Significance
Precise manipulation of Hedgehog pathway activity holds great value for biological research and clinical applications, but pathway agonists amenable to engineering have been lacking. We selected a nanobody that potentially targets the conformational changes of the Hedgehog receptor Patched1 and demonstrated that this nanobody potently activates the pathway in vivo. This nanobody can serve as the basis for mechanistic studies of Hedgehog pathway activation and for potential therapeutic applications. Our method may further apply to the investigation of other related transporters in the Resistance-Nodulation-Division superfamily.
Keywords: hedgehog, cryo-EM, nanobody, Patched, transporter
Abstract
Activation of the Hedgehog pathway may have therapeutic value for improved bone healing, taste receptor cell regeneration, and alleviation of colitis or other conditions. Systemic pathway activation, however, may be detrimental, and agents amenable to tissue targeting for therapeutic application have been lacking. We have developed an agonist, a conformation-specific nanobody against the Hedgehog receptor Patched1 (PTCH1). This nanobody potently activates the Hedgehog pathway in vitro and in vivo by stabilizing an alternative conformation of a Patched1 “switch helix,” as revealed by our cryogenic electron microscopy structure. Nanobody-binding likely traps Patched in one stage of its transport cycle, thus preventing substrate movement through the Patched1 sterol conduit. Unlike the native Hedgehog ligand, this nanobody does not require lipid modifications for its activity, facilitating mechanistic studies of Hedgehog pathway activation and the engineering of pathway activating agents for therapeutic use. Our conformation-selective nanobody approach may be generally applicable to the study of other PTCH1 homologs.
Hedgehog signaling functions in embryonic tissue patterning and in postembryonic regulation of tissue homeostasis and regeneration. The postembryonic regenerative activities of the Hedgehog pathway clearly suggest potential therapeutic benefits of pathway activation. The only modality of pathway modulation tested clinically, however, is inhibition, with clear benefits for patients suffering from malignancies whose initiation and growth depend on pathway-activating mutations in the primary cells of the tumor, such as medulloblastoma (1) and basal cell carcinoma (2).
The apparent lack of clinical interest in pathway-activating therapies, despite availability of potent small-molecule pathway activators (3), is likely due to the expectation that such systemic treatments may cause overgrowth of mesenchyme and potential initiation or exacerbation of fibrosis in multiple organs (4, 5). These dangerous side effects might be avoided by restricting pathway activation to specific cell types.
A pathway agonist conjugated to targeting agents would fulfill this purpose, but the native Hedgehog protein is difficult to engineer for cell-type specificity. Mature Hedgehog protein contains two lipid modifications (6, 7), including a cholesteryl moiety on its carboxy-terminus (8) and a palmitoyl adduct on its amino terminus (9), which is especially critical for signaling activity. The requirement for lipid modification in signaling poses a challenge for large-scale production, storage, and further derivatization for tissue targeting. Other synthetic or genetically encoded peptides that could easily be conjugated to targeting agents are currently lacking.
Recent insights into the mechanism of Hedgehog signal transduction suggest a route to remedying this deficiency. The primary receptor for Hedgehog is Patched1 (PTCH1), which maintains pathway quiescence by suppressing Smoothened (SMO), a downstream G-protein–coupled receptor (GPCR)-like protein (10–12). When bound to Hedgehog, PTCH1 is inactivated, permitting SMO to become active and trigger downstream signaling events (12–15). Mechanistically, the activation of SMO requires binding of a sterol, likely entering the 7TM bundle from the inner leaflet of the plasma membrane (16–21). PTCH1 is proposed to prevent SMO activation by transporting sterols from the inner leaflet of the plasma membrane, thereby limiting SMO access to activating sterols (22–25). A hydrophobic conduit coursing through the PTCH1 extracellular domain is required for this transport activity (24), and Hedgehog blocks this conduit and inactivates PTCH1 by inserting its essential amino-terminal palmitoyl adduct (23–26).
Transporters typically act by moving through a repeated cycle of conformational changes. If PTCH1 transport function employs such a conformational cycle, an agent that preferentially binds and stabilizes a specific PTCH1 conformation would be expected to disrupt its conformational cycle and transport activity, thus permitting activation of SMO. Such an agent thus may serve as a pathway modulator that could make lipid modifications dispensable and may shed light on conformational changes that occur during the PTCH1 working cycle.
Results
Development of a Conformation-Specific Nanobody that Activates Pathway.
Nanobodies are single-chain antibody fragments (27) that have frequently been used to stabilize specific GPCR protein conformations (28) and are amenable to genetic engineering. We have chosen as a starting point a synthetic yeast display library to select for conformation-specific nanobodies against PTCH1 (29). To select conformation-specific nanobodies, we first introduced conformational bias in PTCH1 by altering three acidic residues buried within its transmembrane domain (D499N, D500N, and E1081Q, termed PTCH1-NNQ). These acidic residues, conserved within the Resistance-Nodulation-Division (RND) transporter family, are required for PTCH1 activity in sterol transport and SMO regulation (17, 24, 30) and are more generally proposed to drive conformational changes in RND transporters in response to cation influx (Fig. 1A). We thus reasoned that alteration of these residues in PTCH1 might affect the relative representation of its conformational states.
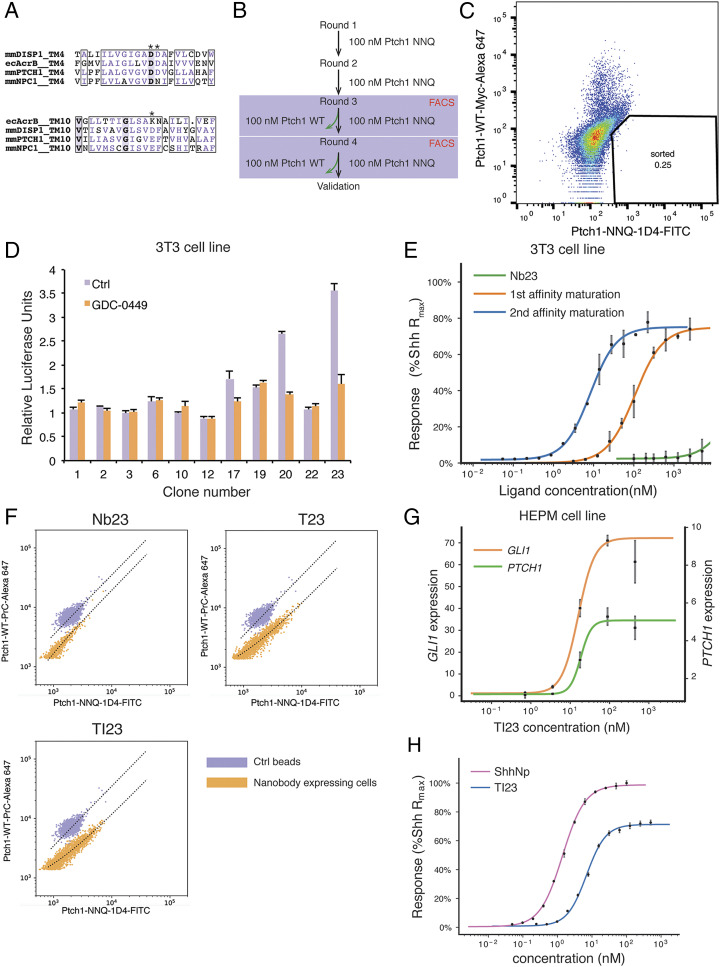
Fig. 1.
Selection of conformation-selective nanobodies. (A) Alignment of transmembrane 4 and 10 from different RND transporters. The charged residues are marked by asterisks. (B) Flowchart of the steps for nanobody selection. The yeast library was first enriched with magnetic-activated cell sorting for clones that bind to the PTCH1-NNQ variant, and then the population that prefers the NNQ variant was selected in FACS using PTCH1-NNQ and PTCH1-WT with different fluorescent labels. (C) Yeast cells stained with PTCH1-NNQ (fluorescein isothiocyanate [FITC] label) and PTCH1-WT (Alexa 647 label) are shown in the FACS plot. (Lower Right Quadrant) Cells that prefer the NNQ variant to the WT variant. Due to more nonspecific binding to the Alexa 647 fluorophore than to the FITC fluorophore, the double-positive population shifts toward the Upper Left Quadrant. (D) Nanobodies expressed and purified in E. coli were tested on Hedgehog-responsive 3T3 cells with a Gli-dependent luciferase reporter. GDC-0449, a pathway antagonist, is a control showing that nanobodies 17, 20, and 23 display weak activation in this assay. (E) Initial nanobody sequences of clones 17, 20, and 23 were mutagenized and selected in yeast display to obtain higher affinity clones (affinity maturation). After two rounds of affinity maturation, the new nanobody variant, named TI23, exhibited an EC50 of 8.6 nM in 3T3 cells, close to that of the native Hedgehog ligand. (F) The TI23 clone resulting from two rounds of affinity maturation showed a preference for binding to the PTCH1-NNQ variant. Yeast cells expressing Nb23, T23, or TI23 were incubated with a mixture of 1:1 protein C-tagged PTCH1-WT and 1D4-tagged PTCH1-NNQ proteins and then stained with antibodies against protein C tag or 1D4 tag. OneComp beads were used as a control for nonselective binding, as these beads bind to the constant region of the kappa chain and do not discriminate between different antibodies used for staining. (G) In the human mesenchymal cell line HEPM, TI23 activated Hedgehog response and induced transcription of the pathway targets GLI1 (EC50 = 16.0 nM) and PTCH1 (EC50 = 18.5 nM), as assayed by qPCR. (H) In NIH 3T3 cells, ShhNp and TI23 are titrated in a Gli-luciferase assay. EC50 for ShhNp and TI23 are determined to be 1.4 and 6.9 nM, respectively. Error bars represent SD, and all data points represent the mean of a triplicate.
We used purified PTCH1-NNQ variant protein for selection of nanobody clones from the yeast display library. After several rounds of enrichment for PTCH1-NNQ–binding yeast clones, we selected nanobodies that preferentially bind to PTCH1-NNQ versus wild-type PTCH1, using Fluorescence-Activated Cell Sorting (FACS) and wild-type and NNQ PTCH1 proteins labeled with antibodies coupled to different fluorophores (Fig. 1B). Yeast cells expressing preferentially bound nanobodies form a population off the diagonal of the FACS plot (Fig. 1C). After selecting nanobody-expressing yeast cells in the NNQ-preferring population, 15 unique clones were identified by sequencing, of which 3 were discarded because they bind directly to the antibody used during selection (SI Appendix, Fig. S1 A and B). As PTCH1 and PTCH1-NNQ differ only in the acidic residues in the transmembrane domain, differences in nanobody binding most likely derive from differences in conformational states between PTCH1 and PTCH1-NNQ.
Stabilization of a specific PTCH1 conformation would be expected to inactivate its transport activity and permit downstream response in the Hedgehog pathway. We therefore tested the activity of purified nanobody proteins on 3T3-Light2 cells, using a Gli-dependent luciferase assay. Clones 17, 20, and 23 showed weak activation effects (Fig. 1D). We enhanced signaling potency through two rounds of affinity maturation, first by selection from an error-prone PCR library (SI Appendix, Fig. S1 C and D) and then from a library targeting the complementarity-determining regions (CDRs) using one-pot mutagenesis (31) (SI Appendix, Fig. S1 E and F). The first round of affinity maturation yielded a series of nanobody clones deriving from clone 23 (“Nb23”), with H105R, G106R substitutions in CDR3 and several variant residues at G50 in CDR2. Among these variants, only the G50T substitution (named “T23”) could be expressed for purification from Escherichia coli. T23 showed better potency in Gli-dependent luciferase assays than its Nb23 parent (Fig. 1E) and was used as the starting sequence for a second round of affinity maturation, in which all CDR residues were systematically randomized in one-pot mutagenesis. After selection based on PTCH1 binding, Y102I in CDR3 was enriched, as well as T77N, an unintended substitution (SI Appendix, Fig. S1G). This variant, named “TI23,” was purified for further characterization, and it exhibited greater potency in pathway activation than its T23 parent (Fig. 1E). All of the nanobody variants showed preferential binding for PTCH1-NNQ, as revealed by two-color staining of yeast cells expressing these variants (Fig. 1F; also see SI Appendix, Fig. S1H). TI23 also strongly activated the human Hedgehog pathway targets GLI1 and PTCH1 at low nanomolar concentrations when tested in a cell line derived from human embryonic palatal mesenchymal (HEPM) (32) (Fig. 1G). In comparison with ShhNp, TI23 exhibited similar potency, but consistently lower efficacy. The maximum response induced by TI23 is ∼75% of that from ShhNp, suggesting that TI23 is a partial agonist (Fig. 1H).
Structure of the PTCH1::TI23 Complex.
To determine the conformational effects of TI23 binding to PTCH1, we prepared the PTCH1::TI23 complex for structure determination by cryogenic electron microscopy (cryo-EM). The complex was clearly visualized in cryo-EM micrographs (SI Appendix, Fig. S2A) with well-fitted contrast transfer function parameters (SI Appendix, Fig. S2B) and two-dimensional class averages (SI Appendix, Fig. S2C). Three-dimensional reconstruction of a cryo-EM dataset yielded a high quality map (Fig. 2A; procedure in SI Appendix, Fig. S2D) at a resolution of 3.4 Å (SI Appendix, Fig. S2F and Table S1). All 12 transmembrane (TM) helices and two major extracellular domains (ECDs) were resolved (Fig. 2A), and an atomic model of the PTCH1::TI23 complex was built based on this map and the previously determined murine PTCH1 structure (24). Most of the intracellular sequence was unresolved, and not modeled, except for two transverse helices preceding TM1 and TM7 (Fig. 2B).
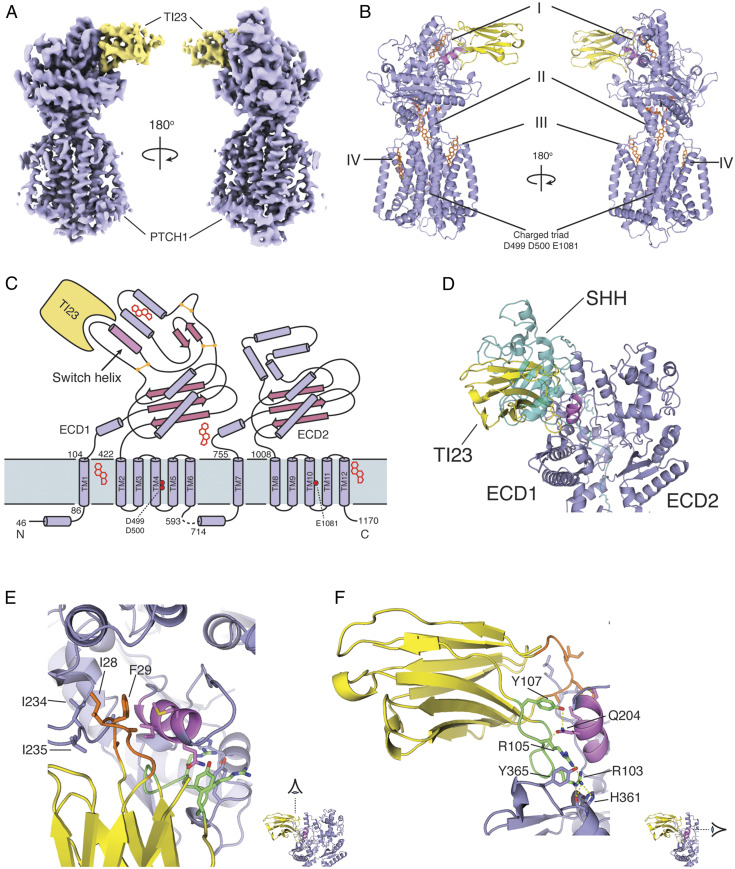
Fig. 2.
Overview of mouse PTCH1::TI23 complex structure. (A) The cryo-EM map of the PTCH1::TI23 complex shows clear features of the proteins. PTCH1, violet; TI23, yellow. (B) Protein model of the complex with PTCH1 and TI23 colored as in A. Lipid-like densities found in the map were modeled in sites I through IV. (C) Schematic view of PTCH1 showing the secondary structure elements and the relative positions of TI23 and the lipid-like densities. The key helix involved in the conformational change is highlighted as “switch helix.” (D) The binding site of TI23 on PTCH1 overlaps with that of SHH (teal). The switch helix, highlighted in violet, is sandwiched by CDR1 and CDR3 of TI23. (E and F) The interactions between TI23 CDRs and PTCH1 are shown in detail. CDR1, orange; CDR3, green; switch helix, violet. The hydrophobic interactions from CDR1 are viewed from above the membrane in E, whereas the hydrogen bond interactions from CDR3 are viewed from the ECD2 side of PTCH1 protein in F.
Sterol-like densities were identified in multiple sites: one in a pocket at the distal tip of ECD1 (farthest from the membrane, density I), one in the cavity proposed as part of the transport conduit (II), and two more at the periphery of the transmembrane domain (III and IV) (Fig. 2B). The density in site II is especially well resolved, and its unusual “Y” shape strongly suggests that it is glyco-diosgenin (GDN), the detergent used during sample preparation (SI Appendix, Fig. S3B). The other sterol-like densities are also most likely GDN, but only the steroidal moiety of GDN, digitogenin, was resolved and modeled.
The nanobody interacts only with ECD1 of PTCH1, as shown in the schematic drawing (Fig. 2C). The binding site of TI23 overlaps with that of SHH, but SHH interacts with both ECD1 and ECD2 (Fig. 2D). The CDR1 and CDR3 loops of the TI23 nanobody contact a short helix in the PTCH1 ECD1 (the “switch helix,” highlighted in Fig. 2C) from different angles. CDR1 interacts with PTCH1 by inserting the hydrophobic residues I28 and F29 into the hydrophobic pocket at lipid site I (Fig. 2E), whereas CDR3 primarily forms a hydrogen bond network with other residues on the surface of PTCH1 (Fig. 2F).
Although TI23 interacts exclusively with ECD1, we noted significant improvement in the resolution of side chains within the transmembrane domain. Of particular interest, the charged residue triad within TM4 and TM10 that was altered for selection of TI23 is better resolved than in most of the other published PTCH1 structures. We thus note that TM4 and TM10 in the PTCH1::TI23 complex associate with each other via a salt bridge between H1085 and D499, whereas, in the SHH-bound PTCH1 structure, this interaction is disrupted (SI Appendix, Fig. S3D). This nanobody-associated change in transmembrane domain side-chain interactions suggests potential allostery between the ECD and the transmembrane domain.
The overall structure of the PTCH1::TI23 complex is similar to the unbound murine PTCH1 structure, with a root mean square deviation of 0.955 Å of the Cα carbon atoms over 910 residues. Both ECD1 and ECD2 display some conformational differences in the complex. One minor difference is a rotation of ECD2 around its connection to the TM domain by ∼5° toward ECD1 as compared to PTCH1 alone (Fig. 3A). A more marked difference is the rotation by ∼32° of the distal end of the switch helix within ECD1 toward the membrane in a manner suggestive of a flipped switch (Fig. 3 A, Inset). We refer to conformations of the switch helix in PTCH1 alone and in the PTCH1::TI23 complex as poses 1 and 2, respectively (Fig. 3 A, Inset). These two alternative poses of the switch helix are present but have gone largely unremarked in other structures of PTCH1 determined under various conditions. For example, in the ternary complex of a single native Shh ligand bound to two human PTCH1 molecules (23), PTCH1 from chain A, the molecule whose sterol conduit is occluded by interaction with the N-terminal palmitoyl moiety of the SHH ligand, adopts pose 2, whereas PTCH1 from chain B adopts pose 1 (23). Indeed, in all published structures of PTCH1, the switch helix adopts one or the other of these two poses (22, 23, 25, 26, 33), suggesting that these poses represent discrete alternative conformations preferentially populated within the PTCH1 activity cycle (Fig. 3B). It is noteworthy that, in the best-resolved SHH-PTCH1 structure (33), the switch helix in the extracellular domain adopts pose 1 while the salt bridge between H1085 and D499 in the transmembrane domain is broken. The PTCH1::TI23 complex, in contrast, adopts the alternative conformation in both of these sites (SI Appendix, Fig. S3). These changes are consistent with allostery between the charged residues in the transmembrane domain and the switch helix in the extracellular domain. None of the other PTCH1 structures have clearly resolved side chains for the charged residues in TM4 and TM10, precluding further comparison.
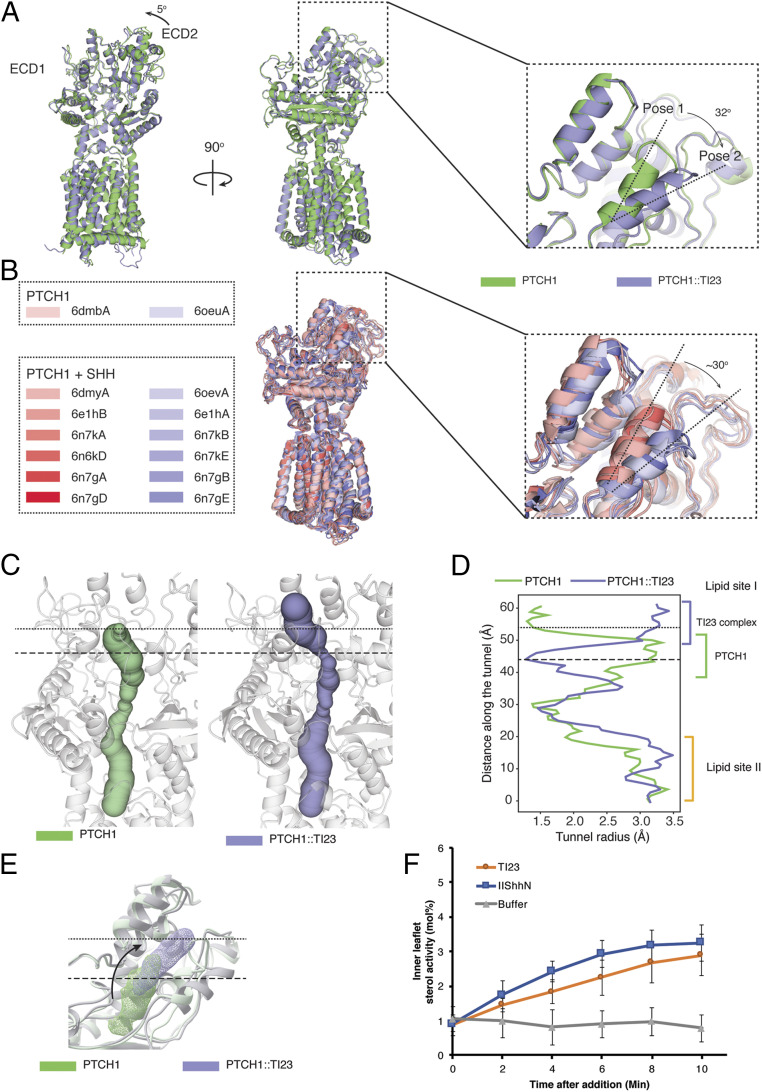
Fig. 3.
Conformational change induced by TI23. (A) Overlay of the structures of murine PTCH1 alone (PDB ID 6mg8) or in complex with TI23 shows two major changes in the extracellular domain. The extracellular domain 2 between TM7 and TM8 turns around 5°, pivoting on its connection to the transmembrane domain. A short helix (the switch helix) in extracellular domain 1 rotates ∼32° toward the membrane. The conformation in PTCH1 alone and in the complex is referred to as pose 1 and 2, respectively. (B) Other published PTCH1 structures also fall into pose 1 and 2 categories. In this overlay of other PTCH1 structures, pose 1-like structures are shown in shades of red, and pose 2-like structures in shades of blue. (C) The rotation of the switch helix alters the shape of the cavity within the extracellular domain. In the PTCH1 structure, the conduit is capped at the end, as indicated by the dotted line, whereas in the TI23-bound structure, the end of the conduit is wide open to the exterior and the lower part is throttled, as marked by the dashed line. (D) The radii at different points along the conduit are plotted here, with the altered parts marked with two horizontal lines. TI23 binding opens the upper end of the conduit but closes the lower part of lipid site I. (E) Position of the lipid-like density in site I changes with TI23 binding. The rotation of the switch helix may push the bound substrate outward while closing down the entry route. (F) In Ptch1−/− mouse embryonic fibroblasts transfected with PTCH1, plasma membrane inner leaflet (IPM) cholesterol activity increased immediately after adding purified TI23 or Hedgehog ligand (ShhN). Hedgehog ligand caused a slightly faster increase in IPM cholesterol activity, which plateaued after ∼6 min. This may reflect the difference in efficacy of these two ligands, as TI23 induces an ∼75% maximum pathway activity at saturating concentration in Gli-dependent luciferase assays. In the control conditions, cholesterol activity did not change over the period of the assay. At the end point (t = 10), cholesterol activity in the TI23 or ShhN group is significantly higher than that of the buffer-treated group (one-way ANOVA with Dunnet’s correction for multiple comparison, P < 0.0001). Error bars represent SD. For ShhN or TI23, n = 10. For buffer-only control, n = 5.
Effects of the Switch Helix on the Sterol Conduit.
These structural rearrangements alter the shape of the transport conduit as assessed by the Caver program (Fig. 3C). The region of the conduit encompassing sterol I in murine PTCH1 thus is seen to be dramatically constricted in the conduit of the PTCH1::TI23 complex, and the conduit in the PTCH1::TI23 complex also acquires a distal opening to the exterior (Fig. 3D). In parallel with this change in conduit shape, the bound sterol-like density shifts from a more proximal enclosed cavity to a more distal position with an opening to the exterior (Fig. 3E). This concerted proximal constriction and distal expansion results primarily from rotation of the switch helix. If PTCH1 activity is, like other RND family members, driven by a chemiosmotic gradient, the conformational change identified here may form part of a defined sequence that results in directional movement of substrates within the transport conduit.
The best-studied bacterial RND homolog, AcrB, also undergoes a series of conformational transitions that affect the substrate conduit, similar in principle although distinct in detail from that of PTCH1. By analysis with the Caver program, a lower and an upper site in AcrB open and close alternatively to enforce directional movement of substrates (SI Appendix, Fig. S4A) (34) whereas only a single upper site has been identified from PTCH1 structures (SI Appendix, Fig. S4B).
The TI23 nanobody appears to stabilize pose 2 of the PTCH1 switch helix. If PTCH1-mediated transport of sterols away from the inner leaflet indeed depend on the dynamic changes in the shape of the conduit associated with switch-helix movement, TI23 binding may lock PTCH1 in a state that is incompatible with sterol movement. To test this idea, we utilized a solvatochromic fluorescent sterol sensor, microinjected into cells to permit ratiometric measurement of sterol available for sensor binding within the inner leaflet of the membrane (35). This sensor previously revealed that available sterol decreases sharply with PTCH1 activity and that PTCH1 inactivation by Shh ligand causes a return to normal sterol availability (24). Similar to the effect of the Shh ligand addition, we noted that TI23 addition reversed the PTCH1-mediated reduction in cholesterol activity (Fig. 3F).
In Vivo Activation of the Hedgehog Pathway.
A small protein such as a nanobody (∼12 kDa) might be expected to display excellent tissue penetrance and be readily accessible to cells in most tissues. We tested the activity of TI23 by injecting mice intravenously with adeno-associated virus (AAV) engineered to express it. This experiment permits observation of biological effects elicited by sustained nanobody exposure as AAV infection is maintained over several weeks. We monitored lingual epithelium and skin, as these tissues display well-characterized responses to Hedgehog pathway activation (36–38).
The TI23 nanobody augmented Hedgehog pathway activity in the dorsal skin, as indicated by a six-fold increase in Gli1 RNA levels (Fig. 4A). The effect from TI23 is weaker than from ShhN or SAG21k, consistent with the observation that TI23 works as a partial agonist in vitro. We also noted expansion of hair follicles into the dermal adipose layer upon histological examination of dorsal skin in mice infected with AAV encoding TI23 or ShhN, but not a control nanobody (Nb4), indicating hair follicle entry into the anagen phase of the hair cycle (Fig. 4B). Consistent with accelerated entry into anagen, we noted faster hair regrowth on the dorsal skin after shaving (Fig. 4C).
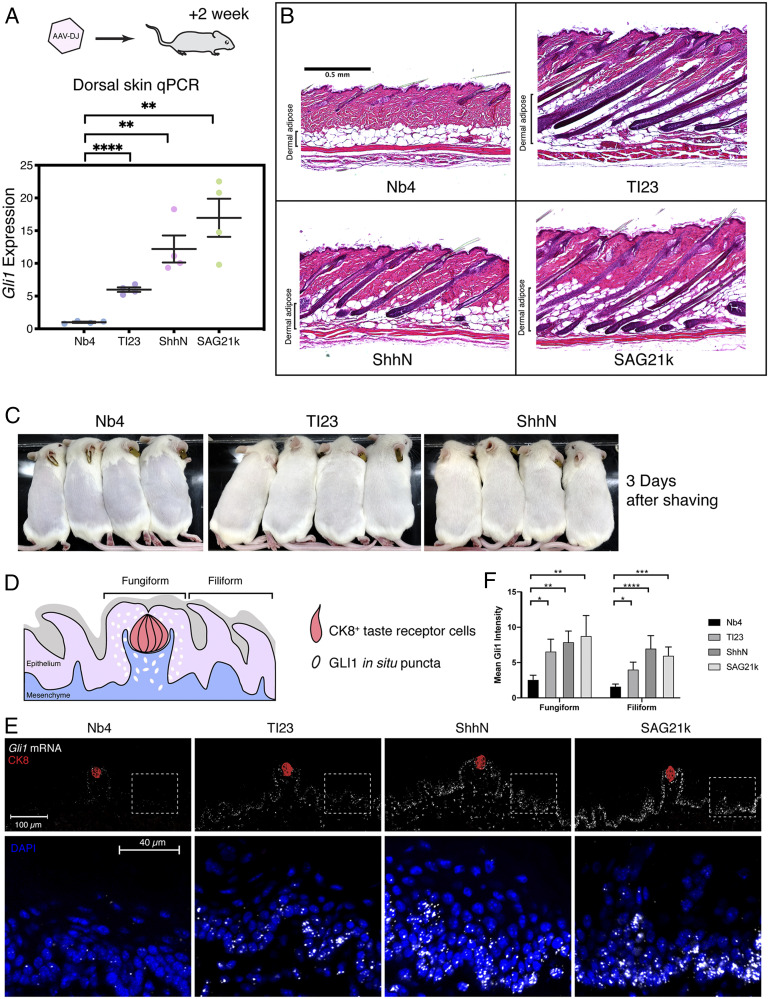
Fig. 4.
Validation of TI23 activity in the skin. (A) Mice were injected with AAV-DJ or treated with small-molecule SAG21k for 2 wk before collecting skin for histology analysis. Gli1 expression (relative to Hprt1) was activated in the dorsal skin of animals receiving TI23, ShhN, or the small-molecule SAG21k, suggesting that TI23 activated the Hedgehog pathway in the skin. Mean and SEM were plotted. (B) Histology of the dorsal skin suggests that hair follicles in the control group were in the quiescent telogen phase, whereas hair follicles grew and invaded the adipocyte layer in with TI23, ShhN, or SAG21k treatment, indicating induction of anagen. (C) Hair regrowth observed 2 wk after virus injection is much accelerated in TI23- or ShhN-treated animals as compared to the control group, suggesting that these hair follicles are in active anagen phase. (D) Schematic view of the dorsal tongue surface. The cells with active Hedgehog pathway response under physiological conditions are primarily located within the fungiform papillae. (E) TI23 induced Gli1 expression in lingual epithelial cells located in the fungiform and filiform papillae, as indicated by in situ hybridization using RNAScope. Tissues were collected from animals 2 wk after injection of AAV-DJ encoding the control nanobody (Nb4), TI23, or ShhN. With the pathway agonists TI23, ShhN, or the small-molecule SAG21k, the expression of Gli1 increased in both fungiform papillae containing taste receptor cells (Ck8+, red) and filiform papillae, as shown in the Insets. For each group, n = 4. (F) The mean fluorescence intensity of Gli1 is compared among regions of fungiform and filiform papillae. One-way ANOVA with Tukey’s multiple comparison suggests that TI23, ShhN, or SAG21k significantly increased Gli1 levels compared to the control conditions. *P < 0.05; **P < 0.005; ***P < 0.0005; ****P < 0.0001. For fungiform regions, n = 5, 3, 4, and 4 for Nb4, TI23, ShhN, and SAG21k, respectively. For filiform regions, n = 5, 4, 3, and 5 for Nb4, TI23, ShhN, and SAG21k, respectively.
We also examined Gli1 messenger RNA by fluorescence in situ hybridization as an indicator of pathway activation in lingual epithelium. Hedgehog pathway activity is limited to the cells surrounding the CK8+ taste receptor cells in untreated animals (Fig. 4D). In ShhN- or TI23-virus–injected mice, the range of Hedgehog pathway activity, as indicated by Gli1 expression, expanded dramatically as compared to the animals that received the control virus (Fig. 4 E and F). A similar expansion of Gli1 expression was also noted in mice given SAG21k (Fig. 4 E and F), a small-molecule Hedgehog agonist that activates SMO (3).
Discussion
The therapeutic applications of Hedgehog pathway modulation have focused primarily on pathway antagonists, and inhibition of the Hedgehog pathway has proven efficacious in the treatment of cancers driven by excessive Hedgehog pathway activity directly in primary cells of the tumor (1, 2). In contrast to promoting tumor growth, however, pathway activity recently has been found to suppress cancer growth and progression when it occurs in stromal cells rather than in primary cells (39), particularly in cancers of endodermal organs, such as bladder carcinoma and colon and pancreatic adenocarcinoma (4, 39–43). Pathway activation may also confer therapeutic benefits in regeneration of taste receptor cells of the tongue (37), which are often lost or diminished in chemotherapy patients, in protection or recovery from diseases such as colitis (43), reduction of tissue overgrowth in prostatic hypertrophy (44), or acceleration of bone healing in diabetes (45).
Despite these potential benefits, pathway activation in clinical settings is hindered by the lack of means to target specific tissues. Available Hedgehog pathway agonists are all hydrophobic in nature, including small-molecule members of the SAG family, certain oxysterols, and purmorphamine, all of which target SMO (46), and the lipid-modified Hedgehog protein or its derivatives, which target PTCH1. Our conformation-selective PTCH1-directed nanobody TI23 represents a class of potent, more hydrophilic agonists, which, unlike the native Hedgehog protein, does not require hydrophobic modification for activity. TI23 furthermore has the potential to be engineered for targeting by fusion to an antibody or other agent with tissue or cell-type specificity. These engineered variants may avoid pleiotropic effects from systemic pathway activation and be better suited for clinical applications.
TI23 not only is a promising candidate for further pharmaceutical development, but also provides insight into the PTCH1 transport mechanism. Directional movement of substrate through a transporter protein implies conformational change, but the identification of such conformational transitions for transporters is a nontrivial challenge. Our conformation-specific nanobody approach allowed us to identify two distinct conformations associated with poses 1 and 2 of the PTCH1 switch helix. The changes in shape of the transport conduit associated with these poses suggest peristaltic movement as a potential mechanism for directed substrate movement. As PTCH1 is distinct from the well-characterized RND transporter AcrB in both its preferred substrate and its extracellular domain structure (47), it is not surprising that the conformational transitions of these proteins differ. Indeed, given these differences, the apparent similarity in peristaltic movement of the substrate conduit in both proteins seems quite remarkable.
In principle, TI23 binding to PTCH1 would be expected to induce a conformational change similar to that of PTCH1-NNQ. As the altered residues in NNQ are buried in the transmembrane domain, whereas TI23 binds to the extracellular domain, the most parsimonious explanation is allostery between the two domains. In bacterial transporters, a charged triad in TM4 and TM10 conducts protons across the membrane to extract energy from a chemiosmotic gradient. In PTCH1, two distinct states of this triad of charged residues have now been observed. In the SHH-bound structure, D513 and E1095 are close to each other, and their negative charges may be stabilized by a bound cation, whereas in the TI23-bound structure, these two residues are far apart and most likely not interacting with any cations. This difference is consistent with the potential effect of NNQ alterations on cation binding, as the lack of charge neutralization in PTCH1-NNQ would be expected to greatly weaken the cation interaction.
One interesting aspect of our TI23 nanobody is that it works as a partial agonist, whereas the PTCH1-NNQ variant exhibits little activity in cells. One explanation for this difference may be that the nanobody may tolerate a small degree of conformational flexibility, thus permitting a low level of PTCH1 transport activity. Indeed, in the local resolution map, resolution of the nanobody region is much worse than the rest of the protein, suggesting substantial structural heterogeneity. Further in vitro evolution to improve structural stability of the nanobody may augment its efficacy to activate the pathway.
Our conformation-selective nanobody approach may be generalizable to the study of other transporters, in particular other members of the RND family. In mammals, this family includes the NPC1 cholesterol transport protein (48) and other PTCH-like proteins, such as PTCHD1, disruption of which is strongly associated with autism (49, 50). For other transporters, mutations that disrupt function may do so by biasing the normal conformational landscape without uniquely stabilizing any one conformation. Selection of nanobodies that preferentially bind such mutants may enable capture of sparsely populated yet critical conformations, expanding the repertoire of experimentally accessible states for structural and functional studies and providing pharmacologic agents with the potential to be targeted to specific cell types or tissue compartments.
Materials and Methods
Culture and treatment of NIH 3T3 fibroblasts, 293 cells, and other cell types is described in SI Appendix, Materials and Methods. Nanobody was selected from a yeast display library and then further mutagenized according to the procedure in SI Appendix, Materials and Methods. The evolved nanobody sequence is packaged into AAV for in vivo delivery. Virus production and analysis of target gene expression in tissues are provided in SI Appendix, Materials and Methods. Quantification of cellular cholesterol after nanobody treatment is given in SI Appendix, Materials and Methods. All procedures using animals were performed under Institutional Animal Care and Use Committee-approved protocol at Stanford University.
Supplementary Material
Acknowledgments
We thank D. Asarnow, L. Feng, and R. Mann for help and discussions during manuscript preparation and P. Lovelace, S. Weber, and the Stanford Stem Cell Institute FACS Core for their support of FACS cell sorting and analysis. Portions of this work were supported by NIH Grants R01GM102498 (to P.A.B.), DP5OD023048 (to A.M.), R01GM098672 (to Y.C.), and R35GM122530 (to W.C.). A.M. acknowledges support from the Pew Charitable Trusts.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2011560117/-/DCSupplemental.
Data Availability.
Structure data have been deposited in the Protein Data Bank (accession code 7K65) and Electron Microscope Data Bank (accession code EMD-22689) (51). All study data are included in the article and supporting information.
References
- 1.Rudin C. M., et al. , Treatment of medulloblastoma with hedgehog pathway inhibitor GDC-0449. N. Engl. J. Med. 361, 1173–1178 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Von Hoff D. D., et al. , Inhibition of the Hedgehog pathway in advanced basal-cell carcinoma. N. Engl. J. Med. 361, 1164–1172 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Brunton S. A., et al. , Potent agonists of the Hedgehog signaling pathway. Bioorg. Med. Chem. Lett. 19, 4308–4311 (2009). [DOI] [PubMed] [Google Scholar]
- 4.Lee J. J., et al. , Stromal response to Hedgehog signaling restrains pancreatic cancer progression. Proc. Natl. Acad. Sci. U.S.A. 111, E3091–E3100 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horn A., et al. , Hedgehog signaling controls fibroblast activation and tissue fibrosis in systemic sclerosis. Arthritis Rheum. 64, 2724–2733 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Taylor F. R., et al. , Enhanced potency of human Sonic hedgehog by hydrophobic modification. Biochemistry 40, 4359–4371 (2001). [DOI] [PubMed] [Google Scholar]
- 7.Mann R. K., Beachy P. A., Novel lipid modifications of secreted protein signals. Annu. Rev. Biochem. 73, 891–923 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Porter J. A., Young K. E., Beachy P. A., Cholesterol modification of hedgehog signaling proteins in animal development. Science 274, 255–259 (1996). [DOI] [PubMed] [Google Scholar]
- 9.Chamoun Z., et al. , Skinny hedgehog, an acyltransferase required for palmitoylation and activity of the hedgehog signal. Science 293, 2080–2084 (2001). [DOI] [PubMed] [Google Scholar]
- 10.Stone D. M., et al. , The tumour-suppressor gene patched encodes a candidate receptor for Sonic Hedgehog. Nature 384, 129–134 (1996). [DOI] [PubMed] [Google Scholar]
- 11.Ingham P. W., Taylor A. M., Nakano Y., Role of the Drosophila patched gene in positional signalling. Nature 353, 184–187 (1991). [DOI] [PubMed] [Google Scholar]
- 12.Chen Y., Struhl G., Dual roles for patched in sequestering and transducing Hedgehog. Cell 87, 553–563 (1996). [DOI] [PubMed] [Google Scholar]
- 13.Goodrich L. V., Milenković L., Higgins K. M., Scott M. P., Altered neural cell fates and medulloblastoma in mouse patched mutants. Science 277, 1109–1113 (1997). [DOI] [PubMed] [Google Scholar]
- 14.Fuse N., et al. , Sonic hedgehog protein signals not as a hydrolytic enzyme but as an apparent ligand for Patched. Proc. Natl. Acad. Sci. U.S.A. 96, 10992–10999 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ingham P. W., McMahon A. P., Hedgehog signaling in animal development: Paradigms and principles. Genes Dev. 15, 3059–3087 (2001). [DOI] [PubMed] [Google Scholar]
- 16.Cooper M. K., et al. , A defective response to Hedgehog signaling in disorders of cholesterol biosynthesis. Nat. Genet. 33, 508–513 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Myers B. R., Neahring L., Zhang Y., Roberts K. J., Beachy P. A., Rapid, direct activity assays for Smoothened reveal Hedgehog pathway regulation by membrane cholesterol and extracellular sodium. Proc. Natl. Acad. Sci. U.S.A. 114, E11141–E11150 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luchetti G., et al. , Cholesterol activates the G-protein coupled receptor Smoothened to promote Hedgehog signaling. eLife 5, e20304 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deshpande I., et al. , Smoothened stimulation by membrane sterols drives Hedgehog pathway activity. Nature 571, 284–288 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qi X., et al. , Cryo-EM structure of oxysterol-bound human Smoothened coupled to a heterotrimeric Gi. Nature 571, 279–283 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang P., et al. , Structural basis of Smoothened activation in Hedgehog signaling. Cell 174, 312–324.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gong X., et al. , Structural basis for the recognition of sonic hedgehog by human Patched1. Science 361, eaas8935 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Qi X., Schmiege P., Coutavas E., Li X., Two Patched molecules engage distinct sites on Hedgehog yielding a signaling-competent complex. Science 362, eaas8843 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y., et al. , Structural basis for cholesterol transport-like activity of the hedgehog receptor patched. Cell 175, 1352–1364.e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qian H., et al. , Inhibition of tetrameric Patched1 by sonic Hedgehog through an asymmetric paradigm. Nat. Commun. 10, 2320 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qi X., Schmiege P., Coutavas E., Wang J., Li X., Structures of human Patched and its complex with native palmitoylated sonic hedgehog. Nature 560, 128–132 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamers-Casterman C., et al. , Naturally occurring antibodies devoid of light chains. Nature 363, 446–448 (1993). [DOI] [PubMed] [Google Scholar]
- 28.Muyldermans S., Nanobodies: Natural single-domain antibodies. Annu. Rev. Biochem. 82, 775–797 (2013). [DOI] [PubMed] [Google Scholar]
- 29.McMahon C., et al. , Yeast surface display platform for rapid discovery of conformationally selective nanobodies. Nat. Struct. Mol. Biol. 25, 289–296 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taipale J., Cooper M. K., Maiti T., Beachy P. A., Patched acts catalytically to suppress the activity of Smoothened. Nature 418, 892–897 (2002). [DOI] [PubMed] [Google Scholar]
- 31.Wrenbeck E. E., et al. , Plasmid-based one-pot saturation mutagenesis. Nat. Methods 13, 928–930 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hwang R. F., et al. , Inhibition of the hedgehog pathway targets the tumor-associated stroma in pancreatic cancer. Mol. Cancer Res. 10, 1147–1157 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qi C., Di Minin G., Vercellino I., Wutz A., Korkhov V. M., Structural basis of sterol recognition by human hedgehog receptor PTCH1. Sci. Adv. 5, w6490 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seeger M. A., et al. , Structural asymmetry of AcrB trimer suggests a peristaltic pump mechanism. Science 313, 1295–1298 (2006). [DOI] [PubMed] [Google Scholar]
- 35.Liu S. L., et al. , Orthogonal lipid sensors identify transbilayer asymmetry of plasma membrane cholesterol. Nat. Chem. Biol. 13, 268–274 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paladini R. D., Saleh J., Qian C., Xu G. X., Rubin L. L., Modulation of hair growth with small molecule agonists of the hedgehog signaling pathway. J. Invest. Dermatol. 125, 638–646 (2005). [DOI] [PubMed] [Google Scholar]
- 37.Lu W. J., et al. , Neuronal delivery of Hedgehog directs spatial patterning of taste organ regeneration. Proc. Natl. Acad. Sci. U.S.A. 115, E200–E209 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Castillo-Azofeifa D., et al. , Sonic hedgehog from both nerves and epithelium is a key trophic factor for taste bud maintenance. Development 144, 3054–3065 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roberts K. J., Kershner A. M., Beachy P. A., The stromal niche for epithelial stem cells: A template for regeneration and a brake on malignancy. Cancer Cell 32, 404–410 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gerling M., et al. , Stromal Hedgehog signalling is downregulated in colon cancer and its restoration restrains tumour growth. Nat. Commun. 7, 12321 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rhim A. D., et al. , Stromal elements act to restrain, rather than support, pancreatic ductal adenocarcinoma. Cancer Cell 25, 735–747 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shin K., et al. , Hedgehog signaling restrains bladder cancer progression by eliciting stromal production of urothelial differentiation factors. Cancer Cell 26, 521–533 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee J. J., et al. , Control of inflammation by stromal Hedgehog pathway activation restrains colitis. Proc. Natl. Acad. Sci. U.S.A. 113, E7545–E7553 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lim A., Shin K., Zhao C., Kawano S., Beachy P. A., Spatially restricted Hedgehog signalling regulates HGF-induced branching of the adult prostate. Nat. Cell Biol. 16, 1135–1145 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tevlin R., et al. , Pharmacological rescue of diabetic skeletal stem cell niches. Sci. Transl. Med. 9, eaag2809 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carney T. J., Ingham P. W., Drugging Hedgehog: Signaling the pathway to translation. BMC Biol. 11, 37 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zwama M., Yamaguchi A., Molecular mechanisms of AcrB-mediated multidrug export. Res. Microbiol. 169, 372–383 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Carstea E. D., et al. , Niemann-pick C1 disease gene: Homology to mediators of cholesterol homeostasis. Science 277, 228–231 (1997). [DOI] [PubMed] [Google Scholar]
- 49.Noor A. et al.; Autism Genome Project Consortium , Disruption at the PTCHD1 locus on Xp22.11 in autism spectrum disorder and intellectual disability. Sci. Transl. Med. 2, 49ra68 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marshall C. R., et al. , Structural variation of chromosomes in autism spectrum disorder. Am. J. Hum. Genet. 82, 477–488 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y., et al. , Hedgehog receptor Patched (PTCH1) in complex with conformation selective nanobody TI23. Electron Microscopy Data Bank. https://www.ebi.ac.uk/pdbe/entry/emdb/ EMD-22689. Deposited 21 September 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Structure data have been deposited in the Protein Data Bank (accession code 7K65) and Electron Microscope Data Bank (accession code EMD-22689) (51). All study data are included in the article and supporting information.