Abstract
Despite the threat to human health posed by some single-stranded RNA viruses, little is understood about their assembly. The goal of this work is to introduce a new tool for watching an RNA genome direct its own packaging and encapsidation by proteins. Contrast variation small-angle X-ray scattering (CV-SAXS) is a powerful tool with the potential to monitor the changing structure of a viral RNA through this assembly process. The proteins, though present, do not contribute to the measured signal. As a first step in assessing the feasibility of viral genome studies, the structure of encapsidated MS2 RNA was exclusively detected with CV-SAXS and compared with a structure derived from asymmetric cryo-EM reconstructions. Additional comparisons with free RNA highlight the significant structural rearrangements induced by capsid proteins and invite the application of time-resolved CV-SAXS to reveal interactions that result in efficient viral assembly.
Keywords: plus-stranded RNA virus, RNA virus, RNA folding, RNA structure, bacteriophage MS2, virus assembly, virus structure, X-ray scattering, small-angle X-ray scattering (SAXS), contrast variation SAXS
Viruses exploit many strategies to encapsidate the genome that codes for its container. The exact packaging process depends on the nature and form of the genetic material as well as the container or capsid. For some dsDNA viruses, there is clear structural separation between container and message; the genome is pumped into the preformed protein capsid by ATP-activated motor proteins (1). The packaged genome, under high pressure, uniformly fills the capsid (2). In contrast, viruses with single-stranded RNA genomes appear to exploit RNA–protein interactions to facilitate encapsidation. The few examples of spatially resolved, encapsidated ssRNA genomes suggest that the genetic material is not uniformly distributed, often asymmetric and with higher density at the protein interface (3–8). The distinct packaging strategies are at least partially because of the very different biophysics of DNA and RNA (9, 10).
Because ssRNA viruses are prevalent in human disease, there is significant interest in revealing their assembly mechanisms and packaged structures. Fortunately, a fair amount is known about RNA structure from work on other biological RNAs. These polymers contain self-complementary regions and fold on themselves to form series of short, base-paired duplexes joined by a wide range of non–base-paired regions, including junctions, bulges, or loops. Many of these latter motifs serve as protein-binding sites. In viral genomic RNA, recent work suggests that some RNA motifs that bind proteins, so-called packaging signals, are essential for viral assembly (11–18). Their presence may explain why ssRNA viruses selectively pack their own genome relative to other cellular RNAs. Thus, the prevailing picture of how ssRNA genomes are encapsidated by proteins in viruses may be best viewed as a cooperative, specific protein-RNA folding process, as opposed to a sequential assembly of protein capsid followed by genome insertion (19). The self-assembly of ssRNA viruses can therefore be viewed as a process that exploits the delicate balance between RNA–RNA interactions, RNA–protein interactions and, of course, protein–protein interactions (20, 21).
From this vantage point, the packaging of some ssRNA genomes can be recast as an RNA folding problem. In the past 10–20 years, significant progress has been made toward defining the rules for RNA folding, largely in response to the growing appreciation of the role of RNA conformational dynamics in biology (22). Many experiments have provided details about RNA folding as it interacts with counterions (23–26), small ligands (27–30), and even proteins (31–33). Time-resolved experiments have been particularly useful at revealing the dynamics and transiently populated states of RNA. When compared with simulations or models, these studies elucidate the principles that direct folding of small to moderately sized (<1 kb) RNAs (24).
Of the many biophysical tools applied to study the dynamic restructuring of RNAs, solution small-angle X-ray scattering (or SAXS) is particularly useful for revealing large-scale, time-dependent conformational changes. SAXS has been recently applied to monitor the changing structures that accompany capsid self-assembly, providing new information about the underlying mechanisms for the simplest systems (34–37). These works are informative but confounded by SAXS' sensitivity to all components present: both capsid components and nucleic acid. In fact, most of the information extracted was related to the number, and/or conformation of protein subunits. Thus, past kinetic studies of virus assembly by SAXS primarily reported the assembly of the capsid. Fortunately, contrast variation (CV-) SAXS can be used to extract the structures of individual components of complexes, when these components have different electron densities. The nucleic acid genome, for example, can be selectively detected even when encapsidated. The high X-ray flux available at synchrotron sources has already enabled time-resolved CV-SAXS studies of nucleic acid structuring by proteins in other systems (38). Applying this technique to study viral assembly could reveal valuable information about the role of RNA in the assembly process.
The goal of this work is to assess the feasibility of time-resolved CV-SAXS studies of virus assembly. Here we report static CV-SAXS studies of bacteriophage MS2, a model system. Bacteriophage MS2 is an Escherichia coli phage from the leviviridae family with an ssRNA genome of 3569 nucleotides, and a T = 3 icosahedral capsid. This container is composed of 180 copies of the capsid protein and a maturation protein. MS2 is commonly used as model system to study +ssRNA viral assembly. As such, it has been widely studied both biochemically (39–44) and structurally, using SAXS (45–48), as well as electron microscopy (4–8). Contrast variation studies of bacteriophage MS2 have been accomplished using small-angle neutron scattering (46) which more readily yields information about the protein conformation within the MS2 capsid. In these studies, structural information about the encapsidated RNA was inferred, not directly measured. Furthermore, the ability to perform these measurements with X-rays exploits the higher signals available from synchrotron sources relative to neutron sources. Recent asymmetric cryo-EM reconstructions have produced electron density models of the encapsidated RNA for comparison with our findings (5–7). The MS2 system is an excellent candidate for future time-resolved studies, as recent work suggests that, as a result of specific interactions with capsid proteins, its assembly pathways are orderly (49–52). Thus, it is an ideal candidate to benchmark the performance of contrast variation SAXS in virus self-assembly experiments.
Results and Discussion
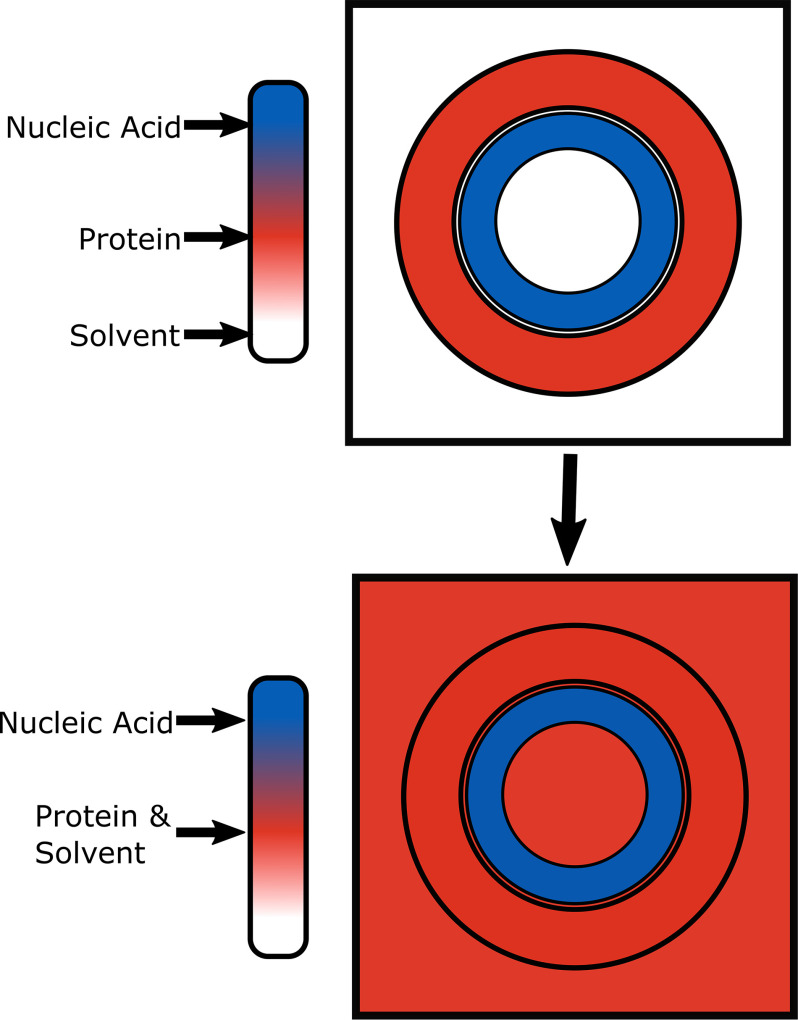
With the goal of directly measuring the encapsidated viral RNA, we performed CV-SAXS on bacteriophage MS2, as well as a recombinantly produced MS2 virus-like particle (VLP) devoid of viral RNA. Contrast variation SAXS, illustrated in Fig. 1 and described in full detail in Ref. 53, exclusively detects the nucleic acid component of a protein-RNA complex, enabling structural studies of the RNA genome contained within the fully assembled virus and potentially throughout the assembly or disassembly process.
Figure 1.
Illustration of the principle of contrast variation SAXS. In CV-SAXS the electron density of the solvent is increased so that it matches that of one of the components of a multicomponent system. In this figure, electron density is represented as color. In a protein–nucleic acid complex the electron density of the solvent can be increased (schematically shown as color changed from white to red to blue) by the addition of sucrose until it matches the electron density of the protein (red). Under this matched condition any scattering signal that contains a contribution from the protein component blends into the background, e.g. disappears. Only the scattering from the denser nucleic acid is detected.
Time-resolved CV-SAXS has been successfully implemented in studies of DNA unwrapping from protein (histone) cores in nucleosome core particles (38). However, in nucleosome core particles, the nucleic acid surrounds the proteins, whereas the opposite is true for viruses. We first address the feasibility of CV-SAXS for particles where the nucleic acid is enclosed by protein. Previous work from Zipper et al. (54) suggests that contrast variation works equally well in either geometry. They performed a limited set of contrast variation SAXS on bacteriophages fr and R17. They extrapolated the radial electron density distribution for the phages but did not thoroughly explore the contrast matched condition. The use of high intensity X-ray sources and sophisticated SAXS data analysis tools make it worthwhile to revisit this approach.
Contrast variation on VLPs
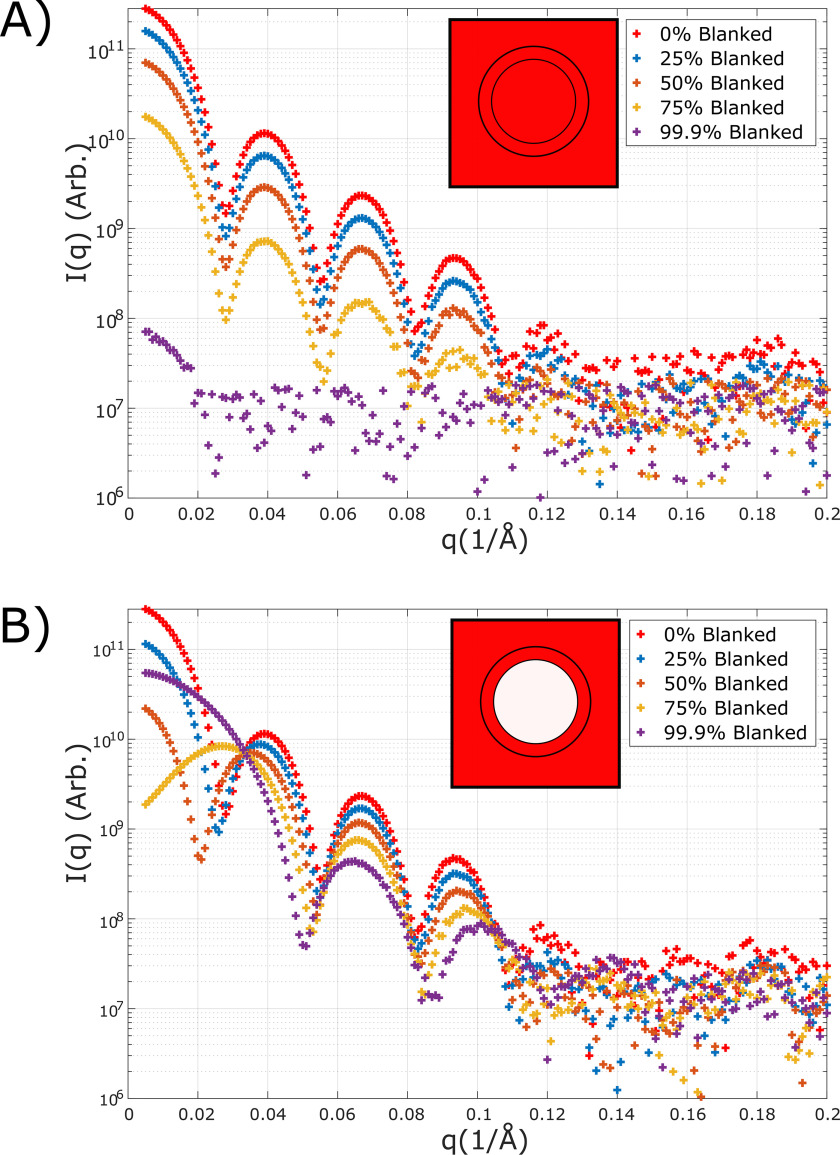
To establish the feasibility of CV-SAXS in virus-like systems, we first performed control experiments on the empty capsid, the MS2 VLP. These studies directly address two primary concerns of CV-SAXS in these systems. The first is that the capsid may be impermeable to the contrast agent, resulting in different electron densities on its inside and outside. As described in Ref. 53, we use high concentrations of sucrose to effect the dramatic changes in solvent electron density required to match the protein density. If the capsid is impermeable to sucrose, the solvent inside the capsid would have an excess negative electron density relative to the solvent outside. Because scattering depends only on density differences, which can be positive or negative, a signal would result. This effect is illustrated using a simple model that treats the capsid as a spherical shell. An analytical solution (see “Experimental procedures”) is used to obtain scattering profiles where the density inside the sphere is different from the density outside. Spheres are particularly amenable to characterization using SAXS; their dimensions are encoded in the positions of (multiple) extrema in the scattering profiles. In particular, the positions of the minima reflect the size of the particle. Fig. 2 shows the predicted behavior of the two cases, a permeable versus nonpermeable capsid, as the (outer) solvent electron density is raised. Here, the expected intensity of the scattered X-rays (I on the y axis) is plotted against the momentum transfer q, defined to be q = (4π sin(θ)/λ), where λ is the X-ray wavelength and 2θ is the scattering angle. Noise was added to simulate realistic experimental conditions. In the former case, the overall signal decreases without significant change in the shape of the scattering profile, assessed through the positions of the extrema. In contrast, for the latter case of a nonpermeable capsid, the positions of the maxima and minima of the scattering profile change, and the signal intensity never drops substantially. A second concern is that sucrose could create osmotic pressures that swell, disrupt, or cause other structural modifications of the capsid. These changes are harder to predict; however, any significant alterations would manifest through a change in the positions of the extrema. The absence of a significant change in the scattering profile of the VLP through the contrast series would suggest that the structure of the capsid is not altered (at the resolution of our measurements) by osmotic effects.
Figure 2.
A and B, the predicted scattering profiles of an empty protein capsid (modeled as a spherical shell) at different contrast conditions in the case that it is permeable (A) or nonpermeable (B) to the contrast variation agent. The % blanked refers to the contrast match between solvent and solute. Perfect contrast matching corresponds to 100% blanked (the line would be difficult to display on a logarithmic axis; we show 99.9% as an alternative). In the inset, the color designates density as in Fig. 1. The curve colors correspond to the fractions quoted in the box. These models suggest that the permeability of the capsid can be readily evaluated from the shape changes in a contrast series. For the desired case of a permeable capsid (top plot), the signal drops dramatically as the contrast match is achieved (purple curve). An impermeable capsid (bottom plot) has a distinctly different signature.
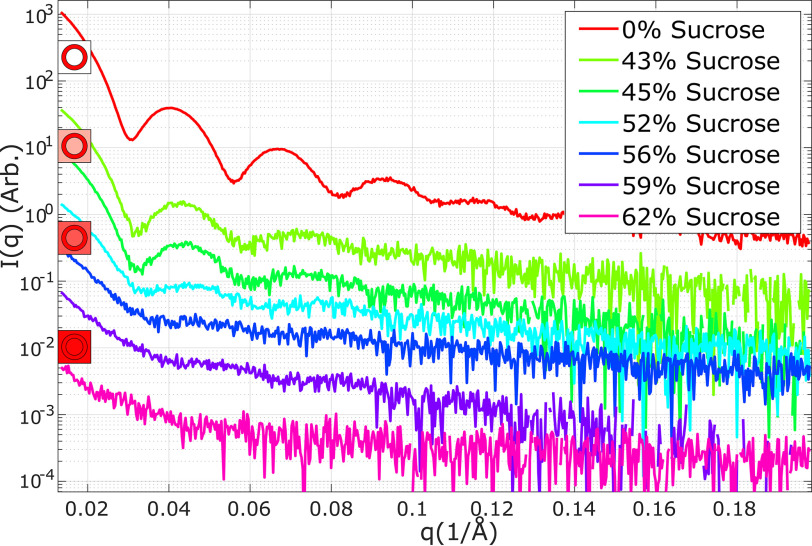
Experimentally determined SAXS scattering profiles of the MS2 VLP, acquired in solutions containing different amounts of sucrose, are shown in Fig. 3. As the protein contrast is reduced by increasing the sucrose concentration, the intensity of the scattering profile decreases, but no change in shape is detected: The positions of the minima remain constant despite the change in solvent electron density. Thus, the capsid appears permeable to sucrose and its structure remains roughly constant. With a further increase in sucrose concentration, the characteristic scattering features of the capsid disappear, and we conclude that it is rendered as transparent as possible. These measurements suggest that an ∼60% (w/w) sucrose solution represents the match condition. Some signal remains, likely because of impurities or the presence of a small amount of mRNA encapsidated during VLP expression. With the knowledge that the capsid can be (largely) blanked we proceed to measure the scattering from the encapsidated RNA inside the WT bacteriophage MS2.
Figure 3.
SAXS profiles of empty MS2 capsids (VLPs) as a function of increasing sucrose in the buffer. The log of the scattering intensity is plotted as a function of q. Curves are offset to aid visualization. At low added sucrose percentages there is no significant change in the position of minima. In conjunction with the models of Fig. 2, this result suggests that the capsid is permeable and therefore can be blanked. Near ∼60% added sucrose there is a significant change in the scattering profile as the signal merges into the noise; this sucrose concentration appears to contrast match, or cancel the protein scattering. The mismatch at low q can be attributed to impurities in the sample; however, we note that even small signals appear amplified when displayed on a logarithmic scale.
Contrast variation on WT MS2
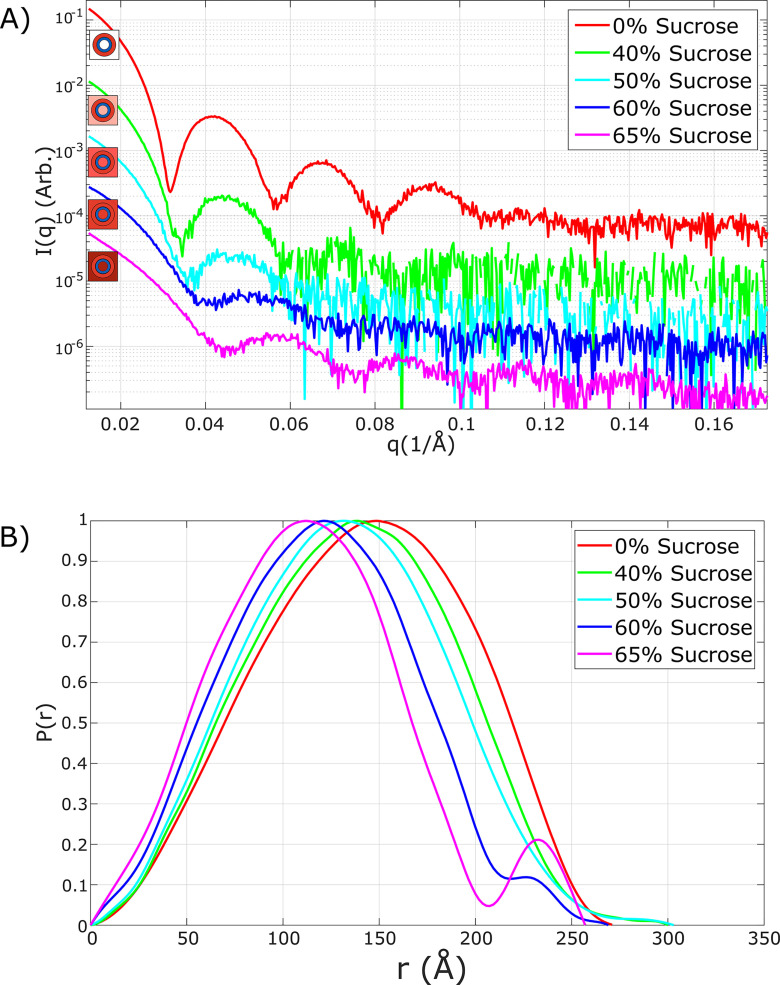
As a first step toward measuring the scattering from the genomic RNA encapsidated within WT MS2 phage particles, we acquired SAXS profiles of the full particle in solutions containing different sucrose concentrations. Measurements acquired at different solution contrast levels allow us to monitor both protein and nucleic acid components as their scattering strengths are varied. Fig. 4A shows SAXS profiles acquired at sucrose concentrations between 0 and 65% (w/w) in discrete steps. It is interesting to compare the shape of the profiles shown here with those of Fig. 2. In the WT MS2 the signal from the RNA becomes apparent; minima in the scattering profile shift to higher q values as the protein contrast is reduced. This trend is consistent with an increased signal from density within the capsid (smaller radius), relative to the capsid itself. The coincident decrease in the depth of the minima can be explained by a loss of spherical symmetry of the molecule (55). We note that the curves change qualitatively above 60% added sucrose. When coupled with the above measurements of the empty capsid, it appears that the “match point,” where the sucrose density equals that of the protein, is around 60%. At higher concentrations, the protein signal reappears, but with lower electron density than the sucrose-rich solvent.
Figure 4.
A and B, SAXS profiles (A) and computed pair distance distribution functions P(r) (B) of WT bacteriophage MS2 as a function of added sucrose, at various concentrations. The observed changes are consistent with an increased contribution from the RNA core (relative to the protein contribution) as the solution contrast increases. All curves are normalized to enable comparison and curves in part (A) are offset to aid in visualization. Beyond the match point, near (but likely just below) 60% added sucrose the contribution from the protein shell reappears as the second peak in pair distance distribution.
Pair distance distributions functions (P(r)) can be computed from the scattering curves and used to interpret the effects of the changing contrast on the signal from the various components of the phage. This formalism displays information from SAXS profiles in real space; as opposed to momentum transfer (q), the x axis in these plots shows real space distances in angstroms. The pair distance distribution functions shown in Fig. 4B were computed using GNOM (56) (ATSAS). All curve features remain robust against variations in selection of parameters for the P(r) computation, the q range, and maximum particle dimension, Dmax. With increasing sucrose concentration, the maximum of the P(r) curve shifts toward a smaller radius. When the sucrose concentration exceeds the match point of about 60%, a local minimum appears above 200 Å, which likely indicates that the solvent density exceeds that of the protein. Under this condition, the signal from the capsid re-emerges, and creates a negative interference term with the RNA. In this case, the second peak above 225 Å could reflect capsid autocorrelation, and the minima above 200 Å could indicate the interference of the signals from RNA (positive contrast) and protein (negative contrast). In any case, all the above measurements are consistent with a match point near 60% sucrose for both the empty and full capsid. Under this condition, the SAXS profile from the native phage most closely represents the scattering from just the encapsidated RNA. Although the contrast match is imperfect, we can nevertheless extract low-resolution structural features about this RNA, for comparison with other measurements on the RNA, and as a proof of principle of the method.
Comparison to EM structure
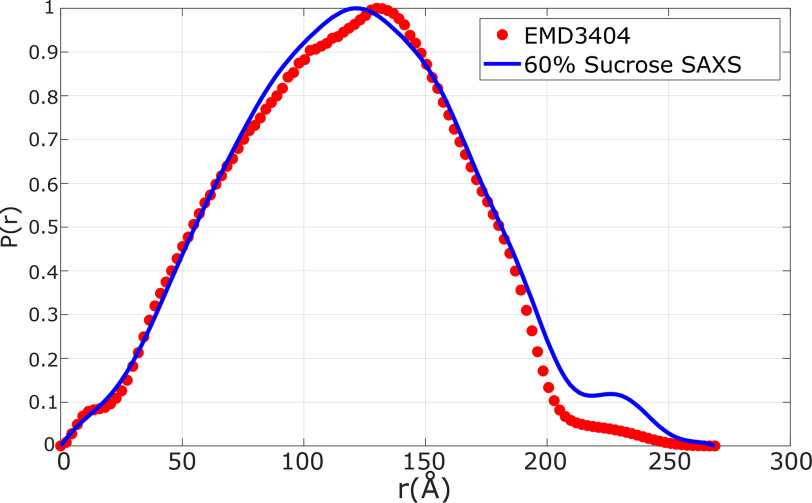
To assess the power and validity of CV-SAXS applied to viruses and virus-like particles, we compare our contrast-matched data (blue curve from Fig. 4) to RNA density derived from a recent cryo-EM study of the MS2 phage by Koning et al. (7). To make this comparison, we computed the pair distance distribution from the electron density map EMD-3404 for voxels above the recommended contour level. Fig. 5 compares the pair distance distribution functions derived from the SAXS data in 60% sucrose (blue curve) with the one computed from this EM density map. The derived distributions are quite similar. Because the spatial resolution of SAXS is lower than that of EM, some of the features in the blue curve are understandably smeared out. Furthermore, the cryo-EM density map accounts for only 95% of the RNA. This loss is attributed to the flexibility near one of the ends of the RNA molecule. In contrast, SAXS retains sensitivity to all the RNA present. Finally, the slight variations at the largest distances may result from incomplete blanking from the capsid shell, e.g. the exact contrast variation point might be 59%, not 60% as measured.
Figure 5.
Comparison of the measured P(r) derived from CVSAXS with one computed from the EM map EMD-3404. The similarity of the two distributions suggests the validity of CV-SAXS for measurements of encapsidated genomes and highlights its potential as a valuable technique for time resolved studies of viral assembly.
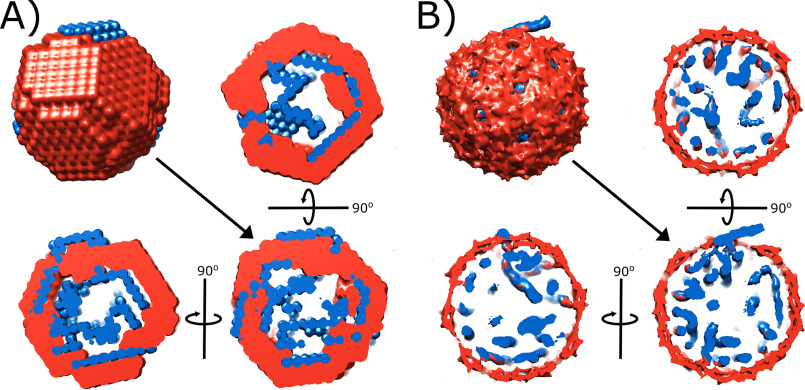
Although it is simplest to interpret data derived from a single molecular component, such as the RNA as described above, several computational tools are available to interpret SAXS data of multicomponent systems acquired under different contrast conditions. To explore this approach, we performed ab initio multiphase reconstruction of the SAXS data using the MONSA algorithm from ATSAS (57). With MONSA we exploit measurements at many different contrast values to obtain bead models for both the protein and the RNA phases of this complex. Results of these multiphase reconstructions are shown in Fig. 6, adjacent to the EM density map of bacteriophage MS2 (EMD-3403 and 3404) (7). Orthogonal cross-sections are shown to help visualize the structure. All molecules were rendered in UCSF Chimera (58).
Figure 6.
A and B, comparison of multicomponent reconstruction of MS2 obtained from SAXS (MONSA) (A), and the models derived from EM (EMD-3404/3403) (B). Protein is shown in red and RNA is shown in blue. The full reconstruction and three orthogonal cross-sections are shown for each case. Although the spatial resolution obtained through SAXS is lower and the reconstruction is not unique, SAXS data are much simpler to acquire than asymmetric cryo-EM reconstructions. Similar structural features are captured by both methods, including a small, protruding piece of RNA that may reflect the position of the maturation protein.
As with all SAXS reconstructions, the solution is neither unique nor high resolution, but it accurately captures several characteristic features present in the cryo-EM model. Specifically, both models suggest that the RNA is localized close to the capsid shell and the genome displays an asymmetry reflecting the position of a maturation protein (top of model). This agreement provides additional confidence in the validity of the contrast variation method as applied to viruses.
Comparison with Mg2+-induced compaction
The contrast variation method offers new opportunities for structural studies of viral genomes. Time-resolved approaches can be applied to follow the genome structure as it folds during assembly. Despite the lack of fully detailed structural information, even low-resolution studies can distinguish different models by measuring the extent and global structure(s) of the RNA; very large changes in conformation are required to compress a long RNA into a capsid. Distinct theoretical models, shown as cartoons in Ref. 59, could be readily distinguished as their global structural signatures are quite distinct. Four different models are discussed in that work and each would present a unique signature. The first, a nucleation-elongation model, would be distinguished by a rapid compaction of RNA, which precedes protein binding. The second, micellar condensation, is most likely consistent with a gradual condensation of RNA as it is slowly condensed by protein. The third, an RNA antenna model, would be distinguished by extended RNA structures as protein binds locally. Finally, a packaging signal model would likely display discrete folding steps because of the cooperative nature of the packaging. When coupled with modeling, the key distinct features (and kinetics) of these different models would be easily distinguished.
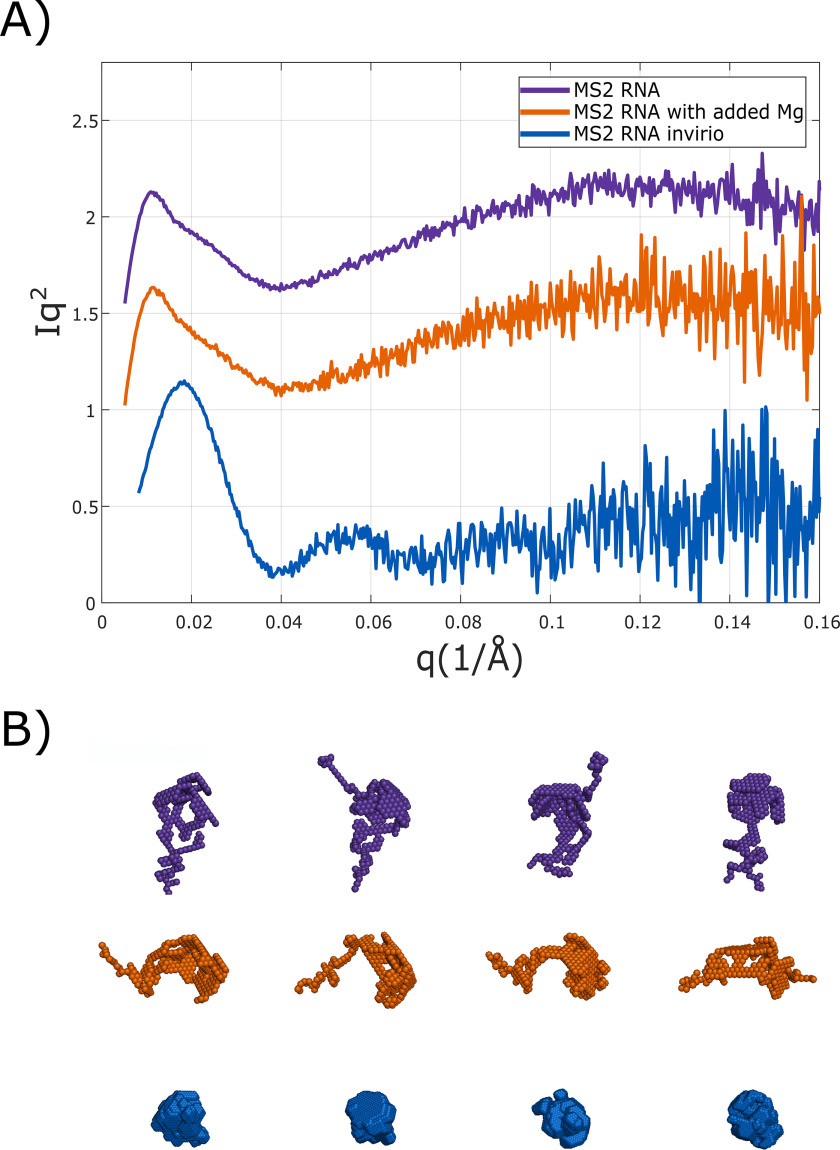
Finally, the global compaction and folding of viral RNAs differs from typical structural of catalytic RNAs. Many of the latter RNAs fold to compact structures following the addition of divalent Mg, which aids in screening the large negative backbone charge and directs catalytic RNAs toward structures that engage tertiary contacts to secure compact states. Fig. 7 shows SAXS profiles and dummy atom reconstructions of free MS2 RNA in a solution containing 150 mm NaCl (purple, top curve) and following the addition of Mg2+ (orange, middle curve) at concentrations that fold many catalytic RNAs compared with the MS2 RNA in virio (blue, bottom curve). Here, scattering profiles are displayed as Kratky plots of Iq2 versus q. This representation of the SAXS profiles emphasizes compaction and is useful in studies of RNA folding, where, in the absence of Mg2+, the molecule assumes more extended states. Dummy atom reconstructions of free MS2 RNA were performed through DAMMIF (60) (ATSAS). Multiple reconstructions are shown for each of the models. Although these reconstructions are not unique, they are useful in exemplifying the changes the RNA undergoes. SAXS studies of other functional RNAs reveal large changes upon the addition of Mg2+ (23, 25). For this RNA, the addition of divalent ions does not lead to a large change in structure or Rg. In contrast, large differences in the SAXS signal between encapsidated (lower, blue curve) and free RNA are striking. This change is also accompanied by a collapse in the radius of gyration from 169 to 95 Å within the capsid, consistent with measurements of MS2 in solution by Zipper et al. (61), the spatial distribution of MS2 (46), and the change in genome size measured in other viruses (62). Folding of the viral RNA seems to require the protein. Perhaps this is by design; the architecture of this RNA has evolved to fold with capsid proteins so that it can assemble efficiently.
Figure 7.
A and B, Kratky plots (Iq2 versus q) of encapsidated MS2 RNA and free MS2 RNA with and without 1 mm MgCl2 (A) and the corresponding bead model reconstructions (B). Kratky curves are offset to facilitate comparison. Although the addition of Mg2+ ions causes many physiologically relevant RNAs to fold, a significant structural change is not detected in this viral RNA. Added protein, on the other hand, initiates a large structural change in the RNA, highlighting the importance and specificity of RNA–protein interactions to genome encapsidation.
Conclusion
In conclusion, these studies not only underscore the important role of protein–RNA interactions in compacting/packaging RNA (as suggested by others) but present a unique strategy for watching this process as it occurs, using CV-SAXS. There is much to learn about the folding of viral RNAs by the proteins that they encode (59). The method, demonstrated here, should be readily transferrable to other nonenveloped viruses and may be useful in unraveling the mechanism of novel antiviral drugs that target assembly, a topic of great current importance.
Experimental procedures
Sample preparation
Escherichia coli bacteriophage MS2 (ATCC® 15597-B1TM) was propagated in Escherichia coli (Migula) Castellani and Chalmers (ATCC® 15597TM) in the recommended growth media. After overnight growth, E. coli were pelleted by precipitation, and the supernatant was collected. PEG and NaCl were added to final concentrations of 10% w/v and 0.5 m, respectively. After incubation for an hour, the bacteriophages were precipitated by centrifugation. The precipitate was resuspended in buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl), filtered, and further purified by gel filtration chromatography though a Superdex 200 column. Contrast variation samples were then dialyzed overnight into the appropriate sucrose concentration. This was done to ensure complete matching between buffer and sample.
MS2 capsid protein (CP) was produced recombinantly in E. coli (BL21). A plasmid containing the MS2 CP sequence was produced de novo by ATUM. Transformed E. coli were grown in LB-Lennox until mid-log phase, after which expression was induced with 1 mg/ml of isopropyl 1-thio-β-d-galactopyranoside. Protein expression continued for 4 h. Afterward E. coli were pelleted by centrifugation, and the pellet was stored at −20°C. The pellet was thawed, resuspended in buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl), sonicated, and clarified by centrifugation. MS2 CP was then purified as self-assembled VLPs using the same protocol as WT MS2. UV absorbance ratios suggest that the VLPs contain a small amount of mRNA. Because of the exploratory nature of the blanking procedure, sucrose was added to samples with a positive displacement pipette.
MS2 RNA was purchased from Sigma and frozen. After thawing, it was buffer exchanged into 50 mm Tris-HCl, pH 7.5, 150 mm by serial concentration. Lastly it was annealed by heating to 95°C for 5 min and then cooled with ice. Magnesium was added immediately before measurement with a pipette.
SAXS data collection
SAXS data were collected at the BioCAT sector of the Advanced Photon Source, in two separate studies. Protocols for acquiring contrast variation SAXS data are detailed in Ref. 53. During the first SAXS data were acquired on MS2 VLP using the standard BioCAT equilibrium setup. Contrast variation data were acquired on MS2 WT in a second beamtime, using a custom built set up that employed coaxial-sheathed continuous flow. In both cases data were collected on a Pilatus 3M detector. Samples were suspended in 50 mm Tris-HCl, pH 7.5, 150 mm NaCl, with added sucrose to enable CV-SAXS. Data reduction was performed using the RAW package (63). Further analysis was performed with the ATSAS suite (64).
Contrast variation shell modeling
The possible effects of permeability on an empty capsid were calculated using the theoretical form factor formula for a core-shell model.
In the above equations, are respectively the outermost and innermost radii of a shell, and are the excess electron density above the solvent for both the shell and core, respectively. In the case of a permeable capsid, was held at zero as was reduced. In the case of the nonpermeable capsid, was reduced by the same amount as . To further simulate experimental conditions, the theoretical SAXS intensity was convoluted with a gaussian function to simulate experimental broadening. Lastly a random noise background was added to simulate the level of signal to noise seen as contrast is reduced.
Data availability
Data will be made available upon request to the corresponding author, Lois Pollack, lp26@cornell.edu.
Supplementary Material
Acknowledgments
We gratefully acknowledge experimental assistance from Srinivas Chakravarthy and useful discussions and manuscript review from Pollack Laboratory group members. This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. Use of the Pilatus 3 1M detector was provided by NIGMS, National Institutes of Health Grant 1S10OD018090-01. Molecular graphics images were produced using the UCSF Chimera package from the Computer Graphics Laboratory, University of California, San Francisco (supported by National Institutes of Health Grant P41 RR-01081).
Author contributions—J. S. E. data curation; J. S. E. formal analysis; J. S. E. investigation; J. S. E. methodology; J. S. E. writing-original draft; L. P. conceptualization; L. P. supervision; L. P. funding acquisition; L. P. project administration; L. P. writing-review and editing.
Funding and additional information—This work was supported by the National Science Foundation Award DBI-1930046 (to L. P.), the National Institutes of Health Grant T32GM008267 (to J. S. E.), and the Colman Fellowship program. This project was supported by NIGMS, National Institutes of Health, Grant 9 P41 GM103622. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- ssRNA
- single-stranded RNA
- SAXS
- small-angle X-ray scattering
- CV-SAXS
- contrast variation SAXS
- VLP
- virus-like particle
- P(r)
- pair distance distributions functions
- CP
- capsid protein.
References
- 1. Aksyuk A. A., and Rossmann M. G. (2011) Bacteriophage assembly. Viruses 3, 172–203 10.3390/v3030172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Duda R. L., Ross P. D., Cheng N., Firek B. A., Hendrix R. W., Conway J. F., and Steven A. C. (2009) Structure and energetics of encapsidated DNA in bacteriophage HK97 studied by scanning calorimetry and cryo-electron microscopy. J. Mol. Biol. 391, 471–483 10.1016/j.jmb.2009.06.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. van den Worm S. H. E., Koning R. I., Warmenhoven H. J., Koerten H. K., and van Duin J. (2006) Cryo electron microscopy reconstructions of the leviviridae unveil the densest icosahedral RNA packing possible. J. Mol. Biol. 363, 858–865 10.1016/j.jmb.2006.08.053 [DOI] [PubMed] [Google Scholar]
- 4. Koning R., Van Den Worm S., Plaisier J. R., Van Duin J., Abrahams J. P., and Koerten H. (2003) Visualization by cryo-electron microscopy of genomic RNA that binds to the protein capsid inside bacteriophage MS2. J. Mol. Biol. 332, 415–422 10.1016/S0022-2836(03)00846-5 [DOI] [PubMed] [Google Scholar]
- 5. Dai X., Li Z., Lai M., Shu S., Du Y., Zhou Z. H., and Sun R. (2017) In situ structures of the genome and genome-delivery apparatus in a single-stranded RNA virus. Nature 541, 112–116 10.1038/nature20589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dent K. C., Thompson R., Barker A. M., Hiscox J. A., Barr J. N., Stockley P. G., and Ranson N. A. (2013) The asymmetric structure of an icosahedral virus bound to its receptor suggests a mechanism for genome release. Structure 21, 1225–1234 10.1016/j.str.2013.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Koning R. I., Gomez-Blanco J., Akopjana I., Vargas J., Kazaks A., Tars K., Carazo J. M., and Koster A. J. (2016) Asymmetric cryo-EM reconstruction of phage MS2 reveals genome structure in situ. Nat. Commun. 7, 12524 10.1038/ncomms12524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Toropova K., Basnak G., Twarock R., Stockley P. G., and Ranson N. A. (2008) The three-dimensional structure of genomic RNA in bacteriophage MS2: Implications for assembly. J. Mol. Biol. 375, 824–836 10.1016/j.jmb.2007.08.067 [DOI] [PubMed] [Google Scholar]
- 9. Chen H., Meisburger S. P., Pabit S. A., Sutton J. L., Webb W. W., and Pollack L. (2012) Ionic strength-dependent persistence lengths of single-stranded RNA and DNA. Proc. Natl. Acad. Sci. U. S. A. 109, 799–804 10.1073/pnas.1119057109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Drozdetski A. V., Tolokh I. S., Pollack L., Baker N., and Onufriev A. V. (2016) Opposing effects of multivalent ions on the flexibility of DNA and RNA. Phys. Rev. Lett. 117, 028101 10.1103/PhysRevLett.117.028101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bunka D. H. J., Lane S. W., Lane C. L., Dykeman E. C., Ford R. J., Barker A. M., Twarock R., Phillips S. E. V., and Stockley P. G. (2011) Degenerate RNA packaging signals in the genome of satellite tobacco necrosis virus: Implications for the assembly of a T = 1 capsid. J. Mol. Biol. 413, 51–65 10.1016/j.jmb.2011.07.063 [DOI] [PubMed] [Google Scholar]
- 12. Stockley P. G., Rolfsson O., Thompson G. S., Basnak G., Francese S., Stonehouse N. J., Homans S. W., and Ashcroft A. E. (2007) A simple, RNA-mediated allosteric switch controls the pathway to formation of a T = 3 viral capsid. J. Mol. Biol. 369, 541–552 10.1016/j.jmb.2007.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dykeman E. C., Stockley P. G., and Twarock R. (2013) Packaging signals in two single-stranded RNA viruses imply a conserved assembly mechanism and geometry of the packaged genome. J. Mol. Biol. 425, 3235–3249 10.1016/j.jmb.2013.06.005 [DOI] [PubMed] [Google Scholar]
- 14. Rolfsson Ó., Middleton S., Manfield I. W., White S. J., Fan B., Vaughan R., Ranson N. A., Dykeman E., Twarock R., Ford J., Cheng Kao C., and Stockley P. G. (2016) Direct evidence for packaging signal-mediated assembly of bacteriophage MS2. J. Mol. Biol. 428, 431–448 10.1016/j.jmb.2015.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stockley P. G., White S. J., Dykeman E., Manfield I., Rolfsson O., Patel N., Bingham R., Barker A., Wroblewski E., Chandler-Bostock R., Weiß E. U., Ranson N. A., Tuma R., and Twarock R. (2016) Bacteriophage MS2 genomic RNA encodes an assembly instruction manual for its capsid. Bacteriophage 6, e1157666 10.1080/21597081.2016.1157666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schneidman-Duhovny D., Hammel M., Tainer J. A., and Sali A. (2013) Accurate SAXS profile computation and its assessment by contrast variation experiments. Biophys. J. 105, 962–974 10.1016/j.bpj.2013.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Patel N., Dykeman E. C., Coutts R. H. A., Lomonossoff G. P., Rowlands D. J., Phillips S. E. V., Ranson N., Twarock R., Tuma R., and Stockley P. G. (2015) Revealing the density of encoded functions in a viral RNA. Proc. Natl. Acad. Sci. U. S. A. 112, 2227–2232 10.1073/pnas.1420812112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shakeel S., Dykeman E. C., White S. J., Ora A., Cockburn J. J. B., Butcher S. J., Stockley P. G., and Twarock R. (2017) Genomic RNA folding mediates assembly of human parechovirus. Nat. Commun. 8, 1–11 10.1038/s41467-016-0009-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Twarock R., and Stockley P. G. (2019) RNA-mediated virus assembly: Mechanisms and consequences for viral evolution and therapy. Annu. Rev. Biophys. 48, 495–514 10.1146/annurev-biophys-052118-115611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Patel N., Wroblewski E., Leonov G., Phillips S. E. V., Tuma R., Twarock R., and Stockley P. G. (2017) Rewriting nature's assembly manual for a ssRNA virus. Proc. Natl. Acad. Sci. U. S. A. 114, 12255–12260 10.1073/pnas.1706951114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Perlmutter J. D., and Hagan M. F. (2015) Mechanisms of virus assembly. Annu. Rev. Phys. Chem. 66, 217–239 10.1146/annurev-physchem-040214-121637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ganser L. R., Kelly M. L., Herschlag D., and Al-Hashimi H. M. (2019) The roles of structural dynamics in the cellular functions of RNAs. Nat. Rev. Mol. Cell Biol. 20, 474–489 10.1038/s41580-019-0136-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Das R., Kwok L. W., Millett I. S., Bai Y., Mills T. T., Jacob J., Maskel G. S., Seifert S., Mochrie S. G. J., Thiyagarajan P., Doniach S., Pollack L., and Herschlag D. (2003) The fastest global events in RNA folding: Electrostatic relaxation and tertiary collapse of the tetrahymena ribozyme. J. Mol. Biol. 332, 311–319 10.1016/S0022-2836(03)00854-4 [DOI] [PubMed] [Google Scholar]
- 24. Plumridge A., Katz A. M., Calvey G. D., Elber R., Kirmizialtin S., and Pollack L. (2018) Revealing the distinct folding phases of an RNA three-helix junction. Nucleic Acids Res. 46, 7354–7365 10.1093/nar/gky363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Russell R., Millett I. S., Tate M. W., Kwok L. W., Nakatani B., Gruner S. M., Mochrie S. G. J., Pande V., Doniach S., Herschlag D., and Pollack L. (2002) Rapid compaction during RNA folding. Proc. Natl. Acad. Sci 99, 4266–4271 10.1073/pnas.072589599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Welty R., Pabit S. A., Katz A. M., Calvey G. D., Pollack L., and Hall K. B. (2018) Divalent ions tune the kinetics of a bacterial GTPase center rRNA folding transition from secondary to tertiary structure. RNA 24, 1828–1838 10.1261/rna.068361.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Felletti M., and Hartig J. S. (2017) Ligand-dependent ribozymes. Wiley Interdiscip. Rev. RNA 8, e1395 10.1002/wrna.1395 [DOI] [PubMed] [Google Scholar]
- 28. Rieder R., Lang K., Graber D., and Micura R. (2007) Ligand-induced folding of the adenosine deaminase A-riboswitch and implications on riboswitch translational control. ChemBioChem. 8, 896–902 10.1002/cbic.200700057 [DOI] [PubMed] [Google Scholar]
- 29. Buck J., Fürtig B., Noeske J., Wöhnert J., and Schwalbe H. (2007) Time-resolved NMR methods resolving ligand-induced RNA folding at atomic resolution. Proc. Natl. Acad. Sci. U. S. A. 104, 15699–15704 10.1073/pnas.0703182104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lee M.-K., Gal M., Frydman L., and Varani G. (2010) Real-time multidimensional NMR follows RNA folding with second resolution. Proc. Natl. Acad. Sci. U. S. A. 107, 9192–9197 10.1073/pnas.1001195107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Welty R., Rau M., Pabit S., Dunstan M. S., Conn G. L., Pollack L., and Hall K. B. (2020) Ribosomal protein L11 selectively stabilizes a tertiary structure of the GTPase center rRNA domain. J. Mol. Biol. 432, 991–1007 10.1016/j.jmb.2019.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ha T., Zhuang X., Kim H. D., Orr J. W., Williamson J. R., and Chu S. (1999) Ligand-induced conformational changes observed in single RNA molecules. Proc. Natl. Acad. Sci. U. S. A. 96, 9077–9082 10.1073/pnas.96.16.9077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rodgers M. L., Tretbar U. S., Dehaven A., Alwan A. A., Luo G., Mast H. M., and Hoskins A. A. (2016) Conformational dynamics of stem II of the U2 snRNA. RNA 22, 225–236 10.1261/rna.052233.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kler S., Asor R., Li C., Ginsburg A., Harries D., Oppenheim A., Zlotnick A., and Raviv U. (2012) RNA encapsidation by SV40-derived nanoparticles follows a rapid two-state mechanism. J. Am. Chem. Soc. 134, 8823–8830 10.1021/ja2110703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chevreuil M., Law-Hine D., Chen J., Bressanelli S., Combet S., Constantin D., Degrouard J., Möller J., Zeghal M., and Tresset G. (2018) Nonequilibrium self-assembly dynamics of icosahedral viral capsids packaging genome or polyelectrolyte. Nat. Commun. 9, 3071 10.1038/s41467-018-05426-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kler S., Wang J. C. Y., Dhason M., Oppenheim A., and Zlotnick A. (2013) Scaffold properties are a key determinant of the size and shape of self-assembled virus-derived particles. ACS Chem. Biol. 8, 2753–2761 10.1021/cb4005518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Asor R., Selzer L., Schlicksup C. J., Zhao Z., Zlotnick A., and Raviv U. (2019) Assembly reactions of hepatitis B capsid protein into capsid nanoparticles follow a narrow path through a complex reaction landscape. ACS Nano. 13, 7610–7626 10.1021/acsnano.9b00648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chen Y., Tokuda J. M., Topping T., Meisburger S. P., Pabit S. A., Gloss L. M., and Pollack L. (2017) Asymmetric unwrapping of nucleosomal DNA propagates asymmetric opening and dissociation of the histone core. Proc. Natl. Acad. Sci. U. S. A. 114, 334–339 10.1073/pnas.1611118114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rolfsson Ó., Toropova K., Ranson N. A., and Stockley P. G. (2010) Mutually induced conformational switching of RNA and coat protein underpins efficient assembly of a viral capsid. J. Mol. Biol. 401, 309–322 10.1016/j.jmb.2010.05.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sugiyama T., Hebert R. R., and Hartman K. A. (1967) Ribonucleoprotein complexes formed between bacteriophage MS2 RNA and MS2 protein in vitro. J. Mol. Biol. 25, 455–463 10.1016/0022-2836(67)90198-2 [DOI] [PubMed] [Google Scholar]
- 41. Peabody D. S. (1993) The RNA binding site of bacteriophage MS2 coat protein. EMBO J. 12, 595–600 10.1002/j.1460-2075.1993.tb05691.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Grahn E., Moss T., Helgstrand C., Fridborg K., Sundaram M., Tars K., Lago H., Stonehouse N. J., Davis D. R., Stockley P. G., and Liljas L. (2001) Structural basis of pyrimidine specificity in the MS2 RNA hairpin-coat-protein complex. RNA 7, 1616–1627 [PMC free article] [PubMed] [Google Scholar]
- 43. Perkett M. R., Mirijanian D. T., and Hagan M. F. (2016) The allosteric switching mechanism in bacteriophage MS2. J. Chem. Phys. 145, 035101 10.1063/1.4955187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dika C., Duval J. F. L., Ly-Chatain H. M., Merlin C., and Gantzer C. (2011) Impact of internal RNA on aggregation and electrokinetics of viruses: Comparison between MS2 phage and corresponding virus-like particles. Appl. Environ. Microbiol. 77, 4939–4948 10.1128/AEM.00407-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kuzmanovic D. A., Elashvili I., Wick C., O'Connell C., and Krueger S. (2006) The MS2 coat protein shell is likely assembled under tension: A novel role for the MS2 bacteriophage A protein as revealed by small-angle neutron scattering. J. Mol. Biol. 355, 1095–1111 10.1016/j.jmb.2005.11.040 [DOI] [PubMed] [Google Scholar]
- 46. Kuzmanovic D. A., Elashvili I., Wick C., O'Connell C., and Krueger S. (2003) Bacteriophage MS2: Molecular weight and spatial distribution of the protein and RNA components by small-angle neutron scattering and virus counting. Structure 11, 1339–1348 10.1016/j.str.2003.09.021 [DOI] [PubMed] [Google Scholar]
- 47. Nguyen T. H., Easter N., Gutierrez L., Huyett L., Defnet E., Mylon S. E., Ferri J. K., and Viet N. A. (2011) The RNA core weakly influences the interactions of the bacteriophage MS2 at key environmental interfaces. Soft Matter 7, 10449–10456 10.1039/c1sm06092a [DOI] [Google Scholar]
- 48. Mylon S. E., Rinciog C. I., Schmidt N., Gutierrez L., Wong G. C. L., and Nguyen T. H. (2010) Influence of salts and natural organic matter on the stability of bacteriophage MS2. Langmuir 26, 1035–1042 10.1021/la902290t [DOI] [PubMed] [Google Scholar]
- 49. Garmann R. F., Goldfain A. M., and Manoharan V. N. (2019) Measurements of the self-assembly kinetics of individual viral capsids around their RNA genome. Proc. Natl. Acad. Sci. U. S. A. 116, 22485–22490 10.1073/pnas.1909223116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bleckley S., and Schroeder S. J. (2012) Incorporating global features of RNA motifs in predictions for an ensemble of secondary structures for encapsidated MS2 bacteriophage RNA. RNA 18, 1309–1318 10.1261/rna.032326.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rolfsson O., Toropova K., Morton V., Francese S., Basnak G., Thompson G. S., Homans S. W., Ashcroft A. E., Stonehouse N. J., Ranson N. A., and Stockley P. G. (2008) RNA packing specificity and folding during assembly of the bacteriophage MS2. Comput. Math. Methods Med. 9, 339–349 10.1080/17486700802168445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Basnak G., Morton V. L., Rolfsson Ó., Stonehouse N. J., Ashcroft A. E., and Stockley P. G. (2010) Viral genomic single-stranded RNA directs the pathway toward a T = 3 capsid. J. Mol. Biol. 395, 924–936 10.1016/j.jmb.2009.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tokuda J. M., Pabit S. A., and Pollack L. (2016) Protein–DNA and ion–DNA interactions revealed through contrast variation SAXS. Biophys. Rev. 8, 139–149 10.1007/s12551-016-0196-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zipper P., Kratky O., Herrmann R., and Hohn T. (1971) An X-ray small angle study of the bacteriophages fr and R17. Eur. J. Biochem. 18, 1–9 10.1111/j.1432-1033.1971.tb01206.x [DOI] [PubMed] [Google Scholar]
- 55. Sun Y., Roznowski A. P., Tokuda J. M., Klose T., Mauney A., Pollack L., Fane B. A., and Rossmann M. G. (2017) Structural changes of tailless bacteriophage ΦX174 during penetration of bacterial cell walls. Proc. Natl. Acad. Sci. U. S. A. 114, 13708–13713 10.1073/pnas.1716614114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Svergun D. I. (1992) Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 25, 495–503 10.1107/S0021889892001663 [DOI] [Google Scholar]
- 57. Tuukkanen A. T., Kleywegt G. J., and Svergun D. I. (2016) Resolution of ab initio shapes determined from small-angle scattering. IUCrJ 3, 440–447 10.1107/S2052252516016018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., and Ferrin T. E. (2004) UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- 59. Dragnea B. (2019) Watching a virus grow. Proc. Natl. Acad. Sci. U. S. A. 116, 22420–22422 10.1073/pnas.1915986116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Franke D., and Svergun D. I. (2009) DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Crystallogr. 42, 342–346 10.1107/S0021889809000338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zipper P., Folkhard W., and Clauwaert J. (1975) A small-angle X-ray scattering investigation on the structure of the RNA from bacteriophage MS2. FEBS Lett. 56, 283–287 10.1016/0014-5793(75)81110-0 [DOI] [PubMed] [Google Scholar]
- 62. Gopal A., Egecioglu D. E., Yoffe A. M., Ben-Shaul A., Rao A. L. N., Knobler C. M., and Gelbart W. M. (2014) Viral RNAs are unusually compact. PLoS One 9, e105875 10.1371/journal.pone.0105875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hopkins J. B., Gillilan R. E., and Skou S. (2017) BioXTAS RAW: Improvements to a free open-source program for small-angle X-ray scattering data reduction and analysis. J. Appl. Crystallogr. 50, 1545–1553 10.1107/S1600576717011438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Franke D., Petoukhov M. V., Konarev P. V., Panjkovich A., Tuukkanen A., Mertens H. D. T., Kikhney A. G., Hajizadeh N. R., Franklin J. M., Jeffries C. M., and Svergun D. I. (2017) ATSAS 2.8: A comprehensive data analysis suite for small-angle scattering from macromolecular solutions. J. Appl. Cryst. 50, 1212–1225 10.1107/S1600576717007786 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available upon request to the corresponding author, Lois Pollack, lp26@cornell.edu.