Abstract
Successful DNA replication requires carefully regulated mechanisms to overcome numerous obstacles that naturally occur throughout chromosomal DNA. Scattered across the genome are tightly bound proteins, such as transcription factors and nucleosomes, that are necessary for cell function, but that also have the potential to impede timely DNA replication. Using biochemically reconstituted systems, we show that two transcription factors, yeast Reb1 and Tbf1, and a tightly positioned nucleosome, are strong blocks to the strand displacement DNA synthesis activity of DNA polymerase δ. Although the block imparted by Tbf1 can be overcome by the DNA-binding activity of the single-stranded DNA-binding protein RPA, efficient DNA replication through either a Reb1 or a nucleosome block occurs only in the presence of the 5'-3' DNA helicase Pif1. The Pif1-dependent stimulation of DNA synthesis across strong protein barriers may be beneficial during break-induced replication where barriers are expected to pose a problem to efficient DNA bubble migration. However, in the context of lagging strand DNA synthesis, the efficient disruption of a nucleosome barrier by Pif1 could lead to the futile re-replication of newly synthetized DNA. In the presence of FEN1 endonuclease, the major driver of nick translation during lagging strand replication, Pif1-dependent stimulation of DNA synthesis through a nucleosome or Reb1 barrier is prevented. By cleaving the short 5' tails generated during strand displacement, FEN1 eliminates the entry point for Pif1. We propose that this activity would protect the cell from potential DNA re-replication caused by unwarranted Pif1 interference during lagging strand replication.
Keywords: DNA replication, DNA polymerase, DNA polymerase δ, DNA helicase, Pif1, nucleosome, transcription factors, Reb1, Tbf1, FEN1
Impeded DNA replication can lead to replication fork stalling and, potentially, genomic instability (1, 2). Yet many barriers must be overcome during DNA replication, such as DNA secondary structures and tightly bound proteins. During DNA replication, nucleosomes are recycled from the parental strand and redeposited randomly on the two daughter strands (3–8), thus ensuring efficient restoration of the proper epigenetic landscape. As lagging strand replication proceeds in the opposite direction of the replication fork, rebinding of transcription factors and reassembly of nucleosomes would form barriers to the lagging strand DNA polymerase δ (Pol δ). Indeed, nucleosomes appear to block Pol δ DNA synthesis both in vivo (9) and in vitro (10), and possibly limit strand displacement synthesis to prevent excessive re-replication, whereas still allowing primer removal. However, in yeast long 5'-flaps can arise during Okazaki fragment maturation and be extended by the 5'-3' DNA helicase Pif1, necessitating the endonuclease activity of the nuclease/helicase Dna2 to cleave the extended flap (11, 12). The flaps extended by Pif1 in vivo can reach lengths of hundreds (13) to thousands (14) of nucleotides, suggesting that during this process Pif1 may displace nucleosomes assembled on downstream Okazaki fragments. This possibility remains to be tested using reconstituted systems.
Interestingly, genome wide analysis of Okazaki fragment junctions showed that their position correlates with the position of the binding sites of the general transcription factors Abf1, Reb1, and Rap1 (9), consistent with these being a barrier to the progression of lagging strand DNA synthesis. Indeed, we showed that in vitro a single Rap1 tightly bound to a high-affinity DNA-binding site is a strong block to the strand displacement DNA synthesis activity of yeast DNA polymerase (Pol) and DNA replication through a single or an array of DNA-bound Rap1 molecules requires the helicase activity of Pif1 (15). It remains to be tested whether other transcription factors are also a strong block to DNA synthesis by Pol δ, and, if so, which ones impart a requirement for the activity of the Pif1 helicase for efficient DNA synthesis by Pol δ.
Here we show that Saccharomyces cerevisiae Reb1 and Tbf1 bound to dsDNA are both strong polar blocks to the strand displacement DNA synthesis activity of Pol δ. However, binding of the ssDNA-binding protein RPA to a 5'-ssDNA flap of the displaced strand is sufficient to stimulate strand displacement DNA synthesis by Pol δ through the block imparted by a bound Tbf1. The same is not true for Reb1, which remains a strong block even in the presence of RPA and requires the activity of the Pif1 helicase for its removal. These findings suggest that a subset of DNA-bound transcription factors may necessitate the activity of the Pif1 helicase for efficient progression of replication. We also show that a positioned nucleosome is a strong block to both strand displacement DNA synthesis by Pol δ and FEN1-mediated nick translation, requiring both the ssDNA-binding protein RPA and Pif1 helicase for through-replication. In the presence of the FEN1 nuclease, Pif1 no longer stimulates DNA synthesis through a nucleosome, as FEN1 removes the entry point for the helicase. On the other hand, whereas RPA bound to a 5'-flap only moderately inhibited FEN1, it was sufficient to promote Pif1 unwinding through a protein block, even in the presence of FEN1. We propose that one function of FEN1 during Okazaki fragment maturation is to prevent the potentially deleterious activity of Pif1.
Results
General transcription factors are strong replication barriers that differ in their requirement for Pif1 to allow through-replication
In vitro primer extension assays were performed with DNA substrates that contain a 1-nt gap to monitor successful base incorporation by the polymerase, and a downstream duplex region with a centrally positioned sequence recognition motif to direct binding of each transcription factor tested. The substrates also contain a 25-nt long 5' poly(dT) flap that prevents PCNA from sliding off the substrate, whereas also providing a binding site for RPA and an entry point for Pif1 5'-3' helicase. The scheme for the assay is depicted in Fig. 1A. Briefly, PCNA is loaded on the DNA substrate by RFC, followed by the binding of the specific transcription factor being tested, in the presence or absence of the single-stranded DNA-binding protein RPA. DNA synthesis is initiated by the addition of Pol δ and dNTPs and the helicase when tested. The formation of the extension products from the labeled primer is monitored over time.
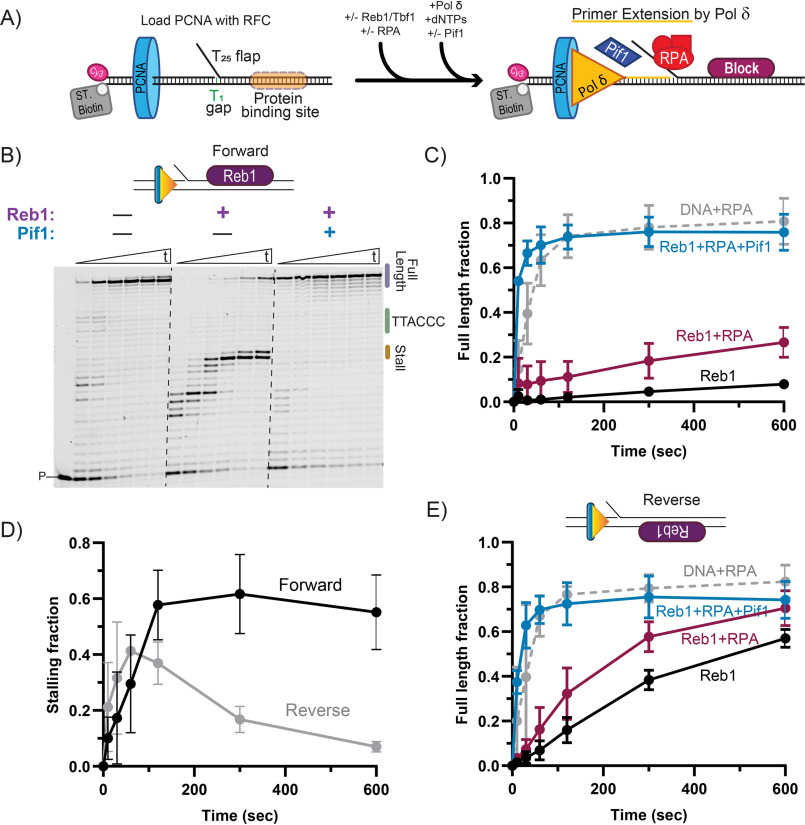
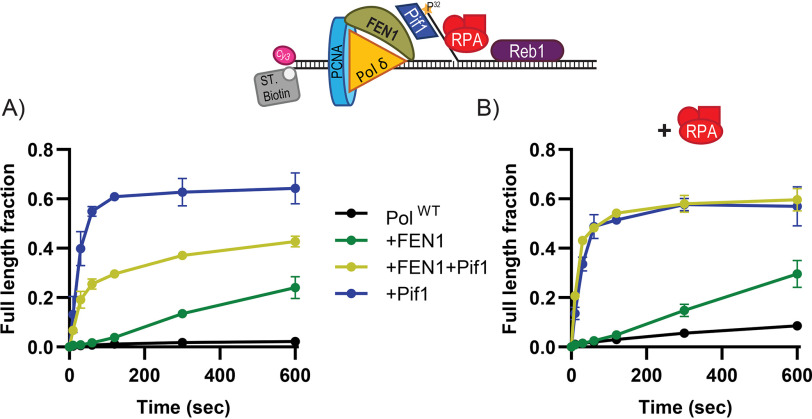
Figure 1.
The activity of the Pif1 helicase is required for DNA synthesis through a Reb1 block. A, schematic of primer extension assays performed with Reb1 or Tbf1 blocks. B, representative sequence gels of primer extension assays performed using Pol δDV in the presence of RPA with and without Reb1 or Pif1, monitored over time (10 sec, 30 sec, 1, 2, 5, and 10 min). Dashed lines have been added for visibility. C, quantification of full-length product formation for the substrate with the forward orientation of the Reb1 logo, performed using Pol δDV with and without Reb1, RPA, and Pif1. D, quantification of the fraction of stalling products (forward: +15-16 nt; reverse: +7-8 nt) formed by Pol δDV in the presence of RPA and the Reb1 binding logo either in the forward or reverse orientations. E, same as in B, but for a DNA substrate with the Reb1 binding logo on the opposite strand (“reverse” substrate). In C–E, the error bars are the mean ± S.D. from 3 independent replicates.
WT Pol δ is stalled by Reb1 bound to the downstream duplex (Fig. S1A). Because of its exonuclease activity, the enzyme idles, cyclically cleaving and resynthesizing DNA close to the block, and results in a distribution of stalled products, which makes their analysis difficult. Thus, we used an exonuclease-deficient variant of Pol δ (Pol δDV) to better quantify stall sites and fractions of extension products. Fig. 1, B and C, show representative primer extension assays and the quantifications of the fraction of full-length DNA synthesis products, respectively. The presence of Reb1 bound to the downstream duplex inhibits formation of full-length extension products, with most of the DNA synthesis stalling 2-3 nt prior to the Reb1 recognition sequence motif (Fig. 1, B, central panel, and D, forward orientation). Thus, Reb1 is a strong block to the strand displacement DNA synthesis activity of Pol δDV.
Importantly, whereas binding of RPA to the 5'-flap of the substrate stimulates DNA synthesis by Pol δ (Fig. S1B), this stimulation is not sufficient to allow replication through the block imparted by the DNA-bound Reb1 (Fig. 1, B and C). Furthermore, Reb1 bound in a reverse orientation relative to the directionality of DNA synthesis still significantly delays Pol δDV (Fig. 1E and Fig. S1C). However, when bound in the reverse orientation Reb1 is a weaker replication block compared with the forward orientation, as indicated by both the larger fraction of full-length DNA synthesis products (Fig. 1E) and by the transient accumulation of the intermediate stalling products (Fig. 1D). Independent of orientation, it is only with the addition of the 5'-3' DNA helicase Pif1 that efficient DNA replication occurs past the Reb1 block, consistent with Pif1 removing the DNA-bound Reb1 downstream of the advancing polymerase.
The requirement of Pif1 for efficient DNA synthesis by Pol δ past the Reb1 block is similar to what we reported for Rap1 (15), and is consistent with the activity of a 5'-3' helicase being required whenever DNA replication must proceed timely and efficiently through a strong protein barrier. However, this observation raises the question of whether any protein tightly bound to DNA is a block to DNA synthesis and, thus, requires the helicase activity of Pif1 for its removal. To address this question we tested whether this conclusion would also hold true for the general transcription factor Tbf1 that tightly binds to DNA (16).
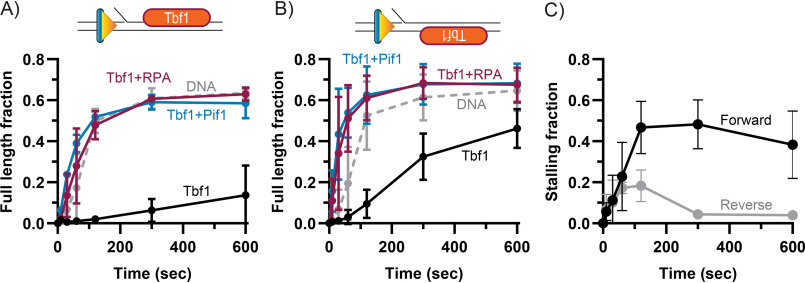
Similar to what we observed for Reb1, the DNA-bound Tbf1 is also a block to the strand displacement DNA synthesis activity of Pol δDV, with one of the DNA-bound orientations displaying a higher ability to impede DNA synthesis (Fig. 2). However, in stark contrast to the Reb1 block, binding of RPA to the 5'-flap of the substrate and its stimulation of DNA synthesis is enough to allow replication past the Tbf1 block, to the same extent as Pif1 stimulated DNA synthesis (Fig. 2). These findings suggest that, in the presence of abundant RPA, not all proteins that are tightly bound to DNA would pose a significant barrier to DNA replication and require the activity of a helicase for their removal.
Figure 2.
RPA is sufficient to stimulate DNA synthesis through a Tbf1 block. A, quantification of full-length product formation during primer extension assays performed using Pol δDV with and without Tbf1, RPA, and Pif1. B, same as A, but for a DNA substrate with the Tbf1 binding logo on the opposite strand (reverse substrate). C, quantification of the fraction of stalling products (forward: +13-16 nt; reverse: +7-10) formed by Pol δDV in the absence of RPA and with Tbf1 binding logo either in the forward or reverse orientations. In A–C, the error bars are the mean ± S.D. from 3 independent replicates.
DNA synthesis by DNA polymerase δ through a nucleosome requires Pif1
Next, we tested whether a tightly positioned nucleosome would block DNA replication, and whether the helicase activity of Pif1 would be needed to remove or reposition the nucleosome to allow DNA synthesis to proceed. For this, we designed a DNA substrate containing the strong nucleosome positioning 601 Widom sequence (17) placed 20 bp from a nick, followed by either a short (3 bp) or long (50 bp) tail to monitor potential sliding of the nucleosome toward the end of the substrate (Fig. 3A). As the DNA substrate is Cy3 labeled prior to nucleosome assembly, it is important that for our experiments all free DNA is assembled into nucleosomes to avoid the contribution from unimpeded DNA synthesis on bare DNA. As shown in Fig. S2A, nucleosome assembly was complete, as no free DNA was detected following nucleosome assembly. On these DNA substrates, Pol δWT showed limited strand displacement synthesis activity, even in the presence of RPA, and failed to reach the nucleosome (Fig. S2B). Therefore, exonuclease-deficient Pol δDV was again used to precisely quantify the polymerase stall sites.
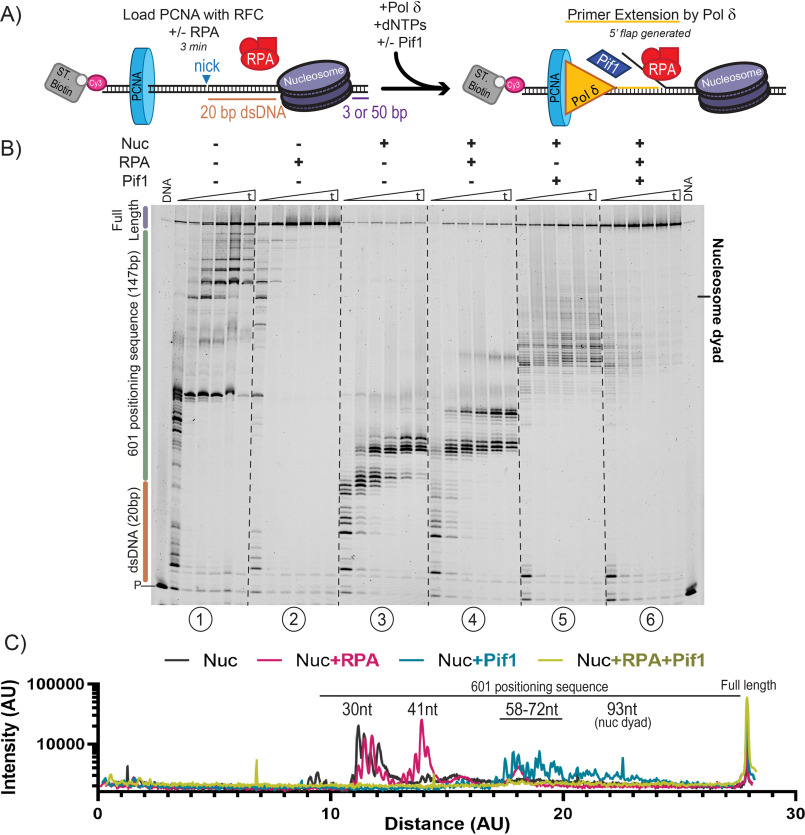
Figure 3.
Pif1 disrupts nucleosomes to allow through-replication. A, schematic of primer extension assays performed on DNA substrates with a nucleosome assembled on a 601 Widom sequence placed 20 bp from a nick. B, representative sequencing gels of primer extension assays performed using Pol δDV with or without a nucleosome, RPA, and/or Pif1 and monitored over time (1, 2, 5, 10, 20, and 30 min). C, lane plots of the band intensity of the 30-min time point for primer extension assays on nucleosome-bound substrates in B.
On a nucleosome-free substrate, Pol δDV carries out strand displacement DNA synthesis as indicated by formation of both intermediate and full-length DNA synthesis products (Fig. 3B, panel 1). Again, binding of RPA to the growing 5'-flap formed by Pol δDV stimulated strand displacement DNA synthesis, as indicated by the extension of all the intermediate products to full-length (Fig. 3B, panel 2, and Fig. S2C). However, on a nucleosome-bound substrate, few full-length products are formed, and Pol δDV is clearly halted at the front edge of the nucleosome (Fig. 3B, panel 3). Importantly, RPA does not stimulate DNA synthesis through the nucleosome, although it promotes synthesis about 8 nt further into it (Fig. 3B, panels 3 and 4). These data are presented quantitatively by monitoring the rate of accumulation of full-length DNA synthesis products (Fig. S2C) and by quantifying the band intensities of all the DNA synthesis products for each reaction condition at 30 min (Fig. 3C).
Next, we tested whether the unwinding activity of Pif1 would be sufficient to remove the stably positioned nucleosome and allow through-replication. Surprisingly, Pif1 was able to disrupt the stably bound nucleosome and promote through-replication, especially in the presence of RPA (Fig. 3B, panels 5 and 6). In the absence of RPA, Pif1 stimulates about 10% formation of full-length product (Fig. S2C), and DNA synthesis by Pol δDV stalls just before the nucleosome dyad (Fig. 3B, panel 5). On the other hand, the combined unwinding activity of Pif1 and binding of RPA to the growing 5' ssDNA flap resulted in the near elimination of stalling just prior to the nucleosome dyad (Fig. 3C, panel 6), and in a rate of accumulation of full-length DNA synthesis product that is similar to the one observed for naked DNA (Fig. S2C).
The stimulation of DNA synthesis through a nucleosome by Pif1 raises the question of whether the nucleosome is simply being removed as Pif1 unwinds the duplex DNA ahead of the polymerase or, because of the linear nature of the DNA substrate used, the nucleosome is being pushed off the end of the DNA. To test this latter possibility, we generated an otherwise identical DNA substrate that contains an extended region of 50 bp rather than 3 bp past the nucleosome positioning sequence. We reasoned that if the nucleosome was pushed downstream by Pif1 along the longer DNA, synthesis should stall further into the duplex, resulting in an offset of the position of the intermediate DNA synthesis products. In the absence of Pif1 and independent of RPA, Pol δDV stalled at the same positions on either substrate (Fig. S3A, panels 1-4). These data indicate that the nucleosome is properly positioned at the same location on both DNA substrates and that the polymerase itself cannot push the nucleosome. Independent of the downstream length past the 601 sequence, DNA synthesis by Pol δDV in the presence of Pif1 stalled at similar positions (Fig. S3A, panels 5-8). We take this as an indication that the nucleosome is not being pushed downstream by Pif1 but, rather, it is removed during unwinding. The mechanism by which this happens remains to be elucidated.
FEN1 nuclease prevents Pif1 stimulation of DNA replication through a protein block
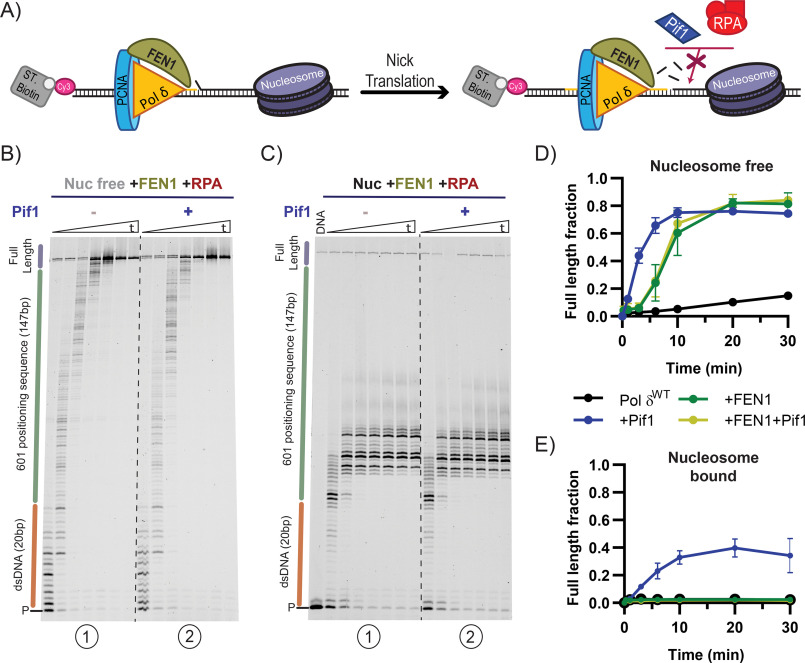
The results in the previous sections suggest that the ability of the Pif1 helicase to efficiently disrupt tightly bound proteins and nucleosomes should allow unperturbed DNA replication through most protein barriers. However, this activity of Pif1 may need to be controlled to avoid potentially deleterious consequences during DNA replication, such as excessive flap generation and unwarranted removal of nucleosomes. Nucleosome disruption during lagging strand replication could lead to loss of epigenetic information or futile re-replication of downstream Okazaki fragments. During Okazaki fragment maturation, FEN1 travels along with Pol δ to promote nick translation: a repetitive cycle of short-range strand displacement by Pol δ and flap cleavage by FEN1, as depicted in Fig. 3A (18). Thus, we sought to test whether the activity of the FEN1 nuclease would be sufficient to remove the entry point for Pif1 and thereby prevent excessive replication through barriers during nick translation.
To measure nick translation under physiologically relevant conditions, primer extension assays were performed with WT Pol δ. On naked DNA, FEN1 allows Pol δWT to efficiently synthesize full-length DNA products (Fig. 4, B, panel 1, and D). However, on a nucleosome-bound substrate, nick translation was blocked at the edge of the nucleosome (Fig. 4C, panel 1), thus preventing synthesis of full-length products (Fig. 4E). These findings are in agreement with previous results (10), and show that nick translation is halted by a strongly positioned nucleosome.
Figure 4.
FEN1 endonuclease activity prevents Pif1 stimulation of replication through a nucleosome. A, schematic of nick translation performed during primer extension assay involving FEN1. B and C, representative primer extension assays using Pol δWT and FEN1 with or without Pif1 on nucleosome-free (B) or nucleosome-bound (C) substrates, monitored over time (15 sec, 1, 2, 5, 10, 20, and 30 min). Dashed lines are added for visibility. D and E, quantification of full-length product formation of reactions with or without FEN1 and Pif1 on nucleosome-free (D) or nucleosome-bound (E) substrates. The error bars are the mean ± S.D. from 3 independent replicates.
Next, we performed the same assays in the presence of the Pif1 helicase. Without FEN1 present, Pif1 promotes replication by Pol δWT through a nucleosome (Fig. 4E and Fig. S2B). However, the situation is quite different when FEN1 is present in the reaction. FEN1 prevents Pif1 from stimulating Pol δWT DNA synthesis even on a nucleosome-free substrate (Fig. 4, B and C, compare panels 1 and 2). Nick translation by FEN1 and Pol δWT is characterized by many intermediate products as Pol δWT cyclically polymerizes a few nucleotides and idles to allow FEN1 cleavage of the short displaced flap. This patterning of DNA products was unaffected by the presence of Pif1. Furthermore, Pif1 had no effect on the rate of full-length product formation in reactions that contained FEN1 (Fig. 4, D and E). We interpret these results as indicative of FEN1 preventing Pif1 from acting on the DNA substrate by continuously cleaving the 5'-flap that is used as an entry point by Pif1. Thus, when Pol δ synthesizes DNA from a nick, the catalytic activity of FEN1 is sufficient to prevent stimulation of DNA synthesis by Pif1.
FEN1 effectively prevented Pif1 stimulation of DNA synthesis through a nucleosome. However, during Okazaki fragment processing, long flaps can form (14) and become inhibitory to FEN1 cleavage if bound by RPA (11). Thus, we tested the ability of FEN1 to protect against Pif1 stimulation on a substrate with a long 5'-flap in the presence or absence of RPA. To test this, we used the DNA substrate that contains a Reb1-binding site in the downstream dsDNA (Fig. 5) and all the experiments were performed in the presence of Reb1 to slow full-length product formation. Interestingly, FEN1 has a slight stimulatory effect on DNA synthesis through Reb1, regardless of the absence (Fig. 5A) or presence (Fig. 5B) of RPA. This was unexpected because in the presence of RPA, FEN1 was expected to be inhibited and, thus, not be able to stimulate Pol δWT. To test whether RPA inhibits FEN1 in our system, the 5'-flap was radiolabeled and cleavage by FEN1 was monitored directly (Fig. S3B). FEN1 cleaves at the base of the 5'-flap; however, at the low RPA concentration used in our reactions RPA only partially inhibits FEN1. Although our findings may appear to be at odds with the commonly accepted idea in the field that RPA inhibits FEN1, the RPA concentration dependence of FEN1 inhibition showed that a large excess of RPA relative to the DNA is needed for significant inhibition to occur (19, 20). Regardless, we found that RPA has a significant impact in promoting Pif1 unwinding over FEN1 cleavage. Without RPA, FEN1 and Pif1 compete for access to the 5'-flap and results in full-length product formation being intermediate of what achieved with Pif1 or FEN1 alone (Fig. 5A). However, in the presence of RPA, reactions containing Pif1 behave identically whether or not FEN1 is present (Fig. 5B), consistent with the finding that RPA inhibition of FEN1 was amplified by Pif1 (11). This result suggests that weak inhibition of FEN1 by RPA bound to a long flap is sufficient to favor Pif1 binding and unwinding, and suggests that for FEN1 to be effective in cleaving the 5' entry point for Pif1 it must do so before a long flap can form and interact with RPA.
Figure 5.
RPA bound to a pre-formed 5'-flap is sufficient to bypass FEN1 inhibition and favor the Pif1-dependent stimulation of DNA synthesis through a protein barrier. A, quantification of primer extension assays performed using Pol δWT with a Reb1-bound substrate with a preformed 5'-flap, the presence or absence of FEN1 and Pif1. B, same as A, but in the presence of RPA. The error bars are the mean ± S.D. from 3 independent replicates.
Together, these results reveal that the cyclical actions of Pol δWT and FEN1 during nick translation are sufficient to prevent Pif1 from acting on the DNA substrate. However, the presence of a long RPA bound 5'-flap blocks FEN1 from cleaving the 5'-flap and protecting the substrate from Pif1 entry.
Discussion
In this work, we showed that two transcription factors from S. cerevisiae, Reb1 and Tbf1, are strong blocks to DNA replication by Pol δ. Removal of the block imparted by Reb1 requires the activity of the Pif1 helicase to allow progression of DNA synthesis, similar to what we had observed for Rap1 (15). Unlike Rap1, which forms a closed complex on DNA (21), crystal structures of Schizosaccharomyces pombe Reb1 suggest that Reb1 binds DNA without forming a closed complex (22), yet we show here that it still forms a significant block to DNA synthesis that can only be removed by the activity of a helicase. On the other hand, whereas Tbf1 also forms a strong barrier to Pol δ DNA synthesis, the binding of RPA to the ssDNA flap of the substrate is all that is required to overcome the Tbf1 block and stimulate the DNA-synthesis activity of the polymerase. Thus, we conclude that in the absence of additional factors Rap1, Reb1, and Tbf1 are all strong barriers to the intrinsic strand displacement DNA-synthesis activity of Pol δ, independent of their DNA-binding affinity or specific configuration adopted on DNA. Interestingly, different additional factors are needed for their removal; either the simple binding of RPA and its stimulation of DNA synthesis or the ATP-dependent DNA-unwinding activity of an accessory helicase, such as Pif1. Although it remains to be determined how the DNA-binding affinity and/or specific DNA binding modes of transcription factors may contribute to defining their ability to act as a barrier to DNA replication, our data strongly suggest that not all transcription factors that can block Pol δ DNA synthesis will be a block to DNA replication when RPA is abundant.
During DNA replication, nucleosomes are removed from in front of the replication fork and redeposited behind the fork, randomly distributed between the two daughter strands (3–8). This process of nucleosome recycling deposits nucleosomes directly in the path of lagging strand replication. It is possible that nucleosomes may serve as a barrier to prevent excessive re-replication by obstructing Pol δ strand displacement synthesis during lagging strand DNA synthesis. Indeed, in vivo isolated Okazaki fragments were found to display length periodicity similar to micrococcal nuclease-treated nucleosome arrays (9). Furthermore, genome wide alignment of Okazaki fragment junctions to nucleosome positioning was highly correlative (9). A similar phenomenon was observed in an in vitro bidirectional replication system in which replication of chromatin resulted in short lagging strand Okazaki fragments similar to lengths seen in vivo (100-300 nt), whereas on bare DNA, much longer and heterogeneous lengths were observed (10). Although in our assay WT Pol δ was not able to perform sufficient strand displacement DNA synthesis to reach the nucleosome, its exonuclease-deficient variant did and showed that a nucleosome is indeed a strong barrier to DNA synthesis by Pol δ.
Here we show that, in addition to removing tightly bound transcription factors, the Pif1 helicase stimulates efficient DNA replication through a nucleosome in the presence of RPA. Because Pif1 disrupts nucleosomes that are highly stable when positioned with the 601 Widom sequence (17), we would expect Pif1 to displace naturally bound nucleosomes as well. Furthermore, our data strongly suggest that Pif1 ejects a nucleosome rather than pushing and/or repositioning it along the DNA. This may be due to the strong interaction of the nucleosome with the optimal configuration of the artificially selected 601 sequence (17, 23), which makes sliding out of position less favorable (24). In cells, nucleosome assembly on random genomic DNA and the activity of nucleosome remodelers (23, 25–27) may favor sliding and, in this context, the nucleosomes may be pushed along DNA as the Pif1 helicase unwinds the duplex to allow replication.
The activity of the Pif1 helicase is implicated in multiple processes involving DNA synthesis, such as processing of long flaps together with the Dna2 helicase/nuclease during Okazaki fragment maturation (11, 12, 14, 28, 29) and bubble migration during break-induced replication (30, 31). In the latter, the sister chromatid is used as a template to copy the genetic information via strand displacement DNA synthesis from the invading 3' end (32, 33). Thus, nucleosomes and select transcription factors on the sister chromatid must be removed to circumvent their strong blocking activity on DNA synthesis. In this scenario, we propose that one of the functions of the Pif1 helicase during break-induced replication is to remove protein barriers to facilitate DNA replication. Our data show that when coupled to DNA synthesis the helicase activity of Pif1 can deal with any protein obstacle we have tested so far.
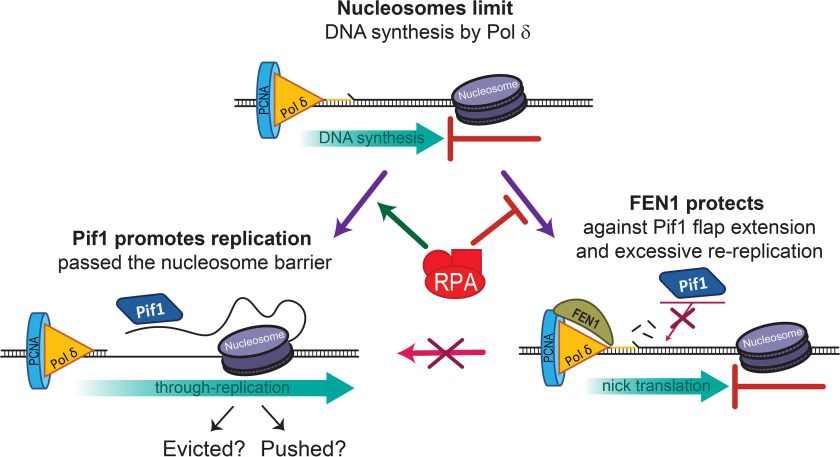
Although the activity of Pif1 is clearly important for long flap processing (11, 12, 14, 28, 29), the stimulation of DNA synthesis and efficient removal of protein barriers raises the question whether in general Pif1 is actively prevented from acting during Okazaki fragment maturation. First, based on our observation that Pif1 efficiently removes protein barriers in front of Pol δ, we would not expect the strong genome wide correlation of the positions of transcription-binding sites and nucleosomes with the position of Okazaki fragment junctions (9). Second, during lagging strand DNA synthesis, uncontrolled activity of Pif1 at the Okazaki fragments would lead to extensive DNA re-synthesis, which would be uneconomical for the cell. Consistent with these arguments, we provided experimental evidence that shows that FEN1 nuclease could play a protective role in preventing unwarranted Pif1-stimulated re-replication. Our data shows that a nucleosome is a block to FEN1-mediated nick translation, consistent with a recent study (10). In addition, FEN1 suppresses the Pif1-dependent stimulation of DNA synthesis on naked or nucleosomal DNA. By repetitively cleaving short flaps generated by the strand displacement activity of Pol δ, FEN1 removes the 5' entry point for Pif1. On the other hand, whereas RPA bound to a long pre-formed flap only moderately inhibits FEN1 cleavage, this inhibition is sufficient to allow Pif1 access to the flap and promote Pif1-stimulated synthesis through a protein block. Fig. 6 depicts our current model for how the interplay between FEN1 activity, RPA binding, Pif1 unwinding, and stimulation of DNA synthesis allows DNA replication past a protein barrier, whereas at the same time protecting the cell from excessive re-replication. Our findings emphasize the importance of FEN1 during lagging strand replication in limiting Pif1 activity and, thus, preventing re-replication and potential epigenetic loss.
Figure 6.
Model of FEN1 and nucleosome protection from re-replication during Okazaki fragment maturation. Predominantly, traveling along with Pol δ, FEN1 will cleave short flaps formed by Pol δ strand displacement activity, removing Pif1's access point for unwinding. However, a delay in endonucleolytic cleavage by FEN1 could result in an extended 5'-flap that could bind RPA and promote Pif1 unwinding. Pif1 unwinding could cause replication through a downstream nucleosome and cause potential loss of epigenetic information or re-replication of downstream Okazaki fragments.
Experimental procedures
Proteins and DNA substrates
S. cerevisiae DNA polymerase δ, Pol δWT, and the exonuclease-defective Pol δDV (D520V) were purified from a yeast overexpression system as described previously (15). S. cerevisiae RPA (34), PCNA (35), RFC (36), and FEN1 (37) were purified as previously described. S. cerevisiae Pif1 was purified as previously described (38). The coding sequences of full-length Reb1 and Tbf1 were PCR amplified from S. cerevisiae genomic DNA from strain S288C, cloned into pGEX-6p-1 and overexpressed in BL21 cells. The cells were grown in LB media, induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside, and grown overnight at 16 °C. The pellet was resuspended in lysis buffer A (50 mm sodium phosphate, 400 mm NaCl, 0.5 mm EDTA, 10% (v/v) glycerol, 1 mm DTT, and 1 mm phenylmethylsulfonyl fluoride) and lysed by sonication. The cell lysate was centrifuged at 12,000 rpm for 30 min at 4 °C and batch bound to glutathione-S-transferase (GST) beads pre-equilibrated with buffer A overnight at 4 °C. Following washing, protein-bound beads were packed in a column and eluted with elution buffer B (20 mm Tris, pH 7.3, 400 mm NaCl, 1 mm EDTA, 10% (v/v) glycerol, 5 mm DTT, and 25 mm GSH). The GST tag was cleaved during overnight dialysis in dialysis buffer C (20 mm Tris, pH 7.3, 200 mm NaCl, 10% (v/v) glycerol, 1 mm EDTA, 5 mm DTT) with 3C protease. Dialyzed and cleaved Reb1 was loaded on a heparin column pre-equilibrated with buffer C, washed with buffer C containing 350 mm NaCl, and eluted in buffer C containing 450-600 mm NaCl. Purified Reb1 was concentrated, dialyzed into storage buffer D (20 mm HEPES, pH 7.4, 400 mm NaCl, 1 mm EDTA, 1 mm DTT, 40% (v/v) glycerol), and stored at −80 °C. Tbf1 was purified in the same way as Reb1, although it eluted from the heparin column in buffer C containing 300-450 mm NaCl.
Reb1 and Tbf1 DNA substrates were purchased from Integrated DNA Technologies (Coralville, IA) and their sequences are listed in Table S1. The 3' biotinylated DNA template strand was annealed to a 5' fluorescently labeled primer and unlabeled top strand in the presence of 20 mm HEPES, pH 8.0, 100 mm NaCl, 2 mm MgCl2. The reaction was heated to 95 °C and allowed to slowly reach room temperature. To generate the DNA substrate containing the 601 Widom sequence (17) a DNA sequence containing a NtBbvCI nickase site 20 bp upstream from the 601 Widom sequence was synthesized into pUC57 by GenScript. Using this plasmid as a template, the DNA substrate was generated by PCR using Phusion polymerase (New England Biolabs) and a 5' biotinylated and fluorescently labeled forward primer and either an unlabeled or a 5' biotinylated reverse primer. The PCR product was purified using the QIAquick PCR Purification Kit from Qiagen (Hilden, Germany). Purified PCR product was nicked using NtBbvCI nickase from New England Biolabs (Ipswich, MA) at 37 °C and purified again. Nicked and purified 601 DNA substrate was quantified by measuring the absorbance at 260 nm on a Varian Cary-100 spectrophotometer. Human octamers were purchased from EpiCypher (Durham, NC). Nucleosome in vitro assembly was performed using salt dialysis similar to as previously described (39) with a 1.8-fold excess of human octamer to DNA. Briefly, the DNA and human octamer were combined and dialyzed sequentially for 50 min in dialysis buffer (10 mm Tris, pH 7.5, 1 mm EDTA, 1 mm DTT) with the following salt (NaCl) concentrations: 2, 1.8, 1.6, 1, 0.8, 0.6, 0.4, 0.2, 0.1, and 0.05 m. The sample was collected and stored at 4 °C.
Primer extension assays
Primer extension assays were carried out in Buffer TM (20 mm Tris-HCl, pH 7.8, 8 mm MgAc2, 1 mm DTT, 0.1 mg/ml of BSA) with 50 mm NaCl for Reb1 and Tbf1 assays and 100 mm NaCl for nucleosome assays. Replication assays were performed with 20 nm of the DNA substrate. A standard loading protocol was followed (35) using the following final concentrations of each component. RFC (20 nm) and PCNA (20 nm) were allowed to react with the biotinylated DNA substrate in the presence of streptavidin (20 nm) and ATP (1 mm) for 2 min at 30 °C, followed by 30 s incubation of RPA (40 nm) and Reb1 (40 nm) or Tbf1 (80 nm), where mentioned. The reactions were started with the addition of Pol δ (20 nm) and dNTPs (100 μm), with or without Pif1 (40 nm) or FEN1 (20 nm). Use of Pol δDV or Pol δWT is stated in the figure legends. At the indicated times, the reactions were stopped by the addition of 80 mm EDTA and 0.08% SDS and incubated at 55 °C for 10 min. After the addition of formamide (50% final) and bromphenol blue, the samples were heated at 95 °C for 2 min and analyzed on a 12% denaturing polyacrylamide gel, pre-run for 1.5 h in 0.5× TBE. The gels were scanned using a Typhoon 9400 Variable Mode Imager (GE Healthcare), monitoring the Cy3 fluorescence of the labeled primer. Accumulation of the full-length product was quantified using ImageQuant; the background was subtracted using the rubber-band option in ImageQuant and the intensity of full-length product was normalized to the intensity of the lane. The reported values in the figures are the mean ± S.D. from three independent replicates. Reactions performed with the 601 substrate were performed similarly, except the concentrations of the proteins were doubled. Furthermore, the DNA from each time point was ethanol precipitated prior to loading on an 8% denaturing polyacrylamide gel that was pre-run for 1.5 h in 1× TBE.
FEN1-cleavage assay
The FEN1-cleavage assays were performed on the Reb1 forward substrate in the presence of 200 nm RPA when tested. The 5'-flap of the top strand was radiolabeled with [γ-32P]ATP using T4-PNK from New England Biolabs (Ipswich, MA). The assay was performed similarly to primer extension assays on Reb1 substrates using Pol δWT and FEN1 but in the absence of dNTPs to prevent DNA synthesis. Samples were run on 12% gels that were dried and developed on a PhosphorImager.
Data availability
All data are contained within the manuscript.
Supplementary Material
Acknowledgments
We thank Dr. Saurabh Singh, Dr. Paolo De Bona, and Carrie Stith for help in producing some of the proteins, and members of the Burgers' group for discussions. We thank Drs. Lohman, Vindigni, and Mosammaparast for suggestions during this project.
This article contains supporting information.
Author contributions—M. A. S., P. M. B., and R. G. conceptualization; M. A. S. data curation; M. A. S. formal analysis; M. A. S. and R. G. investigation; M. A. S. and R. G. visualization; M. A. S., P. M. B., and R. G. methodology; M. A. S., P. M. B., and R. G. writing-original draft; P. M. B. and R. G. supervision; P. M. B. and R. G. funding acquisition; P. M. B. and R. G. writing-review and editing; R. G. resources; R. G. project administration.
Funding and additional information—This work was supported by the National Institutes of Health Grants GM098509 (to R. G.) and GM118129 (to P. B.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- Pol δ
- polymerase δ
- dsDNA
- double-stranded DNA
- ssDNA
- single-stranded DNA
- nt
- nucleotide
- PCNA
- proliferating cell nuclear antigen
- RFC
- replication factor C.
References
- 1. Patel D. R., and Weiss R. S. (2018) A tough row to hoe: when replication forks encounter DNA damage. Biochem. Soc. Trans. 46, 1643–1651 10.1042/BST20180308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gaillard H., García-Muse T., and Aguilera A. (2015) Replication stress and cancer. Nat. Rev. Cancer 15, 276–289 10.1038/nrc3916 [DOI] [PubMed] [Google Scholar]
- 3. Groth A. (2009) Replicating chromatin: a tale of histones. Biochem. Cell Biol. 87, 51–63 10.1139/O08-102 [DOI] [PubMed] [Google Scholar]
- 4. Annunziato A. T. (2015) The fork in the road: histone partitioning during DNA replication. Genes 6, 353–371 10.3390/genes6020353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Prado F., and Maya D. (2017) Regulation of replication fork advance and stability by nucleosome assembly. Genes 8, 49 10.3390/genes8020049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gan H., Serra-Cardona A., Hua X., Zhou H., Labib K., Yu C., and Zhang Z. (2018) The Mcm2-Ctf4-Polα axis facilitates parental histone H3-H4 transfer to lagging strands. Mol. Cell 72, 140–151 10.1016/j.molcel.2018.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Huang H., Strømme C. B., Saredi G., Hödl M., Strandsby A., González-Aguilera C., Chen S., Groth A., Dinshaw., and Patel D. J. (2015) A unique binding mode enables MCM2 to chaperone histones H3–H4 at replication forks. Nat. Struct. Mol. Biol. 22, 618–626 10.1038/nsmb.3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Clemente-Ruiz M., González-Prieto R., and Félix P. (2011) Histone H3K56 acetylation, CAF1, and Rtt106 coordinate nucleosome assembly and stability of advancing replication forks. PLoS Genet. 7, e1002376 10.1371/journal.pgen.1002376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Smith D. J., and Whitehouse I. (2012) Intrinsic coupling of lagging-strand synthesis to chromatin assembly. Nature 483, 434–438 10.1038/nature10895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Devbhandari S., Jiang J., Kumar C., Whitehouse I., and Remus D. (2017) Chromatin constrains the initiation and elongation of DNA replication. Mol. Cell 65, 131–141 10.1016/j.molcel.2016.10.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rossi M. L., Pike J. E., Wang W., Burgers P. M. J., Campbell J. L., and Bambara R. A. (2008) Pif1 helicase directs eukaryotic Okazaki fragments toward the two-nuclease cleavage pathway for primer removal. J. Biol. Chem. 283, 27483–27493 10.1074/jbc.M804550200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pike J. E., Burgers P. M. J., Campbell J. L., and Bambara R. A. (2009) Pif1 helicase lengthens some Okazaki fragment flaps necessitating Dna2 nuclease/helicase action in the two-nuclease processing pathway. J. Biol. Chem. 284, 25170–25180 10.1074/jbc.M109.023325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu B., Hu J., Wang J., and Kong D. (2017) Direct visualization of RNA-DNA primer removal from Okazaki fragments provides support for flap cleavage and exonucleolytic pathways in eukaryotic cells. J. Biol. Chem. 292, 4777–4788 10.1074/jbc.M116.758599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rossi S. E., Foiani M., and Giannattasio M. (2018) Dna2 processes behind the fork long ssDNA flaps generated by Pif1 and replication-dependent strand displacement. Nat. Commun. 9, 1–11 10.1038/s41467-018-07378-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Koc K. N., Singh S. P., Stodola J. L., Burgers P. M., and Galletto R. (2016) Pif1 removes a Rap1-dependent barrier to the strand displacement activity of DNA polymerase δ. Nucleic Acids Res. 44, 3811–3819 10.1093/nar/gkw181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Koering C. E., Fourel G., Binet-Brasselet E., Laroche T., Klein F., and Gilson E. (2000) Identification of high affinity Tbf1p-binding sites within the budding yeast genome. Nucleic Acids Res. 28, 2519–2526 10.1093/nar/28.13.2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lowary P. T., and Widom J. (1998) New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 276, 19–42 10.1006/jmbi.1997.1494 [DOI] [PubMed] [Google Scholar]
- 18. Stodola J., and Burgers P. (2016) Resolving individual steps of Okazaki-fragment maturation at a millisecond timescale. Nat. Struct. Mol. Biol. 23, 402–408 10.1038/nsmb.3207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Henry R. A., Balakrishnan L., Tan Ying-Lin S., Campbell J. L., and Bambara R. A. (2010) Components of the secondary pathway stimulate the primary pathway of eukaryotic Okazaki fragment processing. J. Biol. Chem. 285, 28496–28505 10.1074/jbc.M110.131870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kao H. I., Veeraraghavan J., Polaczek P., Campbell J. L., and Bambara R. A. (2004) On the roles of Saccharomyces cerevisiae Dna2p and Flap endonuclease 1 in Okazaki fragment processing. J. Biol. Chem. 279, 15014–15024 10.1074/jbc.M313216200 [DOI] [PubMed] [Google Scholar]
- 21. König P., Giraldo R., Chapman L., and Rhodes D. (1996) The crystal structure of the DNA-binding domain of yeast RAP1 in complex with telomeric DNA. Cell 85, 125–136 10.1016/S0092-8674(00)81088-0 [DOI] [PubMed] [Google Scholar]
- 22. Jaiswal R., Choudhury M., Zaman S., Singh S., Santosh V., Bastia D., and Escalante C. R. (2016) Functional architecture of the Reb1-Ter complex of Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. U.S.A. 113, E2267–E2276 10.1073/pnas.1525465113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Struhl K., and Segal E. (2013) Determinants of nucleosome positioning. Nat. Struct. Mol. Biol. 20, 267–273 10.1038/nsmb.2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Niina T., Brandani G. B., Tan C., and Takada S. (2017) Sequence-dependent nucleosome sliding in rotation-coupled and uncoupled modes revealed by molecular simulations. PLoS Comput. Biol. 13, e1005880 10.1371/journal.pcbi.1005880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mueller-Planitz F., Klinker H., and Becker P. B. (2013) Nucleosome sliding mechanisms: new twists in a looped history. Nat. Struct. Mol. Biol. 20, 1026–1032 10.1038/nsmb.2648 [DOI] [PubMed] [Google Scholar]
- 26. Park Y. J., Chodaparambil J. V., Bao Y., McBryant S. J., and Luger K. (2005) Nucleosome assembly protein 1 exchanges histone H2A-H2B dimers and assists nucleosome sliding. J. Biol. Chem. 280, 1817–1825 10.1074/jbc.M411347200 [DOI] [PubMed] [Google Scholar]
- 27. Längst G., and Becker P. B. (2001) ISWI induces nucleosome sliding on nicked DNA. Mol. Cell 8, 1085–1092 10.1016/S1097-2765(01)00397-5 [DOI] [PubMed] [Google Scholar]
- 28. Budd M. E., Reis C. C., Smith S., Myung K., and Campbell J. L. (2006) Evidence suggesting that Pif1 helicase functions in DNA replication with the Dna2 helicase/nuclease and DNA polymerase δ. Mol. Cell Biol. 26, 2490–2500 10.1128/MCB.26.7.2490-2500.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Stith C., Sterling J., Resnick M., Gordenin D., and Burgers P. (2008) Flexibility of eukaryotic Okazaki fragment maturation through regulated strand displacement synthesis. J. Biol. Chem. 283, 34129–34140 10.1074/jbc.M806668200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wilson M. A., Kwon Y., Xu Y., Chung W.-H., Chi P., Niu H., Mayle R., Chen X., Malkova A., Sung P., and Ira G. (2013) Pif1 helicase and Pold promote recombination-coupled DNA synthesis via bubble migration. Nature 502, 393–396 10.1038/nature12585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Saini N., Ramakrishnan S., Elango R., Ayyar S., Zhang Y., Deem A., Ira G., Haber J. E., Lobachev K. S., and Malkova A. (2013) Migrating bubble during break-induced replication drives conservative DNA synthesis. Nature 502, 389–392 10.1038/nature12584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Malkova A., Ivanov E. L., and Haber J. E. (1996) Double-strand break repair in the absence of RAD51 in yeast: a possible role for break-induced DNA replication. Proc. Natl. Acad. Sci. U.S.A. 93, 7131–7136 10.1073/pnas.93.14.7131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Malkova A., Naylor M. L., Yamaguchi M., Ira G., and Haber J. E. (2005) RAD51-dependent break-induced replication differs in kinetics and checkpoint responses from RAD51-mediated gene conversion. Mol. Cell Biol. 25, 933–944 10.1128/MCB.25.3.933-944.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Henricksen L. A., Umbricht C. B., and Wold M. S. (1994) Recombinant replication protein A expression, complex formation, and functional characterization. J. Biol. Chem. 269, 11121–11132 [PubMed] [Google Scholar]
- 35. Eissenberg J. C., Ayyagari R., Gomes X. V., and Burgers P. M. J. (1997) Mutations in yeast proliferating cell nuclear antigen define distinct sites for interaction with DNA polymerase δ and DNA polymerase ε. Mol. Cell Biol. 17, 6367–6378 10.1128/mcb.17.11.6367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gomes X. V., Gary S. L., and Burgers P. M. J. (2000) Overproduction in Escherichia coli and characterization of yeast replication factor C lacking the ligase homology domain. J. Biol. Chem. 275, 14541–14549 10.1074/jbc.275.19.14541 [DOI] [PubMed] [Google Scholar]
- 37. Gomes X. V., and Burgers P. M. J. (2000) Two modes of FEN1 binding to PCNA regulated by DNA. EMBO J. 19, 3811–3821 10.1093/emboj/19.14.3811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Singh S. P., Koc K. N., Stodola J. L., and Galletto R. (2016) A monomer of Pif1 unwinds double-stranded DNA and it is regulated by the nature of the non-translocating strand at the 3′-end. J. Mol. Biol. 428, 1053–1067 10.1016/j.jmb.2016.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Luger K., Rechsteiner T. J., and Richmond T. J. (1999) Preparation of nucleosome core particles from recombinant histones. Methods Enzymol. 304, 3–19 10.1016/s0076-6879(99)04003-3 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the manuscript.