Abstract
Coronaviruses (CoVs) are causing a number of human and animal diseases because of their zoonotic nature such as Middle East respiratory syndrome (MERS), severe acute respiratory syndrome (SARS) and coronavirus disease 2019 (COVID-19). These viruses can infect respiratory, gastrointestinal, hepatic and central nervous systems of human, livestock, birds, bat, mouse, and many wild animals. The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a newly emerging respiratory virus and is causing CoVID-19 with high morbidity and considerable mortality. All CoVs belong to the order Nidovirales, family Coronaviridae, are enveloped positive-sense RNA viruses, characterised by club-like spikes on their surfaces and large RNA genome with a distinctive replication strategy. Coronavirus have the largest RNA genomes (~26–32 kilobases) and their expansion was likely enabled by acquiring enzyme functions that counter the commonly high error frequency of viral RNA polymerases. Non-structural proteins (nsp) 7–16 are cleaved from two large replicase polyproteins and guide the replication and processing of coronavirus RNA. Coronavirus replicase has more or less universal activities, such as RNA polymerase (nsp 12) and helicase (nsp 13), as well as a variety of unusual or even special mRNA capping (nsp 14, nsp 16) and fidelity regulation (nsp 14) domains. Besides that, several smaller subunits (nsp 7– nsp 10) serve as essential cofactors for these enzymes and contribute to the emerging “nsp interactome.” In spite of the significant progress in studying coronaviruses structural and functional properties, there is an urgent need to understand the coronaviruses evolutionary success that will be helpful to develop enhanced control strategies. Therefore, it is crucial to understand the structure, function, and interactions of coronaviruses RNA synthesizing machinery and their replication strategies.
Keywords: Coronaviruses, Human, Emerging, Control, Replication
1. Introduction
Coronaviruses (CoVs) are major threats to humans and vertebrate species. They can infect human, livestock, birds, bat, mouse and many other wild animals with the respiratory, gastrointestinal, hepatic and central nervous system infections [[1], [2], [3]]. The current classification of coronaviruses recognizes 39 species in 27 subgenera, five genera and two subfamilies that belong to the family Coronaviridae, suborder Cornidovirineae, order Nidovirales and realm Riboviria [4,5]. Alternatively, coronaviruses are divided into four genera on the basis of genetic and serologic properties; Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus in the subfamily Coronavirinae [[6], [7], [8], [9]]. While CoVs can infect many hosts [10], the coronaviruses infecting humans are all belonging to either alpha- or beta- CoVs. The outbreaks of severe acute respiratory syndrome (SARS), Middle East respiratory syndrome (MERS) and coronavirus disease 2019 (COVID-19) have shown the potential for transmission of newly emerging CoVs from animal to human and human to human [5,11,12]. The SARS-CoV proteins consist of two large polyproteins: ORF1a and ORF1ab (which cleavage proteolytically to shape 16 non-structural proteins) (Table 1 ). While accessory proteins have been found to be dispensable for in vitro viral replication, others have been shown to play a significant role in in vivo virus-host interactions [13]. Comparatively, the SARS-CoV-2 lacks the hemagglutinin esterase gene found in other human coronavirus (hCoV) HKU1, a lineage A betacoronavirus [[14], [15]]. It has been suggested that spike protein, envelope protein, membrane protein, nucleocapsid protein, 3CL protease, papain such as protease, RNA polymerase [16], and helicase protein are viable antiviral drug targets. The CoV outbreaks are highly likely to be unavoidable in the future due to climate and ecology changes, and increased human-animal interactions. Thus, the development of effective therapies and vaccines against CoVs is urgently needed.
Table 1.
| Nonstructural Protein (nsp) | Function |
|---|---|
| nsp 1 | Promotes cellular mRNA degradation and blocks host cell translation, results in blocking innate immune response |
| nsp 2 | No known function, binds to prohibitin proteins |
| nsp 3 | Large, multi-domain transmembrane protein, activities include: • Ubl1 and Ac domains, interact with N protein • ADRP activity, promotes cytokine expression • PLPro/Deubiquitinase domain, cleaves viral polyprotein and blocks host innate immune response • Ubl2, NAB, G2M, SUD, Y domains, unknown functions |
| nsp 4 | Potential transmembrane scaffold protein, important for proper structure of DMVs |
| nsp 5 | Mpro, cleaves viral polyprotein |
| nsp 6 | Potential transmembrane scaffold protein |
| nsp 7 | Forms hexadecameric complex with nsp8, may act as processivity clamp for RNA polymerase |
| nsp 8 | Forms hexadecameric complex with nsp7, may act as processivity clamp for RNA polymerase; may act as primase |
| nsp 9 | RNA binding protein phosphatase |
| nsp 10 | Cofactor for nsp16 and nsp14, forms heterodimer with both and stimulates ExoN and 2-O-MT activity |
| nsp 12 | Replication enzyme (RNA-dependent RNA polymerase) |
| nsp 13 | RNA helicase, 5′ triphosphatase |
| nsp 14 | N7 MTase and 3′–5′ exoribonuclease, ExoN; N7 MTase adds 5′ cap to viral RNAs, ExoN activity is important for proofreading of viral genome |
| nsp 15 | Viral endoribonuclease, NendoU |
| nsp 16 | 2′-O-MT; shields viral RNA from MDA5 recognition |
2. Virion properties and genome organization (with main focus on the nsps)
Coronaviruses are enveloped, 80–220 nm in size, pleomorphic but mostly spherical, and carry characteristic and large (20 nm long) club-shaped spikes (trimers spike protein). The combination of nucleocapsid (N) protein with the genomic RNA forms the helical nucleocapsid that is surrounded by a viral membrane (M) proteins which are composed of icosahedral structures. Some coronaviruses often have a second peripheral short (5 nm long) spikes (hemagglutinin-esterase (HE) protein), which is a peculiar feature of certain betacoronaviruses. Coronaviruses genome is linear positive-sense, infectious, single-stranded RNA 5′ capped and 3′ polyadenylated, the biggest known non-segmented RNA viral genomes (27.6–31 kb). However, the overall organization of the genomes is similar [6]. Maintaining such a large CoV genome may be linked to the unique features of the CoV replication transcription complex (RTC), which contains many RNA processing enzymes such as the non-structural protein 14's (nsp14's) 3′–5′ exoribonuclease. The 3′–5′ exoribonuclease is unique to CoVs in all RNA viruses and is likely to provide an RTC proofreading function [[16], [17], [18]]. The major virion proteins include a nucleocapsid protein (N, 50–60 kDa) and several envelope proteins; the spike glycoprotein trimer (S, 180–220 kDa per monomer), a triple-spanning transmembrane protein (M, 23–35 kDa) and a minor transmembrane protein (E, 9–12 kDa), which together with the M protein is essential for coronavirus virion assembly and budding. Cellular immune responses are generated primarily against the S and N proteins. The 5′-terminal two thirds of the genome include two open reading frames (ORFs), 1a and 1b, that together encode all non-structural proteins for the formation of the RTC, whereas the 3′- proximal third encodes the structural and accessory proteins [19]. ORF1a encodes polyprotein (pp) 1a containing nsp1-11, while ORF1a and ORF1b together produce pp1ab containing nsp1-16 through a (−1) ribosomal frameshift overreading the stop codon of ORF1a [20]. In general, SARS-CoV-2 has a total of 11 genes with 11 open reading frames (ORFs); ORF1ab, ORF2 (Spike protein), ORF3a, ORF4 (Envelope protein), ORF5 (Membrane protein), ORF6, ORF7a, ORF7b, ORF8, ORF9 (Nucleocapsid protein), and ORF10 [14]. Coronaviruses are unique among Nidoviruses because their genomes encode variable numbers of accessory proteins (four or five in majority; eight in the SARS coronaviruses) that are valueless during virus replication in vitro, but they improve the virus fitness in vivo.
3. Non-structural proteins (nsps) of coronaviruses
The enzymatic activities and functional domains of CoVs nsps are expected to be conserved between the various genera of CoVs, suggesting their significance in viral replication [6,21]. Apart from these nsps with established functions, there are other nsps whose biological functions and roles remain to be explored throughout the CoV life cycle. This review considers comprehensive analysis of the nsp of CoVs and to critically assess their functionalities among well-known coronaviruses.
3.1. Coronavirus nsp1
CoV nsp1's cumulative knowledge has confirmed the symmetric features and disparate mechanisms among various CoVs to block expression of the host gene and antagonise innate immune responses that can provide perspective into the expanding repertoire of new viral immune evasion strategies. Furthermore, despite the lack of obvious primary sequence homology within CoVs, there was also a significant correlation between the nsp1 of various CoVs belonging to different genera, inferring their evolutionary linkage and role in the adaptation of CoVs to different host species. Previous studies reported that the nsp1 proteins of SARS-CoV can promote host mRNA degradation, suppress host gene expression [[22], [23], [24]] and block host translational machinery function by binding to the ribosome small subunit [22]. Likewise, SARS-CoV nsp1 has a novel b-barrel structure mixed with helixes based on Nuclear magnetic resonance (NMR) analysis [25]. These studies suggest that coronavirus nsp1 is a major virulence and pathogenicity factor [22,23,26]. Overall, CoV nsp1, with its intriguing properties and characteristics, is an exciting avenue for future research that could potentially lead to the discovery of novel players and pathways of host gene regulation. The fact that nsp1 of various CoVs share a similar biological role to inhibit host gene expression using different modes of action has also posed some important questions about the effect of these functions and divergent mechanisms on the virulence and pathogenesis of emerging human CoVs.
3.2. Coronavirus nsp2
The nsp2 protein is an interesting target for genetic studies, as it has been reported that engineered mutations that eliminate cleavage at CS1 between nsp1 and nsp2 produce infectious virus [27], suggesting that nsp2 either maintains role in un-cleaved form or has a viral replication feature that can be dispensed with. Still, it is not known whether nsp2, as a mature protein or as a component of the polyprotein coronavirus, is essential for viral replication. Reverse genetic deletion of the MHV and SARS-CoV polyprotein nsp2 domains enabled the recovery of infectious mutants with growth deficiencies and RNA synthesis, and demonstrated intact polyprotein processing, including cleavage at engineered chimeric nsp1/3 cleavage sites. SARS-CoV holds the most similar general structure and sizes of nsps 1, 2, and 3 to betacoronaviruses [6,21]. However, there are also major variations between the MHV and SARS-CoV nsps 1, 2, and 3. The identification or resemblance between MHV and SARS-CoV nsp1 and nsp2 is very minimal [28,29]. Previous studies showed that nsp2 and nsp3 potentially originated as precursor proteins until they are transformed into mature nsp2 and nsp3 products. Because of the large size of this nsp2-3 precursor, previous studies identified it as 290-kDa [30] or 250-kDa [31] based on sodium dodecyl sulphate (SDS) polyacrylamide gel electrophoresis (PAGE). In addition, subsequent studies showed that the MHV and SARS-CoV replicase polyproteins nsp2 domains are not required for viral replication [32]. These findings indicate that the coronavirus polyprotein has significant structural and functional flexibility and that ORF1 encodes at least one and perhaps number of protein domains, which may be devoted to functions other than those of the product [32]. Reverse genetics studies were carried out to establish mutant MHV and SARS-CoV nsp2 knockout, the rescue viruses did not replicate, yet processed other replicase proteins correctly [32]. These findings put the basis for studies of replicase protein involvement in host pathogenesis, virus-cell interactions, and virus complementation and approaches to the development of stably attenuated animal and human coronaviruses [32].
3.3. Coronavirus nsp3
Nsp3 is the biggest multi-domain protein produced by coronaviruses, with different domain structure and organization in CoV genera. The individual coronaviruses may have 10 to 16 domains, 8 domains and two conserved transmembrane regions [33]. The nsp3 multidomain plays various roles in CoV infection; it releases nsp1, nsp2, and itself from the polyproteins and binds to form the replication/transcription complex with other viral nsps as well as RNA. Nsp3 acts on host protein post-translation modifications to antagonise the host's innate immune response (by de-MARylation, de-PARylation (possibly), deubiquitination, or deISGylation). Recent studies have shown that the biochemical characterization of SARS-CoV-2's deubiquitinating and deISGylating behaviours are closer to that of its counterpart in MERS-CoV than that of SARS-CoV. SARS-CoV-2 papain-like protease (PLpro) deISGylating activity appeared to be the most dominant of its diverse proteolytic functions and appeared to be species-specific [34]. Additionally, in host cells, nsp3 itself is changed, namely by N-glycosylation of the domain 3Ecto. Nsp3 may also interact with host proteins (such as RCHY1) to promote survival of viruses. Nsp3 was also identified as the largest non-structural protein of CoVs based on a high rate of positively selected mutation sites as the major selective target for driving evolution in CoVs [35]. The papain-like protease domain(s) releases nsp3 from polyprotein, which is (are) part of nsp3 itself [36]. Nsp3 plays major roles in the CoV life cycle; it can act as a scaffold protein to interact with itself and to bind to other viral nsps or host proteins [[37], [38], [39], [40]]. Nsp3 is essential for the formation of RTC, which in association with modified host ER membranes may result in formation of convoluted membranes (CMs) and double-membrane (DMVs) [[41], [42], [43], [44], [45], [46]]. Speculating why coronaviruses retain many essential functions in one protein is interesting, while nsp3 protein shows high-rate genetic diversity during CoV evolution. Ultimately, increased research into the structure and function of nsp3 is required to get a more complete understanding of this protein.
3.4. Coronavirus nsp4
Nsp4 is a transmembrane protein, 500 amino acid residues in length, and is the only protein of the viral polyprotein produced by both PLpro and Mpro after processing. Nsp4 has four transmembrane helices and a conserved cytosolic C-terminal domain throughout the Nidovirales [47], but only the nsp4 part of the C-terminal appears to be retained in the Nidovirales however, deletion of the C-terminal domain resulted in slightly reduction in growth [48]. It was also shown that nsp4 interacts with nsp2 in a two-hybrid yeast system [37] and in cells with other nsp4 molecules [45]. SARS-CoV nsp4 is an important component for viral double-membrane vesicle formation [43]. Studies on intracellular expression have shown a biological interaction between the carboxyl-terminal region of MHV (betacoronavirus) nsp3 and nsp4 [45], and full-length co-expression of SARS-CoV nsp3 and nsp4 results in comprehensive membrane pairing, where the paired membranes are kept at the same distance as the authentic DMVs [43].
3.5. Coronavirus nsp5
Coronavirus nsp5 is one of three parts of the coronavirus replicase machinery, together with the nsp12 polymerase and nsp13 helicase regions, that is preserved all over the Nidovirales [49]. Nsp5 is regarded as the main protease (Mpro), a protease similar to chymotrypsin related to the enteroviral 3C protease. It belongs to the endopeptidase's family C30 and is responsible for cleavage within polyprotein 1a/1 ab at 11 sequence specific sites. The resultant "mature" protein products (nsp4- 16) are assembled into replication complex components [36,50]. Nsp5 can be divided into three domains based on both structure and sequence characteristics that are conserved in all coronaviruses, Nidovirales and several other RNA viruses that share a similar processing scheme for polyproteins; a two-domain active region (I and II) and a third domain (III) play a role in nsp5 dimerization [36]. Previous study based on interactome analysis revealed that nsp12 and nsp14 can interact directly with nsp5 [51], and nsp14 and 16 can also interact indirectly with nsp5 as part of nsp10-14-16 complex [38,[51], [52], [53]]. Overall, this indicates that nsp5 plays a critical role in both RNA replication and in the formation of DMV, possibly by releasing nsp4 and nsp6 proteolytically. To date, nsp3, nsp5, nsp10, nsp12, nsp14 and nsp16 are the only proteins where temperature sensitive mutations have been discovered [[54], [55], [56]]. Nsp10 can interact directly with nsp5 [38], and paradoxically, both nsp10 and nsp3 mutations inhibit Mpro activity [56,57].
3.6. Coronavirus nsp6
While most nsp6 coronavirus proteins are predicted to contain seven transmembrane regions by TMHMM2.0 [58], only six of these functions as membrane-spanning helices. Nsp6 has six regions of transmembrane, with both termini on the cytosolic side of the membrane [[59], [60]]. Nsp6 over-expression disturbs intracellular membrane trafficking [[59], [60]], resulting in an accumulation of single membrane vesicles around the complex of microtubules [43]. It has also been demonstrated that SARS-CoV nsp6 interacts with nsp2, nsp8, nsp9 and accessory protein 9b by two-hybrid yeast assays [37]. It is interesting that both the 4Endo and 6Endo domains are just as well conserved in coronaviruses, as is the Mpro catalytic domain. Mapping the structural variations of the SARS-CoV-2 genome and selection trends, there were two mutations affecting non-structural protein 6 (nsp6) and Open Reading Frame10 (ORF 10) and associated with virus-host interaction, mainly cellular autophagy induced by viruses [61].
3.7. Coronavirus nsp7
The SARS-CoV (betacoronavirus) nsp7 protein structure (83-amino acid) was determined using both NMR [62] and X-ray crystallography with a hexadecameric supercomplex consisting of recombinant nsp7 and nsp8 [63]. Reverse-genetic studies aimed at particular residues within SARS-CoV nsp7 verified the significance of this protein for the virus replication [64], even though the effect of single point mutations was less than predicted based on the in vitro biochemical characterization of the RNA-binding properties of protein complexes containing nsp7. The nsp7-fold includes four helices with quiet different position and spatial orientation, suggesting that the protein's configuration is mainly affected by the interaction with nsp8 [65].
3.8. Coronavirus nsp8 and nsp7– nsp8 complexes
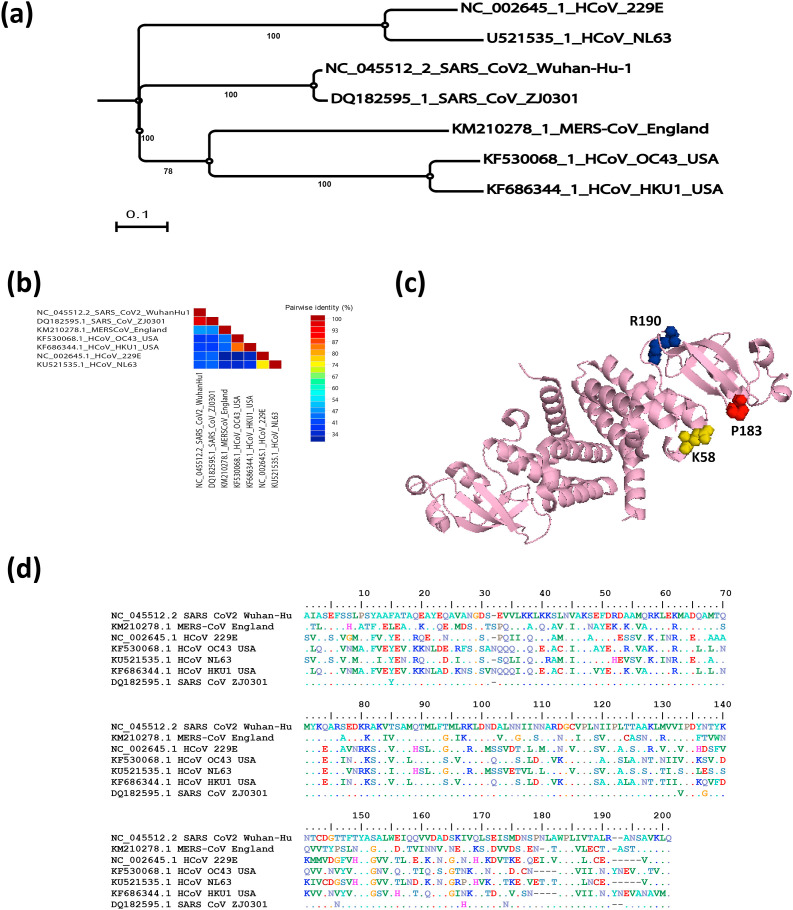
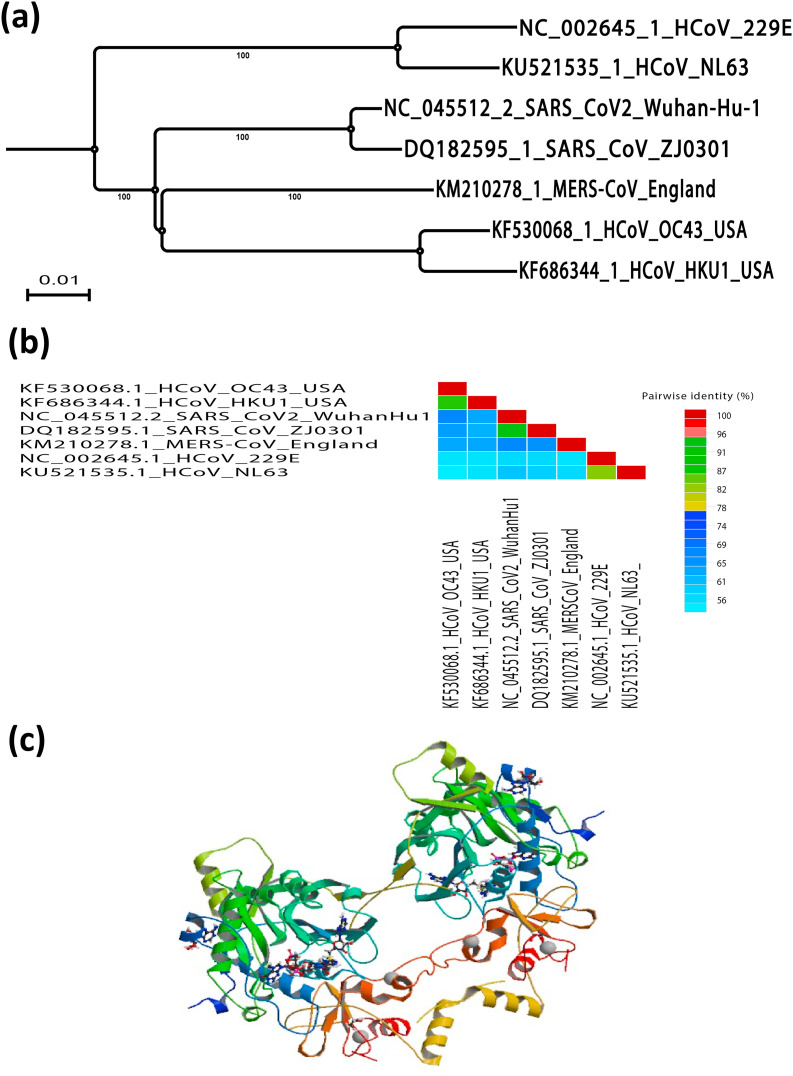
Initially, the 200-amino-acid-long nsp8 subunit took centre stage due to two reports, the first describing a fascinating hexadecameric structure consisting of eight copies of each of nsp7 and nsp8 [63], and the second revealing a nsp8-specific "secondary" RNA polymerase activity [66] involved in the CoV RNA synthesis process. Although the structures of feline coronavirus (FCoV; alphacoronavirus 1) nsp7 and nsp8 were found to mimic their SARS-CoV (betacoronavirus) counterparts, two copies of nsp7 and one copy of nsp8 forming a heterotrimer were found to be assembled into a very different higher-order complex [67]. SARS-CoV nsp8 was found to adopt two different conformations inside the nsp7– nsp8 hexadecamer. The phylogenetic relationship and similarity percentage of SARS-CoV-2 in relation with other human coronaviruses is shown in Fig. 1 a and b, respectively. These have been named "golf club" and "bent shaft golf club" [63], with the golf club's globular head considered a new fold. Biochemistry and reverse genetics studies pointed to an important role in RNA synthesis for SARS-CoV nsp8 residues K58, P183, and R190 (Fig. 1c), replacing which was lethal to SARS-CoV whereas P183 and R190 residues were presumed to be involved in interactions with nsp12, while K58 may be critical for interactions with nsp8–RNA [64]. The amino acid sequence alignment for nsp8 of SARS-CoV-2 compared to other human coronaviruses is shown in Fig. 1d. It has been reported that SARS-CoV nsp8 is an interaction partner of many other viral proteins (including nsp2, nsp3 and nsp5 to nsp16) based on yeast two-hybrid and glutathione S-transferase (GST) pull-down assays, although most of these interactions remain to be verified in the infected cell [68].
Fig. 1.
SARS-CoV-2 nsp8 evolutionary changes in compared to other human coronaviruses. (a) Phylogenetic tree construction by the neighbour joining method was performed using MEGA X software, with bootstrap values being calculated from 1000 trees using amino acid sequences of nsp8 (b) Pairwise identity % plot of nsp8 CoVs amino acid sequences performed using SDT program, (c) 3D crystal structure of the nsp7- nsp8 complex of SARS-CoV-2 (PDB ID: 6YHU) and (d) Multiple amino acid sequence alignment for nsp8 of SARS-CoV-2 compared to other human coronaviruses.
3.9. Coronavirus nsp9
CoV nsp9 subunit is the second replicase cleavage product after nsp5 based on obtaining crystal structures, is approximately 110 amino acids long [69,70]. The dimerization of the nsp9's biologically active form may be capable of binding nucleic acids in a non-sequential manner, with an apparent preference for single-stranded RNA [[69], [70], [71]]. The nsp9 protein function still so far unknown however, site directed mutagenesis studies within nsp9 revealed that point mutations within nsp9 can block CoV replication [72,73] that suppose its role during viral pathogenesis. Mutations in nsp9 were also found to lead to increase the pathogenesis of SARS-CoV (betacoronavirus) in mice model infected with a mouse-adapted strain of virus (MA-15) [74]. Further studies are required to describe how the nsp9 dimerization and mutagenesis may affect interactions with other replicase subunits, such as nsp8 and nsp12-RdRp. Nsp8 and nsp12 have been identified as interaction partners for nsp9 [68,70,75] and colocalize on membranous replication organelles with nsp9 [76].
3.10. Coronavirus nsp10
The nsp10 subunit protein (139 residues; SARS-CoV) is one of the most conserved CoV proteins and is believed to serve as an essential multifunctional replication factor. Nsp10 was shown to be dimerized, as well as interact with nsp1, nsp7, nsp14, and nsp16 using yeast two-hybrid assay. These interactions were confirmed by coimmunoprecipitation and/or GST pull-down assays [37,38,51,77]. Nsp10 ′s interactions with nsp14 and nsp16 and possibly other subunits of the viral replication complex can be a target for the development of antiviral compounds against pathogenic coronaviruses [78]. The significant role of nsp10 in replication was asserted from the MHV (betacoronavirus) temperature-sensitive mutant phenotype in which a nsp10 mutation was responsible for a deficiency in the synthesis of minus-strand RNA [54]. Moreover, the protein was involved in the regulation of polyprotein processing [57]. Based on biochemical and structural tests, nsp10 protein has been found to bind two strongly affinated Zn2+ ions, indicating the presence of two zinc finger motifs [79]. Similarly, nsp10 exhibited a weak affinity for single- and double-stranded RNA and DNA, proposing that protein might act as part of a larger RNA-binding complex. Nsp10 interacts with nsp14 and nsp16 and controls their respective activities ExoN and ribose-2 ′-O-MTase (2′-O-MTase) based on the recent biochemical studies [52,80].
3.11. Coronavirus nsp11
Inside the polyprotein, coronavirus nsp10 is accompanied by a short peptide of highly variable sequence mapping the genomic RNA region where the ribosomal frameshift signal leading to the replicase enzyme cluster being translated into open reading frame 1b is located. Depending on the CoV species, nsp11 consists of 13–23 residues. In SARS-CoV (betacoronavirus), nsp11 is a 13-residue peptide which is very small cleavage product processed from the C-terminus of polyprotein 1a (pp1a) at the nsp10/11junction, however processing of nsp11 has not been demonstrated in infected cells. The structure of the un-cleaved nsp10–11 polypeptide showed some differences in oligomerization and crystal packing, but little difference in the core nsp10 structure [81]. Thus, nsp11 more likely forms part of an essential translation reading frame shift mechanism and is unlikely to significantly influence the function of nsp10. The N-terminal sequence of nsp11 (encoded between the nsp10/11 junction and the ORF1a/1b frameshift site) in the pp1ab frameshift component is equivalent to the N-terminal portion of nsp12 subunit.
3.12. Coronavirus nsp12
Nsp12 has at least two domains, the recently described, "nidovirus-wide conserved domain with nucleotidyl transferase activity" (nidovirus RdRp-associated nucleotidyltransferase (NiRAN)) [82] and the canonical RdRp domain C-terminal [83]. The nsp12-coding sequence contains the ribosomal frameshift site ORF1a/1b, and a programmed −1 frameshifting event drives the translation of ORF1b to produce the polyprotein pp1ab that contains nsp12. CoVs' nsp12-RdRp is a primary drug target, which can be inhibited within the host cell without any toxic side effects. Nucleoside analogues are an important class of antiviral drug candidates able to target the viral RdRps but attempts to use them to inhibit CoV replication have so far not been very successful [1,84]. Ribavirin, a guanosine analogue with a wide range of antiviral activity commonly used against various RNA viruses due to its mechanism in the induction of lethal mutagenesis by increasing the RdRp error rate, inhibition of viral mRNA capping and reduction of viral RNA synthesis by cellular enzyme inhibition (inosine monophosphate dehydrogenase (IMPDH)), which decreases the availability of intracellular GTP [17,85,86]. In spite of ribavirin was used to treat small numbers of SARS and MERS infected patients [87], in vitro and vivo studies with different CoVs and infected cell cultures [84,[88], [89], [90]] established its poor activity and strongly suggested that ribavirin does not target the CoV RdRp directly or is targeted (itself) by the nsp14-ExoN activity [17]. It will be important to better understand the structure and function of nsp12-RdRp that will be helpful to develop new strategies that will reduce the impact of drug resistance-inducing mutations, which are a common problem when targeting rapidly evolving RNA viruses.
3.13. Coronavirus nsp13
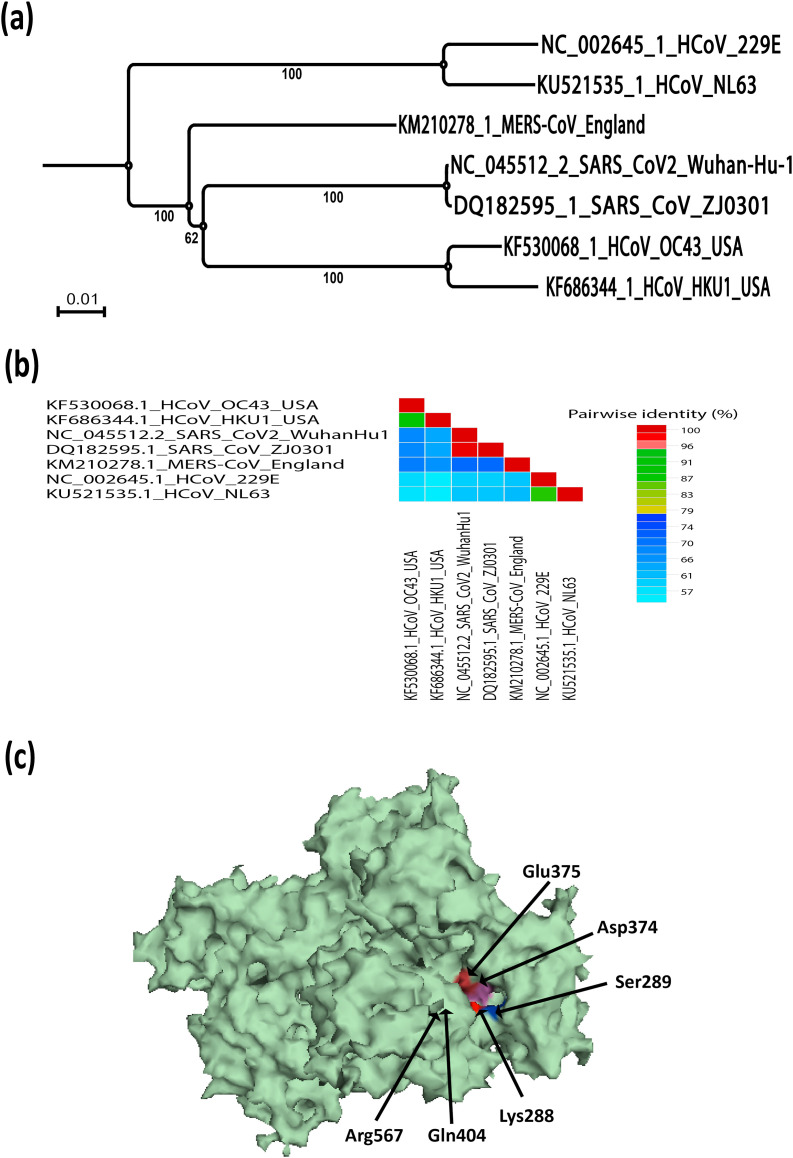
The CoV genome encodes two replicase polyproteins pp1a and pp1ab to support effective replication, which is processed proteolytically into 16 non-structural proteins (nsps) [21,91,92] that assemble into the membrane-associated replication-transcription complexes (RTCs), to drive viral genome replication and translation. The RNA-dependent RNA polymerase (nsp12) and the helicase (nsp13) are main components of RTC [28,29,64]. Positive stranded RNA viruses with a genome greater than 7 kb have been shown to encode helicases [93,94] that are classified into six super-families (SF1-SF6) and participate in almost every aspect of nucleic acid metabolism [95]. Whatever their functional diversity, all helicases contain core domains which hydrolyse NTPs and have accessory domains or inserts of different functions, such as assisting in the catalytic activity or interacting with other protein partners [93,94]. Bioinformatic analysis revealed that CoV nsp13 belongs to the superfamily SF1, including Rep, UvrD, PcrA, RecD, Pif1, Dda, Upf1-like helicases and various + RNA virus helicases [83] and exhibits multiple enzymatic activities, which include hydrolysis of NTPs and dNTPs, unwinding of DNA and RNA duplexes with 5′-3′ directionality and the RNA 5′-triphosphatase activity [[96], [97], [98]]. CoV helicase is one of the three most conserved evolutionary proteins in nidoviruses [99] and is thus an important target for drug development [100]. Physically, CoV's RNA-dependent RNA polymerase (RdRP, nsp12) might interacts with nsp13 and improve its relaxing activity [101,102]. In silico prediction for SARS-CoV-2, nsp13 is about 596 amino acids (located in polyprotein orf1ab). SARS-CoV-2 nsp13's overall structure adopted a triangular pyramid shape and included five domains similar to SARS and MERS. Among these, two “RecA-like” domains, 1A (261–441 a.a) and 2A (442–596 a.a), and 1B domain (150–260 a.a) forming the triangular base, while N-terminal Zinc binding domain (ZBD) (1–99 a.a) and stalk domain (100–149 a.a), which connects ZBD and 1B domain, are arranged at the apex of the pyramid [103]. It has shown that small molecules capable of inhibiting the NTPase activity through interference with ATP binding [103]. The phylogenetic relationship and similarity % of SARS-CoV-2 nsp13 in relation with other human coronaviruses is shown in Fig. 2 a and b, respectively. The SARS-CoV-2 nsp13 identified similar retained active site residues of the NTPase including Lys288, Ser289, Asp374, Glu375, Gln404 and Arg567 similar to SARS-CoV nsp13 [103] (Fig. 2c). All of these residues were clustered together in the cleft between domain 1A and 2B at the base, while the docking grid was formed by locating bound ADP of crystallised yeast Upf1 and identifying top hits [103].
Fig. 2.
SARS-CoV-2 nsp13 evolutionary changes in compared to other human coronaviruses. (a) Phylogenetic tree construction by the neighbour joining method was performed using MEGA X software, with bootstrap values being calculated from 1000 trees using amino acid sequences of nsp13 (b) Pairwise identity % plot of nsp13 CoVs amino acid sequences performed using SDT program, (c) 3D crystal structure of the nsp13 of SARS-CoV-2 (PDB ID: 6JYT).
3.14. Coronavirus nsp14
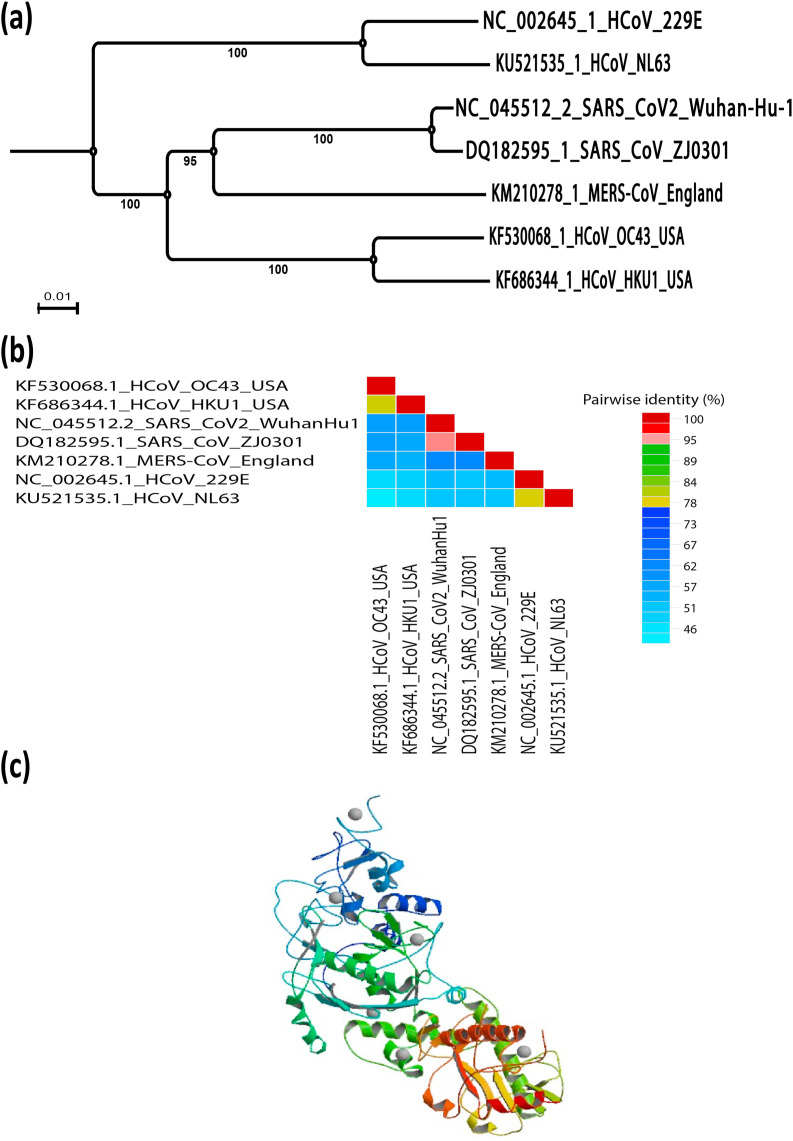
Coronavirus nsp14 plays a crucial role in viral RNA synthesis and has a bifunctional through its N-terminal exonuclease (ExoN) domain and C-terminal part [6,104]. The N-terminal exonuclease (ExoN) domain is thought to promote the fidelity of CoV RNA synthesis while the C-terminal part carries an AdoMet-dependent guanosine N7-MTase activity [6,104]. The phylogenetic relationship and similarity percentage of SARS-CoV-2 nsp14 compared to other human coronaviruses is shown in Fig. 3 a and b, respectively. X-ray structure for nsp14 demonstrated functionally important interactions between the N-terminal (ExoN) and C-terminal (N7-MTase) domains, with three ExoN α-helices maintaining the core of the N7-MTase substrate-binding pocket [105]. Reverse genetics studies confirmed that the specific role of N7-MTase activity during virus replication whereas the SARS-CoV N7-MTase has been shown to methylate 5′ cap structures sequentially independently using a variety of RNAs and to be active on cap analogues and GTP [106]. Alanine scanning mutagenesis has identified a number of 10 primary residues for enzymatic activity within the N7-MTase domain [105]. Similarly, two clusters of residues essential to MTase activity have been identified; the first cluster (nsp14 residues 331–336) corresponds to the AdoMet-binding site's DXGXPXA motif and correlates to 3H-labeled AdoMet binding. The second cluster (nsp14 residues 414 and 428) forms a constricted pocket that holds the cap structure (GpppA) between two β-strands (β1 and β2) and helix 1, placing the guanine's N7 position close to AdoMet based on the analysis of the X-ray structure of a complex SARS-CoV nsp10/nsp14 [107] (Fig. 3c). Drugability of the nsp14 N7-MTase has been explored using a small set of MTase inhibitors previously documented [52,108,109]. The nsp14 N7-MTase is an obvious prospect for antiviral strategies, particularly since it demonstrates a variety of features distinctive from MTases host cell [110].
Fig. 3.
SARS-CoV-2 nsp14 evolutionary changes in compared to other human coronaviruses. (a) Phylogenetic tree construction by the neighbour joining method was performed using MEGA X software, with bootstrap values being calculated from 1000 trees using amino acid sequences of nsp14 (b) Pairwise identity % plot of nsp14 CoVs amino acid sequences performed using SDT program, (c) 3D crystal structure of the nsp14- nsp10 complex of SARS-CoV-2(PDB ID: 5C8U).
3.15. Coronavirus non-structural protein 15 (nsp15; endoribonuclease)
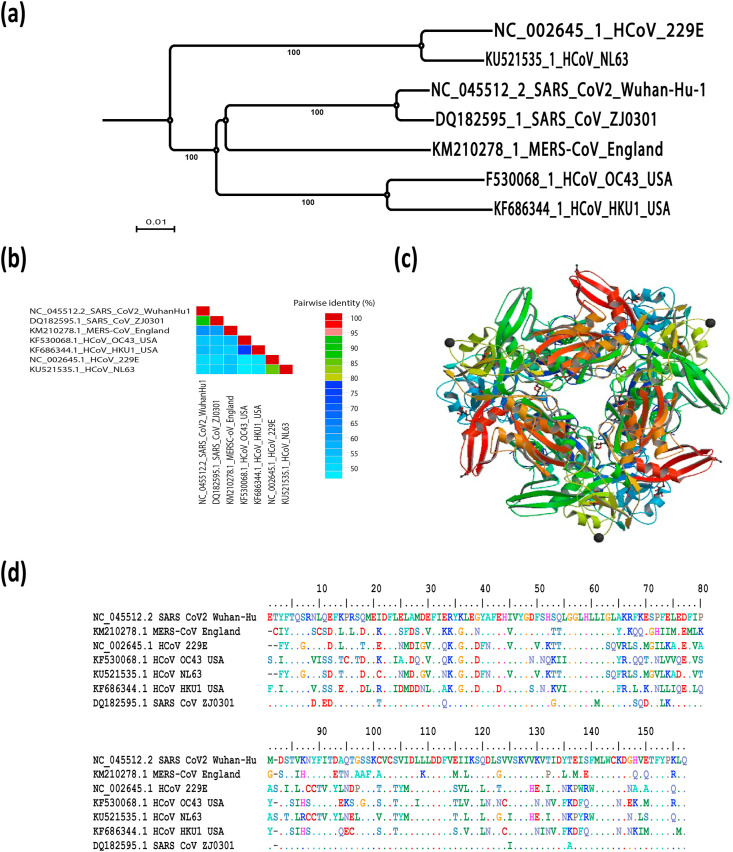
Coronavirus non-structural protein 15 (nsp15), a highly conserved portion of nidovirus with endoribonuclease activity, acts in combination with the viral replication complex to restrict the access of viral dsRNA to host dsRNA sensors that means nsp15 is not required for viral RNA synthesis but acts to mediate evasion of host dsRNA sensors [111]. Nsp15 is a key component of coronavirus pathogenesis that highlighted in nsp15 mutant viruses; CoVs that include a mutation in nsp15, whether render nsp15 unstable or deactivate endoribonuclease function, enhance the IFN production dependent on MDA5, and activate host dsRNA sensors. Therefore, mutant nsp15 viruses can elicit cell apoptosis and demonstrate lower macrophage replication [111]. The phylogenetic relationship and similarity percentage of SARS-CoV-2 nsp15 in relation with other human coronaviruses is shown in Fig. 4 a and b, respectively. Functional genomics analysis showed that nsp15 comprises a cellular endoribonucleases domain with distant similarity. Nsp15, called NendoU, endoribonuclease is highly preserved among vertebrate nidoviruses (coronaviruses and arteriviruses) [6]. Structural and functional studies revealed that the SARS-CoV nsp15 create oligomers to cleave RNA molecules with a preference for uridylates at the 3′-end [[112], [113], [114], [115]]. Previous studies reported that coronavirus nsp15 overexpression can antagonise the innate immune responses, but there was no direct evidence to suggest that in case of viral infection it can counteracts the innate immunity [116]. 3D crystal structure of the nsp15 of SARS-CoV-2 (PDB ID: 6VWW) and the amino acid sequence alignment for nsp8 of SARS-CoV-2 compared to other human coronaviruses (HCoVs) is shown in Fig. 4d and c, respectively. Nsp15 can act as a "gatekeeper" for sequestration of viral dsRNA within complex replication and away from host dsRNA sensors. Previous reports suggested that nsp15 could be part of a viral RNA decay pathway due to increased accumulation of viral dsRNA in cells infected with nsp15 mutant viruses [117]. Further studies are needed to fully elucidate the mechanisms used by nsp15 to potentially hide or degrade viral RNA and ultimately prevent host dsRNA sensors from activating and evaluate the nsp15-mediated dsRNA cleavage in the virus infection context.
Fig. 4.
SARS-CoV-2 nsp15 evolutionary changes in compared to other human coronaviruses. (a) Phylogenetic tree construction by the neighbour joining method was performed using MEGA X software, with bootstrap values being calculated from 1000 trees using amino acid sequences of nsp15, (b) Pairwise identity % plot of nsp15 CoVs amino acid sequences performed using SDT program, (c) 3D crystal structure and (d) the multiple amino acid sequence alignment for of the nsp15 of SARS-CoV-2 (PDB ID: 6VWW) compared to other human coronaviruses.
3.16. Coronavirus nsp16 2′-O-methyl transferase
The existence of the 2′-O-methyl transferase (2′-O-MTase) domain in CoV nsp16 was identified using bioinformatics tools [6,118] that illustrated a model containing a conserved K–D–K–E catalytic tetrad characteristic of AdoMet-dependent 2′-O-MTases and conserved AdoMet-binding site [118]. The FCoV (alphacoronavirus 1) nsp16 protein showed specific interaction with cap-0-containing RNAs and responsible for the transfer of a methyl group from AdoMet to the 2′-O position of the first N7-methylated substrate nucleotide [119]. The phylogenetic relationship and similarity % of SARS-CoV2 nsp15 in relation with other human coronaviruses is shown in Fig. 5 a and b. The nsp16 amino acid sequence is highly conserved throughout the entire CoV family and suggests similar structural domains and functional activities that illustrated in the structural similarities between SARS-CoV and MERS-CoV nsp16/nsp 10 complexes, suggest mutations that would maintain or modify activity in the viral family, leading to similar phenotypic mutants [120,121]. Therefore, antiviral therapies that target nsp16/nsp10's behaviour and function may also be successful against SARS-CoV and HCoV 229E, as well as emerging viruses such as MERS-CoV, PEDV [120,121]. RNA-cap methyltransferase (nsp16) may be regarded as key for antiviral drug development against SARS-CoV-2 [122], while no effective inhibitors or licenced medicines currently exist that can be used as targets for the production of antivirals.
Fig. 5.
SARS-CoV-2 nsp16 evolutionary changes in compared to other human coronaviruses. (a) Phylogenetic tree construction by the neighbour joining method was performed using MEGA X software, with bootstrap values being calculated from 1000 trees using amino acid sequences of nsp16, (b) Pairwise identity % plot of nsp16 CoVs amino acid sequences performed using SDT program and (c) 3D crystal structure of the nsp16 of SARS-CoV-2 (PDB ID: 7BQ7).
Surprisingly, the nsp10 residues included in the nsp10/nsp16 interaction are highly conserved within the CoV family and it has recently been shown that nsp10 of various CoVs (FCoV, MHV, SARS-CoV, MERS-CoV) are functionally compatible in stimulating activity of nsp16 2′-O-MTase [123]. Thereby, compounds or peptides that block such mechanism can have broad-spectrum anti-CoV effects, a hypothesis that has been explored and supported using synthetic peptides that imitate the nsp10 interface and in vitro suppress nsp16 2′-O-MTase activity [123,124]. Previous studies stated that deleting the nsp16 coding sequence can ablate RNA synthesis and viral replication; similar to deleting up stream components (Exonuclease- nsp14 and N-terminal zinc binding domain endoribonuclease- nsp15) [125]. While the nsp16 exhibited an increased type I IFN sensitivity as in case of MHV (betacoronavirus) and 229E mutants [126], the SARS-CoV mutant virus failed to induce further type IFN either in vitro or in vivo [120]. 3D crystal structure of the nsp16 of SARS-CoV-2 (PDB ID: 6VWW) and the amino acid sequence alignment for nsp8 of SARS-CoV-2 compared to other human coronaviruses (HCoVs) is shown in Fig. 5c and d, respectively.
Similarly, following the nsp16 mutant virus challenge compared to wild type virus; there was no significant change in the induction magnitude or kinetics of interferon stimulated genes (ISGs) including IFIT1 and MDA5 [120]. Without functional nsp16, both in vitro and in vivo infections for SARS-CoV (betacoronavirus), HCoV 229E (alphacoronavirus), and MHV (betacoronavirus) are significantly attenuated as a result of increasing the viral RNA recognition by host sensor molecules as well as the effector responses of the IFIT family of ISGs [120]. Identifying these viral/host interactions allows the development of new therapies against the virus, while also enhancing the effectiveness of the existing immune response [120].
4. Conclusions
Accumulated knowledge around CoV nsps' has revealed conserved functions and divergent mechanisms among various CoVs to block expression of the host gene and antagonise innate immune responses that provide insight into the expanding repertoire of new immune evasion virus strategies. Furthermore, given the lack of apparent primary sequence homology between nsps of CoVs, it is important to highlight the functional similarities between them that will help to understand their evolutionary association and the adaptation of CoVs to specific host species. There is no doubt that further characterization of the "nsp interactome" within the CoV-infected cell will provide more clues about how specific functions are switched on and off or modulated. Understanding these mechanisms will not only highlight their critical roles in the virus replication cycle but may also exposed some key druggable targets to propose novel therapeutics.
Author contributions
All the authors contributed equally to the work reported. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financed by International Foundation for Science (IFS), Sweden (project No. I-3-B-6270-1) and The Organisation of Islamic Cooperation's Standing Committee on Scientific and Technological Cooperation (COMSTECH). Additionally, this study was supported by the Biotechnology and Biological Sciences Research Council (BBSRC) (BB/M008681/1 and BBS/E/I/00001852) and British Council (172710323 and 332228521). The funding sources had no role in the study design, collection, or analysis of the data, writing of the manuscript, or in the decision to submit the manuscript for publication.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Chu C.K., Gadthula S., Chen X., Choo H., Olgen S., Barnard D.L., Sidwell R.W. Antiviral activity of nucleoside analogues against SARS-coronavirus (SARS-coV) Antivir. Chem. Chemother. 2006;17:285–289. doi: 10.1177/095632020601700506. [DOI] [PubMed] [Google Scholar]
- 2.Wang L.F., Shi Z., Zhang S., Field H., Daszak P., Eaton B.T. Review of bats and SARS. Emerg. Infect. Dis. 2006;12(12):1834–1840. doi: 10.3201/eid1212.060401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ge X.Y., Li J.L., Yang X.L., Chmura A.A., Zhu G., Epstein J.H., Mazet J.K., Hu B., Zhang W., Cheng Peng C., Zhang Y., Luo C., Tan B., Wang N., Zhu Y., Crameri G., Zhang S., Wang L., Daszak P., Shi Z. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503:535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siddell S.G., Walker P.J., Lefkowitz E.J., Mushegian A.R., Adams M.J., Dutilh B.E., Gorbalenya A.E., Harrach B., Harrison R.L., Junglen S., Knowles N.J., Kropinski A.M., Krupovic M., Kuhn J.H., Nibert M., Rubino L., Sabanadzovic S., Sanfaçon H., Simmonds P., Varsani A., Zerbini F.M., Davison A.J. Additional changes to taxonomy ratified in a special vote by the international committee on taxonomy of viruses (october 2018) Arch. Virol. 2019;164(3):943–946. doi: 10.1007/s00705-018-04136-2. [DOI] [PubMed] [Google Scholar]
- 5.Coronaviridae Study Group of the International Committee on Taxonomy of Viruses The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5(4):536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Snijder E.J., Bredenbeek P.J., Dobbe J.C., Thiel V., Ziebuhr J., Poon L.L., Guan Y., Rozanov M., Spaan W.J.M., Gorbalenya A.E. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003;331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Groot R.J., Cowley J.A., Enjuanes L., Faaberg K.S., Perlman S., M Rottier P.J., Snijder E.J., Ziebuhr J., Gorbalenya A.E. In: Virus Taxonomy. Ninth Report of the International Committee on Taxonomy of Viruses. King A.M.Q., Adams M.J., Carstens E.B., Lefkowitz E.J., editors. Elsevier Academic Press; Amsterdam: 2012. Order Nidovirales; pp. 785–795. [Google Scholar]
- 8.Woo P.C., Lau S.K., Lam C.S., Lau C.C.Y., L Tsang A.K., Lau J.H.N., BaI R., Teng J.L.L., Tsang C.C.C., Wang M., Zheng B., Chan K., Yuen K. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012;86(7):3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adams M.J., Lefkowitz E.J., King A.M., Harrach B., Harrison R.L., Knowles N.J., Kropinski A.M., Krupovic M., Kuhn J.H., Mushegian A.R., Nibert M.L., Sabanadzovic S., Sanfaçon H., Siddell S.G., Simmonds P., Varsani A., Zerbini F.M., Orton R.J., Smith D.B., Gorbalenya A.E., Davison A.J. 50 years of the international committee on taxonomy of viruses: progress and prospects. Arch. Virol. 2017;162:1441–1446. doi: 10.1007/s00705-016-3215-y. [DOI] [PubMed] [Google Scholar]
- 10.Fehr A.R., Perlman S. Springer; New York: 2015. Coronaviruses: an Overview of Their Replication and Pathogenesis. Coronaviruses; p. 1e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17(3):181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Y., Liu Q., Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J. Med. Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu D.X., Fung T.S., Chong K.K.-L., Shukla A., Hilgenfeld R. Accessory proteins of SARS-CoV and other coronaviruses. Antivir. Res. 2014;109:97–109. doi: 10.1016/j.antiviral.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan J.F.-W., Kok K.-H., Zhu Z., Chu H., To K.K.-W., Yuan S., Yuen K.-Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microb. Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calligari P., Bobone S., Ricci G., Bocedi A. Molecular investigation of SARS-CoV-2 proteins and their interactions with antiviral drugs. Viruses. 2020;12:445. doi: 10.3390/v12040445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eckerle L.D., Becker M.M., Halpin R.A., Li K., Venter E., Lu X., Scherbakova S., Graham R.L., Baric R.S., Stockwell T.B., Spiro D.J., Denison M.R. Infidelity of SARS-CoV NSP14-exonuclease mutant virus replication is revealed by complete genome sequencing. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith E.C., Blanc H., Surdel M.C., Vignuzzi M., Denison M.R. Coronaviruses lacking exoribonuclease activity are susceptible to lethal mutagenesis: evidence for proofreading and potential therapeutics. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ogando N.S., Ferron F., Decroly E., Canard B., Posthuma C.C., Snijder E.J. The curious case of the nidovirus exoribonuclease: its role in RNA synthesis and replication fidelity. Front. Microbiol. 2019;10:1813. doi: 10.3389/fmicb.2019.01813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brian D.A., Baric R.S. Coronavirus genome structure and replication. Curr. Top. Microbiol. Immunol. 2005;287:1–30. doi: 10.1007/3-540-26765-4_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brierley I., Digard P., Inglis S.C. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell. 1989;57:537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thiel V., Ivanov K.A., Putics A., Hertzig T., Schelle B., Bayer S., Weissbrich B., Snijder E.J., Rabenau H., Doerr H.W., Gorbalenya A.E., Ziebuhr J. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 2003;84(9):2305–2315. doi: 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- 22.Kamitani W., Narayanan K., Huang C., Lokugamage K., Ikegami T., Ito N., Kubo H., Makino S. Severe acute respiratory syndrome coronavirus NSP1 protein suppresses host gene expression by promoting host mRNA degradation. Proc. Natl. Acad. Sci. U.S.A. 2006;103(34):12885–12890. doi: 10.1073/pnas.0603144103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Züst R., Cervantes-Barragán L., Kuri T., Blakqori G., Weber F., Ludewig B., Thiel V. Coronavirus non-structural protein 1 is a major pathogenicity factor: implications for the rational design of coronavirus vaccines. PLoS Pathog. 2007;3(8):e109. doi: 10.1371/journal.ppat.0030109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Narayanan N., Huang C., Makino S. SARS coronavirus accessory proteins. Virus Res. 2008;133:113–121. doi: 10.1016/j.virusres.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Almeida M.S., Johnson M.A., Herrmann T., Geralt M., Wuthrich K. Novel -barrel fold in the nuclear magnetic resonance structure of the replicase nonstructural protein 1 from the severe acute respiratory syndrome coronavirus. J. Virol. 2007;81(7):3151–3161. doi: 10.1128/JVI.01939-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wathelet M.G., Orr M., Frieman M.B., Baric R.S. Severe acute respiratory syndrome coronavirus evades antiviral signaling: role of NSP1 and rational design of an attenuated strain. J. Virol. 2007;81:11620–11633. doi: 10.1128/JVI.00702-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Denison M.R., Yount B., Brockway S.M., Graham R.L., Sims A.C., Lu X., Baric R.S. Cleavage between replicase proteins p28 and p65 of mouse hepatitis virus is not required for virus replication. J. Virol. 2004;78(11):5957–5965. doi: 10.1128/JVI.78.11.5957-5965.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prentice E., Jerome W.G., Yoshimori T., Mizushima N., Denison M.R. Coronavirus replication complex formation utilizes components of cellular autophagy. J. Biol. Chem. 2004;279:10136–10141. doi: 10.1074/jbc.M306124200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prentice E., McAuliffe J., Lu X., Subbarao K., Denison M.R. Identification and characterization of severe acute respiratory syndrome coronavirus replicase proteins. J. Virol. 2004;78:9977–9986. doi: 10.1128/JVI.78.18.9977-9986.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Denison M.R., Spaan W.J., van der Meer Y., Gibson C.A., Sims A.C., Prentice E., Lu X.T. The putative helicase of the coronavirus mouse hepatitis virus is processed from the replicase gene polyprotein and localizes in complexes that are active in viral RNA synthesis. J. Virol. 1999;73:6862–6871. doi: 10.1128/jvi.73.8.6862-6871.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schiller J.J., Kanjanahaluethai A., Baker S.C. Processing of the coronavirus mhv-jhm polymerase polyprotein: identification of precursors and proteolytic products spanning 400 kilodaltons of ORF1a. Virology. 1998;242:288–302. doi: 10.1006/viro.1997.9010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Graham R.L., Sims A.C., Brockway S.M., Baric R.S., Denison M.R. The NSP2 replicase proteins of murine hepatitis virus and severe acute respiratory syndrome coronavirus are dispensable for viral replication. J. Virol. 2005;79:13399–13411. doi: 10.1128/JVI.79.21.13399-13411.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neuman B.W. Bioinformatics and functional analyses of coronavirus nonstructural proteins involved in the formation of replicative organelles. Antivir. Res. 2016;135:97–107. doi: 10.1016/j.antiviral.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Freitas B.T., Durie I.A., Murray J., Longo J.E., Miller H.C., Crich D., Hogan R.J., Tripp R.A., Pegan S.D. Characterization and noncovalent inhibition of the deubiquitinase and deISGylase activity of SARS-CoV-2 papain-like protease. ACS Infect Dis. acsinfecdis. 2020 doi: 10.1021/acsinfecdis.0c00168. [DOI] [PubMed] [Google Scholar]
- 35.Forni D. Extensive positive selection drives the evolution of nonstructural proteins in lineage C betacoronaviruses. J. Virol. 2016;90:3627–3639. doi: 10.1128/JVI.02988-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ziebuhr J., Snijder E.J., Gorbalenya A.E. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 2000;81:853–879. doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]
- 37.von Brunn A., Teepe C., Simpson J.C., Pepperkok R., Friedel C.C., Zimmer R., Roberts R., Baric R., Haas J. Analysis of intraviral protein-protein interactions of the SARS coronavirus ORFeome. PloS One. 2007;2:e459. doi: 10.1371/journal.pone.0000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Imbert I., Snijder E.J., Dimitrova M., Guillemot J.C., Lecine P., Canard B. The SARS-Coronavirus PLnc domain of NSP3 as a replication/transcription scaffolding protein. Virus Res. 2008;133:136–148. doi: 10.1016/j.virusres.2007.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pfefferle S., Schöpf J., Kögl M., Friedel C.C., Müller A.A., Carbajo-Lozoya J., Stellberger T., von Dall'Armi E., Herzog P., Kallies S., Niemeyer D., Ditt V., Kuri T., Züst R., Pumpor K., Hilgenfeld R., Schwarz F., Zimmer R., Steffen I., Weber F., Thiel V., Herrler G., Thiel H.J., Schwegmann-Wessels C., Pöhlmann S., Haas J., Drosten C., von Brunn A. The SARS-coronavirus-host interactome: identification of cyclophilins as target for pan-coronavirus inhibitors. PLoS Pathog. 2011;7(10) doi: 10.1371/journal.ppat.1002331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma-Lauer Y., Carbajo-Lozoya J., Hein M.Y., Müller M.A., Deng W., Lei J., Meyer B., Kusov Y., von Brunn B., Bairad D.R., Hünten S., Drosten C., Hermeking H., Leonhardt H., Mann M., Hilgenfeld R., von Brunn A. p53 down-regulates SARS coronavirus replication and is targeted by the SARS-unique domain and PLpro via E3 ubiquitin ligase RCHY1. Proc. Natl. Acad. Sci. U.S.A. 2016;113:5192–5201. doi: 10.1073/pnas.1603435113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Snijder E.J., van der Meer Y., Zevenhoven-Dobbe J., Onderwater J.J., van der Meulen J., Koerten H.K., Mommaas A.M. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J. Virol. 2006;80(12):5927–5940. doi: 10.1128/JVI.02501-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Hemert M.J., de Wilde A.H., Gorbalenya A.E., Snijder E.J. The in vitro RNA synthesizing activity of the isolated arterivirus replication/transcription complex is dependent on a host factor. J. Biol. Chem. 2008;283:16525–16536. doi: 10.1074/jbc.M708136200. [DOI] [PubMed] [Google Scholar]
- 43.Angelini M.M., Akhlaghpour M., Neuman B.W., Buchmeier M.J. Severe acute respiratory syndrome coronavirus nonstructural proteins 3, 4, and 6 induce doublemembrane vesicles. mBio. 2013;4 doi: 10.1128/mBio.00524-13. e00524–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Knoops K., Kikkert M., Worm S.H., Zevenhoven-Dobbe J.C., van der Meer Y., Koster A.J., Mommaas A.M., Snijder E.J. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6:e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hagemeijer M.C., Rottier P.J., de Haan C.A. Biogenesis and dynamics of the coronavirus replicative structures. Viruses. 2012;4:3245–3269. doi: 10.3390/v4113245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Wilde A.H., Raj V.S., Oudshoorn D., Bestebroer T.M., van Nieuwkoop S., Limpens R.W.A.L., Posthuma C.C., van der Meer Y., Bárcena M., Haagmans B.L., Snijder E.J., van den Hoogen B.G. MERS-coronavirus replication induces severe in vitro cytopathology and is strongly inhibited by cyclosporin A or interferon-α treatment. J. Gen. Virol. 2013;94(8):1749–1760. doi: 10.1099/vir.0.052910-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oostra M., te Lintelo E.G., Deijs M., Verheije M.H., Rottier P.J., de Haan C.A. Localization and membrane topology of coronavirus nonstructural protein 4: involvement of the early secretory pathway in replication. J. Virol. 2007;81:12323–12336. doi: 10.1128/JVI.01506-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sparks J.S., Lu X., Denison M.R. Genetic analysis of Murine hepatitis virus NSP4 in virus replication. J. Virol. 2007;81:12554–12563. doi: 10.1128/JVI.01257-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lauber C., Goeman J.J., Parquet Mdel C., Nga P.T., Snijder E.J., Morita K., Gorbalenya A.E. The footprint of genome architecture in the largest genome expansion in RNA viruses. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hilgenfeld R. From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J. 2014;281:4085–4096. doi: 10.1111/febs.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pan J., Peng X., Gao Y., Li Z., Lu X., Chen Y., Ishaq M., Liu D., Dediego M.L., Enjuanes L., Guo D. Genome-wide analysis of protein-protein interactions and involvement of viral proteins in SARS-CoV replication. PloS One. 2008;3:e3299. doi: 10.1371/journal.pone.0003299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bouvet M., Debarnot C., Imbert I., Selisko B., Snijder E.J., Canard B., Decroly E. In vitro reconstitution of SARS-coronavirus mRNA cap methylation. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Decroly E., Debarnot C., Ferron F., Bouvet M., Coutard B., Imbert I., Gluais L., Papageorgiou N., Sharff A., Bricogne G., Ortiz-Lombardia M., Lescar J., Canard B. Crystal structure and functional analysis of the SARS-coronavirus RNA cap 2’-O-methyltransferase NSP10/NSP16 complex. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sawicki S.G., Sawicki D.L., Younker D., Meyer Y., Thiel V., Stokes H., Siddell S.G. Functional and genetic analysis of coronavirus replicase-transcriptase proteins. PLoS Pathog. 2005;1:e39. doi: 10.1371/journal.ppat.0010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Al-Mulla H.M., Turrell L., Smith N.M., Payne L., Baliji S., Zust R., Thiel V., Baker S.C., Siddell S.G., Neuman B.W. Competitive fitness in coronaviruses is not correlated with size or number of double-membrane vesicles under reduced-temperature growth conditions. mBio. 2014;5 doi: 10.1128/mBio.01107-13. e01107–e01113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stokes H.L., Baliji S., Hui C.G., Sawicki S.G., Baker S.C., Siddell S.G. A new cistron in the murine hepatitis virus replicase gene. J. Virol. 2010;84:10148–10158. doi: 10.1128/JVI.00901-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Donaldson E.F., Graham R.L., Sims A.C., Denison M.R., Baric R.S. Analysis of murine hepatitis virus strain A59 temperature-sensitive mutant TS-LA6 suggests that NSP10 plays a critical role in polyprotein processing. J. Virol. 2007;81:7086–7098. doi: 10.1128/JVI.00049-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Krogh A., Larsson B., von Heijne G., Sonnhammer E.L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 59.Oostra M., Hagemeijer M.C., van Gent M., Bekker C.P., te Lintelo E.G., Rottier P.J., de Haan C.A. Topology and membrane anchoring of the coronavirus replication complex: not all hydrophobic domains of NSP3 and NSP6 are membrane spanning. J. Virol. 2008;82(24):12392–12405. doi: 10.1128/JVI.01219-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cottam E.M., Maier H.J., Manifava M., Vaux L.C., Chandra-Schoenfelder P., Gerner W., Britton P., Ktistakis N.T., Wileman T. Coronavirus NSP6 proteins generate autophagosomes from the endoplasmic reticulum via an omegasome intermediate. Autophagy. 2011;7(11):1335–1347. doi: 10.4161/auto.7.11.16642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Benvenuto D., Angeletti S., Giovanetti M., Bianchi M., Pascarella S., Cauda R., Ciccozzi M., Cassone A. Evolutionary analysis of SARS-CoV-2: how mutation of Non-Structural Protein 6 (NSP6) could affect viral autophagy. J. Infect. 2020;81(1):e24–e27. doi: 10.1016/j.jinf.2020.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peti W., Johnson M.A., Herrmann T., Neuman B.W., Buchmeier M.J., Nelson M., Joseph J., Page R., Stevens R.C., Kuhn P., Wüthrich K. Structural genomics of the severe acute respiratory syndrome coronavirus: nuclear magnetic resonance structure of the protein NSP7. J. Virol. 2005;79:12905–12913. doi: 10.1128/JVI.79.20.12905-12913.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhai Y., Sun F., Li X., Pang H., Xu X., Bartlam M., Rao Z. Insights into SARS-CoV transcription and replication from the structure of the NSP7-NSP8 hexadecamer. Nat. Struct. Mol. Biol. 2005;12:980–986. doi: 10.1038/nsmb999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Subissi L., Posthuma C.C., Collet A., Zevenhoven-Dobbe J.C., Gorbalenya A.E., Decroly E., Snijder E.J., Canard B., Imbert I. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc. Natl. Acad. Sci. U.S.A. 2014;111:3900–3909. doi: 10.1073/pnas.1323705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Johnson M.A., Jaudzems K., Wuthrich K. NMR structure of the SARS-CoV non-structural protein 7 in solution at pH 6.5. J. Mol. Biol. 2010;402:619–628. doi: 10.1016/j.jmb.2010.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Imbert I., Guillemot J.C., Bourhis J.M., Bussetta C., Coutard B., Egloff M.P., Ferron F., Gorbalenya A.E., Canard B. A second, non-canonical RNA-dependent RNA polymerase in SARS coronavirus. EMBO J. 2006;25(20):4933–4942. doi: 10.1038/sj.emboj.7601368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xiao Y., Ma Q., Restle T., Shang W., Svergun D.I., Ponnusamy R., Sczakiel G., Hilgenfeld R. Nonstructural proteins 7 and 8 of feline coronavirus form a 2:1 heterotrimer that exhibits primer-independent RNA polymerase activity. J. Virol. 2012;86:4444–4454. doi: 10.1128/JVI.06635-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.von Brunn A., Teepe C., Simpson J.C., Pepperkok R., Friedel C.C., Zimmer R., Roberts R., Baric R., Haas J. Analysis of intraviral protein-protein interactions of the SARS coronavirus ORFeome. PloS One. 2007;2(5):e459. doi: 10.1371/journal.pone.0000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Egloff M.P., Ferron F., Campanacci V., Longhi S., Rancurel C., Dutartre H., Snijder E.J., Gorbalenya A.E., Cambillau C., Canard B. The severe acute respiratory syndrome-coronavirus replicative protein NSP9 is a single-stranded RNA-binding subunit unique in the RNA virus world. Proc. Natl. Acad. Sci. U.S.A. 2004;101:3792–3796. doi: 10.1073/pnas.0307877101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sutton G., Fry E., Carter L., Sainsbury S., Walter T., Nettleship J., Berrow N., Owens R., Gilbert R., Davidson A., Siddell S., Poon L.L., Diprose J., Alderton D., Walsh M., Grimes J.M., Stuart D.I. The NSP9 replicase protein of SARS-coronavirus, structure and functional insights. Structure. 2004;12:341–353. doi: 10.1016/j.str.2004.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ponnusamy R., Moll R., Weimar T., Mesters J.R., Hilgenfeld R. Variable oligomerization modes in coronavirus non-structural protein 9. J. Mol. Biol. 2008;383:1081–1096. doi: 10.1016/j.jmb.2008.07.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen B., Fang S., Tam J.P., Liu D.X. Formation of stable homodimer via the C-terminal alpha-helical domain of coronavirus nonstructural protein 9 is critical for its function in viral replication. Virology. 2009;383:328–337. doi: 10.1016/j.virol.2008.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Miknis Z.J., Donaldson E.F., Umland T.C., Rimmer R.A., Baric R.S., Schultz L.W. Severe acute respiratory syndrome coronavirus NSP9 dimerization is essential for efficient viral growth. J. Virol. 2009;83:3007–3018. doi: 10.1128/JVI.01505-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Frieman M., Yount B., Agnihothram S., Page C., Donaldson E., Roberts A., Vogel L., Woodruff B., Scorpio D., Subbarao K., Baric R.S. Molecular determinants of severe acute respiratory syndrome coronavirus pathogenesis and virulence in young and aged mouse models of human disease. J. Virol. 2012;86(2):884–897. doi: 10.1128/JVI.05957-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brockway S.M., Clay C.T., Lu X.T., Denison M.R. Characterization of the expression, intracellular localization, and replication complex association of the putative mouse hepatitis virus RNA-dependent RNA polymerase. J. Virol. 2003;77:10515–10527. doi: 10.1128/JVI.77.19.10515-10527.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bost A.G., Carnahan R.H., Lu X.T., Denison M.R. Four proteins processed from the replicase gene polyprotein of mouse hepatitis virus colocalize in the cell periphery and adjacent to sites of virion assembly. J. Virol. 2000;74(7):3379–3387. doi: 10.1128/jvi.74.7.3379-3387.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Brockway S.M., Lu X.T., Peters T.R., Dermody T.S., Denison M.R. Intracellular localization and protein interactions of the gene 1 protein p28 during mouse hepatitis virus replication. J. Virol. 2004;78:11551–11562. doi: 10.1128/JVI.78.21.11551-11562.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bouvet M., Lugari A., Posthuma C.C., Zevenhoven J.C., Bernard S., Betzi S., Imbert I., Canard B., Guillemot J.C., Lécine P., Pfefferle S., Drosten C., Snijder E.J., Decroly E., Morelli X. Coronavirus NSP10, a critical co-factor for activation of multiple replicative enzymes. J. Biol. Chem. 2014;289:25783–25796. doi: 10.1074/jbc.M114.577353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Matthes N., Mesters J.R., Coutard B., Canard B., Snijder E.J., Moll R., Hilgenfeld R. The non-structural protein NSP10 of mouse hepatitis virus binds zinc ions and nucleic acids. FEBS Lett. 2006;580:4143–4149. doi: 10.1016/j.febslet.2006.06.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bouvet M., Imbert I., Subissi L., Gluais L., Canard B., Decroly E. RNA 3’-end mismatch excision by the severe acute respiratory syndrome coronavirus non-structural protein NSP10/NSP14 exoribonuclease complex. Proc. Natl. Acad. Sci. U.S.A. 2012;109:9372–9377. doi: 10.1073/pnas.1201130109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bhardwaj K., Sun J., Holzenburg A., Guarino L.A., Kao C.C. RNA recognition and cleavage by the SARS coronavirus endoribonuclease. J. Mol. Biol. 2006;361:243–256. doi: 10.1016/j.jmb.2006.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lehmann K.C., Gulyaeva A., Zevenhoven-Dobbe J.C., Janssen G.M., Ruben M., Overkleeft H.S., van Veelen P.A., Samborskiy D.V., Kravchenko A.A., Leontovich A.M., Sidorov I.A., Snijder E.J., Posthuma C.C., Gorbalenya A.E. Discovery of an essential nucleotidylating activity associated with a newly delineated conserved domain in the RNA polymerase-containing protein of all nidoviruses. Nucleic Acids Res. 2015;43:8416–8434. doi: 10.1093/nar/gkv838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gorbalenya A.E., Koonin E.V., Donchenko A.P., Blinov V.M. Coronavirus genome: prediction of putative functional newdomains in the non-structural polyprotein by comparative amino acid sequence analysis. Nucleic Acids Res. 1989;17:4847–4861. doi: 10.1093/nar/17.12.4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ikejiri M., Saijo M., Morikawa S., Fukushi S., Mizutani T., Kurane I., Maruyama T. Synthesis and biological evaluation of nucleoside analogues having 6-chloropurine as anti- SARS-CoV agents. Bioorg. Med. Chem. Lett. 2007;17:2470–2473. doi: 10.1016/j.bmcl.2007.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Crotty S., Maag D., Arnold J.J., Zhong W., Lau J.Y., Hong Z., Andino R., Cameron C.E. The broad spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nat. Med. 2000;6:1375–1379. doi: 10.1038/82191. [DOI] [PubMed] [Google Scholar]
- 86.Crotty S., Hix L., Sigal L.J., Andino R. Poliovirus pathogenesis in a new poliovirus receptor transgenic mouse model: age‐dependent paralysis and a mucosal route of infection. J. Gen. Virol. 2002;83:1707. doi: 10.1099/0022-1317-83-7-1707. [DOI] [PubMed] [Google Scholar]
- 87.Zumla A., Chan J.F., Azhar E.I., Hui D.S., Yuen K.Y. Coronaviruses—drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016;15:327–347. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Barnard D.L., Day C.W., Bailey K., Heiner M., Montgomery R., Lauridsen L., Winslow S., Hoopes J., Li J.K., Lee J., Carson D.A., Cottam H.B., Sidwell R.W. Enhancement of the infectivity of SARS-CoV in BALB/c mice by IMP dehydrogenase inhibitors, including ribavirin. Antivir. Res. 2006;71:53–63. doi: 10.1016/j.antiviral.2006.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Falzarano D., de Wit E., Rasmussen A.L., Feldmann F., Okumura A., Scott D.P., Brining D., Bushmaker T., Martellaro C., Baseler L., Benecke A.G., Katze M.G., Munster V.J., Feldmann H. Treatment with interferon-alpha2b and ribavirin improves outcome in MERS CoV- infected rhesus macaques. Nat. Med. 2013;19:1313–1317. doi: 10.1038/nm.3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pyrc K., Bosch B.J., Berkhout B., Jebbink M.F., Dijkman R., Rottier P., van der Hoek L. Inhibition of human coronavirus NL63 infection at early stages of the replication cycle. Antimicrob. Agents Chemother. 2006;50:2000–2008. doi: 10.1128/AAC.01598-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gorbalenya A.E., Enjuanes L., Ziebuhr J., Snijder E.J. Nidovirales: evolving the largest RNA virus genome. Virus Res. 2006;117:17–37. doi: 10.1016/j.virusres.2006.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Subissi L., Imbert I., Ferron F., Collet A., Coutard B., Decroly E., Canard B. SARS-CoV ORF1b-encoded nonstructural proteins 12–16: replicative enzymes as antiviral targets. Antivir. Res. 2014;101:122–130. doi: 10.1016/j.antiviral.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Singleton M.R., Dillingham M.S., Wigley D.B. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 94.Fairman-Williams M.E., Guenther U.P., Jankowsky E. SF1 and SF2 helicases: family matters. Curr. Opin. Struct. Biol. 2010;20:313–324. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jankowsky E., Fairman M.E. RNA helicases – one fold for many functions. Curr. Opin. Struct. Biol. 2007;17:316–324. doi: 10.1016/j.sbi.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 96.Adeyemi A.O., Lazarus H. Biochemical characterization of Middle East respiratory syndrome coronavirus helicase. mSphere. 2016;1(5) doi: 10.1128/mSphere.00235-16. e00235-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ivanov K.A., Thiel V., Dobbe J.C., van der Meer Y., Snijder E.J., Ziebuhr J. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J. Virol. 2004;78:5619–5632. doi: 10.1128/JVI.78.11.5619-5632.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ivanov K.A., Ziebuhr J. Human coronavirus 229E nonstructural protein 13: characterization of duplex-unwinding, nucleoside triphosphatase, and RNA 5′-triphosphatase activities. J. Virol. 2004;78:7833–7838. doi: 10.1128/JVI.78.14.7833-7838.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ziebuhr J. The coronavirus replicase. Curr. Top. Microbiol. Immunol. 2005;287:57–94. doi: 10.1007/3-540-26765-4_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lehmann K.C., Hooghiemstra L., Gulyaeva A., Samborskiy D.V., Zevenhoven-Dobbe J.C., Snijder E.J., Gorbalenya A.E., Posthuma C.C. Arterivirus NSP12 versus the coronavirus NSP16 2’-O-methyltransferase: comparison of the C-terminal cleavage products of two nidovirus pp1ab polyproteins. J. Gen. Virol. 2015;96:2643–2655. doi: 10.1099/vir.0.000209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Adedeji A.O., Marchand B., Te Velthuis A.J., Snijder E.J., Weiss S., Eoff R.L. Mechanism of nucleic acid unwinding by SARS-CoV helicase. PloS One. 2012;7 doi: 10.1371/journal.pone.0036521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Adedeji A.O., Singh K., Calcaterra N.E., DeDiego M.L., Enjuanes L., Weiss S., Sarafianos S.G. Severe acute respiratory syndrome coronavirus replication inhibitor that interferes with the nucleic acid unwinding of the viral helicase. Antimicrob. Agents Chemother. 2012;56:4718–4728. doi: 10.1128/AAC.00957-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hao W., Wojdyla J.A., Zhao R., Han R., Das R., Zlatev I., Manoharan M., Wang M., Cui S. Crystal structure of Middle East respiratory syndrome coronavirus helicase. PLoS Pathog. 2017;13(6) doi: 10.1371/journal.ppat.1006474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Minskaia E., Hertzig T., Gorbalenya A.E., Campanacci V., Cambillau C., Canard B., Ziebuhr J. Discovery of an RNA virus 3’-5’exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc. Natl. Acad. Sci. U.S.A. 2006;103:5108–5113. doi: 10.1073/pnas.0508200103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chen Y., Cai H., Pan J., Xiang N., Tien P., Ahola T., Guo D. Functional screen reveals SARS coronavirus nonstructural protein NSP14 as a novel cap N7 methyltransferase. Proc. Natl. Acad. Sci. U.S.A. 2009;106:3484–3489. doi: 10.1073/pnas.0808790106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jin X., Chen Y., Sun Y., Zeng C., Wang Y., Tao J., Wu A., Yu X., Zhang Z., Tian J., Guo D. Characterization of the guanine-N7 methyltransferase activity of coronavirus NSP14 on nucleotide GTP. Virus Res. 2013;176:45–52. doi: 10.1016/j.virusres.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ma Y., Wu L., Shaw N., Gao Y., Wang J., Sun Y., Lou Z., Yan L., Zhang R., Rao Z. Structural basis and functional analysis of the SARS coronavirus NSP14-NSP10 complex. Proc. Natl. Acad. Sci. U.S.A. 2015;112:9436–9441. doi: 10.1073/pnas.1508686112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.He R., Adonov A., Traykova-Adonova M., Cao J., Cutts T., Grudesky E., Deschambaul Y., Berry J., Drebot M., Li X. Potent and selective inhibition of SARS coronavirus replication by aurintricarboxylic acid. Biochem. Biophys. Res. Commun. 2004;320:1199–1203. doi: 10.1016/j.bbrc.2004.06.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sun Y., Wang Z., Tao J., Wang Y., Wu A., Yang Z., Wang K., Shi L., Chen Y., Guo D. Yeast-based assays for the high-throughput screening of inhibitors of coronavirus RNA cap guanine-N7-methyltransferase. Antivir. Res. 2014;104:156–164. doi: 10.1016/j.antiviral.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ferron F., Decroly F., Selisko B., Canard B. The viralRNA capping machinery as a target for antiviral drugs. Antivir. Res. 2012;96:21–31. doi: 10.1016/j.antiviral.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Deng X., Hackbart M., Mettelman R.C., O'Brien A., Mielech A.M., Yi G., Kao C.C., Baker S.C. Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. Proc. Natl. Acad. Sci. U.S.A. 2017;114(21):4251–4260. doi: 10.1073/pnas.1618310114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bhardwaj K., Liu P., Leibowitz J.L., Kao C.C. The coronavirus endoribonuclease NSP15 interacts with retinoblastoma tumor suppressor protein. J. Virol. 2012;86(8):4294–4304. doi: 10.1128/JVI.07012-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ricagno S., Egloff M.P., Ulferts R., Coutard B., Nurizzo D., Campanacci V., Cambillau C., Ziebuhr J., Canard B. Crystal structure and mechanistic determinants of SARS coronavirus non-structural protein 15 define an endoribonuclease family. Proc. Natl. Acad. Sci. U.S.A. 2006;103:11892–11897. doi: 10.1073/pnas.0601708103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Xu X., Zhai Y., Sun F., Lou Z., Su D., Xu Y., Zhang R., Joachimiak A., Zhang X.C., Bartlam M., Rao Z. New antiviral target revealed by the hexameric structure of mouse hepatitis virus nonstructural protein NSP15. J. Virol. 2006;80:7909–7917. doi: 10.1128/JVI.00525-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nedialkova D.D., Ulferts R., van den Born E., Lauber C., Gorbalenya A.E., Ziebuhr J., Snijder E.J. Biochemical characterization of arterivirus nonstructural protein 11 reveals the nidovirus-wide conservation of a replicative endoribonuclease. J. Virol. 2009;83:5671–5682. doi: 10.1128/JVI.00261-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Frieman M., Ratia K., Johnston R.E., Mesecar A.D., Baric R.S. Severe acute respiratory syndrome coronavirus papain-like protease ubiquitin-like domain and catalytic domain regulate antagonism of IRF3 and NF-κB signaling. J. Virol. 2009;83:6689–6705. doi: 10.1128/JVI.02220-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kindler E., Gil-Cruz C., Spanier J., Li Y., Wilhelm J., Rabouw H.H., Züst R., Hwang M., V'kovski P., Stalder H., Marti S., Habjan M., Cervantes-Barragan L., Elliot R., Karl N., Gaughan C., Kuppeveld F.J.M., Silverman R.H., Keller M., Ludewig B., Bergmann C.C., Ziebuhr J., Weiss S.R., Kalinke U., Thiel V. Early endonuclease-mediated evasion of RNA sensing ensures efficient coronavirus replication. PLoS Pathog. 2017;13(2) doi: 10.1371/journal.ppat.1006195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.von Grotthuss M., Wyrwicz L.S., Rychlewski L. mRNA cap-1 methyltransferase in the SARS genome. Cell. 2003;113:701–702. doi: 10.1016/S0092-8674(03)00424-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Decroly E., Imbert I., Coutard B., Bouvet M., Selisko B., Alvarez K., Gorbalenya A.E., Snijder E.J., Canard B. Coronavirus nonstructural protein 16 is a cap-0 binding enzyme possessing (nucleoside-2’O)- methyltransferase activity. J. Virol. 2008;82:8071–8084. doi: 10.1128/JVI.00407-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Menachery V.D., Eisfeld A.J., Schäfer A., Josset L., Sims A.C., Proll S., Fan S., Li C., Neumann G., Tilton S.C., Chang J., Gralinski L.E., Long C., Green R., Williams C.M., Weiss J., Matzke M.M., Webb-Robertson B., Schepmoes A.A., Shukla A.K., Metz T.O., Smith R.D., Waters K.M., Katze M.G., Kawaoka Y., Baric R.S. Pathogenic influenza viruses and coronaviruses utilize similar and contrasting approaches to control interferon-stimulated gene responses. mBio. 2014;5(3) doi: 10.1128/mBio.01174-14. e01174-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Menachery V.D., Yount B.L., Josset L., Gralinski L.E., Scobey T., Agnihothram S., Katze M.G., Baric R.S. Attenuation and restoration of severe acute respiratory syndrome coronavirus mutant lacking 2′-o-methyltransferase activity. J. Virol. 2014;88:4251–4264. doi: 10.1128/JVI.03571-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O'Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., Tummino T.A., Hüttenhain R., Kaake R.M., Richards A.L., Tutuncuoglu B., Foussard H., Batra J., Haas K., Modak M., Kim M., Haas P., Polacco B.J., Braberg H., Fabius J.M., Eckhardt M., Soucheray M., Bennett M.J., Cakir M., McGregor M.J., Li Q., Meyer B., Roesch F., Vallet T., Mac Kain A., Miorin L., Moreno E., Naing Z.Z.C., Zhou Y., Peng S., Shi Y., Zhang Z., Shen W., Kirby I.T., Melnyk J.E., Chorba J.S., Lou K., Dai S.A., Barrio-Hernandez I., Memon D., Hernandez-Armenta C., Lyu J., Mathy C.J.P., Perica T., Pilla K.B., Ganesan S.J., Saltzberg D.J., Rakes R., Liu X., Rosenthal S.B., Calviello L., Venkataramanan S., Liboy-Lugo J., Lin Y., Huang X.P., Liu Y., Wankowicz S.A., Bohn M., Safari M., Ugur F.S., Koh C., Savar N.S., Tran Q.D., Shengjuler D., Fletcher S.J., O'Neal M.C., Cai Y., J Chang J.C., Broadhurst D.J., Klippsten S., Sharp P.P., Wenzell N.A., Kuzuoglu-Ozturk D., Wang H.Y., Trenker R., Young J.M., Cavero D.A., Hiatt J., Roth T.L., Rathore U., Subramanian A., Noack J., Hubert M., Stroud R.M., Frankel A.D., Rosenberg O.S., Verba K.A., Agard D.A., Ott M., Emerman M., Jura N., von Zastrow M., Verdin E., Ashworth A., Schwartz O., d'Enfert C., Mukherjee S., Jacobson M., Malik H.S., Fujimori D.G., Ideker T., Craik C.S., Floor S.N., Fraser J.S., Gross J.D., Sali A., Roth B.L., Ruggero D., Taunton J., Kortemme T., Beltrao P., Vignuzzi M., García-Sastre A., Shokat K.M., Shoichet B.K., Krogan N.J. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wang Y., Sun Y., Wu A., Xu S., Pan R., Zeng C., Jin X., Ge X., Shi Z., Ahola T., Che Y., Guo D. Coronavirus NSP10/NSP16 methyltransferase can be targeted by NSP10-derived peptide in vitro and in vivo to reduce replication and pathogenesis. J. Virol. 2015;89:8416–8427. doi: 10.1128/JVI.00948-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ke M., Chen Y., Wu A., Sun Y., Su C., Wu H., Jin X., Tao J., Wang Y., Ma X., Pan J., Guo D. Short peptides derived from the interaction domain of SARS coronavirus nonstructural protein NSP10 can suppress the 2’-O-methyltransferase activity of NSP10/NSP16 complex. Virus Res. 2012;167:322–328. doi: 10.1016/j.virusres.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Almazan F., Dediego M.L., Galan C., Escors D., Alvarez E., Ortego J., Sola I., Zuñiga S., Alonso S., Moreno J.L., Nogales A., Capiscol C., Enjuanes L. Construction of a severe acute respiratory syndrome coronavirus infectious cDNA clone and a replicon to study coronavirus RNA synthesis. J. Virol. 2006;80:10900–10906. doi: 10.1128/JVI.00385-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zust R., Cervantes-Barragan L., Habjan M., Maier R., Neuman B.W., Ziebuhr J. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat. Immunol. 2011;12:137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]