Significance
Antibiotic resistance can spread from person to person, a phenomenon exemplified by the rapid global spread of novel resistance determinants. Antibiotic resistance can also “spill over” between family members or between a hospital and its surrounding community, such that antibiotic use in one population selects for resistance that is transmitted into the other population. In theory, antibiotic use in one population could spill over into its neighbors, making it difficult to understand to what degree a population’s own behavior, such as its antibiotic use, determines its level of resistance. Here we use theoretical modeling and observational data to quantify spillover, finding that even modest interactions between populations can lead to substantial sharing of resistance levels.
Keywords: antibiotic resistance, antibiotic stewardship, Streptococcus pneumoniae, Escherichia coli
Abstract
Antibiotic use is a key driver of antibiotic resistance. Understanding the quantitative association between antibiotic use and resulting resistance is important for predicting future rates of antibiotic resistance and for designing antibiotic stewardship policy. However, the use–resistance association is complicated by “spillover,” in which one population’s level of antibiotic use affects another population’s level of resistance via the transmission of bacteria between those populations. Spillover is known to have effects at the level of families and hospitals, but it is unclear if spillover is relevant at larger scales. We used mathematical modeling and analysis of observational data to address this question. First, we used dynamical models of antibiotic resistance to predict the effects of spillover. Whereas populations completely isolated from one another do not experience any spillover, we found that if even 1% of interactions are between populations, then spillover may have large consequences: The effect of a change in antibiotic use in one population on antibiotic resistance in that population could be reduced by as much as 50%. Then, we quantified spillover in observational antibiotic use and resistance data from US states and European countries for three pathogen–antibiotic combinations, finding that increased interactions between populations were associated with smaller differences in antibiotic resistance between those populations. Thus, spillover may have an important impact at the level of states and countries, which has ramifications for predicting the future of antibiotic resistance, designing antibiotic resistance stewardship policy, and interpreting stewardship interventions.
Antibiotic resistance is a major threat to public health (1). Outpatient antibiotic use, which accounts for ∼80% of human antibiotic use (2, 3), is considered a principal driver of antibiotic resistance in the community (4). Understanding the relationship between use and resistance is important because it allows accurate predictions of the future of antibiotic resistance and goal-oriented antibiotic stewardship policy. The use–resistance association has been previously characterized in many ecological studies at the level of US states (5–7) and European countries (8, 9). However, antibiotic resistance is a complex, temporally dynamic phenomenon (10–13), and many factors complicate the use–resistance association, making what should be an “obvious” connection sometimes difficult to identify and quantify (11). Even when detected, observed use–resistance associations are sometimes weaker than might be expected (7). One factor that could account for the difficulty in detecting use–resistance associations in ecological studies and for the apparent weakness of such associations is “spillover.”
Spillover is a consequence of the fact that antibiotic-resistant and -susceptible bacteria can be transmitted from person to person. Thus, one person’s risk of an antibiotic resistant infection depends on their own antibiotic use (14, 15) as well as the rates of antibiotic use among their contacts (16). For example, one person’s use of antibiotics increases the risk of an antibiotic-resistant infection among their family members (17–20). As another example, hospitalized patients with no recent antibiotic use can have a higher risk of resistance than people in the community with high antibiotic use (21) because antibiotic use and resistance in other hospitalized patients are high.
Spillover is important for three reasons. First, it means antibiotic resistance is not merely a localized problem. It is well understood that new resistance determinants can emerge in one geography and spread globally (22, 23), but the role of spillover in determining the levels of resistance in a given locale is not well quantified. To what degree, for example, can one US state expect that its antibiotic resistance levels are due to antibiotic use within its borders, rather than in surrounding states? Second, spillover makes it difficult to design antibiotic stewardship interventions and understand their results. For example, if antibiotic use in one hospital changes, resistance might not change as expected because of spillover from the community or other hospitals into that hospital’s patients. Finally, spillover makes it difficult to interpret the results of controlled antibiotic interventions, such as the effect of mass drug administration on antibiotic resistance (24, 25), when the intervention and control populations are not wholly epidemiologically separate.
The effect of spillover should scale with the amount of interaction between populations. If two populations do not interact at all, then antibiotic use in one population cannot affect resistance in the other. However, if two populations liberally exchange bacteria, then the rates of antibiotic resistance in the two populations will be very similar, regardless of whether their rates of antibiotic use differ greatly.
The effects of spillover also depend on population sizes. For example, two large populations will have most interactions within themselves, rather than between each other. Spillover should therefore be most pronounced when considering small populations and become less important for large populations. As mentioned above, a single individual’s risk of resistance is modulated by antibiotic use in their family or in their healthcare facility. Spillover is also observed at the level of hospitals, as the level of resistance in one hospital appears to be affected by resistance levels in nearby hospitals as well as by antibiotic use rates in the surrounding communities (26–28). Presumably, when examining ever larger populations, such as US Census tracts (29), US states, or European countries, the effect of spillover will become less important. However, the relationship between population size and spillover effects is not well understood.
We hypothesized that US states and European countries, which are large populations with relatively independent public health policies, may be subject to substantially lower levels of antibiotic resistance spillover than family- or hospital-sized populations. This hypothesis, if true, would mean that individual states or countries could act as independent “laboratories” of antibiotic use and resistance. If not, it means that outpatient antibiotic resistance policy must be national or international in order to achieve its full effect. To evaluate this hypothesis, we first use mathematical models of antibiotic use and resistance to make quantitative predictions about the effect of spillover between populations as a function of their amount of mutual interaction. Then, we search for signals of spillover in observational data of antibiotic use and resistance in US states and European countries.
Methods
Dynamical Models of Antibiotic Resistance.
To examine how interactions between populations could theoretically affect the association between antibiotic use and resistance, we used the within-host neutrality (WHN) mathematical model presented by Davies et al. (30) and described in SI Appendix, Supplemental Methods. Briefly, the model predicts the prevalence ρ of antibiotic resistance that results from an antibiotic use rate τ in a single, well-mixed population. To verify that conclusions drawn from the WHN model are not specific to the model structure, we also repeated all analyses with the “D-types” model of use and resistance (31). We selected these two models because they demonstrate coexistence between sensitive and resistant strains at equilibrium over a wide parameter space. Parameter values and simulation methodology for both models are in SI Appendix, Supplemental Methods. In the simulations, antibiotic use is measured as monthly treatments per capita, and resistance is measured as the proportion of colonized hosts carrying resistant strains.
To conceptually frame and clarify the question of spillover, we simulated an antibiotic stewardship intervention experiment using a structured host population approach inspired by Blanquart et al. (32). We considered pairs of an intervention population with antibiotic use rate τint and a control population with use rate τcont. To determine how spillover affects the intervention’s measured outcome, we modulated the proportion ε of each population’s contacts that support bacterial transmission that are in the other population. For ε = 0%, the populations are completely separate. For ε = 50%, contacts across populations are just as likely as contacts within populations (SI Appendix, Supplemental Methods). We varied ε between 0 and 50%, and we varied the difference in use Δτ = τcont – τint between 0 and 0.15 treatments per person per month while keeping the average use 0.5 × (τcont + τint) fixed at 0.125, reflecting the range of antibiotic use rates in the original model presentations. We quantified the use–resistance association between the two populations as Δρ/Δτ, the absolute difference in resistance divided by the difference in antibiotic use. Because the simulations suggested that Δρ/Δτ is a predictable function of the degree of population mixing, we considered only this functional form for the use–resistance association.
Spatial Antibiotic Use–Resistance Simulation.
To explore how spillover as simulated in the two-population model would manifest in cross-sectional use–resistance data from multiple populations, we ran a second simulation that incorporates the effects of varying interactions across a network of populations. We randomly placed 50 theoretical populations in a square grid, one length unit on a side, and randomly assigned each population an antibiotic use rate τ drawn from a uniform distribution. We then assigned each population i an antibiotic resistance prevalence ρi according to a weighted average of all populations’ use rates according to . The constant β is the slope of the use–resistance line; we use β = 1 for this simulation. The weights wij decline exponentially with the distance between populations: , where dij is the Euclidean distance between the two populations and d0 is a scaling parameter. For , there is no interaction between the populations, and each population’s resistance rate is determined by its own use rate. As d0 increases, the amount of spillover increases.
To evaluate spillover in this simulation, we measured the association between population pairs’ ranked interactions (closer distance means higher interaction) and use–resistance associations Δρ/Δτ using the nonparametric correlation (Spearman’s ρ). To quantify the effect of spillover, we compared the median use–resistance associations among the populations that are in the top and bottom deciles of ranked interactions.
Observational Data.
We examined antibiotic use and resistance for three pathogen-antibiotic combinations: Streptococcus pneumoniae and macrolides, S. pneumoniae and β-lactams, and Escherichia coli and quinolones. We considered these three combinations because they are the subject of many modeling (30, 31) and empirical studies (5, 14).
Observational data were drawn from three sources. First, we used MarketScan (33) and ResistanceOpen (34) as previously described (7). The MarketScan data include outpatient pharmacy antibiotic prescription claims for 62 million unique people during 2011 to 2014. ResistanceOpen includes antibiotic resistance data collected during 2012 to 2015 from 230 hospitals, laboratories, and surveillance units in 44 states. Second, we used the QuintilesIMS Xponent database (35) and the US Centers for Disease Control and Prevention’s National Healthcare Safety Network (NHSN) (36). The Xponent data includes state-level data on US quinolone use during 2011 to 2014. NHSN includes state-level data on quinolone resistance among E. coli catheter-associated urinary tract infections during 2011 to 2014. Third, we used the European Centre for Disease Prevention and Control’s (ECDC) European Surveillance of Antimicrobial Consumption Network (ESAC-Net) antimicrobial consumption database (37) and European Antimicrobial Resistance Surveillance Network (EARS-Net) Surveillance Atlas of Infectious Disease (38) for 2011 to 2015. The ESAC-Net data include country-level outpatient antibiotic use data provided by World Health Organization and ministries of health from member countries. The EARS-Net data include country-level resistance data. In the observational data, we quantified antibiotic use as yearly treatments per capita and resistance as the proportion of collected isolates that were nonsusceptible. We excluded the S. pneumoniae resistance to β-lactams in US states from the analysis because, in previous work using the same primary datasets, the point estimate for the use–resistance relationship was negative (7). Further details about preparation of these data sources and their availability are in SI Appendix, Supplemental Methods.
Use–Resistance Associations by Interactions.
To test the theoretical prediction that the same difference in antibiotic use will be associated with smaller differences in antibiotic resistance when two populations (US states or European countries) have stronger interactions, we tested whether population pairs’ use–resistance associations are inversely correlated with their interpopulation interactions as measured using transportation data. In other words, we tested whether stronger-interacting pairs have weaker use–resistance associations.
Similar to the approach in the simulations, we quantified the use–resistance association as the percentage point difference (i.e., absolute risk difference) in resistance (proportion of nonsusceptible isolates) divided by the difference in antibiotic use. We then quantified interactions using intercounty commuting statistics from the US Census (39) for US states and using intercountry passenger flight data from Eurostat (40) for European countries. Rather than trying to infer a precise mathematical relationship between transportation statistics and epidemiological contacts, we used a nonparametric approach: We assumed that pairs of populations with relatively little interpopulation transportation also have relatively few interpopulation contacts, but we infer only the rank ordering of interpopulation contacts, not their magnitudes. Specifically, we first obtained the matrix of the number of counts (workers in the commuting data and passengers in the flight data) from each population to every other population. The diagonal matrix entries are the number of workers who live and work in the same US state or the number of intracountry European flight passengers. We then converted this matrix of counts into a matrix of the proportion of counts moving from one population to another (i.e., divided each row by its sum), then symmetrized the resulting matrix by taking the elementwise average of the matrix and its transpose, and finally converted the resulting values into ranks. We assumed that intrapopulation interactions outnumber interpopulation interactions, and so we set diagonal entries, which represent within-population interactions, to the highest rank.
Like in the spatial simulation, we measured the association between ranked interactions and use–resistance associations using the nonparametric correlation (Spearman’s ρ). We computed CIs using the jackknife method to account for correlations between population pairs (e.g., the use–resistance relationship between populations A and B is not independent of the relationship between A and C). We tested for statistical significance using the Mantel test with 999 permutations. To quantify the effect of spillover, we used an approach similar to that in the spatial simulation, comparing the median use–resistance associations among the top and bottom deciles of ranked interactions.
In a separate analysis, we also tested whether the use–resistance association is weaker in adjacent pairs of populations, which presumably have more cross-population contacts, compared to nonadjacent populations (SI Appendix, Supplemental Methods).
Simulations and observational analyses were made using R (Version 3.6.0) (41). The Mantel test used the vegan package (42). Multiple hypotheses were accounted for using the Benjamini–Hochberg false discovery rate.
Results
In simulations of two populations, representing intervention and control groups, interactions between the two populations attenuated the effect of the intervention (Fig. 1). With increasing interaction strength, the same intervention, that is, the same difference in antibiotic use between the populations, was associated with a smaller difference in antibiotic resistance. The difference in resistance between populations increases with the difference in antibiotic use but decreases with increasing interaction strength. Thus, spillover between populations attenuates the measured use–resistance association.
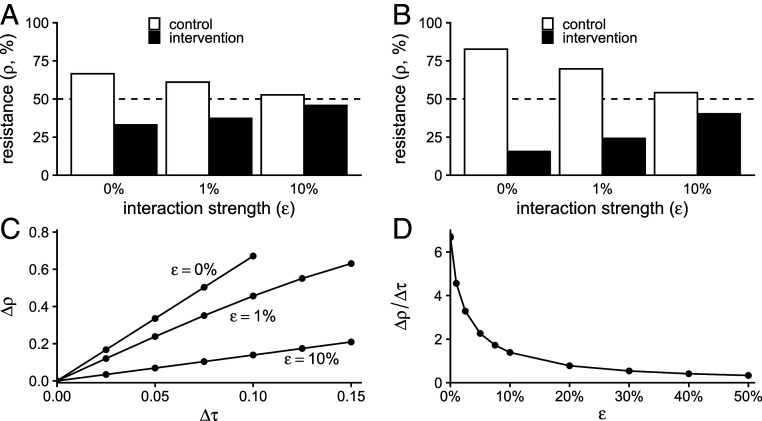
Fig. 1.
Simulated interactions between populations attenuate the effect of interventions. (A) Results of simulations of the two-population WHN model for a modest intervention (difference in antibiotic use between populations Δτ = 0.05 monthly treatments per capita; average of control and intervention treatment rates is 0.125). As interaction strength (ε, horizontal axis) increases, the difference in antibiotic resistance between the two populations decreases. The dashed line shows the resistance level in a population with a mean treatment rate of 0.125. (B) The same pattern holds for a stronger intervention (Δτ = 0.1, same average treatment rate). (C) The difference in resistance between populations (Δρ, vertical axis) increases with the difference in antibiotic use (Δτ, horizontal axis), but the rate of increase is lower with increasing interaction ε. (D) In the WHN model, the use–resistance relationship (Δρ/Δτ, vertical axis) declines exponentially with the interaction strength ε (Δτ fixed at 0.10).
The precise relationship between ε, the proportion of each population’s contacts that are in the other population, and the attenuation of the use–resistance association depended on the choice of mathematical model (SI Appendix, Fig. S1 and Table S1). For ε = 1%, the use–resistance declined by ∼30% in the WHN model and more than 60% in the D-types model. In other words, the models predict that as few as 1% of contacts need to be across populations, rather than within populations, to cause the observed effect of an antibiotic stewardship intervention to shrink by one-third, or even half, compared to an idealized situation with zero interaction between the populations.
In simulations of interacting multiple populations arranged randomly on a grid, increased interaction strength led to a weaker observed use–resistance association in terms of both the association’s slope and its variance (Fig. 2). Spillover could be detected as an inverse correlation between population pairs’ interaction strength and their use–resistance associations. For example, when the interaction strength was sufficiently strong (d0 = 0.075) to reduce the observed, cross-sectional use–resistance association by ∼50%, there was an inverse correlation (Spearman’s ρ = −0.17) between interaction rank and use–resistance associations, and population pairs in the highest decile of interactions had use–resistance associations 47% weaker than for those pairs in the lowest-interacting decile (SI Appendix, Table S2).
Fig. 2.
Simulated pairs of populations with stronger interactions have weaker use–resistance associations. (A) Each filled circle represents a simulated population. When spillover is weak (i.e., d0, measured in arbitrary length units, is small), populations fall along an idealized use–resistance association (dashed black line). As spillover is increased, the cross-sectional use–resistance association becomes weaker and has greater variance. Gray lines show the simple linear regression best fit. (B) Each open circle represents a pair of the populations shown in the corresponding facet in A. For amounts of spillover, all pairs have the same use–resistance association Δρ/Δτ (i.e., 1, the slope of the dashed black line in A). For stronger spillover, there is substantial variance in the associations. For nonzero spillover (d0 > 10−6), stronger-interacting pairs have weaker use–resistance associations (Spearman’s ρ; SI Appendix, Table S2). The red line shows the robust regression best fit. The vertical axis is truncated for visual clarity.
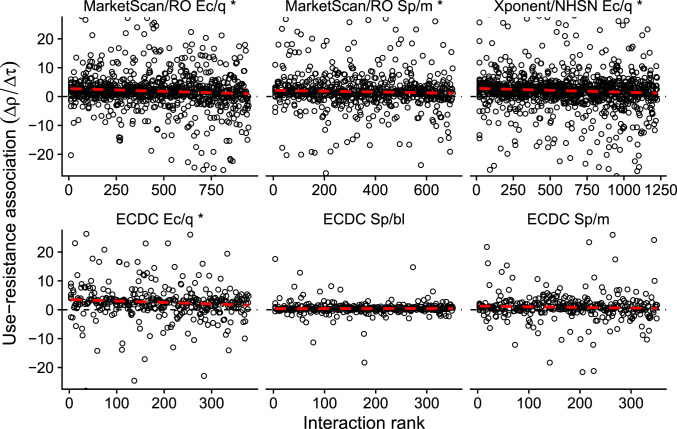
To test whether spillover is important at the scale of US states or European countries, we use empirical use–resistance associations between pairs of populations in six combinations of pathogen species, antibiotic class, and data source (Fig. 3). We reasoned that if spillover is relevant at these scales, then pairs of states or countries with stronger interactions would have detectably weaker use–resistance associations. Using a rank-ordered estimate of interactions drawn from US commuting and European airline passenger flows, we found that, in four of six dataset/pathogen/antibiotic combinations, the nonparametric association between increased interpopulation interactions and decreased use–resistance associations was statistically significant (i.e., a negative correlation; Fig. 4 and SI Appendix, Table S3). The weakest significant result was for S. pneumoniae and macrolides in the MarketScan/ResistanceOpen dataset (Spearman’s ρ = −0.07, 95% jackknife CI of −0.18 to 0.03; P = 0.028, Mantel test), and the strongest was for E. coli and quinolones in the Xponent/NHSN dataset (ρ = −0.13, 95% jackknife CI of −0.25 to −0.007; P = 0.001). The correlation for S. pneumoniae and macrolides in the ECDC data has a point estimate suggesting spillover but was not statistically significant, while the correlation for S. pneumoniae and β-lactams in the ECDC data had the opposite sign of that expected by the simulations. An analysis of use–resistance associations by populations’ physical adjacency did not yield statistically significant results (SI Appendix, Figs. S2 and S3 and Table S4).
Fig. 3.
Use–resistance relationships across US states and European countries. Each point represents antibiotic use and resistance in a US state (Top) or European country (Bottom). Lines show the simple linear regression best fit. Gray areas show the 95% CI. Ec/q: E. coli and quinolones. Sp/m: S. pneumoniae and macrolides. Sp/bl: S. pneumoniae and β-lactams. RO: ResistanceOpen.
Fig. 4.
Use–resistance associations by ranked interaction. Each point represents the use–resistance association in a pair of US states (Top) or European countries (Bottom), the same pairs as shown in Fig. 3, rank ordered by increasing interpopulation interaction as inferred from transportation data. For visual clarity, the horizontal axes are truncated to exclude outliers. The dashed red line is a visual illustration of how increasing interaction is correlated with decreasing use–resistance associations (robust regression; compare SI Appendix, Tables S3 and S4). The asterisk (*) indicates a statistically significant association between increased interaction and decreased use–resistance relationship (SI Appendix, Table S3). Ec/q: E. coli and quinolones. Sp/m: S. pneumoniae and macrolides. Sp/bl: S. pneumoniae and β-lactams. RO: ResistanceOpen.
Finally, to quantify the effect of increased interactions on the observed use–resistance associations, we compared the use–resistance associations in pairs of populations within the lowest decile of interactions against those in the highest decile, using the same approach as for the spatial simulation (SI Appendix, Table S5). In five of six dataset/pathogen/antibiotic combinations, the sign of the point estimate for the difference in use–resistance associations was consistent with spillover, with a weaker association among pairs of populations with greater interactions. In those five cases, the point estimates ranged from a 18% reduction up to a 75% reduction in use–resistance associations among the highest-interacting pairs of populations, compared to the lowest-interacting populations.
Discussion
We used theoretical models to show that interactions between two populations can attenuate the observed use–resistance association. In simulations, the quantitative relationship between interpopulation interactions and the attenuation of the use–resistance association was dependent on the theoretical model used. However, we found that, in two models of the use–resistance association, having on the order of 1% of interactions that can support bacterial transmission between a control and intervention population was sufficient to attenuate the observed effect of theoretical stewardship intervention by 50%, relative to a situation where the two populations were completely isolated. These theoretical results suggest that even small numbers of interactions could lead to substantial spillover. We furthermore found that, in simulations, spillover led to apparently weaker cross-sectional use–resistance relationships across multiple populations and that pairs of populations with stronger interpopulation interactions tended to have weaker measured use–resistance associations.
When examining observational antibiotic use and resistance data from US states and European countries and using transportation data to estimate the relative ranking of epidemiological contacts between those populations, we found a correlation between increased interactions and attenuated use–resistance associations. Pairs of populations in the highest decile of interpopulation interactions, that is, those most subject to spillover, had use–resistance associations on the order of 50% weaker than pairs in the lowest decile of interactions. The two pathogen/antibiotic dataset combinations with data not indicative of spillover, namely, S. pneumoniae and β-lactams and macrolides in the ECDC data, may not have shown the same signal as other cases because the smaller number of populations in those cases (27 versus 28 to 50 in the other cases) led to insufficient statistical power, because air transportation data are not an appropriate proxy for epidemiological interactions (SI Appendix, Fig. S3), because the available data were insufficiently accurate, or potentially because the biology or epidemiology of S. pneumoniae resistance in these cases is somehow different and does not exhibit spillover.
These theoretical and empirical results suggest that spillover is relevant at the level of US states and European countries. This finding has important ramifications. First, attempts to attribute changes in a population’s level of antibiotic resistance to changes in that population’s rates of antibiotic use may lead to inaccurate conclusions unless use and resistance in surrounding populations are accounted for. Second, state- or country-level antibiotic stewardship pilot studies may substantially underestimate the potential reduction in antibiotic resistance that would follow from a reduction in antibiotic use if that reduction were implemented at a larger scale. Spillover does not necessarily mean that the intervention led to a smaller global benefit but, rather, that the benefit of an intervention might have spilled over into neighboring, unmeasured populations. Third, mass drug administration trials may lead to elevated levels of antibiotic resistance in the control populations if those populations are not entirely separated from the intervention population. Finally, spillover can at least partly explain why use–resistance associations at the level of US states or European countries are sometimes difficult to detect and, when they are detected, are sometimes weaker than expected (5, 7, 11). Thus, theoretical models of antibiotic use and resistance that treat US states or European countries as epidemiologically independent populations will not accurately represent the dynamics of resistance (32).
Our study has several limitations. First, we interpreted the theoretical results and ecological data as if the association between antibiotic use and resistance were causal and deterministic. However, decreases in the use of an antibiotic may not necessarily lead to declines in resistance to that antibiotic in a target pathogen (12, 43–45). We do not address coresistance and cross selection (46, 47), and we assumed that resistance equilibrates on a timescale comparable to an intervention. Previous research has shown that resistance among E. coli, S. pneumoniae, N. gonorrhoeae, and other organisms can respond to changes in antibiotic use on the timescale of months (48–51), but the expected delay between a perturbation to antibiotic use and the resulting change in resistance remains a subject of active study (13, 48, 52, 53). Nevertheless, the use of ecological data was essential to addressing our hypothesis, as data from multiple controlled, state- or country-wide experiments are not available.
Second, our analyses attributed all differences in antibiotic resistance between populations to differences in use across those populations and to interactions between them. In fact, antibiotic resistance is associated with factors beyond antibiotic use (6, 54), and those factor are likely spatially correlated. In other words, closely interacting populations might have more similar use–resistance associations because they tend to be more similar with respect to other determinants of antibiotic use. Our estimates of the correlation between interpopulation interactions and the attenuation of use–resistance relationships may therefore be overestimates. A more careful quantification of the relative roles of spillover versus other spatially correlated determinants of resistance is required.
Third, our analysis only considered pairs of populations, when, in fact, spillover is happening between all pairs of populations in our analysis simultaneously. We used the pairs approach because it allowed for a simple theoretical model and a straightforward comparison of theory with the observational data. However, more sophisticated approaches that account for the network of spillover interactions will likely lead to more refined characterizations of spillover.
Finally, analyses based on administrative entities like US states or European countries, although logistically attractive “laboratories” of antibiotic stewardship, will always be difficult to interpret because administrative entities average over important dimensions of population structure like age (55), sexual networks (56), and race/ethnicity (57). Thus, use–resistance associations measured across states and countries may be different from those that appear among geographically proximate populations with dissimilar antibiotic use rates, such as the sexes (58) and racial/ethnic groups (59). The ideal data source for studying antibiotic use and resistance would be linked records of individual-level antibiotic use, antibiotic resistance, geography, and behavior, but these types of datasets are not generally available.
We suggest four lines of investigation that could refine our understanding about the role of spillover at levels of US states and European countries. First, further mathematical modeling studies with more realistic structuring of the host population might articulate more detailed theoretical expectations about the relationship between intervention scale and spillover. For example, models could be parameterized with epidemiological information about individuals’ contacts and travel patterns, as has been done for other infectious diseases (60). Second, meta-analysis of existing studies of use–resistance relationships (5, 61, 62), both experimental and observational, could potentially determine the empirical relationship between intervention population size and the importance of spillover. This kind of meta-analysis might reveal that populations other than US states are feasible “laboratories” for resistance: It may be that cities, daycares, schools, workplaces, or even families represent the optimal trade-off between maximizing logistical feasibility and minimizing spillover. Third, prospective cohort studies could track the transmission of bacteria between different types of populations, providing direct estimates of the magnitude of interpopulation interactions relevant to bacterial transmission. Finally, future experimental outpatient antibiotic stewardship interventions should make careful and deliberate decisions about the sizes and interconnectedness of the populations they target. We hope that a better understanding of spillover will improve predictions about the future of antibiotic resistance, the formulation of stewardship policy, the design of stewardship interventions and antibiotic administration trials, and theoretical models of resistance.
Supplementary Material
Acknowledgments
We thank Dr. Stephen M. Kissler and the anonymous peer reviewers for helpful comments on the manuscript. The views and opinions of the authors expressed herein do not necessarily state or reflect those of the ECDC. The accuracy of the authors’ statistical analysis and the findings they report are not the responsibility of ECDC. ECDC is not responsible for conclusions or opinions drawn from the data provided. ECDC is not responsible for the correctness of the data and for data management, data merging, and data collation after provision of the data. ECDC shall not be held liable for improper or incorrect use of the data. This work was supported by NIH (Grant U54GM088558 to M.L.). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2013694117/-/DCSupplemental.
Data Availability.
Data and code to reproduce results have been deposited in Zenodo (DOI: 10.5281/zenodo.3909812).
References
- 1.Review on Antimicrobial Resistance , “Tackling drug-resistant infections globally: Final report and recommendations” (2016). https://amr-review.org/. Accessed 28 October 2020.
- 2.Public Health England , “English surveillance programme for antimicrobial utilisation and resistance (ESPAUR)” (Report no. 2014362, Public Health England, London, UK 2014).
- 3.Public Health Agency of Sweden , National Veterinary Institute, “Consumption of antibiotics and occurrence of antibiotic resistance in Sweden" (Report no. 16124, Public Health Agency of Sweden and National Veterinary Institute, Solna/Uppsala, Sweden, 2016).
- 4.US Centers for Disease Control and Prevention , Antibiotic Resistance Threats in the United States, 2013, (US Centers for Disease Control and Prevention, 2013). [Google Scholar]
- 5.Bell B. G., Schellevis F., Stobberingh E., Goossens H., Pringle M., A systematic review and meta-analysis of the effects of antibiotic consumption on antibiotic resistance. BMC Infect. Dis. 14, 13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.MacFadden D. R., McGough S. F., Fisman D., Santillana M., Brownstein J. S., Antibiotic resistance increases with local temperature. Nat. Clim. Chang. 8, 510–514 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olesen S. W., et al. , The distribution of antibiotic use and its association with antibiotic resistance. eLife 7, e39435 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goossens H., Ferech M., Vander Stichele R., Elseviers M.; ESAC Project Group , Outpatient antibiotic use in Europe and association with resistance: A cross-national database study. Lancet 365, 579–587 (2005). [DOI] [PubMed] [Google Scholar]
- 9.van de Sande-Bruinsma N. et al.; European Antimicrobial Resistance Surveillance System Group; European Surveillance of Antimicrobial Consumption Project Group , Antimicrobial drug use and resistance in Europe. Emerg. Infect. Dis. 14, 1722–1730 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Livermore D. M., The 2018 Garrod Lecture: Preparing for the black swans of resistance. J. Antimicrob. Chemother. 73, 2907–2915 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Turnidge J., Christiansen K., Antibiotic use and resistance–Proving the obvious. Lancet 365, 548–549 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Arason V. A., et al. , Clonal spread of resistant pneumococci despite diminished antimicrobial use. Microb. Drug Resist. 8, 187–192 (2002). [DOI] [PubMed] [Google Scholar]
- 13.Lipsitch M., The rise and fall of antimicrobial resistance. Trends Microbiol. 9, 438–444 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Costelloe C., Metcalfe C., Lovering A., Mant D., Hay A. D., Effect of antibiotic prescribing in primary care on antimicrobial resistance in individual patients: Systematic review and meta-analysis. BMJ 340, c2096 (2010). [DOI] [PubMed] [Google Scholar]
- 15.Harbarth S., Harris A. D., Carmeli Y., Samore M. H., Parallel analysis of individual and aggregated data on antibiotic exposure and resistance in gram-negative bacilli. Clin. Infect. Dis. 33, 1462–1468 (2001). [DOI] [PubMed] [Google Scholar]
- 16.Lipsitch M., Measuring and interpreting associations between antibiotic use and penicillin resistance in Streptococcus pneumoniae. Clin. Infect. Dis. 32, 1044–1054 (2001). [DOI] [PubMed] [Google Scholar]
- 17.Hannah E. L., et al. , Drug-resistant Escherichia coli, rural Idaho. Emerg. Infect. Dis. 11, 1614–1617 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kalter H. D., et al. , Risk factors for antibiotic-resistant Escherichia coli carriage in young children in Peru: Community-based cross-sectional prevalence study. Am. J. Trop. Med. Hyg. 82, 879–888 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Samore M. H., et al. , High rates of multiple antibiotic resistance in Streptococcus pneumoniae from healthy children living in isolated rural communities: Association with cephalosporin use and intrafamilial transmission. Pediatrics 108, 856–865 (2001). [DOI] [PubMed] [Google Scholar]
- 20.Gottesman B.-S., Low M., Almog R., Chowers M., Quinolone consumption by mothers increases their children’s risk of acquiring quinolone-resistant bacteriuria. Clin. Infect. Dis. 71, 532–538 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Lipsitch M., Samore M. H., Antimicrobial use and antimicrobial resistance: A population perspective. Emerg. Infect. Dis. 8, 347–354 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moellering R. C., Jr, NDM-1–A cause for worldwide concern. N. Engl. J. Med. 363, 2377–2379 (2010). [DOI] [PubMed] [Google Scholar]
- 23.Wang R., et al. , The global distribution and spread of the mobilized colistin resistance gene mcr-1. Nat. Commun. 9, 1179 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bogoch I. I., Utzinger J., Lo N. C., Andrews J. R., Antibacterial mass drug administration for child mortality reduction: Opportunities, concerns, and possible next steps. PLoS Negl. Trop. Dis. 13, e0007315 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doan T. et al.; MORDOR Study Group , Macrolide resistance in MORDOR I–A cluster-randomized trial in Niger. N. Engl. J. Med. 380, 2271–2273 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cooper B. S., et al. , Methicillin-resistant Staphylococcus aureus in hospitals and the community: Stealth dynamics and control catastrophes. Proc. Natl. Acad. Sci. U.S.A. 101, 10223–10228 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.MacFadden D. R., Fishman D. N., Hanage W. P., Lipsitch M., The relative impact of community and hospital antibiotic use on the selection of extended-spectrum beta-lactamase-producing Escherichia coli. Clin. Infect. Dis. 69, 182–188 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knight G. M., et al. , Quantifying where human acquisition of antibiotic resistance occurs: A mathematical modelling study. BMC Med. 16, 137 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klevens R. M., et al. , Outpatient antibiotic prescribing in Massachusetts, 2011–2015. Open Forum Infect. Dis. 6, ofz169 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davies N. G., Flasche S., Jit M., Atkins K. E., Within-host dynamics shape antibiotic resistance in commensal bacteria. Nat. Ecol. Evol. 3, 440–449 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lehtinen S., et al. , Evolution of antibiotic resistance is linked to any genetic mechanism affecting bacterial duration of carriage. Proc. Natl. Acad. Sci. U.S.A. 114, 1075–1080 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blanquart F., Lehtinen S., Lipsitch M., Fraser C., The evolution of antibiotic resistance in a structured host population. J. R. Soc. Interface 15, 20180040 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Truven Health MarketScan Database, Commercial Claims and Encounters. Ann Arbor, MI, 2015. https://marketscan.truvenhealth.com/marketscanportal/. Accessed 28 October 2020.
- 34.MacFadden D. R., et al. , A platform for monitoring regional antimicrobial resistance, using online data sources: ResistanceOpen. J. Infect. Dis. 214, S393–S398 (2016). [DOI] [PubMed] [Google Scholar]
- 35.US Centers for Disease Control and Prevention , Patient safety atlas–Outpatient antibiotic use. https://gis.cdc.gov/grasp/PSA/indexAU.html. Accessed 25 October 2018.
- 36.US Centers for Disease Control and Prevention , Patient safety atlas–Antibiotic resistance. https://gis.cdc.gov/grasp/PSA/AboutTheData.html. Accessed 25 October 2018.
- 37.European Centre for Disease Prevention and Control , Antimicrobial consumption database. https://www.ecdc.europa.eu/en/antimicrobial-consumption/surveillance-and-disease-data/database. Accessed 25 October 2018.
- 38.European Centre for Disease Prevention and Control , Surveillance atlas of infectious diseases. https://atlas.ecdc.europa.eu/public/index.aspx. Accessed 25 October 2018.
- 39.Census U.S., 2011-2015 5-Year ACS commuting flows. https://www.census.gov/data/tables/2015/demo/metro-micro/commuting-flows-2015.html. Accessed 18 February 2020.
- 40.Eurostat, Air transport measurement. https://ec.europa.eu/eurostat/cache/metadata/en/avia_pa_esms.htm. Accessed 21 May 2020.
- 41.R Core Team , R: A language and environment for statistical computing, Version 3.6.0 (R Core Team, Vienna, 2018). https://www.r-project.org/.
- 42.Oksanen J., et al. , vegan: Community Ecology Package (R package Version 2.5-6). https://CRAN.R-project.org/package=vegan. Accessed 1 September 2019.
- 43.Hennessy T. W., et al. , Changes in antibiotic-prescribing practices and carriage of penicillin-resistant Streptococcus pneumoniae: A controlled intervention trial in rural Alaska. Clin. Infect. Dis. 34, 1543–1550 (2002). [DOI] [PubMed] [Google Scholar]
- 44.Sundqvist M., et al. , Little evidence for reversibility of trimethoprim resistance after a drastic reduction in trimethoprim use. J. Antimicrob. Chemother. 65, 350–360 (2010). [DOI] [PubMed] [Google Scholar]
- 45.Enne V. I., Livermore D. M., Stephens P., Hall L. M., Persistence of sulphonamide resistance in Escherichia coli in the UK despite national prescribing restriction. Lancet 357, 1325–1328 (2001). [DOI] [PubMed] [Google Scholar]
- 46.Pouwels K. B., et al. , Association between use of different antibiotics and trimethoprimresistance: Going beyond the obvious crude association. J. Antimicrob. Chemother. 73, 1700–1707 (2018). [DOI] [PubMed] [Google Scholar]
- 47.Tedijanto C., Olesen S. W., Grad Y. H., Lipsitch M., Estimating the proportion of bystander selection for antibiotic resistance among potentially pathogenic bacterial flora. Proc. Natl. Acad. Sci. U.S.A. 115, E11988–E11995 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Olesen S. W., et al. , Azithromycin susceptibility among Neisseria gonorrhoeae isolates and seasonal macrolide use. J. Infect. Dis. 219, 619–623 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dagan R., et al. , Seasonality of antibiotic-resistant streptococcus pneumoniae that causes acute otitis media: A clue for an antibiotic-restriction policy? J. Infect. Dis. 197, 1094–1102 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun L., Klein E. Y., Laxminarayan R., Seasonality and temporal correlation between community antibiotic use and resistance in the United States. Clin. Infect. Dis. 55, 687–694 (2012). [DOI] [PubMed] [Google Scholar]
- 51.Blanquart F., Lehtinen S., Fraser C., An evolutionary model to predict the frequency of antibiotic resistance under seasonal antibiotic use, and an application to Streptococcus pneumoniae. Proc. Biol. Sci. 284, 20170679 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dingle K. E. et al.; Modernising Medical Microbiology Informatics Group , Effects of control interventions on Clostridium difficile infection in England: An observational study. Lancet Infect. Dis. 17, 411–421 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McCormick A. W., et al. , Geographic diversity and temporal trends of antimicrobial resistance in Streptococcus pneumoniae in the United States. Nat. Med. 9, 424–430 (2003). [DOI] [PubMed] [Google Scholar]
- 54.Collignon P., Beggs J. J., Walsh T. R., Gandra S., Laxminarayan R., Anthropological and socioeconomic factors contributing to global antimicrobial resistance: A univariate and multivariable analysis. Lancet Planet. Health 2, e398–e405 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Mossong J., et al. , Social contacts and mixing patterns relevant to the spread of infectious diseases. PLoS Med. 5, e74 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Garnett G. P., et al. , Sexual mixing patterns of patients attending sexually transmitted diseases clinics. Sex. Transm. Dis. 23, 248–257 (1996). [DOI] [PubMed] [Google Scholar]
- 57.Newman M. E. J., Mixing patterns in networks. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 67, 026126 (2003). [DOI] [PubMed] [Google Scholar]
- 58.Hicks L. A., et al. , US outpatient antibiotic prescribing variation according to geography, patient population, and provider specialty in 2011. Clin. Infect. Dis. 60, 1308–1316 (2015). [DOI] [PubMed] [Google Scholar]
- 59.Olesen S. W., Grad Y. H., Racial/ethnic disparities in antimicrobial drug use, United States, 2014–2015. Emerg. Infect. Dis. 24, 2126–2128 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Charu V., et al. , Human mobility and the spatial transmission of influenza in the United States. PLoS Comput. Biol. 13, e1005382 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schechner V., Temkin E., Harbarth S., Carmeli Y., Schwaber M. J., Epidemiological interpretation of studies examining the effect of antibiotic usage on resistance. Clin. Microbiol. Rev. 26, 289–307 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.O’Brien K. S., et al. , Antimicrobial resistance following mass azithromycin distribution for trachoma: A systematic review. Lancet Infect. Dis. 19, E14–E25 (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data and code to reproduce results have been deposited in Zenodo (DOI: 10.5281/zenodo.3909812).