Abstract
Traditional clinicopathological indices are insufficient in predicting the prognosis of patients diagnosed with oral and oropharyngeal squamous cell carcinoma (OSCC/OPSCC). Notably, autophagy and long non-coding RNAs (lncRNAs) regulate the development and progression of various types of cancer. The present study aimed to assess the association between autophagy-related lncRNAs and the prognosis of patients diagnosed with OSCC/OPSCC. Gene sequencing and clinicopathological data of patients with OSCC/OPSCC were downloaded from The Cancer Genome Atlas database, while gene set functional classification was downloaded from the Gene Set Enrichment Analysis database. Out of the 413 transcriptome data samples and 402 clinicopathological data samples retrieved, a total of nine autophagy-related lncRNAs, including PTCSC2, AC099850.3, LINC01963, RTCA-AS1, AP002884.1, UBAC2-AS1, AL512274.1, MIR600HG and AL354733.3, were screened. This was geared towards establishing a signature through gene co-expression network, univariate and Least Absolute Shrinkage and Selection Operator Cox regression analyses. Based on this signature, the patients were subdivided into a high-risk group and a low-risk group. Kaplan-Meier survival analysis revealed that the overall survival of the high-risk group was significantly lower than that of the low-risk group. Furthermore, principal components analysis demonstrated that the patients diagnosed with OSCC/OPSCC could be distinguished into low-survival and high-survival groups according to the signature. Univariate and multivariate Cox regression analyses of clinicopathological data and the signature revealed that the signature could potentially be used as an independent prognostic factor for OSCC/OPSCC. In addition, reverse transcription-quantitative PCR analysis of clinical samples demonstrated the validity of the signature. In summary, the present study revealed that the signature based on autophagy-related lncRNAs potentially acts as an independent prognostic indicator for patients with OSCC/OPSCC. Furthermore, it promotes research on targeted diagnosis and treatment of patients diagnosed with OSCC/OPSCC.
Keywords: oral squamous cell carcinoma, oropharyngeal squamous cell carcinoma, autophagy, long non-coding RNAs, survival, prognosis, signature
Introduction
Head and neck squamous cell carcinoma (HNSCC) is the sixth most common malignant tumor worldwide (1). Notably, the main types of HNSCC are oral and oropharyngeal squamous cell carcinoma (OSCC/OPSCC), which severely affect the quality of life of patients and endanger their lives (2). Recent studies have demonstrated that the development of OSCC/OPSCC is closely associated with the changes of key genes (3–5). Therefore, it is necessary to identify novel gene-level biomarkers that predict the prognosis of OSCC/OPSCC.
Autophagy, a mechanism maintaining the stability of the internal environment and the balance of protein metabolism in cells, regulates the development and progression of various types of cancer (6). In the early stage, autophagy removes abnormal organelles and proteins in normal cells to prevent the generation of the stress response, protect the genes from being damaged and inhibit the occurrence of tumors; however, autophagy also maintains the survival of tumor cells in the late stage of tumor development under certain conditions, such as hypoxia or nutrient deficiency, thereby sustaining proliferation and progression of tumor cells (7). By investigating autophagy-related molecules, the present study aimed to identify novel biomarkers for prognostic prediction of OSCC/OPSCC.
Abnormal expression of long non-coding RNAs (lncRNAs) regulates the proliferation, apoptosis and migration of tumor cells, implying that lncRNAs can potentially be used as important biomarkers and therapeutic targets (8). Nonetheless, lncRNAs in OSCC/OPSCC represent an important subject which has not been sufficiently investigated. In the present study, The Cancer Genome Atlas (TCGA) database was used to evaluate the potential value of autophagy-related lncRNAs as a prognostic tool for patients with OSCC/OPSCC. Through TCGA data analysis, nine autophagy-related lncRNAs associated with the overall survival (OS) of patients with OSCC/OPSCC were identified, and their independent associations were further verified. Subsequently, the nine lncRNAs were integrated into an independent signature, and it was revealed that their predictive performance for the prognosis of patients with OSCC/OPSCC was higher than that of clinicopathological features (age, sex, grade, T stage, N stage and TNM comprehensive stage). This demonstrated the potential of autophagy-related lncRNAs as biomarkers to predict the prognosis of patients with OSCC/OPSCC.
Materials and methods
Acquisition of autophagy-related lncRNAs and clinicopathological data of patients with OSCC/OPSCC
The level 3 RNA-seq and clinicopathological data (project ID: TCGA-HNSC) of patients diagnosed with OSCC/OPSCC were extracted from TCGA (https://portal.gdc.cancer.gov/), while the autophagy gene set (accession number: M10281) was obtained from the Molecular Signatures Database v7.0 (https://www.gsea-msigdb.org/gsea/msigdb/index.jsp). Subsequently, the function of lncRNAs was explored based on the hypothesis that co-expressed genes are more likely to be functionally related (9). Spearman correlation analysis was carried out on the expression of lncRNAs and autophagy genes through the ‘limma’ (version 3.11; http://bioconductor.org/packages/limma/) package of R software (version 3.6.1; R foundation) and lncRNAs with high correlation (|correlation coefficient (cor)|>0.4; P<0.001) with autophagy genes were identified as autophagy-related lncRNAs.
Identification of a prognostic multi-lncRNA signature
First, univariate Cox regression analysis was applied to explore lncRNAs associated with the prognosis of patients (P<0.01). Thereafter, Least Absolute Shrinkage and Selection Operator (LASSO) Cox regression analysis was used to optimize the prognostic multi-lncRNA signature (10). Afterwards, the present study substituted the expression of lncRNAs in patients with OSCC/OSPCC into the Cox model, and using the LASSO Cox regression coefficient, the risk score of each patient was calculated. Subsequently, based on the median risk score, the patients were subdivided into a high-risk group and a low-risk group.
The Kaplan-Meier method and log-rank test were performed to evaluate differences in OS between patients in the high- and low-risk groups. The risk score distribution, survival status of patients with OSCC/OPSCC and expression profiles of prognostic lncRNAs were visualized through images. R software (version 3.6.1; R foundation) was used for statistical calculations and data plotting.
Independence of multi-lncRNA signature in predicting prognosis of patients with OSCC/OPSCC
The independent associations of the multi-lncRNA signature-based risk score and clinicopathological factors (age, sex, TNM stage and pathological stage) with the prognosis of patients with OSCC/OPSCC were determined through univariate and multivariate Cox regression analysis using R software. Furthermore, a receiver operating characteristic (ROC) curve was used to evaluate the accuracy of these factors in predicting the prognosis of OSCC/OPSCC.
Kruskal-Wallis with Dunn's post hoc test was used to further explore the relationship between the expression levels of the nine autophagy-related lncRNAs and the clinicopathological characteristics of patients with OSCC/OPSCC.
Principal components analysis (PCA) and gene set enrichment analysis (GSEA)
Furthermore, PCA was used to test the differentiation of patients in low- and high-risk groups. GSEA (https://www.gsea-msigdb.org/gsea/index.jsp) was used to investigate the functions of autophagy-related genes in low- and high-risk groups. Statistical calculations and data plotting were done using R software (version 3.6.1; R foundation).
Validation of the clinical OSCC/OPSCC specimens by reverse transcription-quantitative PCR (RT-qPCR)
To further validate the findings from TCGA, RT-qPCR was performed to detect the expression levels of the nine autophagy-related lncRNAs in OSCC/OPSCC samples (n=55). All patients (30 men, 25 women; median age, 46 years; age range, 35–68 years) received lesion excision between January 2015 and October 2018 at the First Affiliated Hospital of Nanchang University (Nanchang, China) where they were pathologically diagnosed with OSCC/OPSCC. Patients with distant metastasis, multiple primary cancers or non-first surgery were excluded from the present study. The collection of the tumor samples from patients with OSCC/OPSCC conformed to the Declaration of Helsinki and current legislation. In addition, clinicopathological information of patients was captured by an investigator through interviews and medical records. The present study was approved by the Ethics Committee of First Affiliated Hospital of Nanchang University (approval no. 2019B0017), and all patients provided written informed consent.
Total RNA was isolated from the tissue samples using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The GoScript Reverse Transcription kit (cat. no. PRA5000; Promega Corporation) and GoTaq® qPCR Master Mix of Power SYBR® Green (Promega Corporation) was used to synthesize cDNA and for RT-qPCR detection. RT-qPCR was performed using the GeneAmp® PCR System 9700 (Applied Biosystems; Thermo Fisher Scientific, Inc.). Additionally, relative fold change results were calculated using the 2−ΔΔCq method (11). (The thermocycling conditions were as follows: 95°C for 10 sec, followed by 40 cycles of 95°C for 5 sec and 60°C for 30 sec. RNA expression were normalized to GAPDH. The following primer sequences were used: PTCSC2 forward 5′-CCCTAAGCCCACCGACTTTT-3′ and reverse, 5′-GGGTGCACTGGGTTTAGACA-3′; AC099850.3 forward 5′-CGTCTTTCACCCAGCCTCTT-3′ and reverse, 5′-AAAGCAGGAACCCCTCTGTG-3′; LINC01963 forward 5′-CCCGGTGTAGGGTAAATGCA-3′ and reverse, 5′-ATTGGCCACTCCCGGATTTT-3′; RTCA-AS1 forward 5′-CCGAGGTGCCGACTTTTAGA-3′ and reverse, 5′-CCACCCAGGTTCACATCTCA-3′; AP002884.1 forward 5′-TACGCTAAACTGCCTGGCAA-3′ and reverse, 5′-GTTGTGCAGCAGAGTTGTGG-3′; UBAC2-AS1 forward 5′-TGAAACGATGGCGGTCAGAA-3′ and reverse, 5-TCAGGTCCTCAGGATGCAGA-3′; AL512274.1 forward 5′-AACACAGTGAGCGAGTCAGG-3′ and reverse, 5′-CAACCTCAGAGCAGAACCGT-3′; MIR600HG forward 5′-GCCAGTCTGACGTGAACAGA-3′ and reverse, 5′-GCGCTTCTATCAGGCCATCT-3′; AL354733.3 forward 5′-TCCCCAGGCCTCAAAAATCC-3′ and reverse, 5′-TCTGCTGTCGACTTTCGCTT-3′; GAPDH forward 5′-GGAAGCTTGTCATCAATGGAAATC-3′ and reverse, 5′-TGATGACCCTTTTGGCTCCC-3′.
Statistical analysis
Statistical differences were calculated using R software (version 3.6.1; R foundation) and SPSS Statistics software (version 20; IBM Corp.). All experiments were performed in triplicate. For non-parametric analysis, Kruskal-Wallis with Dunn's post hoc test was used. Survival curves were plotted using the Kaplan-Meier method, and differences were analyzed via the log-rank test. The regression analysis of univariate and multivariate Cox proportional hazards analysis was completed using the ‘survival’ package (version 2.36-10, http://cran.r-project.org/web/packages/survival) of R software. Results are presented in the form of hazard ratio and corresponding 95% confidence interval (CI). P<0.05 was considered to indicate a statistically significant difference.
Results
Autophagy-related lncRNAs and clinicopathological data in patients with OSCC/OPSCC
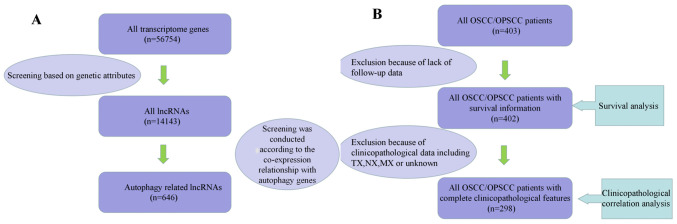
A total of 646 autophagy-related lncRNAs, 402 patients with survival information and 298 patients with complete clinicopathological information were identified (Fig. 1).
Figure 1.
Process of data selection. Screening process of (A) lncRNA data and (B) clinicopathological data. lncRNA, long non-coding RNA; OSCC/OPSCC, oral and oropharyngeal squamous cell carcinoma.
Identification of a prognostic autophagy-related lncRNA signature in patients with OSCC/OPSCC
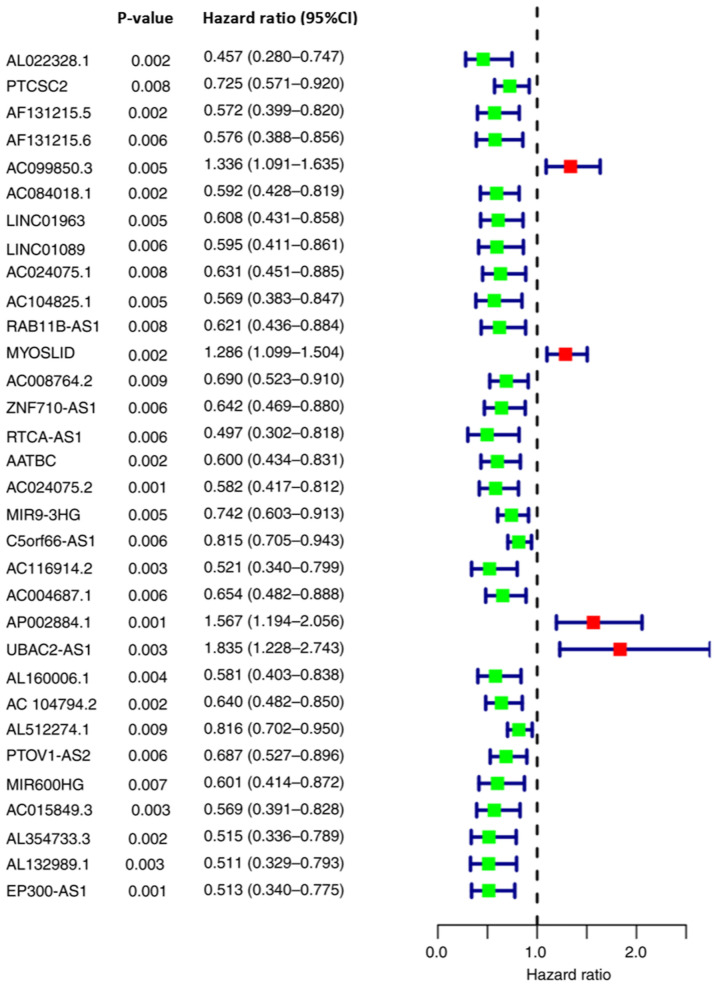
Using univariate Cox regression analysis, 32 autophagy-related lncRNAs associated with the OS of patients with OSCC/OPSCC were identified (Fig. 2).
Figure 2.
Autophagy-related long non-coding RNAs affecting overall survival of patients. 95% CI, 95% confidence interval.
The 32 autophagy-related lncRNAs were further screened using LASSO Cox regression analysis. Among these, nine lncRNAs, including PTCSC2, AC099850.3, LINC01963, RTCA-AS1, AP002884.1, UBAC2-AS1, AL512274.1, MIR600HG and AL354733.3, were incorporated into the Cox model. Subsequently, the expression levels of the aforementioned nine lncRNAs and the LASSO Cox regression coefficients were integrated to establish the signature (Table I). Based on the signature, the risk scores of patients with OSCC/OPSCC (n=371; among the 402 patients, 31 patients with no detectable target lncRNA expression were excluded) were calculated and subdivided into a high-risk group (n=185) and a low-risk group (n=186) according to the median value (1.057).
Table I.
Least Absolute Shrinkage and Selection Operator Cox proportional hazard model of the nine autophagy-related lncRNAs.
| lncRNA ID | Coefficient | Hazard ratio | 95% Confidence interval | P-value |
|---|---|---|---|---|
| PTCSC2 | −0.310 | 0.733 | 0.557–0.966 | 0.027 |
| AC099850.3 | 0.270 | 1.309 | 1.064–1.611 | 0.011 |
| LINC01963 | −0.483 | 0.617 | 0.421–0.904 | 0.013 |
| RTCA-AS1 | −0.526 | 0.591 | 0.341–0.958 | 0.021 |
| AP002884.1 | 0.392 | 1.480 | 1.089–2.010 | 0.012 |
| UBAC2-AS1 | 0.387 | 1.473 | 1.035–2.312 | 0.022 |
| AL512274.1 | −0.178 | 0.837 | 0.710–0.986 | 0.034 |
| MIR600HG | 0.3798 | 1.462 | 1.057–2.334 | 0.030 |
| AL354733.3 | −0.777 | 0.460 | 0.277–0.762 | 0.002 |
lncRNA, long non-coding RNA.
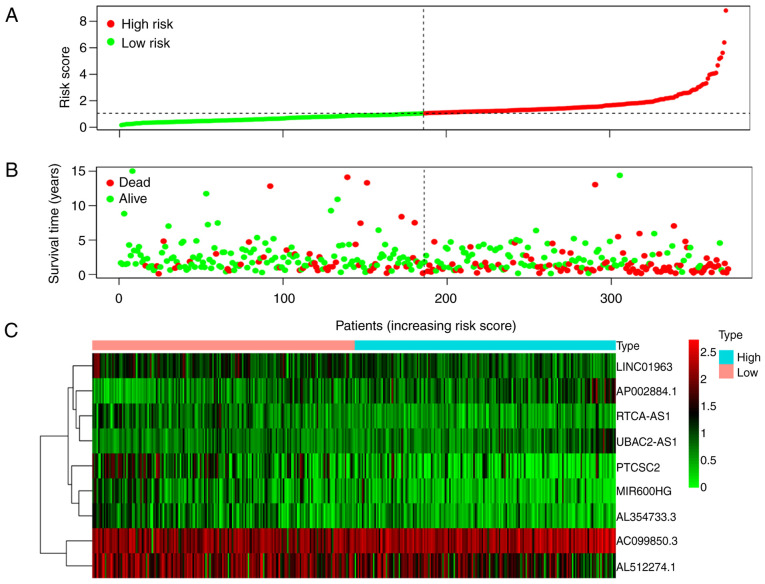
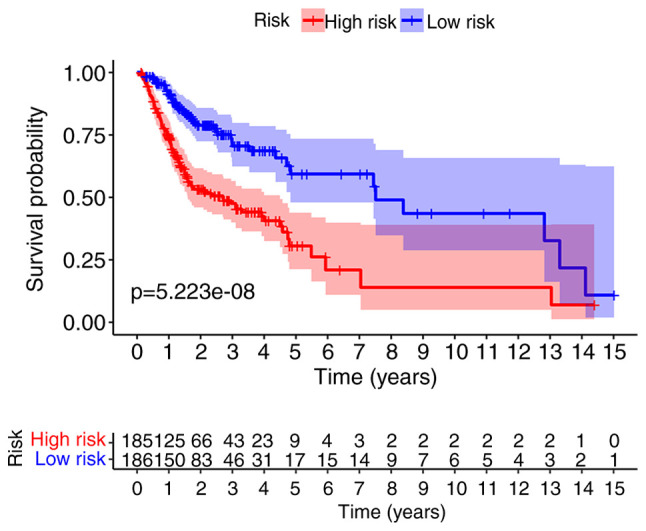
The Kaplan-Meier curve demonstrated that the OS of the high-risk group was significantly poorer than that of low-risk patients (P=5.223×10−8; Fig. 3). The risk score distribution, survival status of patients with OSCC/OPSCC, and expression profiles of the nine prognostic lncRNAs are shown in Fig. 4. It was revealed that the rate of mortality among patients with high-risk scores was higher compared with that in patients with low-risk scores. The expression levels of AC099850.3, AP002884.1 and UBAC2-AS1 were higher in high-risk patients compared with in patients with low-risk. The expression levels of AL512274.1, PTCSC2, LINC01963, RTCA-AS1, AL354733.3 and MIR600HG were lower in high-risk patients compared with in patients with low-risk.
Figure 3.
Kaplan-Meier plot of overall survival according to the risk score.
Figure 4.
Prognostic value of the nine-lncRNA signature in patients with oral and oropharyngeal squamous cell carcinoma. (A) Risk distribution map of patients arranged according to the risk score, and divided into low-risk and high-risk groups according to the median risk score. (B) Distribution of survival status revealing that patients in the high-risk group had higher mortality and a shorter survival time than those in the low-risk group. (C) Heat map of lncRNA expression revealing that the expression levels of AC099850.3, AP002884.1, UBAC2-AS1 and MIR600HG were higher in high-risk patients compared with in patients with low-risk, while the expression levels of AL512274.1, PTCSC2, LINC01963, RTCA-AS1 and AL354733.3 were lower in the high-risk patients compared with in patients with low-risk. lncRNA, long non-coding RNA.
In order to determine the role of the nine lncRNAs, we extracted and displayed the correlation analysis results between these nine lncRNAs and autophagy-related genes (Fig. S1; Table SI). All of the nine lncRNAs were positively correlated with autophagy genes. PTCSC2 exhibited the highest correlation coefficient (cor=0.614), while MIR600HG had the most co-expression genes (n=20).
Association between the nine-lncRNA signature and prognosis in patients with OSCC/OPSCC
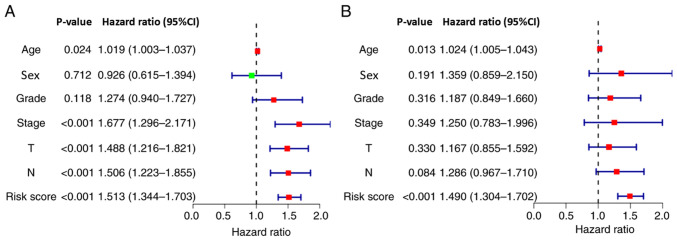
The results of univariate Cox regression analysis demonstrated that the nine-lncRNA signature-based risk score, age, T stage, N stage and TNM comprehensive stage were significantly associated with patient survival (Fig. 5A). Furthermore, multivariate Cox regression analysis revealed that the signature-based risk score and age were independent factors associated with the OS of patients with OSCC/OPSCC (Fig. 5B). Due to the absence of M1 in the case data, the M stage was not included in the analysis.
Figure 5.
Independent testing of the association between the nine-long non-coding RNA signature and prognosis of patients. (A) Univariate Cox regression analysis demonstrated that the signature-based risk score (P<0.001), age (P=0.024), T stage (P<0.001), N stage (P<0.001) and TNM comprehensive stage (P<0.001) were significantly associated with patient survival. (B) Multivariate Cox regression analysis revealed that the signature-based risk score (P<0.001) and age (P=0.013) were independent factors associated with patient survival.
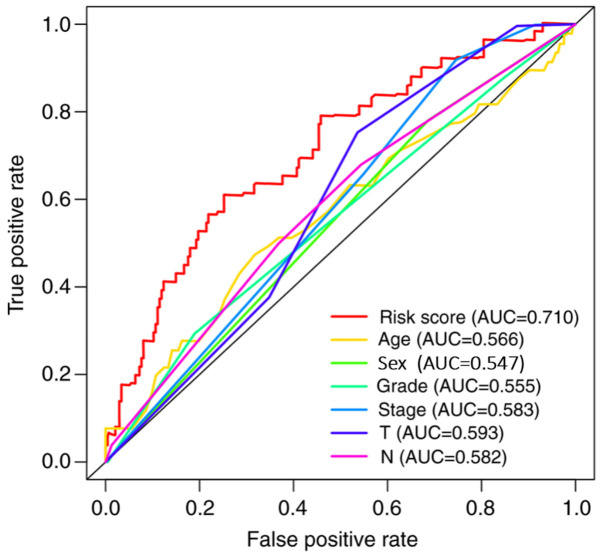
The prediction capability of the signature was further analyzed using ROC curves, and the area under the curve value was 0.710, which was higher than that of the other clinicopathological characteristics (Fig. 6).
Figure 6.
Receiver operating characteristic curve analysis of the nine-long non-coding RNA signature and other clinicopathological characteristics for predicting the overall survival of patients. Compared with other clinicopathological characteristics, the signature-based risk score had a higher AUC value. AUC, area under the curve.
Relationship between nine autophagy-related lncRNAs and clinicopathological factors in patients with OSCC/OPSCC
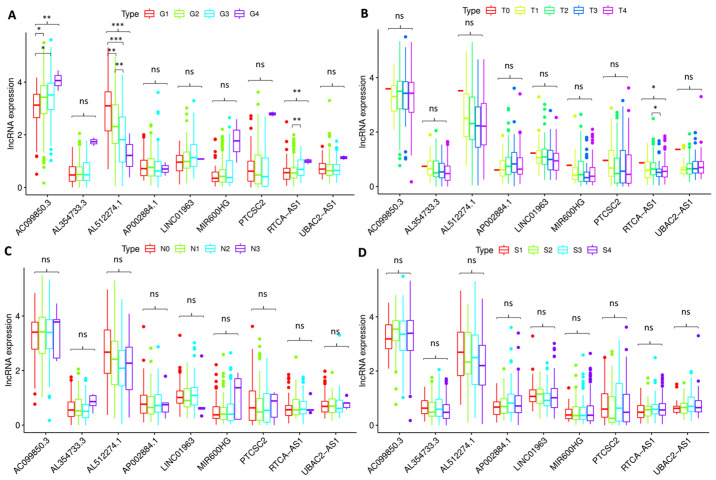
A Kruskal-Wallis test was used to investigate the association between the expression levels of the nine autophagy-related lncRNAs and the clinicopathological characteristics of the patients (Fig. 7). The results demonstrated that AC099850.3 (P<0.01), AL512274.1 (P<0.001) and RTCA-AS1 (P<0.01) were significantly associated with the pathological grade (Fig. 7A), and RTCA-AS1 was significantly associated with T stage (P<0.05; Fig. 7B). However, none of the lncRNAs were significantly associated with the clinical stage or N stage of the patients (Fig. 7C and D).
Figure 7.
Association between the expression levels of the nine lncRNAs and clinicopathological characteristics of the patients. The expression levels of (A) AC099850.3 (P<0.01), AL512274.1 (P<0.001) and RTCA-AS1 (P<0.01) were significantly associated with the pathological grade. (B) RTCA-AS1 expression (P<0.05) was significantly associated with T stage. None of the lncRNAs were significantly associated with the (C) N stage and (D) clinical stage of the patients. *P<0.05, **P<0.01 and ***P<0.001. lncRNA, long non-coding RNA; ns, not significant; G1, well differentiated; G2, moderately differentiated; G3, poorly differentiated; G4, undifferentiated; T, T stage; N, N stage; S, TNM comprehensive stage; S1, stage I; S2, stage II; S3, stage III; S4, stage IV.
Autophagy status of low- and high-risk groups
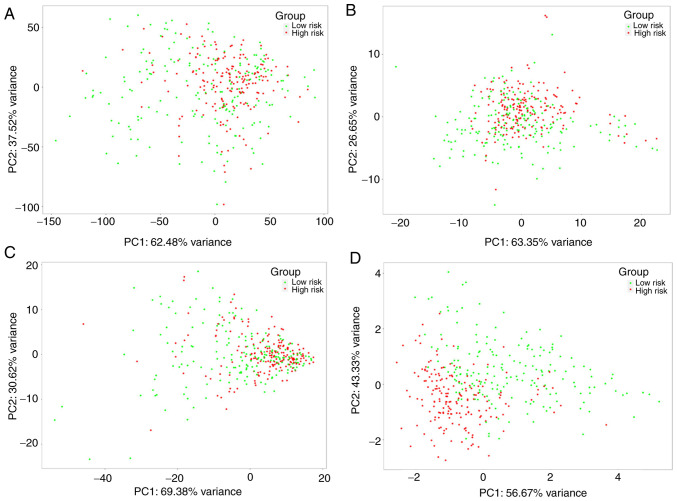
PCA was conducted to investigate differences in the expression of the nine autophagy-related lncRNAs, autophagy-related lncRNAs, autophagy-related genes and all genes between low- and high-risk groups. The patients in the two groups were mixed when the expression of all the genes was used as the spatial indicator. The patients in the two groups were separated gradually when the index was optimized gradually from all genes to autophagy-related genes, autophagy-related lncRNAs and the nine autophagy-related lncRNAs (Fig. 8), suggesting that the signature established in the present study exhibited an improved discriminative power for prognostic prediction of patients with OSCC/OPSCC compared with other gene indices.
Figure 8.
Principal components analysis between low- and high-risk groups. The patients in the two groups were separated gradually when the index was optimized gradually from (A) all genes to (B) autophagy-related genes, (C) autophagy-related lncRNAs and (D) the nine autophagy-related lncRNAs. lncRNA, long non-coding RNA.
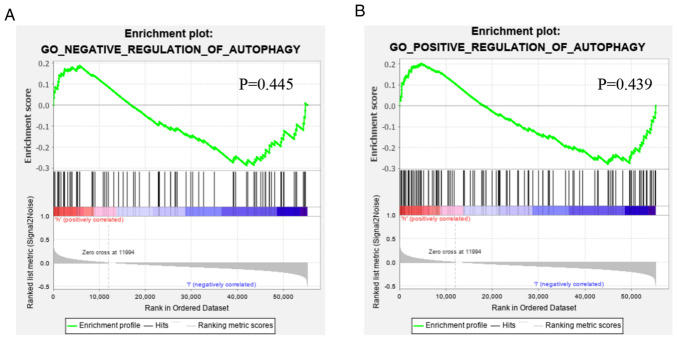
Functional annotation was further performed by GSEA, and the results demonstrated that the difference in gene function between low- and high-risk patients were not significantly enriched in the positive or negative regulation gene sets of autophagy (Fig. 9), indicating that, based on the overall increase or decrease of autophagy, determining the prognosis of the patients was not tenable.
Figure 9.
Gene Set Enrichment Analysis of the difference of the autophagy status between patients in the low- and high-risk groups. The difference in gene function between low- and high-risk patients were not significantly enriched in the (A) negative or (B) positive regulation gene sets of autophagy. GO, Gene Ontology.
RT-qPCR validation
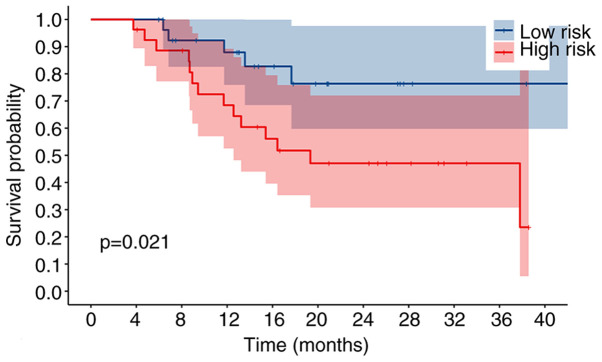
To assess the validity and reliability of the bioinformatics results, the expression levels of the nine autophagy-related lncRNAs were detected by RT-qPCR in 55 patients with OSCC/OPSCC. Based on the results of RT-qPCR and risk scores of the signature, the patients were divided into a high-risk group and a low-risk group. Kaplan-Meier analysis revealed that the OS of the high-risk group was significantly lower than that of the low-risk group (P=0.021; Fig. 10), and univariate and multivariate Cox regression analysis demonstrated the potential of the signature as an independent prognostic factor for patients with OSCC/OPSCC (Fig. 11), confirming the credibility of the signature.
Figure 10.
Kaplan-Meier plot of overall survival based on reverse transcription-quantitative PCR analysis of patients. The overall survival of the high-risk group was significantly lower than that of the low-risk group (P=0.021).
Figure 11.
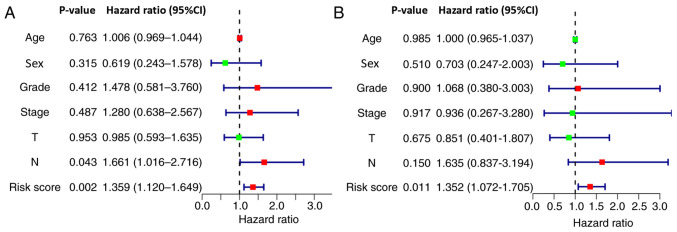
Independent assessment of the association between the nine-long non-coding RNA signature and prognosis of patients at the First Affiliated Hospital of Nanchang University. (A) Univariate Cox regression analysis demonstrated that the signature-based risk score (P=0.002) and N stage (P=0.043) were significantly associated with patient survival. (B) Multivariate Cox regression analysis revealed that the signature-based risk score (P=0.011) was an independent factor associated with patient survival.
Discussion
Autophagy is an important material catabolism process in cells, is caused by hypoxia, peroxidation, drug and DNA damage, and regulates cell self-renewal and homeostasis. According to previous studies, autophagy can be a double-edged sword in tumor development due to its ability to kill tumor cells during tumor development (12). However, it also protects tumor cells from being damaged (13).
At present, more than 40 autophagy-related genes have been identified in yeast and mammals (14). Being highly conserved between species, these genes regulate the occurrence and degree of intracellular autophagy via complex regulatory networks, and they are also implicated in the pathophysiology of diseases (15).
Targeted regulation of the autophagy level has become a subject of research focus in the treatment of cancer and other diseases (16). In the complex regulatory network of intracellular genes, regulating the expression of proteins associated with autophagy is a basic method to regulate autophagy (17).
Previous studies have asserted that lncRNAs, such as lncRNA-HOTAIR (18–21) and lncRNA-MALAT1 (22–25), serve a pivotal role in regulating the expression of autophagy-related proteins through positive or negative effects on mRNAs. Therefore, understanding and utilizing the role of lncRNAs in the regulation of autophagy will advance the clinical diagnosis and treatment of tumors.
With the advent of high throughput RNA sequencing and advanced computer technology, lncRNAs can continuously be investigated. Nevertheless, due to wide expression, complexity of the relationship and functional diversity, research on the functions of most lncRNAs has not matured (26). Bioinformatics analysis is an effective method for exploring the functions of lncRNAs, and provides a basis for subsequent experimental studies (27,28).
Using Cox regression analysis and the Kaplan-Meier method, the present study identified nine autophagy-related lncRNAs which significantly influenced the prognosis of patients with OSCC/OPSCC. Based on the aforementioned findings, a nine-lncRNA signature with satisfactory performance was developed. As a result, the present study further classified the patients with OSCC/OPSCC into a high-risk group and a low-risk group with significantly different prognosis.
By reviewing related literature and the genes exhibiting co-expression relationships with IncRNAs, the functions of the nine lncRNAs in the signature were explored.
Zhou et al (29) revealed that high expression levels of AC099850.3 were closely associated with decreased survival of patients diagnosed with tongue cancer. The present study revealed that AC099850 exhibited the greatest positive co-expression correlation with eukaryotic translation initiation factor 2 alpha kinase 4 (EIF2AK4). Notably, EIF2AK4 belongs to the family of protein kinases that phosphorylates the subunit of eukaryotic translation initiation factor 2 (EIF2) in response to various stress stimuli (30). To activate autophagy-related gene expression in response to cellular stress, activation of the EIF2AK4-EIF2A-activating transcription factor 4 signaling pathway is essential (31).
Zhu et al (32) reported that UBAC2-AS1 was a potential therapeutic target and a prognostic biomarker of clear cell kidney carcinoma. Furthermore, Chen et al (33) suggested that UBAC2-AS1 might be implicated in adipogenesis by acting as a competing endogenous RNA or being co-expressed with its targets. The present study demonstrated that UBAC2-AS1 exhibited the greatest positive co-expression correlation with a suppressor of Ty homolog-5, which has been demonstrated to be a novel tumor-specific human telomerase reverse transcriptase promoter-binding protein in colon cancer cells (34).
According to Song et al (35), MIR600HG is a potential prognostic biomarker in predicting the survival of patients diagnosed with pancreatic ductal adenocarcinoma. The present study demonstrated a positive correlation in the co-expression of MIR600HG and ubiquitin-specific protease 30 (USP30). Furthermore, USP30 potentially reverses depolarization-induced PTEN induced kinase 1-parkin RBR E3 ubiquitin protein ligase-dependent mitophagy and has rapidly emerged as a potential therapeutic target in Parkinson's disease (36).
It was also identified that the PTCSC2, LINC01963, AP002884.1, RTCA-AS1, AL512274.1 and AL354733.3 genes have not been studied in detail. Additionally, a significant positive co-expression relationship was identified between PTCSC2 and Xeroderma pigmentosum group A (XPA). Notably, as a key subunit implicated in the nucleotide excision repair (NER) system, XPA protein is a central organizer in the NER signaling pathway which identifies DNA damage and recruits other NER proteins to DNA lesions (37). A study by Ge et al (38) demonstrated that XPA potentially promotes cell-protective autophagy in a DNA repair-independent manner by enhancing the activation of poly(ADP-ribose) polymerase 1 (PARP1) in melanoma cells resistant to cisplatin.
It was revealed that LINC01963 harbored the greatest positive co-expression correlation with tuberous sclerosis 2 (TSC2), while AL354733.3 had the greatest positive co-expression correlation with tuberous sclerosis 1 (TSC1). By suppressing mTOR signaling, studies have revealed that TSC1 and TSC2 potentially induce autophagy (39,40).
AP002884.1 and fasciculation as well as elongation zeta/zygin1 (FEZ1) had a significant positive co-expression relationship. FEZ1 potentially acts as an adaptor of cargo transport and may be a scaffold protein; the complexes of FEZ1 formed with unc-51 like autophagy activating kinase 1, short coiled-coil protein, RAB3 GTPase activating protein catalytic subunit 1 or RAB3 GTPase activating non-catalytic protein subunit 2 have been demonstrated to be associated with autophagy (41).
RTCA-AS1 was markedly positively correlated with chromatin-modifying protein 4 (CHMP4)A. Notably, CHMP4A, CHMP4C and CHMP2B belong to the family of chromatin-modifying protein/charged multivesicular body protein. They are components of the endosomal sorting complex required for transport III involved in the formation of endocytic multivesicular bodies (42). CHMP4A expression is associated with the recurrence of ovarian cancer (43), whereas CHMP4C regulates radiation resistance in non-small cell lung cancer (44).
AL512274.1 was significantly positively co-expressed with mitogen-activated protein kinase 3 (MAPK3). Previous studies have identified MAPK3 to be specifically implicated in the control of cell proliferation, differentiation and autophagy (45,46).
To the best of our knowledge, except for AC099850.3, the other eight lncRNAs have not been reported in previous studies on OSCC/OPSCC, implying that these lncRNAs represent a potential target for the treatment of OSCC/OPSCC. In addition, their biological roles in the autophagy of OSCC/OPSCC will be a focus in future studies.
The present investigation had some limitations. For instance, the present study was based on profiles of high-throughput RNA-sequencing and data analysis, and therefore, lacked validation in a large clinical sample. A multi-center, large sample, longitudinal study with diverse clinical, radiographic and histopathologic factors is required to further demonstrate the reliability of the present study. Furthermore, the roles of the nine autophagy-related lncRNAs deserve further in vitro and in vivo investigation.
In conclusion, based on nine autophagy-related lncRNAs, a signature that demonstrated the capability to predict the prognosis of patients diagnosed with OSCC/OPSCC was developed. Using this signature, patients with a higher risk of mortality can be predicted, and therefore more priority in treatment should be given to these patients.
Supplementary Material
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science Foundation of China (grant no. 81860477), Jiangxi Provincial Key R&D Plan (grant no. 20181ACG70009) and Special Fund for Postgraduate Innovation in Jiangxi Province (grant no. YC2020-B025).
Availability of data and materials
The datasets used can be obtained from TCGA (https://portal.gdc.cancer.gov). The RT-qPCR data generated in the present study is available from the corresponding author upon reasonable request.
Authors' contributions
QJ and JQ conceived and designed the present study. QJ performed the bioinformatics analysis. DX analyzed the data. QJ and DX drafted the initial manuscript. QJ and FS performed tissue tests and followed up patients. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
For the use of human samples, the present study was approved by the Institutional Ethics Committee of First Affiliated Hospital of Nanchang University (Nanchang, China; approval no. 2019B0017), and written informed consent was provided by all patients prior to the study start.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Chaturvedi AK, Anderson WF, Lortet-Tieulent J, Curado MP, Ferlay J, Franceschi S, Rosenberg PS, Bray F, Gillison ML. Worldwide trends in incidence rates for oral cavity and oropharyngeal cancers. J Clin Oncol. 2013;31:4550–4559. doi: 10.1200/JCO.2013.50.3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Falzone L, Lupo G, La Rosa GRM, Crimi S, Anfuso CD, Salemi R, Rapisarda E, Libra M, Candido S. Identification of novel MicroRNAs and their diagnostic and prognostic significance in oral cancer. Cancers (Basel) 2019;11:610. doi: 10.3390/cancers11050610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakagaki T, Tamura M, Kobashi K, Koyama R, Fukushima H, Ohashi T, Idogawa M, Ogi K, Hiratsuka H, Tokino T, Sasaki Y. Profiling cancer-related gene mutations in oral squamous cell carcinoma from Japanese patients by targeted amplicon sequencing. Oncotarget. 2017;8:59113–59122. doi: 10.18632/oncotarget.19262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bavle RM, Venugopal R, Konda P, Muniswamappa S, Makarla S. Molecular classification of oral squamous cell carcinoma. J Clin Diagn Res. 2016;10:ZE18–ZE21. doi: 10.7860/JCDR/2016/19967.8565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu WK, Coffelt SB, Cho CH, Wang XJ, Lee CW, Chan FK, Yu J, Sung JJ. The autophagic paradox in cancer therapy. Oncogene. 2012;31:939–953. doi: 10.1038/onc.2011.295. [DOI] [PubMed] [Google Scholar]
- 7.Kimmelman AC. The dynamic nature of autophagy in cancer. Genes Dev. 2011;25:1999–2010. doi: 10.1101/gad.17558811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu T, Yu C, Ni H, Liang W, Yan H, Jin W. Expression of the long non-coding RNA H19 and MALAT-1 in growth hormone-secreting pituitary adenomas and its relationship to tumor behavior. Int J Dev Neurosci. 2018;67:46–50. doi: 10.1016/j.ijdevneu.2018.03.009. [DOI] [PubMed] [Google Scholar]
- 9.Stuart JM, Segal E, Koller D, Kim SK. A gene-coexpression network for global discovery of conserved genetic modules. Science. 2003;302:249–255. doi: 10.1126/science.1087447. [DOI] [PubMed] [Google Scholar]
- 10.Tibshirani R. Regression shrinkage and selection via the lasso. J R Statist Soc B. 1996;58:267–288. [Google Scholar]
- 11.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 12.Nassour J, Radford R, Correia A, Fuste JM, Schoell B, Jauch A, Shaw RJ, Karlseder J. Autophagic cell death restricts chromosomal instability during replicative crisis. Nature. 2019;565:659–663. doi: 10.1038/s41586-019-0885-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poillet-Perez L, Xie X, Zhan L, Yang Y, Sharp DW, Hu ZS, Su X, Maganti A, Jiang C, Lu W, et al. Autophagy maintains tumour growth through circulating arginine. Nature. 2018;563:569–573. doi: 10.1038/s41586-018-0697-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 15.Shibutani ST, Yoshimori T. A current perspective of autophagosome biogenesis. Cell Res. 2014;24:58–68. doi: 10.1038/cr.2013.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levy JMM, Towers CG, Thorburn A. Targeting autophagy in cancer. Nat Rev Cancer. 2017;17:528–542. doi: 10.1038/nrc.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chandra V, Bhagyaraj E, Parkesh R, Gupta P. Transcription factors and cognate signalling cascades in the regulation of autophagy. Biol Rev Camb Philos Soc. 2016;91:429–451. doi: 10.1111/brv.12177. [DOI] [PubMed] [Google Scholar]
- 18.Bhan A, Hussain I, Ansari KI, Kasiri S, Bashyal A, Mandal SS. Antisense transcript long noncoding RNA (lncRNA) HOTAIR is transcriptionally induced by estradiol. J Mol Biol. 2013;425:3707–3722. doi: 10.1016/j.jmb.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang L, Zhang X, Li H, Liu J. The long noncoding RNA HOTAIR activates autophagy by upregulating ATG3 and ATG7 in hepatocellular carcinoma. Mol Biosyst. 2016;12:2605–2612. doi: 10.1039/C6MB00114A. [DOI] [PubMed] [Google Scholar]
- 20.Yang Y, Jiang C, Yang Y, Guo L, Huang J, Liu X, Wu C, Zou J. Silencing of lncRNA-HOTAIR decreases drug resistance of non-small cell lung cancer cells by inactivating autophagy via suppressing the phosphorylation of ULK1. Biochem Biophys Res Commun. 2018;497:1003–1010. doi: 10.1016/j.bbrc.2018.02.141. [DOI] [PubMed] [Google Scholar]
- 21.Bao X, Ren T, Huang Y, Sun K, Wang S, Liu K, Zheng B, Guo W. Knockdown of long non-coding RNA HOTAIR increases miR-454-3p by targeting Stat3 and Atg12 to inhibit chondrosarcoma growth. Cell Death Dis. 2017;8:e2605. doi: 10.1038/cddis.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.YiRen H, YingCong Y, Sunwu Y, Keqin L, Xiaochun T, Senrui C, Ende C, XiZhou L, Yanfan C. Long noncoding RNA MALAT1 regulates autophagy associated chemoresistance via miR-23b-3p sequestration in gastric cancer. Mol Cancer. 2017;16:174. doi: 10.1186/s12943-017-0743-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo D, Ma J, Yan L, Li T, Li Z, Han X, Shui S. Down-Regulation of Lncrna MALAT1 Attenuates neuronal cell death through suppressing Beclin1-Dependent autophagy by regulating Mir-30a in cerebral ischemic stroke. Cell Physiol Biochem. 2017;43:182–194. doi: 10.1159/000480337. [DOI] [PubMed] [Google Scholar]
- 24.Guo X, Wu X, Han Y, Tian E, Cheng J. lncRNA MALAT1 protects cardiomyocytes from isoproterenol-induced apoptosis through sponging miR-558 to enhance ULK1-mediated protective autophagy. J Cell Physiol. 2019;234:10842–10854. doi: 10.1002/jcp.27925. [DOI] [PubMed] [Google Scholar]
- 25.Wang S, Yu W, Luo X, Chen J, Deng F. MALAT1/miR-204/LC3-II: A potential regulated axis of autophagy in myocardial ischemia-reperfusion injury. Int J Cardiol. 2019;277:222. doi: 10.1016/j.ijcard.2018.08.075. [DOI] [PubMed] [Google Scholar]
- 26.Tang YH, He GL, Huang SZ, Zhong KB, Liao H, Cai L, Gao Y, Peng ZW, Fu SJ. The long noncoding RNA AK002107 negatively modulates miR-140-5p and targets TGFBR1 to induce epithelial-mesenchymal transition in hepatocellular carcinoma. Mol Oncol. 2019;13:1296–1310. doi: 10.1002/1878-0261.12487. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Kapusta A, Feschotte C. Volatile evolution of long noncoding RNA repertoires: Mechanisms and biological implications. Trends Genet. 2014;30:439–452. doi: 10.1016/j.tig.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao K, Wang M, Kang H, Wu A. A prognostic five long-noncoding RNA signature for patients with rectal cancer. J Cell Biochem. 2019 doi: 10.1002/jcb.29549. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 29.Zhou RS, Zhang EX, Sun QF, Ye ZJ, Liu JW, Zhou DH, Tang Y. Integrated analysis of lncRNA-miRNA-mRNA ceRNA network in squamous cell carcinoma of tongue. BMC Cancer. 2019;19:779. doi: 10.1186/s12885-019-5983-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bretin A, Carriere J, Dalmasso G, Bergougnoux A, B'Chir W, Maurin AC, Muller S, Seibold F, Barnich N, Bruhat A, et al. Activation of the EIF2AK4-EIF2A/eIF2α-ATF4 pathway triggers autophagy response to Crohn disease-associated adherent-invasive Escherichia coli infection. Autophagy. 2016;12:770–783. doi: 10.1080/15548627.2016.1156823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.B'Chir W, Maurin AC, Carraro V, Averous J, Jousse C, Muranishi Y, Parry L, Stepien G, Fafournoux P, Bruhat A. The eIF2α/ATF4 pathway is essential for stress-induced autophagy gene expression. Nucleic Acids Res. 2013;41:7683–7699. doi: 10.1093/nar/gkt563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu H, Lu J, Zhao H, Chen Z, Cui Q, Lin Z, Wang X, Wang J, Dong H, Wang S, Tan J. Functional long noncoding RNAs (lncRNAs) in clear cell kidney carcinoma revealed by reconstruction and comprehensive analysis of the lncRNA-miRNA-mRNA regulatory network. Med Sci Monit. 2018;24:8250–8263. doi: 10.12659/MSM.910773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen K, Xie S, Jin W. Crucial lncRNAs associated with adipocyte differentiation from human adipose-derived stem cells based on co-expression and ceRNA network analyses. Peerj. 2019;7:e7544. doi: 10.7717/peerj.7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen R, Zhu J, Dong Y, He C, Hu X. Suppressor of Ty homolog-5, a novel tumor-specific human telomerase reverse transcriptase promoter-binding protein and activator in colon cancer cells. Oncotarget. 2015;6:32841–32855. doi: 10.18632/oncotarget.5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song J, Xu Q, Zhang H, Yin X, Zhu C, Zhao K, Zhu J. Five key lncRNAs considered as prognostic targets for predicting pancreatic ductal adenocarcinoma. J Cell Biochem. 2018;119:4559–4569. doi: 10.1002/jcb.26598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kluge AF, Lagu BR, Maiti P, Jaleel M, Webb M, Malhotra J, Mallat A, Srinivas PA, Thompson JE. Novel highly selective inhibitors of ubiquitin specific protease 30 (USP30) accelerate mitophagy. Bioorg Med Chem Lett. 2018;28:2655–2659. doi: 10.1016/j.bmcl.2018.05.013. [DOI] [PubMed] [Google Scholar]
- 37.Camenisch U, Dip R, Schumacher SB, Schuler B, Naegeli H. Recognition of helical kinks by xeroderma pigmentosum group A protein triggers DNA excision repair. Nat Struct Mol Biol. 2006;13:278–284. doi: 10.1038/nsmb1061. [DOI] [PubMed] [Google Scholar]
- 38.Ge R, Liu L, Dai W, Zhang W, Yang Y, Wang H, Shi Q, Guo S, Yi X, Wang G, et al. Xeroderma pigmentosum group A promotes autophagy to facilitate cisplatin resistance in melanoma cells through the activation of PARP1. J Invest Dermatol. 2016;136:1219–1228. doi: 10.1016/j.jid.2016.02.693. [DOI] [PubMed] [Google Scholar]
- 39.Inoki K, Li Y, Zhu T, Wu J, Guan KL. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol. 2002;4:648–657. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- 40.Zhang J, Kim J, Alexander A, Cai S, Tripathi DN, Dere R, Tee AR, Tait-Mulder J, Di Nardo A, Han JM, et al. A tuberous sclerosis complex signalling node at the peroxisome regulates mTORC1 and autophagy in response to ROS. Nat Cell Biol. 2013;15:1186–1196. doi: 10.1038/ncb2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Teixeira MB, Alborghetti MR, Kobarg J. Fasciculation and elongation zeta proteins 1 and 2: From structural flexibility to functional diversity. World J Biol Chem. 2019;10:28–43. doi: 10.4331/wjbc.v10.i2.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsang HT, Connell JW, Brown SE, Thompson A, Reid E, Sanderson CM. A systematic analysis of human CHMP protein interactions: Additional MIT domain-containing proteins bind to multiple components of the human ESCRT III complex. Genomics. 2006;88:333–346. doi: 10.1016/j.ygeno.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 43.Barlin JN, Jelinic P, Olvera N, Bogomolniy F, Bisogna M, Dao F, Barakat RR, Chi DS, Levine DA. Validated gene targets associated with curatively treated advanced serous ovarian carcinoma. Gynecol Oncol. 2013;128:512–517. doi: 10.1016/j.ygyno.2012.11.018. [DOI] [PubMed] [Google Scholar]
- 44.Li K, Liu J, Tian M, Gao G, Qi X, Pan Y, Ruan J, Liu C, Su X. CHMP4C disruption sensitizes the human lung cancer cells to irradiation. Int J Mol Sci. 2015;17:18. doi: 10.3390/ijms17010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cagnol S, Chambard JC. ERK and cell death: Mechanisms of ERK-induced cell death-apoptosis, autophagy and senescence. FEBS J. 2010;277:2–21. doi: 10.1111/j.1742-4658.2009.07366.x. [DOI] [PubMed] [Google Scholar]
- 46.Ellington AA, Berhow MA, Singletary KW. Inhibition of Akt signaling and enhanced ERK1/2 activity are involved in induction of macroautophagy by triterpenoid B-group soyasaponins in colon cancer cells. Carcinogenesis. 2006;27:298–306. doi: 10.1093/carcin/bgi214. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used can be obtained from TCGA (https://portal.gdc.cancer.gov). The RT-qPCR data generated in the present study is available from the corresponding author upon reasonable request.