Abstract
Rationale: Platinum-based chemotherapy is one of treatment mainstay for patients with advanced lung squamous cell carcinoma (LUSC) but it is still a “one-size fits all” approach. Here, we aimed to investigate the predictive and monitoring role of circulating cell-free DNA (cfDNA) profiling for the outcome of first-line chemotherapy in patients with advanced LUSC.
Methods: Peripheral blood samples of 155 patients from a phase IV trial and 42 cases from an external real-world cohort were prospectively collected. We generated a copy number variations-based classifier via machine learning algorithm to integrate molecular profiling of cfDNA, named RESPONSE SCORE (RS) to predict the treatment outcome. To monitor the treatment efficacy, cfDNA samples collected at different time points were subjected to an ultra-deep sequencing platform.
Results: The results showed that patients with high RS showed substantially higher objective response rate than those with low RS in training set (P < 0.001), validation set (P < 0.001) and real-world cohort (P = 0.019). Furthermore, a significant difference was observed in both progression-free survival (training set, P < 0.001; validation set: P < 0.001; real-world cohort: P = 0.019) and overall survival (training set, P < 0.001; validation set: P = 0.037) between high and low RS group. Notably, variant allele frequency (VAF) calculated from an ultra-deep sequencing platform significantly reduced in patients experienced a complete or partial response after 2 cycles of chemotherapy (P < 0.001), while it significantly increased in these of non-responder (P < 0.001). Moreover, VAF undetectable after 2 cycles of chemotherapy was correlated with markedly better objective response rate (P < 0.001) and progression-free survival (P < 0.001) than those with detectable VAF.
Conclusions: These findings indicated that the RS, a circulating cfDNA sequencing-based stratification index, could help to guide first-line chemotherapy in advanced LUSC. The change of VAF is valuable to monitor the treatment response.
Keywords: Non-small-cell lung cancer, cell-free DNA, chemotherapy, machine learning
Introduction
Lung squamous cell carcinoma (LUSC) is a common histological type of non-small-cell lung cancer (NSCLC) 1, 2. Unlike lung adenocarcinoma, most of LUSC does not harbor targetable driver mutations. Even though some of LUSC had driver mutations, targeted therapies are rarely used in this setting 3, 4. Recently, immunotherapy targeted programmed cell death 1 (PD-1) and its ligand (PD-L1) has shifted the treatment landscape in patients with advanced LUSC, but only ~20% of them got response to anti-PD-(L)1 monotherapy without biomarker selection 5-8. Therefore, platinum-based chemotherapy still plays an important role in the treatment for patients with LUSC as either front line or second ling setting 9. However, platinum-based chemotherapy in LUSC is still a “one-size fits all” approach. Although we endeavored to investigate single or combined molecular features to predict chemotherapy response, to date, none of them had been successfully implemented in clinical practice 9, 10. Recently, next generation sequencing (NGS) characterizes alterations in genome and demonstrated that tumor mutation burden (TMB) was associated with response to checkpoint inhibitors 11, 12. Furthermore, other genomic signature was found to be correlated with molecular targeted agents 13, which showed the potent for identifying efficacy predictors to chemotherapy via genetic profiling.
Circulating cell-free DNA (cfDNA) is a potential surrogate for the genomic profiling of tumor 14-17. Several publications have reviewed the clinical application of cfDNA in NSCLC, and suggested cfDNA as an alternative predictor for targeted therapy and immunotherapy 14, 18-20. Indeed, it is feasible for real-time monitoring of tumor relapse or disease progression 21-23. Moreover, our previous study in a limited number of NSCLC patients found that those with objective response to first-line chemotherapy have distinct mutational landscape of cfDNA when compared with non-responders 24, indicating cfDNA profiling might be a potential marker to guide chemotherapy in NSCLC.
To further investigate the predictive value of cfDNA profiling for doublet chemotherapy in patients with advanced LUSC, we conducted this biomarker exploratory analysis in patients from a randomized phase IV trial (named LIPUSU, NCT02996214). From the training cohort, we developed a copy number variations (CNV)-based classifier, named RESPONSE SCORE (RS, details are listed in the Methods: Definition and Algorithm of RESPONSE SCORE), via machine learning algorithm to integrate molecular profiling. Our results showed that patients with high RS showed significantly superior objective response rate (ORR), progression-free survival (PFS) and overall survival (OS) than those with low RS in training, validation set and an external real-world cohort. Notably, change of variant allele frequency (VAF) of common mutations could monitor response and might help to identify early disease progression of chemotherapy.
Methods
Patients' enrollment and sample collection
Eligible patients were consecutively enrolled from a randomized phase IV trial, named LIPUSU. The purpose of LIPUSU is to investigate the efficacy and safety of paclitaxel liposome injection plus cisplatin compared with gemcitabine plus cisplatin as first-line therapy in patients with advanced LUSC. Details of the study design and patient eligibility criteria were summarized in https://clinicaltrials.gov/ (NCT02996214). Briefly, enrolled patients were randomly assigned to receive up to 6 cycles of paclitaxel liposome (175 mg/m2) plus cisplatin at a dose of 75 mg/m2 (LP) on day 1, every 21 days, or gemcitabine 1000 mg/m2 (on day 1 and 8, every 21 days) plus cisplatin at a dose of 75 mg/m2 (GP), on day 1, every 21 days. Blood samples were collected at the baseline and two cycle of chemotherapy or disease progression. The study protocol was approved by the institutional ethics committee of each participating medical center. All patients signed informed consent forms before the initiation of any study-related procedure. We also adopted an external real-word cohort from online data to further validate the finding in this study.
DNA extraction and sequencing
Peripheral blood cells and plasma were separated by centrifugation at 1600×g for 10 min. Supernatant plasma was transferred to a 2 milliliter (mL) centrifuge tube and centrifuged at 16,000×g for 10 min. MagMAXTM Cell-Free DNA isolation kit (Life Technologies, California, USA) was utilized to extract cfDNA in the plasma according to the instruction. TIANGEN whole blood DNA kit (TIANGEN, Beijing, China) was used to extract DNA from peripheral blood cells according to the manufacturer's instructions. Further details of sequencing and bioinformatic analyses were provided in Supplementary Methods.
Definition and Algorithm of RESPONSE SCORE
To better predict the treatment response, we identified a set of genes to generate CNV-based classifier, named RESPONSE SCORE (RS). The criteria for the included genes were: (i) common driver mutations with frequency ≥ 2%; (ii) genes showed predictive value with P < 0.1 in univariate analysis from our cohort; (iii) genes potentially associated with efficacy of chemotherapy (e.g. ERCC1/2, BRAC1/2, RRM1, etc.) in previous studies; (iv) genes potentially correlated with the transport, metabolism and resistance of chemotherapeutic agents in previous publications; (v) genes involved in several biological processes associated with cancer cell survival, growth and apoptosis such as DNA replication, transcription and damage repair, cell cycle, immune response pathways and so on. The detailed algorithm of RS calculation was summarized in Supplemental Materials.
Statistical analysis
Both Wilcoxon signed rank test and t test were applied for comparison of CNV and mutation frequency between defined patients' groups. Correlations between RS high and low group were analyzed using the chi-squared or Fisher's exact test for categorical variables. The continuous variables were analyzed by ANOVA and Tukey's multiple comparison tests. Mann-Whitney U tests or Kruskal-Wallis rank sum tests were used for comparisons of continuous variables across multiple groups. The Kaplan-Meier curve with log-rank test was used to test the significance of differences between two groups. All the diagrams were drawn with R packages including ComplexHeatmap and ClusterProfile. Circos-0.69-6 was used to generate circos plots for CNV distributions. All statistical analyses were conducted using GraphPad PRISM 6.0 and the SPSS statistical software, version 22.0 (SPSS Inc., Chicago, IL, USA). P < 0.05 was considered statistically significant.
Results
Baseline characteristics of included patients
Totally, 155 patients with advanced LUSC were identified and their blood samples at baseline and cycle 2 treatment were prospectively collected (Figure 1 and Figure S1). Baseline characteristics were summarized in Table S1. In brief, 151 (97.4%) of them were male and 129 (83.2%) had Eastern Cooperative Oncology Group (ECOG) performance status of 1. Most of patients had smoking history (96.1%). 80 patients received LP and 75 received GP. In LP group, there were 1, 47, 18 and 14 patients' that experienced complete response (CR), partial response (PR), stable disease (SD) and disease progression (PD) to first-line treatment, respectively. In GP group, 45, 10 and 20 patients experienced PR, SD and PD. Median PFS and OS were 153 and 341 days in LP group, 154 and 384 days in GP group, respectively.
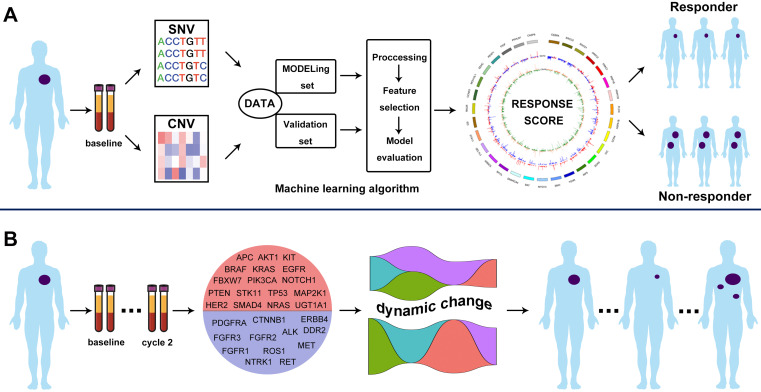
Figure 1.
Schematic illustration of the overall investigation. A. Machine learning algorithm to generate CNV-based RS for response prediction via integrating cfDNA molecular features; B. ICP-based dynamic change of VAF as baseline and cycle 2 treatment monitored the treatment response.
Mutational landscape of cfDNA and its association with treatment response
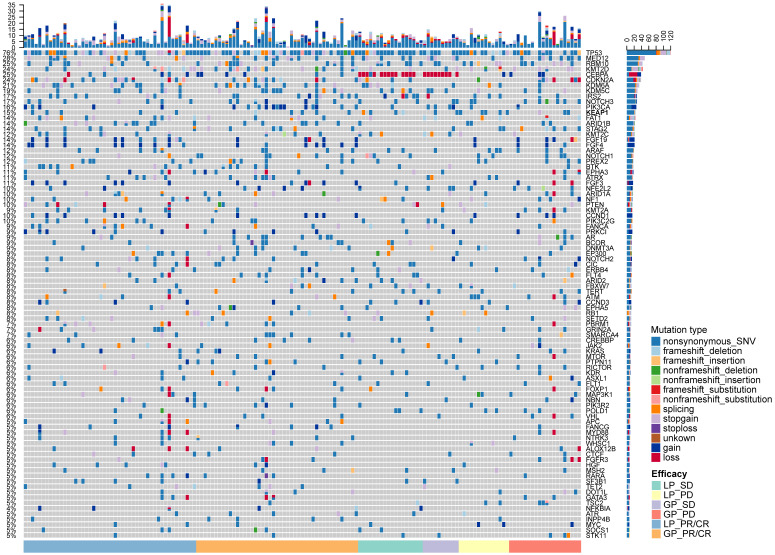
We identified 106 common genetic alterations with mutational frequency ≥ 2% (Figure 2). The most common genetic alteration was TP53 (76.1%, 118/155). We listed the SNV and CNV landscape of LP and GP group in Figure S2-5. Overall, the mutational landscape was analogous between patients with CR/PR and SD/PD (Figure 2). The median TMB was 6.5 and 7.6 mutations/Mb in LP and GP group, respectively. While we used different cutoffs of TMB, it did not show any predictive value for both ORR and PFS (cutoff of TMB 25th, 50th and 75th: ORR, P = 0.793, P = 0.760, P = 0.880, respectively; PFS, P = 0.9267, P = 0.7128, P = 0.5887, respectively; Figure S6A-F). Subgroup analysis showed distinct cutoffs of TMB was also not associated with ORR and PFS in both LP (cutoff of TMB 25th, 50th and 75th: ORR, P = 0.598, P = 0.999, P = 0.598, respectively; PFS, P = 0.9701, P = 0.8431, P = 0.7685, respectively; Figure S7A-F) and GP group (cutoff of TMB 25th, 50th and 75th: ORR, P = 0.448, P = 0.925, P = 0.912, respectively; PFS, P = 0.6805, P = 0.5232, P = 0.8230, respectively; Figure S7G-L). Of note, when we investigated the predictive value of each prevalent gene alteration (frequency ≥ 5%), we found no individual gene alterations showed association with the outcome of chemotherapy.
Figure 2.
The mutational landscape of included patients. Upper panel: The frequency of listed driver genes. Middle panel: The matrix of mutations in a selection of frequently mutated genes. Columns represent samples. Right panel: The total number of patients harboring mutations in each gene. LP, paclitaxel liposome plus cisplatin; GP, gemcitabine plus cisplatin; CR, complete response; PR, partial response; SD, stable disease; PD, disease progression.
Generation of CNV-based RS for response prediction
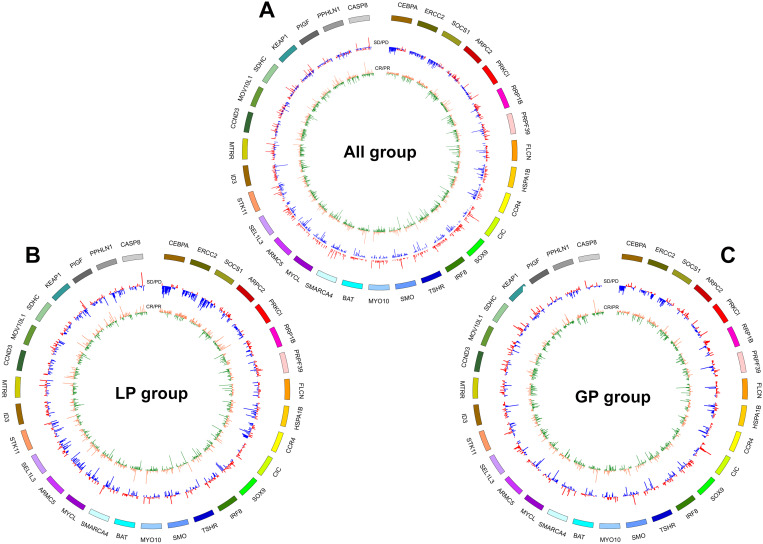
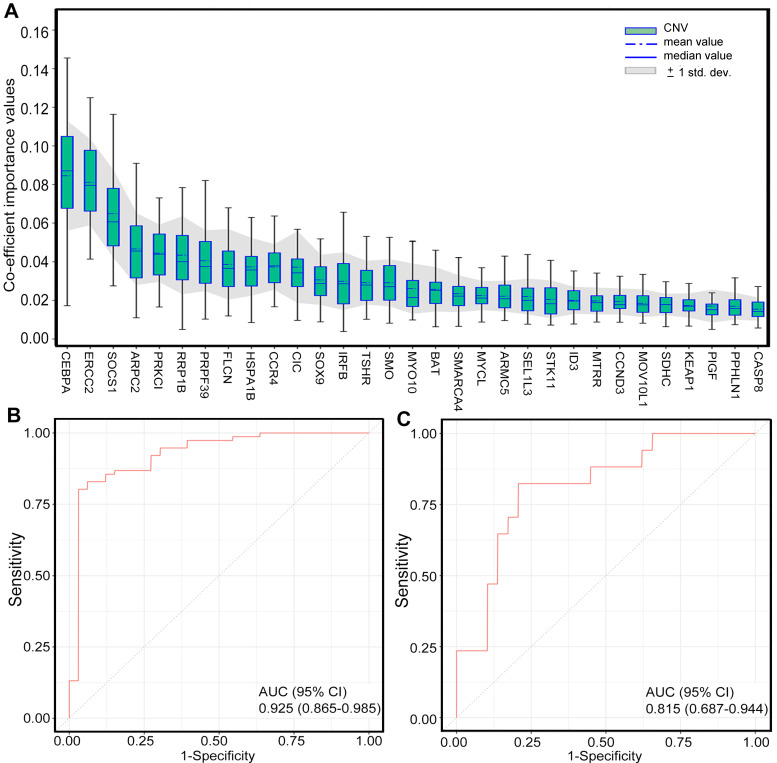
To identify the patients who might benefit from chemotherapy, we generated a CNV-based classifier, named RS (Figure 1A). Firstly, we focused on the potential impact of cfDNA concentrations. The results showed that baseline cfDNA concentrations had no significant difference between patients with CR/PR and SD/PD in all, LP and GP group (P > 0.05, P > 0.05, P > 0.05, respectively; Figure S8A-C). The median PFS was also similar among patients with different baseline cfDNA concentrations in three groups (P = 0.143, P = 0.656, P = 0.103, respectively; Figure S8D-F). Secondly, we found that the fraction of circulating tumor DNA (ctDNA) also had no significant difference between patients with CR/PR and SD/PD in all, LP and GP group (P > 0.05, P > 0.05, P > 0.05, respectively; Figure S9A-C). Moreover, the median PFS was also comparable among patients with different fraction of ctDNA in three groups (P = 0.975, P = 0.869, P = 0.834, respectively; Figure S9D-F). Finally, we excluded the potential impact of maximum VAF of SNV and CNV on therapeutic response (Figure S10). Following our above-mentioned defined criteria and strict algorithm, we identified CNV pattern of 31 genes including CASP8, PPHLN1, PIGF, KEAP1, SDHC, MOV10L1, CCND3, MTRR, ID3, STK11, SEL1L3, ARMC5, MYCL, SMARCA4, BAT, MYO10, SMO, TSHR, IRFB, SOX9, CIC, CCR4, HSPA1B, FLCN, PRPF39, RRP1B, PRKCI, ARPC2, SOCS1, ERCC2 and CEBPA. The results showed obviously different distribution between patients with CR/PR and SD/PD in all, LP and GP group (Figure 3A-C). These genes had different co-efficient importance values in this predictive model (Figure 4A) and the sum of co-efficient importance values based on the selected features for each sample was its individual RS. Receiver operator characteristic (ROC) curve analysis indicated that RS, the developed predictor in this study, could effectively distinguish patients with CR/PR from these with SD/PD in both training set [area under the ROC curve (AUC) = 0.925, Figure 4B] and validation set (AUC = 0.815, Figure 4C).
Figure 3.
CNV pattern of 31 genes including CASP8, PPHLN1, PIGF, KEAP1, SDHC, MOV10L1, CCND3, MTRR, ID3, STK11, SEL1L3, ARMC5, MYCL, SMARCA4, BAT, MYO10, SMO, TSHR, IRFB, SOX9, CIC, CCR4, HSPA1B, FLCN, PRPF39, RRP1B, PRKCI, ARPC2, SOCS1, ERCC2, CEBPA showed obviously distinct distribution between patients with CR/PR and SD/PD in all (A), LP (B) and GP (C) group. From inside to out of each circus plot: the first circle represents the CNVs of patients in SD and PD group (orange represents amplification, green represents loss or deletion); the second circle represents the CNVs of patients in PR and CR group (red represents amplification, blue represents loss or deletion). Outermost circle represents the chromosomes. CR, complete response; PR, partial response; SD, stable disease; PD, disease progression. LP, paclitaxel liposome plus cisplatin; GP, gemcitabine plus cisplatin.
Figure 4.
Generation of CNV-based RS for response prediction. A. Different co-efficient importance values in this model via selecting features with the best accuracy score in the ensemble or LASSO supervised method; B. Receiver operator characteristic curve analysis result in training set; C. Receiver operator characteristic curve analysis result in validation set. CNV, copy number variation; AUC, area under the ROC curve. Feature selection (Fig. A) was carried out with two steps. First, several statistical methods were utilized to evaluate the difference between two groups of samples in training set for each feature, including deviation, mutual information, AUC and p-values of Chi-Square test, Wilcoxon rank sum test, ANOVA and Student's t test, after which features with significantly different signal in at least four of criteria mentioned above were selected. Then, the method of LASSO was conducted to select features with the best accuracy score.
Relationship between RS and treatment outcomes
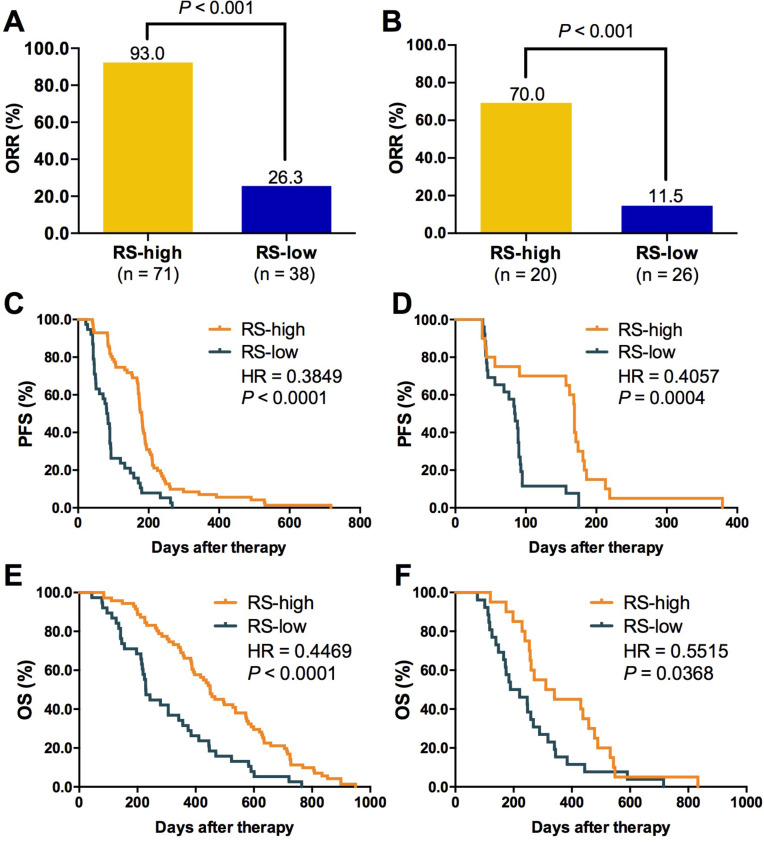
We then evaluated the relationship between RS and outcomes of first-line chemotherapy. The cutoff of RS was defined as the numerical value that showed the best accuracy and AUC in distinguishing patients with different treatment response. Therefore, all the patients in this study were divided into high or low RS group. As shown in Figure 5, patients with high RS showed markedly higher ORR than those with low RS in both training (93.0% vs. 26.3%, P < 0.001; Figure 5A) and validation set (70.0% vs. 11.5%, P < 0.001; Figure 5B). A significant difference was also observed in PFS (training set: HR = 0.38, P < 0.0001, Figure 5C; validation set: HR = 0.41, P = 0.0004, Figure 5D) and OS (training set: HR = 0.45, P < 0.0001, Figure 5E; validation set: HR = 0.55, P = 0.0368, Figure 5F) between two groups. Subgroup analysis in training set indicated that RS was a suitable predictor for both LP and GP group (Figure S11). However, RS could not distinguish the ORR (Figure S12A-B) and PFS (Figure S12C-D) of LP from GP in training and validation set.
Figure 5.
Relationship between RS and treatment outcomes. A. ORR comparison between RS high and low group in training set; B. ORR comparison between RS high and low group in validation set; C. Kaplan-Meier curve of PFS comparison between RS high and low group in training set; D. Kaplan-Meier curve of PFS comparison between RS high and low group in validation set; E. Kaplan-Meier curve of OS comparison between RS high and low group in training set; F. Kaplan-Meier curve of OS comparison between RS high and low group in validation set. RS, RESPONSE SCORE; ORR, objective response rate; PFS, progression-free survival; HR, hazard ratio. Unpaired student t test were applied for comparison of response rate between RS high and low groups. The Kaplan-Meier curve with log-rank test was used to test the significance of differences between two groups.
Validation of predictive value of RS in a real-word cohort
Considering the potential impact of different histology (e.g. LUSC vs. LUAD) and chemotherapeutic regimens, we further survey the universal significance of RS for predicting first-line chemotherapy outcomes in advanced NSCLC. We evaluated its predictive value in an external real-world cohort from previous publication24. 42 patients with advanced NSCLC received docetaxel plus cisplatin/carboplatin as first-line treatment (Figure S13A). The results showed that high RS was also correlated with significantly better ORR (54.5% vs. 15.0%, P = 0.019; Figure S13B) and PFS (HR = 0.42, P = 0.0023; Figure S13C) than those with low RS. These results suggested RS might be served as a universal predictor for first-line platinum-based doublet chemotherapy in advanced NSCLC and further investigation with large sample size is warranted.
Change of VAF monitored the treatment response
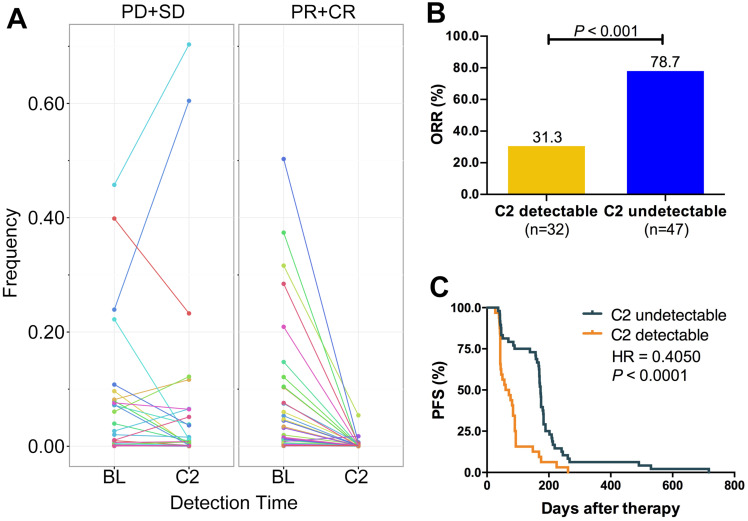
Several studies revealed that cfDNA dynamics could predict the treatment response of targeted therapies or immune checkpoint inhibitors. Here, we designed the Panel 2 covering 29 prevalent tumor related driver genes (Table S3) to explore whether changes of VAF in cfDNA could monitor chemotherapy response (Figure 1B). We collected eligible blood samples from 79 cases at baseline and cycle 2 treatment. Patients of responder (CR+PR) experienced a significant decrease of VAF while patients of non-responder (SD+PD) experienced an increase at cycle 2 treatment (Figure 6A). Moreover, patients of VAF undetectable at cycle 2 had significantly higher ORR (78.7% vs. 31.3%, P < 0.001; Figure 6B) and longer PFS (HR = 0.41, P < 0.0001; Figure 6C) than those of VAF detectable. Subgroup analysis showed that reduction of VAF was associated with durable clinical benefit in both LP (Figure S14A) and GP (Figure S14D) group. VAF undetectable at cycle 2 was correlated with substantially better ORR and PFS in both LP (ORR: 87.0% vs. 31.3%, P < 0.001, Figure S14B; PFS: HR = 0.36, P < 0.0001, Figure S14C) and GP group (ORR: 70.8% vs. 31.3%, P = 0.014, Figure S13E; PFS: HR = 0.46, P = 0.0089, Figure S14F) than those with detectable VAF. These findings suggested that changes of VAF in cfDNA could monitor the response to first-line chemotherapy in patients with advanced LUSC.
Figure 6.
ICP-based dynamic change of VAF monitored the treatment response. A. ICP-based change of VAF between CR/PR and SD/PD at baseline and cycle 2 treatment. B. ORR comparison between VAF detectable and undetectable at cycle 2 treatment; C. Kaplan-Meier curve of PFS comparison between VAF detectable and undetectable at cycle 2 treatment. ORR, objective response rate; PFS, progression-free survival; BL, baseline; C2, cycle 2 treatment; CR, complete response; PR, partial response; SD, stable disease; PD, disease progression. Paired student t test were applied for the dynamic change of cfDNA VAF between baseline and C2 detection time. Unpaired student t test were applied for comparison of response rate between C2 detectable and undetectable groups. The Kaplan-Meier curve with log-rank test was used to test the significance of differences between two groups.
Discussion
The current study comprehensively investigated the predictive value of cfDNA profiling for first-line platinum-based chemotherapy in patients with advanced LUSC. Our genetic analysis indicated that no single gene alternations were associated with outcome of chemotherapy and TMB could also not predict therapeutic response in patients with advanced LUSC. Alternatively, we generated a CNV-based classifier (RS) via machine learning algorithm to integrate cfDNA molecular profiling. We found that patients with high RS showed significantly superior ORR, PFS and OS than those with low RS in both training and validation set. We also validated these findings in an external real-world cohort. Notably, patients with objective response experienced a significant decrease of VAF after 2 cycles of treatment. Moreover, VAF undetectable at cycle 2 treatment was correlated with significantly better ORR and PFS than those with detectable VAF.
cfDNAs are derived from dying cells, detectable in plasma and are typically short DNA fragments (average length of 120-160 bp). In spite of the ambiguous biology of cfDNA, its clinical application (e.g. prediction or monitoring of treatment response, relapse, drug resistance, prognosis, etc.) has been extensively investigated 25. Using cfDNA as a predictor for therapeutic response in NSCLC has been investigated in many previous studies and majority of them focused on cfDNA levels 26, 27. However, a recent large-scale study found that the baseline cfDNA concentration did not validate its predictive value for outcome of systemic therapy in NSCLC 28. Meanwhile, dynamic changes in plasma cfDNA also did not correlate with radiologic response 28, suggesting that cfDNA concentration could not serve as a predictor of systemic therapy. Consistently, our results also found that the baseline cfDNA level cannot distinguish patients with CR/PR from those with SD/PD. Collectively, these results recommended that future studies on the predictive value of cfDNA should shift from its concentration or dynamics.
Single gene alterations as predictor for first-line chemotherapy in NSCLC went through a tortuous course and most of them finally failed 9. Our findings also showed that there were no frequent genetic mutations associated with the outcomes of chemotherapy in patients with advanced LUSC, suggesting that individual gene alterations showed very limited and inconsistent value for predicting outcomes of first-line chemotherapy in advanced LUSC. To improve the predictive power, we developed a CNV-based classifier (RS) via integrating cfDNA profiling in this study. CNV is considered as one of the major types of genome aberrations that contribute to tumorigenesis, maintenance and progression 29. Previous studies indicated that CNV pattern in cfDNA could act as a surrogate of primary tumor in various solid tumors 30, 31. Moreover, Louise et al. reported that CNV-based classification from circulating tumor cells could distinguish chemosensitive from chemorefractory cases with an accuracy of 83.3% in small cell lung cancer 32. In the current study, we integrated 31 frequent genes CNV as RS. As we mentioned above, most of them were involved in the carcinogenesis (e.g. KEAP1, SMARCA4, MYCL, SOX9, STK11), cell cycle regulation (e.g. CCND3, SEL1L3), DNA replication, transcription and damage repair (e.g. PPHLN1, MOV10L1, ID3, CIC, FLCN, PRPF39, RRP1B, ARPC2, CEBPA), immune response pathways (e.g. IRF8, HSPA1B, CCR4, SOCS1), chemotherapeutic drug transport and metabolism (e.g. ERCC2, MTRR, PIGF, SDHC, TSHR) in NSCLC. Although the association of each individual gene with treatment response was limited, a robust correlation between RS and chemotherapeutic response was observed. Patients with high RS had significantly better ORR, PFS and OS than those with low RS. Taken together, these findings indicated that this 31-genes CNV-based RS could be utilized to guide first-line chemotherapy in patients with advanced LUSC.
The genetic profiling of cfDNA also exhibited promising results for monitoring the efficacy of systemic therapy. Mok T et al. found that patients with circulating EGFR mutation clearance at cycle 3 had longer PFS and OS, suggesting that dynamic change of blood-based EGFR status could be a useful predictive marker 22. Several recent publications reported that plasma cfDNA profiling could also predict response to immune checkpoint inhibitors 23, 33-35. However, to date, no biomarkers have been developed to monitor the chemotherapy response in advanced LUSC. Our previous studies reported that the dynamic changes of TP53 mutational burden might have monitoring value for the efficacy of first-line chemotherapy in advanced NSCLC. To improve its reliability, we optimized a small panel that included 29 frequent tumor related driver genes with an ultra-deep sequencing to increase the sensitivity of mutation detection. The result showed that change of VAF could effectively monitor the treatment response. Interestingly, Diehn et al. reported that cancer personalized profiling by deep sequencing (CAPP-seq) circulating tumor DNA (ctDNA) analysis could assess a response earlier than radiographic approaches and identify molecular residual disease after definitive therapy in patients with lung cancer 36, 37. These findings suggested that the application of customized panel of cfDNA sequencing could monitor the clinical benefits of chemotherapy.
There are several limitations that should be acknowledged. First, although the number of patients in the phase IV trial is large enough (n = 536), only 155 cases were included in this biomarker research, which may lead to the potential selection bias. However, when we compared the baseline features of two cohorts, we did not observe the obvious differences regarding to the baseline features including age, gender and ECOG PS. Second, we have utilized an external cohort to investigate the universal significance of RS for predicting first-line chemotherapy outcomes in advanced NSCLC regardless of histology. Due to the accessibility of sequencing data and clinical information for previous publication, only 42 cases were included, which is relatively small. A large prospective study is warranted in the future to validate the predictive efficacy of RS. Third, we only identified the CNV profile from cfDNA, whether it could well represent the CNV features in primary tumor cohort of LUSC remains future investigation. Last but not least, immunotherapy based combination therapy is the standard of care for advanced LUSC. Only to investigate the biomarkers to predict chemotherapy is less clinical significant nowadays in the era of immunotherapy. Nevertheless, platinum-based chemotherapy still plays an important role in the treatment for patients with LUSC considering the accessibility and price of immunotherapy in some areas of China. A substantial number of patients still need chemotherapy and are the potential population who benefit this biomarker analysis.
In summary, the current study indicated that cfDNA profiling is correlated with therapeutic response to first-line chemotherapy in patients with advanced LUSC. CNV-based RS showed potential value in predicting therapeutic effects, and change of VAF is valuable to monitor treatment response. These findings support the feasibility for utilization of cfDNA profiling to guide first-line chemotherapy in patients with advanced LUSC, and worth further validation in large scale population.
Supplementary Material
Supplementary figures and tables.
Acknowledgments
This study was sponsored by Nanjing Luye Pharmaceutical Co. Ltd, Nanjing, China, and supported in part by grants from the National Natural Science Foundation of China (No. 81772467, 81871865, 81874036 and 81972167). The Backbone Program of Shanghai Pulmonary Hospital (No. FKGG1802), “Shuguang Program” supported by Shanghai Education Development Foundation and Shanghai Municipal Education Commission (No. 16SG18), Shanghai Pujiang Talent Plan (No. 2019PJD048), Shanghai Key disciplines of Respiratory (No. 2017ZZ02012), and Shanghai Major Diseases Multidisciplinary Cooperation Diagnosis and Treatment Construction Project.
Author Contributions
Tao Jiang, Shengxiang Ren, and Caicun Zhou designed this study. All of the listed authors collected the blood samples and clinical data. Tao Jiang, Shengxiang Ren, Henghui Zhang, Lihong Wu, Lu Fang, Dandan Liang, Yi Zhao, and Caicun Zhou performed the statistical analyses. Tao Jiang, Shengxiang Ren, and Caicun Zhou drafted the manuscript. Caicun Zhou provided critical comments, suggestions and revised the manuscript. All authors read and approved the final version of the manuscript. Beijing Genecast Biotechnology Co. conducted the next-generation sequencing of all included samples.
Supporting information
The data sets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- cfDNA
cell-free DNA
- CI
confidence interval
- CNV
copy number variations
- CR
complete response
- DCR
disease control rate
- ECOG PS
Eastern Cooperative Oncology Group performance status
- HR
hazard ratios
- ICP
individually customized panel
- LUSC
lung squamous cell carcinoma
- NGS
next generation sequencing
- NSCLC
non-small-cell lung cancer
- ORR
objective response rate
- OS
overall survival
- ROC
Receiver operator characteristic
- RS
RESPONSE SCORE
- PD
progressive disease
- PD-1
programmed cell death 1
- PD-L1
programmed cell death ligand 1
- PR
partial response
- SD
stable disease
- TMB
tumor mutation burden
- VAF
variant allele frequency
References
- 1.Gandara DR, Hammerman PS, Sos ML, Lara PN Jr. and Hirsch FR. Squamous cell lung cancer: from tumor genomics to cancer therapeutics. Clin Cancer Res. 2015;21:2236–43. doi: 10.1158/1078-0432.CCR-14-3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen Z, Fillmore CM, Hammerman PS, Kim CF, Wong KK. Non-small-cell lung cancers: a heterogeneous set of diseases. Nat Rev Cancer. 2014;14:535–46. doi: 10.1038/nrc3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reck M, Rabe KF. Precision diagnosis and treatment for advanced non-small-cell lung cancer. N Engl J Med. 2017;377:849–861. doi: 10.1056/NEJMra1703413. [DOI] [PubMed] [Google Scholar]
- 4.Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553:446–454. doi: 10.1038/nature25183. [DOI] [PubMed] [Google Scholar]
- 5.Rittmeyer A, Barlesi F, Waterkamp D, Park K, Ciardiello F, von Pawel J. et al. Atezolizumab versus docetaxel in patients with previously treated non-small-cell lung cancer (OAK): a phase 3, open-label, multicentre randomised controlled trial. Lancet. 2017;389:255–265. doi: 10.1016/S0140-6736(16)32517-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borghaei H, Paz-Ares L, Horn L, Spigel DR, Steins M, Ready NE. et al. Nivolumab versus Docetaxel in advanced nonsquamous non-small-cell lung cancer. N Engl J Med. 2015;373:1627–39. doi: 10.1056/NEJMoa1507643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brahmer J, Reckamp KL, Baas P, Crino L, Eberhardt WE, Poddubskaya E. et al. Nivolumab versus Docetaxel in advanced squamous-cell non-small-cell lung Cancer. N Engl J Med. 2015;373:123–35. doi: 10.1056/NEJMoa1504627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ribas A, Wolchok JD. Cancer immunotherapy using checkpoint blockade. Science. 2018;359:1350–1355. doi: 10.1126/science.aar4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olaussen KA, Postel-Vinay S. Predictors of chemotherapy efficacy in non-small-cell lung cancer: a challenging landscape. Ann Oncol. 2016;27:2004–2016. doi: 10.1093/annonc/mdw321. [DOI] [PubMed] [Google Scholar]
- 10.Ren SX, Li AW, Zhou SW, Zhang L, Wang YS, Li B. et al. Individualized Chemotherapy in advanced NSCLC patients based on mRNA levels of BRCA1 and RRM1. Chin J Cancer Res. 2012;24:226–31. doi: 10.1007/s11670-012-0226-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hellmann MD, Ciuleanu TE, Pluzanski A, Lee JS, Otterson GA, Audigier-Valette C. et al. Nivolumab plus Ipilimumab in lung cancer with a high tumor mutational burden. N Engl J Med. 2018;378:2093–2104. doi: 10.1056/NEJMoa1801946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Samstein RM, Lee CH, Shoushtari AN, Hellmann MD, Shen R, Janjigian YY. et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet. 2019;51:202–206. doi: 10.1038/s41588-018-0312-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu J, Zhong H, Wu J, Chu T, Zhang L, Li H. et al. Circulating DNA-based sequencing guided anlotinib therapy in non-small cell lung cancer. Adv Sci (Weinh) 2019;6:1900721. doi: 10.1002/advs.201900721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wan JC, Massie C, Garcia-Corbacho J, Mouliere F, Brenton JD, Caldas C, Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer. 2017. [DOI] [PubMed]
- 15.Tong L, Ding N, Tong X, Li J, Zhang Y, Wang X. et al. Tumor-derived DNA from pleural effusion supernatant as a promising alternative to tumor tissue in genomic profiling of advanced lung cancer. Theranostics. 2019;9:5532–5541. doi: 10.7150/thno.34070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang L, Ni J, Zhu Y, Pang B, Graham P, Zhang H. et al. Liquid biopsy in ovarian cancer: recent advances in circulating extracellular vesicle detection for early diagnosis and monitoring progression. Theranostics. 2019;9:4130–4140. doi: 10.7150/thno.34692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hlady RA, Zhao X, Pan X, Yang JD, Ahmed F, Antwi SO. et al. Genome-wide discovery and validation of diagnostic DNA methylation-based biomarkers for hepatocellular cancer detection in circulating cell free DNA. Theranostics. 2019;9:7239–7250. doi: 10.7150/thno.35573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang T, Ren S, Zhou C. Role of circulating-tumor DNA analysis in non-small cell lung cancer. Lung Cancer. 2015;90:128–34. doi: 10.1016/j.lungcan.2015.09.013. [DOI] [PubMed] [Google Scholar]
- 19.Siravegna G, Marsoni S, Siena S, Bardelli A. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol. 2017. [DOI] [PubMed]
- 20.Iijima Y, Hirotsu Y, Amemiya K, Ooka Y, Mochizuki H, Oyama T. et al. Very early response of circulating tumour-derived DNA in plasma predicts efficacy of nivolumab treatment in patients with non-small cell lung cancer. Eur J Cancer. 2017;86:349–357. doi: 10.1016/j.ejca.2017.09.004. [DOI] [PubMed] [Google Scholar]
- 21.Dawson SJ, Tsui DW, Murtaza M, Biggs H, Rueda OM, Chin SF. et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. 2013;368:1199–209. doi: 10.1056/NEJMoa1213261. [DOI] [PubMed] [Google Scholar]
- 22.Mok T, Wu YL, Lee JS, Yu CJ, Sriuranpong V, Sandoval-Tan J. et al. Detection and dynamic changes of EGFR mutations from circulating Tumor DNA as a predictor of survival outcomes in NSCLC patients treated with first-line intercalated erlotinib and chemotherapy. Clin Cancer Res. 2015;21:3196–203. doi: 10.1158/1078-0432.CCR-14-2594. [DOI] [PubMed] [Google Scholar]
- 23.Hofman P, Heeke S, Alix-Panabieres C, Pantel K. Liquid biopsy in the era of immuno-oncology: is it ready for prime-time use for cancer patients? Ann Oncol. 2019;30:1448–1459. doi: 10.1093/annonc/mdz196. [DOI] [PubMed] [Google Scholar]
- 24.Jiang T, Li X, Wang J, Su C, Han W, Zhao C. et al. Mutational Landscape of cfDNA identifies distinct molecular features associated with therapeutic response to first-line platinum-based doublet chemotherapy in patients with advanced NSCLC. Theranostics. 2017;7:4753–4762. doi: 10.7150/thno.21687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bennett CW, Berchem G, Kim YJ, El-Khoury V. Cell-free DNA and next-generation sequencing in the service of personalized medicine for lung cancer. Oncotarget. 2016;7:71013–71035. doi: 10.18632/oncotarget.11717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar S, Guleria R, Singh V, Bharti AC, Mohan A, Das BC. Plasma DNA level in predicting therapeutic efficacy in advanced nonsmall cell lung cancer. Eur Respir J. 2010;36:885–92. doi: 10.1183/09031936.00187909. [DOI] [PubMed] [Google Scholar]
- 27.Pan S, Xia W, Ding Q, Shu Y, Xu T, Geng Y. et al. Can plasma DNA monitoring be employed in personalized chemotherapy for patients with advanced lung cancer? Biomed Pharmacother. 2012;66:131–7. doi: 10.1016/j.biopha.2011.11.022. [DOI] [PubMed] [Google Scholar]
- 28.Li BT, Drilon A, Johnson ML, Hsu M, Sima CS, McGinn C. et al. A prospective study of total plasma cell-free DNA as a predictive biomarker for response to systemic therapy in patients with advanced non-small-cell lung cancers. Ann Oncol. 2016;27:154–9. doi: 10.1093/annonc/mdv498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharp AJ, Locke DP, McGrath SD, Cheng Z, Bailey JA, Vallente RU. et al. Segmental duplications and copy-number variation in the human genome. Am J Hum Genet. 2005;77:78–88. doi: 10.1086/431652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heitzer E, Ulz P, Belic J, Gutschi S, Quehenberger F, Fischereder K. et al. Tumor-associated copy number changes in the circulation of patients with prostate cancer identified through whole-genome sequencing. Genome Med. 2013;5:30. doi: 10.1186/gm434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shaw JA, Page K, Blighe K, Hava N, Guttery D, Ward B. et al. Genomic analysis of circulating cell-free DNA infers breast cancer dormancy. Genome Res. 2012;22:220–31. doi: 10.1101/gr.123497.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carter L, Rothwell DG, Mesquita B, Smowton C, Leong HS, Fernandez-Gutierrez F. et al. Molecular analysis of circulating tumor cells identifies distinct copy-number profiles in patients with chemosensitive and chemorefractory small-cell lung cancer. Nat Med. 2017;23:114–119. doi: 10.1038/nm.4239. [DOI] [PubMed] [Google Scholar]
- 33.Guibert N, Jones G, Beeler JF, Plagnol V, Morris C, Mourlanette J. et al. Targeted sequencing of plasma cell-free DNA to predict response to PD1 inhibitors in advanced non-small cell lung cancer. Lung Cancer. 2019;137:1–6. doi: 10.1016/j.lungcan.2019.09.005. [DOI] [PubMed] [Google Scholar]
- 34.Lee JH, Long GV, Menzies AM, Lo S, Guminski A, Whitbourne K. et al. association between circulating tumor DNA and pseudoprogression in patients with metastatic melanoma treated with anti-programmed cell death 1 antibodies. JAMA Oncol. 2018;4:717–721. doi: 10.1001/jamaoncol.2017.5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jensen TJ, Goodman AM, Kato S, Ellison CK, Daniels GA, Kim L. et al. genome-wide sequencing of cell-free DNA identifies copy-number alterations that can be used for monitoring response to immunotherapy in cancer patients. Mol Cancer Ther. 2019;18:448–458. doi: 10.1158/1535-7163.MCT-18-0535. [DOI] [PubMed] [Google Scholar]
- 36.Newman AM, Bratman SV, To J, Wynne JF, Eclov NC, Modlin LA. et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. 2014;20:548–54. doi: 10.1038/nm.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chaudhuri AA, Chabon JJ, Lovejoy AF, Newman AM, Stehr H, Azad TD. et al. Early detection of molecular residual disease in localized lung cancer by circulating tumor DNA profiling. Cancer Discov. 2017;7:1394–1403. doi: 10.1158/2159-8290.CD-17-0716. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures and tables.