Abstract
Multidrug-resistant (MDR; resistance to ≥3 antimicrobial classes) members of the Pasteurellaceae family may compromise the efficacy of therapies used to prevent and treat bovine respiratory disease (BRD) in feedlot cattle. This study examined the prevalence of multidrug resistance in strains of Mannheimia haemolytica and Pasteurella multocida collected from BRD cattle mortalities in North America. Isolates of M. haemolytica (n = 147) and P. multocida (n = 70) spanning 69 Alberta feedlots from 2011 to 2016 and two United States feedlots from 2011 to 2012 were examined for antimicrobial resistance (AMR) in association with integrative and conjugative elements (ICEs). Overall, resistance was high in both bacterial species with an increase in the prevalence of MDR isolates between 2011 and 2016. Resistance to >7 antimicrobial drugs occurred in 31% of M. haemolytica and 83% of P. multocida isolates. Resistance to sulfadimethoxine, trimethoprim/sulfamethoxazole, neomycin, clindamycin oxytetracycline, spectinomycin, tylosin, tilmicosin, and tulathromycin was most common. Although >80% of strains harbored three or more ICE-associated genes, only 12% of M. haemolytica and 77% of P. multocida contained all six, reflecting the diversity of ICEs. There was evidence of clonal spread as P. multocida and M. haemolytica isolates with the same pulsed-field gel electrophoresis profile from the United States in 2011 were isolated in Alberta in 2015–2016. This work highlights that MDR strains of Pasteurellaceae containing ICEs are widespread and may be contributing to BRD therapy failure in feedlot cattle. Given the antimicrobial resistance gene profiles identified, these MDR isolates may be selected for by the use of macrolides, tetracyclines, and/or in-feed supplements containing heavy metals.
Keywords: antimicrobial resistance, bovine respiratory disease, Mannheimia haemolytica, Pasteurella multocida, integrative and conjugative elements, Pasteurellaceae
Introduction
Bovine respiratory disease (BRD) continues to be the principal health problem for beef production, contributing significantly to economic loss through mortality (1% of death loss) and morbidity (up to 80%) (Hilton, 2014). Multiple viral [bovine herpesvirus type 1 (BHV-1), parainfluenza-3 virus (PI3), bovine viral diarrhea virus (BVDV), and bovine respiratory syncytial virus (BRSV)] and bacterial agents (Mannheimia haemolytica, Pasteurella multocida, Histophilus somni, and Mycoplasma bovis) are implicated in BRD, resulting in a disease complex comprised of numerous forms of pneumonia with varied pathologies. These include bronchopneumonia and fibrinous pneumonia typically associated with P. multocida and M. haemolytica, respectively (Booker et al., 2008); both species are members of the Pasteurellaceae family.
Because it is a complex infection, prevention and management of BRD is challenging. While vaccination is used to protect against viral infections, antimicrobials are the primary tool used to address bacterial agents. As a result, attempts to prevent or treat BRD contribute significantly to the volume of antimicrobials used in beef production. Frequently this is in the form of injectable metaphylaxis used in cattle at high risk of developing respiratory infection upon arrival at feedlots. However, targeted antimicrobial treatment is also used for cattle diagnosed with any ailment due to bacterial infection, which can include cattle exhibiting symptoms of respiratory illness. Additionally, frequent in-feed use of antimicrobials like chlortetracycline and tylosin to prevent or reduce the incidence of liver abscesses can also contribute to bacterial drug exposure in feedlots. Although the use of antimicrobials in the beef industry is necessary to support production and animal health it carries an associated risk of promoting the development and/or selection of antimicrobial-resistant bacteria. This selection pressure may be higher for pathogenic bacteria that are the primary targets of antimicrobial therapy, such as the BRD-associated species, as they are likely to be more frequently exposed to drugs than commensal species.
Multidrug resistance development in bacterial pathogens is a growing concern as microbial agents resistant to multiple drugs can be co-selected by the use of different drug classes. Interim standard definitions (Kwon et al., 2012) have defined multidrug-resistant strains as those resistant to at least one agent in three or more antimicrobial categories and extensively drug-resistant (XDR) strains as those non-susceptible to at least one agent in all but two or fewer antimicrobial categories. For the purpose of this dataset we identify strains as XDR when they are resistant to greater than seven of the nine antimicrobial classes tested, although we did not test all possible drug classes within this panel.
Since the report of a MDR strain of P. multocida harboring 12 antimicrobial resistance genes (ARGs) in Michael et al. (2012a), multidrug resistance has been repeatedly documented in Pasteurellaceae isolates recovered from cattle (Michael et al., 2012a; Lubbers and Hanzlicek, 2013; Klima et al., 2016; Rainbolt et al., 2016; Snyder et al., 2017). Many of the antimicrobial ARGs in these isolates are associated with integrative and conjugative elements (ICEs) (Michael et al., 2012a, b; Eidam et al., 2015; Klima et al., 2016). ICEs are self-transmissible, mobile genetic elements (MGEs) that can contain cassettes of accessory genes enhancing host survivability, including genes associated with environmental adaptation (e.g., ARGs, heavy metal resistance, or phage resistance genes), metabolism (sucrose degradation genes), and fitness (e.g., bacteriocin, nitrogen fixation, biofilm formation, and DNA repair genes) (Armshaw and Pembroke, 2013). Although ICE transfer can be similar to that of plasmids, they differ in that they integrate into the host chromosome and are maintained during cell division (Johnson and Grossman, 2015). Notably, ICEs identified in Pasteurellaceae isolates contain toxin-antitoxin systems that promote their persistence in bacterial populations irrespective of any selective pressure in their environment (Klima et al., 2016).
The ability of ICEs to accumulate and transfer ARGs between Pasteurellaceae spp. may promote the spread of antimicrobial resistance (AMR) in beef cattle production. To address the potential development and dissemination of MDR BRD pathogens within the North American cattle production system, a better understanding of the prevalence of multidrug resistance and its association with ICE is required. This study examines MDR strains of M. haemolytica and P. multocida originating from BRD cattle mortalities as part of three longitudinal studies spanning 2011–2016. The purpose was to describe the diversity of multidrug resistance found in these BRD isolates collected over geographic and temporal distances, and to examine genetic associations between observed multidrug resistance and existing ICEs previously described in these species.
Materials and Methods
Sample Collection, Isolation, and Antimicrobial Susceptibility Testing of M. haemolytica and P. multocida
Samples used for this study originated from three longitudinal projects that collected post-mortem lung tissue samples (n = 755) from cattle BRD mortalities between 2011–2012 (study 1), 2014–2015 (study 2), and 2015–2016 (study 3) (Table 1). Deceased animals were identified, and samples collected based on gross pathological evidence of infectious pneumonia. Infected tissues were excised aseptically from the perimeter of pneumonic lesions, collected in sterile containers, and transported to diagnostic laboratories for processing. Samples from study 1 consisted of fresh tissue stored at 4°C for a period no longer than 10 days prior to bacterial isolation (Klima et al., 2014b). In studies 2 and 3, when delivery time was projected to exceed 72 h, tissues were frozen and stored at −20°C until processed (Anholt et al., 2017; Stanford et al., 2020). Metadata for the animals sampled were provided from submitting veterinary practices and included diagnosis and gross pathology for each case. Drug use data, in the form of how many treatments each animal received, was provided from all veterinary practices and all but one practice also specified the specific antimicrobials used. In total, seven of the 33 feedlots sampled were represented in more than one study (i.e., each were represented in both study 2 and study 3) (Supplementary Table S1).
TABLE 1.
Characteristics of each longitudinal study; the number of isolates screened using antimicrobial susceptibility testing (AST), percentage of multidrug-resistant (MDR) isolates detected, and number of isolates from each study used for subsequent analyses.
| Surveillance study 1 |
Surveillance study 2 |
Surveillance study 3 |
Total |
|
| Year(s) study was performed | 2011–2012 | 2014–2015 | 2015–2016 | – |
| No. of feedlots sampleda | ||||
| Canada | 10 | 17 | 12 | 31 |
| United States | 2 | 0 | 0 | 2 |
| No. animals sampled in the study | 68 | 528 | 159 | 755 |
| No. of isolates recovered | ||||
| M. haemolytica | 55 | 238 | 35 | 408 |
| P. multocida | 8 | 85 | 34 | 140 |
| Percentage of isolates found to be MDRb (No. of strains screened with AST) | ||||
| M. haemolytica | 67.3 (55) | 55 (238) | 81.8 (33) | – |
| P. multocida | 50 (8) | 70.5 (85) | 90.9 (33) | – |
| No. of MDR isolates included in this analysis | ||||
| M. haemolytica | 40 | 80 | 27 | 147 |
| P. multocida | 6 | 34 | 30 | 70 |
| Total | 46 | 114 | 57 | 217 |
aValues include feedlots where MDR strains were recovered. Only three feedlots were represented in more than one study, both sampled in study 2 and study 3 (Supplementary Table S1).bMDR = an isolate expressing resistance to ≥3 antimicrobial classes.
All lung tissue samples were processed for the isolation of M. haemolytica and P. multocida and tested for antimicrobial susceptibility with broth microdilution using a commercially available bovine/porcine BOPO6F panel (Sensititre; Trek Diagnostic Systems, Cleveland, OH, United States) as previously described for M. haemolytica (Klima et al., 2014a) and as per the manufacturer for P. multocida. The minimum inhibitory concentrations (MICs) were assigned as outlined in the Clinical and Laboratory Standards document M31-A3 (Clinical and Laboratory Standards Institute [CLSI], 2008) for those antimicrobials with described breakpoints. Antimicrobials that lacked breakpoints were not assigned a susceptibility designation, with the exception of those antimicrobials where populations exhibited a bimodal distribution with high MICs and isolates possessed the corresponding resistant determinant (Supplementary Table S2). Methods used for species identification and serotyping are detailed in Klima et al. (2014b). Isolates expressing resistance to three or more antimicrobial drugs were classified as MDR and selected for further analysis in this study. Longitudinal study 1 incorporated Canadian and American feedlot sites, while studies 2 and 3 only included feedlots in Canada. The number of feedlots sampled, the number of strains isolated per bacterial species, the percentage of multidrug resistance observed in each study, and the number of MDR isolates included in this analysis are listed in Table 1. In total, 147 M. haemolytica and 70 P. multocida isolates were genotyped using pulsed-field gel electrophoresis (PFGE) with 95% similarity as a cut-off as per Klima et al. (2011) and ICE-associated genes and ARGs screened using PCR as described below.
PCR Assays
Three multiplex PCR assays were designed to improve the efficiency of ARG screening. Primers were designed to target genes in ICEPmu1 (Michael et al., 2012b) using MPprimer v.1.5 (Shen et al., 2010). The RR1-MPLEX assay targeted five ARGs (floR, strB, aphA1, strA, and sul2) in an accessory gene region of ICEPmu1 termed resistance region 1. The RR2-MPLEX assay amplified four ARGs [tet(H), blaOXA–2, aadA25, and aadB] in an accessory gene region of ICEPmu1 termed resistance region 2 as well as a multicopper oxidase gene (mco) located between resistance region 1 and 2. The MCRLD-MPLEX targeted three macrolide ARGs [msr(E), mph(E), and erm(42)] dispersed across both ICEPmu1 regions 1 and 2. Five genes that are part of the core functional regions of ICEPmu1, including a hypothetical protein gene (Pmu_02680), an integrase gene (Pmu_02700), a relaxase gene (Pmu_02890), a transposase gene (Pmu_03510), and a binding protein gene (Pmu_03540), were also screened for by individual PCR assays as previously described (Klima et al., 2014b).
Multiplex PCR Conditions
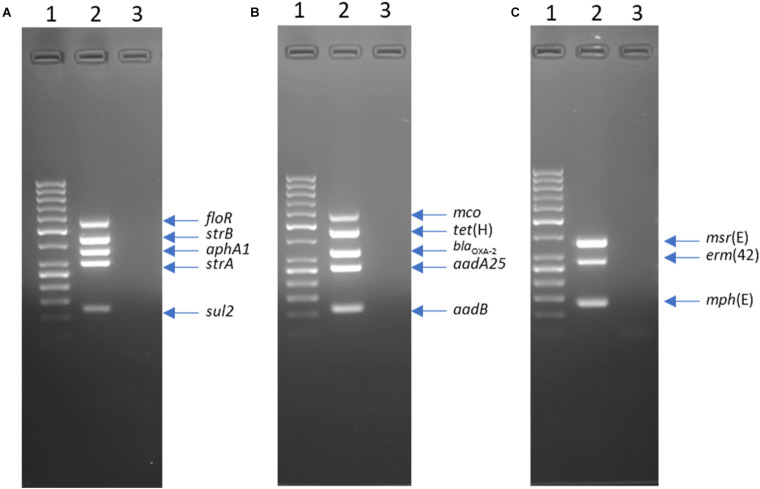
Template DNA was generated using a 1 μL loop of bacterial culture that was suspended in 100 μL of TE buffer (10 mM Tris–HCl, 1 mM EDTA; pH 8.00) and lysed at 98°C for 5 min. The lysate was centrifuged at 13,000 × g for 5 min and 2 μL of supernatant was used as template DNA. For RR1-MPLEX, RR2-MPLEX, and MCRLD-MPLEX, PCR mixtures contained 1X Qiagen Multiplex PCR Master Mix (Qiagen Inc., Mississauga, ON, Canada), 0.5 × Qiagen Q-Solution, varying concentrations of each associated primer as listed in Supplementary Table S3, and 2 μL of template DNA in a total volume of 50 μL. All PCRs were performed as follows: 95°C for 15 min; 35 cycles of 94°C for 30 s, 62°C for 45 s, and 72°C for 1 min; with a final extension at 72°C for 10 min. Following PCR, 20 μL of product was resolved by electrophoresis in a 2.0% (w/v) agarose gel alongside a 50 bp DNA ladder (GeneRuler; Thermo Fisher Scientific, Burlington, ON, Canada) and visualized using ethidium bromide (Figure 1). Representative PCR products from 12 M. haemolytica and P. multocida isolates were sequenced to confirm the accuracy of amplified fragments.
FIGURE 1.
Agarose gel electrophoresis of products from multiplex PCR RR1-MPLEX (A) RR2-MPLEX, (B) and MCRLD-MPLEX (C). Lanes: 1, GeneRuler 50 bp DNA Ladder; 2, Mannheimia haemolytica strain L024A (accession LFXX00000000); 3, negative control.
Statistical Analysis
Fisher’s exact test and Cramer’s V were used in R v. 3.5.1 with the vcd package v. 4.1–4 to determine the strength of associations between categorical (nominal) variables for MDR M. haemolytica and P. multocida isolates. Multidrug resistance profile, study, feedlot, country, submitting veterinary practice, diagnosis, profile of antimicrobial treatment, individual drug treatment, individual resistance phenotypes, and ARG presence were included in the analysis. Associations between the presence/absence of ARGs and antimicrobial resistance phenotype in M. haemolytica and P. multocida were assessed using Pearson’s correlation coefficient in R. All P-values were corrected for multiple comparisons using the Benjamini-Hochberg method. Cramer’s V values close to 0 indicate little association between variables while those close to 1 indicate a strong association.
Results and Discussion
Antimicrobial Susceptibility Testing of M. haemolytica and P. multocida Isolates
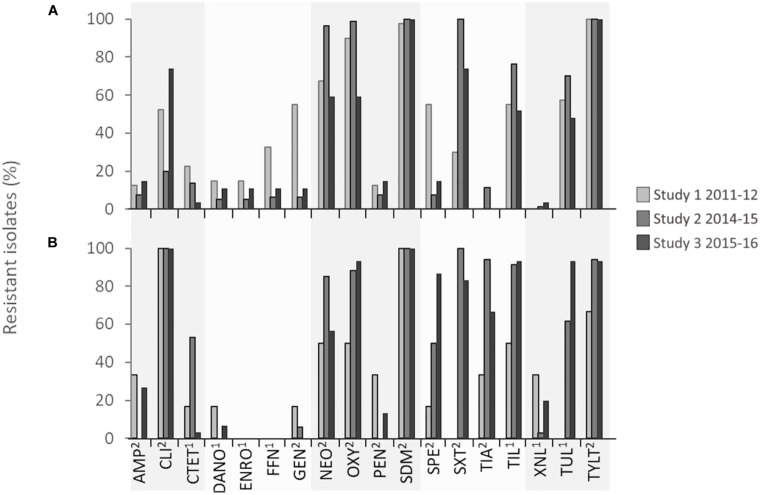
Pasteurella multocida isolates showed very high levels of resistance (>75% of isolates within each study) to neomycin, oxytetracycline, spectinomycin, tiamulin, tilmicosin, tulathromycin, tylosin, and trimethoprim/sulfamethoxazole, and complete resistance (100% of isolates) to clindamycin and sulfadimethoxine in all three studies (Figure 2 and Table 2). With the exception of clindamycin and sulfadimethoxine, the frequency of drug resistance fluctuated over time, but an increase in the percentage of isolates resistant to oxytetracycline, spectinomycin, tilmicosin, and tulathromycin occurred from 2011 to 2016. M. haemolytica isolates were completely resistant (100% of isolates) to tylosin in all years and to sulfadimethoxine from 2014 to 2016. Overall, high levels of resistance (>75% of isolates within a study) to neomycin, oxytetracycline, trimethoprim/sulfamethoxazole, and sulfadimethoxine were observed in M. haemolytica isolates. However, this varied by year, with the highest levels of resistance to florfenicol, gentamicin, and spectinomycin observed in 2011–2012 and to neomycin, oxytetracycline, trimethoprim/sulfamethoxazole, and macrolides in 2014–2015. Although a trend toward higher resistance to some antimicrobials was observed over time in both P. multocida and M. haemolytica isolates, there was considerable temporal and/or geographical variation in resistance to most antimicrobials. Of the antimicrobials included for susceptibility testing, only macrolides (tilmicosin, tulathromycin, tylosin), tetracyclines (chlortetracycline, oxytetracycline), β-lactams (ceftiofur), phenicols (florfenicol), and fluoroquinolones (danofloxacin, enrofloxacin) were used frequently in feedlots (Brault et al., 2019). As a result, the resistance observed to aminoglycosides (gentamicin, neomycin, spectinomycin), lincosamides (clindamycin), and pleuromutilins (tiamulin) was unexpected.
FIGURE 2.
Percentage of (A) Mannheimia haemolytica and (B) Pasteurella multocida isolates in each surveillance study that were resistant to each antimicrobial. 1Minimum inhibitory concentration (MIC) breakpoints defined by Clinical Laboratory Standards Institute (CLSI) guidelines. 2MIC determined by bimodal distribution of MIC coupled with the presence of associated antimicrobial resistance genes. 2Ampicillin, AMP; 2Clindamycin, CLI; 1Chlortetracycline, CTET; 1Danofloxacin, DANO; 1Enrofloxacin, ENRO; 1Florfenicol, 2FFN; Gentamicin, GEN; 2Neomycin, NEO; 2Oxytetracycline, OXY; 2Penicillin, PEN; 2Trimethoprim/sulfamethoxazole, SDM; 2Spectinomycin, SPE; 2Sulfadimethoxine, SXT; 2Tiamulin, TIA; 1Tilmicosin, TIL; 1Ceftiofur, XNL; 1Tulathromycin, TUL; 2Tylosin tartrate, TYLT.
TABLE 2.
Multidrug resistance profiles and associated pulsotypes for Mannheimia haemolytica and Pasteurella multocida isolates collected from bovine respiratory disease mortalities.
| No. drugs in MDR profile | No. drug classes isolates are resistant to | Drugs isolates are resistant to | Pulsotype | M. haemolytica | P. multocida |
| 3 | 3 | CLI, OXY, SDM | |||
| 59 | 1 | ||||
| 61 | 1 | ||||
| 3 | 3 | CLI, SDM, TIA | |||
| 57 | 1 | ||||
| 3 | 3 | CLI, SDM, TYLT | |||
| 5 | 1 | ||||
| 9 | 1 | ||||
| 19 | 1 | ||||
| 38 | 1 | ||||
| 49 | 1 | ||||
| 3 | 3 | OXY, SDM, TYLT | |||
| 25 | 1 | ||||
| 26 | 1 | ||||
| 4 | 3 | CLI, NEO, SDM, SXT | |||
| 61 | 1 | ||||
| 3 | CLI, SDM, SXT, TYLT | ||||
| 4 | 1 | ||||
| 15 | 1 | ||||
| 26 | 1 | ||||
| 27 | 1 | ||||
| 29 | 1 | ||||
| 37 | 1 | ||||
| 42 | 2 | ||||
| 45 | 1 | ||||
| 4 | 4 | NEO, OXY, SDM, TYLT | |||
| 1 | 1 | ||||
| 12 | 2 | ||||
| 21 | 2 | ||||
| 26 | 1 | ||||
| 29 | 1 | ||||
| 39 | 1 | ||||
| 4 | 4 | CLI, SDM, TIA, TYLT | |||
| 58 | 1 | ||||
| 4 | 3 | AMP, CLI, PEN, SDM, XNL | |||
| 55 | 1 | ||||
| 4 | 3 | CLI, GEN, NEO, SDM, SXT | |||
| 63 | 1 | ||||
| 4 | 5 | CLI, NEO, OXY, SDM, TYLT | |||
| 35 | 1 | ||||
| 4 | 4 | CLI, SDM, TIL, TUL, TYLT | |||
| 61 | 1 | ||||
| 4 | 4 | NEO, OXY, SDM, SXT, TYLT | |||
| 2 | 1 | ||||
| 3 | 1 | ||||
| 21 | 7 | ||||
| 22 | 1 | ||||
| 29 | 1 | ||||
| 32 | 3 | ||||
| 36 | 1 | ||||
| 6 | 5 | CLI, NEO, OXY, SDM, SXT, TYLT | |||
| 21 | 1 | ||||
| 22 | 1 | ||||
| 32 | 2 | ||||
| 6 | 5 | CLI, NEO, OXY, SDM, TIL, TYLT | |||
| 57 | 1 | ||||
| 6 | 4 | FFN, NEO, SDM, SXT, TUL, TYLT | |||
| 15 | 1 | ||||
| 6 | 4 | NEO, OXY, SDM, SXT, TIL, TYLT | |||
| 21 | 2 | ||||
| 30 | 1 | ||||
| 6 | 4 | NEO, OXY, SDM, TIL, TUL, TYLT | |||
| 21 | 1 | ||||
| 6 | 3 | OXY, SDM, SXT, TIL, TUL, TYLT | |||
| 23 | 3 | ||||
| 7 | 5 | CLI, CTET, NEO, OXY, SDM, SXT, TYLT | |||
| 21 | 1 | ||||
| 7 | 5 | CLI, CTET, NEO, OXY, SDM, TIL, TYLT | |||
| 54 | 1 | ||||
| 7 | 4 | CLI, CTET, OXY, SDM, TIL, TUL, TYLT | |||
| 61 | 1 | ||||
| 7 | 6 | CLI, FFN, NEO, OXY, SDM, TIL, TYLT | |||
| 12 | 1 | ||||
| 7 | 6 | CLI, NEO, OXY, SDM, SXT, TIA, TYLT | |||
| 26 | 1 | ||||
| 7 | 5 | CLI, NEO, OXY, SDM, SXT, TIL, TYLT | |||
| 21 | 1 | ||||
| 26 | 1 | ||||
| 7 | 4 | CTET, GEN, OXY, SDM, SPE, TUL, TYLT | |||
| 12 | 1 | ||||
| 7 | 3 | CTET, GEN, OXY, SPE, TIL, TUL, TYLT | |||
| 12 | 1 | ||||
| 7 | 4 | NEO, OXY, SDM, SPE, SXT, TIL, TYLT | |||
| 32 | 1 | ||||
| 8 | 4 | NEO, OXY, SDM, SXT, TIL, TUL, TYLT | |||
| 6 | 1 | ||||
| 8 | 1 | ||||
| 15 | 1 | ||||
| 17 | 1 | ||||
| 21 | 20 | ||||
| 22 | 1 | ||||
| 24 | 1 | ||||
| 26 | 1 | ||||
| 28 | 1 | ||||
| 29 | 1 | ||||
| 32 | 8 | ||||
| 33 | 1 | ||||
| 34 | 1 | ||||
| 40 | 1 | ||||
| 53 | 1 | ||||
| 8 | 5 | CLI, NEO, OXY, SDM, TIL, TUL, TYLT | |||
| 21 | 1 | ||||
| 61 | 1 | ||||
| 8 | 5 | CLI, NEO, OXY, SDM, SPE, TIL, TUL, TYLT | 1 | ||
| 61 | |||||
| 8 | 5 | CLI, NEO, OXY, SDM, SXT, TIL, TUL, TYLT | |||
| 16 | 1 | ||||
| 26 | 1 | ||||
| 32 | 2 | ||||
| 8 | 5 | CLI, NEO, SDM, SXT, TIA, TIL, TUL, TYLT | |||
| 61 | 1 | ||||
| 8 | 5 | CLI, OXY, SDM, SPE, SXT, TIL, TUL, TYLT | |||
| 61 | 2 | ||||
| 8 | 6 | CLI, OXY, SDM, SPE, TIA, TIL, TUL, TYLT | |||
| 61 | 1 | ||||
| 8 | 5 | CLI, OXY, SDM, SXT, TIA, TIL, TUL, TYLT | |||
| 61 | 1 | ||||
| 8 | 5 | CTET, GEN, OXY, SDM, SPE, SXT, TUL, TYLT | |||
| 48 | 1 | ||||
| 8 | 4 | CTET, NEO, OXY, SDM, SXT, TIL, TUL, TYLT | |||
| 21 | 2 | ||||
| 8 | 4 | NEO, OXY, SDM, SPE, SXT, TIL, TUL, TYLT | |||
| 15 | 1 | ||||
| 8 | 5 | NEO, OXY, SDM, SXT, TIA, TIL, TUL, TYLT | |||
| 21 | 1 | ||||
| 32 | 2 | ||||
| 9 | 6 | AMP, CLI, NEO, OXY, PEN, SDM, SXT, TIL, TYLT | |||
| 21 | 1 | ||||
| 9 | 6 | AMP, CLI, NEO, OXY, PEN, SDM, SXT, XNL, TYLT | |||
| 20 | 1 | ||||
| 9 | 6 | CLI, CTET, NEO, OXY, SDM, SXT, TIA, TIL, TYLT | |||
| 31 | 1 | ||||
| 61 | 1 | ||||
| 62 | 1 | ||||
| 64 | 1 | ||||
| 65 | 1 | ||||
| 68 | 1 | ||||
| 70 | 1 | ||||
| 72 | 2 | ||||
| 74 | 1 | ||||
| 9 | 6 | CLI, CTET, NEO, SDM, SXT, TIA, TIL, TUL, TYLT | |||
| 61 | 1 | ||||
| 9 | 5 | CLI, CTET, OXY, SDM, SXT, TIA, TIL, TUL, TYLT | |||
| 61 | 1 | ||||
| 9 | 5 | CLI, NEO, OXY, SDM, SPE, SXT, TIL, TUL, TYLT | |||
| 61 | 1 | ||||
| 9 | 6 | CLI, OXY, SDM, SPE, SXT, TIA, TIL, TUL, TYLT | |||
| 61 | 4 | ||||
| 9 | 6 | CTET, NEO, OXY, SDM, SXT, TIA, TIL, TUL, TYLT | |||
| 21 | 1 | ||||
| 32 | 1 | ||||
| 9 | 5 | DANO, ENRO, GEN, OXY, SDM, SPE, SXT, TUL, TYLT | |||
| 14 | 1 | ||||
| 10 | 6 | AMP, CLI, NEO, OXY, SDM, SPE, SXT, TIL, TUL, TYLT | |||
| 61 | 2 | ||||
| 10 | 5 | CLI, CTET, GEN, NEO, OXY, SDM, SPE, TIL, TUL, TYLT | |||
| 52 | 1 | ||||
| 10 | 6 | CLI, CTET, NEO, OXY, SDM, SXT, TIA, TIL, TUL, TYLT | |||
| 21 | 1 | ||||
| 61 | 1 | ||||
| 10 | 6 | CLI, CTET, OXY, SDM, SPE, SXT, TIA, TIL, TUL, TYLT | |||
| 61 | 1 | ||||
| 10 | 6 | CLI, FFN, GEN, NEO, OXY, SDM, SPE, TIL, TUL, TYLT | |||
| 12 | 2 | ||||
| 32 | 1 | ||||
| 41 | 1 | ||||
| 48 | 1 | ||||
| 10 | 6 | CLI, NEO, OXY, SDM, SPE, SXT, TIA, TIL, TUL, TYLT | |||
| 21 | 1 | ||||
| 61 | 10 | ||||
| 62 | 1 | ||||
| 66 | 1 | ||||
| 69 | 1 | ||||
| 70 | 1 | ||||
| 72 | 3 | ||||
| 73 | 1 | ||||
| 10 | 7 | CLI, OXY, SDM, SPE, SXT, TIA, TIL, XNL, TUL, TYLT | |||
| 61 | 1 | ||||
| 11 | 7 | AMP, CLI, DANO, OXY, SDM, SPE, SXT, TIA, TIL, TUL, TYLT | |||
| 61 | 1 | ||||
| 11 | 6 | AMP, DANO, ENRO, GEN, OXY, PEN, SDM, SPE, TIL, TUL, TYLT | |||
| 13 | 1 | ||||
| 39 | 1 | ||||
| 11 | 6 | CLI, CTET, FFN, GEN, NEO, OXY, SDM, SPE, TIL, TUL, TYLT | |||
| 7 | 1 | ||||
| 26 | 1 | ||||
| 46 | 1 | ||||
| 11 | 6 | CLI, CTET, GEN, NEO, OXY, SDM, SPE, SXT, TIL, TUL, TYLT | |||
| 12 | 1 | ||||
| 11 | 7 | CLI, CTET, GEN, OXY, SDM, SPE, SXT, TIA, TIL, XNL, TYLT | |||
| 61 | 1 | ||||
| 11 | 6 | CLI, CTET, NEO, OXY, SDM, SPE, SXT, TIA, TIL, TUL, TYLT | |||
| 61 | 1 | ||||
| 62 | 1 | ||||
| 67 | 1 | ||||
| 71 | 1 | ||||
| 11 | 6 | CLI, FFN, GEN, NEO, OXY, SDM, SPE, SXT, TIL, TUL, TYLT | |||
| 12 | 1 | ||||
| 15 | 1 | ||||
| 47 | 1 | ||||
| 11 | 7 | CLI, NEO, OXY, SDM, SPE, SXT, TIA, TIL, XNL, TUL, TYLT | |||
| 61 | 1 | ||||
| 12 | 7 | AMP, CLI, DANO, GEN, NEO, OXY, PEN, SDM, SPE, TIL, XNL, TYLT | |||
| 56 | 1 | ||||
| 12 | 8 | AMP, CLI, DANO, OXY, PEN, SDM, SPE, SXT, TIA, TIL, TUL, TYLT | |||
| 61 | 1 | ||||
| 12 | 7 | AMP, CLI, NEO, OXY, SDM, SPE, SXT, TIA, TIL, XNL, TUL, TYLT | |||
| 61 | 1 | ||||
| 12 | 6 | AMP, CTET, DANO, ENRO, GEN, OXY, PEN, SDM, SPE, TIL, TUL, TYLT | |||
| 18 | 1 | ||||
| 13 | 6 | AMP, CLI, CTET, GEN, NEO, OXY, PEN, SDM, SXT, TIL, XNL, TUL, TYLT | |||
| 32 | 1 | ||||
| 13 | 7 | AMP, CLI, DANO, ENRO, GEN, OXY, PEN, SDM, SPE, SXT, TIL, TUL, TYLT | |||
| 18 | 1 | ||||
| 13 | 7 | AMP, CLI, NEO, OXY, PEN, SDM, SPE, SXT, TIA, TIL, XNL, TUL, TYLT | |||
| 60 | 1 | ||||
| 61 | 2 | ||||
| 15 | 8 | AMP, CLI, CTET, DANO, ENRO, FFN, GEN, NEO, OXY, PEN, SDM, SPE, TIL, TUL, TYLT | |||
| 10 | 1 | ||||
| 15 | 8 | AMP, CLI, DANO, ENRO, FFN, GEN, NEO, OXY, PEN, SDM, SPE, SXT, TIL, TUL, TYLT | |||
| 7 | 1 | ||||
| 11 | 1 | ||||
| 26 | 1 | ||||
| 32 | 1 | ||||
| 16 | 8 | AMP, CLI, CTET, DANO, ENRO, FFN, GEN, NEO, OXY, PEN, SDM, SPE, SXT, TIL, TUL, TYLT | |||
| 21 | 1 | ||||
| 32 | 1 | ||||
| 50 | 1 | ||||
| Total | 147 | 70 |
The markedly higher resistance to clindamycin, tiamulin, and ceftiofur in P. multocida isolates compared to M. haemolytica is difficult to explain (Figure 2). Although not frequently administered to cattle, clindamycin and tiamulin use is common in swine production where P. multocida is an important cause of pneumonia and atrophic rhinitis (Davies et al., 2003). Consequently, it is possible that these phenotypes could originate from other food production sectors where P. multocida is a pathogen frequently targeted with these therapeutics.
There has been recent emergence of pleuromutilin (tiamulin) and lincosamide (clindamycin) resistance in Brachyspira hyodysenteriae, an agent of swine dysentery worldwide that has been associated with lnu(C) (lincosamide resistance), rplC and tva(A) (pleuromutilin resistance), and various point mutations in the 23S rRNA gene (macrolide, lincosamide, and pleuromutilin resistance) (Card et al., 2018). These mechanisms have not been examined here, but cross-resistance has been documented within the MLSB (macrolides, lincosamides and streptogramins B) group of antimicrobials, with the rRNA methylase gene erm(42) shown to confer strong resistance to clindamycin in P. multocida in vitro (Desmolaize et al., 2011). Correlation between clindamycin and macrolide resistance in P. multocida isolates in the present study was not 100% as six clindamycin-resistant isolates were susceptible to all macrolides tested. Likewise, five tiamulin-resistant isolates were susceptible to clindamycin indicating that further characterization is needed to determine what is contributing to the resistance observed.
Why moderate levels of resistance to ceftiofur (<30% of isolates) were observed in P. multocida isolates, but not in M. haemolytica, is also unclear as this antimicrobial is used in both beef and pork production for the treatment and control of respiratory infections. Historically, in Canada, ceftiofur (a third-generation cephalosporin) has been used less frequently than tetracyclines, florfenicol, or macrolides to treat BRD-associated infections in cattle (Brault et al., 2019). Consequently, ceftiofur is still potentially highly efficacious against BRD agents with <1% of Pasteurellaceae isolates collected from the United States and Canada expressing resistance (Klima et al., 2014b; Anholt et al., 2017; Timsit et al., 2017; Kadlec et al., 2018; Woolums et al., 2018). However, recent increased resistance in Pasteurellaceae spp. to the previously mentioned and more commonly used drugs (Klima et al., 2014b; Anholt et al., 2017; Timsit et al., 2017; Kadlec et al., 2018; Woolums et al., 2018), may lead to an increase in the employment of ceftiofur for BRD therapy. This is concerning, because in addition to being an important veterinary therapy, ceftiofur is classified in Canada as an antimicrobial of very high importance in human medicine (category I) (Government of Canada, 2009) and can select for extended-spectrum β-lactamase-producing Enterobacteriaceae (Mollenkopf et al., 2017). Therefore, the development of cross-resistance to other cephalosporins because of ceftiofur use in veterinary medicine could potentially negatively affect human therapies.
Interestingly, although some florfenicol resistance was observed in M. haemolytica, no resistance to this drug was seen in P. multocida. Similar to tiamulin, florfenicol is licensed for use in both swine and cattle for the treatment of bacterial pneumonia, although it may be used more frequently in cattle with only 7 to 13% of swine farms in Canada reporting injectable florfenicol use in growing-finishing pigs between 2013 and 2016 (Government of Canada, 2018).
Multidrug Resistance in M. haemolytica and P. multocida
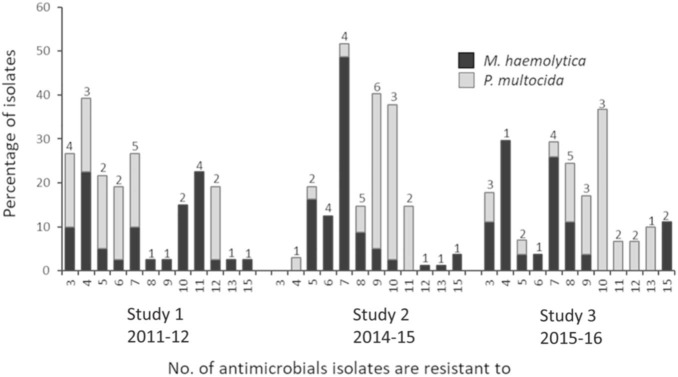
Possible XDR M. haemolytica and P. multocida isolates were found in all three studies (Figure 3 and Table 2). Of the 104 unique multidrug resistance profiles identified across both species, three M. haemolytica isolates from two different feedlots in Canada, were resistant to 16 antimicrobials spanning nine antimicrobial classes (ampicillin, clindamycin, chlortetracycline, danofloxacin, enrofloxacin, florfenicol, gentamicin, neomycin, oxytetracycline, penicillin, sulfadimethoxine, spectinomycin, tilmicosin, trimethoprim/sulfamethoxazole, tulathromycin, and tylosin). From 2011 to 2016 the prevalence of M. haemolytica isolates resistant to >7 antimicrobials fell from 50 to 26%. In contrast, the incidence of P. multocida isolates resistant to >7 antimicrobials increased from 17% in 2011 to 87% in 2016. It is unclear why there was an increase in the number of P. multocida resistant to a large number of drugs, but not in M. haemolytica; particularly since the overall level of multidrug resistance in M. haemolytica was highest at 86% in 2016. The variability in the AMR profiles and the lack of correlation of these profiles with genotype (see below) suggests that horizontal gene transfer rather than clonal dissemination is contributing to the complexity of multidrug resistance in Pasteurellaceae spp. in cattle. It is possible that the rate of transfer and overall stability of the ARGs present in M. haemolytica ICEs could differ from that of P. multocida, accounting for some of the variability in prevalence and AMR patterns seen between the species. Some ICEs have been shown to be active in the host immediately before becoming quiescent. Having not integrated directly into the host genome, these elements are not yet under repression from their ICE-encoded regulators and therefore can rapidly spread to other bacterial cells (Johnson and Grossman, 2015). Although speculation at this point, it is possible that the kinetics behind the transfer and integration rates of ICEs could differ among species.
FIGURE 3.
Multidrug resistance in Mannheimia haemolytica and Pasteurella multocida isolates. Numbers above bars indicate the number of unique antimicrobial susceptibility testing profiles within each multidrug resistance category.
Co-Isolation of M. haemolytica and P. multocida
Of the 946 lung tissue samples collected, only 20 animals were culture positive for both MDR M. haemolytica and P. multocida. Of these cases, two animals harbored isolates of M. haemolytica and P. multocida that exhibited identical multidrug resistance profiles, with one pair of isolates resistant to clindamycin, chlortetracycline, neomycin, oxytetracycline, sulfadimethoxine, trimethoprim/sulfamethoxazole, tiamulin, tilmicosin, tylosin and the other resistant to clindamycin, neomycin, oxytetracycline, sulfadimethoxine, spectinomycin, trimethoprim/sulfamethoxazole, tiamulin, tilmicosin, tulathromycin, and tylosin. The remaining 18 animals with co-infections harbored strains of M. haemolytica and P. multocida that had different multidrug resistance profiles.
Detection of ICE and Antimicrobial Resistance Genes in M. haemolytica and P. multocida
Since the discovery of ICEPmu1 in P. multocida (Michael et al., 2012a), and ICEMh1 in M. haemolytica (Eidam et al., 2015), multiple ICEs have been documented in Pasteurellaceae, many of which are associated with possible XDR (Klima et al., 2016; Cameron et al., 2018; Kadlec et al., 2018; Stanford et al., 2020). All have a similar backbone with regions of functional genes associated with conjugation, recombination and regulation grouped together and in similar orientation. As a result, much of the diversity observed occurs due to the presence or absence, and varied composition, of two accessory gene cassettes that can harbor ARGs, heavy metal resistance genes and other virulence factors.
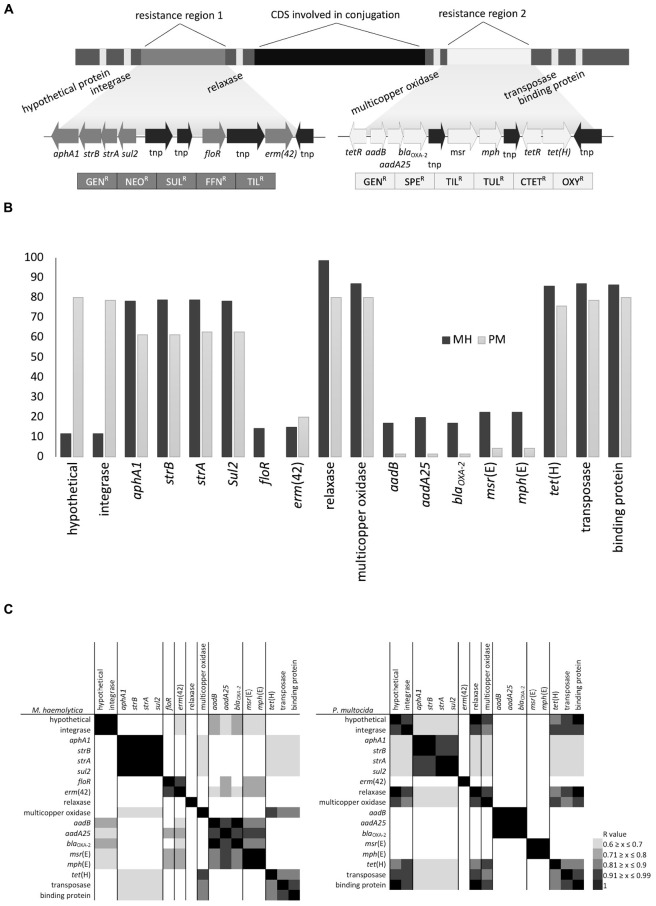
As part of this study, PCR was used to screen for the presence of core and accessory genes identified in ICEPmu1. At the time of the study design, ICEPmu1 was one of the largest ICEs described in Pasteurellaceae, with the highest number of accessory genes, therefore its coding sequence was used to design all PCR primers and included five genes present within the functional gene regions of the ICE, and twelve ARGs and a multicopper oxidase gene present in two accessory gene cassettes known to be resistance regions (Figure 4A). The core genes targeted represented the full length of ICEPmu1 with the relaxase gene (Pmu_02890) from the central region, a hypothetical protein gene (Pmu_02680) and integrase gene (Pmu_02700) located on the left terminus and a transposase gene (Pmu_03510) and binding protein gene (Pmu_03540) located on the right terminus.
FIGURE 4.
(A) Schematic representation of gene arrangement in ICEPmu1 originating from Pasteurella multocida, (B) percentage of isolates positive for antimicrobial resistance genes (ARGs) and ICE-associated genes, and (C) heatmaps displaying the association between ARGs and ICE-associated genes based on Pearson’s correlation coefficient.
The majority (>80%) of M. haemolytica and P. multocida isolates harbored both the relaxase and multicopper oxidase genes located in the center of ICEPmu1 along with the transposase and single-stranded DNA-binding protein genes on the right terminus. However, although 80% of P. multocida isolates carried the hypothetical and integrase genes present on the left flank of ICEPmu1, less than 10% of M. haemolytica isolates were PCR positive for both of these genes indicating that differences may occur between species on the left flank of potential ICEs. The high prevalence of the multicopper oxidase gene in both species suggests that this gene is either closely linked with the functional backbone and not easily lost or is maintained due to selection pressure.
Although ICEs from P. multocida have been shown to transfer to M. haemolytica in vitro (Michael et al., 2012b) and strong similarities between ICEs from species within the Pasteurellaceae family have been previously reported (Beker et al., 2018; Bhatt et al., 2018; Klima et al., 2019), only two of the multidrug resistance profiles identified in this study were present in both species (Table 2). As well, the prevalence of individual ARGs was either high (60–80% of isolates) or low (0–20% of isolates) within the isolates screened (Figure 4B). There were, however, strong correlations between the presence of multiple ARGs in both species (Figure 4C) suggesting that regardless of abundance, some of these genes could be linearly arranged or linked in patterns that reflect gene arrangements within the resistance gene regions in ICEPmu1. This is significant because linked genes typically transfer together and are more likely to be co-selected by the antimicrobials to which they confer resistance. In the case of M. haemolytica, aphA1, strB, strA, and sul2, aadB and blaOXA–2, and msr(E) and mph(E), appeared to be linked based on Pearson’s correlation coefficient (r = 1, P ≤ 0.05). In P. multocida, linkages between aphA1 and strB, strA and sul2, aadB, aadA25, blaOXA–2, and msr(E) and mph(E) were also evident (r = 1, P ≤ 0.05). Similar ARG linkages to those seen here were also reported from the recent sequence analysis of over 1,000 MDR M. haemolytica genomes (Clawson et al., 2016).
Integrative and conjugative elements are thought to acquire accessory genes through the presence of insertion sequences, transposons, and specific recombinases within the ICE genome (Armshaw and Pembroke, 2013). Weaker positive correlations were observed (Pearson correlation r = 0.6 to 0.99, P ≤ 0.05) in this group of isolates that reflect ARGs adjacent to one another on ICEPmu1 and that are flanked by transposases [i.e., aphaA1/strB/strA/sul2 and floR and erm(42), or aadB/blaOXA–2/aadA25 and msr(E)/mph(E)]. It is likely that ARGs located next to transposases are part of insertion sequences and are independently mobile within these cassettes. If some genes are acquired or lost as groups while others are individually mobile, this could account for both the similarity in patterns and the overall diversity of multidrug resistance profiles observed in Pasteurellaceae ICEs. Previous work in our lab has shown that ARGs within ICEMhL044A (Klima et al., 2016) lacked post-conjugal stability, particularly floR and erm(42), which we have observed to be easily lost in recipient strains (C.L. Klima and T.A. McAllister, unpublished data). The instability of ARGs within existing ICEs would be positive from the standpoint of limiting AMR dissemination and spread. Many ICEs in Pasteurellaceae contain toxin-antitoxin systems that promote stability of the element within populations (Klima et al., 2016). However, if some ARGs in ICEs can be independently lost from their conserved functional regions, strategies to enhance ARG loss and not ICE loss could be developed to mitigate AMR in existing populations.
Phenotype vs. Antimicrobial Resistance Genes Present in M. haemolytica and P. multocida
Although strong correlations were observed between AMR phenotype and ARGs, with the exception of florfenicol/floR in M. haemolytica isolates, none were completely correlated (Table 3). This may be due to other ARGs contributing to the phenotypes observed, the ARGs are inactive, or the breakpoints used for susceptibility testing need to be re-evaluated. The narrow-spectrum β–lactamase gene, blaOXA–2, was the only ARG screened in association with β-lactam resistance (ampicillin, penicillin, ceftiofur). Previously reported in ICEPmu1, blaOXA–2 is inactive in both host and recipient strains of P. multocida and M. haemolytica post-conjugation, but active when transferred into Escherichia coli (Michael et al., 2012b). Varied activity of blaOXA–2 based on bacterial host species has been observed before with the gene conferring extended-spectrum β-lactamase activity in Acinetobacter baumannii, an opportunistic nosocomial pathogen, but producing a narrow-spectrum antibiotic resistance pattern in E. coli (Antunes et al., 2014). Given the low concordance of blaOXA–2 with β-lactam resistance (<50%) in our study, its occurrence in 12.9% of susceptible M. haemolytica isolates suggests that it is inactive in these strains. Phenotypic resistance to β-lactams in M. haemolytica has been previously reported to be associated with blaROB–1 (Klima et al., 2014b) and more recently with blaROB–2 (Kadlec et al., 2018). It is possible that either of these genes could be contributing to the β-lactam resistance observed in the current study.
TABLE 3.
Associations between antimicrobial resistance phenotype and antimicrobial resistance gene (ARG) presence in Mannheimia haemolytica (MH) and Pasteurella multocida (PM) isolates collected post-mortem from bovine respiratory disease mortalities.
| Resistance phenotype/ARG | No. isolates with resistance phenotype MH| PM | % of the resistant isolates with ARGa MH| PM | % of the susceptible isolates with ARG MH| PM |
| AMP/blaOXA–2 | 15 | 10 | 54 | 10 | 12.9 | 0 |
| PEN/blaOXA–2 | 15 | 6 | 53.4 | 16.7 | 12.9 | 0 |
| XNL/blaOXA–2 | 2 | 9 | 0 | 11.2 | 17.3 | 0 |
| CTET/tet(H) | 21 | 20 | 90.5 | 85 | 85 | 72 |
| OXYT/tet(H) | 131 | 61 | 96.2* | 86.9 | 0 | 0 |
| FFN/floR | 21 | 0 | 100** | 0 | 0 | 0 |
| SDM/sul2 | 146 | 70 | 79.5 | 62.9 | 0 | 0 |
| SXT/sul2 | 112 | 59 | 83.1 | 67.8 | 65.8 | 36.4 |
| SPE/aadA25 | 32 | 44 | 90.7** | 2.3 | 0 | 0 |
| SPE/aadB | 32 | 44 | 78.2* | 2.3 | 0 | 0 |
| NEO/strA | 120 | 49 | 96.7** | 85.8* | 0 | 9.6 |
| NEO/strB | 120 | 49 | 96.7** | 85.8* | 0 | 4.8 |
| NEO/aphA1 | 120 | 49 | 96.7** | 85.8* | 0 | 4.8 |
| GENT/strA | 30 | 3 | 70 | 33.4 | 81.2 | 64.2 |
| GENT/strB | 30 | 3 | 70 | 33.4 | 81.2 | 62.7 |
| GENT/ aphA1 | 30 | 3 | 70 | 33.4 | 81.2 | 62.7 |
| GENT/aadB | 30 | 3 | 83.4* | 33.4 | 0 | 0 |
| GENT/aadA25 | 30 | 3 | 96.7** | 33.4 | 0 | 0 |
| TUL/msr(E) | 92 | 49 | 33.7 | 4.1 | 3.7 | 4.8 |
| TUL/mph(E) | 92 | 49 | 33.7 | 4.1 | 3.7 | 4.8 |
| TUL erm(42) | 92 | 49 | 22.9 | 8.2 | 1.9 | 47.7 |
| TIL/msr(E) | 97 | 62 | 29.9 | 4.9 | 8 | 0 |
| TIL/mph(E) | 97 | 62 | 29.9 | 4.9 | 8 | 0 |
| TIL/erm(42) | 97 | 62 | 22.7 | 22.6 | 0 | 0 |
| TYLT/msr(E) | 147 | 64 | 22.5 | 4.7 | 0 | 0 |
| TYLT/mph(E) | 147 | 64 | 22.5 | 4.7 | 0 | 0 |
| TYLT/erm(42) | 147 | 64 | 15 | 21.9 | 0 | 0 |
| CLIND/msr(E) | 57 | 70 | 42.2 | 4.3 | 10 | 0 |
| CLIND/mph(E) | 57 | 70 | 42.2 | 4.3 | 10 | 0 |
| CLIND/erm(42) | 57 | 70 | 38.6 | 20 | 0 | 0 |
a Based on significant (P < 0.05) Pearson’s correlation coefficient; *r > 0.8, **r > 0.9.
The tetracycline efflux gene tet(H) was found in 90.5% of M. haemolytica and 85% of P. multocida tetracycline-resistant isolates. Interestingly, the association between tet(H) and tetracycline resistance (oxytetracycline, chlortetracycline) was significant in M. haemolytica expressing oxytetracycline resistance, but not chlortetracycline resistance. A large number (>70%) of M. haemolytica and P. multocida isolates were susceptible to chlortetracycline even though they carried tet(H), indicating that tet(H) does not always confer resistance to chlortetracycline. The prevalent resistance to oxytetracycline is not unexpected as it was administered as a therapeutic to animals surveyed here. Given that resistance to oxytetracycline was not 100% correlated with tet(H) it is possible that an alternative tetracycline resistance gene is conferring the phenotype observed. Although tet(H) has been the most commonly reported tetracycline resistance gene in North American M. haemolytica and P. multocida isolates, tet(M) and tet(B) have been identified in P. multocida from the United States (Hansen et al., 1996), tet(G) in M. haemolytica from Germany (Kehrenberg et al., 2001) and P. multocida from Taiwan (Wu et al., 2003), and tet(L) in M. haemolytica and P. multocida isolates from Belgium (Kehrenberg et al., 2005).
In terms of unresolved macrolide, sulfonamide, and sulfonamide/trimethoprim resistance, it is likely that ARGs other than those screened conferred resistance. Although sul2 was present in many of the sulfadimethoxine- and trimethoprim/sulfamethoxazole-resistant isolates, it was absent in at least 20% of these isolates. We did not screen for the dihydrofolate reductase genes, dfrA20 (Kehrenberg and Schwarz, 2005) or dfrA14 (Niemann et al., 2019), that have been previously linked to trimethoprim resistance in P. multocida. Macrolide resistance was one of the most frequently observed phenotypes in both M. haemolytica and P. multocida, but the ARGs commonly affiliated with these phenotypes, [i.e., erm(42), msr(E), mph(E)], were not present in the majority of resistant isolates. Using functional libraries, we were unable to identify novel ARGs in strains here that lacked erm(42), msr(E), or mph(E) (C.L. Klima and T.A. McAllister, unpublished data), suggesting that macrolide resistance is likely associated with mutations in the 23S rRNA gene that can confer high-level resistance to macrolides in both M. haemolytica and P. multocida (Olsen et al., 2015). There were many incidences of clindamycin resistance where erm(42) was not present, likely indicating that an alternative mechanism is contributing to clindamycin resistance.
Although aadB and aadA25 are both commonly present in spectinomycin-resistant M. haemolytica, aadA25 was detected in 90.7% of these isolates and was more strongly associated with the spectinomycin-resistant phenotype than aadB. Neither gene was highly prevalent (<3%) in the 62.8% of P. multocida that were spectinomycin-resistant, but mutations in both the 16S rRNA and rpsE genes have been reported to confer high-level resistance to spectinomycin in P. multocida (Kehrenberg and Schwarz, 2007). Incidentally, aadA25 was strongly associated with gentamicin resistance in M. haemolytica, and identified in 96.7% of gentamicin-resistant isolates. Although the strA, strB, and aphA1 genes were all present in M. haemolytica and P. multocida neomycin–and gentamicin-resistant isolates, they were more often (>85%) associated with neomycin resistance.
Despite the fact that multiple ARGs that typically confer resistance to aminoglycosides were detected, no single gene was found to be 100% associated with any of the resistance phenotypes. As a result, it is probable that another aminoglycoside resistance gene is also contributing to the resistance profiles seen; possibly the newly described aadA31 (Cameron et al., 2018) or aadA15 (Clawson et al., 2016) genes that have been recently reported in M. haemolytica.
Relationships Between Multidrug Resistance Profiles and Metadata Categories
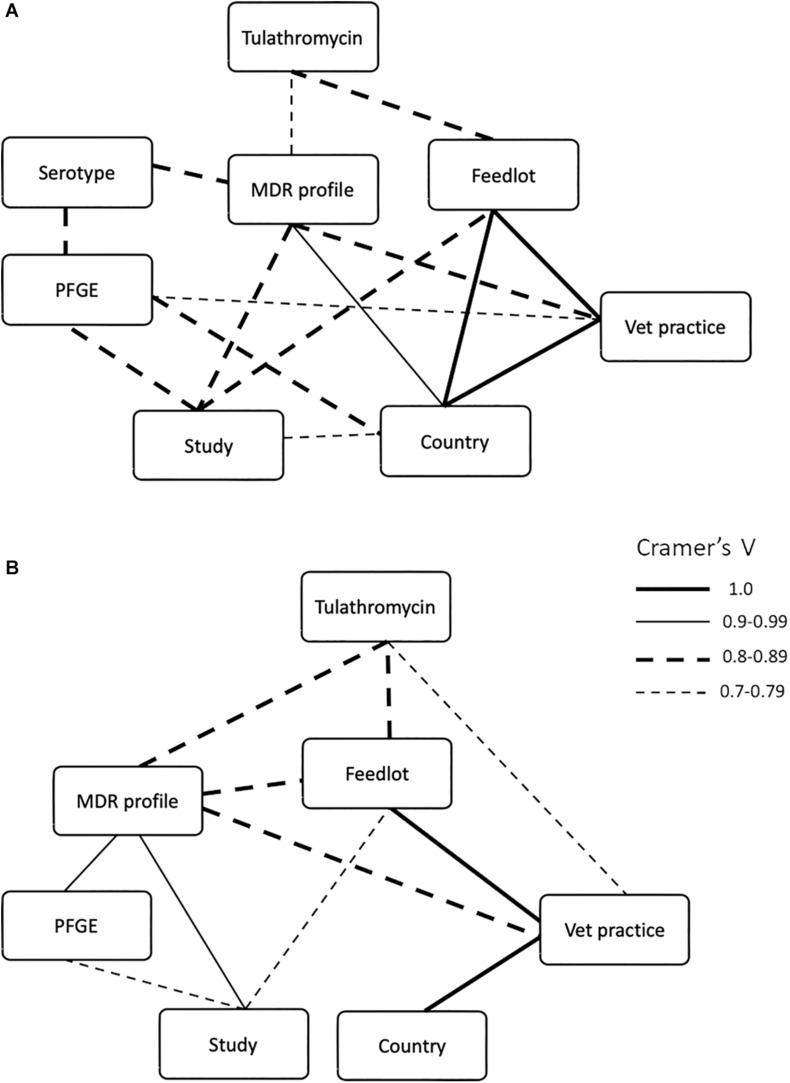
Metadata, including feedlot, country, submitting veterinary clinic, diagnosis, multidrug resistance profile, PFGE profile, serotype (for M. haemolytica), and treatment, were collected for samples from all three studies and the strength of association among these nominal (categorical) data was determined using Cramer’s V (Figure 5). Because these isolates were collected from naturally occurring mortality cases and are specifically the MDR subset (representing anywhere from 0 to 100% of isolates collected depending on location), there is a skew in the number of strains recovered from the different feedlots examined (Supplementary Table S1). Consequently, the statistical analyses used were designed to examine data of this type, specifically non-parametric nominal data. As might be expected of bacterial populations existing in closed environments, strong associations were observed in both bacterial species between multidrug resistance profile and any factor associated with the origin of the isolates, including year, country of isolate origin, feedlot, or the veterinary practice that collected the sample. For M. haemolytica, the multidrug resistance profile was strongly associated with study (Cramer’s V = 0.841, P = 0.0004), country of origin (Cramer’s V = 0.961, P = 0.0004), veterinary practice (Cramer’s V = 0.850, P = 0.0004), serotype (Cramer’s V = 0.845, P = 0.0166), and the use of tulathromycin (Cramer’s V = 0.796, P = 0.0004) (Figure 5A). For P. multocida, the multidrug resistance profile was strongly associated with the study (Cramer’s V = 0.915, P = 0.0007), feedlot (Cramer’s V = 0.891, P = 0.0007), veterinary practice (Cramer’s V = 0.887, P = 0.0007), PFGE profile (Cramer’s V = 0.917, P = 0.0007) and the use of tulathromycin (Cramer’s V = 0.803, P = 0.0092) (Figure 5B). Although not exclusive, most M. haemolytica isolates were collected from cattle with a diagnosis of fibrinous pneumonia (67%), while P. multocida isolates were most often collected from bronchopneumonia (40%) cases (Supplementary Table S4).
FIGURE 5.
Significant (Cramer’s V ≥ 0.7, P ≤ 0.05) associations between multidrug resistance profiles of (A) Mannheimia haemolytica and (B) Pasteurella multocida isolates with feedlot, country of origin, submitting veterinary clinic, diagnosis, PFGE profile, serotype (for M. haemolytica), and treatment. Weight of the lines correspond to Cramer’s V values.
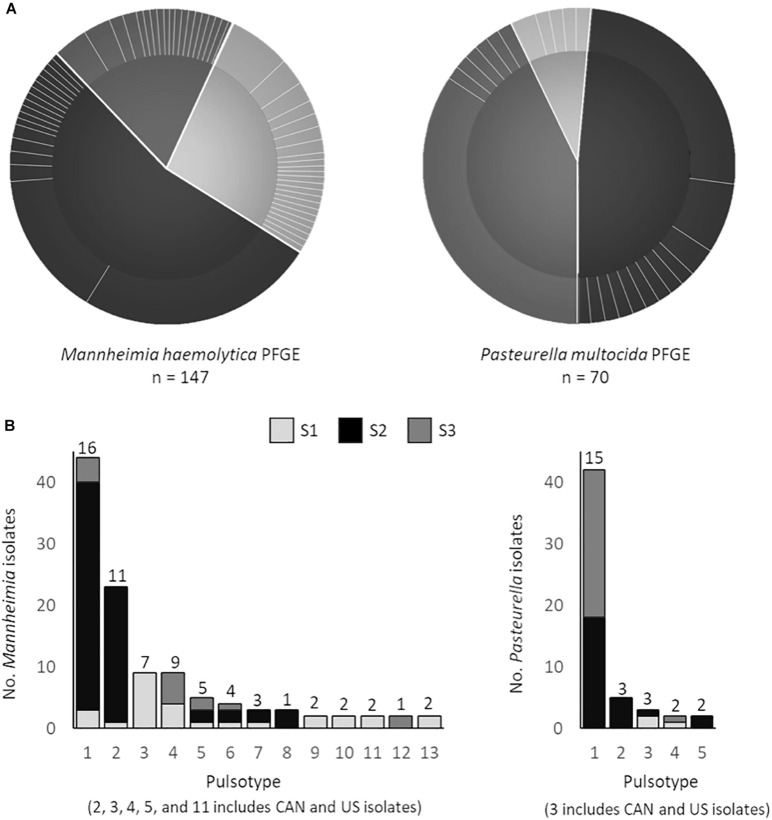
The PFGE data, which reflects overall genetic content rather than only multidrug resistance profile highlight less geographic-based differences than the correlation statistics indicate. Of the 21 unique pulsotypes identified in P. multocida, a single pulsotype was observed in 60% of all isolates originating from 13 feedlots. Furthermore, of the 53 unique pulsotypes in M. haemolytica, two pulsotypes accounted for 45% of all M. haemolytica isolates, from 13 to 7 feedlots, respectively (Figure 6A). Five of the thirteen M. haemolytica pulsotypes found in multiple locations were detected in both the United States and Canada (Figure 6B). None of the P. multocida pulsotypes were shared between the United States and Canada, however, the total number of isolates examined was half that of M. haemolytica. The three M. haemolytica pulsotypes found in all three studies and the three pulsotypes of P. multocida that span two studies, indicate that MDR Pasteurellaceae are widely disseminated within the cattle populations investigated. However, whole genome sequence analysis will be required to determine if the same strains are present across these times and distances.
FIGURE 6.
(A) Pulsed-field gel electrophoresis (PFGE) profiles of Mannheimia haemolytica (SalI) and Pasteurella multocida (ApaI) isolates. Pie charts depict the number of unique pulsotypes (>95% similarity) per study (study 1 M. haemolytica = 40, P. multocida = 6; study 2 M. haemolytica = 80, P. multocida = 34; study 3 M. haemolytica = 27, P. multocida = 34) and the proportion of isolates with each pulsotype. (B) PFGE profiles that contain strains from multiple locations. Numbers above each bar represent the number of unique antimicrobial susceptibility profiles within each unique PFGE profile.
Of note, there was no association between pulsotype (PFGE) and multidrug resistance profile (Figure 5), indicating that isolates with similar genotypes may contain MGEs that carry different ARGs. As a result, genotype based on PFGE is likely not a good metric to track the spread of AMR associated with MGEs. Whole genome sequencing is becoming standard practice for surveillance studies, but where funds or infrastructure for genomics analysis is limited, diagnostics that trace the movement of unique MGEs may be more helpful in assessing AMR spread compared to strain typing.
Multidrug Resistance and M. haemolytica Serotype
An association was found between multidrug resistance profile and serotype in the M. haemolytica isolates (Cramer’s V = 0.8453, P = 0.0082; Figure 5A), with 91.8% of the isolates being serotype 1 (Supplementary Table S5). Although the correlation between serovar and PFGE profile indicates that serovar is linked with genotype (Figure 5A), an overlap in the multidrug resistance profile between serotype 1 and serotype 6 isolates (five out of 45 multidrug resistance profiles) suggests that MGEs likely transfer between serovars and that genotype is not a strong correlate of AMR profile.
Of the three serovars of M. haemolytica commonly found in cattle, serotype 2 is considered less virulent and is often isolated from the nasopharynx of healthy cattle (Klima et al., 2014a). Although not typically targeted by antimicrobial therapy, serotype 2 strains were resistant to six and 10 drugs, respectively. This suggests that serotype 2 strains could act as a reservoir of ARGs in healthy cattle populations even though they are not directly implicated in BRD. As serotype 2 strains are considered to be more prevalent in healthy cattle, they may make a larger contribution to the respiratory tract resistome than do serotypes 1 and 6.
Antimicrobial Resistance in M. haemolytica and P. multocida Associated With Antimicrobial Use
Of the veterinary clinics that submitted samples, all but one was willing to provide therapeutic drug administration data. The remaining clinic was willing to provide how many times an animal was treated but not the specific drugs or dosages used. No information was available from any of the feedlots regarding the antimicrobials that were administered infeed. However, it is reasonable to assume, given standard industry practices at the time of these studies, that the majority of these animals had either tylosin and/or chlortetracycline included in their diets. Although previous work has shown a relationship between the frequency of antimicrobial exposure and the amount of AMR observed in M. haemolytica and P. multocida (Magstadt et al., 2018), no strong associations were observed between multidrug resistance and the number of treatments each animal received (M. haemolytica; Cramer’s V = 0.396, P = 0.048; P. multocida; Cramer’s V = 0.3307, P > 0.05). In fact, there were multiple cases where isolates resistant to four or five antimicrobials were recovered from animals treated up to four times, whereas isolates resistant to 12 to 15 antimicrobials were recovered from cattle that received only one to three antimicrobial treatments (Table 4). There was also no strong association between the individual antimicrobial used and the year of sample collection (Cramer’s V < 0.5).
TABLE 4.
Percentage (No.) of cattle that had been treated multiple times as a result of bovine respiratory disease (BRD).
| Percentage of BRD cases by number of antimicrobials treated (No. animals) |
|||||||
| Bacterial species isolated | None | Once | Twice | Three | Four | Five | Six |
| Mannheimia haemolytica | 33 (48) | 39 (58) | 17 (25) | 7 (11) | 3 (4) | 1 (1) | – |
| Pasteurella multocida | 34 (24) | 21 (15) | 11 (8) | 7 (5) | 13 (9) | 10 (7) | 3 (2) |
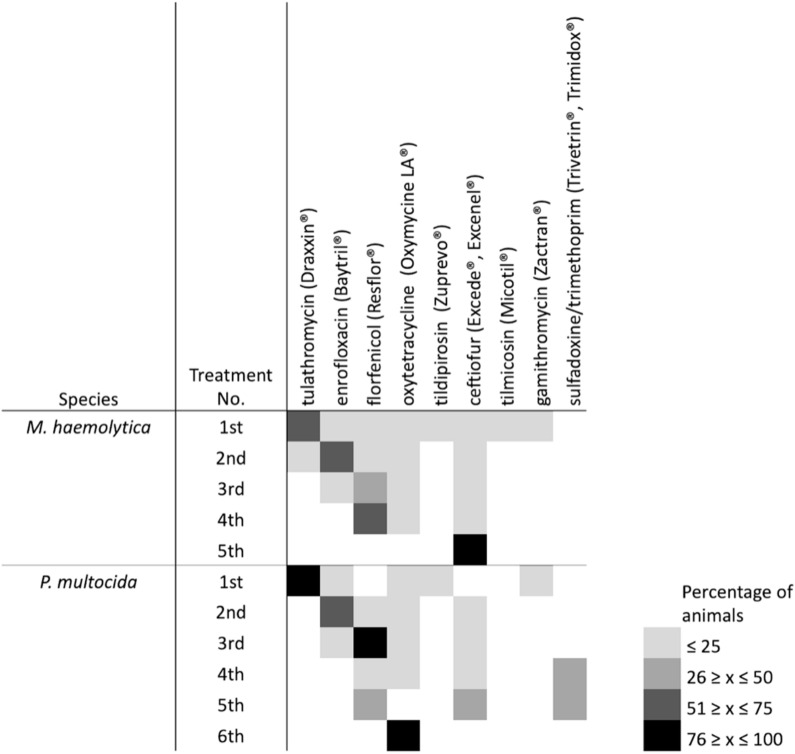
Over 65% of mortalities sampled were from cattle that had received at least one antimicrobial treatment, with the majority being treated only once (Table 5). Tulathromycin, enrofloxacin, florfenicol, ceftiofur and oxytetracycline were the antimicrobials most commonly administered (Figure 7), with tulathromycin most often given as a first-line treatment, enrofloxacin second, and florfenicol third. Of the cattle that were treated multiple times, ceftiofur, florfenicol, or oxytetracycline were most frequently used. The aforementioned antimicrobials were most commonly administered to cattle from which M. haemolytica was isolated, while a combination of trimethoprim-sulfadoxine or oxytetracycline were most often administered to cattle from which P. multocida was isolated. No associations were found between gross pathology (i.e., bronchointerstitial pneumonia vs. fibrinous pleuritis) and the antimicrobials administered, suggesting that any form of respiratory illness, regardless of its presentation, is likely managed similarly, among different feedlots.
TABLE 5.
Percentage (No.) of Mannheimia haemolytica and Pasteurella multocida isolates from bovine respiratory disease mortalities that are multidrug resistant by number of antimicrobial treatments the animal received.
| Percentage of isolates (no. isolates) with drug count of multidrug resistance profile |
||||||||||||||
| Species | Treatment No. | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 15 | 16 |
| M. haemolytica | None | 4 (2) | 15 (7) | 19 (9) | 15 (7) | 17 (8) | 2 (1) | 2 (1) | 8 (4) | 13 (6) | – | – | – | 6 (3) |
| First | 3 (2) | 7 (4) | 9 (5) | 5 (3) | 52 (30) | 10 (6) | 2 (1) | 5 (3) | 3 (2) | – | 2 (1) | 2 (1) | – | |
| Second | 8 (2) | 12 (3) | 8 (2) | – | 36 (9) | 12 (3) | 12 (3) | 4 (1) | 4 (1) | – | – | 4 (1) | – | |
| Third | – | 18 (2) | – | – | 18 (2) | 9 (1) | 9 (1) | – | – | 9 (1) | 9 (1) | 27 (3) | – | |
| Fourth | – | 25 (1) | – | 50 (2) | 25 (1) | – | – | – | – | – | – | – | – | |
| Fifth | 100 (1) | – | – | – | – | – | – | – | – | – | – | – | – | |
| P. multocida | None | – | 4 (1) | 4 (1) | 4 (1) | 13 (3) | 21 (5) | 25 (6) | 17 (4) | 8 (2) | 4 (1) | – | ||
| First | – | 7 (1) | 7 (1) | – | – | 7 (1) | 27 (4) | 33 (5) | 7 (1) | 13 (2) | – | |||
| Second | 25 (2) | – | 13 (1) | – | – | – | 13 (1) | 38 (3) | – | – | 13 (1) | |||
| Third | – | – | – | – | – | – | 20 (1) | 40 (2) | – | – | 40 (2) | |||
| Fourth | 11 (1) | – | – | – | – | – | 11 (1) | 56 (5) | 22 (2) | – | – | |||
| Fifth | – | – | – | – | – | – | 43 (3) | 43 (3) | 14 (1) | – | – | |||
| Sixth | – | – | – | – | – | – | – | 50 (1) | 50 (1) | – | – | |||
FIGURE 7.
Heatmap of the antimicrobial treatment data for animals examined post-mortem after having died of bovine respiratory disease. Box color indicates overall percentage of animals that received an antimicrobial based on the data that was provided by veterinary clinics.
For MDR M. haemolytica isolates, strong associations were observed between multidrug resistance profile and usage of ceftiofur (Cramer’s V = 0.7172, P = 0.0423) and tulathromycin (Cramer’s V = 0.7019, P = 0.0004), with weaker associations found between multidrug resistance profile and the use of florfenicol (Cramer’s V = 0.6765, P = 0.0288). Strong associations were only observed in MDR P. multocida with tulathromycin use (Cramer’s V = 0.8025, P = 0.0129).
Perspectives on Antimicrobial Resistance in Beef Cattle
A key function of AMR surveillance is to assess risks associated with antimicrobial use in food production settings. However, beef production systems vary widely. Feedlots can range significantly in size (3,000 to 100,000 head), how animals are procured (auction vs. direct sourced), the risk category of animals that operators are willing to purchase, the distance cattle are shipped, drug regimens used to prevent and treat disease, and how animals are commingled and moved within the feedlot environment. As a result, the information obtained here cannot effectively address how antimicrobial use may be affecting AMR development and spread in all types of beef production.
What the data do support is that possible XDR M. haemolytica and P. multocida are prevalent in mortality cases associated with BRD in North America. Despite not being frequently observed in BRD cases prior to Lubbers and Hanzlicek (2013), the AMR profiles are both diverse and persistent. The increased multidrug resistance observed during the 7 years of study, in combination with the abundance of ICE-associated genes present in M. haemolytica and P. multocida, suggests that ICE carrying ARGs are likely contributing to treatment failures in cases of respiratory infection in North American cattle. The linkage between the use of tulathromycin, MDR, and ICEs likely reflects the extensive metaphylactic use of this antimicrobial in Canadian feedlots (Brault et al., 2019) Further examination is warranted to determine if ICEs are arising independently through recombination with plasmids and other MGEs, or if the majority of the AMR observed is due to a common element that is transferred and then modified through gene loss or uptake dependent upon selective pressures. The selective pressures promoting ICE spread are also not known. The diversity in both AMR phenotype and the ARGs detected indicate that there is likely more genetic diversity than what has been previously described in ICE from Pasteurellaceae spp.
Conclusion
This study suggests that between 2011 and 2016 there was a significant increase in the prevalence of MDR isolates of M. haemolytica and P. multocida associated with BRD. Furthermore, the AMR profiles observed were diverse and persistent over time. Given the presence of multiple genes associated with both functional gene coding regions and accessory gene cassettes previously reported in ICEs from Pasteurellaceae, it is likely that similar ICEs are abundant in the populations examined. The complexity of the multidrug resistance profiles and the lack of correlation with genotype suggests that horizontal gene transfer is contributing to the spread of multidrug resistance in Pasteurellaceae spp. in cattle. Interestingly, the occurrence of resistance to more than seven antimicrobials in M. haemolytica decreased over time but increased in P. multocida indicating that the dynamics of multidrug resistance spread may differ amongst species that contribute to BRD. There was no correlation with number of antimicrobial treatments individual animals received and the number or types of drugs to which isolates were resistant. A multicopper oxidase gene was prevalent among the isolates and may be a target of selection pressure and play a role in ICE maintenance and spread in feedlots. To address concerns over multidrug resistance spread in beef production in North America, more information regarding the diversity, prevalence, and host range of ICEs in Pasteurellaceae is required. To gain a greater understanding of ICEs within these populations, including how they are developing and spreading will likely require combined efforts amongst industry, veterinarians, producers, and researchers.
Ethics Statement
The animal study was reviewed and approved by Lethbridge Research Centre Animal Care Committee. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author Contributions
CK and TM designed the experiment BR, CB, SH, NA, RM, KS, and TM provided the study samples and metadata. SC, CC, and YN supported the technical aspects of the study and carried out experiments. CK carried out the experiments and wrote the manuscript. CK and DH analyzed the data. All authors revised and approved the manuscript to be published.
Conflict of Interest
CB is part owner and managing partner of Feedlot Health Management Services Ltd. and Southern Alberta Veterinary Services Ltd. SH is an employee of Feedlot Health Management Services Ltd. RM is owner of POV Inc. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Susanne Trapp and Yidong Graham for technical support.
Footnotes
Funding. The project was funded in part by the Beef Cattle Research Council and the Alberta Beef Producers (ANH.09.16). Additional funds were provided by the Agriculture and Agri-Food Canada Growing Forward II, the Government of Canada AMR Genomic Research and Development Initiative, and by a grant provided by the Alberta Livestock and Meat Agency (2015R025) to investigate ICE associated with BRD. Funders had no input into the design, analysis, writing of the manuscripts or the overall conclusion that were derived from the study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.606438/full#supplementary-material
References
- Anholt R. M., Klima C., Allan N., Matheson-Bird H., Schatz C., Ajitkumar P., et al. (2017). Antimicrobial susceptibility of bacteria that cause bovine respiratory disease complex in Alberta, Canada. Front. Vet. Sci. 4:207. 10.3389/fvets.2017.00207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antunes N. T., Lamoureaux T. L., Toth M., Stewart N. K., Frase H., Vakulenko S. B. (2014). Class D β-lactamases: are they all carbapenemases? Antimicrob. Agents Chemother. 58 2119–2125. 10.1128/AAC.02522-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armshaw P., Pembroke J. T. (2013). “Integrative conjugative elements (ICEs) of the SXT/R391 group as vehicles for acquisition of resistance determinants, stable maintenance and transfer to a wide range of enterobacterial pathogens,” in Microbial Pathogens and Strategies for Combating Them: Science, Technology and Education, ed. Méndez-Vilas A. (Badajoz: Formatex Research Center; ), 439–446. [Google Scholar]
- Beker M., Rose S., Lykkebo C. A., Douthwaite S. (2018). Integrative and conjugative elements (ICEs) in Pasteurellaceae species and their detection by multiplex PCR. Front. Microbiol. 9:1329. 10.3389/fmicb.2018.01329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt K., Timsit E., Rawlyk N., Potter A., Liljebjelke K. (2018). Integrative Conjugative Element ICEHs1 encodes for antimicrobial resistance and metal tolerance in Histophilus somni. Front. Vet. Sci. 5:153. 10.3389/fvets.2018.00153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booker C. W., Abutarbush S. M., Morley P. S., Jim G. K., Pittman T. J., Schunicht O. C., et al. (2008). Microbiological and histopathological findings in cases of fatal bovine respiratory disease of feedlot cattle in Western Canada. Can. Vet. J. 49 473–481. [PMC free article] [PubMed] [Google Scholar]
- Brault S., Hannon S., Gow S., Warr B., Withell J., Song J., et al. (2019). Antimicrobial use on 36 beef feedlots in Western Canada: 2008-2012. Front. Vet. Sci. 6:329. 10.3389/fvets.2019.00329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron A., Klima C. L., Ha R., Gruninger R. J., Zaheer R., Mcallister T. A. (2018). A novel aadA aminoglycoside resistance gene in bovine and porcine pathogens. mSphere 3:e00568-17. 10.1128/mSphere.00568-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Card R. M., Stubberfield E., Rogers J., Nunez-Garcia J., Ellis R. J., Abuoun M., et al. (2018). Identification of a new antimicrobial resistance gene provides fresh insights into pleuromutilin resistance in Brachyspira hyodysenteriae, aetiological agent of swine dysentery. Front. Microbiol. 9:1183. 10.3389/fmicb.2018.01183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clawson M. L., Murray R. W., Sweeney M. T., Apley M. D., Dedonder K. D., Capik S. F., et al. (2016). Genomic signatures of Mannheimia haemolytica that associate with the lungs of cattle with respiratory disease, an integrative conjugative element, and antibiotic resistance genes. BMC Genomics 17:982. 10.1186/s12864-016-3316-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute [CLSI] (2008). Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Collected From Animals; Approved Standard. CLSI Document M31-A3, 3rd Edn Wayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- Davies R. L., Maccorquodale R., Baillie S., Caffrey B. (2003). Characterization and comparison of Pasteurella multocida strains associated with porcine pneumonia and atrophic rhinitis. J. Med. Microbiol. 52 59–67. 10.1099/jmm.0.05019-0 [DOI] [PubMed] [Google Scholar]
- Desmolaize B., Rose S., Wilhelm C., Warrass R., Douthwaite S. (2011). Combinations of macrolide resistance determinants in field isolates of Mannheimia haemolytica and Pasteurella multocida. Antimicrob. Agents Chemother. 55 4128–4133. 10.1128/AAC.00450-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eidam C., Poehlein A., Leimbach A., Michael G. B., Kadlec K., Liesegang H., et al. (2015). Analysis and comparative genomics of ICEMh1, a novel integrative and conjugative element (ICE) of Mannheimia haemolytica. J. Antimicrob. Chemother. 70 93–97. 10.1093/jac/dku361 [DOI] [PubMed] [Google Scholar]
- Government of Canada (2009). Categorization of Antimicrobial Drugs Based on Importance in Human Medicine [Online]. Available online at: https://www.canada.ca/en/health-canada/services/drugs-health-products/veterinary-drugs/antimicrobial-resistance/categorization-antimicrobial-drugs-based-importance-human-medicine.html (accessed on 12 September 2020) [Google Scholar]
- Government of Canada (2018). Canadian Integrated Program for Antimicrobial Resistance Surveillance (CIPARS) 2016 annual report [Online]. Ottawa, ON: Public Health Agency of Canada. [Google Scholar]
- Hansen L. M., Blanchard P. C., Hirsh D. C. (1996). Distribution of tet(H) among Pasteurella isolates from the United States and Canada. Antimicrob. Agents Chemother. 40 1558–1560. 10.1128/AAC.40.6.1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton W. M. (2014). BRD in 2014: where have we been, where are we now, and where do we want to go? Anim. Health Res. Rev. 15 120–122. 10.1017/S1466252314000115 [DOI] [PubMed] [Google Scholar]
- Johnson C. M., Grossman A. D. (2015). Integrative and conjugative elements (ICEs): what they do and how they work. Annu. Rev. Genet. 49 577–601. 10.1146/annurev-genet-112414-055018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadlec K., Watts J. L., Schwarz S., Sweeney M. (2018). Plasmid-located extended-spectrum β-lactamase gene blaROB-2 in Mannheimia haemolytica. J. Antimicrob. Chemother. 74 851–853. 10.1093/jac/dky515 [DOI] [PubMed] [Google Scholar]
- Kehrenberg C., Catry B., Haesebrouck F., De Kruif A., Schwarz S. (2005). tet(L)-mediated tetracycline resistance in bovine Mannheimia and Pasteurella isolates. J. Antimicrob. Chemother. 56 403–406. 10.1093/jac/dki210 [DOI] [PubMed] [Google Scholar]
- Kehrenberg C., Salmon S. A., Watts J. L., Schwarz S. (2001). Tetracycline resistance genes in isolates of Pasteurella multocida, Mannheimia haemolytica, Mannheimia glucosida and Mannheimia varigena from bovine and swine respiratory disease: intergeneric spread of the tet(H) plasmid pMHT1. J. Antimicrob. Chemother. 48 631–640. 10.1093/jac/48.5.631 [DOI] [PubMed] [Google Scholar]
- Kehrenberg C., Schwarz S. (2005). dfrA20, A novel trimethoprim resistance gene from Pasteurella multocida. Antimicrob. Agents Chemother. 49 414–417. 10.1128/AAC.49.1.414-417.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehrenberg C., Schwarz S. (2007). Mutations in 16S rRNA and ribosomal protein S5 associated with high-level spectinomycin resistance in Pasteurella multocida. Antimicrob. Agents Chemother. 51 2244–2246. 10.1128/AAC.00229-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klima C., Alexander T., Read R., Gow S., Booker C., Hannon S., et al. (2011). Genetic characterization and antimicrobial susceptibility of Mannheimia haemolytica isolated from the nasopharynx of feedlot cattle. Vet. Microbiol. 149 390–398. 10.1016/j.vetmic.2010.11.018 [DOI] [PubMed] [Google Scholar]
- Klima C. L., Alexander T. W., Hendrick S., Mcallister T. A. (2014a). Characterization of Mannheimia haemolytica isolated from feedlot cattle that were healthy or treated for bovine respiratory disease. Can. J. Vet. Res. 78 38–45. [PMC free article] [PubMed] [Google Scholar]
- Klima C. L., Zaheer R., Cook S. R., Booker C. W., Hendrick S., Alexander T. W., et al. (2014b). Pathogens of bovine respiratory disease in North American feedlots conferring multidrug resistance via integrative conjugative elements. J. Clin. Microbiol. 52 438–448. 10.1128/JCM.02485-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klima C. L., Cook S. R., Zaheer R., Laing C., Gannon V. P., Xu Y., et al. (2016). Comparative genomic analysis of Mannheimia haemolytica from bovine sources. PLoS One 11:e0149520. 10.1371/journal.pone.0149520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klima C. L., Holman D. B., Ralston B. J., Stanford K., Zaheer R., Alexander T. W., et al. (2019). Lower respiratory tract microbiome and resistome of bovine respiratory disease mortalities. Microb. Ecol. 78 446–456. 10.1007/s00248-019-01361-3 [DOI] [PubMed] [Google Scholar]
- Kwon K. H., Hwang S. Y., Moon B. Y., Park Y. K., Shin S., Hwang C. Y., et al. (2012). Occurrence of antimicrobial resistance and virulence genes, and distribution of enterococcal clonal complex 17 from animals and human beings in Korea. J. Vet. Diagn. Invest. 24 924–931. 10.1177/1040638712455634 [DOI] [PubMed] [Google Scholar]
- Lubbers B. V., Hanzlicek G. A. (2013). Antimicrobial multidrug resistance and coresistance patterns of Mannheimia haemolytica isolated from bovine respiratory disease cases–a three-year (2009-2011) retrospective analysis. J. Vet. Diagn. Invest. 25 413–417. 10.1177/1040638713485227 [DOI] [PubMed] [Google Scholar]
- Magstadt D. R., Schuler A. M., Coetzee J. F., Krull A. C., O’connor A. M., Cooper V. L., et al. (2018). Treatment history and antimicrobial susceptibility results for Mannheimia haemolytica, Pasteurella multocida, and Histophilus somni isolates from bovine respiratory disease cases submitted to the Iowa State University Veterinary Diagnostic Laboratory from 2013 to 2015. J. Vet. Diagn. Invest. 30 99–104. 10.1177/1040638717737589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael G. B., Kadlec K., Sweeney M. T., Brzuszkiewicz E., Liesegang H., Daniel R., et al. (2012a). ICEPmu1, an integrative conjugative element (ICE) of Pasteurella multocida: analysis of the regions that comprise 12 antimicrobial resistance genes. J. Antimicrob. Chemother. 67 84–90. 10.1093/jac/dkr406 [DOI] [PubMed] [Google Scholar]
- Michael G. B., Kadlec K., Sweeney M. T., Brzuszkiewicz E., Liesegang H., Daniel R., et al. (2012b). ICEPmu1, an integrative conjugative element (ICE) of Pasteurella multocida: structure and transfer. J. Antimicrob. Chemother. 67 91–100. 10.1093/jac/dkr411 [DOI] [PubMed] [Google Scholar]
- Mollenkopf D. F., Stull J. W., Mathys D. A., Bowman A. S., Feicht S. M., Grooters S. V., et al. (2017). Carbapenemase-producing Enterobacteriaceae recovered from the environment of a swine farrow-to-finish operation in the United States. Antimicrob. Agents Chemother. 61: e01298-16. 10.1128/AAC.01298-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemann L., Feudi C., Eichhorn I., Hanke D., Muller P., Brauns J., et al. (2019). Plasmid-located dfrA14 gene in Pasteurella multocida isolates from three different pig-producing farms in Germany. Vet. Microbiol. 230 235–240. 10.1016/j.vetmic.2019.01.016 [DOI] [PubMed] [Google Scholar]
- Olsen A. S., Warrass R., Douthwaite S. (2015). Macrolide resistance conferred by rRNA mutations in field isolates of Mannheimia haemolytica and Pasteurella multocida. J. Antimicrob. Chemother. 70 420–423. 10.1093/jac/dku385 [DOI] [PubMed] [Google Scholar]
- Rainbolt S., Pillai D. K., Lubbers B. V., Moore M., Davis R., Amrine D., et al. (2016). Comparison of Mannheimia haemolytica isolates from an outbreak of bovine respiratory disease. Vet. Microbiol. 182 82–86. 10.1016/j.vetmic.2015.10.020 [DOI] [PubMed] [Google Scholar]
- Shen Z., Qu W., Wang W., Lu Y., Wu Y., Li Z., et al. (2010). MPprimer: a program for reliable multiplex PCR primer design. BMC Bioinform. 11:143. 10.1186/1471-2105-11-143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder E., Credille B., Berghaus R., Giguere S. (2017). Prevalence of multi drug antimicrobial resistance in isolated from high-risk stocker cattle at arrival and two weeks after processing. J. Anim. Sci. 95 1124–1131. 10.2527/jas.2016.1110 [DOI] [PubMed] [Google Scholar]
- Stanford K., Zaheer R., Klima C., Mcallister T., Peters D., Niu Y. D., et al. (2020). Antimicrobial resistance in members of the bacterial bovine respiratory disease complex isolated from lung tissue of cattle mortalities managed with or without the use of antimicrobials. Microorganisms 8:288. 10.3390/microorganisms8020288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timsit E., Hallewell J., Booker C., Tison N., Amat S., Alexander T. W. (2017). Prevalence and antimicrobial susceptibility of Mannheimia haemolytica, Pasteurella multocida, and Histophilus somni isolated from the lower respiratory tract of healthy feedlot cattle and those diagnosed with bovine respiratory disease. Vet. Microbiol. 208 118–125. 10.1016/j.vetmic.2017.07.01 [DOI] [PubMed] [Google Scholar]
- Woolums A. R., Karisch B. B., Frye J. G., Epperson W., Smith D. R., Blanton J., et al. (2018). Multidrug resistant Mannheimia haemolytica isolated from high-risk beef stocker cattle after antimicrobial metaphylaxis and treatment for bovine respiratory disease. Vet. Microbiol. 221 143–152. 10.1016/j.vetmic.2018.06.005 [DOI] [PubMed] [Google Scholar]
- Wu J. R., Shieh H. K., Shien J. H., Gong S. R., Chang P. C. (2003). Molecular characterization of plasmids with antimicrobial resistant genes in avian isolates of Pasteurella multocida. Avian. Dis. 47 1384–1392. 10.1637/z7035 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.