Abstract
The exponential increase in cases and mortality of coronavirus disease (COVID-19) has called for a need to develop drugs to treat this infection. Using in silico and molecular docking approaches, this study investigated the inhibitory effects of Pradimicin A, Lamivudine, Plerixafor and Lopinavir against SARS-CoV-2 Mpro. ADME/Tox of the ligands, pharmacophore hypothesis of the co-crystalized ligand and the receptor, and docking studies were carried out on different modules of Schrodinger (2019-4) Maestro v12.2. Among the ligands subjected to ADME/Tox by QikProp, Lamivudine demonstrated drug-like physico-chemical properties. A total of five pharmacophore binding sites (A3, A4, R9, R10, and R11) were predicted from the co-crystalized ligand and the binding cavity of the SARS-CoV-2 Mpro. The docking result showed that Lopinavir and Lamivudine bind with a higher affinity and lower free energy than the standard ligand having a glide score of −9.2 kcal/mol and −5.3 kcal/mol, respectively. Plerixafor and Pradimicin A have a glide score of −3.7 kcal/mol and −2.4 kcal/mol, respectively, which is lower than the co-crystallized ligand with a glide score of −5.3 kcal/mol. Molecular dynamics confirmed that the ligands maintained their interaction with the protein with lower RMSD fluctuations over the trajectory period of 100 nsecs and that GLU166 residue is pivotal for binding. On the whole, present study specifies the repurposing aptitude of these molecules as inhibitors of SARS-CoV-2 Mpro with higher binding scores and forms energetically stable complexes with Mpro.
Communicated by Ramaswamy H. Sarma
Keywords: Protease inhibitor, SARS-CoV-2, COVID-19, lamivudine, lopinavir, docking, Mpro
1. Introduction
The emergence of COVID-19 has put the whole world in a standstill for more than 60 days. The disease originated from Wuhan (Hubei, China) in December 2019 (Liu et al., 2020) and spread to the rest of the world, and was then declared a global pandemic in February 2020. COVID-19 is caused by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), a member of the coronavirus family. They are the largest known RNA viruses made up of positive single-stranded RNA and are subdivided into alpha, beta, gamma and delta coronaviruses (Shereen et al., 2020; Huynh et al., 2012). This set of viruses have been documented to have a zoonotic origin from bats, mice or domestic animals and are linked with severe respiratory illness in humans (Ye et al., 2020; Moore and June, 2020). The SARS-CoV was first implicated as the causative agent of the severe acute respiratory syndrome outbreaks between 2002/2003 in the Guangdong area of China (Zhong et al., 2003; Zhong, 2004), and the Middle East respiratory coronavirus (MERS-CoV) in 2012 (de Groot et al., 2013). SARS-CoV-2, another variant of this devastating virus family has forced the human race to be on lockdown as a preventive measure to avoid disease transmission (contraction and spreading of the disease). The disease symptoms include but are not limited to fever, dry cough, sore throat and difficulty breathing. As of 29 May 2020, there are 5 701337 confirmed cases and 357 688 mortalities due to this alarming infection have been reported from 216 countries (WHO, 2020).
Unfortunately, there is neither a specific drug nor vaccine approved for the treatment or modulation of immune response against this infection, yet the infection and mortality rates are steadily increasing. Angiotensin-converting enzyme-2 (ACE2) receptor in the host was identified as one of the receptors that the spike protein of SARS-CoV-2 interact with during infection, and was explored as a potential target for treatment of COVID-19. Chloroquine and its derivative Hydroxychloroquine were the first drugs to be repurposed for use in the treatment of COVID-19 (Omar et al., 2020). Although these drugs were potent in inhibiting SARS-CoV, and showed no serious health effects when used for treatment of malaria and autoimmune diseases (Vincent et al., 2005), they are now red flagged for use in COVID-19 (Mehra et al., 2020). The efficacy of these drugs (Colson et al., 2020) together with Lopinavir/Ritonavir (Lim et al., 2020), and Remdesivir (Wang et al., 2020) are currently undergoing investigation and clinical trials. However, the World Health Organization (WHO) have suspended the use of Chloroquine and Hydroxychloroquine after they were reported to have serious side effects including cardiac toxicity (Mehra et al., 2020). As the world is gradually easing their lockdown restrictions, there are concerns about the possibility of the infections increasing exponentially, hence there is an urgent need to identify and develop drug candidates and vaccines for the treatment and containment of SARS-CoV-2 infections. Notably, traditional drug discovery and development pipeline are time-consuming, very costly and often associated with high clinical failure (Aruleba, 2018). Therefore, employing computational drug discovery tools for this raging infectious agent is of paramount importance. In silico techniques can augment the opportunities for designing, verifying activity, and developing new drug and vaccine candidates within a short space of time. In this regard, bioinformatic tools have been utilized extensively in many interesting studies; and recently for the SARS-CoV-2 (Elmezayen et al., 2020; Gupta et al., 2020; Bhattacharya et al., 2020).
The virus main protease (Mpro) is one of the best characterized drug targets because its inhibition blocks viral replication (Zhang et al., 2020). Holistically, the Mpro is highly promising as no human protease has a similar cleavage pathway, thus making it a non-probable toxic agent (Zhang et al., 2020). The interaction of this enzyme and several compounds as inhibitors have shown promising results (Khaerunnisa et al., 2020; Muralidharan et al., 2020; Ton et al., 2020). Furthermore, Mpro is highly conserved among the coronavirus family showing 40–44% of sequence homology (Muralidharan et al., 2020). Inhibiting the virus replication and maturation in the host cells by blockade of the enzymatic activity of the SARS-CoV-2 Mpro could potentially give the patients a fighting chance. Hence, we have employed in silico molecular docking to explore whether Lamivudine, Pradimicin A, Plerixafor, and Lopinavir could inhibit the Mpro of the SARS-CoV-2 and used molecular dynamics simulation to validate the stable and strong binding interaction between the protein and the ligands with the aim of providing structural basis for rational drug discovery.
2. Materials and methods
2.1. Data selection
The SARS-CoV-2 Mpro receptor with Protein Database (PDB) ID: 6W63 and the potential ligands: Pradimicin A (PubChem ID: 5479145), Lamivudine (PubChem ID: 60825), Plerixafor (PubChem ID: 65015), and Lopinavir (PubChem ID: 92727) were used in this study. The ligands were downloaded from NCBI PubChem database in 3D conformer (SDF) format at (https://www.ncbi.nlm.nih.gov/pccompound) while the crystallized 3D structure of the receptor in complex with co-crystallized ligand: X77, PDB ID:6W66 was retrieved from protein data bank at http://www.rcsb.org/pdb/home/home.do. The Mpro selection is as a result of the presence of an inhibitor ligand (X77) at the active site. The antiviral drug, Lopinavir served as control in this study.
2.2. Adme/tox analysis
To determine whether the ligands can easily gain access to the target site in Mpro after entering the blood stream, the QikProp module in the Schrodinger-2019-4 software package was used to evaluate the 3D molecular structures of the ligands’ pharmacokinetic properties. With respect to Lipinski’s rule of five, the absorption, distribution, metabolism, excretion and toxicity (ADME/T) properties were predicted (Ligprep and Macromodel, 2011).
2.3. Ligand preparation
These compounds were further subjected to ligand preparation (LigPrep) prior to docking. LigPrep is a robust collection of tools designed to prepare high quality, all-atom 3D structures for large numbers of drug-like molecules, starting with 2D or 3D structures in spatial data file (SDF) or Maestro format (Release, 2017). In order to generate a single, low-energy, 3D structure with correct chiralities for each successfully processed input structure, the ligands were subjected to preparation by LigPrep tool. This tool is also capable of generating multiple structures from input structure with various ionization states, tautomers, stereochemistries, and ring conformations, and eliminate molecules using specified criteria. The ligands for this study were also prepared by optimizing geometries through OPLS_2005 force field and Ionization of possible state were generated at pH 7.0 ± 2.0 by Epik (Greenwood et al., 2010). Desalt and generate tautomers were also selected and the stereoisomer computation was checked to retain specific chiralities (vary other chiral centers) and to generate at most 32 conformations per ligand.
2.4. Receptor preparation
Excellent docking studies require both accurate software and starting structures (Fadaka et al., 2020). Experimentally-derived structures can waste time and resources or produce false result if not corrected. Schrödinger’s Protein Preparation Wizard is designed to ensure structural correctness at the onset of a project, equipping them with a high-confidence structure ideal for use with a wide variety of modeling applications (Sastry et al., 2013). Because the typical structure file of the receptor obtained from the PDB is not suitable for immediate use in molecular modeling calculations, the protein preparation wizard was used to prepare the receptor (PDB ID: 6W63). Following the previously reported method (Fadaka et al., 2019), the Protein Preparation Wizard was used for the following: remove alternate conformation, remove HetAtoms from the protein structure, add hydrogen atoms, correct missing or incorrectly specified residues, detect and correct valence violations, optimize protonation state and hydrogen positions.
2.5. Generation of pharmacophore hypothesis (model)
An energy-optimized pharmacophore (e-pharmacophore) model combines the advantages of structure-based and ligand-based drug-design theories, and can be used to rapidly screen ligands based on pharmacophore properties (Salam et al., 2009; Negi et al., 2014). Using the ‘Develop Pharmacophore hypothesis’ option in Maestro v12.2 tasks view mode, pharmacophore sites were generated from the receptor with redocked co-crystalized ligand (Mpro-X77) complex, preserving a maximum of seven pharmacophore features as default. Pharmacophore chemical properties include: hydrogen-bond acceptor (A), represented as vectors, hydrogen-bond donor (D) as projected points, aromatic ring (R) as ring, positive ionizable (P), and negative ionizable (N) (Veeramachaneni et al., 2015). Explicit matching was required in the e-pharmacophore approach for generation of the most energetically favorable site. The hypothesis settings were configured to treat atoms as projected points with a radii scaling factor of 0.50 and limit excluded volume shell to 5.0 Å.
2.6. Docking study
Glide uses a hierarchical series of filters to search for possible locations of the ligand in the active-site region of the receptor. This is done by ligand ranking via high-throughput virtual screening (HTVS). Three modes of sampling ligand conformational and positional degrees of freedom are available to determine the optimal ligand orientation relative to a rigid protein receptor geometry (Repasky et al., 2007). Flexible ligand docking with Glide, optionally includes ligand constraints or ligand molecular similarities. Docking grid of the receptor’s active site was detected using the PDB file of the coordinates with the receptor grid generation tool in maestro v12.2. This site defines the area around the active site in term of co-ordinates x, y and z. The receptor grid box resolution was centered at coordinates −20.57, 18.10, and −26.99 corresponding to x, y and z-axis, respectively. Docking and calculations were run in the extra precision (XP) mode of Glide (Schrödinger, 2018).
2.7. Prime MM-GBSA calculation
The free binding energies of ligand docked complexes were computed using the molecular mechanic-generalized Born surface area (MM-GBSA) (Huang et al., 2006) with specific parameters. Based on the docking score and MM/GBSA binding-free energy, Jin et al. (Jin et al., 2011) developed correlation model between docking scores or calculated binding-free energies and experimental pIC50 values. The Prime module in Maestro was employed to calculate the MM-GBSA energy of Glide XP docked complex. The OPLS_2005 force field in conjunction with GBSA continuum model (Yu et al., 2006) was used to calculate energies of complexes of ligands following the equation reported by Lyne et al. (Lyne et al., 2006) and according to our previous study (Fadaka et al., 2020).
2.8. Molecular dynamic simulation
The docked complexes were subjected to MD simulation using Desmond module of Schrödinger software with OPLS 2005 force field. The protein–ligand complex was bounded with a predefined TIP3P water model (Jorgensen et al., 1983) in orthorhombic box. The volume of the box was minimized and the overall charge of the system was neutralized by adding Na+ and Cl- ions. The temperature and pressure were kept constant at 310 Kelvin and 1.01325 bar using Nose–Hoover thermostat (Hoover, 1985) and Martyna–Tobias–Klein barostat (Zhong, 2004) methods. The simulations were performed using NPT ensemble by considering number of atoms, pressure and timescale. During simulations, the long–range electrostatic interactions were calculated using Particle–Mesh–Ewald method (Essmann et al., 1995). RMSD plots for the backbone atoms for both the protein and ligand bound protein were generated to decipher the relative stability of the ligand in the binding pocket of the Mpro. The results were analyzed and visualized by simulation interaction diagram and MS-MD trajectory analysis.
3. Results
3.1. Ligands’ structures
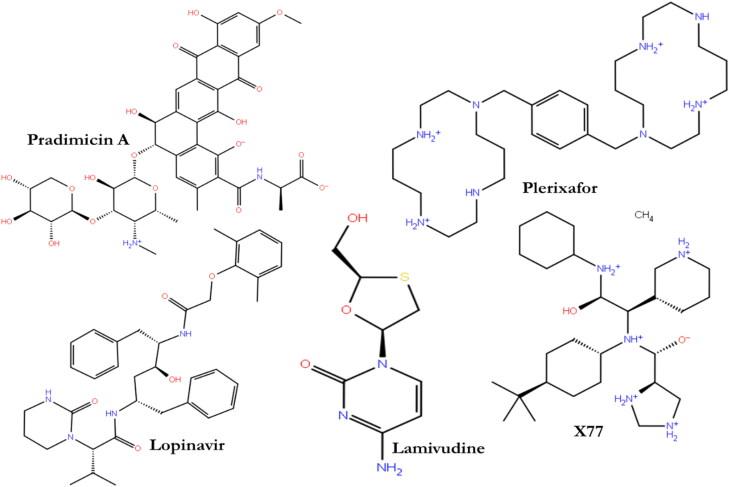
The chemical 2D structures of the four ligands (Figure 1) were obtained from PubChem database. In essence, the database is a public repository composed of numerous validated chemical entities and their biological activities (Kim et al., 2016).
Figure 1.
2D structures of proposed Mpro ligands.
3.2. Pharmacokinetic and toxicological properties prediction
Based on the Lipinski’s rules of five, the pharmacokinetic properties (ADME) of Pradimicin A, Lamivudine, Plerixafor, and Lopinavir were calculated using the QikProp module in the Schrodinger-2019-4-software package. Here, Lipinski stated that a drug/compound would be orally bioavailable if it follows the following criteria such as molecular weight < 500 amu, Hydrogen bond acceptor sites < 10, Hydrogen bond donor sites < 5, and Lipophilicity value LogP ≤ 5. The result of the present study showed that only lamivudine satisfied Lipinski’s rules, which indicated good oral bioavailability (Table 1).
Table 1.
Docking results with pharmacological properties of the studied ligands.
| Ligand | Molecule | ROF | MW | QPlogKhsa | PSA |
|---|---|---|---|---|---|
| Lopinavir | 92727 | 1 | 628.8 | 0.562 | 130.95 |
| Pradimicin A | 5479145 | 3 | 840.78 | −1.137 | 335.54 |
| Plerixafor | 65015 | 2 | 502.78 | 0.375 | 64.555 |
| Lamivudine | 60825 | 0 | 229.26 | −0.758 | 100.229 |
| X77 | 145998279 | 1 | 479.75 | −0.059 | 82.45 |
ROF: Lipinski’s Rule of Five; M.W: Molecular Weight of compounds (130.0 to 725.0 g/mol); QPlogKhsa: Prediction of binding to human serum albumin (−1.5 to 1.5); and PSA: polar surface area.
3.3. Docking study
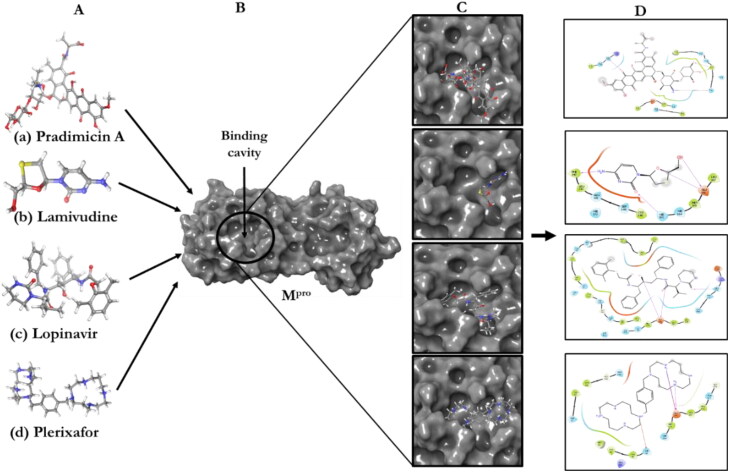
Extra precision docking was used to investigate favorable interactions between the selected ligands and the SARS-CoV-2 Mpro. The redocked X77 replicated the binding pose and orientation of the crystal ligand, and indicated that it was successful in mimicking the native pose (Figure 2). Using the same docking approach, the Mpro receptor was docked with the four ligands (Figure 1). Pradimicin A, Lamivudine, and Plerixafor were considered for docking studies. An FDA-approved anti-viral drug Lopinavir was also docked to correlate the docking scores and interactions (Figure 3).
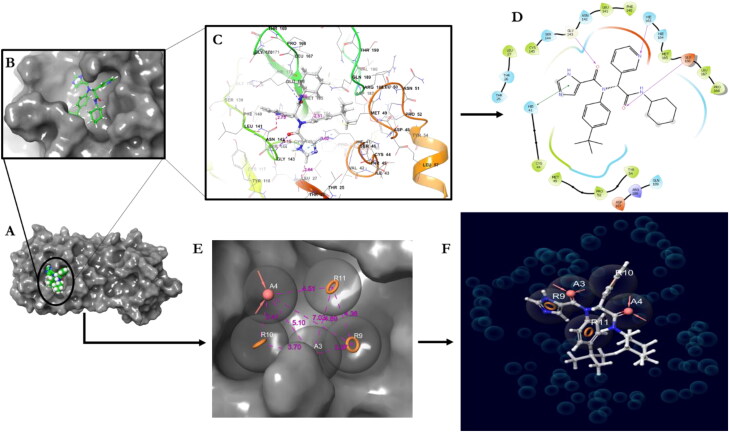
Figure 2.
Validation of the docking algorithm of Mpro (6W63) and the co-crystalized ligand (X77) and the Ephamacophore hypothesis. A) The receptor; B) pose of the redocked co-crystalized ligand; C) amino acid residues of the binding cavity D) 2D of ligand interaction; E) The pharmacophore model; F) Co-crystalized ligand modeled pharmacophore.
Figure 3.
Molecular docking study of the ligands to the receptor. A) Ligands; B) Receptor; C) ligand-receptor complex (binding poses); D) 2D interactions.
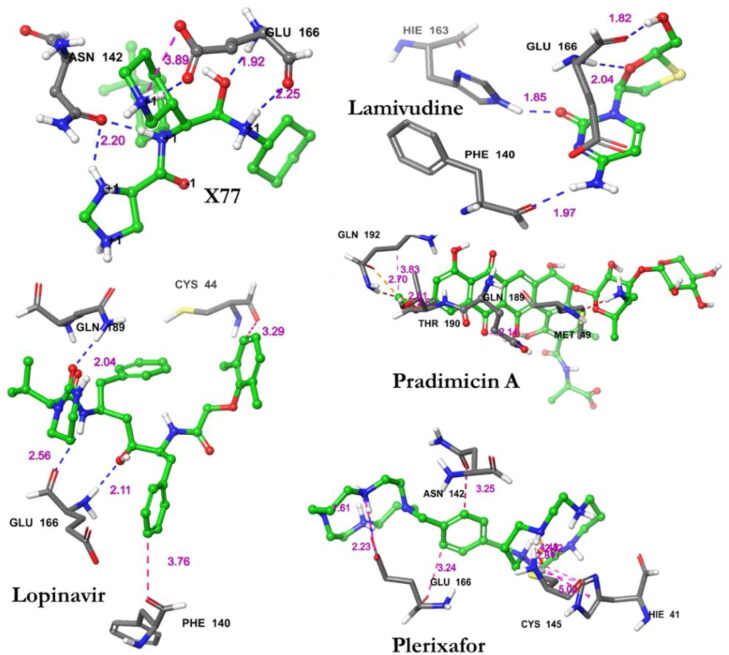
The results revealed that Pradimicin A was properly positioned into the binding cavity constructed by polar amino acids: GLN192 THR190 GLN189 ASN142 THR45 SER46 THR26 THR25 THR24; hydrophobic amino acids: ALA191 PRO168 LEU167 MET165 CYS44 MET49; and charged amino acids: GLU166 with binding energy of −2.4 kcal/mol (Figure 3(A)). The binding parameters of the ligands together with the number of hydrogen bonds and the residues within the distance of 4 Ao are presented in Table 2. From the 2D interaction result, CYS44 exhibited bi-hedral H-bonding with hydroxyl group of oxane ring (tetrahydropyran) of Pradimicin A and other oxygen group showed H-bonding with GLN189 (Figure 3(A)).
Table 2.
Binding interactions of the ligands with the active site of SARS-CoV-2 main protease (PDB ID: 6W63) along with their binding scores (kcal/mol).
| Ligand | Glide Gscore | Dock score | ΔGbind (kcal/mol) | Hydrophobic residues | (No) of H-bonds (4 Å) |
|---|---|---|---|---|---|
| Co-crystalized ligand | −5.3 | −5.3 | −48.60 | PRO168, LEU167, MET165, PHE140, LEU141, CYS145, CYS44, MET49, PRO52, TRY54, LEU27 | (3) 2GLU166, GLY143 |
| Pradimicin A | −2.4 | −1.4 | −45.90 | ALA191, PRO168, LEU167, MET165, CYS44, MET49 | (3) 2Cys44, GLN189 |
| Plerixafor | −3.7 | −2.4 | −75.80 | LEU141, PHE140, CYS145, MET165, LEU167 | (3) 2GLU166 ASN142, CYS145 |
| Lamivudine | −5.3 | −5.3 | −34.90 | LEU141, PHE140, CYS145, MET165, LEU167 | (4) 2GLU166, PHE140 HIE 163 |
| Lopinavir | −9.2 | −9.2 | −85.80 | PRO168, LEU167, MET165, PHE140, LEU141, CYS145, VAL42, CYS44, TYR54, PRO52, MET49 | (3) 2GLU166, GLN189 |
Lamivudine was properly positioned into the binding cavity constructed by polar amino acids: ASN 142 SER144, hydrophobic amino acids: LEU141 PHE140 CYS145 MET165 LEU167, charged amino acids: GLU166; with binding energy −5.3 kcal/mol. The hydroxyl and Oxo groups of lamivudine exhibited bi-hedral H-bonding with GLU166. The amine group and keto groups of the ring structure were also involved in hydrogen bonding with PHE140 (Figures 3(B) and Ye et al., 2020and Table 2).
Lopinavir was properly docked into the binding cavity of the receptor as constructed by polar amino acids: GLN192 THR190 GLN189 ASN142 SER144 THR25 THR45 SER46; hydrophobic amino acids: PRO168 LEU167 MET165 PHE140 LEU141 CYS145 VAL42 CYS44 TYR54 PRO52 MET49; Charged amino acids: ASP187 GLU166; and glycine: GLY143 with binding energy of −9.2 kcal/mol (Figure 3). Among the residues of Lopinavir and Mpro, three H-bonds where involved between the residues GLU166 and GLN189 and the interacting atoms of Lopinavir (Figure 4 and Table 2).
Figure 4.
Specific interactions observed between the residues of Mpro (PDB ID: 6W63) and the studied ligands within specific distances (4 Å).
Plerixafor interacted with the binding cavity constructed by polar amino acids: ASN142 THR25 THR169; hydrophobic amino acids: CYS145 LEU27 MET49 CYS44 VAL42 MET165 LEU167 PRO168; charged amino acids: GLU166 and glycine amino acids: GLY170 with a binding energy of −3.7 kcal/mol. Two amine groups showed bilateral H-bonding with GLU166 and other bonds with CYS145 and ASN142 (Figure 3(D) and Figure 4).
3.3.1. Calculation of prime MM-GBSA
To predict the binding mode and binding free energy (ΔGbind), the Prime MM-GBSA simulation was calculated for Mpro–ligands and Mpro–co-crystalized ligand complexes utilizing Maestro v12.2 (Table 2). The ΔGbind (kcal/mol) of all the interactions were reported in order to understand the binding affinities between the ligands and the receptor to provide an insight into the stability of the ligands in the active site of Mpro using the molecular mechanics-generalized Born surface area (MMGBSA) method. We computationally estimated the binding energy of the co-crystalized ligand to be −48.56 kcal/mol. The binding energy of lopinavir (−85.83 kcal/mol) to the Mpro appears to be the lowest among the ligands estimated followed by plerixafor (−75.78 kcal/mol). Pradimicin and lamivudine showed relatively higher (−45.89 and −34.94 kcal/mol, respectively) when compared to the co-crystalized ligand. The binding free energy determination, based on Prime MM-GBSA, established the stability of Mpro-ligands complexes.
3.4. Molecular dynamic analysis
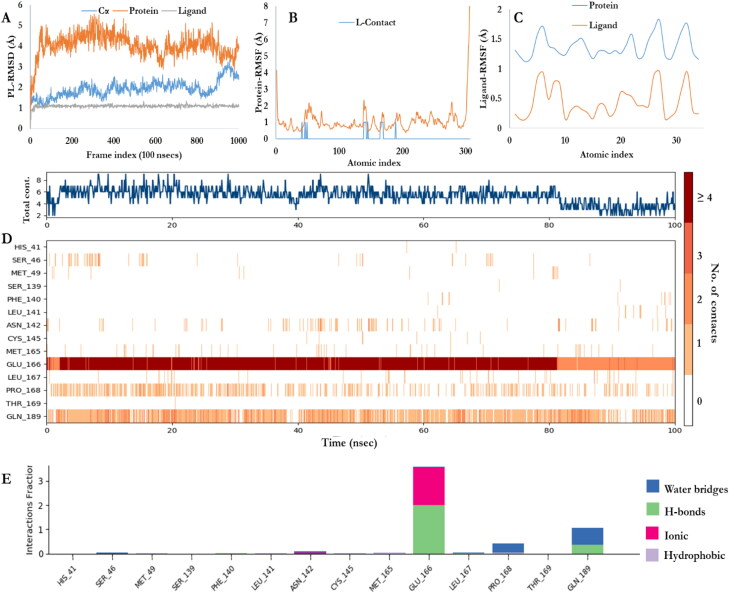
The simulation interaction diagram of Schrodinger suit was used to analyze and survey the protein-ligand interactions for the desmond molecular dynamic trajectory carried out on Mpro and the studied ligands’ complexes. The stability of Mpro, co-crystalized ligand and test ligands complexes were evaluated through 100 nanoseconds (nsec) molecular dynamics simulations (Figures 5–de Groot et al., 2013). Methods comprising virtual screening, molecular docking, and molecular dynamics (MD) simulation are a widely used method for the exploration of novel inhibitors against a target protein (Jani and Dalafave, 2012). MD simulation provides information regarding the time dependent behavior of any molecular system by integrating Newton’s laws of motion (Mandlik and Singh, 2016). The stability of the docked complexes were determined by performing an MD simulation as previously described (Al-Shabib et al., 2018; AlAjmi et al., 2018). The protein-ligand root mean square deviation (PL-RMSD) was used to measure the scaler distance between Mpro and the ligands throughout the trajectory (100 nsecs) and also to evaluate the evolvement of the Cα protein backbone as well as the protein and ligand RMSD over the course of 100 nsecs period. All the protein frames were initially aligned on the reference frame backbone and then the RMSDs were calculated based the Cα, the protein and the ligands. Monitoring the RMSD of Mpro provided insights into the structural conformation throughout the 100 nsecs. Furthermore, it was used to evaluate the equilibration of the complexes and its fluctuation directly to simulation within the sum of thermal average.
Figure 5.
MD simulation of Mpro-Co-crystalized ligand. A) Backbone RMSD of the co-crystalized Mpro-ligand; B; The RMSF plot of the docked complex. The fluctuations indicate the flexibility in the docked complex; C) The Ligand Root Mean Square Fluctuation (L-RMSF). Useful for characterizing changes in the ligand atom positions; D) Mpro-co-crystalized ligand contacts; E) Timeline representation of the interactions and contacts (H-bonds, Hydrophobic, and water bridges).
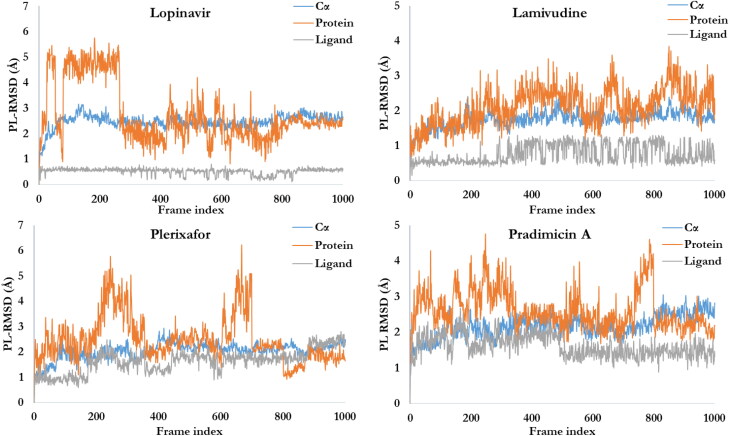
Figures 5(A) and 6 shows the PL-RMSD of the co-crystalized ligand (X77) and studied ligands, respectively, in the active site of Mpro over the 100 nsecs simulation. This illustrates the super positioning or fitting as well as measuring the RMSD using the protein and ligand’s heavy atoms. The ligand RMSD shows the stability of X77 as well as the studied ligands with respect to the protein binding pocket that is, the internal fluctuation within the ligands’ structures is low. MD simulation of the docked complexes protein backbone RMSD plots indicate the stability of the ligands in the active site of the Mpro. The ligands’ backbone RMSD plots indicate that all the four ligands are comparable to the co-crystalized ligand and maintained their interactions with Mpro.
Figure 6.
The RMSD plots of ligands-Mpro. The ligands appear to maintain their stability within the binding pocket as they show lower RMSD fluctuations over the 100 nsec simulation period.
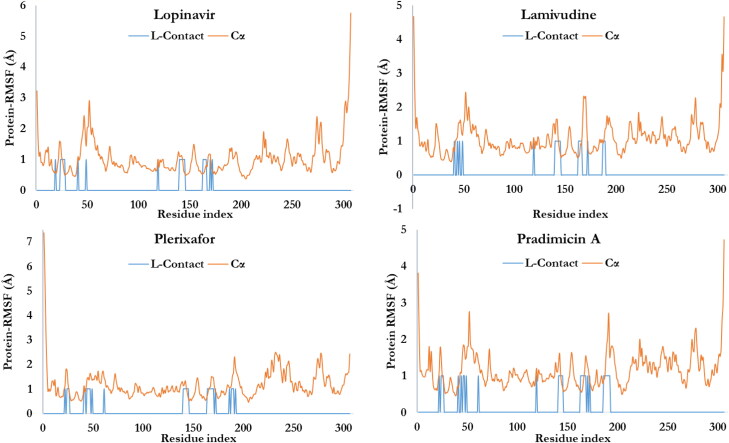
The characterization of the local fluctuation of the protein was further investigated using the protein root mean square fluctuation (P-RMSF) as depicted in Figures 5(B) and 7. The peaks indicate the areas of the protein that fluctuate the most during the simulation period with respect to residue location (the X-axis). Additionally, the ligands’ contacts were shown in bars. This corresponds to the protein residues interacting with the ligands. 31 residues of the Mpro were involved in the contacts of pradimicin A and plerixafor. With respect to 14 contacts with the X77, lamivudine and lopinavir interacted with 19 and 21 Mpro residues, respectively. Figure 5(C) shows the Ligand RMSF which explains how the ligands fragment interact with Mpro and the entropic role in their binding events. Most of the atoms of the X77 (Figure 5(C)), plerixafor and pradimicin A (not shown) showed a high degree of fluctuation due to the fact that the atoms are solvent exposed and are able to rotate easily around the atom bonds and interact with water. Atoms of lamivudine and lopinavir (not shown) are buried deeply in the binding pocket of Mpro with less chance to rotate freely and thus, have lower fluctuation.
Figure 7.
The root mean square fluctuation (RMSF) of the.
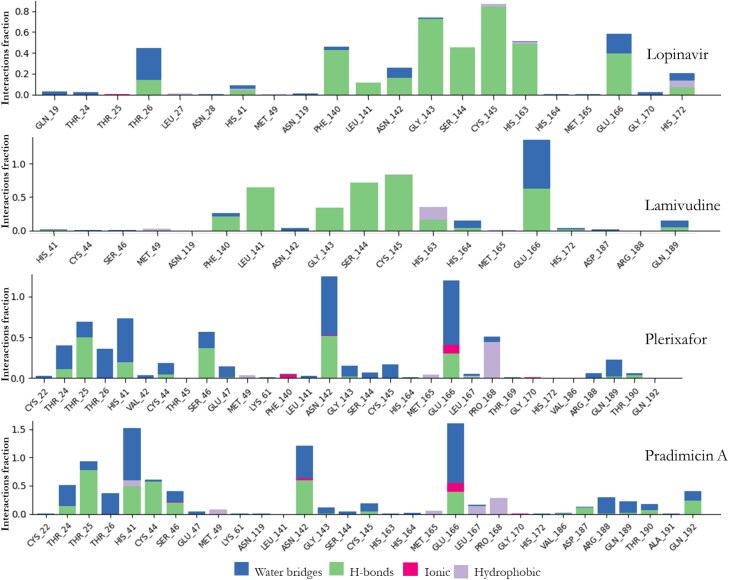
The timeline of the contacts of the residues of the Mpro to X77 over the 100 nsecs period as well as the time of contact (X-axis), the specific residues (Y-axis) and number of contacts (color intensity) were shown in Figure 5(D). The Mpro-ligands interaction was monitored throughout the simulation as presented in Figures 5(E) and 8 for the co-crystalized ligand as well as the studied ligands, respectively. These interactions or the amount of times the ligands made contact with Mpro were categorized into four types: Hydrogen Bonds, Hydrophobic, Ionic and Water Bridges (Ojo et al., 2020). The result of the Mpro-ligands complexes were compared with Mpro-co-crystalized ligand and observed that the contacts made with Mpro were three namely; Hydrogen Bonds, Hydrophobic, and Water Bridges. GLU166 was maintained almost throughout the period of simulation with hydrogen bond and water bridges. In comparison with the four ligands used in this study, several amino acids were constantly in contact with Mpro but the residues ASN142, GLY143, GLN189, GLN192, and GLN166 were significantly in contact with Mpro. Residues SER46 and ASN142 were significantly common to Lopinavir, Pradimicin A, and Plerixafor maintained 90%contacts Mpro over the period of the simulation time.
Figure 8.
Protein-Ligand contacts of Mpro and all the ligands with their respective bond interactions.
4. Discussion
Computational biology entails in silico investigation which is an essential arm of biotechnology focused at enhancing a deeper insight of biomolecular interactions in order to address cellular disease pathogenesis whilst having immense contribution towards design and development of possible therapeutic candidates (Abdullahi et al., 2017). Indeed, this technique have assisted in identifying lead compounds for various diseases (Aruleba et al., 2018; Shanmuga Priya et al., 2018; Fadaka et al., 2019). On the other hand, the emergence and sporadic spread of COVID-19 has called for the urgent identification of novel drug and/or repurposing of existing available chemotherapeutic agents against this pandemic. The virus polyprotein encodes two proteases, the 3-C-like protease (main protease) and a papain-like protease, both are vital targets for drug discovery platforms against coronaviruses (Kandeel and Al-Nazawi, 2020). Accordingly, the Mpro is the most probable antiviral candidate due to its crucial role in self-maturation and ensuing development of polyproteins (Muralidharan et al., 2020). Hence, we explored the molecular implications of blocking the activity of Mpro using compounds (Pradimicin A, Lamivudine, Plerixafor and Lopinavir) that have been shown to possess pharmacological activity against viruses.
In view of the aforementioned, we redocked the 6W63 with its co-crystalized ligand using the Glide XP docking protocol. With the receptor grid and extra precision option in the precision mode, X77 was docked into the active site of the SARS-CoV-2 Mpro receptor. The Mpro-X77 redocked complex was used as an input for the e-pharmacophore to generate a hypothesis. The initial number for pharmacophore site generation was set to default; however, five pharmacophore sites were predicted, and the final hypothesis consisted of three aromatic rings (R9, R10, and R11) and two H-bond acceptors (A3 and A4) as shown in Figure 2(E). The quantitative structure-activity relationship (QSAR) of this enzyme (Mpro) also known as novel 3C-like protease (3CLpro) was developed using a multi-linear regression (MLR) based 2D-QSAR model for the identification of important structural features responsible for the inhibition of Mpro in SARS-CoV-2 infections using eight significant descriptors classified as topological, 2D atom pair, functional group count, and atom entered fragment descriptors (Kumar and Roy, 2020). The study reported that molecules or ligands consisting of features such as O-S fragments and thiophene rings could contribute to higher Mpro enzyme inhibitory activity, and the presence of pyridine rings, methylene groups connected with the highly electronegative atoms, imides (including thioimides) group, and N–O fragment may lower the inhibitory activity for 3CLpro enzyme. Based on this context, the features that negatively influence the inhibitory activity against Mpro are not contained in the studied ligands’ structures and thus the ligands may have high Mpro enzyme inhibitory activities.
Pharmacophore-based screening was performed against the ligands in order to determine Mpro inhibitors with desired chemical features and to compensate for the QSAR due to limited dataset in this study. The generated hypothesis can be used in a virtual-based screening study to generate additional inhibitors of Mpro if they satisfy a minimum match of four sites on the generated e-pharmacophore hypothesis.
Subsequently, molecular docking using the Glide molecular tool revealed that Lopinavir bound to the active site with the highest affinity and lowest free energy with a Glide score of −9.2 kcal/mol; suggesting that it could be a vital inhibitor of protease activity of coronavirus (Ye et al., 2020). Following Lopinavir in terms of excellent binding affinity for Mpro is Lamivudine; having a glide score of −5.3 kcal/mol. Earlier studies have shown this compound to be a potent anti-viral agent and it has been proposed for COVID-19 treatment due to its ability to impair DNA synthesis (Lai et al., 2020). Jarvis et al. (Jarvis and Faulds, 1999) highlighted Lamivudine as an excellent inhibitor of hepatitis B virus (HBV) replication and suppresses viral replication in HIV patients. Interestingly, Lamivudine fulfilled the five Lipinski’s rule in our study. In light of the anti-viral activity of Plerixafor, we repurposed it for the management of COVID-19. Plerixafor had a glide score of −3.7 kcal/mol against the SARS-CoV-2 Mpro, a score lower than the co-crystallized ligand. Nevertheless, this compound is an antagonist to C-X-C chemokine receptor type-4 (CXCR4) (Keating, 2011), with anti-HIV activities. It inhibits the attachment of the virus to the host cell. Amongst all the docked Mpro-ligand complexes, the Mpro-Pradimicin A complex had the lowest binding affinity of −2.4 kcal/mol. However, it has been shown to possess inhibitory activities against virus infection/replication. Importantly, the anti-viral mechanism of action of Pradimicin A is through binding to mannose residue (Tanabe-Tochikura et al., 1990) and recent studies have documented anti-HIV (Balzarini et al., 2007) and anti-coronavirus (Van der Meer et al., 2007) properties for this compound.
Finally, docking of the ligands with the Mpro revealed their binding modes. To account for the flexibility of Mpro and ligands and to evaluate their binding affinity with Mpro, the MD simulation of the docked complexes were carried out at three different time points (1.2 nsecs, 10 nsecs and 100 nsecs) in order to avoid false positive result of the stability result (only the 100 nsecs was reported in this study). Binding mode analysis revealed that the binding modes obtained after MD simulation were more or less similar to that obtained after docking. The presence of a large number of H bond acceptors, H bond donors as well as hydrophobic groups in the ligands account for the stability of the ligand inside the binding pocket of Mpro. Based on the RMSD of the ligand-protein complexes, it was confirmed that ligands maintained their interaction with Mpro with lower root mean square fluctuations.
Globally, various research institutions and hospitals are on a quest to identify suitable drug targets to stop/manage SARS-CoV-2. However, using the traditional methods for identification of drug targets for COVID-19 might not be viable as the disease is already crippling the health care resources. Hence, anti-viral drugs are repurposed for immediate attention. This have proven to be the most expensive exercise as the drugs did not behave as hypothesized. As such, Bioinformatics could serve as a tool to screen potential drug activity before use. Accordingly, computational drug design and repurposing has laid a foundation for researchers to investigate various chemotherapeutic agents towards identifying potent drug candidates for clinical trials.
5. Conclusion
Our study revealed that all the antiviral drugs had satisfactory binding affinity for the SARS-CoV-2 Mpro. Amongst all the inhibitors, Lopinavir had the best binding score while pradimicin A had the lowest binding score for the SARS-CoV-2 Mpro. These results were comparable after we used molecular dynamics simulation to validate the stability of all the ligand-receptor complexes. The ligands bound steadily to Mpro active site and restricted the target movement. GLU166 was found to be an important residue in the active site of Mpro necessary for binding. Furthermore, Lamivudine warrants an in-depth investigation taking into account its binding affinity for Mpro and fulfilment of the Lipinski’s rule of five. It is worth noting that this is the first report that indicated the inhibitory effect of Pradimicin A and Plerixafor against the Mpro, and could serve as potential drug candidates for COVID-19 treatment. Overall, these data can be utilized in drug repurposing pipelines in the continued search for SARS-CoV-2 therapy with high efficacy. The QSAR of these ligands alongside other dataset is recommended for further analysis in order to provide the structure features against the inhibitory activities of the SARS-CoV-2 main protease (Mpro).
Acknowledgements
We would like to acknowledge the National Integrated Cyberinfrastructure system, Center for High Performance Computing (CHPC), Department of Science and Technology, Republic of South Africa for the license to the Lengau cluster and other modules under the Schrodinger suit. Additionally, we appreciate Dr. Samuel Egieyeh, Department of Pharmacy, University of the Western Cape for the time and guidance he invested in this paper to validate its accuracy.
Authors’ contributions
All authors have made significant contributions to the submission of the article. A.O.F, R.T.A, and N.R.S.S conceived the concept and the design of the study, A.K, A.M, and M.M supervised and provided the necessary supports and software required for analysis. The analysis and data interpretations were done collaboratively by all the authors while AOF, RTA, and NRSS prepared the initial draft and also substantially revised the manuscript. A.K, A.M, and M.M thoroughly revised the manuscript. Finally, all authors read and approved the submitted version of the manuscript for publication.
Disclosure statement
The authors declare no competing interests.
Funding
This is not applicable
References
- Abdullahi, M., Olotu, F. A., & Soliman, M. E. (2017). Dynamics of allosteric modulation of lymphocyte function associated antigen-1 closure-open switch: unveiling the structural mechanisms associated with outside-in signaling activation . Biotechnology Letters, 39(12), 1843–1851. 10.1007/s10529-017-2432-0 [DOI] [PubMed] [Google Scholar]
- AlAjmi, M. F., Rehman, M. T., Hussain, A., & Rather, G. M. (2018). Pharmacoinformatics approach for the identification of Polo-like kinase-1 inhibitors from natural sources as anti-cancer agents. International Journal of Biological Macromolecules, 116, 173–181. 10.1016/j.ijbiomac.2018.05.023 [DOI] [PubMed] [Google Scholar]
- Al-Shabib, N. A., Khan, J. M., Malik, A., Alsenaidy, M. A., Rehman, M. T., AlAjmi, M. F., Alsenaidy, A. M., Husain, F. M., & Khan, R. H. (2018). Molecular insight into binding behavior of polyphenol (rutin) with beta lactoglobulin: Spectroscopic, molecular docking and MD simulation studies. Journal of Molecular Liquids, 269, 511–520. [Google Scholar]
- Aruleba, R. T. (2018). Identification and structural bioinformatics of druggable proteins in schistosoma species. University of Zululand. [Google Scholar]
- Aruleba, R. T., Adekiya, T. A., Oyinloye, B. E., & Kappo, A. P. (2018). Structural studies of predicted ligand binding sites and molecular docking analysis of Slc2a4 as a therapeutic target for the treatment of cancer. International Journal of Molecular Sciences , 19(2), 386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balzarini, J., Van Laethem, K., Daelemans, D., Hatse, S., Bugatti, A., Rusnati, M., Igarashi, Y., Oki, T., & Schols, D. (2007). Pradimicin A, a carbohydrate-binding nonpeptidic lead compound for treatment of infections with viruses with highly glycosylated envelopes, such as human immunodeficiency virus. Journal of Virology, 81(1), 362–373. 10.1128/JVI.01404-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya, M., Sharma, A. R., Patra, P., Ghosh, P., Sharma, G., Patra, B. C., Lee, S. S., & Chakraborty, C. (2020). Development of epitope-based peptide vaccine against novel coronavirus 2019 (SARS-COV-2): Immunoinformatics approach. Journal of Medical Virology, 92(6), 618–631. 10.1002/jmv.25736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colson, P., Rolain, J.-M., Lagier, J.-C., Brouqui, P., & Raoult, D. (2020). Chloroquine and hydroxychloroquine as available weapons to fight COVID-19. International Journal of Antimicrobial Agents, 55(4), 105932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot, R. J., Baker, S. C., Baric, R. S., Brown, C. S., Drosten, C., Enjuanes, L., Fouchier, R. A. M., Galiano, M., Gorbalenya, A. E., Memish, Z. A., Perlman, S., Poon, L. L. M., Snijder, E. J., Stephens, G. M., Woo, P. C. Y., Zaki, A. M., Zambon, M., & Ziebuhr, J. (2013). Commentary: Middle East respiratory syndrome coronavirus (MERS-CoV): Announcement of the Coronavirus Study Group. Journal of Virology, 87(14), 7790–7792. 10.1128/JVI.01244-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmezayen, A. D., Al-Obaidi, A., Şahin, A. T., & Yelekçi, K. (2020). Drug repurposing for coronavirus (COVID-19): in silico screening of known drugs against coronavirus 3CL hydrolase and protease enzymes. Journal of Biomolecular Structure and Dynamics, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essmann, U., Perera, L., Berkowitz, M. L., Darden, T., Lee, H., & Pedersen, L. G. (1995). A smooth particle mesh Ewald method. The Journal of Chemical Physics, 103(19), 8577–8593. [Google Scholar]
- Fadaka, A. O., Bakare, O. O., Sibuyi, N. R. S., & Klein, A. (2020). Gene expression alterations and molecular analysis of CHEK1 in solid tumors. Cancers, 12(3), 662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadaka, A. O., Pretorius, A., & Klein, A. (2019). Functional Prediction of Candidate MicroRNAs for CRC Management Using in Silico Approach. International Journal of Molecular Sciences , 20(20), 5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadaka, A. O., Pretorius, A., & Klein, A. (2019). MicroRNA assisted gene regulation in colorectal cancer. International Journal of Molecular Sciences, 20(19), 4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadaka, A. O., Sibuyi, N. R. S., Madiehe, A. M., & Meyer, M. (2020). Computational insight of dexamethasone against potential targets of SARS-CoV-2. Journal of Biomolecular Structure and Dynamics, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood, J. R., Calkins, D., Sullivan, A. P., & Shelley, J. C. (2010). Towards the comprehensive, rapid, and accurate prediction of the favorable tautomeric states of drug-like molecules in aqueous solution. Journal of Computer-Aided Molecular Design, 24(6-7), 591–604. 10.1007/s10822-010-9349-1 [DOI] [PubMed] [Google Scholar]
- Gupta, M. K., Vemula, S., Donde, R., Gouda, G., Behera, L., & Vadde, R. (2020). In-silico approaches to detect inhibitors of the human severe acute respiratory syndrome coronavirus envelope protein ion channel. Journal of Biomolecular Structure and Dynamics, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoover, W. G. (1985). Canonical dynamics: Equilibrium phase-space distributions. Physical Review. A, General Physics, 31(3), 1695–1697. 10.1103/physreva.31.1695 [DOI] [PubMed] [Google Scholar]
- Huang, N., Kalyanaraman, C., Bernacki, K., & Jacobson, M. P. (2006). Molecular mechanics methods for predicting protein-ligand binding. Physical Chemistry Chemical Physics: PCCP, 8(44), 5166–5177. 10.1039/b608269f [DOI] [PubMed] [Google Scholar]
- Huynh, J., Li, S., Yount, B., Smith, A., Sturges, L., Olsen, J. C., Nagel, J., Johnson, J. B., Agnihothram, S., Gates, J. E., Frieman, M. B., Baric, R. S., & Donaldson, E. F. (2012). Evidence supporting a zoonotic origin of human coronavirus strain NL63. Journal of Virology, 86(23), 12816–12825. 10.1128/JVI.00906-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jani, K. S., & Dalafave, D. (2012). computational Design of Targeted Inhibitors of polo-Like Kinase 1 (plk1). Bioinformatics and Biology Insights, 6, BBI. S8971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis, B., & Faulds, D. (1999). Lamivudine. Drugs, 58(1), 101–141. [DOI] [PubMed] [Google Scholar]
- Jin, M., Shepardson, N., Yang, T., Chen, G., Walsh, D., & Selkoe, D. J. (2011). Soluble amyloid beta-protein dimers isolated from Alzheimer cortex directly induce Tau hyperphosphorylation and neuritic degeneration . Proceedings of the National Academy of Sciences of the United States of America, 108(14), 5819–5824. 10.1073/pnas.1017033108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen, W. L., Chandrasekhar, J., Madura, J. D., Impey, R. W., & Klein, M. L. (1983). Comparison of simple potential functions for simulating liquid water. The Journal of Chemical Physics, 79(2), 926–935. [Google Scholar]
- Kandeel, M., & Al-Nazawi, M. (2020). Virtual screening and repurposing of FDA approved drugs against COVID-19 main protease. Life Sciences, 251, 117627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating, G. M. (2011). Plerixafor: a review of its use in stem-cell mobilization in patients with lymphoma or multiple myeloma. Drugs, 71(12), 1623–1647. 10.2165/11206040-000000000-00000 [DOI] [PubMed] [Google Scholar]
- Khaerunnisa, S., Kurniawan, H., Awaluddin, R., Suhartati, S., & Soetjipto, S. (2020). Potential inhibitor of COVID-19 main protease (Mpro) from several medicinal plant compounds by molecular docking study. Prepr. doi10. 20944/preprints202003. 0226. v1 2020:1–14.
- Kim, S., Thiessen, P. A., Bolton, E. E., Chen, J., Fu, G., Gindulyte, A., Han, L., He, J., He, S., Shoemaker, B. A., Wang, J., Yu, B., Zhang, J., & Bryant, S. H. (2016). PubChem substance and compound databases. Nucleic Acids Research, 44(D1), D1202–D1213. 10.1093/nar/gkv951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, V., & Roy, K. (2020). Development of a simple, interpretable and easily transferable QSAR model for quick screening antiviral databases in search of novel 3C-like protease (3CLpro) enzyme inhibitors against SARS-CoV diseases. SAR and QSAR in Environmental Research, 31(7), 511–516. [DOI] [PubMed] [Google Scholar]
- Lai, C.-C., Shih, T.-P., Ko, W.-C., Tang, H.-J., & Hsueh, P.-R. (2020). Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and corona virus disease-2019 (COVID-19): the epidemic and the challenges. International Journal of Antimicrobial Agents, 55(3), 105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligprep, M., & Macromodel, G. (2011). QikProp. Schrodinger, LLC. [Google Scholar]
- Lim, J., Jeon, S., Shin, H.-Y., Kim, M. J., Seong, Y. M., Lee, W. J., Choe, K.-W., Kang, Y. M., Lee, B., & Park, S.-J. (2020). Case of the index patient who caused tertiary transmission of COVID-19 infection in Korea: The application of lopinavir/ritonavir for the treatment of COVID-19 infected pneumonia monitored by quantitative RT-PCR. Journal of Korean Medical Science, 35(7), e79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., Gayle, A. A., Wilder-Smith, A., & Rocklöv, J. (2020). The reproductive number of COVID-19 is higher compared to SARS coronavirus. Journal of Travel Medicine, 27(2), 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyne, P. D., Lamb, M. L., & Saeh, J. C. (2006). Accurate prediction of the relative potencies of members of a series of kinase inhibitors using molecular docking and MM-GBSA scoring. Journal of Medicinal Chemistry, 49(16), 4805–4808. 10.1021/jm060522a [DOI] [PubMed] [Google Scholar]
- Mandlik, V., & Singh, S. (2016). Molecular docking and molecular dynamics simulation study of inositol phosphorylceramide synthase–inhibitor complex in leishmaniasis: Insight into the structure based drug design. F1000Research, 5, 1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehra, M. R., Desai, S. S., Ruschitzka, F., & Patel, A. N. (2020). Hydroxychloroquine or chloroquine with or without a macrolide for treatment of COVID-19: A multinational registry analysis. The Lancet. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Moore, J. B., & June, C. H. (2020). Cytokine release syndrome in severe COVID-19. Science (New York, N.Y.).), 368(6490), 473–474. 10.1126/science.abb8925 [DOI] [PubMed] [Google Scholar]
- Muralidharan, N., Sakthivel, R., Velmurugan, D., & Gromiha, M. M. (2020). Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 Protease against COVID-19. Journal of Biomolecular Structure and Dynamics, 1–6. [DOI] [PubMed] [Google Scholar]
- Negi, B., Raj, K. K., Siddiqui, S. M., Ramachandran, D., Azam, A., & Rawat, D. S. (2014). In vitro antiamoebic activity evaluation and docking studies of metronidazole-triazole hybrids. Chemmedchem, 9(11), 2439–2444. 10.1002/cmdc.201402240 [DOI] [PubMed] [Google Scholar]
- Ojo, O. A., Aruleba, R. T., Adekiya, T. A., Sibuyi, N. R. S., Ojo, A. B., Ajiboye, B. O., Oyinloye, B. E., Adeola, H. A., & Fadaka, A. O. (2020). Deciphering the interaction of puerarin with cancer macromolecules: An in silico investigation. Journal of Biomolecular Structure and Dynamics, 1–12. [DOI] [PubMed] [Google Scholar]
- Omar, S., Bouziane, I., Bouslama, Z., & Djemel, A. (2020). In-Silico Identification of Potent Inhibitors of COVID-19 Main Protease (Mpro) and Angiotensin Converting Enzyme 2 (ACE2) from Natural Products: Quercetin. Hispidulin, and Cirsimaritin Exhibited Better Potential Inhibition than Hydroxy-Chloroquine Against COVID-19 Main Protease Active Site and ACE2.
- Release, S. (2017). 2: LigPrep, Schrödinger. New York, NY: LLC. [Google Scholar]
- Repasky, M. P., Shelley, M., & Friesner, R. A. (2007). Flexible ligand docking with Glide. Curr Protoc Bioinformatics Chapter 8:Unit 8.12. [DOI] [PubMed]
- Salam, N. K., Nuti, R., & Sherman, W. (2009). Novel method for generating structure-based pharmacophores using energetic analysis. Journal of Chemical Information and Modeling, 49(10), 2356–2368. 10.1021/ci900212v [DOI] [PubMed] [Google Scholar]
- Sastry, G. M., Adzhigirey, M., Day, T., Annabhimoju, R., & Sherman, W. (2013). Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. Journal of Computer-Aided Molecular Design, 27(3), 221–234. 10.1007/s10822-013-9644-8 [DOI] [PubMed] [Google Scholar]
- Schrödinger, L. (2018). Schrödinger Release 2018-1: Maestro. Schrödinger, LLC. [Google Scholar]
- Shanmuga Priya, A., Balakrishnan, S. b., Veerakanellore, G. B., & Stalin, T. (2018). In-vitro dissolution rate and molecular docking studies of cabergoline drug with β-cyclodextrin. Journal of Molecular Structure, 1160, 1–8. [Google Scholar]
- Shereen, M. A., Khan, S., Kazmi, A., Bashir, N., & Siddique, R. (2020). COVID-19 infection: Origin, transmission, and characteristics of human coronaviruses. Journal of Advanced Research, 24, 91–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe-Tochikura, A., Tochikura, T. S., Yoshida, O., Oki, T., & Yamamoto, N. (1990). Pradimicin A inhibition of human immunodeficiency virus: Attenuation by mannan. Virology, 176(2), 467–473. [DOI] [PubMed] [Google Scholar]
- Ton, A. T., Gentile, F., Hsing, M., Ban, F., & Cherkasov, A. Rapid identification of potential inhibitors of SARS‐CoV‐2 main protease by deep docking of 1.3 billion compounds. Molecular Informatics (2020). [DOI] [PMC free article] [PubMed]
- Van der Meer, F., de Haan, C., Schuurman, N., Haijema, B., Verheije, M., Bosch, B., Balzarini, J., & Egberink, H. (2007). The carbohydrate-binding plant lectins and the non-peptidic antibiotic pradimicin A target the glycans of the coronavirus envelope glycoproteins. The Journal of Antimicrobial Chemotherapy, 60(4), 741–749. 10.1093/jac/dkm301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeramachaneni, G. K., Raj, K. K., Chalasani, L. M., Bondili, J. S., & Talluri, V. R. (2015). High-throughput virtual screening with e-pharmacophore and molecular simulations study in the designing of pancreatic lipase inhibitors. Drug design. Drug Design, Development and Therapy, 9, 4397–4412. 10.2147/DDDT.S84052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent, M. J., Bergeron, E., Benjannet, S., Erickson, B. R., Rollin, P. E., Ksiazek, T. G., Seidah, N. G., & Nichol, S. T. (2005). Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virology Journal, 2, 69. 10.1186/1743-422X-2-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, M., Cao, R., Zhang, L., Yang, X., Liu, J., Xu, M., Shi, Z., Hu, Z., Zhong, W., & Xiao, G. (2020). Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Research, 30(3), 269–271. 10.1038/s41422-020-0282-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, X.-T., Luo, Y.-L., Xia, S.-C., Sun, Q.-F., Ding, J.-G., Zhou, Y., Chen, W., Wang, X.-F., Zhang, W.-W., Du, W.-J., Ruan, Z.-W., & Hong, L. (2020). Clinical efficacy of lopinavir/ritonavir in the treatment of Coronavirus disease 2019. European Review for Medical and Pharmacological Sciences, 24(6), 3390–3396. 10.26355/eurrev_202003_20706 [DOI] [PubMed] [Google Scholar]
- Ye, Z.-W., Yuan, S., Yuen, K.-S., Fung, S.-Y., Chan, C.-P., & Jin, D.-Y. (2020). Zoonotic origins of human coronaviruses. International Journal of Biological Sciences, 16(10), 1686–1697. 10.7150/ijbs.45472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Z., Jacobson, M. P., & Friesner, R. A. (2006). What role do surfaces play in GB models? A new-generation of surface-generalized born model based on a novel gaussian surface for biomolecules. Journal of Computational Chemistry, 27(1), 72–89. 10.1002/jcc.20307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., Lin, D., Sun, X., Curth, U., Drosten, C., Sauerhering, L., Becker, S., Rox, K., & Hilgenfeld, R. (2020). Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science (New York, N.Y.).), 368(6489), 409–412. 10.1126/science.abb3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, N. (2004). Management and prevention of SARS in China. Philosophical Transactions of the Royal Society of London. Series B: Biological Sciences, 359(1447), 1115–1116. 10.1098/rstb.2004.1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, N. S., Zheng, B. J., Li, Y. M., Poon, L. L. M., Xie, Z. H., Chan, K. H., Li, P. H., Tan, S. Y., Chang, Q., Xie, J. P., Liu, X. Q., Xu, J., Li, D. X., Yuen, K. Y., Peiris, J. S. M., & Guan, Y. (2003). Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People's Republic of China, in February, 2003. The Lancet, 362(9393), 1353–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]