Abstract
A primary limitation in the use of pluripotent stem cell-derived cardiomyocytes (PSC-CMs) for both patient health and scientific investigation is the failure of these cells to achieve full functional maturity. In vivo, cardiomyocytes undergo numerous adaptive structural, functional and metabolic changes during maturation. By contrast, PSC-CMs fail to fully undergo these developmental processes, instead remaining arrested at an embryonic stage of maturation. There is thus a significant need to understand the biological processes underlying proper CM maturation in vivo. Here, we discuss what is known regarding the initiation and coordination of CM maturation. We postulate that there is a critical perinatal window, ranging from embryonic day 18.5 to postnatal day 14 in mice, in which the maturation process is exquisitely sensitive to perturbation. While the initiation mechanisms of this process are unknown, it is increasingly clear that maturation proceeds through interconnected regulatory circuits that feed into one another to coordinate concomitant structural, functional and metabolic CM maturation. We highlight PGC1α, SRF and the MEF2 family as transcription factors that may potentially mediate this cross-talk. We lastly discuss several emerging technologies that will facilitate future studies into the mechanisms of CM maturation. Further study will not only produce a better understanding of its key processes, but provide practical insights into developing a robust strategy to produce mature PSC-CMs.
Graphical Abstract
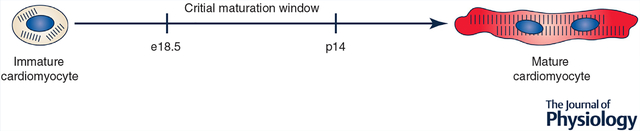
Here, we postulate that there is a critical window, ranging from embryonic day 18.5 to postnatal day 14 in mice, in which interconnected regulatory circuits enable coordinated, concomitant structural, functional and metabolic cardiomyocyte maturation.
There is tremendous enthusiasm in the application of pluripotent stem cell (PSC) technology to cardiovascular medicine. PSCs (including embryonic stem cells (ESCs) and induced PSCs (iPSCs)) can be expanded and subsequently differentiated into cardiomyocytes (CMs) in vitro. Because adult CMs are non-proliferative and difficult to obtain from human patients, PSC-derived CMs (PSC-CMs) serve as the most viable approach to generating large quantities of CMs ex vivo (Batalov & Feinberg, 2015). Among many potential uses, PSC-CMs have been proposed for application in regenerative medicine, drug and toxicity screening, and disease modelling (Passier et al. 2006; du Pré et al. 2013; Wang et al. 2013; Chen et al. 2016; Youssef et al. 2016). Given the burden of cardiovascular disease (Benjamin et al. 2017) and the need for improved cardiac cellular models (Gwathmey et al. 2009), PSC-CMs offer potential solutions to significant unmet health needs in the United States.
Extensive analysis has led to an improved understanding of regulatory pathways in early cardiogenesis. This in turn has allowed for the development of improved and optimized protocols, inspired by in vivo development, to differentiate PSCs to CMs in vitro (Kattman et al. 2011; Burridge et al. 2012; Mummery et al. 2012; Uosaki et al. 2012; Cao et al. 2013; Lian et al. 2013). Despite these successes, however, use of PSC-CMs has been limited due to the failure of these cells to mature to a fully adult phenotype in vitro. In vivo, mature CMs demonstrate increased size, polyploidization, development of well-formed sarcomeres, improved calcium handling, t-tubules, large numbers of mitochondria, primarily oxidative metabolism, and other characteristic features (Robertson et al. 2013; Yang et al. 2014). However, PSC-CMs fail to show these same characteristics, and instead resemble mid-to-late embryonic CMs, even following extended culture (Beqqali et al. 2006; Cao et al. 2008; Snir et al. 2009; Davis et al. 2011; Robertson et al. 2013; Keung et al. 2014; Yang et al. 2014; Uosaki et al. 2015; Veerman et al. 2015). These differences significantly impede further research and clinical use of PSC-CMs.
As with advances in PSC-CM differentiation, improved maturation of PSC-CMs will likely require deeper understanding of CM maturation as it occurs during development in vivo. To date, the regulatory mechanisms underlying CM maturation remain an open area of investigation. The developing heart is a highly dynamic environment, characterized by changes in mechanical forces, electrical coupling, extracellular matrix composition, oxygen and cytokine gradients, and many others (Robertson et al. 2013; Yang et al. 2014); it is unclear which, if any, of these properties serves as upstream regulators or downstream effects of others. As many facets of maturation are conserved between species, animal models (including rodent as well as large animal, such as sheep, models) have been a primary tool for understanding regulation of CM maturation in vivo. Knowledge from these animal studies has been further supplemented by in vitro studies seeking to recapitulate aspects of the native milieu (Scuderi & Butcher, 2017). Though no single ex vivo method has captured the full complexity of in vivo maturation nor generated fully mature CMs, each has furthered our knowledge of the biological underpinnings of CM maturation.
In this review, we aim to summarize potential regulatory mechanisms linking together the multiple facets of CM maturation. As our focus is on in vivo maturation, we primarily consider studies utilizing animal models. While in vivo human data regarding maturation is limited, where appropriate, we also comment on informative results from in vitro studies utilizing human PSC-CMs. We postulate a postnatal ‘critical window’ for maturation, in which interconnected positive feedback circuits regulate concomitant structural, functional, metabolic and transcriptomic CM maturation. We discuss the implications of this critical window in the generation of mature PSC-CMs. We lastly look forward to new technologies that may enable further studies of regulatory mechanisms in CM maturation.
Dynamics of CM maturation
The temporal dynamics of CM maturation have been comprehensively reviewed elsewhere (Robertson et al. 2013; Galdos et al. 2017). Nevertheless, we briefly summarize some of the most important aspects here, focusing particularly on the rodent model. In the perinatal period, CMs undergo significant structural changes as they transition from small, round cells with disorganized features to large, cylindrical cells with highly organized components. Beginning at embryonic day 18.5 (e18.5) and continuing through the first weeks of birth, CMs increase in cell length and length-to-width ratio while displaying longer and more aligned myofibrils (Hirschy et al. 2006). From e19.5 to postnatal day 7 (p7), several sarcomeric proteins (including myosin heavy chain, cardiac troponin T, and cardiac troponin I) undergo isoform switching to enable more efficient contraction (Siedner et al. 2003; Yin et al. 2015). CMs fully cease proliferation around p3–p7 (Porrello et al. 2011; Notari et al. 2018) and subsequently undergo hypertrophic growth with polyploidization and multinucleation (Leu et al. 2001; Liu et al. 2010). From an electrophysiological perspective, ion channel activity and localization changes significantly postnatally (Liu et al. 2002; Harrell et al. 2007). Maturation of cell-cell and cell-ECM junctions, particularly at the intercalated disc, occur during the first week of birth (Wu et al. 2002; Hirschy et al. 2006). T-tubulation, which enables efficient excitation-contraction coupling, initiates at approximately p6 but continues through 2—3 weeks of birth (Sedarat et al. 2000). Lastly, CMs undergo several adaptive metabolic changes. While mitochondrial remodelling begins as early as e13.5 (Hom et al. 2011), significant improvements in mitochondrial number, size, distribution and internal organization occur in the peri- and postnatal periods (Hallman, 1971; Smolich et al. 1989; Marin-Garcia et al. 2000; Vega et al. 2015). Likewise, from p0 to p7, CMs undergo a transition from primarily glycolytic metabolism to one reliant on oxidative phosphorylation, and the primary energy source transitions from glucose to fatty acids (Marsh & Marsh, 1991; Itoi & Lopaschuk, 1993; Lopaschuk & Jaswal, 2010). Taken as a whole, these adaptive processes enable mature myocytes to meet their highly energetic demands. While many key changes occur up through the first week of birth, CM maturation progressively continues, with CMs reaching their maximal volume at approximately 3 months of age (Leu et al. 2001).
Species-specific differences in CM maturation
The hearts of different species have notably different force generation demands; correspondingly adult myocytes show phenotypic differences across species, particularly in terms of contractile and electrophysiological properties (Milani-Nejad & Janssen, 2015). While variations in maturation dynamics between species remain relatively unstudied, some prominent differences have been observed. For example, in mice, heart rate increases from approximately 150—190 beats per minute (bpm) at e12.5 to 245 bpm at e19.5 and eventually reaches a mature heart rate of 310–840 bpm (Yu et al. 2008). In contrast, the human fetal heart rate is 120–160 bpm, and progressively slows during maturation to achieve an adult heart rate of 60–100bpm(Pildner von Steinburg etal. 2013).As a result, the shape of the rodent action potential is different from that of the human action potential, with a notably shorter duration and lack of a prominent plateau phrase. The differences are further reflected in expression dynamics of various isoforms of contractile proteins. While isoform switching is observed across species in CM maturation, the specific switches depend on each particular species’ cardiac contraction/relaxation demands. One particularly well-known example of this phenomenon is myosin heavy chain (MHC) isoform switching. In rodent ventricles, MHC transitions from the β-isoform to the α-isoform over the course of maturation, enabling improved contractile velocity. In human ventricles, the β-isoform increases over maturation, enabling improved contractile economy and lower tension cost (Milani-Nejad & Janssen, 2015). T-tubulation dynamics also show species differences. Unlike in smaller mammals, primitive t-tubules may be observed in fetal development in large mammals such as sheep, cows, rhesus monkeys and humans (Kim et al. 1992). Lastly, nucleation dynamics also demonstrate significant species variations in maturation. For example, while terminal differentiation of CMs to multinucleated, quiescent cells occurs largely postnatally in rodents, this process may be up to 75% complete prior to birth in sheep (Burrell et al. 2003; Jonker et al. 2007). While binucleation occurs prenatally in humans, as in sheep, adult human hearts present with a notably smaller number of binucleated myocytes (25–60%) compared to both rodents and sheep (~90% binucleated) (Botting et al. 2012). It is estimated that human CMs reach an adult-like state by 10 years of age (Takamatsu et al. 1983; Peters et al. 1994).
Further investigation is required to understand the mechanisms underlying these species-specific differences in CM maturation dynamics and adult CM properties. Intriguingly, comparative gene expression analysis indicates that transcriptomic changes across maturation are broadly conserved in mouse and humans (Uosaki & Taguchi, 2016). On the other hand, xenogenic transplantations of PSC-CMs typically yield only partially mature myocytes, suggesting significant environmental or regulatory differences between species with regards to maturation (Dai et al. 2007; Laflamme et al. 2007; Shiba et al. 2012; Chong et al. 2014; Liu et al. 2018). It is possible that while gene trends remain similar across species, species-specific gene dosages or relative gene expression levels contribute to species differences in maturation. Moreover, these species-specific effects may be regulated through post-translational processes. Identifying a mechanism underlying species-specific differences in CM maturation remains a significant unanswered question in cardiac developmental biology.
Initiation of CM maturation
To date, the factor(s) responsible for initiating CM maturation remain unknown. Birth, which is accompanied by significant changes in haemodynamics, oxygenation and biochemical milieu (Rudolph, 1970; Dawes et al. 1980; Teitel et al. 1987), has frequently been thought of as a trigger for maturation. During birth, the closure of shunts in fetal circulation results in significant changes in cardiac load and output (Agata et al. 1991; Schubert et al. 2013), which in turn promotes structural, contractile and force generation improvements in CMs (Barbera et al. 2000; Ruan et al. 2015). Oxygenation of arterial blood doubles (Teitel et al. 1987), leading to metabolic and mitochondrial maturation through reduction in HIF1a signalling (Breckenridge et al. 2013; Neary et al. 2014). The timely regulation of these changes is critical; for example, in pediatric patients, when the fetal-to-neonatal transition in circulation does not occur (often called ‘persistent fetal circulation’), myocardial maturation may become delayed (Hines, 2013). In several studies conducted in both humans and other animals, premature birth was found to be associated with abnormal CM hypertrophy and nucleation and premature cell cycle cessation (Bensley et al. 2010, 2018; Aye et al. 2017). As a caveat, however, patients in these studies were often treated antenatally and/or postnatally with corticosteroids, which may in turn affect CM maturation (Agnew et al. 2017).
Despite these observations, the idea of birth as a triggering event for CM maturation has been contested (Jonker et al. 2015). Indeed, many facets of maturation appear to be initiated in utero, including increase in myocyte length and volume, myofibril length and number, sarcomeric isoform switching, cessation of cell cycle, aerobic metabolism, and other changes (Kim et al. 1992; Burrell et al. 2003; Siedner et al. 2003; Hirschy et al. 2006; Jonker et al. 2007, 2015; Porter et al. 2011; Baker & Ebert, 2013; Yin et al. 2015). Moreover, disruption of the intrauterine environment, particularly in the period prior to birth, can lead to retardation of CM maturation (Bubb et al. 2007). Thus, it appears that the initiation of CM maturation occurs in the period prior to birth (e.g. e16.5–e18.5 in mice, gestational days 110–130 of 147 in sheep, pregnancy weeks 28–32 in human), perhaps in anticipation of significant changes in cardiac function in the fetal-to-neonatal transition.
The evolving hormonal and neuroendocrine environment of the fetus may influence these maturation phenomena. For example, in mice, endogenous glucocorticoid production increases from e15.5 and peaks at e17.5; correspondingly, maternal plasma glucocorticoid levels peak at approximately e16–e17 (Rog-Zielinska et al. 2015). This spike in glucocorticoids has been shown to mediate CM myofibrillar structure, oxygen consumption, ion channel expression and calcium handling, with many of these processes mediated in a PGC1α-dependent manner (Rog-Zielinska et al. 2013, 2015). Similarly, in rodents, thyroid hormone secretion initiates at approximately e17.5, and mediates transcriptional effects on CM contractile function, among other effects (Li et al. 2014). Other hormones that have been implicated in CM maturation include IGF1 and NRG1 (Rupert & Coulombe, 2017).
Evidence for a critical perinatal window in maturation
While the initiation of CM maturation is still unknown, recent data strongly demonstrate the critical importance of the perinatal cardiac environment in regulating CM maturation. For example, we recently demonstrated that transplantation of ESC-CMs into the neonatal rat heart at p0–p3 led to generation of CMs that were structurally, functionally and transcriptomically indistinguishable from adult CMs (Cho et al. 2017). Similarly, Kadota et al. (2017) have shown that full maturation of neonatal rat ventricular CMs (NRVCMs) is possible following transplantation in the neonatal heart. On the other hand, results of same-species transplantation experiments in adult hearts have been more equivocal. For example, some studies have observed significant and near-complete maturation of transplanted cells by structural and electrophysiological analysis, including primary cells (Klug et al. 1996; Gojo et al. 1997; Roell et al. 2002; Rubart et al. 2003) and PSC-CMs (Didié et al. 2013). On the other hand, other studies have shown only partial and limited maturation even following extended transplantation, with failure to achieve full adult size and structure (Leor et al. 1996; Watanabe et al. 1998; Reinecke et al. 1999; Müller-Ehmsen et al. 2002; Christoforou et al. 2010; Shiba et al. 2016). Likewise, we have observed that transplantation of ESC-CMs at p14 resulted in limited maturation with incomplete sarcomere alignment (Cho et al. 2017). In addition to these transplantation experiments, other studies have demonstrated that the proliferation-to-hypertrophy transition (Anatskaya et al. 2010) and mitochondrial/metabolic maturation (Gong et al. 2015) processes are exquisitely sensitive to perturbation up to approximately 2—3 weeks after birth in rodents. Based on these data, we believe that the perinatal time period (which we define as e18.5–p14 in rodents) may represent a critical window for CM maturation, analogous to the perinatal regenerative window. Perturbations to normal developmental phenomena during this time period may lead to an immature CM phenotype.
Critical window for maturation in PSC-CMs
Intriguingly, a similar critical window for maturation may be observed in PSC-CM differentiation. In vitro, PSC-CMs mature through the first 20 days of culture before undergoing maturation arrest (Uosaki et al. 2015). At this time, they structurally, functionally and transcriptomically resemble fetal CMs (Robertson et al. 2013; Galdos et al. 2017), though they display numerous aberrant regulatory networks (Uosaki et al. 2015). While it is thought that long term culture improves maturation of PSC-CMs (Kamakura et al. 2013; Kuppusamy et al. 2015; Dias et al. 2018), recent analyses have suggested that even at 1 year of culture, PSC-CMs continue to resemble late embryonic/early fetal CMs (DeLaughter et al. 2016). We hypothesize that, analogous to in vivo development, PSC-CMs are receptive to signalling cues prompting them to undergo maturation during an early critical window. Perturbation during this window (e.g. due to stresses induced by cell culture) leads to failure of complete maturation.
This hypothesis is supported by transplantation studies. For example, Kadota et al. (2017). observed that transplantation of early human PSC-CMs at day 5 of differentiation into neonatal rat heart led to significantly improved maturation over PSC-CMs transplanted at days 18–20 of differentiation. Similarly, we have observed complete maturation of mouse ESC-CMs transplanted into neonatal heart at days 5–7 of differentiation (Cho et al. 2017), but incomplete maturation of the same cells when transplanted at day 14+ of differentiation (authors’ unpublished data). We summarize these results in Fig. 1. It is possible that the results of these transplantation studies are confounded by difficulties in handling/dissociating late stage PSC-CMs from culture or their poor retention in vivo. Interestingly, however, Ronaldson-Bouchard et al. (2018) recently described a similar phenomenon in an ex vivo bioreactor system in which iPSC-CMs were subjected to various electrical simulation regimes. They found that an intensity training-based simulation protocol resulted in significant force generation, calcium handling and ultrastructural maturation in early stage (day 12) iPSC-CMs but only limited maturation in late stage (day 28) iPSC-CMs.
Figure 1. Summary of transplantation experiments for PSC-CM maturation.
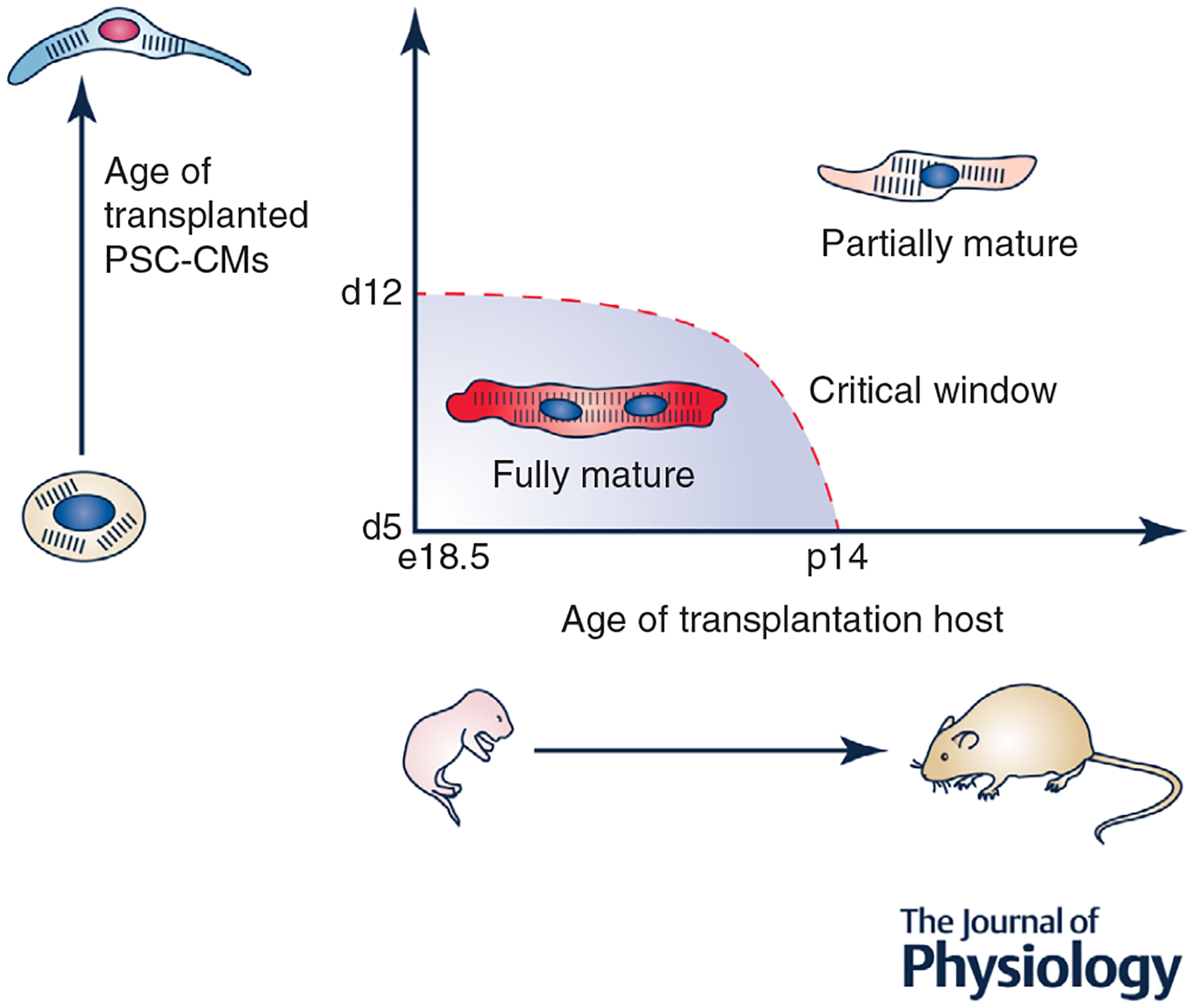
When early PSC-CMs are transplanted in vivo during the perinatal period, they achieve full structural, functional and transcriptomic maturity. However, when either late PSC-CMs are transplanted or an older host is used, only partial maturation occurs. These results support the existence of a critical window for CM maturation both in vitro and in vivo.
These results have significant research implications for the use of PSC-CMs. In particular, interventions designed to improve the maturation of PSC-CMs may be limited if they use cells that have already passed the critical window and have undergone subsequent maturation arrest. Likewise, early stage PSC-CMs may be most optimal for regenerative medicine therapies to ensure that transplanted cells achieve full maturity in their transplanted niche.
Regulation of coordinated maturation processes
While a number of studies have aimed to identify factors regulating various individual aspects of CM maturation (Galdos et al. 2017)., it is still unclear how maturation is coordinated. A number of individual maturation-related processes have been identified, as discussed in previous sections (e.g. structural, functional, metabolic, cell cycle maturation). However, a major open question is whether these processes are independently regulated or, if they are co-regulated, whether they are organized hierarchically or interdependently. To date, it has been difficult to individually manipulate each maturation-related process in vivo to observe the effect on other processes. In vitro studies, typically using PSC-CM models, have facilitated this type of perturbation study. Intriguingly, several studies have hinted at significant co-regulation of maturation processes. As an example, fatty acid treatment of PSC-CMs not only results in expected improvements in PSC-CM metabolic maturation, but also leads to improvements in sarcomeric gene expression and structure, calcium handling and cell cycle inhibition (Correia et al. 2017; Mills et al. 2017). This in turn suggests potential regulation of structural, functional and cell cycle maturation through a metabolic mechanism. In Table 1, we summarize studies in which perturbation of one facet of CM maturation results in novel observations of improvements in other maturation-related processes. What emerges from these data supports the notion of maturation as composed of intertwined regulatory circuits that feed into one another to allow concomitant structural and functional maturation (Fig. 2).
Table 1.
In vitro studies demonstrate co-regulated circuits in maturation
| Perturbation | Primary response | Secondary effect | Reference |
|---|---|---|---|
| Structural | |||
| Anisotropic ECM micropatterning |
|
|
(Ribeiro et al. 2015; Lyra-Leite et al. 2017) |
| 3D CM aggregate formation |
|
|
(Correia et al. 2018) |
| Functional | |||
| Auxotonic contraction of engineered heart tissue (EHT) |
|
|
(Ulmer et al. 2018) |
| Electrical stimulation with increasing intensity |
|
|
(Ronaldson-Bouchard et al. 2018) |
| Metabolic | |||
| Differentiation with fatty acids and galactose |
|
|
(Correia et al. 2017) |
| Palmitate treatment |
|
|
(Mills et al. 2017) |
| Glucose deprivation following differentiation |
|
|
(Nakano et al. 2017) |
| Cell cycle | |||
| Mitomycin C treatment |
|
|
(Zhou et al. 2017) |
Here, we describe several in vitro studies of myocyte maturation in which one pathway of maturation (e.g. structural, functional, metabolic, cell cycle) was perturbed experimentally. We describe putative primary and secondary effects of the intervention. This table is not an exhaustive list of tissue engineered approaches to improved CM maturation; for more comprehensive reviews on that topic, please see Zhu et al. (2014) and Scuderi & Butcher (2017). Instead, we compile studies in which perturbation of one maturation pathway led to previously undescribed changes to other maturation-related processes, suggesting potential co-regulation between pathways.
Figure 2. Cross-talk between processes involved in CM maturation.
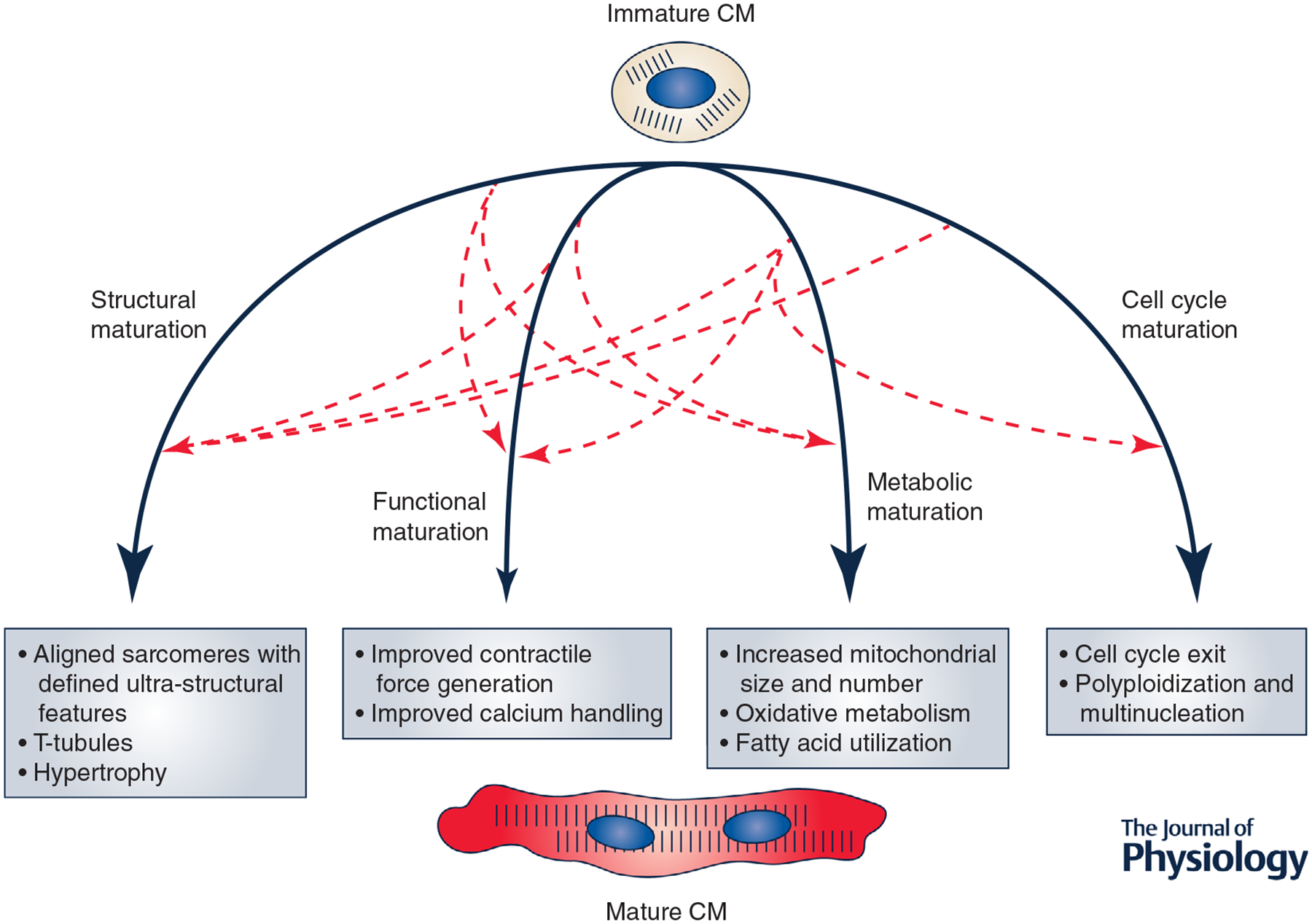
During maturation, CMs undergo significant changes in structure, function, metabolism and cell cycle, among other processes. Evidence from various in vitro studies suggests that these processes may function in an interdependent manner, allowing for coordinated CM maturation.
How the interplay between the various facets of CM maturation is regulated remains unknown. It is still unclear whether circuits are co-regulated through direct means, for example a common upstream transcriptional mechanism, or through indirect methods. Moreover, while in vitro methods have been highly informative in terms of demonstrating potential co-regulation, the numerous differences between experimental methodologies, as well as potential divergence from in vivo biology, have made it difficult to identify common mechanisms responsible for coordination of maturation. We anticipate that new studies will shine further light on this question.
Nevertheless, given the tight coordination of the maturation regulatory network, we predict the existence of factors that must be upstream of and simultaneously directly regulate multiple facets of maturation. Here we highlight three factors that may fit this description: PGC1α, SRF and MEF2. We do not intend this to be an exhaustive list, but rather highlight these factors because they have previously been implicated in multiple maturation circuits in vivo. These factors are known to interact with one another and have been implicated as key nodes in the CM maturation regulatory network (Xu et al. 2009; Schlesinger et al. 2011). Moreover, PGC1α and SRF in particular are dysregulated in PSC-CM maturation and may contribute to maturation arrest (Uosaki et al. 2015). While it is possible that these factors may exhibit species-specific regulatory patterns, the largely conserved nature of these factors suggests they may play a significant role in CM maturation across species (Uosaki & Taguchi, 2016).
PGC1α
Peroxisome proliferator-activated receptor γ (PPARγ) coactivator-1α (PGC1α) is a critical regulator of mitochondrial biogenesis in a variety of tissues (Finck & Kelly, 2006) and plays a role in metabolic regulation in the heart in development and disease (Duncan & Finck, 2008). In addition to PPARg, PGC1α binds to a large number of nuclear receptor and non-nuclear receptor transcription factors to mediate chomatin remodelling and gene transcription (Finck & Kelly, 2006). In the mouse fetal heart, PGC1α is a target of glucocorticoid activity (Rog-Zielinska et al. 2015) and is expressed beginning at e15.5 (Lai et al. 2008). Critically, PGC1α has been implicated in mediating the metabolic switch away from glycolysis to oxidative phosphorylation and fatty acid metabolism (Lehman & Kelly, 2002). Over-expression of PGC1α at birth leads to a dramatic increase in mitochondrial volume density and size (Russell et al. 2004). Developmentally, PGC1α has partial redundancy with PGC1b; indeed, while knockout of either alone leads to a minimal phenotype under physiological conditions, the double knockout is lethal shortly after birth (Lai et al. 2008). PGC1α/b−/− mice show severe defects in mitochondrial number and size, and demonstrate a failure to transition from anaerobic glycolysis to oxidative metabolism with fatty acid utilization. Interestingly, PGC1α may have effects on CM sarcomeric structure and function as well. For example, the same study showed that PGC1α/b−/− mice display CMs with significant disarray or even absence of sarcomeric structure. In an in vitro study of fetal myocytes, PGC1α siRNA-mediated knockdown eliminated myofibril maturation induced by glucocorticoid treatment (Rog-Zielinska et al. 2015). Likewise, PGC1α knockdown in PSC-CMs led to decreased CM beat rate, altered action potential and a failure of sarcomeric integrity (Birket et al. 2013). Currently, it is thought that PGC1α mediates its effects on sarcomeric organization and contractile function indirectly through energetic/metabolic regulation (Birket et al. 2013; Rog-Zielinska et al. 2015), though studying maturation-specific direct targets of PGC1α is an area of ongoing investigation.
SRF
Serum response factor (SRF) has been implicated in key processes in mesoderm formation and muscle development (Arsenian et al. 1998), and is essential to cardiomyocyte differentiation and maturation (Parlakian et al. 2004; Dirkx et al. 2013). CM-specific deletion of SRF leads to embryonic lethality between e10.5 and e13.5 in mice, and is characterized by failure of chamber maturation and disruption of the CM contractile apparatus (Parlakian et al. 2004; Balza & Misra, 2006). SRF also mediates cardiac function postnatally. For example, Zhang et al. (2001) generated a CM-specific SRF mutant with impaired binding of SRF to target binding sites. These mice died within 12 days of birth and demonstrated significant dilated cardiomyopathy. SRF disruption in adult mice similarly leads to dilated cardiomyopathy and heart failure-induced death, with significant defects in CM structural integrity and contractile function (Parlakian et al. 2005). It is increasingly recognized that SRF forms a key node in the cardiac transcription network, and may regulate a range of CM processes including contraction, conduction, growth/apoptosis, miRNA regulation, and others (Schlesinger et al. 2011; Schueler et al. 2012). Intriguingly, SRF may mediate its effects in a stage-specific manner, and may play a particularly critical role in perinatal CM maturation. In a recent study, CRISPR/Cas9-driven knockdown of SRF at p1 led to CM defects in cell size, sarcomeric structure, and T-tubulation, as well as gene-expression changes related to mitochondrial biogenesis and oxidative metabolism (Guo et al. 2018a). SRF may regulate these latter processes in a multifactorial way - both directly through target gene binding and indirectly through disruption of overall CM cytoarchitecture (Schlesinger et al. 2011; Guo et al. 2018a).
MEF2 family
The myocyte enhancer factor 2 family (MEF2) consists of a family of transcription factors responsible for regulating a range of processes in cardiac development and differentiation. The full dynamics of MEF2 isoform expression is outside the scope of this review and may be found elsewhere (Desjardins & Naya, 2016). Nevertheless, we summarize by noting that expression of individual isoforms of MEF2 initiates between e7.5 and e8.5 in mice; postnatally, MEF2A, MEF2C and MEF2D are expressed (Iida et al. 1999). In the perinatal period, these MEF2 isoforms may have non-overlapping and even potentially antagonistic function (Desjardins & Naya, 2017). In particular, MEF2A and MEF2D appear to be required for cell-cycle inhibition and activation of sarcomeric gene expression, while MEF2C performs the opposite function. MEF2A knockout in vivo leads to death within the first week of life, with mice exhibiting significant myofibrillar disarray, mitochondrial disorganization, and failure to activate mature gene expression patterns (Naya et al. 2002). By contrast, the MEF2D knockout has no phenotype under physiological conditions, though these mice display attenuated hypertrophy and remodelling following application of cardiac stressors (Kim et al. 2008). Intriguingly, while MEF2C has primarily been implicated in early cardiac development (Lin et al. 1997), it may regulate mitochondrial function and oxidative metabolism during the perinatal period (Desjardins & Naya, 2017).
Emerging technologies for studying CM maturation
Elucidating the CM maturation regulatory network remains a major area of investigation. It is being appreciated that this network is extraordinarily complex, comprising not only transcription factors (such as those discussed above) but other regulatory molecules such as microRNAs (Kuppusamy et al. 2015; Lee et al. 2015; White et al. 2016; Alfar et al. 2018) and long non-coding RNAs (Touma et al. 2016), as well as epigenetic regulation (Schlesinger et al. 2011). Here, we survey two major scientific tools that we believe will influence ongoing research in maturation: CASAAV and transcriptomics.
CASAAV
In vivo analysis of factors regulating maturation has been limited, owing not only to the challenge of generating mouse models for a large number of candidates, but also due to confounding results from secondary effects of heart failure in knockdown models. The recent development of the CRISPR/Cas9/AAV (CASAAV)-based somatic mutagenesis platform may facilitate future in vivo loss-of-function studies (VanDusen et al. 2017; Guo et al. 2018b). In this system, an AAV9 vector delivers guide RNAs (under a ubiquitous promoter) and Cre (under a CM-specific promoter) to Cre-dependent Cas9-P2A-GFP knock-in mice. Thus, knockdown of target genes is done in a CM-specific mosaic pattern, enabling the study of cell autonomous effects of knockdown without confounding secondary effects. Moreover, the use of the CRISPR/Cas9 system enables rapid testing of many target genes. Thus far, this system has been used to study the cell autonomous effects of a variety of genes in cardiac maturation, including junctophilin-2 (Guo et al. 2018b), GATA4/6 (Prendiville et al. 2015) and SRF (Guo et al. 2018a).
Transcriptomics
In addition to novel methods for performing in vivo studies such as the CASAAV system, improving technologies in the field of transcriptomics (particularly RNA-sequencing (RNA-seq)) will enable an improved understanding of regulatory networks in CM maturation. To date, transcriptomic analysis has been used to: elucidate stage-specific regulatory networks guiding cardiac development and maturation (Uosaki et al. 2015); identify nucleosome and histone-modifying genes in maturation (van den Berg et al. 2015); identify the role of Let-7 family of microRNAs in guiding CM maturation through metabolic switch (Kuppusamy et al. 2015); and identify miR-200c as a regulator of mature ion channel expression and calcium handling (Poon et al. 2018). In concert with chromatin immunoprecipitation-sequencing (ChIP-seq), transcriptomic analyses have also been used to develop a stronger understanding of epigenetic dynamics of maturation (Sim et al. 2015; Gilsbach et al. 2018). Lastly, transcriptomics has provided a powerful tool to benchmark the maturation status of in vitro-generated CM tissues (Kuppusamy et al. 2015; Uosaki et al. 2015; van den Berg et al. 2015; DeLaughter et al. 2016).
Single cell RNA-seq (scRNA-seq) represents a major opportunity for a further understanding of CM maturation. DeLaughter et al. (2016) performed a seminal study in which they generated scRNA-seq libraries for > 1,200 cardiac cells from various developmental time points ranging from e9.5 to p21. They subsequently analyzed the developmental dynamics of CMs. As expected, they observed distinct stage-specific progression of maturation, with a notable transition from e14.5 to e18.5/p0 representing the initiation of perinatal maturation. Crucially, however, while transition states were observable on bulk, there was significant heterogeneity of maturation state at any given discrete time point. In particular, between e18.5 and p3, individual cells spanned an overlapping spectrum of maturation states before converging to a final mature phenotype at p21. These results indicate that maturation may be best viewed at the level of the single cell which, upon receiving the appropriate cues, proceeds through maturation at its own unique rate before reaching maturity (Fig. 3A).
Figure 3. scRNA-seq enables improved understanding of CM maturation.

A, scRNA-seq data of CMs at various stages of development reveals that while maturation proceeds in a stage-specific manner, individual CMs proceed heterogeneously across the maturation trajectory before converging on the final mature phenotype. B, scRNA-seq profiles may enable more precise benchmarking of PSC-CM maturation. By comparing individual PSC-CMs to in vivo CM data, their position along the maturation trajectory can be ascertained and used to quantify a maturation score for single cells. This approach has enabled us to identify, for example, that PSC-CMs transplanted in vivo achieve a maturation score greater than those cultured in vitro and comparable with adult CMs (authors’ unpublished data).
The use of scRNA-seq may additionally provide a powerful method for benchmarking the precise maturation state of PSC-CMs (Fig. 3B). Indeed, to date, one of the primary challenges in PSC-CM research is the lack of a consensus metric or metrics to precisely quantify maturation state, particularly with reference to physiological maturation in vivo. scRNA-seq is a particularly useful tool as it integrates information from the range of phenomena perturbed in maturation (e.g. sarcomeric, electrophysiological, metabolic, cell cycle and other changes). By comparing scRNA-seq profiles of PSC-CMs to the inferred trajectory of in vivo CM maturation, the maturation state of PSC-CMs can be quantified in a biologically meaningful manner. This analysis may facilitate more comparable and reproducible studies of CM maturation, and may allow further biological insight to be gleaned from studies using PSC-CMs as model systems.
Conclusion
The biology of CM maturation remains a fast-moving and highly exciting area of research, with emerging technologies offering new opportunities for insight. Here, we aim to emphasize the perinatal period as a critical window for maturation, consisting of interconnected regulatory modules guiding concommitant structural and functional maturation of CMs. We believe that new breakthroughs in understanding CM maturation can be leveraged towards improving patient health.
Acknowledgements
We thank Renjun Zhu for providing helpful comments and feedback in preparing this manuscript.
Funding
NICHD/NIH (R01HD086026), AHA and The Magic that Matters Fund.
Biography
Suraj Kannan is an MD/PhD student at Johns Hopkins University. His career goal is to help translate regenerative medicine therapies to alleviate cardiovascular disease. Chulan Kwon is an Associate Professor of Medicine at Johns Hopkins University Heart & Vascular Institute and Institute for Cell Engineering. His lab focuses on studying the molecular underpinnings of cardiomyocyte maturation with the goal of improving the maturation of pluripotent-stem cell derived cardiomyocytes for regenerative medicine and disease modelling.

Footnotes
This review was presented at the symposium ‘2018 Gordon Research Conference on Cardiac Regulatory Mechanisms’, which took place at Colby-Sawyer College, New London, NH, USA, 3–8 June 2018.
Competing interests
The authors declare no competing interests with regard to this manuscript.
References
- Agata Y, Hiraishi S, Oguchi K, Misawa H, Horiguchi Y, Fujino N, Yashiro K & Shimada N (1991). Changes in left ventricular output from fetal to early neonatal life. J Pediatr 119, 441–445. [DOI] [PubMed] [Google Scholar]
- Agnew EJ, Agnew EJ, Ivy JR, Stock SJ & Chapman KE (2017). Glucocorticoids, antenatal corticosteroid therapy and fetal heart maturation. J Mol Endocrinol 61, R61–R73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfar EA, El-Armouche A & Guan K (2018). MicroRNAs in cardiomyocyte differentiation and maturation. Cardiovasc Res 114, 779–781. [DOI] [PubMed] [Google Scholar]
- Anatskaya OV, Sidorenko NV, Beyer TV & Vinogradov AE (2010). Neonatal cardiomyocyte ploidy reveals critical windows of heart development. Int J Cardiol 141, 81–91. [DOI] [PubMed] [Google Scholar]
- Arsenian S, Weinhold B, Oelgeschlä Ger M, Rü Ther U & Nordheim A (1998). Serum response factor is essential for mesoderm formation during mouse embryogenesis. EMBO J 17, 6289–6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aye CYL, Lewandowski AJ, Lamata P, Upton R, Davis E, Ohuma EO, Kenworthy Y, Boardman H, Wopperer S, Packham A, Adwani S, McCormick K, Papageorghiou AT & Leeson P (2017). Disproportionate cardiac hypertrophy during early postnatal development in infants born preterm. Pediatr Res 82, 36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker CN & Ebert SN (2013). Development of aerobic metabolism in utero: requirement for mitochondrial function during embryonic and foetal periods. OA Biotechnol 2, 1–7. [Google Scholar]
- Balza RO & Misra RP (2006). Role of the serum response factor in regulating contractile apparatus gene expression and sarcomeric integrity in cardiomyocytes. J Biol Chem 281, 6498–6510. [DOI] [PubMed] [Google Scholar]
- Barbera A, Giraud GD, Reller MD, Maylie J, Morton MJ & Thornburg KL (2000). Right ventricular systolic pressure load alters myocyte maturation in fetal sheep. Am J Physiol Regul Integr Comp Physiol 279, R1157–R1164. [DOI] [PubMed] [Google Scholar]
- Batalov I & Feinberg AW (2015). Differentiation of cardiomyocytes from human pluripotent stem cells using monolayer culture. Biomark Insights 10, 71–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin EJ et al. , American Heart Association Statistics Committee and Stroke Statistics Subcommittee (2017). Heart Disease and Stroke Statistics—2017 Update: A Report From the American Heart Association. Circulation 135, e146–e603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensley JG, Moore L, De Matteo R, Harding R & Black MJ (2018). Impact of preterm birth on the developing myocardium of the neonate. Pediatr Res 83, 880–888. [DOI] [PubMed] [Google Scholar]
- Bensley JG, Stacy VK, De Matteo R, Harding R & Black MJ (2010). Cardiac remodelling as a result of pre-term birth: Implications for future cardiovascular disease. Eur Heart J 31, 2058–2066. [DOI] [PubMed] [Google Scholar]
- Beqqali A, Kloots J, Ward-van Oostwaard D, Mummery C & Passier R (2006). Genome-wide transcriptional profiling of human embryonic stem cells differentiating to cardiomyocytes. Stem Cells 24, 1956–1967. [DOI] [PubMed] [Google Scholar]
- Birket MJ, Casini S, Kosmidis G, Elliott DA, Gerencser AA, Baartscheer A, Schumacher C, Mastroberardino PG, Elefanty AG, Stanley EG & Mummery CL (2013). PGC-1α and reactive oxygen species regulate human embryonic stem cell-derived cardiomyocyte function. Stem Cell Reports 1, 560–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botting K, Wang K, Padhee M, McMillen I, Summers-Pearce B, Rattanatray L, Cutri N, Posterino G, Brooks D & Morrison J (2012). Early origins of heart disease: Low birth weight and determinants of cardiomyocyte endowment. Clin Exp Pharmacol Physiol 39, 814–823. [DOI] [PubMed] [Google Scholar]
- Breckenridge RA, Piotrowska I, Ng KE, Ragan TJ, West JA, Kotecha S, Towers N, Bennett M, Kienesberger PC, Smolenski RT, Siddall HK, Offer JL, Mocanu MM, Yelon DM, Dyck JRB, Griffin JL, Abramov AY, Gould AP & Mohun TJ (2013). Hypoxic regulation of hand1 controls the fetal-neonatal switch in cardiac metabolism. PLoS Biol 11, 4–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubb KJ, Cock ML, Black MJ, Dodic M, Boon WM, Parkington HC, Harding R & Tare M (2007). Intrauterine growth restriction delays cardiomyocyte maturation and alters coronary artery function in the fetal sheep. J Physiol 578, 871–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrell JH, Boyn AM, Kumarasamy V, Hsieh A, Head SI & Lumbers ER (2003). Growth and maturation of cardiac myocytes in fetal sheep in the second half of gestation. Anat Rec - Part A Discov Mol Cell Evol Biol 274, 952–961. [DOI] [PubMed] [Google Scholar]
- Burridge PW, Keller G, Gold JD & Wu JC (2012). Production of de novo cardiomyocytes: Human pluripotent stem cell differentiation and direct reprogramming. Cell Stem Cell 10, 16–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao F, Wagner RA, Wilson KD, Xie X, Fu J-D, Drukker M, Lee A, Li RA, Gambhir SS, Weissman IL, Robbins RC & Wu JC (2008). Transcriptional and functional profiling of human embryonic stem cell-derived cardiomyocytes. PLoS One 3, e3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao N, Liang H, Huang J, Wang J, Chen Y, Chen Z & Yang HT (2013). Highly efficient induction and long-term maintenance of multipotent cardiovascular progenitors from human pluripotent stem cells under defined conditions. Cell Res 23, 1119–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen IY, Matsa E & Wu JC (2016). Induced pluripotent stem cells: At the heart of cardiovascular precision medicine. Nat Rev Cardiol 13, 333–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho GS, Lee DI, Tampakakis E, Murphy S, Andersen P, Uosaki H, Chelko S, Chakir K, Hong I, Seo K, Chen HSV, Chen X, Basso C, Houser SR, Tomaselli GF, O’Rourke B, Judge DP, Kass DA & Kwon C (2017). Neonatal transplantation confers maturation of PSC-derived cardiomyocytes conducive to modeling cardiomyopathy. Cell Rep 18, 571–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong JJH, Yang X, Don CW, Minami E, Liu YW, Weyers JJ, Mahoney WM, Van Biber B, Cook SM, Palpant NJ, Gantz JA, Fugate JA, Muskheli V, Gough GM, Vogel KW, Astley CA, Hotchkiss CE, Baldessari A, Pabon L, Reinecke H, Gill EA, Nelson V, Kiem HP, Laflamme MA & Murry CE (2014). Human embryonic-stem-cell-derived cardiomyocytes regenerate non-human primate hearts. Nature 510, 273–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoforou N, Oskouei BN, Esteso P, Hill CM, Zimmet JM, Bian W, Bursac N, Leong KW, Hare JM & Gearhart JD (2010). Implantation of mouse embryonic stem cell-derived cardiac progenitor cells preserves function of infarcted murine hearts. PLoS One 5, e11536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correia C, Koshkin A, Duarte P, Hu D, Carido M, Sebastião MJ, Gomes-Alves P, Elliott DA, Domian IJ, Teixeira AP, Alves PM & Serra M (2018). 3D aggregate culture improves metabolic maturation of human pluripotent stem cell derived cardiomyocytes. Biotechnol Bioeng 115, 630–644. [DOI] [PubMed] [Google Scholar]
- Correia C, Koshkin A, Duarte P, Hu D, Teixeira A, Domian I, Serra M & Alves PM (2017). Distinct carbon sources affect structural and functional maturation of cardiomyocytes derived from human pluripotent stem cells. Sci Rep 7, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai W, Field LJ, Rubart M, Reuter S, Hale SL, Zweigerdt R, Graichen RE, Kay GL, Jyrala AJ, Colman A, Davidson BP, Pera M & Kloner RA (2007). Survival and maturation of human embryonic stem cell-derived cardiomyocytes in rat hearts. J Mol Cell Cardiol 43, 504–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RP, van den Berg CW, Casini S, Braam SR & Mummery CL (2011). Pluripotent stem cell models of cardiac disease and their implication for drug discovery and development. Trends Mol Med 17, 475–484. [DOI] [PubMed] [Google Scholar]
- Dawes BYGS, Johnston BM & Walker DW (1980). Relationship of arterial pressure and heart rate in fetal, new-born, and adult sheep. J Physiol 405–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLaughter DM, Bick AG, Wakimoto H, McKean D, Gorham JM, Kathiriya IS, Hinson JT, Homsy J, Gray J, Pu W, Bruneau BG, Seidman JG & Seidman CE (2016). Single-cell resolution of temporal gene expression during heart development. Dev Cell 39, 480–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjardins C & Naya F (2016). The function of the MEF2 family of transcription factors in cardiac development, cardiogenomics, and direct reprogramming. J Cardiovasc Dev Dis 3, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjardins CA & Naya FJ (2017). Antagonistic regulation of cell-cycle and differentiation gene programs in neonatal cardiomyocytes by homologous MEF2 transcription factors. J Biol Chem 292, 10613–10629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias TP, Pinto SN, Santos JI, Fernandes TG, Fernandes F, Diogo MM, Prieto M & Cabral JMS (2018). Biophysical study of human induced pluripotent stem cell-derived cardiomyocyte structural maturation during long-term culture. Biochem Biophys Res Commun 499, 611–617. [DOI] [PubMed] [Google Scholar]
- Didié M, Field LJ, Didié M, Christalla P, Rubart M & Muppala V (2013). Parthenogenetic stem cells for tissue- engineered heart repair Find the latest version: Technical advance Parthenogenetic stem cells for tissue-engineered heart repair. J Clin Invest 123, 1285–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirkx E, da Costa Martins PA & De Windt LJ (2013). Regulation of fetal gene expression in heart failure. Biochim Biophys Acta - Mol Basis Dis 1832, 2414–2424. [DOI] [PubMed] [Google Scholar]
- Duncan JG & Finck BN (2008). The PPARα-PGC-1α axis controls cardiac energy metabolism in healthy and diseased myocardium. PPAR Res 2008, 253817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- du Pré BC, Doevendans PA & van Laake LW (2013). Stem cells for cardiac repair: an introduction. J Geriatr Cardiol 10, 186–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finck BN & Kelly DP (2006). PGC-1 coactivators: Inducible regulators of energy metabolism in health and disease. J Clin Invest 116, 615–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galdos FX, Guo Y, Paige SL, Vandusen NJ, Wu SM & Pu WT (2017). Cardiac regeneration: Lessons from development. Circ Res 120, 941–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilsbach R, Schwaderer M, Preissl S, Grüning BA, Kranzhöfer D, Schneider P, Nührenberg TG, Mulero-Navarro S, Weichenhan D, Braun C, Dreßen M, Jacobs AR, Lahm H, Doenst T, Backofen R, Krane M, Gelb BD & Hein L (2018). Distinct epigenetic programs regulate cardiac myocyte development and disease in the human heart in vivo. Nat Commun 9, 391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gojo S, Kitamura S, Hatano O, Takakusa A, Hashimoto K, Kanegae Y & Saito I (1997). Transplantation of genetically marked cardiac muscle cells. J Thorac Cardiovasc Surg 113, 10–18. [DOI] [PubMed] [Google Scholar]
- Gong G, Song M, Csordas G, Kelly DP, Matkovich SJ & Dorn GW (2015). Parkin-mediated mitophagy directs perinatal cardiac metabolic maturation in mice. Science 350, aad2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Jardin BD, Zhou P, Sethi I, Akerberg BN, Toepfer CN, Ai Y, Li Y, Ma Q, Guatimosim S, Hu Y, Varuzhanyan G, Vandusen NJ, Zhang D, Chan DC, Yuan G-C, Seidman CE, Seidman JG & Pu WT (2018a). Hierarchical and stage-specific regulation of cardiomyocyte maturation by serum response factor. Nat Commun 9, 3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Vandusen NJ, Zhang L, Gu W, Sethi I, Jardin BD, Ai Y, Zhang D, Chen B, Yuan G, Song L, Pu WT, Biology C, De Minas UF, Horizonte B, City I & City I (2018b). Platform for rapid dissection of cardiac myocyte gene function. Circ Res 120, 1874–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwathmey JK, Tsaioun K & Hajjar RJ (2009). Cardionomics: a new integrative approach for screening cardiotoxicity of drug candidates. Expert Opin Drug Metab Toxicol 5, 647–660. [DOI] [PubMed] [Google Scholar]
- Hallman M (1971). Changes in mitochondrial respiratory chain proteins during perinatal development. Biochim Biophys Acta 360–372. [DOI] [PubMed] [Google Scholar]
- Harrell MD, Harbi S, Hoffman JF, Zavadil J & Coetzee WA (2007). Large-scale analysis of ion channel gene expression in the mouse heart during perinatal development. Genomics, 273–283. [DOI] [PubMed] [Google Scholar]
- Hines MH (2013). Neonatal cardiovascular physiology. Semin Pediatr Surg 22, 174–178. [DOI] [PubMed] [Google Scholar]
- Hirschy A, Schatzmann F, Ehler E & Perriard JC (2006). Establishment of cardiac cytoarchitecture in the developing mouse heart. Dev Biol 289, 430–441. [DOI] [PubMed] [Google Scholar]
- Hom JR, Quintanilla RA, Hoffman DL, de Mesy Bentley KL, Molkentin JD, Sheu SS & Porter GA (2011). The permeability transition pore controls cardiac mitochondrial maturation and myocyte differentiation. Dev Cell 21, 469–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida K, Hidaka K, Takeuchi M, Nakayama M, Yutani C, Mukai T & Morisaki T (1999). Expression of MEF2 genes during human cardiac development. Tohoku J Exp Med 187, 15–23. [DOI] [PubMed] [Google Scholar]
- Itoi T & Lopaschuk GD (1993). The contribution of glycolysis, glucose oxidation, lactate oxidation, and fatty acid oxidation to ATP production in isolated biventricular working hearts from 2-week-old rabbits. Pediatr Res 34, 735–741. [DOI] [PubMed] [Google Scholar]
- Jonker SS, Louey S, Giraud GD, Thornburg KL & Faber JJ (2015). Timing of cardiomyocyte growth, maturation, and attrition in perinatal sheep. FASEB J 29, 4346–4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonker SS, Zhang L, Louey S, Giraud GD, Thornburg KL & Faber JJ (2007). Myocyte enlargement, differentiation, and proliferation kinetics in the fetal sheep heart. J Appl Physiol 102, 1130–1142. [DOI] [PubMed] [Google Scholar]
- Kadota S, Pabon L, Reinecke H & Murry CE (2017). In vivo maturation of human induced pluripotent stem cell-derived cardiomyocytes in neonatal and adult rat hearts. Stem Cell Reports 8, 278–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamakura T, Makiyama T, Sasaki K, Yoshida Y, Wuriyanghai Y, Chen J, Hattori T, Ohno S, Kita T, Horie M, Yamanaka S & Kimura T (2013). Ultrastructural maturation of human-induced pluripotent stem cell-derived cardiomyocytes in a long-term culture. Circ J 77, 1307–1314. [DOI] [PubMed] [Google Scholar]
- Kattman SJ, Witty AD, Gagliardi M, Dubois NC, Niapour M, Hotta A, Ellis J & Keller G (2011). Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell 8, 228–240. [DOI] [PubMed] [Google Scholar]
- Keung W, Boheler KR & Li RA (2014). Developmental cues for the maturation of metabolic, electrophysiological and calcium handling properties of human pluripotent. Stem Cell Res Ther 5, 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Kim D, Lee I, Rah B, Sawa Y & Schaper J (1992). Human fetal heart development after mid-term: Morphometry and ultrastructural study. J Mol Cell Cardiol 24, 949–965. [DOI] [PubMed] [Google Scholar]
- Kim Y, Phan D, Van Rooij E, Wang DZ, McAnally J, Qi X, Richardson JA, Hill JA, Bassel-Duby R & Olson EN (2008). The MEF2D transcription factor mediates stress-dependent cardiac remodeling in mice. J Clin Invest 118, 124–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug MG, Soonpaa MH, Koh GY & Field LJ (1996). Genetically selected cardiomyocytes from differentiating embryonic stem cells form stable intracardiac grafts. J Clin Invest 98, 216–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuppusamy KT, Jones DC, Sperber H, Madan A, Fischer KA, Rodriguez ML, Pabon L, Zhu W-Z, Tulloch NL, Yang X, Sniadecki NJ, Laflamme MA, Ruzzo WL, Murry CE & Ruohola-Baker H (2015). Let-7 family of microRNA is required for maturation and adult-like metabolism in stem cell-derived cardiomyocytes. Proc Natl Acad Sci U S A 112, E2785–E2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laflamme MA, Chen KY, Naumova AV, Muskheli V, Fugate JA, Dupras SK, Reinecke H, Xu C, Hassanipour M, Police S, O’Sullivan C, Collins L, Chen Y, Minami E, Gill EA, Ueno S, Yuan C, Gold J & Murry CE (2007). Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat Biotechnol 25, 1015–1024. [DOI] [PubMed] [Google Scholar]
- Lai L, Leone TC, Zechner C, Schaeffer PJ, Kelly SM, Flanagan DP, Medeiros DM, Kovacs A & Kelly DP (2008). Transcriptional coactivators PGC-lα and PGC-lβ control overlapping programs required for perinatal maturation of the heart. Genes Dev 22, 1948–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DS, Chen JH, Lundy DJ, Liu CH, Hwang SM, Pabon L, Shieh RC, Chen CC, Wu SN, Yan YT, Lee ST, Chiang PM, Chien S, Murry CE & Hsieh PCH (2015). Defined microRNAs induce aspects of maturation in mouse and human embryonic-stem-cell-derived cardiomyocytes. Cell Rep 12, 1960–1967. [DOI] [PubMed] [Google Scholar]
- Lehman JJ & Kelly DP (2002). Transcriptional activation of energy metabolic switches in the developing and hypertrophied heart. Clin Exp Pharmacol Physiol 29, 339–345. [DOI] [PubMed] [Google Scholar]
- Leor J, Patterson M, Quinones MJ, Kedes LH & Kloner RA (1996). Transplantation of fetal myocardial tissue into the infarcted myocardium of rat. A potential method for repair of infarcted myocardium? Circulation 94, II332–336. [PubMed] [Google Scholar]
- Leu M, Ehler E & Perriard J (2001). Characterisation of postnatal growth of the murine heart. Anat Embryol 204, 217–224. [DOI] [PubMed] [Google Scholar]
- Li M, Iismaa SE, Naqvi N, Nicks A, Husain A & Graham RM (2014). Thyroid hormone action in postnatal heart development. Stem Cell Res 13, 582–591. [DOI] [PubMed] [Google Scholar]
- Lian X, Zhang J, Azarin SM, Zhu K, Hazeltine LB, Bao X, Hsiao C, Kamp TJ & Palecek SP (2013). Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/β-catenin signaling under fully defined conditions. Nat Protoc 8, 162–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q, Schwarz J, Bucana C & Olson EN (1997). Control of mouse cardiac morphogenesis and myogenesis by transcription factor MEF2C. Science 276, 1404–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Yasui K, Opthof T, Ishiki R, Lee JK, Kamiya K, Yokota M & Kodama I (2002). Developmental changes of Ca2+ handling in mouse ventricular cells from early embryo to adulthood. Life Sci 71, 1279–1292. [DOI] [PubMed] [Google Scholar]
- Liu Y-W Chen B, Yang X, Fugate JA, Kalucki FA, Futakuchi-Tsuchida A, Couture L, Vogel KW, Astley CA, Baldessari A, Ogle J, Don CW, Steinberg ZL, Seslar SP, Tuck SA, Tsuchida H, Naumova AV, Dupras SK, Lyu MS, Lee J, Hailey DW, Reinecke H, Pabon L, Fryer BH, MacLellan WR, Thies RS & Murry CE (2018). Human embryonic stem cell - derived cardiomyocytes restore function in infarcted hearts of non-human primates. Nat Biotechnol 36, 597–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Yue S, Chen X, Kubin T & Braun T (2010). Regulation of cardiomyocyte polyploidy and multinucleation by cyclinG1. Circ Res 106, 1498–1506. [DOI] [PubMed] [Google Scholar]
- Lopaschuk GD & Jaswal JS (2010). Energy metabolic phenotype of the cardiomyocyte during development, differentiation, and postnatal maturation. J Cardiovasc Pharmacol 56, 130–140. [DOI] [PubMed] [Google Scholar]
- Lyra-Leite DM, Andres AM, Petersen AP, Ariyasinghe NR, Cho N, Lee JA, Gottlieb RA & McCain ML (2017). Mitochondrial function in engineered cardiac tissues is regulated by extracellular matrix elasticity and tissue alignment. Am J Physiol Heart Circ Physiol 313, H757–H767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin-Garcia J, Ananthakrishnan R & Goldenthal MJ (2000). Heart mitochondrial DNA and enzyme changes during early human development. Mol Cell Biochem 210, 47–52. [DOI] [PubMed] [Google Scholar]
- Marsh R & Marsh DR (1991). Glycolysis production is predominant source of myocardial immediately after birth ATP. Am J Physiol 1698–1705. [DOI] [PubMed] [Google Scholar]
- Milani-Nejad N & Janssen PM (2015). Small and large animal models in cardiac contraction research: advantages and disadvantages. Pharmacol Ther 141, 235–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills RJ, Titmarsh DM, Koenig X, Parker BL, Ryall JG, Quaife-Ryan GA, Voges HK, Hodson MP, Ferguson C, Drowley L, Plowright AT, Needham EJ, Wang QD, Gregorevic P, Xin M, Thomas WG, Parton RG, Nielsen LK, Launikonis BS, James DE, Elliott DA, Porrello ER & Hudson JE (2017). Functional screening in human cardiac organoids reveals a metabolic mechanism for cardiomyocyte cell cycle arrest. Proc Natl Acad Sci U S A 114, E8372–E8381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller-Ehmsen J, Whittaker P, Kloner RA, Dow JS, Sakoda T, Long TI, Laird PW & Kedes L (2002). Survival and development of neonatal rat cardiomyocytes transplanted into adult myocardium. J Mol Cell Cardiol 34, 107–116. [DOI] [PubMed] [Google Scholar]
- Mummery CL, Zhang J, Ng E, Elliott DA, Elefanty AG & Kamp TJ (2012). Differentiation of Human ES and iPS cells to cardiomyocytes: a methods overview. Circ Res 111, 344–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano H, Minami I, Braas D, Pappoe H, Wu X, Sagadevan A, Vergnes L, Fu K, Morselli M, Dunham C, Ding X, Stieg AZ, Gimzewski JK, Pellegrini M, Clark PM, Reue K, Lusis AJ, Ribalet B, Kurdistani SK, Christofk H, Nakatsuji N & Nakano A (2017). Glucose inhibits cardiac muscle maturation through nucleotide biosynthesis. Elife 6, 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naya FJ, Black BL, Wu H, Bassel-Duby R, Richardson JA, Hill JA & Olson EN (2002). Mitochondrial deficiency and cardiac sudden death in mice lacking the MEF2A transcription factor. Nat Med 8, 1303–1309. [DOI] [PubMed] [Google Scholar]
- Neary MT, Ng KE, Ludtmann MHR, Hall AR, Piotrowska I, Ong SB, Hausenloy DJ, Mohun TJ, Abramov AY & Breckenridge RA (2014). Hypoxia signaling controls postnatal changes in cardiac mitochondrial morphology and function. J Mol Cell Cardiol 74, 340–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notari M, Ventura-Rubio A, Bedford-Guaus SJ, Jorba I, Mulero L, Navajas D, Martí M & Raya Á (2018). The local microenvironment limits the regenerative potential of the mouse neonatal heart. Sci Adv, 4, eaao5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parlakian A, Charvet C, Escoubet B, Mericskay M, Molkentin JD, Gary-Bobo G, De Windt LJ, Ludosky MA, Paulin D, Daegelen D, Tuil D & Li Z (2005). Temporally controlled onset of dilated cardiomyopathy through disruption of the srf gene in adult heart. Circulation 112, 2930–2939. [DOI] [PubMed] [Google Scholar]
- Parlakian A, Tuil D, Hamard G, Hentzen D, Concordet J, Li Z & Daegelen D (2004). Targeted inactivation of serum response factor in the developing heart results in myocardial defects and embryonic lethality. Mol Cell Biol 24, 5281–5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passier R, Denning C & Mummery C (2006). Cardiomyocytes from human embryonic stem cells. Stem Cells Handb Exp Pharmacol 1, 101–122. [PubMed] [Google Scholar]
- Peters NS, Severs NJ, Rothery SM, Lincoln C, Yacoub MH & Green CR (1994). Spatiotemporal relation between gap junctions and fascia adherens junctions during postnatal development of human ventricular myocardium. Circulation 90, 713–725. [DOI] [PubMed] [Google Scholar]
- Pildner von Steinburg S, Boulesteix A-L, Lederer C, Grunow S, Schiermeier S, Hatzmann W, Schneider K-TM & Daumer M (2013). What is the “normal” fetal heart rate? PeerJ 1, e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon ENY, Hao B, Guan D, Jun Li M, Lu J, Yang Y, Wu B, Wu SCM, Webb SE, Liang Y, Miller AL, Yao X, Wang J, Yan B & Boheler KR (2018). Integrated transcriptomic and regulatory network analyses identify microRNA-200c as a novel repressor of human pluripotent stem cell-derived cardiomyocyte differentiation and maturation. Cardiovasc Res 114, 894–906. [DOI] [PubMed] [Google Scholar]
- Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, Olson EN & Sadek HA (2011). Transient regenerative potential of the neonatal mouse heart. Science 331, 1078–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter GA, Hom JR, Hoffman DL, Quintanilla RA, Bentley KLdM & Sheu SS (2011). Bioenergetics, mitochondria, and cardiac myocyte differentiation. Prog Pediatr Cardiol 31, 75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prendiville TW, Guo H, Lin Z, Zhou P, Stevens SM, He A, VanDusen N, Chen J, Zhong L, Wang DZ, Gao G & Pu WT (2015). Novel roles of GATA4/6 in the postnatal heart identified through temporally controlled, cardiomyocyte-specific gene inactivation by adeno-associated virus delivery of Cre recombinase. PLoS One 10, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinecke H, Zhang M, Bartosek T & Murry CE (1999). A study in normal and injured rat hearts. Circulation 100, 193–202. [DOI] [PubMed] [Google Scholar]
- Ribeiro AJS, Ang Y-S, Fu J-D, Rivas RN, Mohamed TMA, Higgs GC, Srivastava D & Pruitt BL (2015). Contractility of single cardiomyocytes differentiated from pluripotent stem cells depends on physiological shape and substrate stiffness. Proc Natl Acad Sci U S A 112, 12705–12710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson C, Tran DD & George SC (2013). Concise review: Maturation phases of human pluripotent stem cell-derived cardiomyocytes. Stem Cells 31, 829–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roell W, Lu ZJ, Bloch W, Siedner S, Tiemann K, Zia Y, Stoecker E, Fleischmann M, Bohlen H, Stehle R, Kolossov E, Brem G, Addicks K, Pfitzer G, Welz A, Hescheler J & Fleischmann BK (2002). Cellular cardiomyoplasty improves survival after myocardial injury. Circulation 105, 2435–2441. [DOI] [PubMed] [Google Scholar]
- Rog-Zielinska EA, Craig MA, Manning JR, Richardson RV, Gowans GJ, Dunbar DR, Gharbi K, Kenyon CJ, Holmes MC, Hardie DG, Smith GL & Chapman KE (2015). Glucocorticoids promote structural and functional maturation of foetal cardiomyocytes: A role for PGC-1α. Cell Death Differ 22, 1106–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rog-Zielinska EA, Thomson A, Kenyon CJ, Brownstein DG, Moran CM, Szumska D, Michailidou Z, Richardson J, Owen E, Watt A, Morrison H, Forrester LM, Bhattacharya S, Holmes MC & Chapman KE (2013). Glucocorticoid receptor is required for foetal heart maturation. Hum Mol Genet 22, 3269–3282. [DOI] [PubMed] [Google Scholar]
- Ronaldson-Bouchard K, Ma SP, Yeager K, Chen T, Song LJ, Sirabella D, Morikawa K, Teles D, Yazawa M & Vunjak-Novakovic G (2018). Advanced maturation of human cardiac tissue grown from pluripotent stem cells. Nature 556, 239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan J-L, Tulloch NL, Saiget M, Paige SL, Razumova MV, Regnier M, Tung KC, Keller G, Pabon L, Reinecke H & Murry CE (2015). Mechanical stress promotes maturation of human myocardium from pluripotent stem cell-derived progenitors. Stem Cells 33, 2148–2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubart M, Pasumarthi KBS, Nakajima H, Soonpaa MH, Nakajima HO & Field LJ (2003). Physiological coupling of donor and host cardiomyocytes after cellular transplantation. Circ Res 92, 1217–1224. [DOI] [PubMed] [Google Scholar]
- Rudolph AM (1970). The changes in the circulation after birth. Their importance in congenital heart disease. Circulation 41, 343–359. [DOI] [PubMed] [Google Scholar]
- Rupert CE & Coulombe KLK (2017). IGF1 and NRG1 enhance proliferation, metabolic maturity, and the force-frequency response in hESC-derived engineered cardiac tissues. Stem Cells Int 2017, 7648409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell LK, Mansfield CM, Lehman JJ, Kovacs A, Courtois M, Saffitz JE, Medeiros DM, Valencik ML, McDonald JA & Kelly DP (2004). Cardiac-specific induction of the transcriptional coactivator peroxisome proliferator-activated receptor γ coactivator-1α promotes mitochondrial biogenesis and reversible cardiomyopathy in a developmental stage-dependent manner. Circ Res 94, 525–533. [DOI] [PubMed] [Google Scholar]
- Schlesinger J, Schueler M, Grunert M, Fischer JJ, Zhang Q, Krueger T, Lange M, Tönjes M, Dunkel I & Sperling SR (2011). The cardiac transcription network modulated by Gata4, Mef2a, Nkx2.5, Srf, histone modifications, and microRNAs. PLoS Genet 7, e1001313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert U, Müller M, Norman M & Abdul-Khaliq H (2013). Transition from fetal to neonatal life: Changes in cardiac function assessed by speckle-tracking echocardiography. Early Hum Dev 89, 803–808. [DOI] [PubMed] [Google Scholar]
- Schueler M, Zhang Q, Schlesinger J, Tönjes M & Sperling SR (2012). Dynamics of Srf, p300 and histone modifications during cardiac maturation in mouse. Mol Biosyst 8, 495–503. [DOI] [PubMed] [Google Scholar]
- Scuderi GJ & Butcher J (2017). Naturally engineered maturation of cardiomyocytes. Front Cell Dev Biol 5, 1–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedarat F, Xu L, Moore ED & Tibbits GF (2000). Colocalization of dihydropyridine and ryanodine receptors in neonate rabbit heart using confocal microscopy. Am J Physiol Heart Circ Physiol 279, H202–H209. [DOI] [PubMed] [Google Scholar]
- Shiba Y, Fernandes S, Zhu WZ, Filice D, Muskheli V, Kim J, Palpant NJ, Gantz J, Moyes KW, Reinecke H, Van Biber B, Dardas T, Mignone JL, Izawa A, Hanna R, Viswanathan M, Gold JD, Kotlikoff MI, Sarvazyan N, Kay MW, Murry CE & Laflamme MA (2012). Human ES-cell-derived cardiomyocytes electrically couple and suppress arrhythmias in injured hearts. Nature 489, 322–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba Y, Gomibuchi T, Seto T, Wada Y, Ichimura H, Tanaka Y, Ogasawara T, Okada K, Shiba N, Sakamoto K, Ido D, Shiina T, Ohkura M, Nakai J, Uno N, Kazuki Y, Oshimura M, Minami I & Ikeda U (2016). Allogeneic transplantation of iPS cell-derived cardiomyocytes regenerates primate hearts. Nature 538, 388–391. [DOI] [PubMed] [Google Scholar]
- Siedner S, Krüger M, Schroeter M, Metzler D, Roell W, Fleischmann BK, Hescheler J, Pfitzer G & Stehle R (2003). Developmental changes in contractility and sarcomeric proteins from the early embryonic to the adult stage in the mouse heart. J Physiol 548, 493–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim CB, Ziemann M, Kaspi A, Harikrishnan KN, Ooi J, Khurana I, Chang L, Hudson JE, El-Osta A & Porrello ER (2015). Dynamic changes in the cardiac methylome during postnatal development. FASEB J 29, 1329–1343. [DOI] [PubMed] [Google Scholar]
- Smolich JJ, Walker AM, Campbell GR & Adamson TM (1989). Left and right ventricular myocardial morphometry in fetal, neonatal, and adult sheep. Am J Physiol Heart Circ Physiol 257, H1–H9. [DOI] [PubMed] [Google Scholar]
- Snir M, Kehat I, Gepstein A, Coleman R, Livne E, Gepstein L, Snir M, Kehat I, Gepstein A, Coleman R, Itskovitz-eldor J, Livne E, Gepstein L, Itskovitz-eldor J & Livne E (2009). Assessment of the ultrastructural and proliferative properties of human embryonic stem cell-derived cardiomyocytes. Am J Physiol Heart Circ Physiol 285, H2355–H2363. [DOI] [PubMed] [Google Scholar]
- Takamatsu T, Nakanishi K, Fukuda M & Fujita S (1983). Cytofluorometric nuclear DNA-determinations in infant, adolescent, adult and aging human hearts. Histochemistry 77, 485–494. [DOI] [PubMed] [Google Scholar]
- Teitel DF, Iwamoto HS & Rudolph AM (1987). Effects of birth-related events on central blood flow patterns. Pediatr Res 22, 557–566. [DOI] [PubMed] [Google Scholar]
- Touma M, Kang Z, Zhao Y, Cass AA, Gao F, Biniwale R, Coppola G, Xiao X, Reemtsen B & Wang Y (2016). Decoding the long noncoding RNA during cardiac maturation: a roadmap for functional discovery. Circ Cardiovasc Genet 9, 395–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmer BM, Stoehr A, Schulze ML, Patel S, Gucek M, Mannhardt I, Funcke S, Murphy E, Eschenhagen T & Hansen A (2018). Contractile work contributes to maturation of energy metabolism in hiPSC-derived cardiomyocytes. Stem Cell Reports 10, 834–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uosaki H, Andersen P, Shenje LT, Fernandez L, Christiansen SL & Kwon C (2012). Direct contact with endoderm-like cells efficiently induces cardiac progenitors from mouse and human pluripotent stem cells. PLoS One 7, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uosaki H, Cahan P, Lee DI, Wang S, Miyamoto M, Fernandez L, Kass DA & Kwon C (2015). Transcriptional landscape of cardiomyocyte maturation. Cell Rep 13, 1705–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uosaki H & Taguchi YH (2016). Comparative gene expression analysis of mouse and human cardiac maturation. Genomics, Proteomics Bioinforma 14, 207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg CW, Okawa S, Chuva de Sousa Lopes SM, van Iperen L, Passier R, Braam SR, Tertoolen LG, del Sol A, Davis RP & Mummery CL (2015). Transcriptome of human foetal heart compared with cardiomyocytes from pluripotent stem cells. Development 142, 3231–3238. [DOI] [PubMed] [Google Scholar]
- VanDusen NJ, Guo Y, Gu W & Pu WT (2017). CASAAV: A CRISPR-based platform for rapid dissection of gene function in vivo. Curr Protoc Mol Biol 2017, 31.11.1–31. 11.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veerman CC, Kosmidis G, Mummery CL, Casini S, Verkerk AO & Bellin M (2015). Immaturity of human stem-cell-derived cardiomyocytes in culture: fatal flaw or soluble problem? Stem Cells Dev 24, 1035–1052. [DOI] [PubMed] [Google Scholar]
- Vega RB, Horton JL & Kelly DP (2015). Maintaining ancient organelles: mitochondrial biogenesis and maturation. Circ Res 116, 1820–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WE, Chen X, Houser SR & Zeng C (2013). Potential of cardiac stem/progenitor cells and induced pluripotent stem cells for cardiac repair in ischaemic heart disease. Clin Sci 125, 319–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe E, Smith DM, Delcarpio JB, Sun J, Smart FW, Van Meter CH & Claycomb WC (1998). Cardiomyocyte transplantation in a porcine myocardial infarction model. Cell Transplant 7, 239–246. [DOI] [PubMed] [Google Scholar]
- White MC, Pang L & Yang X (2016). MicroRNA-mediated maturation of human pluripotent stem cell-derived cardiomyocytes: Towards a better model for cardiotoxicity? Food Chem Toxicol 98, 17–24. [DOI] [PubMed] [Google Scholar]
- Wu JC, Sung HC, Chung TH & DePhilip RM (2002). Role of N-cadherin- and integrin-based costameres in the development of rat cardiomyocytes. J Cell Biochem 84, 717–724. [DOI] [PubMed] [Google Scholar]
- Xu XQ, Soo SY, Sun W & Zweigerdt R (2009). Global expression profile of highly enriched cardiomyocytes derived from human embryonic stem cells. Stem Cells 27, 2163–2174. [DOI] [PubMed] [Google Scholar]
- Yang X, Pabon L & Murry CE (2014). Engineering adolescence: maturation of human pluripotent stem cell-derived cardiomyocytes. Circ Res 114, 511–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Z, Ren J & Guo W (2015). Sarcomeric protein isoform transitions in cardiac muscle: A journey to heart failure. Biochim Biophys Acta - Mol Basis Dis 1852, 47–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youssef AA, Ross EG, Bolli R, Pepine CJ, Leeper NJ & Yang PC (2016). The promise and challenge of induced pluripotent stem cells for cardiovascular applications. JACC Basic to Transl Sci 1, 510–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Leatherbury L, Tian X & Lo CW (2008). Cardiovascular assessment of fetal mice by in utero echocardiography. Ultrasound Med Biol 34, 741–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Chai J, Azhar G, Sheridan P, Borras AM, Furr MC, Khrapko K, Lawitts J, Misra RP & Wei JY (2001). Early postnatal cardiac changes and premature death in transgenic mice overexpressing a mutant form of serum response factor. J Biol Chem 276, 40033–40040. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Wang L, Liu Z, Alimohamadi S, Yin C, Liu J & Qian L (2017). Comparative gene expression analyses reveal distinct molecular signatures between differentially reprogrammed cardiomyocytes. Cell Rep 20, 3014–3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu R, Blazeski A, Poon E, Costa KD, Tung L & Boheler KR (2014). Physical developmental cues for the maturation of human pluripotent stem cell-derived cardiomyocytes. Stem Cell Res Ther 5, 117. [DOI] [PMC free article] [PubMed] [Google Scholar]


