Abstract
Purpose
Our research aimed to investigate the expression level of circ_0002232, which is transcribed from PTEN, and find out the association of circ_0002232/miR-92a-3p/PTEN network in acute myeloid leukemia (AML).
Methods
Circ_0002232 expression in 115 AML patients and 48 controls was detected by using real-time quantitative PCR. The diagnostic value of circ_0002232 expression was evaluated by receiver operating characteristic curve. Kaplan–Meier curves were used to analyse the impact of circ_0002232 for overall survival. Associated network of circ_0002232 was predicted by using interaction prediction websites.
Results
Compared with controls, circ_0002232 was notably low-expressed in AML (P<0.001). According to the result of receiver operating characteristic curve, circ_0002232 expression could distinguish AML patients from controls (P<0.001). There were significant differences in patients’ age (P=0.004), FAB classifications (P=0.036), white blood cell count (P=0.041) and platelet count (P=0.021) between low-expressed circ_0002232 group and high-expressed circ_0002232 group. Moreover, there was a positive correlation between circ_0002232 expression and patients’ age (Pearson r=0.256, P=0.0057). Interestingly, we found that patients in low-expressed circ_0002232 group had better overall survival both in whole AML (P=0.030) and non-APL AML (P=0.014). Remarkably, the expression of circ_0002232 was positively correlated with PTEN (Spearman r=0.678, P<0.001). Furthermore, there was a negative correlation in AML between circ_0002232 and miR-92a-3p (Spearman r=−0.301, P=0.016), miR-92a-3p and PTEN (Spearman r=−0.324, P=0.034). Interaction prediction websites revealed that circ_0002232 might affect the expression of PTEN and the process of AML through sponging miR-92a-3p.
Conclusion
Circ_0002232, one of the circRNAs transcribed from PTEN, was remarkably down-regulated in AML and could act as a promising biomarker for the diagnosis of AML. In addition, there might be a potential association network of circ_0002232/miR-92a-3p/PTEN in AML.
Keywords: circular RNAs, circ_0002232, miR-92a-3p, PTEN, acute myeloid leukemia
Introduction
Acute myeloid leukemia (AML), the most common malignant myeloid disease in adults, is characterized by loss of differentiation of blasts (myeloid progenitor cell) and clonal amplification in the peripheral blood and bone marrow.1,2 It had a poor prognosis in the past.2 Cytogenetics analyses play a crucial role to identify subgroups of AML with different outcomes.3 Meanwhile, identifying molecular genetic markers also helps to divide AML patients into different groups and refine their prognosis.3
In recent years, non-coding RNAs have increasingly caught researchers’ attention. A wide variety of studies have shown that non-coding RNAs participate in the process of controlling cell differentiation through regulating expression of the gene.4
Circular RNAs (circRNAs) are an emerging class of non-coding RNAs and are characterized by having covalent binding between the 3ʹ and 5ʹ ends which are generated by the mechanism of reverse splicing.5 Due to the conserved characteristic across species and tissue, circRNAs have been found to be ideal diagnostic and prognostic biomarkers for disease, especially cancer.6 For example, according to Xia et al, their study indicated that high-expressed of circ_0067934 in esophageal cancer was related with poor proliferation. Up-regulated expression of circ_0067934 was an unfavorable factor for esophageal squamous cell carcinoma.7 Shao et al revealed that circ_0014717 expression significantly decreased in gastric carcinoma. The level of its expression was related to tumor staging and distal metastasis. Due to the stable expression of circ_0014717, it had been regarded as an ideal biomarker for clinical detection of gastric cancer.8
Moreover, circular RNAs, which have also been named as competing endogenous RNAs (ceRNAs), could regulate cell biological function through acting as miRNA sponges.5 Actually, circRNAs play an essential regulatory role in diseases through interacting with disease-related miRNAs.9 For example, Weng et al illustrated that over-expressed ciRs-7 acted as miRNA sponge to abolish the tumor suppressive effect of miR-7 and promoted tumorgenesis in colorectal cancer.10 Circ_FBLIM1 had been found to serve as ceRNA and regulate the process of hepatocellular cancer through binding with miR-346. This process promoted the progression of hepatocellular cancer.11 But there are few studies focused on the diagnostic and prognostic value of circular RNAs or their function acting as ceRNA in malignant hematonosis.
Phosphatase and tensin homolog deleted on chromosome 10 (PTEN) serves as a classic tumor suppressor.12 It mainly participates the homeostasis of the phosphatidylinositol 3 kinase (PI3K)/AKT pathway.12 And losing the suppressive function of PTEN plays an essential role in the occurrence of cancer. PTEN has been found to be down-expressed in several solid cancers, like prostate cancer and breast cancer.13,14 Furthermore, some research illustrated that the expression of PTEN transcript was remarkably lower in AML than controls and inactivation of PTEN promoted AML progression.15,16
To our knowledge, PTEN-dependent circular RNAs have seldom been studied in cancer, let alone AML. Circ_0002232 is one of circRNAs of PTEN. The purpose of this research was to analyse circ_0002232 expression in AML and to investigate its clinical relevance. We wanted to find whether it could serve as a biomarker for diagnosis and prognosis of AML and reveal the potential ceRNA network behind it.
Materials and Methods
Patients and Samples
A total of 163 samples, including 48 controls and 115 de novo AML patients, were provided by the Affiliated People’s Hospital of Jiangsu University. This study was approved by Human Research Ethics Committee of the Affiliated People’s Hospital of Jiangsu University and was conducted in accordance with the Declaration of Helsinki. Patients involved in this study were clearly diagnosed and classified according to guidelines of World Health Organization (WHO) and French-American-British (FAB) criteria.17,18 To refine the group, the total 115 AML samples we used included 98 non-acute promyelocytic leukemia AML (non-APL AML) samples and 51 normal cytogenetic AML (CN-AML) samples. All of the enrolled patients have to meet the following conditions: 1) age ≥ 18 years old; 2) patients were diagnosed with AML for the first time; 3) patients had not received any form of treatment; and 4) patients had complete pathological data in our hospital. 5) participatants signed informed consent and patients who failed to meet above criteria were excluded from the study. Bone marrow (BM) specimen was collected after every participant signed informed consent. Extraction of bone marrow mononuclear cells (BMNCs) was conducted by using Lymphocyte Separation Medium (TBD sciences corporation, Tianjin, China). The vital clinical and laboratory features of these patients are listed in Table 1 and demographic data of controls are shown in Supplemental File 2
Table 1.
Comparison of clinical and laboratory characteristics between AML patients with low and high circ_0002232 expression
| Patient’s Parameters | Low (n=87) | High (n=28) | P value |
|---|---|---|---|
| Sex, male/female | 58/29 | 15/13 | 0.261 |
| Median age, years (range) | 54 (21–81) | 64 (20–88) | 0.004* |
| Median WBC, ×109/L (range) | 14.9 (0.3–528.0) | 34.50 (1.1–207.5) | 0.041* |
| Median hemoglobin, g/L (range) | 78 (34–144) | 82 (42–119) | 0.618 |
| Median platelets, ×109/L (range) | 34 (3–415) | 52 (9–382) | 0.021* |
| BM blasts, % (range) | 48.50 (1.00–109.00) | 30.00 (6.50–81.00) | 0.566 |
| CR(+/-) | 38/37 | 8/15 | 0.235 |
| FAB | 0.036* | ||
| M0 | 0 (0%) | 0 (0%) | |
| M1 | 4 (4.9%) | 0 (0%) | |
| M2 | 39 (48.1%) | 6 (26.1%) | |
| M3 | 14 (17.3%) | 2 (8.7%) | |
| M4 | 17 (21%) | 9 (39.1%) | |
| M5 | 7 (8.6%) | 6 (26.1%) | |
| M6 | 0 (0%) | 0 (0%) | |
| Cytogenetic abnormalities’ classification | 0.279 | ||
| Favorable | 24 (27.6%) | 3 (11.1%) | |
| Intermediate | 52 (59.8%) | 20 (74.1%) | |
| Poor | 9 (10.3%) | 3 (11.1%) | |
| No data | 2 (2.3%) | 1 (3.7%) | |
| Cytogenetic abnormalities | 0.350 | ||
| Normal | 40 (46%) | 12 (44.4%) | |
| t (8;21) | 9 (10.3%) | 1 (3.7%) | |
| t (15;17) | 14 (16.1%) | 2 (7.4%) | |
| +8 | 2 (2.3%) | 3 (11.1%) | |
| Complex | 8 (9.2%) | 3 (11.1%) | |
| Others | 11 (12.6%) | 5 (18.5%) | |
| No data | 2 (2.3%) | 1 (3.7%) | |
| Gene mutation | |||
| CEBPA (+/-) | 10/64 | 0/18 | 0.201 |
| NPM1(+/-) | 7/67 | 0/18 | 0.338 |
| FLT3-ITD (+/-) | 11/63 | 1/17 | 0.448 |
| C-KIT (+/-) | 4/70 | 1/17 | 1.000 |
| N/K-RAS(+/-) | 3/60 | 2/11 | 0.200 |
| IDH1/2 (+/-) | 0/74 | 1/17 | 0.196 |
| DNMT3A (+/-) | 5/69 | 1/17 | 1.000 |
| U2AF1 (+/-) | 1/73 | 1/17 | 0.355 |
| SRSF2(+/-) | 1/62 | 0/13 | 1.000 |
Notes: Prognostic group standard according to chromosomal abnormalities: Karyotype associated with favorable prognosis include t(8;21)(q22;q22), inv(16)(p13q22)/t(16;16)(p13;q22); Karyotype associated with intermediate prognosis include t(15;17)(q22;q12), normal cytogenesis, +8; Karyotype associated with adverse prognosis include complex karyotype consisting of ≥3 abnormalities, t(6;9)(p23;q34), abnormal 11q23 excluded t(9:11), del(5q), −5, del(7q),-7, t(9:22). *Indicated statistical significance (P<0.05).
Abbreviations: WBC, white blood cell; BM blast, bone marrow blast; FAB, French-American-British criteria.
RNA Isolation and Reverse Transcription
The process of isolating total RNA from BMNCs was conducted by using Trizol reagent (Thermo Fisher Scientific, Waltham, MA, USA). Reverse transcription mixture contains 2μg of total RNA from each sample, 10mM of dNTPs, 10μM of random hexamers, 80U of RNase inhibitor, and 200U of reverse transcriptase (Thermo Fisher Scientific, Waltham, MA, USA). The reverse transcript system was incubated at 25°C for 10 min, at 42°C for 60 min and stored at −20°C.
Real-Time Quantitative PCR
The expression of circ_0002232, miR-92a-3p and PTEN was detected by real-time quantitative PCR (RQ-PCR) with specific primers listed in supplemental File 1. The PCR reaction systems of detecting circ_0002232 and PTEN were SYBR Premix Ex Taq II (TaKaRa, Japan) and the reaction system of detecting the expression of miR-92a-3p was miScript SYBR green PCR kit (Qiagen, Duesseldorf, Germany). 7500 Thermocycler (Thermo Fisher Scientific, Waltham, MA, USA) was used to perform reaction system. A housekeeping gene (ABL) was used to calculate the quantity of circ_0002232 and PTEN. And the quantity of miR-92a-3p was valued by U6. Relative expression level of circ_0002232, miR-92a −3p and PTEN were calculated by using 2−ΔΔCT formula.
Gene Mutation Detection
Mutations of gene NPM1, N/K-RAS, DNMT3A, c-KIT, U2AF1, IDH1/2 and SRSF2 were detected by High Resolution Melting analysis.19–22 Direct DNA sequencing was used to detect mutations of gene CEBPA and FLT3-ITD.
Bioinformatics and Statistical Analysis
Micro RNAs which might bind with circ_0002232 were predicted by circRNA-miRNA interaction prediction websites, miRanda (http://miranda.org.uk) and RNAhybrid (https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid/submission.html). Target genes of miR-92a-3p were predicted by miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/php/index.php), miRDB (http://www.mirdb.org) and TargetScan (http://www.targetscan.org/vert_72/).
Statistical analysis was conducted by using spss software version 22.0. The diagnostic value of circ_0002232 expression was evaluated by receiver operating characteristic (ROC) curve and area under the ROC curve (AUC). The differences of categorical variables between the two groups were analysed by using Pearson Chi-square analysis or Fisher exact test and the differences of continuous variables were evaluated by using Mann–Whitney U-test. To explore prognostic potential of circ_0002232, Kaplan-Meier curves were used to analyse the impact of circ_0002232 for overall survival (OS) and Cox regression analysis was used to assess its independent prognostic value. Spearman correlation analysis was used respectively to examine the correlationship among the expression of circ_0002232, miR-92a-3p and PTEN. P value less than or equal to 0.05 (two-sided) was considered statistically significant in all analyses.
Results
Circ_0002232 Expression in AML and Controls
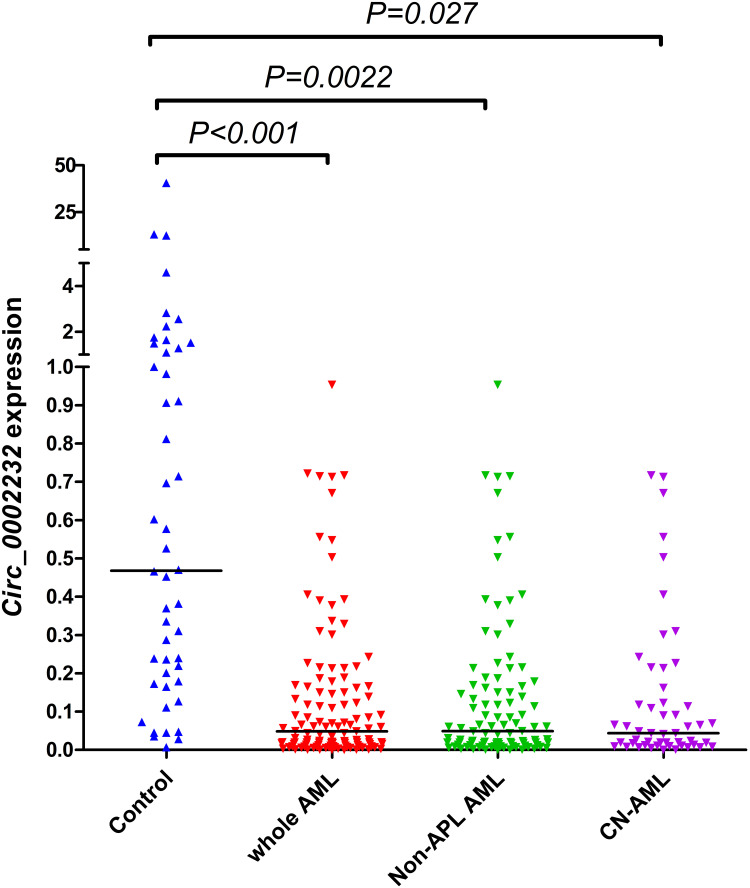
In our experiment, the expression level of circ_0002232 in de novo AML (median 0.0486, range 0.000215–0.953) was notably decreased compared with that in controls (median 0.468, range 0.00693–40.518; P<0.001, Figure 1). In addition, compared with controls, circ_0002232 expression level was remarkably down-regulated in non-APL AML patients (P=0.0022, Figure 1) and was low-expressed in CN-AML patients (P=0.027, Figure 1).
Figure 1.
Relative expression level of circ_0002232 in controls and AML. The expression of circ_0002232 in controls, whole AML, non-APL AML and CN-AML patients was measured by using RQ-PCR. Each dot represents a single sample and horizontal line represents the median level of expression.
Differentiating Ability of Circ_0002232 Expression
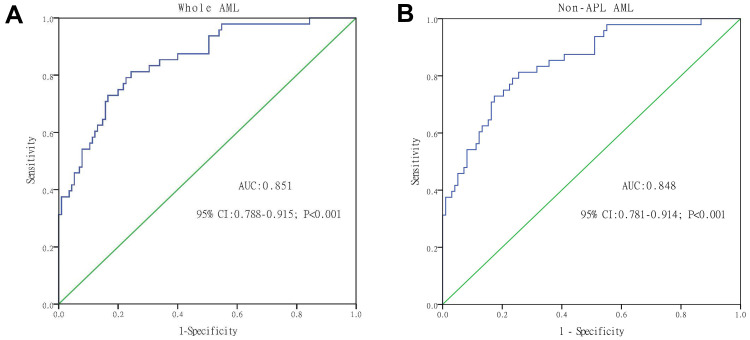
The capacity of circ_0002232 expression to distinguish AML patients from controls was analysed by ROC curve (AUC:0.851, 95% CI:0.788–0.915, P<0.001, Figure 2A). It indicated that cric_00022232 could act as a significant marker in differentiating between AML patients and controls. In addition, the remarkable significance was found in distinguishing non-APL AML patients from controls (AUC:0.848, 95% CI:0.781–0.914, P<0.001, Figure 2B).
Figure 2.
ROC curve analysis of circ_0002232 for distinguishing AML patients from controls: (A) Whole AML; and (B) non-APL AML.
Clinical and Laboratory Characteristics of AML Patients
For the purpose of exploring the relationship between clinical parameters and circ_0002232 expression, we obtained the cut-off value, which had the maximum sum of sensitivity and specificity according to ROC curve analysis, and divided 115 AML patients into low-expressed group (circ_0002232low) and high-expressed group (circ_0002232high). Hence, we used 0.165 as the cut-off value, whose sensitivity was 0.813 and specificity was 0.757. There were no significant discrepancies between the two groups in sex, hemoglobin, BM blasts, complete remission (CR), cytogenetic abnormalities and nine gene mutations (P>0.05, Table 1).
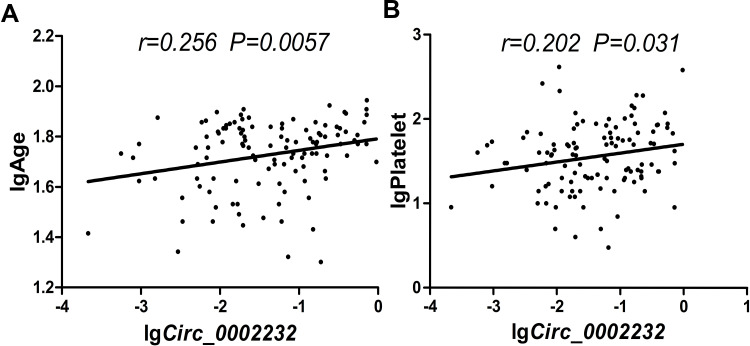
However, remarkable differences were observed in FAB classifications (P=0.036), white blood cell (WBC) count (P=0.041) and platelet count (P=0.021) between circ_0002232low and circ_0002232high groups. Age of the patients in circ_0002232low group was notably younger than those in circ_0002232high group (P=0.004). Moreover, we found that there was a positive correlation between circ_0002232 expression and patients’ age (Pearson r=0.256, P=0.0057, Figure 3A). And a positive correlation between the expression of circ_0002232 and platelet count was also found (Pearson r=0.202, P=0.031, Figure 3B).
Figure 3.
Pearson correlation analysis: (A) correlationship between patients’ age and circ_0002232 expression in AML; and (B) correlationship between platelet count and circ_0002232 expression in AML.
Correlation Between Circ_0002232 Expression and Patients’ Clinical Outcome
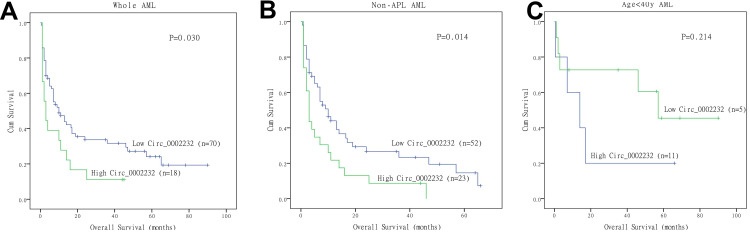
Survival analysis included 88 AML patients and excluded 27 patients who failed to follow up. Median follow-up time of included patients was 8 months, which ranged from 1 month to 90 months. According to Kaplan-Meier analysis, circ_0002232low group had significantly longer OS compared with circ_0002232high group in whole AML (P=0.030, Figure 4A). In non-APL AML, patients in low-expressed circ_0002232 group tended to have better prognosis (P=0.014, Figure 4B). However, in low age group (age<40y), we found that patients with high circ_0002232 expression tended to have better OS, but it was not statistically significant (P=0.214, Figure 4C).
Figure 4.
Kaplan-Meier analysis showed the differences in overall survival between circ_0002232low and circ_0002232high group: (A) overall survival among whole AML; (B) overall survival among non-APL AML; and (C) overall survival among AML (age<40y).
Univariate analysis, including age (≤60y or >60y), WBC count (≥30×109/L or <30×109/L), cytogenetic abnormalities’ classification, circ_0002232 expression, RAS mutation, U2AF1 mutation with P<0.05, showed that expression of circ_0002232 could be used as a valuable factor for AML patients’ prognosis. However, according to multivariate analysis, expression of circ_0002232 could not act as an independent factor for OS (P=0.727) among AML patients (Table 2).
Table 2.
Univariate and multivariate analyses of prognostic variables for overall survival in whole AML patients
| Variables | Overall Survival | |||
|---|---|---|---|---|
| Univariate Analysis | Multivariate Analysis | |||
| HR (95% CI) | P value | HR (95% CI) | P value | |
| Age | 2.532 (1.534–4.179) | <0.001 | 1.310 (0.731–2.346) | 0.364 |
| WBC | 3.021 (1.829–4.990) | <0.001 | 2.277 (1.302–3.981) | 0.004* |
| Cytogenetic abnormalities’ classifications | 2.076 (1.503–2.867) | <0.001 | 1.962 (1.306–2.948) | 0.001* |
| Circ_0002232 expression | 1.817 (1.024–3.223) | 0.041 | 0.874 (0.409–1.866) | 0.727 |
| FLT3-ITD mutation | 0.881 (0.397–1.954) | 0.755 | – | – |
| NPM1 mutation | 1.697 (0.672–4.287) | 0.263 | – | – |
| CEBPA mutation | 0.891 (0.380–2.093) | 0.791 | – | – |
| c-KIT mutation | 0.586 (0.142–2.416) | 0.460 | – | – |
| N/K-RAS mutation | 2.753 (1.071–7.081) | 0.036 | 3.000 (1.155–7.795) | 0.024* |
| IDH1/2 mutation | 5.328 (0.697–40.737) | 0.107 | – | – |
| DNMT3A mutation | 1.644 (0.652–4.147) | 0.292 | – | – |
| U2AF1 mutation | 4.609 (1.073–19.801) | 0.040 | 1.593 (0.202–12.584) | 0.659 |
| SRSF-2 mutation | 2.610 (0.353–19.303) | 0.347 | – | – |
Notes: Prognostic variables included WBC (≥30×109 vs <30×109/L), patients’ age (≤60 vs >60 years), cytogenetic abnormalities’ classifications (favorable vs intermediate vs poor), circ_0002232 expression level (Low vs High), and gene mutations (mutant vs wild-type). Variables with P<0.05 in univariate analysis were included into multivariate analysis. *Indicated statistical significance (P<0.05).
Abbreviations: HR, hazard ratio; CI, confidence interval; WBC, white blood cell.
Correlation Between Expression of Circ_0002232 and PTEN in AML
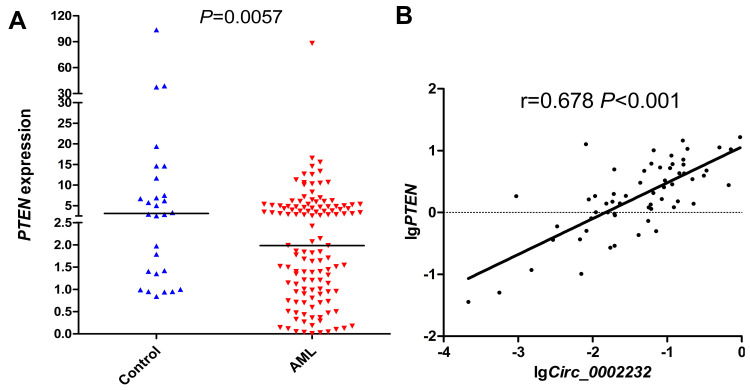
The expression of PTEN in AML (median 1.984, range 0.00701–88.0896) was remarkably down-regulated compared with controls (median 3.330, range 0.842–103.788) (P=0.0057, Figure 5A). Furthermore, the expression of circ_0002232 was positively correlated with its parental gene PTEN (Spearman r=0.678, P<0.001, Figure 5B).
Figure 5.
(A) Relative expression level of PTEN in controls and whole AML. (B) Spearman correlation analysis between the expression of PTEN and circ_0002232 in AML.
Potential Interaction Network of Circ_0002232/miR-92a-3p/PTEN
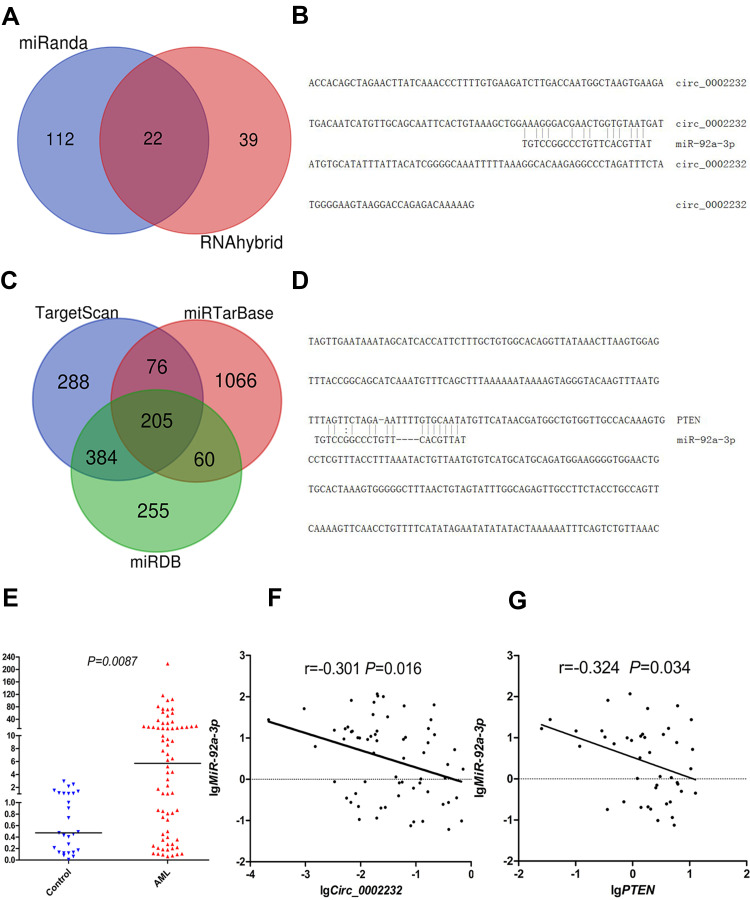
CircRNA-miRNA interaction prediction websites were used to predict miRNAs which might bind with circ_0002232. Through searching literature, we finally choose miR-92a-3p (Figure 6A and B). The expression of miR-92a-3p was detected in controls and AML patients. MiR-92a-3p was notably up-expressed in AML (median 6.215, range 0.0610–218.199) compared with controls (median 0.472, range 0.00815–2.964) (P=0.0087, Figure 6E). Spearman correlation analysis revealed that circ_0002232 expression was negatively correlated with miR-92a-3p expression in AML (Spearman r=−0.301, P=0.016, Figure 6F). Moreover, prediction websites showed the potential binding sites between miR-92a-3p and PTEN (Figure 6C and D). According to the result of Spearman analysis, miR-92a-3p had negative correlation with PTEN (Spearman r=−0.324, P=0.034, Figure 6G).
Figure 6.
(A) Venn results of microRNAs which could bind with circ_0002232 predicted by miRanda and RNAhybrid. (B) Binding sites between circ_0002232 and miR-92a-3p predicted by miRanda. (C) Venn results of genes which could bind with miR-92a-3p predicted by TargetScan, miRTarBase and miRDB. (D) Binding sites between miR-92a-3p and PTEN predicted by miRTarBase. (E) Relative expression level of miR-92a-3p in controls and AML. (F) Spearman correlation analysis between the expression of circ_0002232 and miR-92a-3p in AML. (G) Spearman correlation analysis between the expression of miR-92a-3p and PTEN in AML.
Discussion
CircRNAs, known as a novel category of non-coding RNAs, exist widely in mammalian cells.23 They have been considered as ideal biomarkers for disease because of their conservative feature across species. There are a few studies concentrated on the role of circRNAs in hematological malignancies. For instance, circ_0004277 expression had been reported to be down-regulated in AML. And the expression of circ_0004277 tended to be up-regulated when the patients had complete remission and down-regulated again when they relapsed. Circ_0004277 expression changed dynamically with process of AML, which proved that it could be used as AML biological marker.24
As far as we know, this is the first report focused on the expression of the circular RNA transcribed from PTEN in AML. In this study, circ_0002232 expression in AML was notably down-regulated compared with that in controls. The same results were found in groups of non-APL AML and CN-AML. According to ROC curve analysis, circ_0002232 could act as a valuable marker to identify AML patients and control groups.
Identifying the relationship between the expression of circ_0002232 and clinical characteristics, we found that the expression level of circ_0002232 was positively correlated with platelet count. Circ_0002232low group tended to have lower platelet count. There have already been several reports focused on the abnormal platelet count and dysfunction in AML.25 Low platelet count was associated with poor prognosis and recovery of platelet was concerned with relapse-free survival rate after chemotherapy in AML.26,27 Moreover, circ_0002232low group also tended to have lower hemoglobin and higher percentage of blast compared with circ_0002232 high expression group. This means circ_0002232low group have more severe myelosuppression and more serious infiltration in BM. Hence, low expression of circ_0002232 is an adverse factor of AML.
Because PTEN, parental gene of circ_0002232, plays a role of tumor suppressor in many diseases, at the beginning of the experiment we proposed that the circular RNA transcribed from PTEN might also have tumor suppressive effect and down-regulation of circ_0002232 might lead to shorter overall survival time. Unexpectedly, results of Kaplan-Meier analysis revealed that OS of patients with low-expressed circ_00002232 were longer than that of patients with high-expressed circ_00002232 in whole AML. This result was obviously in contrast with our initial prospect.
However, our study indicated that patients in circ_0002232high group were significantly older than those in circ_0002232low group. Pearson analysis was used to confirm this result, which revealed that the circ_0002232 expression was positively correlated with patients’ age. Age is an important risk factor for AML. Survival time of AML patients tends to decrease with increased age.28,29 We suppose that it may help us to understand this conflicting result. High-expressed circ_0002232 group tends to have poor prognosis because of the increase of age.
Then according to the expression level of circ_0002232, we divided the patients (age<40y) into two groups and compared the differences in survival time. The result in the subgroup under 40 showed that circ_0002232high group tended to have better OS compared with circ_0002232low group. This result confirmed our conjecture. But due to the limitation of our experiment size, this result was not statistically significant. In the future, additional experiments are needed to enlarge sample size and identify the relationship between circ_0002232 expression and OS in different age subgroups.
Moreover, the phenomenon of circRNAs acting as miRNA sponges in regulating proliferation, metastasis and relapse of gastrointestinal cancer have been reported in some studies.5 In this research, prediction websites revealed that there were potential binding sites among circ_0002232, miR-92a-3p and PTEN. MiR-92a-3p expression has been revealed to be up-regulated in several solid cancers including breast cancer and brain glioma.30,31 According to our experiment, miR-92a-3p expression of AML patients was obviously up-regulated compared with controls and was negatively correlated with the expression of circ_0002232.
Furthermore, high-expressed miR-92a have been found to regulate colorectal cell migration and invasion by suppressing the expression of PTEN.32 Alteration of miR-92a also promoted its effect on metastatic behavior of nasopharyngeal carcinoma cell by targeting PTEN.33 Notably, we found that the expression level of miR-92a-3p was also negatively correlated with PTEN in AML. Hence, we proposed that circ_0002232 might affect the process of AML and the expression of PTEN through sponging miR-92a-3p. In the future, we plan to design more experiments to explore the mechanism of this pathway in AML.
Conclusion
Our experiment revealed circ_0002232, one of circRNAs of PTEN, was remarkably down-regulated in AML and could act as a promising biomarker for the diagnosis of AML. In addition, there might be a potential ceRNA interaction network of circ_0002232/miR-92a-3p/PTEN in AML.
Funding Statement
This study was supported by National Natural Science foundation of China (81970156, 81970118), Medical Innovation Team of Jiangsu Province (CXTDB2017002), Zhenjiang Clinical Research Center of Hematology (AA2018009), and Jiangsu Provincial “innovative & entrepreneurial talent team” Program. Sponsors of fundings were not involved in any stages from study design to submission of the paper for publication.
Abbreviations
PTEN, phosphatase and tensin homolog; circRNAs, circular RNAs; ceRNA, competing endogenous RNAs; AML, acute myeloid leukemia; APL, acute promyelocytic leukemia; CN-AML, cytogenetic normal AML; RQ-PCR, real-time quantitative PCR; BM, bone marrow; BMNCs, BM mononuclear cells; FAB classification, French–American–British classification; WHO criteria, World Health Organization criteria; ROC, receiver operating characteristic; AUC, area under the ROC curve; CI, confidence interval; OS, overall survival; CR, complete remission; WBC, white blood cell; Lg, logarithm based on 10.
Data Sharing Statement
The datasets used during the current study are available from the corresponding author on reasonable request.
Ethics Approval and Informed Consents
This study was approved by Human Research Ethics Committee of the Affiliated People’s Hospital of Jiangsu University and was conducted in accordance with the Declaration of Helsinki. All patients signed informed consent to participate in our research.
Author Contributions
All authors contributed to data analysis, drafting or revising the article, agreed on the journal to which the article will be submitted, gave final approval of the version to be published, and agreed to be accountable for all aspects of the work.
Disclosure
The authors declare that they have no competing interests.
References
- 1.Short NJ, Rytting ME, Cortes JE. Acute myeloid leukaemia. Lancet. 2018;392(10147):593–606. doi: 10.1016/S0140-6736(18)31041-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Medinger M, Passweg JR. Acute myeloid leukaemia genomics. Br J Haematol. 2017;179(4):530–542. doi: 10.1111/bjh.14823 [DOI] [PubMed] [Google Scholar]
- 3.Valk PJ, Verhaak RG, Beijen MA, et al. Prognostically useful gene-expression profiles in acute myeloid leukemia. N Engl J Med. 2004;350(16):1617–1628. doi: 10.1056/NEJMoa040465 [DOI] [PubMed] [Google Scholar]
- 4.Wei JW, Huang K, Yang C, Kang CS. Non-coding RNAs as regulators in epigenetics (Review). Oncol Rep. 2017;37(1):3–9. doi: 10.3892/or.2016.5236 [DOI] [PubMed] [Google Scholar]
- 5.Li X, Yang L, Chen LL. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71(3):428–442. doi: 10.1016/j.molcel.2018.06.034 [DOI] [PubMed] [Google Scholar]
- 6.Qu S, Yang X, Li X, et al. Circular RNA: a new star of noncoding RNAs. Cancer Lett. 2015;365(2):141–148. doi: 10.1016/j.canlet.2015.06.003 [DOI] [PubMed] [Google Scholar]
- 7.Xia W, Qiu M, Chen R, et al. Circular RNA has_circ_0067934 is upregulated in esophageal squamous cell carcinoma and promoted proliferation. Sci Rep. 2016;6:35576. doi: 10.1038/srep35576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shao YF, Li JY, Lu RD, et al. Global circular RNA expression profile of human gastric cancer and its clinical significance. Cancer Med. 2017;6(6):1173–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu L, Wang J, Khanabdali R, Kalionis B, Tai XT, Xia SJ. Circular RNAs: isolation, characterization and their potential role in diseases. RNA Biol. 2017;14(12):1715–1721. doi: 10.1080/15476286.2017.1367886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weng W, Wei Q, Toden S, et al. Circular RNA ciRS-7-A promising prognostic biomarker and a potential therapeutic target in colorectal cancer. Clin Cancer Res. 2017;23(14):3918–3928. doi: 10.1158/1078-0432.CCR-16-2541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bai N, Peng E, Qiu X, et al. circFBLIM1 act as a ceRNA to promote hepatocellular cancer progression by sponging miR-346. J Exp Clin Cancer Res. 2018;37(1):172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen L, Guo D. The functions of tumor suppressor PTEN in innate and adaptive immunity. Cell Mol Immunol. 2017;14(7):581–589. doi: 10.1038/cmi.2017.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wise HM, Hermida MA, Leslie NR. Prostate cancer, PI3K, PTEN and prognosis. Clin Sci (Lond). 2017;131(3):197–210. doi: 10.1042/CS20160026 [DOI] [PubMed] [Google Scholar]
- 14.Koboldt DC, Fulton RS, McLellan MD, et al. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, Gao L, Luo X, et al. Epigenetic silencing of microRNA-193a contributes to leukemogenesis in t(8;21) acute myeloid leukemia by activating the PTEN/PI3K signal pathway. Blood. 2013;121(3):499–509. doi: 10.1182/blood-2012-07-444729 [DOI] [PubMed] [Google Scholar]
- 16.Zayed R, Eltaweel M, Botros S, Zaki M. MN1 and PTEN gene expression in acute myeloid leukemia. Cancer Biomark. 2017;18(2):177–182. doi: 10.3233/CBM-160235 [DOI] [PubMed] [Google Scholar]
- 17.Bennett JM, Catovsky D, Daniel MT, et al. Proposed revised criteria for the classification of acute myeloid leukemia. A report of the French-American-British Cooperative Group. Ann Intern Med. 1985;103(4):620–625. doi: 10.7326/0003-4819-103-4-620 [DOI] [PubMed] [Google Scholar]
- 18.Arber DA, Orazi A, Hasserjian R, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127(20):2391–2405. doi: 10.1182/blood-2016-03-643544 [DOI] [PubMed] [Google Scholar]
- 19.Lin J, Yao DM, Qian J, et al. Recurrent DNMT3A R882 mutations in Chinese patients with acute myeloid leukemia and myelodysplastic syndrome. PLoS One. 2011;6(10):e26906. doi: 10.1371/journal.pone.0026906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin J, Yao DM, Qian J, et al. IDH1 and IDH2 mutation analysis in Chinese patients with acute myeloid leukemia and myelodysplastic syndrome. Ann Hematol. 2012;91(4):519–525. doi: 10.1007/s00277-011-1352-7 [DOI] [PubMed] [Google Scholar]
- 21.Yang X, Qian J, Sun A, et al. RAS mutation analysis in a large cohort of Chinese patients with acute myeloid leukemia. Clin Biochem. 2013;46(7–8):579–583. doi: 10.1016/j.clinbiochem.2012.12.022 [DOI] [PubMed] [Google Scholar]
- 22.Qian J, Yao DM, Lin J, et al. U2AF1 mutations in Chinese patients with acute myeloid leukemia and myelodysplastic syndrome. PLoS One. 2012;7(9):e45760. doi: 10.1371/journal.pone.0045760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dong Y, He D, Peng Z, et al. Circular RNAs in cancer: an emerging key player. J Hematol Oncol. 2017;10(1):2. doi: 10.1186/s13045-016-0370-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li W, Zhong C, Jiao J, et al. Characterization of hsa_circ_0004277 as a new biomarker for acute myeloid leukemia via circular RNA profile and bioinformatics analysis. Int J Mol Sci. 2017;18(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qian X, Wen-jun L. Platelet changes in acute leukemia. Cell Biochem Biophys. 2013;67:1473–1479. doi: 10.1007/s12013-013-9648-y [DOI] [PubMed] [Google Scholar]
- 26.Zhang QY, Dai KC, Bi LX, et al. Pretreatment platelet count predicts survival outcome of patients with de novo non-M3 acute myeloid leukemia. PeerJ. 2017;5:e4139. doi: 10.7717/peerj.4139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamazaki E, Kanamori H, Itabashi M, et al. Hyper-recovery of platelets after induction therapy is a predictor of relapse free survival in acute myeloid leukemia. Leuk Lymphoma. 2017;58(1):104–109. doi: 10.1080/10428194.2016.1190969 [DOI] [PubMed] [Google Scholar]
- 28.Appelbaum FR, Gundacker H, Head DR, et al. Age and acute myeloid leukemia. Blood. 2006;107(9):3481–3485. doi: 10.1182/blood-2005-09-3724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abelson S, Collord G, Ng SWK, et al. Prediction of acute myeloid leukaemia risk in healthy individuals. Nature. 2018;559(7714):400–404. doi: 10.1038/s41586-018-0317-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cun J, Yang Q. Bioinformatics-based interaction analysis of miR-92a-3p and key genes in tamoxifen-resistant breast cancer cells. Biomed Pharmacother. 2018;107:117–128. doi: 10.1016/j.biopha.2018.07.158 [DOI] [PubMed] [Google Scholar]
- 31.Song H, Zhang Y, Liu N, et al. miR-92a-3p exerts various effects in glioma and glioma stem-like cells specifically targeting CDH1/β-catenin and Notch-1/Akt signaling pathways. Int J Mol Sci. 2016;17(11):1799. doi: 10.3390/ijms17111799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang G, Zhou H, Xiao H, Liu ZL, Tian HP, Zhou T. MicroRNA-92a functions as an oncogene in colorectal cancer by targeting PTEN. Dig Dis Sci. 2014;59(1):98–107. doi: 10.1007/s10620-013-2858-8 [DOI] [PubMed] [Google Scholar]
- 33.Zhang H, Cao H, Xu D, Zhu K. MicroRNA-92a promotes metastasis of nasopharyngeal carcinoma by targeting the PTEN/AKT pathway. Onco Targets Ther. 2016;9:3579–3588. [DOI] [PMC free article] [PubMed] [Google Scholar]