Abstract
High-altitude pulmonary edema occurs most frequently in non-acclimatized low landers on exposure to altitude ≥2500 m. High-altitude pulmonary edema is a complex condition that involves perturbation of signaling pathways in vasoconstrictors, vasodilators, anti-diuretics, and vascular growth factors. Genetic variations are instrumental in regulating these pathways and evidence is accumulating for a role of epigenetic modification in hypoxic responses. This review focuses on the crosstalk between high-altitude pulmonary edema-associated genetic variants and transcription factors, comparing high-altitude adapted and high-altitude pulmonary edema-afflicted subjects. This approach might ultimately yield biomarker information both to understand and to design therapies for high-altitude adaptation.
Keywords: genetics, epigenetics, high-altitude adaptation
Evolution and physiological adaptation have permitted survival at the highest topographically elevated regions of the world.1–5 Reduced air pressure at high-altitude decreases the partial pressure of inspired oxygen, affecting lungs, brain, heart, and blood and can lead to a spectrum of high-altitude disorders including high-altitude pulmonary edema (HAPE), acute mountain sickness, and high-altitude cerebral edema.6,7 This review focuses on HAPE. HAPE is a consequence of hypoxic pulmonary vasoconstriction leading to increased pulmonary arterial pressure and capillary stress failure.8–11 HAPE victims have lower arterial oxygen saturation (SaO2) and higher heart rate, pulmonary vascular resistance, and pulmonary vascular resistance index12,13 than do unaffected sojourners to altitude. Clinically, HAPE is characterized by dyspnea, elevated body temperature, pink frothy sputum, tachypnea, tachycardia, persistent cough, and cyanosis.14–17 Chest X-rays and CT scans show increased lung vascular markings and patchy shadows.18,19 There have been great strides in understanding the clinical and physiological mechanisms of HAPE that has led to the discovery of successful treatments. Information on genetic contributions to this disorder has also grown rapidly over the last decade, partly to the development and implementation of newer genetic techniques.
Candidate gene approaches, advanced techniques such as Next-Generation Sequencing and Genome-Wide Association Studies have led to association of multiple genetic variants with high-altitude adaptation or maladaptation.20–26 The majority of these genes belong to multiple, frequently related pathways. These include the renin–angiotensin–aldosterone system, apelin signaling, nitric oxide (NO) signaling, and hypoxia-induced signaling. These pathways regulate vasoactive molecules including angiotensin II (ANG II), apelin, NO, aldosterone, and beta-adrenergics.27–31 In addition to genetic variation, epigenetics plays prominent role in HAPE and other diseases.32–35 DNA methylation, acetylation, histone modifications/chromatin remodeling, and post translational RNA regulations are increasingly being recognized as mediators of crosstalk between genes and environment36,37 paving the way to epigenetic-based therapeutics.38,39 Interestingly, the Encyclopedia of DNA elements project consortium has mapped active transcription sites with an aim to identify the functional elements in the human genome.40 Genetic variants that alter the binding site of transcription factors (TFs) are increasingly being identified and are associated with epigenetics.41,42 It is now known that most of the genome is likely regulatory, and TFs play a crucial role in its recognition and defining the function that may again be in favor or against normal physiological signaling.43 It is here that the relevance of TFs in diseases becomes integral. In fact, last few years have seen increasing efforts being put in to understanding the TF-mediated gene regulatory mechanisms. These efforts also highlighted the synchronization between the TFs and methylation; a synchronization that works in tandem to regulate and define a function.35,44 Altered TF binding results in differential gene expression which brings out phenotypic differences in associated diseases.35,45,46 This review will attempt to integrate genetics, TFs, and the molecular regulation of vascular homeostasis in HAPE. We first summarize the signaling pathways, the associated genetic variants, and the signaling molecules, followed by allele-specific transcriptional regulation by TFs. The overall purpose is to highlight the cellular crosstalk between HAPE-associated genetic variants and the TFs. We expect this will ultimately lead to biomarker identification and to focus on development of improved therapeutics.
Renin–angiotensin–aldosterone system-mediated allele-specific TF binding and salt sensitivity contribute to vascular dysfunction
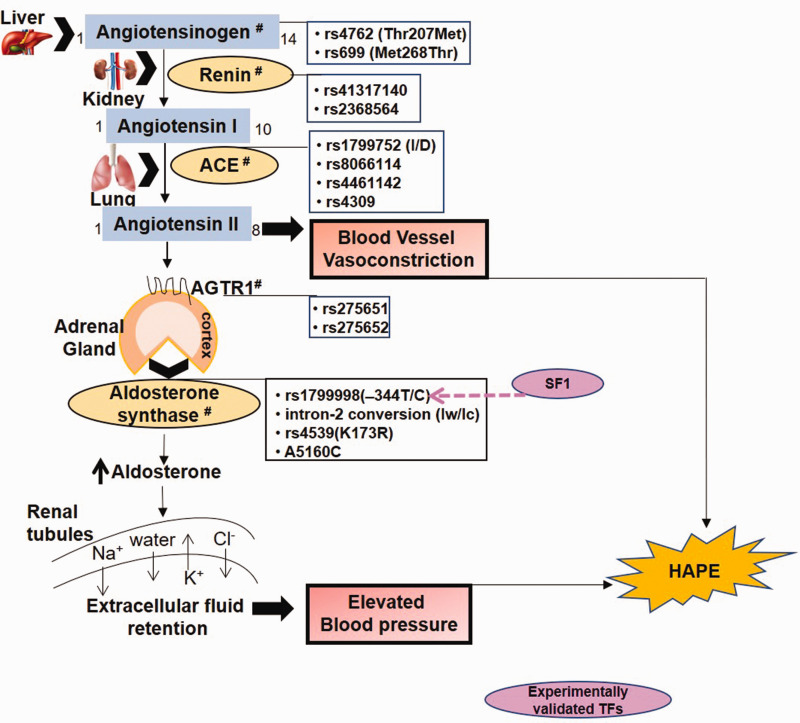
The renin–angiotensin–aldosterone system (RAAS) involves renin-mediated conversion of angiotensinogen (AGT) to a decapeptide, angiotensin I (Fig. 1). The latter is converted into the vasoconstrictive, octapeptide ANG II by angiotensin-1 converting enzyme (ACE). In addition to vasoconstriction, ANG II, through the ANG II receptor (AGTR1), stimulates the adrenal cortex to secrete aldosterone, a major minerlocorticoid hormone, under the regulation of aldosterone synthase enzyme. Aldosterone acts on the mineralocorticoid receptors on the renal duct cells to increase the extracellular fluid retention via sodium chloride reabsorption and acts as an anti-diuretic, thereby regulating blood pressure and blood volume.47,48
Fig. 1.
The renin–angiotensin–aldosterone pathway regulates blood pressure and electrolyte balance in the body.
ACE: angiotensin-1 converting enzyme; AGTR1: angiotensin II receptor; SF1: steroidogenic transcription factor.
Note: Hash (#) represents HAPE-associated genetic variants.
Disturbance of the RAAS is attributed, in part, to genetic variants in at least five critical genes namely REN, AGT, ACE, AGTR1, and CYP11B2.20,49–53 Of these, the major genetic variants that have been associated with HAPE are rs4762 (Thr207Met) and rs699 (Met268Thr) of AGT,53,54 the insertion/deletion (indel) alleles (I/D, 287bp alu repeat sequence), rs8066114 and rs4461142 of ACE,20,50,51,53,55 rs275651 and rs275652 of AGTR1,56 and –344T/C and intron 2 conversion of CYP11B2 (Fig. 1 and Table 1).48,57 Whereas in case of the healthy native population, the homozygotes of major alleles of these genes such as the homozygotes rs4762CC of AGT, I/I of ACE1,20,58 and –344TT of CYP11B248 were associated with HA adaptation, but not without conflicts especially on ACE1 I/D polymorphism.13,59 These allelic variations in health and disease are also substantiated with varied circulating levels of the respective protein or enzyme levels. For example, the variants associated with increased circulating levels; where ACE, aldosterone, and sodium levels were increased in HAPE patients.20,50,60 Here, the D allele was associated with elevated activity of ACE; likewise, –344C allele of CYP11B2 was associated with elevated levels of aldosterone. Interestingly, however, ethnicity-based differences were also observed for few of the genes but in sealand population, such as the CYP11B2 –344T/C polymorphism associated with increased salt sensitivity in the Japanese,61 as the subjects with TT genotype displayed inappropriately higher aldosterone levels and systolic blood pressure in response to high salt intake. In addition to high-altitude adaptation and maladaptation, RAAS genetic variants have been extensively investigated in hypertension and cardiovascular diseases.52,62,63
Table 1.
Distribution of few significant SNPs in healthy controls and patients of HAPE.
| S no. | Gene | rs ID | Allele |
P-Value |
Significant genetic models | ||
|---|---|---|---|---|---|---|---|
| HAPE-p vs HAPE-f | HAPE-p vs HL | HL vs HAPE-f | |||||
| 1 | AGT | rs69955 | A/G | 0.05 | – | – | Co-dominant and additive model |
| rs476256 | G/A | – | 0.024 | 0.03 | Co-dominant and additive model | ||
| 2 | ACE | I/D20,50,53 | I/D | ≤0.05 | – | – | Co-dominant and additive model |
| rs806611457 | C/G | 0.04; 0.03 | – | – | Additive model; Dominant model | ||
| rs446114257 | T/C | 0.03 | – | – | Dominant model | ||
| 3 | AGTR1 | rs27565156 | T/A | 0.017 | – | – | Additive model |
| rs27565256 | T/G | 0.016 | – | – | Additive model | ||
| 4. | CYP11B2 | –344T/C48 | T/C | – | – | <0.0001 | Additive model |
| intron 2 conversion57 | intron 2 conversion | – | 0.03 | – | Co-dominant model | ||
| 5 | APLN | rs3761581 25 | T/G | 0.0027 | 3.9E-05 | – | Co-dominant and additive model |
| rs223531225 | C/T | 1.0E-06 | 1.2E-06 | – | Co-dominant and additive model | ||
| rs311575725 | C/G | 0.0032 | 0.04 | – | Co-dominant and additive model | ||
| 6 | APLNR | rs1154437425 | G/A | 0.004 | – | – | Co-dominant and additive model |
| rs228262325 | A/G | 0.013 | 1.0E-07 | 4.5E-05 | Co-dominant and additive model | ||
| 7 | NOS3 | rs179998321,25,80 | G/T | 0.03 | 1.2E-05 | – | Co-dominant and additive model |
| rs783025 | A/C | 1.6E-05 | 3.0E-06 | – | Co-dominant and additive model | ||
| 4b/4a21,25,80 | b/a | 0.0003 | 9.0E-07 | – | Co-dominant and additive model | ||
| Gene |
rs ID |
Allele |
Minor allele predominance |
||||
| 8 | EPAS1 | rs56721780104 | G/C | Absence of minor allele C in other world population except Tibetans | |||
| rs13419896105 | G/A | Predominance of A allele in Tibetans and Sherpas | |||||
| rs149594770106 | T/A | Absence of minor allele A in other world population except Tibetans | |||||
| 9 | EGLN1 | rs186996510107 | G/C | Absence of minor allele C in other world population except Tibetans | |||
| rs12097901107 | C/G | Absence of minor allele G in other world population except Tibetans | |||||
|
P-Value HAPE-p vs HAPE-f |
Significant genetic models | ||||||
| rs153866445 | T/C | P < 0.008 | Co-dominant and additive model | ||||
| rs47920045 | G/A/C | ||||||
| rs248672945 | C/G/T | ||||||
| rs279087945 | A/C | ||||||
| rs48090245 | T/C | ||||||
| rs248673645 | C/T | ||||||
| rs97325245 | A/G | ||||||
HAPE-p: HAPE-patients; HAPE-f: HAPE-free controls; HL: high-landers.
Variants of RAAS pathway are predicted to change the respective protein's physicochemical properties, secondary structure, and solvent accessibility, which in turn may affect the binding of a molecule with its target.64 Under the given conditions, allele-specific binding of TFs may amend gene expression, leading to phenotypic differences in several diseases.42,46 Of the several genetic variants of RAAS, the CYP11B2 variant –344T/C appears significant because of its interaction with a TF, namely steroidogenic TF (SF1).65 Stronger binding of SF1 to variant –344C, as confirmed by electrophoretic mobility shift assay (EMSA),65 was associated with elevated aldosterone levels in HAPE and hypertension.59,66 However, as conveyed already elsewhere above, this gene represents variations from population to populations, such as the T allele was also associated with SF1 factor and with high aldosterone levels and blood pressure.67,68 Such conflicts were attributed to sampling and genetic differences between the populations.68 Under such circumstances, it is possible that a TF may strongly bind to one allele at a given locus but may also bind indirectly and weakly to other allele. Furthermore, at any given time more than one, TFs are attracted to a particular allelic locus,35,69,70 though due to specificity, only one TF will bind to the allelic locus and other TFs will hang along the first TF.45 Likewise, altered binding of p300, a histone acetyl transferase and HDACs, a histone deacetylase at ACE I/D polymorphism may control ACE levels by differentially regulating histone acetylation and deacetylation.71 It is vital to note that most TFs do not work alone, instead these form homotypic and heterotypic interactions. Such interactions, which are abundant in the system, comprise of TF–TF, TF–nucleosome, and such other combinations. Thus, there are ways to interact and collaborate, such as the cooperative binding and the synergistic regulation. Now, these combinations/attractions could be dimeric, trimeric, and higher-order.72 It also defines the various types or classes of TFs and thereby the mechanisms that have been so very well elucidated in recent times. However, it is not the intention of this review to interpret these interactions. It is pretty obvious from these findings that physiological traits are vulnerable to the complex human system.
Allele-dependent control of apelin and NO signaling
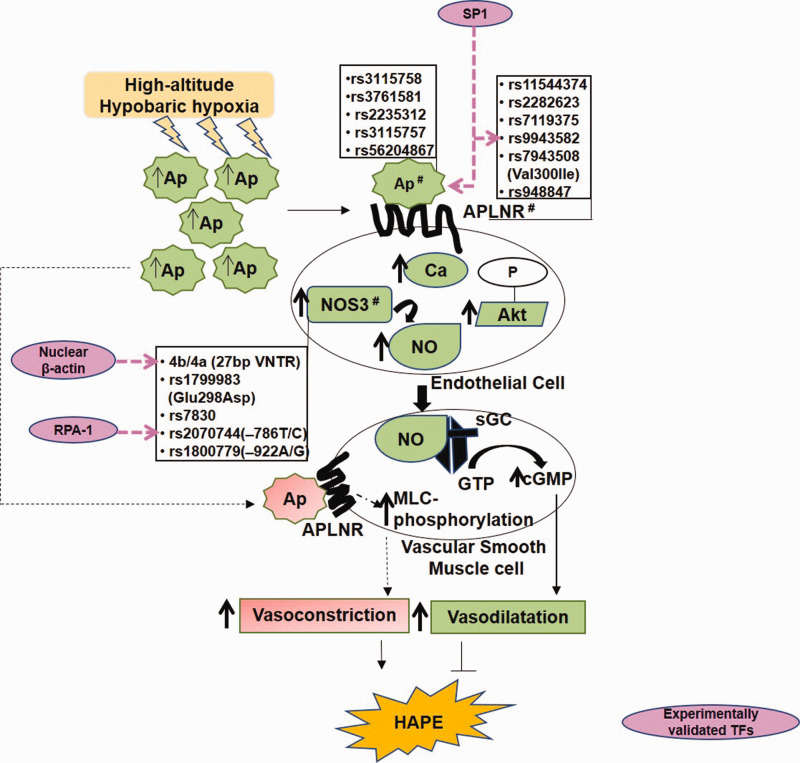
G-protein-coupled apelin receptors (APJ) and NO signaling play crucial role in maintaining pulmonary vascular homeostasis.25,73 Its malfunction is associated with several diseases including HAPE, pulmonary arterial hypertension, and cardiovascular diseases.25,74 X-linked apelin (APLN) gene encodes different variants of apelin peptide, differing in number of amino acid residues (apelin-36, -31, -28, -13), which upon binding to the vascular endothelial APJ possesses hypotensive as well as angiogenic activity (Fig. 2). Apelin phosphorylates serine/threonine-specific protein kinase B (Akt), raises intracellular calcium levels, and facilitates the production of NO, a potent vasodilator, generated by the endothelial NO synthase (NOS3) protein.57 Apelin-mediated angiogenesis is a consequence of its phosphorylation (ERKs and Akt), leading to the proliferation of endothelial cells and the formation of new blood vessels.75 Apelin also mediates vasoconstriction by stimulating myosin light chain phosphorylation in vascular smooth muscle cells.76
Fig. 2.
The apelin and nitric oxide signaling system. Apelin mediates nitric oxide-mediated smooth muscle relaxation via Akt and calcium signaling (Solid lines). Apelin mediates vascular smooth muscle cells contraction via myosin light chain (MLC) phosphorylations (dashed lines).
Ap: Apelin; APLNR: Apelin receptor; Ca: calcium; NOS3: nitric oxide synthase; NO: nitric oxide; sGC: soluble guanylate cyclase; GTP: guanosine triphosphate; cGMP: cyclic guanosine monophosphate; SP1: specificity protein 1; RPA1: replication protein 1; VNTR: variable number tandem repeat; MLC: myosin light chain; HAPE: high-altitude pulmonary edema; TF: transcription factor.
Note: Hash (#) represents HAPE-associated genetic variants.
Interestingly, the expression of Apelin and NOS3 as well as circulating levels of apelin 13 and NO when downregulated contribute to impaired vasodilation in HAPE patients.25 This decrease in the levels of vasodilators was associated with several genetic variants such as Apelin rs3761581, rs2235312, and rs3115757; apelin receptor (APLNR) rs11544374 and rs2282623; and NOS3 rs1799983, 4b/4a, and rs7830 (Fig. 2 and Table 1). The apelin–APJ polymorphisms also were reported with low apelin 13 levels and the risk of hypertension.77 The risk alleles include Apelin rs3115757C, rs56204867C, and rs3761581A.78 Beside polymorphisms, in vitro functional assays viz luciferase assay and real-time PCR, revealed allele-specific transcriptional regulation of apelin/APJ pathway by TFs such as TF specificity protein 1 (SP1).79 Likewise, electrophoretic mobility shift assay and chromatin immunoprecipitation confirmed SP1 binding to the APLNR polymorphism rs9943582 (–154G/A), specifically to G allele and thereby upregulating its expression. In addition to APLNR, apelin is also a direct transcriptional target of SP1 (Fig. 2). Thus, transcriptional upregulation of both, Apelin and APLNR, in response to SP1 enhances apelin–APJ signaling. This relates to blood pressure disorder, progression of atherosclerosis, and increased susceptibility to brain infarction.79 Interestingly, of the several TFs, SP1, signal transducer and activator of transcription 3 (STAT3), and Activating transcription factor 4 (ATF4) are known to control transcriptional regulation of apelin and APLNR.80,81
NOS has three isoforms, two constitutive, i.e. neuronal (NOS1) and endothelial (NOS3), and one inducible (NOS2), and all isoforms produce NO, which promotes cyclic guanosine monophosphate-mediated vascular smooth muscle relaxation by activating guanylate cyclase.21,27 Differential NO levels were associated with NOS3 polymorphism at high-altitude.21,82,83 Low NO levels were associated with haplotype of NOS3 bearing heterozygotes, i.e. GTbaAGTC of the polymorphisms 894G/T, 4b/4a (27bp repeat), –922A/G and –786T/C in HAPE patients, while the homozygous haplotype GG/bb of G894T and 4b/4a polymorphisms was associated with elevated NO levels in HA natives.21,25,61,81,83 Three NOS3 polymorphisms namely 894G/T, 4b/4a, and –786T/C are the most studied and validated (Fig. 2). Beginning with rs1799983 (894G/T), it encodes protease-sensitive NOS3 Glu298Asp variant (894T) that associates with decreased NO levels in HAPE patients.25,82,84,85 The reduced levels of NO in turn cause hypoxia-mediated pulmonary vasoconstriction72,86 and are inversely related to increased levels of an endogenous NOS inhibitor, asymmetric dimethylarginine.87 On the contrary, the 894G allele associates with increased NO levels and adaptation and acclimatization in several different high-altitude populations like the Ladhakis, the Chinese from Qinghai-China, the Han recruits traveling to the Lhasa plateau, and the Quechua of the Andean population.25,82,83 Likewise, the NOS3 4b/4a intron 4 variant that expresses five or four copies of the 27bp variable number tandem repeat is pertinent. The 4b allele associates with elevated NO levels and NOS3 expression in high-altitude adaptation,82 while the 4a allele associates with low NO and NOS3 expression in HAPE and hypertension.21,88 Of the several nuclear TFs bound to 27bp repeats as revealed through biotin–streptavidin pull down assay, mass spectrometry, and luciferase reporter assay,89 β-actin (Fig. 2) upregulated NOS3 expression in the presence of 4b allele.90 With β-actin's specificity for 27bp repeats and its role in the associated NOS3 transcriptional regulation, perhaps β-actin works here like a TF.
On the contrary, in vitro studies such as northern blot showed endothelial cells with 4b allele displayed higher levels of a 27bp small interference RNA, leading to decreased NOS3 expression compared to the 4a allele.91 In case of the NOS3 –786T/C promoter polymorphism, the protective –786T allele enhances NOS3 transcription efficiency as compared to its risk –786C allele;89 whereas, specific binding of replication protein A1 to its risk –786C allele decreased NOS3 transcription.92 This could be related to risk –786C allele-dependent decreased NO levels in HA adaptation. Surprisingly, in relation to –786T/C polymorphism, 4b/4a plays a contrasting cis-acting role in NOS3 regulation. Here, both the protective 4b and risk 4a alleles decreased the NOS3 transcription efficiency in the presence of the protective –786T allele.89 On the other hand, both the 4b and 4a alleles increased the NOS3 transcription efficiency in the presence of the risk C allele of NOS3 –786T/C promoter polymorphism. This could be attributed to the various TFs that are attracted to these allele-specific sites (Fig. 2).
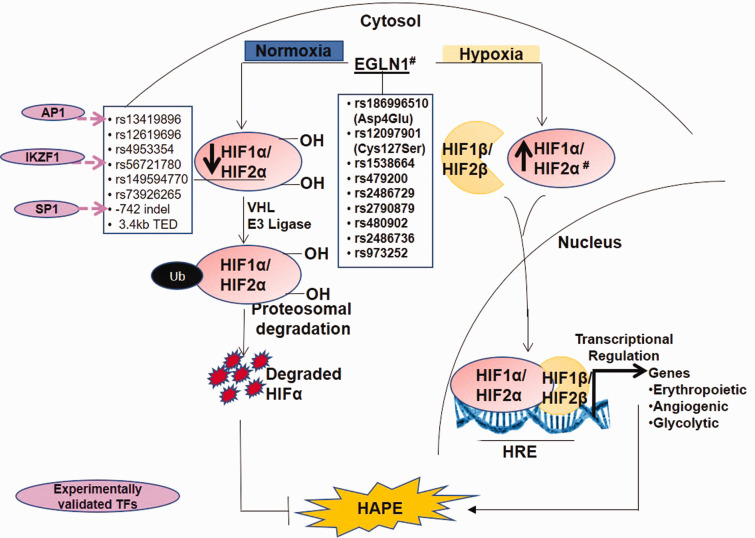
Differential expression of the hypoxia-inducible factor-signaling in the presence of TFs
Expression and activity of hypoxia-inducible factor (HIF)-1, a key oxygen-sensitive TF, increases exponentially with decrease in cellular oxygen.93,94 The HIF signaling is depicted in Fig. 3. HIF-1 consists of an oxygen-sensitive HIF-1α subunit and a constitutively expressed HIF-1β subunit. HIF-1 drives transcriptional activation of numerous genes involved in vascular homeostasis, erythropoiesis, angiogenesis, and glycolysis.95,96 Although HIF-1α subunit shares 48% sequence homology with HIF-2α subunit (encoded by the endothelial Per/ARNT/Sim domain protein-1 (EPAS-1)) and both bind to the same consensus sequence, hypoxia response element, however, HIF1 and HIF2 mediate different responses to hypoxia.97,98 Another molecule in this system, EGLN1, encoding prolyl hydroxylase 2 (PHD2), negatively regulates the activity of HIF-1α by hydroxylation of its two prolines.97 Additionally, PHD2/HIF-2α axis regulates pulmonary arterial pressure in vivo by antagonistically regulating vasoconstrictor, Endothelin 1, and the vasodilator, apelin-mediated signaling.99 However, which of the two molecules is activated more at any given time will depend on the overall pathways influenced. The expression of PHD2 is regulated under hypobaric environment.45 Over the years, the polymorphisms in these genes have found relevance (Fig. 3 and Table 1).
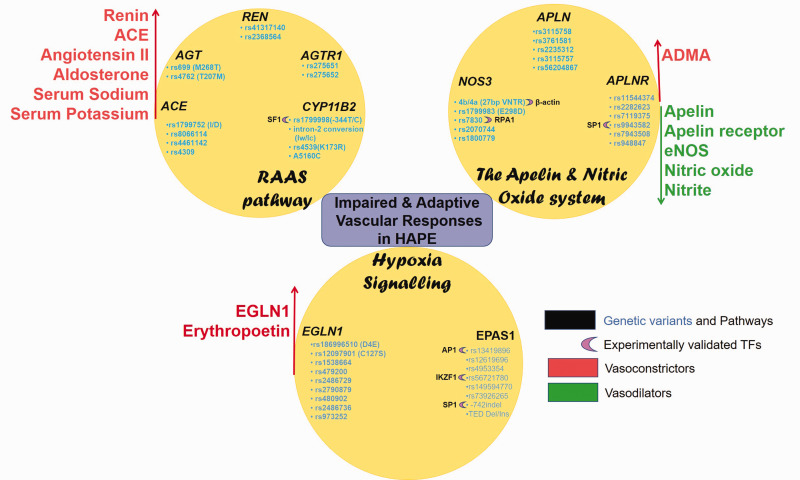
Fig. 4.
Genetic variants and associated TFs in pathways of interest control vascular homeostasis by regulating the levels of vasodilators and vasoconstrictors.
Ren: renin; AGT: angiotensinogen; ACE: angiotensin converting enzyme; AGTR1: angiotensin receptor; CYP11B2: aldosterone synthase; SF1: steroidogenic transcription factor; APLN: apelin; APLNR: apelin receptor; NOS3: endothelial nitric oxide synthase; RPA1: replication protein A1; SP1: specificity protein 1; AP1: activator protein 1; ADMA: asymmetric dimethylarginine; EGLN1: Egl nine homolog 1; EPAS1: endothelial Per/ARNT/Sim domain protein-1; IKZF1: IKAROS family zinc finger 1; RAAS: Renin–angiotensin–aldosterone system.
Fig. 3.
The hypoxia signaling pathway involves EGLN1-mediated hydroxylation and VHL-dependent ubiquitination of HIF-1α/2α subunits leading to its proteosomal degrardation under normoxic conditions. However, under hypobaric hypoxic conditions, HIF-1α/2α subunits are stabilized, which complexes with HIF-β subunit and drives transcriptional regulation of numerous genes involved in vascular homeostasis.
HIF: hypoxia-inducible factor; HRE: hypoxia response element; VHL: Von Hippel-Lindau; Ub: ubiquitin; indel: insertion–deletion; 3.4kb TED: Tibetan-enriched 3.4kb deletion; D4E: aspartate4glutamate; C127S: cysteine127serine; IKZF1: IKAROS family zinc finger 1; SP1: specificity protein 1; AP1: activator protein 1; EGLN1: Egl nine homolog 1; HAPE: high-altitude pulmonary edema; TF: transcription factor.
Note: Hash (#) represents high-altitude adaptation/maladaptation associated genetic variants.
Several independent groups have shown association of the polymorphisms in EPAS1/HIF2α and EGLN1 with high-altitude adaptation in Tibetans.100–103 Interestingly, two EPAS1 promoter polymorphisms, rs56721780 and an indel positioned at –886 and –742 upstream of its TSS, regulate EPAS1 by allele-specific TF binding (Fig. 3). Here, the rs56721780C allele decreased the binding of EPAS1 transcriptional repressor, IKAROS family zinc finger 1 as confirmed by EMSA. While the 40bp insertion at –742 indel increased the binding of its transcriptional activator SP1. Thus, both the rs56721780C allele and the –742 insertion increased EPAS1 levels as revealed by luciferase reporter assays. This modification has been associated with higher birth weight and embryonic development in Tibetan newborns.104 Similarly, another EPAS1 polymorphism in intron 1, namely rs13419896, regulates EPAS1 transcription.105 The rs13419896A allele that was associated with high-altitude adaptation in Tibetans and Sherpas, has also been reported to bind to the TF Activator protein-1 (AP1) that upregulated the EPAS1 levels. Contrary to this, evidence of diminished EPAS1 activity at high-altitude has also been reported.106 For example, another Tibetan-enriched EPAS1 rs149594770A allele weakens its TF-binding capacity as well as its promoter activity as seen by EMSA and luciferase reporter assay.106 This weakened EPAS1 activity was attributed to the relatively low hemoglobin level in Tibetans.100,106
Among the other members of HIF signaling, EGLN1 polymorphisms have been investigated (Fig. 3 and Table 1). Several EGLN1 variants and its haplotypes are reported to associate with decreased SaO2 levels, increased pulmonary arterial systolic pressure (PASP), and circulating EGLN1 levels in HAPE.45 Specifically, two key exonic polymorphisms, rs186996510 (Asp4Glu) and rs12097901 (Cys127Ser), contribute to high-altitude adaptation in Tibetans,107 albeit at a lower frequency in highland Andeans.108 Interestingly, these two missense variants exhibit low Km for oxygen and promote increased HIF-1α degradation under hypoxic conditions.107 Abrogation of HIF-mediated responses like erythropoiesis prevents polycythemia in Tibetans. In fact, EGLN1 gain-of-function mutants in combination with EPAS1 polymorphisms also associated with decreased hemoglobin concentration in Tibetan highlanders.109 Surprisingly, another study reported the EGLN1 Tibetan haplotype, D4E/C127S, as loss-of-function mutant, which increases HIF-1α-mediated respiration and NO levels.110 Although TFs have so far not been reported in regard to EGLN1 variants, however, we have found few important TFs associating with EGLN1 variants (unpublished). Therefore, altered binding of TFs to EGLN1 variants (Fig. 3) may regulate HIF-signaling under the hypobaric hypoxic environment at high-altitude.
Gene to drug interactions in HAPE
Drug responses increasingly are being seen to be influenced by genetic polymorphisms. Nifedipine pharmacokinetics are influenced by genetic polymorphisms in cytochrome P450 (CYP) 3A5.111 Sildenafil protects against the development of altitude-induced hypoxemia, pulmonary hypertension, and improves gas exchange.112 Sildenafil response is influenced by NOS3 genetic polymorphisms in several diseases including pulmonary hypertension.113 The polymorphism in the β-adrenergic receptor 2 (ADRB2) gene affects therapeutic responses to beta agonists. The ADRB2 Arg16Gly loss-of-function polymorphism in particular dictates responses in heart patients.114 Renin polymorphisms predict the effects of thiazide diuretics, genetic variants in patients with decompensated heart influenced furosemide drug response.115 While mineralocorticoid receptor gene variation influences dexamethasone-induced stress response.116 Such insights from pharmacogenomics research will help to characterize genetic determinants effecting drug response to HAPE; paving way to personalized medicines. Of consequence, research on the role of TFs in influencing drug response is increasing.117,118 For instance, one mechanism of action of aminophylline involves recruitment and activation of HDACs,119 allele-specific HDACs binding to genetic variants may alter drug response in HAPE. Likewise, acetazolamide controls transcriptional activation of several TFs namely, AP-1, HIF, Heat shock factor (HSF), Nuclear factor-kappa B (NF-κB), Nuclear factor erythroid 2-related factor 2 (NRF2), Tumor suppresor p53 (p53), and STAT3.120 Also, the corticosteroid action involves binding to several TFs including AP-1 or NF-κB as well recruitment of HDACs.121 Thus, recent advances surely encourage to envisage equally greater role for the various TFs in conjunction with the genetic variants in shaping a physiological path.
Conclusion
We conclude that genetic variants and TFs play a pivotal role in the regulation of HAPE. Allele-specific binding of TFs seems to play a critical role in determining the causal role and may be contributed by select loci. This observation is an outcome of understanding of several of the pertinent physiological pathways. Evidences of cellular role of allele-specific TF binding in regulating these effects are wanting. As of now, none of the genetic marker has diagnostic potential, unless tested in a larger sample size. Investigations on association of genetic polymorphisms with predicted TFs under hypobaric hypoxic conditions would shed light on the physiological processes thereby advancing the development of diagnostics and therapeutics.
Future perspectives
In addition to genetics, epigenetic influences play a significant role in the regulation of physiological functions and human health. With newer information on disease-associated genetic variants, more extensive studies on polymorphisms-mediated changes in putative TF-binding motifs, chromatin structure, chromatin states, methylation, and gene expression will further elucidate the mechanisms underlying the disease. Additionally, pharmacogenomics research in turn will greatly enhance the life-expectancy or survival of the fittest under the given extreme environment.
Acknowledgements
We thank the Director of CSIR-Institute of Genomics and Integrative Biology, Delhi, for the infrastructure.
Author contributions: N.C. contributed to designing the concept, writing, and correcting the manuscript. T.P. contributed the write-up and designing the figures. J.H.N. conceived, designed and supervised the project, contributed to writing, and correcting the manuscript. M.A.Q.P. conceived, designed and supervised the project, contributed to human sample collection, analyses, writing, and correcting the manuscript.
Conflict of interest: The author(s) declare that there is no conflict of interest.
Funding: This work is majorly supported by CSIR, India (grant numbers MLP1401 and BSC0123) and in part by CMREF, USA (grant number GAP0119) and ICMR (fellowship to NC. and EMS to Q.P., ref 74/6/2015-Pers/EMS).
References
- 1.Beall CM. High-altitude adaptations. Lancet 2003; 362: s14–s15. [DOI] [PubMed] [Google Scholar]
- 2.Qadar Pasha MA, Newman JH. High-altitude disorders: pulmonary hypertension: pulmonary vascular disease: the global perspective. Chest 2010; 137: 13S–19S. [DOI] [PubMed] [Google Scholar]
- 3.Simonson TS, Wagner PD. Oxygen transport adaptations to exercise in native highland populations. Exp Physiol 2015; 100: 1231–1232. [DOI] [PubMed] [Google Scholar]
- 4.West JB. Oxygen conditioning: a new technique for improving living and working at high altitude. Physiology 2016; 31: 216–222. [DOI] [PubMed] [Google Scholar]
- 5.Julian CG, Moore LG. Human genetic adaptation to high altitude: evidence from the Andes. Genes 2019; 10: 150–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peacock AJ. Oxygen at high altitude. BMJ 1998; 317: 1063–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hackett PH, Roach RC. High-altitude illness. N Engl J Med 2001; 345: 107–114. [DOI] [PubMed] [Google Scholar]
- 8.West JB. Oxygen enrichment of room air to relieve the hypoxia of high altitude. Respir Physiol 1995; 99: 225–232. [DOI] [PubMed] [Google Scholar]
- 9.Swenson ER, Maggiorini M, Mongovin S, et al. Pathogenesis of high-altitude pulmonary edema: inflammation is not an etiologic factor. JAMA 2002; 287: 2228–2235. [DOI] [PubMed] [Google Scholar]
- 10.Sartori C, Allemann Y, Scherrer U. Pathogenesis of pulmonary edema: learning from high-altitude pulmonary edema. Respir Physiol Neurobiol 2007; 159: 338–349. [DOI] [PubMed] [Google Scholar]
- 11.Scherrer U, Rexhaj E, Jayet PY, et al. New insights in the pathogenesis of high-altitude pulmonary edema. Prog Cardiovasc Dis 2010; 52: 485–492. [DOI] [PubMed] [Google Scholar]
- 12.Maggiorini M, Mélot C, Pierre S, et al. High-altitude pulmonary edema is initially caused by an increase in capillary pressure. Circulation 2001; 103: 2078–2083. [DOI] [PubMed] [Google Scholar]
- 13.Hotta J, Hanaoka M, Droma Y, et al. Polymorphisms of renin-angiotensin system genes with high-altitude pulmonary edema in Japanese subjects. Chest 2004; 126: 825–830. [DOI] [PubMed] [Google Scholar]
- 14.Bärtsch P. High altitude pulmonary edema. Respiration 1997; 64: 435–443. [DOI] [PubMed] [Google Scholar]
- 15.Gluecker T, Capasso P, Schnyder P, et al. Clinical and radiologic features of pulmonary edema. Radiographics 1999; 19: 1507–1531. [DOI] [PubMed] [Google Scholar]
- 16.Maggiorini M. High altitude-induced pulmonary oedema. Cardiovasc Res 2006; 72: 41–50. [DOI] [PubMed] [Google Scholar]
- 17.Milledge JS, West JB, Schoene RB. High altitude medicine and physiology, 4th ed London, UK: CRC Press, 2007, pp. 480. [Google Scholar]
- 18.West JB, Mathieu-Costello O. High altitude pulmonary edema is caused by stress failure of pulmonary capillaries. Int J Sports Med 1992; 13: S54–S58. [DOI] [PubMed] [Google Scholar]
- 19.Bartsch P, Mairbaurl H, Maggiorini M, et al. Physiological aspects of high-altitude pulmonary edema. J Appl Physiol 2005; 98: 1101–1110. [DOI] [PubMed] [Google Scholar]
- 20.Qadar Pasha MA, Khan AP, Kumar R, et al. Angiotensin converting enzyme insertion allele in relation to high altitude adaptation. Ann Hum Genet 2001; 65: 531–536. [DOI] [PubMed] [Google Scholar]
- 21.Ahsan A, Mohd G, Norboo T, et al. Heterozygotes of NOS3 polymorphisms contribute to reduced nitrogen oxides in high-altitude pulmonary edema. Chest 2006; 130: 1511–1519. [DOI] [PubMed] [Google Scholar]
- 22.Stobdan T, Karar J, Qadar Pasha MA. High altitude adaptation: genetic perspectives. High Alt Med Biol 2008; 9: 140–147. [DOI] [PubMed] [Google Scholar]
- 23.Bigham AW, Wilson MJ, Julian CG, et al. Andean and Tibetan patterns of adaptation to high altitude. Am J Hum Biol 2013; 25: 190–197. [DOI] [PubMed] [Google Scholar]
- 24.Beall CM. Adaptation to high altitude: phenotypes and genotypes. Annu Rev Anthropol 2014; 43: 251–272. [Google Scholar]
- 25.Mishra A, Kohli S, Dua S, et al. Genetic differences and aberrant methylation in the apelin system predict the risk of high-altitude pulmonary edema. Proc Natl Acad Sci 2015; 112: 6134–6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moore LG. Human genetic adaptation to high altitudes: current status and future prospects. Quatern Int 2017; 461: 4–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beall CM, Laskowski D, Strohl KP, et al. Pulmonary nitric oxide in mountain dwellers. Nature 2001; 414: 411–412. [DOI] [PubMed] [Google Scholar]
- 28.Pandey P, Qadar Pasha MA. Oxidative stress at high altitude: genotype–phenotype correlations. Adv Genomics Genet 2014; 4: 29–43. [Google Scholar]
- 29.Mishra A, Mohammad G, Norboo T, et al. Lungs at high-altitude: genomic insights into hypoxic responses. J Appl Physiol 2015; 119: 1–5. [DOI] [PubMed] [Google Scholar]
- 30.Richalet JP. Physiological and clinical implications of adrenergic pathways at high altitude. Adv Exp Med Biol 2016; 903: 343–356. [DOI] [PubMed] [Google Scholar]
- 31.Swenson ER, Bartsch P. High-altitude pulmonary edema. Compr Physiol 2012; 2: 2753–2573. [DOI] [PubMed] [Google Scholar]
- 32.Alam P, Saini N, Qadar Pasha MA. MicroRNAs: an apparent switch for high-altitude pulmonary edema. MicroRNA 2015; 4: 158–167. [DOI] [PubMed] [Google Scholar]
- 33.Blissenbach B, Nakas CT, Krönke M, et al. Hypoxia-induced changes in plasma micro-RNAs correlate with pulmonary artery pressure at high altitude. Am J Physiol Lung Cell Mol Physiol 2017; 314: L157–L164. [DOI] [PubMed] [Google Scholar]
- 34.Julian CG. Epigenomics and human adaptation to high altitude. J Appl Physiol 2017; 123: 1362–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lambert SA, Jolma A, Campitelli LF, et al. The human transcription factors. Cell 2018; 172: 650–665. [DOI] [PubMed] [Google Scholar]
- 36.Chen LJ, Wei SY, Chiu JJ. Mechanical regulation of epigenetics in vascular biology and pathobiology. J Cell Mol Med 2013; 17: 437–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Turunen MP, Aavik E, Ylä-Herttuala S. Epigenetic regulation in vascular cells. Curr Opin Lipidol 2013; 24: 438–443. [DOI] [PubMed] [Google Scholar]
- 38.Ti D, Li M, Fu X, et al. Causes and consequences of epigenetic regulation in wound healing. Wound Repair Regen 2014; 22: 305–312. [DOI] [PubMed] [Google Scholar]
- 39.Davis FM, Gallagher K. Epigenetic mechanisms in monocytes/macrophages regulate inflammation in cardiometabolic and vascular disease. Arterioscler Thromb Vasc Biol 2019; 39: 623–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 2012; 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Y, Walavalkar NM, Dozmorov MG, et al. Identification of breast cancer associated variants that modulate transcription factor binding. PLoS Genet 2017; 13: e1006761–e1006782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wagih O, Merico D, Delong A, et al. Allele-specific transcription factor binding as a benchmark for assessing variant impact predictors. BioRxiv 2018, pp. 253427–253463. [Google Scholar]
- 43.Kellis M, Wold B, Snyder MP, et al. Defining functional DNA elements in the human genome. Proc Natl Acad Sci 2014; 111: 6131–6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Héberlé É, Bardet AF. Sensitivity of transcription factors to DNA methylation. Essays Biochem 2019; 63: 727–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mishra A, Mohammad G, Thinlas T, et al. EGLN1 variants influence expression and SaO2 levels to associate with high-altitude pulmonary oedema and adaptation. Clin Sci 2012; 124: 479–489. [DOI] [PubMed] [Google Scholar]
- 46.Cavalli M, Pan G, Nord H, et al. Allele-specific transcription factor binding to common and rare variants associated with disease and gene expression. J Hum Genet 2016; 135: 485–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paul M, Poyan Mehr A, Kreutz R. Physiology of local renin-angiotensin systems. Physiol Rev 2006; 86: 747–803. [DOI] [PubMed] [Google Scholar]
- 48.Rajput C, Arif E, Vibhuti A, et al. Predominance of interaction among wild-type alleles of CYP11B2 in Himalayan natives associates with high-altitude adaptation. Biochem Biophys Res Commun 2006; 348: 735–740. [DOI] [PubMed] [Google Scholar]
- 49.Rigat B, Hubert C, Alhenc-Gelas F, et al. An insertion/deletion polymorphism in the angiotensin I-converting enzyme gene accounting for half the variance of serum enzyme levels. J Clin Invest 1990; 86: 1343–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Charu R, Stobdan T, Ram RB, et al. Susceptibility to high altitude pulmonary oedema: role of ACE and ET-1 polymorphisms. Thorax 2006; 61: 1011–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bigham AW, Kiyamu M, León-Velarde F, et al. Angiotensin-converting enzyme genotype and arterial oxygen saturation at high altitude in Peruvian Quechua. High Alt Med Biol 2008; 9: 167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nejatizadeh A, Shekari M, Stobdan T, et al. Genetic susceptibility of angiotensin-I converting enzyme and G-protein β3-subunit gene polymorphisms to essential hypertension. Genet Third Millennium 2010; 8: 2133–2144. [Google Scholar]
- 53.Stobdan T, Ali Z, Khan AP, et al. Polymorphisms of renin–angiotensin system genes as a risk factor for high-altitude pulmonary oedema. J Renin Angiotensin Aldosterone Syst 2011; 12: 93–101. [DOI] [PubMed] [Google Scholar]
- 54.Srivastava S, Bhagi S, Kumari B, et al. Association of polymorphisms in angiotensin and aldosterone synthase genes of the renin–angiotensin–aldosterone system with high-altitude pulmonary edema. J Renin Angiotensin Aldosterone Syst 2012; 13: 155–160. [DOI] [PubMed] [Google Scholar]
- 55.Wang QQ, Yu L, Huang GR, et al. Polymorphisms of angiotensin converting enzyme and nitric oxide synthase 3 genes as risk factors of high-altitude pulmonary edema: a case-control study and meta-analysis. Tohoku J Exp Med 2013; 229: 255–266. [DOI] [PubMed] [Google Scholar]
- 56.Jin T, Ren Y, Zhu X, et al. Angiotensin II receptor 1 gene variants are associated with high-altitude pulmonary edema risk. Oncotarget 2016; 7: 77117–77123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ahsan A, Charu R, Qadar Pasha MA, et al. eNOS allelic variants at the same locus associate with HAPE and adaptation. Thorax 2004; 59: 1000–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Droma Y, Hanaoka M, Basnyat B, et al. Adaptation to high altitude in Sherpas: association with the insertion/deletion polymorphism in the angiotensin-converting enzyme gene. Wilderness Environ Med 2008; 19: 22–29. [DOI] [PubMed] [Google Scholar]
- 59.Kumar R, Qadar Pasha MA, Khan AP, et al. Renin angiotensin aldosterone system and ACE I/D gene polymorphism in high-altitude pulmonary edema. Aviat Space Envir Md 2004; 75: 981–983. [PubMed] [Google Scholar]
- 60.Barker KR, Conroy AL, Hawkes M, et al. Biomarkers of hypoxia, endothelial and circulatory dysfunction among climbers in Nepal with AMS and HAPE: a prospective case–control study. J Travel Med 2016; 23: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Iwai N, Kajimoto K, Tomoike H, et al. Polymorphism of CYP11B2 determines salt sensitivity in Japanese. Hypertension 2007; 49: 825–831. [DOI] [PubMed] [Google Scholar]
- 62.Kumar R, Nejatizadeh A, Arif E, et al. Multi-locus interactions of vascular homeostasis genes in essential hypertension: a gender-based study. Clin Chim Acta 2009; 405: 87–93. [DOI] [PubMed] [Google Scholar]
- 63.Ellis KL, Palmer BR, Frampton CM, et al. Genetic variation in the renin–angiotensin–aldosterone system is associated with cardiovascular risk factors and early mortality in established coronary heart disease. J Hum Hypertens 2013; 27: 237–244. [DOI] [PubMed] [Google Scholar]
- 64.Singh KD, Karthikeyan M. Combined sequence and sequence-structure-based methods for analyzing RAAS gene SNPs: a computational approach. J Recept Sig Transduct Res 2014; 34: 513–526. [DOI] [PubMed] [Google Scholar]
- 65.Bassett MH, Zhang Y, Clyne C, et al. Differential regulation of aldosterone synthase and 11beta-hydroxylase transcription by steroidogenic factor-1. J Mol Endocrinol 2002; 28: 125–135. [DOI] [PubMed] [Google Scholar]
- 66.Tamaki S, Iwai N, Tsujita Y, et al. Genetic polymorphism of CYP11B2 gene and hypertension in Japanese. Hypertension 1999; 33: 266–270. [DOI] [PubMed] [Google Scholar]
- 67.Davies E, Holloway CD, Ingram MC, et al. Aldosterone excretion rate and blood pressure in essential hypertension are related to polymorphic differences in the aldosterone synthase gene CYP11B2. Hypertension 1999; 33: 703–707. [DOI] [PubMed] [Google Scholar]
- 68.Lim PO, Macdonald TM, Holloway C, et al. Variation at the aldosterone synthase (CYP11B2) locus contributes to hypertension in subjects with a raised aldosterone-to-renin ratio. J Clin Endocrinol Metab 2002; 87: 4398–4402. [DOI] [PubMed] [Google Scholar]
- 69.Porter AH, Johnson NA, Tulchinsky AY. A new mechanism for Mendelian dominance in regulatory genetic pathways: competitive binding by transcription factors. Genetics 2017; 1: 101–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Deplancke B, Alpern D, Gardeux V. The genetics of transcription factor DNA binding variation. Cell 2016; 3: 538–554. [DOI] [PubMed] [Google Scholar]
- 71.Song R, Van Buren T, Yosypiv IV. Histone deacetylases are critical regulators of the renin-angiotensin system during ureteric bud branching morphogenesis. Pediatr Res 2010; 67: 573–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Reiter F, Wienerroither S, Stark A. Combinatorial function of transcription factors and cofactors. Curr Opin Genet Dev 2017; 43: 73–81. [DOI] [PubMed] [Google Scholar]
- 73.Beall CM, Laskowski D, Erzurum SC. Nitric oxide in adaptation to altitude. Free Radic Biol Med 2012; 52: 1123–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Berger MM, Hesse C, Dehnert C, et al. Hypoxia impairs systemic endothelial function in individuals prone to high-altitude pulmonary edema. Am J Respir Crit Care Med 2005; 172: 763–767. [DOI] [PubMed] [Google Scholar]
- 75.Cheng J, Luo X, Huang Z, et al. Apelin/APJ system: a potential therapeutic target for endothelial dysfunction-related diseases. J Cell Physiol 2019; 234: 12149–12160. [DOI] [PubMed] [Google Scholar]
- 76.Hashimoto T, Kihara M, Ishida J, et al. Apelin stimulates myosin light chain phosphorylation in vascular smooth muscle cells. Arter Thromb Vasc Biol 2006; 26: 1267–1272. [DOI] [PubMed] [Google Scholar]
- 77.Gupta MD, Girish MP, Shah D, et al. Biochemical and genetic role of apelin in essential hypertension and acute coronary syndrome. Int J Cardiol 2016; 223: 374–378. [DOI] [PubMed] [Google Scholar]
- 78.Zhang M, Peng F, Lin L, et al. Association study of apelin-APJ system genetic polymorphisms with incident metabolic syndrome in a Chinese population: a case-control study. Oncotarget 2019; 10: 3807–3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hata J, Matsuda K, Ninomiya T, et al. Functional SNP in an Sp1-binding site of AGTRL1 gene is associated with susceptibility to brain infarction. Hum Mol Genet 2007; 16: 630–639. [DOI] [PubMed] [Google Scholar]
- 80.O'Carroll AM, Lolait SJ, Howell GM. Transcriptional regulation of the rat apelin receptor gene: promoter cloning and identification of an Sp1 site necessary for promoter activity. J Mol Endocrinol 2006; 36: 221–235. [DOI] [PubMed] [Google Scholar]
- 81.Jeong K, Oh Y, Kim SJ, et al. Apelin is transcriptionally regulated by ER stress-induced ATF4 expression via a p38 MAPK-dependent pathway. Apoptosis 2014; 19: 1399–1410. [DOI] [PubMed] [Google Scholar]
- 82.Ahsan A, Norboo T, Baig MA, et al. Simultaneous selection of the wild-type genotypes of the G894T and 4B/4A polymorphisms of NOS3 associate with high-altitude adaptation. Ann Hum Genet 2005; 69: 260–267. [DOI] [PubMed] [Google Scholar]
- 83.Wang P, Ha AY, Kidd KK, et al. A variant of the endothelial nitric oxide synthase gene (NOS3) associated with AMS susceptibility is less common in the Quechua, a high altitude native population. High Alt Med Biol 2010; 11: 27–30. [DOI] [PubMed] [Google Scholar]
- 84.Droma Y, Hanaoka M, Basnyat B, et al. Genetic contribution of the endothelial nitric oxide synthase gene to high altitude adaptation in Sherpas. High Alt Med Biol 2006; 7: 209–220. [DOI] [PubMed] [Google Scholar]
- 85.Sofowora G, Dishy V, Xie HG, et al. In-vivo effects of Glu298Asp endothelial nitric oxide synthase polymorphism. Pharmacogenet Genom 2001; 11: 809–814. [DOI] [PubMed] [Google Scholar]
- 86.Busch T, Bartsch P, Pappert D, et al. Hypoxia decreases exhaled nitric oxide in mountaineers susceptible to high-altitude pulmonary edema. Am J Respir Crit Care Med 2001; 163: 368–373. [DOI] [PubMed] [Google Scholar]
- 87.Ali Z, Mishra A, Kumar R, et al. Interactions among vascular-tone modulators contribute to high altitude pulmonary edema and augmented vasoreactivity in highlanders. PLoS One 2012; 7: e44049–e44057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kumar R, Kohli S, Mishra A, et al. Interactions between the genes of vasodilatation pathways influence blood pressure and nitric oxide level in hypertension. Am J Hypertens 2014; 28: 239–247. [DOI] [PubMed] [Google Scholar]
- 89.Wang J, Dudley D, Wang XL. Haplotype-specific effects on endothelial NO synthase promoter efficiency: modifiable by cigarette smoking. Arterioscler Thromb Vasc Biol 2002; 22: e1–e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ou H, Shen YH, Utama B, et al. Effect of nuclear actin on endothelial nitric oxide synthase expression. Arterioscler Thromb Vasc Biol 2005; 25: 2509–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang MX, Zhang C, Shen YH, et al. Biogenesis of short intronic repeat 27-nucleotide small RNA from endothelial nitric-oxide synthase gene. J Biol Chem 2008; 283: 14685–14693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Miyamoto Y, Saito Y, Nakayama M, et al. Replication protein A1 reduces transcription of the endothelial nitric oxide synthase gene containing a–786T→ C mutation associated with coronary spastic angina. Hum Mol Genet 2000; 9: 2629–2637. [DOI] [PubMed] [Google Scholar]
- 93.Semenza GL. Regulation of mammalian O2 homeostasis by hypoxia-inducible factor 1. Annu Rev Cell Dev Biol 1999; 15: 551–578. [DOI] [PubMed] [Google Scholar]
- 94.Ratcliffe PJ, O'Rourke JF, Maxwell PH, et al. Oxygen sensing, hypoxia-inducible factor-1 and the regulation of mammalian gene expression. J Exp Biol 1998; 201: 1153–1162. [DOI] [PubMed] [Google Scholar]
- 95.Maxwell PH. Hypoxia-inducible factor as a physiological regulator. Exp Physiol 2005; 90: 791–797. [DOI] [PubMed] [Google Scholar]
- 96.Rey S, Semenza GL. Hypoxia-inducible factor-1-dependent mechanisms of vascularization and vascular remodelling. Cardiovasc Res 2010; 86: 236–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ratcliffe PJ. HIF-1 and HIF-2: working alone or together in hypoxia?. J Clin Invest 2007; 117: 862–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Koh MY, Powis G. Passing the baton: the HIF switch. Trends Biochem Sci 2012; 37: 364–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kapitsinou PP, Rajendran G, Astleford L, et al. The endothelial prolyl-4-hydroxylase domain 2/hypoxia-inducible factor 2 axis regulates pulmonary artery pressure in mice. Mol Cell Biol 2016; 36: 1584–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Beall CM, Cavalleri GL, Deng L, et al. Natural selection on EPAS1 (HIF2α) associated with low hemoglobin concentration in Tibetan highlanders. Proc Natl Acad Sci 2010; 107: 11459–11464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Simonson TS, Yang Y, Huff CD, et al. Genetic evidence for high-altitude adaptation in Tibet. Science 2010; 329: 72–75. [DOI] [PubMed] [Google Scholar]
- 102.Xu S, Li S, Yang Y, et al. A genome-wide search for signals of high-altitude adaptation in Tibetans. Mol Biol Evol 2010; 28: 1003–1011. [DOI] [PubMed] [Google Scholar]
- 103.Yi X, Liang Y, Huerta-Sanchez E, et al. Sequencing of 50 human exomes reveals adaptation to high altitude. Science 2010; 329: 75–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Xu XH, Huang XW, Qun L, et al. Two functional loci in the promoter of EPAS1 gene involved in high-altitude adaptation of Tibetans. Sci Rep 2014; 4: 7465–7473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hanaoka M, Droma Y, Basnyat B, et al. Genetic variants in EPAS1 contribute to adaptation to high-altitude hypoxia in Sherpas. PloS One 2012; 7: e50566–e50574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Peng Y, Cui C, He Y, et al. Down-regulation of EPAS1 transcription and genetic adaptation of Tibetans to high-altitude hypoxia. Mol Biol Evol 2017; 34: 818–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lorenzo FR, Huff C, Myllymäki M, et al. A genetic mechanism for Tibetan high-altitude adaptation. Nat Genet 2014; 46: 951–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Heinrich EC, Wu L, Lawrence ES, et al. Genetic variants at the EGLN1 locus associated with high-altitude adaptation in Tibetans are absent or found at low frequency in highland Andeans. Ann Hum Genet 2019; 83: 171–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tashi T, Reading NS, Wuren T, et al. Gain-of-function EGLN1 prolyl hydroxylase (PHD2 D4E: C127S) in combination with EPAS1 (HIF-2α) polymorphism lowers hemoglobin concentration in Tibetan highlanders. J Mol Med 2017; 95: 665–670. [DOI] [PubMed] [Google Scholar]
- 110.Song D, Li LS, Arsenault PR, et al. Defective Tibetan PHD2 binding to p23 links high altitude adaption to altered oxygen sensing. J Biol Chem 2014; 289: 14656–14665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Haas DM, Quinney SK, Clay JM, et al. Nifedipine pharmacokinetics are influenced by CYP3A5 genotype when used as a preterm labor tocolytic. Am J Perinatol 2013; 30: 275–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Richalet JP, Gratadour P, Robach P, et al. Sildenafil inhibits altitude-induced hypoxemia and pulmonary hypertension. Am J Resp Crit Care 2005; 171: 275–281. [DOI] [PubMed] [Google Scholar]
- 113.Muniz JJ, Lacchini R, Rinaldi TO, et al. Endothelial nitric oxide synthase genotypes and haplotypes modify the responses to sildenafil in patients with erectile dysfunction. Pharmacogenomics J 2013; 13: 189–196. [DOI] [PubMed] [Google Scholar]
- 114.Huang J, Li C, Song Y, et al. ADRB2 polymorphism Arg16Gly modifies the natural outcome of heart failure and dictates therapeutic response to β-blockers in patients with heart failure. Cell Discov 2018; 4: 57–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.de Denus S, Rouleau JL, Mann DL, et al. A pharmacogenetic investigation of intravenous furosemide in decompensated heart failure: a meta-analysis of three clinical trials. Pharmacogenomics J 2017; 17: 192–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Van Leeuwen N, Kumsta R, Entringer S, et al. Functional mineralocorticoid receptor (MR) gene variation influences the cortisol awakening response after dexamethasone. Psychoneuroendocrinology 2010; 35: 339–349. [DOI] [PubMed] [Google Scholar]
- 117.Hanson C, Cairns J, Wang L, et al. Computational discovery of transcription factors associated with drug response. Pharmacogenomics J 2016; 16: 573–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Shah F, Medvedev A, Wassermann AM, et al. The identification of pivotal transcriptional factors mediating cell responses to drugs with drug-induced liver injury liabilities. Toxicol Sci 2017; 162: 177–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ranjani R, Vinotha AS. A prospective randomized controlled study: theophylline on oxidative stress and steroid sensitivity in chronic obstructive pulmonary disease patients. Int J Pharm Investig 2017; 7: 119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Noma N, Fujii G, Miyamoto S, et al. Impact of acetazolamide, a carbonic anhydrase inhibitor, on the development of intestinal polyps in min mice. Int J Mol Sci 2017; 18: 851–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Adcock IM, Ito K, Barnes PJ. Glucocorticoids: effects on gene transcription. Proc Am Thorac Soc 2004; 1: 247–254. [DOI] [PubMed] [Google Scholar]