Abstract
The protocol here presents a single experiment designed to introduce a trainee to multiple advanced bench and analysis techniques, including high dimensional cytometry, profiling cell signaling networks, functional assays with primary human tissue, and single cell analysis with machine learning tools. The trainee is expected to have only minimal lab experience and is not required to have any prior training in flow cytometry, immunology, or data science. This protocol aims to introduce the advanced research areas with a design that is robust enough that novice trainees will succeed, flexible enough to allow some project customization, and fundamental enough that the skills and knowledge gained will provide a template for future experiments. For advanced users, the updated phospho-flow protocol and the established controls, best practices, and expected outcomes presented here also provide a framework for adapting these tools in new areas with unexplored biology.
Keywords: Mass cytometry, signaling, phospho-flow, training
INTRODUCTION:
Phospho-specific flow cytometry or “phospho-flow” protocols were initially developed two decades ago (Krutzik, Irish, Nolan, & Perez, 2004; Krutzik & Nolan, 2003). At its core, phospho-flow takes advantage of three major advances that have reshaped cytometry since: 1) expansion of the number of per-cell measurements, 2) movement away from cell line models to the use of primary human tissues, and 3) an explosion in quantitative and computational analysis. Perhaps because it exists at the intersection of these advancing areas, phospho-flow has been highly impactful in basic and clinical research (Irish & Doxie, 2014; Rogne & Tasken, 2013). For example, phospho-flow has revealed risk stratifying cell biology in leukemia (Good et al., 2018; Irish et al., 2004; Levine et al., 2015), lymphoma (Irish et al., 2010; Myklebust et al., 2017), ovarian cancer (Gonzalez et al., 2018), and brain tumors (Nalin Leelatian et al., 2019). In non-malignant cells, phospho-flow has identified cell type specific signaling in common and rare immune cells, including B cells (Bendall et al., 2014; Huse et al., 2019; Irish, Czerwinski, Nolan, & Levy, 2006; Polikowsky, Wogsland, Diggins, Huse, & Irish, 2015) and T cells (Kalland, Oberprieler, Vang, Tasken, & Torgersen, 2011), and has provided insight into stem cell biology and the action of drugs (Bodenmiller et al., 2012; Krutzik, Crane, Clutter, & Nolan, 2008; Qin et al., 2020) and natural product discovery (Earl et al., 2018). The protocols have also been updated as new technologies emerged, such as mass cytometry (Bandyopadhyay, Fisher, Malkova, & Oh, 2017; N. Leelatian, Diggins, & Irish, 2015). Phospho-flow remains a cutting-edge approach that draws in trainees looking to make an impact in their chosen fields. For the same reasons, modern phospho-flow protocols can be overwhelming to new users who are typically unfamiliar with the core elements. Thus, there is a need to introduce new users from diverse fields to the core techniques of phospho-flow without overwhelming them with jargon and field-specific biology.
The primary goal of this protocol is to familiarize a new lab member with stimulating and measuring intracellular protein phosphorylation using human peripheral blood mononuclear cells and a set of well-characterized cytokines. The basis of this protocol comes from now well-established mass cytometry protocols (N. Leelatian et al., 2015), but could be replaced with those of spectral or traditional fluorescence flow cytometry. The protocol concludes with data analysis – an increasingly essential aspect of training new cytometry users (Diggins, Ferrell, & Irish, 2015; Saeys, Gassen, & Lambrecht, 2016)– and introduces a wide range of tools, including t-SNE (Amir el et al., 2013), UMAP (Becht et al., 2018), FlowSOM (Van Gassen et al., 2015b), and MEM (Diggins, Gandelman, Roe, & Irish, 2018; Diggins, Greenplate, Leelatian, Wogsland, & Irish, 2017). In addition to the analysis protocol, an R script and example data files are included so the user can walk through an automated analysis. This protocol was designed with cytometry novices in mind and emphasizes key principles of phospho-specific flow cytometry experimental design, teaches users to recognize major cell subsets in human blood, and explores fundamental cytokines and phospho-protein signaling events. The protocol is also designed to include both well-known signaling events and an optional “wildcard” that allows new users to bring in their interests or elements of a potential project. Ultimately, this introductory experiment is designed to demystify cutting-edge cytometry techniques that may initially seem daunting to beginners.
This phospho-flow protocol will instruct a flow cytometry novice in the basics of design and execution of a robust experiment that assesses signaling at the single cell level in primary human peripheral blood mononuclear cells (PBMCs). This benchtop protocol is accompanied by a supplementary protocol designed to offer a primer in high dimensional data analysis and mass cytometry specific data processing steps. These two protocols address the perceived difficulty of these each technique by simplifying the protocols, teaching the basics of each, and providing a reliable output. By executing these protocols, one will obtain a pilot mass cytometry dataset of healthy human blood, useful for learning immunology basics if they are not already known by the trainee. The trainee will also obtain signaling data, which can be compared to that of fellow trainees and existing literature to assess experiment execution. There are two protocols in this document. Basic Protocol 1 guides the user through a basic phospho-flow experiment, inducing signaling in healthy human peripheral blood with three different stimuli. These cells are then stained with a basic immunophenotyping panel and a signaling panel to phenotype cells in the sample and assess the effects of the stimuli used. These data can be analyzed in whatever way the user sees fit. Support Protocol 1 is provided for those who do not have an existing mass cytometry data analysis pipeline, and includes a manual gating scheme for data pre-processing as well as an R script for automated analysis of the data generated in Basic Protocol 1.
Significance:
Phospho-specific flow cytometry has been an especially impactful technique in clinical research. For trainees, although the phospho-flow protocol can be daunting, it provides an outstanding introduction to a range of essential quantitative cell biology techniques. This protocol updates established phospho-flow protocols with recent technique advances and a complete data analysis script in R. This protocol thus aims to increase the effective use of single cell signaling measurements and to provide a primer for a “first training experience” in quantitative cytometry labs.
Basic Protocol 1: Phospho-protein stimulation and mass cytometry data collection
Supplementary Protocol 1: Semi-automated analysis of signaling mass cytometry data in R.
BASIC PROTOCOL 1
Phospho-protein stimulation and mass cytometry data collection
In this protocol, the user will stimulate signaling in live human PBMCs using cytokines IL-4 and IFNγ as well as one other stimuli of their choice for 15 minutes, then fix the cells with paraformaldehyde and stain with a panel of mass cytometry antibodies designed to identify the major cell subsets in human blood as well as induced signaling events. This protocol is based on long-standing workflows in the flow cytometry community (Krutzik & Nolan, 2003). If multiple trainees are conducting the experiment in parallel with biologically identical samples (PBMCs from the same donor), near identical results should be observed across replicates, both in sample phenotyping and signaling results. It may be useful to have a seasoned technician perform the protocol in parallel with the trainees(s) to provide a benchmark for experimental outcomes. Prior protocols include preparation and cryopreservation of PBMCs (N. Leelatian et al., 2015) and other cell types form human tissue (N. Leelatian et al., 2017). As was done here, the usual precautions taken with human-derived materials should be observed, including best practices to avoid potential blood borne pathogen exposure and collection of samples in accordance with local and institutional guidelines of ethics, informed consent, and in accordance with the Declaration of Helsinki. Chemical hazards include paraformaldehyde and methanol; care should be taken to avoid inhalation or skin exposure to these reagents.
MATERIALS
Reagents, solutions, and samples:
Healthy human PBMCs cryopreserved in 90% FBS/10% DMSO (Nicholas et al., 2016)
Antibodies cocktail (see Reagents and Solutions for preparation)
Cell-ID™ Intercalator-Rh—500 μM (Fluidigm 201103A)
Cell-ID™ Intercalator-Ir—125 μM (Fluidigm 201192A)
EQ™ Four Element Calibration Beads (Fluidigm 201078)
RPMI with L-Glutamine
RPMI with L-Glutamine, 10% FBS, and antibiotic
PBS without Ca2+, Mg2+
DNase (Sigma Aldrich 12352100, reconstituted and aliquoted per manufacturer directions)
1% w/v BSA in PBS without Ca2+, Mg2+, sterile filtered
Ice cold methanol, stored at −20°C or lower
16% paraformaldehyde (Fisher Scientific 30525-89-4)
Ultra-pure water
15 mL screw top Falcon tubes
5 mL Falcon 2052 tubes, also known as FACS tubes (Fisher Scientific 30525-89-4)
Filter top FACS tubes (Fisher Scientific 352235)
Parafilm
Trypan blue
IL-4 20 ng/mL final concentration
IFN-γ 20 ng/mL final concentration
Trainee selected stimulation: suggested conditions include 10 mM final concentration H2O2, 20 ng/mL final concentration IL-6, or 20 ng/mL final concentration IL-2.
A note on cytokine preparation: Working stocks of cytokines should be 50X and made from a concentrated stock of “500X” that is different for each stim. For the final stimulation of cells in a 490 uL volume you will add 10 uL of working stock to obtain a 500 uL final reaction volume.
A note on H2O2 concentration: 10 mM H2O2 will strongly stimulate most cell types and is a natural secondary messenger produced by leukocytes that inhibits the catalytic cysteine of protein tyrosine phosphatases (Reth, 2002). A lower dose of 3.3 mM H2O2 in combination with stimulation through the B cell antigen receptor can be used to robustly and selectively activate B cells (Irish et al., 2006; Irish et al., 2010; Polikowsky et al., 2015). Pervanadate may be substituted as a mechanistically similar irreversible inhibitor (Bendall et al., 2011).
Hardware and instruments:
Centrifuge capable of spinning at 200xg and 800xg at room temperature
Rotors for 15 mL falcon tubes and 5 mL FACS tubes
37°C water bath
Incubator at 37°C, 5% CO2
Fridge at 4°C
−20°C or −80°C storage space
Benchtop vortex
Microscope and hemocytometer, or other method for cell counting
Aspiration setup with trap
Sink or vessel for decanting
Helios mass cytometer
Steps and annotations
Stimulation and extracellular stain
-
1
Warm media (RPMI with L-Glutamine, 10% FBS, and antibiotic) in 37°C water bath
-
2
Remove cryopreserved samples from liquid nitrogen.
-
3
Thaw samples rapidly (~60 seconds) in 37°C water bath and dilute in >10 mL of media in a 15 mL Falcon tube
-
4
Centrifuge samples at 200xg for 5 minutes at room temperature.
-
5
Aspirate supernatant and suspend cells in 1–2 mL media for counting.
-
6
Rest cells in 37°C incubator for 15 minutes with 1x DNase in serum-free media
-
7
While cells are resting, count viable nucleated cells using a hemocytometer (M. R. Green & Sambrook, 2019)
-
8
After 15 min, spin cells again at 200xg for 5 minutes and resuspend in 1–2 mL media
-
9
Rest cells again in 37°C incubator for 15 minutes in serum-free media
-
10
Centrifuge cells at 200xg for 5 minutes
-
11
Aspirate supernatant and re-suspend each sample in PBS at ~2×106 cells/mL
-
12
Add rhodium intercalator to 1μM and vortex
-
13
Incubate 5 minutes at room temperature
-
14
Add 2 mL PBS+BSA
-
15
Spin at 200xg for 5 min at room temperature
-
16
Aspirate supernatant and resuspend cells in 3 mL media
-
17Split cells into 6 FACS tubes by pipetting 490 uL of cells in media into each tube
- Vortex cells gently before pipetting to ensure equal distribution of cells
-
18
Volume in each tube should be exactly 490 uL before adding cytokines
-
19Add cytokines and other stimulations to desired concentration (see materials list above)
- Add 10 uL PBS to control tube
-
20
After 15 minutes of stimulation, fix cells by adding 55 uL 16% PFA to each tube to a final concentration of 1.6% and vortex.
-
21
Incubate for 10 min at room temperature
-
22
Spin down cells at 800xg for 5 minutes
-
23
Remove supernatant by decanting and then vortex.
-
24
Wash cells with 1 mL PBS+BSA.
-
25
Remove as much supernatant as possible by decanting and then vortexing.
-
26Transfer 35 uL of cells to a new tube containing 15 uL of extracellular antibody cocktail
- Add PBS+BSA if pellet volume is <35 uL
-
27
Vortex and incubate cells with antibodies for 30 minutes at room temperature
-
28
Add 1 mL PBS+BSA, spin down cells at 800xg for 5 minutes
-
29
Remove supernatant by decanting and vortex.
-
30
Add 1 mL PBS and spin down cells at 800xg for 5 minutes
-
31
Remove supernatant by decanting and then vortexing.
-
32
Add 1mL ice cold (≤ −20 °C) methanol to each tube and parafilm the top of the tube and transfer to freezer
-
33Samples must remain in methanol at −20°C for a minimum of 20 minutes to permeabilize cells before intracellular staining.
- Samples can remain at −20°C for weeks, or at −80°C for months, without apparent degradation of staining.
Intracellular Staining
-
34
Remove tubes from freezer and remove parafilm
-
35
Add 1 mL PBS and spin down the cells at 800xg for 5 minutes
-
36
Remove supernatant by decanting and vortexing.
-
37
Add 1 mL PBS+BSA and vortex well. Then, spin down cells at 800xg for 5 minutes
-
38
Remove as much supernatant as possible by decanting and vortexing.
-
39Transfer 35 uL of resuspended pellet to a new tube containing 15 uL of intracellular staining master mix
- Add PBS/BSA if pellet volume is <35 uL
-
40
Vortex and incubate cells with antibodies for 30’ at room temperature
-
41
Add 1 mL PBS+BSA and spin down cells at 800xg for 5 minutes
-
42
Remove supernatant by decanting and then vortex.
-
43
Add 1 mL PBS and spin down cells at 800xg for 5 minutes
-
44
Remove supernatant by decanting and then vortex.
-
45
Add 1 mL pre-diluted iridium intercalator (125 nM in 1.6% PFA in PBS)
-
46
Vortex and parafilm the top of the tubes
-
47
Incubate cells at least 20 minutes at room temperature or overnight at 4°C
Data acquisition
-
48
Add 1 mL PBS to each tube and spin down cells at 800xg for 5 minutes
-
49
Remove supernatant by decanting and then vortex.
-
50
Add 1 mL ddH2O and spin down cells at 800xg for 5 minutes
-
51
Remove supernatant by decanting and then vortex.
-
52
Resuspend cells in 1x beads in ddH2O at 0.5 ×106 cells per mL
-
53Pass resuspended cells and beads through filtered FACS tube and acquire on Helios mass cytometer
- If event rate is >500 events/second, pause acquisition and add 1X beads
- Aim to acquire 100,000 events per tube
-
54
After collection, normalize files in the Fluidigm software, opting to remove beads, and analyze
SUPPORT PROTOCOL 1
Analysis of signaling mass cytometry data
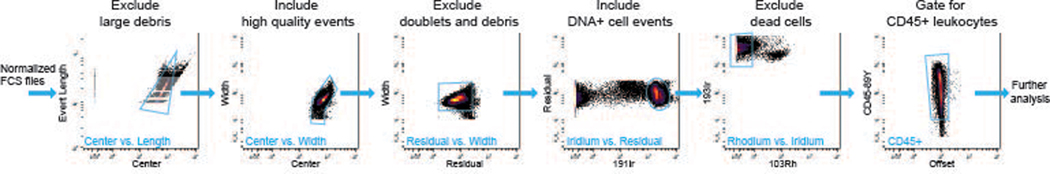
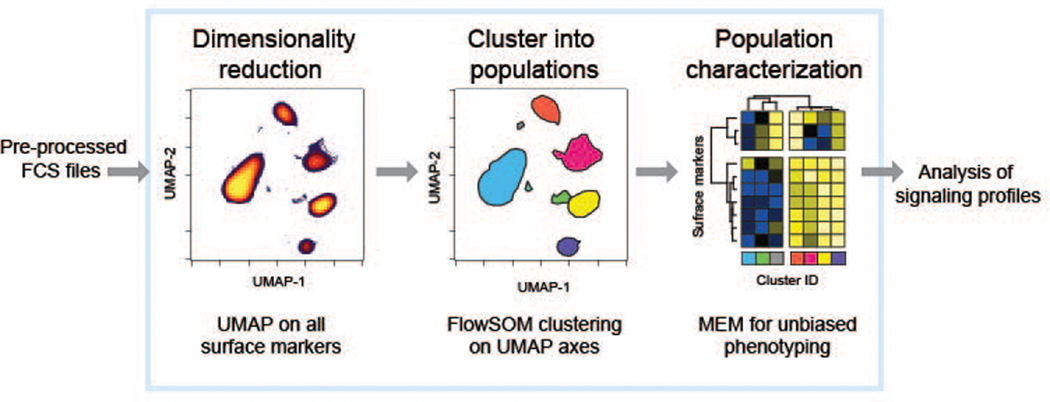
This support protocol provides a framework for high-dimensional, unsupervised analysis of the data generated in Basic Protocol 1. Pre-processing steps including normalization and manual, clean-up gating were completed prior to analysis of the data in R. Manual, clean-up gating is typically accomplished in a GUI such as Cytobank (Kotecha, Krutzik, & Irish, 2010), FCS Express (De Novo Software, Glendale, CA), or FlowJo (Becton, Dickinson & Company, Franklin Lakes, NJ). Automated options may also be used (Aghaeepour et al., 2018; Li et al., 2017). Clean-up gating follows the strategy illustrated in Figure 1 to remove atypical events from the normalized data files. This gating scheme is based on literature from the manufacturer of mass cytometry systems (“Recommendations for Use of Gaussian Discrimination Parameters,” 2016), and should be applicable to all data generated on third generation mass cytometry (Helios, Fluidigm Inc., South San Francisco, CA) systems. Following data pre-processing, data are exported to FCS and analyzed in the R programming language using an R Markdown (RMD) script that includes dimensionality reduction using UMAP (Becht et al., 2018; McInnes, 2018), clustering using FlowSOM (Van Gassen et al., 2015a), and cluster characterization using MEM (Diggins et al., 2017), following the schematic depicted in Figure 1. The unsupervised, automated workflow implemented in the RMD is modular, as visualized in Figure 2, meaning advanced users can modify this pipeline to utilize alternative algorithms for the steps mentioned prior, i.e. dimensionality reduction using t-SNE (Amir el et al., 2013).
Figure 1.
Mass cytometry data are traditionally manually reviewed to prepare for automated analysis. This figure demonstrates representative gating for manual cleanup of normalized FCS files using a GUI. Panels are to be followed stepwise, with each subsequent gate removing one or more forms of debris or computationally undesirable cell events. More details of this gating scheme can be found in Support Protocol 1.
Figure 2.
A semi-automated analysis workflow introduces trainees to multidimensional analysis and computational tools. Manually pre-processed FCS files are read into the R pipeline, symbolized here by a large light box, where multidimensional data is reduced to two dimensions using UMAP for ease of visualization and simplification of subsequent clustering using FlowSOM. The population clusters identified by FlowSOM are then phenotypically characterized by MEM, and these cluster IDs along with the UMAP coordinates are encoded into a new FCS file, which can be exported and analyzed by any means desired.
Materials:
A GUI for biaxial gating of mass cytometry data, such as FlowJo, FCS express, or Cytobank
R version 3.6.2 for your operating system with Bioconductor/flowCore
R Studio (version 1.2 used here)
R Tools PC version Rtools35, or X11 Quartz for MAC users
RMD from https://github.com/cytolab/roe_phospho_flow
Normalized fcs files generated in Basic Protocol 1
Steps and annotations
Manual data cleanup.
-
1
Open your normalized FCS files in the GUI of choice
-
2
Verify that the scales of all Gaussian parameters and event length are on an archsinh scale
-
3
Plot Center vs. Event Length and draw a gate as indicated in Figure 1, excluding large pieces of debris and computational doublets.
-
4
Using the population gated in step 3, plot Center vs. Width and gate as indicated in Figure 3 for high quality, Gaussian normal events.
-
5
Using the population obtained in step 4, plot Residual vs. Width and draw a gate as indicated in Figure 1, excluding those events with high residual (doublets) or low width (debris).
-
6
Using the population obtained in step 5, plot Ir191 intercalator vs. residual and draw a gate as indicated in Figure 1, for DNA positive cell events. Take care to include the population highest for iridium intercalator as this may represent dividing cells.
-
7
Using the population obtained in step 6, plot Ir193 intercalator vs. live/dead (103Rh intercalator) and draw a gate as indicated in Figure 3, excluding dead cells.
-
8
For the data generated in Basic Protocol 1, and most immune cell datasets, gate also for CD45+ cells from the Live Cell population obtained in step 7.
-
9
Export CD45+ cells to FCS, and proceed with R analysis.
Figure 3.
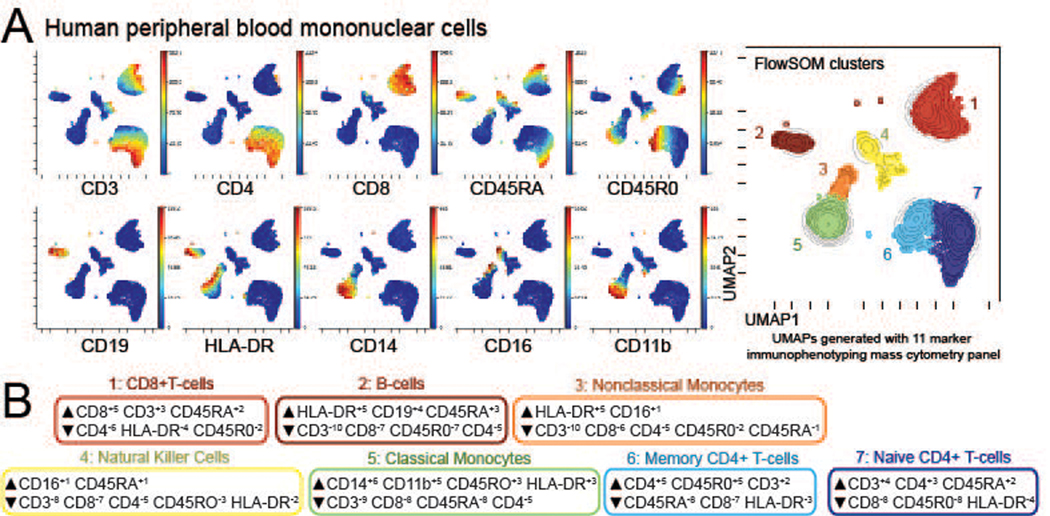
An 11-antibody core panel is sufficient for automated analysis to resolve major immune cell types in blood. This figure demonstrates expected phenotyping results from Protocol 1, using a minimal mass cytometry panel to subset healthy human blood mononuclear cells. In A, the large plot shows dots color-coded by FlowSOM cluster ID, overlaid with contours indicating cell density. Each smaller plot shows the same dots, with color indicating relative marker expression. This marker expression is summarized in the MEM labels in part B, with the numbered boxes labelled with an expert defined cell population name based on the MEM labels. Numbers in part B correspond to numbers next to cell ‘islands’ in part A, which represent the most common expected cell types in human PBMCs.
Unsupervised Analysis in R
-
10
Install R, RStudio, and Rtools35/X11 Quartz, as is appropriate for your operating system
-
11
Install packages by opening and running 00_install_tools.RMD script in RStudio
-
12
Open 01_data_analysis.RMD script in RStudio
-
13
Follow the directions in the RMD to analyze data
-
14
Analyzed data are exported to FCS with analysis results included (UMAP1, UMAP2, FlowSOM cluster ID) and can be visualized in a GUI of your choice
REAGENTS AND SOLUTIONS:
Antibody cocktail preparation: See Table 1 for antibody cocktail information, including antibody clones and volumes for staining. This table includes 10% extra total cocktail volume, in the form of additional PBS+BSA, to allow for pipetting error. Add PBS+BSA to cocktail tube before adding antibodies, and vortex each antibody tube well before pipetting into the cocktail. Vortex the final cocktail well before staining cells. Antibodies listed in Table 1 are available conjugated to the indicated metal isotopes from Fluidigm, Inc.
Table 1:
Antibody cocktail preparation.
| Antibody cocktail | |||||
|---|---|---|---|---|---|
| Isotope Tag | Target [site] (clone) | uL/stain | Stains | Ex total | In Total |
| 89Y | CD45 (HI30) | 0.5 | 4 | 2 | 0 |
| 142Nd | CD19 (2H7) | 0.25 | 4 | 1 | 0 |
| 144Nd | p-PLCγ2 [Y759] (K86.689.37) | 0.25 | 4 | 0 | 1 |
| 145Nd | CD4 (RP-T4) | 0.25 | 4 | 1 | 0 |
| 146Nd | CD8a (RPA-T8) | 0.25 | 4 | 1 | 0 |
| 148Nd | CD14 (RMO52) | 0.5 | 4 | 2 | 0 |
| 150Nd | p-STAT5 [Y694] (47) | 0.25 | 4 | 0 | 1 |
| 153Eu | p-STAT1 [Y701] (4a) | 0.25 | 4 | 0 | 1 |
| 158Gd | p-STAT3 [Y705] (4/P-Stat3) | 0.25 | 4 | 0 | 1 |
| 165Ho | CD16(3G8) | 0.5 | 4 | 2 | 0 |
| 169Er | CD45RA (HI100) | 0.25 | 4 | 1 | 0 |
| 170Er | CD3 (UCHT1) | 0.25 | 4 | 1 | 0 |
| 174Yb | HLA-DR (L243) | 0.25 | 4 | 1 | 0 |
| 176Lu | CD56 (CMSSB) | 0.25 | 4 | 1 | 0 |
| 175Lu | p-STAT6 [Y641] (18/P-Stat6) | 0.25 | 4 | 0 | 1 |
| 209Bi | CD11b (ICRF44) | 0.25 | 4 | 1 | 0 |
| Total antibody volume (μL) | 14 | 5 | |||
| PBS+BSA to add* (μL) | 52 | 61 | |||
| Total volume cocktail (μL) | 66 | 66 | |||
| Cocktail volume per tube to be stained (μL) | 15 | 15 | |||
| Cell volume per tube to be stained (μL) | 35 | 35 | |||
COMMENTARY
BACKGROUND INFORMATION:
Phospho-specific Flow Cytometry Reveals Cell Signaling Profiles
Signaling governs both intrinsic and extrinsic functions of cells, guiding development and directing a cell’s behavior as it responds to events in its environment. Changes in how individual cells respond to stimulation is central to many human diseases, such as cancer and auto-immunity (Irish et al., 2010; O’Gorman et al., 2015). Single cell analysis of signaling has significant potential for understanding this in the laboratory, as cell subpopulations within a primary human sample—such as human blood—can be identified on the basis of surface markers and further distinguished by their phenotypic signaling potential and biological response profiles.
Mass Cytometry Provides Extensive Opportunities for Multiplexing of Measurements
Advances in single-cell biology have enabled measurements of more than 40 protein features on millions of cells in a variety of sample types using cytometry by time-of-flight, also known as mass cytometry (Ornatsky et al., 2008). This technology offers a unique opportunity to multiplex signaling measurements with surface marker analysis by flow cytometry, enabling deep phenotyping of cells while simultaneously characterizing cell response to stimuli at the single cell level. This has led to impressive data sets (Bendall et al., 2011; Bodenmiller et al., 2012), but this quantity of data has necessitated the development of new high-dimensional, often automated, data analysis strategies.
Unsupervised Analysis to Characterize Cell Populations and Reveal Signaling Outcomes
A multitude of algorithms have been developed to tackle high-dimensional single cell data (Nowicka et al., 2017), and a set of common themes have emerged in analysis pipelines for mass cytometry data (Pedersen & Olsen, 2019). After vendor-provided normalization to metal bead standards (Finck et al., 2013), raw data is manually gated by an expert for live, intact, single cell events to generate flow cytometry standard (FCS) files ready for further computational analysis. This analysis consists of dimensionality reduction for ease of visualization, clustering of cells into populations of similar phenotype, and then characterization of these populations to look for changes in signaling patterns or other functional readouts across cell populations and treatment conditions.
CRITICAL PARAMETERS:
The reproducibility of this protocol hinges on accuracy in pipetting, sample and reagent quality, and timing of all steps involving live cells. Inaccuracies in pipetting during cell staining will introduce variability in surface marker measurement, and, more distressingly, to detection of protein phosphorylation events, hindering analysis of data generated. Similarly, timing and sample quality are critical for accurate, robust signaling responses to stimulation. Cells not permitted adequate time to return to (low) basal signaling levels following the trauma of thaw will exhibit high basal signaling, masking responses to stimuli. Conversely, poor quality, dying samples will signal little, if at all. The duration of stimulation before fixation is also important, as any variation at this step will alter the cell’s protein phosphorylation signature in possibly unpredictable ways.
TROUBLESHOOTING:
See Table 2 for potential protocol problems and solutions.
Table 2:
Troubleshooting Guide for Mass Cytometry Phospho-flow
| No intracellular staining observed | Cells were not adequately permeabilized | Permeabilize cells in methanol overnight |
| Surface marker appears to be absent | Antibody is faulty or was omitted from cocktail | Repeat ensuring antibody is included in cocktail |
| Large number of dead cells | Sample was poorly cryopreserved or thawed incorrectly | Ensure that thawing is rapid (<1 minute) and sample is diluted at least 1:10 in warm media |
| Few cells | Decanting was done incorrectly, or cells not adequately recovered after methanol permeabilization | Vortex sample well during rehydration after methanol. Decant assertively, without wobbling |
| Poor antibody staining | Cells may be of poor quality, over-fixed, or antibody cocktail may have been prepared incorrectly | Samples with >20% Trypan blue positive cells are likely low quality. |
| Globally low or unexpected signaling patterns | Cells were not adequately fixed, or stimulation timing was incorrect | Ensure fixation reagent is fresh |
| High signaling in control sample | Cells were not rested a full 30 minutes before stimulation, or were rested in serum containing media | Check media and resting time |
| Absence of specific expected signals | Stimulation reagent is faulty or stimulation timing was incorrect | Include a positive control, such as H2O2 to determine which is faulty |
UNDERSTANDING RESULTS:
Figure 3 illustrates phenotyping of a healthy human PBMC sample using the protocol above. Major cell populations expected are present, including CD4+ and CD8+ T-cells, B-cells, classical and non-classical monocytes, and natural killer cells (Maecker, McCoy, & Nussenblatt, 2012). CD4+ T-cells, an abundant population, are further subdivided into naïve and memory cells based on CD45RA and R0 expression. However, there is poor resolution of rare cell types, such as dendritic cells, which did not have identifying antibodies in the panel and are thus not apparent to the automated analysis tools, and further subtyping of existing populations, such as T-cells, is not straightforward using a minimal antibody panel, as shown here. Figure 4 part A demonstrates expected signaling results obtained with this protocol, including know responses such as IFNγ > p-STAT1 (D. S. Green, Young, & Valencia, 2017) and IL-4 > p-STAT6 (Wills-Karp & Finkelman, 2008). In this experiment, the trainee selected high dose 10 mM hydrogen peroxide as their wildcard stimuli, which resulted in phosphorylation of STAT1 and PLCγ. However, not all cell types responded to peroxide as expected (N. Leelatian et al., 2015), suggesting a less than optimal stimulation. In part B of Figure 4, the reproducibility of this protocol is demonstrated. Here, two trainees thawed aliquots of PBMC from the same donor and followed the protocol above, resulting in near identical signaling results in theirs samples. These data can be found on Flow Repository at https://flowrepository.org/experiments/2564, or along with the RMD for analysis at https://github.com/cytolab/roe_phospho_flow.
Figure 4,F.

The phospho-flow protocol produces reliable signaling results and allows for trainee customization. This figure shows the expected signaling outcome from Protocol 1. In part A, one iteration is shown, with heatmaps indicating the arcsinh transformed ration of median protein phosphorylation compared to the unstimulated condition. Each row represents a cell population as defined in Figure 3, and each column represents a stimulation condition. Five different phosphorylated protein readouts are shown here. In this experiment, the trainee selected peroxide as their third condition. In part B, the results of two trainees performing the protocol in parallel are shown. In part C, a variety of ‘wildcard’ stimuli and their most robust readouts are shown.
TIME CONSIDERATIONS:
Stimulation and extracellular stain requires four hours. Intracellular stain and intercalation requires one hour. If done on the same day, these portions require six hours total. Data acquisition requires one hour of instrument setup and 20 minutes per sample to be acquired. The entire protocol could be completed in one eight to ten-hour day, but it is preferable to divide up the experiment to allow time for errors. Data analysis a dynamic process, but manual gating and R analysis as described in Supplementary Protocol 1 takes one to two hours.
ACKNOWLEDGEMENTS:
We thank all the trainees who helped test this protocol and generate the data presented here. This work was supported by research grants: National Institutes of Health/National Cancer Institute (NIH/NCI R01 CA226833, U54 CA217450) and the Vanderbilt-Ingram Cancer Center (VICC, P30 CA68485).
Footnotes
CONFLICTS OF INTEREST:
Jonathan M. Irish was a co-founder and was a board member of Cytobank Inc. and received research support from Incyte Corp, Janssen, and Pharmacyclics. The authors declare that there are no other conflicts of interest.
WORKS CITED
- Aghaeepour N, Simonds EF, Knapp D, Bruggner RV, Sachs K, Culos A, . . . Nolan GP (2018). GateFinder: projection-based gating strategy optimization for flow and mass cytometry. Bioinformatics, 34(23), 4131–4133. doi: 10.1093/bioinformatics/bty430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir el AD, Davis KL, Tadmor MD, Simonds EF, Levine JH, Bendall SC, . . . Pe’er D. (2013). viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol, 31(6), 545–552. doi: 10.1038/nbt.2594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay S, Fisher DAC, Malkova O, & Oh ST (2017). Analysis of Signaling Networks at the Single-Cell Level Using Mass Cytometry. Methods Mol Biol, 1636, 371–392. doi: 10.1007/978-1-4939-7154-1_24 [DOI] [PubMed] [Google Scholar]
- Becht E, McInnes L, Healy J, Dutertre CA, Kwok IWH, Ng LG, . . . Newell EW (2018). Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol. doi: 10.1038/nbt.4314 [DOI] [PubMed] [Google Scholar]
- Bendall SC, Davis KL, Amir el AD, Tadmor MD, Simonds EF, Chen TJ, . . . Pe’er D. (2014). Single-cell trajectory detection uncovers progression and regulatory coordination in human B cell development. Cell, 157(3), 714–725. doi: 10.1016/j.cell.2014.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendall SC, Simonds EF, Qiu P, Amir el AD, Krutzik PO, Finck R, . . . Nolan GP (2011). Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science, 332(6030), 687–696. doi: 10.1126/science.1198704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenmiller B, Zunder ER, Finck R, Chen TJ, Savig ES, Bruggner RV, . . . Nolan GP (2012). Multiplexed mass cytometry profiling of cellular states perturbed by small-molecule regulators. Nat Biotechnol, 30(9), 858–867. doi: 10.1038/nbt.2317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diggins KE, Ferrell PB Jr., & Irish JM (2015). Methods for discovery and characterization of cell subsets in high dimensional mass cytometry data. Methods, 82, 55–63. doi: 10.1016/j.ymeth.2015.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diggins KE, Gandelman JS, Roe CE, & Irish JM (2018). Generating Quantitative Cell Identity Labels with Marker Enrichment Modeling (MEM). Curr Protoc Cytom, 83, 10 21 11–10 21 28. doi: 10.1002/cpcy.34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diggins KE, Greenplate AR, Leelatian N, Wogsland CE, & Irish JM (2017). Characterizing cell subsets using marker enrichment modeling. Nat Methods, 14(3), 275–278. doi: 10.1038/nmeth.4149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl DC, Ferrell PB Jr., Leelatian N, Froese JT, Reisman BJ, Irish JM, & Bachmann BO (2018). Discovery of human cell selective effector molecules using single cell multiplexed activity metabolomics. Nat Commun, 9(1), 39. doi: 10.1038/s41467-017-02470-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finck R, Simonds EF, Jager A, Krishnaswamy S, Sachs K, Fantl W, . . . Bendall SC (2013). Normalization of mass cytometry data with bead standards. Cytometry A, 83(5), 483–494. doi: 10.1002/cyto.a.22271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez VD, Samusik N, Chen TJ, Savig ES, Aghaeepour N, Quigley DA, . . . Fantl WJ (2018). Commonly Occurring Cell Subsets in High-Grade Serous Ovarian Tumors Identified by Single-Cell Mass Cytometry. Cell Rep, 22(7), 1875–1888. doi: 10.1016/j.celrep.2018.01.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good Z, Sarno J, Jager A, Samusik N, Aghaeepour N, Simonds EF, . . . Davis KL (2018). Single-cell developmental classification of B cell precursor acute lymphoblastic leukemia at diagnosis reveals predictors of relapse. Nat Med, 24(4), 474–483. doi: 10.1038/nm.4505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green DS, Young HA, & Valencia JC (2017). Current prospects of type II interferon gamma signaling and autoimmunity. J Biol Chem, 292(34), 13925–13933. doi: 10.1074/jbc.R116.774745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green MR, & Sambrook J. (2019). Estimation of Cell Number by Hemocytometry Counting. Cold Spring Harb Protoc, 2019(11), pdb prot097980. doi: 10.1101/pdb.prot097980 [DOI] [PubMed] [Google Scholar]
- Huse K, Wogsland CE, Polikowsky HG, Diggins KE, Smeland EB, Myklebust JH, & Irish JM (2019). Human Germinal Center B Cells Differ from Naive and Memory B Cells in CD40 Expression and CD40L-Induced Signaling Response. Cytometry A, 95(4), 442–449. doi: 10.1002/cyto.a.23737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irish JM, Czerwinski DK, Nolan GP, & Levy R. (2006). Kinetics of B cell receptor signaling in human B cell subsets mapped by phosphospecific flow cytometry. J Immunol, 177(3), 1581–1589. doi: 10.4049/jimmunol.177.3.1581 [DOI] [PubMed] [Google Scholar]
- Irish JM, & Doxie DB (2014). High-dimensional single-cell cancer biology. Curr Top Microbiol Immunol, 377, 1–21. doi: 10.1007/82_2014_367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irish JM, Hovland R, Krutzik PO, Perez OD, Bruserud O, Gjertsen BT, & Nolan GP (2004). Single cell profiling of potentiated phospho-protein networks in cancer cells. Cell, 118(2), 217–228. doi: 10.1016/j.cell.2004.06.028 [DOI] [PubMed] [Google Scholar]
- Irish JM, Myklebust JH, Alizadeh AA, Houot R, Sharman JP, Czerwinski DK, . . . Levy R. (2010). B-cell signaling networks reveal a negative prognostic human lymphoma cell subset that emerges during tumor progression. Proc Natl Acad Sci U S A, 107(29), 12747–12754. doi: 10.1073/pnas.1002057107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalland ME, Oberprieler NG, Vang T, Tasken K, & Torgersen KM (2011). T cell-signaling network analysis reveals distinct differences between CD28 and CD2 costimulation responses in various subsets and in the MAPK pathway between resting and activated regulatory T cells. J Immunol, 187(10), 5233–5245. doi: 10.4049/jimmunol.1101804 [DOI] [PubMed] [Google Scholar]
- Kotecha N, Krutzik PO, & Irish JM (2010). Web-based analysis and publication of flow cytometry experiments. Curr Protoc Cytom, Chapter 10, Unit10 17. doi: 10.1002/0471142956.cy1017s53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krutzik PO, Crane JM, Clutter MR, & Nolan GP (2008). High-content single-cell drug screening with phosphospecific flow cytometry. Nat Chem Biol, 4(2), 132–142. doi: 10.1038/nchembio.2007.59 [DOI] [PubMed] [Google Scholar]
- Krutzik PO, Irish JM, Nolan GP, & Perez OD (2004). Analysis of protein phosphorylation and cellular signaling events by flow cytometry: techniques and clinical applications. Clin Immunol, 110(3), 206–221. doi: 10.1016/j.clim.2003.11.009 [DOI] [PubMed] [Google Scholar]
- Krutzik PO, & Nolan GP (2003). Intracellular phospho-protein staining techniques for flow cytometry: monitoring single cell signaling events. Cytometry A, 55(2), 61–70. doi: 10.1002/cyto.a.10072 [DOI] [PubMed] [Google Scholar]
- Leelatian N, Diggins KE, & Irish JM (2015). Characterizing Phenotypes and Signaling Networks of Single Human Cells by Mass Cytometry. Methods Mol Biol, 1346, 99–113. doi: 10.1007/978-1-4939-2987-0_8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leelatian N, Doxie DB, Greenplate AR, Sinnaeve J, Ihrie RA, & Irish JM (2017). Preparing Viable Single Cells from Human Tissue and Tumors for Cytomic Analysis. Curr Protoc Mol Biol, 118, 25C 21 21–25C 21 23. doi: 10.1002/cpmb.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leelatian N, Sinnaeve J, Mistry AM, Barone SM, Diggins KE, Greenplate AR, . . . Irish JM (2019). High risk glioblastoma cells revealed by machine learning and single cell signaling profiles. bioRxiv. [Google Scholar]
- Levine JH, Simonds EF, Bendall SC, Davis KL, Amir el AD, Tadmor MD, . . . Nolan GP (2015). Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis. Cell, 162(1), 184–197. doi: 10.1016/j.cell.2015.05.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Shaham U, Stanton KP, Yao Y, Montgomery RR, & Kluger Y. (2017). Gating mass cytometry data by deep learning. Bioinformatics, 33(21), 3423–3430. doi: 10.1093/bioinformatics/btx448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maecker HT, McCoy JP, & Nussenblatt R. (2012). Standardizing immunophenotyping for the Human Immunology Project. Nat Rev Immunol, 12(3), 191–200. doi: 10.1038/nri3158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnes L. (2018). UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. Retrieved from https://umap-learn.readthedocs.io/en/latest/
- Myklebust JH, Brody J, Kohrt HE, Kolstad A, Czerwinski DK, Walchli S, . . . Levy R. (2017). Distinct patterns of B-cell receptor signaling in non-Hodgkin lymphomas identified by single-cell profiling. Blood, 129(6), 759–770. doi: 10.1182/blood-2016-05-718494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas KJ, Greenplate AR, Flaherty DK, Matlock BK, Juan JS, Smith RM, . . . Kalams SA (2016). Multiparameter analysis of stimulated human peripheral blood mononuclear cells: A comparison of mass and fluorescence cytometry. Cytometry A, 89(3), 271–280. doi: 10.1002/cyto.a.22799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowicka M, Krieg C, Crowell HL, Weber LM, Hartmann FJ, Guglietta S, . . . Robinson MD (2017). CyTOF workflow: differential discovery in high-throughput high-dimensional cytometry datasets. F1000Res, 6, 748. doi: 10.12688/f1000research.11622.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Gorman WE, Hsieh EW, Savig ES, Gherardini PF, Hernandez JD, Hansmann L, . . . Davis MM (2015). Single-cell systems-level analysis of human Toll-like receptor activation defines a chemokine signature in patients with systemic lupus erythematosus. J Allergy Clin Immunol, 136(5), 1326–1336. doi: 10.1016/j.jaci.2015.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornatsky OI, Lou X, Nitz M, Schafer S, Sheldrick WS, Baranov VI, . . . Tanner SD (2008). Study of cell antigens and intracellular DNA by identification of element-containing labels and metallointercalators using inductively coupled plasma mass spectrometry. Anal Chem, 80(7), 2539–2547. doi: 10.1021/ac702128m [DOI] [PubMed] [Google Scholar]
- Pedersen CB, & Olsen LR (2019). Analysis of Mass Cytometry Data. Methods Mol Biol, 1989, 267–279. doi: 10.1007/978-1-4939-9454-0_17 [DOI] [PubMed] [Google Scholar]
- Polikowsky HG, Wogsland CE, Diggins KE, Huse K, & Irish JM (2015). Cutting Edge: Redox Signaling Hypersensitivity Distinguishes Human Germinal Center B Cells. J Immunol, 195(4), 1364–1367. doi: 10.4049/jimmunol.1500904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X, Sufi J, Vlckova P, Kyriakidou P, Acton SE, Li VSW, . . . Tape CJ (2020). Cell-type-specific signaling networks in heterocellular organoids. Nat Methods. doi: 10.1038/s41592-020-0737-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recommendations for Use of Gaussian Discrimination Parameters. (2016). In Fluidigm I (Ed.). Cytoforum. [Google Scholar]
- Reth M. (2002). Hydrogen peroxide as second messenger in lymphocyte activation. Nat Immunol, 3(12), 1129–1134. doi: 10.1038/ni1202-1129 [DOI] [PubMed] [Google Scholar]
- Rogne M, & Tasken K. (2013). Cell signalling analyses in the functional genomics era. N Biotechnol, 30(3), 333–338. doi: 10.1016/j.nbt.2013.01.003 [DOI] [PubMed] [Google Scholar]
- Saeys Y, Gassen SV, & Lambrecht BN (2016). Computational flow cytometry: helping to make sense of high-dimensional immunology data. Nat Rev Immunol, 16(7), 449–462. doi: 10.1038/nri.2016.56 [DOI] [PubMed] [Google Scholar]
- Van Gassen S, Callebaut B, Van Helden MJ, Lambrecht BN, Demeester P, Dhaene T, & Saeys Y. (2015a). FlowSOM: Using self-organizing maps for visualization and interpretation of cytometry data. Cytometry A, 87(7), 636–645. doi: 10.1002/cyto.a.22625 [DOI] [PubMed] [Google Scholar]
- Van Gassen S, Callebaut B, Van Helden MJ, Lambrecht BN, Demeester P, Dhaene T, & Saeys Y. (2015b). FlowSOM: Using self-organizing maps for visualization and interpretation of cytometry data. Cytometry A. doi: 10.1002/cyto.a.22625 [DOI] [PubMed] [Google Scholar]
- Wills-Karp M, & Finkelman FD (2008). Untangling the complex web of IL-4- and IL-13-mediated signaling pathways. Sci Signal, 1(51), pe55. doi: 10.1126/scisignal.1.51.pe55 [DOI] [PMC free article] [PubMed] [Google Scholar]