Significance
Diabetic kidney disease (DKD) is the most common cause of chronic and end-stage renal failure in the world. In a genetically susceptible host, poor metabolic control contributes to DKD development. The epigenome integrates signals from sequence variations and environmental alterations. We performed genome-wide DNA methylation association analysis in one of the best-characterized kidney disease cohorts: The Chronic Renal Insufficiency Cohort study. Complex computational integration analysis indicated the key role of genetic variations in DNA methylation. Our analysis highlighted loci, where methylation and gene-expression changes likely mediate the genotype effect on kidney disease development. Functional annotation of high-confidence genes suggested the causal role of inflammation, specifically, complement activation and apoptotic cell clearance in kidney disease development.
Keywords: methylation quantitative trait loci (mQTL), multitrait colocalization analysis (moloc), epigenetics, multiomics integration analysis, chronic kidney disease
Abstract
Poor metabolic control and host genetic predisposition are critical for diabetic kidney disease (DKD) development. The epigenome integrates information from sequence variations and metabolic alterations. Here, we performed a genome-wide methylome association analysis in 500 subjects with DKD from the Chronic Renal Insufficiency Cohort for DKD phenotypes, including glycemic control, albuminuria, kidney function, and kidney function decline. We show distinct methylation patterns associated with each phenotype. We define methylation variations that are associated with underlying nucleotide variations (methylation quantitative trait loci) and show that underlying genetic variations are important drivers of methylation changes. We implemented Bayesian multitrait colocalization analysis (moloc) and summary data-based Mendelian randomization to systematically annotate genomic regions that show association with kidney function, methylation, and gene expression. We prioritized 40 loci, where methylation and gene-expression changes likely mediate the genotype effect on kidney disease development. Functional annotation suggested the role of inflammation, specifically, apoptotic cell clearance and complement activation in kidney disease development. Our study defines methylation changes associated with DKD phenotypes, the key role of underlying genetic variations driving methylation variations, and prioritizes methylome and gene-expression changes that likely mediate the genotype effect on kidney disease pathogenesis.
More than 800 million people worldwide suffer from chronic kidney disease (CKD) (1). Despite the important clinical needs, there is no curative therapy for CKD. Current treatments mostly rely on improving blood pressure and blood glucose control. New therapies that target novel causal pathways are desperately needed.
The role of immune cells and inflammation in diabetic kidney disease (DKD) development remains controversial (2, 3). DKD is traditionally considered a nonimmune-mediated kidney disease (2). Genome-wide association analysis studies highlighted the role of podocytes and proximal tubules in kidney disease development (4, 5). On the other hand, human kidney gene-expression studies have reproducibly indicated a correlation between immune cells, certain cytokine levels, and disease severity (6–8). The lack of genetic support for inflammation in CKD led to the notion that inflammation might be a secondary phenomenon, and targeting such a pathway could be futile for DKD.
Metabolic factors—such as diabetes, obesity, aging, and intrauterine nutritional environment—play critical roles in CKD development (9, 10). Intrauterine nutritional deprivation or periods of hyperglycemia will increase kidney disease risk, even after several decades of good metabolic control, a phenomenon called “metabolic memory or programming” (11–15). Epigenetic changes have been proposed to mediate this long-lasting effect of nutritional environment, as epigenome editing enzymes require intermediates of cellular metabolism (such as acetyl and methyl groups) for histone and DNA modifications; thus, nutrient availability can directly influence the epigenome (16). Epigenetic modifications are maintained during cell division, therefore, the epigenome can serve as a long-term environmental footprint.
Methylome-wide associated studies (MWAS) have been performed to characterize methylation changes in CKD (17, 18). Studies from the Diabetes Control and Complications Trial (DCCT) group identified changes around the thioredoxin-interacting protein (TXNIP) gene in subjects with diabetes (14). Changes in CpG methylation associated with kidney function were also identified in blood samples of Pima Indian subjects (19). The largest MWAS study included subjects from the Atherosclerosis Risk in Communities (ARIC) and Framingham Heart Study (FHS) cohorts and identified signals with genome-wide significance. A large number of differentially methylated regions have been reported in recent studies that analyzed microdissected human kidney tubule samples (20, 21). Critical limitations of these studies are the lack of replication and the lack of examining the contribution of underlying genetic variations to MWAS signals.
The heritability of kidney function was estimated to be around 30 to 50% (22, 23). Recently, published large population-based genome-wide association studies (GWAS) have identified hundreds of variants showing genome-wide significant association with estimated glomerular filtration rate (eGFR) (24–27). GWAS studies highlighted important differences in the genetic architecture of different kidney disease traits, such as albuminuria, eGFR, and kidney function decline (4, 28). Despite the success of the GWAS mapping, genes, pathways, and cell types explaining CKD heritability are poorly understood. Almost all identified GWAS variants (>90%) are in the noncoding region of the genome. Expression of quantitative trait (eQTL) studies have been powerful to annotate disease-driving genetic variations to prioritize disease-causing genes. Our initial integration of CKD GWAS and kidney eQTL data were able to prioritize likely causal genes for 20% of the GWAS loci (5). These initial studies highlighted the key role of the proximal tubules and endolysosomal trafficking in kidney disease pathogenesis. eQTL analysis relies on detecting genotype-driven differences in gene expression at baseline condition; however, it is possible that regulatory region-driven gene-expression differences only manifest upon an external stimulus (19, 21, 29). As cells constantly respond to external stimuli, it is difficult to catalog all context-dependent changes determined by underlying genetic variation. Integration of epigenetic information might be able to capture such gene regulatory logic, and therefore can define context-dependent expression changes and improve GWAS target identification.
Here, we adopted a comprehensive approach by integrating epigenetic and genetic signals to identify novel disease-driving pathways and therapeutic targets. We analyzed subjects with varying degrees of kidney disease from one of the largest and best phenotyped CKD cohorts: The Chronic Renal Insufficiency Cohort (CRIC) (30, 31). Given new developments in immune therapeutics, we focused on blood immune cells. We found that underlying genetic variations play important roles in modulating the epigenome-disease association, indicating that epigenetic variations can be used to prioritize GWAS loci. Bayesian integration and summary data-based Mendelian randomization analysis of methylation QTL (mQTL) and eQTL datasets suggested the causal role of inflammation, specifically, apoptotic cell clearance and complement activation in CKD, which could open new avenues for therapeutics development for this devastating condition.
Results
Cohort Characteristics.
Baseline demographic and clinical characteristics of the participants are described in Dataset S1. The study used a subsample of the entire CRIC cohort. To reduce disease heterogeneity, we selected only subjects with diabetes (SI Appendix, Fig. S1). The baseline kidney function (eGFR) and its distribution followed the pattern observed in the CRIC cohort (SI Appendix, Fig. S2A). The mean baseline eGFR at the time of enrollment was 44 mL/min/1.73 m2 (Dataset S1). We also enriched the population for subjects with progressive kidney disease by selecting subjects with the fastest eGFR decline and matched control samples based on their baseline characteristics, such as age, race, and gender (Materials and Methods and SI Appendix, Figs. S2 B and C and S3). The mean eGFR slope of the fast progressor DKD group was −3.97 mL/min/1.73 m2/y, based on close to 8 y of follow-up data. Hemoglobin A1c, a measure of glycemic control, was 8.06% ± 1.76%. The mean 24-h urine albumin was 1.27 ± 2.18 g (Dataset S1). Although this design has its own limitation, it allowed us to analyze patients with diabetes and also subjects with progressive kidney function decline, as a large proportion of subjects in the CRIC study had stable kidney function.
Methylome-Wide Association Analysis for DKD Phenotypes.
DKD has different phenotypic manifestations, such as albuminuria, eGFR, and the rate of kidney function decline. We analyzed the relationship between methylation levels and DKD-associated phenotypes using the cross-sectional design. We first investigated the association between glycemic control (HgbA1c) and methylation levels of 866,836 cytosines (CpGs), interrogated by the Illumina Human MethylationEPIC BeadChip, in 473 whole-blood samples obtained at the time of enrollment (Fig. 1A). After data cleaning and normalization, we used linear regression to characterize the association between methylation and HgbA1c. The final model included batch effect, age, sex, genetic background, hypertension, and cell heterogeneity as covariates and cytosine methylation (M values) as outcome. The associations between HgbA1c and methylation changes (differentially methylated probes, DMPs) across the genome are shown in Fig. 1B. One probe, cg19693031, with P = 6.22E-14 (SI Appendix, Figs. S4 A and B and S5 A and B), located in the promoter region of the TXNIP gene (SI Appendix, Fig. S4C) passed the stringent Bonferroni-corrected genome-wide significance value (P < 6.42E-08), while 110 passed the discovery significance threshold (two-sided P < 5E-05) (Dataset S2).
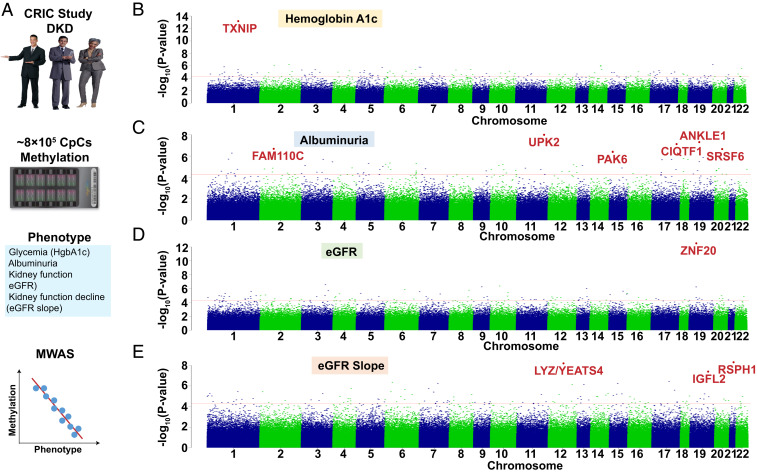
Fig. 1.
Methylation changes associated with DKD phenotypes (glycemia, albuminuria, eGFR, and eGFR slope) in whole-blood samples of the CRIC study participants. (A) Study schematics. Methylation levels of ∼800,000 loci measured by Illumina Human MethylationEPIC arrays were used to analyze the associations with DKD phenotypes (glycemia, albuminuria, eGFR, and eGFR slope) using a linear regression model. (B) Manhattan plot. The y axis is the −log10 of the association P value of hemoglobin A1c and methylation. The x axis represents the genomic location of the CpG probes. n = 473 samples. (C) Manhattan plot. The y axis is the −log10 of P value of albuminuria (24 h) and methylation. The x axis represents the genomic location of the CpG probes. n = 473 samples. (D) Manhattan plot. The y axis is the −log10 of P value of kidney function (baseline eGFR) and methylation. The x axis represents the genomic location of the CpG probes. n = 473 samples. (E) Manhattan plot. The y axis is the −log10 of P value of kidney function decline (eGFR slope) and methylation. The x axis represents the genomic location of the CpG probes. n = 410 samples.
Next, we performed methylome-wide association analysis for albuminuria, an important manifestation of DKD. Six probes showed significant association with albuminuria (false-discovery rate [FDR] < 0.05) (Fig. 1C and SI Appendix, Fig. S5 C and D), while 73 probes passed the discovery significance threshold (Dataset S3). The top locus was around Uroplakin 2 (UPK2), a urothelial specific gene.
Methylome-wide association analysis for baseline kidney function (eGFR) identified 99 DMPs at the discovery significance threshold (Fig. 1D and Dataset S4). One CpG site, cg17944885, passed the most stringent, Bonferroni-corrected P-value threshold (SI Appendix, Fig. S5 E and F). This top DMP located close to the 3′ region of zinc finger protein 20 (ZNF20), a transcription factor with unknown function (32) (Fig. 1D and SI Appendix, Fig. S6A). The methylation of cg17944885 showed an observable association with eGFR, but not with HgbA1c and albuminuria (SI Appendix, Fig. S6 B–G). Sensitivity analysis was conducted to test the robustness of our results (SI Appendix, Fig. S7) and indicated lack of measurable influence of smoking (33), age (34), and body mass index (35).
Defining future kidney function decline is one of the most important clinical question. We next examined the association between methylation changes and future kidney function decline using two different models. First, we used a conditional logistic regression model comparing 410 subjects using a stratified design (Materials and Methods) by matching for age, race, gender, baseline eGFR, duration of diabetes, and glycemic control (36). The second analysis used a linear regression model by adjusting for age, batch effect, top 10 genetic PCs, hypertension, cell proportions, hemoglobin A1c, and urinary albumin to creatinine ratio. The logistic regression model identified 9 CpG sites (SI Appendix, Fig. S8 and Dataset S5), while the linear regression model identified 111 probes at a discovery significance threshold (P < 5E-05) (Dataset S6). Three DMPs (cg16408865, cg15507486, and cg01491004) passed the most stringent Bonferroni-corrected P-value cutoff (6.42E-08) (Fig. 1E and SI Appendix, Fig. S5 G and H). The distribution of the P values across the whole genome is shown in Fig. 1E. For example, the methylation of cg02713581 (SI Appendix, Fig. S9A) showed significant negative association with kidney function decline (two-sided P = 5.63E-07) (SI Appendix, Fig. S9 B and C). Sensitivity analysis, performed to test the robustness of our results, indicated the lack of measurable influence of smoking and body mass index (SI Appendix, Figs. S10 and S11). Quality-control metrics, such as inflation coefficient, statistical significance, and correlation strength association (SI Appendix, Fig. S5) further supported our conclusions. In summary, we defined genome-wide methylation changes associated with four DKD phenotypes: Glycemia, albuminuria, eGFR, and eGFR decline.
Cytosine Methylation Changes Can Be Replicated in External Cohorts.
We next examined the overlap between methylation changes associated with different DKD phenotypes. Consistent with earlier epigenetic studies, methylation changes were mostly specific to the analyzed phenotypes. Glycemic control showed the greatest overlap with other DKD phenotypes, indicating the potential role of glycemia in driving epigenetic changes and phenotype development (Fig. 2A). In our review of the literature, no prior studies have analyzed methylation changes associated with albuminuria and kidney function decline. A previous study by Chen et al. (14) identified important association between methylation at the TXNIP locus and hyperglycemic metabolic memory in patients with type 1 diabetes in the DCCT cohort. Our data further support the broad association between the TXNIP locus methylation and glycemia in patients with type 2 diabetes and diverse genetic background (14, 37).
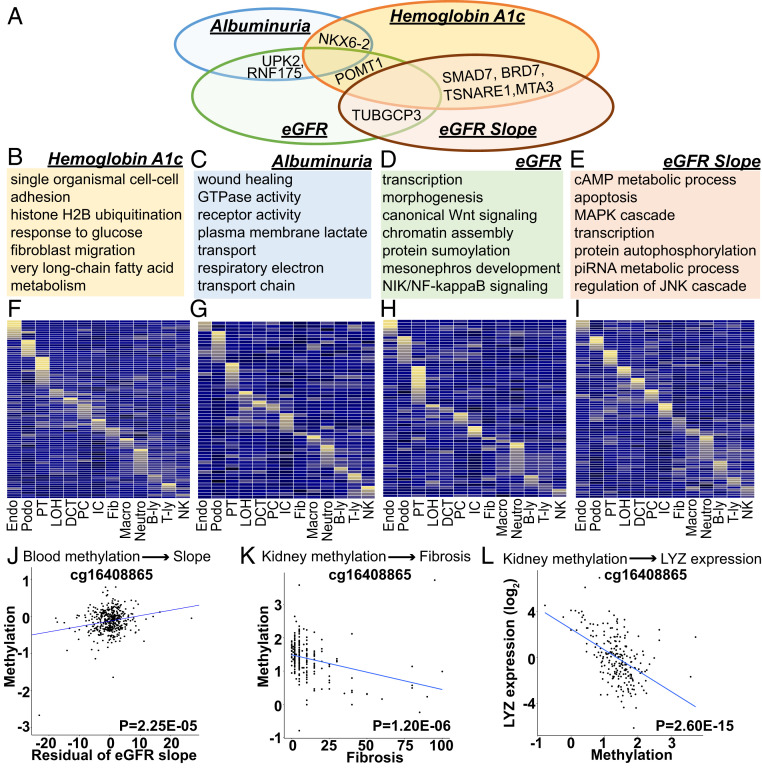
Fig. 2.
Functional annotation of DKD phenotype-associated methylation changes. (A) Overlap of loci that showed methylation association with multiple DKD phenotypes (as denoted by the closest genes to the methylation change). (B–E) Functional annotation (gene ontology) of genes showing association with DKD phenotypes in MWAS. (F–I) Adult kidney single-cell expression enrichment of MWAS identified genes (as defined by DMP proximity) (41). Each row represents one gene and each column represents one cell type. B-ly, B-lymphocytes; DCT, distal convoluted tubule; Endo, endothelia; fib, fibroblast; IC, intercalated cells of the collecting duct; LOH, loop of Henle; macro, macrophages; neutro, neutrophils; NK, natural killer cells; PC, principal cells of the collecting duct; Podo, podocyte; PT, proximal tubule; T-ly, T lymphocytes. Mean expression values of the genes were calculated in each cluster. The color scheme is based on z-score distribution. Yellow indicates higher expression while blue indicates low expression. (J) Correlation between the methylation of cg1640885 and kidney function decline (residual of eGFR slope) (SI Appendix) in blood samples of CRIC study participants. The x axis is the residual of eGFR slope and y axis is M value (methylation). n = 410 samples. (K) Correlation between the methylation of cg1640885 and degree of fibrosis in microdissected human kidney samples (Dataset S13). The x axis is the percent fibrosis (0–100%) and y axis is M value (methylation). n = 227 samples. (L) Correlation between the methylation of cg1640885 and expression of LYZ in microdissected human kidney samples. The x axis is normalized M value (M) and y axis is normalized gene expression [log2(TPM); transcripts per million]. n = 227 samples.
Despite multiple studies have analyzed the association between eGFR and methylation patterns in peripheral blood mononuclear cells (PBMCs), no consistent or validated DMPs have been reported. Here, we used the summary statistics data from the ARIC and FHS studies (29) that included mixed diabetic and nondiabetic cohorts, the Pima Indian cohort (19) that included subjects with early DKD, and the Veterans Aging Cohort Study (VACS) that included subjects with HIV and kidney disease (38). In addition, microdissected kidney tubule-specific methylation and kidney function datasets were available from the Susztak laboratory Biobank (7, 21) (Dataset S7).
There was a significant, direction-consistent association between the methylation levels of cg17944885 and eGFR in the ARIC and FHS studies with two-sided P = 1.61E-07 and 2.03E-17, respectively (Dataset S8). The methylation of this CpG site showed an association with baseline eGFR in subjects with diabetes of Pima Indian heritage (two-sided P = 3.01E-04) (19), and patients with HIV at the VACS cohort (two-sided P = 2.5E-05) (Dataset S8) (38). Furthermore, the methylation levels of cg17944885 showed significant association with kidney disease (fibrosis) (two-sided P = 4.70E-03) (21) in microdissected human kidney tubule samples (Dataset S7). Our results indicated that phenotype-specific methylation changes could be successfully replicated in different cohorts, and even in different tissue types.
Functional Annotation of DKD Phenotype-Associated Loci.
Most current cytosine methylation models proposed that methylation of promoter or enhancer regions can alter transcription factor binding, leading to quantitative changes in transcript levels. Gene regulatory region annotation (promoter and enhancer, and so forth) for PBMCs was generated by combining multiple histone chromatin immunoprecitation data (ChiP-seq) by ChromHMM (39, 40). Compared to all probes present on the EPIC arrays, eGFR-associated DMPs were enriched in regions annotated as enhancers and promoters (SI Appendix, Fig. S12A). Slope-associated DMPs were enriched in promoter and transcribed regions in PBMCs (SI Appendix, Fig. S12B). Similar enrichment analysis using human kidney tissue indicated that DMPs were more likely to be located in regions annotated as promoters or enhancers in human kidneys. Comparing regulatory annotations of different organs, we found that DMP-enriched enhancers showed kidney and blood specificity (SI Appendix, Fig. S12).
To understand the potential functional role of DKD phenotype-associated methylation changes, we performed gene ontology-based functional annotation. We found that methylation changes associated with glycemia showed enrichment around genes involved in glucose and fatty acid metabolism (Fig. 2B and Dataset S9). Methylome association analysis for albuminuria identified changes around the vicinity of genes associated with wound healing and small GTPase functions (Fig. 2C and Dataset S10). Methylome-wide association for eGFR showed enrichment for transcription and development including kidney development (Fig. 2D and Dataset S11). Annotation of loci associated with kidney function decline showed enrichment for transcription, MAPK and JNK cascades (Fig. 2E and Dataset S12).
Next, we examined cell-type expression of genes associated with DKD phenotypes. We used single-cell gene-expression datasets that we generated earlier by profiling whole kidney samples (41). Our results indicated that the closest genes of kidney phenotype-associated DMPs showed important cell-type–specific expression. Several genes expressed in kidney epithelial and endothelial cells; others showed important immune cell-specific expression (Fig. 2 F–I). Overall, the results indicated that DKD-associated methylation changes affected a variety of cell types.
The methylation levels of cg16408865 that associated with kidney function decline in blood samples (Fig. 2J), also strongly associated with kidney fibrosis in microdissected human kidney tissue samples (Dataset S13), indicating that changes observed in blood samples could be relevant for kidney tissue samples as well (Fig. 2K). For example, the methylation of cg16408865 strongly correlated with LYZ expression in microdissected human kidney samples (Dataset S13) (Fig. 2L). Overall, our results indicated that DKD-associated methylation changes showed enrichment in cell-type–specific regulatory regions in blood and kidney cells and altered phenotype-specific pathways.
Genetically Driven Methylation Changes.
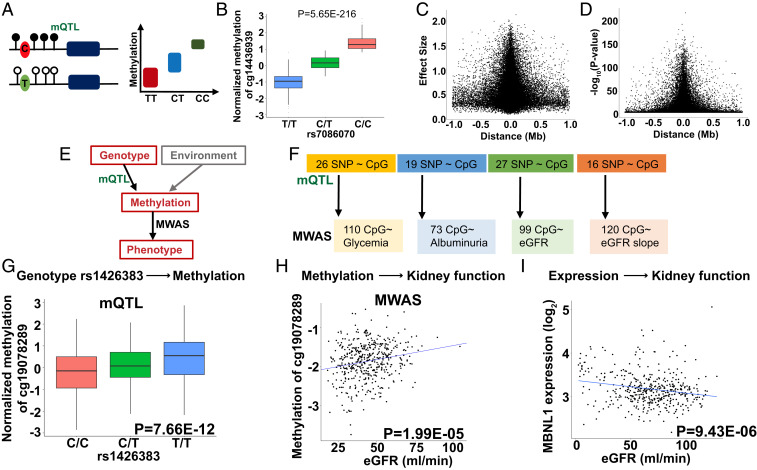
To understand the contribution of genetic variations to methylation variations, we analyzed the association of genetic variations and local methylation changes (mQTL) (Fig. 3A) (42). We interrogated the association of 6,177,888 SNPs and methylation levels of 836,828 CpG sites in 473 blood samples using a linear regression model. The mQTL analysis was limited to SNPs located within ± 1-Mb (cis) window of each queried CpG site (Materials and Methods). We identified 171,732 CpG as significant mQTLs (CpG site that regulated by at least one SNP) at FDR < 0.05 and 123,541,191 significant SNP-CpG pairs. For example, the underlying nucleotide variant (rs7086070) had a robust effect on the local DNA methylation level of cg14436939 (P = 5.65E-216) (Fig. 3B). The effect size (Fig. 3C) and significance (Fig. 3D) of the lead SNP on each mCpG decreased for SNPs further away from the transcription start site, suggesting that genetic variations in promoter regions have larger effects on methylation levels. Our results replicated the significant SNP-CpG associations described earlier with the fraction of true positives (π1) being around 0.92 to 0.94 (Dataset S14) [using the threshold criteria of 1E-14 established by Gaunt et al. (43)].
Fig. 3.
Genotype-driven methylation changes (mQTL) contribute to MWAS signals. (A) Schematic representation of mQTL; genotype-methylation association study. DNA methylation changes regulated by a nearby SNP. (B) An example of mQTL. Higher allele dosage C of the rs7086070 variant is associated with higher methylation levels of cg14436939 (P = 5.65E-216). The x axis shows the allelic dosage of rs7086070; y axis shows normalized methylation (INT-transformed M value). Center lines show the medians; box limits indicate the 25th and 75th percentiles; whiskers extend to the fifth and 95th percentiles; outliers are represented by dots. (C) The effect size (y axis) of the best mSNPs (the lead mQTL) decreases as the distance (x axis) from the transcription start site increases. n = 473 samples. (D) The strength of association (y axis) of the best mSNPs (the lead mQTL) decreases as the distance (x axis) from the transcription start site increases. n = 473 samples. (E) Both genetic variations and environmental changes contribute to methylation variation. Genotype-associated methylation changes can be uncovered by mQTL analysis, while MWAS analysis detects methylation and phenotype association. (F) Differentially methylated probes identified in DKD MWAS studies. The number of DMPs influenced by underlying genetic variants, identified by mQTL analysis. (G) Genotype and methylation association of rs1426383 variant and the methylation of cg19078289 identified by mQTL analysis of blood samples of CRIC participants. The x axis shows allele dosage of rs1426383; y axis shows normalized methylation (INT-transformed M value). Center lines show the medians; box limits indicate the 25th and 75th percentiles; whiskers extend to the 5th and 95th percentiles; outliers are represented by dots. n = 473 samples. (H) The methylation level of cg19078289 correlate with kidney function (eGFR) in CRIC study participants. The x axis: eGFR (mL/min); y axis: normalized methylation (INT-transformed M value), n = 473 samples. (I) Correlation of MBNL1 expression and kidney function in 433 microdissected human kidney samples (Dataset S16) (5). The x axis: eGFR (mL/min); y axis: expression (log2FPKM), n = 433 samples.
Given the robust genotype-driven signals on methylation levels, we next examined the potential role of genetic variation in our MWAS analysis (Fig. 3E). Most prior studies have failed to incorporate genotype analysis into MWAS studies. To ascertain the contribution of genetic variations in our MWAS results, we overlapped our MWAS and mQTL signals. We found that 22% of the identified MWAS signals could be driven by underlying genetic variations. For example, 26 of the 110 identified methylation associated with glycemia showed significant associations with underlying genetic variations (Fig. 3F and Dataset S15).
As an example, we show the muscleblind-like protein 1 (MBNL1) locus (SI Appendix, Fig. S13A). The genotype of rs1426383 (C or T) showed association with the cytosine methylation of this locus cg19078289 (mQTL) (Fig. 3G). The methylation variations of cg19078289 associated with eGFR in our MWAS analysis (Fig. 3H and SI Appendix, Fig. S13B), suggesting that the underlying genetic variations likely contributed to the detected MWAS signal. To further prove the association between genetic variants and kidney function (eGFR), we examined the eGFR GWAS study. This SNP (rs1426383) showed a nominally significant association with eGFR (two-sided P = 1.70E-05) (28), further substantiating the role of underlying genetic variants driving the MWAS association. Furthermore, MBNL1 expression in microdissected human kidney tubule samples correlated with eGFR (Fig. 3I) and fibrosis (SI Appendix, Fig. S13C) (P = 9.43E-06 and P = 2E-16, respectively) (Dataset S16). Future studies shall define the functional role of MBNL1 in the kidney.
In summary, our results indicated that underlying genetic variation play an important role in influencing local methylation and downstream gene-expression levels, and likely also contributed to the MWAS signals.
Integration of Genetically Driven Methylation and Gene-Expression Changes with GWAS Signals Can Prioritize Genes for Kidney Dysfunction.
As our results indicated the critical role for genetic variations influencing the association between methylation and disease state, we therefore systematically investigated whether we could identify kidney function-associated genetic loci that are also associated with methylation and gene-expression changes, and disease state (Fig. 4A). As genotypes do not suffer from reverse causation, such analysis can further prioritize methylation changes that are causally linked to disease development.
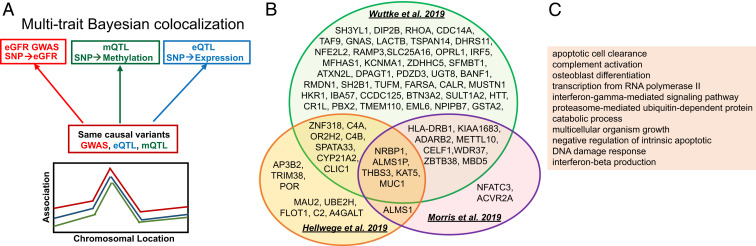
Fig. 4.
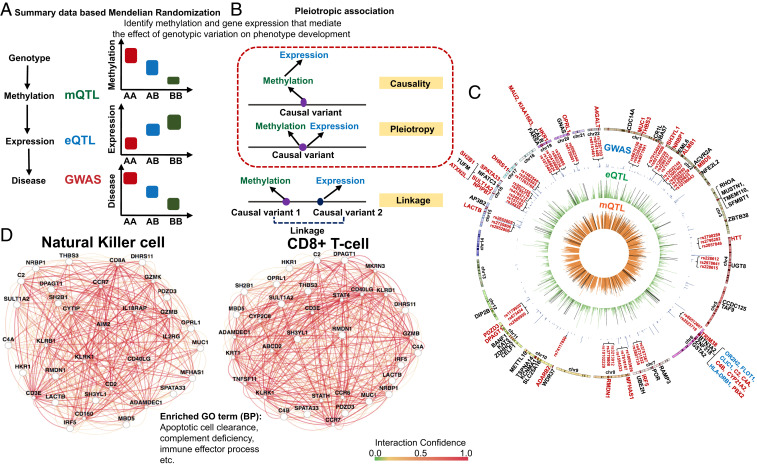
Bayesian moloc (GWAS, mQTL, and eQTL) to identify genetic loci associated with kidney function, methylation, and gene-expression changes. (A) Illustration of the Bayesian framework for moloc analysis. The analysis will test for different scenarios, such as whether the causal variants associated with kidney function (GWAS), methylation (mQTL), and gene expression (eQTL) are shared. (B) We identified 71 protein-coding genes (85 total), where disease state (GWAS), methylation levels (mQTL), and gene expression (eQTL) share causal genetic variants. Each circle represents summary data from a specific GWAS study (28, 45, 46). (C) Functional enrichment (gene ontology) analysis results of the 71 protein-coding genes, where causal genetic variants for kidney function, methylation, and gene expression are shared.
First, we used a Bayesian statistical framework established in the multiple traits colocalization (moloc) analysis (44). We analyzed loci from three recently published large multiethnic studies that examined genotype and kidney function (eGFR) correlations (GWAS) (28, 45, 46). We identified, genetic variants that showed association with methylation levels at 267 CpG loci. The expression of 85 genes (71 protein-coding genes and 14 noncoding genes), were associated with genetic and epigenetic changes (probability of moloc abc_PP ≥ 0.8) (Datasets S17–S19). We observed strong consistency between the different GWAS cohorts and larger GWAS studies [such as CKDGen (28)] identified more loci. Several genes were prioritized by multiple GWAS studies, including nine genes (five protein-coding genes) identified by all GWAS/mQTL/eQTL integrations (Fig. 4B). A couple of putative CKD risk genes—such as NRBP1, ALMS1P, MUC1, and METTL10—have been identified earlier by CKD GWAS and kidney compartment gene expression (eQTL) integration studies (5, 28, 47). Nine of the 71 protein-coding genes, which located within the major histocompatibility complex (MHC) regions, such as HLA-DRB1 and C4B, C4A, and C2 (Dataset S20), need further validation due to the complex genetic architecture of this region (48).
Functional enrichment analysis of the 267 significant moloc-prioritized CpG sites indicated enrichment in enhancer and promoter regions in PBMC and kidney samples (SI Appendix, Fig. S14). Gene ontology analysis indicated that genes prioritized for kidney function were enriched for inflammation, specifically, apoptotic cell clearance, complement activation, and IFN signaling (Fig. 4C and Dataset S21). Taken together, our Bayesian moloc integration highlighted genetic signals, where methylation, gene expression, and phenotype variations were driven by the same genetic variants, and prioritized 267 methylation sites and 85 likely causal kidney disease risk genes.
Summary Data-Based Mendelian Randomization to Define Genetic Variations Driving Methylation and Gene-Expression Changes.
Next, we narrowed the moloc-identified loci by performing summary data-based Mendelian randomization (SMR) (49) analysis to understand whether the effect of genetic variants on phenotype development is mediated by gene-expression changes via cytosine methylation (Fig. 5A). SMR tests three scenarios: Causality, where the effect of a genetic variant on transcription is mediated by methylation; pleiotropy, where a genetic variant has direct effects on both methylation and transcription; and linkage, where two or more distant genetic variants in linkage disequilibrium (LD) affecting methylation and transcription independently (Fig. 5B). We further complemented the SMR analysis with a HEIDI test (heterogeneity in dependent instruments) to distinguish causality and pleiotropy from linkage (50). Our analytical framework included pleiotropic association tests in three directions, including methylation to transcription, methylation to phenotype, and transcription to phenotype (Materials and Methods) (28, 45, 46).
Fig. 5.
SMR to define genetic variations driving methylation and gene-expression changes. (A) We used SMR to distinguish scenarios, where the effect of a genetic variant on transcription is mediated by methylation. (B) Pleiotropic association test was conducted to distinguish causality, pleiotropy, and linkage. Causality: The effect of a genetic variant on transcription is mediated by methylation. Pleiotropy: A genetic variant has direct effects on both methylation and transcription. Linkage: Genetic variants in LD affecting methylation and transcription independently. (C) The moloc and SMR results are shown on a Circos plot. Negative log10 of P value of inner (orange ring) mQTL (blood), middle (green ring) eQTL [blood; GTEx V7 (53)], and GWAS (blue ring) (28). Only the lead SNP for each mCpG or eGene, and significant GWAS SNPs (two-sided PEWAS < 5E-08) were plotted. Significant moloc regions that covered 1,142 eGFR significant GWAS variants and 267 CpG sites were highlighted by the black lines in the green ring and orange ring, respectively. Eighty-five genes were highlighted in dark red color and bars on the outmost chromosomes ring. Seventy-one protein-coding genes were labeled with their gene names. Nine protein-coding genes located within MHC regions were highlighted in blue color. Gene names of the 31 high fidelity CKD risk genes and 2 to 3 representative SNPs in the center of regions showing significant pleiotropic associations were highlighted in dark red. (D) Pathway analysis performed by GIANT (51), indicating that high-fidelity CKD causal genes were enriched for immune function, including apoptotic cell clearance, and also strongly coexpressed in NK cells and CD8+ cells.
The SMR analysis narrowed the 85 moloc-prioritized genes into 40 high-confidence likely causal genes (31 protein-coding genes and 9 noncoding genes), where the effect of genetic variants on phenotype development was mediated by methylation and gene-expression changes (Fig. 5C and Datasets S22–S24). Pleiotropic associations of CKD GWAS and mQTL data highlighted 102 CpG loci (Dataset S25). In these regions, we observed methylation changes likely driven by GWAS variants. We observed an attenuation of effect sizes of genetic variants on methylation and gene expression toward kidney function (eGFR), further supporting that genetic variations are the key drivers of methylation changes (SI Appendix, Fig. S15).
Gene ontology analysis of the 31 high-fidelity protein-coding genes identified in our multitrait integration analysis showed enrichment for immune response, specifically, positive regulation of apoptotic cell clearance and regulation of complement activation (Dataset S26). Pathway analysis, performed using genome scale integration analysis of gene networks in tissues (GIANT) (51), indicated that high-fidelity CKD causal genes were enriched for immune function and apoptotic cell clearance, and also strongly coexpressed in NK cells and CD8+ cells (Fig. 5D and Datasets S27 and S28). We confirmed the cell-type–specific gene enrichment by analyzing adult human kidney single-cell RNA-sequencing (RNA-seq) data (SI Appendix, Fig. S16) (52). To further explore the functional role of inflammation, apoptotic cell clearance and the complement system, we investigated gene-expression changes in microdissected human kidney samples. We observed positive association between expression of complement components, such as C3, C6, and C7, and kidney disease severity (SI Appendix, Fig. S17). Similarly, expression of genes in the apoptotic clearance pathway, such as TREM2, CCL2, and CD300LF were higher in microdissected human diabetic CKD samples (SI Appendix, Fig. S18), further supporting the role of these pathways in kidney disease development.
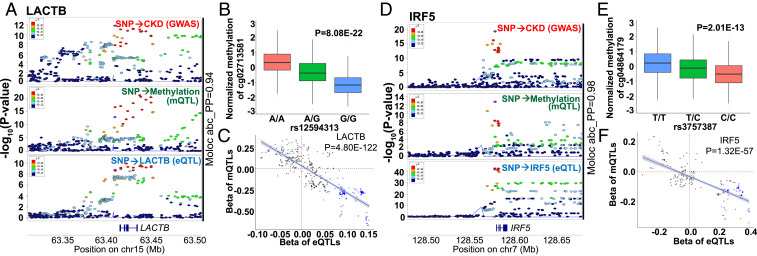
For example, we observed that, on chromosome 15, the eGFR-associated GWAS variants were also the causal variants for methylation changes and for the expression of the Serine β-lactamase-like protein (LACTB) (Fig. 6A). We identified 53 moloc signals (abc_PP range from 93 to 94%) (Dataset S29) for the LACTB. We found that the eGFR-associated genetic variant rs12594313 influenced the methylation levels of the nearby CpG, cg02713581 (Fig. 6B). The same variant (rs12594313) was also associated with the expression levels of LACTB (P = 3.92E-10) in blood samples (53). The distribution of statistical associations of the SMR tests (conducted in three directions) for variants located within ±100 kb of rs12594313 is shown in SI Appendix, Fig. S19A. Effect sizes, of mQTLs on cg02713581 and eQTLs on LACTB for variants located within ±100 kb around rs4775622 were significantly correlated (Fig. 6C), supporting that methylation changes in this region will influence the expression of LACTB. Finally, this association was not limited to blood samples, as LACTB expression positively correlated with kidney function (eGFR) and negatively correlated with kidney structure damage (fibrosis) in 433 human kidney tubule samples (SI Appendix, Fig. S19C and Dataset S16) (5). This correlation was direction-consistent with the effect size (T2P analysis) estimated in the SMR analysis (Dataset S22). Interestingly, this genotype-driven methylation signal was also observed in the (eGFR slope) MWAS study (SI Appendix, Fig. S9 B and C), further supporting the functional role of genetic variations in driving methylation changes.
Fig. 6.
Functional annotation supports the causal roles of LACTB and IRF5 in kidney disease development. (A) LocusZoom plot of eGFR GWAS, blood mQTLs on cg02713581, and blood eQTLs on LACTB. The x axis is the chromosomal location; the y axis is the strength (negative log10 of the association P value) of association between trait and genotype. The moloc signal of this region was observed (abc_PP = 94%). The y axis shows −log10(P value) of association tests (by linear regression). Each data point represents a single SNP. SNPs located within ±100 kb around rs4775622 (the purple data point) were illustrated. The moloc signal of this region was observed (abc_PP = 94%). (Top) GWAS associations (genotype and eGFR); (Middle) mQTLs (genotype and methylation value of cg02713581); (Bottom) eQTLs (genotype and expression of LACTB). The sample size was n = 765,348 in GWAS; n = 473 in mQTLs in whole blood; n = 369 in eQTLs in whole blood (from GTEx V7). r2 was used to show the degree of LD of variants. (B) The association of genetic variant rs12594313 and CpG methylation (cg02713581) [PmQTL(rs12594313) = 8.08E-22] in human blood samples. n = 473 samples. (C) The effect sizes of mQTLs on cg02713581 and eQTLs on LACTB of variants located within ±100 kb of rs4775622 significantly correlated. Each data point represents a single SNP: if the P values of eQTL or mQTL were <0.05, the data points are shown as a “cross”; if the P values of both eQTL and mQTL were <0.05, the data points are shown in blue. The sample size is n = 473 for mQTLs in whole blood; n = 369 for eQTLs in whole blood (from GTEx). (D) LocusZoom plot of eGFR GWAS, blood mQTLs on cg04864179, and blood eQTLs on IRF5. The x axis is the chromosomal location; the y axis is the strength (negative log10 of the association P value) of the association between trait and genotype. Each data point represents a single SNP. SNPs located within ±100 kb around rs3757387 (the purple data point) are illustrated. The moloc signal of this region was observed (abc_PP = 98%). (Top) GWAS association (genotype and eGFR); (Middle) mQTLs (genotype and the methylation values of cg04864179); (Bottom) eQTLs (genotype and expression of IRF5). The sample size was n = 765,348 in GWAS; n = 473 in whole-blood mQTL; n = 369 in whole-blood eQTL (GTEx V7). r2 was used to show the degree of LD of variants. (E) Association of genetic variant rs3757387 and the methylation of cg04864179 [two-sided PmQTL(rs3757387) = 2.01E-13] in human blood samples (n = 473 samples). (F) The effect sizes of mQTLs on cg04864179 and eQTLs on IRF5 variants located within ±100 kb of rs3757387 significantly negatively correlated. Each data point represents a single SNP: If the P values of eQTL or mQTL were <0.05, the data points are shown as a "cross"; if the P values of both eQTL and mQTL were <0.05, the data points are shown in blue. The sample size is n = 473 for mQTLs in whole blood; n = 369 for eQTLs in whole blood (from GTEx V7).
Another example is the IRF5 region, where methylation and gene-expression changes mediated the effect of genetic variants on phenotype development. On chromosome 7, we identified 48 moloc signals (abc_PP∼98%) (Dataset S30) for IRF5. For example, centered around the eGFR GWAS SNP of rs3757387, a moloc signal was observed (Fig. 6D). We found that genetic variant rs3757387 influenced the methylation levels of the nearby CpG cg04864179 in human blood samples (Fig. 6E). The same variant (rs3757387) also influenced the expression levels of IRF5. Histone modification tracks illustrated that this CpG site cg04864179 located in an enhancer region both in kidney and PBMCs (SI Appendix, Fig. S20). The distribution of the statistical associations of SMR tests for variants within ±100 kb of rs3757387 is shown in SI Appendix, Fig. S19B. Effect sizes, of mQTLs on cg04864179 methylation and eQTLs on IRF5 expression for variants within ±100 kb of rs3757387, were significantly correlated (Fig. 6F), supporting the causal role of methylation changes of cg0486179 affecting the expression of IRF5 in blood samples. Furthermore, the expression of IRF5 negatively correlated with kidney function (eGFR) and positively correlated with kidney structure damage (fibrosis) in microdissected human kidney tubule samples (SI Appendix, Fig. S19D), which was also direction-consistent with the association between genetically driven IRF5 expression changes and kidney function (eGFR) variations estimated by the SMR analysis (Dataset S22). Overall, our stringent analysis indicated that IRF5 as a high-fidelity causal gene for kidney function and could likely explain the association between rs3757387 and kidney function.
To conclude, the Bayesian moloc analysis highlighted a core set of methylation changes and gene-expression variations that originated from kidney function-associated genetic loci. SMR narrowed these regions, where the genetic variants drive gene-expression changes via methylation variations leading to phenotype development.
Discussion
Here, we performed an integrative genetic and epigenetic analysis to identify novel causal pathways for diabetic CKD. We took a multipronged approach that included the evaluation of the association between methylation levels and DKD associated traits (MWAS). We defined genetically driven methylation changes (mQTL). Finally, using moloc and SMR analyses, we identified methylation and gene-expression changes that likely mediated the genotype effect on kidney disease development.
We believe that this study that analyzes multiple phenotypic manifestations of DKD, such as glycemia, albuminuria, kidney function, and kidney function decline, is unique. We defined trait-specific methylation patterns. Glycemia-associated methylation showed the greatest overlap with DKD phenotypes, indicating the potential role of glycemia in other traits. It is interesting to note that the MWAS analysis for glycemic control identified methylation changes around TXNIP. Methylation changes in this region have previously shown association with glycemic metabolic memory and kidney disease in the DCCT cohort (14). TXNIP encodes for thioredoxin-interacting protein that plays an important role in redox homeostasis and a physiologic regulator of peripheral glucose uptake into fat and muscle in human (54–57). We identified a single CpG cg17944885, whose methylation levels correlated with eGFR in our study and could be validated in multiple studies that analyzed blood or kidney samples. Despite the consistent associations, the functional role of this methylation change remains to be established, as it is not located on the gene regulatory element in blood and kidney samples. It is possible that this is a regulatory region during development or plays a functional role in a rare cell type, that was not captured by bulk epigenome and expression analysis. Future single-cell expression and epigenome analysis shall examine the functional role of cg17944885. Our analysis for albuminuria identified methylation changes around UPK2. Uroplakins cover urothelial apical surfaces. Mice with null mutation of Upk2 are often born with congenital kidney disease (58).
We generated a new mQTL database to understand the association between genotype and methylation changes. This dataset indicates that a considerable portion of methylation changes associated with kidney function (in MWAS) is driven by underlying genetic variations. This is best illustrated by the chromosome 15 locus; genetic variant rs12594313 influences the methylation of its nearby CpG cg02713581, whose methylation variations show the association with kidney function decline in the eGFR slope-MWAS study. Furthermore, the genetic locus not only controlled the methylation of this CpG site, but also altered the expression of LACTB. LACTB encodes the Serine β-lactamase-like protein that is involved in mitochondrial phospholipid metabolism (59). Our results indicate that it will be critical to integrate genetically driven methylation signals into future MWAS studies to differentiate genetically and environmentally driven methylation differences.
Given the critical role of genetic variations driving methylation changes, here we used kidney function-associated genetic variations (from GWAS) to identify methylation changes that likely mediate phenotype development. We demonstrate that the integration of epigenetic signals can significantly improve our understanding of kidney disease pathogenesis driven by GWAS variants (28, 45, 46). The present data indicate that the effects of variants on methylation are widespread and can even be observed in the absence of eQTL effects. Changes in mQTL likely play an important role in how cells with different genotypes respond to external stimuli. We narrowed the colocalization regions with SMR associations across methylation, transcription, and complex traits in our analysis to identify changes that mediate the genotype effect on gene expression via DNA methylation.
The current integrative analysis highlighted that genetic variants influence methylation and expression levels of multiple genes are known to play important roles in the immune system and inflammation. Specifically, genes associated with the clearance of apoptotic cells and complement pathways have been identified as the putative kidney disease risk genes. While further studies are needed to examine the role of C2 and C4 as these genes located in the MHC regions with high genetic complexity, coding mutations in the complement pathway have been shown to cause rare forms of kidney diseases (60, 61). Increased expression and activation of the complement pathway have also been observed in diabetic CKD (62, 63). The causal role of complement activation in diabetic CKD has been debated, however, as complement activation has traditionally been considered as a secondary phenomenon (64). As genotypes do not suffer from reverse causation, our SMR analysis suggests a causal role for complement in diabetic CKD.
Our studies also highlighted the potential role for IRF5. IRF5 is an IFN-responsive transcription factor that could also play a role in clearance of apoptotic cells in macrophages (65, 66). Blocking IRF5 in macrophages may help to treat a wide range of inflammatory diseases and could be an important new therapeutic target for CKD. Gene ontology analysis of our integrative study strongly supports the role of immune cells in kidney disease development. Specifically, our network analysis showed enrichment for NK and CD8+ T cells in kidney disease development.
There are two important limitations of the study, such as the use of only diabetic CKD samples. It seems that eGFR-associated methylation changes are shared in multiple studies, indicating that they are likely linked to common CKD mechanisms rather than specific diseases. This is further supported by the current work as the top eGFR-associated DMP could be validated in mixed CKD, diabetic CKD, and HIV-associated CKD cohorts. We also acknowledge that the stratified design originally aimed to identify signals for kidney function decline is an important limitation of the presented work.
To conclude, our work defined distinct cytosine methylation changes associated with different DKD phenotypes, the key role of underlying genetic variations driving methylation variations, identified methylation changes that mediate the genotype effect of kidney disease development, and illustrated how methylome variations can be used to prioritize genes for kidney disease pathogenesis.
Materials and Methods
Study Population.
For the study population, 1,394 participants with DKD and phenotype records were selected from the 3,668 CRIC study participants. The best linear unbiased predictor modeling was used to adjust eGFR slope (67). Two-hundred-fifty CRIC study subjects with diabetes with adjusted eGFR slope > −2.85 (fast-progressor group) and 250 matched participants with diabetes, but with slower kidney function decline (the adjusted eGFR slope < −2.85; slow-progressor group) were selected for our study (SI Appendix, Figs. S1 and S2B). The pairmatch function in the optmatch package in R was used for matching (68). We applied a distance matrix that combined a caliper on an estimated propensity score with a rank based Mahalanobis distance (69). Strata pairs were matched for age, hemoglobin A1c, baseline eGFR, logarithm of urine albumin, gender, race, and days with diabetes (self-reported) (SI Appendix, Figs. S2C and S3). After combining with good-quality genotype data, 473 and 410 subjects were used in our MWAS analyses with HgbA1c, albuminuria, kidney function, and functional decline, respectively.
MWAS.
To account for cell heterogeneity of whole-blood samples, cell compositions were estimated using a reference-based approach, such as the CIBERSORT algorithm implemented in the EpiDISH package (70–72). We generated estimated cell counts for B cells, TCD4+ cells, TCD8+ cells, NK cells, monocytes, and granulocytes, and used them in the regression models. To identify methylation changes associated with hemoglobin A1c and albuminuria, we used M values as outcome, hemoglobin A1c, and albuminuria as independent variables, respectively, and batch effect, age, sex, genetic background, hypertension, and cell heterogeneity as covariates. For kidney function MWAS analysis, we used linear mixed-effect models with batch effect (chip number) as the random effect, age, top 10 PCs of genetic background, hypertension, and imputed cell counts of B cells, TCD4+ cells, TCD8+ cells, NK cells, monocytes, and granulocytes as fixed effects to generate residuals of M values. Baseline eGFR was then adjusted for age, sex, top 10 PCs of genetic background, and hemoglobin A1c by linear regression. Last, we used residuals of baseline eGFR (eGFR adjusted for sample constitution difference) as the outcome and residuals of M values as the independent variable to examine DNA methylation associated with baseline eGFR. Four-hundred-ten subjects (1:1 stratified sampling with 205 strata as illustrated in SI Appendix, Fig. S2B) with longitudinal eGFR records and good-quality genotype data were used in the kidney function decline MWAS analysis. Accounting for the 1:1 stratified design, we first used conditional logistic regression to assess the relationship between kidney function decline rate (fast/slow) and DNA methylation without adjusting for additional covariates. To avoid the strict stratification-induced power reduction (based on the QQ-plot, as illustrated in SI Appendix, Fig. S8B), we further directly performed a linear regression model using M value as dependent variable and eGFR slope as independent variables by adjusting for baseline eGFR, age, sex, batch effect, top 10 genetic PCs, hypertension, blood cell proportions, hemoglobin A1c, and urinary albumin to creatinine ratio. Similarly, these covariates were selected by backward stepwise procedure (SI Appendix). All analyses were performed using R (3.4.3). The mixed-effect model analysis was performed using lmer in the lme4 package.
mQTL Mapping.
Cis-mQTL (referred to as mQTL herein) mapping was conducted on blood samples from 473 CRIC study participants. We inverse normal-transformed M values (INT-transformed M values) and implemented probabilistic estimation of expression residuals (PEER) (73) on the INT-transformed M values using age, batch effect, top 10 PCs of genetic background, hypertension, and whole-blood cell subtype proportions as covariates. We performed PEER analysis by including different numbers of factors (k = 5–50) at intervals of 5 to optimize for mQTL discovery. To identify epigenome-wide associations between SNPs and DNA methylation, an additive linear model was fitted to test if the number of alleles (coded as 0, 1, and 2) correlated with DNA methylation (INT-transformed M values) at each site, including covariates for age, chip number, top 10 PCs of genetic background, hypertension, imputed whole-blood cell-type proportions, and different numbers of PEER factors using the R package MatrixEQTL (74). We calculated mQTLs for all SNPs within ±1 Mb of the queried methylation probe. mQTLs with Benjamini–Hochberg FDR < 0.05 (by MatrixEQTL) were used to select the number of PEER factors that could maximize the identified mCpGs (CpG sites that significantly regulated by at least one SNP). Twenty PEER factors were included in our final mQTL mapping model (SI Appendix, Fig. S21). We next implemented FastQTL (75) to estimate significance of the top associated variant per CpG by setting adaptive permutation as “-permute 10000”. The β-distribution–adjusted empirical P values were used to calculate q using Storey’s q method (76), and a q threshold ≤ 0.05 was applied to identify mCpGs. We finally defined a nominal P threshold, Pt, as the empirical P of the CpG closest to the 0.05 FDR threshold. Pt was used to calculate a nominal Pt for each mCpG based on the beta distribution model (from FastQTL) of the minimum P distribution f(Pmin) obtained from the permutations for the CpG sites. For each mCpG, variants with a nominal P below the CpG-level cutoff were regarded as significant mSNPs (SNPs that significantly regulate at least one CpG site).
moloc Analysis.
To estimate the posterior probabilities of whether DNA methylation and gene expression in whole-blood and kidney disease share common genetic causal variants in a given region, we performed multiple-trait-colocalization analysis using moloc (44) with summary data of GWAS and our mQTL and eQTL data of whole blood from GTEx (V7) (53). moloc computes the evidence supporting the 15 all possible effect configurations of sharing of SNPs among kidney disease risk, gene-expressions levels, and methylations in a genomic region (44). By specifying the prior probabilities and using the association evidence of the data, moloc outputs the posterior probability that the SNPs in a genomic region are associated with all three traits (methylation, gene expression, and phenotype) (Fig. 4A). Summary data from eGFR-associated GWAS studies from Hellwege et al. (46), Wuttke et al. (28), and Morris et al. (45) were used. GWAS variants associated with eGFR at genome-wide significance (P < 5E-08) were selected. To avoid the inflation caused by GWAS variants representing the same signal, we performed LD pruning using swiss (https://github.com/statgen/swiss). Variants in LD r2 ≥ 0.8 with the lead SNP at each locus were removed. Regions within ±100 kb of each pruned GWAS variant overlapping with eGenes (genes significantly regulated by an SNP) were selected for further moloc analysis. Summary data of blood mQTL (here), blood eQTL [GTEx V7 (53)], and SNPs within 100 kb of the GWAS SNPs were used to calculate the posterior probability. All available eGene-mCpGs-GWAS triplets were tested in each region. In the moloc results, abc_PP represents the posterior probability that all three traits are associated and share causal variants. We used abc_PP ≥ 0.8 as the threshold of moloc.
SMR Analysis and HEIDI Test.
We focused on defining causal effects of regions showing significant association in the moloc analysis. An SMR&HEIDI test was used to test potential causal effects with the publicly available software SMR (49). Using genetic variants as possible instruments, SMR can be used to calculate a potential causal relationship between any two traits. We conducted SMR&HEIDI tests in three directions (Fig. 5A), including methylation to transcription (M2T), methylation to phenotype (M2P), and transcription to phenotype (T2P) analyses. First, to identify target genes for the CpG sites, we tested the associations between each mCpG and its neighboring genes (within ±1 Mb of each mCpG), using the top associated mQTL as the instrumental variable (M2T analysis). We used a Bonferroni-corrected P-value threshold to obtain the genes that showed pleiotropic associations of transcription and methylation. For example, we adopted 2.4E-04 (i.e., 0.05/208 as cutoff for PSMR in M2T analysis) as 208 CpG sites were identified in the moloc step, while using GWAS data from the Wuttke et al. dataset (28). Next, we narrowed functionally relevant CpG sites by testing the associations of each mCpG with eGFR (the phenotype) with the top associated mQTL as instrumental variable (M2P analysis). Similarly, 208 CpG sites were identified, when integrated with GWAS data (28) in the moloc step, Bonferroni-corrected P-value cutoff 2.4E-04 (0.05/208) was used. We obtained the PSMR threshold similarly, while combing with other GWAS studies (45, 46). We prioritized the trait-associated eGenes by conducting association test between each eGene and eGFR, using the top associated eQTL (T2P analysis) as the instrumental variable. For example, eGenes were identified as functionally relevant by two-sided PSMR < 7E-04 (i.e., 0.05/71, where 71 was the number of eGenes in moloc regions), while combining with eGFR GWAS data from Wuttke et al. (28). We further performed the HEIDI test to reject the hypothesis that the association detected by the SMR test is due to linkage (not rejected by the HEIDI test at two-sided PHEIDI ≥ 0.01) (Fig. 5B).
Data Access.
Genotype data are available from https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000524.v1.p1 with the dbGaP Study accession no.: phs000524.v1.p1. The clinical records for CRIC samples are available from https://clinicaltrials.gov/ct2/show/NCT00304148?term=CRIC+study. The MWAS and mQTL data are available via the CRIC study and a searchable public website https://zenodo.org/record/4148467#.X5ohRy1VZR0. Summary data of eQTL in whole blood samples were available via GTEx Portal https://gtexportal.org/home/.
Ethics Approval and Consent to Participate.
The CRIC study protocol was approved by the institutional review boards at each of the primary sites and all participants provided written informed consent. The specific human research review committees included: 1) University of Pennsylvania Office of Regulatory Affairs, Philadelphia, PA; 2) The Johns Hopkins University School of Medicine, Office of Human Subjects Research Institutional Review Boards, Baltimore, MD; 3) University of Maryland Institutional Review Board, Baltimore, MD; 4) Case Western Reserve University, University Hospitals, Case Medical Center Institutional Review Board for Human Investigation, Cleveland, OH; 5) MetroHealth System Institutional Review Board, Cleveland, OH; 6) Cleveland Clinic Foundation Institutional Review Board, Cleveland, OH; 7) University of Michigan Medical School Institutional Review Board, Ann Arbor, MI; 8) St. John Hospital and Medical Center Institutional Review Board, Grosse Pointe Woods, MI; 9) University of Illinois at Chicago Office of the Protection of Research Subjects, Chicago, IL; 10) Tulane University Health Science Center Human Research Protection Program Institutional Review Boards, New Orleans, LA; and 11) Kaiser Permanente of Permanente of Northern California, Kaiser Foundation Research Institute Institutional Review Board, Oakland, CA. All participants provided written informed consent.
Consent for Publication.
Consent for publication was obtained from the CRIC Publication committee.
Supplementary Material
Acknowledgments
This work in the K.S. laboratory has been supported by National Institute of Health Grants R01 DK087635, DK076077, and DK105821 and in the H. Li laboratory by Grant R01 GM129781. Funding for the Chronic Renal Insufficiency Cohort study was obtained under a cooperative agreement from National Institute of Diabetes and Digestive and Kidney Diseases (Grants 5U01DK060990, 5U01DK060984, 5U01DK06102, 5U01DK061021, 5U01DK061028, 5U01DK60980, 5U01DK060963, and 5U01DK060902). CRIC is supported partially by Tulane Centers of Biomedical Research Excellence for Clinical and Translational Research in Cardiometabolic Diseases P20 GM109036, NIGMS/NIH. The authors thank the Diabetes Research Center (P30-DK19525) at the University of Pennsylvania for the services.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. R.N. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2005905117/-/DCSupplemental.
Data Availability.
The raw genotype, methylation and clinical information contain personally identifiable information. Therefore, they are available via the following restricted access. Genotype data are available from https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000524.v1.p1 with the dbGaP Study accession no.: phs000524.v1.p1. The clinical records for CRIC samples are available from https://clinicaltrials.gov/ct2/show/NCT00304148?term=CRIC+study. The MWAS and mQTL data are available via the CRIC study and a searchable public website https://zenodo.org/record/4148467#.X5ohRy1VZR0.
References
- 1.Alicic R. Z., Rooney M. T., Tuttle K. R., Diabetic kidney disease: Challenges, progress, and possibilities. Clin. J. Am. Soc. Nephrol. 12, 2032–2045 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boumpas D. T., Chrousos G. P., Wilder R. L., Cupps T. R., Balow J. E., Glucocorticoid therapy for immune-mediated diseases: Basic and clinical correlates. Ann. Intern. Med. 119, 1198–1208 (1993). [DOI] [PubMed] [Google Scholar]
- 3.Silverstein D. M., Inflammation in chronic kidney disease: Role in the progression of renal and cardiovascular disease. Pediatr. Nephrol. 24, 1445–1452 (2009). [DOI] [PubMed] [Google Scholar]
- 4.Salem R. M. et al.; SUMMIT Consortium, DCCT/EDIC Research Group, GENIE Consortium , Genome-wide association study of diabetic kidney disease highlights biology involved in glomerular basement membrane collagen. J. Am. Soc. Nephrol. 30, 2000–2016 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qiu C., et al. , Renal compartment-specific genetic variation analyses identify new pathways in chronic kidney disease. Nat. Med. 24, 1721–1731 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woroniecka K. I., et al. , Transcriptome analysis of human diabetic kidney disease. Diabetes 60, 2354–2369 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beckerman P., et al. , Human kidney tubule-specific gene expression based dissection of chronic kidney disease traits. EBioMedicine 24, 267–276 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kang H. M., et al. , Defective fatty acid oxidation in renal tubular epithelial cells has a key role in kidney fibrosis development. Nat. Med. 21, 37–46 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Winnicki E., et al. , Use of the kidney failure risk equation to determine the risk of progression to end-stage renal disease in children with chronic kidney disease. JAMA Pediatr. 172, 174–180 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saely C., et al. , Type 2 diabetes, chronic kidney disease, and mortality in patients with established cardiovascular disease. J. Am. Coll. Cardiol. 71, A1841 (2018). [Google Scholar]
- 11.Kaelin W. G. Jr, McKnight S. L., Influence of metabolism on epigenetics and disease. Cell 153, 56–69 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beckerman P., Ko Y.-A., Susztak K., Epigenetics: A new way to look at kidney diseases. Nephrol. Dial. Transplant. 29, 1821–1827 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sayyed S. G., et al. , Progressive glomerulosclerosis in type 2 diabetes is associated with renal histone H3K9 and H3K23 acetylation, H3K4 dimethylation and phosphorylation at serine 10. Nephrol. Dial. Transplant. 25, 1811–1817 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Chen Z., et al. , Epigenomic profiling reveals an association between persistence of DNA methylation and metabolic memory in the DCCT/EDIC type 1 diabetes cohort. Proc. Natl. Acad. Sci. U.S.A. 113, E3002–E3011 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Boer I. H. et al.; Diabetes Control and Complications Trial/Epidemiology of Diabetes Interventions and Complications Study Research Group , Long-term renal outcomes of patients with type 1 diabetes mellitus and microalbuminuria: An analysis of the diabetes control and complications trial/epidemiology of diabetes interventions and complications cohort. Arch. Intern. Med. 171, 412–420 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park J., et al. , Functional methylome analysis of human diabetic kidney disease. JCI Insight 4, 128886 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greally J. M., Drake A. J., The current state of epigenetic research in humans: Promise and reality. JAMA Pediatr. 171, 103–104 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Krupinski J., et al. , DNA methylation in stroke. Update of latest advances. Comput. Struct. Biotechnol. J. 16, 1–5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qiu C., et al. , Cytosine methylation predicts renal function decline in American Indians. Kidney Int. 93, 1417–1431 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gluck C., et al. , Kidney cytosine methylation changes improve renal function decline estimation in patients with diabetic kidney disease. Nat. Commun. 10, 2461 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ko Y.-A., et al. , Cytosine methylation changes in enhancer regions of core pro-fibrotic genes characterize kidney fibrosis development. Genome Biol. 14, R108 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fox C. S., et al. , Genomewide linkage analysis to serum creatinine, GFR, and creatinine clearance in a community-based population: The Framingham Heart Study. J. Am. Soc. Nephrol. 15, 2457–2461 (2004). [DOI] [PubMed] [Google Scholar]
- 23.Sandholm N., et al. , The genetic landscape of renal complications in type 1 diabetes. J. Am. Soc. Nephrol. 28, 557–574 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Köttgen A., et al. , Multiple loci associated with indices of renal function and chronic kidney disease. Nat. Genet. 41, 712–717 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gudbjartsson D. F., et al. , Association of variants at UMOD with chronic kidney disease and kidney stones-role of age and comorbid diseases. PLoS Genet. 6, e1001039 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parsa A., et al. , Genome-wide association of CKD progression: The chronic renal insufficiency cohort study. J. Am. Soc. Nephrol. 28, 923–934 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mueller P. W., et al. , Genetics of Kidneys in Diabetes (GoKinD) study: A genetics collection available for identifying genetic susceptibility factors for diabetic nephropathy in type 1 diabetes. J. Am. Soc. Nephrol. 17, 1782–1790 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wuttke M. et al.; Lifelines Cohort Study; V. A. Million Veteran Program , A catalog of genetic loci associated with kidney function from analyses of a million individuals. Nat. Genet. 51, 957–972 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chu A. Y., et al. , Epigenome-wide association studies identify DNA methylation associated with kidney function. Nat. Commun. 8, 1286 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feldman H. I. et al.; Chronic Renal Insufficiency Cohort (CRIC) Study Investigators , The Chronic Renal Insufficiency Cohort (CRIC) study: Design and methods. J. Am. Soc. Nephrol. 14(7, suppl. 2)S148–S153 (2003). [DOI] [PubMed] [Google Scholar]
- 31.Lash J. P. et al.; Chronic Renal Insufficiency Cohort (CRIC) Study Group , Chronic Renal Insufficiency Cohort (CRIC) study: Baseline characteristics and associations with kidney function. Clin. J. Am. Soc. Nephrol. 4, 1302–1311 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sheng X., et al. , MTD: A mammalian transcriptomic database to explore gene expression and regulation. Brief. Bioinform. 18, 28–36 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsaprouni L. G., et al. , Cigarette smoking reduces DNA methylation levels at multiple genomic loci but the effect is partially reversible upon cessation. Epigenetics 9, 1382–1396 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Horvath S., DNA methylation age of human tissues and cell types. Genome Biol. 14, R115 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wahl S., et al. , Epigenome-wide association study of body mass index, and the adverse outcomes of adiposity. Nature 541, 81–86 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li W., Christiansen L., Hjelmborg J., Baumbach J., Tan Q., On the power of epigenome-wide association studies using a disease-discordant twin design. Bioinformatics 34, 4073–4078 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Albao D. S., et al. , Methylation changes in the peripheral blood of Filipinos with type 2 diabetes suggest spurious transcription initiation at TXNIP. Hum. Mol. Genet. 28, 4208–4218 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Chen J., et al. , Epigenetic associations with estimated glomerular filtration rate among men with human immunodeficiency virus infection. Clin. Infect. Dis. 70, 667–673 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ernst J., Kellis M., ChromHMM: Automating chromatin-state discovery and characterization. Nat. Methods 9, 215–216 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kundaje A. et al.; Roadmap Epigenomics Consortium , Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park J., et al. , Single-cell transcriptomics of the mouse kidney reveals potential cellular targets of kidney disease. Science 360, 758–763 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mailman M. D., et al. , The NCBI dbGaP database of genotypes and phenotypes. Nat. Genet. 39, 1181–1186 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gaunt T. R., et al. , Systematic identification of genetic influences on methylation across the human life course. Genome Biol. 17, 61 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Giambartolomei C. et al.; CommonMind Consortium , A Bayesian framework for multiple trait colocalization from summary association statistics. Bioinformatics 34, 2538–2545 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morris A. P., et al. , Trans-ethnic kidney function association study reveals putative causal genes and effects on kidney-specific disease aetiologies. Nat. Commun. 10, 29 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hellwege J. N., et al. , Mapping eGFR loci to the renal transcriptome and phenome in the VA Million Veteran Program. Nat. Commun. 10, 3842 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu X., et al. , Molecular insights into genome-wide association studies of chronic kidney disease-defining traits. Nat. Commun. 9, 4800 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumasaka N., Knights A. J., Gaffney D. J., Fine-mapping cellular QTLs with RASQUAL and ATAC-seq. Nat. Genet. 48, 206–213 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu Z., et al. , Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet. 48, 481–487 (2016). [DOI] [PubMed] [Google Scholar]
- 50.Wu Y., et al. , Integrative analysis of omics summary data reveals putative mechanisms underlying complex traits. Nat. Commun. 9, 918 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Greene C. S., et al. , Understanding multicellular function and disease with human tissue-specific networks. Nat. Genet. 47, 569–576 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Young M. D., et al. , Single-cell transcriptomes from human kidneys reveal the cellular identity of renal tumors. Science 361, 594–599 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Consortium G.; GTEx Consortium , Human genomics. The genotype-tissue expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science 348, 648–660 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Muoio D. M., TXNIP links redox circuitry to glucose control. Cell Metab. 5, 412–414 (2007). [DOI] [PubMed] [Google Scholar]
- 55.Shalev A., Minireview: Thioredoxin-interacting protein: Regulation and function in the pancreatic β-cell. Mol. Endocrinol. 28, 1211–1220 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schulze P. C., et al. , Hyperglycemia promotes oxidative stress through inhibition of thioredoxin function by thioredoxin-interacting protein. J. Biol. Chem. 279, 30369–30374 (2004). [DOI] [PubMed] [Google Scholar]
- 57.Sheth S. S., et al. , Thioredoxin-interacting protein deficiency disrupts the fasting-feeding metabolic transition. J. Lipid Res. 46, 123–134 (2005). [DOI] [PubMed] [Google Scholar]
- 58.Jenkins D., et al. , Mutation analyses of Uroplakin II in children with renal tract malformations. Nephrol. Dial. Transplant. 21, 3415–3421 (2006). [DOI] [PubMed] [Google Scholar]
- 59.Keckesova Z., et al. , LACTB is a tumour suppressor that modulates lipid metabolism and cell state. Nature 543, 681–686 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Abrera-Abeleda M. A., et al. , Allelic variants of complement genes associated with dense deposit disease. J. Am. Soc. Nephrol. 22, 1551–1559 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bu F., et al. , High-throughput genetic testing for thrombotic microangiopathies and C3 glomerulopathies. J. Am. Soc. Nephrol. 27, 1245–1253 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wada T., Nangaku M., Novel roles of complement in renal diseases and their therapeutic consequences. Kidney Int. 84, 441–450 (2013). [DOI] [PubMed] [Google Scholar]
- 63.RASTOGI P & Obediat M , Mon-024 unusual presentation of dense deposit disease. Kidney Int. Rep. 4, S313–S314 (2019). [Google Scholar]
- 64.Flyvbjerg A., The role of the complement system in diabetic nephropathy. Nat. Rev. Nephrol. 13, 311–318 (2017). [DOI] [PubMed] [Google Scholar]
- 65.Banga J., et al. , Inhibition of IRF5 cellular activity with cell-penetrating peptides that target homodimerization. Sci. Adv. 6, eaay1057 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Seneviratne A. N., et al. , Interferon regulatory factor 5 controls necrotic core formation in atherosclerotic lesions by impairing efferocytosis. Circulation 136, 1140–1154 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Robinson G. K., That BLUP is a good thing: The estimation of random effects. Stat. Sci. 6, 15–32 (1991). [Google Scholar]
- 68.Hansen B. B., Klopfer S. O., Optimal full matching and related designs via network flows. J. Comput. Graph. Stat. 15, 609–627 (2007). [Google Scholar]
- 69.Rosenbaum P. R., “Constructing matched sets and strata.” in Observational Studies (Springer, 2002), pp. 295–331. [Google Scholar]
- 70.Newman A. M., et al. , Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 12, 453–457 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Teschendorff A. E., Relton C. L., Statistical and integrative system-level analysis of DNA methylation data. Nat. Rev. Genet. 19, 129–147 (2018). [DOI] [PubMed] [Google Scholar]
- 72.Teschendorff A. E., Breeze C. E., Zheng S. C., Beck S., A comparison of reference-based algorithms for correcting cell-type heterogeneity in Epigenome-Wide Association Studies. BMC Bioinformatics 18, 105 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stegle O., Parts L., Piipari M., Winn J., Durbin R., Using probabilistic estimation of expression residuals (PEER) to obtain increased power and interpretability of gene expression analyses. Nat. Protoc. 7, 500–507 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shabalin A. A., Matrix eQTL: Ultra fast eQTL analysis via large matrix operations. Bioinformatics 28, 1353–1358 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ongen H., Buil A., Brown A. A., Dermitzakis E. T., Delaneau O., Fast and efficient QTL mapper for thousands of molecular phenotypes. Bioinformatics 32, 1479–1485 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Storey J. D., A direct approach to false discovery rates. J. R. Stat. Soc. Series B Stat. Methodol. 64, 479–498 (2002). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw genotype, methylation and clinical information contain personally identifiable information. Therefore, they are available via the following restricted access. Genotype data are available from https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000524.v1.p1 with the dbGaP Study accession no.: phs000524.v1.p1. The clinical records for CRIC samples are available from https://clinicaltrials.gov/ct2/show/NCT00304148?term=CRIC+study. The MWAS and mQTL data are available via the CRIC study and a searchable public website https://zenodo.org/record/4148467#.X5ohRy1VZR0.