Significance
The chloroviruses are unusual because they are predicted to encode most, if not all, of the machinery to synthesize the glycans attached to their major capsid proteins. Here we show that two of the virus-encoded proteins A064R and A061L are functionally active. A064R has three domains: The first two are GTs and the third domain is a methyltransferase. A061L has a methyltransferase activity. The action of these two enzymes produce the fragment 2,3-di-O-methyl-α-l-Rha-(1→2)-β-l-Rha, which is part of the complex N-linked glycan attached to the virus capsid protein. A064R domain 2 is a member of a new GT family. This provides direct evidence that the synthesis of PBCV-1 glycans are accomplished with virus-encoded enzymes.
Keywords: glycosyltransferases, methyltransferases, multi domain protein, chloroviruses, N-glycan
Abstract
Paramecium bursaria chlorella virus-1 (PBCV-1) is a large double-stranded DNA (dsDNA) virus that infects the unicellular green alga Chlorella variabilis NC64A. Unlike many other viruses, PBCV-1 encodes most, if not all, of the enzymes involved in the synthesis of the glycans attached to its major capsid protein. Importantly, these glycans differ from those reported from the three domains of life in terms of structure and asparagine location in the sequon of the protein. Previous data collected from 20 PBCV-1 spontaneous mutants (or antigenic variants) suggested that the a064r gene encodes a glycosyltransferase (GT) with three domains, each with a different function. Here, we demonstrate that: domain 1 is a β-l-rhamnosyltransferase; domain 2 is an α-l-rhamnosyltransferase resembling only bacterial proteins of unknown function, and domain 3 is a methyltransferase that methylates the C-2 hydroxyl group of the terminal α-l-rhamnose (Rha) unit. We also establish that methylation of the C-3 hydroxyl group of the terminal α-l-Rha is achieved by another virus-encoded protein A061L, which requires an O-2 methylated substrate. This study, thus, identifies two of the glycosyltransferase activities involved in the synthesis of the N-glycan of the viral major capsid protein in PBCV-1 and establishes that a single protein A064R possesses the three activities needed to synthetize the 2-OMe-α-l-Rha-(1→2)-β-l-Rha fragment. Remarkably, this fragment can be attached to any xylose unit.
Posttranslational glycosylation is a common event in eukaryotic cells, and it has been estimated that ≥50% of cellular proteins are glycosylated. Eukaryotic protein glycosylation usually takes place in the endoplasmic reticulum (ER) and Golgi apparatus (1, 2). Likewise, structural proteins of many viruses that infect eukaryotes, such as rhabdoviruses, poxviruses, and paramyxoviruses, are glycosylated (3). Virus glycoprotein glycans are typically N-linked to Asn via N-acetylglucosamine (GlcNAc), whereas O-linked glycosylation also occurs but is less frequent (4). Most viruses studied to date use host-encoded GTs and glycosidases to add and remove sugar residues from viral N-linked glycoproteins either cotranslationally or shortly after translation of the protein (5–8). As a result, virus glycoproteins are transported to the host plasma membrane where progeny viruses acquire their glycoprotein(s) coats by budding through the membrane.
However, glycosylation of the major capsid protein (MCP) of the plaque-forming chloroviruses (family Phycodnaviridae) differs from this scenario. Chloroviruses encode most, if not all, of the machinery for MCP glycosylation. Furthermore, all experimental results indicate that this process occurs in the cytoplasm rather than in the ER and Golgi (3, 9). The prototype chlorovirus PBCV-1 has a 330-kb genome that is predicted to code for ∼410 proteins (10); at least, 17 of these genes encode enzymes that manipulate sugars at different levels. Its MCP (also referred to as Vp54) is coded by gene a430l and has a predicted molecular mass of 48,165 Da, which increases to 53,790 Da as a result of N-glycosylation (11–13).
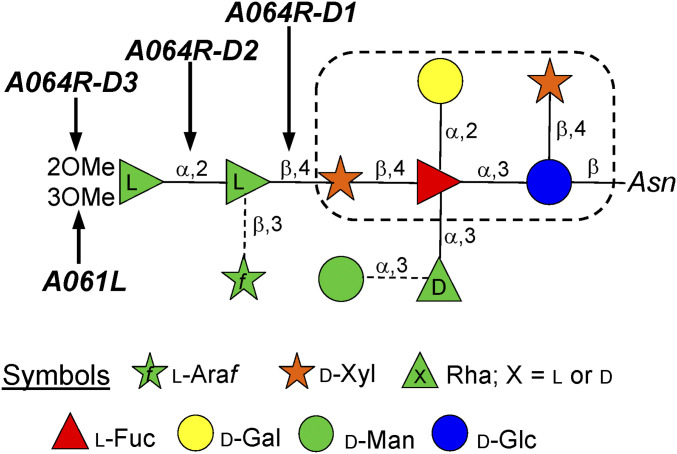
Vp54 has four glycosylation sites, and the predominant oligosaccharide is a nonasaccharide (Fig. 1) (14) with several unusual structural features: 1) It is not linked to a typical Asn-X-(Thr/Ser) sequon; 2) it is attached to the protein by a β-glucose linkage, which is rare in nature and has only been reported in glycoproteins from a few organisms (15–18); 3) it is highly branched; 4) fucose (Fuc) is substituted at all available positions; 5) it contains two Rha residues with opposite configurations plus a terminal l-Rha capped with two O-methyl groups; and 6) two monosaccharides, arabinose (Ara) and mannose (Man), occur as nonstoichiometric substituents, resulting in four glycoforms. These N-glycan structures are unique and do not resemble any known eukaryotic, bacterial or archaeal glycans. Interestingly, all chloroviruses analyzed to date shared a conserved core region (Fig. 1) that is further substituted with other monosaccharides, whose pattern depends on the chlorovirus (19–21).
Fig. 1.
N-glycan structure isolated from the chlorovirus PBCV-1 (adapted from 14) indicating the activities of the GTs described in this study. The dotted box defines the motif of the conserved core region common to all chloroviruses studied to date. Glycosidic linkages denoted with a broken line indicate non stoichiometric substituents, creating four different glycoforms.
PBCV-1 encodes seven putative sugar manipulating enzymes with most of them predicted to be involved in the synthesis of its N-glycan(s) as disclosed by bioinformatic analyses (3, 9): A064R (638 amino acids), A111/114R (860 aa), A219/222/226R (677 aa), A473L (517 aa), A546L (396 aa), A071R (354 aa), and A075L (280 aa). However, this number is less than the 10 sugars present in the N-glycan attached to the MCP. Several possibilities could explain this discrepancy: 1) One or more of the seven putative GTs might have multiple functional domains, making this restricted repertoire of enzymes sufficient to synthesize these structures; 2) PBCV-1 genes encode enzymes that do not resemble GTs in the databases and, hence, they are not recognized during query searches; 3) a host-encoded GT(s) could contribute to the glycosylation process.
Among the genes mentioned above, the protein coded by gene a064r is of interest because it is found in PBCV-1 but not in other chloroviruses where the glycan structures are known (19–21). Also, experimental evidence shows that isolated spontaneous mutants (or antigenic variants) of PBCV-1 with mutations in a064r have truncated glycan structures (9, 11). A combination of genetic and structural analyses suggested that the A064R protein has three putative domains of ∼200 amino acids each (SI Appendix, Fig. S1). Domain 1 (A064R-D1) was predicted to encode a β-l-rhamnosyltransferase and domain 3 (A064R-D3) was predicted to encode a S-adenosyl-l-methionine-dependent methyltransferase (SAM)-dependent methyltransferase that decorates two positions in the terminal α-l-Rha unit (Speciale 2019). The A064R domain 2 (A064R-D2) was hypothesized to encode an α-l-rhamnosyltransferase, despite only resembling bacterial proteins with unknown functions, thus, suggesting that it could be a new GT family.
Here we provide evidence that the predictions about the three A064R domains are correct with the exception of A064R-D3, which adds only one methyl group and not two as proposed. It methylates O-2 of the α-l-Rha residue, while O-3 methylation is accomplished by another virus-encoded protein A061L.
Results
A064R-D1: β-l-Rhamnosyltransferase Activity.
A064R-D1, amino acid residues 1–212 in the PBCV-1 A064R protein (SI Appendix, Fig. S1A) was originally predicted to be a GT in subfamily 34 (GT34; retaining) (22). Thereafter, A064R-D1 was expressed as a recombinant protein and crystallized (22) confirming it to be a member of the GT34 subfamily and preferring uridine 5′-diphosphate (UDP)-glucose as a donor substrate. However, this study occurred before the PBCV-1 glycan structure(s) was known, and this first hypothesis was later refuted by Speciale et al. (9) who predicted that A064R-D1 added l-Rha to d-Xyl.
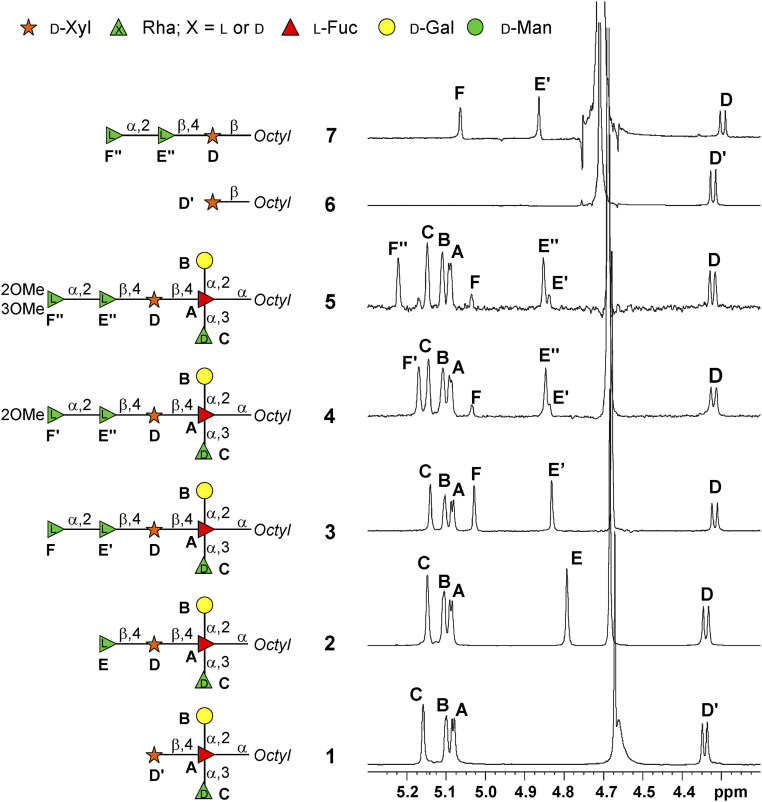
Here, we biochemically evaluated this prediction by using the tetrasaccharide 1 (Fig. 2) as the acceptor substrate. This oligosaccharide is a simplified truncated version of the N-glycan from PBCV-1 in which the d-Xyl is available for glycan elongation. In contrast to the natural glycan, the Fuc in 1 is capped at the reducing end with an octyl group (Fig. 2 and SI Appendix, NMR Characterization, Figs. S2 and S3, and Table S1), facilitating the monitoring of the reaction via high-pressure liquid chromatography (HPLC) and the purification of the products. UDP-β-l-Rha was used as the donor, and the reaction was monitored by either the HPLC or by the UDP-Glo assay. When the reaction mixture included Mn2+, the HPLC profile (Fig. 3A) showed the formation of pentasaccharide 2 while 1 simultaneously decreased. NMR analysis of 2 revealed an additional signal at 4.79 parts per million (ppm), labeled E, in the anomeric region (Fig. 2). Comparison of the heteronuclear single quantum correlation (HSQC) spectrum of 2 (SI Appendix, Fig. S4) with that of 1 identified the three common residues: namely, A (2,3,4-substitued α-l-Fuc), B (terminal α-d-Gal), and C (terminal α-d-Rha). D The β-d-Xyl unit showed some chemical shift variations because of glycosylation at O-4 as inferred by the downfield displacement of the corresponding carbon chemical shift (79.4 ppm, SI Appendix, Table S2) compared to the reference (70.7 ppm as found in 1, SI Appendix, Table S1).
Fig. 2.
(600 MHz and 310 K, D2O) 1H NMR spectra detailing the anomeric region of the oligosaccharides 1–7, drawn as a cartoon next to each spectrum. Each monosaccharide unit is labeled with a capital letter, and the full NMR attribution of each compound is reported in the SI Appendix. The peak at ca. 4.7 ppm is the residual solvent signal.
Fig. 3.
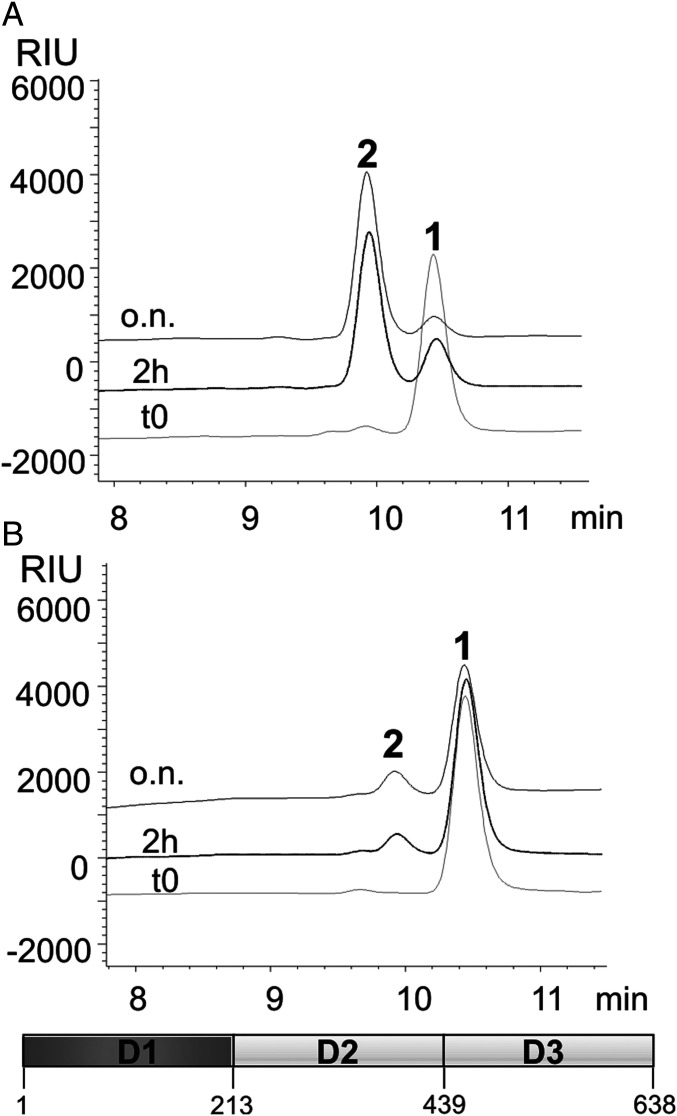
HPLC chromatographic profiles obtained from the reaction of the A064R-D1 enzyme (schematic at the bottom indicating its length in aa) with tetrasaccharide 1 (acceptor) and UDP-β-l-Rha (donor) in the presence of (A) Mn2+ and (B) with EDTA and without cations. Peak labels refer to oligosaccharide structures in Fig. 2.
As expected, the newly added residue in 2 was a β-l-Rha (E) linked at position 4 of d-Xyl (D) as proven by the diagnostic correlations found in both heteronuclear multiple bond correlation (HMBC) and T-ROESY spectra (SI Appendix, Figs. S4 and S5B). The β-configuration of this anomeric center was inferred by the diagnostic nuclear Overhauser effect (NOE) contacts between the anomeric proton H-1 of E with its H-3 and H-5 protons and confirmed by the 1JC,H value (160.3 Hz).
To demonstrate the importance of the cation, the same reaction was repeated without Mn2+ along with ethylenediaminetetraacetic acid (EDTA); the HPLC profile (SI Appendix and Fig. 3B) revealed that the enzyme required Mn2+ to function. Together, these results establish that A064R-D1 acts as a Mn2+-dependent β-l-rhamnosyltransferase, able to form the β-l-Rha-(1→4)-β-d-Xyl linkage. Moreover, the complete conversion of the substrate attests to a lack of a relevant hydrolytic activity versus the donor under the conditions tested, similar to what is observed for the other enzymes (A064R-D2 and A064R-D1D2).
These results were confirmed by the bioluminescent UDP-Glo assay in which the specificity of the enzyme for Mn2+ and Mg2+ was tested, disclosing the enzyme’s affinity for Mn2+ (SI Appendix, Fig. S6A), in agreement with previous studies (22). The nature of the acceptor and donor substrates was also investigated, leading to further information (SI Appendix, Fig. S6B): 1) The enzyme did not recognize UDP-α-d-Glc, in agreement with our previous prediction and contrary to previous studies (22); and 2) the enzyme recognized d-Xyl alone as an acceptor, albeit with a lower affinity with respect to acceptor 1.
A064R-D2: α-l-Rhamnosyltransferase Activity.
We previously predicted that A064R-D2 adds the second l-Rha to the first l-Rha (3, 9). To test this prediction, three versions of the second domain were designed by changing the boundaries, cloned and tested (SI Appendix, Fig. S1A). The first construct (215 aa; named D2) was based on bioinformatic studies. The other two constructs included 33 additional amino acids at the C terminus and differed in length of the N-terminal region: D2L (aa 191–438) and D2L2 (aa 213–438).
C-terminal elongation was investigated because the antigenic variant CME6 (9) has a functional D2 domain longer than the bioinformatic prediction. The first 17 amino acids of the N-terminal region were omitted in the D2L2 construct because they were predicted to form a flexible loop and, therefore, unlikely required for activity.
The three proteins were screened for activity using pentasaccharide 2 (Fig. 2) as an acceptor and UDP-β-l-Rha as a donor with or without cations (Mg2+ or Mn2+). Interestingly, the construct initially developed from the bioinformatic studies (D2) was not active (SI Appendix, Fig. S7A), while the other two D2L and D2L2 produced the hexasaccharide 3 (SI Appendix, Fig. S7 B and C).
The same reaction occurred with the D2L enzyme after five days of storage at 4 °C indicating that the enzyme was still active (SI Appendix, Fig. S7D). 1H-NMR spectroscopic investigation showed that 3 had an additional anomeric signal (5.03 ppm, labeled F) compared to 2 (Fig. 2). Comparison of the HSQC spectra of 2 and 3 (SI Appendix, Fig. S8A) identified l-Fuc (A), d-Gal (B), α-d-Rha (C), and d-Xyl (D), while the chemical shifts of β-l-Rha (E′) were slightly different due to its glycosylation at C-2, (78.1 ppm instead of 71.8 ppm, SI Appendix, Table S3). The new unit F was a terminal α-l-Rha; its anomeric signal showed only one correlation in the total correlation spectroscopy (TOCSY) pattern (SI Appendix, Fig. S8B) ascribed to the H-1/H-2 cross peak, whereas the connections up to the methyl group H-6 (1.30 ppm) were visible from H-2, while the C-5 chemical shift value at 69.7 ppm supported the α-configuration of this residue (23).
These results establish that A064R-D2 is an inverting GT, that forms the α-l-Rha-(1→2)-l-Rha linkage. Moreover, amino acids 191–212 at the N terminus do not contribute to the functionality of the protein, but the 33 extra amino acids at the C terminus in D2L2 and D2L are crucial for activity. It is possible that these residues are involved in some dynamic interaction that enables the proper protein folding or confers stability. The reaction was cation independent as confirmed by adding EDTA to the mixture (SI Appendix, Fig. S7E).
Two additional experiments were performed to learn more about the substrate specificity of A064R-D2. First, the octyl xyloside 6 (Fig. 2) was used as the acceptor with the D2L construct, and no reaction occurred (SI Appendix, Fig. S7F). Next, the reaction was performed using l-Rha monosaccharide as the acceptor. In this case, the reaction could not be monitored via HPLC, but NMR analysis after overnight incubation revealed the formation of a α-l-Rha-(1→2)-l-Rha disaccharide. The 1H-NMR spectrum of the product differed from those of the reagents Rha and UDP-Rha (SI Appendix, Fig. S9A), and the HSQC spectrum (SI Appendix, Fig. S9B) displayed two anomeric signals at 1H/13C 5.22/93.7 ppm and 4.96/103.3 ppm related to the two-linked α-l-Rha at the reducing end (R) and to the terminal α-Rha (T), respectively (SI Appendix, Table S4). The NMR analysis (SI Appendix, Fig. S9 B and C) confirmed the identity of the disaccharide. In particular, the downfield shift of the C-2 carbon signal of residue R (to 80.3 ppm), confirmed its glycosylation and showed that the enzyme was able to link α-l-Rha at O-2 to another l-Rha unit. A detailed inspection of the spectra detected trace amounts of disaccharide α-l-Rha-(1→2)-β-l-Rha, as well.
Thus, D2L recognizes free Rha as an acceptor. Based on the structure of 2, which has a β-Rha, we hypothesize that the β-anomer of Rha is the substrate of the enzyme not the α-anomer. Once formed, the reducing end of the disaccharide is converted by mutarotation into the predominant α-form, while some of the free α-Rha replaces the β-form that was consumed. This β-form then reacts with the enzyme and additional UDP-β-l-Rha to give the disaccharide, and this process continues up to the total consumption of the monosaccharide. This hypothesis needs to be validated by future work.
Because A064R-D2 resembles several proteins with unknown functions in the GenBank, we predict that these homologs are likely to be undiscovered GTs. Thus, A064R-D2 is a new cation-independent inverting GT that can interact with different Rha-containing molecules.
A064R-D1D2 Activity.
To confirm our previous findings and expand our understanding about A064R-D1 and A064R-D2 activities, two additional experiments were performed. We made a recombinant protein, A064R-D1D2 (aa 1–438), similar to that encoded by the antigenic variant CME6 virus (9), comprising D1 and D2L active domains.
The first reaction used tetrasaccharide 1 as an acceptor and two equivalents of UDP-l-Rha and Mn2+; formation of hexasaccharide 3 was almost complete after 2 h (SI Appendix, Fig. S10A).
The other acceptor used was monosaccharide 6 (Fig. 2) which disclosed that A064R-D1D2 worked also with a different d-Xyl source. The chromatographic profile after 2 h showed that 6 was replaced with trisaccharide 7 (SI Appendix, Fig. S10B), whose identity was confirmed by NMR analysis (Fig. 2 and SI Appendix, Fig. S11, chemical shifts in SI Appendix, Table S5), namely, that it was 6 with the α-l-Rha-(1→2)-β-l-Rha disaccharide attached to O-4 of the xylose unit as expected.
A064R-D1 and A064R-D2 Bioinformatic Analysis.
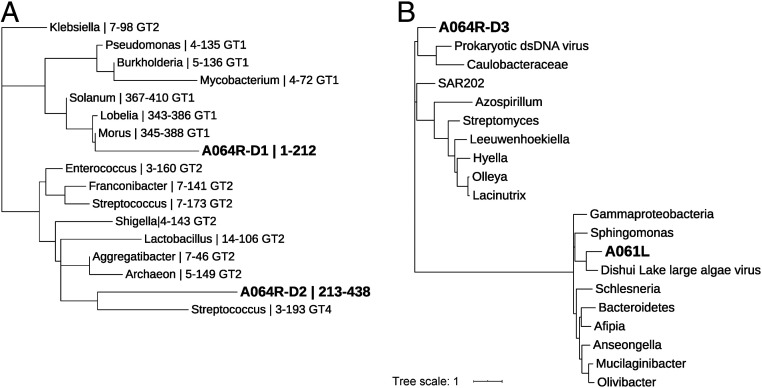
Phylogenetic analysis of annotated GT domains from characterized rhamnosyltransferases revealed that the A064R-D1 was more like members in the GT1 family rather than the GT2 family (Fig. 4A, detailed list of the organisms used in SI Appendix, Table S6). However, a previous crystal structure of A064R-D1 indicated a structural fold similar to catalytic domains of retaining glycosyltransferases in the GT-A family (characterized by a N-terminal Rossmann-like fold) (22), although the amino acid sequence similarity was very low (less than 14% for equivalent Cα atoms), further validating its separation from other characterized rhamnosyltransferase proteins. An A064R-D1 pBLAST query search shows the highest sequence similarity to hypothetical proteins from bacteria and a galactosyl transferase domain from Burkholderia. Interestingly, A064R-D1 shares an ancestral node of origin with eukaryotic homologs (SI Appendix, Fig. S12A).
Fig. 4.
Maximum-likelihood phylogenetic trees of some PBCV-1 encoded proteins involved in glycan synthesis. Phylogenetic trees are shown for GT-associated domains from (A) characterized rhamnosyltransferases and (B) selected methyltransferases. (A) Tree construction is based on GT-associated domains of virus-encoded rhamnosyltransferases A064R-D1, A064R-D2, and a variety of annotated rhamnosyltransferases from CAZy. (B) Tree construction is based on methyltransferase homologs of virus-encoded proteins A064R-D3 and A061L. The branch lengths indicate evolutionary distance and are labeled with (A) organism, residue range of GT domain, and GT family (A); and organism (B). PBCV-1 encoded proteins are in bold. The list of the organisms used along with their UniProtKB gene accession nos. is reported in SI Appendix, Table S6.
Interestingly, A064R-D2 is the most unusual of the three A064R domains because it least resembles a recognizable GT. Its lack of sequence homology with annotated rhamnosyltransferases (<30% with only 15% coverage) makes it an outlier in a phylogenetic analysis that groups more closely with other GT2 family members rather than GT1s in the phylogenetic tree (Fig. 4A). We used BLASTp to further evaluate A064R-D2 (aa 213–439, SI Appendix, Fig. S1A). This domain shares limited homology with several hypothetical and uncharacterized proteins from bacteria, including one GT-family protein from Lacipirellula parvula (31.36% identities with 95% coverage) and two homologs in Pithovirus: a hypothetical protein and a GT10 fucosyltransferase (>45% and >35% identities covering the C-terminal domain, respectively). The majority of A064R-D2 homologs are of bacterial origin (SI Appendix, Fig. S12B).
A064R-D3: SAM-Dependent Methyltransferase Activity.
A064R-D3 (aa 439–638) is predicted to have SAM-dependent methyltransferase activity. An A064R-D3 pBLAST query search shows the highest sequence similarity to several hypothetical proteins from bacteria and many class I SAM-dependent methyltransferases of both bacteria and virus origins (SI Appendix, Fig. S13A). Therefore, we previously hypothesized that it might add the two methyl groups to the terminal l-Rha of the glycan (9). To address this hypothesis, we used the complete A064R protein because the third domain by itself did not form a soluble recombinant protein.
Hence, A064R was incubated with 1 as the acceptor and with two equivalents of UDP-β-l-Rha with Mn2+ and Mg2+ as the cation requirement of this domain was unknown. Due to the labile nature of SAM, it was not possible to add two equivalents with certainty, thus, we repeatedly added SAM until no increase in new products occurred as judged by HPLC.
The reaction produced two products (SI Appendix, Fig. S14A). The less abundant one was hexasaccharide 3 as established by its retention time and by NMR analysis of the mixture. The NMR analysis of the most abundant product 4 (Fig. 2 and SI Appendix, Fig. S14 B and C and Table S7) revealed that methylation only occurred at O-2 of the terminal α-l-Rha residue not at both O-2 and O-3 as hypothesized.
1H-NMR and HSQC spectra of 4 (Fig. 2 and SI Appendix, Fig. S14 B and C) revealed the same residues found in 3 with H-1 of E (here, labeled E″) slightly shifted and a new anomeric signal at 5.17 ppm, labeled as F′, which instead replaced F. The anomeric proton of F′ had the TOCSY pattern typical of a Rha unit, α-configured based on its C-5 value (69.5 ppm), and methylated at O-2 based on the diagnostic value of C-2 (81.2 ppm). This finding was confirmed from the NOE contact between H-2 of F′ with the methyl group at 3.45 ppm in the T-ROESY spectrum, which also reported that F′ was linked to O-2 of E'' (SI Appendix, Fig. S14C).
This reaction established that the third domain of A064R only added one methyl group to the C-2 hydroxyl group of the terminal α-l-Rha, leaving open the question about what enzyme methylated the C-3 hydroxyl group.
A061L: Methyltransferase Activity.
To determine the enzyme responsible for adding a methyl group to O-3 of the terminal α-l-Rha residue, we screened the PBCV-1 genome for another gene encoding an O-methyltransferase. This led to the discovery of the gene a061l (SI Appendix, Fig. S1A). Phylogenetically distinct from A064R-D3 (Fig. 4B), the closest homolog of A061L is a macrocin-O-methyltransferase from Dishui Lake large algae virus 1. An A061L pBLAST query search shows the highest sequence similarity to several well-characterized class I SAM-dependent methyltransferases from bacteria and, although more distantly related, from cyanobacteria and algae (SI Appendix, Fig. S13B). Moreover, it had a structural homology of 90% of the sequence modeled with 100% confidence with the O-methyltransferase NovP from Streptomyces spheroids.
Recombinant A061L was tested for activity using hexasaccharides 3 or 4 as acceptors, SAM as a donor of the methyl group, and with both Mg2+ and Mn2+. The two acceptors differed only in the presence of the methyl group on O-2 of the terminal l-Rha (Fig. 2). A061L only methylated hexasaccharide 4, the substrate with the terminal l-Rha already possessing a methyl group at O-2 (SI Appendix, Fig. S15). No reaction occurred with the unmethylated hexasaccharide 3. The structure of product 5 was inferred via NMR spectroscopic analysis (SI Appendix, Fig. S16 and Table S8) which confirmed the presence of two methyl groups (at O-2 and O3) on the terminal l-Rha unit.
This last experiment demonstrated that the virus-encoded A061L added a methyl group to the O-3 of the terminal l-Rha only when it was already methylated at O-2. However, it remains unclear if it has a cation requirement. Additional experiments will be performed to better define all of the characteristics of this enzyme.
Modeling the Two Methyltransferases.
The methyltransferase activity of A064R-D3 was supported by bioinformatic information; the protein has 31.5% identity with 91% coverage (3e−19 confidence value) with a protein coded by a prokaryotic dsDNA virus sp. (UniProtKB gene accession no. A0A516L7Y9). This viral gene encodes a putative 8-demethyl-8-α-l-rhamnosyl tetracenomycin-C2′-O-methyltransferase (Fig. 4B).
Moreover, a protein structure prediction by Phyre2 analysis chose the SAM/metal-dependent O-methyltransferase MycE from Micromonospora griseorubida to predict the 3D structure of A064R-D3 based on 178 residues (89% of the sequence modeled with 100% confidence). The choice by Phyre2 to use this particular methyltransferase is consistent with our results.
Previously, crystal structures were determined for MycE bound to the product S-adenosyl-l-homocysteine (SAH) and magnesium, the first structure of a natural product sugar methyltransferase in complex with its natural substrate (24). The structure of A064R-D3 (aa 446–638) is predicted to accurately trace the catalytic methyltransferase domain of MycE (aa161–399) from residues 161–340 situated in the C terminus comprising an α/β-sandwich with a seven-stranded β-sheet core sandwiched by three α-helices on the front face and two on the back. In agreement with other class I methyltransferases, A064R-D3 possesses a series of conserved motifs shared among these proteins, named motifs I–VI (25).
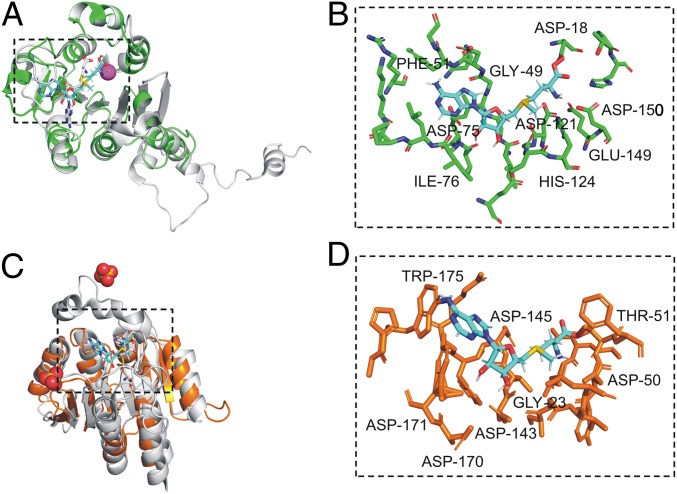
Notably, A064R-D3 residues 43–51 (Ile43, Ile44, Glu45, Ile46, Gly47, Ile48, Gly49, Asp50, and Phe51) resemble motif I, a nine-residue amino acid block with the consensus sequence (V/I/L)(L/V)(D/E)(V/I)G(G/C)G(T/P)G. This nine-residue structure contains the glycine-rich ‘‘GxGxG’’ signature sequon, a SAM-binding motif found in almost all SAM-dependent methyltransferases. Prediction of the A064R-D3 SAM-binding sites was corroborated by the enzyme-ligand structure model (Fig. 5 A and B). Residues positioned within 5 Å of the SAM ligand are organized into three major regions: 46–51, 74–81, and 121–124. The predicted SAM-binding site for A064R-D3 is located within the loops at the C-terminal end of the central β-sheet as usually occurs for methyltransferases. Prediction models detect coordination of a metal ion by homologous residues Asp121, Glu149, and Asp150 in accord with the metal dependence of activity observed in MycE (Asp275, Glu303, and Asp304). These residues are strictly conserved among the MycE homologs, implying that all homologs are metal dependent. The SAM amino acid moiety contacts Asp18 through the carboxyl group and with Asp121 and Asp150 through the amino group. As for the metal ligands, the residues that contact SAM are highly conserved. Protein sequence alignment of A064R-D3 and MycE displays homology in critical functional residues (SI Appendix, Fig. S17). In total, A064R-D3 has 12 of the 23 SAM-binding residues with MycE. Furthermore, active site residues Tyr208 and His278 determined in MycE have homology with A064R-D3 residues Phe51 and His124. The single-residue substitution Y208F MycE resulted in reduced but not abolished enzyme activity (24).
Fig. 5.
Stereoviews showing superpositions of methyltransferases A064R-D3 and A061L with structural homologs. (A) Superposition of predicted A064R-D3 (green) complexed with SAM (cyan) with crystalized SAM/metal-dependent sugar O-methyltransferase MycE chain E (gray) from Micromonospora griseorubida in complex with Mg ion (magenta) and SAH (gray). (B) Magnified view of the predicted A064R-D3 SAM-binding site. (C) Superposition of predicted A061L (orange) complexed with SAM (cyan) with crystalized SAM/metal-dependent sugar O-methyltransferase NovP chain A (gray) from S. spheroids in complex with sulfate ion (red and yellow) and SAH (gray). (D) Magnified view of the predicted A061L SAM-binding site. Residues represented by stick models are predicted to be positioned within 5 Å of the SAM ligand and have the equivalent residue position of ligand-binding amino acids in characterized sugar O-methyltransferase homologs. Labeled residues are predicted to bind SAM or metal ion via hydrogen bonding or participate in polar contact. Bound SAM and SAH are represented by stick models.
Phyre2 analysis of the SAM/metal dependent sugar O-methyltransferase NovP from S. spheroides allowed the prediction of the 3D structure of A061L based on 188 residues (modeled with 100% confidence). The A061L sequence contains recognizable motifs I and II that are typical of SAM-dependent methyltransferases (SI Appendix, Fig. S18). Motif I lies between β1 and α4 and forms the expected interactions with the amino acyl portion of SAM via Glu21 and Gly23, equivalent to residues Glu92 and Gly94 in NovP. Motif II, defined as an acidic loop, which lies at the C-terminal end of β2, forms the expected interactions with the ribose hydroxyls groups via Asp50, equivalent to residue Asp122 in NovP. The active center of NovP contains a strictly conserved metal-binding site composed of three Asp residues: Asp196, Asp223, and Asp224 (26). A further conserved Asp198 likely acts as the general base that initiates the methyl transfer reaction. Using in silico docking, we generated models of the A061L-SAM complex that are consistent with this mechanism (Fig. 5 C and D). A061L possesses a homologous three Asp sequence (Asp143, Asp170, and Asp171) and a general base Asp145, the putative general base of the reaction, is well conserved in A061L structural homologs. We propose this residue initiates the methyl transfer reaction by deprotonating the C-3 hydroxyl group of the l-Rha unit.
Discussion
To date, the evidence indicates that chloroviruses decorate their MCP with atypical N-glycans (19–21) and that the viral genome encodes most, if not all, of the enzymes responsible for synthesizing these glycans. Results in this paper support this hypothesis by defining the activity of protein A064R, one of the GTs encoded by chlorovirus PBCV-1, along with the function of the A061L protein encoded by another gene a061l.
A064R (638 aa) has three functional domains of ∼200 aa each. The first two are GTs, and the third is a methyltransferase that methylates O-2 of the terminal α-l-Rha residue. The finding that the third domain only transfers one methyl group prompted the search for a second virus-encoded methyltransferase, which led to the identification of A061L. This enzyme completes the methylation of the terminal α-l-Rha by adding a methyl group to its O-3 position but only after the O-2 position has been methylated.
From a structural viewpoint, a064r encodes a rare protein because it has three functional transferase activities: two different rhamnosyltransferases and one methyltransferase. To date, few enzymes with multiple GT activities have been identified, and most of them only have two functions. The oldest is KpsC, an enzyme with two GT activities that are involved in the biosynthesis of the Escherichia coli K5 polysaccharide (27). More recently, Clarke et al. (28) discovered an O2a polymerase (WbbM) in Klebsiella pneumoniae that possesses two domains, a galactopyranosyltransferase resembling known GT8 family enzymes and a galactofuranosyltransferase defining a previously unrecognized family (GT111). Only a few other multidomain enzymes have been identified with more than one GT active site: WbdA mannosyltransferase involved in the synthesis of the E. coli O9, O9a, and O8 lipopolysaccharide O antigens, which is recognized as a bifunctional α-(1→2)-, α-(1→3)-mannosyltransferase in serotype O9a, while its counterpart in serotype O8 is a trifunctional mannosyltransferase (α-[1→2], α-[1→3], and β-[1→2]) (29, 30). Analyses on the O-antigenic polysaccharide produced by some isolates of Raoultella terrigena and K. pneumoniae identified another trifunctional GT protein (WbbB), which is able to generate a polysaccharide of [4)-α-Rha-(1→3)-β-GlcNAc-(1→] repeating units capped with a nonreducing terminal residue of β-linked 3-deoxy-d-manno-oct-2-ulosonic acid (β-Kdo) (31). Other bifunctional microbial GTs are those involved in the biosynthesis of glycosaminoglycans and glycosaminoglycan-like polysaccharides, such as those involved in synthesis of hyaluronan (32), including one from chlorovirus PBCV-1 (33), chondroitin (34), and heparosan (35), along with KpsC from E. coli K5, mentioned above. The dGT1 enzyme from Streptococcus parasanguinis is a bifunctional protein involved in the biosynthesis of the serine-rich repeat protein adhesin Fimbriae associate protein (Fap1), which is essential for glycosylation of Fap1 (36). Thus, A064R is part of a restricted pool of GTs with three activities with the qualifier that the third domain is a methyltransferase.
The results in this paper lead to several significant conclusions: 1) PBCV-1 and probably other chloroviruses encode multifunctional proteins involved in glycan synthesis of their MCP glycoproteins; 2) the PBCV-1 protein A064R possesses a domain (D2) that may represent a new GT family; 3) the A064R protein and probably other chlorovirus encoded GTs are likely to be soluble due to the lack of N-terminal signal peptides that target them to the ER or the Golgi. This last conclusion suggests they may be easier to express and purify.
Beyond the significance of our findings to chlorovirus biology, we can imagine employing this protein or its individual domains for other purposes, e.g., the A064R-D1D2 protein could be used to produce a different class of rhamnolipid biosurfactants, having the rhamnobiose unit attached to an alkyl xyloside. Rhamnolipids are primarily produced from some strains of Pseudomonas (37) and have multiple uses and properties as active agents in skin reepithelialization in wound healing (38) or used in the cosmetics field for the treatment of wrinkles (39) or to alleviate and/or prevent immunological activities associated with autoimmune diseases (38).
In conclusion, the results described herein provide direct evidence that the synthesis of the PBCV-1 N-glycan, or, at least, part of it, is accomplished with enzymes encoded by the virus itself. This finding is particularly relevant as it subverts the dogma that all viruses uses host enzymes to glycosylate their proteins.
Materials and Methods
All recombinant proteins were expressed in E. coli cells. The A064R full-length protein was produced with a C-terminal 6xHis-tag (SI Appendix, Fig. S1 A and B) using the pET23a vector as described (40). Protein purification to homogeneity was performed using the Probond nickel-chelating resin (ThermoFisher, code R801-01), following the manufacturer’s protocol.
A064R domains (SI Appendix, Fig. S1 A and B) were cloned and expressed in the pGEX-6P1 vector as described (40) obtaining fusion proteins with the glutathione S-transferase (GST) tag at the N terminus, that were purified using glutathione Sepharose 4B (GE Healthcare, code no. 17–0756-01). The glutathione-Sepharose-bound proteins were used for the enzymatic reactions. Cleavage was achieved for A064R-D1 only by using Prescission protease (GE Healthcare, code no. 27–0843-01) on the column, and the released protein was concentrated using Amicon Ultra-4 (Millipore). The released domain was used for the UDP-Glo assay.
Purity of full-length A064R-6xHis protein and of the GST-fusion domains was determined by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS/PAGE) and Coomassie blue staining (SI Appendix, Fig. S1C).
A061L protein (SI Appendix, Fig. S1) was cloned and expressed in pGEX-5X-1 expression vector (GE Healthcare), producing a GST tag at the N terminus of the target protein. The recombinant protein was purified on glutathione Sepharose as before and used for enzymatic reactions (more details in SI Appendix, Materials and Methods).
All eluted proteins were resolved by SDS/PAGE with Coomassie brilliant blue staining solution (41).
Synthesis of Donor and Acceptor Substrates.
The UDP-β-l-Rha substrate (donor) was synthesized by using an enzymatic tool developed in our laboratory and then purified via HPLC. The synthesis of 1 and 6 is described in the SI Appendix (Synthesis, SI Appendix, Figs. S19–S21) and followed protocols previously reported (42).
Enzymatic Reactions.
All of the reactions were performed at 25 °C in phosphate buffer saline ([PBS]; Sigma-Aldrich, code no. P4417, composition: 0.01-M phosphate buffer, 0.0027-M potassium chloride, and 0.137-M sodium chloride, pH 7.4) by using the enzyme still attached to the resin through its GST tag or its His tag in the case of the whole A064R. In a typical experiment, the reaction volume was about 150 μL and included 50 μL of resin (glutathione Sepharose 4B GE Healthcare or Probond nickel-chelating resin) with ∼250 μg of protein adsorbed unless otherwise specified.
All enzymatic reactions were screened via HPLC, using a C18 column (Phenomenex Kinetex 5 μ, 2,50 × 4.60 mm, code no. 00G-4601-E0) and 70% methanol as an eluent (flow rate at 0.8 mL/min) by injecting 10 μL of the crude reaction after centrifugation.
For A064R-D1, the reaction was performed by using 1 (1.5 mM) (Fig. 2) as an acceptor and UDP-β-l-Rha (1.5 mM) as a donor in the presence of Mn2+ (2 mM). In order to verify the cation dependence of the enzyme, the reaction was repeated by adding EDTA (1 mM) to the protein 30 min prior to the addition of all of the other reagents except the cation.
For A064R-D2, three different constructs were produced and tested: D2 (191–405 aa), D2L (191–438 aa), and D2L2 (213–438 aa). All three constructs were tested with 2 (1.5 mM) as an acceptor and UDP-β-l-Rha (1.5 mM) and with or without cations (Mg2+ and Mn2+, 2 mM). To exclude any cation dependency, a reaction with D2L was also tested by adding EDTA (0.5 mM), keeping the other reagents unchanged. The D2L reaction was repeated replacing acceptor 2 with l-Rha monosaccharide (1.5 mM).
The resin with A064R-D1D2 was suspended in PBS buffer and incubated with 1 (1.5-mM UDP-β-l-Rha (3.6 mM) and Mn2+ (2 mM). In the second reaction, 6 (1.5 mM) was used instead of 1, while all other components were kept unchanged.
The resin with the full-length A064R attached was suspended with 1 (1.5 mM), UDP-β-l-Rha (3.4 mM), the precursor for the methyltransferases SAM (CAS no. 86867–01-8; code no. A7007, Microtech), and with both Mn2+ and Mg2+ (2 mM each).
As for A061L, two reactions were performed by varying the nature of the acceptor. In the first reaction, 3 (1 mM) was used as an acceptor along with an excess of SAM (ca. 2 mM), Mn2+ and Mg2+ (2 mM each); in the second reaction, 3 was replaced with 4.
Compounds Isolation.
The crude reaction solutions were centrifuged using spin columns with collection tubes (code no. H6787, Sigma-Aldrich) to remove the resin, and, then, the pure products were recovered by C18 Sep-Pak cartridges (pore size 125 Å, particle size 55–105 μm, code no. WAT051910, Waters) as detailed below.
The Sep-Pak cartridge was activated with 10 mL of ethanol, 4 mL of acetonitrile, and 10 mL of water. After being loaded with the supernatant, the following elution was performed: 20 mL of water, 15 mL of acetonitrile/water 1:4, 5 mL of acetonitrile, and 20 mL of ethanol. All fractions were dried and analyzed via NMR spectroscopy.
Bioluminescent Assay: UDP-Glo Kit.
Two UDP standard curves were prepared in a 96-well plate, performing a serial twofold dilution as described in the UDP-Glo kit (code no. V6961, Promega), obtaining 12 solutions at different UDP concentrations. One reaction contained Mn2+ the other contained Mg2+ at the same concentration (40 mM). Thereafter, 25 μL of each solution was transferred to a second 96-well plate, and 25 μL of UDP detection reagent (provided by the UDP-Glo kit) was added; the solutions were incubated at 25 °C for 1 h, and luminescence was measured with a luminometer.
Reactions that involved the A064R-D1 enzyme, 1 as an acceptor and UDP-β-l-Rha as a donor in separate phosphate buffer solutions (one with Mg2+ and the other with Mn2+), were performed maintaining a constant ratio of UDP-l-Rha/1 (2:1 ratio; 2.470 and 1.250 nmol, respectively), while the GT amount decreased from the first solutions to the last where GT was absent. Similar to the UDP standard curves, the solutions at different GT concentrations were obtained using twofold serial dilutions: 100 μL of the GT solution (5 μL of GT 150 μg/mL; 5-μL Mn2+/or Mg2+, 40 mM; 4.75-μL PBS 20×; 85.25 μL of water) was added in the first well followed by the addition of 50-μL buffer (composed of Mn2+/Mg2+, 40 mM; PBS 20×, and H2O) to the additional 11 wells. Then, 15 μL from each well was transferred into 12 wells of another 96-well plate, and 10 μL of the solution containing UDP-l-Rha (3.3 mM) and 1 (1 mM) was added. The reactions were incubated at 25 °C for 1 h, after that 25 μL of the UDP-detection buffer was added to each well, and the solutions were kept at the same temperature for one additional hour prior to measuring luminescence. Two replicas for each experiment were performed. The same strategy was used for the following two reactions, in which the activated sugar donor or the acceptor substrate were changed, maintaining the 2:1 ratio as before: in one, the UDP-l-Rha was substituted with the UDP-D-Glc (10 mM), and in the other, the d-Xyl monosaccharide (1 mM) was used instead of structure 1. Experiments were performed in the presence of both bivalent cations.
Luminescence measurements were performed with a Synergy HT multimode microplate reader (BioTek instrument) using a 96-well microplate with standard 128 × 86-mm geometry with an integration time of 1.0 s. Luminescence was measured by a low-noise photomultiplier detector through an empty filter position in the emission filter wheel. Data analysis and the construction of the graphs were achieved with Excel Software.
NMR Spectroscopy.
All NMR experiments of the oligosaccharides from 1 to 7 in Fig. 2 were recorded in D2O at 310 K on a Bruker DRX-600 MHz (1H: 600-MHz and 13C: 150-MHz) instrument equipped with a cryoprobe except for the Rha disaccharide for which a temperature of 298 K was used. All chemical shifts are referred to internal acetone (1H 2.225 and 13C 31.45 ppm).
The set of 2D spectra (correlation spectroscopy, TOCSY, T-ROESY, and HSQC) were measured for each substrate, except for 3, 7, and the α-l-Rha-(1→2)-l-Rha disaccharide for which only the HSQC and the TOCSY experiments were acquired and for structures 1 and 2 where HMBC was also recorded.
Homonuclear experiments were recorded using 512 free induction decays (FIDs) of 2,048 complex data points, setting 24 scans per FID for all experiments, whereas for structures 3 and 4, 32 scans per FID were set in order to obtain a better signal to noise ratio. Mixing time of 100 ms was applied for TOCSY and 300 ms for T-ROESY spectra acquisitions. 1H-13C heteronuclear experiments were acquired with 512 FIDs of 2,048 complex points with 40–80 scans per FID, depending on the sample abundance. Standard Bruker software Topspin 3.1 was used to process and analyze all spectra. NMR operating conditions used for the characterization of the synthetic intermediates of 1 and 6 are reported in the SI Appendix.
Phylogenetic Analyses.
Multiple sequence alignment and phylogenetic reconstructions were performed using the Environment for Tree Exploration3 (ETE3) v3.1.1 program (43) as implemented on the GenomeNet (https://www.genome.jp/tools/ete/).
Phylogenetic trees were constructed using maximum likelihood and neighbor-joining approaches using PhyML v20160115 ran with model parameters: -f m–pinv e -o tlr–nclasses 4–bootstrap 100–alpha e (44). The branch supports are the χ2-based parametric values return by the approximate likelihood ratio test. Maximum-likelihood annotation was generated by Interactive Tree of Life (iTOL) (45).
Viral proteins were used as queries to search the publicly available NCBI nonredundant sequence database using BLASTp and hits filtered based on general parameters excluding chloroviruses.
The characterized GT domains were identified using UniProt (46) and HHpred (47) from selected rhamnosyltransferases in the CAZy database.
Protein Modeling.
PBCV-1 A064R-D3 and A061L structure predictions were constructed by Phyre2 (48). Molecular docking of viral proteins complexed with SAM were performed using PatchDock (49), an algorithm based on shape complementarity principles. Protein complexes were inputted into the molecular modeling system, Chimera where 3D protein models were constructed. SAM-binding residues were generated by SAMbinder (50, 51). Final drawings and residue analysis were prepared with molecular graphics system PyMol, a molecular graphics system, Version 1.2r3pre, Schrödinger, LLC.
Supplementary Material
Acknowledgments
This work was funded in part by Mizutani Foundation Grant 180047 (C.D.C.), the National Sciences and Engineering Research Council of Canada (T.L.L.), the Canadian Glycomics Network (T.L.L.), NSF Grant 1736030 (J.L.V.E.), and the NSF Graduate Research Fellowship Program Grant 2505060195001 (E.N.). Alberta Innovates Technology Future is thanked for a studentship for S.L.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2016626117/-/DCSupplemental.
Data Availability.
All study data are included in the article and supporting information and the UniProt Knowledgebase (UniProtKB), https://www.uniprot.org/ (accession no. A0A516L7Y9).
References
- 1.Nagashima Y., von Schaewen A., Koiwa H., Function of N-glycosylation in plants. Plant Sci. 274, 70–79 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Varki A., Biological roles of glycans. Glycobiology 27, 3–49 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Etten J. L., et al. , Chloroviruses have a sweet tooth. Viruses 9, 88 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bagdonaite I., Wandall H. H., Global aspects of viral glycosylation. Glycobiology 28, 443–467 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doms R. W., Lamb R. A., Rose J. K., Helenius A., Folding and assembly of viral membrane proteins. Virology 193, 545–562 (1993). [DOI] [PubMed] [Google Scholar]
- 6.Olofsson S., Hansen J. E. S., Host cell glycosylation of viral glycoproteins–A battlefield for host defence and viral resistance. Scand. J. Infect. Dis. 30, 435–440 (1998). [DOI] [PubMed] [Google Scholar]
- 7.Hunter E., “Virus assembly” in Fields Virology, Knipe D. M., et al., Eds. (Wolters Kluwer/Lippincott Williams & Wilkins, Philadelphia, ed. 5, 2007), pp. 141–168. [Google Scholar]
- 8.Vigerust D. J., Shepherd V. L., Virus glycosylation: Role in virulence and immune interactions. Trends Microbiol. 15, 211–218 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Speciale I., et al. , Glycan structures of chlorovirus PBCV-1 major capsid protein antigenic variants help identify virus-encoded glycosyltransferases. J. Biol. Chem. 294, 5688–5699 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Etten J. L., Agarkova I. V., Dunigan D. D., Chloroviruses. Viruses 12, E20 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graves M. V., Bernadt C. T., Cerny R., Van Etten J. L., Molecular and genetic evidence for a virus-encoded glycosyltransferase involved in protein glycosylation. Virology 285, 332–345 (2001). [DOI] [PubMed] [Google Scholar]
- 12.Nandhagopal N., et al. , The structure and evolution of the major capsid protein of a large, lipid-containing DNA virus. Proc. Natl. Acad. Sci. U.S.A. 99, 14758–14763 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Castro C., et al. , Structure of the chlorovirus PBCV-1 major capsid glycoprotein determined by combining crystallographic and carbohydrate molecular modeling approaches. Proc. Natl. Acad. Sci. U.S.A. 115, E44–E52 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Castro C., et al. , Structure of N-linked oligosaccharides attached to chlorovirus PBCV-1 major capsid protein reveals unusual class of complex N-glycans. Proc. Natl. Acad. Sci. U.S.A. 110, 13956–13960 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wieland F., Heitzer R., Schaefer W., Asparaginylglucose: Novel type of carbohydrate linkage. Proc. Natl. Acad. Sci. U.S.A. 80, 5470–5474 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mengele R., Sumper M., Drastic differences in glycosylation of related S-layer glycoproteins from moderate and extreme halophiles. J. Biol. Chem. 267, 8182–8185 (1992). [PubMed] [Google Scholar]
- 17.Schreiner R., Schnabel E., Wieland F., Novel N-glycosylation in eukaryotes: Laminin contains the linkage unit beta-glucosylasparagine. J. Cell Biol. 124, 1071–1081 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gross J., et al. , The Haemophilus influenzae HMW1 adhesin is a glycoprotein with an unusual N-linked carbohydrate modification. J. Biol. Chem. 283, 26010–26015 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Castro C., et al. , N-linked glycans of chloroviruses sharing a core architecture without precedent. Angew. Chem. Int. Ed. 55, 654–658 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quispe C. F., et al. , Characterization of a new chlorovirus type with permissive and non-permissive features on phylogenetically related algal strains. Virology 500, 103–113 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Speciale I., Agarkova I., Duncan G. A., Van Etten J. L., De Castro C., Structure of the N-glycans from the chlorovirus NE-JV-1. Ant. van Leeu 110, 1391–1399 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y., Xiang Y., Van Etten J. L., Rossmann M. G., Structure and function of a chlorella virus-encoded glycosyltransferase. Structure 15, 1031–1039 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bock K., Pedersen S., Carbon-13 nuclear magnetic resonance spectroscopy of monosaccharides. Adv. Carbohydr. Chem. Biochem. 41, 27–65 (1983). [Google Scholar]
- 24.Akey D. L., et al. , A new structural form in the SAM/metal-dependent o-methyltransferase family: MycE from the mycinamicin biosynthetic pathway. J. Mol. Biol. 413, 438–450 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liscombe D. K., Louie G. V., Noel J. P., Architectures, mechanisms and molecular evolution of natural product methyltransferases. Nat. Prod. Rep. 29, 1238–1250 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Gómez García I., et al. , The crystal structure of the novobiocin biosynthetic enzyme NovP: The first representative structure for the TylF O-methyltransferase superfamily. J. Mol. Biol. 395, 390–407 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rigg G. P., Barrett B., Roberts I. S., The localization of KpsC, S and T, and KfiA, C and D proteins involved in the biosynthesis of the Escherichia coli K5 capsular polysaccharide: Evidence for a membrane-bound complex. Microbiology 144, 2905–2914 (1998). [DOI] [PubMed] [Google Scholar]
- 28.Clarke B. R., et al. , A bifunctional O-antigen polymerase structure reveals a new glycosyltransferase family. Nat. Chem. Biol. 16, 450–457 (2020). [DOI] [PubMed] [Google Scholar]
- 29.Greenfield L. K., et al. , Biosynthesis of the polymannose lipopolysaccharide O-antigens from Escherichia coli serotypes O8 and O9a requires a unique combination of single- and multiple-active site mannosyltransferases. J. Biol. Chem. 287, 35078–35091 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greenfield L. K., et al. , Domain organization of the polymerizing mannosyltransferases involved in synthesis of the Escherichia coli O8 and O9a lipopolysaccharide O-antigens. J. Biol. Chem. 287, 38135–38149 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams D. M., et al. , Single polysaccharide assembly protein that integrates polymerization, termination, and chain-length quality control. Proc. Natl. Acad. Sci. U.S.A. 114, E1215–E1223 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jing W., DeAngelis P. L., Dissection of the two transferase activities of the Pasteurella multocida hyaluronan synthase: Two active sites exist in one polypeptide. Glycobiology 10, 883–889 (2000). [DOI] [PubMed] [Google Scholar]
- 33.DeAngelis P. L., Jing W., Graves M. V., Burbank D. E., Van Etten J. L., Hyaluronan synthase of chlorella virus PBCV-1. Science 278, 1800–1803 (1997). [DOI] [PubMed] [Google Scholar]
- 34.Sobhany M., Kakuta Y., Sugiura N., Kimata K., Negishi M., The chondroitin polymerase K4CP and the molecular mechanism of selective bindings of donor substrates to two active sites. J. Biol. Chem. 283, 32328–32333 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chavaroche A. A., van den Broek L. A., Boeriu C., Eggink G., Synthesis of heparosan oligosaccharides by Pasteurella multocida PmHS2 single-action transferases. Appl. Microbiol. Biotechnol. 95, 1199–1210 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang H., et al. , New helical binding domain mediates a glycosyltransferase activity of a bifunctional protein. J. Biol. Chem. 291, 22106–22117 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abdel-Mawgoud A. M., Lépine F., Déziel E., Rhamnolipids: Diversity of structures, microbial origins and roles. Appl. Microbiol. Biotechnol. 86, 1323–1336 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stipcevic T., Piljac A., Piljac G., Enhanced healing of full-thickness burn wounds using di-rhamnolipid. Burns 32, 24–34 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Piljac T., Piljac G., “Use of rhamnolipids in wound healing, treating burn shock, atherosclerosis, organ transplants, depression, schizophrenia and cosmetics.” US Patent 7,262,171 B1 (2007).
- 40.Piacente F., et al. , Characterization of a UDP-N-acetylglucosamine biosynthetic pathway encoded by the giant DNA virus Mimivirus. Glycobiology 24, 51–61 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Smith B. J., Methods in Molecular Biology (Humana Press, Clifton, NJ, 1984), vol. 1. [Google Scholar]
- 42.Lin S., Lowary T. L., Synthesis of the highly branched hexasaccharide core of chlorella virus N-linked glycans. Chemistry 24, 16992–16996 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Huerta-Cepas J., Serra F., Bork P., ETE 3: Reconstruction, analysis, and visualization of phylogenomic data. Mol. Biol. Evol. 33, 1635–1638 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guindon S., et al. , New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321 (2010). [DOI] [PubMed] [Google Scholar]
- 45.Letunic I., Bork P., Interactive tree of life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 47, W256–W259 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.UniProt Consortium , UniProt: A worldwide hub of protein knowledge. Nucleic Sci. Res. 47, D506–D515 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zimmermann L., et al. , A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J. Mol. Biol. 430, 2237–2243 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Kelley L. A., Mezulis S., Yates C. M., Wass M. N., Sternberg M. J. E., The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schneidman-Duhovny D., Inbar Y., Nussinov R., Wolfson H. J., PatchDock and SymmDock: Servers for rigid and symmetric docking. Nucleic Acids Res. 33, W363-7 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Agrawal P., Mishra G., Raghava G. P. S., SAMbinder: A web server for predicting S-adenosyl-L-methionine binding residues of a protein from its amino acid sequence. Front. Pharmacol. 10, 1690 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Neuhaus J.-M., Sticher L., Meins F. Jr, Boller T., A short C-terminal sequence is necessary and sufficient for the targeting of chitinases to the plant vacuole. Proc. Natl. Acad. Sci. U.S.A. 88, 10362–10366 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and supporting information and the UniProt Knowledgebase (UniProtKB), https://www.uniprot.org/ (accession no. A0A516L7Y9).