Borém A., Alzate-Marin A.L., Fagundes S.D.A., Silva M.G.D.M.E., De Barros E.G., and Moreira M.A.. 2005. Allelic relationships for genes that confer resistance to angular leaf spot in common bean. Euphytica 145:237–245. doi: 10.1007/s10681-005-1258-3 [DOI] [Google Scholar]
- Caixeta E.T., Borém A., de Azevedo Fagundes S., Niestche S., de Barros E.G., and Moreira M.A.. 2003. Inheritance of angular leaf spot resistance in common bean line BAT 332 and identification of RAPD markers linked to the resistance gene. Euphytica 134:297–303. doi: 10.1023/B:EUPH.0000004948.41083.1f [DOI] [Google Scholar]
- Castellanos G., Jara C., and Mosquera G.. 2016. Phaeoisariopsis griseola: Managing the fungus in the laboratory In: Bean pathogens: Practical guide for lab and greenhouse work. 2nd ed. Publ. 420. CIAT, Cali, Colombia. [Google Scholar]
- Chacón M.I., Jara C., Castellanos G., Posso C.E., Buruchara R. Cuasquer , J.B., and Pastor-Corrales M.A.. 1997. Genetic diversity and relation between common bean angular leaf spot fungus isolates from Africa and South America: Genetic improvement implications. Annu. Rep. Bean Improv. Coop. 40:127–128. [Google Scholar]
- Chataika B., Bokosi J., Kwapata M., Chirwa R., Mwale V., Mnyenyembe P., and Myers J.. 2010. Performance of parental genotypes and inheritance of Angular Leaf Spot (Phaeosariopsis griseola) resistance in the common bean (Phaseolus vulgaris). Afr. J. Biotechnol. 9:4398–4406. [Google Scholar]
- Chilagane L.A., Tryphone G.M., Protas D., Kweka E., Kusolwa P.M., and Nchimbi-Msolla S.. 2013. Incorporation of resistance to angular leaf spot and bean common mosaic necrosis virus diseases into adapted common bean (Phaseolus vulgaris L.) genotype in Tanzania. Afr. J. Biotechnol. 12:4343–4350. doi: 10.5897/AJB2012.9651 [DOI] [Google Scholar]
- CIAT 1995. Bean Program annual report 1994. CIAT, Cali, Colombia. [Google Scholar]
- CIAT 1996. Bean Program annual report 1995. CIAT, Cali, Colombia. [Google Scholar]
- CIAT 1997. Bean Program annual report 1997. CIAT, Cali, Colombia. [Google Scholar]
- CIAT 1999. Meeting demand for beans for sub-Saharan Africa in sustainable ways: Annual report Project IP-2. CIAT, Cali, Colombia. [Google Scholar]
- CIAT 2005. Bean improvement for the tropics: Annual report. CIAT, Cali, Colombia. [Google Scholar]
- CIAT 2006. Bean improvement for the tropics: Annual report. CIAT, Cali, Colombia. [Google Scholar]
- Cichy K.A., Porch T.G., Beaver J.S., Cregan P., Fourie D., Glahn R.P., et al. 2015. A Phaseolus vulgaris diversity panel for Andean bean improvement. Crop Sci. 55:2149–2160. doi: 10.2135/cropsci2014.09.0653 [DOI] [Google Scholar]
- Correa F.J., and Saettler A.W.. 1987. Angular leaf spot of red kidney beans in Michigan. Plant Dis. 71:915–918. doi: 10.1094/PD-71-0915 [DOI] [Google Scholar]
- Corrêa R.X., Good-God P.I.V., Oliveira M.L.P., Nietsche S., Moreira M.A., and Barros E.G.D.E.. 2001. Inheritance of resistance to the common bean angular leaf spot and identification of molecular markers flanking the resistance locus. (In Portuguese, with English abstract.) Fitopatol. Bras. 26:27–32. [Google Scholar]
- Correa-Victoria F.J., Pastor-Corrales M.A., and Saettler A.W.. 1989. Angular leaf spot. In: Schwartz H.F. and Pastor-Corrales M.A., editors, Bean production problems in the tropics. CIAT, Cali, Colombia: p. 59–75. [Google Scholar]
- Crous P.W., Liebenberg M.M., Braun U., and Groenewald J.Z.. 2006. Re-evaluating the taxonomic status of Phaeoisariopsis griseola, the causal agent of angular leaf spot of bean. Stud. Mycol. 55:163–173. doi: 10.3114/sim.55.1.163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ddamulira G., Mukankusi C., Edema R., Sseruwagi P., and Gepts P.. 2014a. Identification of new sources of resistance to angular leaf spot among Uganda common bean landraces. Can. J. Plant Breed. 2:55–65. [Google Scholar]
- Ddamulira G., Mukankusi C., Ochwo-Ssemakula M., Edema R., Sseruwagi P., and Gepts P.. 2014b. Distribution and Variability of Pseudocercospora griseola in Uganda. J. Agric. Sci. 6:16–29. [Google Scholar]
- Ddamulira G., Mukankusi C., Ochwo-Ssemakula M., Edema R., Sseruwagi P., and Gepts P.. 2015. Gene pyramiding improved resistance to angular leaf spot in common bean. Am. J. Exp. Agric. 9:1–12. [Google Scholar]
- de Carvalho G.A., Paula Junior. T.J., Alzate-Marin A.L., Nietsche S., Barros E.G., and Moreira M.A.. 1998. Inheritance of resistance of the Andean bean line AND-277 to race 63-23 of Phaeoisariopsis griseola and identification of a RAPD marker linked to the resistance gene. Fitopatol. Bras. 23:482–485. [Google Scholar]
- de Jesus W.C. Junior, do Vale F.X. R.,Coelho R.R.,Hau B.,Zambolim L., Costa L.C., and Bergamin Filho A.. 2001. Effects of angular leaf spot and rust on yield loss of Phaseolus vulgaris. Phytopathology 91:1045–1053. doi: 10.1094/PHYTO.2001.91.11.1045 [DOI] [PubMed] [Google Scholar]
- de Paula T.J. Júnior, Vieira R.F., Teixeira H,Lobo Júnior M., and Wendland A.. 2015. Doenças do feijoeiro In: Carneiro J.E.D.S., et al., editors, Feijão do plantio à colheita. Fed. Univ. Viçosa, Viçosa, Brazil: p. 270–299. [Google Scholar]
- del Río L.E., Lamppa R.S., Gross P.L., Brolley B., and Prischmann J.. 2003. Identification of Colletotrichum lindemuthianum race 73 in Manitoba, Canada. Can. J. Plant Pathol. 25:104–107. doi: 10.1080/07060660309507055 [DOI] [Google Scholar]
- Faleiro F.G., Nietsche S., Ragagnin V.A., Borém A., Moreira M.A., and Barros E.G.. 2001. Resistance of common bean to rust and angular leaf spot under greenhouse conditions. (In Portuguese, with English abstract.) Fitopatol. Bras. 26:1995–1998. [Google Scholar]
- Faleiro F.G., Ragagnin V.A., Schuster I., Corrêa R.X., Good-God P.I., Brommonshenkel S.H., et al. 2003. Mapping rust, anthracnose and angular leaf spot resistance genes in common bean using RAPD markers. (In Portuguese, with English abstract.) Fitopatol. Bras. 28:59–66. doi: 10.1590/S0100-41582003000100009 [DOI] [Google Scholar]
- Fang D.D. 2015. Molecular breeding In: Fang D.D. and Percy R.G., editors, Cotton. 2nd ed. Agron. Monogr. 57; ASA, CSSA, and SSSA, Madison, WI: p. 255–288. doi: 10.2134/agronmonogr57.2013.0027 [DOI] [Google Scholar]
- Ferreira C.F., Borém A., Carvalho G.A., Nietsche S., Paula T.J. Jr., Barros E.G., and Moreira M.A.. 2000. Inheritance of angular leaf spot resistance in common bean and identification of a RAPD marker linked to a resistance gene. Crop Sci. 40:1130–1133. doi: 10.2135/cropsci2000.4041130x [DOI] [Google Scholar]
- Gonçalves-Vidigal M.C., Cruz A.S., Garcia A., Kami J., Vidigal Filho P.S., Sousa L.L., et al. 2011. Linkage mapping of the Phg-1 and Co-14 genes for resistance to angular leaf spot and anthracnose in the common bean cultivar AND 277. Theor. Appl. Genet. 122:893–903. doi: 10.1007/s00122-010-1496-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves-Vidigal M.C., Cruz A.S., Lacanallo G.F., Vidigal Filho P.S., Sousa L.L., Pacheco C.M.N.A., et al. 2013. Co-segregation analysis and mapping of the anthracnose Co-10 and angular leaf spot Phg-ON disease-resistance genes in the common bean cultivar Ouro Negro. Theor. Appl. Genet. 126:2245–2255. doi: 10.1007/s00122-013-2131-8 [DOI] [PubMed] [Google Scholar]
- Gonçalves-Vidigal M.C., and Kelly J.D.. 2006. Inheritance of anthracnose resistance in the common bean cultivar Widusa. Euphytica 151:411–419. doi: 10.1007/s10681-006-9164-x [DOI] [Google Scholar]
- Gonçalves-Vidigal M.C., Vidigal Filho P.S., Medeiros A.F., and Pastor-Corrales M.A.. 2009. Common bean landrace Jalo Listras Pretas is the source of a New Andean anthracnose resistance gene. Crop Sci. 49:133–138. doi: 10.2135/cropsci2008.01.0004 [DOI] [Google Scholar]
- Guzman P., Gilbertson R.L., Nodari R.O., Johnson W.C., Temple S.R., Mandala D., et al. 1995. Characterization of variability in the fungus Phaeoisariopsis griseola suggests coevolution with the common bean (Phaseolus vulgaris). Phytopathology 85:600–607. doi: 10.1094/Phyto-85-600 [DOI] [Google Scholar]
- Jiang G.L. 2013. Molecular markers and marker-assisted breeding in plants. In: Anderson S.B, editor, Plant breeding from laboratories to fields. IntechOpen, London. doi: 10.5772/52583 [DOI] [Google Scholar]
- Keller B., Manzanares C., Jara C., Lobaton J.D., Studer B., and Raatz B.. 2015. Fine-mapping of a major QTL controlling angular leaf spot resistance in common bean (Phaseolus vulgaris L.). Theor. Appl. Genet. 128:813–826. doi: 10.1007/s00122-015-2472-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly J.D., Afanador L., and Cameron L.S.. 1994. New races of Colletotrichum lindemuthianum in Michigan and implications in dry bean resistance breeding. Plant Dis. 78:892–894. doi: 10.1094/PD-78-0892 [DOI] [Google Scholar]
- Kijana R., Abang M., Edema R., Mukankusi C., Buruchara R., and Kivu S.. 2017. Prevalence of angular leaf spot disease and sources of resistance in common bean in eastern Democratic Republic of Congo. Afr. Crop Sci. J. 25:109–122. doi: 10.4314/acsj.v25i1.8 [DOI] [Google Scholar]
- Landeras E., Trapiello E., Braña M., and González A.J.. 2017. Occurrence of angular leaf spot caused by Pseudocercospora griseola in Phaseolus vulgaris in Asturias, Spain. Spanish J. Agric. Res. 15:e10SC03. doi: 10.5424/sjar/2017153-10798 [DOI] [Google Scholar]
- Leitich R.K. 2016. Mapping of angular leaf spot disease hotspot areas in western Kenya towards its management. Am. J. Appl. Sci. Res. 2:75–81. [Google Scholar]
- Liebenberg M.M., and Pretorius Z.A.. 1997. A review of angular leaf spot of common bean (Phaseolus vulgaris L). Afr. Plant Prot. 3:81–106. [Google Scholar]
- Lobaton J.D., Miller T., Gil J., Ariza D., de la Hoz J.F., Soler A., et al. 2018. Re-sequencing of common bean identifies regions of inter-gene pool introgression and provides comprehensive resources for molecular breeding. Plant Genome 11:170068. doi: 10.3835/plantgenome2017.08.0068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López C.E., Acosta I.F., Jara C., Pedraza F., Gaitan-Solis E., Gallego G., et al. 2003. Identifying resistance gene analogs associated with resistances to different pathogens in common bean. Phytopathology 93:88–95. doi: 10.1094/PHYTO.2003.93.1.88 [DOI] [PubMed] [Google Scholar]
- Mahuku G.S., Henríquez M.A., Montoya C., Jara C., Teran H., and Beebe S.. 2011. Inheritance and development of molecular markers linked to angular leaf spot resistance genes in the common bean accession G10909. Mol. Breed. 28:57–71. doi: 10.1007/s11032-010-9461-x [DOI] [Google Scholar]
- Mahuku G.S., Henríquez M.A., Munõz J., and Buruchara R.A.. 2002a. Molecular markers dispute the existence of the Afro-Andean group of the bean angular leaf spot pathogen, Phaeoisariopsis griseola. Phytopathology 92:580–589. doi: 10.1094/PHYTO.2002.92.6.580 [DOI] [PubMed] [Google Scholar]
- Mahuku G.S., Iglesias A.M., and Jara C.. 2009. Genetics of angular leaf spot resistance in the Andean common bean accession G5686 and identification of markers linked to the resistance genes. Euphytica 167:381–396. doi: 10.1007/s10681-009-9897-4 [DOI] [Google Scholar]
- Mahuku G.S., Jara C., Cajiao C., and Beebe S.. 2003. Sources of resistance to angular leaf spot (Phaeoisariopsis griseola) in common bean core collection, wild Phaseolus vulgaris and secondary gene pool. Euphytica 130:303–313. doi: 10.1023/A:1023095531683 [DOI] [Google Scholar]
- Mahuku G.S., Jara C., Cuasquer J.B., and Castellanos G.. 2002b. Genetic variability within Phaeoisariopsis griseola from Central America and its implications for resistance breeding of common bean. Plant Pathol. 51:594–604. doi: 10.1046/j.1365-3059.2002.00742.x [DOI] [Google Scholar]
- Mahuku G.S., Montoya C., Henríquez M.A., Jara C., Teran H., and Beebe S.. 2004. Inheritance and characterization of angular leaf spot resistance gene present in common bean accession G 10474 and identification of an AFLP marker linked to the resistance gene. Crop Sci. 44:1817–1824. doi: 10.2135/cropsci2004.1817 [DOI] [Google Scholar]
- Melzer M.S., and Boland G.J.. 2001. First report of angular leaf spot caused by Phaeoisariopsis griseola on bean in Ontario, Canada. Plant Dis. 85:919. doi: 10.1094/PDIS.2001.85.8.919D [DOI] [PubMed] [Google Scholar]
- Miklas P.N., Kelly J.D., Beebe S.E., and Blair M.W.. 2006. Common bean breeding for resistance against biotic and abiotic stresses: From classical to MAS breeding. Euphytica 147:105–131. doi: 10.1007/s10681-006-4600-5 [DOI] [Google Scholar]
- Miller T., Gepts P., Kimmo S., Arunga E., Chilagane L.A., Nchimbi-Msolla S., et al. 2018. Alternative markers linked to the Phg-2 angular leaf spot resistance locus in common bean using the Phaseolus genes marker database. Afr. J. Biotechnol. 17:818–828. doi: 10.5897/AJB2018.16493 [DOI] [Google Scholar]
- Namayanja A., Buruchara R., Mahuku G., Rubaihayo P., Kimani P., Mayanja S., and Eyedu H.. 2006. Inheritance of resistance to angular leaf spot in common bean and validation of the utility of resistance linked markers for marker assisted selection outside the mapping population. Euphytica 151:361–369. doi: 10.1007/s10681-006-9158-8 [DOI] [Google Scholar]
- Nay M.M., Buendia H.F., Portilla A.E., Studer B., and Raatz B.. 2018. Angular leaf spot resistance: GWAS of field and greenhouse screenings in Colombia. Annu. Rep. Bean Improv. Coop. 61:5–6. [Google Scholar]
- Ng’ayu-Wanjau B.N. 2013. Breeding for durable resistance to angular leaf spot (Pseudocercospora griseola) in common bean (Phaseolus vulgaris) in Kenya. Ph.D. diss., Univ. KwaZulu-Natal, Durban, Republic of South Africa. [Google Scholar]
- Nietsche S., Borém A., Assis De Carvalhos G., De Paula Júnior T.J., Fortes Ferreira C., Gonçalves De Barros E., and Alves Moreira M.. 2001. Genetic diversity of Phaeoisariopsis griseola in the state of Minas Gerais, Brazil. Euphytica 117:77–84. doi: 10.1023/A:1004096421990 [DOI] [Google Scholar]
- Nietsche S., Borém A., Carvalho G.A., Rocha R.C., Paula T.J., De Barros E.G., and Moreira M.A.. 2000. RAPD and SCAR markers linked to a gene conferring resistance to angular leaf spot in common bean. J. Phytopathol. 148:117–121. doi: 10.1046/j.1439-0434.2000.00479.x [DOI] [Google Scholar]
- Oblessuc P.R., Baroni R.M., Garcia A.A.F., Chioratto A.F., Carbonell S.A.M., Camargo L.E.A., and Benchimol L.L.. 2012. Mapping of angular leaf spot resistance QTL in common bean (Phaseolus vulgaris L.) under different environments. BMC Genet. 13:50. doi: 10.1186/1471-2156-13-50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oblessuc P.R., Matiolli C.C., Chiorato A.F., Camargo L.E.A., Benchimol-Reis L.L., and Melotto M.. 2015. Common bean reaction to angular leaf spot comprises transcriptional modulation of genes in the ALS10.1 QTL. Front. Plant Sci. 6:152. doi: 10.3389/fpls.2015.00152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oblessuc P.R., Perseguini J.M.K.C., Baroni R.M., Chiorato A.F., Carbonell S.A.M., Mondego J.M.C., et al. 2013. Increasing the density of markers around a major QTL controlling resistance to angular leaf spot in common bean. Theor. Appl. Genet. 126:2451–2465. doi: 10.1007/s00122-013-2146-1 [DOI] [PubMed] [Google Scholar]
- Okii D., Tukamuhabwa P., Tusiime G., Talwana H., Odong T., Mukankusi C., et al. 2017. Agronomic qualities of genetic pyramids of common bean developed for multiple-disease-resistance. Afr. Crop Sci. J. 25:457–472. doi: 10.4314/acsj.v25i4.5 [DOI] [Google Scholar]
- Opio F., Ugen M.A., Kyamanywa S., David S., and Mugisa-Mutetikka M.. 2001. Beans In: Mukiibi J.K., editor, Agriculture in Uganda: Crops II. Fountain Publ., Kampala, Uganda: p. 162–191. [Google Scholar]
- Pastor-Corrales M.A. 1996. Traditional and molecular confirmation of the coevolution of beans and pathogens in Latin America. Annu. Rep. Bean Improv. Coop. 39:46–47. [Google Scholar]
- Pastor-Corrales M.A., and Aime M.C.. 2004. Differential cultivars and molecular markers segregate isolates of Uromyces appendiculatus into two distinct groups that correspond to the gene pools of their common bean hosts. Phytopathology 94:82.18943823 [Google Scholar]
- Pastor-Corrales M.A., and Jara C.E.. 1995. The evolution of Phaeoisariopsis griseola with the common bean in Latin America. Fitopatol. Colomb. 1:15–24. [Google Scholar]
- Pastor-Corrales M.A., Jara C., and Singh S.P.. 1998. Pathogenic variation in, sources of, and breeding for resistance to Phaeoisariopsis griseola causing angular leaf spot in common bean. Euphytica 103:161–171. doi: 10.1023/A:1018350826591 [DOI] [Google Scholar]
- Pastor-Corrales M.A., Kelly J.D., Steadman J.R., Lindgren D.T., Stavely J.R., and Coyne D.P.. 2007. Registration of six great northern bean germplasm lines with enhanced resistance to rust and bean common mosaic and necrosis potyviruses. J. Plant Reg. 1:77–79. doi: 10.3198/jpr2005.12.0517crg [DOI] [Google Scholar]
- Pastor-Corrales M.A., Rayapati J., Osorno J.M., Kelly J.D., Wright E.M., Brick M.A.G., et al. 2010. Reaction of common bean cultivars to two new races of the rust pathogen from Michigan and North Dakota. Annu. Rep. Bean Improv. Coop. 53:64–65. [Google Scholar]
- Pereira R., de Souza E.A., Barcelos Q.L., and Abreu Â.F.B.. 2013. Evaluation of resistance in common bean genotypes to the causal agent of angular leaf spot. Annu. Rep. Bean Improv. Coop. 56:33–34. [Google Scholar]
- Pereira R., Souza E.A., Barcelos Q.L, Abreu Â.F.B., and Librelon S.S.. 2015. Aggressiveness of Pseudocercospora griseola strains in common bean genotypes and implications for genetic improvement. Genet. Mol. Res. 14:5044–5053. doi: 10.4238/2015.May.12.7 [DOI] [PubMed] [Google Scholar]
- Perseguini J.M.K.C., Oblessuc P.R., Rosa J.R.B.F., Gomes K.A., Chiorato A.F., Carbonell S.A.M., et al. 2016. Genome-wide association studies of anthracnose and angular leaf spot resistance in common bean (Phaseolus vulgaris L.). PLoS One 11:e0150506. doi: 10.1371/journal.pone.0150506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragagnin V.A., De Souza T.L.P.O., Sanglard D.A., Arruda K.M.A., Costa M.R., Alzate-Marin A.L., et al. 2009. Development and agronomic performance of common bean lines simultaneously resistant to anthracnose, angular leaf spot and rust. Plant Breed. 128:156–163. doi: 10.1111/j.1439-0523.2008.01549.x [DOI] [Google Scholar]
- Ramalho M.A.P., Abreu Â.F.B., Santos J.D., and Nunes J.A.R.. 2012. Aplicações da genética quantitativa no melhoramento de plantas autógamas. Fed. Univ. Lavras, Lavras, Brazil. [Google Scholar]
- Rava Seijas C.A., Sartorato A., and Porto de Carvalho J.R.. 1985. Yield losses in dry bean (Phaseolus vulgaris L.) caused by angular leaf spot (Isariopsis griseola Sacc.). Annu. Rep. Bean Improv. Coop. 28:5–6. [Google Scholar]
- Sanglard D.A., Ribeiro C.A.G., Balbi B.P., Arruda K.M.A., De Barros E.G., and Moreira M.A.. 2013. Characterization of the angular leaf spot resistance gene present in common bean cultivar Ouro Negro. J. Agric. Sci. 5:19–23. [Google Scholar]
- Sartorato A. 2004. Pathogenic variability and genetic diversity of Phaeoisariopsis griseola isolates from two counties in the state of Goias, Brazil. J. Phytopathol. 152:385–390. doi: 10.1111/j.1439-0434.2004.00858.x [DOI] [Google Scholar]
- Sartorato A., and Alzate-Marin A.L.. 2004. Analysis of the pathogenic variability of Phaeoisariopsis griseola in Brazil. Annu. Rep. Bean Improv. Coop. 47:235–236. [Google Scholar]
- Sartorato A., Nietsche S., Barros E.G., and Moreira M.A.. 1999a. Inheritance of angular leaf spot resistance and RAPD markers linked to disease resistance gene in common beans. Annu. Rep. Bean Improv. Coop. 42:21–22. [Google Scholar]
- Sartorato A., Nietsche S., Barros E.G., and Moreira M.A.. 1999b. SCAR marker linked to angular leaf spot resistance gene in common bean. Annu. Rep. Bean Improv. Coop. 42:23–24. [Google Scholar]
- Sartorato A., Nietsche S., Barros E., Moreira M.A., Sartorato A., Nietsche S., et al. 2000. RAPD and SCAR markers linked for resistance gene to angular leaf spot in common beans. Fitopatol. Bras. 25:637–642. [Google Scholar]
- Schmutz J., McClean P.E., Mamidi S., Wu G.A., Cannon S.B., Grimwood J., et al. 2014. A reference genome for common bean and genome-wide analysis of dual domestications. Nat. Genet. 46:707–713 doi: 10.1038/ng.3008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch C.L., Crous P.W., Groenewald J.Z., Boehm E.W.A., Burgess T.I., de Gruyter J., et al. 2009. A class-wide phylogenetic assessment of Dothideomycetes. Stud. Mycol. 64:1–15. doi: 10.3114/sim.2009.64.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz H.F., Correa F., Pineda P., Otoya M.M., and Katherman M.J.. 1981. Dry bean yield losses caused by Ascochyta, angular, and white leaf spots in Colombia. Plant Dis. 65:494–496. doi: 10.1094/PD-65-494 [DOI] [Google Scholar]
- Schwartz H.F., and Pastor-Corrales M.A.. 1989. Bean production problems in the tropics. CIAT, Cali, Colombia. [Google Scholar]
- Schwartz H.F., Pastor-Corrales M.A., and Singh S.P.. 1982. New sources of resistance to anthracnose and angular leaf spot of beans (Phaseolus vulgaris). Euphytica 31:741–754. doi: 10.1007/BF00039213 [DOI] [Google Scholar]
- Schwartz H.F., Steadman J.R., Hall R., and Forster R.L.. 2005. Compendium of bean diseases. 2nd ed. APS Press, St. Paul, MN. [Google Scholar]
- Semagn K., Bjørnstad Å., and Ndjiondjop M.N.. 2006. An overview of molecular methods for plants. Afr. J. Biotechnol. 5:2540–2568. [Google Scholar]
- Silva K.J.D.E., De Souza E.A., Sartorato A., and De Souza Freire C.N.. 2008. Pathogenic variability of isolates of Pseudocercospora griseola, the cause of common bean angular leaf spot, and its implications for resistance breeding. J. Phytopathol. 156:602–606. doi: 10.1111/j.1439-0434.2008.01413.x [DOI] [Google Scholar]
- Singh S.P. 1999. Production and utilization. In: S.P. Singh, editor, Common bean improvement in the twenty-first century. Springer, Dordrecht, the Netherlands: p. 1–24. doi: 10.1007/978-94-015-9211-6_1 [DOI] [Google Scholar]
- Singh S.P., and Schwartz H.F.. 2010. Breeding common bean for resistance to diseases: A review. Crop Sci. 50:2199–2223. doi: 10.2135/cropsci2009.03.0163 [DOI] [Google Scholar]
- Solarte D. 2014. Validación y aplicación de marcadores moleculares SNPs en el mejoramiento de frijol común (Phaseolus vulgaris L.) para resistencia a mancha angular (Phaeoisariopsis griseola Sacc.). Univ. Nac Colombia, Sede Palmira, Colombia. [Google Scholar]
- Souza T.L.P.O., Dessaune S.N., Sanglard D.A., Moreira M.A., and de Barros E.G.. 2011. Characterization of the rust resistance gene present in the common bean cultivar Ouro Negro, the main rust resistance source used in Brazil. Plant Pathol. 60:839–845. doi: 10.1111/j.1365-3059.2011.02456.x [DOI] [Google Scholar]
- Souza T.L.P.O., Gonçalves-Vidigal M.C., Raatz B., Mukankusi C.M., Abreu Â.F.B., Melo L.C., and Pastor-Corrales M.A.. 2016. Major loci controlling resistance to the angular leaf spot of common bean. Annu. Rep. Bean Improv. Coop. 59:49–50. [Google Scholar]
- Stenglein S.A., and Balatti P.A.. 2006. Genetic diversity of Phaeoisariopsis griseola in Argentina as revealed by pathogenic and molecular markers. Physiol. Mol. Plant Pathol. 68:158–167. doi: 10.1016/j.pmpp.2006.10.001 [DOI] [Google Scholar]
- Stenglein S., Ploper L.D., Vizgarra O., and Balatti P.. 2003. Angular leaf spot: A disease caused by the fungus Phaeoisariopsis griseola (Sacc.) Ferraris on Phaseolus vulgaris L. Adv. Appl. Microbiol. 52:209–243. doi: 10.1016/S0065-2164(03)01009-8 [DOI] [PubMed] [Google Scholar]
- St. Clair D.A. 2010. Quantitative disease resistance and quantitative resistance loci in breeding. Annu. Rev. Phytopathol. 48:247–268. doi: 10.1146/annurev-phyto-080508-081904 [DOI] [PubMed] [Google Scholar]
- Teixeira F.F., Bosco dos Santos J., Ramalho M.A.P., Abreu Â.F.B., Teixeira Guimarães C., and de Oliveira A.C.. 2005. QTL mapping for angular leaf spot in common bean using microsatellite markers. Crop Breed. Appl. Biotechnol. 5:272–278. doi: 10.12702/1984-7033.v05n03a03 [DOI] [Google Scholar]
- Thompson H.J., McGinley J.N., Neil E.S., and Brick M.A.. 2017. Beneficial effects of common bean on adiposity and lipid metabolism. Nutrients 9:998. doi: 10.3390/nu9090998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tock A.J., Fourie D., Walley P.G., Holub E.B., Soler A., Cichy K.A., et al. 2017. Genome-wide linkage and association mapping of halo blight resistance in common bean to Race 6 of the globally important bacterial pathogen. Front. Plant Sci. 8:1170. doi: 10.3389/fpls.2017.01170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Schoonhoven A., and Pastor-Corrales M.A.. 1987. Standard system for the evaluation of bean germplasm. CIAT, Cali, Colombia. [Google Scholar]
- Viguiliouk E., Blanco Mejia S., Kendall C.W., and Sievenpiper J.L.. 2017. Can pulses play a role in improving cardiometabolic health? Evidence from systematic reviews and meta-analyses. Ann. N. Y. Acad. Sci. 1392:43–57. doi: 10.1111/nyas.13312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagara I.N., Mwang’ombe A.W., Kimenju J.W., Buruchara R.A., Jamnadass R., and Majiwa P.A.O.. 2004. Genetic diversity of Phaeoisariopsis griseola in Kenya as revealed by AFLP and group-specific primers. J. Phytopathol. 152:235–242. doi: 10.1111/j.1439-0434.2004.00836.x [DOI] [Google Scholar]
- Wagara I.N., Mwang’ombe A.W., Kimenju J.W., Buruchara R.A., and Kimani P.M.. 2011. Reaction of selected common bean genotypes to physiological races of Phaeoisariopsis griseola occurring in Kenya. Afr. Crop Sci. J. 19:343–355. [Google Scholar]
- Wortmann C.S., Kirby R.A., Eledu C.A., and Allen D.J.. 1998. Atlas of common bean (Phaseolus vulgaris L.) production in Africa. CIAT, Cali, Colombia. [Google Scholar]
- Zuiderveen G.H., Padder B.A., Kamfwa K., Song Q., and Kelly J.D.. 2016. Genome-wide association study of anthracnose resistance in Andean beans (Phaseolus vulgaris). PLoS One 11:e0156391. doi: 10.1371/journal.pone.0156391 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.