Significance
Mutation of CHD7 causes multiple developmental disorders, including CHARGE syndrome, in which conotruncal anomalies are the most prevalent form of heart defects. Our study provides mouse genetic evidence demonstrating that CHD7 cell-autonomously regulates cardiac neural crest cell (cNCC) development to promote normal conotruncal morphogenesis, clarifying a long-standing controversy. Furthermore, through comprehensive in vitro and in vivo analyses, we reveal that CHD7 acts in cNCCs through both nucleosome remodeling and recruiting histone modifying enzymes to target loci. Our study provides fundamental insights regarding the etiology of CHD7-related congenital cardiovascular disorders. Importantly, our data suggest that patients carrying truncation mutations versus missense mutations will likely display different molecular alterations; these patients might therefore require personalized therapeutic interventions.
Keywords: CHARGE syndrome, cardiac neural crest cells, cardiovascular development, nucleosome remodeling, CHD7
Abstract
CHD7 encodes an ATP-dependent chromatin remodeling factor. Mutation of this gene causes multiple developmental disorders, including CHARGE (Coloboma of the eye, Heart defects, Atresia of the choanae, Retardation of growth/development, Genital abnormalities, and Ear anomalies) syndrome, in which conotruncal anomalies are the most prevalent form of heart defects. How CHD7 regulates conotruncal development remains unclear. In this study, we establish that deletion of Chd7 in neural crest cells (NCCs) causes severe conotruncal defects and perinatal lethality, thus providing mouse genetic evidence demonstrating that CHD7 cell-autonomously regulates cardiac NCC development, thereby clarifying a long-standing controversy in the literature. Using transcriptomic analyses, we show that CHD7 fine-tunes the expression of a gene network that is critical for cardiac NCC development. To gain further molecular insights into gene regulation by CHD7, we performed a protein–protein interaction screen by incubating recombinant CHD7 on a protein array. We find that CHD7 directly interacts with several developmental disorder-mutated proteins including WDR5, a core component of H3K4 methyltransferase complexes. This direct interaction suggested that CHD7 may recruit histone-modifying enzymes to target loci independently of its remodeling functions. We therefore generated a mouse model that harbors an ATPase-deficient allele and demonstrates that mutant CHD7 retains the ability to recruit H3K4 methyltransferase activity to its targets. Thus, our data uncover that CHD7 regulates cardiovascular development through ATP-dependent and -independent activities, shedding light on the etiology of CHD7-related congenital disorders. Importantly, our data also imply that patients carrying a premature stop codon versus missense mutations will likely display different molecular alterations; these patients might therefore require personalized therapeutic interventions.
The ATP-dependent chromatin remodeling factors regulate genomic functions and numerous biological processes through the modulation of chromatin structure. Mutations in these factors are associated with a large number of pathologies (1–3). Based on protein sequence features, chromatin remodelers are classified into four major families: SWI/SNF, ISWI, CHD, and INO80 (1–3). CHD7 belongs to the CHD (Chromodomain Helicase DNA-binding) family of enzymes and heterozygous mutations of CHD7 account for ∼70% of CHARGE (Coloboma of the eye, Heart defects, Atresia of the choanae, Retardation of growth/development, Genital abnormalities, and Ear anomalies) syndrome cases (4–12). In addition, CHD7 mutations also have been identified in idiopathic hypogonadotropic hypogonadism (IHH), Kallmann syndrome (KS), autism spectrum disorders (ASD), DiGeorge syndrome, and nonsyndromic patients with congenital heart defects (13–18).
Congenital heart defects are a major factor contributing to the high mortality rate of infants with CHARGE syndrome and these malformations affect more than 75% of patients (4–12). Among CHARGE patients with inborn heart anomalies, more than 50% display conotruncal defects, which include pharyngeal arch (PA) artery (PAA) patterning defects, tetralogy of Fallot (TOF), double-outlet right ventricle (DORV), overriding aorta, and interrupted aortic arch type B (IAA-B), among others (5, 6, 10). This spectrum of malformations highly resembles those caused by abnormal development of cardiac neural crest cells (cNCCs) in animals (19–24), consistent with the hypothesis that the clinical features of CHARGE may result from abnormal development of NCCs (25). NCCs are a group of multipotent stem-like cells that arise from the dorsal neural tube through epithelial-to-mesenchymal transition after the neural tube folds (19–24). cNCCs are formed between the otic placode and the posterior border of the third somite (19–24). In mouse embryos between embryonic day (E) 9.0 to E10.5, cNCCs invade the newly formed PAs 3, 4, and 6, where they populate and serve as the primary source for PA mesenchyme. A large portion of PA cNCCs differentiates into vessel smooth muscle cells (SMCs) during remodeling of PAAs into aortic arch arteries. A subgroup of PA NCCs continues to migrate into cardiac outflow tract (OFT) cushions and participate in separating the common OFT into the aorta and pulmonary trunk.
While CHARGE patients display conotruncal defects similar to those caused by cNCC dysfunction in animal models, the function of CHD7 in cNCCs is still unclear and even controversial. On the one hand, the critical role of CHD7 in the regulation of NCCs is well supported by studies using several model systems. In murine Joma1.3 NC stem cells and human induced pluripotent stem cells, CHD7 promotes the formation of migratory NCCs (26, 27). Moreover, knocking-down CHD7 expression in zebrafish and Xenopus leads to abnormalities in NCC-derived structures (27–29). On the other hand, the importance of CHD7 in NCCs for cardiovascular development has been challenged by two independent mouse studies. In a genetic rescue experiment using a Chd7 gene-trap model (Chd7xk), restoring expression of CHD7 in NCCs failed to rescue the abnormal PAA phenotype in Chd7xk/+ embryos (30). In a conditional gene knockout study, in which Chd7 was specifically inactivated in NCCs using the Wnt1-Cre driver (31), Wnt1-Cre;Chd7loxp/loxp embryos showed severe craniofacial defects, confirming an essential role for CHD7 in craniofacial NCCs. However, these embryos did not show any heart or PAA defects (31). Therefore, whether CHD7 has essential cell-autonomous roles in mammalian cNCCs and whether cNCC dysfunction acts as a major contributing factor to the cardiovascular defects in CHARGE remains unknown.
In this study, we used an improved NCC Cre driver, Wnt1-Cre2 (32), to delete Chd7 in NCCs. Mutant embryos displayed severe conotruncal defects resembling those observed in CHARGE patients, providing mouse genetic evidence to support the essential cell-autonomous role of Chd7 in cNCCs. We further show that CHD7 regulates the epigenetic status of target loci through both nucleosome remodeling-dependent and -independent activities. This study greatly advances our knowledge of cNCC development and provides a better understanding of the developmental and molecular basis of the cardiovascular defects associated with CHD7 mutations.
Results
Deletion of Chd7 in NCCs Led to Severe Conotruncal Defects and Perinatal Lethality.
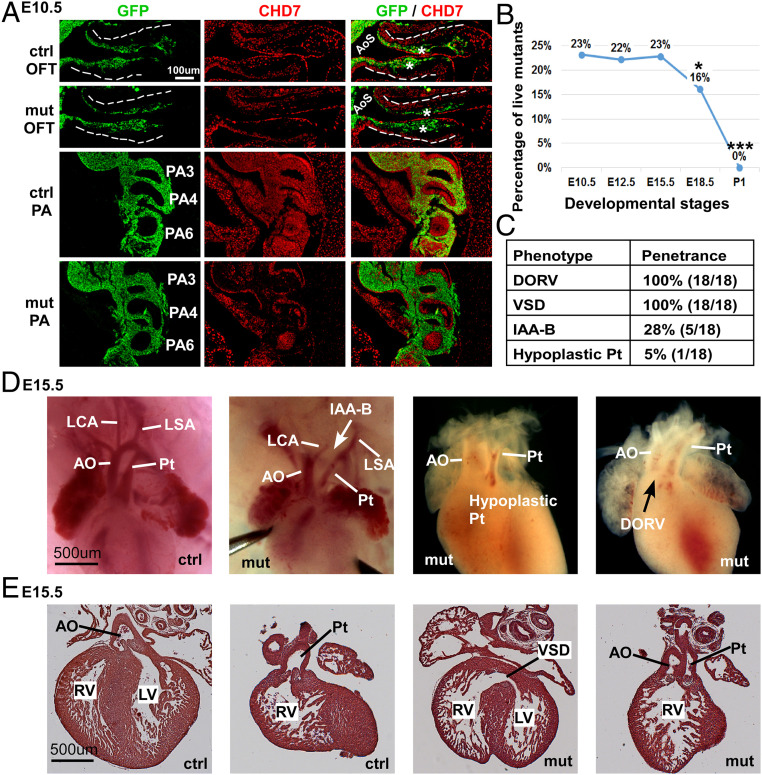
To determine the role of CHD7 in cNCCs, we used the Wnt1-Cre2 driver (32) to specifically delete Chd7 in NCCs. To distinguish NCCs from other cell types, we used the R26mTmG reporter allele, which expresses membrane-tethered GFP in CRE-recombinase+ cells (33). At E9.5, ∼30% of GFP+ cells (NCCs) still expressed CHD7 (SI Appendix, Fig. S1 A and B). However, by E10.5 and at E11.5, less than 5% of GFP+ cells were CHD7+ (Fig. 1A and SI Appendix, Fig. S1 C–E), suggesting that NCC-expression of CHD7 was efficiently inactivated starting from E10.5. Notably, the deletion of Chd7 in NCCs led to perinatal lethality (Fig. 1B and Dataset S1), demonstrating that the expression of Chd7 in NCCs is required for mammalian embryo viability. Morphological examination of mutant embryos (Wnt1-Cre2;Chd7loxp/loxp or Wnt1-Cre2;Chd7loxp/loxp;R26mtmg/+) at E15.5 and E16.5 revealed that all mutant animals displayed the DORV and ventricular septal defect (VSD) (Fig. 1 C–E). About 30% of mutant embryos showed IAA-B and 5% showed a hypoplastic pulmonary trunk. Given that these anomalies resemble the inborn conotruncal defects observed in CHARGE patients (5, 6, 12, 34–37), our data support the idea that these patient malformations arise, at least in part, as a result of impaired CHD7 function in NCCs. In addition to assessing cardiovascular defects, we also examined the craniofacial structure of E17.5 mutant embryos (SI Appendix, Fig. S2). Consistent with a previous report (31), deletion of Chd7 in NCCs led to smaller frontal bone, maxilla, and mandible (which are all derivatives of NCCs), thereby providing further evidence to support a critical role for CHD7 in craniofacial NCCs.
Fig. 1.
Deletion of Chd7 in NCCs led to severe conotruncal defects. (A) Immunostaining of control (Ctrl; Wnt1-Cre2;Chd7+/+;R26mtmg/+) and mutant (mut; Wnt1-Cre2;Chd7loxp/loxp;R26mtmg/+) embryos at E10.5 with antibodies against GFP (green) and CHD7 (red). Areas of the OFT and PA regions are shown. The dashed lines outline the OFT. OFT cushions are indicated with an asterisk (*). Broader views of the same embryos are shown in SI Appendix, Fig. S1C. (B) Percentage of live mutant (Wnt1-Cre2;Chd7loxp/loxp) animals, resulting from crossing Wnt1-Cre2;Chd7loxp/+ and Chd7loxp/loxp mice. Data were obtained from at least four litters at each of the indicated developmental stages. *P < 0.05; ***P < 0.001, χ2 test. (C) Summary of heart defects observed in 18 mutant embryos (Wnt1-Cre2;Chd7loxp/loxp) at E15.5. Pt, pulmonary trunk. (D) Whole-mount examination revealing conotruncal defects in mutant embryos at E15.5. AO, aorta; LCA, left carotid artery; LSA, left subclavian artery. (E) Section examination showing the DORV defect in a mutant embryo at E15.5. LV, left ventricle; RV, right ventricle.
Chd7 Mutant Embryos Displayed Severe Defects in Postmigratory cNCCs.
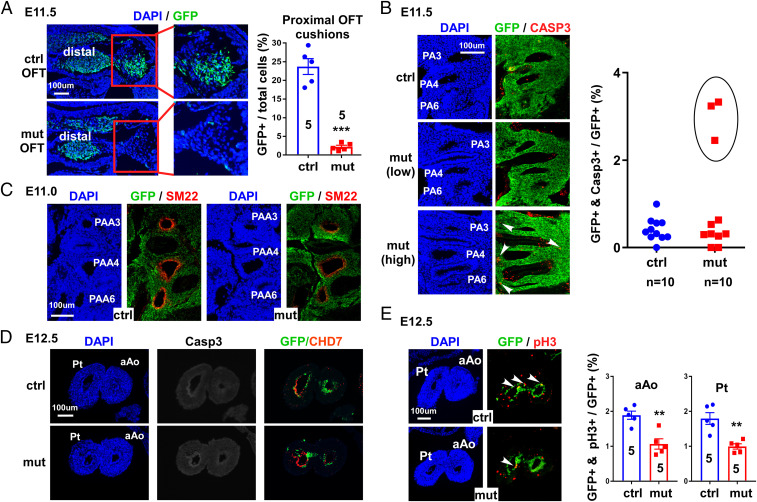
To assess how Chd7-deletion affects cNCC function, we performed a detailed examination of mutant embryos at E10.5. Interestingly, we did not observe any morphological or cellular defects at this stage (SI Appendix, Figs. S3 and S4), suggesting that the conotruncal defects in mutant embryos are unlikely caused by impaired migration of cNCCs into PAs. However, at E11.5, the number of NCCs in the proximal OFT cushions was dramatically reduced in mutants compared to controls (Fig. 2A). We did not observe significantly reduced cell proliferation in the dorsal neural tube, PA regions and OFT cushions of mutant embryos at E11.5 (SI Appendix, Figs. S5–S7). We therefore examined whether this reduction in NCCs resulted from increased apoptosis. No increased cell death was observed in dorsal neural tube and OFT cushions of mutant embryos (SI Appendix, Figs. S5–S7). In the PA region, we found that 30% of mutant embryos showed increased cell death, as judged by cleaved-Caspase 3 (CASP3) staining (Fig. 2B). It is possible that, in these embryos, expression of CHD7 was more efficiently inactivated than in embryos showing no increase in apoptosis. Interestingly, all mutant embryos examined (n = 5) displayed the same OFT cushion defect, including three embryos with normal cell apoptosis. These results suggest that the reduced number of cNCCs in mutant proximal OFT cushions is due to impaired invasion of cNCCs, rather than reduced cell proliferation or increased cell death.
Fig. 2.
Chd7 mutant embryos showed severe cellular defects, starting from E11.0. (A) Sections covering the OFT region from control (Ctrl; Wnt1-Cre2;Chd7+/+;R26mtmg/+) and mutant (mut; Wnt1-Cre2;Chd7loxp/loxp;R26mtmg/+) embryos (E11.5) were immunostained with an anti-GFP antibody (green). Total nuclei were visualized with DAPI (blue). The boxed regions show the proximal OFT cushions. Quantification of GFP+ cells (cNCCs) in control and mutant embryos is shown (Right). All examined mutant embryos (n = 5) displayed a similar phenotype. (B) Sections of control and mutant embryos at E11.5 were immunostained with an antibody against the apoptosis marker CASP3 (red). Sections covering the PA3–6 region are shown. Arrowheads indicate examples of GFP and CASP3–double-positive nuclei. Quantification of CASP3+ (apoptotic) cells is shown on the right. In mutant samples, seven showed (“low”) apoptosis levels (Middle) that were indistinguishable from control samples (Top). The remaining three mutant samples (circled in the quantification chart) showed increased (“high”) cell death (Bottom). (C) Sections of control and mutant embryos at E11.0 were immunostained with antibodies against the smooth muscle cell marker SM22 (red) and GFP (green). The same results were obtained from five pairs of control and mutant embryos. (D) Cross sections of pulmonary trunk (Pt) and ascending aorta (aAo) from control and mutant hearts at E12.5 were immunostained with antibodies against GFP (green), cleaved CASP3 (white), and CHD7 (red). No cell death was observed in either sample. (E) Same as D, except that samples were stained with an antiphospho histone H3 (pH3) to detect proliferating cells. Arrowheads indicate examples of GFP and pH3–double-positive nuclei. As shown by the quantification of the immunostainings (Right), mutant embryos show reduced cell proliferation in both the pulmonary trunk and ascending aorta regions. **P < 0.01, ***P < 0.001, unpaired two-tailed Student’s t test.
We also observed a reduction in the expression of SM22, an SMC marker, in mutant cNCCs (Fig. 2C), suggesting that Chd7-deletion also impairs the differentiation of cNCCs into SMCs. To confirm this phenotype more quantitatively, we isolated GFP+ cells from the trunk regions (PA3–6) of E11.5 embryos and monitored the expression of three SMC differentiation markers—namely SM22, Acta2, and Cnn1—using qRT-PCR (SI Appendix, Fig. S8). Expression of all these SMC markers was significantly reduced, confirming our SM22 immunostaining result. To further reveal how deletion of Chd7 impacts NCC derivatives, we examined samples at E12.5. We did not observe cell death in cNCC derivatives in aorta and pulmonary trunk walls (Fig. 2D), while cell proliferation was significantly decreased in mutant samples (Fig. 2E). Interestingly, at this stage, expression of CHD7 was already undetectable in the majority of NCC derivatives (GFP+) in control embryos (Fig. 2 D and E and SI Appendix, Fig. S9). Therefore, the reduced cell proliferation in mutant NCC derivatives at E12.5 is, most likely, an epigenetically inherited defect resulting from earlier alterations in PA cNCCs.
CHD7 Regulates a Complex Gene Network in cNCCs.
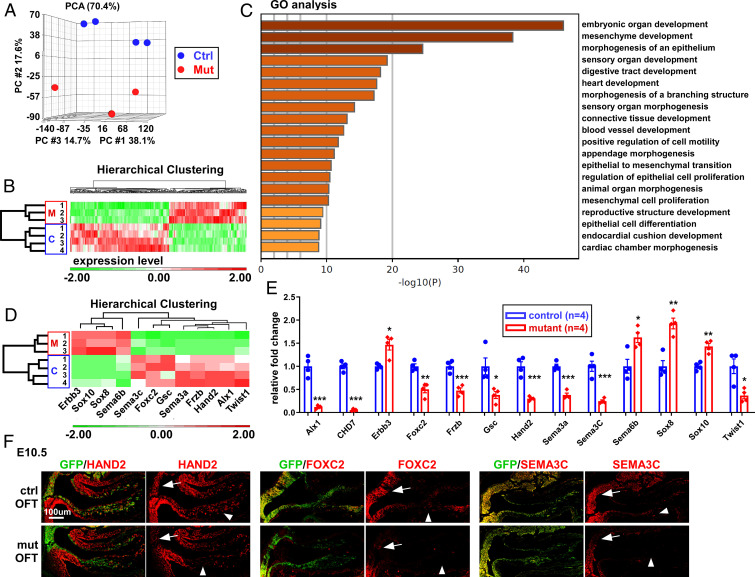
To better understand the molecular basis for the conotruncal defects observed in embryos after deletion of Chd7 in cNCCs, we examined the gene-expression profile of cNCCs isolated from control (Wnt1-Cre2;Chd7+/+;R26mtmg/+) and mutant (Wnt1-Cre2;Chd7loxp/loxp;R26mtmg/+) embryos at E10.5, using mRNA-sequencing (mRNA-seq) (Fig. 3 and SI Appendix, Fig. S10). Principal component analysis (PCA) of these profiles showed a clear separation of mutant and control groups, suggesting that deletion of Chd7 led to significant changes in gene expression (Fig. 3A). Mutant samples did not appear to cluster as close as the control samples in PCA, suggesting that their mRNA profiles were more variable. This might reflect variations in Chd7-deletion efficacy by the Cre recombinase in cNCC subpopulations. Alternatively, this might simply reflect the significant variability seen between CHARGE patients, including siblings with identical mutations (38, 39). This could explain, at least in part, why only six genes reached the threshold by the false-discovery rate (FDR < 0.05) (Dataset S2). In addition, this might also result from the relatively “small” number of replicates used (n = 4 for control and n = 3 for mutants) in comparison to the number of transcripts examined (∼20,000) and the relatively moderate effect of Chd7-deletion on gene expression. Indeed, the expression of nearly 90% of these genes was altered within 2.5-fold, suggesting that the primary role of CHD7 in cNCCs is fine-tuning target gene expression rather than switching genes on and off. We therefore pondered this factor and used the P value to set the threshold (P < 0.05) for selection of candidate genes [as used by other groups examining the impact of Chd7-deletion on transcriptomes (40–42)]. The expression of 316 genes was significantly altered by at least 50% in mutant samples (Fig. 3B and Dataset S2). Among these, 177 were down-regulated and 139 were up-regulated. We next performed gene ontology (GO) term enrichment analysis using Metascape (43) (Fig. 3C and Dataset S3). Strikingly, the deregulated genes were predominantly associated with developmental GO terms, such as “embryonic organ development” or “mesenchyme development,” fully consistent with the idea that Chd7 is required for normal development of NCC-derived mesenchymal cells in the PA and OFT regions. For example, genes including Hand1, Hand2, Sema3A, Sema3C, Twist1, and Foxc2 are all well-established critical regulators of mesenchymal cell development. Furthermore, GO terms involved in the development of the cardiovascular system and other organs/tissues (such as digestive tract, cartilage, skeleton, and connective tissue) were also highly enriched (Fig. 3C and Dataset S3). Thus, these analyses provide a transcriptional explanation for the NCC differentiation defects seen upon Chd7-deletion.
Fig. 3.
CHD7 regulates a complex gene network in cNCCs. (A) PCA of mRNA-seq data obtained from cNCCs (GFP+) cells isolated from the PA3–6 regions of control (Wnt1-Cre2;Chd7+/+;R26mtmg/+) and mutant (Wnt1-Cre2;Chd7loxp/loxp;R26mtmg/+) embryos (E10.5). (B) Hierarchical clustering of the 316 genes showing significantly altered expression in mutant samples (P < 0.05, unpaired two-tailed Student’s t test). The z-normalized intensities (average = 0 and SD = 1) were used to show changes in expression levels as a result of Chd7 deletion. Up-regulated genes are represented in red and down-regulated genes are represented in green. The scale (expression level) indicates the range of color intensities used to represent the range of changes in gene expression. Changes of twofold and above are represented by the darkest green tone, while changes of negative twofold and below are represented by the darkest red tone. (C) GO term enrichment analysis of the above-mentioned 315 genes. The top 20 GO terms are shown (for full list, see Dataset S3). This chart shows that genes which were deregulated upon Chd7 deletion are predominantly associated with developmental GO terms, including many pathways linked to cardiovascular development. (D) Hierarchical clustering focusing on 12 genes that are both deregulated in Chd7 mutant cells and are known to be critical for NCC development. (E) qRT-PCR validating the deregulation of these 12 genes and confirming Chd7 deletion, using total RNA from cNCCs of control and mutant embryos (E10.5). The RNA samples used in qRT-PCR were prepared independently from those used for mRNA-seq. *P < 0.05; **P < 0.01; ***P < 0.001, unpaired two-tailed Student’s t test. (F) Immunostaining was performed to verify reduced expression (at the protein level) of HAND2, FOXC2, and SEMA3C in the OFT of mutant (mut; Wnt1-Cre2;Chd7loxp/loxp;R26mtmg/+) compared to control (ctrl; Wnt1-Cre2;Chd7+/+;R26mtmg/+) embryos at E10.5. Arrows indicate NCCs in the aortic sac region while arrowheads indicate NCCs in OFT cushions.
Indeed, among the 316 CHD7-regulated genes, 12 are known to be important for NCC development (Fig. 3D). We confirmed that their expression was significantly altered in mutant cNCCs using qRT-PCR (Fig. 3E). Moreover, we validated that Hand2, Foxc2, and Sema3C were reduced at the protein level in mutant cNCCs (Fig. 3F). Therefore, these results suggest that CHD7 regulates a core gene network in cNCCs and that the abnormal expression of these genes most likely contributes to the conotruncal defects observed in CHARGE patients.
CHD7 Directly Regulates Genes That Are Critical for cNCC Development.
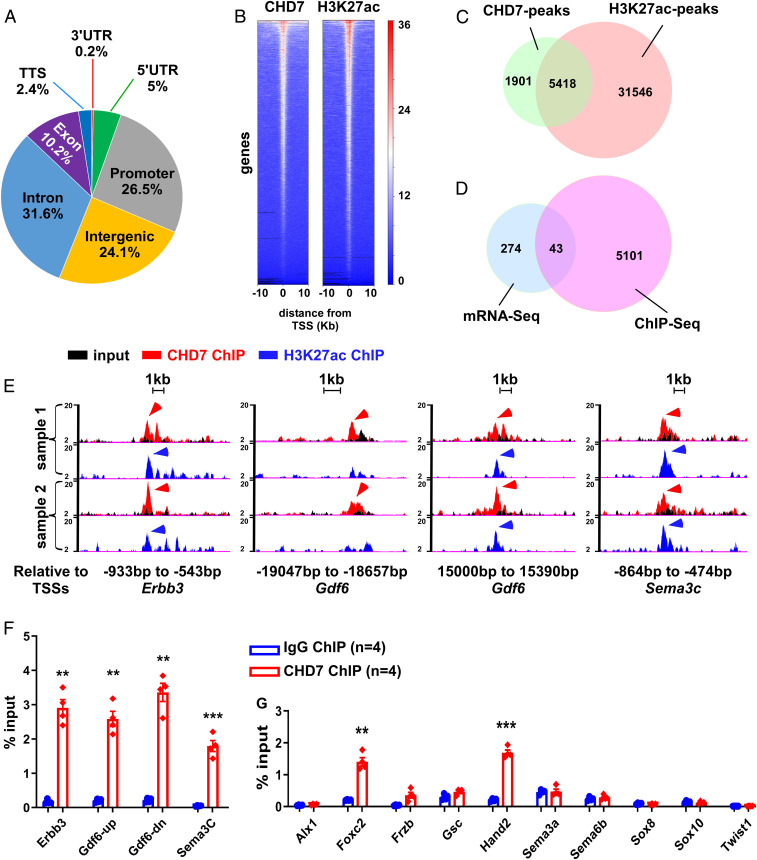
To identify CHD7 direct target loci, we performed chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq) using the PA3–6 regions (after removal of the primitive heart chambers and neural tubes) of wild-type embryos (E10.5). ChIPs against CHD7 and histone H3 Lysine 27 acetylation (H3K27ac), a mark for active enhancers/promoters (44), were performed in parallel using biological duplicate samples. A total of 7,319 peaks and 36,964 peaks were associated with CHD7 and H3K27ac, respectively (Datasets S4 and S5). The distribution of CHD7-peaks in the genome of NCCs is shown in Fig. 4 A and B. About 75% of CHD7-associated peaks overlapped with H3K27ac (Fig. 4C and Dataset S6), suggesting that CHD7 preferentially localizes to active enhancer or promoter regions, as observed in other cell types (45, 46); 5,418 CHD7-peaks were within 20 kb upstream or downstream of transcription start sites (TSSs) of 5,144 genes (Dataset S7). However, only 43 genes showed a significantly altered expression upon Chd7-deletion, as judged from our mRNA-seq data (Fig. 4D and Dataset S8), suggesting that these genes are direct targets of CHD7.
Fig. 4.
CHD7 directly regulates genes that are critical for cNCC development. (A) Pie chart showing the distribution of CHD7 binding peaks in the genome. UTR, untranslated region. (B) ChIP-seq density heat maps for CHD7 (Left) and H3K27ac (Right) within ±10 kb of TSSs of genes. The deepTools plotHeatmap tool was used to create the heatmap for scores associated with the ChIP genomic regions. The red and white tiles indicate that a larger number of reads were mapped to the TSS of the represented genes, whereas the blue tiles indicate no or a smaller number of reads were mapped to these genes. The numbers refer to the relative signal intensity. (C) Venn diagram showing the number of overlapping and nonoverlapping CHD7 and H3K27ac ChIP-seq peaks. (D) Venn diagram showing that 43 genes that are deregulated upon Chd7 deletion, also contain CHD7-ChIP peaks within ±20 kb of their TSSs. (E) The promoters of Erbb3 and Sema3C, and the enhancer regions of Gdf6 are shown as examples of CHD7 and H3K27ac overlapping or nonoverlapping ChIP-seq peaks. Arrowheads indicate peaks that were called by the HOMER software. (F) Validation of CHD7 binding to the cis-regulatory elements of Gdf6, Errb3, and Sema3C shown in E, by ChIP-qPCR using GFP+ cells isolated from the PA3–6 regions of wild-type (Wnt1-Cre2;Chd7+/+;R26mtmg/+) embryos (E10.5). (G) Assessment of CHD7 binding to the promoters of the remaining 10 NCC critical genes (identified in Fig. 3D) by ChIP-qPCR, as indicated. The positions of genomic elements that were tested are shown in Dataset S11. **P < 0.01; ***P < 0.001, unpaired two-tailed Student’s t test.
We verified the binding of CHD7 to several of the 43 genes identified above. We focused on Gdf6 (Bmp13), Erbb3, and Sema3C (Fig. 4E). The latter two genes are known to be important for normal NCC development (47–49) and Gdf6 is a member of the BMP cytokines, which are well-recognized for their roles in regulating cNCC development (50). In addition, Gdf6 mutations have been associated with inborn heart defects (51). Using qRT-PCR, we first verified that the expression of Gdf6 was significantly altered in Chd7 mutant cNCCs (SI Appendix, Fig. S11), as observed for other genes examined in Fig. 3E. We next confirmed all of the tested CHD7 peaks identified by ChIP-seq through ChIP-qPCR using NCCs (GFP+) isolated from E10.5 wild-type (Wnt1-Cre2;Chd7+/+;R26mTmG/+) embryos (Fig. 4F).
To test whether these CHD7-bound sites were indeed bona fide enhancers, we cloned these fragments into a modified pREP4-luc reporter vector (52) and performed reporter assays. All of the tested elements significantly enhanced expression of the luciferase reporter compared to the empty vector control, suggesting that they indeed act as enhancers (SI Appendix, Fig. S12).
To complement these approaches, we also used cNCCs purified from wild-type embryos (E10.5) to directly examine the potential association of CHD7 with the promoters of the 10 remaining CHD7-downstream genes listed in Fig. 3E. The use of purified cNCCs allowed us to clearly detect association of CHD7 with the promoters of Foxc2 and Hand2 (Fig. 4G). Thus, our data collectively establish that CHD7 directly regulates a set of genes that are central to NCC development, thereby providing a transcriptional basis for CHARGE patient heart malformations. We also noticed that expression of Sox8, Sox10, and Sema6B were all significantly increased in mutant samples, and yet these genes could not be associated with any detectable CHD7-binding peak. Therefore, this suggests that CHD7 likely impacts these genes (and additional targets) indirectly.
CHD7 Interacts with Many Disease-Mutated Proteins, Including WDR5.
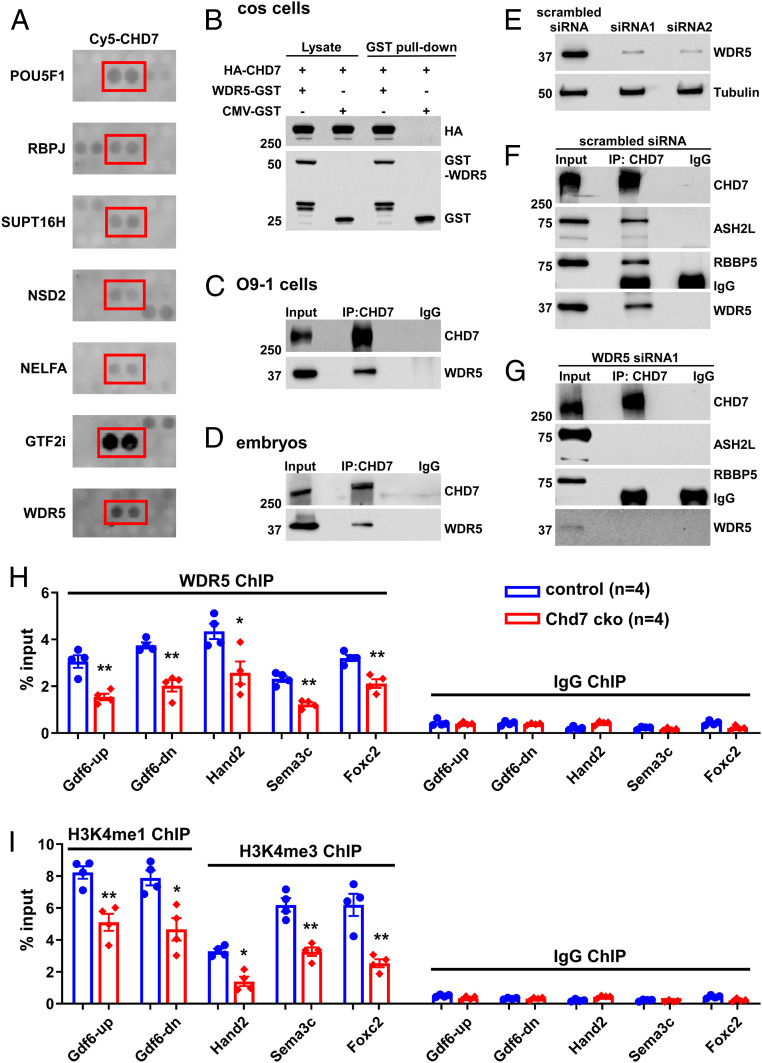
To gain further molecular insights into gene regulation by CHD7 and into the etiology of CHD7-related disorders, we used a protein array to screen for CHD7 interactors and retrieved 303 nuclear proteins, including known interactors, such as POU5F1 (OCT4), RBPJ, and SUPT16H (53, 54) (Fig. 5A and Dataset S9). Of note, this screen revealed that CHD7 interacts with multiple factors encoded by disease genes that cause congenital heart defects, such as WDR5 (Table 1 and Dataset S10). In addition, CHD7 interacts with NSD2 and NELFA, which are also referred to as Wolf-Hirschhorn syndrome candidate genes. This syndrome shares clinical features with CHARGE, such as congenital heart defects and coloboma of the eye (55). We also identify the Williams-Beuren syndrome-deleted GTF2I as a strong CHD7 interactor and confirmed this interaction using coimmunoprecipitation (co-IP) (Fig. 5A and SI Appendix, Fig. S13). Interestingly, Williams-Beuren syndrome patients also show partial phenotypical overlaps with CHARGE patients, some of which also display TOF and VSD (56). Hence, these findings suggest that many of these interactions are specific while potentially implicating CHD7 in previously unanticipated pathways and human disorders.
Fig. 5.
CHD7 interacts with and recruits WDR5 to CHD7-target loci. (A) Examples of positive CHD7 interactors from the protein–protein array screen, including known (POU5F1 and RBPJ) and newly identified (including WDR5) direct interactors. (B) COS cells were transfected with different combinations of plasmids encoding HA-CHD7 or WDR5-GST. CMV-GST was included as a control, as indicated on top. Transfections were followed by GST-pull down and Western blot analyses. (C and D) Protein extracts from O9-1 cells (C) or from the trunk region of wild-type embryos at E10.5 (D) were subjected to IP using an anti-CHD7 antibody or a control preimmune IgG. (E–G) Efficient knockdown of WDR5 expression in O9-1 cells (using two independent siRNAs) was assessed by Western blot. Tubulin was used as a protein loading control (E). IP using an anti-CHD7 antibody or a control preimmune IgG were performed using protein extracts from O9-1 cells treated with a scrambled siRNA control (F) or siRNA1 (G). IP of CHD7 and potential co-IP of ASH2L, RBBP5, and WDR5 were assessed by Western blot. The same results were obtained when using siRNA2 (SI Appendix, Fig. S15B). (H and I) ChIP-qPCR was performed using GFP+ cells (cNCCs) isolated from the PA3–6 regions of control (Wnt1-Cre2;Chd7+/+;R26mtmg/+) and mutant (cko, Wnt1-Cre2;Chd7loxp/loxp;R26mtmg/+) embryos at E10.5 to examine association of WDR5 with the indicated promoters/enhancers (H). In addition, H3K4me1 was examined for the enhancers of Gdf6, and H3K4me3 was examined for the promoters of Foxc2, Hand2, and Sema3C (I). *P < 0.05; **P < 0.01, unpaired two-tailed Student’s t test.
Table 1.
CHD7-interacting proteins that are mutated in patients with syndromic and nonsyndromic congenital heart defects
| CHD7-interacting proteins | Associated diseases |
| AFF4 | CHOPS syndrome |
| DGCR8 | Velocardiofacial and DiGeorge syndrome |
| GTF2I | Williams-Beuren syndrome |
| NELFA | Wolf-Hirschhorn syndrome |
| NSD2 | Wolf-Hirschhorn syndrome |
| RPS6KA3 | Coffin-Lowry syndrome |
| RXRA | Congenital heart anomalies, dilated cardiomyopathy, systemic lupus erythematosus |
| SMAD2 | Congenital heart anomalies |
| SMC1A | Cornelia de Lange syndrome; Wiedemann-Steiner syndrome |
| UBE2B | Congenital heart anomalies |
| WDR5 | Congenital heart anomalies |
CHD7 Interacts with H3K4 Methyltransferase Complexes through WDR5 to Epigenetically Regulate Target Promoters/Enhancers in cNCCs.
To further shed light on the basic molecular mechanisms by which CHD7 regulates transcription, we chose to focus on the histone methyltransferase (HMTase) complex subunit WDR5 (Fig. 5A), since CHD7 functions have previously been linked to histone methylation (57, 58). WDR5 is a core component of H3K4 methyltransferase complexes (59–62) and it has been proposed that CHD7 is recruited to its target loci through the binding of methylated H3K4 (H3K4me), a histone modification associated with active transcription (59–62). However, the detection of a direct interaction between CHD7 and WDR5 in our screen suggested that CHD7 itself might regulate H3K4 methylation at CHD7-target loci. Therefore, we further examined the CHD7–WDR5 interaction and its role in cNCCs. We confirmed the CHD7–WDR5 interaction in COS cells, O9-1 NCCs, and mouse embryos (Fig. 5 B–D). We mapped this interaction to the first putative SANT-SLIDE region (amino acids 1535–1852) of CHD7 using co-IP (SI Appendix, Fig. S14). Additional co-IP experiments using the PA3–6 region of E10.5 embryos show that CHD7 associates with the WDR5 partners ASH2L and RBBP5 (SI Appendix, Fig. S15A), the other two core components of H3K4 methyltransferase complexes (59–62). However, knocking down WDR5 expression disrupted the interaction of CHD7 with ASH2L and RBBP5, implying that WDR5 is required to bridge the interaction of CHD7 with the two proteins (Fig. 5 E–G and SI Appendix, Fig. S15B). Collectively, our data suggest that CHD7 interacts with HMTase complexes through a direct interaction with WDR5.
To address the biological significance of this interaction at CHD7-target genes, we purified cNCCs (GFP+) from the PA3–6 regions of control (Wnt1-Cre2;Chd7+/+;R26mtmg/+) and mutant (Wnt1-Cre2;Chd7loxp/loxp;R26mtmg/+) embryos at E10.5 and performed ChIP-qPCR using antibodies against WDR5 and H3K4 methylation marks that are catalyzed by WDR5-containing complexes, namely H3K4 mono- and trimethylation (H3K4me1 and H3K4me3, respectively). Deletion of Chd7 significantly reduced association of WDR5 with the CHD7-target promoters/enhancers tested (Fig. 5H), and we also observed a significant decrease in H3K4 methylation at these sites (Fig. 5I). Therefore, our data suggest that CHD7 is important for loading WDR5/HMTase complexes onto its target promoters/enhancers and, consequently, for the deposition of H3K4 methylation. These results were unexpected since CHD7 had so far been thought to function downstream of H3K4 methylation.
CHD7 Regulates cNCC and Embryo Development through Both ATP-Dependent and -Independent Activities.
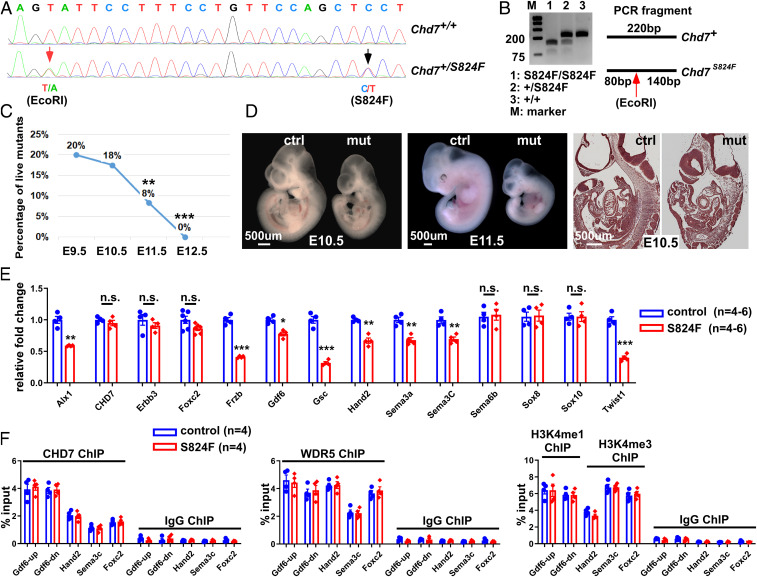
Given that CHD7 is believed to function primarily as a nucleosome remodeling ATPase (63), we tested whether we could separate its remodeling activity from the regulation of H3K4 methylation. We therefore generated a mouse line in which the S824F mutation was knocked into the endogenous Chd7 allele using the CRISPR/Cas9 technique (Fig. 6 A and B). The mouse S824F mutation corresponds to the human S834F mutation, which is a pathological allele identified in CHARGE, IHH, and KS patients (13, 64). This single point mutation within the first chromodomain of CHD7 abolishes both its ATPase and nucleosome remodeling activities (63). We first examined the impact of this mutation on development. All Chd7S824F/S824F embryos died between E10.5 and E12.5 (Fig. 6C). Beginning at E10.5, mutant embryos displayed a gross developmental delay and their growth was arrested at the ∼35 somite stage at E11.5 (Fig. 6D). Interestingly, one-third of the mutant embryos survived to E11.5, whereas no embryos with homozygous Chd7-null alleles survived to this stage (30, 52, 65, 66). These observations are consistent with the S824F allele being less severe than a null allele, as it may still be able to carry out CHD7 ATP-independent functions.
Fig. 6.
CHD7 regulates embryo development and target genes through ATP-dependent and -independent activities. (A) The S824F allele was knocked into the endogenous Chd7 locus through CRISPR/Cas9. Successful knockin was confirmed by genomic DNA sequencing of the founder mouse. The red arrow indicates a silent mutation that generates an EcoRI site (introduced to facilitate genotyping), while the black arrow indicates the C-to-T mutation that generates the S824F mutation. (B) PCR products from mice with different genotypes were digested with EcoRI. (C) Percentage of live mutant embryos (Chd7S824F/S824F) from the self-cross of Chd7S824F/+ mice. **P < 0.01; ***P < 0.001, χ2 test. (D) Control (Ctrl, Chd7+/+) and mutant (mut, Chd7S824F/S824F) embryos at E10.5 (Left) and 11.5 (Center) were subjected to whole mount and section examination (Right). (E) Total RNA was isolated from GFP+ cells of the PA3–6 regions of control (Wnt1-Cre2;Chd7+/+;R26mtmg/+) and mutant (Wnt1-Cre2;Chd7S824F/S824F;R26mtmg/+) embryos (25 to 35 somites). qRT-PCR was performed to examine expression of the indicated genes. n.s., no significant difference; *P < 0.05; **P < 0.01; ***P < 0.001, unpaired two-tailed Student’s t test. (F) GFP+ cells were isolated from the PA3–6 regions of control (ctrl, Wnt1-Cre2;Chd7+/+;R26mtmg/+) and mutant (Wnt1-Cre2;Chd7S824f/S824F;R26mtmg/+) embryos followed by ChIP-qPCR using antibodies against CHD7, WDR5, H3K4me1 (for Gdf6up and Gdf6-dn), and H3K4me3 (for Hand2, Sema3C, and Foxc2), as indicated. A preimmune IgG was used as a control.
Prior to assessing the effect of the S824F mutation on histone methylation, we investigated its impact on the expression of CHD7 target genes in cNCCs. To this end, we crossed Wnt1-Cre2;Chd7S824F/+ male mice with Chd7S824/+;R26mTmG/mTmG female mice to obtain mutant (Wnt1-Cre2;Chd7S824f/S824F;R26mTmG/+) and control (Wnt1-Cre2;Chd7+/+;R26mTmG/+) embryos with 25 to 35 somites and isolated cNCCs (GFP+ cells) from their trunk regions between PA3 and PA6. We then analyzed the expression of the 13 CHD7-regulated genes we tested above (Fig. 3 and SI Appendix, Fig. S11). Expression of eight of these genes was significantly impaired in Chd7S824f/S824F cNCCs (Fig. 6E). Collectively, this set of data demonstrates that the nucleosome remodeling activity of CHD7 is essential for mouse embryo development, and in particular, for the regulation of genes that are important for cNCC functions. Interestingly, the S824F mutation did not significantly alter the expression of the five remaining CHD7-regulated genes, suggesting that proper regulation of these genes does not require CHD7 ATP-dependent activities and that ATP-independent functions were sufficient. Among the eight genes whose expression was altered by the S824F mutation, the expression of Alx1, Hand2, Sema3a, and Sema3C in Chd7S824f/S824F cNCCs was significantly different from the expression of these genes in Chd7 knockout cNCCs (comparing Fig. 3E to Fig. 6E and SI Appendix, Fig. S16). This indicates that the impact of the S824F mutation on expression of these genes was less severe than that of Chd7-deletion. These data are fully consistent with our observation that Chd7S824f/S824F embryos displayed weaker phenotypes than Chd7-null embryos. Collectively, these data suggest that CHD7-regulated genes show different requirements for CHD7 ATP-dependent and -independent functions. In addition, these results further support the idea that patients with CHD7 mutations likely suffer from different molecular alterations depending on the type of mutation they carry.
Finally, we tested whether the promotion of H3K4 methylation by CHD7 relies on its ATP-dependent activity. We examined whether CHD7 (S824F) could still recruit WDR5/HMTases to CHD7 target loci. Importantly, we first confirmed that neither CHD7 expression nor its interaction with WDR5 were reduced by the S824F mutation (SI Appendix, Fig. S17). ChIP analysis using cNCCs (GFP+) from mutant and control embryos demonstrated that mutant CHD7 (S824F) still binds to its target elements at levels identical to wild-type CHD7 (Fig. 6F). Furthermore, the S824F mutant is still capable of targeting H3K4 methylation to CHD7-regulated loci (Fig. 6F). Thus, these data uncover an ATP-independent function for CHD7.
Discussion
The present study provides mouse genetic evidence to support essential cell-autonomous functions for CHD7 in the regulation of mammalian cNCC development, thereby clarifying a long-standing controversy in the literature. We characterized the morphological, cellular, and molecular defects in the conotruncal regions of Chd7 mutant embryos through combined developmental, genetic, molecular, and biochemical approaches. Our data reveal that CHD7 regulates its target genes through both nucleosome remodeling and recruiting H3K4 methyltransferase activity to its target loci.
Deletion of Chd7 in NCCs Led to Severe Cardiovascular Defects and Perinatal Lethality.
Previous studies have suggested that CHD7 is critical for the proper development of NCCs (26–29), yet its essential role in mammalian cNCCs was not supported by published genetic rescue or conditional gene inactivation studies (30, 31). Notably, it has recently been shown that the widely utilized Wnt1-Cre driver (67) [which was used in Chd7 genetic studies mentioned above (30, 31)] leads to ectopic expression of WNT1 from the Wnt1-Cre transgene (32). While this study only examined ectopic expression in the brain region (32), it is likely that WNT1 is also ectopically expressed in cNCCs (since the Wnt1-Cre transgene is activated in nearly all NCCs). Our study using the improved Wnt1-Cre2 line (32) suggests that in previous studies, ectopically expressed WNT1 in NCCs (in Wnt1-Cre;Chd7loxp/loxp embryos) may have rescued the conotruncal defects caused by the Chd7-deletion. Interestingly, this implies a potential interaction between WNT signaling and Chd7. This hypothesis is further supported by bioinformatic Ingenuity Pathway Analysis, which indicates that Wnt signaling is one of the top potential upstream regulators of CHD7 target genes (SI Appendix, Fig. S18), suggesting that CHD7 may mediate the activity of WNT cytokines in cNCCs. However, the CHD7–WNT interaction might be more central to cardiovascular than to craniofacial development, since ectopic expression of WNT1 in NCCs was not sufficient to rescue the craniofacial defects in Wnt1-Cre;Chd7loxp/loxp embryos (31). Alternatively, CHD7 might simply carry out other central WNT-independent functions in craniofacial development.
In addition to the potential involvement of the WNT–CHD7 interaction, there are several other possibilities that might also account for the different cardiovascular phenotypes reported in the present vs. the previous study (31). First, we cannot formally rule out that Wnt1-Cre and Wnt1-Cre2 have subtle spatial or temporal differences in recombination that cannot be detected by reporter analysis. This might also result in delayed deletion of Chd7 and these subtle differences might lead to the different cardiovascular phenotype. Second, the two studies used two different conditional Chd7 alleles. In the previous study (31), the second exon of Chd7 was floxed, while in the present study the third exon was floxed. Although we cannot fully exclude the possibility that the two different alleles have caused the different results on cardiovascular development, this explanation is highly unlikely, as deletion of either exon leads to a null allele (31, 68). Third, the difference might be due to using different mouse genetic backgrounds. In the previously published study, mice were maintained on C57bl6/129 hybrid background (31), while in this study we have maintained our lines on the 129S6 congenic background. Understanding how WNT and CHD7 functions are linked in cNCCs and whether the activity of CHD7 is modified by genetic/environmental factors warrants further investigation.
Cellular Basis for the Conotruncal Defects Caused by Deletion of Chd7 in NCCs.
Deletion of Chd7 in NCCs leads to severe conotruncal defects, including the DORV, IAA-B, and hypoplastic pulmonary trunk (Fig. 1), all of which have been observed in CHARGE patients (5, 6, 12, 34–37). Our observations support the idea that abnormal development of NCCs significantly contributes to the cardiovascular clinical features of CHARGE patients. Interestingly, inactivation of Chd7 in the anterior mesoderm can result in similar defects (68), suggesting that mesodermal expression of CHD7 is also important for conotruncal development. Therefore, the cardiovascular phenotypes of CHARGE patients are likely determined by the combined defects in multiple cell populations.
In contrast to the hypoplastic PAA4 defect observed in Chd7xk/+ embryos (30), we found that the initial formation of PAAs 4–6 was not impaired by deletion of Chd7 at E10.5 (SI Appendix, Fig. S3). Therefore, the IAA-B observed in Wnt1-Cre2;Chd7loxp/loxp mutants is unlikely caused by the abnormal formation of PAAs at earlier stages. Instead, 30% of mutant embryos displayed a significant increase in NCC apoptosis in the PA regions (Fig. 2), and this percentage correlates well with 28% of mutant embryos displaying an IAA-B at E15.5 (Fig. 1). Therefore, we speculate that the IAA-B phenotype is primarily caused by increased cell death in the PAs. Interestingly, increased cell death and impaired SMC differentiation were observed throughout PAAs 3–6 on both the left and right sides, and yet IAA-B was the only defect identified in mutant great arteries. Given that IAA-B is caused by abnormal development of the left PAA4, our result is consistent with the idea that development of left PAA4 is particularly sensitive to abnormalities in NCCs.
We did not observe reduced length of the OFT in mutant embryos nor abnormal cardiac looping, suggesting that the etiology of the DORV caused by deletion of Chd7 is different from that resulting from NCC-ablation (69). Instead, only a few NCCs are able to invade into the proximal OFT cushions in mutant embryos at E11.5 (Fig. 2). NCCs in the proximal cushions are critical for OFT septation and alignment with ventricles (19–24). Therefore, the dramatically reduced invasion of cNCCs into proximal OFT cushions likely acts as the major contributor to the DORV and the VSD in mutant embryos. At the molecular level, Sema3C is required for NCCs to invade proximal OFT cushions (70) and Sema3C is a direct downstream target of CHD7. Therefore, reduced expression of Sema3C may be a major molecular cause for the OFT alignment defect in Chd7-deletion embryos and in CHARGE patients.
While CHD7 appears to directly regulate a rather small number of genes in NCCs, a closer inspection reveals that many CHD7 targets are in fact critical to NCC development (Fig. 4). Nonetheless, it is also possible that this rather low number of direct targets also reflects the difficulty of assigning which distant CHD7-bound enhancer regulates a given promoter. For example, CHD7 binds to the Sox9 enhancer that is ∼250 kb away from its TSS (71).
CHD7 Regulates Target Genes through ATP-Dependent and -Independent Functions.
CHD7 is a SNF2-family ATPase that is highly conserved among mammals, with mouse CHD7 being 95% identical and 97% similar to human CHD7 (72). Through knocking the catalytically inactive S824F allele into the Chd7 endogenous loci, we show that the CHD7 ATP-dependent activity is essential for mammalian embryo development. The deregulation of multiple CHD7 target genes in these mutant embryos indicates that CHD7 regulates gene expression, in part through its nucleosome remodeling activity.
We also noticed that Chd7S824F/S824F embryos displayed weaker phenotypes compared to all reported Chd7-null embryos. Approximately 30% of Chd7S824F/S824F embryos survived to E11.5, whereas no Chd7-null embryos survived beyond E10.5 (30, 52, 65, 66). The fact that a milder phenotype was observed with the S824F allele is consistent with a weaker impact on the transcription of some genes, since the mutated CHD7 protein is still capable of performing ATP-independent functions. Of note, in humans the corresponding S834F heterozygous mutation also leads to a mild phenotype for a CHARGE patient (13), or “only” leads to reproductive organ disorders (IHH and KS) in two other patients (14). This hypothesis is consistent with the more general observation that truncating/stop codon mutations in CHD7 are typically found in patients with CHARGE, while patients with IHH (who displays less severe phenotypes) predominantly carry missense mutations (73). Hence, our data provide a molecular explanation for the phenotypical differences observed between patients with CHD7 mutations.
The present study reveals a nucleosome-remodeling independent function for CHD7, namely direct recruitment of H3K4 methyltransferase activity to target promoters/enhancers. H3K4 methylation is particularly relevant to congenital heart diseases; in nonsyndromic patients with inborn heart defects, mutations in genes involved in the modification of H3K4 are significantly enriched (15). Furthermore, a mouse study confirmed that H3K4 methylation is essential for normal heart development (74). While CHD7 has initially been proposed to bind the H3K4me mark (59–62), our findings strongly suggest that CHD7 can also directly recruit an H3K4me writer to target elements. The dual activities of CHD7 may represent an efficient mechanism to coordinate nucleosome remodeling and H3K4 methylation at these target loci. The mutual interaction between the CHD7 nucleosome remodeler and histone methylation machinery could form a positive feedback loop to stabilize epigenetic states at target elements.
It has been hypothesized that CHD7 may interact with H3K4 methyltransferase complexes via CHD8 (58). However, through an unbiased approach using purified proteins, we found that CHD7 directly interacts with WDR5 in vitro (Fig. 5A). Consistent with this, our co-IP data also indicate that this interaction does not require CHD8 in cells (SI Appendix, Fig. S19). Hence, our data point to a more direct link to explain the phenotypical overlap between CHARGE and Kabuki syndromes. In addition to WDR5, our screen revealed that CHD7 interacts with other chromatin-modifying enzymes (Dataset S9). It is therefore likely that CHD7 carries out additional ATP-independent functions through the targeting of other chromatin-modifying activities to specific genomic loci. This also implies that patients bearing missense mutations (such as CHD7 S834F) will most likely exhibit different molecular alterations compared to patients bearing a truncation mutation. In addition, CHD8, one of the most frequently mutated proteins in ASD, also interacts with H3K4 methyltransferase (58). We thus speculate that, contrary to what was suggested by Chd8 knockout studies, ASD patients carrying point mutations might not necessarily benefit from H3K4 demethylase inhibitors since point-mutated CHD8 proteins will most likely still be able to recruit H3K4me. By extension, our conclusions may apply to other disorders that are either caused by stop codons or missense mutations, if the mutated protein carries out other functions. In other words, our data suggest that patients with the same syndrome may require personalized therapeutic interventions based on the type of mutations they carry, despite displaying similar phenotypes.
CHD7 Is Implicated in a Wider Range of Physiological Processes and Diseases than Previously Anticipated.
The discovery of CHD7 ATP-independent functions is consistent with the observation that, in patients, some mutations do not significantly impact the catalytic functions of this remodeling ATPase (63). It is therefore likely that, instead, some human disorders result from the disruption of protein–protein interactions. In some cases, disruption of the interaction between CHD7 and other proteins might not be the consequence of mutations in CHD7 itself, but might result from mutations in CHD7-interacting partners. Consistent with this hypothesis, mutation in several genes encoding for CHD7-interaction partners (such as NSD2, NELFA, and GTF2I) lead to syndromic diseases with phenotypes overlapping with CHARGE syndrome (see discussion above). Given that 10 to 30% of CHARGE patients do not display a mutation in CHD7 (4–11), sequencing the genes encoding CHD7 candidate interactors identified here might be informative for determining the etiology of CHARGE in these patients. Similarly, since our data unexpectedly link CHD7 to a wider range of pathologies, it may also be informative to assess potential mutations in CHD7 in idiopathic cases for these newly linked pathologies.
In summary, our study provides mouse genetic evidence to support an essential cell-autonomous role of CHD7 in cardiac NCCs. Our study also demonstrates that a single point mutation in the Chd7 gene is sufficient to cause severe developmental defects and embryonic lethality in mammals. In addition, determining the CHD7 interactome allowed us to identify WDR5 as a direct CHD7-interacting protein and to reveal that CHD7 also regulates mammalian cNCC development through recruiting HMTase(s) to its target elements. This implies that patients harboring a CHD7 mutation may require personalized therapeutic interventions, as a function of their mutation. Last but not least, the CHD7 interactome also suggests that CHD7 is likely implicated in an even wider range of physiological processes and human diseases than previously anticipated. Importantly, we now provide a molecular framework of direct candidate interactors to investigate known or new CHD7 functions, as well as the molecular etiology of CHD7-associated diseases or phenotypes.
Materials and Methods
Detailed materials and methods are provided in SI Appendix, Detailed Materials and Methods.
Mouse Works.
This study conforms to the Guide for the Care and Use of Laboratory Animals (75). All protocols were approved by the Institutional Animal Care and Use Committee in the University of Alabama at Birmingham. Euthanasia of mice was achieved through inhalation of CO2 followed by cervical dislocation. The S824F allele was knocked into the Chd7 endogenous locus through CRISPR/Cas9 performed by the University of Alabama at Birmingham Transgenic Core Facility.
Tissue Treatment, Immunostaining, and Biochemical Assays.
Procedures for tissue treatment and immunostaining were performed as previously described (52, 76). Biochemical assays, including Western blot, GST-pull down, and co-IP, were performed following standard protocols with modifications detailed in SI Appendix, Detailed Materials and Methods.
Subcloning, Cell Culture, and Transfection.
Subcloning was performed using standard PCR amplification and cloning procedures. The O9-1 cells were purchased from Millipore (SCC049) and cNCCs were isolated as described previously (77, 78). Transfections were performed using Lipofectamine 2000 (ThermoFisher) or PureFection (System Biosciences).
mRNA-Seq, ChIP-qPCR, and ChIP-Seq.
GFP+ cNCCs were isolated from E10.5 embryos. These cells were used for mRNA-seq, ChIP-qPCR, and ChIP-seq.
Protein–Protein Interaction Screen.
The full-length CHD7 protein was purified as previously described (63), subsequently labeled with Cy5 and incubated with the “ProtoArray Human Protein Microarray” (ThermoFisher) according the manufacturer’s instructions.
Statistic Tests.
All statistic tests were performed using the Prism 8 software. A χ2 test was performed in Fig. 1B. P < 0.05 was considered as significant. For other datasets, Shapiro–Wilk (n = 3, 4) and Kolmogorov–Smirnov (n ≥ 5) tests were used to determine the normality of data. All data followed normal distribution except for cell apoptosis data of mutant samples in Fig. 2B. For data following normal distribution, an unpaired two-tailed Student’s t test was used to compare control and mutant samples. Data are shown as mean ± SE, with P < 0.05 considered as significant.
Supplementary Material
Acknowledgments
We thank Dr. S. Astrof (Rutgers University) for suggestions on isolation of neural crest cells; Drs. R. Serra, A. Javed, and H. Seo (University of Alabama at Birmingham, UAB) for examination of the craniofacial defects; members in K.J.’s and K.B.’s laboratories for suggestions on this project; the UAB Transgenic & Genetically Engineered Models Core for making the CHD7 (S824F) mouse line using CRISPR/Cas9; the UAB Genomics Core for performing deep-seq and Sanger sequencing; and the UAB Comprehensive Flow Cytometry Facility for sorting neural crest cells. This work was supported by National Institutes of Health Grants 2R01HL095783 and R56 HL130711 (to K.J.); American Heart Association Grant 19TPA34890006 (to K.J.); and Universitätsklinikum Giessen und Marburg Projekt 5/2016 (to K.B.) with additional support from TRR81/3.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2005222117/-/DCSupplemental.
Data Availability.
The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession nos. GSE149855 and GSE149860) (79, 80).
References
- 1.Hota S. K., Bruneau B. G., ATP-dependent chromatin remodeling during mammalian development. Development 143, 2882–2897 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clapier C. R., Iwasa J., Cairns B. R., Peterson C. L., Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat. Rev. Mol. Cell Biol. 18, 407–422 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Runge J. S., Raab J. R., Magnuson T., Epigenetic regulation by ATP-dependent chromatin-remodeling enzymes: SNF-ing out crosstalk. Curr. Top. Dev. Biol. 117, 1–13 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vissers L. E., et al. , Mutations in a new member of the chromodomain gene family cause CHARGE syndrome. Nat. Genet. 36, 955–957 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Bergman J. E., et al. , CHD7 mutations and CHARGE syndrome: The clinical implications of an expanding phenotype. J. Med. Genet. 48, 334–342 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Corsten-Janssen N., et al. , The cardiac phenotype in patients with a CHD7 mutation. Circ. Cardiovasc. Genet. 6, 248–254 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Basson M. A., van Ravenswaaij-Arts C., Functional insights into chromatin remodelling from studies on CHARGE syndrome. Trends Genet. 31, 600–611 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Micucci J. A., Sperry E. D., Martin D. M., Chromodomain helicase DNA-binding proteins in stem cells and human developmental diseases. Stem Cells Dev. 24, 917–926 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Ravenswaaij-Arts C., Martin D. M., New insights and advances in CHARGE syndrome: Diagnosis, etiologies, treatments, and research discoveries. Am. J. Med. Genet. C. Semin. Med. Genet. 175, 397–406 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corsten-Janssen N., Scambler P. J., Clinical and molecular effects of CHD7 in the heart. Am. J. Med. Genet. C. Semin. Med. Genet. 175, 487–495 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Pauli S., Bajpai R., Borchers A., CHARGEd with neural crest defects. Am. J. Med. Genet. C. Semin. Med. Genet. 175, 478–486 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Zentner G. E., Layman W. S., Martin D. M., Scacheri P. C., Molecular and phenotypic aspects of CHD7 mutation in CHARGE syndrome. Am. J. Med. Genet. A. 152A, 674–686 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delahaye A., et al. , Familial CHARGE syndrome because of CHD7 mutation: Clinical intra- and interfamilial variability. Clin. Genet. 72, 112–121 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Kim H. G., et al. , Mutations in CHD7, encoding a chromatin-remodeling protein, cause idiopathic hypogonadotropic hypogonadism and Kallmann syndrome. Am. J. Hum. Genet. 83, 511–519 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zaidi S., et al. , De novo mutations in histone-modifying genes in congenital heart disease. Nature 498, 220–223 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanka M., Tangsinmankong N., Loscalzo M., Sleasman J. W., Dorsey M. J., Complete DiGeorge syndrome associated with CHD7 mutation. J. Allergy Clin. Immunol. 120, 952–954 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Gamsiz E. D., et al. , Discovery of rare mutations in autism: Elucidating neurodevelopmental mechanisms. Neurotherapeutics 12, 553–571 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gonçalves C. I., et al. , High frequency of CHD7 mutations in congenital hypogonadotropic hypogonadism. Sci. Rep. 9, 1597 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hutson M. R., Kirby M. L., Neural crest and cardiovascular development: A 20-year perspective. Birth Defects Res. C Embryo Today 69, 2–13 (2003). [DOI] [PubMed] [Google Scholar]
- 20.Snider P., Olaopa M., Firulli A. B., Conway S. J., Cardiovascular development and the colonizing cardiac neural crest lineage. ScientificWorldJournal 7, 1090–1113 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keyte A., Hutson M. R., The neural crest in cardiac congenital anomalies. Differentiation 84, 25–40 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keyte A. L., Alonzo-Johnsen M., Hutson M. R., Evolutionary and developmental origins of the cardiac neural crest: Building a divided outflow tract. Birth Defects Res. C Embryo Today 102, 309–323 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Plein A., Fantin A., Ruhrberg C., Neural crest cells in cardiovascular development. Curr. Top. Dev. Biol. 111, 183–200 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Lin C. J., Lin C. Y., Chen C. H., Zhou B., Chang C. P., Partitioning the heart: Mechanisms of cardiac septation and valve development. Development 139, 3277–3299 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siebert J. R., Graham J. M. Jr, MacDonald C., Pathologic features of the CHARGE association: Support for involvement of the neural crest. Teratology 31, 331–336 (1985). [DOI] [PubMed] [Google Scholar]
- 26.Li W., et al. , Brg1 governs distinct pathways to direct multiple aspects of mammalian neural crest cell development. Proc. Natl. Acad. Sci. U.S.A. 110, 1738–1743 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bajpai R., et al. , CHD7 cooperates with PBAF to control multipotent neural crest formation. Nature 463, 958–962 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Balow S. A., et al. , Knockdown of fbxl10/kdm2bb rescues chd7 morphant phenotype in a zebrafish model of CHARGE syndrome. Dev. Biol. 382, 57–69 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Asad Z., et al. , Rescue of neural crest-derived phenotypes in a zebrafish CHARGE model by Sox10 downregulation. Hum. Mol. Genet. 25, 3539–3554 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Randall V., et al. , Great vessel development requires biallelic expression of Chd7 and Tbx1 in pharyngeal ectoderm in mice. J. Clin. Invest. 119, 3301–3310 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sperry E. D., et al. , The chromatin remodeling protein CHD7, mutated in CHARGE syndrome, is necessary for proper craniofacial and tracheal development. Dev. Dyn. 243, 1055–1066 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lewis A. E., Vasudevan H. N., O’Neill A. K., Soriano P., Bush J. O., The widely used Wnt1-Cre transgene causes developmental phenotypes by ectopic activation of Wnt signaling. Dev. Biol. 379, 229–234 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muzumdar M. D., Tasic B., Miyamichi K., Li L., Luo L., A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Blake K. D., Prasad C., CHARGE syndrome. Orphanet J. Rare Dis. 1, 34 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanlaville D., Verloes A., CHARGE syndrome: An update. Eur. J. Hum. Genet. 15, 389–399 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Wyse R. K., al-Mahdawi S., Burn J., Blake K., Congenital heart disease in CHARGE association. Pediatr. Cardiol. 14, 75–81 (1993). [DOI] [PubMed] [Google Scholar]
- 37.Hsu P., et al. , CHARGE syndrome: A review. J. Paediatr. Child Health 50, 504–511 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Jongmans M. C., et al. , CHARGE syndrome: The phenotypic spectrum of mutations in the CHD7 gene. J. Med. Genet. 43, 306–314 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lalani S. R., et al. , Spectrum of CHD7 mutations in 110 individuals with CHARGE syndrome and genotype-phenotype correlation. Am. J. Hum. Genet. 78, 303–314 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.He D., et al. , Chd7 cooperates with Sox10 and regulates the onset of CNS myelination and remyelination. Nat. Neurosci. 19, 678–689 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feng W., et al. , Chd7 is indispensable for mammalian brain development through activation of a neuronal differentiation programme. Nat. Commun. 8, 14758 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marie C., et al. , Oligodendrocyte precursor survival and differentiation requires chromatin remodeling by Chd7 and Chd8. Proc. Natl. Acad. Sci. U.S.A. 115, E8246–E8255 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou Y., et al. , Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Creyghton M. P., et al. , Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U.S.A. 107, 21931–21936 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schnetz M. P., et al. , Genomic distribution of CHD7 on chromatin tracks H3K4 methylation patterns. Genome Res. 19, 590–601 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schnetz M. P., et al. , CHD7 targets active gene enhancer elements to modulate ES cell-specific gene expression. PLoS Genet. 6, e1001023 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kodo K., et al. , Regulation of Sema3c and the interaction between cardiac neural crest and second heart field during outflow tract development. Sci. Rep. 7, 6771 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Plein A., et al. , Neural crest-derived SEMA3C activates endothelial NRP1 for cardiac outflow tract septation. J. Clin. Invest. 125, 2661–2676 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Erickson S. L., et al. , ErbB3 is required for normal cerebellar and cardiac development: A comparison with ErbB2-and heregulin-deficient mice. Development 124, 4999–5011 (1997). [DOI] [PubMed] [Google Scholar]
- 50.Scholl A. M., Kirby M. L., Signals controlling neural crest contributions to the heart. Wiley Interdiscip. Rev. Syst. Biol. Med. 1, 220–227 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bejiqi R., Retkoceri R., Bejiqi H., Zeka N., Klippel–Feil syndrome associated with congential heart disease presentaion of cases and a review of the curent literature. Open Access Maced. J. Med. Sci. 3, 129–134 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu Y., et al. , CHD7 interacts with BMP R-SMADs to epigenetically regulate cardiogenesis in mice. Hum. Mol. Genet. 23, 2145–2156 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oliviero G., et al. , The variant polycomb repressor complex 1 component PCGF1 interacts with a pluripotency sub-network that includes DPPA4, a regulator of embryogenesis. Sci. Rep. 5, 18388 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Engelen E., et al. , Sox2 cooperates with Chd7 to regulate genes that are mutated in human syndromes. Nat. Genet. 43, 607–611 (2011). [DOI] [PubMed] [Google Scholar]
- 55.Rutherford E. L., Lowery L. A., Exploring the developmental mechanisms underlying Wolf-Hirschhorn syndrome: Evidence for defects in neural crest cell migration. Dev. Biol. 420, 1–10 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.De Rubens Figueroa J., Rodríguez L. M., Hach J. L., Del Castillo Ruíz V., Martínez H. O., Cardiovascular spectrum in Williams-Beuren syndrome: The Mexican experience in 40 patients. Tex. Heart Inst. J. 35, 279–285 (2008). [PMC free article] [PubMed] [Google Scholar]
- 57.Gervais L., et al. , Stem cell proliferation is kept in check by the chromatin regulators kismet/CHD7/CHD8 and Trr/MLL3/4. Dev. Cell 49, 556–573.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schulz Y., et al. , CHARGE and Kabuki syndromes: A phenotypic and molecular link. Hum. Mol. Genet. 23, 4396–4405 (2014). [DOI] [PubMed] [Google Scholar]
- 59.Eissenberg J. C., Shilatifard A., Histone H3 lysine 4 (H3K4) methylation in development and differentiation. Dev. Biol. 339, 240–249 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang Q. J., Liu Z. P., Histone methylations in heart development, congenital and adult heart diseases. Epigenomics 7, 321–330 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Black J. C., Van Rechem C., Whetstine J. R., Histone lysine methylation dynamics: Establishment, regulation, and biological impact. Mol. Cell 48, 491–507 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Justin N., De Marco V., Aasland R., Gamblin S. J., Reading, writing and editing methylated lysines on histone tails: New insights from recent structural studies. Curr. Opin. Struct. Biol. 20, 730–738 (2010). [DOI] [PubMed] [Google Scholar]
- 63.Bouazoune K., Kingston R. E., Chromatin remodeling by the CHD7 protein is impaired by mutations that cause human developmental disorders. Proc. Natl. Acad. Sci. U.S.A. 109, 19238–19243 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Janssen N., et al. , Mutation update on the CHD7 gene involved in CHARGE syndrome. Hum. Mutat. 33, 1149–1160 (2012). [DOI] [PubMed] [Google Scholar]
- 65.Hurd E. A., et al. , Loss of Chd7 function in gene-trapped reporter mice is embryonic lethal and associated with severe defects in multiple developing tissues. Mamm. Genome 18, 94–104 (2007). [DOI] [PubMed] [Google Scholar]
- 66.Alavizadeh A., et al. , The Wheels mutation in the mouse causes vascular, hindbrain, and inner ear defects. Dev. Biol. 234, 244–260 (2001). [DOI] [PubMed] [Google Scholar]
- 67.Danielian P. S., Muccino D., Rowitch D. H., Michael S. K., McMahon A. P., Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr. Biol. 8, 1323–1326 (1998). [DOI] [PubMed] [Google Scholar]
- 68.Payne S., et al. , A critical role for the chromatin remodeller CHD7 in anterior mesoderm during cardiovascular development. Dev. Biol. 405, 82–95 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yelbuz T. M., et al. , Shortened outflow tract leads to altered cardiac looping after neural crest ablation. Circulation 106, 504–510 (2002). [DOI] [PubMed] [Google Scholar]
- 70.Feiner L., et al. , Targeted disruption of semaphorin 3C leads to persistent truncus arteriosus and aortic arch interruption. Development 128, 3061–3070 (2001). [DOI] [PubMed] [Google Scholar]
- 71.Mondal T., et al. , Sense-antisense lncRNA pair encoded by locus 6p22.3 determines neuroblastoma susceptibility via the USP36-CHD7-SOX9 regulatory axis. Cancer Cell 33, 417–434.e7 (2018). [DOI] [PubMed] [Google Scholar]
- 72.Bosman E. A., et al. , Multiple mutations in mouse Chd7 provide models for CHARGE syndrome. Hum. Mol. Genet. 14, 3463–3476 (2005). [DOI] [PubMed] [Google Scholar]
- 73.Balasubramanian R., Crowley W. F. Jr, Reproductive endocrine phenotypes relating to CHD7 mutations in humans. Am. J. Med. Genet. C. Semin. Med. Genet. 175, 507–515 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ang S. Y., et al. , KMT2D regulates specific programs in heart development via histone H3 lysine 4 di-methylation. Development 143, 810–821 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.National Research Council , Guide for the Care and Use of Laboratory Animals (National Academies Press, Washington, DC, ed. 8, 2011). [Google Scholar]
- 76.Peng Y., et al. , Sema6D acts downstream of bone morphogenetic protein signalling to promote atrioventricular cushion development in mice. Cardiovasc. Res. 112, 532–542 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang X., Astrof S., Neural crest cell-autonomous roles of fibronectin in cardiovascular development. Development 143, 88–100 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang X., Astrof S., Isolation of mouse cardiac neural crest cells and their differentiation into smooth muscle cells. Bio Protoc. 7, e2530 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yan S., et al. , Distribution of CHD7 and H3K27ac in cardiac nerual crest cells. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE149855. Deposited 5 May 2020.
- 80.Yan S., et al. , CHD7 downstream genes in mouse cardiac neural crest cells https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE149860. Deposited 5 May 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession nos. GSE149855 and GSE149860) (79, 80).