Abstract
Background
COVID-19 is an infectious disease caused by a novel positive-sense single-stranded RNA coronavirus called as SARS-CoV-2. This viral disease is known to infect the respiratory system, eventually leading to pneumonia. Crystallographic studies of the viral structure reveal its mechanism of infection as well as active binding sites and the druggable targets as scope for treatment of COVID-19.
Hypothesis
The role of tea polyphenols in prophylaxis and treatment of COVID-19 was established in this study.
Study design
Molecular docking interactions of tea polyphenols with some of the possible binding sites of SARS-CoV-2 were performed.
Materials and methods
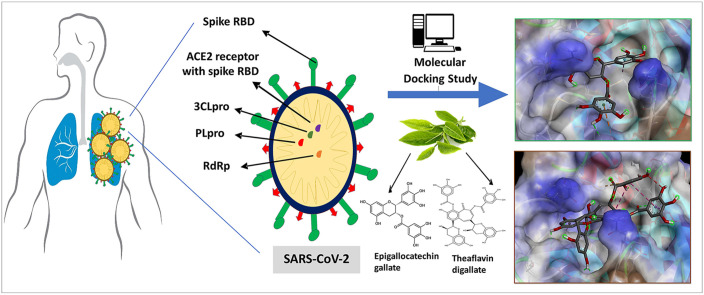
From various studies on the SARS-CoV-2 reported in the literature, we chose possible drug targets (Chymotrypsin-like protease, RNA dependant RNA polymerase, Papain like protease, Spike RBD and ACE2 receptor with spike RBD) which are vital proteins. These receptors were docked against two tea polyphenols, Epigallocatechin gallate (EGCG) from green tea and Theaflavin digallate (TF3) from black tea. These polyphenols have been previously reviewed for their antiviral activities, especially against single-stranded RNA viruses. Two antiviral drugs, Remdesivir and Favipiravir were studied for comparative docking results.
Results
A comparative study of docking scores and the type of interactions of EGCG, TF3 with the possible targets of COVID-19 showed that the tea polyphenols had good docking scores with significant in-silico activity.
Conclusion
These results can provide a lead in exploring both the tea polyphenols in prophylaxis as well as treatment of COVID-19.
Keywords: COVID-19, Molecular docking, Tea polyphenols, ECCG, Theaflavins
Graphical abstract
Highlights
-
•
Tea polyphenols are known for their broad-spectrum antiviral activity.
-
•
EGCG and theaflavin digallate were docked against SARS-CoV-2 receptors.
-
•
Polyphenols docked to the receptors with good affinity showing several interactions.
-
•
The affinity of polyphenols was higher than Remdesivir and Favipiravir against RdRp.
-
•
MD Simulation validated the stability of the docked complex.
List of abbreviations
- 3CLpro
Chymotrypsin-like protease
- ACE2
Angiotensin-converting enzyme 2
- EC
Epicatechin
- ECG
Epicatechin-3-gallate
- EGC
Epigallocatechin
- EGCG
Epigallocatechin Gallate
- GC
Gallocatechin
- H-bond
Hydrogen Bond
- Mpro
Main protease
- NAMD
Nanoscale Molecular Dynamics Simulation
- Nsp
Non-structural proteins
- PDB
Protein Data Bank
- PLpro
Papain like protease
- RBD
Receptor-binding domain
- RdRp
RNA dependant RNA polymerase
- TF
Theaflavin
- TF2A
Theaflavin-3-gallate
- TF2B
Theaflavin-3′-gallate
- TF3
Theaflavin-3,3′-digallate
- VMD
Visual Molecular Dynamics
1. Introduction
The crystallographic structure of SARS-CoV-2 reveals that the virus is a positive-sense single-stranded RNA virus with four major structural proteins, the spike protein, envelope protein, membrane protein, nucleocapsid protein and some non-structural proteins (Nsp). A glycoprotein known as hemagglutinin esterase exists on the structure of β-strands [1]. The S protein further has two subunits S1 and S2. S1 helps the virus in binding to the host cell with its receptor-binding domain (RBD) whereas the S2 subunit is known to perform membrane fusion to complete the infection process [2]. The S protein is known to target specifically the angiotensin-converting enzyme 2 (ACE2) receptor on the host cell with the RBD. Various studies performed on the structural features of SARS-CoV-2 help us in identifying the major druggable targets. The Main protease or Chymotrypsin-like protease (3CLpro) performs a vital function in the viral maturation step. It cleaves the non-structural proteins (Nsp) at 11 different sites which results in the formation of Nsp4-Nsp16. Since these Nsps perform important roles at some step in the viral replication cycle, 3CLpro is the most important drug target for COVID-19. Papain-like protease (PLpro) is another important enzyme, and hence a drug target that performs a function similar to 3CLpro. It generates Nsp1-Nsp3 by cleaving at different sites [3]. It is also known to suppress the immune system of the host cell, facilitating the infection process. RNA dependant RNA polymerase (RdRp) has a crucial role in the replication and transcription of the viral genome. Looking at the indispensable role of RdRp, it is yet another target for antiviral drugs. The spike RBD is the part that attaches the ACE2 receptor and initiates the infection. An alteration with the protein structure is, therefore, a possible mechanism of inhibition, making ACE2 receptor another drug target. Other binding sites of SARS-CoV-2 have also been explored in literature and attempts to discover their inhibitors have been made [4,5]
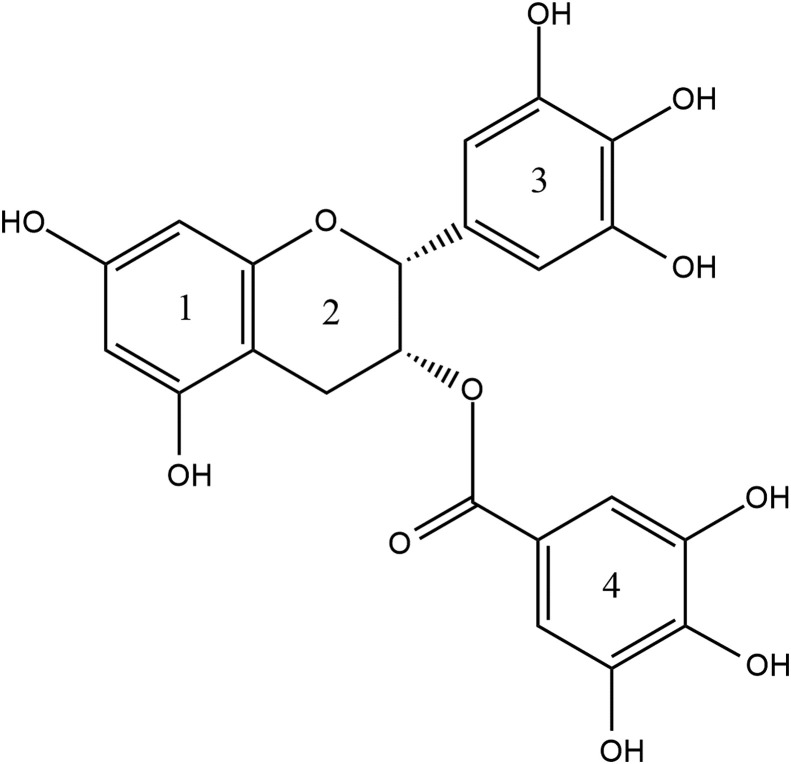
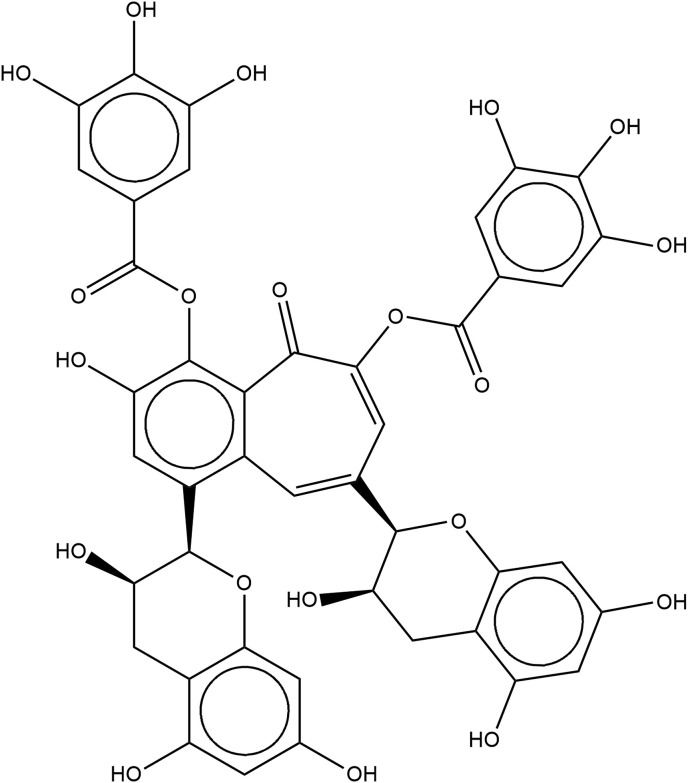
Management of COVID-19 is still a challenge for healthcare workers around the world. Although techniques have been developed for effective diagnosis [6], no drug has been yet approved for treatment. Tea polyphenols are bioactive dietary nutraceuticals. Catechins, the most abundant phytoconstituents found in green tea and theaflavins, the polyphenols found in black tea are active phytochemicals. They display a vast array of bioactive properties like antitumorigenic, anti-inflammatory, antibacterial, antioxidative, antiproliferative apart from their antiviral effects [7,8]. Their antiviral activity has already been reported in numerous single-stranded RNA viruses hinting at the application of tea polyphenols as potential treatment methods for COVID-19 [9]. There are also some reports of these molecules showing activity in SARS-CoV-2 [[10], [11], [12]]. Bhardwaj et al. performed molecular docking studies of tea polyphenols on the main protease of SARS-CoV-2 and discussed their potential as inhibitors of 3CLpro [13]. The catechins found in green tea are catechin, gallocatechin (GC), epicatechin (EC), epicatechin-3-gallate (ECG), epigallocatechin (EGC) and epigallocatechin-3-gallate (EGCG). Out of these, EGCG is found in the highest quantity in green tea, and also researched extensively as compared to the other catechins owing to its antiviral properties [14] (see Fig. 1 ). The theaflavins found in black tea are theaflavin (TF1), theaflavin-3-gallate (TF2a), theaflavin-3′-gallate (TF2b) and theaflavin-3,3-digallate (TF3). TF3 (see Fig. 2 ) has particularly shown better antiviral activity than the other theaflavins [15]. In this study, we analysed interactions of two prime phytoconstituents present in tea, EGCG from green tea and TF3 from black tea with known binding sites of SARS-CoV-2 to anticipate the probable use of these polyphenols as a treatment method in COVID-19.
Fig. 1.
Structure of epigallocatechin gallate (EGCG).
Fig. 2.
Structure of theaflavin digallate (TF3).
Both the tea polyphenols are generally regarded as safe by the FDA, hence there are no dose-dependent side effects. A cup of green tea of roughly 250 mL is expected to have around 50–100 mg of EGCG. Although no clear upper limit exists for the consumption of EGCG, a study estimated that a dose of 800 mg or higher per day can cause liver damage [16]. Similarly, there are no distinct dose restrictions for theaflavins. However, in one study 700 mg of Theaflavins showed side-effects, indicating that 700 mg is a safe amount, but not necessarily an upper limit [17]. A study performed in mice showed that toxic effects were visible at a dosage of 562 mg/kg [18].
To study the exact interaction of EGCG and TF3 with the possible binding sites of SARS-CoV-2, we performed molecular docking studies. Molecular docking is an in-silico method of predicting the most stable orientation of a particular ligand (which is generally a small molecule) in which it will bind to an active site of the receptor. There are two parameters to evaluate a better docking, docking score and the type of interaction. A better docking score is the one with more negative binding energy. Interactions can be non-covalent bonds like hydrogen bonding (H-bonding), Van der Walls interactions, pi-pi-stacking and in cases some ionic bonds. Remdesivir and Favipiravir were chosen as antiviral drugs to be tested specifically the RNA polymerase inhibiting activity. Molecular Dynamics Simulation was performed with the best-docked complex among all the receptors. The drug-likeliness of these molecules is also discussed.
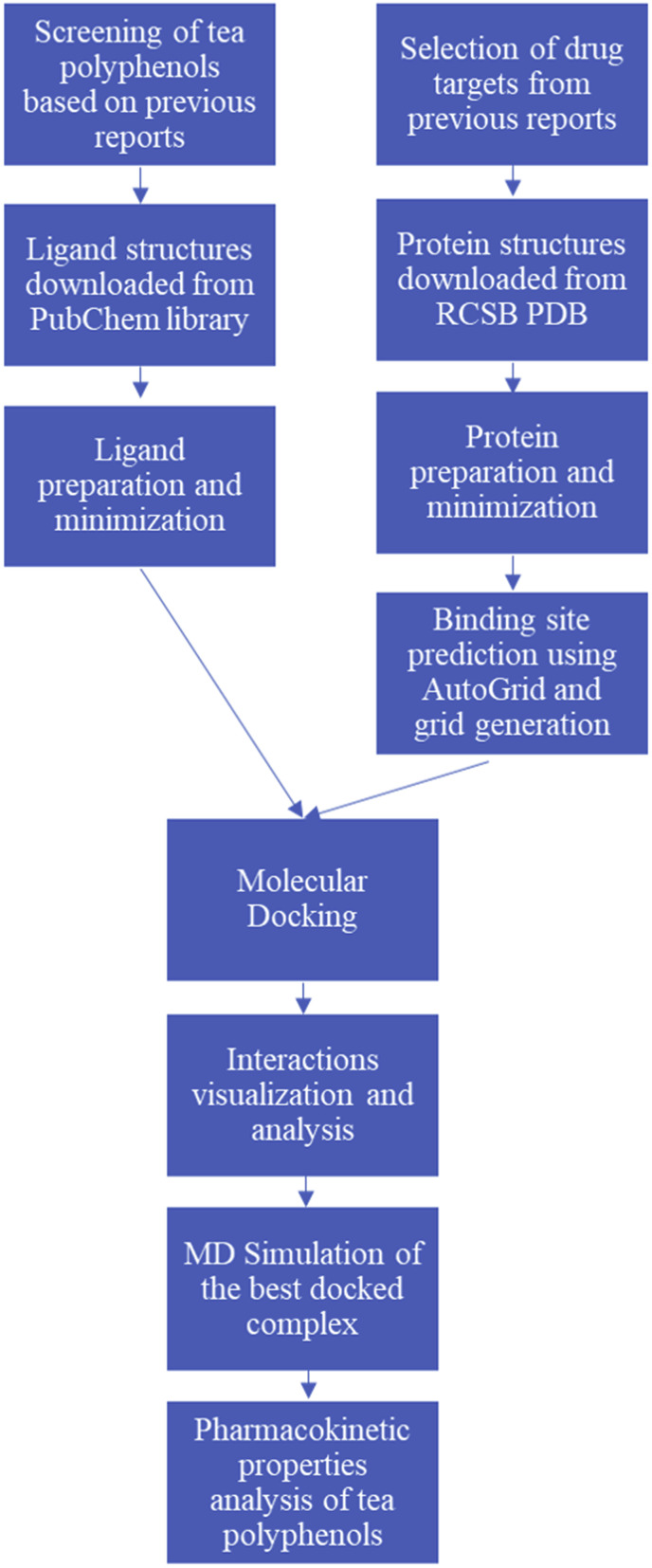
2. Materials and methods
2.1. Obtaining protein structures and small molecules
The ligand 3-D structures were obtained from PubChem in the form of PubChem CIDs. The PubChem CIDs of the ligands were 65064, 136825043, 3652, 121304016, 492405 for EGCG, TF3, Remdesivir and Favipiravir respectively. The 3-D protein structures of the receptors were downloaded from the RCSB Protein Data Bank (PDB) as.pdb files. The PDB IDs of 3CLpro, Spike RBD, PLpro, RdRp and ACE2 receptor with spike RBD were 6LU7, 6XE1, 6W9C, 6M71, 6M0J respectively. These protein structures were imported in USCF Chimera version 1.14 [19].
2.2. Ligand preparation
The PubChem CIDs of EGCG, TF3, Remdesivir and Favipiravir were used as direct input to UCSF Chimera version 1.14 software. From their respective CID's the structure was built using the structure builder option of Chimera. The structures obtained in 3-D format were checked for the number of hydrogens and saved as separate.pdb files.
2.3. Receptor preparation
After importing the receptor structures as.pdb files, the non-standard residues, native ligands and the water molecules if present in the structure were removed. The protein structures were initially prepared by structural minimization with 200 steepest descent steps with a step size of 0.02 Å and 20 conjugate gradient steps with 0.02 Å step size. Hydrogen atoms and charges were added using the Dunbrack 2010 rotamer library [20]. The charges were computed with the help of ANTECHAMBER using AM1-BCC charge method [21]. This minimized structure was saved as a.pdb file.
2.4. Binding site prediction
Binding sites were predicted using AutoGridFR, a platform for the prediction of docking sites on protein receptors. It is part of the AutoDockFR suite [22]. Crucial amino acids participating in ligand interaction cited in the primary citation of the receptor and previous reports were considered in the selection of the docking site. The grid coordinates generated with AutoGridFR were used for molecular docking.
2.5. Molecular docking
EGCG, TF3 and HCQ were docked with all the receptors chosen. Remdesivir and Favipiravir are well known as RNA polymerase inhibitors, hence were docked only with RdRp. The protein structure and ligands which were prepared in USCF Chimera were imported as.pdb files. Molecular docking studies were performed in PyRx version 1.14 using AutoDock Vina web service by the National Biomedical Computation Resources [23]. The docking grid was created with the help of coordinates found by AutoGridFR. A total of 8 binding modes were generated. Out of these modes, the binding mode with the best docking score was saved as a.pdb file and was used for further evaluation. This ligand-receptor complex was then minimized again using the same parameters used to prepare the receptor. The interactions formed between the receptors and the ligands were visualized using Discovery Studio Visualizer v.20.
2.6. Mutagenesis analysis
To understand the importance of amino acids composing the binding pocket, the active site was mutated and the polyphenols were re-docked to the mutated protein structures. The three-dimensional structure was visualized using PyMOL and point mutations at amino acids in the active sites were carried out using the mutagenesis wizard [24,25]. The stability of the mutant protein structure was evaluated using the DynaMut web server [26]. The ligands were docked with these mutated protein structures using the procedure described earlier for the wild protein structures.
2.7. Molecular dynamics simulation
To validate the docking results, molecular dynamics simulation of the complex was performed. The docked protein-ligand complex was simulated using the Nanoscale Molecular Dynamics Simulation (NAMD) software. The best-docked pose of the ligand was used to generate ligand topology files with the help of the CHARMM-GUI web-server using the input generator for the NAMD feature [[27], [28], [29]]. The server employs the CHARMM force field to the ligand. The ligand file was then imported into Visual Molecular Dynamics (VMD), a software supporting molecular visualization. The protein structure file was build using the automatic psf builder feature of VMD. The ligand and protein files were prepared using a VMD script to form a single system. The system was solvated in VMD using the solvate feature. The system was simulated for 1.2 ns at 310 K in the NPT ensemble.
2.8. Pharmacokinetic properties of tea polyphenols
The pharmacokinetic properties were evaluated using the Swiss ADME and the pkCSM web server [30,31]. The entire methodology is summarized in Fig. 3 .
Fig. 3.
Flowchart depicting the process of in-silico study.
3. Results and discussion
3.1. Binding sites prediction
The amino acid residues composition of the binding sites and their grid coordinates for the receptors is summarized in Table S1 in the supplementary information. The binding site for 3CLpro was chosen based on the native ligand N3 which is established as a potential inhibitor of the protein. For all other receptors, there are no reported inhibitors and hence the docking site is not reported in the literature. The binding site with the highest active site score as reported by AutoGridFR was chosen for molecular docking.
3.2. Docking results of tea polyphenols and other drugs
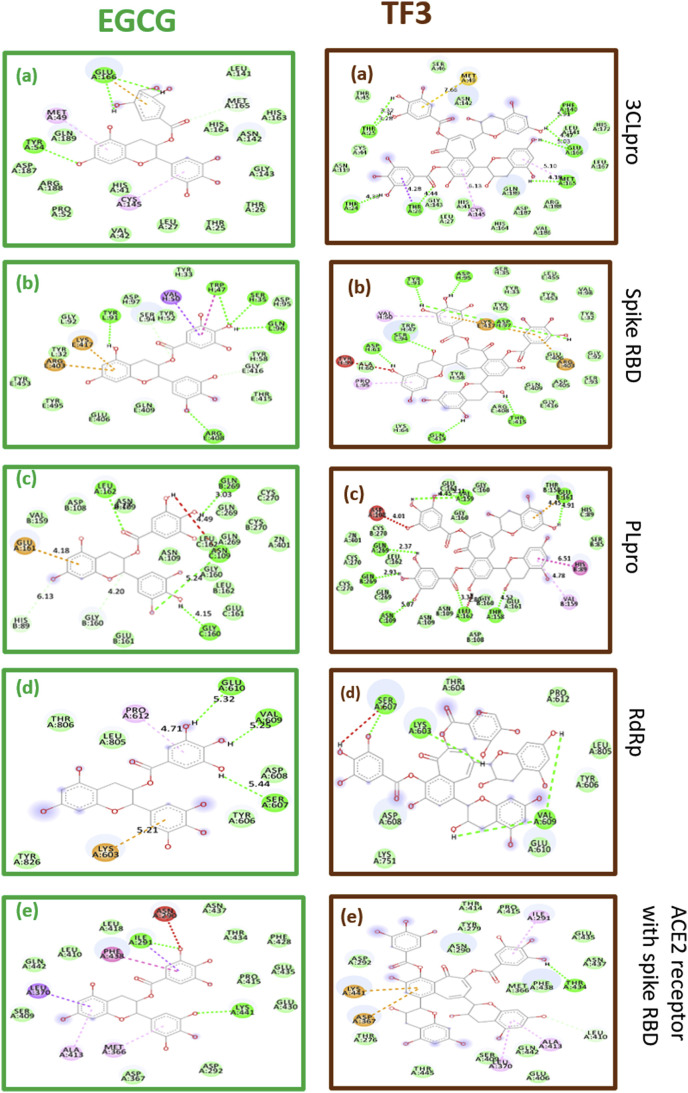
As observed from Table 1 , the docking scores for both the tea polyphenols with different receptors of SARS-CoV-2 are encouraging. The reported docking score for N3, the native ligand of 3CLpro is −7.0 kcal/mol [10], whereas the observed scores for tea polyphenols were −8.3 kcal/mol and −8.4 kcal/mol for EGCG and TF3 respectively. For the 3CLpro receptor, EGCG and TF3 exhibited a combination of hydrogen bonding, van der Waals and other hydrophobic interactions in the docked complex. EGCG formed 2 hydrogen bonds and a pi-anion bond with GLU166 residue of the A chain. GLU residues are generally functionally important in isomerases [32]. The same residue was found to form 2 hydrogen bonds with galloyl moieties of TF3. MET49 also was observed to form Pi-alkyl bond with EGCG and a Pi-sulphur bond with TF3. CYS145, which is usually important in transferases, was observed to form Pi-alkyl bonds with both the tea polyphenols. Unsurprisingly, these amino acid residues are also reported to form interactions with N3 in various reports [[33], [34], [35]]. Apart from them, a lot of other residues were observed to be forming van der Waals interactions with the polyphenols as mentioned in table no 2.
Table 1.
Docking results with SARS-CoV-2 receptors.
| Sr. no | Receptor with PDB IDs | Ligand | Docking score (kcal/mol) | No of H-bonds | Amino acid residues forming H-bond with their length in Å |
|---|---|---|---|---|---|
| 1 | 3CLpro (6LU7) | EGCG | −8.3 | 3 | GLU166 (3.86, 3.33), TYR54 (6.79) |
| TF3 | −8.4 | 8 | THR25 (3.32, 3.28), PHE140 (5.91, 4.47), GLU166 (5.03), THR26 (4.44), THR24 (4.33) | ||
| 2 | Spike RBD (6XE1) | EGCG | −9.7 | 5 | TYR91 (4.76), ARG408 (4.49), GLN96 (4.87), SER35 (5.19), TRP47 (5.26) |
| TF3 | −11.6 | 8 | TYR91 (4.65), ASP95 (3.84), ASP97 (6.45, 5.05), THR415 (4.71), GLN414 (4.22), ASP61 (2.93), SER94 (4.22) | ||
| 3 | PLpro (6W9C) | EGCG | −8.9 | 4 | LEU162 (3.27), B:GLN269 (3.03), ASN109 (5.24), GLY160 (4.15) |
| TF3 | −11.3 | 8 | A:GLN269 (2.37), B:GLN269 (293), ASN109 (5.07), LEU162 (3.32), THR158 (4.52), GLU161 (4.91), VAL159 (4.45, 5.11) | ||
| 4 | RdRp (6M71) | EGCG | −5.7 | 3 | GLU610 (5.32), VAL609 (5.25), SER607 (5.44) |
| TF3 | −6.0 | 4 | SER607 (4.40), LYS603 (3.43), VAL609 (4.90, 4.75) | ||
| Remdesivir | −5.0 | 2 | LYS603(4.36), GLU610 (4.74) | ||
| Favipiravir | −4.8 | 3 | ARG750 (3.86, 4.63), ASP608 (4.39) | ||
| 5 | ACE2 receptor with spike RBD (6M0J) | EGCG | −8.5 | 2 | LYS441 (5.36), ILE291 (4.17) |
| TF3 | −8.0 | 1 | THR434 (4.09) |
Table 2.
Interacting residues of SARS-CoV-2 receptors with tea polyphenols.
| Sr. no | Receptor with PDB IDs | Ligand | Interacting residues |
|
|---|---|---|---|---|
| Polar Interactions | Hydrophobic interactions | |||
| 1 | 3CLpro | EGCG | GLN189, ASP187, ARG188, PRO52, HIS41, VAL42, LEU27, THR25, THR26, GLY142, ASN142, HIS164, HIS163, MET165, LEU141 | MET49, CYS145, GLU166 |
| TF3 | THR45, SER46, ASN142, LEU141, HIS172, LEU167, ARG188, GLN189, ASP187, VAL186, HIS164, HIS41, GLY143, LEU27, ASN119, CYS44 | MET49, THR26, CYS145, MET165 | ||
| 2 | Spike RBD | EGCG | GLY92, TYR32, TYR453, TYR495, GLU406, GLN409, THR415, TYR58, ASP95, TYR33, ASP97, TYR52 | ARG403, LYS417, VAL50, TRP47, GLY416, SER94 |
| TF3 | SER35, LEU455, TYR33. TYR52, TYR453, VAL98, TYR32, GLY92, GLU406, SER93, ASP405, GLN409, GLY416, ARG408, LYS64, TYR58, ALA60, TRP47 | LYS417, ARG403, VAL50 | ||
| 3 | PLpro | EGCG | VAL159, ASP108, ASN109, B:CYS270, C:CYS270, GLN269, LEU162, ASN109, GLN269, GLY160, LEU162, GLU161, GLU161, | GLU161, GLY160, HIS89 |
| TF3 | CYS270, CYS270, GLN269, LEU162, CYS270, A:GLY160, C:GLY160, GLU161, THR158, HIS89, SER85, GLU161, GLY160, ASP108, ASN109, A:ASN109, B:ASN109, GLN269 | HIS89, VAL159, GLU161 | ||
| 4 | RdRp | EGCG | THR806, LEU805, TYR826, TYR606, ASP608 | PRO612, LYS603 |
| TF3 | THR604, PRO612, LEU805, TYR606, GLU610, LYS751, ASP608 | - | ||
| Remdesivir | TYR828, PRO612, VAL609, SER607, ASP608 | TYR606, LEU805, LYS603 | ||
| Favipiravir | SER754, SER607, THR604, LYS751 | ARG750, ASP608 | ||
| 5 | ACE2 receptor with spike RBD | EGCG | LEU418, LEU410, GLN442, SER409, ASP367, ASP292, GLU430, PRO415, GLU435, PHE428, THR434, ASN437 | PHE438, LEU370, ALA413, MET366, ILE291 |
| TF3 | THR414, PRO415, TYR279, ASN290, ASP292, THR276, THR445, SER409, GLU406, GLN442, MET336, PHE438, ASN437, DLU4435 | LYS441, ASP367, ILE291, ALA413, LEU370 | ||
The spike RBD is a possible druggable target due to its role in viral attachment. The tea polyphenols showed significantly high docking scores of −9.7 kcal/mol and −11.6 kcal/mol. EGCG was observed to form 5 and TF3 was found to form as many as 8 hydrogen bonds. TYR91 amino acid residue of the L chain was observed to form hydrogen bonds with both the polyphenols. Pi-cation bonds were formed by residues LYS417 and ARG403 of the E chain with EGCG and TF3, VAL50 was another amino acid residue which was seen interaction with both polyphenols. It formed a Pi-Sigma bond with EGCG and a Pi-alkyl bond with TF3.
EGCG and TF3 showed docking scores of −8.9 kcal/mol and −11.3 kcal/mol with the PLpro. 3 amino acid residues, namely LEU162 from chain A, GLN269 of chain B and ASN109 of chain C, were observed to form hydrogen bonds with both the polyphenols. GLU161 of chain B formed a hydrogen bond with TF3 whereas that of chain A formed a Pi-anion bond with EGCG. Several amino acid residues were observed to form van der Waals and hydrophobic interactions with both the polyphenols.
RdRp inhibitor drugs like Remdesivir and Favipiravir are some of the antiviral drugs being repurposed and are currently in clinical trials [36]. Remdesivir has also been approved by the US FDA for emergency use. A study of how the tea polyphenols performed as compared to these drugs is interesting. With RdRp, the docking scores of EGCG and TF3 were better than both the antiviral drugs. EGCG formed 3 hydrogen bonds with GLU610, VAL609 and SER607 of the chain A. GLU106 was also observed to form a hydrogen bond with Remdesivir whereas SER607 formed a hydrogen bond with TF3. A graphical representation of the docking score is added in the supplementary section (Fig. S1). The interactions formed by tea polyphenols, especially EGCG were better than those formed by Remdesivir and Favipiravir as observed from the interaction diagrams in Fig. 4 and Fig. 5 .
Fig. 4.
Docking interactions of EGCG and TF3 with druggable targets of SARS-CoV-2. The first column represents EGCG and the second column represents TF3. The rows present: a) 3Clpro b) spike RBD 3) PLpro 4) RdRp 5) ACE2 receptor with spike RBD. The dotted green line indicates hydrogen bonding, amino acids in green colour indicate can der Waals interactions, orange dotted line represents pi-anion or pi-cation interaction, the pink dotted line represents a pi-alkyl bond, the purple dotted line indicates pi-sigma or pi-pi bond, yellow dotted line represent a pi-sulphur bond, a cyan dotted line indicates a halogen bond and red dotted line represents unfavourable interaction.
Fig. 5.
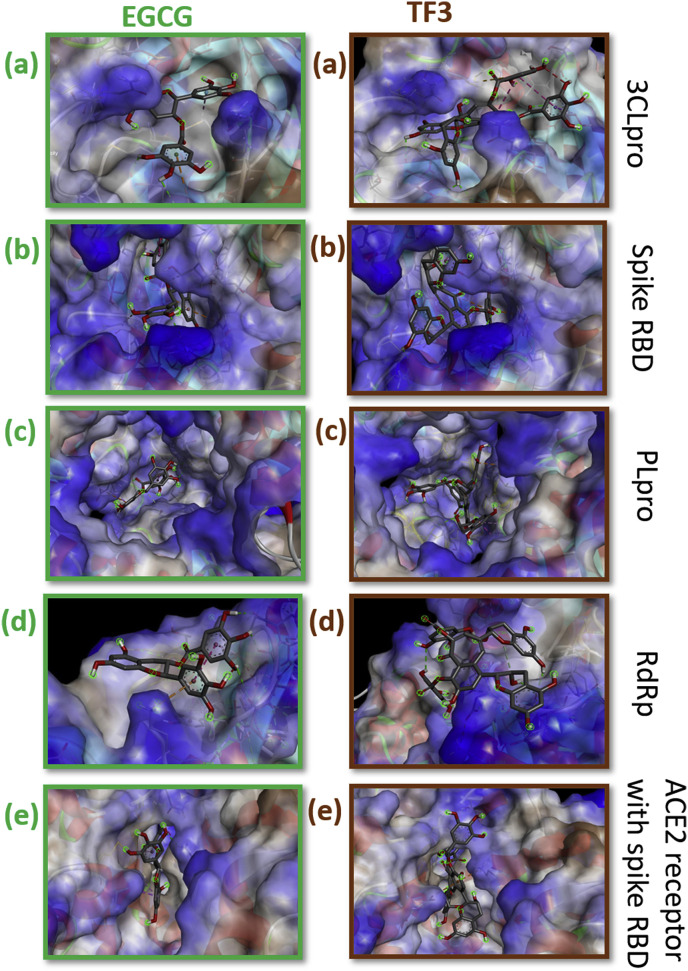
3-D docked poses of EGCG and TF3 with druggable targets of SARS-CoV-2. The first column represents EGCG and the second column represents TF3. The rows present: a) 3Clpro b) spike RBD 3) PLpro 4) RdRp 5) ACE2 receptor with spike RBD. The dotted green line indicates hydrogen bonding, amino acids in green colour indicate can der Waals interactions, orange dotted line represents pi-anion or pi-cation interaction, the pink dotted line represents a pi-alkyl bond, the purple dotted line indicates pi-sigma or pi-pi bond, yellow dotted line represent a pi-sulphur bond, a cyan dotted line indicates a halogen bond and red dotted line represents unfavourable interaction.
The polyphenols also showed a considerable binding affinity towards complex formed at the viral entry, consisting of ACE2 receptor and the spike RBD. ILE291 residue formed a hydrogen bond with EGCG and Pi-alkyl bond with TF3. LEU370 residue formed Pi-Sigma bond with EGCG and a Pi-alkyl bond with TF3. Several residues were involved in van der Waals interactions as mentioned in Table 2 .
For all the receptors studied, both the polyphenols exhibited good docking scores with various types of interactions. Except for RdRp, the docking scores of EGCG and TF3 were consistently higher than −8.0 kcal/mol, in some cases even higher than −11 kcal/mol which is a good indicator. The interactions observed are a mix of hydrogen bonding, van der Waals interactions, Pi-Pi stacking, Pi-alkyl bonds and Pi-sigma bond with multiple amino acid residues interacting with the polyphenols and are represented in Fig. 4. For RdRp, although the absolute scores of EGCG and TF3 were −5.7 kcal/mol and −6 kcal/mol respectively, these scores were better than those for Remdesivir and Favipiravir, which are RNA polymerase inhibitors already in clinical trials for repurposing in COVID-19. Both the polyphenols also showed a better binding affinity toward the receptors than HCQ. The broad-spectrum activity of naturally derived phytoconstituents is evident from the results in the present study.
3.3. Mutagenesis analysis
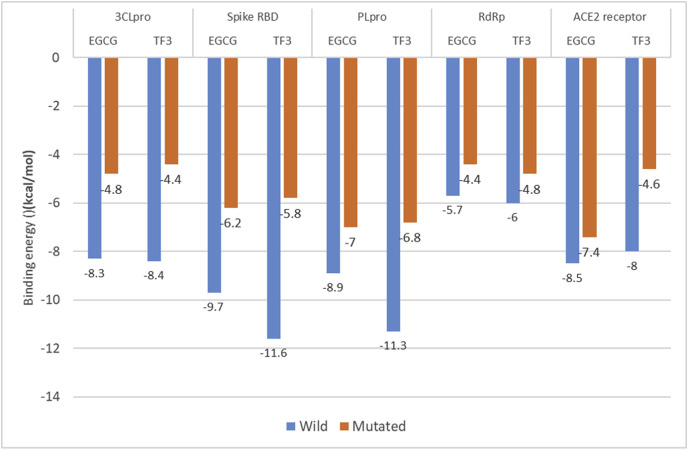
The amino acids that form the binding pocket and participate in interactions are crucial in terms of the binding affinity of the tea polyphenols. When these amino acids were mutated, the protein stability changed. Protein structure stability prediction results showed that the mutations were destabilizing with a marked shift in the Δ(ΔG) value between the wild and mutated protein structures. On docking the tea polyphenols with mutated proteins, the docking score was significantly compromised. This could be because the polyphenols could not form interactions with the mutated active site as formed with wild structures. This also explains the significance of the amino acids comprising the wild-type active site in the activity of tea polyphenols. Moreover, the tea polyphenols also exhibit a targeted activity towards the amino acids and any alteration can weaken the interactions reducing the binding affinity. The following figure no. 6 shows the variation between the docking scores.
Fig. 6.
Comparison of docking scores of wild and mutated structures.
3.4. Molecular dynamics simulation
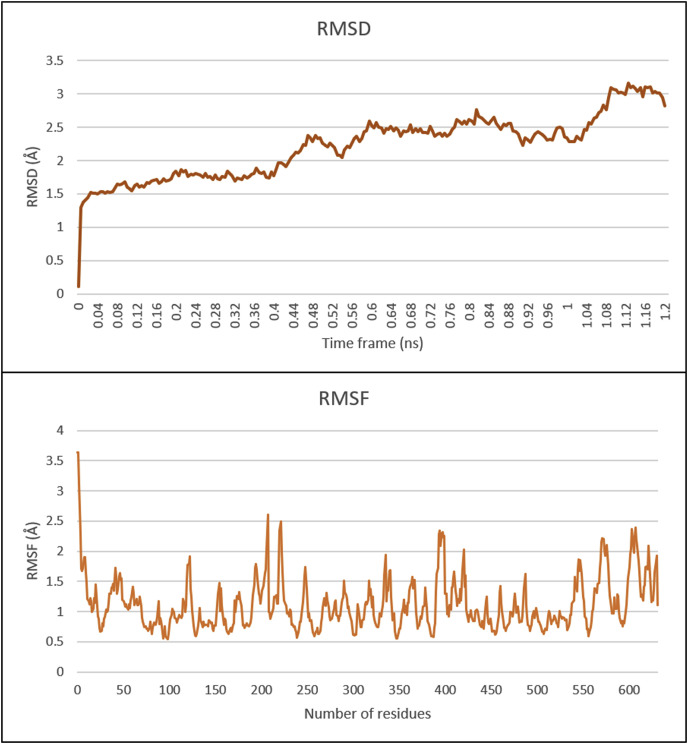
To validate the docking scores, the best docking model was simulated. The complex of TF3 with spike RBD had the highest docking score (−11.6 kcal/mol) and multiple amino acid residues were observed to interact with TF3. The protein-ligand complex was simulated for 1.2 ns under NTP ensemble. RMSD and RMSF calculations were performed to analyse the trajectory generated after simulation and represented in Fig. 7 . The RMSD of the protein-ligand complex attained a value of 1.5 Å at 40 ps and a constant value till 0.4 ns with an average of 1.72 Å. The average RMSD value after 0.4 ns until 1 ns was 2.38 Å remaining constant in that time frame. There was an increase in the RMSD to 3.1 Å after 1 ns but dropped down again to 2.8 Å at the end of the simulation. Calculations of RMSF of the backbone atoms to check the mobility of residues to allow conformational changes on forming the complex ligand were also performed. The average RMSF for the TF3-spike RBD complex was 1.11 Å and the fluctuations were not high, contained in the range of 0.5–2.5 Å.
Fig. 7.
RMSD And RMSF analysis of docked complex of TF3 and spike RBD.
On basis of RMSD and RMSF analyses, it can be concluded that the docked protein-ligand complex of TF3 with spike RBD was stable and the docking result could be validated.
3.5. Pharmacokinetic properties analysis
The physicochemical properties of these molecules were studied and summarized in Table 3 .
Table 3.
Physicochemical properties of tea polyphenols and the drugs evaluated in this study.
| Ligand | EGCG | TF3 |
|---|---|---|
| Molecular weight | 458.37 | 868.8 |
| Lipinski Rule Violations | 2 | 3 |
| Intestinal Absorption (%) | 58.48 | 43.2 |
| BBB permeability (log BB) | −2.238 | −3.559 |
| Fraction unbounded (Fu) | 0.314 | 0.388 |
| Total clearance (log mL/min/kg) | 0.51 | 0.357 |
| Hepatotoxicity | No | No |
| Oral rat acute toxicity (mol/kg) | 2.634 | 2.482 |
| Oral rat chronic toxicity (log mg/kg of bw/day) | 4.077 | 9.52 |
The number of hydrogen bond donors and acceptors is high in both the polyphenols violating the Lipinski's rule of 5. TF3 also exceeds the ideal molecular weight of less than 500. The tea polyphenols need structural optimization to increase the drug likeliness to be considered as strong drug candidates. However, the absorption of these polyphenols is acceptably high, and these molecules also do not pass the Blood-brain barrier. These polyphenols show no hepatotoxicity. The LD50 values of EGCG and TF3 are 2.634 and 2.482 respectively. The clearance of these polyphenols is also faster with a rate of 0.51 and 0.357 log mL/min/kg for EGCG and TF3 respectively.
4. Conclusions and future perspective
In the present study, we performed molecular docking studies of EGCG and TF3 with possible active binding sites of SARS-CoV-2. The docking scores and the interactions of tea polyphenols with these receptors are a good basis to further explore the application of these natural phytochemicals in COVID-19. Tea polyphenols showed good docking scores with all the receptors of SARS-CoV-2 studied here. The polyphenols also showed better docking scores than Remdesivir and Favipiravir when docked with RdRp. MD simulation studies also confirmed the stability of the TF3-spike RBD complex on basis of RMSD and RMSF values. These results suggest the potential activity of tea polyphenols in the treatment of COVID-19. The use of tea phytoconstituents over synthetic drugs is an exciting treatment option since they are safer, and a higher dose is feasible. However, the bulkiness of these polyphenols and violations of Lipinski's rules could be hindrances in their development. Oral bioavailability is a concern for these polyphenols for which to overcome their derivatives are being developed. EGCG derivatives are being tested for enhancement in physicochemical properties and better efficacy.
Among the derivatives, esters derivatives like palmitates and stearates have shown better antiviral activity [37,38]. These derivatives have to be further explored in the application of COVID-19. The in-silico results presented in the study should provide a lead in evaluating the broad-spectrum antiviral activity of the tea polyphenols in the treatment of COVID-19.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author contributions
Mr. Susmit Mhatre has performed all the docking studies. He analysed and interpreted the results. He has written the original manuscript and made all the tables and figures. Mr. Shivraj Naik designed the manuscript and revised the final draft. Dr. Vandana Patravale planned the manuscript as well as administered and supervised the entire work.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Molecular graphics and analyses were performed with UCSF Chimera, developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, with support from NIH P41-GM103311. We would also like to thank Dr. Sreeranjini Pulakkat for her efforts in revising the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.compbiomed.2020.104137.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Hilgenfeld R. From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J. 2014;281:4085–4096. doi: 10.1111/febs.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Du L., He Y., Zhou Y., Liu S., Zheng B., Jiang S. The spike protein of SARS-CoV - a target for vaccine and therapeutic development. Nat. Rev. Microbiol. 2009;7:226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harcourt B.H., Jukneliene D., Kanjanahaluethai A., Bechill J., Severson K.M., Smith C.M., Rota P.A., Baker S.C. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J. Virol. 2004;78:13600–13612. doi: 10.1128/JVI.78.24.13600-13612.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Azeez S.A., Alhashim Z.G., Al Otaibi W.M., Alsuwat H.S., Ibrahim A.M., Almandil N.B., Borgio J.F. State-of-the-art tools to identify druggable protein ligand of SARS-CoV-2. Arch. Med. Sci. 2020;16:497–507. doi: 10.5114/aoms.2020.94046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borgio J.F., Alsuwat H.S., Al Otaibi W.M., Ibrahim A.M., Almandil N.B., Al Asoom L.I., Salahuddin M., Kamaraj B., AbdulAzeez S. State-of-the-art tools unveil potent drug targets amongst clinically approved drugs to inhibit helicase in SARS-CoV-2. Arch. Med. Sci. 2020;16:508–518. doi: 10.5114/aoms.2020.94567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abbas A., Abdelsamea M.M., Gaber M.M. Classification of COVID-19 in chest X-ray images using DeTraC deep convolutional neural network. Appl. Intell. 2020 doi: 10.1007/s10489-020-01829-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang C.S., Landau J.M. Effects of tea consumption on nutrition and health. J. Nutr. 2000;130:2409–2412. doi: 10.1093/jn/130.10.2409. [DOI] [PubMed] [Google Scholar]
- 8.Chacko S.M., Thambi P.T., Kuttan R., Nishigaki I. Beneficial effects of green tea: a literature review. Chin. Med. 2010;5:13. doi: 10.1186/1749-8546-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mhatre S., Srivastava T., Naik S., Patravale V. Antiviral activity of green tea and black tea polyphenols in prophylaxis and treatment of COVID-19: a review. Phytomedicine. 2020:153286. doi: 10.1016/j.phymed.2020.153286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghosh R., Chakraborty A., Biswas A., Chowdhuri S. Evaluation of green tea polyphenols as novel corona virus (SARS CoV-2) main protease (Mpro) inhibitors – an in silico docking and molecular dynamics simulation study. J. Biomol. Struct. Dyn. 2020:1–13. doi: 10.1080/07391102.2020.1779818. 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Menegazzi M., Campagnari R., Bertoldi M., Crupi R., Di Paola R., Cuzzocrea S. Protective effect of epigallocatechin-3-gallate (EGCG) in diseases with uncontrolled immune activation: could such a scenario Be helpful to counteract COVID-19? Int. J. Mol. Sci. 2020;21:5171. doi: 10.3390/ijms21145171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adem S., Eyupoglu V., Sarfraz I., Rasul A. Identification of potent COVID-19 main protease (mpro) inhibitors from natural polyphenols: an in silico strategy unveils a hope against CORONA. 2020. [DOI] [PMC free article] [PubMed]
- 13.Bhardwaj V.K., Singh R., Sharma J., Rajendran V., Purohit R., Kumar S. Identification of bioactive molecules from tea plant as SARS-CoV-2 main protease inhibitors. J. Biomol. Struct. Dyn. 2020:1–10. doi: 10.1080/07391102.2020.1766572. 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song J.-M., Lee K.-H., Seong B.-L. Antiviral effect of catechins in green tea on influenza virus. Antivir. Res. 2005;68:66–74. doi: 10.1016/j.antiviral.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 15.Imbert I., Guillemot J.-C., Bourhis J.-M., Bussetta C., Coutard B., Egloff M.-P., Ferron F., Gorbalenya A.E., Canard B. A second, non-canonical RNA-dependent RNA polymerase in SARS coronavirus. EMBO J. 2006;25:4933–4942. doi: 10.1038/sj.emboj.7601368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu J., Webster D., Cao J., Shao A. The safety of green tea and green tea extract consumption in adults – results of a systematic review. Regul. Toxicol. Pharmacol. 2018;95:412–433. doi: 10.1016/j.yrtph.2018.03.019. [DOI] [PubMed] [Google Scholar]
- 17.Frank K., Patel K., Lopez G., Willis B. Theaflavins research analysis. 2019. https://examine.com/supplements/theaflavins/
- 18.Kar K., Mohanta P.K., Popli S.P., Dhawan B.N. Inhibition of passive cutaneous anaphylaxis by compounds of Camellia sinensis. Planta Med. 1981;42:75–78. doi: 10.1055/s-2007-971549. [DOI] [PubMed] [Google Scholar]
- 19.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 20.Shapovalov M.V., Dunbrack R.L. A smoothed backbone-dependent rotamer library for proteins derived from adaptive kernel density estimates and regressions. Structure. 2011;19:844–858. doi: 10.1016/j.str.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J., Wang W., Kollman P.A., Case D.A. Automatic atom type and bond type perception in molecular mechanical calculations. J. Mol. Graph. Model. 2006;25:247–260. doi: 10.1016/j.jmgm.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 22.Ravindranath P.A., Forli S., Goodsell D.S., Olson A.J., Sanner M.F. AutoDockFR: advances in protein-ligand docking with explicitly specified binding site flexibility. PLoS Comput. Biol. 2015;11 doi: 10.1371/journal.pcbi.1004586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trott O., Olson A.J., Vina AutoDock. Improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Janson G., Zhang C., Prado M.G., Paiardini A. PyMod 2.0: improvements in protein sequence-structure analysis and homology modeling within PyMOL. Bioinformatics. 2017;33:444–446. doi: 10.1093/bioinformatics/btw638. [DOI] [PubMed] [Google Scholar]
- 25.Sonawane P., Patel K., Vishwakarma R.K., Singh S., Khan B.M. In Silico mutagenesis and docking studies of active site residues suggest altered substrate specificity and possible physiological role of Cinnamoyl CoA Reductase 1 (Ll-CCRH1) Bioinformation. 2013;9:224–232. doi: 10.6026/97320630009224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodrigues C.H., Pires D.E., Ascher D.B. DynaMut: predicting the impact of mutations on protein conformation, flexibility and stability. Nucleic Acids Res. 2018;46:W350–W355. doi: 10.1093/nar/gky300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jo S., Kim T., Iyer V.G., Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 28.Lee J., Cheng X., Swails J.M., Yeom M.S., Eastman P.K., Lemkul J.A., Wei S., Buckner J., Jeong J.C., Qi Y., Jo S., Pande V.S., Case D.A., Brooks C.L., MacKerell A.D., Klauda J.B., Im W. CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J. Chem. Theor. Comput. 2016;12:405–413. doi: 10.1021/acs.jctc.5b00935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Phillips J.C., Hardy D.J., Maia J.D.C., Stone J.E., Ribeiro J.V., Bernardi R.C., Buch R., Fiorin G., Hénin J., Jiang W., McGreevy R., Melo M.C.R., Radak B.K., Skeel R.D., Singharoy A., Wang Y., Roux B., Aksimentiev A., Luthey-Schulten Z., Kalé L.V., Schulten K., Chipot C., Tajkhorshid E. Scalable molecular dynamics on CPU and GPU architectures with NAMD. J. Chem. Phys. 2020;153 doi: 10.1063/5.0014475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pires D.E.V., Blundell T.L., Ascher D.B. pkCSM: predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J. Med. Chem. 2015;58:4066–4072. doi: 10.1021/acs.jmedchem.5b00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7:42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holliday G.L., Mitchell J.B.O., Thornton J.M. Understanding the functional roles of amino acid residues in enzyme catalysis. J. Mol. Biol. 2009;390:560–577. doi: 10.1016/j.jmb.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 33.Mahanta S., Chowdhury P., Gogoi N., Goswami N., Borah D., Kumar R., Chetia D., Borah P., Buragohain A.K., Gogoi B. Potential anti-viral activity of approved repurposed drug against main protease of SARS-CoV-2: an in silico based approach. J. Biomol. Struct. Dyn. 2020:1–10. doi: 10.1080/07391102.2020.1768902. 0. [DOI] [PubMed] [Google Scholar]
- 34.Huynh T., Wang H., Luan B. In silico exploration of the molecular mechanism of clinically oriented drugs for possibly inhibiting SARS-CoV-2’s main protease. J. Phys. Chem. Lett. 2020;11:4413–4420. doi: 10.1021/acs.jpclett.0c00994. [DOI] [PubMed] [Google Scholar]
- 35.Elmezayen A.D., Al-Obaidi A., Şahin A.T., Yelekçi K. Drug repurposing for coronavirus (COVID-19): in silico screening of known drugs against coronavirus 3CL hydrolase and protease enzymes. J. Biomol. Struct. Dyn. 2020:1–13. doi: 10.1080/07391102.2020.1758791. 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lythgoe M.P., Middleton P. Ongoing clinical trials for the management of the COVID-19 pandemic. Trends Pharmacol. Sci. 2020;41:363–382. doi: 10.1016/j.tips.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hsu S. Compounds derived from epigallocatechin-3-gallate (EGCG) as a novel approach to the prevention of viral infections. Inflamm. Allergy - Drug Targets. 2015;14:13–18. doi: 10.2174/1871528114666151022150122. [DOI] [PubMed] [Google Scholar]
- 38.Zhao M., Jiang J., Zheng R., Pearl H., Dickinson D., Fu B., Hsu S. A proprietary topical preparation containing EGCG-stearate and glycerin with inhibitory effects on herpes simplex virus: case study. Inflamm. Allergy - Drug Targets. 2012;11:364–368. doi: 10.2174/187152812803251033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.