Significance
In this study, we posited that the metabolic and mitogenic activities of enterokine FGF19 are reflections of different thresholds of FGF19-induced FGF receptor (FGFR) dimerization. To test our hypothesis, we engineered three FGF19 variants that have progressively reduced FGFR dimerization capacity. We show that these variants have progressively reduced cell proliferative/tumorigenic activity but maintain FGF19WT-like metabolic activities. These observations substantiate our model in which FGF signaling specificity is regulated by different thresholds in FGF-induced FGFR dimer stability and longevity, and provide a simple stratagem to engineer safer agonists of FGF19—or indeed any other FGF—for the treatment of a range of metabolic diseases.
Keywords: FGF19, FGFR4 dimerization, structure-based drug design, tumorigenic activity, metabolic activity
Abstract
As a physiological regulator of bile acid homeostasis, FGF19 is also a potent insulin sensitizer capable of normalizing plasma glucose concentration, improving lipid profile, ameliorating fatty liver disease, and causing weight loss in both diabetic and diet-induced obesity mice. There is therefore a major interest in developing FGF19 as a therapeutic agent for treating type 2 diabetes and cholestatic liver disease. However, the known tumorigenic risk associated with prolonged FGF19 administration is a major hurdle in realizing its clinical potential. Here, we show that nonmitogenic FGF19 variants that retain the full beneficial glucose-lowering and bile acid regulatory activities of WT FGF19 (FGF19WT) can be engineered by diminishing FGF19’s ability to induce dimerization of its cognate FGF receptors (FGFR). As proof of principle, we generated three such variants, each with a partial defect in binding affinity to FGFR (FGF19ΔFGFR) and its coreceptors, i.e., βklotho (FGF19ΔKLB) or heparan sulfate (FGF19ΔHBS). Pharmacological assays in WT and db/db mice confirmed that these variants incur a dramatic loss in mitogenic activity, yet are indistinguishable from FGF19WT in eliciting glycemic control and regulating bile acid synthesis. This approach provides a robust framework for the development of safer and more efficacious FGF19 analogs.
FGF19, FGF21, and FGF23 comprise the endocrine acting FGF19 subfamily within the FGF family of ligands (1). FGF19 is a postprandial hormone that is expressed and released from enterocytes in response to bile acid (BA), then enters the portal venous circulation (2). Upon reaching the liver and gallbladder, FGF19 inhibits new hepatic BA biosynthesis by suppressing transcription of Cyp7A1 (the rate-limiting enzyme in cholesterol catabolism) and stimulating gallbladder filling, thus providing negative feedback (2–5). Recent studies have extended the metabolic functions of FGF19 to stimulation of hepatic protein and glycogen synthesis and to inhibition of gluconeogenesis (6, 7).
FGF19 exerts its physiological effects by binding, dimerizing, and thereby activating FGF receptor 4 (FGFR4), its principal cognate FGF receptor and the most abundantly expressed FGFR isoform in liver (8). FGF19-FGFR4 signaling requires an obligatory interaction with βKlotho (KLB), a single-pass transmembrane protein that is predominantly expressed in liver, fat, and hypothalamus (9–11). Mechanistically, KLB promotes FGF19-FGFR4 binding by simultaneously grasping FGF19 by its C tail and FGFR4 by its Ig domain 3 (D3); in so doing, KLB enforces FGF19-FGFR4 proximity and confers complex stability (11–14). However, dimerization of the endocrine FGF19-FGFR4 complex and, hence, signaling also requires heparan sulfate (HS), the glycan moiety of ubiquitously expressed HS proteoglycans, as an additional coreceptor (9). The precise nature of the FGF19-FGFR4-KLB-HS quaternary dimer is currently unknown. However, by analogy to paracrine FGF-FGFR signaling, we recently proposed that HS induces formation of a symmetric 2:2:2:2 FGF19-FGFR4-KLB-HS quaternary dimer (9, 14).
Fgf15 (the murine ortholog of Fgf19), Fgfr4, or KLB-knockout mice all present with an elevated BA pool, revealing an intimate functional linkage between FGF19, FGFR4, and KLB in the physiological regulation of BA homeostasis (15, 16). However, pharmacological studies in type 2 diabetic (T2D) mice have shown that—similar to FGF21—FGF19 can also improve glucose, cholesterol, and lipid triglycerides, as well as enhance energy expenditure and reduce body weight via insulin-dependent (acute treatment) and independent (chronic treatment) mechanisms (6, 7, 17, 18). Like FGF21, FGF19 exerts its beneficial pharmacological effects by binding and activating the “c” splice isoform of FGFR1 (FGFR1c) in adipose tissue and hypothalamus in a KLB-dependent fashion (8, 19, 20). Unlike FGF19, FGF21 shows exclusive specificity for FGFR1c and is completely devoid of mitogenic activity (8, 21–23).
The striking pharmacological properties of FGF19 have brought this ligand to the forefront as a promising target for the potential treatment of diabetes, nonalcoholic steatohepatitis (NASH), cholestatic liver disease, and BA metabolism disorders. However, a major safety concern has emerged from the fact that prolonged treatment with FGF19 induces hepatocellular carcinoma (24–27). For this reason, there has been an intensive search for nonmitogenic FGF19 variants that preserve its beneficial metabolic activities (26, 28–32).
We recently proposed that FGF signaling specificity is regulated by different thresholds in FGF-induced FGFR dimer stability and longevity (33, 34). Specifically, via the use of engineered FGF1 variants with weakened HS or receptor binding abilities, we showed that a proliferative FGF response requires strong FGFR dimerization and a correspondingly persistent intracellular signal, whereas a metabolic response can be evoked by relatively weak dimerization and transient signals (33). Given these observations, we hypothesized that nonmitogenic FGF19 molecules might be engineered by weakening FGF19’s ability to dimerize its cognate FGFRs. Here, we describe the generation of a series of three FGF19 partial agonists, termed FGF19ΔFGFR, FGF19ΔHBS, and FGF19ΔKLB, each with a progressively reduced dimerization potential. We show that the mitogenic activity of these mutants is progressively lost, while their metabolic activities remain unchanged. Notably, RNA-sequencing (RNA-seq) analysis of mice treated with FGF19ΔKLB, which exhibits the weakest dimerizing potential, reveals that intracellular signals involved in hepatocellular carcinogenesis are dramatically attenuated, while pathways mediating BA biosynthesis remain fully intact. Thus, fine tuning of receptor dimerization and downstream signaling thresholds is a practical approach toward engineering safer agonists of FGF19—or any other FGF—for the treatment of a range of metabolic diseases.
Results
Engineering FGF19 Variants with Reduced FGFR Dimerization Capacity.
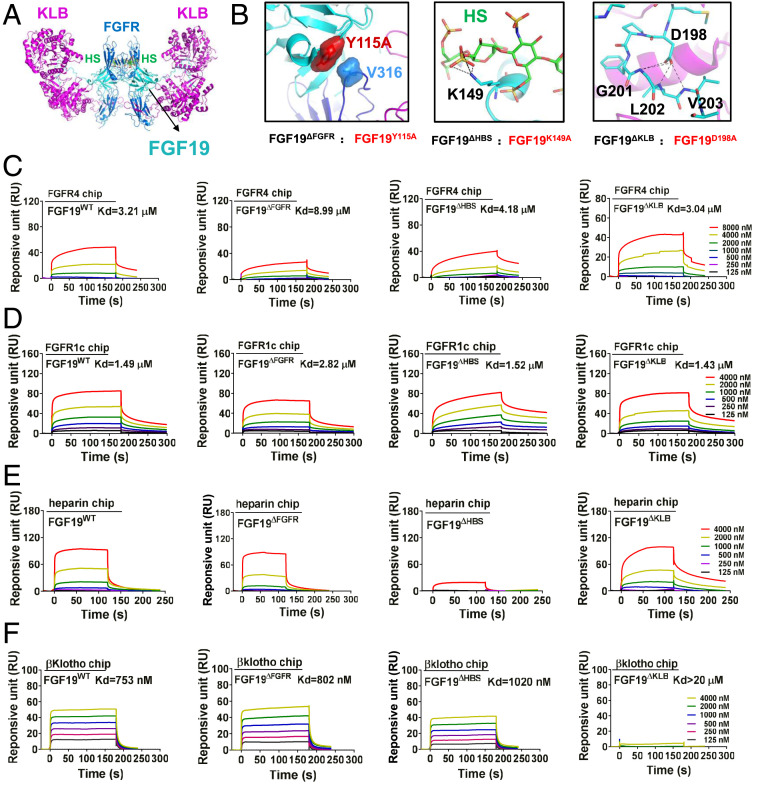
We recently showed that endocrine FGFs require both Klotho and HS as coreceptors to dimerize and activate their cognate FGFRs (14). Based on our structural and biochemical data, we constructed a 2:2:2:2 FGF19-FGFR1c-KLB-HS dimer model (Fig. 1A) and used this as a guide to design three FGF19 mutants (termed FGF19ΔFGFR, FGF19ΔHBS, and FGF19ΔKLB, respectively) with anticipated diminished capacities to induce FGFR dimerization. FGF19ΔFGFR, FGF19ΔHBS, and FGF19ΔKLB have partial defects in FGFR, HS, or KLB binding, respectively. FGF19ΔFGFR carries a Tyr-115-Ala mutation predicted to eliminate hydrophobic interactions between Tyr-115 of FGF19 and Val-316, located within the D3 domain of FGFR1c (Fig. 1 B, Left); FGF19ΔHBS harbors a Lys-149-Ala mutation, which maps to the atypical HS binding site of FGF19 and is predicted to impair HS binding (Fig. 1 B, Center); and FGF19ΔKLB harbors an Asp-198-Ala mutation, which affects KLB binding by abrogating intramolecular hydrogen bonds within the FGF19 C tail (Fig. 1 B, Right) (13). As a negative control, we generated a C-terminally truncated FGF19 (FGF19ΔC-tail) lacking the very last 25 amino acids of FGF19WT, which is predicted to be completely incapable of KLB binding and, hence, FGFR dimerization.
Fig. 1.
Design and characterization of FGF19 variants. (A) A hypothetical 2:2:2:2 FGF19-FGFR1c-KLB-HS dimeric complex was constructed by superimposing structures of FGF19 alone (PDB ID code: 2P23) and of KLB in complex with the C-terminal tail of FGF19 (FGF19CT) (PDB ID code: 6NFJ) onto a dimeric model of a FGF23-FGFR1c-αKlotho-HS dimer (PDB ID code: 5W21). FGF19, FGFR1c, and KLB are shown as cyan, light blue, and purple cartoons, respectively; HS is shown as sticks with carbon, nitrogen, oxygen, and sulfur colored in green, blue, red, and yellow, respectively. (B) Close-up view of interfaces between FGF19 and FGFR (Left), FGF19 and HS (Center), and FGF19 and KLB (Right). In Left, Tyr-115 of FGF19 and Val-316 of FGFR1c are shown as surfaces to highlight their hydrophobic interaction. At Center, the side chain of Lys-149 of FGF19 which engages in hydrogen bonds with HS is shown as sticks. Right shows multiple intramolecular hydrogen bonds mediated by Asp-198 that facilitate FGF19CT conformation, thereby contributing to KLB binding. In Center and Right, black dashed lines denote hydrogen bonds. (C–F) Representative SPR sensorgrams of binding of FGF19WT and FGF19 variants to the ligand-binding domains of FGFR4 (C) and FGFR1 (D), and to HS (E) and KLB (F). Equilibrium dissociation constants (Kd values) were derived from saturation binding curves.
We first assessed the binding affinities of these mutants with respect to FGFR, HS, or KLB via surface plasmon resonance (SPR) spectroscopy, using FGF19WT as a positive control. Consistent with our predictions, the binding affinities of FGF19ΔFGFR with either FGFR1 or FGFR4 were much weaker than the other mutants (Fig. 1 C and D and SI Appendix, Fig. S1 A and B). However, FGF19ΔHBS exhibited a major loss in HS binding capacity but was fully capable of binding KLB and FGFR4 (Fig. 1E). Finally, FGF19ΔKLB had a markedly diminished KLB binding affinity, but its binding to HS and FGFR4 was preserved (Fig. 1F and SI Appendix, Fig. S1C). As expected, FGF19ΔC-tail completely failed to bind KLB (SI Appendix, Fig. S1D).
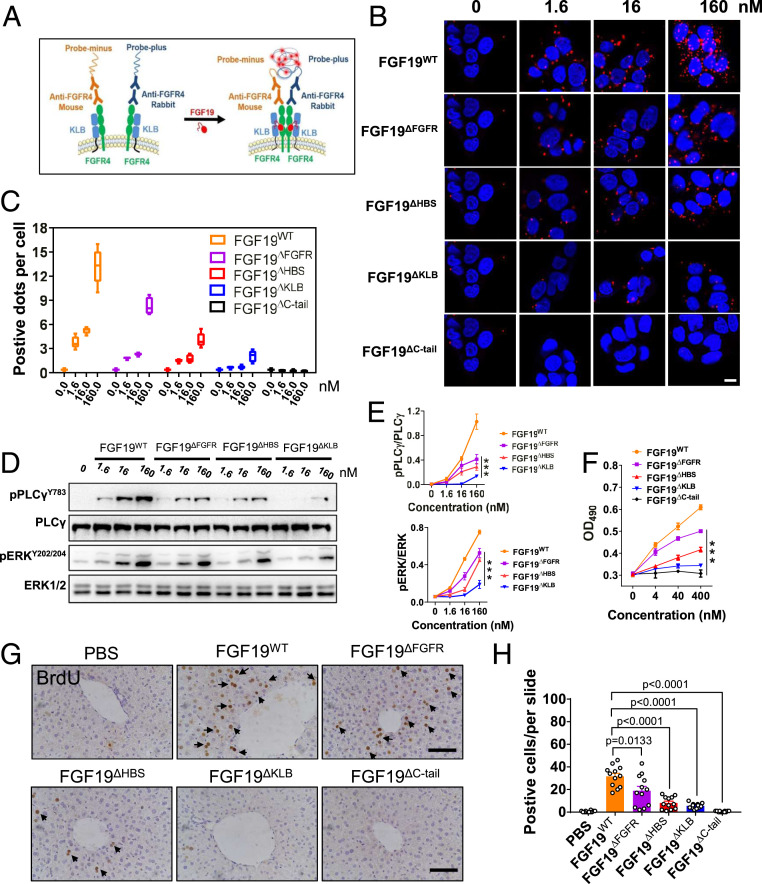
We next devised a proximity ligation assay (PLA) to measure the abilities of FGF19ΔFGFR, FGF19ΔHBS, or FGF19ΔKLB to promote FGFR4 dimerization in situ (Fig. 2A). Human HepG2 hepatoma cells (which naturally express KLB and FGFR4) were exposed to increasing concentrations of FGF19ΔFGFR, FGF19ΔHBS, or FGF19ΔKLB, followed by incubation with primary mouse anti-FGFR4 and rabbit anti-FGFR4 antibodies and then oligonucleotide-labeled secondary antibodies (PLA probes). FGF19WT and FGF19ΔC-tail served as positive and negative controls, respectively; FGFR4 dimerization was visualized by confocal microscopy. Qualitative and semiquantitative analyses of images showed that the three variants incurred successively greater reductions in their ability to promote FGFR4 dimerization, in the order FGF19WT > FGF19ΔFGFR > FGF19ΔHBS > FGF19ΔKLB (Fig. 2 B and C). FGF19ΔC-tail completely failed to induce FGFR4 dimerization, reflecting the mandatory role of KLB in supporting FGF19-FGFR4 binding and dimerization.
Fig. 2.
FGFR4 dimerization and downstream signaling activation induced by FGF19WT and its variants. (A) Schematic illustration of the PLA. (B) PLA analysis of FGFR4 dimerization strength induced by FGF19WT and its variants at 0, 1.6, 16, and 160 nM. (Scale bar, 10 μm.) (C) Quantification of PLA (B) by counting the number of positive dots per nucleus. (D) Immunoblots showing dose-dependent activation of PLCγ and the MAPK pathway (ERK1/2) by FGF19WT, FGF19ΔHBS, FGF19ΔFGFR, and FGF19ΔKLB in the human hepatoma cell line HepG2. (E) Quantification of Western blot in D by densitometric analysis. Data from three independent measurements are presented as mean ± SEM; ***P < 0.001 vs. FGF19WT. (F) In vitro proliferative activity determined by a MTT assay using HepG2 cells. Data from three independent measurements are presented as mean ± SEM; ***P < 0.001 vs. FGF19WT. (G) Representative images of BrdU immunostained livers from C57BL/6J mice after 7 d of treatment with PBS, FGF19WT, or FGF19 variants; (Scale bar, 100 μm.) (H) Semiquantitative analysis of BrdU-positive hepatocytes was performed on three images of randomly selected areas from four different mice for each treatment group. Data are presented as mean values ± SEM (n = 12); a value of P < 0.05 was considered to be statistically significant.
To corroborate these findings functionally, we measured the activation levels of PLCγ and ERK signaling pathways in HepG2 cells as readouts for FGFR dimerization in response to increasing concentrations of WT or mutant ligands. Consistent with our PLA data, all three FGF19 mutants were impaired in their ability to induce PLCγ and ERK1/2 phosphorylation (Fig. 2 D and E). The extent of impairment correlated with the observed graded reduction in dimerization abilities seen in PLA assays (Fig. 2B). As a further measure of receptor dimerization capacity, we examined the time course of ERK1/2 activation by FGF19WT and FGF19ΔKLB (the least dimerizing mutant) in HepG2 cells. As shown in SI Appendix, Fig. S2 A and B, stimulation with FGF19WT led to activation of ERK1/2 as early as 2 min, and the signal continued to intensify until 15 min. By 30 min, the signal intensity waned but still remained above that observed in buffer-treated cells. In contrast, in FGF19ΔKLB-treated cells, a weaker activation of ERK1/2 was observed at all time points tested compared to FGF19WT-treated cells, and the signal completely dissipated by 30 min (SI Appendix, Fig. S2 A and B). These data were recapitulated using a rat hepatoma cell line (H4IIE). Specifically, Western blotting and quantitative homogeneous time-resolved fluorescence (HTRF) assays measuring ERK phosphorylation showed a successive decrease in signaling capacities of FGF19 variants in the order FGF19ΔFGFR > FGF19ΔHBS > FGF19ΔKLB (SI Appendix, Fig. S2 C and D).
Dramatically Reduced Proliferative Activity of FGF19ΔHBS and FGF19ΔKLB.
The extent of impairment in the abilities of FGF19ΔFGFR, FGF19ΔHBS, and FGF19ΔKLB to induce FGFR4 dimerization/activation resulted in corresponding reductions in the ability of these variants to promote HepG2 cell proliferation (Fig. 2F). A similar trend in the proliferative activity of these variants was observed using H4IIE cells (SI Appendix, Fig. S2E). Notably, these proliferative activities were not significantly impacted by the tyrosine kinase inhibitor AZD4547 (SI Appendix, Fig. S2F), which primarily inhibits FGFR1-FGFR3 and, to a much lesser extent, FGFR4 (35), implying that FGFR4 mediates the proliferative activity of FGF19.
We corroborated these cell-based data via an in vivo BrdU labeling experiment in which we measured the proliferative activity of FGF19WT and its variants in C57BL/6J mouse liver tissue following one week of administration. Consistent with our cell-based data, the BrdU signal was successively diminished in blots from livers of animals treated with FGF19ΔFGFR, FGF19ΔHBS, and FGF19ΔKLB (Fig. 2 G and H). FGF19ΔC-tail failed to elicit any proliferative activity in cultured cells or in mice, implying that the mitogenic activity of FGF19 is also βKlotho-dependent (Fig. 2 F–H). This finding was further validated by the inability of FGF19WT and all its variants to induce mitogenic activity in kidney tissue, which is βKlotho-deficient (SI Appendix, Fig. S3).
FGF19ΔFGFR, FGF19ΔHBS, and FGF19ΔKLB Are Indistinguishable from FGF19WT in Suppressing BA Synthesis.
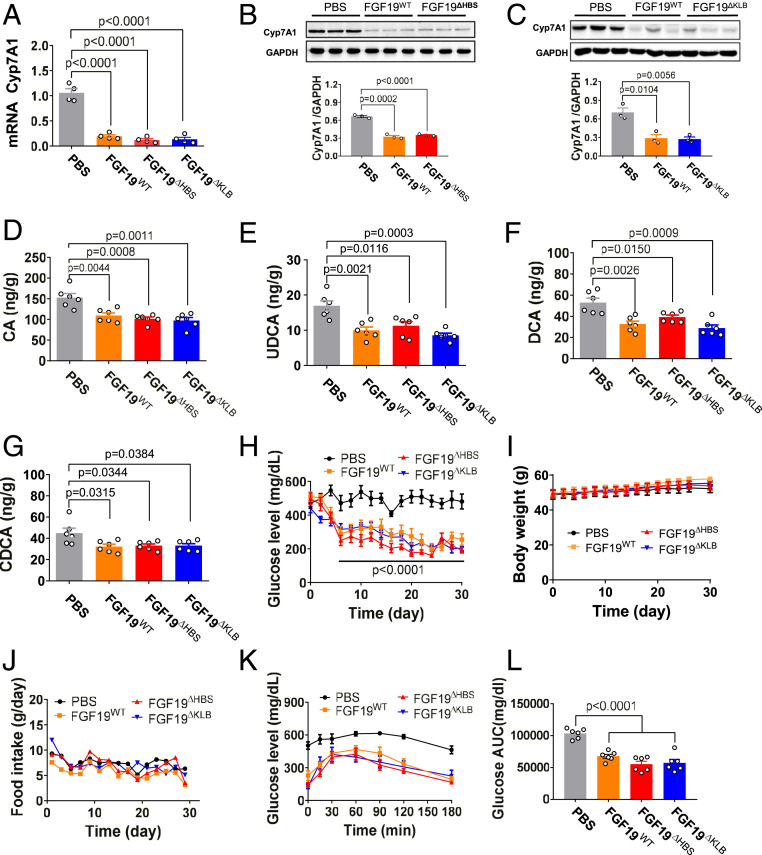
To establish whether FGF19ΔFGFR, FGF19ΔHBS, and FGF19ΔKLB retain the BA regulatory activity of FGF19WT, we administered a single dose of each these proteins into normal C57BL/6J mice and measured hepatic expression of Cyp7A1 (a key readout for BA synthesis) 4 h postinjection. In all cases, reductions in hepatic expression of Cyp7A1 were indistinguishable from mice treated with FGF19WT (Fig. 3A and SI Appendix, Fig. S4A). The results from this short-term treatment were recapitulated in a T2D mouse model (db/db) in which mice were chronically treated with the identical engineered ligands. Hepatic expression of Cyp7A1 (Fig. 3 B and C) and BA levels (Fig. 3 D–G) in liver tissues were reduced to the same extent as in mice treated with FGF19WT. As expected, the negative control (FGF19ΔC-tail) lacked any ability to regulate BA synthesis (SI Appendix, Fig. S4A). We conclude that the BA regulatory activities of FGF19ΔFGFR, FGF19ΔHBS, and FGF19ΔKLB are unimpaired with respect to their WT counterpart.
Fig. 3.
FGF19ΔHBS and FGF19ΔKLB have FGF19WT-like BA and glucose regulatory activities. C57BL/6J or db/db mice were injected with either FGF19WT or its variants (all at 21 nmol/kg of body weight), or with PBS as a control. (A) Real-time PCR analysis of Cyp7A1 mRNA levels in C57BL/6J liver extracts 4 h after injection. Data are presented as mean values ± SEM (n = 4); a value of P < 0.05 was considered to be statistically significant. (B and C) Protein expression (Upper) and quantification (Lower) of Cyp7A1 determined by Western blotting in extracts from livers of db/db mice after chronic administration for 1 mo. Data are presented as mean values ± SEM (n = 3); a value of P < 0.05 was considered to be statistically significant. (D–G) Tissue levels of CA (D), UDCA (E), DCA (F), and CDCA (G) in livers of db/db mice treated for 1 mo. Data are presented as mean values ± SEM (n = 6); a value of P < 0.05 was considered to be statistically significant. (H–J) Blood glucose (H), body weight (I), and food intake (J) in ad libitum-fed db/db mice after chronic daily treatment with FGF19WT, FGF19ΔHBS, or FGF19ΔKLB. (K and L) A GTT done after chronic treatment of db/db mice (K) and the accompanying integrated AUC (L) for changes in blood glucose. Data are presented as mean values ± SEM (n = 6); a value of P < 0.05 was considered to be statistically significant.
FGF19ΔFGFR, FGF19ΔHBS, and FGF19ΔKLB Are Indistinguishable from FGF19WT in Exerting Glycemic Control.
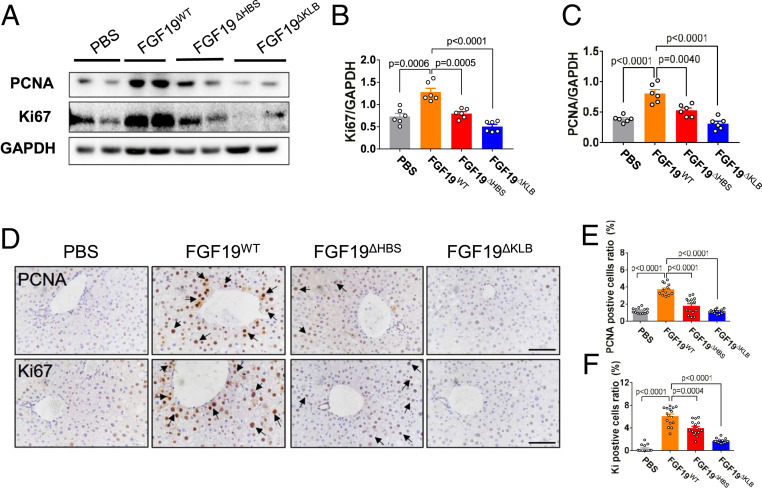
We next compared the glucose-lowering activities of FGF19ΔFGFR, FGF19ΔHBS, and FGF19ΔKLB in db/db mice treated daily with the corresponding ligands for 1 mo; db/db mice treated with FGF19WT and FGF19ΔC-tail served as positive and negative controls, respectively. All three variants were comparable to FGF19WT in normalizing blood glucose levels, with no significant effect on food intake or body weight (Fig. 3 H–J and SI Appendix, Fig.S4 B and C). Additionally, blood glucose levels of db/db mice chronically treated with any of the three variants remained low throughout a glucose tolerance test (GTT) (Fig. 3 K and L and SI Appendix, Fig. S4 D and E). These data show that despite their loss of mitogenic activity, FGF19 variants retain their capacity to exert glycemic control. As expected, administration of FGF19ΔC-tail lacked any ability to lower glucose levels (SI Appendix, Fig. S4 B–E). Consistent with the results obtained in chronically treated healthy C57BL/6J mice, chronic treatment of db/db mice with FGF19ΔKLB did not cause any overt signs of proliferation in liver (Fig. 4). However, consistent with our in vivo BrdU labeling results obtained from C57BL/6J mouse livers, chronic administration of FGF19ΔFGFR induced a substantial increase in the number of hyperproliferating hepatocytes, albeit to a lesser extent than FGF19WT (SI Appendix, Fig. S5).
Fig. 4.
Long-term treatment of db/db mice with FGF19ΔKLB does not promote hepatic cell proliferation. Ad libitum-fed db/db mice were i.p. administrated with FGF19WT, FGF19ΔHBS, FGF19ΔKLB (all at 21 nmol/kg of body weight), or a buffer (PBS) control daily for 1 mo. (A–C) Protein expression of PCNA and Ki67 in liver tissues as determined by Western blotting (A) and data quantitated using ImageJ software (B and C). Data are presented as mean values ± SEM (n = 6); a value of P < 0.05 was considered to be statistically significant. (D) Representative immunohistochemical staining of PCNA and Ki67 in the livers of db/db mice. (Scale bars, 100 μm.) (E and F) Semiquantitation of PCNA and Ki67-positive hepatocytes was performed on 2–3 images of randomly selected areas from six different mice for each treatment group. Data are presented as mean values ± SEM (n = 15); a value of P < 0.05 was considered to be statistically significant.
We also compared the glucose-lowering activity of FGF19ΔKLB with that of M70 (NGM282), a nonmitogenic FGF19 variant that is currently undergoing a phase II clinical trial for the treatment of NASH (36). M70 has a five amino acid (24PLAFS28) truncation and a A30S/G31S/H33L triple substitution in its N terminus (31). These modifications are thought to abolish M70’s mitogenic activity by further skewing its specificity toward FGFR4. However, our structural data suggest that in addition to participating in FGFR binding specificity, the N termini of endocrine FGFs are also directly involved in FGFR dimerization (14). Indeed, analysis of M70 via PLA showed that M70 has a dramatically diminished ability to promote FGFR4 dimerization (SI Appendix, Fig. S6 A and B). These data imply that the nonmitogenic character of M70 is also attributable to its reduced FGFR dimerization capacity. As previously reported (31), we found that M70 possess BA regulatory activity comparable to that of FGF19WT when injected intraperitoneally (i.p.) into C57BL/6J mice (SI Appendix, Fig. S6C). However, in contrast to FGF19WT and FGF19ΔKLB, injection i.p. of M70 (at the same dosage as FGF19ΔKLB, 21 nmol/kg of body weight) did not reduce blood glucose or food intake in db/db mice (SI Appendix, Fig. S6 D and E). Moreover, in a GTT, M70 failed to exert any glycemic control (SI Appendix, Fig. S6 F and G). We surmise that the combination of an N-terminal truncation and a triple substitution disproportionately affects M70’s ability to induce FGFR1c dimerization relative to FGFR4 dimerization. Consequently, activation of the adipose-tissue resident FGFR1c pathway is below the required “signaling threshold” for the glucose-lowering effect.
FGF19ΔKLB has Reduced Ability to Up-Regulate Genes/Pathways Involved in Hepatocarcinogenesis.
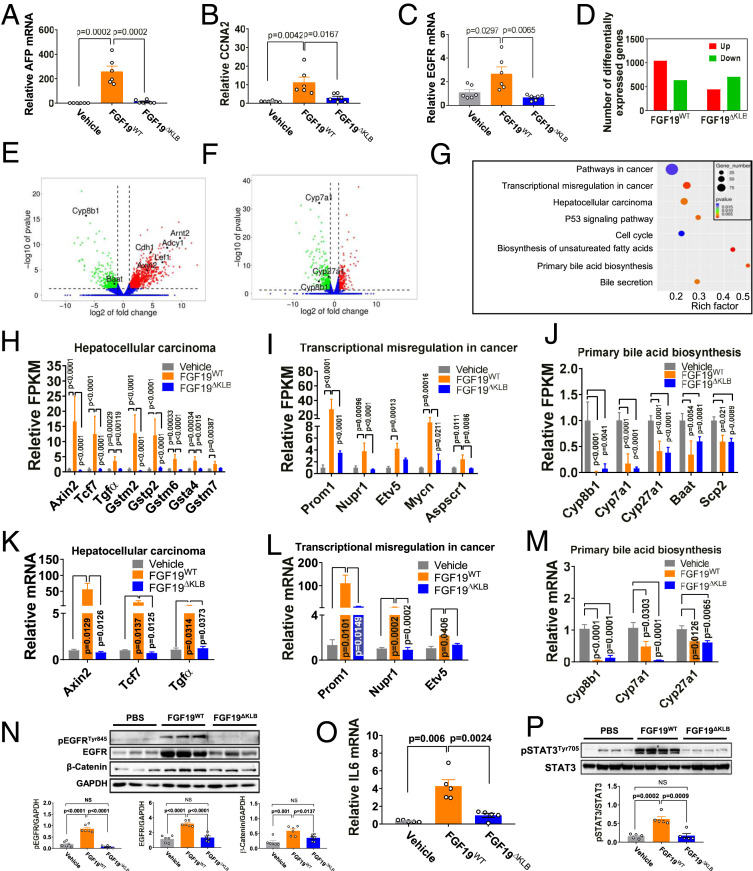
Consistent with previous reports (25, 28), we did not observe any increase in the expression of known hepatocellular carcinoma biomarkers such as AFP, EGFR, and CCNA2 following a 4-wk treatment of db/db mice with FGF19WT or FGF19ΔKLB (SI Appendix, Fig. S7). To better assess the mitogenic risk of FGF19WT and FGF19ΔKLB, we extended the duration of this treatment to 12 wk. Under these conditions, FGF19WT did lead to an up-regulation of AFP, CCNA2, and EGFR, whereas FGF19ΔKLB had no such effect (Fig. 5 A–C). To corroborate these data, at the conclusion of the 12-wk treatment period, we excised the db/db mouse livers and subjected extracts to RNA-seq analysis. Volcano plots of RNA-seq data revealed a conspicuous difference in the number of differentially up-regulated genes between mice receiving FGF19ΔKLB and those receiving FGF19WT (Fig. 5 D–F). Specifically, compared to a buffer-treated control, both FGF19WT (Fig. 5E) and FGF19ΔKLB (Fig. 5F) inhibited expression of a comparable number of genes, most notably those involved in BA synthesis such as Cyp7A1, Cyp27A1, and Cyp8B1. In contrast, the number of up-regulated genes—in particular those involved in cancer-related pathways—was significantly less in the FGF19ΔKLB-treated group relative to the FGF19WT-treated group (Fig. 5 E and F). This differential gene expression pattern was confirmed by gene ontology (GO) pathway analysis (Fig. 5G). Notably, FGF19WT caused up-regulation of multiple genes belonging to EGFR/TGFα and Wnt/Axin2/TCF7 signaling pathways that are involved in hepatocarcinogenesis (Fig. 5H) and transcriptional misregulation in cancer (Fig. 5I), whereas FGF19ΔKLB did not. In contrast, principal BA biosynthesis genes were down-regulated to a similar extent after treatment with either FGF19WT or FGF19ΔKLB (Fig. 5J). Differential expression of genes identified via RNA-seq was confirmed by RT-PCR analyses (Fig. 5 K–M). Finally, to functionally validate the RNA-seq and RT-PCR data, we measured the levels of phosphorylated EGFR (pEGFR), total EGFR, and total β-Catenin—a key downstream effector of Wnt signaling pathway—in livers of db/db mice after 12 wk of treatment with FGF19WT or FGF19ΔKLB. Relative to a buffer-treated control group, we observed increased levels of pEGFR, EGFR, and β-Catenin in the livers of FGF19WT-treated mice, but not in FGF19ΔKLB-treated mice (Fig. 5N). These data corroborate the RNA-seq data and collectively show that FGF19WT enhances EGFR and Wnt signaling pathways, whereas FGF19ΔKLB does not.
Fig. 5.
Heterogeneous signals in the dissected functions of FGF19ΔKLB. Ad libitum-fed db/db mice were i.p. administered with FGF19WT or FGF19ΔKLB (both at 21 nmol/kg of body weight) daily for 12 wk; mice treated with PBS buffer served as controls. (A–C) Real-time PCR analysis of hepatic expression levels of AFP (A), CCNA2 (B), and EGFR (C). Data are presented as mean values ± SEM (n = 6); a value of P < 0.05 was considered to be statistically significant. (D–J) Transcriptome analyses of liver tissues. (D) Numbers of differentially expressed genes (red column: numbers of up-regulated genes; green column: numbers of down-regulated genes). (E and F) Volcano plot of genes in livers of FGF19WT (E) or FGF19ΔKLB (F) treated animals (red: genes up-regulated >twofold; green: genes down-regulated >twofold [P < 0.05]; blue: unchanged genes); (G) GO pathway analysis of the differentially expressed genes between vehicle, FGF19WT, and FGF19ΔKLB obtained by edgeR analysis. (H–J) FPKM of selected gene expression in cancer-related pathways (H and I) and in primary BA biosynthesis (J). Data are presented as mean values ± SEM (n = 3); P values were obtained by edgeR analysis, and a value of P < 0.05 was considered to be statistically significant. (K–M) Real-time PCR analysis of gene expression levels in pathways involved in hepatocellular carcinoma (K), transcriptional misregulation in cancer (L) and primary BA biosynthesis (M). Data are presented as mean values ± SEM (n = 6). A value of P < 0.05 was considered to be statistically significant. (N) Representative Western blot analysis (Upper) and densitometric quantification (Lower) of phosphorylated EGFR (pEGFR), total EGFR, and β-Catenin expression in liver tissues from db/db mice. Data are presented as mean values ± SEM (n = 6); a value of P < 0.05 was considered to be statistically significant. (O and P) db/db mice received a single i.p. injection of 42 nmol/kg FGF19WT or FGF19ΔKLB; mice treated with PBS buffer served as controls. (O) Real-time PCR analysis of hepatic expression levels of IL-6. (P) Representative Western blot analysis of pSTAT3/STAT3 expression in livers (Upper) and their densitometric quantification (Lower). Data are presented as mean values ± SEM (n = 5); a value of P < 0.05 was considered to be statistically significant.
The interleukin-6 (IL-6)/signal transducer and activator of transcription 3 (STAT3) signaling pathway has been shown to mediate FGF19-driven hepatocarcinogenesis (27). Accordingly, we measured IL-6 expression and STAT3 activation in liver tissues of db/db mice following administration of FGF19WT and FGF19ΔKLB. Indeed, hepatic mRNA levels of IL-6 in FGF19WT-treated db/db mice were significantly up-regulated, as reflected in an increase in STAT3 activation (Fig. 5 O and P). In contrast, FGF19ΔKLB failed to enhance IL-6 expression and STAT3 activation (Fig. 5 O and P), implying that the inability of FGF19ΔKLB to activate the IL-6/STAT3 signaling pathway may account for its noncarcinogenic property. Importantly, the BA synthesis inhibitory, glucose lowering, and insulin sensitizing activities of FGF19ΔKLB persisted throughout the 12-wk treatment period (SI Appendix, Fig. S8). We conclude that FGF19ΔKLB carries little risk of hepatic proliferation and, hence, holds significant potential as a safer and more efficacious molecule compared to its parent (FGF19WT) for use in the treatment of cholestatic liver disease and T2D.
Discussion
In this study, we harnessed our insights into the mechanism of the FGF19-FGFR-KLB-HS quaternary complex to engineer three FGF19 mutants that have diminished FGFR dimerization abilities and, hence, a reduced propensity to cause unwanted carcinogenesis (Figs. 1 and 2). We found that the three mutant FGF19 proteins we produced lay on a declining gradient in their ability to induce hepatocellular proliferation while retaining the full metabolic effects of the parent molecule (Figs. 2–5). These data validate our working model that the pleiotropic effects of FGFs can be dissected by tuning FGFR dimerization strength (33).
Besides M70 (NGM282), there is a significant list of nonmitogenic FGF19 variants in the literature (26, 29, 30). However, the molecular basis of the nonmitogenic properties of these variants has not been defined. Notably, Li and coworkers engineered another nonmitogenic FGF19 chimera by replacing the N-terminal 38WGDPI42 motif and HS binding regions in FGF19WT with the corresponding residues of FGF21 (26, 29). These modifications were introduced to restrict the specificity of the chimera to FGFR1c, thereby eliminating the chimera’s ability to engage and activate liver resident FGFR4 and, hence, its hepatocarcinogenic activity but retaining glucose lowering effects via adipose tissue resident FGFR1c (26, 29). It is also highly plausible that just as with M70, these modifications within the N terminus and HS binding regions of the chimera diminish its receptor dimerization potential and that this is the underlying cause for the nonmitogenic character of this chimera.
Preclinical and clinical studies have consistently validated robust effects of nonmitogenic FGF19 and FGF21 molecules for the alleviation of dyslipidemia and NAFLD in mice, monkeys, and humans. However, the antidiabetic effects of FGF19 and FGF21 variants seen in mice have not been extended to humans (30, 36–38). The reason for this is unclear, but it may be related to species differences in the molecular pathways or target organs that regulate lipid and glucose metabolism.
A number of other studies have reported ligands engineered so as to modulate receptor–ligand complex stability or geometry with a view to exerting control over the extent of receptor dimerization regulating the strength of downstream signals (39–42). Notably, Ho et al. recently described a mechanism-based stem cell factor (SCF) partial agonist that impaired c-Kit receptor dimerization, thus truncating downstream signaling amplitude (39). This SCF partial agonist retained therapeutic efficacy while exhibiting no anaphylactic off-target effects. The engineered mutations reduced the c-Kit binding affinity of this SCF variant below the threshold necessary for the activation of mast cells that give rise to an allergic and/or anaphylactic response. However, the remaining c-Kit binding affinity of these variants still enabled it to activate hematopoietic progenitors and, hence, to exert radioprotective effects (39).
Taken together, the experiments described here reinforce the emerging concept put forth by us and others that proteins with pleiotropic functions can be modified to retain one or more individual and desirable functions while eliminating those that are adverse. In the case of FGFs, this involves changing their binding affinity with their cognate-binding partners (i.e., receptor and coreceptor). Such relatively straightforward manipulations provide an unprecedented opportunity to engineer a repertoire of biologics with unique properties for use in both basic and translational research.
Materials and Methods
Expression and Purification of Recombinant FGF19WT, FGF19 Variants, and FGFRs.
A DNA fragment encoding the mature form of the WT human FGF19 gene (Arg23-Lys216, termed FGF19WT) was subcloned into the bacterial expression vector pET28a. A Fast Mutagenesis System Kit (TRANS) was used to introduce single mutations (Y115A, K149A, or D198A) into the FGF19WT expression vector, resulting in FGF19ΔFGFR, FGF19ΔHBS, and FGF19ΔKLB. A C-terminally truncated FGF19 (FGF19ΔC-tail, Arg23-Pro191) construct, which lacked 25 amino acids (βKlotho binding region) at the C terminus of FGF19WT, was generated by PCR. Complementary DNA (cDNA) fragments encoding the ligand binding regions (D2-D3 domains) of human FGFR1c (residues 142–365) and human FGFR4 (residues 144–355) were subcloned into pET-28a. Constructs were transformed into Escherichia coli BL21 (DE3). Protein expression was induced with 1 mM isopropyl β-d-1-thiogalactopyranoside at 37 °C for 4 h and the cells were collected by centrifugation. FGF19WT, FGF19 variants, and ligand-binding regions of FGFR1c and FGFR4 were refolded in vitro from isolated bacterial inclusion bodies using published protocols (11, 43). Briefly, refolded FGF19WT and its variants containing an N-terminal histidine tag were purified by nickel affinity column (HisTrap HP) and size exclusion chromatography (HiLoad 16/600 Superdex 75 column) with an AKTA purifier (GE Healthcare). Refolded FGFR1c and FGFR4 were purified using a heparin affinity column, followed by size exclusion chromatography (HiLoad 16/600 Superdex 75). Protein concentrations were determined by Nanodrop.
SPR Spectroscopy.
SPR experiments were performed on a BIAcore T200 system (GE Healthcare) in HBS-EP buffer (10 mM Hepes-NaOH, pH 7.4, 150 mM NaCl, 3 mM EDTA, 0.005% Surfactant P20). FGFR and βklotho chips were prepared by immobilizing ligand-binding regions of FGFR1c, FGFR4, or recombinant human βklotho (R&D Systems) onto CM5 biosensor chips using an amine coupling kit (GE Healthcare) as previously described (43). Briefly, purified FGFR1c, FGFR4, or βklotho samples were passed over the activated chip in Na acetate buffer, pH 5.5 and immobilized to 200–480 response units (RU). Chip surfaces were blocked using 1 M ethanolamine-HCl, pH 8.5. Heparin chips were prepared by immobilizing biotinylated heparin (Sigma-Aldrich) on flow channels of a research grade streptavidin chip (GE Healthcare). The control flow channel was left blank.
After chip preparation, increasing concentrations of FGF19WT or its variants in HBS-EP buffer were injected over the chip at 50 μL/min for 180 s. After each injection, HBS-EP buffer was passed over the chip for 180 s to monitor dissociation. Sensor chip surfaces were regenerated with 1.5 M NaCl in 10 mM Hepes-NaOH, pH 7.5 or 2.0 M NaCl in 10 mM sodium acetate, pH 4.5. For each injection, nonspecific responses from the control flow channel were subtracted from responses recorded for the FGF19 mutant flow channel. Data were processed with BIAEvaluation software, and equilibrium dissociation constants (Kd) were calculated from fitted saturation binding curves.
PLA.
HepG2 cells cultured in 12-well plates (1.5 × 105 per well) were starved in Dulbecco’s Modified Eagle Medium (DMEM) without fetal bovine serum (FBS) for 8 h and stimulated with 1.6, 16, or 160 nM FGF19WT or its variants for 15 min. After treatment, samples were rinsed three times with PBS and fixed with precooled 4% paraformaldehyde for 30 min. Samples were deposited on glass slides, blocked with 5% BSA for 1 h in a 37 °C incubator and incubated with a mixture of anti-FGFR4 primary antibody (Abcam, ab41948 and Santa Cruz Biotechnology, sc-136988) in a humidity chamber at 4 °C overnight. PLA reactions were completed according to the manufacturer’s instructions (DUO92101, Sigma). Briefly, slides were incubated with diluted PLUS and MINUS PLA probes in a preheated humidity chamber for 1 h at 37 °C, ligase was added to ligate the two probes for 30 min, and the slides were incubated with amplification solution for 100 min. Finally, slides were mounted with a coverslip using Duolink PLA Mounting Medium containing DAPI for 15 min and viewed in a confocal microscope. The resulting data were analyzed to determine the total number of PLA signals per cell.
Comparison of In Vitro Signaling Capacity of FGF19WT and Its Variants.
For analysis of in vitro signaling induced by FGF19WT and its variants, HepG2 or H4IIE cells were cultured in six-well plates, starved in DMEM without FBS overnight, and stimulated with different doses (1.6, 16, or 160 nM) of FGF19WT or its variants for 15 min. The cells were snap-frozen in liquid nitrogen, lysed in a buffer containing phosphatase and protease inhibitors, and processed for Western blot analysis using antibodies against phosphorylated PLCγ, total PLGγ, phospho-44/42 mitogen-activated protein kinase (ERK1/2), and total ERK1/2.
HTRF.
HTRF assays were performed as reported previously (44). Briefly, H4IIE cells cultured in 96-well tissue culture plates were starved in DMEM without FBS overnight and stimulated with different concentrations of FGF19WT or its variants for 15 min. Cell culture media were removed and the cells incubated in 50 μL of lysis buffer for at least 30 min at room temperature with shaking. Following homogenization by pipetting up and down, 16 µL of each cell lysate was transferred to a 384-well small volume assay plate. Four microliters of premixed antibody solutions contained in detection buffer was added to each well, the plate sealed with a plate sealer and incubated for 4 h at room temperature with shaking. A plate reader (TECAN Spark 10M) was set up for Eu3+ Cryptate and fluorescence emission read at two different wavelengths (665 nm and 620 nm). Curves were constructed by plotting the HTRF ratio versus log [protein] concentrations. The fluorescence signal intensity is directly proportional to the phospho-ERK1/2 (Thr202/Tyr204) content of the sample.
MTT Assays.
For in vitro mitogenicity assays, HepG2 or H4IIE cells (5 × 103 per well) were cultured in 96-well plates in a 5% CO2 incubator at 37 °C. Cells were starved for 12 h in DMEM without FBS, stimulated with 0, 4, 40, and 400 nM FGF19WT or its variants for 48 h, and treated with MTT (5.0 mg/mL) for 4 h. Culture medium was carefully aspirated, and 150 μL of DMSO was added to each well to fully dissolve MTT. Viable cells were quantitated by measuring the A490 using a microplate reader. To identify potential FGFRs isoforms mediating mitogenic activity of FGF19WT, HepG2 cell were pretreated with AZD4547 (40 nM, Selleckchem, S2801) before stimulation with FGF19 or its variants; the MTT assay was then performed using the protocol described above.
Animals and Animal Welfare.
Male db/db (C57BLKS/J-leprdb/leprdb) mice and male C57BL/6J mice were purchased from the Model Animal Research Center of Nanjing University. All animals were acclimatized to our laboratory environment before use and housed in a specific pathogen-free animal facility in a controlled environment (22 ± 2 °C, 50–60% humidity, 12-h light/dark cycle) and given free access to food and water. All protocols used in our studies were approved by the Animal Care and Use Committee of Wenzhou Medical University, China.
In Vivo Hepatocyte BrdU Labeling.
Six- to eight-week-old male C57BL/6J mice were randomized and injected i.p. with 25 mg/kg 5-bromo-2-deoxyuridine (BrdU, Sigma) twice daily for six consecutive days. During this period, mice were treated with either PBS alone or with 21 nmol/kg FGF19WT or its variants. Liver tissues were collected and fixed in 4% paraformaldehyde prior to paraffin embedding, sectioning, and evaluation by light microscopy. Sections of all collected tissues were stained to visualize BrdU incorporation as a marker of proliferation activity as previous described (26).
Measurement of BA Regulatory Activity of FGF19 and Its Variants in Mice.
To evaluate the acute effects of FGF19WT and its variants on BA, 8-wk-old male C57BL/6J mice received a single i.p. injection of PBS, FGF19WT, or its variants at a dose of 21 nmol/kg. Four hours after administration, Cyp7A1 mRNA levels in liver tissues were determined via a qRT-PCR. To analyze chronic effects of FGF19WT and its variants on BA biosynthesis, liver tissues were collected from db/db mice treated for 1 mo with either PBS, FGF19WT, FGF19ΔHBS, or FGF19ΔKLB (21 nmol/kg) (n = 6). Hepatic Cyp7A1 expression levels were determined by Western blot analysis using an anti-Cyp7A1 antibody. Hepatic bile acid concentrations, including cholic acid (CA), ursodeoxycholic acid (UDCA), deoxycholic acid (DCA), and chenodeoxycholic acid (CDCA), were determined by mass spectrometry as described previously (45).
Glucose Regulatory Activity of FGF19 and Its Variants in db/db Mice.
Twelve-week-old db/db mice were randomized and injected i.p. with PBS, FGF19WT, or its variants (21 nmol/kg body weight daily). Glucose levels were measured on alternate days using a glucose meter (Roche Diagnostics). Db/db mice treated with PBS served controls. Body weight and food intake were monitored during chronic administration experiments. After treatment for 4–12 wk, a GTT was performed. Briefly, after overnight fasting, db/db mice were challenged with a glucose solution (2.0 g/kg body weight), blood samples were collected at 0, 15, 30, 60, 120, and 180 min, and blood glucose levels were determined as described above. The area under the curve (AUC) was calculated by the trapezoid rule for the glucose tolerance curve using GraphPad Prism 7 software (GraphPad Software).
In Vivo Activation of IL-6/STAT3 Signaling Pathway.
Twelve-week-old db/db mice were injected i.p. with 42 nmol/kg FGF19WT or FGF19ΔKLB. Two hours later, liver tissues were excised and hepatic IL-6 mRNA levels were determined via a qRT-PCR. To analyze the effects of FGF19WT and FGF19ΔKLB on STAT3 activation, phosphorylated STAT3 (pSTAT3) and total STAT3 expression levels in the liver tissues were determined by Western blot analysis using anti-pSTAT3 and anti-STAT3 antibodies.
RNA Extraction, cDNA Synthesis, and qRT-PCR.
Total RNA samples were extracted from liver tissues with TRIzol reagent (Thermo Fisher Scientific) and purified using an RNAeasy Mini Kit (Qiagen). Total RNA was reverse-transcribed into cDNA with the One-Step gDNA Removal Kit (TransGen Biotech). qRT-PCRs were done using the ChamQ Universal SYBR qPCR Master Mix (Vazyme) with specific primers (listed in SI Appendix, Table S1) using a Step One Plus Real-Time PCR system (Applied Biosystems Quant Studio 3). Expression levels of the β-actin gene were used as endogenous controls to normalize for differences in the amount of total RNA added to each reaction.
Western Blot Analysis.
HepG2 cells, H4IIE cells, or liver tissues were homogenized in RIPA lysis buffer (25 mM Tris, pH 7.6, 150 mM NaCl, 1% Nonidet P-40, 1% sodium deoxycholate, 0.1% SDS) supplemented with protease and phosphatase inhibitors (all from Thermo Fisher Scientific). Protein concentrations were determined using a BCA Kit (Protein Assay Kit, Beyotime Biotechnology). Equal quantities of soluble protein (40 μg) were separated using 8–12% SDS/PAGE and electro-transferred onto a nitrocellulose membrane. Blots were probed with antibodies against phosphorylated PLCγ (Cell Signaling Technology, 14008), total PLGγ (Cell Signaling Technology, 5690), phospho-44/42 mitogen-activated protein kinase (ERK1/2) (Cell Signaling Technology, 4370S), total ERK1/2 (Cell Signaling Technology, 4695S), PCNA (Santa Cruz Biotechnology, sc25280), Ki67 (Abcam, ab15580), Cyp7A1 (Abcam, ab65596), phosphorylated EGFR (Cell Signaling Technology, 2231), total EGFR (Cell Signaling Technology, 9561), and β-Catenin (Cell Signaling Technology, 8480). Immunereactive bands were detected by incubation with secondary antibody (Santa Cruz Biotechnology) conjugated with horseradish peroxidase and visualized using enhanced chemiluminescence (ECL) reagents (Bio-Rad). Densitometric analyses of immunoblots were done using ImageJ software (version 1.38e, NIH).
Immunohistochemical Analyses.
After deparaffinization and rehydration, paraffin sections (5.0 mm) were subjected to immunohistochemical staining with primary antibodies against BrdU (Abcam, ab8152), PCNA (Santa Cruz Biotechnology, sc-25280), or Ki67 (Abcam, ab15580) overnight at 4 °C. After washing, sections were incubated with horseradish peroxidase conjugated secondary antibody against mouse or rabbit and developed using a 3,3-diaminobenzidine developing system (Vector Laboratories), counterstained with hematoxylin, and observed under light microscopy.
RNA-seq Analysis.
RNA-seq analysis was performed as previous described (46). Three independent RNA samples per treatment group were subjected to RNA-seq. Briefly, liver tissues of db/db mice treated with PBS, FGF19WT, or FGF19ΔKLB for 12 wk were collected. Total RNA was extracted using the TRIzol reagent (Invitrogen) and analyzed using a Bio-analyzer 2100 and an RNA 6000 Nano Lab Chip Kit (Agilent) with application of an RNA integrity number >7.0. Cleaved RNA fragments were reverse-transcribed to create a cDNA library in accordance with the protocol for the mRNA Seq sample preparation kit (Illumina), with a resulting average insert size for paired-end libraries of 200 bp (±50 bp). Samples were subjected to paired-end sequencing on an Illumina HiSeq 4000 (LC Sciences) following the vendor’s recommended protocol.
Mapped reads for each sample were assembled using StringTie. All transcriptomes were merged to reconstruct a comprehensive transcriptome using Perl scripts. Following generation of the final transcriptome, StringTie and edgeR were used to estimate the expression levels of all transcripts. StringTie was used to determine the expression level of mRNAs by calculating the fragment per kilobase per million (FPKM). Differentially expressed mRNAs and genes were selected as log2 (fold change) > 1 or log2 (fold change) < −1 and with statistical significance (P < 0.05) by R package.
Pathway analysis was conducted with ggplot2 as previously described (46). Briefly, networks of genes were generated based on their connectivity and then aligned against the KEGG. Enriched genes identified by KEGG analysis were analyzed using Fisher’s exact test and the χ2 test, and P values were corrected and adjusted to obtain the q value. A pathway was considered significant if the q value was <0.05. Raw sequencing data were submitted to Gene Expression Omnibus (GSE148997).
Statistical Analyses.
All results were expressed as mean values ± SEM. Statistical analysis was performed with one-way or two-way ANOVA and a Student t test using the statistical software NASDAQ: SPSS (SPSS Inc.). A value of P < 0.05 was considered to be statistically significant.
Supplementary Material
Acknowledgments
This work was supported by National Key R&D Program of China Grant 2017YFA0506000 (to X.L. and Z.H.), Natural Science Foundation of China Grants 81874323 and 81803415 (to Z.H. and J.N.), Natural Science Foundation of Zhejiang Grants LQ19H300002 and LGF19H30004 (to J.W. and D.W.), Science and Technology Project of Wenzhou Grant Y20180176 (to J.W.), and College Students’ science and technology of Zhejiang Grant 2019R413061 (to J.Z.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2010984117/-/DCSupplemental.
Data Availability.
All study data are included in the article and supporting information.
References
- 1.Beenken A., Mohammadi M., The FGF family: Biology, pathophysiology and therapy. Nat. Rev. Drug Discov. 8, 235–253 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Inagaki T., et al. , Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2, 217–225 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Potthoff M. J., Kliewer S. A., Mangelsdorf D. J., Endocrine fibroblast growth factors 15/19 and 21: From feast to famine. Genes Dev. 26, 312–324 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi M., et al. , Identification of a hormonal basis for gallbladder filling. Nat. Med. 12, 1253–1255 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Song K. H., Li T., Owsley E., Strom S., Chiang J. Y., Bile acids activate fibroblast growth factor 19 signaling in human hepatocytes to inhibit cholesterol 7alpha-hydroxylase gene expression. Hepatology 49, 297–305 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Potthoff M. J., et al. , FGF15/19 regulates hepatic glucose metabolism by inhibiting the CREB-PGC-1α pathway. Cell Metab. 13, 729–738 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kir S., et al. , FGF19 as a postprandial, insulin-independent activator of hepatic protein and glycogen synthesis. Science 331, 1621–1624 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kurosu H., et al. , Tissue-specific expression of betaKlotho and fibroblast growth factor (FGF) receptor isoforms determines metabolic activity of FGF19 and FGF21. J. Biol. Chem. 282, 26687–26695 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu X., et al. , Co-receptor requirements for fibroblast growth factor-19 signaling. J. Biol. Chem. 282, 29069–29072 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Lin B. C., Wang M., Blackmore C., Desnoyers L. R., Liver-specific activities of FGF19 require Klotho beta. J. Biol. Chem. 282, 27277–27284 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Goetz R., et al. , Molecular insights into the klotho-dependent, endocrine mode of action of fibroblast growth factor 19 subfamily members. Mol. Cell. Biol. 27, 3417–3428 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu X., et al. , C-terminal tail of FGF19 determines its specificity toward Klotho co-receptors. J. Biol. Chem. 283, 33304–33309 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuzina E. S., et al. , Structures of ligand-occupied β-Klotho complexes reveal a molecular mechanism underlying endocrine FGF specificity and activity. Proc. Natl. Acad. Sci. U.S.A. 116, 7819–7824 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen G., et al. , α-Klotho is a non-enzymatic molecular scaffold for FGF23 hormone signalling. Nature 553, 461–466 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu C., et al. , Elevated cholesterol metabolism and bile acid synthesis in mice lacking membrane tyrosine kinase receptor FGFR4. J. Biol. Chem. 275, 15482–15489 (2000). [DOI] [PubMed] [Google Scholar]
- 16.Ito S., et al. , Impaired negative feedback suppression of bile acid synthesis in mice lacking betaKlotho. J. Clin. Invest. 115, 2202–2208 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alvarez-Sola G., et al. , Fibroblast growth factor 15/19 (FGF15/19) protects from diet-induced hepatic steatosis: Development of an FGF19-based chimeric molecule to promote fatty liver regeneration. Gut 66, 1818–1828 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Fu L., et al. , Fibroblast growth factor 19 increases metabolic rate and reverses dietary and leptin-deficient diabetes. Endocrinology 145, 2594–2603 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Perry R. J., et al. , FGF1 and FGF19 reverse diabetes by suppression of the hypothalamic-pituitary-adrenal axis. Nat. Commun. 6, 6980 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lan T., et al. , FGF19, FGF21, and an FGFR1/beta-Klotho-activating antibody act on the nervous system to regulate body weight and glycemia. Cell Metab. 26, 709–718.e3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang C., et al. , Differential specificity of endocrine FGF19 and FGF21 to FGFR1 and FGFR4 in complex with KLB. PLoS One 7, e33870 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu X., et al. , FGF19-induced hepatocyte proliferation is mediated through FGFR4 activation. J. Biol. Chem. 285, 5165–5170 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Agrawal A., et al. , Molecular elements in FGF19 and FGF21 defining KLB/FGFR activity and specificity. Mol. Metab. 13, 45–55 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miura S., et al. , Fibroblast growth factor 19 expression correlates with tumor progression and poorer prognosis of hepatocellular carcinoma. BMC Cancer 12, 56 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nicholes K., et al. , A mouse model of hepatocellular carcinoma: Ectopic expression of fibroblast growth factor 19 in skeletal muscle of transgenic mice. Am. J. Pathol. 160, 2295–2307 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu X., et al. , Separating mitogenic and metabolic activities of fibroblast growth factor 19 (FGF19). Proc. Natl. Acad. Sci. U.S.A. 107, 14158–14163 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou M., Yang H., Learned R. M., Tian H., Ling L., Non-cell-autonomous activation of IL-6/STAT3 signaling mediates FGF19-driven hepatocarcinogenesis. Nat. Commun. 8, 15433 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou M., et al. , Separating tumorigenicity from Bile acid regulatory activity for endocrine hormone FGF19. Cancer Res. 74, 3306–3316 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Ge H., et al. , Characterization of a FGF19 variant with altered receptor specificity revealed a central role for FGFR1c in the regulation of glucose metabolism. PLoS One 7, e33603 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DePaoli A. M., et al. , FGF19 analog as a surgical factor mimetic that contributes to metabolic effects beyond glucose homeostasis. Diabetes 68, 1315–1328 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Luo J., et al. , A nontumorigenic variant of FGF19 treats cholestatic liver diseases. Sci. Transl. Med. 6, 247ra100 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Zhou M., et al. , Engineered fibroblast growth factor 19 reduces liver injury and resolves sclerosing cholangitis in Mdr2-deficient mice. Hepatology 63, 914–929 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang Z., et al. , Uncoupling the mitogenic and metabolic functions of FGF1 by tuning FGF1-FGF receptor dimer stability. Cell Rep. 20, 1717–1728 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zinkle A., Mohammadi M., A threshold model for receptor tyrosine kinase signaling specificity and cell fate determination. F1000 Res. 7, F1000 Faculty Rev-872 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gavine P. R., et al. , AZD4547: An orally bioavailable, potent, and selective inhibitor of the fibroblast growth factor receptor tyrosine kinase family. Cancer Res. 72, 2045–2056 (2012). [DOI] [PubMed] [Google Scholar]
- 36.Harrison S. A., et al. , NGM282 for treatment of non-alcoholic steatohepatitis: A multicentre, randomised, double-blind, placebo-controlled, phase 2 trial. Lancet 391, 1174–1185 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Gaich G., et al. , The effects of LY2405319, an FGF21 analog, in obese human subjects with type 2 diabetes. Cell Metab. 18, 333–340 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Geng L., Lam K. S. L., Xu A., The therapeutic potential of FGF21 in metabolic diseases: From bench to clinic. Nat. Rev. Endocrinol. 10.1038/s41574-020-0386-0 (2020). [DOI] [PubMed] [Google Scholar]
- 39.Ho C. C. M., et al. , Decoupling the functional pleiotropy of stem cell factor by tuning c-kit signaling. Cell 168, 1041–1052.e18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim A. R., et al. , Functional selectivity in cytokine signaling revealed through a pathogenic EPO mutation. Cell 168, 1053–1064.e15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swanson M. D., et al. , Engineering a therapeutic lectin by uncoupling mitogenicity from antiviral activity. Cell 163, 746–758 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moraga I., et al. , Tuning cytokine receptor signaling by re-orienting dimer geometry with surrogate ligands. Cell 160, 1196–1208 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Y., et al. , Regulation of receptor binding specificity of FGF9 by an autoinhibitory homodimerization. Structure 25, 1325–1336.e3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ayoub M. A., et al. , Homogeneous time-resolved fluorescence-based assay to monitor extracellular signal-regulated kinase signaling in a high-throughput format. Front. Endocrinol. 5, 94 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li T., et al. , Overexpression of cholesterol 7α-hydroxylase promotes hepatic bile acid synthesis and secretion and maintains cholesterol homeostasis. Hepatology 53, 996–1006 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Halim L., et al. , An atlas of human regulatory T helper-like cells reveals features of Th2-like tregs that support a tumorigenic environment. Cell Rep. 20, 757–770 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and supporting information.