Significance
Nipah virus (NiV) is a zoonotic virus and World Health Organization (WHO) priority pathogen that causes near-annual outbreaks in Bangladesh and India with >75% mortality. This work advances our understanding of transmission of NiV in its natural bat reservoir by analyzing data from a 6-y multidisciplinary study of serology, viral phylogenetics, bat ecology, and immunology. We show that outbreaks in Pteropus bats are driven by increased population density, loss of immunity over time, and viral recrudescence, resulting in multiyear interepizootic periods. Incidence is low, but bats carry NiV across Bangladesh and can shed virus at any time of year, highlighting the importance of routes of transmission to the timing and location of human NiV outbreaks.
Keywords: bats, henipavirus, Nipah virus, Pteropus, disease modeling
Abstract
Nipah virus (NiV) is an emerging bat-borne zoonotic virus that causes near-annual outbreaks of fatal encephalitis in South Asia—one of the most populous regions on Earth. In Bangladesh, infection occurs when people drink date-palm sap contaminated with bat excreta. Outbreaks are sporadic, and the influence of viral dynamics in bats on their temporal and spatial distribution is poorly understood. We analyzed data on host ecology, molecular epidemiology, serological dynamics, and viral genetics to characterize spatiotemporal patterns of NiV dynamics in its wildlife reservoir, Pteropus medius bats, in Bangladesh. We found that NiV transmission occurred throughout the country and throughout the year. Model results indicated that local transmission dynamics were modulated by density-dependent transmission, acquired immunity that is lost over time, and recrudescence. Increased transmission followed multiyear periods of declining seroprevalence due to bat-population turnover and individual loss of humoral immunity. Individual bats had smaller host ranges than other Pteropus species (spp.), although movement data and the discovery of a Malaysia-clade NiV strain in eastern Bangladesh suggest connectivity with bats east of Bangladesh. These data suggest that discrete multiannual local epizootics in bat populations contribute to the sporadic nature of NiV outbreaks in South Asia. At the same time, the broad spatial and temporal extent of NiV transmission, including the recent outbreak in Kerala, India, highlights the continued risk of spillover to humans wherever they may interact with pteropid bats and the importance of limiting opportunities for spillover throughout Pteropus’s range.
Outbreaks of zoonotic diseases are often sporadic, rare events that are difficult to predict, but can have devastating consequences (1). Emerging viral zoonoses of wildlife that have become pandemic include HIV/AIDS, 1918 H1N1 influenza virus, severe acute respiratory syndrome (SARS) coronavirus, and the current COVID-19 pandemic caused by SARS-CoV-2 (2–5). Bats are important hosts for many zoonotic viruses, including Ebola virus, SARS-CoV, SARS-CoV-2, and Nipah virus (NiV); the ecological drivers and transmission dynamics of these viruses in their reservoir hosts are poorly understood (6–12). A better understanding of the transmission dynamics of zoonotic pathogens in their natural reservoirs may help anticipate and prevent outbreaks (10, 13).
NiV is an emerging zoonotic paramyxovirus (genus Henipavirus) that has repeatedly spilled over from bats to cause outbreaks in people and livestock with high case-fatality rates across a broad geographic range. To date, human NiV infections have been identified in India, Bangladesh, Malaysia, Singapore, and the Philippines (14–18). It has caused repeated outbreaks in Bangladesh and India, with a mean case-fatality rate greater than 70% (14, 19, 20). A single genus of frugivorous bats (Pteropus) appears to be the main reservoir for henipaviruses throughout Asia and Australia (21–25). This includes Pteropus medius [formerly Pteropus giganteus (26)], the only pteropid bat present in Bangladesh and India (16, 27–30). NiV has several characteristics that make it a significant threat to human and animal health: 1) Its bat reservoir hosts are widely distributed throughout Asia and occur within dense human and livestock populations, leading to widespread frequent spillover events and outbreaks; 2) it can be transmitted directly to humans by bats or via domestic animals; 3) it can be transmitted from person to person; 4) spillover has repeatedly occurred in highly populous and internationally connected regions; 5) it is associated with high mortality rates in people; and 6) there are currently no commercially available vaccines to prevent infection or drugs to mitigate disease (31–33). As a result, the World Health Organization has listed NiV in its R&D Blueprint as one of the 10 highest-priority pathogens for the development of medical countermeasures due to its potential to cause significant outbreaks (34). In May 2018, an outbreak of NiV encephalitis with a 91% mortality rate occurred in a new location—Kerala, India—more than 1,200 km southwest of previous Indian and Bangladeshi outbreaks (35). A single case was subsequently reported in Kerala in 2019, and while local P. medius populations have been implicated as the local source of infection, the route of spillover in both instances remains unknown (35, 36).
In Malaysia and Bangladesh, consumption of cultivated food resources contaminated with bat excreta, such as mangoes in Malaysia and date palm sap in Bangladesh and northeastern India, have been identified as the predominant cause of spillover to pigs and people, respectively (37). Human outbreaks occur almost annually in Bangladesh, and the seasonal timing (November to April) and spatial distribution of outbreaks coincide with patterns of raw date-palm-sap consumption in a region termed the “Nipah belt” (38). However, there is variability in the geographic locations and number of spillover events, as well as the number and magnitude of human outbreaks that occur each year (39, 40). Spillover has also occurred outside the predominant season and region of date-palm-sap consumption (41). Whereas no human outbreaks have been reported in eastern Bangladesh, despite date-palm-sap harvesting and consumption, human outbreaks have been reported in Kerala, India, where date-palm sap is not cultivated (38). These observations suggest an alternate route of spillover in certain locations and a critical need to understand the mechanisms of underlying viral infection dynamics in bats and the extent of genetic diversity within the virus—each of which may influence the timing, location, and epidemiology of human outbreaks (38).
Previous research on the transmission dynamics of NiV and Hendra virus in Pteropus species (spp.) has produced mixed, and sometimes contradictory, findings. NiV, like Ebola, Marburg, Hendra, and some bat coronaviruses, is associated with seasonal spikes in infection that coincide with annual or semiannual synchronous birth pulses (21, 42–48). Seasonal periods of NiV shedding were observed in Pteropus lylei in Thailand, and seasonal spikes in NiV (or a related henipavirus) seroprevalence coinciding with pregnancy periods were observed in Eidolon dupreanum in Madagascar (49, 50), but not in Pteropus vampyrus or Pteropus hypomelanus in Peninsular Malaysia (25). Hendra virus prevalence in Australian pteropid bats has shown multiyear interepizootic periods, during which little virus can be detected, followed by periods of markedly increased viral shedding (51–53). It has been hypothesized that multiyear periodicity in the incidence of henipavirus infections could arise from a buildup and waning of herd immunity in the reservoir host, with reintroduction of virus via immigration, recrudescence, or viral persistence (11, 54–56).
There is a paucity of data related to henipavirus-associated immune dynamics in free-ranging pteropid bats, including the duration of immunity in adults and juveniles, which limits our understanding of population-level viral dynamics. Experimental infections of Pteropus bats with Hendra virus and NiV show that bats mount an antibody response following infection with Hendra virus and NiV (57–59). Waning of anti-NiV antibodies was observed in recaptured wild Eidolon helvum, a bat related to Pteropus spp., in Madagascar (60). Passive transfer and waning of maternal antibodies also occurred in captive Pteropus species, and, along with loss of immunity in adults, could contribute to the loss of herd immunity in wild populations (61). Some pteropid bat species are migratory, and interconnected colonies form a metapopulation, which could allow for viral reintroductions into susceptible colonies (10, 25, 62, 63). In addition, NiV recrudescence has been observed in wild-caught P. vampyrus and possibly also in E. helvum (64–66). Either of these phenomena could allow NiV to persist regionally during periods of high local immunity. However, no study has yet shown evidence in open, free-ranging bat populations that examines the influence of these factors on NiV transmission dynamics.
Here, we examine the distribution, dynamics, genetic diversity, and underlying drivers of NiV infection in P. medius in Bangladesh to improve our understanding of human outbreak patterns. Specifically, we analyze the spatial, temporal, and demographic variation in serological dynamics and viral shedding in bats over a 6-y period to determine the spatiotemporal drivers and dynamics of virus transmission. We also analyze the movement patterns of individual bats and analyze NiV phylogenetics to understand patterns of spatial mixing and virus strain diversity.
Results
Comparative Nipah Seroprevalence and Virus Infection Study in Bats Inside and Outside the Nipah Belt and Concurrent Longitudinal Bat Study Inside the Nipah Belt.
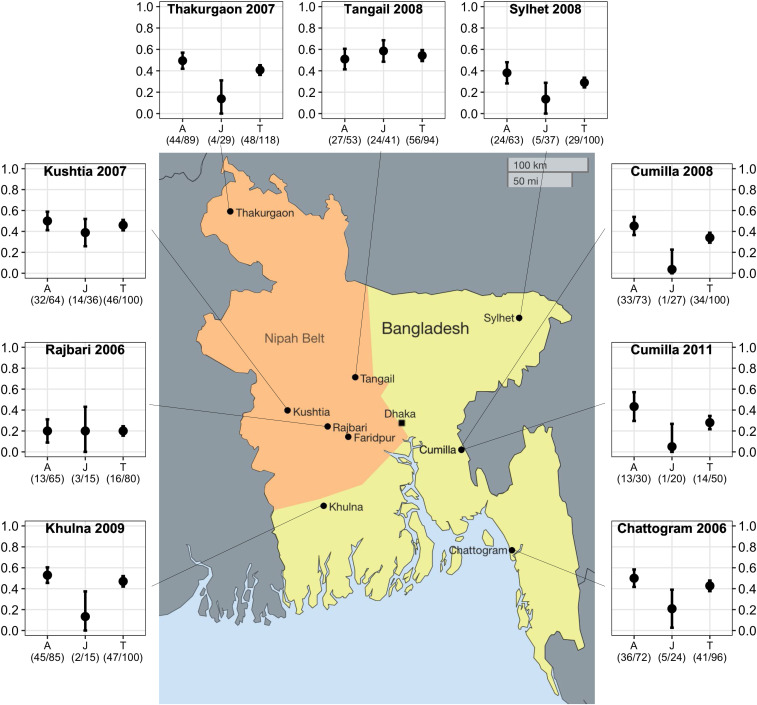
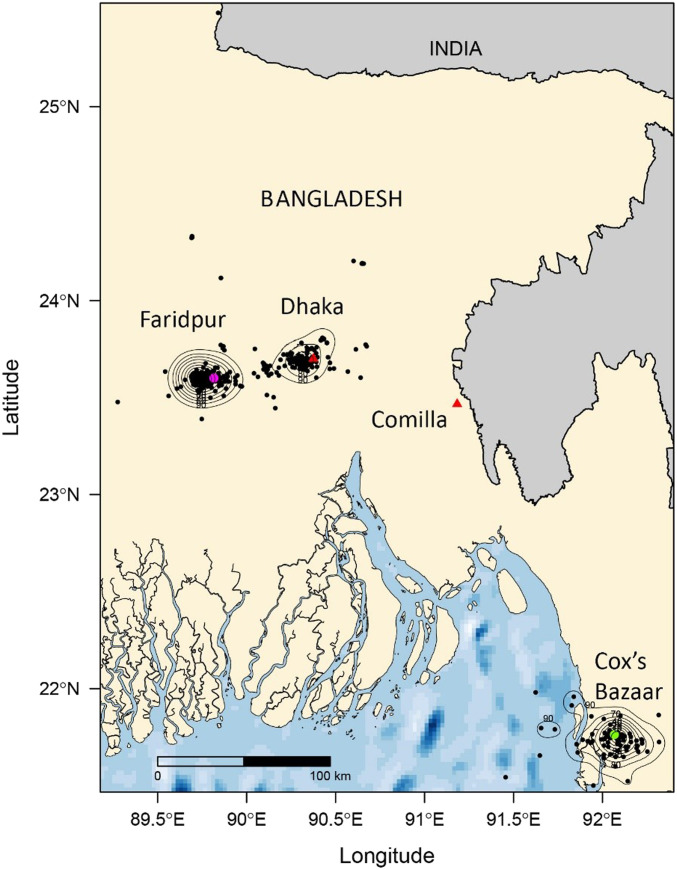
In a cross-sectional spatial study conducted between January 2006 and July 2012, we caught and tested 883 P. medius (∼100 per district) from eight colonies in different districts across Bangladesh. We detected anti-Nipah immunoglobulin G (IgG) antibodies in all colonies (Fig. 1). Seroprevalence varied by location (χ2 = 55.61, P < 0.001). In most locations, adult seroprevalence exceeded juvenile seroprevalence; in Tangail and Rajbari, seroprevalence was similar across ages. Viral detection in individuals was rare; overall, we detected NiV RNA in 11 of 2,088 individuals and in three pooled oropharyngeal samples (representing five bats, but which could not be resolved to an individual) (Table 1). We detected viral RNA in individual bats in Faridpur and Rajbari and from pooled samples from Thakurgaon and roost urine samples from Cumilla. Of the 11 PCR-positive individuals, three had IgG antibodies (SI Appendix, Table S1). We also detected virus in pooled urine collected from tarps placed below bats at roosts associated with human outbreaks in Bhanga and Joypurhat. The viral prevalence in Rajbari in January 2006 was 3.8% (95% CI: 0 to 11%; n = 78). In Faridpur, which is adjacent to Rajbari and where we conducted an intensive longitudinal study (see below), viral prevalence estimates ranged from 0 to 3% (95% CI: 0 to 10%; n = 100 at each of 18 sampling times) (Table 1). NiV RNA was detected in bats from inside (Rajbari, Thakurgaon, and Faridpur) and outside (Cumilla) the Nipah Belt. There was no significant difference between NiV detection rates from individual bats by the two main sample types: urine/urogenital swabs, 0.37% (n = 2,126) and oropharyngeal swabs, 0.15% (n = 1973) (χ2= 1.92 P = 0.17). The estimated detection rate from pooled urine samples, collected from tarps placed underneath roosts) across the entire study was 2.5% (n = 829), which was significantly higher than either sample type collected from individual bats (χ2 = 55.6, P < 0.001).
Fig. 1.
Map showing age-stratified seroprevalence in P. medius colonies, Bangladesh. Bats from eight colonies were sampled and tested for anti-NiV IgG antibodies: four within the “Nipah belt” (orange shaded) and four outside. Seroprevalence of adults (A), juveniles (J), and total seroprevalence (T) are shown with 95% CI error bars. The shaded region represents the “Nipah belt,” where previous human NiV outbreaks have been reported.
Table 1.
PCR detection of NiV RNA in P. medius 2006 to 2012
| Location | Date | Bats tested | Throat swabs tested | Throat swabs positive | Urine tested | Urine positive | Rectal swabs tested | Rectal swabs positive | Bats positive | Prevalence | ±95% CI |
| Spatial study | |||||||||||
| Rajbari | Jan-06 | 99 | 79 | 3 | 78 | 0 | 79 | 1 | 3 | 0.04 | 0.11 |
| Thakurgaon | Mar-07 | 118 | 115 | 3* | 72 | 0 | — | — | unk. | 0.00 | — |
| Kushtia | Aug-07 | 101 | 100 | 0 | 99 | 0 | — | — | 0 | 0.00 | — |
| Tangail | Jun-08 | 100 | 61 | 0 | 77 | 0 | — | — | 0 | 0.00 | — |
| Chattogram | Aug-06 | 115 | 19 | 0 | — | — | — | — | 0 | — | — |
| Cumilla | May-08 | 100 | 0 | 0 | 50 | 0 | — | — | 0 | — | — |
| Sylhet | Sep-08 | 100 | 100 | 0 | 49 | 0 | — | — | 0 | 0.00 | — |
| Khulna | Jan-09 | 100 | 50 | 0 | 80 | 0 | — | — | 0 | 0.00 | — |
| Cumilla | Mar-11 | 50 | 50 | 0 | 50 | 0 | — | — | 0 | 0.00 | — |
| Longitudinal study | |||||||||||
| Faridpur | Jul-07 | 102 | 64 | 0 | 50 | 0 | — | — | 0 | 0.00 | |
| Faridpur | Dec-07 | 101 | N/A | N/A | N/A | — | — | — | 0 | ||
| Faridpur | Apr-08 | 100 | 64 | 0 | 88 | 0 | — | — | 0 | 0.00 | |
| Faridpur | Jul-08 | 100 | 58 | 0 | 74 | 0 | — | — | 0 | 0.00 | |
| Faridpur | Jul-08 | 100 | 98 | 0 | 99 | 0 | — | — | 0 | 0.00 | |
| Faridpur | Feb-09 | 100 | 50 | 0 | 100 | 1 | — | — | 1 | 0.01 | 0.10 |
| Faridpur | May-09 | 101 | 100 | 0 | 99 | 2 | — | — | 2 | 0.02 | 0.10 |
| Faridpur | Aug-09 | 100 | 100 | 0 | 99 | 0 | — | — | 0 | 0.00 | |
| Faridpur | Nov-09 | 100 | 100 | 0 | 82 | 1 | — | — | 1 | 0.01 | 0.11 |
| Faridpur | Feb-10 | 100 | 100 | 0 | 100 | 0 | — | — | 0 | 0.00 | |
| Faridpur | Jun-10 | 100 | 100 | 0 | 100 | 3 | — | — | 3 | 0.03 | 0.10 |
| Faridpur | Sep-10 | 100 | 100 | 0 | 100 | 0 | — | — | 0 | — | |
| Faridpur | Jan-11 | 100 | 100 | 0 | 100 | 0 | — | — | 0 | 0.00 | |
| Faridpur | May-11 | 102 | 102 | 0 | 102 | 1 | — | — | 1 | 0.01 | 0.10 |
| Faridpur | Aug-11 | 100 | 100 | 0 | 100 | 0 | — | — | 0 | — | |
| Faridpur | Dec-11 | 100 | 100 | 0 | 100 | 0 | — | — | 0 | — | |
| Faridpur | Apr-12 | 100 | 78 | 0 | 78 | 0 | — | — | 0 | — | |
| Faridpur | Jul-12 | 100 | 100 | 0 | 100 | 0 | — | — | 0 | — | |
| Faridpur | Nov-12 | 100 | 100 | 0 | 100 | 0 | — | — | 0 | ||
| Total | 2,789 | 2,088 | 6 | 2,126 | 8 | 79 | 1 | 11 | 0.005 | 0.02 | |
| Outbreak investigation | |||||||||||
| Pooled roost urine samples n = no. pos. | |||||||||||
| Bangha | Feb-10 | 19 | 3 | ||||||||
| Joypurhat | Jan-12 | 19 | 16† | ||||||||
| Rajbari | Dec-09 | 35 | 0 | ||||||||
| West Algi | Jan-10 | 31 | 0 | ||||||||
Unk., unknown.
NiV RNA was detected in three pooled oropharyngeal samples, confirmed by sequencing, although confirmation from individual samples could not be made. These data are not used in prevalence estimates.
Detection by qPCR, Ct ranges 20 to 38.
Factors Associated with NiV IgG Serostatus in P. medius.
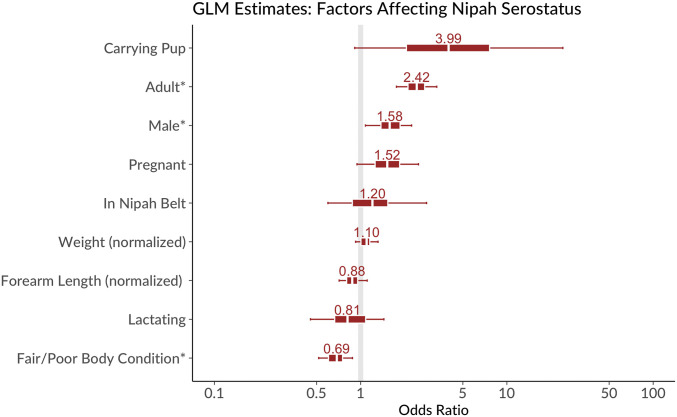
There was no statistical difference between seroprevalence in bats inside the Nipah Belt and outside (95% odds ratio [OR] 1.2, highest posterior density interval [HDPI] 0.47 to 3.1). Adults had higher seropositivity than juveniles (OR 2.4, 1.7 to 3.6 HDPI), and males greater than females (OR 1.6, 1.0 to 2.4 HDPI) (Fig. 2). There was weak evidence that seroprevalence was higher in pup-carrying (OR 4.0, HDPI 0.6 to 34) and pregnant (1.5 times, HDPI 0.85 to 2.8) individuals than other females. Neither mass, forearm length, nor the mass:forearm ratio (a proxy for age) consistently correlated with seropositivity. However, bats with poor body condition (an assessment of pectoral muscle mass by palpation) were less likely to be seropositive (poor/fair body condition OR = 0.69, HDPI 0.49 to 0.96). Finally, serostatus was strongly correlated in mother–pup pairs; 39 of 41 pups with seropositive mothers were seropositive, and 32 of 39 pups with seronegative mothers were seronegative.
Fig. 2.
Results of Bayesian generalized linear model of factors affecting Nipah serostatus in bats in cross-sectional study. Bars indicate ORs and 50% (inner) and 95% (outer) credible intervals for model parameters. Factors with asterisks (*) have 95% CIs that do not overlap one. Model intercept (predicted probability of seropositivity for a juvenile, female bat outside the Nipah belt of mean size and good body condition) was 0.26 (95% CI 0.12 to 0.56).
Longitudinal NiV Serodynamics in P. medius, Faridpur District (2006 to 2012).
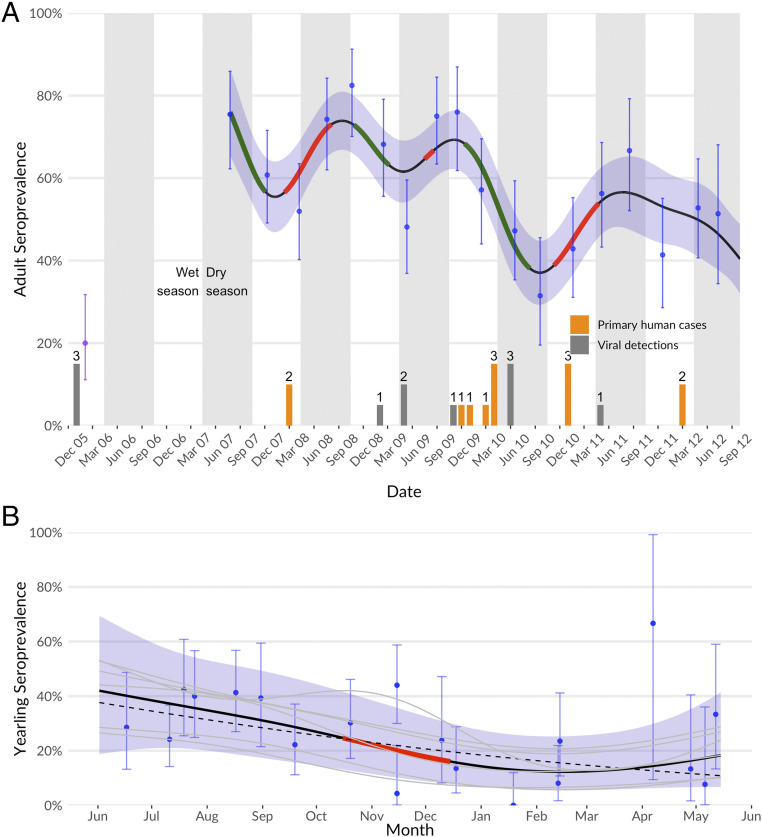
We sampled bats quarterly from a single population in the Faridpur district between 2007 and 2012 (total bats sampled = 2,789). We also microchipped a total of 2,345 bats. We used generalized additive models (GAMs) to characterize changes in NiV seroprevalence over time. There were significant fluctuations in adult (>24 mo) and juvenile (6 to 24 mo) seroprevalence over the 6-y study period (Fig. 3A). Juvenile seroprevalence ranged from 0 to 44% (95% CI: 37 to 51%), and decreased over the first year of life for bats born in each year (“yearlings”), consistent with loss of maternal antibodies in juveniles. A more pronounced decrease occurred from mid-October to mid-December than other parts of the year. However, the GAM indicating this had only slightly better fit (ΔAIC [ΔAkaike information criterion] < 1) than one with a linear decrease over the whole year (Fig. 3B).
Fig. 3.
Serodynamics of the Faridpur bat population, measured and fit to a GAM. (A) Adult seroprevalence over time, with measured values and 95% CI in blue and mean GAM prediction and 95% shown with line and surrounding shaded areas. Point from February 2006 (purple) is shown separately due to ELISA vs. Luminex measure. Periods of significant change (where GAM derivative 95% does not overlap zero) are shown in red (increasing) and green (decreasing). Periods of increase indicate viral-circulation events in the adult population; these do not occur with consistent periodicity or seasonality. Counts of primary human cases from local district (dark gray) and bat viral detections (orange; Table 1) are shown on bottom for comparison. (B) Juvenile seroprevalence during the first year of life (“yearlings”). All years’ measurements are collapsed onto the scale of a single year overlain to show yearling dynamics. Measured values and 95% CIs are shown in blue, and mean and 95% CIs for the GAM model pooled across cohorts are shown with line and surrounded shaded areas. GAM realizations for individual years are in gray and overall effect in black. The period of significant decline in the GAM is shown in red. Juvenile seroprevalence decreases over the course of the year and is not distinguishable from a simple linear decrease (ΔAIC < 1, dotted line).
Adult seroprevalence ranged from 31% (95% CI: 20 to 46%) to 82% (95% CI: 77 to 87%) with three cycles of clear variability over the course of the study (Fig. 3A). We found no evidence of regular seasonal fluctuations; a GAM with annual cyclic terms fit worse than one without (ΔAIC > 10). Viral RNA was detected during periods of increasing, decreasing, and stable seroprevalence.
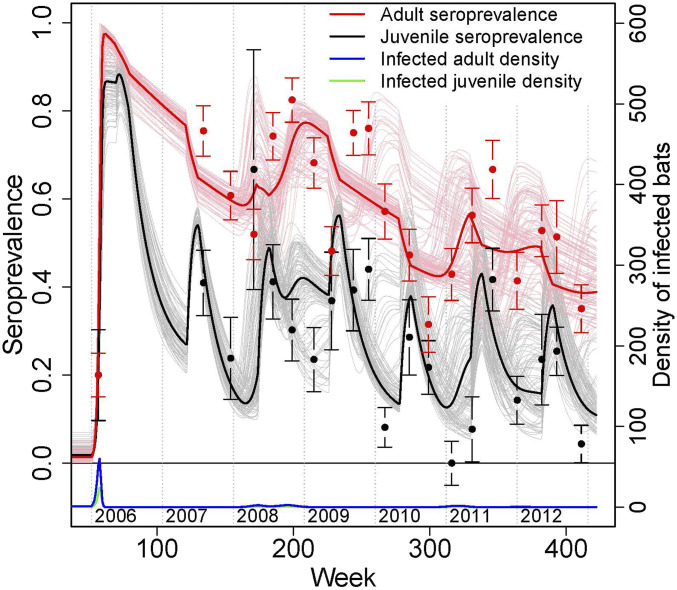
We fitted a series of age-stratified compartmental susceptible–infected–recovered models to examine different biological processes influencing serodynamics, including density- vs. frequency-dependent transmission, recrudescence vs. immigration of infected individuals, and seroreversion (loss of antibodies) in both juveniles and adults (Methods and Fig. 4). Density-dependent models were a far better fit to the data than frequency-dependent models (difference in log-likelihood 10.0; ΔAIC = 20.0), suggesting that movements of bats and fluctuations in colony size alter spatiotemporal variation in the risk of NiV infection in bats. In Faridpur (“Domrakhandi/Khaderdi” in SI Appendix, Fig. S1) during the period of sampling, the roost population declined from ∼300 bats to 185, which decreased transmission potential in the fitted model: R0 in adult bats was estimated to decrease from 3.5 to 2.1 as the number of bats in the colony decreased. As a result, over the 6-y study period, the fitted model predicted that the threshold for herd immunity (i.e., the seroprevalence below which the reproductive ratio Rt > 1) for adults fell from 72% (when bat counts were highest—in 2006) to 52% (when bat counts were lowest).
Fig. 4.
Longitudinal data and fitted model for NiV serological dynamics in adult and juvenile bats. Red and black points show observed data (±1 SE), and solid lines show the fitted model (thick lines show the trajectory for the model with maximum-likelihood parameter estimates; thin lines show realizations for parameter estimates drawn from the estimated distributions) for the fraction of adults and juveniles seropositive for NiV (left axis), and the model-estimated number of infected adult and juvenile bats (bottom and right axes). See Methods for details of model structure.
The fitted model suggests that serodynamics in juveniles were strongly driven by inheritance and loss of maternal antibodies. The estimated duration of maternal antibodies was 17.6 wk (95% CI: 13.7 to 25.0), which was much quicker than the loss of antibodies in adults (290.8 wk, 95% CI: 245.0 to 476.4) (SI Appendix, Table S2). Finally, models with recrudescence fit the data better than models without recrudescence (SI Appendix, Table S2; difference in log-likelihood 32.6; ΔAIC = 65.1), and models with recrudescence fit the data better than models with immigration (ΔAIC = 3.76).
Mark–Recapture and Seroconversion/Seroreversion.
There were 56 recapture events over the study period (SI Appendix, Table S3). Thirty-one bats were recaptured at a nearby roost other than the original capture location. This network of roosts, or “roost complex,” formed a polygon covering ∼80 km2 and included 15 roosts sampled during the longitudinal study (SI Appendix, Fig. S2 A and B). Ten instances of seroconversion (change from IgG-negative to IgG-positive) and nine instances of seroreversion (positive to negative) were observed (SI Appendix, Table S3). The mean time between positive and negative tests in adults (excluding juveniles with maternal antibodies) was 588 d (n = 6) (range: 124 to 1,082 d).
Home Range and Intercolony Connectivity Analysis.
Home-range analysis of satellite telemetry data from 14 bats (mean duration of collar data transmission = 6.25 mo; range = 1 to 25 mo; SI Appendix, Table S4) showed that the majority of bats roosted within 10 km of where the bats were originally collared, in the Faridpur (Nipah belt) colony, and within 7 km from where the bats in the Cox’s Bazaar colony were originally collared (315 km east of Faridpur). The average foraging radius was 18.7 km (SD 21.5 km) for the Faridpur bats and 10.8 km (SD 11.9 km) for the Cox’s Bazaar bats (SI Appendix, Fig. S2). Home range analysis suggests that bats in Faridpur and Cox’s Bazar (separated by approx. 310 km) would have a <5% probability of intermingling (Fig. 5). Home-range size was larger during the wet season than the dry season (2,746 km2 vs. 618 km2) (SI Appendix, Figs. S3 and S4).
Fig. 5.
Satellite telemetry and home-range analysis. Location data from satellite collars (n = 14) placed on 11 bats from Faridpur and 3 bats from Cox’s Bazaar, Chattogram, collected between 2009 and 2011 were used to calculate local and long-range movement patterns and home range for these two groups.
NiV Phylogenetic Analysis.
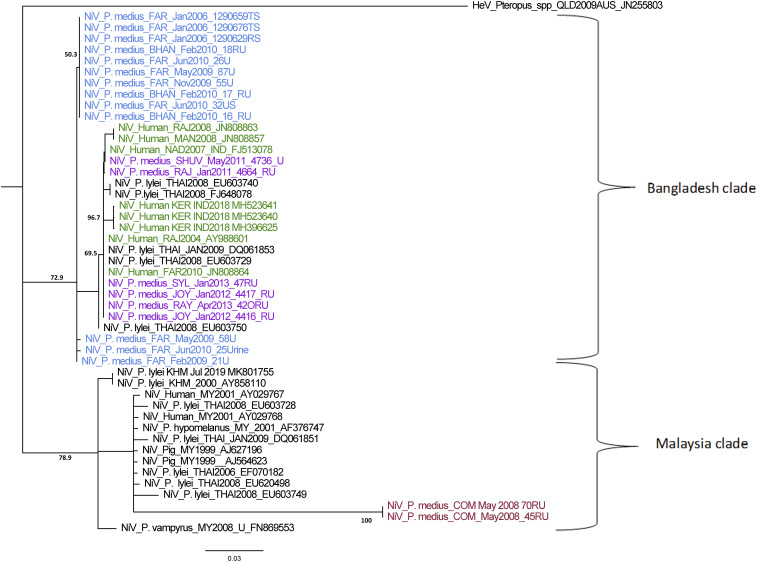
Phylogenetic analysis of NiV sequences from a 224-nt section of the N gene (nt position 12,90 to 1,509 [position ref gb|FJ513078.1| India]) suggests that strains from both India and Malaysia clades are present in bats in Bangladesh (Fig. 6). This finding was supported by an additional analysis of near-whole N gene sequences (∼1,720 nt) from bats, pigs, and humans, including those from a subset of P. medius from this and a more recent study by our group (SI Appendix, Fig. S5) (67). Eleven 224-nt N gene sequences obtained from bats between 2006 and 2012 (all from the Faridpur population) were identical. Overall, the N gene sequences identified from the Faridpur, Rajbari and Bhanga colonies between 2006 and 2011 had 98.21 to 100% shared nucleotide identity. Sequences from Rajbari district obtained 5 y apart (January 2006 and January 2011) had only a single nucleotide difference, resulting in a synonymous substitution (G to A) at position 1,304, which was found in four other bat NiV sequences from this study, as well as in the NiV isolate from P. vampyrus in Malaysia. Five human NiV N gene sequences from various locations within the Nipah belt over the same time period as our bat study show more nucleotide diversity than those from the Faridpur P. medius population. Human sequences throughout Bangladesh and from Kerala, India, all nested within the diversity found in P. medius (Fig. 6). By contrast, the sequences found in P. medius from Cumilla, a location 150 km to the east of Faridpur, showed 80.8 to 82.59% shared nucleotide identity with sequences from P. medius in Faridpur and clustered within the Malaysia group of NiV sequences. The two Cumilla sequences were identical to each other and had up to 87.95% shared nucleotide identity to sequences from P. lylei in Thailand.
Fig. 6.
NiV partial N-gene phylogeny (224 nt). Phylogenetic neighbor-joining tree created in Geneious Prime 2019 using a Tamura-Nei model with 1,000 bootstrap replicates and Hendra virus as an outgroup is shown. Branch lengths are shown as the number of substitutions per site. Sample collection date, location, and GenBank accession numbers are included in the label for each sequence, except P. medius sequences we collected (GenBank accession nos. MK995284–MK995302). Blue labels indicate bat sequences from Faridpur and Bhanga (an outbreak response in Faridpur). Purple sequences are from P. medius from other roosts sampled during the longitudinal study. Red sequences are from P. medius in Cumilla. Green sequences are human NiV sequences from Bangladesh and India.
Discussion
Our findings suggest that NiV circulation occurs in bat populations throughout the country. We observed that virus can be shed by bats at any time of year and that viral dynamics are cyclical, but not annual or seasonal. Our models fit to serological data suggest that these cycles may be driven by demographic and immunological factors; the waning of herd immunity through turnover or individual waning in bat populations allows heightened viral transmission when seroprevalence passes below a critical threshold. Previous studies from Bangladesh suggested that human NiV outbreaks occur primarily within a defined region in western Bangladesh, termed the “Nipah belt,” during a defined season (November through April) (41, 68). These observations raised the question of whether the timing and location of human infections are due solely to differences in the frequency and intensity of date-palm-sap consumption, or whether ecological factors such as the distribution and timing of bat viral infection also play a role (19, 38, 69). Our extensive survey of P. medius, which is common across Bangladesh and throughout the Indian subcontinent, demonstrates that viral circulation within their populations is not limited to the Nipah belt (16, 27, 30).
A number of mechanisms have been proposed for the maintenance of acute viral infections in bat populations, which are often formed of interconnected colonies, including synchronous birthing and subsequent loss of maternal antibodies (11, 43, 45); lowered immunity within pregnant females due to stress; nutritional stress and other factors (47); immigration of infected individuals from other colonies (62, 70, 71); and recrudescence within previously infected individuals (11, 64, 72). However, little is known about how henipaviruses are transmitted among wild bats. Pteropus species are typically gregarious and their roosts, often comprising multiple hardwood trees, and are highly socially structured, with individuals segregated by age, sex, and social dominance (69, 73, 74). Interactions among individuals are often dependent on their grouping, and the intensity of social interactions varies with specific behaviors such as mother–pup interactions, play (juveniles), territorial fighting (adult males), and mating (adults) (74). Our data and previous experiments show that henipaviruses can be shed orally, urogenitally, in feces, and in birthing fluids (59, 75). This suggests that multiple mechanisms for transmission are possible, including mutual grooming, fighting, mating, exposure to excreta or birthing fluids, and ingestion of food contaminated by saliva. Roost size also increases seasonally during mating and birthing periods, which the fitted models suggest would increase transmission, if seroprevalence is below the herd-immunity threshold (30, 73). While P. medius does not roost with other bat species, it does feed with other frugivorous bats, and it’s possible that interspecies viral transmission occurs during feeding (76, 77). In Madagascar, henipavirus antibodies have been detected in multiple species of frugivorous bats, though it is unknown whether the same virus or antigenically related viruses was shared among them (60). While serological evidence suggests that it is possible henipaviruses circulate in other frugivorous bat species, our findings, as well as those of others (16, 59), suggest that in Bangladesh, P. medius is the main natural reservoir for NiV. Henipaviruses other than Nipah may be circulating in P. medius (28). We assumed that the anti-IgG antibodies detected by the serological assays used in this study were specific to NiV, but it is possible that the enzyme-linked immunosorbent assay (ELISA) used in the cross-sectional study may have detected antibodies against unknown henipaviruses, which could elevate NiV seroprevalence estimates. An advantage of the Luminex assay used in the longitudinal study is that we could compare median fluorescent intensity (MFI) values to multiple specific henipaviruses (Nipah, Hendra, and Cedar) and differentiate between specific reactions to NiV and reactions to the other viruses, which could indicate antibodies against an unknown henipavirus. Hendra and Cedar viruses are enzootic in Australian Pteropus spp. and are not known to occur in Bangladesh, so we considered reactions to these viruses NiV-negative results.
Our modeling indicates that NiV is primarily driven by immunity and density-dependent transmission dynamics among bats, with cycles of higher seroprevalence dampening intracolony transmission followed by waning of antibody titers within individuals and death of seropositive individuals. Waning humoral immunity against NiV has been consistently shown in henipavirus studies of African pteropodid bats (56, 60). Our recapture data provided reported evidence of the loss of detectable NiV IgG antibodies in individual free-ranging bats, which supports the fitted model suggesting limited duration individual immunity and the importance of population-level waning immunity. The consistently decreasing seroprevalence that we observed in juveniles suggests that they lose maternal antibodies over their first year (the fitted model estimates after 3 to 5 mo), consistent with other studies of maternal antibodies against henipaviruses in pteropodid bats (47, 56, 61, 65). Our analysis do not support the hypothesis (45) that seasonal pulses of these new seronegative individuals are sufficient to drive new outbreaks because high seroprevalence in adults limited transmission in several years (Fig. 4).
NiV reintroduction into a colony may occur from a persistently infected individual (e.g., via recrudescence) or immigration of an infected individual. Our analyses suggested that recrudescence was a more important driver of transmission dynamics than immigration. Recrudescence of henipavirus infection has been observed for NiV in captive P. vampyrus (64), for henipavirus in captive E. helvum (56, 66), and humans infected by NiV (78) and Hendra virus (79). It is difficult to know from serology alone whether wild-caught seronegative bats had been previously infected. Experimental infections comparing naïve to previously infected P. medius that have sero-reverted would provide a better understanding of how humoral immunity influences individual susceptibility to infection and inform dynamics models attempting to explain viral maintenance within bat populations (60).
Our longitudinal study was limited to a single population of interacting subcolonies and bat populations across Bangladesh likely represent a dynamic metapopulation. Our roost count data and recapture data from microchipped bats showed how roost sizes can fluctuate and that bats shift among local roosts. The fitted model strongly suggested that decreases in local roost counts substantially reduced local transmission potential of NiV. However, a larger study across multiple regional populations would be needed to understand how local shifts in bat colonies impact broader fluctuations in regional populations and spatial patterns of NiV transmission.
Understanding how bat populations connect across landscapes is important for understanding viral maintenance, and studying local and migratory bat movements can provide important ecological information related to viral transmission, including how bats move between different colonies (62, 80). Our satellite telemetry data suggest that P. medius exists as a metapopulation, like other pteropid species (11, 71). The numbers of individuals we collared represents a small sample size; however, they are comparable to other bat satellite telemetry studies of related species, and our data suggest that bat dispersal in Bangladesh may currently be more localized than other species elsewhere. P. medius appear to travel shorter distances and remain within a smaller home range (321.46 and 2,865.27 km2 for two groups) than P. vampyrus in Malaysia (64,000 and 128,000 km2) and Acerodon jubatus in the Philippines, both of which are similarly sized fruit bats (62, 81). Pteropodid bat migration is primarily driven by seasonal food-resource availability (63, 82–84). In Bangladesh, P. medius prefer to roost in human-dominated environments in highly fragmented forests, as opposed to less-populated, intact forested areas, such as in national parks (85). The conversion of land to villages and farmland over recent human history has likely led to increased food availability for P. medius and may have reduced the impetus for long-distance migration (37). This may reflect a similar adaptation to anthropogenic food resources, as observed over the last few decades in Australian Pteropus species (71). Home ranges were significantly smaller during the dry season, which corresponds to winter months and the time when most female bats are pregnant, likely resulting in them flying shorter distances to conserve energy. Genetic analysis of P. medius across Bangladesh has shown that, historically, there has been extensive gene flow and intermixing among populations, and we did observe a few instances of longer-distance flights; however, the movement data indicated that, overall, these bats had much smaller home ranges (80). Less connectivity among bat populations across Bangladesh may influence NiV transmission by creating longer interepizootic periods and larger amplitude fluctuations in population-level immunity in P. medius compared to more migratory species (71). Bat movement and population connectivity may also influence the genetic diversity of NiV found in different locations.
The potential existence of a more transmissible or pathogenic strain of NiV already circulating in bats further underscores the need to strengthen efforts to prevent spillover. While the overall strain diversity among NiV has not been well characterized due to a dearth of isolates, two distinct NiV clades have been described: a Bangladesh clade, that includes sequences identified in India and Bangladesh; and a Malaysian clade, that comprises sequences from Malaysia, Cambodia, The Philippines, and Thailand (18, 67, 86). Our findings of substantially different NiV sequences in Faridpur and Cumilla suggest that viruses from both clades are circulating in Bangladesh. Strains of NiV from these two clades are associated with differences in pathogenesis, epidemiological and clinical profiles in humans and animal models, and observed shedding patterns in bats (49, 87–91). Phenotypic variation in NiV could influence human outbreak patterns by altering transmission to, or pathogenesis in, humans and the likelihood of smaller outbreaks being identified or reported (92). Human-to-human NiV transmission via contact with respiratory and other secretions has been regularly observed in Bangladesh and India, including the recent 2018 outbreak in Kerala (14, 68, 93), whereas transmission among people was not a common feature of the Malaysia outbreak, despite close contact between cases and health care providers (94, 95). NiV cases in Bangladesh have shown more strain diversity than in the Malaysia outbreak, which could be due to greater virus diversity in P. medius (96).
Until now, there have been very few Nipah sequences obtained from P. medius. We found that Nipah N-gene sequences from bats from the Faridpur population were nearly identical over time, compared to variation in N-gene sequences from bats and humans from other locations observed over the same time period (2006 to 2010). This suggests that there may be locally prevalent and stable NiV genotypes that persist within bat colonies. Human NiV genotype diversity is likely a reflection of the diversity of the NiV strains in the local bats that seed outbreaks (10). This is also supported by viral sequences obtained from humans and bats associated with the 2018 NiV outbreak in Kerala, India, where human NiV sequences were most closely related to local P. medius sequences (97).
Connectivity of pteropid bats in Bangladesh with those in Southeast Asia could explain the observed strain diversity in our study. Historical interbreeding of P. medius with pteropid species found in Myanmar, Thailand, and Malaysia and our telemetry findings showing bats are capable of flying hundreds of kilometers could explain the presence of a Malaysia clade NiV sequence in bats from Cumilla (80). NiV Bangladesh strains have also been found in P. lylei in Thailand (98). The N gene of the Cumilla NiV strain differs from those reported from bats in the Nipah belt by 20%, whereas NiV Malaysia and NiV Bangladesh differ by only 6 to 9% and are associated with different clinical profiles. Whole-genome sequence would have allowed for better characterization of the Cumilla strain; however, this was not obtained. Despite the short sequences used in our analysis, the N gene is generally conserved relative to other genes and is representative of the diversity across henipavirus genomes (86). We would expect the rest of the Cumilla viral genome to also be highly divergent, potentially even qualifying it as a different henipavirus species. It is, therefore, plausible that the clinical profile of a 20% divergent Nipah-related virus differs significantly from known strains. Sequence information from an isolated human NiV case in Cumilla has not been reported, so comparison to the sequence we found in bats was not possible (41). Studies linking viral genotype to clinical outcomes in people would provide additional insight into the effect of strain diversity in bats on the potential for larger-scale human outbreaks.
Our study sheds light on the sporadic nature of human NiV outbreaks, with multiyear interepizootic periods in South Asia. PCR results show that overall NiV incidence in P. medius is low, consistent with previous studies of Hendra virus and NiV (25, 47, 52). The fitted model suggests that transmission increases when bat populations have become susceptible through waning immunity (11). In the current study, observed seroprevalence patterns and the fitted model suggest that three periods of transmission occurred over the 6 y of sampling, each of which followed periods of low adult and juvenile seroprevalence. Viral detection in bats has coincided with some human outbreaks, supporting the hypothesis that spillover is a sporadic event (67, 97). In our study periods, low seroprevalence in bats was not always followed by outbreaks in humans. We detected NiV RNA during periods of both increasing and high seroprevalence, consistent with recrudescence, which was strongly supported by the fitted model and has been demonstrated in captive animals (64–66). This likely contributes to the sporadic variation in human outbreaks (e.g., spillover events) from year to year in Bangladesh.
Overall, our results suggest that NiV outbreaks in humans stem from an interaction of four factors: 1) multiyear fluctuations in transmission intensity among bats driven by immunity and colony size/density-dependent transmission; 2) relatively localized bat movements creating spatially variable transmission dynamics; 3) occasional shedding by previously infected bats due to recrudescence; and 4) highly seasonal contact between bats and humans via consumption of raw date-palm sap. The timing of multiple factors involved in driving transmission dynamics needs to align for intracolony NiV transmission events and further align with human behavior and availability of a route of spillover for human outbreaks to occur, as previously hypothesized (99). We further conclude that NiV dynamics in bats combined with the seasonality and specific geography of date-palm-sap consumption in Bangladesh likely explains the sporadic nature of human outbreaks in the region (38).
These findings suggest that human NiV outbreaks in other regions of Bangladesh (and Asia) where Pteropus bats occur are also likely to be sporadic and rare, leading to underreporting or a lack of reporting. This is probably exacerbated by the fact that the clinical syndrome is similar to that of other common infections, such as Japanese encephalitis, malaria, and measles (100). Understanding whether some NiV strains are capable of causing mild or asymptomatic cases will provide important insights about why outbreaks have not been detected in areas such as eastern Bangladesh or other parts of Asia, where host, virus, and potential routes of spillover exist. One reason is that mild or asymptomatic cases would be unlikely to be detected by current surveillance systems. About half of all Nipah outbreaks in Bangladesh between 2007 and 2014 were unreported, suggesting that many cryptic spillover events have occurred (101). The 2018 and 2019 spillover events in Kerala, India, which were linked to local P. medius colonies and which occurred in an area that does not cultivate date-palm sap, further emphasize the point, but raise questions about the mechanism of spillover.
In the last two decades, the world has experienced large epidemics of bat-associated viruses, including Ebola in West Africa and Democratic Republic of the Congo, SARS coronavirus, and SARS-CoV-2. The World Health Organization has listed NiV and other henipaviruses as priority pathogens for vaccine and therapeutic research and development, along with Ebola viruses and coronaviruses. Surveillance for henipaviruses and antibodies in bats and people where they are in close contact will help determine spillover risk; characterize henipavirus genetic diversity; and understand the genetic determinants of NiV transmissibility and pathogenicity among humans. These measures may help target interventions that reduce spillover, substantially improving our ability to reduce the risk of a more transmissible strain of NiV emerging and causing a large-scale epidemic with significant human and animal mortality.
Methods
The study period was between January 2006 and November 2012. The study was conducted under Tufts University Institutional Animal Care and Use Committee protocol G929-07 and International Centre for Diarrhoeal Diseases Research, Bangladesh (ICDDR,B) Animal Experimentation Ethical Committee Protocol 2006-012 with permission from the Forest Department, Government of Bangladesh. Locations were selected based on whether the district had any previously recorded human NiV encephalitis clusters at the time of this study and was therefore inside the Nipah Belt (e.g., Rajbari, Tangail, Thakurgaon, and Kushtia) or whether they had not and were outside the Nipah Belt (e.g., Cumilla, Khulna, Sylhet, and Chattogram). The Thakurgaon study was conducted as part of an NiV outbreak investigation and coincided with ongoing human transmission (102). Between 2006 and 2012, three different studies of P. medius with similar bat-sampling protocols were performed: 1) a cross-sectional spatial study with a single sampling event in each of the eight locations listed above; 2) a longitudinal study of a Faripur bat colony with repeated sampling approximately every 3 mo from July 2007 to November 2012; and 3) a longitudinal study of the Rajbari colony with repeated sampling at a monthly interval between 12 mo period between April 2010 and May 2011. Opportunistic sampling of P. medius was also performed during this time period during NiV outbreak investigations (Bangha, Faridpur [February 2010], Joypurhat [January 2012], Rajbari [December 2009], and West Algi, Faridpur [January 2010]). Bats were captured using mist nets at locations within eight different districts across Bangladesh between January 2006 and December 2012 (Fig. 1).
Capture and Sample Collection.
For the country-wide cross-sectional and Faridpur longitudinal study, ∼100 bats were sampled at each sampling event, which lasted 7 to 10 d. This sample size allowed us to detect at least one exposed bat (IgG antibody-positive) given a seroprevalence of 10% with 95% confidence. Bats were captured using a custom-made mist net of ∼10 m × 15 m suspended between bamboo poles, which were mounted atop trees close to the target bat roost. Catching occurred between 11 PM and 5 AM as bats returned from foraging. To minimize bat stress and chance of injury, nets were continuously monitored, and each bat was extracted from the net immediately after entanglement. Personal protective equipment was worn during capture and sampling, which included dedicated long-sleeve outerwear or Tyvek suits, P100 respirators (3M), safety glasses, nitrile gloves, and leather welding gloves for bat restraint. Bats were placed into cotton pillowcases and held for a maximum of 6 h before being released at the site of capture. Bats were sampled at the site of capture using a field laboratory setup. Bats were anesthetized by using isoflurane gas (103), and blood, urine, oropharyngeal swabs, and wing-membrane biopsies (for genetic studies) were collected. For some sampling periods, rectal swabs were collected, but due to resource constraints, these samples were deemed to likely be lower-yield than saliva and urine for NiV and were discontinued during the study. For each bat sampled, we recorded age, weight, sex, physiologic and reproductive status, and morphometric measurements, as described (27). Bats were classified as either juveniles (approximately 4 to 6 mo—the age by which most pups are weaned) to 2 y old (the age when most Pteropus species reach sexual maturity) or adults (sexually mature) based on body size and the presence of secondary sexual characteristics, pregnancy, or lactation—indicating reproductive maturity (27, 104).
Up to 3.0 mL of blood was collected from the brachial vein and placed into serum tubes with serum clot activator (Vaccutainer). Blood tubes were stored vertically on ice packs in a cold box, and serum was allowed to separate overnight. Serum was drawn from the tube after 24 h, placed in a screw-top cryovial (Corning), and stored in a liquid nitrogen dewar (Princeton Cryogenics). Sterile pediatric swabs with polyester tips and aluminum shafts were used to collect urogenital and rectal samples, and larger polyester swabs with plastic shafts (Fisher) were used to collect oropharyngeal samples. All swabs were collected in duplicate, with one set being placed individually in cryotubes containing lysis buffer (either trireagent or NucliSENS Lysis buffer; BIOMERIEUX) and the second set in viral transport medium (VTM). All tubes were stored in liquid nitrogen in the field and then transferred to a −80 °C freezer.
During each sampling event, pooled urine samples were collected beneath bat roosts using polyethylene sheets (2′ × 3′) distributed evenly under the colony between 3 AM and 6 AM. Urine was collected from each sheet either by using a sterile swab to soak up droplets or a sterile disposable pipette. Swabs or syringed urine from a single sheet were combined to represent a pooled sample. Each urine sample was divided in half and aliquoted into lysis buffer or VTM at an approximate ratio of one part sample to two parts preservative.
Serological and Molecular Assays.
Sera from the cross-sectional survey were heat-inactivated at 56 °C for 30 min, as described (105), prior to shipment to the Center for Infection and Immunity at Columbia University for analysis. Samples were screened for anti-NiV IgG antibodies using an ELISA, as described in ref. 27. Sera from the longitudinal studies were sent to the Australian Animal Health Laboratory and were gamma-irradiated upon receipt. Because of the large sample size and development of a high-throughput multiplex assay of comparable specificity and sensitivity, for these samples, we used a Luminex-based microsphere binding assay to detect anti-Nipah G IgG antibodies reactive to a purified NiV-soluble G-protein reagent, as described (106, 107). Samples resulting in an MFI value of 250 and below are considered negative for other bat species, and previous studies have reported using a threshold of at least three times the mean MFI of negative sera to determine the cutoff (47, 108–110). For this study, MFI values of over 1,000 were considered positive for NiV antibodies, an approach considered appropriate for research purposes for bats.
Total nucleic acids from swabs and urine samples were extracted and complementary DNA was synthesized by using SuperScript III (Invitrogen) according to manufacturer’s instructions. A nested RT-PCR and a real-time assay targeting the N gene were used to detect NiV RNA in samples (111). An RT-qPCR designed to detect the nucleocapsid gene of all known NiV isolates was also utilized (112). Oligonucleotide primers and probe were as described (112). Assays were performed by using AgPath-ID One-StepRT-PCR Reagents (Thermofisher) with 250 nM probe, 50 nM forward, and 900 nM reverse primers. Thermal cycling was 45 °C for 10 min, followed by 45 cycles of 95 °C for 15 s and 60 °C for 45 s. Cutoff values were cycle threshold (CT) ≤ 40 for positive and CT ≥ 45 for negative. Results with CT values between 40 and 45 were deemed indeterminate, i.e., not conclusively positive or negative. Samples with NiV RNA detected by real-time PCR were confirmed by gel electrophoresis and product sequencing.
A subset of NiV-positive samples was processed by high-throughput sequencing (HTS) on the Ion Torrent PGM platform in order to obtain additional NiV genomic sequence. Libraries were prepared according to the manufacturer’s instructions, and 1 million reads were allocated per sample. HTS reads were aligned against host reference databases to remove host background by using the bowtie2 mapper, and host-subtracted reads were primer-trimmed and filtered based on quality, GC content, and sequence complexity. The remaining reads were de novo assembled by using Newbler (Version 2.6) and mapped to the full-length NiV genome. Contigs and unique singletons were also subjected to homology search by using MegaBlast against the GenBank nucleotide database, in case variance in parts of the genome precluded efficient mapping. From these data, N-gene consensus sequences were constructed by using Geneious (Version 7.1) and were used for phylogenetic analyses.
Phylogenetic Analysis.
All P. medius NiV sequences have been submitted to GenBank, and accession numbers are included in Fig. 6. Sequence alignments were constructed by using ClustalW in Geneious Prime software (Geneious Prime 2019.0.3; https://www.geneious.com) (113). Phylogenetic trees of NiV N-gene sequences were constructed by using neighbor-joining algorithms, and figures were constructed in FigTree (Version 1.4.2).
Satellite Telemetry and Home-Range Analysis.
We developed a collar system to attach 12g solar-powered Platform Terminal Transmitters (PTTs) (Microwave Telemetry) to adult bats using commercial nylon feline collars with the buckle removed and 0-gauge nylon suture to attach the PTT to the collar and to fasten the collar around the bat’s neck. Collars were fitted to the bat such that there was enough space to allow for normal neck movement and swallowing, but so that the collar would not slip over the head of the animal (SI Appendix, Fig. S6). PTTs were programmed with a duty cycle of 10 h on and 48 h off. Data were accessed via the Argos online data service (argos-system.org). A total of 14 collars were deployed as follows: February 2009: three males and three females from a colony in Shuvarampur, Faridpur district; February 2011: three males and two females from the same colony; and April 2011 Cox’s Bazaar, three bats from a colony in Cox’s Bazaar, Chattogram district. Bats were selected based on size, such that the total weight of the collar (∼21 g) was less than 3% of the bat’s body mass (SI Appendix, Table S3).
The individual telemetry dataset was combined for each region, and its aggregate utilization distributions (UDs) were computed in R by using package “adehabitatHR” (114). Population-specific home range is represented by the *95% area enclosure of its UD’s volume. The volume of intersection between the colonies quantifies the extent of home-range overlap. To evaluate the potential for contact with the Cox’s Bazaar colony, we calculated the most likely distance moved (“mldm”) for each sampled bat at Faridpur, where the population was more intensively monitored. Movement distance was measured in kilometers with respect to a center location (w) shared by the whole colony. This information was used to predict how likely an animal was to use the landscape.
Statistical Approach–Cross-Sectional Study.
We fit a Bayesian generalized linear model with a logit link and a Bernoulli outcome to identify potential predictors which influenced a bat’s serostatus. We included age, sex, age- and sex-normalized mass and forearm length, mass:forearm ratio, body condition, and whether the bat was pregnant, lactating, or carrying a pup, using weak zero-centered normal priors for all coefficients. We included location of sampling as a group effect (similar to a random effect in a frequentist context) nested within Nipah Belt or non-Nipah Belt regions. We fit the models and performed posterior predictive checks in R 3.4.3, using the rstanarm and rstan packages.
Statistical Approach–Longitudinal Study.
We fit binomial GAMs (115) to the time series of adult and juvenile seroprevalence in the longitudinal study. We included annual, synchronous birthing that occurred between March and April. We assumed that pups weaned from their dams at 3 mo and became independent flyers, and that maternal antibodies waned after 6 mo, at which point pups transitioned into the “juvenile” class (30, 61). We assumed that juveniles became sexually mature at 24 mo and entered the “adult” class based on other pteropid species (30, 47, 116). For juveniles, we modeled the birth cohort of bats as separate random effects in a pooled model of juvenile seroprevalence starting from June of their birth year, June being the earliest month we sampled free-flying juveniles in any cohort. We determined the cohort year of juveniles by using cluster analysis to group individuals by weight, assuming that those in the smallest group were born in the current year and those in the larger group were born the previous year. Of juveniles captured, 92% were yearlings. For adults, we analyzed seroprevalence of adults as a single pool over the entire course of the study. We tested models with and without annual cyclic effects.
Where time series had significant temporal autocorrelation (adults only), we aggregated data by week. We determined periods of significant increase or decrease as those where the 95% CI of the GAM prediction’s derivative did not overlap zero. We fit the models and performed checks in R (Version 3.4.3), using the mgcv package.
To examine the importance of different biological mechanisms in transmission, we fit an age-structured (adult and juvenile) maternally immune (M)–susceptible (S)–infected (I)–recovered (R) model with recrudescence (R to I) and loss of immunity (R to S) to the seroprevalence data on a weekly timescale:
.
We included a class M for the density of juvenile bats with maternal antibodies to allow for the biological possibility that maternal antibodies are lost at a much higher rate than antibodies acquired following infection. The subscripts J refer to juveniles and A to adults; β is the transmission rate; γ is the recovery rate; μ is the mortality rate; τ is the rate of loss of adult immunity; λ is the rate of loss of maternal antibodies (61); Δ is the adult recrudescence rate (R to I); and b is the birth rate (pups join the juvenile population after 5 wk). Juveniles transition to adults after 52 wk. We included terms for loss of antibody in adults (τ, S to R) and viral recrudescence (Δ, R to I) based on previous studies on captive bats that demonstrated the existence of these processes without providing enough data to characterize them precisely (64, 65). We fit this deterministic model to the seroprevalence data by maximum likelihood, which assumes that deviations from the model are due to observation error. We estimated the CIs around maximum-likelihood parameter estimates using likelihood profiles using the profile function in package bbmle in R (Version 3.2.2).
We used model fitting and model comparison to examine the need for several of the biological processes in the model above that could influence NiV dynamics. First, we examined both density- and frequency-dependent transmission by comparing the fit of the model above to one with transmission terms that have population size (NA or NJ) in the denominator. Second, we examined the CIs of the parameters describing viral recrudescence, loss of antibodies in adult bats, and loss of antibodies in juvenile bats. If the confidence bounds for these parameters included zero, then these biological processes are not needed to explain the serological dynamics. Finally, we examined the confidence bounds for parameters describing the loss of maternal and nonmaternal antibodies (τ and λ) to determine if the rate of loss of these two types of immunity were different. We note that this model structure has similarities to a susceptible–infected–latently infected (L)–infected (SILI) model (if latently infected individuals are seropositive), but the model above differs in allowing for the possibility of individuals to transition from the R class back to the S class.
Code Availability.
SIR model code written in R is available upon request.
Supplementary Material
Acknowledgments
We thank Pitu Biswas, Md. Sheikh Gofur, Abdul Hai, and Craig Smith for assistance in the field; Toph Allen, Parviez Hosseini, and Emma Mendelsohn for technical input; Eliza Choi, Yan-Ru Feng, and Lianying Yan for preparing recombinant viral glycoproteins; Jennifer Barr, Vishal Kapoor, Neil Renwick, and Mohammad Ziadur for laboratory diagnostic and technical support; anonymous reviewers for their useful comments; and the Government of Bangladesh for permits and logistical support that made this work possible. This study was funded by NIH National Institute of Allergy and Infectious Diseases (NIAID) Awards AI067549 and U01AI153420 (to J.H.E.) and AI054715 (to C.C.B.); NIH Fogarty International Center Ecology and Evolution of Infectious Diseases (EEID) Award R01 TW005869 (to P.D.); NSF-NIH EEID Award EF-0914866 (to A.M.K.); NIH NIAID Award U19 AI109761 (to W.I.L.); US Agency for International Development PEER Award 226 (to M.S.U.K.); and the US Agency for International Development Emerging Pandemic Threats: PREDICT Program (P.D., J.H.E., Ariful Islam, S.J.A., N.R., C.Z.-T., and K.J.O.). The ICDDR,B is also grateful to the Governments of Bangladesh, Canada, Sweden, and the United Kingdom for providing core/unrestricted support.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2000429117/-/DCSupplemental.
Data Availability.
All molecular sequences are available via GenBank (GenBank accession nos. MK995284–MK995302). The datasets generated during and/or analyzed during the current study are available from the corresponding author on request.
References
- 1.Morse S. S. et al., Prediction and prevention of the next pandemic zoonosis. Lancet 380, 1956–1965 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Bank , World Bank Group Ebola Response Fact Sheet, (World Bank, Washington, DC, 2014). [Google Scholar]
- 3.Keogh-Brown M. R., Smith R. D., The economic impact of SARS: How does the reality match the predictions? Health Policy 88, 110–120 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reid A. H., Fanning T. G., Hultin J. V., Taubenberger J. K., Origin and evolution of the 1918 “Spanish” influenza virus hemagglutinin gene. Proc. Natl. Acad. Sci. U.S.A. 96, 1651–1656 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou P. et al., A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olival K. J. et al., Host and viral traits predict zoonotic spillover from mammals. Nature 546, 646–650 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li W. et al., Bats are natural reservoirs of SARS-like coronaviruses. Science 310, 676–679 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Towner J. S. et al., Isolation of genetically diverse Marburg viruses from Egyptian fruit bats. PLoS Pathog. 5, e1000536 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leroy E. M. et al., Fruit bats as reservoirs of Ebola virus. Nature 438, 575–576 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Plowright R. K. et al., Ecological dynamics of emerging bat virus spillover. Proc. Biol. Sci. 282, 20142124 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Plowright R. K. et al., Transmission or within-host dynamics driving pulses of zoonotic viruses in reservoir-host populations. PLoS Negl. Trop. Dis. 10, e0004796 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Latinne A., et al. , Origin and cross-species transmission of bat coronaviruses in China. bioRxiv:10.1101/2020.05.31.116061 (31 May 2020). [DOI] [PMC free article] [PubMed] [Retracted]
- 13.Karesh W. B. et al., Ecology of zoonoses: Natural and unnatural histories. Lancet 380, 1936–1945 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chadha M. S. et al., Nipah virus-associated encephalitis outbreak, Siliguri, India. Emerg. Infect. Dis. 12, 235–240 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chua K. B. et al., Nipah virus: A recently emergent deadly paramyxovirus. Science 288, 1432–1435 (2000). [DOI] [PubMed] [Google Scholar]
- 16.Hsu V. P. et al., Nipah virus encephalitis reemergence, Bangladesh. Emerg. Infect. Dis. 10, 2082–2087 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paton N. I. et al., Outbreak of Nipah-virus infection among abattoir workers in Singapore. Lancet 354, 1253–1256 (1999). [DOI] [PubMed] [Google Scholar]
- 18.Ching P. K. G., et al. , Outbreak of henipavirus infection, Philippines, 2014. Emerg. Infect. Dis. 21, 328–331 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luby S. P. et al., Recurrent zoonotic transmission of Nipah virus into humans, Bangladesh, 2001-2007. Emerg. Infect. Dis. 15, 1229–1235 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.World Health Organization , Morbidity and Mortality Due to Nipah or Nipah-like Virus Encephalitis in WHO South-East Asia Region, 2001-2018, (World Health Organization, Geneva, Switzerland, 2018). [Google Scholar]
- 21.Field H., Crameri G., Kung N. Y.-H., Wang L.-F., “Ecological aspects of Hendra virus” in Henipavirus: Ecology, Molecular Virology, and Pathogenesis, Lee B., Rota P. A., Eds. (Current Topics in Microbiology and Immunology, Springer, Berlin, Germany, 2012), Vol. vol. 359, pp. 11–23. [DOI] [PubMed] [Google Scholar]
- 22.Reynes J. M. et al., Nipah virus in Lyle’s flying foxes, Cambodia. Emerg. Infect. Dis. 11, 1042–1047 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sendow I. et al., Henipavirus in Pteropus vampyrus bats, Indonesia. Emerg. Infect. Dis. 12, 711–712 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sendow I. et al., Nipah virus in the fruit bat Pteropus vampyrus in Sumatera, Indonesia. PLoS One 8, e69544 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rahman S. A., et al. , Risk factors for Nipah virus infection among pteropid bats, peninsular Malaysia. Emerg. Infect. Dis. 19, 51–60 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mlikovsky J., Correct name for the Indian flying fox (Pteropodidae). Vespertilio 16, 203–210 (2012). [Google Scholar]
- 27.Epstein J. H. et al., Henipavirus infection in fruit bats (Pteropus giganteus), India. Emerg. Infect. Dis. 14, 1309–1311 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anthony S. J. et al., A strategy to estimate unknown viral diversity in mammals. MBio 4, e00598-13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yadav P. D. et al., Detection of Nipah virus RNA in fruit bat (Pteropus giganteus) from India. Am. J. Trop. Med. Hyg. 87, 576–578 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bates P. J. J., Harrison D. L., “Sub-order MEGACHIROPTERA: Family Pteropodidae: Old World fruit bats” in Bats of the Indian Subcontinent, (Harrison Zoological Museum, Kent, England, 1997), pp. 13–15. [Google Scholar]
- 31.Epstein J. H., Field H. E., Luby S., Pulliam J. R. C., Daszak P., Nipah virus: Impact, origins, and causes of emergence. Curr. Infect. Dis. Rep. 8, 59–65 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luby S. P., The pandemic potential of Nipah virus. Antiviral Res. 100, 38–43 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Broder C. C. et al., A treatment for and vaccine against the deadly Hendra and Nipah viruses. Antiviral Res. 100, 8–13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.World Health Organization , WHO Research and Development Blueprint: 2018 Annual Review of Diseases Prioritized under the Research and Development Blueprint, (World Health Organization, Geneva, Switzerland, 2018). [Google Scholar]
- 35.Arunkumar G. et al., Outbreak investigation of Nipah virus disease in Kerala, India, 2018. J. Infect. Dis. 219, 1867–1878 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Mourya D. T. et al., Spatial association between a Nipah virus outbreak in India and Nipah virus infection in Pteropus bats. Clin. Infect. Dis. 69, 378–379 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Altizer S. et al., Food for contagion: Synthesis and future directions for studying host-parasite responses to resource shifts in anthropogenic environments. Philos. Trans. R. Soc. Lond. B Biol. Sci. 373, 20170102 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gurley E. S. et al., Convergence of humans, bats, trees, and culture in Nipah virus transmission, Bangladesh. Emerg. Infect. Dis. 23, 1446–1453 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.World Health Organization , Nipah Virus Outbreaks in the WHO South-East Asia Region, (World Health Organization, Geneva, Switzerland, 2018). [Google Scholar]
- 40.Cortes M. C. et al., Characterization of the spatial and temporal distribution of Nipah virus spillover events in Bangladesh, 2007-2013. J. Infect. Dis. 217, 1390–1394 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chakraborty A. et al., Evolving epidemiology of Nipah virus infection in Bangladesh: Evidence from outbreaks during 2010-2011. Epidemiol. Infect. 144, 371–380 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Amman B. R. et al., Seasonal pulses of Marburg virus circulation in juvenile Rousettus aegyptiacus bats coincide with periods of increased risk of human infection. PLoS Pathog. 8, e1002877 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peel A. J. et al., The effect of seasonal birth pulses on pathogen persistence in wild mammal populations. Proc. Biol. Sci. 281, 20132962 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wacharapluesadee S. et al., Longitudinal study of age-specific pattern of coronavirus infection in Lyle’s flying fox (Pteropus lylei) in Thailand. Virol. J. 15, 38 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hayman D. T. S., Biannual birth pulses allow filoviruses to persist in bat populations. Proc. Biol. Sci. 282, 20142591 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Drexler J. F. et al., Amplification of emerging viruses in a bat colony. Emerg. Infect. Dis. 17, 449–456 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Plowright R. K. et al., Reproduction and nutritional stress are risk factors for Hendra virus infection in little red flying foxes (Pteropus scapulatus). Proc. Biol. Sci. 275, 861–869 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reed Hranac C., Marshall J. C., Monadjem A., Hayman D. T. S., Predicting Ebola virus disease risk and the role of African bat birthing. Epidemics 29, 100366 (2019). [DOI] [PubMed] [Google Scholar]
- 49.Wacharapluesadee S. et al., A longitudinal study of the prevalence of Nipah virus in Pteropus lylei bats in Thailand: Evidence for seasonal preference in disease transmission. Vector Borne Zoonotic Dis. 10, 183–190 (2010). [DOI] [PubMed] [Google Scholar]
- 50.Iehlé C. et al., Henipavirus and Tioman virus antibodies in pteropodid bats, Madagascar. Emerg. Infect. Dis. 13, 159–161 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Field H., Hendra virus in Australian flying foxes: Possible maintenance strategies. J. Clin. Virol. 28S, S87–S108 (2003). [Google Scholar]
- 52.Field H. et al., Hendra virus infection dynamics in Australian fruit bats. PLoS One 6, e28678 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Field H. et al., Spatiotemporal aspects of Hendra virus infection in pteropid bats (Flying-Foxes) in Eastern Australia. PLoS One 10, e0144055 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bjørnstad O. N., Finkenstädt B. F., Grenfell B. T., Dynamics of measles epidemics: Estimating scaling of transmission rates using a time series SIR model. Ecol. Monogr. 72, 169–184 (2002). [Google Scholar]
- 55.Keeling M. J., Grenfell B. T., Disease extinction and community size: Modeling the persistence of measles. Science 275, 65–67 (1997). [DOI] [PubMed] [Google Scholar]
- 56.Peel A. J. et al., Support for viral persistence in bats from age-specific serology and models of maternal immunity. Sci. Rep. 8, 3859 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Williamson M. M. et al., Transmission studies of Hendra virus (equine morbillivirus) in fruit bats, horses and cats. Aust. Vet. J. 76, 813–818 (1998). [DOI] [PubMed] [Google Scholar]
- 58.Middleton D. J. et al., Experimental Nipah virus infection in pteropid bats (Pteropus poliocephalus). J. Comp. Pathol. 136, 266–272 (2007). [DOI] [PubMed] [Google Scholar]
- 59.Halpin K. et al.; Henipavirus Ecology Research Group , Pteropid bats are confirmed as the reservoir hosts of henipaviruses: A comprehensive experimental study of virus transmission. Am. J. Trop. Med. Hyg. 85, 946–951 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brook C. E. et al., Disentangling serology to elucidate henipa- and filovirus transmission in Madagascar fruit bats. J. Anim. Ecol. 88, 1001–1016 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Epstein J. H. et al., Duration of maternal antibodies against canine distemper virus and Hendra virus in pteropid bats. PLoS One 8, e67584 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Epstein J. H. et al., Pteropus vampyrus, a hunted migratory species with a multinational home-range and a need for regional management. J. Appl. Ecol. 46, 991–1002 (2009). [Google Scholar]
- 63.Eby P., Seasonal movements of gray-headed flying-foxes, Pteropus poliocephalus (Chiroptera, Pteropodidae), from 2 maternity Camps in Northern New-South-Wales. Wildl. Res. 18, 547–559 (1991). [Google Scholar]
- 64.Sohayati A. R. et al.; Henipavirus Ecology Research Group , Evidence for Nipah virus recrudescence and serological patterns of captive Pteropus vampyrus. Epidemiol. Infect. 139, 1570–1579 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Baker K. S. et al., Viral antibody dynamics in a chiropteran host. J. Anim. Ecol. 83, 415–428 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Peel A. J. et al., Henipavirus neutralising antibodies in an isolated island population of African fruit bats. PLoS One 7, e30346 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Anderson D. E. et al., Isolation and full-genome characterization of Nipah viruses from bats, Bangladesh. Emerg. Infect. Dis. 25, 166–170 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nikolay B. et al., Transmission of Nipah virus—14 years of investigations in Bangladesh. N. Engl. J. Med. 380, 1804–1814 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hahn M. B. et al., Roosting behaviour and habitat selection of Pteropus giganteus reveals potential links to Nipah virus epidemiology. J. Appl. Ecol. 51, 376–387 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cross P. C., Lloyd-Smith J. O., Johnson P. L. F., Getz W. M., Duelling timescales of host movement and disease recovery determine invasion of disease in structured populations. Ecol. Lett. 8, 587–595 (2005). [Google Scholar]
- 71.Plowright R. K. et al., Urban habituation, ecological connectivity and epidemic dampening: The emergence of Hendra virus from flying foxes (Pteropus spp.). Proc. Biol. Sci. 278, 3703–3712 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang H.-H., Kung N. Y., Grant W. E., Scanlan J. C., Field H. E., Recrudescent infection supports Hendra virus persistence in Australian flying-fox populations. PLoS One 8, e80430 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hall L., Richards G., Eds., Flying Foxes: Fruit and Blossom Bats of Australia, (UNSW Press, Sydney, Australia, ed. 1, 2000), p. 135. [Google Scholar]
- 74.Markus N., Blackshaw J. K., Behaviour of the black flying fox Pteropus alecto: 1. An ethogram of behaviour, and preliminary characterisation of mother-infant interactions. Acta Chiropt. 4, 137–152 (2002). [Google Scholar]
- 75.Halpin K., Young P. L., Field H. E., Mackenzie J. S., Isolation of Hendra virus from pteropid bats: A natural reservoir of Hendra virus. J. Gen. Virol. 81, 1927–1932 (2000). [DOI] [PubMed] [Google Scholar]
- 76.Sudhakaran M. R., Doss P. S., Food and foraging preferences of three pteropodid bats in southern India. J. Threat. Taxa 4, 2295–2303 (2012). [Google Scholar]
- 77.Salah Uddin Khan M. et al., Use of infrared Camera to understand bats’ access to date palm sap: Implications for preventing Nipah virus transmission. EcoHealth 7, 517–525 (2011). [DOI] [PubMed] [Google Scholar]
- 78.Wong K. T., Tan C. T., “Clinical and pathological manifestations of human henipavirus infection” in Henipavirus: Ecology, Molecular Virology, and Pathogenesis, Lee B., Rota P. A., Eds. (Current Topics in Microbiology and Immunology, Springer-Verlag, Berlin, 2012), Vol. vol. 359, pp. 95–104. [DOI] [PubMed] [Google Scholar]
- 79.Wong K. T. et al., Human Hendra virus infection causes acute and relapsing encephalitis. Neuropathol. Appl. Neurobiol. 35, 296–305 (2009). [DOI] [PubMed] [Google Scholar]
- 80.Olival K. J. et al., Population genetics of fruit bat reservoir informs the dynamics, distribution, and diversity of Nipah virus. Mol. Ecol. 29, 970–985 (2020). [DOI] [PubMed] [Google Scholar]
- 81.de Jong C., et al. , Foraging behaviour and landscape utilisattion by the endangered golden-crowned flying fox (Acerodon jubatus), the Philippines. PLoS One 8, e79665 (2013). Correction in: PLoS One10, e0120945 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Numata S., Yasuda M., Okuda T., Kachi N., Noor N. S. M., Temporal and spatial patterns of mass flowerings on the Malay Peninsula. Am. J. Bot. 90, 1025–1031 (2003). [DOI] [PubMed] [Google Scholar]
- 83.Fleming T. H., Eby P., “Ecology of bat migration” in Bat Ecology, Kunz T. H., Fenton M. B., Eds. (The University of Chicago Press, Chicago, IL, 2003), pp. 156–208. [Google Scholar]
- 84.Richter H. V., Cumming G. S., Food availability and annual migration of the straw-colored fruit bat (Eidolon helvum). J. Zool. 268, 35–44 (2006). [Google Scholar]
- 85.Hahn M. B. et al., The role of landscape composition and configuration on Pteropus giganteus roosting ecology and Nipah virus spillover risk in Bangladesh. Am. J. Trop. Med. Hyg. 90, 247–255 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lo M. K. et al., Characterization of Nipah virus from outbreaks in Bangladesh, 2008-2010. Emerg. Infect. Dis. 18, 248–255 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Clayton B. et al., Nipah viruses from Malaysia and Bangladesh: Differences in transmission and pathogenesis. EcoHealth 7, S140–S141 (2011). [Google Scholar]
- 88.Clayton B. A. et al., Transmission routes for Nipah virus from Malaysia and Bangladesh. Emerg. Infect. Dis. 18, 1983–1993 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.DeBuysscher B. L. et al., Comparison of the pathogenicity of Nipah virus isolates from Bangladesh and Malaysia in the Syrian hamster. PLoS Negl. Trop. Dis. 7, e2024 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chong H. T., Hossain M. J., Tan C. T., Differences in epidemiologic and clinical features of Nipah virus encephalitis between the Malaysian and Bangladesh outbreaks. Neurol. Asia 13, 23–26 (2008). [Google Scholar]
- 91.Mire C. E. et al., Pathogenic differences between Nipah virus Bangladesh and Malaysia strains in primates: Implications for antibody therapy. Sci. Rep. 6, 30916 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hegde S. T. et al., Investigating rare risk factors for Nipah virus in Bangladesh: 2001-2012. EcoHealth 13, 720–728 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kumar C. P. G. et al., Infections among contacts of patients with Nipah virus, India. Emerg. Infect. Dis. 25, 1007–1010 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mounts A. W. et al.; Nipah Virus Nosocomial Study Group , A cohort study of health care workers to assess nosocomial transmissibility of Nipah virus, Malaysia, 1999. J. Infect. Dis. 183, 810–813 (2001). [DOI] [PubMed] [Google Scholar]
- 95.Song X. et al., Satellite tracking of post-nesting movements of green turtles Chelonia mydas from the Gangkou Sea Turtle National Nature Reserve, China, 2001. Mar. Turtle Newsl. 97, 8–9 (2002). [Google Scholar]
- 96.Luby S. et al., Recurrent Nipah virus outbreaks in Bangladesh, 2001-2007. Am. J. Trop. Med. Hyg. 77, 273 (2007). [Google Scholar]
- 97.Yadav P. D. et al., Nipah virus sequences from humans and bats during Nipah outbreak, Kerala, India, 2018. Emerg. Infect. Dis. 25, 1003–1006 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wacharapluesadee S. et al., Genetic characterization of Nipah virus from Thai fruit bats (Pteropus lylei). Asian Biomed. 7, 813–819 (2013). [Google Scholar]
- 99.Plowright R. K. et al., Pathways to zoonotic spillover. Nat. Rev. Microbiol. 15, 502–510 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sejvar J. J., The evolving epidemiology of viral encephalitis. Curr. Opin. Neurol. 19, 350–357 (2006). [DOI] [PubMed] [Google Scholar]
- 101.Hegde S. T. et al., Using healthcare-seeking behaviour to estimate the number of Nipah outbreaks missed by hospital-based surveillance in Bangladesh. Int. J. Epidemiol. 48, 1219–1227 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Homaira N. et al., Nipah virus outbreak with person-to-person transmission in a district of Bangladesh, 2007. Epidemiol. Infect. 138, 1630–1636 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jonsson N. N., Johnston S. D., Field H., de Jong C., Smith C., Field anaesthesia of three Australian species of flying fox. Vet. Rec. 154, 664 (2004). [DOI] [PubMed] [Google Scholar]
- 104.Pierson E. D., Rainey W. E., . “The biology of flying foxes of the genus Pteropus: A review” in Pacific Island Flying Foxes: Proceedings of an International Conservation Conference, Wilson D. E., Graham G. L., Eds. (US Department of the Interior Fish and Wildlife Service, Washington, DC, 1992), pp. 1–17. [Google Scholar]
- 105.Daniels P. W., Ksiazek T., Eaton B., Diagnostic tests for Nipah and Hendra viruses. Microbes Infect. 3, 289–295 (2001). [DOI] [PubMed] [Google Scholar]
- 106.Chowdhury S. et al., Serological evidence of henipavirus exposure in cattle, goats and pigs in Bangladesh. PLoS Negl. Trop. Dis. 8, e3302 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bossart K. N. et al., Neutralization assays for differential henipavirus serology using Bio-Plex protein array systems. J. Virol. Methods 142, 29–40 (2007). [DOI] [PubMed] [Google Scholar]
- 108.Breed A. C., Breed M. F., Meers J., Field H. E., Evidence of endemic Hendra virus infection in flying-foxes (Pteropus conspicillatus)—implications for disease risk management. PLoS One 6, e28816 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hayman D. T. S. et al., Evidence of henipavirus infection in West African fruit bats. PLoS One 3, e2739 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hayman D. T. S. et al., Antibodies to henipavirus or henipa-like viruses in domestic pigs in Ghana, West Africa. PLoS One 6, e25256 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wacharapluesadee S., Hemachudha T., Duplex nested RT-PCR for detection of Nipah virus RNA from urine specimens of bats. J. Virol. Methods 141, 97–101 (2007). [DOI] [PubMed] [Google Scholar]
- 112.Feldman K. S. et al., Design and evaluation of consensus PCR assays for henipaviruses. J. Virol. Methods 161, 52–57 (2009). [DOI] [PubMed] [Google Scholar]
- 113.Kearse M. et al., Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Calenge C., The package “adehabitat” for the R software: A tool for the analysis of space and habitat use by animals. Ecol. Modell. 197, 516–519 (2006). [Google Scholar]
- 115.Wood S. N., Generalized Additive Models: An Introduction with R, (CRC Texts in Statistical Science, Chapman & Hall/CRC, Boca Raton, FL, 2006). [Google Scholar]
- 116.Tidemann C. R., Nelson J. E., Life expectancy, causes of death and movements of the grey-headed flying-fox (Pteropus poliocephalus) inferred from banding. Acta Chiropt. 13, 419–429 (2011). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All molecular sequences are available via GenBank (GenBank accession nos. MK995284–MK995302). The datasets generated during and/or analyzed during the current study are available from the corresponding author on request.