Significance
Enterotoxigenic Escherichia coli (ETEC), a leading cause of acute watery diarrheal illness in children, have also been linked to nondiarrheal sequelae including malnutrition. ETEC produce heat-labile toxin (LT) to stimulate the production of cellular cAMP, ultimately modulating ion channels to release salt and water into the intestinal lumen, resulting in diarrhea. Given the importance of cAMP as a “second messenger,” we reasoned that cellular features could be altered by LT to benefit the bacteria. Interestingly, we found that LT-mediated increases in cAMP stimulate production of CEACAMs, a family of cell surface glycoproteins, which then serve as receptors for ETEC. Collectively, our findings may have implications for our understanding of toxin-mediated illness as well as sequelae associated with these important pathogens.
Keywords: enterotoxin, CEACAM, diarrhea
Abstract
The enterotoxigenic Escherichia coli (ETEC) are among the most common causes of diarrheal illness and death due to diarrhea among young children in low-/middle-income countries (LMICs). ETEC have also been associated with important sequelae including malnutrition and stunting, placing children at further risk of death from diarrhea and other infections. Our understanding of the molecular pathogenesis of acute diarrheal disease as well as the sequelae linked to ETEC are still evolving. It has long been known that ETEC heat-labile toxin (LT) activates production of cAMP in the cell, signaling the modulation of cellular ion channels that results in a net efflux of salt and water into the intestinal lumen, culminating in watery diarrhea. However, as LT also promotes ETEC adhesion to intestinal epithelial cells, we postulated that increases in cAMP, a critical cellular “second messenger,” may be linked to changes in cellular architecture that favor pathogen–host interactions. Indeed, here we show that ETEC use LT to up-regulate carcinoembryonic antigenrelated cell adhesion molecules (CEACAMs) on the surface of small intestinal epithelia, where they serve as critical bacterial receptors. Moreover, we show that bacteria are specifically recruited to areas of CEACAM expression, in particular CEACAM6, and that deletion of this CEACAM abrogates both bacterial adhesion and toxin delivery. Collectively, these results provide a paradigm for the molecular pathogenesis of ETEC in which the bacteria use toxin to drive up-regulation of cellular targets that enhances subsequent pathogen–host interactions.
The burden of illness caused by enterotoxigenic Escherichia coli (ETEC) is remarkable, with yearly estimates of hundreds of millions of infections worldwide (1). Although the death rate from acute diarrheal illness has declined appreciably in the past few decades (2), ETEC and other enteric pathogens have recently been recognized as a cause of substantial morbidity in children who survive the acute infection. Increasingly, these organisms have been implicated in postdiarrheal sequelae including growth stunting, malnutrition, and cognitive deficiencies, thereby exacting a tremendous global health burden (3).
ETEC were discovered as a cause of severe cholera-like diarrhea more than five decades ago (4–6), and the ETEC pathovar came to be defined as organisms capable of producing heat-labile (LT) and/or heat-stable (ST) enterotoxins. LT is a heterohexameric protein, similar to cholera toxin (CT), that is composed of a pentameric B subunit that engages host cell receptors to promote uptake of the enzymatically active A subunit. Heat-stable toxins are small peptides of 18 or 19 amino acids that bind to guanylate cyclase C. In the canonical paradigm for ETEC pathogenesis, these organisms adhere to the small intestinal mucosa via plasmid-encoded colonization factors, where they deliver their toxin payloads to cognate receptors on intestinal epithelia (7). Toxin-mediated activation of adenylate cyclase by LT and guanylate cyclase by ST result in respective production of host cAMP and cGMP. Intracellular accumulation of these cyclic nucleotides results in activation of protein kinases that in turn modulate ion channels in enterocyte apical membranes, resulting in a net loss of salt and water into the lumen of the small intestine and ensuing watery diarrheal illness (7).
However, more recent studies have suggested that the molecular pathogenesis of these organisms is considerably more complex than had been appreciated (8, 9). One aspect of ETEC virulence which has not been completely explored is the role played by ETEC toxins in mediating pathogen–host interactions. Interestingly, heat-labile toxin has been shown to promote colonization of the small intestine in animal models of ETEC infection (10, 11) and to enhance bacterial adhesion in vitro (12). While the intracellular signaling pathways involved in LT-stimulated adhesion have been explored (13), the precise mechanism by which LT promotes pathogen–host interactions remains undefined.
Interestingly, E. coli pathovars other than ETEC (14, 15) interact with certain carcinoembryonic antigen-related cell adhesion molecules (CEACAMs), surface glycoproteins belonging to a superfamily of immunoglobulin-like cell adhesion molecules encoded on chromosome 19 (16). Each CEACAM possess an amino terminal Ig-like variable domain and varying numbers of domains homologous to Ig constant region (SI Appendix, Fig. S1A). Data to date suggest that chromosomally encoded type 1 fimbriae (pili), common to virtually all E. coli, engage these glycoprotein molecules, including CEA (CD66e, CEACAM5), as well as NCA (CD66c, CEACAM6) (17–19), through specific lectin-mediated interactions with mannosylated residues (20, 21). We have recently demonstrated that, in addition to plasmid-encoded pathovar specific pili, known as colonization factors, the majority of ETEC express chromosomally encoded type 1 fimbriae, and that these structures play an essential role in virulence (22). We therefore questioned whether CEACAMs expressed in the intestine contribute to the pathogenesis of ETEC. Here we demonstrate that heat-labile toxin stimulates production of CEACAMs that then serve as critical receptors for ETEC type 1 fimbriae, accelerating bacterial adhesion and toxin delivery to host tissues.
Results
Heat-Labile Toxin Induces CEACAM Expression in Intestinal Epithelial Cells.
Of the different CEACAMs, CEACAM1, CEACAM5, and CEACAM6 are normally expressed predominantly in the colon, but also by small intestinal epithelia, while CEACAM7 is largely restricted to the colon (SI Appendix, Table S1 and Fig. S1B). Because CEACAMs have been linked to the pathogenesis of other E. coli pathovars (15, 17–19), and because heat-labile toxin (LT) has been shown to promote bacterial adhesion (12) and intestinal colonization (11), we questioned whether LT could exert its effect in part through modulation of CEACAM expression on the surface of intestinal epithelial cells. Therefore, we treated intestinal epithelial cell monolayers with either wild type LT or a mutant version of LT (mLT) containing an inactivating point mutation (E112K) within the A subunit active site. We found that wild type LT promoted transcriptional activation of each of the CEACAM genes with known expression in the intestine (CEACAM1, CEACAM5, CEACAM6, and CEACAM7) in Caco-2 cells relative to untreated cells or cells treated with mLT (SI Appendix, Fig. S2).
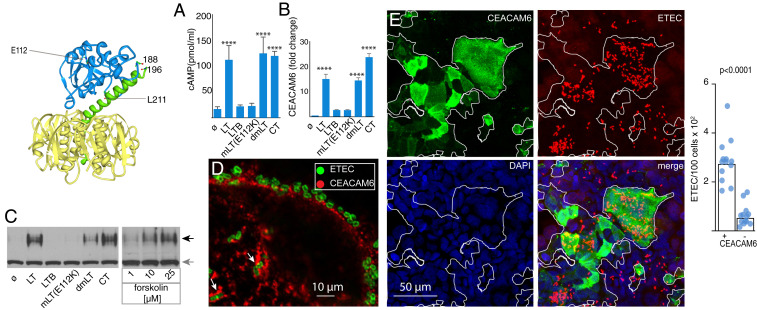
Of the genes significantly up-regulated by LT, CEACAM6 is known to be expressed at low levels in small intestine, the site of ETEC colonization. We therefore performed additional experiments to examine the impact of LT and related toxins, which vary in their ADP ribosylating activity, required for cAMP production in the host. These included cholera toxin (CT), which shares structural and functional features of LT; double mutant (R192G, L211A) LT (dmLT) that retains the ability to induce cAMP (23); mLT; or the catalytically inactive B subunit pentamer of LT (LT-B) that retains the ability to bind to target cells. These studies demonstrated that cAMP production (Fig. 1A) paralleled increases in CEACAM6 transcription (Fig. 1B). Likewise, we observed increased production of the CEACAM6 protein following treatment with active versions of LT, CT, or forskolin, a known activator of adenylyl cyclase (Fig. 1C). Intriguingly, we also observed marked accumulation of CEACAM6 surrounding sites of bacterial attachment (Fig. 1D) and significantly more bacteria attached to epithelial cells exhibiting robust CEACAM6 expression (Fig. 1E), suggesting the possibility of an intimate interaction of ETEC and CEACAM6.
Fig. 1.
Heat-labile toxin stimulates production of CEACAM6 in intestinal epithelia. (Upper Left) Schematic depicts the heat-labile toxin (LT) with the A1, A2, and B subunits depicted in blue, green, and yellow, respectively, with the location of sites mutated in mLT (E112K) and dmLT (R192G, L211A). (A) cAMP activation in target Caco-2 epithelial cells following treatment with LT, LT mutants, the B subunit of LT alone (LTB), or cholera toxin (CT). ø, control baseline untreated cells (****P < 0.0001 by ANOVA comparison to untreated cells). (B) RT-PCR data demonstrating fold change in CEACAM6 expression relative to untreated cells normalized to GAPDH (mean ± SD, n = 3 technical replicates) relative to control (****P < 0.0001 by ANOVA). (C) Immunoblot of CEACAM6 expression following treatment with toxins or forskolin (Right). Black arrow indicates migration of CEACAM6, while gray arrow indicates migration of tubulin loading control. (D) Accumulation of CEACAM6 (red) adjacent to sites of ETEC (green) attachment to Caco-2 cells. (E) Confocal microscopic images showing association of ETEC with areas of CEACAM6 expression on Caco-2 cells. (Right) Bars on the graph indicate geometric means (****P < 0.0001 by Mann–Whitney U two-tailed nonparametric comparison of n = 13 replicates in each group).
CEACAM6 Is Required for Optimal Adhesion to Intestinal Epithelial Cells.
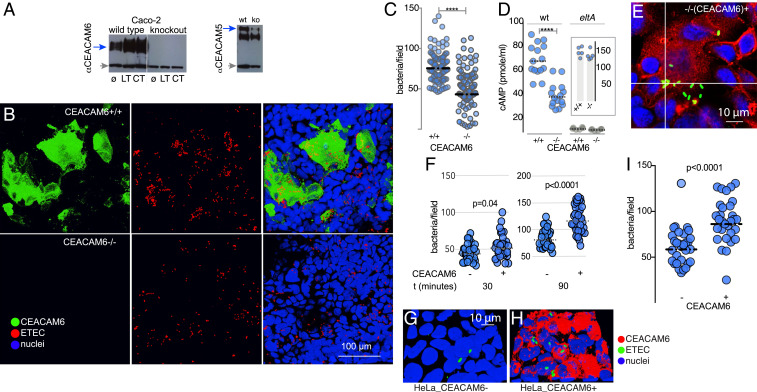
To further evaluate the importance of CEACAM6 in ETEC interactions with target intestinal epithelial cells, we constructed a mutant cell line that lacked CEACAM6 using CRISPR-Cas9 genome editing of intestinal epithelial cells (Fig. 2A). We found that interruption of CEACAM6 led to a marked decrease in bacterial adhesion by wild type ETEC (Fig. 2 B and C). Toxin delivery by ETEC requires intimate contact of the bacteria with epithelial cells, a process that is facilitated by the interaction of multiple adhesins with cognate cellular receptors (24). Accordingly, we found that delivery of heat-labile toxin was diminished in the absence of CEACAM6, as determined by production of cAMP following infection (Fig. 2D), suggesting that CEACAM6 is both stimulated by LT and essential for optimal delivery of this toxin.
Fig. 2.
CEACAM6 facilitates ETEC pathogen–host interactions. (A) CRISPR-Cas9–generated CEACAM6−/− mutant cell line (knockout, ko) does not make CEACAM6 (anti-CEACAM6 immunoblot; Left), but makes CEACAM1 and CEACAM5 (Right). Gray arrows indicate tubulin as a loading control. ø, untreated cells; LT and CT, stimulation with heat-labile or cholera toxin overnight, respectively. (B) Confocal images of wild type H10407 ETEC adherent to CEACAM6+/+ cells (Top) and CEACAM6-deficient cells (−/−; Bottom). (C) CEACAM6 is required for optimal interaction of ETEC with intestinal epithelial cells. (D) CEACAM6 is required for efficient toxin delivery by ETEC. Intracellular cAMP concentrations were determined in CEACAM6+/+ and −/− cells following infection with either wild type (wt) or control LT-negative eltA mutant strain (n = 15 replicates from a total of three independent experiments). (Inset) cAMP responses (in pmol/mL) of CEACAM6+/+ and −/− cells following treatment with forskolin. Symbols represent n = 5 replicates (bars represent geometric mean values; ****P < 0.0001 by Mann–Whitney U two-tailed nonparametric testing). (E and F) Restoration of CEACAM6 expression (red) in CEACAM6−/− mutant cell line G2 restores ETEC (green) adhesion. Graphs depict ETEC adhesion to G2 cells (−/+CEACAM6) expressed as bacteria per field 30 and 90 min after infection. (G–I) Introduction of CEACAM6 into HeLa cells promotes ETEC adhesion.
To further investigate the importance of CEACAM6 to pathogen–host interactions, we complemented the defect in the CEACAM6−/− cells in trans by lentiviral transduction with CEACAM6, restoring the adhesion phenotype (Fig. 2 E and F). Similarly, heterologous expression of CEACAM6 in HeLa cells promoted ETEC adhesion (Fig. 2 G–I), and, among the individual CEACAMs, CEACAM6 expression was most efficient in promoting bacterial adhesion (SI Appendix, Fig. S3 A and B).
Earlier studies demonstrated that E. coli can interact not only with CEACAM6, but also with CEA (CEACAM5) (17–19). While we found that treatment of Caco-2 cells with CEACAM5 siRNA resulted in significant decreases in adhesion, this was not true of CEACAM6−/− cells treated with the same siRNA (SI Appendix, Fig. S4A). Moreover, we found that this siRNA treatment resulted in decreased production of both CEACAM5 and CEACAM6 (SI Appendix, Fig. S4B). In contrast to distinct localization of bacteria within areas of CEACAM6 expression, we found that ETEC were not preferentially distributed to areas of CEACAM5 production (SI Appendix, Fig. S4C). Nevertheless, we observed clear areas of robust CEACAM5 expression in some areas of ETEC attachment (SI Appendix, Fig. S4D). Collectively, these data suggest that, while ETEC can induce and engage CEACAM5, other molecules may be more efficient in facilitating these interactions.
ETEC Interaction with CEACAMs Is Directed by the FimH Adhesin.
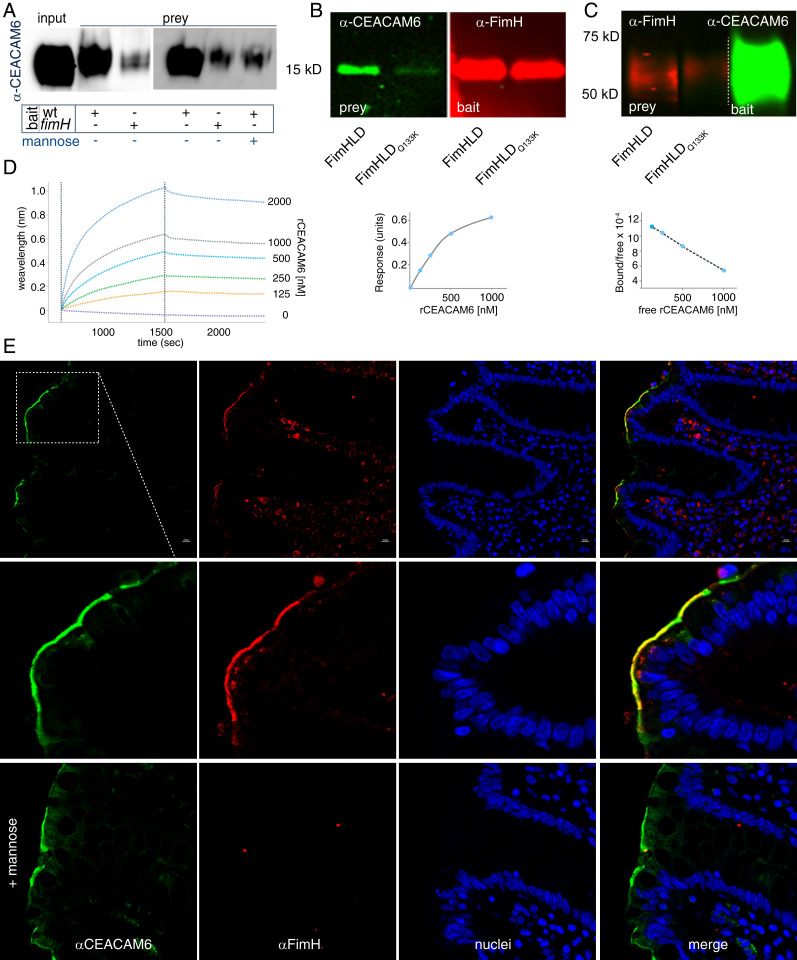
As type 1 fimbriae are highly conserved within the ETEC pathovar and critical for bacterial adhesion and toxin delivery (22), we next examined the interaction of the mannose-binding FimH type 1 tip adhesin with CEACAM6. In the absence of fimH or in the presence of mannose, ETEC binding of CEACAM6 was drastically reduced, suggesting that these interactions are largely dependent on expression and assembly of type 1 fimbriae (Fig. 3A). Type 1 fimbrial interaction with intestinal epithelia is mediated by the mannose-binding lectin domain of FimH (FimHLD) (22). Far Western analyses demonstrated that soluble recombinant CEACAM6 interacted with target FimHLD, but not with FimHLDQ133K containing a point mutation in the mannose binding pocket (Fig. 3B). Likewise, FimHLD, but not FimHLDQ133K, interacted with immobilized target recombinant CEACAM6 molecules (Fig. 3C). We also found that ETEC adhesion to CEACAM6 expressed on HeLa cells was dependent on mannose (SI Appendix, Fig. S5A), and that interaction with CEACAMs was dependent on fimH, but not cfaE, the tip adhesin expressed on colonization factor 1 (CFA/I) fimbriae (SI Appendix, Fig. S5B). In addition, CEACAM6 engaged FimHLD but not FimHLDQ133K with high affinity (Kd ∼ 2.8 × 10−8) in biolayer interferometry experiments using immobilized biotinylated adhesin (Fig. 3D). Finally, confocal imaging of human small intestinal sections incubated with rFimHLD demonstrated distinct colocalization of the adhesin with CEACAM6 expressed on the surface of enterocytes (Fig. 3E), a process that was completely inhibited by the addition of exogenous mannose.
Fig. 3.
CEACAM6 interacts with the FimH tip adhesin of type 1 fimbriae. (A) FimH promotes interaction of CEACAM6 with ETEC. Anti-CEACAM6 immunoblot shows amount of CEACAM6 retained by wild type (wt) and fimH mutant ETEC following incubation of bacteria with recombinant CEACAM6 (rCEACAM6). (B, Left) CEACAM6 far Western blot (green) in which either FimHLD or the FimHLDQ133K mutant protein was used as bait. (Right) Immunoblot (red) shows relative amounts of protein transferred to the blot as bait. (C, Left) FimH far Western blot in which immobilized CEACAM6 is used as bait for FimH or the FimHLDQ133K mutant. (Right) Transferred rCEACAM6 used as bait is shown. (D) Biolayer interferometry studies demonstrate interaction between immobilized rFimH and varying concentrations of rCEACAM6. (E) rFimHLD colocalizes with CEACAM6 on human small intestinal biopsies in a mannose-dependent fashion.
ETEC Interactions with CEACAM6 on Human Small Intestinal Epithelia.
The expression of CEACAMs is known to be altered in gastrointestinal cancers (25, 26) and inversely correlates with enterocyte differentiation (27), and CEA (CEACAM5) serves as a clinical biomarker for advanced colon carcinoma. As ETEC targets the small intestine rather than the colon, and Caco-2 cells are derived from a metastatic colon tumor, we next examined ETEC interactions with CEACAM6 in a contextually relevant ileal enteroid model. When grown as polarized differentiated monolayers, small intestinal enteroids develop a defined apical brush border with distinct microvilli. Transmission electron microscopy of small intestinal enterocytes demonstrated CEACAM6 associated with the microvilli (Fig. 4A) and with the glycocalyx at the tips of the microvilli (Fig. 4B). As with the transformed Caco-2 cell line, we found ETEC in close association with robust CEACAM6 expression on the apical surface of these cells (Fig. 4C) and observed distinct “footprinting” of CEACAM6 surrounding attached bacteria (Fig. 4D). As with Caco-2 cells, wild type, but not LT-negative, bacteria provoked increases in intracellular cAMP (Fig. 4E), and LT led to increased production of CEACAM6 in target enteroids derived from small intestinal stem cells (Fig. 4F). Finally, both LT (Fig. 4G) and CEACAM6 (Fig. 4H) were required for optimal interaction of ETEC with these cells.
Fig. 4.
ETEC–CEACAM interactions on human small intestinal epithelia. (A) CEACAM6 is expressed on the apical surface of small intestinal enteroids. Arrows point to immunogold labeling of CEACAM6 on tips of microvilli (7,500×). (B) CEACAM6 associated with glycocalyx at tips of microvilli (15,000×). (C) ETEC (red) associates with CEACAM6 (green) on the surface of human small intestinal (Hu135D) epithelial cells (enteroids). Image prepared in Volocity from confocal image z-stacks. (D) Confocal microscopy images of “footprinting” of CEACAM6 (green) at sites of bacterial attachment (red) to small intestinal epithelia. Each panel is flanked by representations of the xz and yz planes (Bottom and Right, respectively). (E) ETEC induction of cAMP in polarized enteroid small intestinal monolayers. Shown are data for blood A (135D), B (248D), and O (235D). ø, untreated; PDEi, phosphodiesterase inhibition. (F) LT mediates up-regulation of CEACAM6 in small intestinal epithelia. Shown are experimental replicate data (n = 3) from control cells or cells treated with LT. (Inset) Immunoblot compares CEACAM6 protein expression in treated and untreated cells. (G) Heat-labile toxin promotes adhesion to small intestinal enteroids (shown are three independent experimental replicates, each with five to seven technical replicates, using enteroids derived from a donor belonging to a different ABO blood group). P values reflect nonparametric comparisons by Mann–Whitney U test. (H) CEACAM6 is required for optimal adhesion to small intestinal enteroids [**P = 0.0067 by ANOVA (Kruskal–Wallis) comparison of untreated (ø) or treatment with scramble RNA (sc) and CEACAM6 siRNA]. (Inset) Immunoblot demonstrates CEACAM6 production (top band) in enteroids by the ø, sc, si groups relative to tubulin (bottom band). (I) Human small intestinal biopsies demonstrating CEACAM6 expression on day 2, day 7, and day 30 post infection with ETEC. Control specimen is from the same patient (at day 7) processed without primary anti-CEACAM6 antibody.
As CEACAM5 is also normally expressed at low levels in the small intestine, we also examined the impact of this molecule in small intestinal enteroids. Like CEACAM6, treatment of cells with LT, but not mLT or LT-B, resulted in significant increases in CEACAM5 expression (SI Appendix, Fig. S6A). Interestingly, in contrast to CEACAM6, the absence of CEACAM5 (SI Appendix, Fig. S6B) had only a modest impact on adhesion (SI Appendix, Fig. S6C), and the majority of ETEC bacteria adhering to parental CEACAM5+/+ cells and CEACAM5−/− cells localized to areas of CEACAM6 expression (SI Appendix, Fig. S6D).
Altogether, these data suggested that, while LT enhances the production of multiple CEACAMs with the capacity to engage ETEC, CEACAM6 plays a critical and predominant role in mediating these interactions. To provide additional validation and overcome the limitations inherent to in vitro model systems, we also examined archived biopsy specimens from patients infected with ETEC in Bangladesh. Here, we were able to observe an apparent increase in CEACAM6 expression in small intestinal epithelia following infection (Fig. 4I and SI Appendix, Fig. S7), suggesting that modulation of these molecules also occurs during natural infection of human hosts.
Discussion
In the classical pathogenesis paradigm for ETEC, heat-labile toxin activates adenylate cyclase, resulting in increased production of cAMP and activation of cAMP-dependent protein kinase A (PKA). This kinase in turn phosphorylates membrane ion channels that mediate Na+ and Cl− ion transport, resulting in the net export of salt and water into the intestinal lumen. Notably, however, toxin-driven accumulation of cAMP in target intestinal epithelia is likely to have many other effects, given the importance of this cyclic nucleotide as a second messenger. Importantly, cAMP binding to PKA regulatory subunits results in liberation of the biologically active A1 catalytic subunit for transport into the nucleus, where PKA is free to phosphorylate transcription factors such as the cAMP response element binding protein (CREB) that modulate activation of a host of genes (28, 29). Interestingly, however, promotor regions for CEACAM genes were not represented in earlier genome-wide searches for CREB binding sites (30), suggesting that activation of multiple CEACAMs may not occur directly through cAMP-response elements. Notably, earlier studies of another E. coli pathovar isolated in the setting of inflammatory bowel disease, adherent-invasive E. coli (AIEC), demonstrate that these bacteria exploit cytokine-induced up-regulation of CEACAMs (15, 31). Further studies will be needed to determine whether a common signaling pathway underlies toxin- and cytokine-stimulated up-regulation of CEACAMs.
Intestinal CEACAM expression is normally most abundant in the large intestine (32). Here, however, we demonstrate that heat-labile toxin of ETEC drives transcriptional activation of genes encoding CEACAM molecules expressed in the small intestine, particularly CEACAM6, which then serve as receptors to promote effective engagement of the bacteria mediated by chromosomally encoded type 1 fimbriae. As our previous studies demonstrate that these structures, universally conserved in the ETEC pathovar, are essential for effective bacterial adhesion and toxin delivery (22), the bacteria are effectively modulating the architecture of the small intestinal epithelia to promote pathogen–host interactions.
Increasingly, it has become recognized that ETEC are associated with important sequelae, particularly among young children of low- and middle-income regions, where these pathogens have been linked to malnutrition and stunted growth (33) due to a poorly understood condition known as environmental enteropathy (34) that is characterized by shortened villi and poor nutrient absorption. Although enhanced expression of CEACAMs may benefit the pathogen by accelerating bacterial adhesion and toxin delivery, CEACAM overexpression in the intestine may be deleterious to the host. During normal maturation of enterocytes along the crypt–villus axis of intestinal microvilli, cells ultimately are released from the tips of the microvilli by apoptosis as cells lose connections to the underlying extracellular matrix, a process known as anoikis (35, 36). Notably enhanced expression of CEACAMs, including CEACAMs 5 and 6, has been shown to inhibit anoikis (37–39) and disrupt epithelial architecture, suggesting a possible link to enteropathy observed with ETEC infection.
Collectively, the studies reported here provide a paradigm for ETEC pathogenesis that extends the role of the toxins beyond simple activation of ion channels that result in diarrhea. The data presented here suggest that ETEC use the heat-labile toxin to modulate expression of important receptors, thus effectively modifying the epithelial landscape to benefit the pathogen.
Materials and Methods
Bacterial Strains and Growth Conditions.
Strains used in these studies are outlined in SI Appendix, Table S2. Bacteria were grown from frozen glycerol stocks at 37 °C in Luria Bertani media.
Growth and Maintenance of Gastrointestinal Cell Lines.
A list of all cell lines used in these experiments is included in SI Appendix, Table S3.
Caco-2 Cells.
Caco-2 intestinal epithelial cells (ATCC HTB-37) were obtained directly from ATCC and propagated at 37 °C, 5% CO2, in Eagle’s MEM containing fetal bovine serum (FBS) at a final concentration of 20%. Transfected HeLa cells expressing different CEACAMs were obtained from author B.B.S. and propagated at 37 °C, 5% CO2, in DMEM media containing 10% FBS and 2 mM L-glutamine (Gibco) and 500 µg/mL of G418 disulfate (G8168; Sigma).
Small Intestinal Enteroids.
Biopsy samples obtained from adult patients undergoing routine endoscopy at Washington University School of Medicine, with patient consent and approval of the institutional review board, were processed as previously described and maintained by the Washington University Digestive Diseases Research Core Center (DDRCC) BioSpecimens Core (40). Briefly, as described previously (8), isolated crypt tissues were thawed and resuspended in a total volume of 15 µL of Matrigel (BD Biosciences) and then added to individual wells of a 24-well plate. The suspended cells were incubated at 37 °C with a 1:1 mixture of conditioned media (CM) from the L-WRN cell line (40, 41) and primary culture media (Advanced DEM/F12; Invitrogen) supplemented with 20% FBS, 2 mM L-glutamine, 100 U/mL penicillin, 0.1 mg/mL streptomycin, 10 μM Y-27632 (ROCK inhibitor; Tocris Bioscience, R&D Systems), and 10 μM SB 431542 (TGFBR1 inhibitor; Tocris Bioscience, R&D Systems). All samples were deidentified prior to use in the studies outlined here.
To obtain polarized enteroid monolayers, cells grown in Matrigel were trypsinized, resuspended in growth media, and plated onto Transwell filters (Corning) coated with type IV human collagen (Sigma), then grown for 4 d prior to induction of differentiation by replacement of media with differentiation media (5% L-WRN CM; primary culture medium lacking SB 431542).
Lentivirus Production and Construction of CEACAM6-Expressing Cells.
To generate an expression plasmid for autologous expression of CEACAM6 in HeLa cells, we amplified CEACAM6 full-length cDNA from plasmid RC202454 (Origene) using primers 021518.1 and 021518.2 (SI Appendix, Table S4) and cloned between the BamHI and AgeI sites of pFCIV using Gibson assembly, a lentivirus expression vector that expresses GFP marker from an IRES promoter (Hope Center, Washington University), yielding pFCIV-CEACAM6. Lentivirus was then produced by cotransfecting HEK293T cells with a total of 2 µg of psPAX2 (no. 12260; Addgene), pMD2.G (no. 12259; Addgene), and pFCIV-CEACAM6 plasmids at 2:2:1 ratio in FuGENE HD transfection reagent (Promega). Media was changed following 18 h post transfection. Two days post transfection, the CEACAM6-lentivirus–containing supernatant was collected, flash-frozen, and stored at −80 °C. The lentivirus transduction mixture was prepared by adding 250 µL of CEACAM6-lentivirus supernatant in 750 µL of cell culture medium supplemented with 8 µg/mL Polybrene. Wild type HeLa cells were grown for 24 h after plating to ∼50% confluency, and 250 µL of CEACAM6-lentivirus mixture was added per well of a 12-well plate. The plate was then centrifuged at 1,000 × g for 30 min at 32 °C, then incubated at 37 °C, 5% CO2, for 6 h, after which media was replaced with fresh cell culture media. Transduction efficiency was monitored by fluorescence microscopy for GFP expression. CEACAM6 expression was then confirmed by immunofluorescence microscopy using anti-CEACAM6 monoclonal (clone-9A6, sc-59899; Santa Cruz Biotechnology) and by immunoblotting of cell lysates. The G2 (Caco-2_CEACAM6−/−) cells were also transcomplemented by transduction with CEACAM6-lentivirus, and GFP-positive cells were FACS-sorted (BD FACS Aria II; BD Biosciences) and analyzed for CEACAM6 expression by Western blotting as well as by immunofluorescence microscopy.
Construction of CEACAM-Deficient Cell Lines and RNAi Experiments.
CEACAM6−/− Caco-2 cells were generated at the Genome Engineering and Induced Pluripotent Stem Cell Center at Washington University School of Medicine (http://geic.wustl.edu/) using CRISPR-Cas9 nuclease to induce a targeted double-strand break in exon 2 of the CEACAM6 gene with guide RNA 5′-AGCTGTAACCAATACGATTC-3′. CEACAM5−/− intestinal cells were generated by lentiviral-mediated delivery of a 5′-CTTGTGAATGAAGAAGCAAC-3′ guide RNA. Briefly, a synthetic fragment 5′-acttgaaagtatttcgatttcttggctttatatatcttgtggaaaggacgaaacaccCTTGTGAATGAAGAAGCAACgttttagagctagaaatagcaagttaaaataaggctagtccgttatcaacttg-3′ was cloned by Gibson assembly into plentiCRISPRv2 (42) following linearization of the vector with BsmB1. The resulting plasmid pCEACAM5gRNA was then transfected (FuGENE; Promega) with psPAX2 and pMD2.G into HEK293T cells for propagation of lentiviral particles for transduction into target cells. In RNA inhibition experiments, siRNAs for CEACAM5 and CEACAM6 or control siRNA (SI Appendix, Table S5) were introduced by lipid-mediated transfection (Lipofectamine; Invitrogen).
Purification of Recombinant CEACAM6.
For recombinant CEACAM6 (rCEACAM6) purification, the CEACAM6 cDNA without the native signal peptide sequence was amplified from RC202454 plasmid (Origene) using primers 030218.1 and 030218.3 (SI Appendix, Table S4). The amplicon was then ligated between the EcoRI and AgeI restriction sites of pcDNA3.4_mIgG2-mMxr8a (43) downstream of an IL-2 signal peptide sequence in frame with a C-terminal 6×His tag sequence by Gibson assembly (New England Biolabs), yielding pCDNA3.4-CEACAM6. Expi293 cells (Thermo Fisher) were seeded at 5 × 106 cells per milliliter in Expi293 expression medium (ThermoFisher) supplemented with HyClone Cell Boost 1 (GE) 2% (wt/vol) 1 d prior to transfection with 250 µg of pCDNA3.4-CEACAM6 using HYPE-5 transfection reagent (OzBiosciences). Cells were supplemented with 10 mL, 20 mL, and 30 mL of 2% HyClone cell Boost 1 supplement (GE) on day 2, day 3, and day 4, respectively. Four days after transfection, supernatant was collected, centrifuged at 3,000 × g for 15 min, and purified by metal-affinity column chromatography (HisTrap HP; GE 17–5248-01)
FimHLD and Q133K Expression and Purification.
Recombinant, polyhistidine-tagged lectin domain of the FimH tip adhesin from type I fimbriae (FimHLD-his) and a mannose binding mutant FimHLD:Q133K (Q133K-his) were expressed in BL21 (DE3) pLys and purified by metal-affinity chromatography as previously described (22).
Bacterial Adhesion Assays.
HeLa cells or HeLa cells expressing CEACAM6 were grown as confluent monolayers on Transwell filters, then infected with ETEC H10407 grown under static conditions at a multiplicity of infection of ∼1 bacteria:10 cells (22). After 1 to 2 h of incubation at 37 °C, 5% CO2 cells were washed with prewarmed media and fixed with paraformaldehyde, and cell-associated ETEC were identified using O78 LPS specific anti-O78 polyclonal antibody (1:200) raised in rabbit, followed by fluorescence-tagged goat anti-rabbit Alexa Fluor 594 antibody (1:400). Cell nuclei were counterstained with DAPI, and cell-associated bacteria enumerated by fluorescence microscopy.
Toxin Delivery Assays.
Effective delivery of ETEC heat-labile toxin was determined using cyclic nucleotide assays as previously described (8). Briefly, wild type Caco-2 or G2 CEACAM6-negative cells were grown to confluency in 96-well plates and infected with ETEC H10407. After 2 h of incubation at 5% CO2, 37 °C, wells were washed with tissue culture media and incubated for an additional 2.5 h, and cells were lysed in sample buffer. Intracellular cAMP concentrations were determined by ELISA (cAMP Direct EIA; Arbor Assays) as per manufacturer’s protocol. For small intestinal enteroids, cells were incubated with phosphodiesterase inhibitors (PDEis) vardenafil hydrochloride trihydrate (Y0001647), rolipram (R6520), and cilostazol (C0737; all from Sigma-Aldrich) at a final concentration of 25 µM for 2 h prior to exposure to bacteria.
Quantitative Real-Time PCR.
Quantitative RT PCR was performed as previously described using a ViiA 7 Real-time PCR System (Applied Biosystems), and data were analyzed using ViiA 7 software (v 1.1). Primers used in PCR analysis are listed in SI Appendix, Table S4.
Immunohistochemistry.
Paraffin sections were taken from small intestinal biopsies performed at ICDDRB in Dhaka, Bangladesh, or age-matched healthy controls obtained from a biobank of specimens at Washington University School of Medicine. All sections were deidentified prior to use. Sections mounted on glass slides were treated with xylene to remove paraffin, antigen unmasking was done using heat retrieval procedure (3 min at 18 psi and 125 °C in a pressure cooker), and then sections were blocked with 1% BSA/10% normal goat serum in PBS. Slides were then incubated with mouse monoclonal anti-CEACAM6 antibody (1:1,000; Santa Cruz) overnight at 4 °C, washed, then incubated with biotin-SP AffiniPure goat anti-mouse IgG (1:800; Jackson Labs) for 1 h at room temperature, washed, and further incubated with streptavidin conjugated to HRP (1:1,000; Jackson Labs) for 1 h at room temperature. After washing, slides were developed with Betazoid DAB (3,3′-diaminobenzidine) chromogen kit (Biocare Medical) as per manufacturer’s protocol, washed, and counterstained with hematoxylin.
CEACAM6 Pull-Downs with ETEC.
Bacteria grown at type 1 pili inducing conditions were used to pull down CEACAM6. Briefly, 100 µL of OD600-1.5 bacteria suspension (∼2 × 108 cells) was incubated with purified CEACAM6-his for 1 h at room temperature on a minirotor at 30 rpm (Glas-Col). Following incubation, bacteria were washed with PBS and boiled in 1× Laemmli sample buffer (Bio-Rad) supplemented with β-mercaptoethanol before SDS/PAGE and Western blotting.
Far-Western and Immunoblotting.
In order to examine molecular interactions between CEACAM6 and FimH, either purified rCEACAM6 or purified FimH was separated on 4 to 20% SDS/PAGE gradient minigel and then transferred onto nitrocellulose membrane (Bio-Rad). The nitrocellulose membranes were then blocked with 5% (wt/vol) nonfat dry milk (blotting-grade blocker; Bio-Rad) for 1 h at room temperature, diluted in PBS/0.05% Tween 20 (PBST). The membranes were then washed and incubated with prey proteins (for CEACAM6 blot either with FimHLD or with FimHLD_Q133K, and for FimHLD blot with CEACAM6 in the presence or absence of methyl-D-mannose) for 1 h at room temperature. Subsequently, the membranes were washed 3× with PBST and incubated with rabbit anti-FimH polyclonal (1:1,000) or mouse anti-CEACAM6 monoclonal (1:1,000) at room temperature for 1 h. Following 3× wash with PBST, membranes were incubated with goat anti-rabbit IgG-HRP (1:1,000; Santa Cruz) or goat anti-mouse IgG-HRP (1:1,000; Cell Signaling Technology) for 45 min at room temperature. After 3× wash with PBST, membranes were developed using Clarity western ECL substrate (Bio-Rad), and an image was taken using an Azure c600 imaging system (Azure Biosystems).
Biolayer Interferometry.
Biolayer interferometry was used to evaluate direct molecular interactions between the FimH adhesin and CEACAM6 (Octet Red96; Pall ForteBio). Briefly, prior to immobilization, streptavidin biosensor (ForteBio) surface was equilibrated with PBS for 10 min. Following equilibration, 5 µg/mL of biotinylated FimHLD or FimHLDQ133K was immobilized on biosensor tips and then quenched with biocytin (10 µg/mL) for 5 min and washed with PBS (baseline). These were then incubated with twofold serial dilutions (2,000 to 100 nM) of purified recombinant CEACAM6 or control BSA (2,000 nM) prepared in PBS as analyte. Q133K immobilized biosensors were used for background binding. Fresh biosensors were used every time without any regeneration step. Data were analyzed using Octet data analysis software version 8.2. Binding sensorgrams were aligned at the beginning of the association cycle and following the background subtraction; binding sensorgrams were globally fit to a 1:1 binding model.
Confocal Laser Scanning Microscopy.
Infected or uninfected cell monolayers were fixed with 2% paraformaldehyde for 30 min at 37 °C and then washed 3× with PBS prior to blocking with 1% BSA in PBS for 1 h at room temperature. Primary antibodies raised in mice (SI Appendix, Table S6) were detected using fluorescence-tagged goat anti-mouse IgG Alexa Fluor 594 (1:200), and antibodies raised in rabbit were detected using fluorescence-tagged goat anti-rabbit IgG Alexa Fluor 488 (1:200). Cell nuclei were counterstained with DAPI and mounted on glass slides using Prolong Gold antifade reagent (Invitrogen). Images were captured and analyzed on a Nikon C2 confocal microscope equipped with NIS-Elements AR 5.11.01 software (Nikon).
Transmission Immunoelectron Microscopy.
For localization of CEACAM6 on intestinal epithelium, human ileal stem cell-derived polarized monolayers grown on Transwell filters (0.45-µM pore size; Corning) were fixed in 4% paraformaldehyde (Polysciences) in 100 mM Pipes/0.5 mM MgCl2, pH 7.2, for 1 h at 4 °C. Samples were then embedded in 10% gelatin and infiltrated overnight with 2.3 M sucrose/20% polyvinyl pyrrolidone in Pipes/MgCl2 at 4 °C. Samples were trimmed, frozen in liquid nitrogen, and sectioned with a Leica Ultracut UCT7 cryo-ultramicrotome (Leica Microsystems). Ultrathin sections of 50 nm were blocked with 5% FBS/5% NGS for 30 min and subsequently incubated with mouse anti-CEACAM6 (1:100) for 1 h at room temperature. Following washes in block buffer, sections were incubated with goat anti-mouse IgG+IgM conjugated to 18 nm colloidal gold (1:30; Jackson ImmunoResearch Laboratories) for 1 h. Sections were stained with 0.3% uranyl acetate/2% methyl cellulose and viewed by transmission electron microscope as described above. All labeling experiments were conducted in parallel with controls omitting the primary antibody. These controls were consistently negative at the concentration of colloidal gold-conjugated secondary antibodies used in these studies.
Statistical Analysis.
Statistical analyses were performed using Prism software version 8 (GraphPad). Analysis between two groups was performed by nonparametric Mann–Whitney U (two-tailed) testing. For multiple groups, ANOVA and Kruskal–Wallis testing of multiple comparisons were used to determine statistical significance. Statistical significance was assigned when P values were < 0.05.
Supplementary Material
Acknowledgments
We thank Dr. John Clements of Tulane University for generously providing heat- labile toxin (LT) and mutant versions of LT, Dr. Michael Diamond for providing pFCIV and resources for lentivirus, Drs. Daved Freemont and Chris Nelson for assistance in purification of recombinant CEACAM6, Dr. Wandy Beatty for her expert assistance with electron microscopy, and Dr. Scott Grey-Owen for his helpful insights. This work was supported by funding from the U.S. Department of Veterans Affairs (VA) (I01BX004825); Grants R01AI89894 and R01AI126887 from the National Institute of Allergy and Infectious Diseases (NIAID) of the NIH; Clinical and Translational Sciences Award UL1TR000448 from the NIH; the Digestive Diseases Research Core Center at Washington University School of Medicine; Grant P30 DK52574 from the National Institute of Diabetes and Digestive and Kidney Diseases. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIAID, NIH, or the VA.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2012480117/-/DCSupplemental.
Data Availability.
Original image files have been deposited in Figshare (10.6084/m9.figshare.12137544, 10.6084/m9.figshare.12141033, 10.6084/m9.figshare.121412-28, 10.6084/m9.figshare.12137658, 10.6084/m9.figshare.12137652, 10.6084/m9.figshare.12137655, 10.6084/m9.figshare.12141138, 10.6084/m9.figshare.12137661, 10.6084/m9.figshare.12138297, 10.6084/m9.figshare.12142092, 10.6084/m9.figshare.12140994) (44–54).
References
- 1.Khalil I. A., et al. , Morbidity and mortality due to shigella and enterotoxigenic Escherichia coli diarrhoea: The global burden of disease study 1990-2016. Lancet Infect. Dis. 18, 1229–1240 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walker C. L. F., et al. , Global burden of childhood pneumonia and diarrhoea. Lancet 381, 1405–1416 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.MAL-ED Network Investigators , The MAL-ED study: A multinational and multidisciplinary approach to understand the relationship between enteric pathogens, malnutrition, gut physiology, physical growth, cognitive development, and immune responses in infants and children up to 2 years of age in resource-poor environments. Clin. Infect. Dis. 59 (suppl. 4), S193–S206 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Sack R. B., The discovery of cholera–Like enterotoxins produced by Escherichia coli causing secretory diarrhoea in humans. Indian J. Med. Res. 133, 171–180 (2011). [PMC free article] [PubMed] [Google Scholar]
- 5.Carpenter C. C., et al. , Clinical and physiological observations during an epidemic outbreak of non-vibrio cholera-like disease in Calcutta. Bull. World Health Organ. 33, 665–671 (1965). [PMC free article] [PubMed] [Google Scholar]
- 6.Sack R. B., et al. , Enterotoxigenic Escherichia coli isolated from patients with severe cholera-like disease. J. Infect. Dis. 123, 378–385 (1971). [DOI] [PubMed] [Google Scholar]
- 7.Fleckenstein J. M., et al. , Molecular mechanisms of enterotoxigenic Escherichia coli infection. Microbes Infect. 12, 89–98 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumar P., et al. , Enterotoxigenic Escherichia coli-blood group A interactions intensify diarrheal severity. J. Clin. Invest. 128, 3298–3311 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chakraborty S., et al. , Interrogation of a live-attenuated enterotoxigenic Escherichia coli vaccine highlights features unique to wild-type infection. NPJ Vaccines 4, 37 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allen K. P., Randolph M. M., Fleckenstein J. M., Importance of heat-labile enterotoxin in colonization of the adult mouse small intestine by human enterotoxigenic Escherichia coli strains. Infect. Immun. 74, 869–875 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berberov E. M., et al. , Relative importance of heat-labile enterotoxin in the causation of severe diarrheal disease in the gnotobiotic piglet model by a strain of enterotoxigenic Escherichia coli that produces multiple enterotoxins. Infect. Immun. 72, 3914–3924 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson A. M., Kaushik R. S., Francis D. H., Fleckenstein J. M., Hardwidge P. R., Heat-labile enterotoxin promotes Escherichia coli adherence to intestinal epithelial cells. J. Bacteriol. 191, 178–186 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X., Gao X., Hardwidge P. R., Heat-labile enterotoxin-induced activation of NF-κB and MAPK pathways in intestinal epithelial cells impacts enterotoxigenic Escherichia coli (ETEC) adherence. Cell. Microbiol. 14, 1231–1241 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muenzner P., et al. , Uropathogenic E. coli exploit CEA to promote colonization of the urogenital tract mucosa. PLoS Pathog. 12, e1005608 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barnich N., et al. , CEACAM6 acts as a receptor for adherent-invasive E. coli, supporting ileal mucosa colonization in Crohn disease. J. Clin. Invest. 117, 1566–1574 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gray-Owen S. D., Blumberg R. S., CEACAM1: Contact-dependent control of immunity. Nat. Rev. Immunol. 6, 433–446 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Leusch H. G., et al. , Escherichia coli of human origin binds to carcinoembryonic antigen (CEA) and non-specific crossreacting antigen (NCA). FEBS Lett. 261, 405–409 (1990). [DOI] [PubMed] [Google Scholar]
- 18.Leusch H. G., Drzeniek Z., Hefta S. A., Markos-Pusztai Z., Wagener C., The putative role of members of the CEA-gene family (CEA, NCA an BGP) as ligands for the bacterial colonization of different human epithelial tissues. Zentralbl. Bakteriol. 275, 118–122 (1991). [DOI] [PubMed] [Google Scholar]
- 19.Leusch H. G., Drzeniek Z., Markos-Pusztai Z., Wagener C., Binding of Escherichia coli and Salmonella strains to members of the carcinoembryonic antigen family: Differential binding inhibition by aromatic alpha-glycosides of mannose. Infect. Immun. 59, 2051–2057 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sauter S. L., Rutherfurd S. M., Wagener C., Shively J. E., Hefta S. A., Identification of the specific oligosaccharide sites recognized by type 1 fimbriae from Escherichia coli on nonspecific cross-reacting antigen, a CD66 cluster granulocyte glycoprotein. J. Biol. Chem. 268, 15510–15516 (1993). [PubMed] [Google Scholar]
- 21.Sauter S. L., Rutherfurd S. M., Wagener C., Shively J. E., Hefta S. A., Binding of nonspecific cross-reacting antigen, a granulocyte membrane glycoprotein, to Escherichia coli expressing type 1 fimbriae. Infect. Immun. 59, 2485–2493 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sheikh A., et al. , Highly conserved type 1 pili promote enterotoxigenic E. coli pathogen-host interactions. PLoS Negl. Trop. Dis. 11, e0005586 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clements J. D., Norton E. B., The mucosal vaccine adjuvant LT(R192G/L211A) or dmLT. MSphere 3, e00215-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dorsey F. C., Fischer J. F., Fleckenstein J. M., Directed delivery of heat-labile enterotoxin by enterotoxigenic Escherichia coli. Cell. Microbiol. 8, 1516–1527 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Neumaier M., Paululat S., Chan A., Matthaes P., Wagener C., Biliary glycoprotein, a potential human cell adhesion molecule, is down-regulated in colorectal carcinomas. Proc. Natl. Acad. Sci. U.S.A. 90, 10744–10748 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schölzel S., et al. , Carcinoembryonic antigen family members CEACAM6 and CEACAM7 are differentially expressed in normal tissues and oppositely deregulated in hyperplastic colorectal polyps and early adenomas. Am. J. Pathol. 156, 595–605 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ilantzis C., Jothy S., Alpert L. C., Draber P., Stanners C. P., Cell-surface levels of human carcinoembryonic antigen are inversely correlated with colonocyte differentiation in colon carcinogenesis. Lab. Invest. 76, 703–716 (1997). [PubMed] [Google Scholar]
- 28.Mayr B., Montminy M., Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat. Rev. Mol. Cell Biol. 2, 599–609 (2001). [DOI] [PubMed] [Google Scholar]
- 29.Sassone-Corsi P., The cyclic AMP pathway. Cold Spring Harb. Perspect. Biol. 4, a011148 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang X., et al. , Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc. Natl. Acad. Sci. U.S.A. 102, 4459–4464 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barnich N., Darfeuille-Michaud A., Abnormal CEACAM6 expression in Crohn disease patients favors gut colonization and inflammation by adherent-invasive E. coli. Virulence 1, 281–282 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Fagerberg L., et al. , Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteomics 13, 397–406 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anderson J. D., 4th, et al. , Burden of enterotoxigenic Escherichia coli and shigella non-fatal diarrhoeal infections in 79 low-income and lower middle-income countries: A modelling analysis. Lancet Glob. Health 7, e321–e330 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Korpe P. S., Petri W. A. Jr, Environmental enteropathy: Critical implications of a poorly understood condition. Trends Mol. Med. 18, 328–336 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barker N., Adult intestinal stem cells: Critical drivers of epithelial homeostasis and regeneration. Nat. Rev. Mol. Cell Biol. 15, 19–33 (2014). [DOI] [PubMed] [Google Scholar]
- 36.Taddei M. L., Giannoni E., Fiaschi T., Chiarugi P., Anoikis: An emerging hallmark in health and diseases. J. Pathol. 226, 380–393 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Ordoñez C., Screaton R. A., Ilantzis C., Stanners C. P., Human carcinoembryonic antigen functions as a general inhibitor of anoikis. Cancer Res. 60, 3419–3424 (2000). [PubMed] [Google Scholar]
- 38.Duxbury M. S., Ito H., Ashley S. W., Whang E. E., CEACAM6 cross-linking induces caveolin-1-dependent, Src-mediated focal adhesion kinase phosphorylation in BxPC3 pancreatic adenocarcinoma cells. J. Biol. Chem. 279, 23176–23182 (2004). [DOI] [PubMed] [Google Scholar]
- 39.Ilantzis C., DeMarte L., Screaton R. A., Stanners C. P., Deregulated expression of the human tumor marker CEA and CEA family member CEACAM6 disrupts tissue architecture and blocks colonocyte differentiation. Neoplasia 4, 151–163 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.VanDussen K. L., et al. , Development of an enhanced human gastrointestinal epithelial culture system to facilitate patient-based assays. Gut 64, 911–920 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miyoshi H., Stappenbeck T. S., In vitro expansion and genetic modification of gastrointestinal stem cells in spheroid culture. Nat. Protoc. 8, 2471–2482 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanjana N. E., Shalem O., Zhang F., Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang R., et al. , Mxra8 is a receptor for multiple arthritogenic alphaviruses. Nature 557, 570–574 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sheikh A., Figure 1c original blots. Figshare. 10.6084/m9.figshare.12137544. Deposited 16 April 2020. [DOI] [Google Scholar]
- 45.Sheikh A., Figure 2a. Figshare. 10.6084/m9.figshare.12141033. Deposited 16 April 2020. [DOI]
- 46.Sheikh A., Figure 3. Figshare. 10.6084/m9.figshare.12141228. Deposited 16 April 2020. [DOI] [Google Scholar]
- 47.Sheikh A., Figure 4a.tiff. Figshare. 10.6084/m9.figshare.12137658. Deposited 16 April 2020. [DOI] [Google Scholar]
- 48.Sheikh A., Figure 4b.tiff. Figshare. 10.6084/m9.figshare.12137652. Deposited 16 April 2020. [DOI] [Google Scholar]
- 49.Sheikh A., Figure 4D.tif. Figshare. 10.6084/m9.figshare.12137655. Deposited 16 April 2020. [DOI] [Google Scholar]
- 50.Sheikh A., Figure 4f. Figshare. 10.6084/m9.figshare.12141138. Deposited 16 April 2020. [DOI]
- 51.Sheikh A., Immunoblot of small intestinal enteroids following CEACAM6 siRNA knockdown. untreated cells>scramble siRNA control, CEACAM6 siRNA. Figshare. 10.6084/m9.figshare.12137661. Deposited 16 April 2020. [DOI]
- 52.Sheikh A., Immunnohistochemistry of small intestinal biopsy sections from infected hosts. Figshare. 10.6084/m9.figshare.12138297. Deposited 16 April 2020. [DOI]
- 53.Sheikh A., Supplementary figure 4b. Figshare. 10.6084/m9.figshare.12142092. Deposited 16 April 2020. [DOI]
- 54.Sheikh A., Immunoblot comparison of CEACAM5 expression in small intestinal enteroids following treatment with LT, mLT(E112K), or LT-B subunit. Figshare. 10.6084/m9.figshare.12140994. Deposited 16 April 2020. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Original image files have been deposited in Figshare (10.6084/m9.figshare.12137544, 10.6084/m9.figshare.12141033, 10.6084/m9.figshare.121412-28, 10.6084/m9.figshare.12137658, 10.6084/m9.figshare.12137652, 10.6084/m9.figshare.12137655, 10.6084/m9.figshare.12141138, 10.6084/m9.figshare.12137661, 10.6084/m9.figshare.12138297, 10.6084/m9.figshare.12142092, 10.6084/m9.figshare.12140994) (44–54).