Significance
Bats have been shown to dampen several key upstream pathogen and danger-associated molecular patterns, yet much of the downstream signaling is yet unknown. Here, we identify residues in caspase-1 which are critical for enzymatic activity and have been targeted for inhibition in Pteropus bats. Further, we discover cleavage-site flanking residues which lead to loss of IL-1β cleavage in Myotis bats. Thus, we report an inverse relationship between caspase-1 function and IL-1β cleavage, resulting in a consistent reduction of downstream signaling by the inflammasome across bats within the two suborders. In sum, we confirm that bats have targeted the inflammasome pathway at multiple levels and via heterogeneous strategies to reduce proinflammatory responses, thus mitigating potential immune-mediated tissue damage and disease.
Keywords: bats, AIM2, caspase-1, inflammasome, IL-1β
Abstract
Bats have emerged as unique mammalian vectors harboring a diverse range of highly lethal zoonotic viruses with minimal clinical disease. Despite having sustained complete genomic loss of AIM2, regulation of the downstream inflammasome response in bats is unknown. AIM2 sensing of cytoplasmic DNA triggers ASC aggregation and recruits caspase-1, the central inflammasome effector enzyme, triggering cleavage of cytokines such as IL-1β and inducing GSDMD-mediated pyroptotic cell death. Restoration of AIM2 in bat cells led to intact ASC speck formation, but intriguingly resulted in a lack of caspase-1 or consequent IL-1β activation. We further identified two residues undergoing positive selection pressures in Pteropus alecto caspase-1 that abrogate its enzymatic function and are crucial in human caspase-1 activity. Functional analysis of another bat lineage revealed a targeted mechanism for loss of Myotis davidii IL-1β cleavage and elucidated an inverse complementary relationship between caspase-1 and IL-1β, resulting in overall diminished signaling across bats of both suborders. Thus we report strategies that additionally undermine downstream inflammasome signaling in bats, limiting an overactive immune response against pathogens while potentially producing an antiinflammatory state resistant to diseases such as atherosclerosis, aging, and neurodegeneration.
Bats are placental mammals which uniquely utilize powered flight for locomotion, harbor a diverse viral repertoire, and possess longevity exceptional to their body size. In recent years, bats have been implicated in major outbreaks caused by fatal zoonotic viruses, such as SARS-CoV and MERS-CoV, henipaviruses, filoviruses including Ebola and Marburg virus, and a high likelihood of the currently circulating SARS-CoV-2 (1–4). Ongoing outbreaks with significant mortality and morbidity in human and livestock have driven a targeted search for the originating hosts of these spillover pathogens and pivotal studies have identified bats as significant reservoir and ancestral hosts to more zoonotic diseases per species, against all other mammalian orders (2, 5). Of key interest is the bat innate and adaptive immune system, due to evolutionarily driven or yet undiscovered, altered interactions between the host–pathogen interface, leading to their tolerance of viral diseases.
Genomic and transcriptomic studies have identified disparities between bats and other mammals. Positive selection has been shown in critical innate immune, tumor suppressive, and DNA damage checkpoint genes of bats (6), including NLRP3, TP53, and ATM. Altered natural killer (NK) cell repertoires were found among Pteropus alecto, Myotis davidii, and Rousettus aegyptiacus bats, along with differential contraction of IFN-α genes and expansion of IFN-ω genes in Pteropus, Myotis, and Rousettus bat species (7, 8). Despite mounting genomic evidence that bats have unique alterations in innate immune pathways, experimental confirmation is rare. We recently demonstrated that NLRP3 is dampened in bats as a result of loss-of-function bat-specific isoforms and impaired transcriptional priming (9). The stimulator of IFN genes (STING), a key adaptor to the DNA-sensing cGAS protein, is also exclusively mutated at S358 in bats, resulting in a reduced IFN response to HSV1 (10). We previously reported a complete absence of Absent in melanoma 2 (AIM2)-like receptor (ALR) genes across all available bat genomes from both Yinpterochiroptera and Yangochiroptera suborders (11). As these modifications in bats signify shifts in cell signaling and immune regulation, we thus investigated the loss of the PYHIN or ALR gene family for implications on the bat DNA-sensing inflammasome response.
The ALRs are an essential group of germline-encoded pattern recognition receptors (PRRs) comprising 5 members in humans and 14 members in mice, with the most well studied being AIM2 (12, 13). AIM2 is the prototypical member of the ALR family and was shown to mediate intracellular dsDNA-responsive inflammasome signaling, typically of invading pathogenic origin or aberrant host cytosolic DNA (14, 15). There is extensive diversity across mammalian ALR families and AIM2 is the only member with preserved orthology among species (12, 16). Upon recognizing exposed intracellular DNA, it binds to the major turn of the DNA helix and recruits its adaptor apoptosis-associated speck-like protein containing a CARD (ASC), which forms aggregates (ASC specks) to complex with downstream procaspase-1 (15, 17). The caspase-1 proenzyme undergoes homodimerization and autocleavage for activation, conferring it the ability to bind, cleave, and mature IL-1β, IL-18, and gasdermin D (GSDMD) and trigger pyroptotic cell death (18–22). The mammalian AIM2 is crucial in its role in sensing intracellular foreign DNA, accompanied by a potential for either a pathological or protective inflammatory response in the host (23–25). Yet the consequence of its absence in bats, a unique animal model shown to down-regulate components of its inflammasome pathway, is poorly understood.
Caspase-1, or cysteine aspartic protease 1, is the central inflammasome effector for pyroptosis and cytokine secretion, playing roles in diverse cellular processes, including apoptosis/necrosis, metabolism, mitophagy, and autophagy (26–29). The propeptide consists of a caspase-recruitment domain (CARD), a p20 and p10 polypeptide sequence, and undergoes sequential autoproteolysis at aspartic acid residues into p20/p10 subunits which dimerize to achieve the activated conformation for substrate binding and cleavage (18, 21, 30). It is converged upon by all canonical inflammasome receptors, including NLRP3, NAIP/NLRC4, NLRP1, AIM2, and pyrin, and many other members of the NLR family such as NLRP6, NLRP7, and NLRP12, mediating critical proinflammatory host responses against microbes or autoimmune and autoinflammatory sequelae (31). Further, it is involved in multiple age-related diseases, including amyloid β accumulation in Alzheimer’s disease and cardiac injury during acute myocardial infarction (32, 33). While intensive study of human caspase-1 inhibitors are currently ongoing, given its therapeutic potential, still little is known about the downstream activation of caspase-1 in bats, especially given their altered immune landscape and dampened inflammasome function.
Here, we confirm that genomic loss of AIM2 in bats dismantles the inflammasome adaptor recruitment responsive to dsDNA. Additionally, reconstitution of the human gene in a bat in vitro environment is sufficient to partly restore this intracellular pathway up to ASC level. However, we discovered an absence of the downstream cytokine release or cell-death initiation despite robust ASC speck formation in bat primary macrophages. We identify key residues in the bat caspase-1 responsible for dampened IL-1β cleavage, or altered IL-1β cleavage sites which significantly reduce its processing and maturation in bats. Importantly, we have elucidated multiple levels of disengagement within the bat inflammasome pathway with key implications in their response toward cellular stress, inflammation, and pathogenic detection.
Results
Absence of ASC Speck Induced by DNA Stimulation in Bat Kidney and Immune Cells Is Restored by Human AIM2.
Given the ALR family members, including AIM2, are the only DNA sensors mediating the intracellular sensing of pathogenic and aberrant host DNA to activate the inflammasome, we hypothesized that absence of all ALR genes in bats would result in the inability for bats to trigger inflammasome signaling. Indeed, with exogenous dsDNA ligand PolydA:dT stimulation of bat bone-marrow-derived macrophages (BMDMs), we observed a lack of recruitment of ASC into aggregates (ASC speck), which could be seen in dsDNA-treated murine BMDMs (Fig. 1A). Further, using high-throughput image-based flow cytometry (Imagestream), we observed that the cytosolic ASC remained diffusely distributed in bat cells, unlike their aggregation into a speck-like morphology in mouse macrophages (Fig. 1B).
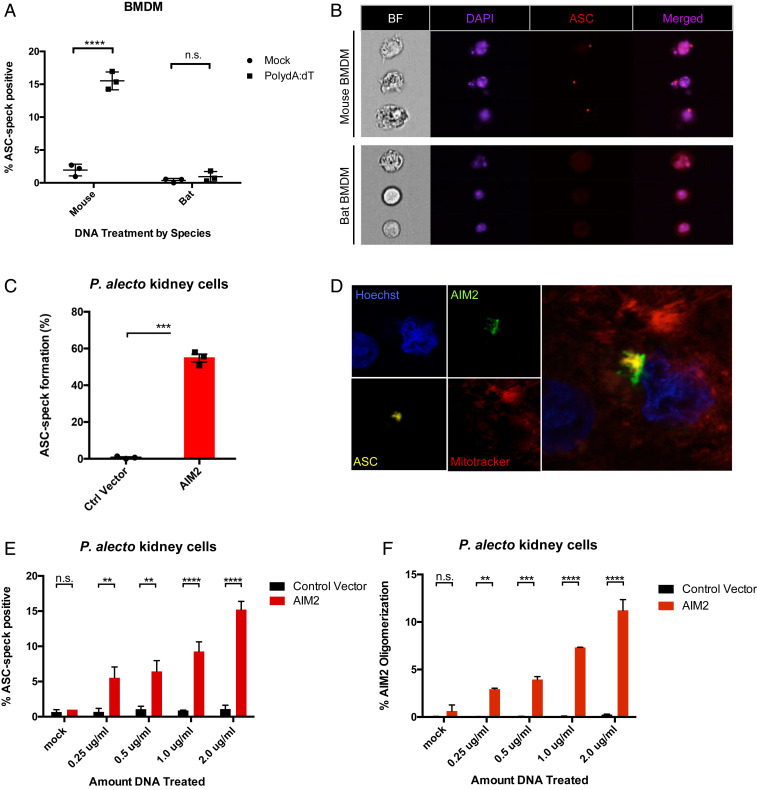
Fig. 1.
Reconstitution of AIM2 restores DNA-triggered ASC speck in bat cells. (A) Measurement of ASC speck formation in mouse and bat BMDMs treated with either vehicle (mock) or transfected dsDNA (PolydA:dT, 1 μg/mL) for 4 h after 3 h LPS (mouse) or CL264 (bat) priming. (B) Single-cell imaging of mouse or bat BMDMs collected on Imagestream to visualize ASC speck aggregation or diffuse intracellular distribution, shown as bright field (BF), DAPI, and ASC signals. (C) ASC speck formation was quantified on Imagestream flow cytometry in P. alecto kidney cells (PaKiS) stably expressing bat ASC-mPlum and transiently expressing human AIM2. (D) Single-plane confocal imaging of transfected PaKiS cells showing ASC speck aggregation and association with cotransfected AIM2. Hoechst 33342 nuclear staining (blue), AIM2-mCitrine (green), ASC-mPlum (yellow), and Mitotracker (red). (E) Retroviral transduction of control vector or AIM2 was performed in ASC-mPlum stably expressing PaKiS cells, and DNA transfected in a dose-curve (mock, 0.25 to 2.0 μg/mL PolydA:dT). Imagestream flow cytometry was performed for triggered ASC speck. (F) AIM2 oligomerization induced in a dose-dependent manner quantified by Imagestream. **P < 0.01, ***P < 0.001, ****P < 0.0001, n.s., nonsignificant; linear regression and two-tailed unpaired t test. Data are presented as mean ± SEM of three biological replicates (A and B) or three independent experiments (C–F).
To investigate whether lack of ASC speck formation was attributable to the absence of gene and protein expression from the ALR family, we generated a human AIM2-mCitrine fusion construct cloned into a mammalian expression vector and rescued the gene in Pteropus alecto kidney-derived (PaKiS) immortalized cells (34). We selected the human AIM2 protein, as AIM2 is the only ALR gene with conserved evolutionary and functional orthology across species (16) and the human ortholog is closest in homology to the only nonfunctional PYHIN peptide fragment identified in bats (Pteronotus parnelli) (11). Transient overexpression of human AIM2 alone was sufficient to restore ASC speck formation in PaKiS cells stably expressing bat ASC (Fig. 1C and SI Appendix, Fig. S1 A and B), resulting in organization into a perinuclear inflammasome complex with colocalization of AIM2 and downstream ASC (Fig. 1D). To evaluate the activation in response to intracellular DNA, lentiviral delivery was used to generate PaKiS cells stably expressing both human AIM2 and bat ASC at low copy number. ASC speck formation was induced with addition of PolydA:dT DNA ligand and increased in a dose-dependent manner in AIM2-positive cells only (12-fold increase at 2.0 μg/mL compared to 0 μg/mL) (Fig. 1E). This was accompanied by increasing detection of AIM2 oligomerization (15-fold), signaling intact sensing of dsDNA by AIM2 and consequent formation of the inflammasome recruitment platform (Fig. 1F). Importantly, interaction of the adaptor ASC with the AIM2 sensor demonstrates the highly conserved nature of bat ASC to retain ability for recruitment to oligomerized human AIM2. This supports our previous observation whereby bat ASC is also conserved with the human ASC in its function and speck properties, including size, density, and shape in response to NLRP3 (9).
Human AIM2 Restores ASC Speck Formation but Not Caspase-1 Activation or IL-1β Release in Bat Macrophages.
As the P. alecto kidney in vitro immortalized cell system lacks classic inflammasome machinery, we examined primary in vitro differentiated bat BMDMs for activation of the inflammasome as a consequence of AIM2 restoration. Transduction of lentivirus carrying a control vector or human AIM2 was performed in P. alecto BMDMs (PaBMDMs), and ASC speck formation was similarly quantified by Imagestream. Only AIM2 reconstituted (AIM2+) bat macrophages treated with PolydA:dT were able to induce endogenous bat ASC aggregation, whereas minimal induction was observed in the mock (vehicle)-treated control vector or AIM2+, or PolydA:dT-treated control vector, conditions (Fig. 2A). Imagestream analysis supported this finding with visible ASC speck in PolydA:dT-treated AIM2+ BMDMs compared to vehicle-only (mock) controls (Fig. 2B). This suggested AIM2-dependent restoration of ASC speck induction in response to the DNA ligands.
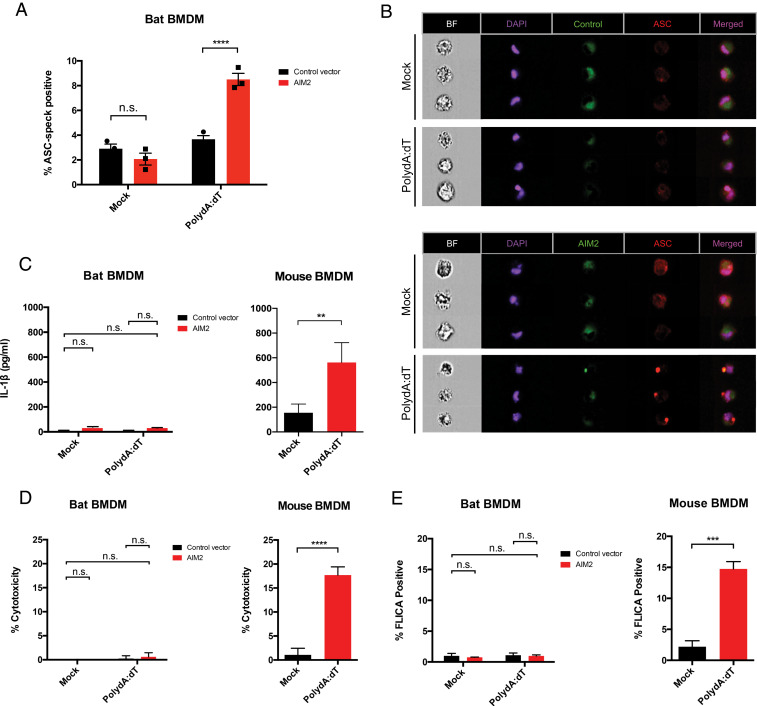
Fig. 2.
Lack of caspase-1 or IL-1β signaling despite ASC speck restoration in AIM2-reconstituted bat macrophages. (A) Mouse or P. alecto BMDMs (PaBMDM) were differentiated in CSF-1 for 5 d and transduced with AIM2 or control vector lentivirus. At 48 h posttransduction, macrophages were primed with CL264 or LPS and treated with vehicle (mock) or DNA (PolydA:dT, 1 μg/mL) for 4 h and ASC speck was quantified via Imagestream. (B) Representative images for PaBMDMs stained for DAPI and anti-PaASC, shown is HsAIM2-mCitrine reconstitution and PaASC speck formation with either mock or PolydA:dT treatment. (C) LDH assay was performed on supernatant to measure cytolytic cell death in PolydA:dT-treated bat control or AIM2+ BMDM (Left) or mouse BMDM (Right). (D) Similarly, IL-1β secretion was quantified in the supernatant of treated bat and mouse BMDMs using in-house bat ELISA protocol as previously published (14) and mouse IL-1β ELISA kit. (E) Fluorescent-labeled inhibitor of caspase activation (FLICA) assay was performed on bat and mouse BMDMs for caspase-1 activation upon DNA treatment, with staining for 1 h and flow cytometry analysis. Statistical analysis was performed using two-way ANOVA with Bonferroni’s multiple comparisons test (A and C–E) and two-tailed unpaired t test (C–E). **P < 0.01, ***P < 0.001, ****P < 0.0001, n.s., nonsignificant. Data are representative of three biological replicates (n = 3) in B or mean ± SEM of three biological replicates (n = 3) in (A, C, and E).
Thus, we next looked for downstream IL-1β cleavage or induction of pyroptosis. Unexpectedly, mature IL-1β was unable to be detected in the supernatant despite ASC speck induction in DNA-treated AIM2-reconstituted bat BMDMs, in contrast to the mouse BMDMs (Fig. 2C). This was accompanied by minimal lytic cell death and low lactate dehydrogenase (LDH) activity levels in the cell supernatant (Fig. 2D). To measure the caspase-1 activity specifically in bat BMDMs, we utilized the 660-YVAD-fmk fluorescent-labeled inhibitor of caspase activation (FLICA) assay with relative specificity for the caspase-1 active site. We observed lack of substrate binding even in DNA-treated and AIM2+ bat BMDMs, compared to the robust activity in treated mouse BMDMs which possess endogenous AIM2 (Fig. 2E). As a previous study similarly identified minimal secretion of IL-1β upon NLRP3 inflammasome activation in bat primary immune cells, we decided to investigate the downstream convergence of both sensing platforms onto caspase-1.
Failure in IL-1β Production Is Due to Substitution of Two Residues in P. alecto Bat Caspase-1.
Lack of downstream activation in the presence of ASC specks can be attributed to either caspase-1, or IL-1β, or both. To dissect the mechanism, we reconstituted the entire AIM2 inflammasome axis in human embryonic kidney (HEK293T) cells, coexpressing AIM2, ASC, caspase-1, and IL-1β. Human genes were used for the upstream components (AIM2 and ASC) to standardize the system in a human cell line. Either control (empty vector), human, or P. alecto caspase-1 (HsCASP1, PaCASP1) proteins were expressed in increasing concentrations and the cell lysates were immunoblotted for cleaved IL-1β p17, representing the product of inflammasome activation. Notably, there was minimal detection of mature bat IL-1β p17 in the cell lysates despite increasing expression of PaCASP1 (Fig. 3A). In contrast, human caspase-1 showed robust cleavage of bat IL-1β, suggesting intact cleavage of bat IL-1β. We observed an overall decrease in pro-PaCASP1 expression in contrast to human protein expression, and an absence of PaCASP1 p32 or other intermediate self-cleavage products. Importantly, a clear reduction in PaCASP1 activity compared to HsCASP1 was observed, which suggested decreased functionality of bat caspase-1.
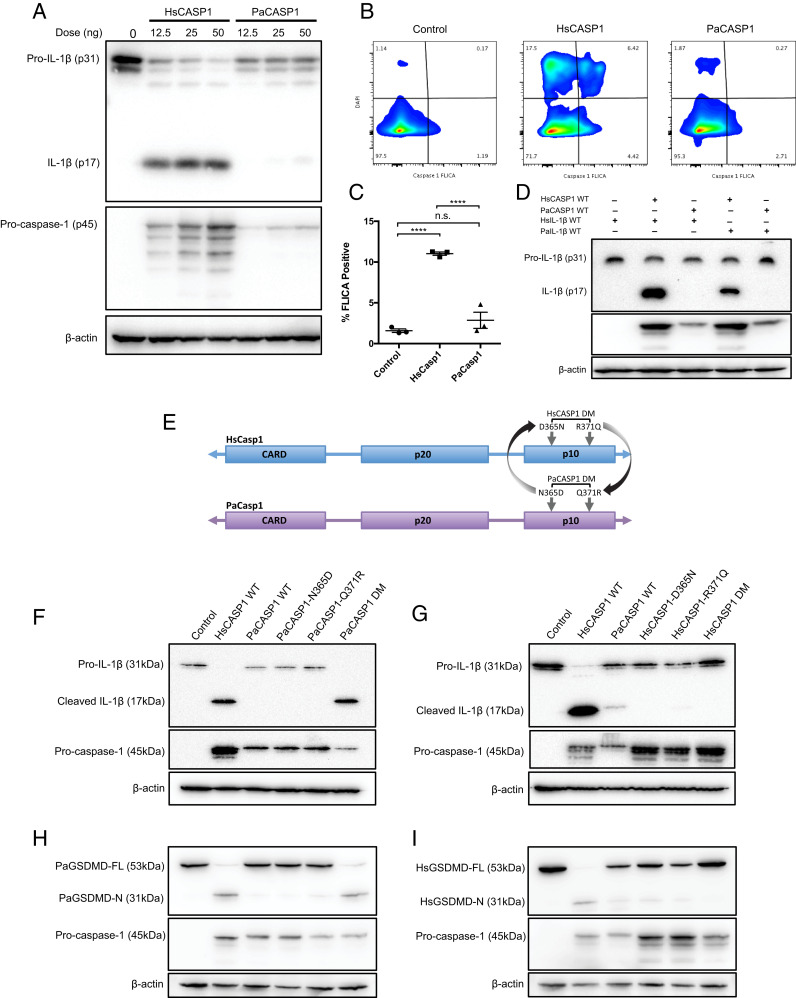
Fig. 3.
Inactive PaCasp1 is rescued by substitution of N365D and Q371R in the p10 domain. (A) The AIM2 inflammasome axis was reconstituted in HEK293T cells using either HsCASP1 or PaCASP1 in a dose-curve. Cell lysates were stained for pro-IL-1β (p31) or mature IL-1β (p17). Procaspase-1 (p45) was assayed to compare expression, and lysates were normalized by β-actin. (B) Cells were reconstituted with the AIM2 inflammasome axis genes and incubated for 48 h, and stained with 660-YVAD-fmk caspase-1 FLICA substrate (Immunochemistry). Flow cytometry was performed to detect caspase-1 activation in control vector, HsCASP1 or PaCASP1 expressing cells, and data analyzed in FlowJo. (C) Quantification of FLICA-positive cells measured via flow cytometry. (D) Immunoblot of cell lysates crossing HsCASP1-3XFLAG with HsIL-1β-HA or PaIL-1β-HA, and PaCasp1-3XFLAG with HsIL-1β-HA or PaIL-1β-HA, coexpressed with human AIM2 and ASC. Cleaved human or bat IL-1β was measured with anti-HA antibody. (E) Schematic of site-derived reverse mutagenesis conducted in the p10 domain of PaCASP1 or HsCASP1. Targeted residues are shown, either single bat N365D or Q371R mutation, or containing both mutations (PaCASP1 DM); and D365N, R371Q, or HsCASP1 DM for the human gene. (F) AIM2 inflammasome axis was reconstituted in HEK283T cells, varying the PaCASP1 for WT, single-mutant N365D, Q371R, or double mutant DM. Cleaved IL-1β p17 was assayed via immunoblotting 48 h posttransfection. (G) Similarly, HsCASP1 WT, D365N, R371Q, or DM was reconstituted, and cleavage of IL-1β was analyzed by immunoblot. Shown are the pro-IL-1β (31 kDa), cleaved IL-1β (17 kda), and human or bat procaspase-1 (45 kDa). (H and I) Similarly, PaGSDMD was coexpressed with PaCASP1 (H) and HsGSDMD with cognate HsCASP1 (I) WT, single-mutants or double-mutant variants, in conjunction with upstream AIM2 inflammasome axis. Cleavage of GSDMD was detected by immunoblotting for the GSDMD-N-2xMyc domain after 48 h incubation in vitro as per D. Data are representative of three independent experiments in A–G. ****P < 0.0001, n.s., nonsignificant. Statistics were performed using unpaired Student’s t test and presented as mean ± SEM of three independent replicates (C).
To confirm the loss of caspase-1 activity, we interrogated PaCASP1 for intact activation by FLICA assay with specificity to caspase-1 to detect for active site binding. Similarly, although HsCASP1 exhibited 10-fold times higher FLICA substrate retention and fluorescence (11.03% ± 0.201), PaCASP1 had minimal detection in activity (2.855% ± 0.991), with levels similar to the control vector (1.58% ± 0.218) (Fig. 3 B and C). To further confirm that loss of inflammasome signaling was at the bat caspase-1 and not IL-1β level, we paired expression of either HsCASP1 or PaCASP1 with human or bat IL-1β. Assay of the lysates for either HsIL-1β or PaIL-1β indicated that HsCASP1, but not PaCASP1, was able to successfully cleave the IL-1β from both species (Fig. 3D). Altogether, this indicates that caspase-1, and not IL-1β, is dampened in the Pteropus bat inflammasome pathway, resulting in failure of IL-1β maturation and cleavage.
Structural studies in human caspase-1 have identified C285 and H237 as essential residues for catalytic activity, among others which form the substrate-binding active site (35, 36). Additional residues within the p10 and p20 fragments have been shown to mediate crucial interactions in dimerization, and mutations result in inability to bind and cleave cytokines for secretion (37). A recent study established that homodimerization of two p20/p10 subunits forms the active conformation of caspase-1, whereby autoprocessing of the caspase-1 p10 fragment is required for GSDMD cleavage and initiation of pyroptosis (22). Given the existing understanding of caspase-1 activation, we analyzed the caspase-1 gene sequence to determine the potential mechanism of its reduced activity in bats. Sequences of available bat caspase-1 genes across Yinpterochiroptera and Yangochiroptera suborders, and 10 nonbat mammalian species were aligned, and phylogenetic analysis by maximum likelihood (PAML) performed to identify lineages and sites acted on by selection pressures (38) (SI Appendix, Fig. S2A). We found one ancestral branch of the bats to have undergone positive selection pressures, and branch-site testing identified greater positive selection pressure exerted on two residue sites in its subsequent branch (SI Appendix, Fig. S2B). Compared to human, mouse, and other included species, the P. alecto and Pteropus vampyrus caspase-1 sequence showed alterations at residue 365 from Asp to Asn, and at 371 from Arg to Gln (red boxes). Both residues are localized within the p10 polypeptide and span five amino acids apart.
To understand the consequence of these substitutions, we performed site-directed reverse mutagenesis of the specific sites, replacing either one, or both residues in the bat with the equivalent human residues, resulting in N365D-only, Q371R-only, or double-mutant (DM) PaCASP1 (Fig. 3E). Likewise, the human HsCASP1 gene was mutated to individually or simultaneously replace both residues with those of Pteropus bat amino acid sites (D365N-only, R371Q-only, and HsCasp1 DM). Only the PaCASP1 DM, but not the single mutants, provoked reversal of the inactive PaCASP1 phenotype to rescue cleavage of IL-1β (Fig. 3F). Conversely for HsCASP1, single mutation of either site to the bat residues was sufficient to abrogate its ability to cleave IL-1β (Fig. 3G). Further, HEK293T cells expressing either HsCASP1 WT or mutant PaCASP1 DM showed increased cellular stress and death morphologically, while HsCASP1 DM and PaCASP1 WT exhibited increased viability regardless of ASC speck formation (SI Appendix, Fig. S3).
Bats also possess the GSDMD gene with conserved pore-forming N-terminal subunits and a caspase-1 recognition site 266FLSD269; yet its function and cleavage potential by bat caspase-1 are still unknown. Thus, PaGSDMD was cloned and compared with HsGSDMD for cleavage by human and P. alecto wild-type (WT) or mutant caspase-1 variants. Similar to the pattern of IL-1β processing by PaCASP1, cleavage of PaGSDMD was undetected in WT PaCASP1, while PaCASP1-DM successfully restored cleavage as observed by the 31-kDa PaGSDMD-N fragment (Fig. 3H). Conversely, HsCASP1-D365N, R371Q, or the combined HsCASP1-DM led to abrogration of GSDMD cleavage (Fig. 3I). To provide the molecular basis of the loss-of-function mutations of D365N and R371Q, we examined the structure of human caspase-1 bound to GSDMD (Protein Data Bank [PDB] code: 6VIE) (39). In the substrate-bound caspase-1 state (p20/p10 dimer), R371 participates in electrostatic interactions with E367 of the opposite p10 subunit, therefore R371Q mutation results in unfavorable interaction across the interface (SI Appendix, Fig. S4). D365 forms part of the dimer interface and may stabilize the homodimeric interactions. As such, similar to R371Q, D365N mutation would weaken the caspase 1 dimer–dimer coordination, leading to inactivation of the enzyme (22, 39, 40). Taken together, our findings demonstrate that both sites in the p10 fragment are necessary for caspase-1 activation by the inflammasome complex, and both IL-1β and GSDMD substrate maturation is dampened in P. alecto bats in a caspase-1-dependent manner.
Complementation between Caspase-1 Activity and IL-1β Cleavage Results in Consistent Inflammasome Dampening across Bats.
Bats, belonging to the order Chiroptera, are the second largest group of mammals with more than 1,000 species. Within the order Chiroptera, Pteropus bats are part of the Pteropodidae family in the suborder Yinpterochiroptera, which are distinct from the suborder Yangochiroptera, containing the rest of the microbat families (41, 42). To better understand if downstream inflammasome dampening is a consistent pattern across bats, we extended our study to include both the Eonycteris spelaea bat (cave nectar bat), also from Yinpterochiroptera, and the M. davidii (David’s myotis) species from the Yangochiroptera suborder. Due to the ability of caspase-1 to signal via multiple upstream sensors, we reconstituted the NLRP3 inflammasome axis in HEK293T cells and expressed either human or relevant species of bat caspase-1 in a dose-dependent manner. We observed that E. spelaea caspase-1 (EsCASP1) retained the ability to cleave P. alecto IL-1β at reduced levels, and M. davidii caspase-1 (MdCASP1) demonstrated intact activity comparable to that of human at low-dose expression (Fig. 4A).
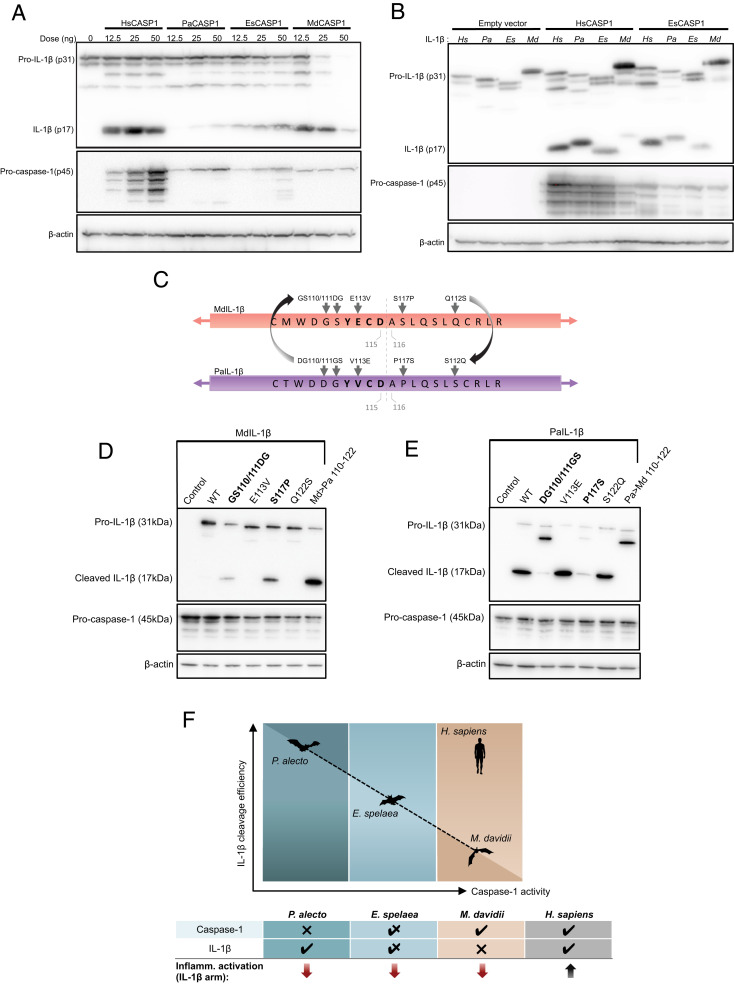
Fig. 4.
Complementary relationship between M. davidii, E. spelaea, and P. alecto caspase-1 and IL-1β. (A) HEK293T cells were transfected with human NLRP3, ASC, pro-IL-1β for inflammasome axis reconstitution, and coexpressed with either H. sapiens, P. alecto, E. spelaea, or M. davidii caspase-1 in increasing concentrations (12.5 to 50 ng/well). Cells were incubated for 48 h and the cell lysate was harvested for Western blot. The amount of mature/cleaved IL-1β (p17) was compared between HsCASP1 and the three bat species and normalized by β-actin. (B) Coexpression of human, P. alecto, E. spelaea, or M. davidii IL-1β in HEK283T with reconstituted human NLRP3 and ASC. Cells were transfected with either empty vector, HsCASP1, or EsCASP1 (as indicated). Cell lysate was immunoblotted for pro-IL-1β (p31) and cleaved IL-1β (p17) with anti-HA, procaspase-1 (p45) with anti-FLAG, and normalized by β-actin. Figures are representative of three independent experiments (A and B). (C) Alignment of full-length IL-1β sequences was performed for H. sapiens, P. alecto, E. spelaea, and M. davidii, and site-directed mutagenesis was performed substituting identified MdIL-1β amino acid residues with equivalent sites from PaIL-1β near the Asp-Ala cleavage site (gray arrows). In bold, cleavage sites YECD and YVCD for MdIL-1β and PaIL-1β, respectively. (D) Full-length WT or mutant MdIL-1β was coexpressed with HsCASP1, AIM2, and ASC and incubated for 48 h; and lysates were assayed for successful cleavage of 17 kDa IL-1β via staining of C-terminal HA-tag. Md → Pa 110 to 122 denotes combined introduction of all identified P. alecto residues expressed by the MdIL-1β mutant protein. (E) Various mutant PaIL-1β was similarly expressed within the AIM2 inflammasome axis and incubated in vitro for 48 h, and various levels of cleaved PaIL-1β were detected by Western blot. (F) Diagram of inverse reciprocal relationship between P. alecto, E. spelaea, and M. davidii caspase-1 activity and IL-1β cleavage efficiency, displayed on the x axis and (Left) y axis, respectively. Top showing increasing pattern of caspase-1 activity (P. alecto < E. spelaea < M. davidii < H. sapiens) is countered by decreasing IL-1β cleavability (M. davidii < E. spelaea < P. alecto < H. sapiens). Table shows vertical summation of either caspase-1 or IL-1β cleavage and resultant function of the downstream inflammasome axis (IL-1β arm).
Next, we reconstituted the NLRP3 inflammasome axis and varied IL-1β of three different bat species in either a HsCASP1, EsCASP1, or MdCASP1 system (Fig. 4B and SI Appendix, Fig. S5A). Interestingly, we observed that M. davidii IL-1β possessed the least capacity for cleavage and P. alecto the highest, in contrast to their respective caspase-1 activity (highest in M. davidii and lowest in P. alecto). We hypothesized that efficient targeting of MdIL-1β for reduced cleavage may occur at or near its cleavage site. Therefore, we aligned the IL-1β amino acid sequence of M. davidii against eight other species of bats (Myotis lucifugus, Eptesicus fuscus, Miniopterus natalensis, Desmodus rotundus, P. alecto, and P. vampyrus, R. aegyptiacus, Hipposideros armiger) and six other model mammalian species (Sus scrofa, Canis lupus, Pan troglodytes, Mus musculus, and Rattus norvegicus) (SI Appendix, Fig. S5B). Among the 15 total species analyzed using branch site modeling in PAML, we discovered a Ser117 residue immediately adjacent to the cleavage site Asp115-Ala116 which was under higher selection in the Myotis branch, which was a proline residue in all except three other species (M. natalensis, S. scrofa, and C. lupus).
We next hypothesized that residues conserved between Homo sapiens, P. alecto, and E. spelaea but differing in M. davidii may be responsible for the impaired cleavability of MdIL-1β. We thus additionally identified G110, S111, E113, and Q122 in MdIL-1β as distinct from equivalent IL-1β residues of the other three species. For a direct comparison of MdIL-1β with PaIL-1β (which is fully cleavable), we performed site-directed mutagenesis replacing each of the respective MdIL-1β residues into their P. alecto counterparts, along with a combined mutation containing all amino acid site substitutions (Fig. 4C). Notably, the MdIL-1β mutant S117P was able to strongly restore IL-1β cleavage, along with a partial restoration by a double mutant GS110/111DG; however, the combined mutant demonstrated strongest cleavage ability (Md > Pa 110 to 112) (Fig. 4D). Conversely, the PaIL-1β mutants DG110/111GS and P117S resulted in defective IL-1β cleavage (Fig. 4E). Further, both residues appear to have a partial effect either in restoration (MdIL-1β) or abrogation (PaIL-1β) of function, whereby the effect was strongest in the combined mutants. Thus, we observe that given a certain level of caspase-1 activity in any of the bat species (P. alecto low, E. spelaea medium, and M. davidii high), the cleavage potential of IL-1β occurs in opposite direction (P. alecto high, E. spelaea medium, and M. davidii low) (Fig. 4F). Taken together, this demonstrates a complementary mechanism whereby full caspase-1 activity is balanced by diminished cleavage potential of IL-1β, and vice versa, resulting in an overall, equivalent dampening of inflammasome signaling across multiple bat species from both suborders.
Discussion
We have confirmed that loss of AIM2/ALRs in bats results in inactive initiation of the inflammasome cascade in response to cytosolic DNA in vitro. Crucially, we reveal another layer of dampening through bat caspase-1, the principal cysteine protease responsible for cleaving inflammatory cytokines such as IL-1β and IL-18. We identified two inactivating alterations N365D and Q371R localized in the p10 sequence of wild-type P. alecto bat caspase-1 which when rescued by substitution of the human residues at equivalent sites, restored caspase-1 enzyme functionality. Simultaneously, when human caspase-1 was replaced with the equivalent bat amino acids, either residue change resulted in abrogation of IL-1β cleavage. Despite retention of caspase-1 activity in other bat species, corresponding reduction in IL-1β cleavage mitigated downstream inflammasome signaling. Thus, we have experimentally validated two additional residues of mammalian caspase-1, which are integral to its activity for substrate maturation, and demonstrated a proof of concept whereby downstream inflammasome activation in bats is dampened through a unique inverse relationship involving bat caspase-1 and IL-1β.
The discovery of dampened caspase-1 in bats has particular significance in their response to infection and immunity. As the classical inflammatory effector of the inflammasome complex, caspase-1 is converged upon by multiple upstream sensors, including NLRP3, NLRP1, AIM2, NLRC4, and others (43–47). These sensors are activated by a diverse array of cell- and pathogen-derived stimuli, including viral and bacterial nucleic acids, flagellin, ATP, and MSU crystals, and reactive oxidative species (ROS). Such signals trigger a systemic activation of alert and defense mechanisms, including pyroptosis, cytokine signaling, and the recruitment of neutrophils and macrophages into the affected tissue area (17, 48, 49). As caspase-1 cleaves cGMP-AMP (cGAMP) synthase (cGAS) to enhance host resistance to DNA viruses, and MAVS and TRIF to abolish IFN signaling (50), it is possible that bats may have evolved other compensatory mechanisms to resist viral pathogenesis by biasing cross-regulation of these pathways (7). Indeed, there is increasing evidence that AIM2 and other inflammasomes oppose type I IFN sensors, including cGAS, STING, and MyD88/IRF7 (51–54). As such, constitutive IFN expression in bats may, in part, be both an outcome and compensatory mechanism of inflammasome dampening, allowing them to mitigate viral pathogenesis. Further, as noncanonical caspase-1 substrates range from cytoskeletal components, enzymes in cell metabolism, and diverse other proteins involved in cellular stress responses and cell death pathway, it is possible that caspase-1 may retain residual baseline activity for regulation of these processes, thus conserving its expression in bats albeit at reduced function. Thus the effect of our findings on these nonimmune substrates would also warrant investigation.
It is notable that both identified caspase-1 residues acted on by high positive selection pressures possess noncharged (Asn, Gln) instead of charged (Asp, Arg) side chains, representing a substantial decrease in capacity for ionic bond formation. Both amino acids are situated within the p10-p10 interface of the caspase-1 (p20p10)2 homodimer and are shown to participate in critical electrostatic interactions across the interface of the active conformation of caspase-1 (SI Appendix, Fig. S4). Loss of these charged residues likely results in weakened caspase-1 dimerization, preventing robust autoprocessing and substrate cleavage. To our knowledge, no prior study has identified either D365 and R371 in α6 as essential for caspase-1 activity in humans or other mammalian species, with early structural studies reporting the interface to only consist of residues 318 to 322 and 386 to 396 (35). Two noncompetitive inhibitors discovered decades earlier, gold thiomalate and auranofin, closely mimic this interface-disruptive mechanism but differ in residue specificity (35). While most widely used caspase-1 inhibitors to date utilize active site-specific mechanisms to impede function (18, 32, 35, 55–57), none of the marketable inhibitors yet allosterically perturb caspase-1 at the p10 dimer–dimer interface despite efforts (58, 59). Thus, our findings may prove to facilitate additional specificity in inhibitor design. Importantly, our findings provide evidence of evolutionary drivers of inhibitory mechanisms in nature informing insight into human caspase-1 and suggest another potential alternative strategy for caspase-1 targeting in human therapeutics.
Positive selection pressure occurred in both caspase-1 residues of the Pteropus bat genus but not in any other bat species. We observed retention of caspase-1 activity by E. spelaea and M. davidii bats from both bat suborders Yinpterochiroptera and Yangochiroptera, which may be explained by a lack of D365 and R371 substitutions in caspase-1 of both bats. Intriguingly, further downstream investigation for IL-1β maturation elucidated an inverse relationship between bat caspase-1 activity and IL-1β cleavage potential, whereby the cleavage of IL-1β within a bat species occurs in the opposite direction to its caspase-1 activity (e.g., in M. davidii, caspase-1 high and IL-1β low). The mechanism by which MdIL-1β cleavage is diminished was shown to occur through the S117 site immediately adjacent to the 115Asp-Ala116 cleavage site, which when restored by mutation to S117P permitted cleavage of the MdIL-1β mutant into its 17-kDa fragment. This was followed to a smaller extent by the double mutant GS110-111DG of MdIL-1β, suggesting that all three residues might play a role in dampening maturation of the cytokine in Myotis bats. We demonstrate that by targeting either caspase-1 or IL-1β, bat species of both suborders possess dampening of important downstream inflammasome components.
Therefore, it is clear that differing strategies have been co-opted by bats to dampen either caspase-1 activation, IL-1β cleavage, or both in a complementary manner, highlighting the importance of this phenotype across bats. Indeed, previous genomic or functional studies have found that different bat species exhibited varying or independent genomic strategies to dampen the AIM2 or NLRP3 inflammasome sensors, which culminated in an equivalent level of loss or reduction of activity (9, 11). However, cleavage of IL-1β alone is not sufficient to generate biological inflammasome functioning, and recent studies have demonstrated requirement of GSDMD for IL-1β and IL-18 secretion and activation of pyroptotic cell death (60–62). Thus, in bat species with intact caspase-1 activity, other significant functions may be retained such as GSDMD-mediated pyroptosis and secretion of IL-18, or activation of IL-37 and inactivation of IL-33, which may confer higher regulatory control of pro/antiinflammatory responses (63–65). As such, multiple other indicators of inflammasome functioning in bats remain unknown and require further investigation in the context of these findings.
In conclusion, we find strong experimental evidence pointing to diminished multisensor inflammasome signaling in bats, suggesting high selection pressures acting not only on single, but multiple levels in this pathway. Given the inflammasome functions at the forefront of innate immune signaling, such alteration of inflammasome signaling in bats has a critical role in viral disease tolerance and asymptomaticity (66). Inflammasome activation has been implicated in multiple coronavirus infections, including MERS-CoV, SARS-CoV, and SARS-CoV-2 (67), possibly affecting the ability of bats to function as a reservoir host. With unique capacity for metabolically costly flight, bats could have adapted to elevated metabolic states by dispensing with this inflammatory arm (68–70). Further, inflammasome suppression improves longevity or prevents age-related decline in mice and promotes longevity in humans (71–75), which is in line with bats’ long-lived mammalian phenotype. Taken together, our study contributes significant mechanistic understanding for strategies targeting inflammasome dampening in bats, offers potential insight in regulation of human inflammation, and further elucidates the ability of bats to harbor and transmit zoonotic pathogens without sustaining detrimental costs of immune activation.
Materials and Methods
Reagents.
Reagents are as previously described (9). Ultrapure LPS-B5, CL264, and Hygromycin B Gold were obtained from InvivoGen. P. alecto ASC-specific monoclonal antibody (mouse IgG2b) was generated by GenScript’s monoclonal antibody service. Rabbit polyclonal anti-ASC (AL177) (human/mouse) was purchased from Adipogen. Goat polyclonal anti-dog IL-1β (ab193852) (cross-reactive to P. alecto) and rabbit polyclonal anti-mouse IL-1β (ab9722) were from Abcam. mAb to β-actin (A2228) was from Sigma-Aldrich and mAb to GFP and variants (including mCitrine) were from Roche (11814460001). Anti-mouse/rabbit/goat horseradish peroxidase (HRP)-conjugated secondary antibodies were from Santa Cruz.
Plasmids.
Generation of expression constructs for NLRP3, ASC-mPlum, IL-1β-HA, and empty vectors (control vectors) are as previously described (34, 76). Human AIM2 was cloned from human peripheral blood mononuclear cells (PBMC) cDNA using Q5 Polymerase (NEB) with AgeI and NotI flanking primers (NUS-IRB reference code H-18–029). AIM2 was digested and ligated into pQCXIH (Clontech) vector containing C-terminal mCitrine or 3× FLAG. Procaspase-1 was cloned from the human pCI-caspase-1 construct (Addgene plasmid 41552) or Omniscript (Qiagen)-generated cDNA of P. alecto spleen. Caspase-1 was inserted into pQCXIH-mCitrine and pQCXIH-3× FLAG vectors. P. alecto or human caspase-1 mutants were generated by overlap extension PCR with primers containing the respective mutations. Similarly, P. alecto, E. spelaea, and M. davidii IL-1β were cloned by PCR of bat spleen cDNA into pQCXIH (Clontech) backbones containing C-terminal HA-tag. P. alecto and M. davidii IL-1β mutants were also generated by overlap extension PCR. Gasdermin D was cloned from human PBMC cDNA and P. alecto spleen cDNA with 2× MYC-tag on the N terminus into pQCXIH (Clontech) backbones containing C- terminal HA-tag. Primer sequences are listed in SI Appendix, Tables S1 and S2. All constructs were prepared with endotoxin-free plasmid maxi-prep kits (Omega Bio-tek).
Cells.
All procedures utilizing animal samples in this study were performed in compliance with all relevant ethical regulations. Capturing and processing of bats (P. alecto) in Australia was approved by the Queensland Animal Science Precinct and University of Queensland Animal Ethics Committee (AEC#SVS/073/16/USGMS) and the Australian Animal Health Laboratory Animal Ethics Committee (AEC#1389 and AEC#1557). Where possible, wild bats with irreparable physical damage (torn wings) already scheduled to be killed were utilized. Processing of bats has been described previously (9, 69). Wild-type C57BL/6 mice were obtained with permission from the Singhealth institutional animal care and use committee. Harvesting and differentiation of bone marrow from P. alecto bats has been described previously and performed according to identical protocols (76, 77). Mouse bone marrow was harvested from C57BL/6 mice and frozen once in liquid nitrogen, thawed, and differentiated over 7 d in 10 ng/mLmacrophage colony-stimulating factor (M-CSF) as described previously (34). GP2-293 retroviral packaging cells were obtained from Clontech. GP2-293, HEK293T, and PaKiS cells were cultured in Dulbecco's modified Eagle medium (DMEM) (Gibco 11965092) medium supplemented with 10% fetal bovine serum (FBS). PaKiS (CVCL_YM14) is a spontaneous-immortalized kidney epithelial cell line with identical origin from the parental primary cells of PakiT03 (RRID: CVCL_DR89) (34). GP2–293 cell culture medium was supplemented with sodium pyruvate and nonessential amino acid (NEAA) cell culture supplement (Life Technologies) during retroviral packaging.
Reconstitution of AIM2 in Bat Macrophages.
Retrovirus was generated by cotransfecting pVSV-G envelope protein with the plasmid containing the gene of interest (AIM2-mCitrine or mCitrine-only) at 1:1 ratio in GP293 cells grown at 70% confluency. Cells were incubated for 48 to 72 h in DMEM containing 10% FBS at 37 °C, and supernatant centrifuged and filtered through 0.45-μm PVDF sterile filters (Millipore). To further concentrate the retrovirus, either 100,000 MW Vivaspin columns (Sartorius) in a benchtop centrifuge or ultracentrifugation at 125,000 × g for 90 min (Optima X, Beckman Coulter) in a SW41-TI rotor was performed to 75 to 100× dilution. Retrovirus was titrated on HEK293T cells and added at multiplicity of infection 5 into PaBMDM media at day 5 of differentiation. Cells were incubated for 48 h, supernatant was removed, and cells were recovered in additional 24 h of PaCSF-1 RPMI with 10% FBS before treatment.
In Vitro dsDNA Stimulation.
P. alecto immortalized kidney cells have been described previously (9). Cells stably expressing P. alecto ASC were transduced with mCitrine-only or AIM2-mCitrine retrovirus generated from GP2-293 cells. Cells were selected with puromycin for 5 d and recovered in 10% FBS DMEM media. AIM2-mCitrine/ASC double-positive cells were sorted by fluorescence activated cell sorting (FACS) using BD FACSAria for medium-to-low expression. Cells were grown to 70% confluency and transfected with increasing doses of PolydA:dT (Invivogen). For BMDM stimulation, differentiated cells were primed with 1 μg/mL CL264 (PaBMDM) or LPS B5 (MmBMDM) (Invivogen) for 3 h, washed with FBS-free RPMI (Gibco), and transfected with 1 mg/mL PolydA:dT using Lipofectamine 3000 (Thermo Fisher) in RPMI for 4 h. Supernatant was collected for LDH assay, IL-1β ELISA, and cells were stained for flow cytometry by Imagestream of FACS.
Imagestream Imaging Flow Cytometry.
Cells were harvested for Imagestream imaging flow cytometry as previously described (9). Briefly, BMDM cells were harvested with 5 mM ethylenediaminetetraacetic acid (EDTA), washed once with phosphate-buffered saline (PBS), fixed in 4% paraformaldehyde, and permeabilized with 0.3% Triton-X + 2% FBS for 10 min at 4 °C. Cells were stained with primary ASC and prelabeled fluorescent anti-mouse antibodies for 1 h at room temperature, with DAPI for 15 min, and then washed twice with PBS and resuspended in FACS buffer. PaKiS were harvested by trypsinization and resuspended in FACS buffer directly. Events on the Imagestream X were acquired using using INSPIRE software on an Amnis ImageStreamX Mk II imaging flow cytometer using 40× magnification. At least 5,000 events were acquired per sample and analyzed with the inbuilt IDEAS software. Cells in focus were gated by brightfield r.m.s. values, single cells by aspect ratio by area values, and intact nuclei using DAPI staining. Double positive cells (AIM2-mCitrine, ASC-mPlum) were gated and analyzed for ASC speck formation plotted via mean fluorescence intensity by max-pixel intensity.
Confocal Microscopy.
PaKiS cells were seeded into 24-well plates containing coverslips (#1.5 thickness). Cells stably expressing P. alecto ASC-mPlum were transiently transfected with human AIM2-mCitrine for 4 h, washed, and incubated for 48 h. Cells were stained for 30 min at 37 °C incubation with working concentration of Mitotracker according to manufacturer’s instructions (Thermo Fisher). Mitotracker probe solution was removed and cells were washed with PBS 2× before fixing with 4% paraformaldehyde. Nuclear staining was performed with DAPI. Coverslips were mounted onto glass slides with Mowiol 4.88 and images acquired on a Leica TCS SP8 machine at 100× resolution. Images were processed using ImageJ 2.0.0 software.
IL-1β ELISA and LDH Release Assay.
Supernatant collected from DNA-treated PaBMDMs was centrifuged to remove debris and frozen once at −80 °C. The supernatant was then measured by a sandwich ELISA protocol as previously described (9). Briefly, purified recombinant PaIL-1β protein was utilized for the standard curve, with goat anti-canine IL-1β primary antibody and rabbit anti-mouse IL-1β antibody used as capture and detection antibodies, respectively. IL-1β in mBMDM supernatants was detected using the BioLegend IL-1β Standard ELISA kit. LDH release assay was performed as previously described using a Cytotoxicity Detection Kit PLUS (LDH) from Roche (9). Calculations were performed as per manufacter’s instructions, with low and high controls included for normalization of individual biological replicates.
Evolutionary Analysis of Mammalian Caspase-1 and IL-1β.
Caspase-1 coding sequences (CDSs) were retrieved from National Center for Biotechnology Information (NCBI) for one armadillo (Dasypus novemcinctus) and many Boreoeutheria species, including Euarchontoglires and Laurasiatheria. Euarchontoglires species include two primates (human and Pan troglodytes), two rodents (rat and mouse), and one tree shrew (Chinese tree shrew, Tupaia belangeri chinensis) (SI Appendix, Table S3). Homologs of caspase-1 in the 15 bat genomes were identified by discontiguous MegaBLAST (BLAST + 2.7.1) with max e-value of 1e-5 and word size of 11. Similarly, IL-1β sequences for M. davidii and eight other species of bats (M. lucifugus, E. fuscus, M. natalensis, D. rotundus, P. alecto, and P. vampyrus, R. aegyptiacus, and H. armiger) and six other model mammalian species (S. scrofa, C. lupus, P. troglodytes, M. musculus, R. norvegicus) were retrieved from NCBI or PCR-cloning and gene sequencing performed on bat cDNA (SI Appendix, Table S4). Alignment of the CDSs was generated by MAFFT (78) and used to plot the phylogeny tree by the maximum-likelihood method with the general-time-reversible (GTR) model and 1,000 bootstrap replicates in PHYML 3.0 software (38). The phylogeny tree and alignment file then served as input for performance of positive selection analysis on CodeML from the PAML package (version 4.9) (79), and branch-site models with relevant branches were marked on the tree. LRTs were performed in different substitution models, including 1) M0 (one‐ratio), M1a (nearly neutral), M2a (positive selection), M3 (discrete), M7 (beta), M8 (beta and ω > 1), and M8a (beta and ω = 1) in site mod; 2) M0 (one‐ratio) and two-ratio model assuming different ω for background and foreground branches in branch mode; and 3) positive selection along specified branches (model A) against a null model (model A null) that allows neutral evolution and negative selection for branch-site mode. Positive selection sites were scored by the Bayes empirical Bayes (BEB) method (80).
Caspase-1 Western Blot and FLICA Assay.
The AIM2 inflammasome axis (HsAIM2, HsASC, human caspase-1, and IL-1β) was reconstituted into HEK293T cells at increasing doses using Fugene 6 (Promega) at 3:1 ratio with total DNA. Cells seeded into 96-well plates (Corning) were incubated for 48 h posttransfection and lysed in lysis buffer (79). cOmplete ULTRA protease inhibitor mixture and PhosSTOP phosphatase inhibitors (Roche) were added to lysis buffer before use. Proteins were separated on 12 to 15% SDS/PAGE gels and transferred onto 0.45-μm polyvinylidene difluoride (PVDF) membrane with a Trans-Blot Turbo transfer system (Biorad). Membranes were blocked in 5% bovine serum albumin (BSA) for 1 h and stained with primary antibody followed by HRP-conjugated secondary antibody. Membranes were developed with Amersham ECL Prime Western blotting detection reagent (GE Healthcare) on a myECL Imager (Thermo Scientific). For FLICA detection, cells were trypsinized, washed once in PBS, and stained with 660-Caspase-1 FLICA substrate (Immunochemistry) with occasional agitation for 1 h. Cells were washed three times with cellular wash buffer (provided), resuspended in PBS with 2% BSA, and analyzed via flow cytometry (LSRFortessa Cell Analyzer, BD Biosciences). Live/dead gating was performed using DAPI and subgated for AIM2-mCitrine and ASC-mPlum positivity, before gated for FLICA-660 positive staining (SI Appendix, Fig. S6). At least 10,000 cell events were collected per replicate and independently analyzed on FlowJo.
Supplementary Material
Acknowledgments
This work was funded by the Singapore National Research Foundation (Grants NRF2012NRF-CRP001–056 to L.-F.W. and NRF2016NRF-NSFC002–013 to L.-F.W.), and a New Investigator’s Grant (to A.T.I.) from the National Medical Research Council of Singapore (NMRC/BNIG/2040/2015). We thank Crameri Research Consulting, J. Meers, H. Field, and Duke-NUS team members for help with collection of bat samples; D. Anderson and M. Wirawan for the critical reading of the manuscript; and P. Rozario for assistance with experiments. We thank A. Bertoletti and A. T. Tan for use of the Amnis ImageStream. We also acknowledge the facilities and technical assistance of the Advanced Bioimaging Core and Flow Cytometry Core at SingHealth Duke-NUS Academic Medical Centre.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2003352117/-/DCSupplemental.
Data Availability.
All study data are included in the article text and SI Appendix.
References
- 1.Li W. et al., Bats are natural reservoirs of SARS-like coronaviruses. Science 310, 676–679 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Drexler J. F. et al., Bats host major mammalian paramyxoviruses. Nat. Commun. 3, 796 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leroy E. M. et al., Fruit bats as reservoirs of Ebola virus. Nature 438, 575–576 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Zhou P. et al., A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olival K. J. et al., Host and viral traits predict zoonotic spillover from mammals. Nature 546, 646–650 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang G. et al., Comparative analysis of bat genomes provides insight into the evolution of flight and immunity. Science 339, 456–460 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou P. et al., Contraction of the type I IFN locus and unusual constitutive expression of IFN-α in bats. Proc. Natl. Acad. Sci. U.S.A. 113, 2696–2701 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kepler T. B. et al., Chiropteran types I and II interferon genes inferred from genome sequencing traces by a statistical gene-family assembler. BMC Genomics 11, 444 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ahn M. et al., Dampened NLRP3-mediated inflammation in bats and implications for a special viral reservoir host. Nat. Microbiol. 4, 789–799 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xie J. et al., Dampened STING-dependent interferon activation in bats. Cell Host Microbe 23, 297–301.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ahn M., Cui J., Irving A. T., Wang L.-F., Unique loss of the PYHIN gene family in bats amongst mammals: Implications for inflammasome sensing. Sci. Rep. 6, 21722 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cridland J. A. et al., The mammalian PYHIN gene family: Phylogeny, evolution and expression. BMC Evol. Biol. 12, 140 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeYoung K. L. et al., Cloning a novel member of the human interferon-inducible gene family associated with control of tumorigenicity in a model of human melanoma. Oncogene 15, 453–457 (1997). [DOI] [PubMed] [Google Scholar]
- 14.Fernandes-Alnemri T., Yu J.-W., Datta P., Wu J., Alnemri E. S., AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature 458, 509–513 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hornung V. et al., AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature 458, 514–518 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brunette R. L. et al., Extensive evolutionary and functional diversity among mammalian AIM2-like receptors. J. Exp. Med. 209, 1969–1983 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rathinam V. A. K. et al., The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat. Immunol. 11, 395–402 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thornberry N. A. et al., A novel heterodimeric cysteine protease is required for interleukin-1 β processing in monocytes. Nature 356, 768–774 (1992). [DOI] [PubMed] [Google Scholar]
- 19.Fantuzzi G., Dinarello C. A., Interleukin-18 and interleukin-1 β: Two cytokine substrates for ICE (caspase-1). J. Clin. Immunol. 19, 1–11 (1999). [DOI] [PubMed] [Google Scholar]
- 20.Shi J. et al., Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 526, 660–665 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Boucher D. et al., Caspase-1 self-cleavage is an intrinsic mechanism to terminate inflammasome activity. J. Exp. Med. 215, 827–840 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang K. et al., Structural mechanism for GSDMD targeting by autoprocessed caspases in pyroptosis. Cell 180, 941–955.e20 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Man S. M., Karki R., Kanneganti T.-D., AIM2 inflammasome in infection, cancer, and autoimmunity: Role in DNA sensing, inflammation, and innate immunity. Eur. J. Immunol. 46, 269–280 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang H. et al., AIM2 inflammasome is critical for influenza-induced lung injury and mortality. J. Immunol. 198, 4383–4393 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choubey D., Panchanathan R., Absent in melanoma 2 proteins in SLE. Clin. Immunol. 176, 42–48 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Denes A., Lopez-Castejon G., Brough D., Caspase-1: Is IL-1 just the tip of the ICEberg? Cell Death Dis. 3, e338 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsuchiya K. et al., Caspase-1 initiates apoptosis in the absence of gasdermin D. Nat. Commun. 10, 2091 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu J. et al., Inflammasome activation leads to Caspase-1-dependent mitochondrial damage and block of mitophagy. Proc. Natl. Acad. Sci. U.S.A. 111, 15514–15519 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suzuki T. et al., Differential regulation of caspase-1 activation, pyroptosis, and autophagy via Ipaf and ASC in Shigella-infected macrophages. PLoS Pathog. 3, e111 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nagar A., DeMarco R. A., Harton J. A., Inflammasome and caspase-1 activity characterization and evaluation: An imaging flow cytometer-based detection and assessment of inflammasome specks and caspase-1 activation. J. Immunol. 202, 1003–1015 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Broz P., Dixit V. M., Inflammasomes: Mechanism of assembly, regulation and signalling. Nat. Rev. Immunol. 16, 407–420 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Flores J. et al., Caspase-1 inhibition alleviates cognitive impairment and neuropathology in an Alzheimer’s disease mouse model. Nat. Commun. 9, 3916 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang X. M. et al., The highly selective caspase-1 inhibitor VX-765 provides additive protection against myocardial infarction in rat hearts when combined with a platelet inhibitor. J. Cardiovasc. Pharmacol. Ther. 22, 574–578 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crameri G. et al., Establishment, immortalisation and characterisation of pteropid bat cell lines. PLoS One 4, e8266 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilson K. P. et al., Structure and mechanism of interleukin-1 β converting enzyme. Nature 370, 270–275 (1994). [DOI] [PubMed] [Google Scholar]
- 36.Walker N. P. et al., Crystal structure of the cysteine protease interleukin-1 β-converting enzyme: A (p20/p10)2 homodimer. Cell 78, 343–352 (1994). [DOI] [PubMed] [Google Scholar]
- 37.Gu Y. et al., Interleukin-1 beta converting enzyme requires oligomerization for activity of processed forms in vivo. EMBO J. 14, 1923–1931 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang Z., PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Liu Z. et al., Caspase-1 engages full-length gasdermin D through two distinct interfaces that mediate caspase recruitment and substrate cleavage. Immunity 53, 106–114.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Datta D., McClendon C. L., Jacobson M. P., Wells J. A., Substrate and inhibitor-induced dimerization and cooperativity in caspase-1 but not caspase-3. J. Biol. Chem. 288, 9971–9981 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsagkogeorga G., Parker J., Stupka E., Cotton J. A., Rossiter S. J., Phylogenomic analyses elucidate the evolutionary relationships of bats. Curr. Biol. 23, 2262–2267 (2013). [DOI] [PubMed] [Google Scholar]
- 42.Hawkins J. A. et al., A metaanalysis of bat phylogenetics and positive selection based on genomes and transcriptomes from 18 species. Proc. Natl. Acad. Sci. U.S.A. 116, 11351–11360 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu H. B., Finlay B. B., The caspase-1 inflammasome: A pilot of innate immune responses. Cell Host Microbe 4, 198–208 (2008). [DOI] [PubMed] [Google Scholar]
- 44.Miao E. A. et al., Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1β via Ipaf. Nat. Immunol. 7, 569–575 (2006). [DOI] [PubMed] [Google Scholar]
- 45.Franchi L. et al., Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1β in salmonella-infected macrophages. Nat. Immunol. 7, 576–582 (2006). [DOI] [PubMed] [Google Scholar]
- 46.Moayeri M. et al., Inflammasome sensor Nlrp1b-dependent resistance to anthrax is mediated by caspase-1, IL-1 signaling and neutrophil recruitment. PLoS Pathog. 6, e1001222 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhong F. L. et al., Germline NLRP1 mutations cause skin inflammatory and cancer susceptibility syndromes via inflammasome activation. Cell 167, 187–202.e17 (2016). [DOI] [PubMed] [Google Scholar]
- 48.Ichinohe T., Lee H. K., Ogura Y., Flavell R., Iwasaki A., Inflammasome recognition of influenza virus is essential for adaptive immune responses. J. Exp. Med. 206, 79–87 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Inoue M., Williams K. L., Gunn M. D., Shinohara M. L., NLRP3 inflammasome induces chemotactic immune cell migration to the CNS in experimental autoimmune encephalomyelitis. Proc. Natl. Acad. Sci. U.S.A. 109, 10480–10485 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Y. et al., Inflammasome activation triggers caspase-1-mediated cleavage of cGAS to regulate responses to DNA virus infection. Immunity 46, 393–404 (2017). [DOI] [PubMed] [Google Scholar]
- 51.Corrales L. et al., Antagonism of the STING pathway via activation of the AIM2 inflammasome by intracellular DNA. J. Immunol. 196, 3191–3198 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nakaya Y., Lilue J., Stavrou S., Moran E. A., Ross S. R., AIM2‐like receptors positively and negatively regulate the interferon response induced by cytosolic DNA. mBio 8, 1–17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu X. et al., Inflammasome activation negatively regulates MyD88-IRF7 type I IFN signaling and anti-malaria immunity. Nat. Commun. 9, 4964 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mayer-Barber K. D., Yan B., Clash of the cytokine titans: Counter-regulation of interleukin-1 and type I interferon-mediated inflammatory responses. Cell. Mol. Immunol. 14, 22–35 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fletcher D. S. et al., A synthetic inhibitor of interleukin-1 β converting enzyme prevents endotoxin-induced interleukin-1 β production in vitro and in vivo. J. Interferon Cytokine Res. 15, 243–248 (1995). [DOI] [PubMed] [Google Scholar]
- 56.Wei Y. et al., The structures of caspases-1, -3, -7 and -8 reveal the basis for substrate and inhibitor selectivity. Chem. Biol. 7, 423–432 (2000). [DOI] [PubMed] [Google Scholar]
- 57.Shahripour A. B. et al., Structure-based design of caspase-1 inhibitor containing a diphenyl ether sulfonamide. Bioorg. Med. Chem. Lett. 11, 2779–2782 (2001). [DOI] [PubMed] [Google Scholar]
- 58.Hardy J. A., Lam J., Nguyen J. T., O’Brien T., Wells J. A., Discovery of an allosteric site in the caspases. Proc. Natl. Acad. Sci. U.S.A. 101, 12461–12466 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Scheer J. M., Romanowski M. J., Wells J. A., A common allosteric site and mechanism in caspases. Proc. Natl. Acad. Sci. U.S.A. 103, 7595–7600 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kanneganti A. et al., GSDMD is critical for autoinflammatory pathology in a mouse model of Familial Mediterranean Fever. J. Exp. Med. 215, 1519–1529 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xiao J. et al., Gasdermin D mediates the pathogenesis of neonatal-onset multisystem inflammatory disease in mice. PLoS Biol. 16, e3000047 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen K. W., Demarco B., Broz P., Beyond inflammasomes: Emerging function of gasdermins during apoptosis and NETosis. EMBO J. 39, e103397 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mantovani A., Dinarello C. A., Molgora M., Garlanda C., Interleukin-1 and related cytokines in the regulation of inflammation and immunity. Immunity 50, 778–795 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cayrol C., Girard J. P., The IL-1-like cytokine IL-33 is inactivated after maturation by caspase-1. Proc. Natl. Acad. Sci. U.S.A. 106, 9021–9026 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bulau A. M. et al., Role of caspase-1 in nuclear translocation of IL-37, release of the cytokine, and IL-37 inhibition of innate immune responses. Proc. Natl. Acad. Sci. U.S.A. 111, 2650–2655 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Swanepoel R. et al., Experimental inoculation of plants and animals with Ebola virus. Emerg. Infect. Dis. 2, 321–325 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tay M. Z., Poh C. M., Rénia L., MacAry P. A., Ng L. F. P., The trinity of COVID-19: Immunity, inflammation and intervention. Nat. Rev. Immunol. 20, 363–374 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shen Y.-Y. et al., Adaptive evolution of energy metabolism genes and the origin of flight in bats. Proc. Natl. Acad. Sci. U.S.A. 107, 8666–8671 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Costantini D., Lindecke O., Pētersons G., Voigt C. C., Migratory flight imposes oxidative stress in bats. Curr. Zool. 65, 147–153 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gutiérrez J. S. et al., Oxidative status and metabolic profile in a long-lived bird preparing for extreme endurance migration. Sci. Rep. 9, 17616 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Camell C. D. et al., Aging induces an Nlrp3 inflammasome-dependent expansion of adipose B cells that impairs metabolic homeostasis. Cell Metab. 30, 1024–1039.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Youm Y. H. et al., Canonical Nlrp3 inflammasome links systemic low-grade inflammation to functional decline in aging. Cell Metab. 18, 519–532 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Marín-Aguilar F. et al., NLRP3 inflammasome suppression improves longevity and prevents cardiac aging in male mice. Aging Cell 19, e13050 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Furman D. et al., Expression of specific inflammasome gene modules stratifies older individuals into two extreme clinical and immunological states. Nat. Med. 23, 174–184 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Christ A. et al., Western diet triggers NLRP3-dependent innate immune reprogramming. Cell 172, 162–175.e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Irving A. T. et al., Optimizing dissection, sample collection and cell isolation protocols for frugivorous bats. Methods Ecol. Evol. 11, 2041–210X.13325 (2019). [Google Scholar]
- 77.Zhou P. et al., Unlocking bat immunology: Establishment of Pteropus alecto bone marrow-derived dendritic cells and macrophages. Sci. Rep. 6, 38597 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Guindon S. et al., New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321 (2010). [DOI] [PubMed] [Google Scholar]
- 79.Sadler A. J., Latchoumanin O., Hawkes D., Mak J., Williams B. R. G., An antiviral response directed by PKR phosphorylation of the RNA helicase A. PLoS Pathog. 5, e1000311 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yang Z., Wong W. S. W., Nielsen R., Bayes empirical bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 22, 1107–1118 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article text and SI Appendix.