Significance
Stem cells in the adult brain undergo a weeks-long differentiation process to become new neurons. During this process, the stem cell continuously senses differentiation cues, which fluctuate over a wide range of physiological timescales. However, how differentiation can proceed reliably despite these fluctuations is poorly understood. We use light-activatable control of the neurogenic β-catenin signal to probe how adult neurogenesis responds to fluctuations in differentiation cues. We discovered that β-catenin dynamics strongly influence cell-fate decisions. Surprisingly, while a sustained signal resulted in neurogenesis, transient signals induced cell death. Our results may provide an explanation for the cell death that is commonly observed during adult neurogenesis and may also illuminate a quality control mechanism for correcting errors during neurogenesis.
Keywords: neural stem cells, neurogenesis, signaling dynamics, β-catenin
Abstract
Stem cells undergo differentiation in complex and dynamic environments wherein instructive signals fluctuate on various timescales. Thus, cells must be equipped to properly respond to the timing of signals, for example, to distinguish sustained signaling from transient noise. However, how stem cells respond to dynamic variations in differentiation cues is not well characterized. Here, we use optogenetic activation of β-catenin signaling to probe the dynamic responses of differentiating adult neural stem cells (NSCs). We discover that, while elevated, sustained β-catenin activation sequentially promotes proliferation and differentiation, transient β-catenin induces apoptosis. Genetic perturbations revealed that the neurogenic/apoptotic fate switch was mediated through cell-cycle regulation by Growth Arrest and DNA Damage 45 gamma (Gadd45γ). Our results thus reveal a role for β-catenin dynamics in NSC fate decisions and may suggest a role for signal timing to minimize cell-fate errors, analogous to kinetic proofreading of stem-cell differentiation.
Cells have evolved to robustly perform a broad range of functions in response to extracellular regulatory signals. For example, within the brain, adult neural stem cells (NSCs) undergo quiescence, self-renewing divisions, glial or neuronal differentiation, or apoptosis (where the majority of cells die prior to full maturation and integration), and this overall process leads to the addition of new neurons to modulate hippocampal circuitry (1, 2). These cellular behaviors are regulated by a spectrum of signals, including specific growth factors, morphogens, cytokines, juxtacrine cues, and the extracellular matrix (3, 4).
Importantly, the brain and other tissues are dynamic systems in which signals known to instruct stem-cell behavior fluctuate over time, and the Wnt/β-catenin pathway is one such cue that regulates the proliferation and neuronal differentiation of neural stem and progenitor cells in the adult hippocampus (5–7). Various studies indicate Wnt activity can vary over a broad range of timescales in the brain and in other organs. On rapid timescales, neuronal activity can modulate Wnt signaling both in vitro (8) and in vivo (9). On an organismal behavior timescale, running modulates Wnt signaling, and by extension neurogenesis, in the adult hippocampus (9). Moreover, the rhythmic production of somites during mouse spinal cord development is regulated by a clock with 2-h periodicity that involves Wnt signaling (10), and oscillatory Wnt signaling in intestinal crypts regulates cell divisions with 12-h periodicity (11). Finally, on a long timescale, Wnt expression becomes down-regulated with age in hippocampal astrocytes, which parallels the age-associated decline in neurogenesis (12). Despite such clear examples where Wnt agonists and antagonists fluctuate on timescales ranging from seconds to years, we have a poor understanding of how time-varying cues can impact differentiation and of how signal disruptions during the differentiation process can affect cell behavior.
New approaches to dynamically perturb cellular signaling on a broad range of timescales could greatly benefit our understanding of how signaling dynamics impact cellular function. Optogenetics has recently emerged as a field in which light—which can readily be varied in intensity, space, and time—is harnessed to precisely modulate cell-signaling pathways. In this approach, light-sensitive proteins are engineered to interface with specific signaling pathways, and the subsequent introduction of such an optogenetic construct into cells renders the signaling pathway responsive to light (13, 14). We recently engineered such an optogenetic method to activate Wnt/β-catenin signaling via blue-light–induced clustering of Cryptochrome 2 (Cry2) fused to the pathway coreceptor LRP6 (15).
Here, we have harnessed optogenetic control of β-catenin signaling as a discovery approach to elucidate how dynamics of a key regulatory signal can impact stem-cell behavior. We found that, while continuous β-catenin activation in cultured neural stem cells lead to robust neuronal differentiation, pulsatile activation yielded frequency-dependent behaviors, where progressively longer period oscillations led to reduced neuronal differentiation and, surprisingly, increased apoptosis. We identified and functionally validated a specific cell-cycle regulator, Growth Arrest and DNA differentiation gamma (Gadd45γ), as a potentially important target in regulating the cell cycle, cell survival, and neuronal differentiation. These results support a model in which continuous exposure to a signal supports strong stem-cell differentiation, whereas premature loss of that signal may instead lead to apoptosis, revealing that distinct NSC behaviors can be driven by dynamic variation in exposure to a single instructive cue.
Results
Optogenetic β-Catenin Enables Optical Dissection of Neurogenesis in Adult Neural Progenitor Cells.
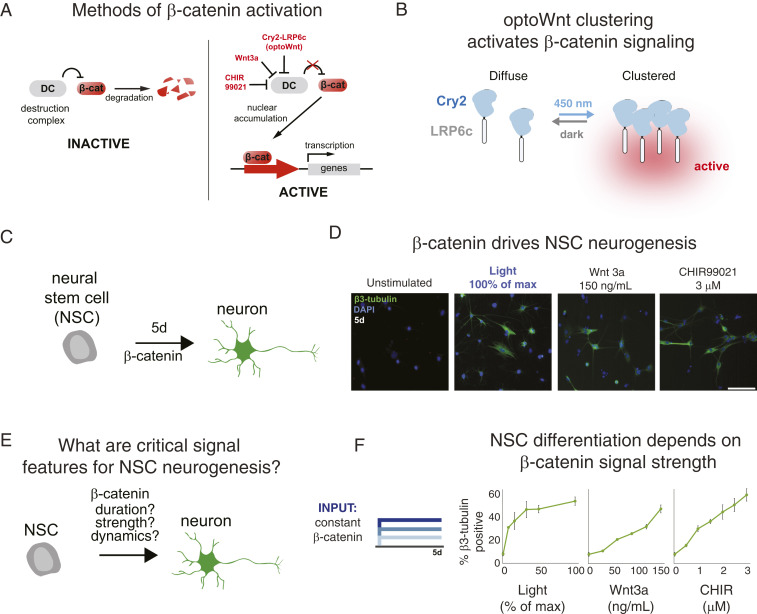
To study how β-catenin signal dynamics drive cellular function, we adapted our previously reported approach (15) (referred to here as optoWnt) to optogenetically manipulate β-catenin activity in cells. In the absence of pathway stimulation, the destruction complex (DC) represses β-catenin through constitutive degradation (Fig. 1A). Our optogenetic Wnt agonist optoWnt is composed of a fusion of the C-terminal domain of the LRP6 receptor (LRP6c) to a truncated Arabidopsis thaliana Cryptochrome 2 (Cry2). Blue-light illumination induces the clustering of Cry2, and the accompanying LRP6c oligomerization [known to be sufficient for Wnt signal transduction (16)] activates downstream β-catenin activity (Fig. 1B) (15). In addition to DC inhibition via LRP6 clustering, the pathway ligand Wnt3a and pharmacological DC inhibition (GSK3β inhibitor CHIR99021, or CHIR) also stabilize β-catenin, resulting in β-catenin accumulation and translocation to the nucleus (Fig. 1A). Previously, we showed that a Cry2-mCherry-LRP6c construct can stimulate β-catenin in NSCs, but only under high-copy-number expression (15). We additionally found here that removal of the fluorescent protein dramatically enhanced light-induced β-catenin activity, enabling strong signal induction from single-copy expression.
Fig. 1.
Optogenetic β-catenin drives neurogenesis in adult hippocampal neural stem cells (NSCs). (A) optoWnt comprises Cry2 fused to the LRP6 endodomain. Blue-light–induced clustering of optoWnt stimulates activation of β-catenin signaling. (B) Analogous to stimulation by Wnt3a or CHIR, optoWnt stimulates β-catenin signaling by inhibiting the destruction complex (DC). DC inhibition stabilizes β-catenin, which acts as a transcriptional activator. (C) NSCs undergo neurogenic differentiation after 5 d of β-catenin activation. (D) Neurogenesis can be achieved using optoWnt, Wnt3a, or CHIR as a pathway inducer. (Scale bar, 500 μm.) (E) Our work examines how signal features such as duration, strength, and dynamics regulate neurogenesis. (F) Increasing signal strength stimulates increased neurogenesis regardless of the method of β-catenin activation. Data represent means ± 1 SE of biological triplicates.
As β-catenin is a strong neurogenic regulator of adult neural stem cells (5), we investigated whether we could control neurogenesis with optoWnt (Fig. 1C). Upon optoWnt expression and illumination over 5 d, we observed strong induction of the neuronal marker β-III-tubulin accompanied by stereotypical morphological changes indicative of neuronal differentiation (Fig. 1D). These high viability cultures could be sustained for at least 15 d, during which neuronal processes continued to elaborate (SI Appendix, Fig. S1). In contrast, cells that expressed optoWnt but received no light stimulation showed minimal staining or morphological changes. Notably, light-induced differentiation was similar in appearance and magnitude to that induced by β-catenin stimulation via either Wnt3a or CHIR addition (Fig. 1D).
Temporal Variation in β-Catenin Directs Distinct Fates in Adult Neural Stem Cells.
Optogenetic control of differentiation offers a tunable strategy to investigate potential dynamic control over cell-fate decisions, and we thus harnessed optoWnt to elucidate how particular features of a differentiation cue—e.g., its strength, dynamics, or duration—may impact cell fate (Fig. 1E). We first examined the effect of signal strength on differentiation and, as anticipated, found that increasing blue-light intensities yielded a monotonic increase in neural stem-cell differentiation when light was applied continuously over 5 d (Fig. 1F). Analogous behavior was found with increased dosing of β-catenin using Wnt3a or CHIR.
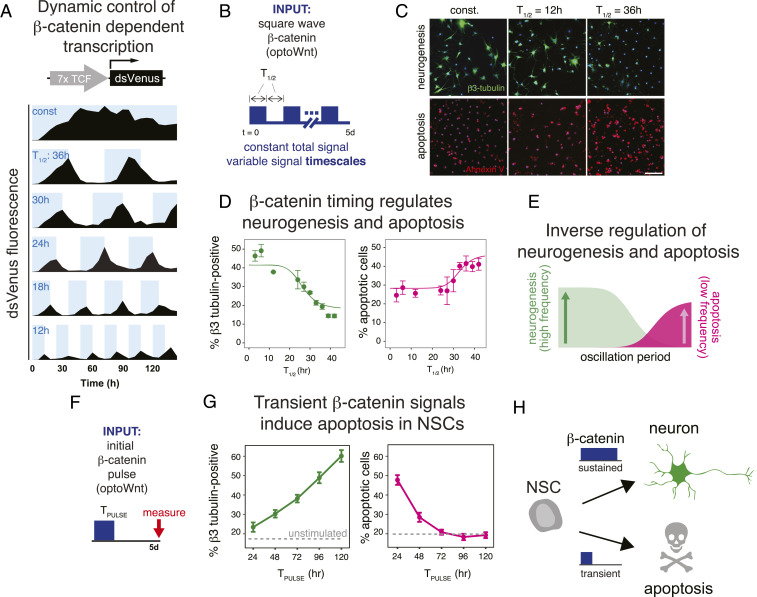
We next asked whether the dynamics of signal presentation could influence cell decisions, analogous to variation in Wnt signaling in a stem-cell niche (5–12). To vary signal timing while holding the total applied signal constant, cells were stimulated with square-wave pulses of varying pulse widths (T1/2) ranging from 1 to 42 h, such that all cells received illumination half of the time but with varying frequencies (Fig. 2 A and B). We first analyzed how square wave light pulses actively drove β-catenin activity within NSCs (Fig. 2A and SI Appendix, Fig. S2) by observing expression of a short half-life fluorescence reporter (17) that we adapted to quantify β-catenin–dependent transcription. Light stimulation and subsequent withdrawal induced the dynamic accumulation followed by degradation of the reporter, where the kinetics of each decay showed no apparent dependence on the prior pulse width or history of activation. Accordingly, the signal decayed to a basal state during the intervals between light activations.
Fig. 2.
β-Catenin signal persistence specifies NSC differentiation or apoptosis. (A) Pulsatile optoWnt stimulation drives dynamic β-catenin–dependent transcription. Blue bars indicate blue-light illumination timing. (B) Square wave stimulation allows interrogation of the role of signal timing while maintaining a constant integrated signal over the duration of the experiment. (C) As square wave T1/2 increases, neurogenesis decreases and apoptosis increases. (Scale bar, 500 μm.) (D) Quantification of neurogenesis and apoptosis in response to all square wave stimuli tested. (E) Data fits from D are overlaid. High-frequency stimulation promoted neurogenesis, while low-frequency stimulation promoted apoptosis. (F) β-Catenin was activated at the beginning of a 5-d differentiation experiment for varying durations (TPULSE), and cell fate was measured at the end of the experiment. (G) Neurogenesis increased monotonically with increasing duration of initial β-catenin signal. In contrast, short-signal durations drove NSCs toward apoptosis, and apoptosis decreased with increasing signal duration. (H) Schematic of results from the experiment depicted in F and G. Data points in D and G represent means ± 1 SE of biological triplicates.
We next examined the effect of dynamic β-catenin pulse trains on differentiation by immunostaining cells for β-III-tubulin after oscillating optoWnt treatment. We observed that, while short β-catenin fluctuations (T1/2 <∼10 h) induced neuronal differentiation at levels similar to continuous stimulation, pulse trains of longer T1/2 duration showed a marked reduction in neurogenesis (Fig. 2 C and D). These results indicated that, despite constant total input (i.e., area under the stimulation curve), differentiation is sensitive to the temporal presentation of β-catenin activity.
To our surprise, cell cultures exposed to the long-timescale fluctuations exhibited morphological features of apoptosis, which we verified via Annexin V staining (Fig. 2 C and D). In particular, short-timescale fluctuations showed little apoptosis above baseline, while square waves of T1/2 >∼20 h induced increasing levels of apoptosis. The inverse relationship between neurogenesis and apoptosis as a function of T1/2 suggested that β-catenin signal timing may drive mutually exclusive cell fates (Fig. 2E).
To better understand the role of β-catenin dynamics in fate choice, we asked whether a simple, initial pulse of β-catenin activity could modulate the neurogenic/apoptotic fate choice. We subjected cells to 5-d stimulation protocols, where an initial optoWnt signal was applied for varying durations, after which the signal was withdrawn for the remainder of the experiment (Fig. 2F). A 24-h pulse of optoWnt yielded a minimal increase in neurogenesis, but with each additional day that optoWnt was applied, a monotonic increase in neurogenesis was observed (Fig. 2 G, Left). Strikingly, while unstimulated cells showed a low level of apoptosis (∼20%), transient 1-d stimulation and withdrawal of optoWnt resulted in almost 50% cell death 4 d after this stimulation (Fig. 2 G, Right). Apoptosis decreased for progressively longer initial optoWnt pulses until reaching near baseline levels for pulses lasting 3 d or more. We obtained qualitatively similar results by repeating this experiment with manual addition of Wnt3a or CHIR, demonstrating that the observed NSC fate responses were not unique to optogenetic stimulation (SI Appendix, Fig. S3).
Notably, CHIR treatment elicited higher levels of both neurogenesis and apoptosis compared to Wnt treatment, which could conceivably be attributed to differences in either the levels or dynamics of induced β-catenin. Collectively, these results show that cell death resulting from dynamic β-catenin is a function of both the initial β-catenin pulse and the duration of signal loss between pulses. Together, our results show that differentiating stem cells can sense and differentially respond to the duration of a β-catenin signal. Sustained signaling is required for differentiation, whereas premature loss of signal diverts differentiating cells toward apoptosis (Fig. 2H).
Gadd45γ Mediates Fate Decisions Driven by β-Catenin Dynamics.
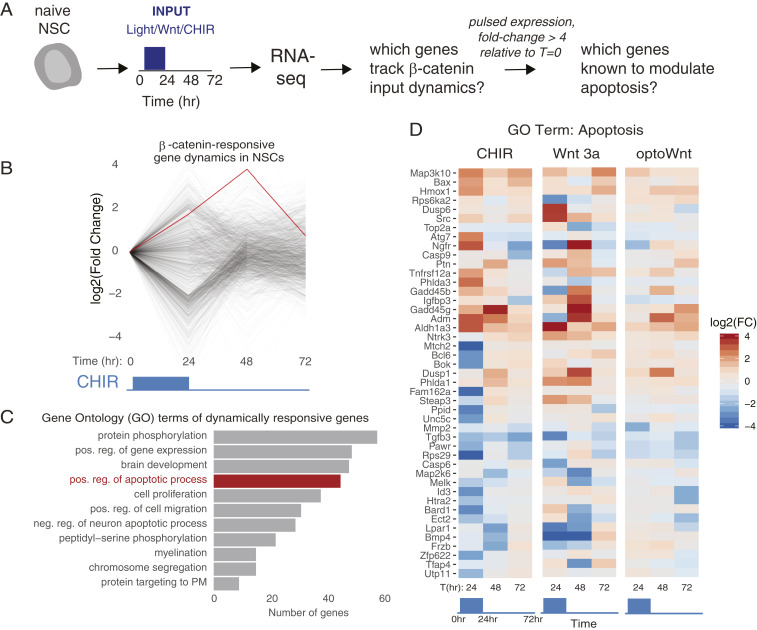
We next investigated the molecular mechanisms by which β-catenin dynamics mediate cell-fate switching. Because β-catenin is a transcriptional regulator and the observed phenotypes appeared on transcriptional timescales, we hypothesized that the apoptotic/neurogenic fate decision resulted from β-catenin–dependent gene regulation. RNA sequencing (RNAseq) analysis was thus performed to identify potential transcriptional targets (Fig. 3A) (18). We exposed NSCs to either no signal, a sustained 72-h signal, or a “withdrawal” signal, whereby cells were stimulated for 24 h with subsequent signal removal. We then harvested and processed cells within each condition at 0, 24, 48, and 72 h after the timing of signal onset. Furthermore, to ensure that results reflected general β-catenin mechanisms, we performed stimulation with all three methods for β-catenin activation (optoWnt, Wnt3a, CHIR). During analysis, we hypothesized that genes that mediated the fate decision between neurogenic or apoptotic phenotypes would follow a dynamic expression pattern either parallel or inverse to β-catenin stimulation, i.e., increasing/decreasing with β-catenin activation and decreasing/increasing upon withdrawal. RNAseq analysis revealed 564 candidate genes that fit this profile (Materials and Methods) (Fig. 3B).
Fig. 3.
Transcriptional profiling reveals genes that track β-catenin input dynamics. (A) Experimental design. (B) Gene expression time courses from genes whose expression changed by more than four-fold in response to a 24-h pulse of β-catenin (from optoWnt, Wnt3a, or CHIR). (C) GO groups that were enriched among the dynamically regulated genes. (D) Genes in the GO group “positive regulation of apoptotic process” were hierarchically clustered based on expression trends over time after a pulse of CHIR treatment. Expression trends for these genes obtained from Wnt3a and optoWnt treatment are also depicted. For further detail, see SI Appendix, Figs. S4 and S5 and Materials and Methods.
We then examined our list of candidate genes for enrichment of functional gene-set modules using Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis (Fig. 3C and SI Appendix, Fig. S4) and found enrichment of several potentially relevant GO categories, including the positive and negative regulation of apoptosis and proliferation (Fig. 3D and SI Appendix, Fig. S5). We searched the literature for known roles for these gene candidates within neurogenesis, neural maintenance, and apoptosis. Notably, growth arrest and DNA-damage–inducible beta (GADD45β) play a role in DNA demethylation during adult neurogenesis (19). However, other members in this family play a role in cell-cycle exit (20), and GADD45γ expression in particular showed strong dynamic regulation in response to multiple stimuli in our RNAseq experiment (Fig. 3D). Thus, GADD45γ was selected for more detailed functional analysis.
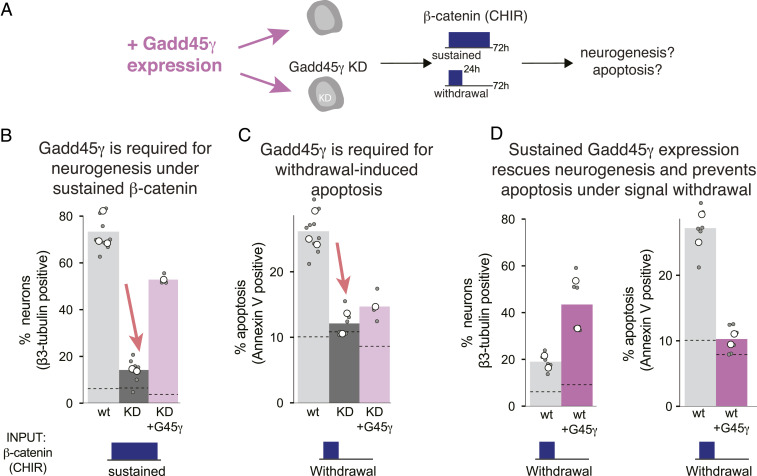
To examine the roles of GADD45γ in fate choice, we disrupted its expression using CRISPR/Cas9 to generate knockdown (KD) NSC lines (SI Appendix, Fig. S6). In addition, GADD45γ was retrovirally overexpressed either in the KD for functional rescue or in wild-type cells (SI Appendix, Fig. S7). We then subjected each cell line to either “sustained” or “withdrawal” β-catenin signal inputs and measured neurogenic or apoptotic cell-fate decisions (Fig. 4A).
Fig. 4.
Gadd45γ mediates β-catenin–dependent fate response in NSCs. (A) The role of Gadd45γ in NSC neurogenesis and apoptosis was determined by probing cell fate within naive NSCs, NSCs engineered for KD of endogenous Gadd45γ, or in either cell line engineered to express exogenous Gadd45γ. Cells were subjected to either sustained or transient β-catenin signaling (3 μm CHIR) and were fixed at 72 h. (B) Loss of neurogenesis under Gadd45γ KD is restored by ectopic Gadd45γ expression. (C) Gadd45γ KD prevents apoptosis that is normally induced by transient β-catenin activation, but apoptosis is only marginally restored with ectopic Gadd45γ expression. (D) Ectopic Gadd45γ expression reduces apoptosis and increases neurogenesis in wild-type (wt) NSCs in response to transient β-catenin stimulation. White circles represent the means of individual experiments. Gray circles represent the means of biological replicates within experiments (each replicate mean obtained from 300 to 3,000 cells). Dashed lines indicate baseline values (no β-catenin stimulus).
We found that Gadd45γ played a strong role in mediating both neuronal differentiation and apoptosis in response to β-catenin dynamics (Fig. 4 and SI Appendix, Fig. S8). First, Gadd45γ was required for neuronal induction. Under sustained CHIR conditions, knockout of Gadd45γ reduced differentiation to baseline levels (Fig. 4B). However, vector-mediated restoration of Gadd45γ expression in the KD NSCs largely rescued the neurogenic response. Gadd45γ was also required to mediate β-catenin withdrawal-induced apoptosis, as Gadd45γ KD NSCs did not show an apoptotic response to β-catenin withdrawal (Fig. 4B). However, the apoptotic phenotype was only partially restored by constitutive overexpression of exogenous Gadd45γ, potentially indicating that dynamic or transient Gadd45γ expression is important for the apoptotic response. Finally, sustained Gadd45γ overexpression in wild-type NSCs reversed the effect of β-catenin withdrawal; i.e., transient β-catenin activation induced strong differentiation and reduced apoptosis to baseline levels (Fig. 4 C and D). Thus, Gadd45γ was necessary to mediate neurogenesis and apoptosis in a manner dependent on β-catenin and Gadd45γ dynamics.
Notably, we performed similar experiments to functionally examine other gene candidates from our RNAseq analysis, including activating transcription factor 3 (ATF3), a protein previously linked with neuronal differentiation, cell-cycle control, and apoptosis. However, both ATF3 knockdown and overexpression had negative effects on progenitor proliferation and apoptosis (SI Appendix, Fig. S9), suggesting that ATF3 likely does not regulate our observed neurogenesis/apoptosis fate decision, so we did not investigate further.
Gadd45γ Blocks the G1/S Transition in NSCs.
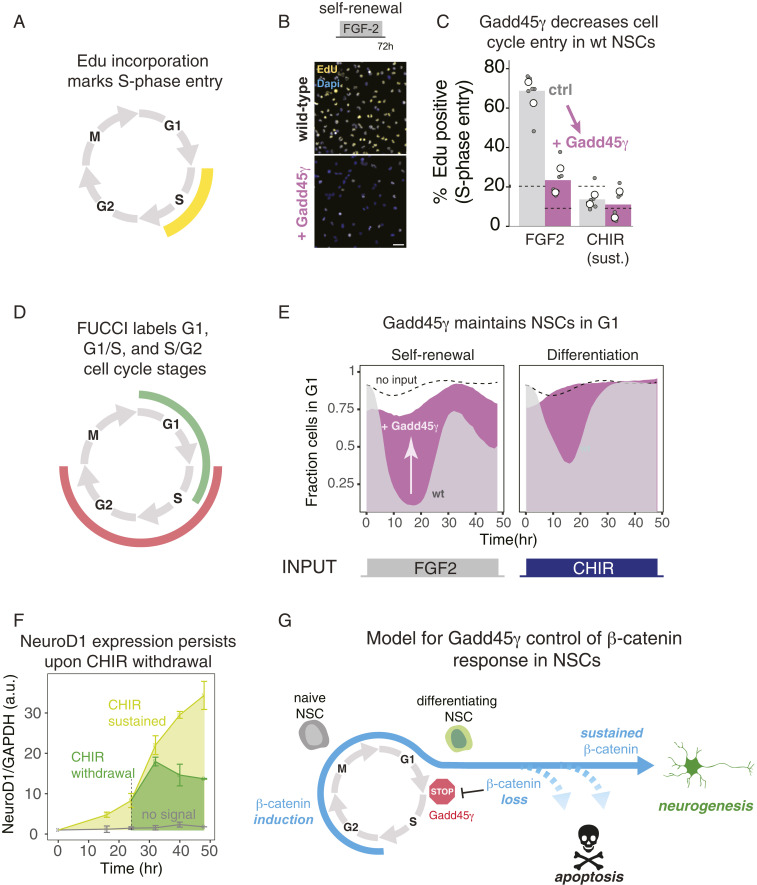
We next asked how Gadd45γ may regulate fate switching during neurogenic differentiation. Because β-catenin can be a mitogenic stimulus, and Gadd45γ is a known cell-cycle regulator, we examined the link between Gadd45γ, cell-cycle regulation, and neurogenesis. Notably, cell-cycle reentry has also been implicated in neuronal apoptosis in neurodegeneration (21–23), further suggesting a link between the cell cycle and the NSC decision to either differentiate or apoptose. Gadd45γ can reportedly block transition between the G1 and S phases of the cell cycle (24–26), and several studies have shown that stem-cell differentiation correlates with the duration of G1 (27–32). Thus, we hypothesized that increased Gadd45γ expression may regulate NSC differentiation by lengthening the G1 phase. To investigate this possibility, we quantified S-phase entry via EdU incorporation by NSCs as a function of Gadd45γ expression (Fig. 5A). As anticipated, wild-type NSCs under self-renewal conditions (20 ng/mL FGF-2) showed strong EdU staining, and S-phase entry was markedly reduced under differentiation (sustained β-catenin) conditions (Fig. 5 B and C). However, Gadd45γ overexpression substantially reduced EdU incorporation under self-renewal conditions, indicating a blockage of cell cycle and confirming Gadd45γ as a strong G1/S checkpoint inhibitor. In contrast, NSCs in which Gadd45γ was knocked down performed similarly to wild-type cells under self-renewal conditions, but showed higher S-phase entry under differentiation conditions, further establishing the role of Gadd45γ in preventing S-phase entry during differentiation (SI Appendix, Fig. S10). Finally, overexpression of Gadd45γ in KD cells restored blockage of the G1/S transition (SI Appendix, Fig. S10).
Fig. 5.
Gadd45γ blocks cell-cycle progression in NSCs. (A) Edu incorporation occurs during DNA synthesis in S phase. (B) Ectopic Gadd45γ expression blocks Edu incorporation and cell-cycle entry in NSCs under both self-renewal conditions (20 ng/mL FGF2) and neurogenic conditions (3 μM CHIR) (B and C). Data in C represent the means of two experiments (white circles), each of which was performed with biological triplicates (gray circles). (Scale bar, 100 μm.) (D) FUCCI reporters are fluorescent indicators that mark a cell’s position in the cell cycle. (E) Live-cell imaging of the FUCCI system in NSCs shows that ectopic Gadd45γ expression blocks cell-cycle entry in response to both self-renewal and neurogenic stimuli. (F) β-Catenin–dependent expression of the early neurogenic factor NeuroD1 persists in NSCs despite withdrawal of the β-catenin stimulus. Data represent quantitation of Western blots (mean ± 1 SE of biological duplicates, see SI Appendix, Fig. S12). (G) Model for how Gadd45γ regulates differential cell-fate control downstream of β-catenin dynamics.
We further examined cell-cycle dynamics using the FUCCI system, a system for visualization of cell-cycle progression that involves expression of fluorescent proteins fused to proteins whose abundance is regulated with the cell-cycle phase (33). With this system, we tracked NSCs transitioning through G1, G1/S, or S/G2/M phases under different experimental conditions in the presence or absence of Gadd45γ overexpression (Fig. 5D). Wild-type NSCs under self-renewal conditions were observed to progress through multiple cell cycles over 48 h, as indicated by the change in the fraction of cells in G1 over time (Fig. 5E). Gadd45γ overexpression markedly reduced the fraction of proliferating cells, consistent with the EdU incorporation data (Fig. 5 B and C).
In wild-type NSCs exposed to sustained β-catenin activation, 50% of cells underwent one round of division before arresting in G1. With Gadd45γ overexpression, however, cells were unable to undergo this round of division, and the initial population of cells not originally in G1 rapidly disappeared, presumably due to completion of the cell cycle and arrest in G1 (Fig. 5D). Cells under withdrawal conditions behaved similarly to differentiating cells, likely because cell-cycle entry occurred during the first 24 h during which the continuous and withdrawal conditions were indistinguishable (SI Appendix, Fig. S11). Collectively, these results definitively establish Gadd45γ as a strong inhibitor of the G1/S transition in NSCs.
Our results suggest a model whereby initial exposure and then premature loss of a differentiation signal may first induce but then release G1/S blockade to initiate cell-cycle reentry, resulting in conflicting cell states resolved via apoptosis. This model predicts that, even under withdrawal conditions, NSCs may continue to express early neurogenic markers despite failing to ultimately differentiate. To test this prediction, we examined the differentiation state of neural stem cells by tracking protein expression of NeuroD1, an early marker and driver of neuronal differentiation (34). Indeed, CHIR withdrawal after 24 h of differentiation resulted in a persistent level of NeuroD1 expression, even after removal of the input stimulus (Fig. 5F and SI Appendix, Fig. S12).
In sum, our results support a model of dynamic regulation of fate decisions, where distinct cell fates are controlled by β-catenin–signaling dynamics (Fig. 5G). Upon β-catenin pathway activation, NSCs complete one round of cell division while up-regulating expression of the G1/S checkpoint inhibitor Gadd45γ. Subsequently, sustained β-catenin and Gadd45γ expression maintain cells in G1 and enable differentiation to occur. However, initially strong induction followed by premature loss of β-catenin removes checkpoint blockade and thereby allows cell-cycle reentry in the differentiating neurons, resulting in conflicting states that cells solve through increased apoptosis.
Discussion
Stem cells are dynamical systems that sense and respond to their environment to promote development or maintain tissue homeostasis. While the signals that regulate important stem-cell programs are known, comparatively less is understood about how the dynamics of signal presentation can regulate cellular behavior. In this report, we used readily controllable optogenetic approaches to investigate stem-cell differentiation and found that the dynamics of β-catenin can regulate distinct cell fates. In the first 24 h, β-catenin can stimulate one round of cell division, after which persistence of the signal induces G1 arrest and neurogenesis, both of which are maintained via Gadd45γ expression. Premature loss of β-catenin, however, can induce apoptosis in differentiating cells, potentially due to inappropriate cell-cycle reentry resulting in part from Gadd45γ loss.
According to our model, the fate of an NSC in response to β-catenin dynamics will depend on 1) the amplitude and duration of β-catenin required to first enter the cell cycle and then to differentiate, 2) the kinetics of apoptosis upon β-catenin withdrawal after cell-cycle entry, and 3) the time window after the first β-catenin signal during which a subsequent signal can rescue the cell from apoptosis. Understanding the signal parameters relevant to each step will provide a high-resolution picture of how dynamic β-catenin signals specify cell fate in differentiating NSCs. We found that Gadd45γ expression alone significantly rescued survival and differentiation even upon Wnt signal withdrawal (Fig. 4D). It will be interesting to examine in future work whether a second pulse of Wnt (alone or in combination with Gadd45γ expression) provides a more complete rescue or whether continuous Wnt stimulation is necessary.
Our results offer insights into the paradoxical observations that Wnt/β-catenin signaling regulates both expansion and differentiation of NSCs (5–7). Previous work in intestinal (35), hair follicle (36), hematopoietic (37), and pluripotent stem cells (38) has shown that the strength and dynamics of β-catenin signals regulate stem-cell-fate decisions. Our data support the model that NSC differentiation requires a strong, sustained β-catenin signal (Fig. 1F) and further suggest that Gadd45γ induction regulates the switch from self-renewal to differentiation at high β-catenin levels, since Gadd45γ knockdown increases proliferation in the presence of high CHIR doses (SI Appendix, Fig. S10). In principle, β-catenin dynamics could also regulate the proliferation/differentiation decision in a regime where short β-catenin pulses activate proliferation but are insufficiently long to stimulate prodifferentiation factors like Gadd45γ. Our data suggest that the duration of such hypothesized pulses must be shorter than 24 h (Fig. 2G), and further work will be required to determine whether such dynamics indeed exist.
While we suspected that signal dynamics may determine cell fate, we were surprised to find that transient β-catenin induced apoptosis. Interestingly, apoptosis has been repeatedly observed as a common feature of neural stem-cell neurogenesis (39–41). Recent intravital imaging of labeled neural stem cells within mouse brains revealed that 60% of stem-cell progeny underwent cell death (39). While the apoptotic mechanisms are poorly understood, apoptosis was prominent within the first 1 to 4 d after cell birth, consistent with results from our in vitro system.
There is a strong precedent for apoptosis occurring upon reinduction of cell-cycle machinery within fully differentiated, postmitotic neurons, both during neuronal development and in adulthood (23, 42). For example, induction of G1 cyclins and cyclin-dependent kinases and subsequent programmed cell death of sympathetic neurons in response to nerve growth factor withdrawal (43), aberrant expression of cell-cycle proteins within neurons in degenerating regions of Alzheimer’s brains (22), and coincidence of cell-cycle reentry and apoptosis upon DNA damage in cultured cortical neurons (21) have been reported. However, to our knowledge, release of cell-cycle arrest and apoptosis in neural stem or progenitor cells at the onset of neuronal fate commitment has not been reported, including within the adult hippocampus. Furthermore, this work establishes a critical role for Gadd45γ—whose analogs protect from apoptosis and down-regulate cell-cycle inhibition to enable neurogenesis within the developing central nervous system in fish (25, 26)—downstream of Wnt signaling in adult neurogenesis. Future work may further investigate how the intersection of two inherently dynamic events—neuronal fate commitment and release of cell-cycle arrest—may contribute to cell death.
While our study focused on the role of Gadd45γ in early NSC decisions to proliferate, differentiate, or apoptose, it is possible that Gadd45γ may also play a separate role in fate specification. A recent study in hematopoietic stem cells (HSCs) found that Gadd45γ not only inhibits proliferation and promotes differentiation, but also biases HSC differentiation toward granulocyte-macrophage progenitors (GMP) at the expense of megakaryocyte-erythroid progenitors (44). Intriguingly, Gadd45γ-expressing HSCs that do not undergo GMP differentiation were more likely to undergo cell death, echoing the findings of our work. However, the extent to which Gadd45γ plays a similar role in adult NSCs is not known.
It is not clear why NSCs have evolved the ability to specify either differentiation or apoptosis through the dynamics of an individual signal. An intriguing possibility is that this regulatory structure could implement a quality control mechanism to safeguard against cells that have not fully differentiated, for example, due to premature loss of the differentiation signal. Such time-based error correction is analogous to kinetic proofreading of cell fate. Kinetic proofreading describes how errors in biomolecular reactions can be minimized through the combination of time delays and rapid removal of reaction errors (45). In the case of neurogenesis, cell-fate behavior analogous to kinetic proofreading may be observed to increase differentiation fidelity through the elimination (apoptosis) of differentiation errors (cells that have not received a sufficient duration of differentiation signal), ensuring that only high-quality neurons (ones receiving sufficiently long differentiation cues) survive the lengthy differentiation process. It will be interesting to explore the prevalence of apparent cell-fate kinetic proofreading across biological systems and to understand its role in organismal homeostasis and in disease.
Materials and Methods
Cell Culture and Viral Packaging.
Cultured rat hippocampal adult NSCs were maintained on polyornithine and laminin (5 μg/mL, Life Technologies) coated polystyrene plates in Dulbecco’s Modified Eagle Medium (DMEM)/F12 (1:1, Life Technologies) high-glucose medium with N-2 supplement (Life Technologies) and stimulated with recombinant human FGF-2 (20 ng/mL, Peprotech). For all differentiation assays, NSCs were cultured in basal medium composed of DMEM/F12 (1:1) high-glucose medium, N-2 supplement, and 0.5% fetal bovine serum (FBS). For viral production, HEK 293Ts were maintained in DMEM, FBS (10%, Life Technologies), and penicillin/streptomycin (1%, Life Technologies). All cells were cultured at 37 °C and 5% CO2. Retroviral and lentiviral production in HEK 293Ts was completed as previously described (46). In short, virus was generated via transient plasmid transfection in HEK 293T cells and was harvested and sterile filtered 48 and 72 h after transfection. Viral particles were purified through ultracentrifugation through a sucrose cushion, resuspended in phosphate-buffered saline, and maintained in −80 °C for long-term storage.
Plasmid Design and Assembly.
The optoWnt design.
OptoWnt was constructed by fusing the LRP6c domain directly to the C terminus of Cry2PHR, but without an intervening mCherry as we previously described (15). This fusion was inserted into the MMLV retroviral CLPIT plasmid backbone between the SfiI and PmeI sites. The CLPIT backbone offers puromycin selection and tetracycline-repressible transgene control.
Gadd45γ and ATF3 overexpression.
pQStrep2-GADD45G was a gift from Konrad Buessow (Helmholz Center for Infection Research, Braunschweig, Germany; Addgene plasmid #31584) (47). pRK-ATF3 was a gift from Yihong Ye (NIH, Bethesda, MD; Addgene plasmid #26115) (48). Full-length human Gadd45γ and ATF3 were introduced individually (CLPIT-hGadd45γ and CLPIT-ATF3) into the CLPIT backbone using Gibson assembly (NEB) between the SfiI and PmeI restriction sites.
CRISPR/Cas9 knockout plasmids.
LentiCRISPRv2 puro was a gift from Brett Stringer (University of Queensland, Brisbane, Australia; Addgene plasmid # 98290) (49). The LentiCRISPRv2 plasmid system was used to build CRISPR/Cas9 knockout vectors specific to rat Gadd45γ. Guide RNAs (gRNAs) specific to the exons of Gadd45γ were designed using Benchling’s CRISPR/Cas9 Guide RNA tool (Benchling biology software retrieved from https://www.benchling.com/). Short annealed gRNAs were ligated into the lentiCRISPRv2 BsmBI site. The lentiCRISPRv2 backbone offers puromycin selection.
FUCCI cell-cycle reporter.
ES-FUCCI was a gift from Pierre Neveu (EMBL, Heidelberg, Germany; Addgene plasmid # 62451) from which the components of the FUCCI plasmids developed for this study were (33). Citrine was fused to the N terminus (1-110) of Gem1 and was introduced into CLPIT using Gibson assembly (NEB) between the SfiI and PmeI restriction sites, maintaining the sites (Citrine-Gem[1-110]). The same technique was used to introduce mCherry fused to the C terminus of hCdt (30-120) into CLPIT (Cherry-hCdt[30-120]).
TdVP reporter.
To quantify β-catenin activity dynamics, an ultra-destabilized Venus fluorescent protein (17) was placed under the control of a 7x TCF promoter.
DNA Transfer: Transfection and Viral Transduction.
Transient transfections of HEK 293Ts for viral production were performed using the calcium phosphate method. Gene transfer into NSCs was performed using viral transduction at a multiplicity of infection (MOI) of 0.1. NSCs were plated in 35-mm polyornithine- and laminin-coated polystyrene plates and transduced with viral particles. Twenty-four hours post infection, puromycin (0.6 μg/mL, Sigma Aldrich) was added for an additional 24 to 48 h to select for positive viral integration at a copy number of ∼1.
Optogenetic and reporter NSCs.
NSCs were transduced with optoWnt for optogenetic assays. For β-catenin dynamics assays, NSCs stably transduced with TdVP were subsequently transduced with optoWnt.
Overexpression and knockdown NSCs.
For overexpression of human Gadd45γ or human ATF3, NSCs were transduced with retroviral vector harboring expression cassettes for hGadd45γ or ATF3. ATF3-transduced cells were puromycin selected. The hGadd45γ-transduced cells were not selected due to the acute antiproliferative and proneurogenic changes associated with hGadd45γ protein expression, but were instead assayed at 24 h post transduction. Protein expression for human hGadd45γ was assessed via Western blot (hGadd45γ, LifeSpan Biosciences) and reached peak expression by 24 h post transduction. To knock down rat Gadd45γ, NSCs were transduced with LentiCRISPRv2 carrying gRNAs to target sites of exon 2 of rat Gadd45γ. Successful gene disruption was measured using the Surveyor Nuclease assay (IDT) and subsequent sequencing of the targeted site. RNA knockdown was assessed with quantitative PCR against the targeted exon site.
FUCCI NSCs.
NSCs were transduced with retroviruses encoding both mCherry h-Cdt and Venus Gem1, each at a MOI of 1. NSCs were puromycin selected for successful transduction. Forty-eight hours post transduction, cells were sorted by fluorescence-activated cell sorting for dual expression of mCherry and Venus, which occurs during the G1/S transition phase of the cell cycle (FUCCI-NSCs). This population was expanded and used for subsequent assays.
Neuronal Differentiation Assay.
NSC differentiation was induced by three methods: the small-molecule GSK3β inhibitor CHIR99021 (Tocris), the Wnt3a ligand (R&D Systems), and the blue-light induction of optoWnt oligomerization. For all assays, optoWnt-transduced NSCs were seeded at ∼20,000 cells per well in polyornithine- and laminin-coated black-walled 96-well plates compatible with microscopy (Greiner µClear, #655087). NSCs were maintained in the “basal” medium (DMEM + N2 supplement and 0.5% FBS) for the duration of the assay. For blue-light stimulation of optoLRP, NSCs seeded in the 96-well plates were placed on custom-built LED illumination devices as described previously (15). In brief, 5-mm blue LEDs were arranged underneath alternating wells. Specific light patterns were controlled by the open-source Arduino electronics platform. Cells were illuminated within a humidified incubator at 37 °C and 5% CO2 for 48 to 144 h.
Post treatment, cells were fixed with 4% paraformaldehyde and immunostained with anti–β3-tubulin (1:1,000, Sigma Aldrich) and DAPI. For apoptotic studies, prior to fixation, cells were stained with Annexin V (ThermoFisher) and imaged before immunostaining. Cell nuclei were counted using CellProfiler (50). Cells that stained positive for either β3-tubulin or Annexin V were counted manually. One-way ANOVA was used to determine statistically relevant differences between experimental conditions.
β-Catenin Activity Fluorescence Reporter Assay.
NSCs were transduced with TdVP retrovirus and optoWnt, each at a MOI of 1 and prepared as described for differentiation assays. β-Catenin activity was induced with blue light, CHIR99021, or Wnt3a ligand. The selected regions of interest (ROIs) within the plate wells were imaged every 3 h over a total of 144 h. Total fluorescence per ROI was quantified with ImageJ (51).
FUCCI System-Based Cell-Cycle Tracking.
Cell cycles in FUCCI-NSCs were monitored over 48 h by fluorescence microscopy. Population cell-cycle changes under various stimuli were tracked over time using image analysis and quantification with MetaMorph (Molecular Devices). Cells were exposed to self-renewal (FGF-2), null (no stimuli), or neurogenic (CHIR99021) stimuli patterns, and all assays were performed in basal medium for cell maintenance.
Microscopy and Image Analysis.
Cells were imaged at 20x on the ImageXpress Micro cellular imaging system (Molecular Devices). Image analysis was performed with MetaExpress (Molecular Devices), CellProfiler, and ImageJ (50).
RNAseq Library Preparation.
NSCs were transduced with optoWnt and treated with either light stimuli, CHIR99021, or Wnt3a ligand for specified stimulation patterns over 0 to 72 h in basal medium. RNA was extracted using RNAeasy kit and protocol (Qiagen). The RNA library was prepared using QuantSeq3 kit (Lexogen) with the low-RNA-input protocol. Sequencing was performed on an Illumina HiSeq in the University of California at Berkeley Vincent Coates Sequencing Facility.
RNAseq, GO, and KEGG Statistical Analysis.
Sequencing reads were aligned using the Bowtie 2 alignment tool (52). Reads were mapped to a Bowtie 2 index built from the Rattus norvegicus Rnor_6.0 reference genome downloaded from the Ensembl database (https://asia.ensembl.org/info/data/ftp/index.html). Alignment was performed using the default preset options in “–local” mode. Aligned reads were counted by processing the output.sam files with the featureCounts (53) software through the Subread package in the R programming language (https://cran.r-project.org/). Within featureCounts, multimapped reads were counted (allowMultiOverlap == TRUE) and were assigned to the feature with the largest number of overlapping bases (largestOverlap == TRUE). Reads were mapped against annotated gene sets from the Rnor_6.0 genome build obtained from the Ensembl database. Feature counts (expressed genes) were analyzed using the edgeR (54) library from the Bioconductor package. Briefly, genes with fewer than 1 count per million in any sample were discarded from analysis. The library sizes of the remaining genes were normalized for each sample using the upper quartile method. Differential gene expression between stimulated cells and unstimulated cells was then calculated using the normalized read counts for each time point of each condition. Genes were filtered for those that exhibited dynamic expression (nonmonotonic) and that had more than four-fold expression change at any given time point. Pathways enriched in the resultant gene sets were identified using GO (55, 56) and KEGG (57) analysis with the DAVID bioinformatics resource (58). Hierarchical clustering was performed by first calculating a distance matrix of gene expression time-course responses to a pulse of CHIR99021. Distance was calculated based on rank using the Spearman method. Clustering was performed using the hclust function with the Ward.D2 algorithm. The resultant gene order from the CHIR99021-stimulated samples was then applied to gene expression data from Wnt3a- and optoWnt-stimulated cells.
Supplementary Material
Acknowledgments
This work was funded by NIH Grants R01NS087253 (D.V.S.) and R35GM138211 (L.J.B.). We thank Sydney Oraskovich for key experimental contributions.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2008509117/-/DCSupplemental.
Data Availability.
RNAseq datasets have been deposited in the Gene Expression Omnibus repository under accession no. GSE155862, and all other data and associated protocols are present in the manuscript (18). Materials are available upon request via a simple material transfer agreement.
References
- 1.Gage F. H., Adult neurogenesis in mammals. Science 364, 827–828 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Kempermann G., et al. , Human adult neurogenesis: Evidence and remaining questions. Cell Stem Cell 23, 25–30 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keung A. J., Kumar S., Schaffer D. V., Presentation counts: Microenvironmental regulation of stem cells by biophysical and material cues. Annu. Rev. Cell Dev. Biol. 26, 533–556 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Discher D. E., Mooney D. J., Zandstra P. W., Growth factors, matrices, and forces combine and control stem cells. Science 324, 1673–1677 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lie D. C., et al. , Wnt signalling regulates adult hippocampal neurogenesis. Nature 437, 1370–1375 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Kalani M. Y., et al. , Wnt-mediated self-renewal of neural stem/progenitor cells. Proc. Natl. Acad. Sci. U.S.A. 105, 16970–16975 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qu Q., et al. , Orphan nuclear receptor TLX activates Wnt/beta-catenin signalling to stimulate neural stem cell proliferation and self-renewal. Nat. Cell Biol. 12, 31–40 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wayman G. A., et al. , Activity-dependent dendritic arborization mediated by CaM-kinase I activation and enhanced CREB-dependent transcription of Wnt-2. Neuron 50, 897–909 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Jang M. H., et al. , Secreted frizzled-related protein 3 regulates activity-dependent adult hippocampal neurogenesis. Cell Stem Cell 12, 215–223 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dequéant M. L., et al. , A complex oscillating network of signaling genes underlies the mouse segmentation clock. Science 314, 1595–1598 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Matsu-Ura T., et al. , Intercellular coupling of the cell cycle and circadian clock in adult stem cell culture. Mol. Cell 64, 900–912 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miranda C. J., et al. , Aging brain microenvironment decreases hippocampal neurogenesis through Wnt-mediated survivin signaling. Aging Cell 11, 542–552 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deisseroth K., Optogenetics: 10 years of microbial opsins in neuroscience. Nat. Neurosci. 18, 1213–1225 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Repina N. A., Rosenbloom A., Mukherjee A., Schaffer D. V., Kane R. S., At light speed: Advances in optogenetic systems for regulating cell signaling and behavior. Annu. Rev. Chem. Biomol. Eng. 8, 13–39 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bugaj L. J., Choksi A. T., Mesuda C. K., Kane R. S., Schaffer D. V., Optogenetic protein clustering and signaling activation in mammalian cells. Nat. Methods 10, 249–252 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Cong F., Schweizer L., Varmus H., Wnt signals across the plasma membrane to activate the beta-catenin pathway by forming oligomers containing its receptors, Frizzled and LRP. Development 131, 5103–5115 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Wang C. C., Bajikar S. S., Jamal L., Atkins K. A., Janes K. A., A time- and matrix-dependent TGFBR3-JUND-KRT5 regulatory circuit in single breast epithelial cells and basal-like premalignancies. Nat. Cell Biol. 16, 345–356 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosenbloom A. B., et al. , Gene Expression Omnibus Entry GSE155862. Gene Expression Omibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE155862. Deposited 7 August 2020.
- 19.Ma D. K., et al. , Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science 323, 1074–1077 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salvador J. M., Brown-Clay J. D., Fornace A. J. Jr, Gadd45 in stress signaling, cell cycle control, and apoptosis. Adv. Exp. Med. Biol. 793, 1–19 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Kruman I. I., et al. , Cell cycle activation linked to neuronal cell death initiated by DNA damage. Neuron 41, 549–561 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Busser J., Geldmacher D. S., Herrup K., Ectopic cell cycle proteins predict the sites of neuronal cell death in Alzheimer’s disease brain. J. Neurosci. 18, 2801–2807 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Konishi Y., Lehtinen M., Donovan N., Bonni A., Cdc2 phosphorylation of BAD links the cell cycle to the cell death machinery. Mol. Cell 9, 1005–1016 (2002). [DOI] [PubMed] [Google Scholar]
- 24.Vairapandi M., Balliet A. G., Hoffman B., Liebermann D. A., GADD45b and GADD45g are cdc2/cyclinB1 kinase inhibitors with a role in S and G2/M cell cycle checkpoints induced by genotoxic stress. J. Cell. Physiol. 192, 327–338 (2002). [DOI] [PubMed] [Google Scholar]
- 25.de la Calle-Mustienes E., Glavic A., Modolell J., Gómez-Skarmeta J. L., Xiro homeoproteins coordinate cell cycle exit and primary neuron formation by upregulating neuronal-fate repressors and downregulating the cell-cycle inhibitor XGadd45-gamma. Mech. Dev. 119, 69–80 (2002). [DOI] [PubMed] [Google Scholar]
- 26.Candal E., Thermes V., Joly J. S., Bourrat F., Medaka as a model system for the characterisation of cell cycle regulators: A functional analysis of ol-Gadd45gamma during early embryogenesis. Mech. Dev. 121, 945–958 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Pauklin S., Vallier L., The cell-cycle state of stem cells determines cell fate propensity. Cell 155, 135–147 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boward B., Wu T., Dalton S., Concise review: Control of cell fate through cell cycle and pluripotency networks. Stem Cells 34, 1427–1436 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dalton S., Linking the cell cycle to cell fate decisions. Trends Cell Biol. 25, 592–600 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Calder A., et al. , Lengthened G1 phase indicates differentiation status in human embryonic stem cells. Stem Cells Dev. 22, 279–295 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Julian L. M., Carpenedo R. L., Rothberg J. L., Stanford W. L., Formula G1: Cell cycle in the driver’s seat of stem cell fate determination. BioEssays 38, 325–332 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Soufi A., Dalton S., Cycling through developmental decisions: How cell cycle dynamics control pluripotency, differentiation and reprogramming. Development 143, 4301–4311 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sladitschek H. L., Neveu P. A., MXS-chaining: A highly efficient cloning platform for imaging and flow cytometry approaches in mammalian systems. PLoS One 10, e0124958 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuwabara T., et al. , Wnt-mediated activation of NeuroD1 and retro-elements during adult neurogenesis. Nat. Neurosci. 12, 1097–1105 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gehart H., Clevers H., Tales from the crypt: New insights into intestinal stem cells. Nat. Rev. Gastroenterol. Hepatol. 16, 19–34 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Choi Y. S., et al. , Distinct functions for Wnt/β-catenin in hair follicle stem cell proliferation and survival and interfollicular epidermal homeostasis. Cell Stem Cell 13, 720–733 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luis T. C., et al. , Canonical wnt signaling regulates hematopoiesis in a dosage-dependent fashion. Cell Stem Cell 9, 345–356 (2011). [DOI] [PubMed] [Google Scholar]
- 38.Marucci L., et al. , β-catenin fluctuates in mouse ESCs and is essential for Nanog-mediated reprogramming of somatic cells to pluripotency. Cell Rep. 8, 1686–1696 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Pilz G. A., et al. , Live imaging of neurogenesis in the adult mouse hippocampus. Science 359, 658–662 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kempermann G., Gast D., Kronenberg G., Yamaguchi M., Gage F. H., Early determination and long-term persistence of adult-generated new neurons in the hippocampus of mice. Development 130, 391–399 (2003). [DOI] [PubMed] [Google Scholar]
- 41.Kuhn H. G., Control of cell survival in adult mammalian neurogenesis. Cold Spring Harb. Perspect. Biol. 7, a018895 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Becker E. B., Bonni A., Cell cycle regulation of neuronal apoptosis in development and disease. Prog. Neurobiol. 72, 1–25 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Freeman R. S., Estus S., Johnson E. M. Jr, Analysis of cell cycle-related gene expression in postmitotic neurons: Selective induction of cyclin D1 during programmed cell death. Neuron 12, 343–355 (1994). [DOI] [PubMed] [Google Scholar]
- 44.Thalheimer F. B., et al. , Cytokine-regulated GADD45G induces differentiation and lineage selection in hematopoietic stem cells. Stem Cell Rep. 3, 34–43 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hopfield J. J., Kinetic proofreading: A new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc. Natl. Acad. Sci. U.S.A. 71, 4135–4139 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ashton R. S., et al. , Astrocytes regulate adult hippocampal neurogenesis through ephrin-B signaling. Nat. Neurosci. 15, 1399–1406 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Büssow K., et al. , Structural genomics of human proteins: Target selection and generation of a public catalogue of expression clones. Microb. Cell Fact. 4, 21 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Q., et al. , ERAD inhibitors integrate ER stress with an epigenetic mechanism to activate BH3-only protein NOXA in cancer cells. Proc. Natl. Acad. Sci. U.S.A. 106, 2200–2205 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stringer B. W., et al. , A reference collection of patient-derived cell line and xenograft models of proneural, classical and mesenchymal glioblastoma. Sci. Rep. 9, 4902 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carpenter A. E., et al. , CellProfiler: Image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7, R100 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schneider C. A., Rasband W. S., Eliceiri K. W., NIH image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liao Y., Smyth G. K., Shi W., featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 54.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ashburner M. et al.; The Gene Ontology Consortium , Gene ontology: Tool for the unification of biology. Nat. Genet. 25, 25–29 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.The Gene Ontology Consortium , Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res. 45, D331–D338 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kanehisa M., Sato Y., Kawashima M., Furumichi M., Tanabe M., KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44, D457–D462 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang W., Sherman B. T., Lempicki R. A., Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNAseq datasets have been deposited in the Gene Expression Omnibus repository under accession no. GSE155862, and all other data and associated protocols are present in the manuscript (18). Materials are available upon request via a simple material transfer agreement.