Abstract
Glioblastoma (GBM) is the most common malignant brain tumor and the most aggressive type of glioma, characterized by strong invasive potential and rapid recurrence despite severe treatment methods, such as maximal tumor resection followed by chemotherapy and radiotherapy. Thrombospondin-1 (THBS1) was first discovered in platelets and subsequent studies have indicated its functions in the development of several cancers, including breast cancer, melanoma, gastric cancer, cervical cancer and GBM. However, to the best of our knowledge, the expression profiles of THBS1 in GBM subtypes remain unknown, and the underlying mechanism by which THBS1 expression is regulated, and its effect on the local immune response in GBM, remains unclear. The present study used public datasets from The Cancer Genome Atlas, the Chinese Glioma Genome Atlas, the Gene Expression Omnibus, the Ivy Glioblastoma Atlas Project, Tumor Immune Estimation Resource, Estimation of STromal and Immune cells in MAlignant Tumor tissues using Expression data and the Human Protein Atlas to investigate the prognostic value of THBS1 and its expression profiles, as well as its correlation with the local immune response in GBM. The results demonstrated that THBS1 was a biomarker of the pathological malignancy of glioma, and predicted the mesenchymal subtype of GBM. Furthermore, DNA methylation of THBS1 may be an important mechanism by which THBS1 expression is regulated in GBM. The hypomethylation or overexpression of THBS1 predicted an unfavorable prognosis in patients with GBM. Additionally, THBS1 was correlated with immune and inflammatory responses in GBM. Thus, the findings of the present study provide insight into the potential value of THBS1 in the treatment of GBM.
Keywords: glioblastoma, bioinformatics analysis, thrombo-spondin-1, prognosis, microenvironment
Introduction
According to the 2016 World Health Organization central nervous system tumor classification, glioblastoma (GBM) is the most common primary malignant brain tumor worldwide and has the lowest 5-year survival rate (<20%) among all malignant brain tumors (1,2). A list of histological and molecular parameters of glioma, including isocitrate dehydrogenase-1 (IDH-1) mutations and 1p/19q codeletion, was generated by the World Health Organization (WHO) in 2016, which has updated the method of diagnosing glioma from histological to molecular methods, improving diagnostic accuracy and providing improved treatment strategies for patients with glioma (1). Based on data from The Cancer Genome Atlas (TCGA) Research Network, GBM was divided into four subtypes: i) Mesenchymal; ii) classical; iii) neural; and iv) proneural, based on different cellular features and genetic contexts (3). However, subsequent studies have demonstrated that the transcriptional subtypes of glioma, defined by clustering based on tumor-intrinsic genes, strongly overlap between the proneural, classical and mesenchymal subtypes; however, the neural subtype was identified as a normal neural lineage contamination (4,5). The molecular characterization of GBM has significantly improved patient stratification and provided insight into novel strategies for treating GBM (1). However, GBM is a cancer with high molecular heterogeneity (1,3) and its tumor microenvironment has been reported to critically affect clinical outcomes and gene expression in tumor tissues (6,7). The tumor microenvironment (TME) consists of local immune cells, mesenchymal cells, endothelial cells, inflammatory factors and the extracellular matrix (ECM), TME contains significant leukocytic infiltrations that convey immunosuppressive activity (ability to block cytotoxic T lymphocytes or natural killer T cell mediated killing of aberrant cells), these include regulatory T cells (Tregs), myeloid-derived suppressor cells, tumor associated macrophages programmed by Th2-type cytokines, and neutrophils and mast cell subtypes that collectively confer cancer cells with a mechanism to escape killing by T cells (8), accounting for the heterogeneity of GBM and elucidating differential responses to oncological therapy (9). Therefore, investigating the molecular events in the GBM microenvironment is necessary for developing targeted therapy (9,10).
Thrombospondins (THBSs) are important components of the ECM and serve important roles in numerous physical and pathological processes, such as cell differentiation, proliferation, migration and fibroblast apoptosis, vascular hemeostasis, immunity and wound healing (11). THBS1, a glycoprotein of the THBS family is comprised of four motifs and was first discovered in platelets; however, subsequent studies have indicated its functions in the development of several cancers, including breast (12), gastric (13), melanoma (14), cervical cancer (15) and GBM (16–18). Numerous studies have reported that THBS1 regulates cell differentiation, proliferation and migration, and the apoptosis of fibroblasts, smooth muscle cells and macrophages (11,15,18,19). Therefore, THBS1 serves an important role in the regulation of various biological processes. The function of THBS1 in the regulation of angiogenesis in GBM has been reported, and similar results have been demonstrated in cervical cancer (15,18). Tumors formed by C6 glioma cells expressing the angiostatic peptide of THBS1 exhibited a strong inhibition of angiogenesis and became more aggressive (20). In the U87 cell line, the knockdown of THBS1 using short hairpin (sh)RNA decreased tumor invasion (21). Overexpressed THBS1 in glioma samples, and its functions in glioma proliferation, have also been reported (21); however, the expression profiles of THBS1 in different molecular subtypes of GBM, and the mechanism of its dysregulation in GBM, have not been fully elucidated. As an ECM factor, THBS1 expression facilitates the induction of TNF-β in monocytes, resulting in the suppression of CD8+ T cell proliferation in glioma (22). Furthermore, CD35+ B cells expressing THBS1 decreased the levels of CD80/86 in dendritic cells, and converted naïve CD4+ T cells to regulatory T cells, thus inhibiting the inflammatory response (19). These results indicate that THBS1 serves multifaceted functional roles in the immune response; however, the relationship between the transcript levels of THBS1 and immune-associated genes and its function in local immune cell infiltration in GBM remain to be fully understood.
The present study comprehensively analyzed THBS1 expression in GBM subtypes and utilized genome-wide methylation data to explore the underlying mechanism by which THBS1 expression is regulated in GBM. Furthermore, a bioinformatics analysis method was utilized to investigate the function of THBS1 in the local immune response in GBM. The prognostic value and immunosuppressive function of THBS1 found in the present study indicate that THBS1 may be a valuable biomarker and therapeutic target of GBM.
Materials and methods
Data resource
In order to ensure that the number of cases in each analysis was adequate, three large public datasets were selected, each of which included >200 patients with glioma. A gene expression array (the AffyU133a array) from the TCGA-GBM dataset (529 GBM tissues and 10 non-tumor tissues) (23) was obtained using the UCSC Xena browser (xenabrowser.net; date accessed 2017-09-08). RNA-sequencing data of 325 glioma tissues [144 GBM tissues and 181 low-grade glioma (LGG) tissues] were downloaded from the Chinese Glioma Genome Atlas (CGGA; cgga.org.cn; date accessed 2017-09-04). Microarray data from the Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/; access no. GSE16011; release date: 25/10/2006), including 8 normal brain tissues, 159 GBM tissues and 117 LGG tissues, were obtained through the R2: Genomics Analysis and Visualization Platform (GPL8542 platform; Affymetrix GeneChip Human Genome U133 Plus 2.0 Array; r2.amc.nl).
THBS1 protein levels in low grade glioma and high grade glioma were evaluated by immunohistochemistry staining based on data from the Human Protein Atlas (HPA; proteinatlas.org) (24). Genes that correlated with THBS1 expression in the TCGA-GBM dataset were calculated using Pearson's correlation coefficient using the online tool GlioVis portal (gliovis.bioinfo.cnio.es; date accessed, 2016-10-31) (25). RNA-seq data with specific tumor anatomic structures in GBM were obtained from the Ivy Glioblastoma Atlas Project (http://glioblastoma.alleninstitute.org; date accessed, 2017-10-20). In each step of the analysis, the patients with detailed genomic and clinical data were included. The genomic and clinical datasets contained data concerning THBS1 mRNA expression of GBM (WHO IV) and LGG, (WHO II, III) GBM subtype information (according to Wang's study) (4,25), overall survival (OS), IDH-1 status, glioma CPG island methylation phenotype (G-CIMP) status and DNA methylation data (data from UCSC Xena browser, DNA methylation profile was measured experimentally using Illumina Infinium Human Methylation 27 platform). Cases without relevant data were eliminated in each analysis.
Bioinformatics analysis
The correlation between THBS1 expression and glioma malignancy was based on the TCGA-GBM, CGGA and GSE16011 datasets. Kaplan-Meier survival curves of patients with GBM from the TCGA-GBM and CGGA datasets were generated and median gene expression value was regarded as the cutoff. Genes that exhibited positive and negative correlations (correlation, |R|>=0.3) with THBS1 in GBM samples from the TCGA-GBM datasets were determined using the GlioVis portal (25). Gene Ontology (GO) and Kyoto Encyclopedia of Gene and Genomic (KEGG) analyses of genes that showed correlations (|R|>=0.3) with THBS1 were performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) bioinformatics resources (version 6.8; david.ncifcrf.gov). GraphPad Prism software (version no. 7; GraphPad Software, Inc.) was used to compare THBS1 expression between different groups. Gene expression heatmaps of the association between THBS1 and inflammatory metagene clusters, Treg signatures (26–28) and THBS1-correlated genes were generated using ClustVis (http://biit.cs.ut.ee/clustvis/; first version online; date accessed 2014-10-31) (7). The Tumor IMmune Estimation Resource database (TIMER; cistrome.shinyapps.io/timer) was used to estimate the abundance of immune infiltrates (29). In the present study the correlation between THBS1 expression and the abundance of immune infiltrates, including tumor purity, B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages and dendritic cells, was analyzed via gene modules in the TIMER database through Spearman correlation as previously described (29). The correlation between THBS1 expression and biomarkers of mesenchymal subtype GBM was evaluated by Pearson correlation coefficient in the present study as described by a previous study (30). The ESTIMATE database (https://bioinformatics.mdanderson.org/estimate/rpackage.html) was used to calculate immune and stromal scores in different malignant tumors by Yoshihara et al (7) and was successfully applied to GBM by Jia et al (9). Therefore, the present study investigated the association between THBS1 expression and immune and stromal scores to investigate the importance of THBS1 in the immune infiltrations of GBM.
Statistical analysis
Kaplan-Meier survival analysis was performed to compare OS data from two cohorts using the log-rank test. Column analysis was performed using GraphPad Prism software (version no. 7; GraphPad Software, Inc.) using unpaired t-tests between 2 groups and one-way ANOVA and Tukey's post-hoc test between >2 groups. Correlation analysis was performed by Pearson's correlation coefficient or Spearman correlation coefficient. Receiver operating characteristic (ROC) curves were generated using SPSS software (version 21.0; IBM Corp.) P<0.05 was considered to indicate a statistically significant difference.
Results
Increased THBS1 expression predicts the pathological malignancy of glioma
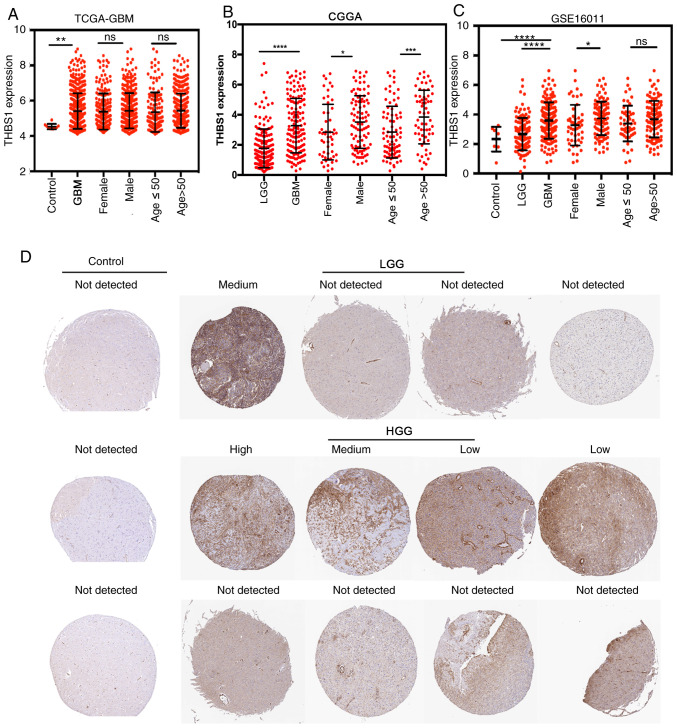
Clinical data of three transcriptome datasets were summarized in Table SI. Analyses of the transcript levels THBS1 expression in glioma demonstrated that THBS1 expression was higher in GBM (WHO IV grade glioma) tissues compared with LGG (WHO II or III grade) and normal brain tissues among three independent glioma datasets, TCGA-GBM (Fig. 1A), CGGA (Fig. 1B) and GSE16011 (Fig. 1C). However, the distribution of THBS1 expression in different sexes (female vs. male) and ages (≤50 vs. >50) was not consistent among the three public datasets (Fig. 1A-C). In the TCGA-GBM database, there was no statistically significant difference in THBS1 expression between different ages and sexes (Fig. 1A). However, in the CGGA and GSE16011 datasets, males exhibited higher THBS1 expression compared with females (Fig. 1B and C). Furthermore, in the CGGA database, older patients with GBM (>50 years) had higher THBS1 expression compared with younger patients (≤50 years) (Fig. 1B). This variable distribution may derive from systematic bias due to different detection methods in the three transcriptome datasets and the limited number of patients with GBM in each group.
Figure 1.
High THBS1 expression is associated with glioma malignancy. In (A) TCGA-GBM, (B) CGGA and (C) GSE16011 datasets, THBS1 expression was associated with the glioma malignancy (GBM has the highest expression) at the transcript level. In the CGGA and GSE16011 datasets, males exhibited higher THBS1 expression than females. Furthermore, in the CGGA dataset, older patients with GBM (>50 years) had higher THBS1 expression compared with younger patients (≤50 years). (D) Data from the Human Protein Atlas demonstrated that THBS1 protein were not detected in glial cells from normal brain tissues. In comparison, among the 12 glioma tissues, weak staining was observed in 2 cases of HGG, moderate staining was found in 1 case of HGG and 1 case of LGG, strong staining was observed in 1 case of HGG and no staining was observed in 4 cases of HGG and 3 cases of LGG. *P<0.05, **P<0.01, ***P<0.001 and ****P<0.0001. THBS1, thrombospondin-1; TCGA, The Cancer Genome Atlas; GBM, glioblastoma; CGGA, Chinese Glioma Genome Atlas; WHO, World Health Organization; HGG, high-grade glioma; LGG, low-grade glioma; NS, not statistically significant.
Immunohistochemical (IHC) staining images from the HPA demonstrated THBS1 protein expression in normal brain and glioma tissues. In comparison, among the 12 glioma tissues, weak IHC staining was observed in 2 cases of high-grade glioma (HGG), moderate staining in 1 case of HGG and 1 case of LGG and strong staining in 1 case of HGG (Fig. 1D). No staining was found in 4 cases of HGG and 3 cases of LGG. Based on the data from the HPA, THBS1 protein levels were not detected in the glial cells of normal brain tissues (Fig. 1D, control group). These results indicated that THBS1 expression was associated with glioma malignancy and that THBS1 expression were detected in glioma.
High THBS1 expression predicts the mesenchymal subtype of GBM
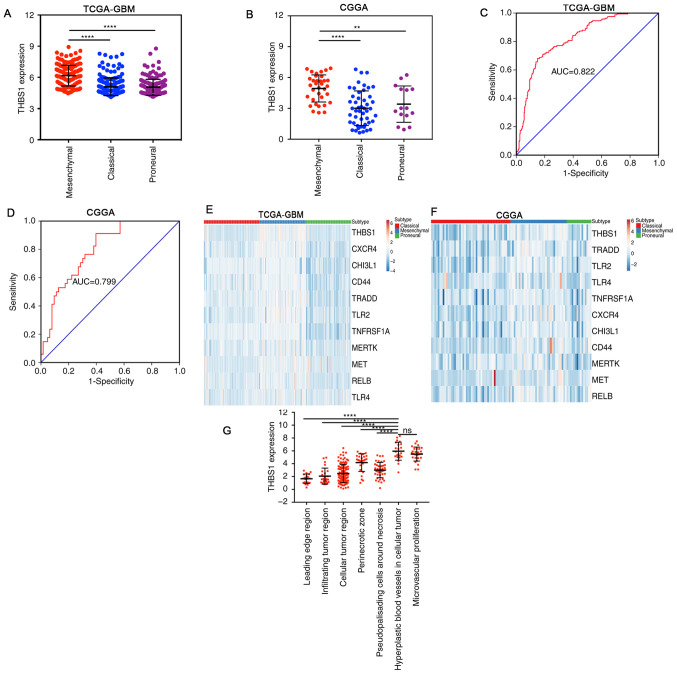
For GBM subtyping the three most recent subtype classification systems (classical, mesenchymal and proneural) were used (4,25). The expression profiles of THBS1 in different GBM subtypes were used using the TCGA-GBM and CGGA-GBM datasets. The results demonstrated that THBS1 expression was highest in the mesenchymal subtype (Fig. 2A and B). ROC curve analysis revealed that THBS1 may be an effective indicator of the mesenchymal subtype of GBM, with area under the curve (AUC) values of 82.2 and 79.9% in the TCGA-GBM (Fig. 2C) and CGGA-GBM datasets (Fig. 2D), respectively. Following this, Pearson's correlation coefficient was used to investigate the association between THBS1 and other well-known mesenchymal biomarkers in the TCGA-GBM and CGGA datasets (Fig. 2E and F) (3,30). Although the correlation coefficients were relatively weak (R<0.5), the results demonstrated that MET, CHI3L1, CD44, TRADD, CXCR4, RELB, TLR4, TNFRSF1A, TLR2 and MERTK were positively correlated with THBS1 expression with statistical significance (Fig. S1). By mining data in the Ivy Glioblastoma Atlas Project, the distribution of THBS1 transcript levels in distinct areas of GBM was investigated, and the results revealed that THBS1 transcript levels were mainly enriched in hyperplastic blood vessels in cellular tumors and in microvascular proliferation, representing characteristic features of the mesenchymal subtype (Fig. 2G). In summary, these results demonstrate that THBS1 is a valuable biomarker of the mesenchymal subtype of GBM.
Figure 2.
High THBS1 expression predicts the mesenchymal subtype of GBM. (A,B) THBS1 expression was higher in the mesenchymal subtype of GBM compared with the other subtypes in (A) TCGA-GBM and (B) CGGA. (C and D) Receiver operating characteristic curves demonstrated high sensitivity in predicting the mesenchymal subtype, with AUC values of ~0.8 in (C) TCGA-GBM and (D) CGGA datasets. (E and F) Comparison of THBS1 expression with other mesenchymal biomarkers. (G) THBS1 expression was enriched in hyperplastic blood vessels in cellular tumors and in microvascular proliferation, as evidence by the Ivy Glioblastoma Atlas Project. **P<0.01 and ****P<0.0001. THBS1, thrombospondin-1; GBM, glioblastoma; TCGA, The Cancer Genome Atlas; CGGA, Chinese Glioma Genome Atlas; NS, not statistically significant; AUC, area under curve.
DNA hypomethylation may participate in the upregulation of THBS1 expression
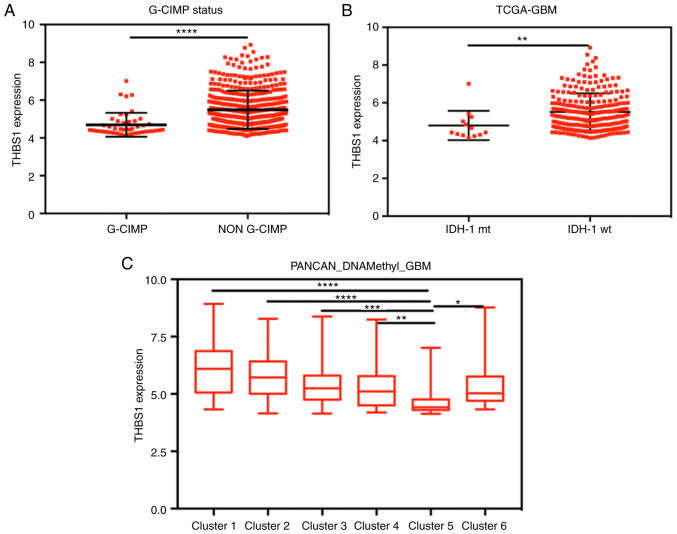
The glioma CpG island methylation phenotype (G-CIMP) results in the hypermethylation of numerous genes in GBM (31). Methylation 27k data was used to explore the association between G-CIMP status, overall GBM DNA methylation, THBS1 DNA methylation and THBS1 expression. Although the correlation coefficients between THBS1 expression and DNA methylation levels were weak (−0.2547 to −0.1685), the results demonstrated that four CpG sites were negatively associated with THBS1 expression with statistical significance (Fig. S2A-D). Furthermore, THBS1 expression was significantly lower in the G-CIMP group compared with the non-G-CIMP group (Fig. 3A).
Figure 3.
High THBS1 expression is negatively correlated with its DNA methylation levels and predicts IDH-1 wild-type GBM. (A) THBS1 expression was significantly lower in the G-CIMP group compared with the non G-CIMP group. (B) IDH-1 mt GBM exhibited lower THBS1 expression compared with IDH-1 wt GBM. (C) THBS1 expression was lowest in the GBM DNA methylation cluster 5 group. *P<0.05, **P<0.01, ***P<0.001 and ****P<0.0001. THBS1, thrombospondin-1; IDH-1, isocitrate dehydrogenase-1; GBM, glioblastoma; G-CIMP, glioma CpG island phenotype; mt, mutant type; wt, wild-type.
The IDH-1 status is an important prognostic indicator of GBM and is highly correlated with CpG island methylation (32). In the present study, IDH-1 mutation type GBM exhibited lower THBS1 expression (Fig. 3B) and higher levels of THBS1 DNA methylation compared with IDH-1 wild-type GBM (Fig. S2E-H). Furthermore, by comparing THBS1 expression in different overall GBM DNA methylation groups, the results indicated that THBS1 expression was lowest in the GBM DNA methylation cluster 5 group (Fig. 3C). Therefore, we hypothesized that the THBS1 DNA methylation status may be an important mechanism in the regulation of THBS1 gene expression.
High expression of THBS1 or its DNA hypomethylation is associated with a poor prognosis in GBM
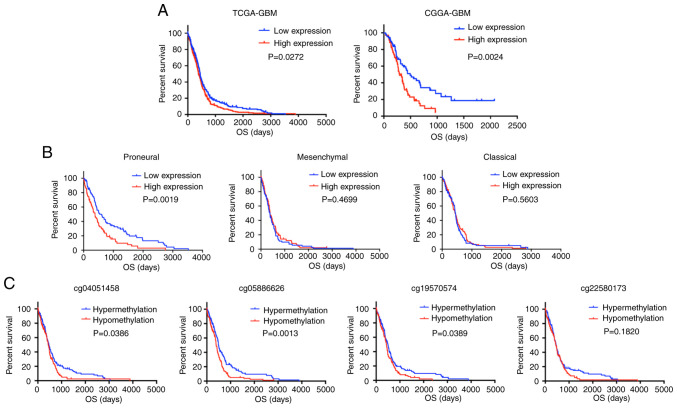
For survival analyses, data were divided into two groups based on the median value of THBS1. A log-rank test of the OS curves demonstrated that high THBS1 expression was associated with unfavorable OS in patients with GBM in the TCGA-GBM (THBS1 expression cutoff, 5.11) and CGGA-GBM (THBS1 expression cutoff, 2.99) datasets (Fig. 4A). As shown previously, THBS1 is an indicator of the mesenchymal subtype of GBM. Therefore, the prognostic value of THBS1 expression in distinct GBM subtypes was further investigated using the TCGA-GBM dataset. The results indicated that high THBS1 expression significantly reduced the OS of patients with the proneural subtype; however, its association with the other subtypes was not significant (Fig. 4B). The methylation analysis demonstrated that hypomethylation levels (β value > the median value) in 3 CpG sites (cg04051458, cg05886626 and cg19570574) were associated with a poor prognosis in GBM (Fig. 4C). These findings indicated that elevated THBS1 expression was an indicator of poor prognosis in patients with GBM.
Figure 4.
High expression of THBS1 or its DNA hypomethylation predicts poor OS in patients with GBM. (A) High THBS1 expression was associated with unfavorable OS in patients GBM from TCGA-GBM and CGGA datasets. (B) High THBS1 expression predicted a poor prognosis of the proneural subtype of GBM in the TCGA-GBM dataset. (C) The DNA hypermethylation of THBS1 predicted favorable OS in patients with GBM from the TCGA-GBM dataset. THBS1, thrombospondin-1; OS, overall survival; TCGA, The Cancer Genome Atlas; CGGA, Chinese Glioma Genome Atlas.
High THBS1 expression is associated with the immune response in GBM
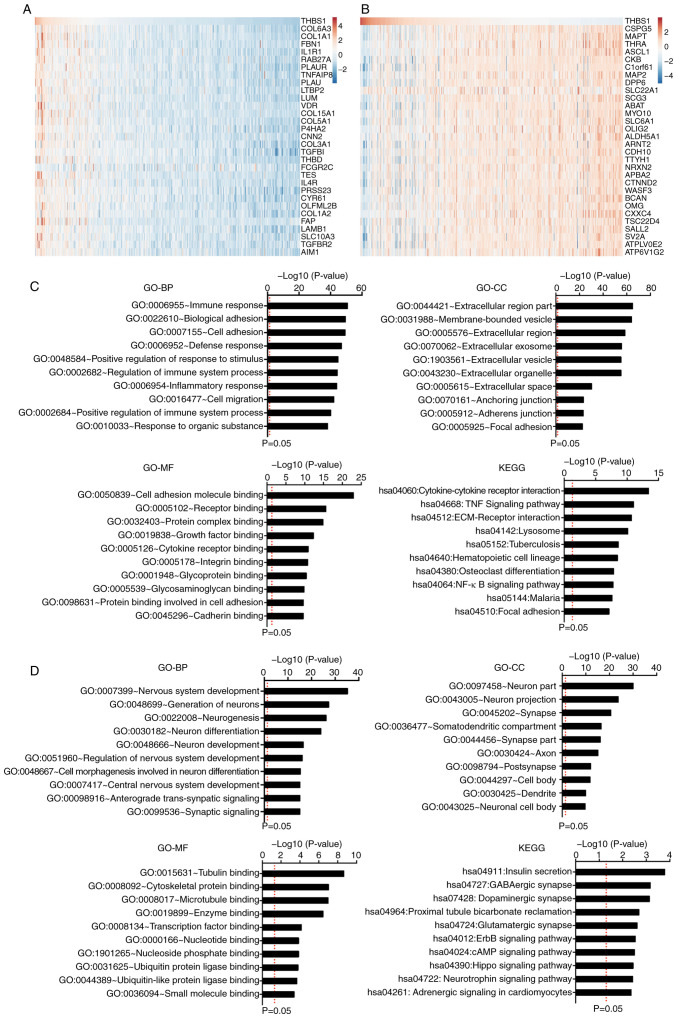
Pearson's correlation coefficient was used to identify genes that exhibited positive and negative correlations (R≥0.3) with THBS1 in the TCGA-GBM dataset using the GlioVis portal. The results discovered 1,040 positively correlated genes and 844 negatively correlated genes. The top 30 genes ranked by Pearson's R value were listed (Fig. 5A and B). Following this, correlated genes were separately uploaded to the online software DAVID to identify GO categories and the KEGG pathway.
Figure 5.
GO and KEGG functional enrichment analyses of THBS1-correlated genes. (A and B) Top 30 genes that correlated with THBS1 expression as ranked by Pearson's R value. (C) GO categories and the KEGG pathway of the genes that positively correlated with THBS1. (D) GO categories and KEGG pathway of the genes that negatively correlated with THBS1. THBS1, thrombospondin-1; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Gene and Genomic; BP, biological processes; CC, cellular component; MF, molecular function.
The top 10 items ranked according to the negative log10 (P-value) of the GO categories and KEGG pathways are displayed (Fig. 5C). The results revealed that the genes which exhibited positive correlations with THBS1 were mainly involved in the immune response. Regarding the biological processes (BP) category, the positively correlated genes were significantly enriched in the ‘immune response’, ‘biological adhesion’, ‘cell adhesion’, ‘defense response’, ‘positive regulation of response to stimulus’, ‘regulation of immune system process’, ‘inflammatory response’, ‘cell migration’, ‘positive regulation of immune system process’ and ‘response to organic substance’. In the cellular component (CC) category, the positively correlated genes were highly enriched in the ‘extracellular region part’, ‘membrane-bounded vesicle’, ‘extracellular region’, ‘extracellular exosome’, ‘extracellular vesicle’, ‘extracellular organelle’, ‘extracellular space’, ‘anchoring junction’, ‘adherence junction’ and ‘focal adhesion’. Furthermore, in the molecular function (MF) category, the positively correlated genes were enriched in ‘cell adhesion molecule binding’, ‘receptor binding’, ‘protein complex binding’, ‘growth factor binding’, ‘cytokine receptor binding’, ‘integrin binding’, ‘glycoprotein binding’, ‘glycosaminoglycan binding’, ‘protein binding involved in cell adhesion’ and ‘cadherin binding’. KEGG pathway analysis showed that the positively correlated genes were mainly enriched in the pathways ‘cytokine-cytokine receptor interaction’, ‘the TNF signaling pathway’, ‘ECM-receptor interaction’, ‘lysosomes’, ‘tuberculosis’, ‘hematopoietic cell lineage’, ‘osteoclast differentiation’, ‘NF-κB signaling pathway’, ‘malaria’ and ‘focal adhesion’. Moreover, genes that exhibited negative correlations with THBS1 were mainly involved in physiological functions, including ‘nervous system development’, ‘neuron part’ and ‘insulin secretion’ (Fig. 5D). These results indicated that THBS1 serves important roles in the immunologic biological processes of GBM.
THBS1 expression is positively correlated with the expression of immune-related genes and is associated with immune cell infiltration levels in GBM
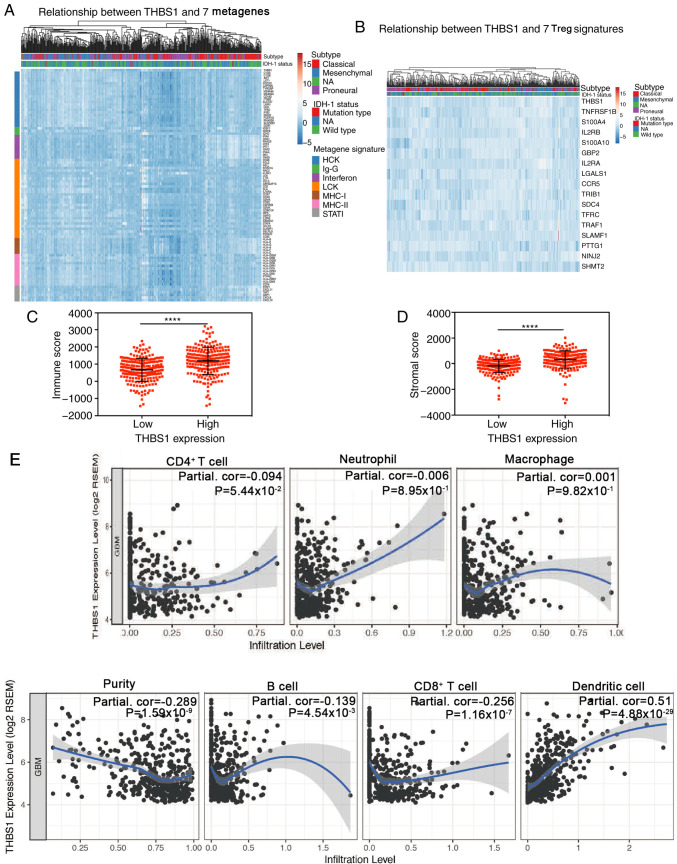
To clarify THBS1-related immunologic biological processes in GBM, 7 inflammatory metagene signatures were selected based on previous studies to explore their association with THBS1 in the TCGA-GBM dataset (26–28). The results indicated that THBS1 was positively correlated with most immune signatures (HCK, MHC-I, MHC-II, STAT1, interferon, LCK and IgG) and negatively correlated with KIRK1, IGSF1, IFI44L and IFIT1 (Fig. 6A; Table I). As GBM is recognized as an immunosuppressive neoplasm, Tregs, are a subpopulation of T cells that regulate the immune system (33). Following this, the association between Treg signatures and THBS1 expression was investigated as described by Wang et al (27). The results demonstrated that THBS1 expression was positively correlated with most Treg signatures (Fig. 6B; Table II). These results indicated that THBS1 overexpression is accompanied by an enhanced immune response and may serve an important role in immunosuppressive processes.
Figure 6.
Correlations between THBS1 expression and that of immune-related genes, immune/stromal scores and tumor-infiltrating lymphocytes in GBM. (A and B) Through Pearson's correlation coefficient, THBS1 expression exhibited significant correlations with most immune-related genes in The Cancer Genome Atlas-GBM dataset and was positively correlated with Treg signatures. (C and D) High THBS1 expression was associated with high immune/stromal scores in GBM. (E) In the Tumor IMmune Estimation Resource database, Spearman correlation demonstrated that THBS1 expression exhibited significant correlations with tumor purity and the infiltrating levels of B cells, CD8+ T cells and dendritic cells. THBS1 expression had a borderline significant correlation with the infiltrating levels of CD4+ T cells (P=0.0544); however, this was not evident in macrophages and neutrophils. ****P<0.0001. THBS1, thrombospondin-1; GBM, glioblastoma; Treg, regulatory T cells.
Table I.
Association between thrombospondin-1 expression and 7 metagene signatures.
| Gene signature | Gene list | Pearson's R | P-value |
|---|---|---|---|
| HCK | C1QB | 0.2248 | 1.738×10−7 |
| C1QA | 0.3056 | 6.788×10−13 | |
| AIF1 | 0.2781 | 7.495×10−11 | |
| LST1 | 0.2023 | 2.739×10−6 | |
| DOCK2 | 0.3436 | 4.231×10−16 | |
| LAPTM5 | 0.3002 | 1.759×10−12 | |
| TYROBP | 0.2312 | 7.550×10−8 | |
| MS4A4A | 0.4597 | 5.112×10−29 | |
| MS4A6A | 0.3133 | 1.642×10−13 | |
| CD163 | 0.4514 | 6.512×10−28 | |
| ITGB2 | 0.3396 | 9.677×10−16 | |
| SLC7A7 | 0.3462 | 2.443×10−16 | |
| LAIR1 | 0.4724 | 9.469×10−31 | |
| HCK | 0.4100 | 7.382×10−23 | |
| TFEC | 0.3583 | 1.811×10−17 | |
| IFI30 | 0.5369 | 7.913×10−41 | |
| MNDA | 0.3904 | 1.047×10−20 | |
| FCER1G | 0.3185 | 6.241×10−14 | |
| RNASE6 | 0.3169 | 8.295×10−14 | |
| SLCO2B1 | 0.3457 | 4.013×10−14 | |
| CCR1 | 0.3426 | 5.154×10−16 | |
| LCK | CD2 | 0.3896 | 1.289×10−20 |
| GZMK | 0.3795 | 1.463×10−19 | |
| GZMA | 0.3817 | 8.675×10−20 | |
| CD3D | 0.3803 | 1.201×10−19 | |
| CD53 | 0.2633 | 7.733×10−10 | |
| LCK | 0.2227 | 2.268×10−7 | |
| ARHGAP15 | 0.2677 | 3.922×10−10 | |
| CCL5 | 0.3790 | 1.642×10−19 | |
| GMFG | 0.3042 | 8.621×10−13 | |
| SELL | 0.2151 | 5.939×10−7 | |
| STAT4 | 0.2212 | 2.773×10−7 | |
| SAMSN1 | 0.3128 | 1.808×10−13 | |
| RAC2 | 0.4862 | 9.818×10−33 | |
| HCLS1 | 0.3191 | 5.485×10−14 | |
| CCR7 | 0.2181 | 4.095×10−7 | |
| PIK3CD | 0.2931 | 6.123×10−12 | |
| CORO1A | 0.2203 | 3.106×10−7 | |
| CD48 | 0.2981 | 2.573×10−12 | |
| IL2RG | 0.3869 | 2.470×10−20 | |
| SH2D1A | 0.1076 | 1.330×10−2 | |
| SLAMF1 | 0.0270 | 5.363×10−1 | |
| IL7R | 0.6121 | 1.132×10−55 | |
| INPP5D | NA | NA | |
| KLRK1 | −0.1006 | 2.070×10−2 | |
| FGL2 | 0.3075 | 4.802×10−13 | |
| IRF8 | 0.1649 | 5.000×10−4 | |
| SELPLG | 0.1510 | 4.927×10−4 | |
| IL10RA | 0.4530 | 4.054×10−28 | |
| SLA | 0.3928 | 5.848×10−21 | |
| CCR2 | 0.4416 | 1.187×10−26 | |
| CSF2RB | 0.5112 | 1.475×10−36 | |
| MHC-II | HLA-DRB1 | 0.2657 | 5.369×10−10 |
| HLA-DRB5 | NA | NA | |
| HLA-DRB3 | NA | NA | |
| HLA-DPA1 | 0.2124 | 8.266×10−7 | |
| HLA-DRA | 0.3009 | 1.558×10−12 | |
| HLA-DQA1 | 0.2889 | 1.260×10−11 | |
| HLA-DQA2 | NA | NA | |
| HLA-DMA | 0.3300 | 6.592×10−15 | |
| HLA-DOA | 0.2709 | 2.374×10−10 | |
| HLA-DRB4 | NA | NA | |
| HLA-DMB | 0.2548 | 2.761×10−9 | |
| HLA-DQB1 | 0.3990 | 1.236×10−21 | |
| HLA-DPB1 | 0.3078 | 4.524×10−13 | |
| HLA-DQB2 | NA | NA | |
| CD74 | 0.2518 | 4.318×10−9 | |
| PTPRC | 0.4077 | 1.344×10−22 | |
| HLA-DOB | 0.1976 | 4.654×10−6 | |
| HLA-DPB2 | NA | NA | |
| STAT1 | STAT1 | 0.1668 | 1.000×10−4 |
| CXCL11 | 0.1265 | 3.564×10−3 | |
| TAP1 | 0.0784 | 7.170×10−2 | |
| GBP1 | 0.2299 | 8.896×10−8 | |
| CXCL9 | 0.2904 | 9.745×10−12 | |
| CXCL10 | 0.1834 | 2.202×10−5 | |
| MHC-I | HLA-E | 0.2868 | 1.777×10−11 |
| HLA-H | NA | NA | |
| HLA-B | 0.3167 | 8.646×10−14 | |
| HLA-J | NA | NA | |
| HLA-F | 0.2961 | 3.668×10−12 | |
| HLA-G | 0.2606 | 1.163×10−9 | |
| HLA-A | 0.2230 | 2.204×10−7 | |
| HLA-C | 0.2885 | 1.353×10−11 | |
| HLA-L | NA | NA | |
| Ig-G | IGSF8 | NA | NA |
| ISLR2 | NA | NA | |
| IGSF21 | NA | NA | |
| IGSF1 | −0.1056 | 1.510×10−2 | |
| IGSF22 | NA | NA | |
| IGDCC3 | NA | NA | |
| IGHD | 0.2129 | 7.721×10−7 | |
| IGSF11 | NA | NA | |
| IGSF5 | NA | NA | |
| IGSF6 | 0.0248 | 5.697×10−1 | |
| Interferon | IFI44 | 0.1342 | 1.984×10−3 |
| DAS1 | 0.0663 | 1.272×10−1 | |
| RSAD2 | 0.0044 | 9.194×10−1 | |
| IFIT1 | −0.1456 | 7.838×10−4 | |
| IFIT3 | 0.0713 | 1.014×10−1 | |
| OAS2 | 0.1404 | 1.205×10−3 | |
| IFI44L | −0.1336 | 2.068×10−3 | |
| MAX1 | 0.0220 | 6.145×10−1 | |
| OAS3 | 0.1000 | 2.142×10−2 |
NA, not acquired.
Table II.
Correlation between thrombospondin-1 and regulatory T cell signatures in The Cancer Genome Atlas-glioblastoma database.
| Gene list | Pearson's R | P-value |
|---|---|---|
| TNFRSF1B | 0.3881 | 1.858×10−20 |
| S100A4 | 0.5349 | 1.791×10−40 |
| IL2RB | 0.4008 | 7.919×10−22 |
| EPSTI1 | NA | NA |
| GBP5 | NA | NA |
| S100A10 | 0.3859 | 3.152×10−20 |
| SLAMF1 | 0.0269 | 5.363×10−1 |
| GBP2 | 0.2880 | 1.455×10−11 |
| IL2RA | 0.5505 | 3.079×10−43 |
| LGALS1 | 0.3885 | 1.691×10−20 |
| CCR5 | 0.4176 | 9.878×10−24 |
| TRIB1 | 0.2944 | 4.877×10−12 |
| LGALS3 | 0.4885 | 1.595×10−18 |
| SDC4 | 0.3449 | 3.205×10−16 |
| TP53INP1 | NA | NA |
| TFRC | 0.2267 | 1.364×10−7 |
| PTTG1 | −0.0297 | 4.952×10−1 |
| TRAF1 | 0.2731 | 1.678×10−10 |
| NINJ2 | 0.1543 | 4.000×10−4 |
| SHMT2 | −0.0093 | 8.301×10−1 |
NA, not acquired.
Immune scores and stromal scores are significantly associated with GBM mesenchymal subtype and predict malignant phenotype of GBM, with high immune scores indicating an unfavorable prognosis for patients with GBM (9). In the present study, THBS1 expression was compared with immune/stromal scores and the results revealed that high THBS1 expression was correlated with high immune/stromal scores (Fig. 6C and D).
Tumor-infiltrating lymphocytes are important predictors of survival in patients with cancer and tumor purity is an important factor that determines glioma prognosis (34,35). Therefore, whether THBS1 expression was correlated with immune infiltration levels in GBM was explored. The correlations between THBS1 expression and numerous immune infiltration levels were assessed with the TIMER database (26). THBS1 expression had significant correlations with tumor purity and the infiltrating levels of B cells, CD8+ T cells and dendritic cells. Additionally, THBS1 expression had a borderline significant correlation with infiltrating levels of CD4+ T cells; however, this was not present in macrophages and neutrophils (Fig. 6E).
Discussion
According to the 2016 WHO central nervous system tumor classification, GBM is the most common malignant brain tumor and is the most aggressive type of glioma, exhibiting highly aggressive features with rapid recurrence following surgery and standard chemotherapy and radiotherapy (1). Investigation of the molecular mechanisms in glioma is necessary and the effect of numerous molecules on the prognosis of glioma have been confirmed, such as IDH-1 mutant type and 1p19q co-deletion status (1). As a glycoprotein, THBS1 was first discovered in platelets and functions in cancers development (12–18). Accumulating evidence has demonstrated that THBS1 regulates cell differentiation, proliferation and migration, and apoptosis of fibroblasts, smooth muscle cells and macrophages (19). However, the function of THBS1 in glioma has not been fully understood.
The present study investigated THBS1 expression in gliomas with different WHO grades and found a significant correlation between THBS1 expression and glioma malignancy, which was consistent with a previous study (21). However, the exact mechanism by which THBS1 expression is regulated in GBM remains unclear. A G-CIMP-positive phenotype correlated with IDH-1 mutations has been demonstrated to have a favorable prognosis in GBM (31). Therefore, in the present study the association between THBS1 expression and its DNA methylation levels was investigated. Notably, the results revealed four CpG sites that correlated with THBS1 expression and that the G-CIMP status was associated with low THBS1 expression. CIMP, in which numerous genes are concordantly methylated, has been confirmed to be associated with OS in certain types of solid tumors, such as hepatocellular carcinoma and ovarian carcinoma, and may indicate different survival outcomes (36,37). Furthermore, it has been reported that CIMP is enriched in the proneural subtype of GBM and is associated with a favorable prognosis (31). The results of the current study indicated that THBS1 expression was associated with the G-CIMP status and DNA methylation. The correlation coefficient between the methylation levels and THBS1 expression was lower in this study (Pearson R <0.5), which indicated that DNA methylation may potentially be a mechanism in regulating THBS1 expression. However, further study is required to confirm this hypothesis. Furthermore, the results demonstrated that elevated THBS1 expression or its DNA hypomethylation reduced OS in patients with GBM. Based on these results, the importance of THBS1 expression in predicting glioma malignancy was revealed and the prognostic value of THBS1 in GBM was further explored. Collectively, the aforementioned results of the present study demonstrate that THBS1 is an effective prognostic indicator of GBM.
Though elevated THBS1 expression in GBM has been reported (13), its expression profile in different GBM subtypes remains unclear. Using Wang's classification (4), the results of the present study demonstrated that THBS1 expression was higher in the mesenchymal subtype compared with the other subtypes. To explore whether THBS1 is an effective biomarker of the mesenchymal subtype, Pearson's correlation coefficient was used in the present study to investigate the correlation between THBS1 and other well-known mesenchymal biomarkers, as described by Yi et al (30). The results of the present study revealed that THBS1 was positively correlated with most mesenchymal biomarkers. The Ivy Glioblastoma Atlas Project analysis reported that THBS1 was enriched in hyperplastic blood vessels in cellular tumor and the microvascular proliferation, which are characteristic features of the mesenchymal subtype (3). Both overexpressed TBHS1 and its intense staining in vascular structures in GBM demonstrated that THBS1 may participate in the progression of angiogenesis and in the formation of mesenchymal characteristics (20).
In glioma, THBS1 expression facilitates the induction of TNF-β in monocytes, suppressing CD8+ T cell proliferation (22). CD35+ B cells with THBS1 expression have been revealed to decrease the levels of CD80/86 in dendritic cells and converted naïve CD4+ T cells to Tregs to inhibit the inflammatory response (19). These results indicate that THBS1 may be a valuable indicator of the immune response; however, the function of THBS1 in the GBM immune microenvironment remains uncertain. The tumor microenvironment, particularly the immune cell components, serves crucial roles in cancer progression and tumor purity has been reported to be an independent factor in determining glioma prognosis (35). Therefore, in the present study the function of THBS1 expression in the GBM immune microenvironment and in local immune infiltration was explored. By mining data from a large microarray and an RNA-seq dataset, THBS1 expression was compared with seven immune metagene signatures. The results revealed that THBS1 expression was positively correlated with the expression of most immune genes at the transcript level.
GBM has been recognized as an immunosuppressive neoplasm (33) and Tregs are vital immunosuppressive cells that serve an important role in suppressing other T cells activities and preventing immune inflammation, and may be involved in GBM immunosuppression (33,38). THBS1 converts latent TGF-β to active TGF-β, which functions in the conversion of Th0 cells to Tregs (19). Elevated THBS1 expression induced Foxp3+ expression, further increasing Treg expression (19). However, whether THBS1 regulates T cells immunosuppressive progression in GBM remains unclear. To clarify its immunosuppressive effects in GBM, correlation analysis was used in the present study to compare THBS1 expression with Treg signatures. The results identified a positive correlation with most Treg signatures. Taken together, the findings of the present study indicated that THBS1 aids in regulating the immune microenvironment in GBM. Therefore, we hypothesize that THBS1 may enhance immunosuppression and promote GBM to escape immune supervision. Furthermore, another important finding of the current study is that THBS1 expression was associated with diverse immune infiltration levels and tumor purity in GBM. By mining data from TIMER (29), the results revealed a positive correlation between THBS1 expression and the infiltration of dendritic cells. However, negative correlations between THBS1 expression and tumor purity, B cells infiltration and CD8+ T cell infiltration were revealed. Furthermore, the results demonstrated that high THBS1 expression predicted high immune/stromal scores, which have been shown to predict the malignant phenotype and an unfavorable prognosis of GBM.
However, even though the present study integrated transcriptional data across different databases, several limitations remain. Firstly, microarrays and sequencing data from different public datasets have inevitably introduced systematic bias, which could be observed in Fig. 1, in which different datasets exhibited variable THBS1 expression according to sex and age. Secondly, as a potential prognostic and diagnostic biomarker in GBM, the validation of protein levels is limited and the exact mechanisms by which THBS1 expression affects survival in patients with GBM remains uncertain, as the results could not confirm that THBS1 affects patient survival by regulating immune infiltration. Thirdly, a bioinformatics analysis of THBS1 expression, methylation levels, immune cell infiltration and patient survival across distinct public databases is indirect evidence and the underlying mechanisms in the regulation of gene expression are complex, involving numerous biological processes. The present study revealed that THBS1 DNA methylation levels were negatively correlated with THBS1 expression with weak correlation coefficients. However, further testing on in vivo and in vitro models combined with sufficient clinical sample examination will be required prior to determining whether THBS1 inhibition may be an effective method for treating GBM. These issues encourage the current authors to investigate this valuable research in the future.
In conclusion, by performing a comprehensive bioinformatics analysis, the present study demonstrated that THBS1 may be a biomarker of glioma malignancy and the GBM mesenchymal subtype. By analyzing the correlations between THBS1 expression and immune signatures, the results reported positive correlations between THBS1 expression and Treg signatures, supporting the conclusion that THBS1 may enhance local immune tolerance in GBM. The value of THBS1 in treating GBM requires further study.
Supplementary Material
Acknowledgements
Not applicable.
Funding
The present study was funded by the Liaonin Provide Special Professor Project (grant no. 3110517003).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
CQ and SO conceptualized and designed the present study. CQ collected the data, performed the statistical analyses and wrote the manuscript. LL, JH, GW and JL participated in the data collection and provided statistical expertise. All authors contributed to data analysis, drafting and revising the manuscript and agree to be accountable for all aspects of the work. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. The 2016 world health organization classification of tumors of the central nervous system: A summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 2.Jaoude DA, Moore JA, Moore MB, Twumasi-Ankrah P, Ablah E, Moore DF., Jr Glioblastoma and increased survival with longer chemotherapy duration. Kans J Med. 2019;12:65–69. doi: 10.17161/kjm.v12i3.11795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Q, Hu B, Hu X, Kim H, Squatrito M, Scarpace L, deCarvalho AC, Lyu S, Li P, Li Y, et al. Tumor evolution of glioma-intrinsic gene expression subtypes associates with immunological changes in the microenvironment. Cancer Cell. 2017;32:42–56.e6. doi: 10.1016/j.ccell.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gill BJ, Pisapia DJ, Malone HR, Goldstein H, Lei L, Sonabend A, Yun J, Samanamud J, Sims JS, Banu M, et al. MRI-localized biopsies reveal subtype-specific differences in molecular and cellular composition at the margins of glioblastoma. Proc Natl Acad Sci USA. 2014;111:12550–12555. doi: 10.1073/pnas.1405839111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Curry JM, Sprandio J, Cognetti D, Luginbuhl A, Bar-ad V, Pribitkin E, Tuluc M. Tumor microenvironment in head and neck squamous cell carcinoma. Semin Oncol. 2014;41:217–234. doi: 10.1053/j.seminoncol.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 7.Yoshihara K, Shahmoradgoli M, Martinez E, Vegesna R, Kim H, Torres-Garcia W, Treviño V, Shen H, Laird PW, Levine DA, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanahan D, Coussens LM. Accessories to the crime: Functions of cells recruited to the tumor microenvironment. Cancer Cell. 2012;21:309–322. doi: 10.1016/j.ccr.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 9.Jia D, Li S, Li D, Xue H, Yang D, Liu Y. Mining TCGA database for genes of prognostic value in glioblastoma microenvironment. Aging (Albany NY) 2018;10:592–605. doi: 10.18632/aging.101415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martinez-Lage M, Lynch TM, Bi Y, Cocito C, Way GP, Pal S, Haller J, Yan RE, Ziober A, Nguyen A, et al. Immune landscapes associated with different glioblastoma molecular subtypes. Acta Neuropathol Commun. 2019;7:203. doi: 10.1186/s40478-019-0803-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miyata Y, Sakai H. Thrombospondin-1 in urological cancer: Pathological role, clinical significance, and therapeutic prospects. Int J Mol Sci. 2013;14:12249–12272. doi: 10.3390/ijms140612249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Byrne GJ, Hayden KE, McDowell G, Lang H, Kirwan CC, Tetlow L, Kumar S, Bundred NJ. Angiogenic characteristics of circulating and tumoural thrombospondin-1 in breast cancer. Int J Oncol. 2007;31:1127–1132. doi: 10.3892/ijo.31.5.1127. [DOI] [PubMed] [Google Scholar]
- 13.Nakao T, Kurita N, Komatsu M, Yoshikawa K, Iwata T, Utsunomiya T, Shimada M. Expression of thrombospondin-1 and Ski are prognostic factors in advanced gastric cancer. Int J Clin Oncol. 2011;16:145–152. doi: 10.1007/s10147-010-0147-5. [DOI] [PubMed] [Google Scholar]
- 14.Trotter MJ, Colwell R, Tron VA. Thrombospondin-1 and cutaneous melanoma. J Cutan Med Surg. 2003;7:136–141. doi: 10.1007/s10227-002-0164-8. [DOI] [PubMed] [Google Scholar]
- 15.Wu MP, Tzeng CC, Wu LW, Huang KF, Chou CY. Thrombospondin-1 acts as a fence to inhibit angiogenesis that occurs during cervical carcinogenesis. Cancer J. 2004;10:27–32. doi: 10.1097/00130404-200401000-00007. [DOI] [PubMed] [Google Scholar]
- 16.Zhang J, Yang W, Zhao D, Han Y, Liu B, Zhao H, Wang H, Zhang Q, Xu G. Correlation between TSP-1, TGF-β and PPAR-γ expression levels and glioma microvascular density. Oncol Lett. 2014;7:95–100. doi: 10.3892/ol.2013.1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tenan M, Fulci G, Albertoni M, Diserens AC, Hamou MF, El Atifi-Borel M, Feige JJ, Pepper MS, Van Meir EG. Thrombospondin-1 is downregulated by anoxia and suppresses tumorigenicity of human glioblastoma cells. J Exp Med. 2000;191:1789–1798. doi: 10.1084/jem.191.10.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kragh M, Quistorff B, Tenan M, Van Meir EG, Kristjansen PE. Overexpression of thrombospondin-1 reduces growth and vascular index but not perfusion in glioblastoma. Cancer Res. 2002;62:1191–1195. [PubMed] [Google Scholar]
- 19.Zhang HP, Wu Y, Liu J, Jiang J, Geng XR, Yang G, Mo L, Liu ZQ, Liu ZG, Yang PC. TSP1-producing B cells show immune regulatory property and suppress allergy-related mucosal inflammation. Sci Rep. 2013;3:3345. doi: 10.1038/srep03345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Fraipont F, Keramidas M, El Atifi M, Chambaz EM, Berger F, Feige JJ. Expression of the thrombospondin 1 fragment 167–569 in C6 glioma cells stimulates tumorigenicity despite reduced neovascularization. Oncogene. 2004;23:3642–3649. doi: 10.1038/sj.onc.1207438. [DOI] [PubMed] [Google Scholar]
- 21.Daubon T, Léon C, Clarke K, Andrique L, Salabert L, Darbo E, Pineau R, Guérit S, Maitre M, Dedieu S, et al. Deciphering the complex role of thrombospondin-1 in glioblastoma development. Nat Commun. 2019;10:1146. doi: 10.1038/s41467-019-08480-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma Y, Qu B, Xia X, Yang L, Kuang Y, Yang T, Cheng J, Sun H, Fan K, Gu J. Glioma-derived thrombospondin-1 modulates cd14+ cell tolerogenic properties. Cancer Invest. 2015;33:152–157. doi: 10.3109/07357907.2015.1010089. [DOI] [PubMed] [Google Scholar]
- 23.Cancer Genome Atlas Research Network, corp-author. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uhlen M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, Zwahlen M, Kampf C, Wester K, Hober S, et al. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol. 2010;28:1248–1250. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- 25.Bowman RL, Wang Q, Carro A, Verhaak RG, Squatrito M. GlioVis data portal for visualization and analysis of brain tumor expression datasets. Neuro Oncol. 2017;19:139–141. doi: 10.1093/neuonc/now247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rody A, Holtrich U, Pusztai L, Liedtke C, Gaetje R, Ruckhaeberle E, Solbach C, Hanker L, Ahr A, Metzler D, et al. T-cell metagene predicts a favorable prognosis in estrogen receptor-negative and HER2-positive breast cancers. Breast Cancer Res. 2009;11:R15. doi: 10.1186/bcr2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang ZL, Li GZ, Wang QW, Bao ZS, Wang Z, Zhang CB, Jiang T. PD-L2 expression is correlated with the molecular and clinical features of glioma, and acts as an unfavorable prognostic factor. Oncoimmunology. 2019;8:e1541535. doi: 10.1080/2162402X.2018.1541535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou Y, Wang X, Huang X, Li XD, Cheng K, Yu H, Zhou YJ, Lv P, Jiang XB. High expression of COPB2 predicts adverse outcomes: A potential therapeutic target for glioma. CNS Neurosci Ther. 2019;26:309–318. doi: 10.1111/cns.13254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, Li B, Liu XS. TIMER: A web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77:e108–e110. doi: 10.1158/0008-5472.CAN-17-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yi L, Tong L, Li T, Hai L, Abeysekera IR, Tao Z, Ma H, Liu P, Xie Y, Li J, et al. Bioinformatic analyses reveal the key pathways and genes in the CXCR4 mediated mesenchymal subtype of glioblastoma. Mol Med Rep. 2018;18:741–748. doi: 10.3892/mmr.2018.9011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, Campos C, Fabius AWM, Lu C, Ward PS, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Balss J, Meyer J, Mueller W, Korshunov A, Hartmann C, von Deimling A. Analysis of the IDH1 codon 132 mutation in brain tumors. Acta Neuropathol. 2008;116:597–602. doi: 10.1007/s00401-008-0455-2. [DOI] [PubMed] [Google Scholar]
- 33.Razavi SM, Lee KE, Jin BE, Aujla PS, Gholamin S, Li G. Immune evasion strategies of glioblastoma. Front Surg. 2016;3:11. doi: 10.3389/fsurg.2016.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pan JH, Zhou H, Cooper L, Huang JL, Zhu SB, Zhao XX, Ding H, Pan YL, Rong L. LAYN is a prognostic biomarker and correlated with immune infiltrates in gastric and colon cancers. Front Immunol. 2019;10:6. doi: 10.3389/fimmu.2019.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang C, Cheng W, Ren X, Wang Z, Liu X, Li G, Han S, Jiang T, Wu A. Tumor purity as an underlying key factor in glioma. Clin Cancer Res. 2017;23:6279–6291. doi: 10.1158/1078-0432.CCR-16-2598. [DOI] [PubMed] [Google Scholar]
- 36.Wu LM, Zhang F, Zhou L, Yang Z, Xie HY, Zheng SS. Predictive value of CpG island methylator phenotype for tumor recurrence in hepatitis B virus-associated hepatocellular carcinoma following liver transplantation. BMC Cancer. 2010;10:399. doi: 10.1186/1471-2407-10-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strathdee G, Appleton K, Illand M, Millan DW, Sargent J, Paul J, Brown R. Primary ovarian carcinomas display multiple methylator phenotypes involving known tumor suppressor genes. Am J Pathol. 2001;158:1121–1127. doi: 10.1016/S0002-9440(10)64059-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Doucette T, Rao G, Rao A, Shen L, Aldape K, Wei J, Dziurzynski K, Gilbert M, Heimberger AB. Immune heterogeneity of glioblastoma subtypes: Extrapolation from the cancer genome atlas. Cancer Immunol Res. 2013;1:112–122. doi: 10.1158/2326-6066.CIR-13-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.