Abstract
The objective of the present study was to investigate the expression levels of toll-like receptor 4 (TLR4) in glioma cells and the mechanisms underlying its regulatory effects on proliferation, migration and apoptosis of glioma cells. A total of three TLR4 silencing short hairpin (sh)RNA plasmids were established, and Lipofectamine® was used to the transfect the human glioma cell line U-87MG. Transfection efficiency was measured via flow cytometry. The interference plasmid exhibiting the largest silencing effect on TLR4 was screened for subsequent experiments using puromycin. Reverse transcription-quantitative PCR and western blot analysis were used to detect the TLR4 gene and protein expression levels, respectively, in stably transfected cells. Flow cytometry measured cell cycle and apoptosis and a wound healing assay was employed to assess the migration ability of transfected cells. The proliferation of transfected cells was detected using Cell Counting Kit-8 assay. TLR4-sh2 exhibited the highest transfection efficiency. Following transfection of U-87MG cells with TLR4-sh2 and negative control (NC) plasmids for 48 h and screening by puromycin, stable transfected cells were named U-87MG-Sh and U-87MG-NC cells respectively. The TLR4 gene and protein expression levels in the U-87MG-Sh cells were significantly lower than in U-87MG and U-87MG-NC cells. The apoptosis rate and the percentage of G0/1 cells were significantly higher, whereas the cell proliferation rate was notably lower, in U-87MG-Sh cells than in the U-87MG-NC and U-87MG cells. The proliferation rate and the cell migration ability of U-87MG-Sh cells were significantly lower than those of U-87MG-NC and U-87MG cells. TLR4 is associated with the proliferation of glioma cells. Inhibition of TLR4 expression levels significantly inhibited proliferation of glioma cells and induced apoptosis. The present study provided insights into the mechanisms associated with the development, progression and invasion ability of glioma cells.
Keywords: glioma, toll-like receptor 4, migration, proliferation, RNA interference
Introduction
Glioma is a type of primary brain cancer, which is characterized by high prevalence and gross invasion and is lethal to human health (1,2). It is estimated that >19,000 new cases of glioma are diagnosed each year, with an age-adjusted average annual incidence rate of 6.24 per 100,000 individuals in the United States between 2007 and 2011 (3). Glioma cells are aggressive and can develop resistance to conventional therapies; treatment options (such as surgery, radiotherapy and chemotherapy) exhibit poor efficiency and the average survival time of patients with high-grade glioma is <1 year (4). With the development of molecular biology, the potential of immunotherapy as a treatment for glioma has been investigated. It is necessary to understand immunotolerance and immunomodulation of glioma to develop an immunotherapy strategy. Toll-like receptors (TLRs) have been identified as key pattern recognition receptors (PRRs). The release of inflammatory mediators and the activation of innate immunity are primarily caused by TLRs, which induce signal transduction via recognition of pathogen-associated molecular patterns (PAMPs) and certain endogenous ligands (5,6). TLRs are also implicated in the pathogenesis of numerous types of immune-associated disease in human (7–9). For example, Tian et al (10) reported that mulberry leaf decreased inflammation and insulin resistance in type 2 diabetic mice via TLRs and the insulin signaling pathway. A total of 10 types of TLR have been identified in humans, among which TLR4 exhibits a significant association with the biological behavior of tumors (11,12). This finding is of interest from a tumor immunity perspective. Although TLRs mediate innate immunity, they are not unique to the innate immune system; in addition to immune cells (monocytes/macrophages, T/B lymphocytes and dendritic cells), they are also expressed in certain non-immune cells (endothelial, smooth muscle and tumor cells) (13). TLR expression levels in tumor cells are important for tumor genesis and development, and is an area of interest for tumor research (14). Among TLRs, TLR4 is more commonly expressed in different types of tumor, such as colon cancer and ovarian carcinoma, compared with other TLRs (15,16). High expression levels of TLR4 accelerate tumor proliferation and mediate tumor immune escape (17,18). Hence, the present study aimed to investigate the association between TLR4 and the development of human glioma, and to provide an experimental basis for understanding of the molecular mechanism underlying proliferation regulation of human glioma cells.
Materials and methods
Reagents and equipment
All-in-One™ quantitative (q)PCR Primer (cat. no. HQP054754) and H-TLR4-short hairpin (sh)RNAs 1–3 (cat. nos. HSH054754-CU6-a, b, c and CSHCTR001-CU6) were purchased from GeneCopoeia, Inc. Lipofectamine® 2000 was obtained from Invitrogen (Thermo Fisher Scientific, Inc.). High-purity Plasmid Mini-Preps kit was obtained from Tiangen Bioctech Co., Ltd.. Minimum Essential Medium (MEM) and bovine serum albumin were obtained from Gibco (Thermo Fisher Scientific, Inc.). RNAiso total RNA extraction reagent, HiScript II 1st Strand complementary cDNA Synthesis kit and qPCR SYBR Green Master mix were acquired from Vazyme Biotech Co., Ltd.. Annexin V-PE/7-AAD kit and Matrigel Matrix Basement Membrane were purchased from BD Biosciences. FC-500 flow cytometer (Beckman Coulter, Inc.), the PCR instrument (Eppendorf) and Mx3000P fluorescence quantitative PCR instrument (Agilent Technologies, Inc.) were also used.
Cell lines
The U-87MG cell line (cat. no. CL-0238) was obtained from Procell Life Science & Technology Co., Ltd. STR profiling, Y-chromosome paint and Q-band assay confirmed that the cell line is male in origin (performed by Procell Life Science & Technology Co., Ltd). Based on current literature, the cell line is likely a glioblastoma of unknown origin (19). The U-87MG cell line had a stable passage in the laboratory, and was cultured in MEM containing 10% FBS, 100 U/ml penicillin and streptomycin. Cells were sustained in an incubator at 37°C with 5% CO2.
RNA interference lentiviral vector construction
GeneCopoeia, Inc. synthesized the interference plasmids [cat. nos. HSH054754-CU6-a,b,c and CSHCTR001-CU6; accession no. NM_003266.3 (Homo sapiens TLR4, mRNA)]. The vector used was psi-U6.1 (GeneCopoeia, Inc.). A total of three shRNAs were designed to interfere with the TLR4 gene. shRNA target sequences are presented in Table I.
Table I.
Short hairpin RNA target sequences.
| Clone name | Symbol | Location | Length, bp | Target sequence (5′-3′) |
|---|---|---|---|---|
| HSH054754-CU6-a(OS397039) | TLR4 | 890 | 21 | GCTCACAATCTTATCCAATCT |
| HSH054754-CU6-b(OS397040) | TLR4 | 678 | 21 | GGTGTGAAATCCAGACAATTG |
| HSH054754-CU6-c(OS397041) | TLR4 | 887 | 21 | GTGGCTCACAATCTTATCCAA |
TLR4, toll-like receptor 4.
Transfection of shRNA-containing plasmids into U-87MG cells
shRNA-containing plasmids (4 µg) were transfected into U-87MG cells, in order to silence the TLR4 gene. For reagent 1, 244 µl MEM (FBS- and antibiotic-free) was used to buffer 6 µl Lipofectamine® 2000. For reagent 2, 240 µl MEM (FBS- and antibiotics-free) was used to dilute 10 µl shRNA plasmids [containing 4 µg TLR4-sh1-3 and negative control (NC) plasmids]. Both reagents were placed at room temperature for 5 min.
U-87MG cells in the logarithmic phase of growth were collected and seeded in 6-well plates at a density of 4×105 cells/well. Cells were transfected until they reached 85–90% confluence. MEM (FBS- and antibiotics-free) was used to wash cells twice, then 2 ml FBS- and antibiotic-free MEM was added into each well. After placing the mixture containing reagents 1 and 2 at room temperature for 20 min, 200 µl of this mixture was added into wells. After 6 h, FBS- and antibiotics-free MEM was replaced with complete culture medium. Following transfection for 48 h, cells were observed under a fluorescence microscope (magnification, ×100). Untreated cells were considered as the blank control group. Cells only treated with Lipofectamine were considered as the liposome group.
Identifying stably transfected cells with puromycin
The TLR4-sh2 plasmid was chosen for subsequent siRNA experiments. After U-87MG cells were transfected with the TLR4-sh2 plasmid and the negative control plasmid for 48 h, the cells were diluted with MEM and cultured in a 6-well plate at a density of 5×104 cells/well. Subsequently, 600 ng/ml puromycin was added into each well. The non-transfected U-87MG cells were used as the blank control group. After the cells were cultured for 14 days at 37°C with 5% CO2, the surviving cells were considered as stably transfected cells. The cells transfected with TLR4-sh2 were named U-87MG-Sh, while the cells transfected with the negative control (NC) plasmid were named U-87MG-NC.
Evaluating transfection efficiency via flow cytometry
Following transfection for 48 h, cells were collected and washed with PBS once. Next, flow cytometry was performed using a FC-500 type flow cytometer (Beckman Coulter, Inc.) to measure green fluorescence in cells. Transfection efficiency was identified as the proportion of cells expressing green fluorescence compared with all observed cells. Green fluorescence was measured using the EXPO32 ADC v1.2 software (Beckman Coulter, Inc.).
Evaluating TLR4 mRNA expression levels via reverse transcription-quantitative (RT-q)PCR
Following transfection for 48 h, cells were collected and washed with PBS twice. Next, based on the one-step method (20), total RNA was extracted by adding 1 ml RNA isolator (Vazyme Biotech Co., Ltd.). As the template for PCR, cDNA was synthesized using the HiScript II First Strand cDNA Synthesis kit according to the manufacturer's protocol (Vazyme Biotech Co., Ltd.). The internal reference was human GAPDH. The fluorescent signal was obtained using SYBR-Green I. Based on the manufacturer's protocol of the qPCR SYBR-Green Master Mix kit (Vazyme Biotech Co., Ltd.), qPCR was performed. The thermocycling conditions were as follows: 95°C for 5 min (pre-denaturation), followed by 40 cycles at 95°C for 10 sec and 60°C for 30 sec, and dissociation at 95°C for 15 sec, 60°C for 1 min and 95°C for 15 sec. For each sample, each experiment was repeated three times. The forward primer for GAPDH was 3′-CACTACCGTACCTGACACCA-5′ and the reverse primer was 3′-ATGTCGTTGTCCCACCACCT-5′. The TLR4 sequence was synthesized by GeneCopoeia, Inc. (cat. no. HQP054754). The relative expression levels of TLR4 mRNA (ΔCq=CqTLR4-CqGAPDH) were calculated using the 2−ΔΔCq method (21). RT-qPCR was used to evaluate the silencing effect of shRNA TLR4 expression levels and to select the interference plasmid with the greatest silencing effect.
Measuring TLR4 protein expression levels via western blotting
U-87MG-Sh, U-87MG-NC and U-87MG cells were collected in the logarithmic phase of growth and washed with cold PBS twice. Total protein was collected by adding cell lysis solution (RIPA:PMSF, 100:1; Beijing Solarbio Science & Technology Co., Ltd.). The protein concentration was evaluated via bicinchoninic acid assay. Protein (50 µg/lane) was separated via 10% SDS-PAGE then transferred to PVDF membranes before being blocked overnight at 4°C with 5% skimmed milk. Next, rabbit-anti-human β-actin (1:4,000; cat. no. ab8227; Abcam) or mouse-anti-human TLR4 (1:500; cat. no. ab22048; Abcam) monoclonal antibodies were added for incubation at room temperature for 1 h. The membrane was washed three times, then incubated at room temperature for 2 h with horseradish peroxidase-conjugated goat anti-mouse and anti-rabbit secondary antibodies (1:20,000; cat. nos. ab205719 and ab205718, respectively; Abcam). The antigen-antibody reaction was visualized by detection using an ECL assay (EMD Millipore). Relative expression levels of TLR4 protein were normalized to those of β-actin using Odyssey v3.0 software (LI-OR Biosciences). A total of three independent repeats were performed.
Flow cytometric analysis of cell cycle
U-87MG-Sh, U-87MG-NC and U-87MG cells were collected in the logarithmic phase of growth and washed with cold PBS twice. Cells were fixed with 70% ethanol at 4°C for 24 h. Cells were washed twice with PBS and cell density was adjusted to 1×107/ml. Then, 100 µl cell suspension was collected and 500 µl propidium iodide including a ribonuclease (BD Biosciences) was added for 30 min at 4°C. Flow cytometry was performed using a FC500 type flow cytometer (Beckman Coulter, Inc.), and the Multicycle AV v6.0 software (Beckman Coulter, Inc.) was used to estimate the percentage of cells in each stage of the cell cycle. The proliferation index (PI) was calculated as the percentage of cells in the S and G2/M phases against all cells, representing cell proliferation activity.
Flow cytometry analysis of apoptosis
U-87MG-Sh, U-87MG-NC and U-87MG cells were collected in the logarithmic phase of growth and washed with cold PBS twice. Then, 1×106 cells were collected and washed with PBS once and suspended in 100 µl 1X binding buffer. Next, cells were incubated on ice with 10 µl Annexin V-PE for 15 min. Then, 380 µl 1X binding buffer and 10 µl 7-AAD were separately added, before being placed on ice for 15 min in the dark, washed and suspended in 1 ml PBS. Flow cytometry (FC500 type flow cytometer; Beckman Coulter, Inc.) was used to evaluate cell apoptosis. Immunofluorescence data and apoptosis rate were measured using the EXPO32 ADC v1.2 software (Beckman Coulter, Inc.).
Measuring cell migration by wound healing assay
On the back of the 6-well culture plate, lines (5/well) at 0.5 cm intervals were drawn. Then, ~5×105 cells (U-87MG-Sh, U-87MG-NC and U-87MG) were seeded into each well and cultivated overnight at 37°C to 100% confluence. The next day, scratches perpendicular to the lines on the back of the 6-well plate were drawn on cells using a 10-µl pipette tip. In order to remove unattached cells, cells were washed with PBS three times and cultured for 24 h at 37°C in serum-free MEM. The cell culture condition was 37°C with 5% CO2, and images were captured at 0 and 24 h using an inverted light microscope (TE2000; Nikon Corporation; magnification, ×100). The area of wound healing was assayed using ImageJ 1.42 software (National Institutes of Health).
Detecting cell viability by cell counting Kit-8 (CCK-8) assay
Monolayer cultured U-87MG-Sh, U-87MG-NC and U-87MG cells were digested at 37°C for 30 sec using 0.25% trypsin. Then, MEM supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) was used to prepare the cell suspension. The cell density was adjusted to 1×104 cells/ml and cells were seeded into 96-well plates before being cultured in the incubator for cell adherence. Following incubation at 37°C with 5% CO2 for 24 h, 20 µl CCK-8 (Wuhan Boster Biological Technology, Ltd.) was added to each well and the plate was incubated for 4 h at 37°C according to the manufacturer's protocol. The 450 nm optical density wavelength was read via a microplate spectrophotometer, calibrated using the blank well. Growth inhibitory rate was calculated as 1-A450 U-87MG-Sh, U-87MG-NC or U-87MG cells/A450 U-87MG cells) ×100%.
Statistical analysis
All data are presented as the mean ± SD (n=3). One-way ANOVA was performed to compare multiple samples, followed by Tukey's post hoc test. Data were analyzed using SPSS software (version 21; IBM Corp.). P<0.05 was considered to indicate a statistically significant difference.
Results
Silencing of TLR4 in U-87MG cells using shRNA plasmid transfection and evaluation of transfection efficiency and TLR4 mRNA expression levels via flow cytometry and fluorescence RT-qPCR
Green fluorescence expression levels in U-87MG cells transfected with H_TLR4-sh1, H_TLR4-sh2, H_TLR4-sh3 and NC plasmid were observed via fluorescence microscope. No green fluorescence was detected in non-transfected cells (blank control) and U-87MG cells in the liposome group.
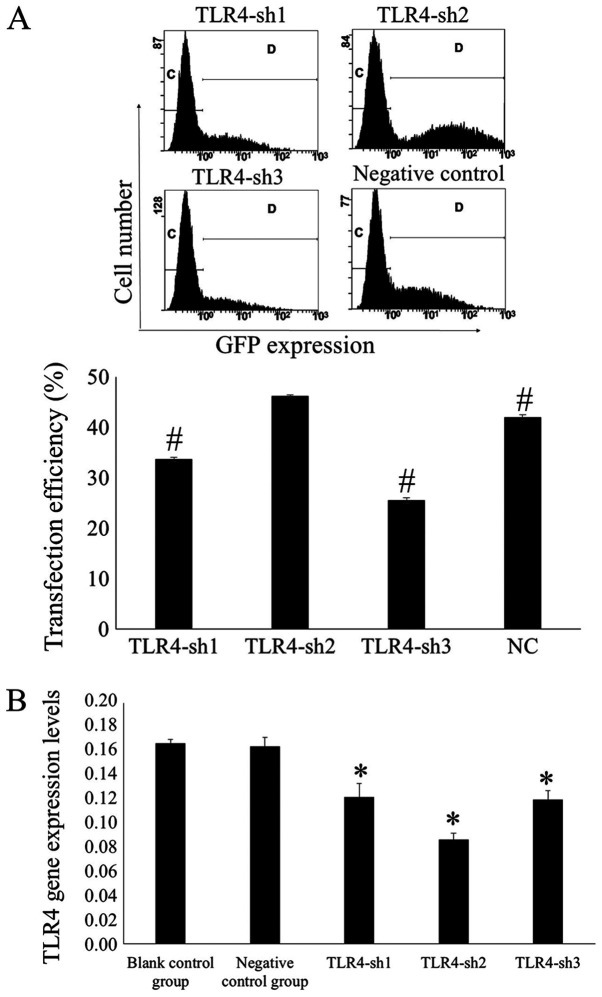
Flow cytometry was used to measure transfection efficiency after 48 h. Results demonstrated that the transfection efficiency was 33.66±0.40, 46.20±0.31, 25.49±0.60 and 41.94±0.55% in the H_TLR4-sh1, H_TLR4-sh2, H_TLR4-sh3 and NC groups, respectively. In the H_TLR4-sh2 group, the transfection efficiency was significantly higher compared with the other groups (P<0.01; Fig. 1A).
Figure 1.
Silencing TLR4 in U-87MG cells using shRNA plasmids. (A) Transfection efficiency was evaluated using flow cytometry. The percentage of cells expressing green fluorescence measured using flow cytometry represents the transfection efficiency. #P<0.01 vs. TLR4-sh2. The letters “C” and “D” represent the gate of flow cytometry. (B) Expression levels of TLR4 mRNA in transfected cells using fluorescence reverse transcription-quantitative PCR. *P<0.05 vs. blank and negative control. n=3. TLR4, toll-like receptor 4; sh, short hairpin RNA.
It was indicated that the TLR4 mRNA expression levels in U-87MG cells transfected with H_TLR4-sh1, H_TLR4-sh2, H_TLR4-sh3 and NC plasmid were 0.1219±0.0116, 0.0867±0.0054, 0.1201±0.0071 and 0.1643±0.0074, respectively, whereas that in the blank control group was 0.1669±0.0029. The TLR4 mRNA expression levels were significantly lower in U-87MG cells transfected with H_TLR4-sh2 than in the other groups (P<0.05; Fig. 1B). This illustrated that H_TLR4-sh2 possessed the largest silencing effect. Hence, H_TLR4-sh2 was selected for subsequent experiments.
TLR4 silencing decreases TLR4 mRNA expression levels
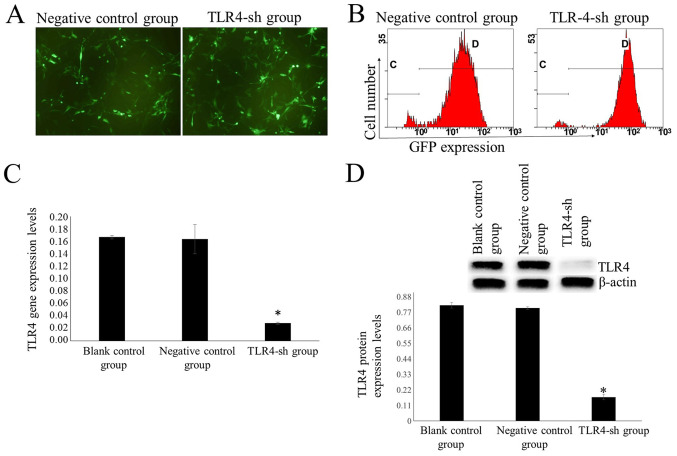
Following treatment with 600 ng/ml puromycin for 14 days, all cells in the blank control group died. A notable number of cells transfected with TLR4-sh2 and NC plasmids were alive (Fig. 2A). Surviving cells represented stably transfected cells. Cells transfected with TLR4-sh2 and NC plasmids were labeled as U-87MG-Sh and U-87MG-NC cells, respectively.
Figure 2.
Identification of stably transfected cells using puromycin. Cells were treated with 600 ng/ml puromycin for 14 days. (A) Observing expression levels of green fluorescence using a fluorescence microscope (magnification, ×100). (B) Flow cytometry was used to evaluate transfection efficiency. Transfection efficiency was 95.05±0.17 and 94.75±0.72% in the H_TLR4-sh2 and NC groups, respectively. The letters “C” and “D” represent the gate of flow cytometry. Measuring expression levels of TLR4 (C) mRNA and (D) protein in transfected cells using fluorescence reverse transcription-quantitative PCR and western blot analysis, respectively. n=3. *P<0.01 vs. U-87MG and U-87MG-NC. TLR4, toll-like receptor 4; sh, short hairpin RNA; NC, negative control; GFP, green fluorescent protein.
Transfection efficiency was measured via flow cytometry. In the TLR4-sh and NC group, transfection efficiency was 95.05±0.17 and 94.75±0.72%, respectively (Fig. 2B). Fluorescence RT-qPCR results demonstrated that the TLR4 mRNA expression levels were significantly lower in the TLR4-sh group than that in NC and blank control groups (P<0.01; Fig. 2C).
TLR4 protein expression levels are decreased in TLR4-sh-transfected cells
Western blot results demonstrated that TLR4 protein expression levels were significantly lower in the TLR4-sh group than in NC and blank control groups (P<0.01; Fig. 2D).
Measuring cell cycle and apoptosis rate of transfected cells via flow cytometry
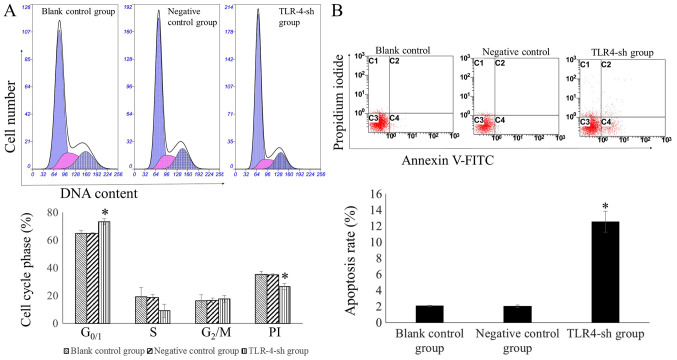
The percentage of cells in the G0/1 phase was significantly higher in the TLR4-sh than that in NC and blank control groups (P<0.01). By contrast, the PI was significantly lower in U-87MG-Sh cells than in U-87MG-NC and U-87MG cells (P<0.01). The percentage of the cells in the G0/1 phase and PI in U-87MG-NC and U-87MG cells exhibited no significant difference (P>0.05; Fig. 3A).
Figure 3.
Evaluation of cell cycle and apoptosis rate of transfected cells using flow cytometry. (A) Cell cycle analysis of transfected, negative control and blank control cells. Propidium iodide staining was used to determine DNA concentration. Multicycle AV software was used to perform fit analysis of the DNA cell cycle and to calculate the percentage of cells in each phase. *P<0.01 vs. blank and negative control. The percentage of G0/1 cells in the U-87MG-Sh group was significantly higher, whereas PI was significantly lower. This indicated that the proliferation activity of the U-87MG-Sh cells decreased significantly. (B) Evaluating cell apoptosis rate using flow cytometry. Measuring apoptosis rates in transfected and negative and blank control cells with flow cytometry. The apoptosis rate in the TLR4-sh group increased significantly. *P<0.01 vs. blank and negative control. TLR4, toll-like receptor 4; sh, short hairpin RNA; PI, proliferation index.
The cell apoptosis rate was significantly higher in U-87MG-Sh cells than in U-87MG-NC and U-87MG cells (P<0.01) whereas no significant difference was observed in the apoptosis rates between U-87MG-NC and U-87MG cells (P>0.05; Fig. 3B).
TLR4 silencing decreases migration ability
Wound healing assay indicated that migration ability of the U-87MG-Sh cells was significantly lower than that of U-87MG-NC and U-87MG cells (Fig. 4).
Figure 4.
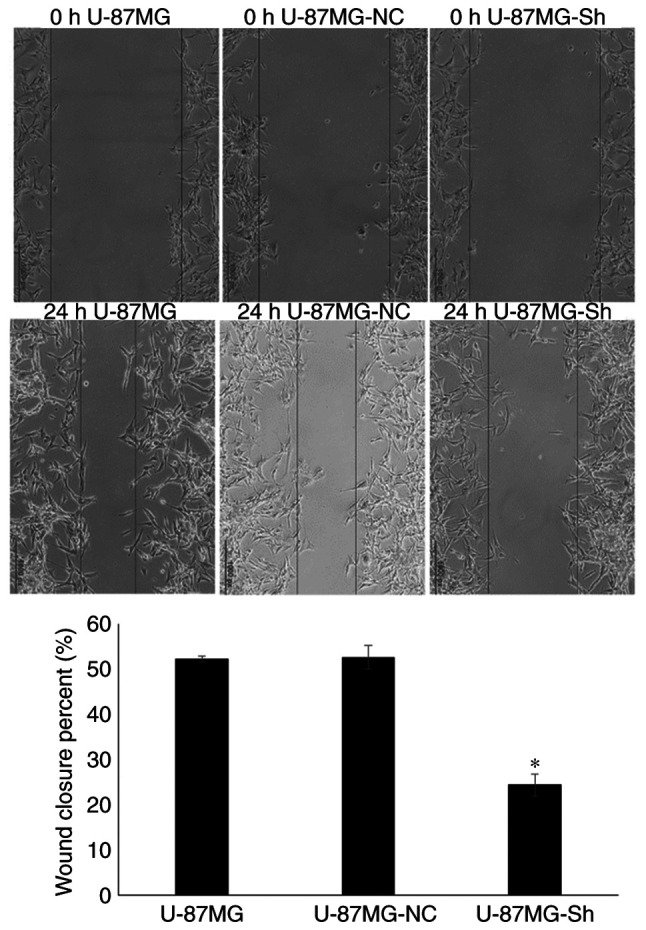
Evaluation of cell migration ability using wound healing assay, viewed under an inverted microscope (magnification, ×100). *P<0.01 vs. U-87MG and U-87MG-NC. NC, negative control; Sh, short hairpin RNA.
TLR4 silencing inhibits cell proliferation
Results of the CCK-8 assay indicated that the proliferation inhibitory rates were 0.40±1.47, 0.31±0.94 and 28.94±1.31% in the blank, NC and TLR4-sh groups, respectively. The proliferation inhibitory rate of TLR4-sh group was significantly higher than that of blank and NC groups (P<0.01) (data not shown).
Discussion
Glioma is a type of primary brain cancer (22), which is characterized by high prevalence and gross invasion and is lethal to human health (1,2). Resistance to conventional therapy (such as surgery, radiotherapy and chemotherapy) and the diffusely aggressive nature of glioma mean overall efficiency of glioma treatment is poor, and the average survival time of patients with high-grade glioma is <1 year (23). With the development of molecular biology, research has focused on immunotherapy against glioma (23). Therefore, it is necessary to determine the characteristics of immunotolerance and immunomodulation of glioma to develop an effective immunotherapy strategy.
TLRs have been investigated as key PRRs in natural immunity and recognize certain highly conserved molecular structures of pathogens (24). Moreover, TLRs are the bridge between innate immunity and acquired immunity and are important to prevent biological pathogenic factor infection (25).
TLRs are type I transmembrane proteins in mammals and they are a family of conserved transmembrane signaling receptors regulating innate immunity. TLRs are a member of the PRR family and are associated with the recognition of PAMPs (26). TLR4 was the first identified human cell TLR homologous to Drosophila, and is therefore the most studied TLR (27). To date, 13 types of TLRs (TLR 1–13) have been identified in the human body (28).
Although TLRs mediate innate immunity, they are not unique to the innate immune system: In addition to immune cells [monocytes/macrophages, T/B lymphocytes and dendritic cells (29)], they are also expressed in certain non-immune cells, such as endothelial (30), smooth muscle (31) and tumor cells (32). TLR expression levels in tumor cells are key for tumorigenesis and progression, and this is an important topic in tumor research (33). Among TLRs, TLR4 is closely associated with tumors; high expression levels of TLR4 accelerate tumor proliferation and mediate tumor immune escape, for example in colon cancer, lung cancer and oral squamous cell carcinoma (34–36). Pandey et al (37) investigated TLR4 polymorphisms and expression level patterns and reported that TLR4 expression levels in different types of solid tumor tissue promote tumor growth, invasion and metastasis. Therefore, TLR4 expression levels in different tumor types may serve as a marker for tumor proliferation, differentiation, metastasis, prognosis and patient survival. Chen et al (38) reported that TLR4 is associated with liver cancer development; the activated TLR4/Nanog oncogenic pathway is associated with the suppression of cytostatic TGF-β signaling and may serve as a therapeutic target for hepatitis C virus-associated hepatocellular carcinoma (HCC). Yao et al (39) reported that the TLR4/STAT3 signaling pathway participates in the carcinogenesis, development, invasion and metastasis of liver cancer. The authors also demonstrated that M2-polarized macrophages facilitate the migration and epithelial-mesenchymal transition of HCC cells via the TLR4/STAT3 signaling pathway, suggesting that TLR4 may be a novel therapeutic target. Szajnik et al (40) demonstrated that the TLR4/MyD88 signaling pathway promotes ovarian cancer development, drug resistance and immune escape. Zhang and Zhang (41) reported that Foxp3 and TLR4 are highly expressed in cervical cancer cells and are associated with clinical stage and lymph node metastasis; Foxp3 and TLR4 may be useful biomarkers for predicting the prognosis of patients and treating cervical cancer. Zhan et al (42) revealed that the molecular factors TLR4 and TLR3 affect the progression of lung cancer by regulating autophagy, which may be a potential treatment strategy for lung cancer. Li et al (43) reported that TLR4 is highly expressed in breast cancer tissue and promotes breast cancer metastasis via the AKT/GSK3β/β-catenin pathway, which is stimulated by lipopolysaccharide. Numerous studies have proved that TLR4 is associated with tumors (44,45), but there are few studies on the association between TLR4 and the occurrence and development of glioma. Hence, the present study aimed to elucidate the association between TLR4 and the occurrence and development of glioma in vitro, and to provide experimental support to further investigate the pathogenesis of glioma.
In the present study, TLR4 gene expression levels in the human glioma cell line U-87MG were silenced by gene interference. The use of only one cell line was a potential limitation of the present study: In future investigations, multiple glioma cell lines should be used to verify the regulatory effect of TLR4 on proliferation. In the present study, a total of three interference plasmids were designed for the human TLR4 gene. After 48 h transfection, flow cytometry detection demonstrated that the transfection and silencing efficiency of TLR4-sh2 was the highest. Therefore, TLR4-sh2 interference plasmids were selected for subsequent experiments. The transfection efficiency of TLR4-sh2 interference and NC plasmid were 95.05±0.17 and 94.75±0.72%, respectively. Results of fluorescence RT-qPCR revealed that the TLR4 gene expression levels in U-87MG-sh cells decreased significantly, which revealed that TLR4 gene silencing in U-87MG cells was successful.
The present study investigated the association between TLR4 and U-87MG cell proliferation. After silencing TLR4 gene expression levels in U-87MG cells via gene interference, flow cytometry indicated that the PI of U-87MG-sh cells declined and the G0/1 phase of the cell cycle extended significantly, demonstrating that TLR4 gene silencing significantly inhibited the proliferation of U-87MG cells. Following TLR4 gene silencing, CCK-8 demonstrated that U-87MG cell proliferation was significantly inhibited, which was consistent with the flow cytometry results. The absence of colony formation assay was a potential limitation of the present study: In future investigations, colony formation assay should be used to verify the CCK-8 results. Following TLR4 gene silencing in U-87MG cells, flow cytometry revealed that the apoptotic rate of U-87MG cells increased significantly, indicating that silencing the TLR4 gene inhibited growth and induced apoptosis in the glioma cell line U-87MG. TLR4 may therefore represent a target for the development of anti-glioma drugs and treatment methods.
TLR4-sh interference plasmids were successfully transfected into U-87MG cells via liposome transfection and stably transfected cells were screened using puromycin to establish a glioma cell model with low TLR4 gene expression levels. The present study demonstrated that the TLR4 gene is associated with proliferation and apoptosis regulation in U-87MG cells. Silencing the TLR4 gene in U-87MG cells significantly inhibited cell proliferation and induced U-87MG cell apoptosis, providing an experimental basis for the development of anti-glioma drugs and therapeutic methods targeting the TLR4 gene. The absence of clinical data (expression levels in tissue, association with patient prognosis) was a limitation of the present study. The molecular mechanisms underlying growth inhibition and cell apoptosis induction of U-87MG cells by silencing the TLR4 gene are complex, necessitating further study in the future.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Excellent Talent Project in Clinical Medicine funded by Hebei provincial government in 2017 [grant no. Jicaishe(2017)46].
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
YL performed the experiments and wrote the manuscript. YJ performed the experiments and statistical analysis. JL, YC and XH performed the experiments. LL designed the study, performed the experiments and revised the manuscript. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Davis ME. Epidemiology and overview of gliomas. Semin Oncol Nurs. 2018;34:420–429. doi: 10.1016/j.soncn.2018.10.001. [DOI] [PubMed] [Google Scholar]
- 2.Ostrom QT, Gittleman H, Stetson L, Virk SM, Barnholtz-Sloan JS. Epidemiology of gliomas. Cancer Treat Res. 2015;163:1–14. doi: 10.1007/978-3-319-12048-5_1. [DOI] [PubMed] [Google Scholar]
- 3.Ostrom QT, Gittleman H, Liao P, Rouse C, Chen Y, Dowling J, Wolinsky Y, Kruchko C, Barnholtz-Sloan J. CBTRUS statistical report: Primary brain and central nervous system tumors diagnosed in the United States in 2007–2011. Neuro Oncol. 2014;16(Suppl 4):iv1–iv63. doi: 10.1093/neuonc/nou327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rasmussen BK, Hansen S, Laursen RJ, Kosteljanetz M, Schultz H, Nørgård BM, Guldberg R, Gradel KO. Epidemiology of glioma: Clinical characteristics, symptoms, and predictors of glioma patients grade I–IV in the the Danish neuro-oncology registry. J Neurooncol. 2017;135:571–579. doi: 10.1007/s11060-017-2607-5. [DOI] [PubMed] [Google Scholar]
- 5.Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34:637–650. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 6.Kumar H, Kawai T, Akira S. Toll-like receptors and innate immunity. Biochem Biophys Res Commun. 2009;388:621–625. doi: 10.1016/j.bbrc.2009.08.062. [DOI] [PubMed] [Google Scholar]
- 7.Hua Z, Hou B. TLR signaling in B-cell development and activation. Cell Mol Immunol. 2013;10:103–106. doi: 10.1038/cmi.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawai T, Akira S. TLR signaling. Semin Immunol. 2007;19:24–32. doi: 10.1016/j.smim.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 9.Seydoux E, Liang H, Dubois Cauwelaert N, Archer M, Rintala ND, Kramer R, Carter D, Fox CB, Orr MT. Effective combination adjuvants engage both TLR and inflammasome pathways to promote potent adaptive immune responses. J Immunol. 2018;201:98–112. doi: 10.4049/jimmunol.1701604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tian S, Wang M, Liu C, Zhao H, Zhao B. Mulberry leaf reduces inflammation and insulin resistance in type 2 diabetic mice by TLRs and insulin signalling pathway. BMC Complement Altern Med. 2019;19:326. doi: 10.1186/s12906-019-2742-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dickinson SE, Wondrak GT. TLR4-directed molecular strategies targeting skin photodamage and carcinogenesis. Curr Med Chem. 2018;25:5487–5502. doi: 10.2174/0929867324666170828125328. [DOI] [PubMed] [Google Scholar]
- 12.Wang YH, Cao YW, Yang XC, Niu HT, Sun LJ, Wang XS, Liu J. Effect of TLR4 and B7-H1 on immune escape of urothelial bladder cancer and its clinical significance. Asian Pac J Cancer Prev. 2014;15:1321–1326. doi: 10.7314/APJCP.2014.15.3.1321. [DOI] [PubMed] [Google Scholar]
- 13.Monlish DA, Bhatt ST, Schuettpelz LG. The role of toll-like receptors in hematopoietic malignancies. Front Immunol. 2016;7:390. doi: 10.3389/fimmu.2016.00390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dajon M, Iribarren K, Cremer I. Toll-like receptor stimulation in cancer: A pro- and anti-tumor double-edged sword. Immunobiology. 2017;222:89–100. doi: 10.1016/j.imbio.2016.06.009. [DOI] [PubMed] [Google Scholar]
- 15.Makkar S, Riehl TE, Chen B, Yan Y, Alvarado DM, Ciorba MA, Stenson WF. Hyaluronic acid binding to TLR4 promotes proliferation and blocks apoptosis in colon cancer. Mol Cancer Ther. 2019;18:2446–2456. doi: 10.1158/1535-7163.MCT-18-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun NK, Huang SL, Chang TC, Chao CC. TLR4 and NFκB signaling is critical for taxol resistance in ovarian carcinoma cells. J Cell Physiol. 2018;233:2489–2501. doi: 10.1002/jcp.26125. [DOI] [PubMed] [Google Scholar]
- 17.Shetab Boushehri MA, Lamprecht A. TLR4-based immunotherapeutics in cancer: A review of the achievements and shortcomings. Mol Pharm. 2018;15:4777–4800. doi: 10.1021/acs.molpharmaceut.8b00691. [DOI] [PubMed] [Google Scholar]
- 18.Dapito DH, Mencin A, Gwak GY, Pradere JP, Jang MK, Mederacke I, Caviglia JM, Khiabanian H, Adeyemi A, Bataller R, et al. Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4. Cancer Cell. 2012;21:504–516. doi: 10.1016/j.ccr.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allen M, Bjerke M, Edlund H, Nelander S, Westermark B. Origin of the U87MG glioma cell line: Good news and bad news. Sci Transl Med. 2016;8:354re3. doi: 10.1126/scitranslmed.aaf6853. [DOI] [PubMed] [Google Scholar]
- 20.Zhou T, Li Y, Yang L, Liu L, Ju Y, Li C. Silencing of ANXA3 expression by RNA interference inhibits the proliferation and invasion of breast cancer cells. Oncol Rep. 2017;37:388–398. doi: 10.3892/or.2016.5251. [DOI] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Ostrom QT, Bauchet L, Davis FG, Deltour I, Fisher JL, Langer CE, Pekmezci M, Schwartzbaum JA, Turner MC, Walsh KM, et al. The epidemiology of glioma in adults: A ‘state of the science’ review. Neuro Oncol. 2014;16:896–913. doi: 10.1093/neuonc/nou087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lim M, Xia Y, Bettegowda C, Weller M. Current state of immunotherapy for glioblastoma. Nat Rev Clin Oncol. 2018;15:422–442. doi: 10.1038/s41571-018-0003-5. [DOI] [PubMed] [Google Scholar]
- 24.Satoh T, Akira S. Toll-like receptor signaling and its inducible proteins. Microbiol Spectr. 2016;4 doi: 10.1128/microbiolspec.MCHD-0040-2016. [DOI] [PubMed] [Google Scholar]
- 25.Steinhagen F, Kinjo T, Bode C, Klinman DM. TLR-based immune adjuvants. Vaccine. 2011;29:3341–3355. doi: 10.1016/j.vaccine.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar V. Toll-like receptors in the pathogenesis of neuroinflammation. J Neuroimmunol. 2019;332:16–30. doi: 10.1016/j.jneuroim.2019.03.012. [DOI] [PubMed] [Google Scholar]
- 27.Medzhitov R, Preston-Hurlburt P, Janeway CA., Jr A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature. 1997;388:394–397. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- 28.Vijay K. Toll-like receptors in immunity and inflammatory diseases: Past, present, and future. Int Immunopharmacol. 2018;59:391–412. doi: 10.1016/j.intimp.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mikulic J, Longet S, Favre L, Benyacoub J, Corthesy B. Secretory IgA in complex with Lactobacillus rhamnosus potentiates mucosal dendritic cell-mediated Treg cell differentiation via TLR regulatory proteins, RALDH2 and secretion of IL-10 and TGF-β. Cell Mol Immunol. 2017;14:546–556. doi: 10.1038/cmi.2015.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang AT, Choi JP, Kotzin JJ, Yang Y, Hong CC, Hobson N, Girard R, Zeineddine HA, Lightle R, Moore T, et al. Endothelial TLR4 and the microbiome drive cerebral cavernous malformations. Nature. 2017;545:305–310. doi: 10.1038/nature22075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yin Q, Jiang D, Li L, Yang Y, Wu P, Luo Y, Yang R, Li D. LPS promotes vascular smooth muscle cells proliferation through the TLR4/Rac1/Akt signalling pathway. Cell Physiol Biochem. 2017;44:2189–2200. doi: 10.1159/000486024. [DOI] [PubMed] [Google Scholar]
- 32.Fu HY, Li C, Yang W, Gai XD, Jia T, Lei YM, Li Y. FOXP3 and TLR4 protein expression are correlated in non-small cell lung cancer: Implications for tumor progression and escape. Acta Histochem. 2013;115:151–157. doi: 10.1016/j.acthis.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 33.Grimmig T, Matthes N, Hoeland K, Tripathi S, Chandraker A, Grimm M, Moench R, Moll EM, Friess H, Tsaur I, et al. TLR7 and TLR8 expression increases tumor cell proliferation and promotes chemoresistance in human pancreatic cancer. Int J Oncol. 2015;47:857–866. doi: 10.3892/ijo.2015.3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiao T, Wu S, Yan C, Zhao C, Jin H, Yan N, Xu J, Wu Y, Li C, Shao Q, Xia S. Butyrate upregulates the TLR4 expression and the phosphorylation of MAPKs and NK-κB in colon cancer cell in vitro. Oncol Lett. 2018;16:4439–4447. doi: 10.3892/ol.2018.9201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li C, Li H, Jiang K, Li J, Gai X. TLR4 signaling pathway in mouse Lewis lung cancer cells promotes the expression of TGF-β1 and IL-10 and tumor cells migration. Biomed Mater Eng. 2014;24:869–875. doi: 10.3233/BME-130879. [DOI] [PubMed] [Google Scholar]
- 36.Ren WH, Zhang LM, Liu HQ, Gao L, Chen C, Qiang C, Wang XL, Liu CY, Li SM, Huang C, et al. Protein overexpression of CIRP and TLR4 in oral squamous cell carcinoma: An immunohistochemical and clinical correlation analysis. Med Oncol. 2014;31:120. doi: 10.1007/s12032-014-0120-7. [DOI] [PubMed] [Google Scholar]
- 37.Pandey N, Chauhan A, Jain N. TLR4 polymorphisms and expression in solid cancers. Mol Diagn Ther. 2018;22:683–702. doi: 10.1007/s40291-018-0361-9. [DOI] [PubMed] [Google Scholar]
- 38.Chen CL, Tsukamoto H, Liu JC, Kashiwabara C, Feldman D, Sher L, Dooley S, French SW, Mishra L, Petrovic L, et al. Reciprocal regulation by TLR4 and TGF-β in tumor-initiating stem-like cells. J Clin Invest. 2013;123:2832–2849. doi: 10.1172/JCI65859. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39.Yao RR, Li JH, Zhang R, Chen RX, Wang YH. M2-polarized tumor-associated macrophages facilitated migration and epithelial-mesenchymal transition of HCC cells via the TLR4/STAT3 signaling pathway. World J Surg Oncol. 2018;16:9. doi: 10.1186/s12957-018-1312-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Szajnik M, Szczepanski MJ, Czystowska M, Elishaev E, Mandapathil M, Nowak-Markwitz E, Spaczynski M, Whiteside TL. TLR4 signaling induced by lipopolysaccharide or paclitaxel regulates tumor survival and chemoresistance in ovarian cancer. Oncogene. 2009;28:4353–4363. doi: 10.1038/onc.2009.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang H, Zhang S. The expression of Foxp3 and TLR4 in cervical cancer: Association with immune escape and clinical pathology. Arch Gynecol Obstet. 2017;295:705–712. doi: 10.1007/s00404-016-4277-5. [DOI] [PubMed] [Google Scholar]
- 42.Zhan Z, Xie X, Cao H, Zhou X, Zhang XD, Fan H, Liu Z. Autophagy facilitates TLR4- and TLR3-triggered migration and invasion of lung cancer cells through the promotion of TRAF6 ubiquitination. Autophagy. 2014;10:257–268. doi: 10.4161/auto.27162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li J, Yin J, Shen W, Gao R, Liu Y, Chen Y, Li X, Liu C, Xiang R, Luo N. TLR4 promotes breast cancer metastasis via Akt/GSK3β/β-catenin pathway upon LPS stimulation. Anat Rec (Hoboken) 2017;300:1219–1229. doi: 10.1002/ar.23590. [DOI] [PubMed] [Google Scholar]
- 44.Kim KH, Jo MS, Suh DS, Yoon MS, Shin DH, Lee JH, Choi KU. Expression and significance of the TLR4/MyD88 signaling pathway in ovarian epithelial cancers. World J Surg Oncol. 2012;10:193. doi: 10.1186/1477-7819-10-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou Q, Wang C, Wang X, Wu X, Zhu Z, Liu B, Su L. Association between TLR4 (+896A/G and +1196C/T) polymorphisms and gastric cancer risk: An updated meta-analysis. PLoS One. 2014;9:e109605. doi: 10.1371/journal.pone.0109605. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.