Abstract
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus is causing a global pandemic, and cases continue to rise. Most infected individuals experience mildly symptomatic coronavirus disease 2019 (COVID-19), but it is unknown whether this can induce persistent immune memory that could contribute to immunity. We performed a longitudinal assessment of individuals recovered from mild COVID-19 to determine whether they develop and sustain multifaceted SARS-CoV-2-specific immunological memory. Recovered individuals developed SARS-CoV-2-specific immunoglobulin (IgG) antibodies, neutralizing plasma, and memory B and memory T cells that persisted for at least 3 months. Our data further reveal that SARS-CoV-2-specific IgG memory B cells increased over time. Additionally, SARS-CoV-2-specific memory lymphocytes exhibited characteristics associated with potent antiviral function: memory T cells secreted cytokines and expanded upon antigen re-encounter, whereas memory B cells expressed receptors capable of neutralizing virus when expressed as monoclonal antibodies. Therefore, mild COVID-19 elicits memory lymphocytes that persist and display functional hallmarks of antiviral immunity.
Keywords: SARS-CoV2, COVID-19, memory B cell, memory T cell, monoclonal antibody, human, vaccine, adaptive immune response
Graphical Abstract
Longitudinal analysis of immune memory following mild COVID-19 elicits memory lymphocytes that persist and display functional hallmarks of antiviral immunity.
Introduction
The rapidly spreading severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) betacoronavirus has infected millions of people and killed hundreds of thousands worldwide in 2020. Infection causes the coronavirus disease 2019 (COVID-19), which ranges in presentation from asymptomatic to fatal. The vast majority of infected individuals experience mild symptoms that do not require hospitalization (Wu and McGoogan, 2020). It is critically important to understand if SARS-CoV-2-infected individuals who recover from mild disease develop functional immune memory cells capable of protection from subsequent SARS-CoV-2 infections, thereby reducing transmission and COVID-19 disease.
Immunological memory is primarily mediated by cells of the adaptive immune system. In response to most acute viral infections, B and T cells that can bind viral proteins through their antigen receptors and become activated, expand, differentiate, and begin secreting effector molecules to help control the infection. Upon resolution of infection, approximately 90% of these virus-specific “effector cells” die, whereas 10% persist as long-lived “memory” cells (Ruterbusch et al., 2020). Immune memory cells can produce a continuous supply of effector molecules, as seen with long-lived antibody-secreting plasma cells (LLPCs). In most cases, however, quiescent memory lymphocytes are strategically positioned to rapidly reactivate in response to re-infection and execute effector programs imprinted upon them during the primary response. Upon re-infection, pathogen-specific memory B cells (MBCs) that express receptors associated with antigen experience and the transcription factor T-bet rapidly proliferate and differentiate into protective immunoglobulin (Ig)G+ antibody-secreting plasmablasts (PBs) (Kim et al., 2019; Knox et al., 2019; Nellore et al., 2019). Reactivated T-bet-expressing memory CD4+ T cells proliferate, “help” activate MBCs, and secrete cytokines (including interferon [IFN]-γ) to activate innate cells (Ruterbusch et al., 2020). Meanwhile, memory CD8+ T cells also secrete cytokines and kill virus-infected cells directly through the delivery of cytolytic molecules (Schmidt and Varga, 2018). These quantitatively and qualitatively enhanced virus-specific memory populations coordinate to quickly clear the virus, thereby preventing disease and reducing the chance of transmission. It is therefore critical to assess the full cadre of SARS-CoV-2-specific immune memory responses to determine whether mild infection induces a lasting, multilayered defense.
To infect cells and propagate, SARS-CoV-2 relies on the interaction between the receptor-binding domain (RBD) of its spike (S) protein and angiotensin converting enzyme 2 (ACE2) on host cells (Hoffmann et al., 2020). Multiple studies have shown that the majority of SARS-CoV-2-infected individuals produce S- and RBD-specific antibodies during the first 2 weeks of the primary response and that RBD-specific monoclonal antibodies can neutralize the virus in vitro and in vivo (Long et al., 2020; Robbiani et al., 2020; Shi et al., 2020). Therefore, RBD-specific antibodies would likely contribute to protection in response to reinfection if maintained in the plasma by LLPCs or rapidly expressed by MBCs.
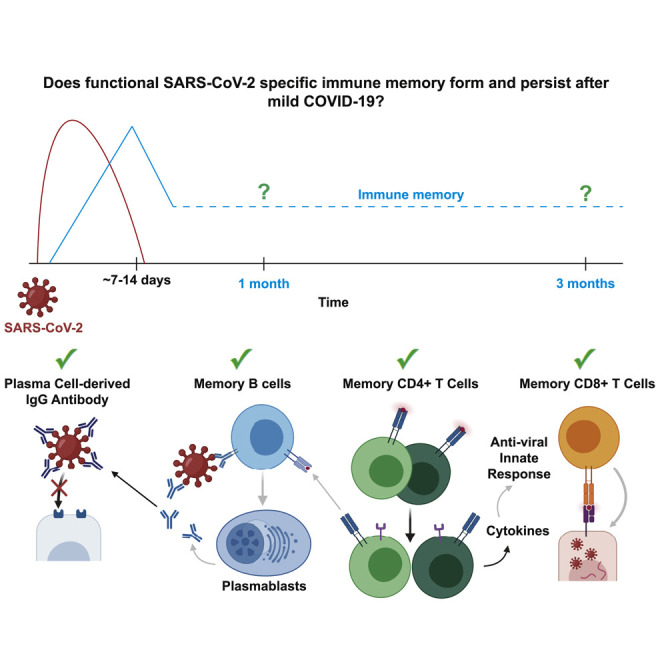
We therefore assessed SARS-CoV-2-specific immune responses at 1 and 3 months post-symptom onset in individuals that had experienced mild COVID-19. We found that a multipotent SARS-CoV-2-specific immune memory response forms and is maintained in recovered individuals for the duration of our study. Furthermore, persistent memory lymphocytes display hallmarks of protective antiviral immunity, including a numerically increased population of virus-specific memory B cells capable of expressing SARS-CoV-2 neutralizing antibodies.
Results
Return to Immune Homeostasis after Mildly Symptomatic COVID-19
To determine whether immune memory cells form after mildly symptomatic COVID-19, we collected plasma and peripheral blood mononuclear cells (PBMCs) from 15 individuals recovered from mild COVID-19 (CoV2+) (UW IRB 00009810). The CoV2+ group had a median age of 47 and reported mild symptoms lasting a median of 13 days (Table 1 ). The first blood sample (Visit 1) was drawn at least 20 days after a positive PCR test for SARS-CoV-2 and a median of 35.5 days post-symptom onset. We expect the primary response to be contracting and early memory populations to be generated at this time point, as viral load is cleared approximately 8 days post-symptom onset (Wölfel et al., 2020). Participants returned for a second blood draw (Visit 2) a median of 86 days post-symptom onset so we could assess the quantity and quality of the long-lived memory populations (Figure 1 A). We compared these samples to samples collected at 2 time points representing a similar sampling interval in a group of 17 healthy controls (HCs). All HCs were considered to have no prior SARS-CoV-2 infection based on having no detectable plasma SARS-CoV-2 RBD- or S-specific antibodies above 3 standard deviations (SDs) of the mean of historical negative (HN) plasma samples drawn prior to 2020 (Figure S1 ).
Table 1.
Study Cohort
| CoV2+ | HCs | |
|---|---|---|
| Number of participantsa | 15 | 17 |
| Age (years) | 47 (28–71) | 42 (24–57) |
| Sex | 27% male, 73% female |
47% male, 53% female |
| Number of symptomsb,c | 5 (1–7) | NAd |
| Symptom duration (days) | 13 (2–31) | NA |
| Time from symptom onset to Visit 1 (days) | 35.5 (19–44) | NA |
| Time from symptom onset to Visit 2 (days) | 86 (73–110) | NA |
| Time from SARS-CoV-2 positive PCR test to Visit 1 (days) | 28 (20–35) | NA |
| Time from SARS-CoV-2 positive PCR test to Visit 2 (days) | 77.5 (64–97) | NA |
| Time from Visit 1 to Visit 2 (days) | 46 (39–69) | 47 (40–61) |
Previously SARS-CoV-2-infected (CoV2+) and HC volunteers were consented and enrolled for this study. Values are reported as the median with the range in parentheses.
Blood drawn from 14 CoV2+ and 13 HCs at Visit 1 and Visit 2. One CoV2+ and 2 HCs were only drawn with Visit 1. Two HCs were only drawn with Visit 2.
All CoV2+ individuals reported symptoms. Nine HCs reported symptoms, and 2 HC had negative SARS-CoV-2 PCR results.
The symptoms surveyed were fever, chills, cough, runny nose, fatigue, muscle ache, and difficulty breathing.
NA = Not applicable.
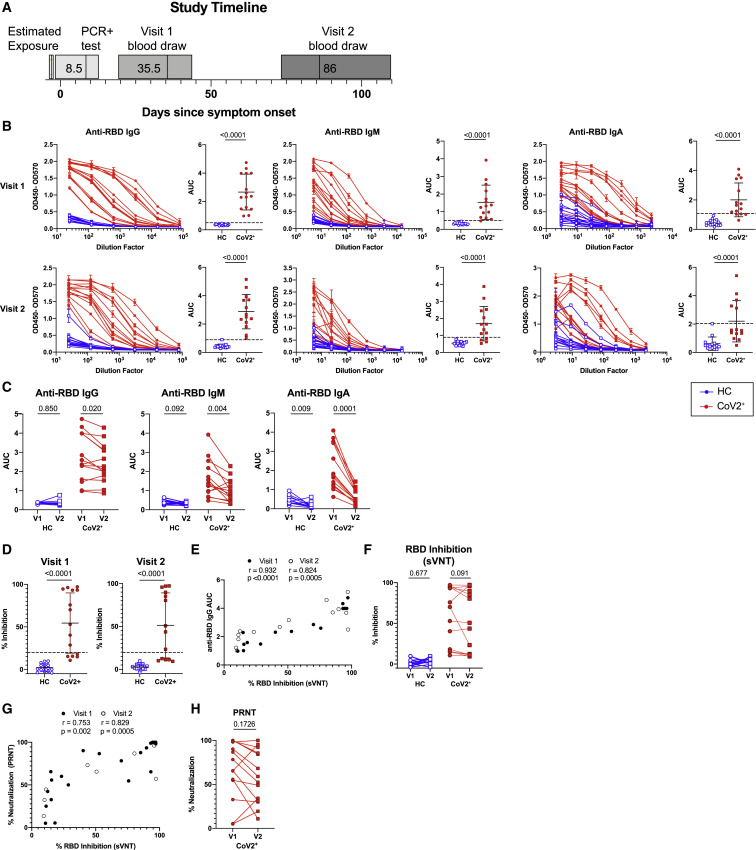
Figure 1.
Mild COVID-19 Induces Persistent, Neutralizing Anti-SARS-CoV-2 IgG Antibody
(A) Study timeline. Range is indicated by box and median indicated by line for each event.
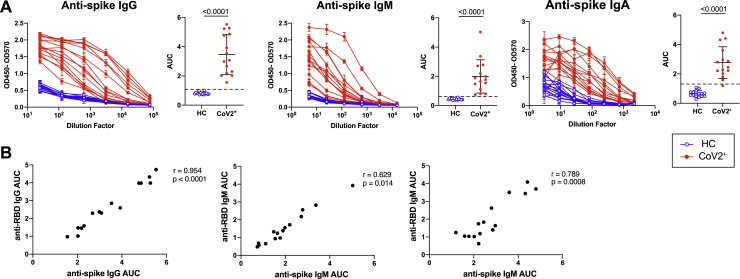
(B) ELISA dilution curves and AUC for anti-RBD IgG (left), IgM (center), and IgA (right) from HC and CoV2+ plasma samples at Visit 1 (V1) and Visit 2 (V2). Dashed line indicates mean + 3 SD of the HC AUC values.
(C) Comparing V1 and V2 AUC in HC and CoV2+ individuals for each antibody isotype. V2 AUC values were normalized to V1 samples run with V2 samples.
(D) Percent inhibition of RBD binding to ACE2 by plasma sVNT at 1:2 plasma dilution.
(E) Spearman correlation between percent RBD inhibition by sVNT at a 1:2 plasma dilution and anti-RBD IgG AUC at both visits.
(F) Percent RBD inhibition at 1:2 plasma dilution at V1 and V2, paired by sample.
(G) Spearman correlation between percent RBD inhibition by sVNT at a 1:2 plasma dilution and percent virus neutralization by PRNT at a 1:160 plasma dilution.
(H) CoV2+ percent virus neutralization by PRNT at a 1:160 plasma dilution normalized and paired as in (C).
Statistics for unpaired data determined by 2-tailed Mann-Whitney tests and, for paired data, by 2-tailed Wilcoxon signed-rank tests. Multiple testing correction significance cutoff at FDR = 0.05 is p value < 0.05. Error bars represent mean and SD.
See also Figures S1 and S4.
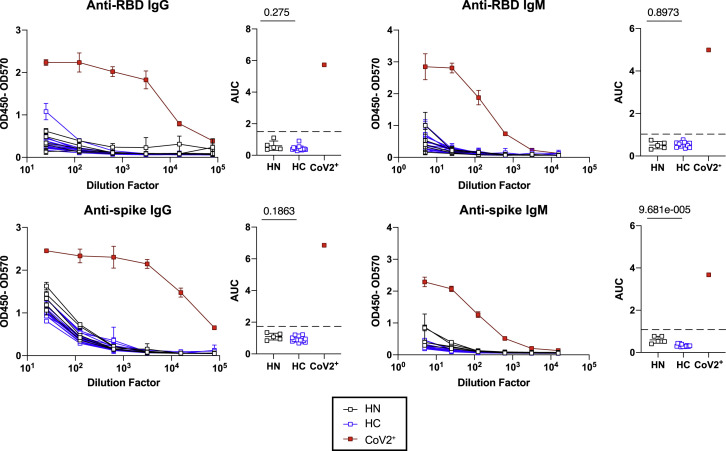
Figure S1.
Healthy Controls Do Not Have SARS-CoV-2 RBD or Spike-Specific Antibodies, Related to Figure 1
ELISA dilution curves and area under the curve (AUC) for anti-RBD and anti-spike IgG (left) and IgM (right) in plasma collected from individuals prior to 2020 and the SARS-CoV-2 pandemic (historical negatives, HN, black), from healthy controls (HC, at Visit 2) and from individuals that tested PCR+ for SARS-CoV-2 (CoV2+, at Visit 1). Dashed line indicates mean + 3 SD of HN AUC values.
Statistics determined by two-tailed Mann-Whitney tests. Multiple testing correction significance cutoff at FDR = 0.05 is p value < 0.05. Error bars represent mean and SD.
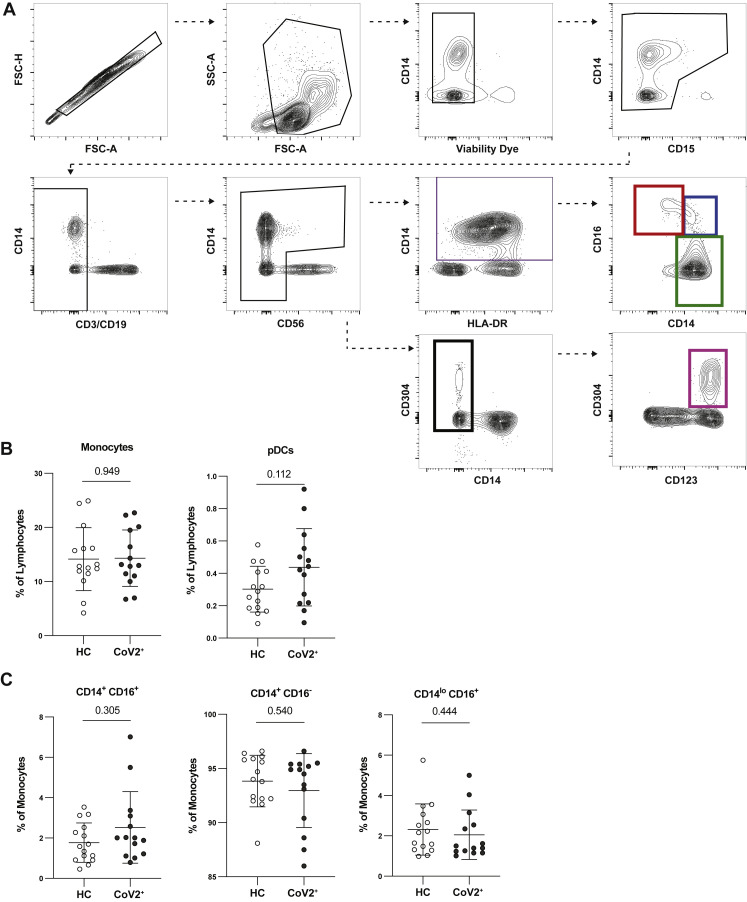
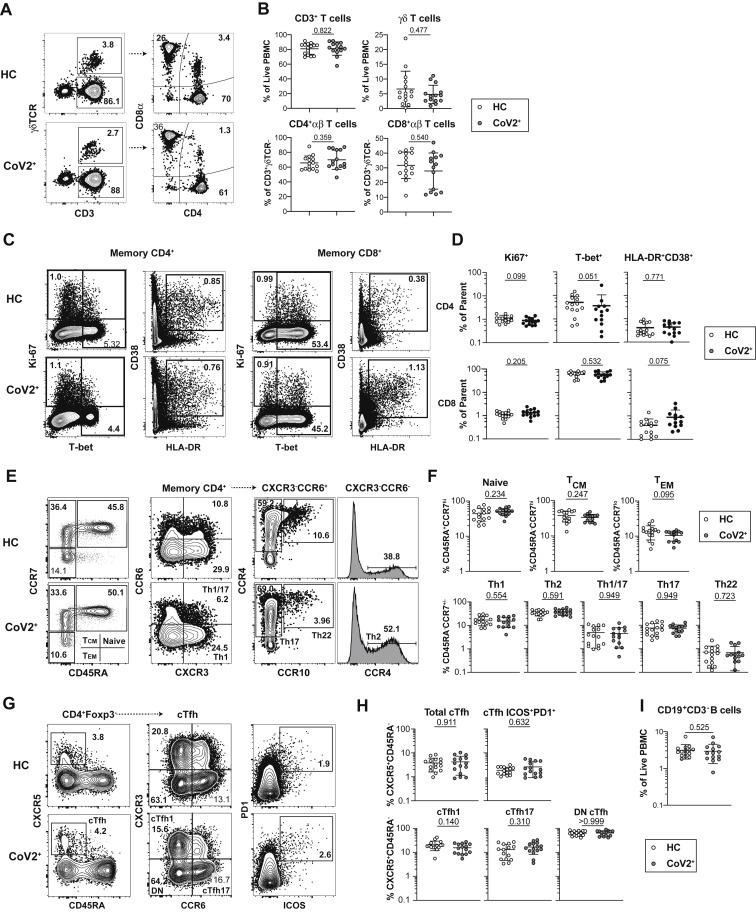
Populations of activated innate and adaptive immune cells expand in the blood during the primary response to SARS-CoV-2 infection (Mathew et al., 2020). When an acute viral infection is cleared, the majority of these highly inflammatory cells either die or become quiescent memory cells such that the proportions and phenotypes of total immune cells are indistinguishable from those seen in pre-infection blood samples. Consistent with resolution of the primary response, we found no differences in frequency of total monocytes, monocyte subsets, or plasmacytoid dendritic cells among PBMCs between CoV2+ and HC individuals (Figure S2 ). We also found no differences in γδ or αβ CD3+ T cell frequencies (CD4+ or CD8+), cell cycle status, expression of molecules associated with activation, migration, function, or proportions of various CD45RA– memory T cell subsets (Figures S3 A–S3H). We also found no differences in frequency of CD19+ B cells (Figure S3I). Together, these data demonstrate that the inflammatory response associated with acute infection had resolved by the Visit 1 time point and that the early immune memory phase had commenced.
Figure S2.
PBMC Innate Populations in CoV2+ Individuals Return to Immune Quiescence by Visit 1, Related to Figure 1
(A) Flow cytometry gating for CD15-CD3-CD19-CD56-HLADR+CD14+ monocytes (purple gate), which were further divided into CD14loCD16+ (red gate), CD14+CD16+ (blue gate), and CD14+CD16- monocytes (green gate), and CD15-CD3-CD19-CD56-CD14-CD304+CD123+ plasmacytoid dendritic cells (pDCs) (pink gate).
(B) Percent monocytes and pDCs of live PBMCs from healthy controls (HC) and previously SARS-CoV-2 infected (CoV2+) individuals.
(C) Percent subsets of monocytes from PBMCs.
Statistics determined by two-tailed Mann-Whitney tests. Multiple testing correction significance cutoff at FDR = 0.05 is p value < 0.05. Error bars represent mean and SD. Data from two experiments.
Figure S3.
Bulk PBMCs Return to Immune Quiescence by Visit 1, Related to Figure 1
(A and B) Representative flow cytometry plots and frequencies of αβ and γδ T cell subsets at Visit 1 (V1) in PBMCs from healthy control (HC) and SARS-CoV-2-recovered (CoV2+) individuals.
(C and D) Representative flow cytometry plots and frequencies of CD4+ and CD8+ T cell effector/activation states (Ki67+, T-bet+, HLA-DR+CD38+) of total non-naive, memory CD45RA+CCR7+/− CD4+ or CD8+ T cells at V1 in HC and CoV2+ PBMCs.
(E and F) Representative flow cytometry plots and frequencies of CD4+ memory and T-helper subsets at V1 in HC and CoV2+ PBMCs.
(G and H) Representative flow cytometry plots and frequencies of cTfh (CXCR5+CD45RA-) and cTfh activation (ICOS+PD-1+) and helper (CXCR3+/−CCR6+/−) subsets at V1 in HC and CoV2+ PBMCs.
(I) Frequency of B cells (CD19+CD3-) at V1 in HC and CoV2+ PBMCs.
Statistics determined by two-tailed Mann-Whitney tests. Multiple testing correction significance cutoff at FDR = 0.05 is p value < 0.05. Error bars represent mean and SD. Data from two experiments.
Mild COVID-19 Induces Persistent, Neutralizing Anti-SARS-CoV-2 IgG Antibody
Humoral immune responses are characterized by a first wave of short-lived, low-affinity antibody-secreting PBs followed by a subsequent germinal center (GC) response that generates high-affinity MBCs and antibody-secreting LLPCs. LLPCs can maintain detectable plasma antibody titers for months to many years, depending upon the specific viral infection (Slifka and Ahmed, 1996). PBs are formed during acute SARS-CoV-2 infection but are no longer present in recovered individuals at approximately 1 month post-symptom onset (Mathew et al., 2020). This suggests that antibodies measured at Visit 1 might include significant contributions from short-lived PBs that are no longer present, but based on calculations of antibody half-life (approximately 23 days; Cohen and Freeman, 1960), only an estimated 6% of PB-derived antibodies present at Visit 1 would be retained at Visit 2. At Visit 2, LLPCs are therefore likely contributing the majority of antibodies and maintaining the level of RBD-specific antibodies in the blood. We therefore examined the levels of SARS-CoV-2-specific IgG, IgM, and IgA antibodies in plasma at Visit 1 and Visit 2 (Ma et al., 2020). At Visit 1, all of CoV2+ individuals had plasma anti-RBD IgG levels 3 SDs above the mean of HCs, as measured by ELISA area under the curve (AUC), in accordance with studies showing 100% seroprevalence by day 14 (Long et al., 2020) (Figure 1B). Additionally, the majority of CoV2+ individuals had anti-RBD IgM and anti-RBD IgA above this negative threshold and possessed IgG, IgM, and IgA anti-spike antibodies above the threshold at Visit 1 as well (Figure S4 A). Levels of anti-RBD and anti-spike binding were highly correlated for all isotypes (Figure S4B). At Visit 2, all CoV2+ individuals maintained anti-RBD IgG levels above the negative threshold, but fewer CoV2+ individuals maintained anti-RBD IgM and IgA (Figure 1B). Anti-RBD IgG levels decreased only slightly among CoV2+ individuals between time points, suggesting the IgG antibody levels were being stabilized by the formation of antibody-secreting LLPCs. Anti-RBD IgM and IgA, however, decreased substantially from Visit 1 to Visit 2, suggesting expression from early PBs that is not maintained by LLPCs (Figure 1C).
Figure S4.
Mild COVID-19 Induces Persistent, Neutralizing Anti-SARS-CoV-2 IgG Antibody, Related to Figure 1
(A) ELISA dilution curves and area under the curve (AUC) for anti-spike IgG (left), IgM (center), and IgA (right) from healthy control (HC) and SARS-CoV-2-recovered (CoV2+) individuals plasma at Visit 1 (V1). Dashed line indicates mean + 3 SD of the HC AUC values.
(B) Spearman correlation of V1 anti-RBD and anti-spike IgG (left), IgM (center), and IgA (right) AUC.
Statistics determined by two-tailed Mann-Whitney tests. Multiple testing correction significance cutoff at FDR = 0.05 is p value < 0.05. Error bars represent mean and SD.
As spike protein, and specifically the RBD, is key for viral entry into the cell, antibodies that target the RBD can be potent inhibitors of infection (Robbiani et al., 2020; Shi et al., 2020). To determine whether CoV2+ individuals form and maintain neutralizing antibodies, we tested for SARS-CoV-2 neutralization indirectly by using a cell-free assay of RBD-ACE2 binding inhibition (surrogate virus neutralization test, sVNT) and directly in a plaque reduction neutralization test (PRNT) (Tan et al., 2020). CoV2+ plasma inhibited RBD binding to ACE2 significantly more than did HC plasma by sVNT, and RBD inhibition correlated strongly with anti-RBD IgG levels at both time points (Figures 1D and 1E). Further, CoV2+ plasma RBD inhibition capacity was maintained from Visit 1 to Visit 2 (Figure 1F). Neutralization by PRNT correlated strongly with RBD inhibition at both time points (Figure 1G) and was similarly maintained between visits (Figure 1H). The majority of CoV2+ plasma achieved at least 50% neutralization by PRNT at a 1:160 dilution at both time points. By the latest time point in our study, the majority of CoV2+ individuals still had better RBD-inhibiting plasma and better neutralizing plasma than did HCs. These data are consistent with the emergence of IgG+ RBD-specific LLPCs that maintain detectable neutralizing anti-SARS-CoV-2 antibody to at least 3 months post-symptom onset.
Mild COVID-19 Induces a Sustained Enrichment of RBD-Specific IgG+ Memory B Cells
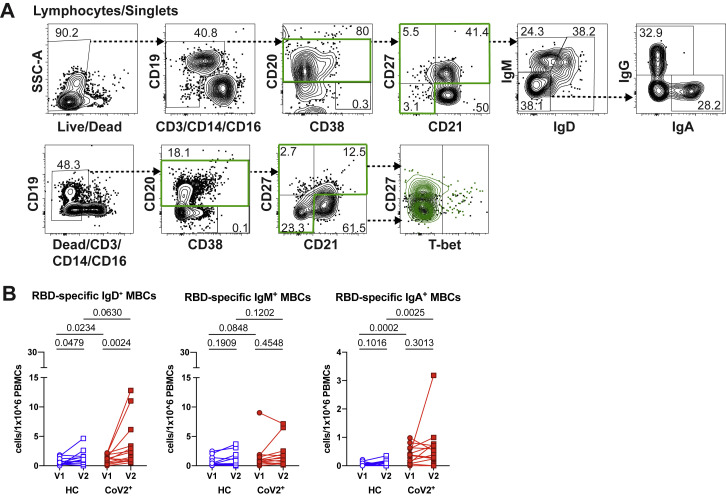
The presence of SARS-CoV-2 neutralizing antibodies 3 months post-symptom onset in CoV2+ individuals suggests that GC-derived LLPCs have formed. GCs also produce long-lived MBCs that play a critical role in the formation of high-affinity antibody-secreting PBs upon antigen re-exposure and can be long-lived. We therefore tested whether SARS-CoV-2-specific MBCs were formed and maintained in CoV2+ individuals throughout the study’s time course. We generated RBD tetramer reagents and used enrichment strategies to identify and phenotype rare RBD-specific B cells that are otherwise undetectable (Krishnamurty et al., 2016). We tested the specificity of our reagent in RBD-immunized mice (Walls et al., 2020) and then used it to identify, enumerate, and phenotype rare, RBD-specific B cells in PBMCs from HC and CoV2+ individuals. Gates used to phenotype RBD-specific B cells were defined on total B cell populations (Figure S5 A).
Figure S5.
Mild COVID-19 Induces a Sustained Enrichment of RBD-Specific IgG+ Memory B Cells, Related to Figure 2
(A) Representative flow cytometry gates for phenotyping RBD-specific B cells from PBMCs in Figure 2 set on total B cells from a healthy control (HC) (surface stain, top; intracellular stain, bottom).
(B) Number of RBD-specific IgD+, IgM+ and IgA+ MBCs (CD20+RBD tetramer+decoy tetramer- CD27+CD21+/CD27+CD21-/CD27-CD21-) from healthy control (HC) and SARS-CoV-2-recovered (CoV2+) PBMCs at Visit 1 (V1) and Visit 2 (V2).
Statistics for unpaired data determined by two-tailed Mann-Whitney tests and, for paired data, by two-tailed Wilcoxon signed-rank tests. Multiple testing correction significance cutoff at FDR = 0.05 is p value < 0.02. Error bars represent mean and SD. Data from two experiments per visit.
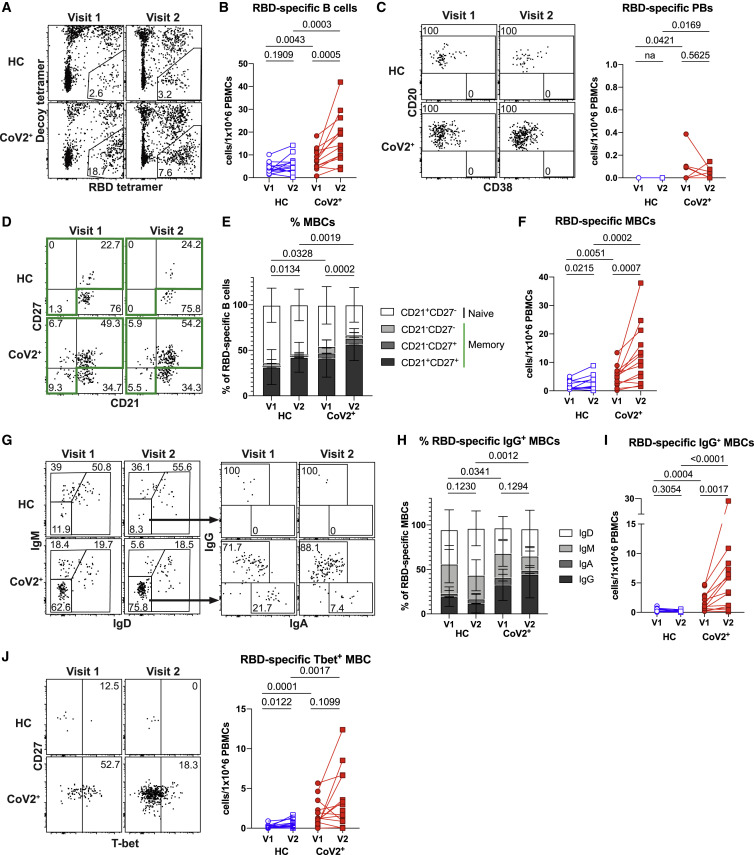
At Visit 1, RBD-specific B cells were expanded in CoV2+ individuals in comparison with HCs, and their numbers in CoV2+ individuals increased significantly from Visit 1 to Visit 2 (Figures 2A and 2B). As expected, we found very few CD20−CD38+ RBD-specific antibody-secreting cells (PBs or PCs) in the blood of HC or CoV2+ individuals at either time point, supporting the idea that LLPCs, likely in the bone marrow, are producing the majority of RBD-specific antibody at Visit 2 (Figure 2C). Expression of CD21 and CD27 distinguishes CD21+CD27− naive B cells from CD21+CD27+ classical MBCs and CD21−CD27+/− activated MBCs (Weisel and Shlomchik, 2017). The majority of RBD-specific B cells in CoV2+ individuals were CD21+CD27+ classical MBCs, whereas the majority of RBD-specific B cells in HC individuals were naive (Figures 2D and 2E). The number of RBD-specific MBCs in CoV2+ samples were on average significantly greater than in HCs and increased even further at Visit 2 (Figure 2F). Class switching of B cell receptors (BCRs) from IgM and IgD to IgG, IgA, or IgE is associated with BCR affinity maturation usually needed to form high-affinity neutralizing antibodies (Weisel and Shlomchik, 2017). CoV2+ RBD-specific MBCs were enriched for IgG+ MBCs, whereas the smaller number of HC RBD-specific MBCs were predominantly unswitched (IgM+ and IgD+) (Figures 2G, 2H, and S5B). Most strikingly, the increased numbers of IgG+ RBD-specific MBCs seen in CoV2+ individuals as compared to HC individuals at Visit 1 were even further enhanced at Visit 2 (Figure 2I). Relatively few of the CoV2+ RBD-specific MBCs expressed IgA, but their number was significantly higher than in HC individuals at both time points (Figure S5B).
Figure 2.
Mild COVID-19 Induces a Sustained Enrichment of RBD-Specific IgG+ Memory B Cells
(A) Representative gating of live CD3–CD14–CD16– cells for SARS-CoV-2 RBD-specific cells (RBD tetramer+decoy tetramer–) from CoV2+ and HC PBMCs at V1 and V2.
(B) Number of RBD-specific B cells (RBD tetramer+decoy tetramer–CD20+) per 1 × 106 PBMCs.
(C) Representative flow cytometry plots and number of RBD-specific PBs (RBD tetramer+decoy tetramer−CD20−CD138hi) (na = could not be calculated because all values 0).
(D) Representative gating of RBD-specific B cells for naive B cells (CD21+CD27−) and MBCs (CD21+CD27+/CD21–CD27+/CD21–CD27– populations outlined in green).
(E) Proportion of RBD-specific B cells that are naive (CD21+CD27−), classical MBCs (CD21+CD27+), or activated MBCs (CD21−CD27+/−), statistics for the proportion that are MBCs.
(F) Number of RBD-specific MBCs (classical and activated).
(G) Representative gating of RBD-specific MBCs for BCR isotype (IgD, IgM, IgA, and IgG) expression.
(H) Proportion of RBD-specific MBCs expressing the BCR isotypes IgD, IgM, IgA, and IgG. Statistics are for the proportion that are IgG+.
(I) Number of RBD-specific IgG+ MBCs.
(J) Representative gating of RBD-specific MBCs for T-bet expression and number of RBD-specific T-bet+ MBCs.
Statistics for unpaired data determined by 2-tailed Mann-Whitney tests and, for paired data, by 2-tailed Wilcoxon signed-rank tests. Multiple testing correction significance cutoff at FDR = 0.05 is p value < 0.04. Error bars represent mean and SD. Data from 2 experiments per visit. See also Figure S5.
An additional measure of antiviral MBC function is the expression of T-bet (Knox et al., 2017a). MBCs that express T-bet are associated with rapid differentiation into secondary PBs that produce high-affinity, viral-specific antibodies during a secondary infection (Knox et al., 2017b). We found on average a higher number of T-bet+ RBD-specific MBCs in CoV2+ individuals than in HCs at Visit 1, and the higher numbers were maintained at Visit 2 (Figure 2J). Together these data demonstrate that IgG+ RBD-specific classical MBCs not only form and persist in response to mild COVID-19, but their numbers continue to increase from 1 to 3 months post-symptom onset.
SARS-CoV-2 Infection Induces Durable, Functional Spike-Reactive Memory CD4+ T Cells
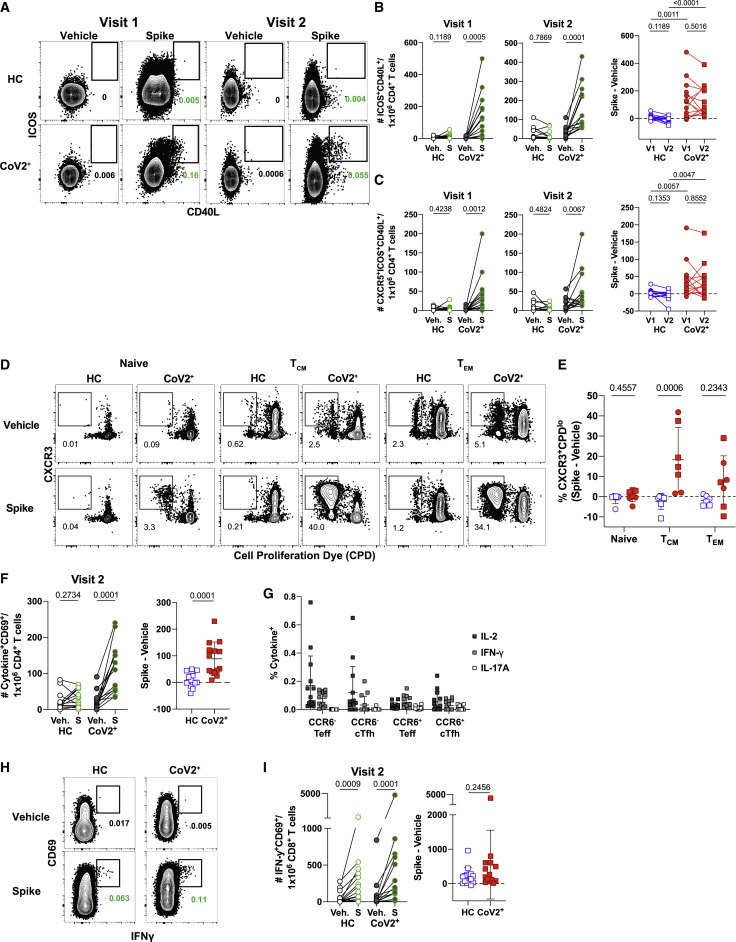
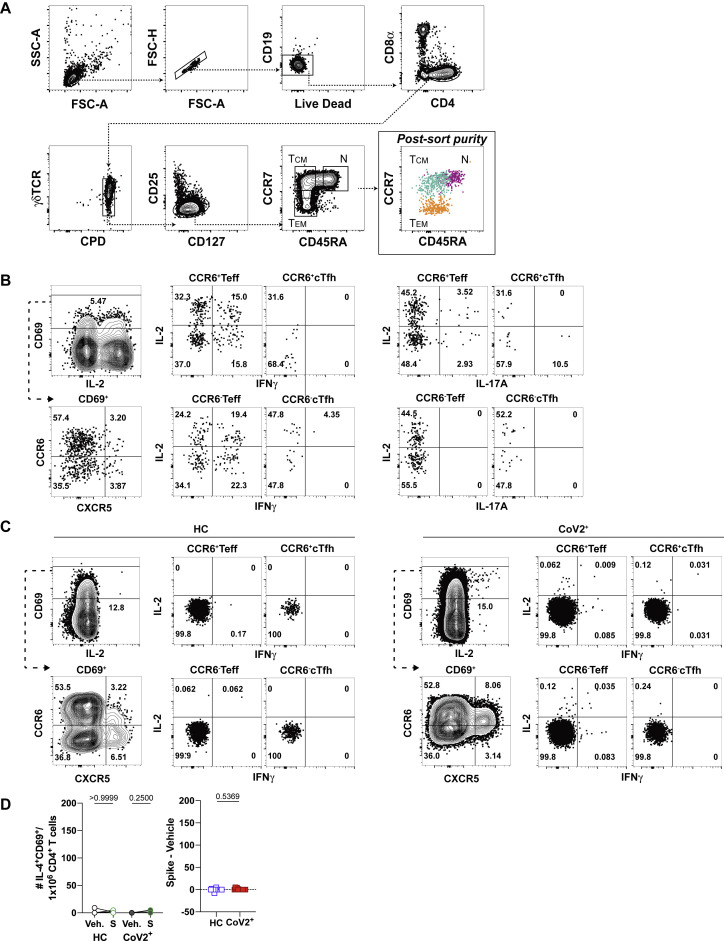
The presence of T-dependent IgG+ RBD-specific MBCs suggested that antigen-specific memory T cell responses were also present in CoV2+ individuals. To enumerate SARS-CoV-2-specific CD4+ memory T cells, total PBMCs from HC or CoV2+ individuals were incubated with vehicle control or spike protein, and activation marker expression was assessed 20 h later (Figure 3 A) (Bentebibel et al., 2013; Reiss et al., 2017). We first focused on the activation-induced expression of members of specific T-B receptor-ligand pairs including ICOS and CD40L. CD4+ T cells from CoV2+ individuals at both Visit 1 and 2 demonstrated enhanced expression of ICOS and CD40L after re-activation with spike protein in comparison with vehicle control (Figure 3B). Small numbers of spike-responsive activated CD4+ T cells could also be found in a few HC individuals, but on average these were not significantly increased across the HC cohort. Furthermore, there were no significant differences in the numbers of responding cells in CoV2+ individuals between Visit 1 and Visit 2, demonstrating that spike-specific memory CD4+ T cells were maintained throughout the study (Figure 3B). In addition, greater numbers of CXCR5-expressing circulating T follicular helper (cTfh) cells (Vinuesa et al., 2016) capable of migrating to B cell follicles were seen in re-stimulated samples from CoV2+ individuals than from HC individuals, and this was maintained between visits (Figure 3C). Together these data demonstrate that SARS-CoV-2-specific memory CD4+ T cells are maintained, and some retain the capacity to provide B cell help, at 3 months post-symptom onset.
Figure 3.
SARS-CoV-2 Infection Induces Durable, Functional Spike-Reactive CD4+ T Cells
(A) Representative flow cytometry plots of ICOS and CD40L expression on antigen-experienced (non-CD45RA+CCR7+) CD4+ T cells 20 h after incubation of PBMCs from HC and CoV2+ individuals at V1 and V2 with vehicle or SARS-CoV-2 spike.
(B) Number of antigen-experienced ICOS+CD40L+CD4+ T cells per 1x106 CD4+ T cells from HC and CoV2+ samples after incubation with vehicle (Veh.) or spike (S) at both time points (right) and calculated number of spike-responsive CD4+ T cells (number after incubation with spike minus number after incubation with vehicle) compared across time points (left).
(C) Number of antigen-experienced CXCR5+ICOS+CD40L+CD4+ T cells (cTfh) per 1 × 106 CD4+ T cells from HC and CoV2+ samples after incubation with Veh. or S at both time points (right) and calculated number of spike-responsive cTfh cells (number after incubation with spike minus number after incubation with vehicle) compared across time points (left).
(D) Representative flow cytometry plots of sorted CD4+ naive (CD45RA+CCR7+), TCM (CD45RA−CCR7+), or TEM (CD45RA−CCR7−) T cells from HC and CoV2+ PBMCs after 5–6 days of co-culture with SARS-CoV-2 spike-protein-pulsed autologous monocytes and measuring proliferation by CPD dilution.
(E) SARS-CoV-2 spike-specific expansion of sorted CD4+ naive T, TCM, and TEM cells from V1 (circles) and V2 (squares) reported as frequency of CXCR3+CPDlo cells after incubation with spike minus frequency after incubation with vehicle.
(F) Number of cytokine-producing, antigen-experienced CD69+CD4+ T cells per 1 × 106 CD4+ T cells after incubation with Veh. or S (right) and calculated number of spike-responsive, cytokine-producing CD4+ T cells (number after incubation with spike minus number after incubation with vehicle) (left).
(G) Frequency of antigen-experienced CD69+CD4+ T cell subsets, CCR6+/− Teff (CXCR5−), and CCR6+/− cTfh (CXCR5+) producing IL-2, IFN-γ, and IL-17A effector cytokines after incubation with spike for 20 h.
(H) Representative flow cytometry plots of CD69 and IFN-γ expression on antigen-experienced CD8+ T cells from HC and CoV2+ PBMCs at V2 after 20 h of incubation with vehicle or SARS-CoV-2 spike.
(I) Number of antigen-experienced IFN-γ+CD69+CD8+ T cells per 1 × 106 CD8+ T cells after 20 h of incubation with Veh. or S (right) and calculated number of spike-responsive CD8+ T cells (number after incubation with spike minus number after incubation with vehicle) (left).
Statistics for unpaired data determined by 2-tailed Mann-Whitney tests and, for paired data, by 2-tailed Wilcoxon signed-rank tests. Multiple testing correction significance cutoff at FDR = 0.05 is p value < 0.05. Error bars represent mean and SD. Data from 2 experiments per visit. See also Figure S6.
Antigen-specific memory CD4+ T cells proliferate to re-seed the memory pool and generate effector cells upon antigen re-exposure (Sallusto et al., 1999). To formally demonstrate that spike-specific CD4+ T cells in CoV2+ individuals were of a memory phenotype prior to re-stimulation, we measured the proliferative capacity of pre-sorted memory or naive subsets in response to spike re-stimulation. For this, we sorted CD45RA+ naive, CD45RA–CCR7+ central memory (TCM) and CD45RA–CCR7– effector memory (TEM) T cells from HC or CoV2+ individuals and cultured them for 5–6 days with autologous CD14+ monocytes and recombinant spike protein (Figure S6 A). We next determined the frequency of cells that proliferated and therefore diluted a cell proliferation dye (CPD) after 5–6 days of culture (Figure 3D). We additionally examined the expression of CCR6+ (associated with IL-17-producing Th17 cells), and CXCR3+ (associated with IFN-γ producing Th1 cells), as Th1 and Th17 subsets have been associated with protection from other respiratory viral infections (Sallusto, 2016). In CoV2+ individuals, a small population of proliferated (CPDlo) CXCR3+CD4+ Th1 cells emerged from sorted naive cells after 5 days of culture, perhaps representing contaminating memory cells, as these were not seen in HC individuals (Figure 3E). Sorted TCM cells from all CoV2+ individuals tested displayed significantly higher frequencies of CXCR3+CPDlo T cells that proliferated in response to spike than did HC samples. Although substantial proliferative responses were observed in TEM cells in some CoV2+ individuals, this was more variable across the CoV2+ group than what was seen in the TCM cells. Together these data demonstrate that, in individuals that have recovered from mild COVID-19, predominantly CXCR3-expressing spike-specific TCM, and in some individuals CXCR3-expressing TEM, persist and have the ability to proliferate and re-populate the memory pool upon antigen re-encounter.
Figure S6.
SARS-CoV-2 Infection Induces Durable, Functional Spike-Reactive CD4+ T Cells, Related to Figure 3
(A) Flow cytometry sorting strategy for naive, T central memory (TCM), and T effector memory (TEM) cells from HC and CoV2+ PBMCs at Visit 1 and Visit 2 before 5-6 days of culture with autologous monocytes and SARS-CoV-2 spike protein or vehicle.
(B) Representative flow cytometry gating on PMA/Ionomycin-activated PBMCs for cytokine expression by antigen experienced (non-CD45RA+CCR7+) CD4+ T cells subset into CCR6+/− T effector cells (Teff, CXCR5-) and circulating T follicular helper cells (cTfh, CXCR5+).
(C) Representative flow cytometry gating on antigen-experienced (non-CD45RA+CCR7+) CD4+ T cells from HC and CoV2+ V2 PBMCs following incubation with SARS-CoV-2 spike for 20 h. Gating on CD69+ CCR6+/− T effector cells (Teff, CXCR5-) and CCR6+/− circulating T follicular helper cells (cTfh, CXCR5+) for IL-2, IFN-γ and IL-17A effector cytokines expression.
(D) Number of IL-4-producing, antigen-experienced CD69+CD4+ T cells per 1x106 CD4+ T cells after incubation with vehicle (Veh.) or SARS-CoV2 spike (S) (left) and calculated number of spike-responsive, cytokine-producing CD4+ T cells (number after incubation with spike minus number after incubation with vehicle)(right).
Statistics determined by two-tailed Mann-Whitney tests. Multiple testing correction significance cutoff at FDR = 0.05 is p value < 0.05. Error bars represent mean and SD. Data from two experiments per visit.
Memory T cells rapidly express a wide variety of cytokines to engage, recruit, or activate innate cells or other adaptive lymphocytes. We next performed a detailed analysis of the cytokine profiles of spike-responsive memory T cells that persisted at Visit 2 in CoV2+ and HC individuals to gain a better understanding of SARS-CoV-2-specific memory cell function. Production of specific cytokines by CD4+ CXCR5+ memory Tfh cells can influence B cell activation and class switching, whereas CD4+ CXCR5− T effector (Teff) cells express high levels of cytokines to engage antiviral programs. Cytokines generated by a variety of T helper subsets were examined (IFN-γ, IL-2, IL-4, IL-17), and staining was confirmed on PMA/ionomycin activated CD69+ cells (representative plots shown in Figure S6B). Activated CD69+ CD4+ memory cells were further subset by CCR6 and CXCR5 expression, as predominant populations of spike-specific CCR6+CXCR5+ cTfh were recently described early after SARS-CoV2 infection (Juno et al., 2020) (Figure S6C). After spike re-stimulation, the CoV2+ samples demonstrated significant numbers of activated, cytokine-producing cells in comparison to vehicle controls, whereas some HC samples exhibited small numbers of activated, cytokine-producing cells (Figure 3F). The cytokine production in the CoV2+ samples was dominated by the expression of IL-2 and IFN-γ, with less frequent IL-17A production and no significant IL-4 production noted (Figures 3G and S6D). Spike-responsive CCR6− subsets expressed higher frequencies of IL-2 and IFN-γ than CCR6+ cells, whereas the few IL-17 producers present expressed CCR6 as expected. Together these data demonstrate that functional, spike-specific memory CXCR5+ cTfh and CXCR5− Teff cells persist 3 months after symptom onset that predominantly make Th1 cytokines, with a small IL-17A-producing population.
Although much recent work has focused on antibodies and B cells, memory CD8+ T cells are uniquely positioned to kill virus-infected cells through their directed expression of cytokines and cytolytic molecules. We therefore interrogated the presence of functional CD8+ memory T cells in the same cytokine reactivation assays and samples described above. Spike-specific memory CD8+ T cells that persisted for 3 months after mild COVID-19 disease could be identified by expression of the activation marker CD69 and expression of the cytokine IFN-γ after incubation with SARS-CoV-2 spike for 20 h (Figure 3H). Unlike CD4+ memory T cells, however, activated cytokine-expressing CD8+ T cells were significantly increased in number over the cells incubated with vehicle in both HC and CoV2+ groups (Figure 3I). Together, these data demonstrate that Th1 CD4+ SARS-CoV-2-specific cTfh and non-Tfh memory T cells are maintained and able to produce effector cytokines after re-stimulation 3 months post-symptom onset in mildly symptomatic COVID-19 individuals. These data further suggest that a population of IFN-γ-producing, cross-reactive CD8+ T cells exist in healthy controls.
SARS-CoV-2-Specific MBCs Can Express Neutralizing Antibodies
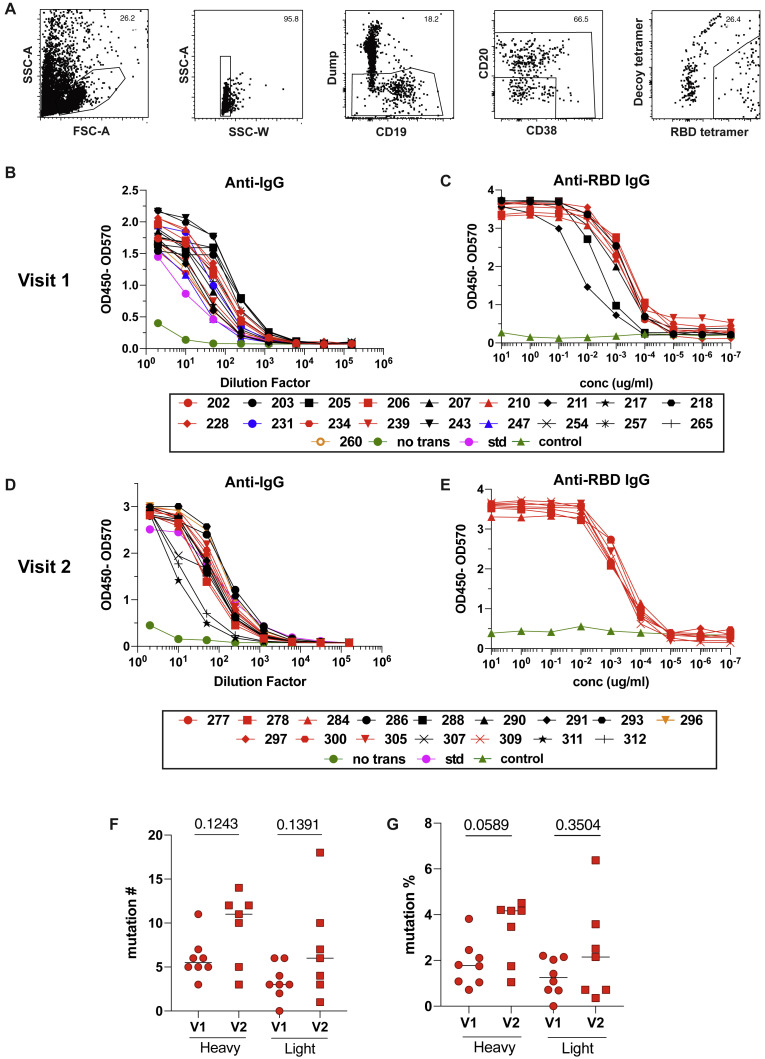
Because SARS-CoV-2 RBD-specific MBC and S-specific CD4+ cTfh were enriched in CoV2+ individuals after 3 months, we asked whether these MBCs had the potential to produce neutralizing antibodies if reactivated by a secondary infection. To this end, we index sorted single RBD-specific B cells and sequenced the BCRs from 3 CoV2+ individuals at both time points (Figures S7 A and 4A). We sequenced all IgG+ RBD-specific classical MBCs (CD21+CD27+), cloned paired heavy and light chain sequences that were obtained, and expressed them as IgG1 monoclonal antibodies. In total, this comprised 19 antibodies from Visit 1 and 16 from Visit 2.
Figure S7.
SARS-CoV-2-Specific MBCs Can Express Neutralizing Antibodies, Related to Figure 4 and Table S1
(A) Gating strategy for sorting RBD-specific B cells.
(B) IgG ELISA to confirm expression of Visit 1 antibodies in transfected cell culture supernatants. Positive control is the kit standard (std) and negative control is supernatant from untransfected cells (no trans, green).
(C) RBD ELISA of purified Visit 1 monoclonal antibodies. Negative control (green) is an irrelevant Plasmodium-specific antibody.
(D) IgG ELISA to confirm expression of Visit 2 antibodies in transfected cell culture supernatants.
(E) RBD ELISA of purified Visit 2 monoclonal antibodies.
(F) Number of mutations in variable regions of RBD-specific monoclonal antibodies.
(G) Mutation frequency of variable regions of RBD-specific monoclonal antibodies.
Statistics determined by two-tailed Mann-Whitney tests. Multiple testing correction significance cutoff at FDR = 0.05 is p value < 0.05.
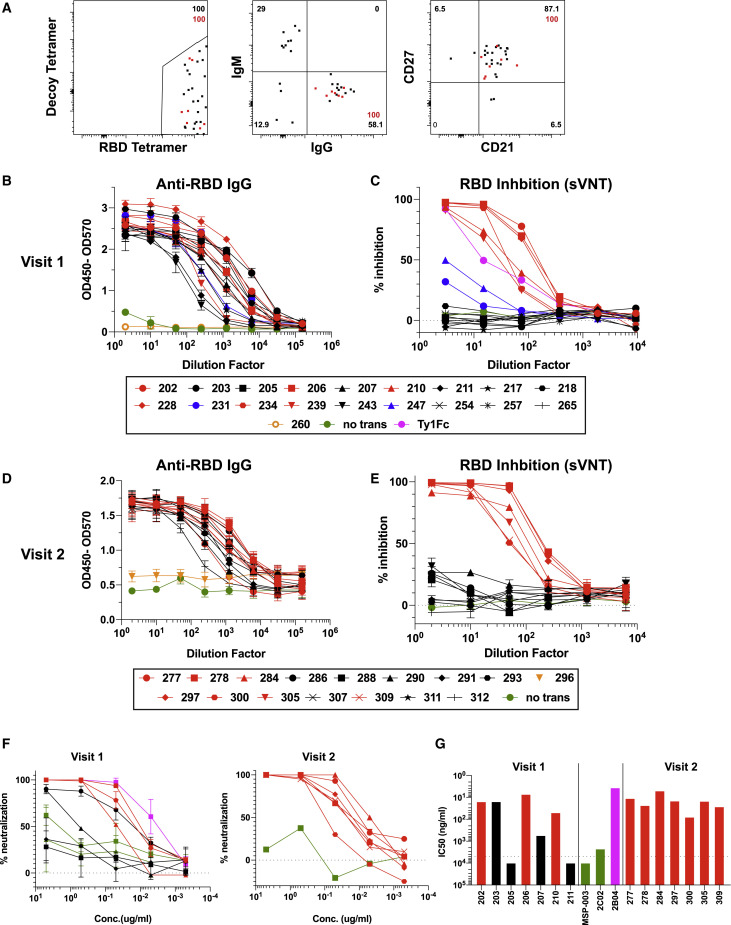
Figure 4.
SARS-CoV-2-Specific MBCs Can Express Neutralizing Antibodies
(A) Representative flow plots of index-sorted RBD-tetramer specific B cells (gating scheme in Figure S7A). BCRs cloned from cells are shown in red.
(B) Anti-RBD ELISA of culture supernatants from cells transfected to express one of the Visit 1 monoclonal antibodies or supernatant from untransfected cells (no trans). Antibodies that did not bind RBD are shown in orange.
(C) Inhibition of RBD binding to ACE2 by Visit 1 monoclonal antibody supernatants measured by sVNT assay, compared to a known RBD-specific neutralizing antibody (Ty1). Red indicates strong inhibitors, blue moderate inhibitors and black non-inhibitors.
(D) Anti-RBD ELISA of culture supernatants from cells transfected to express one of the Visit 2 monoclonal antibodies. Antibodies that did not bind RBD are shown in orange.
(E) Inhibition of RBD binding to ACE2 by Visit 2 monoclonal antibodies measured by sVNT assay. Red indicates strong inhibitors and black non-inhibitors.
(F) Neutralization capacity of purified monoclonal antibodies as measured by PRNT. 2B04 and 2C02 are previously identified strong and weak neutralizing murine antibodies, respectively, and MSP-003 is an irrelevant Plasmodium-specific antibody.
(G) IC50 values calculated from PRNT. Dotted line represents the limit of detection.
See also Figure S7.
These antibodies were first expressed in small scale cultures for screening. Transfection supernatants were assessed for antibody expression by IgG ELISA (Figures S7B and S7D) and specificity by RBD ELISA, where all but 1 antibody at each time point showed strong binding to RBD (Figures 4B and 4D). Antibodies were then assessed for their capacity to inhibit RBD binding to the ACE2 receptor by sVNT assay. Eight of 18 antibodies cloned from subjects at Visit 1 (44%) and 7 of 15 from Visit 2 (47%) were able to inhibit RBD binding to ACE2 (Figures 4C and 4E). Although 2 of the Visit 1 antibodies showed lower levels of inhibition, the others showed levels of inhibition similar to a strongly neutralizing alpaca nanobody (Hanke et al., 2020). These antibodies were then expressed on a larger scale and purified for more detailed analysis. The specificity of the purified antibodies for RBD was again confirmed by ELISA (Figures S7C and S7E), and all Visit 2 inhibitors, the majority of Visit 1 inhibitors, and several non-inhibitors were assayed for SARS-CoV-2 neutralization capacity by PRNT assay. All of the inhibitory antibodies were able to strongly neutralize the virus, with IC50 values ranging from 7.15 to 49.61 ng/mL (Figures 4F and 4G). This was comparable to a previously published strongly neutralizing mouse antibody (2B04) which was included as a positive control (IC50 = 3.6 ng/mL) (Alsoussi et al., 2020). Two of the non-inhibitory antibodies (#205 and #211) were unable to inhibit virus infection as expected, similar to a non-neutralizing mouse antibody (2C02) and an irrelevant Plasmodium-specific human antibody (MSP-003). Interestingly, the other 2 non-inhibitory antibodies tested (#203 and #207) were able to neutralize live virus, with #203 having an IC50 comparable to the strong inhibitors (15.4 ng/mL), suggesting that the sVNT assay does not detect all monoclonal RBD-specific antibodies capable of neutralizing live virus. More than 50% of the antibodies tested showed activity by one or both of these methods.
This set of antibodies utilized a wide variety of heavy and light chains, had all undergone somatic hypermutation (SHM) and were all unique clones (Tables 2 and S1). Somatic hypermutation (SHM) levels in both heavy and light chains increased on average between Visit 1 and Visit 2, but these differences were not statistically significant (Figures S7F and S7G). Thus, RBD-specific MBCs induced by SARS-CoV-2 infection are capable of producing a variety of neutralizing antibodies against the virus and could contribute to protection from a second exposure.
Table 2.
Neutralizing Monoclonal Antibody Information
| mAb ID | Heavy Chain | Light Chain | Heavy Chain Junction AA Sequence | Heavy Chain Mutation # | Light Chain Mutation # |
|---|---|---|---|---|---|
| 202 | IGHV 3-66 | IGLV 1-40 | CARGGEEPLPFDPW | 7 | 0 |
| 203 | IGHV 1-69 | IGLV 1-40 | CARDEAQTTVNTNWFDPW | 11 | 6 |
| 206 | IGHV 3-66 | IGKV 1-39 | CARGDGSYYRAFDYW | 6 | 3 |
| 207 | IGHV 3-23 | IGlV 1-21 | CAKDPGTVTTYEYFQHW | 3 | 6 |
| 210 | IGHV 3-53 | IGKV 1-39 | CARDASSYGIDW | 5 | 3 |
| 228 | IGHV 3-66 | IGKV 1-33 | CARGVKDNIW | 6 | 3 |
| 234 | IGHV 3-53 | IGKV 3-20 | CARAFGGDYMDVW | 5 | 4 |
| 239 | IGHV 3-23 | IGLV 1-40 | CAKAGGRDYYDRSGTLNVGAWNFQHW | 5 | 2 |
| 278 | IGHV 1-46 | IGKV 1-39 | CARANSGSYHYYDYW | 12 | 4 |
| 277 | IGHV 4-59 | IGKV 3-20 | CARSWLRPHNWLDPW | 12 | 18 |
| 284 | IGHV 1-69 | IGKV 1-39 | CAGREKRWFGELNWDDGMDVW | 14 | 6 |
| 297 | IGHV 3-9 | IGKV 1-39 | CAKGHDPFHYYYYGMDVW | 11 | 7 |
| 300 | IGHV 1-69 | IGKV 1-39 | CASVSHYYDGSGYPTGFDPW | 10 | 1 |
| 305 | IGHV 3-53 | IGKV 1-NL1 | CARGPGVIIDW | 3 | 3 |
| 309 | IGHV 3-53 | IGKV 1-12 | CARELSSYYDLW | 5 | 10 |
Heavy and light chain gene usage, somatic hypermutation rate, and VDJ junction sequence of neutralizing antibodies.
Recovered Individuals Formed Multifaceted SARS-CoV-2-Specific Immune Memory
In response to human viral infections such as measles, polio, and hepatitis A, the amount and function of virus-specific IgG+ antibodies, IgG+ MBCs, CD4+ memory T cells, and CD8+ memory T cells have been correlated with long-lived protection (Amanna et al., 2007; Plotkin, 2010). Although we do not yet know which components of immune memory are sufficient for protection from a second exposure to SARS-CoV-2, these components should synergize to form a multi-layered defense.
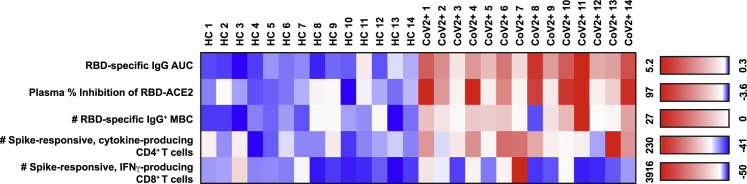
To determine if the CoV2+ individuals in our cohort each formed and maintained multiple immune memory components, we assessed each individual across components of SARS-CoV-2-specific functional immune memory that were significantly different in CoV2+ individuals at Visit 2 and, thus, might influence protection from a second exposure. The HC and CoV2+ individuals were grouped by infection status, and the values for independent metrics of immune memory at Visit 2 were reported in a heatmap (Figure 5 ). Because we do not yet know what a protective threshold is for each of these metrics, we set one SD above the mean of the values from HCs for each metric (row) as a threshold (white on the color scale). This allows us to see which CoV2+ individuals had responses above those detected in HCs. All 14 CoV2+ individuals in our cohort formed and sustained multiple components of immune memory at Visit 2 at levels above those found in HCs. All but one CoV2+ individual (CoV2+ 8) had higher RBD-specific IgG+ antibody titers, improved plasma inhibition of RBD-ACE2 binding, and higher numbers of RBD-specific IgG+ MBCs than HCs. Most CoV2+ individuals (11/14) also maintained higher numbers of spike-responsive CD4+ memory T cells that could rapidly produce effector cytokines (IL-2, IFN-γ, and/or IL-17A) than HCs. A few CoV2+ individuals (5/14) also had higher numbers of spike-responsive, IFN-γ-producing CD8+ memory T cells than did HCs. Together these data demonstrate that all of the CoV2+ individuals in our cohort formed and maintained multiple components of functional, adaptive immune memory.
Figure 5.
Recovered Individuals Formed Multifaceted SARS-CoV-2-Specific Immune Memory
Heatmap of values for independent SARS-CoV2 RBD- or spike-specific immune memory components from each HC and CoV2+ individual at Visit 2. RBD-specific IgG measured by ELISA (AUC). Percent inhibition by sVNT calculated at 1:2 plasma dilution. Number of RBD-specific IgG+ MBCs per 1 × 106 PBMCs. Number of spike-responsive (CD69+), cytokine-producing (IL-2/IFN-γ/IL-17A), antigen-experienced CD4+ T cells calculated by number after 20 h incubation with spike minus number after incubation with vehicle. Number of spike-responsive (CD69+IFN-γ+), antigen-experienced CD8+ T cells calculated by number after 20 h incubation with spike minus number after incubation with vehicle. The color scales are set for each metric (row) with the mean + 1 SD of the HC set to white.
Discussion
Although a vaccine is needed to safely reach herd immunity against SARS-CoV-2, understanding if natural infection induces viral-specific immunological memory that could influence transmission and disease severity is critical to controlling this pandemic. We therefore investigated whether individuals that experienced mild COVID-19 developed and sustained multilayered, functional immune memory. We found that 3 months after mildly symptomatic COVID-19, recovered individuals had formed an expanded arsenal of SARS-CoV-2-specific immune memory cells that exhibited protective antiviral functions. Recovered individuals had increased neutralizing antibodies, IgG+ classical MBCs with BCRs that formed neutralizing antibodies, Th1 cytokine-producing CXCR5+ circulating Tfh and CXCR5− non-Tfh cells, proliferating CXCR3+ CD4+ memory cells, and IFN-γ-producing CD8+ T cells. These components of immune memory have all been associated with protection from other viruses in humans (Ahmed and Gray, 1996; Amanna et al., 2007; Morita et al., 2011). Together, these data demonstrate that all of the recovered individuals in our cohort formed a multifaceted defense, which suggests attenuated virus vaccines are likely to be similarly successful in eliciting a functional immune memory response.
Sustained production of neutralizing IgG+ virus-specific antibodies is a frequent correlate of protection from viral infection (Amanna et al., 2007). Some studies examining the longevity of the antibody response to coronaviruses have suggested that antibodies wane rapidly (Seow et al., 2020; Tang et al., 2011; Wu et al., 2007). Our study, as well as other recent studies, has examined memory time points when only LLPCs, and not short-lived PBs, are thought to be producing circulating antibodies. Together, we demonstrate elevated IgG+ RBD-specific plasma antibodies and neutralizing plasma are generated and maintained at elevated levels for at least 3 months post-SARS-CoV-2 infection (Isho et al., 2020; Perreault et al., 2020; Ripperger et al., 2020; Wajnberg et al., 2020).What level of antibody is needed to contribute to protection and whether that level will be maintained in the long term will require studies of later time points. Although we detected IgA+ RBD-specific antibodies early, the levels had dropped significantly by 3 months, suggesting that the early IgA was derived from short-lived PBs. IgA-producing LLPCs either do not form or are sequestered in a tissue such that antibodies are not secreted into the blood.
Functional virus-specific memory B and T cells are key mediators of protective immune memory (Plotkin, 2010) and, unlike LLPCs, can be directly measured. Although previous studies have described the emergence of SARS-CoV-2-specific MBCs within a month of infection (Grifoni et al., 2020; Juno et al., 2020), (Wilson et al., 2020) we characterized SARS-CoV-2-specific MBCs at 1 and 3 months from symptom onset in the same individuals to determine whether this population is maintained. Our study also analyzed additional attributes of MBCs that have been associated with anti-viral protection. Our study revealed a prominent population of RBD-specific IgG+CD27+CD21+ MBCs, which, in other infections, has been associated with GC derivation, rapid differentiation into antibody-secreting PBs upon re-exposure (Nellore et al., 2019), and effective antiviral responses (Rubtsova et al., 2013). Not only was this population maintained from 1 to 3 months, the numbers increased significantly. We also found that these cells express BCRs capable of neutralizing SARS-CoV-2 when expressed as monoclonal antibodies. Approximately half of the antibodies derived from the IgG+ MBCs analyzed were able to neutralize the virus in vitro. The BCRs all exhibited SHM, and the number of mutations from Visit 1 BCRs was similar to those previously reported from samples obtained at a similar time point (Robbiani et al., 2020). SHM modestly increased in both heavy and light chains from Visit 1 to Visit 2, which could reflect additional affinity maturation in the GC, but further analysis of a larger numbers of samples is needed. Taken together, these data suggest that upon re-exposure with SARS-CoV-2, these individuals will have MBCs that can rapidly generate neutralizing SARS-CoV-2 antibody titers and help control the infection.
Memory CD4+ T cells can help reactivate MBCs through their expression of key molecules associated with T-B interactions including CXCR5, ICOS, CD40L, and a variety of cytokines (Vinuesa et al., 2016). SARS-CoV-2-specific CD4+ memory T cells in recovered individuals in our cohort exhibited the capacity to express all of these molecules and to undergo robust proliferation upon re-exposure to spike protein. Notably, spike-specific CD4+ memory T cells from CoV2+ individuals rapidly displayed increased levels of ICOS and CD40L on CXCR5+ and CXCR5– cells after stimulation as well as expression of Th1- and Th17-associated cytokines. These results are consistent with other recent reports of SARS-CoV-2-specific cTfh cells (Juno et al., 2020), although they detected a high frequency of Th17-like cTfh cells, which could be due to the earlier time point they were examining as Th17 cells can develop into Th1 cells late in an immune response (Lee et al., 2009). The expression of IFN-γ and IL-17 by cTfh cells is notable as these cytokines are associated with class switching to IgG and IgA isotypes, respectively (Hirota et al., 2013; Peng et al., 2002).
We also likely found cross-reactive memory B and T cells in healthy controls. In response to a viral infection, B cells that could recognize a viral antigen, but did not enter a GC, predominantly form IgM+ and IgD+ MBCs which tend to be low affinity and do not rapidly form PBs upon re-exposure, but might be able to recognize a variant of the viral protein (Weisel and Shlomchik, 2017). Because we detected RBD-specific IgM+ and IgD+ MBCs in HCs, we hypothesize that some of these could be cross-reactive MBCs generated in response to a previous human coronavirus infection as recent work suggests (Song et al., 2020). We also found a small number of spike-responsive CD4+ memory T cells in HCs, which other groups have similarly attributed to cross-reactive memory T cells potentially associated with a previous human coronavirus infection (Braun et al., 2020; Grifoni et al., 2020; Sekine et al., 2020; Sette and Crotty, 2020). We also found SARS-CoV-2-specific CD8+ memory T cells in equal numbers in HCs and CoV2+ individuals. This finding suggests cross-reactivity within a population of IFN-γ producing CD8+ memory cells in our HC samples and raises the possibility that our inability to interrogate CD8+ resident memory cells in the lungs could mask the true expansion of this compartment in CoV2+ individuals. How these cross-reactive cells contribute to the SARS-CoV-2 memory response in recovered individuals is difficult to discern without knowledge of each individual’s SARS-CoV-2-specific BCR and TCR repertoires prior to infection. However, we can conclude that mild COVID-19 induces an expanded population of MBCs and CD4+ memory T cells with markers of increased functionality in comparison with the cross-reactive pool present in our controls.
It is currently impossible to perform controlled SARS-CoV-2 reinfection studies to test the protective capacity of the SARS-CoV-2-specific memory lymphocytes we have described in humans. However, previous studies of human coronaviruses have shown protection from homologous rechallenge that correlated with antibody titers (Callow et al., 1990). Although studies of SARS-CoV-2 have confirmed rare second exposures months after the first, they suggest prior exposure can be protective (Abu-Raddad, 2020; To et al., 2020). Additional studies have supported this finding, including evidence from a fishing vessel where 85% of the crew became infected, yet 3 previously exposed individuals with neutralizing antibodies did not get sick (Addetia et al., 2020). More recently, during an outbreak at an overnight camp, none of the attendees that were seropositive (16%) prior to attending the camp tested positive for infection, whereas 91% of the remaining susceptible population tested positive for infection (Pray, 2020). Animal studies provide additional support, as macaques infected with SARS-CoV-2 were protected from rechallenge (Chandrashekar, 2020). Although these studies support the role of immune memory contributing to protection from SARS-CoV-2 re-exposure, future studies will require data on the SARS-CoV-2-specific immune memory compartment prior to re-exposure to assess a correlation to protection.
It is also important to note that different levels of severity of COVID-19 could be associated with different levels of immune memory and subsequent immune protection. We focused on the immune memory response to mild COVID-19, but whether similar memory populations form after severe COVID-19 is still unclear. In one largely histological study of post-mortem tissues from patients that succumbed to severe COVID-19, the lack of GC formation or the generation of CD4+ Tfh lymphocytes required for an optimal immune memory response suggested that forming immune memory could be difficult (Kaneko et al., 2020). However, as these patients died of acute disease, it is impossible to determine if germinal centers were transiently disrupted due to acute inflammation as has been seen in other highly inflammatory diseases like malaria (Keitany, 2016). Although additional studies are needed to determine how long memory to SARS-CoV-2 infection lasts, our work suggests that mild COVID-19 induces persistent, multifaceted immune memory. These functional antiviral memory lymphocytes are poised for a coordinated response to SARS-CoV-2 re-exposure that could contribute to immunity and help to curtail the pandemic.
Limitations of Study
First, our cohort is of relatively small size, preventing us from identifying patient characteristics that correlate with maintained immune memory. We would need a larger cohort with a diverse racial make-up as well as details on disease characteristics to achieve this. Second, we restricted our cohort to individuals recovered from mildly symptomatic COVID-19. Thus, these data do not describe the immune memory response after asymptomatic or severe disease. Studying a larger cohort of individuals with a spectrum of disease severity would allow this extension of our data. Third, we found SARS-CoV-2-specific immune memory was maintained at 3 months post-symptom onset, but we do not know whether SARS-CoV-2-specific immune memory will be maintained for a year or longer. Repeating these studies at later time points will yield these data. Finally, we demonstrate that SARS-CoV-2-specific immune memory is maintained and functional in vitro, but our data cannot say whether these individuals will be protected from disease upon a second exposure and, if they are, which arms of immune memory correlate with protection. Addressing this will require study of individuals before and after a natural re-exposure event. Large epidemiologic studies to determine whether the rate of symptomatic re-infection in previously exposed individuals is different than in naive individuals will also help address whether previous exposure and immune memory generation could be protective from symptomatic re-infection.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-human CD16-BUV395, Clone 3G8 | BD | Cat #563785; RRID: AB_2744293 |
| Anti-human CD15-BUV563, Clone W6D3 | BD | Cat #741417; RRID: AB_2868406 |
| Anti-human CD14-BUV615, Clone M5E2 | BD | Cat #751150; RRID: AB_2875174 |
| Anti-human CD56-BUV737, Clone NCAM16.2 | BD | Cat #612767; RRID: AB_2860005 |
| Anti-human CD31-CD31, Clone WM59 | Biolegend | Cat #303123; RRID: AB_2562179 |
| Anti-human HLADR-BV510, Clone L243 | Biolegend | Cat #307646; RRID: AB_2561948 |
| Anti-human CD11c-BV605, Clone 3.9 | Biolegend | Cat #301635; RRID: AB_2562191 |
| Anti-human CD3-BV650, Clone OKT3 | Biolegend | Cat #317324; RRID: AB_2563352 |
| Anti-human CD19-BV650, Clone HIB19 | Biolegend | Cat #302237; RRID: AB_11126981 |
| Anti-human CD10-BV711, Clone H110a | Biolegend | Cat #312226; RRID: AB_2565876 |
| Anti-human CD123-BV785, Clone 6H6 | Biolegend | Cat #306032; RRID: AB_2566448 |
| Anti-human FCAR-FITC, Clone A59 | Biolegend | Cat #354114; RRID: AB_2728341 |
| Anti-human CD66b-PerCp Cy5.5, Clone G10F5 | Biolegend | Cat #305108; RRID: AB_2077855 |
| Anti-human CD9-PE, Clone HI9a | Biolegend | Cat #312106; RRID: AB_2075892 |
| Anti-human CD163-PE-Cy7, Clone GHI/61 | Biolegend | Cat #333614; RRID: AB_2562641 |
| Anti-human CD41-APC, Clone HIP8 | Biolegend | Cat #303710; RRID: AB_2249385 |
| Anti-human CD304-APC-Fire, Clone 12C2 | Biolegend | Cat #354523; RRID: AB_2715882 |
| Anti-human CD19-FITC, Clone HIB19 | BD | Cat #555412; RRID: AB_395812 |
| Anti-human CD20-P5.5, Clone 2H7 | BD | Cat #560736; RRID: AB_1727451 |
| Anti-human CD3-BV711, Clone UCHT1 | BD | Cat #563725; RRID: AB_2744392 |
| Anti-human CD14-BV711, Clone MɸP9 | BD | Cat #563372; RRID: AB_2744290 |
| Anti-human CD16-BV711, Clone 3G8 | BD | Cat #563127; RRID: AB_2732050 |
| Anti-human IgM-BV510, Clone MHM-88 | BioLegend | Cat #314521; RRID: AB_2561513 |
| Anti-human IgD-PE-Cy7, Clone IA6-2 | BD | Cat #561314; RRID: AB_10642457 |
| Anti-human IgG-BV786, Clone G18-145 | BD | Cat #564230; RRID: AB_2738684 |
| Anti-human CD21-SB600 Clone, HB5 | ThermoFisher | Cat #63-0219-41 |
| Anti-human CD27-BV421, Clone M-T271 | BioLegend | Cat #356418; RRID: AB_2562599 |
| Anti-human CD38-AF700, Clone HIT2 | ThermoFisher | Cat #56-0381-82; RRID: AB_657740 |
| Anti-human IgA-Biotin, Clone IS11-8E10 | Miltenyi | Cat #130-113-474; RRID: AB_2733547 |
| Anti-human CD27-BV711, Clone M-T271 | BioLegend | Cat #356430 |
| Anti-human FCLR5-EF660, Clone 509F6 | ThermoFisher | Cat #50-3078-42; RRID: AB_2574199 |
| Anti-human CD11c-PEDazzle594, Clone 3.9 | BioLegend | Cat #301642; RRID: AB_2564083 |
| Anti-human Tbet-BV421, Clone 4B10 | BioLegend | Cat #644816; RRID: AB_10959653 |
| Anti-human Ki67-BUV395, Clone B56 | BD | Cat #564071; RRID: AB_2738577 |
| Anti-human CD45RA-BUV737, Clone HI100 | BD | Cat #612846; RRID: AB_2870168 |
| Anti-human CXCR5-BV421, Clone RF8B2 | BD | Cat #562747; RRID: AB_2737766 |
| Anti-human CD3-BV510, Clone UCHT1 | BD | Cat #563109; RRID: AB_2732053 |
| Anti-human CD4-BV605, Clone SK3 | BioLegend | Cat #344646; RRID: AB_2734348 |
| Anti-human CCR6-BV650, Clone 11A9 | BD | Cat #563922; RRID: AB_2738488 |
| Anti-human CD19-BV711, Clone SJ25C1 | BD | Cat #563038; RRID: AB_2737970 |
| Anti-human CD14-BV711, Clone M5E2 | BD | Cat #740773; RRID: AB_2740436 |
| Anti-human CD8-BV750, Clone SK1 | BD | Cat #747097; RRID: AB_2871850 |
| Anti-human CCR7-BV785, Clone G043H7 | BioLegend | Cat #353230; RRID: AB_2563630 |
| Anti-human FoxP3-AF488, Clone 259D/C7 | BD | Cat #560047; RRID: AB_1645349 |
| Anti-human ICOS-Percp-eF710, Clone ISA-3 | ThermoFisher | Cat #46-9948-42; RRID: AB_10854730 |
| Anti-human Bcl-6-PE, Clone 7D1 | BioLegend | Cat #358504; RRID: AB_2562152 |
| Anti-human Blimp-1-PE CF594, Clone 6D3 | BD | Cat #565274; RRID: AB_2739147 |
| Anti-human CXCR3-PE Cy5, Clone IC6/CXCR3 | BD | Cat #561731; RRID: AB_10892799 |
| Anti-human T-bet-PE Cy7, Clone 4B10 | BioLegend | Cat #644823; RRID: AB_2561760 |
| Anti-human PD-1-APC, Clone EH12.2H7 | BioLegend | Cat #329908; RRID: AB_940475 |
| Anti-human HLA-DR-APC H7, Clone L243 | BioLegend | Cat #307610; RRID: AB_314688 |
| Anti-human CD3-BUV395, Clone SK7 | BD | Cat #564001; RRID: AB_2744382 |
| Anti-human CXCR3-BV421, Clone BVD2-21C11 | BD | Cat #562930; RRID: AB_2737899 |
| Anti-human CCR7-BV605, Clone G043H7 | BioLegend | Cat #353224; RRID: AB_2561753 |
| Anti-human CD4-BV711, Clone OKT4 | BioLegend | Cat #317440; RRID: AB_2562912 |
| Anti-human CD127-BV786, Clone hIL7Rm21 | BD | Cat #563324; RRID: AB_2738138 |
| Anti-human pan-gdTCR-PerCP/ef710, Clone B1 | BioLegend | Cat #331224; RRID: AB_2738871 |
| Anti-human CCR4-PE/Dazzle594, Clone L291H4 | BioLegend | Cat #359420; RRID: AB_2564095 |
| Anti-human CD25-PE/Cy5, Clone BC96 | BioLegend | Cat #302608; RRID: AB_314278 |
| Anti-human CD8a-PE/Cy7, Clone SK1 | ThermoFisher | Cat #25-0087-42; RRID: AB_10733019 |
| Anti-human CD19-Af700, Clone SJ25C1 | ThermoFisher | Cat #56-0198-42; RRID: AB_2815226 |
| Anti-human CD8a-BV510, Clone SK1 | BioLegend | Cat #344732; RRID: AB_2564624 |
| Anti-human CD19-BV605, Clone SJ25C1 | BioLegend | Cat #363006; RRID: AB_2564128 |
| Anti-human IL-4-FITC, Clone MP4-25D2 | BioLegend | Cat #500806; RRID: AB_315125 |
| Anti-human IL-13-PE, Clone JES10-5A2 | BioLegend | Cat #501903; RRID: AB_315198 |
| Anti-human IL17a-PE/Dazzle594, Clone BL168 | BioLegend | Cat #512336; RRID: AB_2564038 |
| Anti-human IL-22-PE/Cy7, Clone 22URTI | ThermoFisher | Cat #25-7229-42; RRID: AB_10853346 |
| Anti-human IFNg-Af700, Clone 4S.B3 | BioLegend | Cat #502520; RRID: AB_528921 |
| Anti-human pan-gdTCR-APC/Fire75, Clone B1 | BioLegend | Cat #331228; RRID: AB_2650627 |
| Anti-human CCR6-BV650, Clone OKT4 | BioLegend | Cat #317436; RRID: AB_2563050 |
| Anti-human CXCR5-BB515, Clone RF8B2 | BD | Cat #564624; RRID: AB_2738871 |
| Anti-human CCR10-PE, Clone 14305 | R&D | Cat #FAB3478P-100 |
| Anti-human CCR4-PE/Dazzle594, Clone L291H4 | BioLegend | Cat #359420; RRID: AB_2564095 |
| Anti-human CD25-PE/Cy5, Clone BC96 | BioLegend | Cat #302608; RRID: AB_314278 |
| Anti-human CD127-PE/Cy7, Clone hIL7Rm21 | BD | Cat #560822; RRID: AB_2033938 |
| Anti-human CD69-BUV395, Clone FN50 | BD | Cat #564364; RRID: AB_2738770 |
| Anti-human CD40L-FITC, Clone 24-31 | ThermoFisher | Cat #11-1548-42; RRID: AB_10669047 |
| Anti-human ICOS-Percp-eF710, Clone ISA-3 | ThermoFisher | Cat #46-9948-42; RRID: AB_10854730 |
| Anti-human IL-21-PE, Clone eBio3A3-N2 | ThermoFisher | Cat #12-7219-42; RRID: AB_1582260 |
| Anti-human IL-17A-PE Dz594, Clone BL168 | Biolegend | Cat #512336; RRID: AB_2564038 |
| Anti-human IL-4-PE Cy7, Clone MP4-25D2 | BioLegend | Cat #500824; RRID: AB_2126746 |
| Anti-human IL-6-AF647, Clone MQ2-13A5 | BioLegend | Cat #501124; RRID: AB_2810624 |
| Anti-human IL-2-AF700, Clone MQ1-17H12 | BioLegend | Cat #500320; RRID: AB_528929 |
| Anti-human IFNg-APC H7, Clone 4S.B3 | BioLegend | Cat #502530; RRID: AB_10663412 |
| Anti-mouse CD4-FITC, Clone GK1.5 | BD | Cat #553729; RRID: AB_395013 |
| Anti-mouse CD8-FITC, Clone 53.67 | BD | Cat #553030; RRID: AB_394568 |
| Anti-mouse B220-BV711, Clone RA3-6B2 | BD | Cat #563892; RRID: AB_2738470 |
| Anti-mouse CD138-BV605, Clone 281-2 | BD | Cat #563147; RRID: AB_2721029 |
| Anti-mouse CD38-AF700, Clone 90 | ThermoFisher | Cat #56-0381-82; RRID: AB_657740 |
| Anti-mouse Gl7-ef450, Clone GL-7 | ThermoFisher | Cat #48-5902-82; RRID: AB_10870775 |
| Anti-human IgG-HRP | Jackson ImmunoResearch | Cat #109-035-088; RRID: AB_2337584 |
| anti-human IgM-HRP | Southern Biotech | Cat #1020-05; RRID: AB_2794201 |
| anti-human IgA-HRP | Southern Biotech | Cat #2050-05; RRID: AB_2687526 |
| Bacterial and Virus Strains | ||
| SARS-CoV-2 WA-1 | BEI resources | NR-52281 |
| Biological Samples | ||
| Human PBMC | This paper | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Live/Dead Blue-BUV450 | Thermo Fisher | Cat#L34962 |
| Live/dead-ef780 | ThermoFisher | Cat#65-0865-14; RRID: AB_2869553 |
| Streptavidin-BUV395 | BD | Cat#564176; RRID: AB_2869553 |
| Decoy Tetreamer- PE-Cy5 | This paper | N/A |
| RBD Tetramer- PE | This paper | N/A |
| PEI-MAX | Polyscience | Cat#26406 |
| PDADMAC | Sigma Aldrich | Cat#409014 |
| Talon cobalt affinity resin | Takara | Cat#635506 |
| EZ-Link Sulfo-NHS-LC Biotinylation Kit | ThermoFisher | Cat#21435 |
| SA-PE | Agilent | Cat#PJRS301-1 |
| Alexa Fluor 647 Antibody Labeling Kit | ThermoFisher | Cat#A20186 |
| 1X 3,3′,5,5′-Tetramethylbenzidine (TMB) | Invitrogen | Cat#00-4201-56 |
| Avicel RC-591 | FMC | |
| anti-PE magnetic beads | Miltenyi Biotec | Cat#130-048-801 |
| eBioscience FoxP3 Fix/Perm kit | ThermoFisher | Cat#00-5521-00 |
| Recombinant Spike Protein | (Walls et al., 2020) | N/A |
| Recombinant RBD Protein | (Walls et al., 2020) | N/A |
| phorbol 12-myristate 13-acetate | Sigma-Aldrich | Cat#P8139 |
| Ionomycin | Sigma-Aldrich | Cat#I9657 |
| GolgiStop/monensin | Becton Dickinson | Cat#554724 |
| Cytofix/Cytoperm | Becton Dickinson | Cat#554714 |
| Brefeldin A | Sigma-Aldrich | Cat#B6542 |
| Cell Proliferation Dye eFluor 670 | ThermoFisher | Cat#65-0840-85 |
| recombinant human IL-2 | Biolegend | Cat#589104 |
| DreamTaq | Thermo Fisher | Cat#EP0702 |
| FastAP | Thermo Fisher | Cat#EF0651 |
| Exonuclease I | ThermoFisher | Cat#EN0582 |
| Nutridoma-SP | Sigma-Aldrich | Cat#11011375001 |
| Critical Commercial Assays | ||
| Human IgG ELISA Kit | Stemcell | Cat#01994 |
| SMART-Seq v4 | Takara | Cat#634470 |
| In-Fusion Cloning Kit | Takara | Cat#638911 |
| Experimental Models: Cell Lines | ||
| 293T | ATCC | Cat#ACS-4500 |
| Oligonucleotides | ||
| 5′-GGAAGGAAGTCCTGTGCGAGGC-3′, 5′-GGAAGAAGCCCTGGACCAGGC-3′ | (Wardemann and Busse, 2019) | N/A |
| 5′-TCTTGTCCACCTTGGTGTTGCT’-3′ | (Smith, 2009) | N/A |
| 5′-GTTTCTCGTAGTCTGCTTTGCTCA-3′ | (Smith, 2009) | N/A |
| 5′-CACCAGTGTGGCCTTGTTGGCTTG-3′ | (Smith, 2009) | N/A |
| 5′-GTGGTATCAACGCAGAGTACATGGG-3′ | This paper | N/A |
| Recombinant DNA | ||
| SARS-CoV-2 SB | BEI Resources | BEI NR-52422 |
| SARS-CoV-2 S-2P ectodomain trimer | BEI Resources | GenBank: YP_009724390.1, BEI NR-52420; cite PMID 32155444 |
| IgG1 vector | Smith et al., 2009 | N/A |
| IgK vector | Smith et al., 2009 | N/A |
| IgL vector | Smith et al., 2009 | N/A |
| Software and Algorithms | ||
| FlowJo10 | Becton Dickinson | N/A |
| Prism | GraphPad | N/A |
| Geneious Prime | Geneious | N/A |
Resource Availability
Lead Contact
Requests for further information and reagents should be directed to Marion Pepper (mpepper@uw.edu).
Materials Availability
Unique reagents generated in this study will be made available on request from Lead Contact, but we may require a payment and/or a completed Materials Transfer Agreement if there is potential for commercial application.
Data and Code Availability
The published article includes all datasets generated or analyzed during this study and no code was generated.
Experimental Model and Subject Details
Ethics Statement
This study was approved by the University of Washington Institutional Review Board (Gale Lab, IRB 00009810). Informed consent was obtained from all enrolled participants. Samples were de-identified prior to transfer to the Pepper Lab.
Study Participants
The study was conceptualized utilizing a case-control design. Cases and controls were identified from a cross-sectional cohort study that recruited via print and online advertising from the Seattle metropolitan area (Table 1). Cases (CoV2+, n = 15) were selected based on a reported history of a positive SARS-CoV-2 PCR nasal swab. Controls (n = 17) were selected based on having no prior positive SARS-CoV-2 PCR nasal swab and having no detectable SARS-CoV-2 RBD- or S-specific IgG or IgM plasma antibodies (within mean + 3 SD of 5 de-identified plasma samples drawn prior to 2020 generously donated by Wesley C. Van Voorhis). At the time of enrollment, information was collected from all participants regarding recent illness symptoms and severity. All CoV2+ cases reported at least one symptom, but all were classified as mild disease, as none required hospitalization.
Method Details
Peripheral blood mononuclear cell (PBMC) and plasma collection
10-50 mL of venous blood from study volunteers were collected in EDTA tubes and spun at 1500xg for 10 min. Plasma was collected, heat-inactivated at 56°C for 30 min, aliquoted and stored at −80°C. The cellular fraction was resuspended in phosphate buffered saline (PBS) and PBMC were separated from red blood cells using Sepmate PBMC Isolation Tubes (STEMCELL Technologies) according to manufacturer’s instruction and frozen at −80°C before being stored in liquid nitrogen. PBMCs were thawed at 37°C and washed twice before use.
SARS-CoV-2 Protein Production and Purification
Plasmid construction
The SARS-CoV-2 SB (BEI NR-52422) construct was synthesized by GenScript into pcDNA3.1- with an N-terminal mu-phosphatase signal peptide and a C-terminal octa-histidine tag (GHHHHHHHH). The boundaries of the construct are N-328RFPN331 and C-528KKST531. The SARS-CoV-2 S-2P ectodomain trimer (GenBank: YP_009724390.1, BEI NR-52420; cite PMID 32155444) was synthesized by GenScript into pCMV with an N-terminal mu-phosphatase signal peptide and a C-terminal TEV cleavage site (GSGRENLYPQG), T4 fibritin foldon (GGGSGYIPEAPRDGQAYVRKDGEWVLLSTPL), and octa-histidine tag (GHHHHHHHH). The construct contains the 2P mutations (proline substitutions at residues 986 and 987; PMID 28807998) and an 682SGAG685 substitution at the furin cleavage site. A pCAGGS vector containing the spike protein receptor binding domain (RBD) from SARS-CoV-2 (Wuhan-Hu-1 isolate) was generously provided by Florian Krammer (Amanat et al., 2020).
Transient expression
Constructs were produced in Expi293F cells grown in suspension using Expi293F expression medium (Life technologies) at 33°C, 70% humidity, and 8% CO2 rotating at 150 rpm. The cultures were transfected using PEI-MAX (Polyscience) with cells grown to a density of 3.0 million cells per mL and cultivated for 3 days. Supernatants was clarified by centrifugation (5 min at 4000 rcf), addition of PDADMAC solution to a final concentration of 0.0375% (Sigma Aldrich, #409014), and a second spin (5 min at 4000 rcf).
Purification of His-tagged proteins
Proteins were purified from clarified supernatants via a batch bind method where each supernatant was supplemented with 1 M Tris-HCl pH 8.0 to a final concentration of 45 mM and 5 M NaCl to a final concentration of ∼310 mM). Talon cobalt affinity resin (Takara) was added to the treated supernatants and allowed to incubate for 15 min with gentle shaking. Resin was collected using vacuum filtration using a 0.2 μm filter and transferred to a gravity column. The resin was washed with 20 mM Tris pH 8.0, 300 mM NaCl, and the protein was eluted with three column volumes of 20 mM Tris pH 8.0, 300 mM imidazole, 300 mM NaCl. The batch bind process was then repeated and the first and second elutions combined. SDS-PAGE was used to assess purity. Purified S-2P trimer was concentrated to ∼1 mg/mL and dialyzed into 50 mM Tris pH 8, 150 mM NaCl, 0.25% L-histidine, 5% glycerol in a hydrated 10k molecular weight cutoff dialysis cassette (Thermo Scientific). The purified RBD protein was dialyzed into 50 mM Tris pH 7, 185 mM NaCl, 100 mM arginine, 4.5% glycerol, 0.75% w/v CHAPS. Due to inherent instability, S-2P was immediately flash frozen and stored at −80°C.
Tetramer generation
Recombinant trimeric spike and the RBD domain were both biotinylated using an EZ-Link Sulfo-NHS-LC Biotinylation Kit (ThermoFisher), tetramerized with streptavidin-phycoerythrin (SA-PE) (Agilent) and stored in 50% glycerol at −20°C as previously described17. Decoy reagent was generated by tetramerizing an irrelevant biotinylated protein with SA-PE previously conjugated to Alexa Fluor 647 using an Alexa Fluor 647 Antibody Labeling Kit (ThermoFisher).
ELISA
96-well plates (Corning) were coated with 2 ug/mL of recombinant SARS-CoV-2 RBD or trimeric spike protein diluted in PBS and incubated at 4°C overnight. Plates were washed with PBS-T (PBS containing 0.05% Tween-20) and incubated with blocking buffer (PBS-T and 3% milk) for 1 h at room temperature (RT). Plasma, culture supernatants or monoclonal antibodies were serially diluted in dilution buffer (PBS-T and 1% milk) in triplicate, added to plates, and incubated at RT for 2 h. Secondary antibodies were diluted in dilution buffer as follows: anti-human IgG-HRP (Jackson ImmunoResearch) at 1:3000, anti-human IgM-HRP (Southern Biotech) at 1:3000, or anti-human IgA-HRP (Southern Biotech) at 1:1500. Plates were incubated with secondary antibodies for 1 h at RT, then detected with 1X 3,3′,5,5′-Tetramethylbenzidine (TMB) (Invitrogen) and quenched with 1M HCl. Sample optical density (OD) was measured by a spectrophotometer at 450nm and 570nm. CR3022, a human SARS-CoV antibody previously determined to cross-react with SARS-CoV-2 was used as a positive control. IgG in culture supernatants was measured using a Human IgG ELISA Kit (Stemcell) according to the manufacturer’s instructions. Data was analyzed in Prism (GraphPad).
Receptor-binding inhibition assay (sVNT)
sVNT assays were performed as previously described (Tan et al., 2020), Briefly, high-binding 96-well plates (Corning) were coated with 5 ug/mL of recombinant human ACE2-Fc diluted in 100mM carbonate-bicarbonate buffer (pH 9.6) and incubated at 4°C overnight. Plates were washed with PBS-T and incubated with blocking buffer (3% milk in PBS-T) for 1 h at RT. Plasma or monoclonal antibody supernatants were serially diluted in triplicate in dilution buffer (1% milk in PBS-T) and incubated with 18ng of recombinant SARS-CoV-2 RBD-HRP (conjugated using Abcam HRP conjugation kit) for 1 h at 37°C. Blocked plates were washed and incubated with the pre-incubated plasma/antibody and RBD-HRP for 1 h at RT, then detected with TMB and 1M HCl. OD was measured by a spectrophotometer at 450nm and 570nm. RBD-HRP alone and plasma with no RBD-HRP incubation were used as controls. The percent inhibition was calculated as (1 – Sample OD value/Average Negative Control OD value) x 100. Data was analyzed in Prism (GraphPad).
Plaque reduction neutralization test (PRNT)
PRNT assays were performed as previously described (Erasmus et al., 2020). Briefly, heat inactivated plasma was diluted 1:5 followed by four 4-fold serial dilutions and monoclonal antibodies were diluted 1:10 followed by 4 10-fold serial dilution and mixed 1:1 with 600 PFU/mL SARS-CoV-2 WA-1 (BEI resources) in PBS+0.3% cold water fish skin gelatin (Sigma). After 30 min of incubation at 37°C, the plasma/virus mixtures were added to 12 well plates of Vero cells and incubated for 1 h at 37°C, rocking every 15 min. All dilutions were done in duplicate, along with virus only and no virus controls. Plates were then washed with PBS and overlaid with a 1:1 mixture of 2.4% Avicel RC-591 (FMC) and 2X MEM (ThermoFisher) supplemented with 4% heat-inactivated FBS and Penicillin/Streptomycin (Fisher Scientific.) After a 48 h incubation, the overlay was removed, plates were washed with PBS, fixed with 10% formaldehyde (Sigma-Aldrich) in PBS for 30 min at room temp and stained with 1% crystal violet (Sigma-Aldrich) in 20% EtOH. Percent neutralization was calculated as (1 – # sample plaques/# positive control plaques) x 100. Data was analyzed in Prism (GraphPad) and IC50 was calculated by sigmoidal interpolation method.
Cell Enrichment, Stimulations and Flow Cytometry
Immunophenotyping and sorting RBD-specific B cells
Thawed PBMCs were first stained with decoy tetramer and then with RBD tetramer prior to incubation with anti-PE magnetic beads and magnetic bead enrichment (Miltenyi Biotec) as previously described.17 Bound cells were stained with surface antibodies and, if required, were fixed/permeabilized using eBioscience FoxP3 Fix/Perm kit (ThermoFisher) for 30 min, followed by incubation with intracellular antibodies. Stained samples were run on a LSRII flow cytometer and analyzed using FlowJo (Becton Dickinson). Samples with less than 5 RBD-specific MBCs were removed from summary data for proportional phenotyping (surface stain V1: 2, surface stain V2: 1, intracellular stain V1: 4, intracellular stain V2: 3). For B cells sorting experiments, single tetramer-specific B cells were indexed sorted on a FACSAriaII cell sorter and collected in a 96-well PCR plate containing SMART-Seq v4 capture buffer (Takara Bio).
Immunophenotyping of PBMCs
For surface phenotyping, total PBMCs (innate cells) or PBMCs from the negative fraction of the antigen-specific B Cell magnetic columns (for lymphocytes) were washed and incubated with fluorescently conjugated antibodies. Staining for cTfh analyses were performed as follows: chemokine-receptors and transcription factors (40 min, RT), surface antigens (20 min, 4°C). Intracellular staining was performed using eBioscience FoxP3 Fix/Perm kit (ThermoFisher). For detection of intracellular cytokine production, PBMC were stimulated with 50 ng/mL phorbol 12-myristate 13-acetate (Sigma-Aldrich) and 1 μg/mL Ionomycin (Sigma-Aldrich) with 10 μg/mL Brefeldin A (Sigma-Aldrich) and 1x dose GolgiStop/monensin (Becton Dickinson) for 4 h. Permeabilization and fixation was performed using Cytofix/Cytoperm (Becton Dickinson). Intracellular stains were performed for 30 min at 4°C. Flow cytometry analysis of innate immune populations was done on 0.5-1 million PBMCs before fraction isolation. Data was acquired on a Cytek Aurora or BD LSR Fortessa and analyzed using FlowJo10 software (Becton Dickinson).
Ex vivo spike Protein Stimulation of Peripheral Blood T Cells
PBMCs from the negative fraction of antigen-specific B Cell magnetic columns were washed and resuspended to 4x106 cells/mL with complete RPMI with 10mM HEPES (ThermoFisher) supplemented with 10% FBS, 2-Mercaptoethanol, Pen-Strep, and L-Glutamine. Spike-stimulated PBMCs were incubated with 2ug/mL full-length recombinant spike protein resuspended in PBS + 5% glycerol. Unstimulated controls received equivalent volume of PBS + 5% glycerol vehicle. Both conditions were left for 20 h at 37°C, 5%–8% CO2, with addition of 10 μg/mL Brefeldin A (Sigma-Aldrich) and 1x dose GolgiStop/monensin (Becton Dickinson) for the final 5 h to allow for intracellular detection of cytokines. Positive controls were stimulated with PMA/Ionomycin (see above) for 5 h in the presence of Brefeldin-A and Monensin. Staining was performed as follows: chemokine-receptors (40 min, RT), surface antigens and cytokines (20 min, 4°C). Cells were run on the Cytek Aurora and analyzed using FlowJo (Becton Dickenson).
Antigen-specific T cell proliferation
Starting with PBMC from healthy control or CoV2+ individuals, cell proliferation dye (CPD)-labeled, 1.25uM (ThermoFisher5), sorted naive or memory T cell subsets (5 × 104) were co-cultured in round-bottomed 96-well plates with irradiated autologous monocytes (5000 rads, 5 × 104), and provided either full-length recombinant human spike protein (2.5ug/mL) resuspended in 5% PBS-glycerol or vehicle control. Cultures were supplemented with 5U/mL recombinant human IL-2 (Biolegend; 589104). Cellular proliferation was assessed after 5-6 days by flow cytometry as above and analyzed using FlowJo10 (Becton Dickenson). The percentage of CXCR3+CPDlo cells (defined as cells that had undergone 3 or more divisions) represented as Spike - Vehicle is calculated by subtracting the vehicle control proliferation from spike-treated proliferation.
Monoclonal antibody generation
BCR sequencing and cloning
Amplification of cDNA was performed using SMART-Seq v4 (Takara Bio) at half reaction volume for each sorted cell. B cell receptor (BCR) chains were amplified in a multiplex PCR using half reactions of DreamTaq (Thermo Fisher) and 1.25 ul of resulting cDNA with 3' primers for constant regions of IgM, IgA, (5′-GGAAGGAAGTCCTGTGCGAGGC-3′, 5′-GGAAGAAGCCCTGGACCAGGC-3′, ) (Wardemann and Busse, 2019)IgG, IgK, IgL (5′-TCTTGTCCACCTTGGTGTTGCT’-3′, 5′-GTTTCTCGTAGTCTGCTTTGCTCA-3′, 5′-CACCAGTGTGGCCTTGTTGGCTTG-3′, (Smith, 2009)) and a 5′ primer for the template switch sequence (5′-GTGGTATCAACGCAGAGTACATGGG-3′). Thermocycler conditions were 95°C for 2 min, 30 cycles of 95°C for 30 s, 57°C for 30 s and 72°C for 1 min. Resulting PCR products were cleaned using 5 ul of PCR reaction, 1 ul FastAP (Thermo Fisher), and 0.5 ul Exonuclease I (ThermoFisher) for 30 min at 37°C and inactivated at 75°C for 15 min. Sanger sequencing for each purified sample was performed using each 3′ primer from the previous BCR PCR amplification. Sequences were trimmed at Q30 using Geneious and submitted to IMGT/HighV-QUEST for alignment (Alamyar, 2012). Primers were designed using 5′ and 3′ cDNA sequence for In-Fusion Cloning Kit and performed according to manufacturer’s instructions. If a 5′ or 3′ sequence was missing, then the closest matching IMGT germline sequence was used for primer design. Heavy chains were inserted into IgG1 vectors, kappa and lambda chains were cloned into vectors with their respective constant regions (Smith, 2009). Cloned plasmids were sequenced and screened by ensuring sequences of chains matched original cDNA sequence.
Expression and purification
For small scale transfections, 12 well plates of 293T cells at 80% confluence were transiently transfected with 0.5ug each of heavy and light chain vectors using polyethylenimine (PEI). After 16 h, media was removed and replaced with serum-free media. After 3-4 days, supernatants were harvested and cell debris was removed by centrifugation at max speed in a microcentrifuge for 1 min. For large scale transfections, expression vectors containing paired heavy and light chains were transiently transfected into 293T cells using PEI. Expression of recombinant full-length human IgG monoclonal antibodies were carried out in serum-free basal medium (Nutridoma-SP, Sigma-Aldrich). Four days after transfection, cell culture medium was collected and protein was purified using HiTrap™ Protein G HP column (1ml, GE Healthcare). Final IgG proteins were concentrated and buffer exchanged into 1x PBS using Millipore concentrator (30K MWCO). IgG protein concentration is determined by Nanodrop 2000 spectrophotometer.
Quantification and Statistical Analysis
Statistics are described in figure legends and were determined using Prism (Graphpad). All measurements within a group are from distinct samples except technical replicates used in ELISAs as described. Statistical significance of all pairwise comparisons was assessed by two-tailed nonparametric tests; Mann-Whitney for unpaired data and Wilcoxon signed rank tests for paired data. Raw p values are displayed and the adjusted p value significance cutoff calculated from the Benjamini-Hochberg multiple testing correction with FDR = 0.05 for each figure is listed in the corresponding legend.
Acknowledgments
We thank David M. Koelle for PBMC samples used for reagent testing; Florian Krammer for vectors used to express the SARS-CoV-2 spike RBD; Wesley C. Van Voorhis for historical negative plasma samples; Ben Murrell and the CoroNAb consortium for providing the Ty1 antibody; Ali Ellebedy for providing the 2BO4 and 2CO2 antibodies; Mike Murphy and Deleah Pettie for assistance with protein production; Noah Simon for assistance with statistical analyses; Brian Hondowicz for technical help; and the Pepper, Gale, Campbell, Rawlings, and Hammerman labs for helpful discussion and the study volunteers for their participation. This work was supported by the following funding: L.B.R. and L.S. (NIH2T32 AI106677), D.J.C. and P.A.M. (NIH R01AI127726, NIH U19AI125378-S1), J.A.H. (NIH R01AI150178-01S1), H.R.W. (NIH TL1 TR002318), D.J.R. (SCRI, Research Integration Hub Covid-19 Award), and M.P. (NIH U01AI142001-02S1; R01AI118803); a Bill & Melinda Gates Foundation grant to N.P.K. (OPP1156262); BWF #1018486 and COVID Pilot grant to M.P.; and Emergent Ventures Fast Grant to M.P.
Author contributions
M.P., L.B.R., and J.N. conceived the study. K.K.T. assisted in cohort recruitment and visit scheduling. J.R., C.S., E.A.H., L.B.R., J.N., and K.K.T. processed and preserved blood and plasma samples. J.N., L.C., and N.P.K. generated proteins with help from S.W. and B.F., and L.B.R. generated and validated tetramer reagents. L.S., Y.C., J.N., and L.B.R. performed ELISAs, and L.S. and J.N. performed sVNT assays. L.S. analyzed plasma data. H.R.W. and J.H. conceived, performed, and analyzed innate cell phenotyping experiments. L.B.R. and J.N. performed and analyzed antigen-specific B cell flow cytometry and sorting. K.B.P. and P.A.M. conceived, performed, and analyzed T cell experiments with help from M.L.F. C.D.T. and M.H. sequenced and generated mAb plasmids. Y.C. and M.H. expressed and purified mAbs. J.E. and E.A.H. performed PRNT assays. L.B.R, J.N., L.S., K.B.P., P.A.M., and M.P. drafted the manuscript. All authors helped edit the manuscript. M.P. secured funds and supervised the project.
Declaration of Interests
M.P., D.J.R., J.N., C.D.T., Y.C., and L.B.R. have filed a patent under the provisional serial no. 63/063,841. Other authors declare no competing interests.
Published: November 23, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.cell.2020.11.029.
Supplemental Information
References
- Addetia A., Crawford K.H.D., Dingens A., Zhu H., Roychoudhury P., Huang M.-L., Jerome K.R., Bloom J.D., Greninger A.L. Neutralizing Antibodies Correlate with Protection from SARS-CoV-2 in Humans during a Fishery Vessel Outbreak with a High Attack Rate. J. Clin. Microbiology. 2020;58 doi: 10.1128/JCM.02107-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed R., Gray D. Immunological memory and protective immunity: understanding their relation. Science. 1996;272:54–60. doi: 10.1126/science.272.5258.54. [DOI] [PubMed] [Google Scholar]
- Alamyar E., et al. IMGT(®) tools for the nucleotide analysis of immunoglobulin (IG) and T cell receptor (TR) V-(D)-J repertoires, polymorphisms, and IG mutations: IMGT/V-QUEST and IMGT/HighV-QUEST for NGS. Methods in molecular biology (Clifton, N.J.) 882(Chapter. 2012;32:569–604. doi: 10.1007/978-1-61779-842-9_32. [DOI] [PubMed] [Google Scholar]
- Alsoussi W.B., Turner J.S., Case J.B., Zhao H., Schmitz A.J., Zhou J.Q., Chen R.E., Lei T., Rizk A.A., McIntire K.M., et al. A Potently Neutralizing Antibody Protects Mice against SARS-CoV-2 Infection. J. Immunol. 2020;205:915–922. doi: 10.4049/jimmunol.2000583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amanat F., Stadlbauer D., Strohmeier S., Nguyen T.H.O., Chromikova V., McMahon M., Jiang K., Arunkumar G.A., Jurczyszak D., Polanco J., et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020;26:1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amanna I.J., Carlson N.E., Slifka M.K. Duration of humoral immunity to common viral and vaccine antigens. N. Engl. J. Med. 2007;357:1903–1915. doi: 10.1056/NEJMoa066092. [DOI] [PubMed] [Google Scholar]
- Bentebibel S.E., Lopez S., Obermoser G., Schmitt N., Mueller C., Harrod C., Flano E., Mejias A., Albrecht R.A., Blankenship D., et al. Induction of ICOS+CXCR3+CXCR5+ TH cells correlates with antibody responses to influenza vaccination. Sci. Transl. Med. 2013;5:176ra32. doi: 10.1126/scitranslmed.3005191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun J., Loyal L., Frentsch M., Wendisch D., Georg P., Kurth F., Hippenstiel S., Dingeldey M., Kruse B., Fauchere F., et al. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. 2020;587:270–274. doi: 10.1038/s41586-020-2598-9. [DOI] [PubMed] [Google Scholar]
- Callow K.A., Parry H.F., Sergeant M., Tyrrell D.A. The time course of the immune response to experimental coronavirus infection of man. Epidemiol. Infect. 1990;105:435–446. doi: 10.1017/s0950268800048019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrashekar A., et al. SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science. 2020;369:812–817. doi: 10.1126/science.abc4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S., Freeman T. Metabolic heterogeneity of human gamma-globulin. Biochem. J. 1960;76:475–487. doi: 10.1042/bj0760475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erasmus J.H., Khandhar A.P., Walls A.C., Hemann E.A., O’Connor M.A., Murapa P., Archer J., Leventhal S., Fuller J., Lewis T., et al. Single-dose replicating RNA vaccine induces neutralizing antibodies against SARS-CoV-2 in nonhuman primates. bioRxiv. 2020 2020.05.28.121640. [Google Scholar]
- Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell. 2020;181:1489–1501.e15, e1415. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanke L., Vidakovics Perez L., Sheward D.J., Das H., Schulte T., Moliner-Morro A., Corcoran M., Achour A., Karlsson Hedestam G.B., Hällberg B.M., et al. An alpaca nanobody neutralizes SARS-CoV-2 by blocking receptor interaction. Nat. Commun. 2020;11:4420. doi: 10.1038/s41467-020-18174-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota K., Turner J.E., Villa M., Duarte J.H., Demengeot J., Steinmetz O.M., Stockinger B. Plasticity of Th17 cells in Peyer’s patches is responsible for the induction of T cell-dependent IgA responses. Nat. Immunol. 2013;14:372–379. doi: 10.1038/ni.2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–280.e8, e278. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isho B., Abe K.T., Zuo M., Jamal A.J., Rathod B., Wang J.H., Li Z., Chao G., Rojas O.L., Bang Y.M., et al. Evidence for sustained mucosal and systemic antibody responses to SARS-CoV-2 antigens in COVID-19 patients. medRxiv. 2020 doi: 10.1126/sciimmunol.abe5511. 2020.2008.2001.20166553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juno J.A., Tan H.X., Lee W.S., Reynaldi A., Kelly H.G., Wragg K., Esterbauer R., Kent H.E., Batten C.J., Mordant F.L., et al. Humoral and circulating follicular helper T cell responses in recovered patients with COVID-19. Nat. Med. 2020;26:1428–1434. doi: 10.1038/s41591-020-0995-0. [DOI] [PubMed] [Google Scholar]
- Kaneko N., Kuo H.H., Boucau J., Farmer J.R., Allard-Chamard H., Mahajan V.S., Piechocka-Trocha A., Lefteri K., Osborn M., Bals J., et al. Massachusetts Consortium on Pathogen Readiness Specimen Working Group Loss of Bcl-6-Expressing T Follicular Helper Cells and Germinal Centers in COVID-19. Cell. 2020;183:143–157.e13, e113. doi: 10.1016/j.cell.2020.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keitany G.J., et al. Blood Stage Malaria Disrupts Humoral Immunity to the Pre- erythrocytic Stage Circumsporozoite Protein. Cell Reports. 2016;17:3193–3205. doi: 10.1016/j.celrep.2016.11.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C.C., Baccarella A.M., Bayat A., Pepper M., Fontana M.F. FCRL5+ Memory B Cells Exhibit Robust Recall Responses. Cell Rep. 2019;27:1446–1460.e4, e1444. doi: 10.1016/j.celrep.2019.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox J.J., Buggert M., Kardava L., Seaton K.E., Eller M.A., Canaday D.H., Robb M.L., Ostrowski M.A., Deeks S.G., Slifka M.K., et al. T-bet+ B cells are induced by human viral infections and dominate the HIV gp140 response. JCI Insight. 2017;2:92943. doi: 10.1172/jci.insight.92943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox J.J., Kaplan D.E., Betts M.R. T-bet-expressing B cells during HIV and HCV infections. Cell. Immunol. 2017;321:26–34. doi: 10.1016/j.cellimm.2017.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox J.J., Myles A., Cancro M.P. T-bet+ memory B cells: Generation, function, and fate. Immunol. Rev. 2019;288:149–160. doi: 10.1111/imr.12736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurty A.T., Thouvenel C.D., Portugal S., Keitany G.J., Kim K.S., Holder A., Crompton P.D., Rawlings D.J., Pepper M. Somatically Hypermutated Plasmodium-Specific IgM(+) Memory B Cells Are Rapid, Plastic, Early Responders upon Malaria Rechallenge. Immunity. 2016;45:402–414. doi: 10.1016/j.immuni.2016.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.K., Turner H., Maynard C.L., Oliver J.R., Chen D., Elson C.O., Weaver C.T. Late developmental plasticity in the T helper 17 lineage. Immunity. 2009;30:92–107. doi: 10.1016/j.immuni.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q.-X., Liu B.-Z., Deng H.-J., Wu G.-C., Deng K., Chen Y.-K., Liao P., Qiu J.-F., Lin Y., Cai X.-F., et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020;26:845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- Ma H., Zeng W., He H., Zhao D., Jiang D., Zhou P., Cheng L., Li Y., Ma X., Jin T. Serum IgA, IgM, and IgG responses in COVID-19. Cell. Mol. Immunol. 2020;17:773–775. doi: 10.1038/s41423-020-0474-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew D., Giles J.R., Baxter A.E., Oldridge D.A., Greenplate A.R., Wu J.E., Alanio C., Kuri-Cervantes L., Pampena M.B., D’Andrea K., et al. UPenn COVID Processing Unit Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science. 2020;369:eabc8511. doi: 10.1126/science.abc8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita R., Schmitt N., Bentebibel S.E., Ranganathan R., Bourdery L., Zurawski G., Foucat E., Dullaers M., Oh S., Sabzghabaei N., et al. Human blood CXCR5(+)CD4(+) T cells are counterparts of T follicular cells and contain specific subsets that differentially support antibody secretion. Immunity. 2011;34:108–121. doi: 10.1016/j.immuni.2010.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nellore A., Scharer C.D., King R.G., Tipton C.M., Zumaquero E., Fucile C., Mousseau B., Bradley J.E., Macon K., Mi T., et al. Fcrl5 and T-bet define influenza-specific memory B cells that predict long-lived antibody responses. bioRxiv. 2019 doi: 10.1101/643973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng S.L., Szabo S.J., Glimcher L.H. T-bet regulates IgG class switching and pathogenic autoantibody production. Proc. Natl. Acad. Sci. USA. 2002;99:5545–5550. doi: 10.1073/pnas.082114899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perreault J., Tremblay T., Fournier M.-J., Drouin M., Beaudoin-Bussières G., Prévost J., Lewin A., Bégin P., Finzi A., Bazin R. Longitudinal analysis of the humoral response to SARS-CoV-2 spike RBD in convalescent plasma donors. bioRxiv. 2020 doi: 10.1182/blood.2020008367. 2020.2007.2016.206847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotkin S.A. Correlates of protection induced by vaccination. Clin. Vaccine Immunol. 2010;17:1055–1065. doi: 10.1128/CVI.00131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pray I.W., et al. COVID-19 Outbreak at an Overnight Summer School Retreat Wisconsin, July-August 2020. MMWR. Morbidity and mortality weekly. report. 2020;69:1600–1604. doi: 10.15585/mmwr.mm6943a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss S., Baxter A.E., Cirelli K.M., Dan J.M., Morou A., Daigneault A., Brassard N., Silvestri G., Routy J.P., Havenar-Daughton C., et al. Comparative analysis of activation induced marker (AIM) assays for sensitive identification of antigen-specific CD4 T cells. PLoS ONE. 2017;12:e0186998. doi: 10.1371/journal.pone.0186998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripperger T.J., Uhrlaub J.L., Watanabe M., Wong R., Castaneda Y., Pizzato H.A., Thompson M.R., Bradshaw C., Weinkauf C.C., Bime C., et al. Detection, prevalence, and duration of humoral responses to SARS-CoV-2 under conditions of limited population exposure. medRxiv. 2020 2020.08.14.20174490. [Google Scholar]
- Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S., et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubtsova K., Rubtsov A.V., van Dyk L.F., Kappler J.W., Marrack P. T-box transcription factor T-bet, a key player in a unique type of B-cell activation essential for effective viral clearance. Proc. Natl. Acad. Sci. USA. 2013;110:E3216–E3224. doi: 10.1073/pnas.1312348110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruterbusch M., Pruner K.B., Shehata L., Pepper M. In Vivo CD4+ T Cell Differentiation and Function: Revisiting the Th1/Th2 Paradigm. Annu. Rev. Immunol. 2020;38:705–725. doi: 10.1146/annurev-immunol-103019-085803. [DOI] [PubMed] [Google Scholar]
- Sallusto F. Heterogeneity of Human CD4(+) T Cells Against Microbes. Annu. Rev. Immunol. 2016;34:317–334. doi: 10.1146/annurev-immunol-032414-112056. [DOI] [PubMed] [Google Scholar]
- Sallusto F., Lenig D., Förster R., Lipp M., Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401:708–712. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- Schmidt M.E., Varga S.M. The CD8 T Cell Response to Respiratory Virus Infections. Front. Immunol. 2018;9:678. doi: 10.3389/fimmu.2018.00678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine T., Perez-Potti A., Rivera-Ballesteros O., Strålin K., Gorin J.B., Olsson A., Llewellyn-Lacey S., Kamal H., Bogdanovic G., Muschiol S., et al. Karolinska COVID-19 Study Group Robust T Cell Immunity in Convalescent Individuals with Asymptomatic or Mild COVID-19. Cell. 2020;183:158–168.e14, e114. doi: 10.1016/j.cell.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seow J., Graham C., Merrick B., Acors S., Steel K.J.A., Hemmings O., O’Bryne A., Kouphou N., Pickering S., Galao R., et al. Longitudinal evaluation and decline of antibody responses in SARS-CoV-2 infection. medRxiv. 2020 doi: 10.1038/s41564-020-00813-8. 2020.2007.2009.20148429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette A., Crotty S. Pre-existing immunity to SARS-CoV-2: the knowns and unknowns. Nat. Rev. Immunol. 2020;20:457–458. doi: 10.1038/s41577-020-0389-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L., et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- Slifka M.K., Ahmed R. Long-term antibody production is sustained by antibody-secreting cells in the bone marrow following acute viral infection. Ann. N Y Acad. Sci. 1996;797:166–176. doi: 10.1111/j.1749-6632.1996.tb52958.x. [DOI] [PubMed] [Google Scholar]
- Smith K., et al. Rapid generation of fully human monoclonal antibodies specific to a vaccinating antigen. Nature protocols. 2009;4:372–384. doi: 10.1038/nprot.2009.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song G., He W.T., Callaghan S., Anzanello F., Huang D., Ricketts J., Torres J.L., Beutler N., Peng L., Vargas S., et al. Cross-reactive serum and memory B cell responses to spike protein in SARS-CoV-2 and endemic coronavirus infection. bioRxiv. 2020 doi: 10.1038/s41467-021-23074-3. 2020.09.22.308965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan C.W., Chia W.N., Qin X., Liu P., Chen M.I.C., Tiu C., Hu Z., Chen V.C.-W., Young B.E., Sia W.R., et al. A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2-spike protein-protein interaction. Nat. Biotechnol. 2020;38:1073–1078. doi: 10.1038/s41587-020-0631-z. [DOI] [PubMed] [Google Scholar]
- Tang F., Quan Y., Xin Z.-T., Wrammert J., Ma M.-J., Lv H., Wang T.-B., Yang H., Richardus J.H., Liu W., Cao W.C. Lack of peripheral memory B cell responses in recovered patients with severe acute respiratory syndrome: a six-year follow-up study. J. Immunol. 2011;186:7264–7268. doi: 10.4049/jimmunol.0903490. [DOI] [PubMed] [Google Scholar]
- To K., Hung I., Ip J.D., Infectious A.C.C. Coronavirus disease 2019 (COVID-19) reinfectionby a phylogenetically distinct severe acute respiratory syndrome coronavirus 2 strainconfirmed by whole genome sequencing. Clin. Inf. Dis. 2020 doi: 10.1093/cid/ciaa1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinuesa C.G., Linterman M.A., Yu D., MacLennan I.C. Follicular Helper T Cells. Annu. Rev. Immunol. 2016;34:335–368. doi: 10.1146/annurev-immunol-041015-055605. [DOI] [PubMed] [Google Scholar]
- Wajnberg A., Amanat F., Firpo A., Altman D., Bailey M., Mansour M., McMahon M., Meade P., Mendu D.R., Muellers K., et al. SARS-CoV-2 infection induces robust, neutralizing antibody responses that are stable for at least three months. medRxiv. 2020 2020.2007.2014.20151126. [Google Scholar]
- Walls A.C., Fiala B., Schäfer A., Wrenn S., Pham M.N., Murphy M., Tse L.V., Shehata L., O’Connor M.A., Chen C., et al. Elicitation of Potent Neutralizing Antibody Responses by Designed Protein Nanoparticle Vaccines for SARS-CoV-2. Cell. 2020 doi: 10.1016/j.cell.2020.10.043. S0092-8674(20)31450-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisel F., Shlomchik M. Memory B Cells of Mice and Humans. Annu. Rev. Immunol. 2017;35:255–284. doi: 10.1146/annurev-immunol-041015-055531. [DOI] [PubMed] [Google Scholar]
- Wilson P., Stamper C., Dugan H., Li L., Asby N., Halfmann P., Guthmiller J., Zheng N.Y., Huang M., Stovicek O., et al. Distinct B cell subsets give rise to antigen-specific antibody responses against SARS-CoV-2. Res Sq. 2020 rs.3.rs-80476. [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Wu Z., McGoogan J.M. Characteristics of and Important Lessons From the Coronavirus Disease 2019 (COVID-19) Outbreak in China: Summary of a Report of 72 314 Cases From the Chinese Center for Disease Control and Prevention. JAMA. 2020;323:1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- Wu L.P., Wang N.C., Chang Y.H., Tian X.Y., Na D.Y., Zhang L.Y., Zheng L., Lan T., Wang L.F., Liang G.D. Duration of antibody responses after severe acute respiratory syndrome. Emerg. Infect. Dis. 2007;13:1562–1564. doi: 10.3201/eid1310.070576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Raddad L.J., et al. Assessment of the risk of SARS-CoV-2 reinfection in an intense reexposure setting. medRxiv. 2020 doi: 10.1101/2020.08.24.20179457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardemann H., Busse C.E. (2019) Expression Cloning of Antibodies from Single Human B Cells. In: Küppers R. (eds) Lymphoma. Methods in Molecular Biology, vol 1956. Humana Press, New York, NY. 10.1007/978-1-4939-9151-8_5 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The published article includes all datasets generated or analyzed during this study and no code was generated.